Abstract
Background
Genetically predicted leukocyte telomere length (LTL) has been evaluated in several studies of childhood and adult cancer. We test whether genetically predicted longer LTL is associated with germ cell tumours (GCT) in children and adults.
Methods
Paediatric GCT samples were obtained from a Children’s Oncology Group study and state biobank programs in California and Michigan (N = 1413 cases, 1220 biological parents and 1022 unrelated controls). Replication analysis included 396 adult testicular GCTs (TGCT) and 1589 matched controls from the UK Biobank. Mendelian randomisation was used to look at the association between genetically predicted LTL and GCTs and TERT variants were evaluated within GCT subgroups.
Results
We identified significant associations between TERT variants reported in previous adult TGCT GWAS in paediatric GCT: TERT/rs2736100-C (OR = 0.82; P = 0.0003), TERT/rs2853677-G (OR = 0.80; P = 0.001), and TERT/rs7705526-A (OR = 0.81; P = 0.003). We also extended these findings to females and tumours outside the testes. In contrast, we did not observe strong evidence for an association between genetically predicted LTL by other variants and GCT risk in children or adults.
Conclusion
While TERT is a known susceptibility locus for GCT, our results suggest that LTL predicted by other variants is not strongly associated with risk in either children or adults.
Subject terms: Germ cell tumours, Cancer genetics, Paediatric cancer
Background
Telomeres are DNA–protein structures consisting of a repeat sequence (TTAGGG) that cap the ends of a chromosome and function to stabilise and protect them from recombination and deterioration [1]. As part of normal cell division, telomere length shortens over time and when a critical length is reached, the cell will undergo cellular senescence or apoptosis. Thus, telomeres are thought of as a DNA biomarker of ageing and have been associated with many age-related diseases [2–4]. However, cancer cells have been shown to upregulate telomerase (the enzyme responsible for adding new DNA onto telomeres) and bypass cellular senescence, which enables a limitless replication potential [5, 6]. Thus, several studies have suggested the maintenance of telomere length plays a critical role in cancer susceptibility. For example, epidemiological studies that have measured telomere length directly have generally found shorter telomere length to be associated with increased risk for multiple adult cancers including lung cancer, melanoma and chronic lymphocytic leukaemia, although the direction of the association has not been consistent across studies [7–10]. Studies have also shown that telomere length is highly variable across individuals starting at birth and is associated with genetic variation, demographics, disease status and environmental factors (e.g., tobacco smoking, ultraviolet radiation) [11–16].
Germ cell tumours (GCTs) are heterogeneous tumours that arise from the primordial germ cell (PGC) [17]. In children and adolescents, GCTs occur in both males and females in the gonads and in extragonadal locations [18]. In adults, testicular GCTs (TGCTs) are the most common malignancy diagnosed in men between the ages of 15 and 45 years [19]. The aetiology of GCTs in children and adolescents is largely unknown. Studies of adult TGCTs, which arise from the same precursor cell as paediatric GCTs, support a role for inherited variation in risk suggesting genetic factors may play a role in their aetiology [20, 21]. GWAS have found susceptibility loci for TGCTs, including variants in TERT (telomerase reverse transcriptase) which encodes a subunit of the enzyme telomerase [22–25]. This raises the possibility that germline genetic susceptibility in other variants related to telomere length could be associated with risk for germ cell tumours.
Measuring circulating telomeres directly in cancer cells is problematic as it is unclear if disease status is the cause or the consequence of shortened telomere length. Such reverse causality is a potential explanation for the observed mixed results from studies of telomere length and cancer risk. In the past decade, several studies have tried to overcome this limitation by identifying genetic predictors of telomere length to be used in prospective analyses [26–32]. A large-scale genome-wide association study (GWAS) of over 37,000 individuals led to the development of a robust genetic predictor of leukocyte telomere length (LTL) using a set of seven common single-nucleotide polymorphisms (SNPs) that together explain an estimated 1.23% of the variance in LTL [27]. These SNPs are located in ACYP2, TERC, NAF1, TERT, OBFC1, ZNF208 and RTEL1 and have been used as a proxy for telomere length in a number of studies investigating cancer risk [33, 34]. Recently, this study was updated to include over 78,000 individuals and identified an additional thirteen SNPs in or near SENP7, MOB1B, CARMIL1, PRRC2A, TERF2, RFWD3, TERT, RTEL1, PARP1, POT1, ATM and MPHOSPH6 loci for a combination of 20 SNPs that together explain an estimated 1.71% of the variance in LTL [32]. Seventeen of these SNPs have been used as instrumental variables to study the association between genetically predicted LTL and risk for a wide range of cancers, including adult testicular cancer [34].
Mendelian randomisation (MR) is a statistical method that utilises known genetic determinants of the exposure, such as SNPs, as instrumental variables (or proxies for exposures) to examine the causal effect of an exposure (e.g., telomere length) on health outcomes (e.g., cancer risk) without the limitations of confounding, reverse causation, and various biases (e.g., measurement error) [35]. Epidemiological studies have reported associations between genetically predicted LTL and increased risk for multiple types of childhood and adult cancers [30, 36–38], including three previous studies reporting inconsistent associations with adult TGCT [34, 39, 40]. These differences may be due to the inclusion of TERT, the previously identified susceptibility locus for adult TGCT, in the genetic instrument for predicted telomere length [22, 23]. For example, Haycock et al. reported genetically increased telomere length was associated with increased risk for adult TGCT, not including TERT in their genetic predictor (OR [95% CI] = 1.76 [1.02–3.04]; P = 0.040) [39]. In contrast, Gao et al. reported a null association between shorter telomere length and increased risk for adult TGCT including TERT (OR [95% CI] = 1.02 [0.95–1.11]; P = 0.549) [34]. More recently, when Brown et al. removed TERT due to the previously identified TGCT association, they reported a non-significant association between longer telomere length and increased risk for TGCT (OR [95% CI] = 1.07 [1.02, 1.13]; P = 0.121) [40]. Notably, the established association between TERT and the LTL lengthening risk allele is in the opposite direction for the TGCT risk allele, which may explain why removing TERT in the previous studies strengthened the association with the other LTL variants. Genetically predicted LTL has not been evaluated as a risk factor for paediatric and adolescent GCTs to date.
In these analyses, we use MR methods to test whether genetically predicted longer LTL is associated with GCTs in children, adolescents and adults using two separate sets of SNPs that are validated genetic predictors of LTL [27, 32]. Given previous findings, we report associations both with and without variants at the TERT locus. We also explored individual TERT variant associations with paediatric, adolescent, and adult GCTs to determine whether the previously reported association with adult TGCT was also important in other groups of GCT patients [22–25].
Methods
Study participants
Germ cell tumour epidemiology study (GaMETES)
Children and adolescents with GCTs were identified through the Children’s Oncology Group (COG) Childhood Cancer Research Network and invited to participate in a case–parent trio study as detailed previously [41, 42]. Briefly, children and adolescents were eligible if they had a primary diagnosis of GCT between July 1, 2008 and December 31, 2015, were <20 years of age at diagnosis, and had at least one biological parent willing to participate and able to complete a questionnaire in English or Spanish. Parents provided written informed consent for participation in the study. Participants aged 18 years or older provided informed consent for participation in the study. Assent was obtained for children aged 8–17 years. Saliva DNA was collected from children with GCTs and from their biological parents.
Newborn biobank samples
GCT cases and unrelated controls identified through newborn biobanks in California and Michigan were also included in the analysis. Neonatal blood spots (NBS) are routinely collected after each live birth in all states as part of a Newborn Screening Program. Once screening is complete, residual NBS are stored in relevant state biobanks and are available for additional research in several states. Children and adolescents who were later diagnosed with a GCT were identified through linkage to the California and Michigan state cancer registries and vital statistics birth/death files. Participants were selected if they had a primary diagnosis of either an intracranial or extracranial GCT between January 1, 1990 and December 31, 2011 and were <20 years of age at diagnosis. In addition, unaffected controls (matched on birth month and year [i.e., age], sex, and race/ethnicity) were selected. One punch from the stored blood spots measuring 1/8 inch was obtained from the California Biobank Program (CBP) and the Michigan Neonatal Biobank (MNB) for each selected case and control and shipped to the Molecular Epidemiology Laboratory at the University of Minnesota for DNA extraction and genotyping.
All study procedures were approved by the University of Minnesota Institutional Review Board. The California Committee for the Protection of Human Subjects and the Michigan Department of Health and Human Services Institutional Review Board approved the use of the biobank samples.
DNA extraction
We performed automated DNA isolation using an Autopure LS system (Qiagen, Venlo, Netherlands) and a Puregene DNA Purification Kit (Gentra Systems, Minneapolis, MN) according to the manufacturer’s protocols. DNA yield was quantified in triplicate using real-time polymerase chain reaction (ABI 7900 Prism; Applied Biosystems, Thermo Fisher Scientific, Waltham, Massachusetts) (1:10 dilution). Extracted DNA was aliquoted and stored at −20 °C until genotyping.
Genotyping and quality control
The University of Minnesota Genomics Center performed genotyping using the Illumina CoreExome BeadChip (Illumina, San Diego) according to the manufacturer’s specified protocol. Allele cluster definitions for each variant were determined via Illumina’s GenomeStudio Genotyping Module using the intensity data from only those study samples with high-quality data (call rate > 0.98; log R ratio standard deviation < 0.25). A HapMap sample was placed on each 96-sample plate. In addition, blind duplicate samples were distributed among the plates to assess genotyping concordance and detect plate effects. There was no evidence of plate-specific genotype effects. Genvisis (http://www.genvisis.org) was used to identify low quality/contaminated samples (N = 95), samples with sex aneuploidy (N = 30) and trisomy 21 (N = 7). These samples were excluded from all analyses.
Ancestry was inferred by performing a principal components analysis (PCA) as implemented in EIGENSOFT [43, 44] that included HapMap samples as anchors. The first two principal components from the analysis were plotted and samples were assigned an ancestry of European, African American, Asian or Hispanic based on where they clustered relative to the three HapMap anchor populations. A second PCA was run within each of these ancestry designations to obtain principal components to use as covariates.
Genotyping imputation
To extend our genotype analysis, imputation for the paediatric GCT cases and controls was performed using 194,512 haplotypes from the Trans-Omics for Precision Medicine (TOPMed) Imputation Reference panel (version TOPMed-r2; Eagle v2.49 and minimac3.7) accessed through the TOPMed Imputation Server [45–47]. Prior to imputation, variants were removed if: (1) the call rate was less than 98%; (2) missingness differed between cases and controls (P < 1E-7); (3) allele frequencies or missingness differed between males and females (P < 1E-7); (4) significant deviation from Hardy–Weinberg equilibrium was observed in the European founders (P < 1E-7); or (5) significant deviation from expected when imputing the variant from nearby markers using PLINK’s ---mishap test (P < 1E-7). In addition, monomorphic variants and non-SNPs were dropped, leaving 364,538 variants for imputation.
SNP selection
We selected two different sets of independent SNPs that were previously reported as validated predictors of LTL based on results from two large-scale GWAS meta-analyses by Codd et al. and updated by Li et al. [27, 32]. We chose to include results that used the Codd et al. genetic predictor so that we could facilitate comparisons with previously published literature as Li et al. was recently published and has only one applicable published study for comparison [34]. Genotyping and imputation information for each variant is provided in Supplemental Table S1. One variant, MOB1B, had an Rsq = 0.5887 but the minimum Rsq for the remaining variants was 0.8314.
Statistical analysis
Analysis was performed using two methods due to the differences in the study design for the case–parent trio and case–control samples. GaMETES study cases that were not part of a full trio were included in the case–control analysis using controls matched on sex and ancestry from the Geisinger Health System (GHS; dbGaP Study Accession: phs000957.eMERGE_III_MyCode.v1.p1; HumanExome-12v1.1 array). Case–parent trio data were analysed using the transmission disequilibrium test (TDT) as implemented in the PLINK software package [48] (version 1.9), which by design controls for population stratification [49]. Case–control data were analysed using multivariable logistic regression adjusting for sex, principal components (two PCs for European Ancestry, three PCs for Hispanic ancestry, three for Asian ancestry and four PCs for African ancestry), and study site. None of the ancestry-specific PCs were associated with GCT status (we also repeated the analysis without PCs and the results did not change). Analyses were stratified by ancestral population. Effect estimates from the TDT analysis and the ancestry-specific multivariable logistic regression analysis were combined using an inverse-variance-weighted (IVW) meta-analysis using METAL [50]. Individual SNPs were evaluated for heterogeneity across samples using Cochran’s Q-test in METAL [51] and for significance using a Bonferroni-corrected p-value (0.05/number of SNPs in the genetic predictor). Resulting estimates and summary statistics for the LTL associated SNPs from Codd et al. and Li et al. were used for MR analyses using the TwoSampleMR software package [52] in R. We ran the analysis using code provided at https://mrcieu.github.io/TwoSampleMR/. We present IVW, MR Egger, and weighted median estimates but consider IVW as our primary model as is the standard approach and assumes all variants are valid instruments. Individual TERT variant associations were evaluated separately. Odds ratios (OR) and 95% confidence intervals (CI) were calculated and correspond to the telomere lengthening alleles reported in Codd et al. and Li et al. We also evaluated subgroup associations by age at diagnosis (<11 years and 11–19 years), sex (male and female), tumour location (extragonadal, intracranial, testis and ovary), and tumour histology (germinoma, teratoma, yolk sac tumours (YST) and mixed/other).
Sensitivity analyses
To validate the results from the IVW method, various additional MR methods were applied as sensitivity analyses to examine potential instrumental outliers and pleiotropy that would suggest a violation of MR assumptions, including MR Egger, and weighted median regression methods. MR relies on three main assumptions: the instrumental variables are (1) associated with the exposure of interest; (2) not associated with any confounder of the exposure-outcome association and (3) not independently associated with the outcome [53, 54]. MR Egger is more robust for the detection of a causal relationship in the presence of pleiotropy as it relaxes the third MR assumption that the genetic variant is independent of the outcome and it assumes that the correlation between the genetic associations with the exposure (LTL) and the direct effects of the genetic variants on the outcome (GCTs) is zero (referred to as the InSIDE [INstrument Strength Independent of Direct Effect] assumption) and does not require an intercept of zero [55]. Weighted median uses a weighted empirical distribution function of each SNP ratio estimate and provides the median estimate and a consistent estimate of true causal effect if at least 50% of the weight comes from valid instruments [56]. In addition, the MR Egger intercept test was used to detect pleiotropy, Cochran’s Q-statistic to detect heterogeneity with respect to IVW and MR Egger, and leave-one-out analysis and MR-Radial regression were used to identify potentially influential SNPs [57]. When heterogeneity was detected, we removed outliers to avoid violating MR assumptions and repeated both the MR analysis and the sensitivity analysis (Supplemental Table S2).
Replication analyses in UK Biobank testicular germ cell tumours
The UK Biobank is a population-based prospective cohort study that recruited more than 500,000 individuals aged 40–69 years from across the United Kingdom (UK) as detailed previously [58, 59]. We selected unrelated males <50 years of age who were diagnosed with testicular cancer (ICD-10 code C62) between January 1, 1971 and December 31, 2015 and had existing genetic data available. Individuals were excluded from the analysis if they were ≥50 years of age at diagnosis, had sex aneuploidy, or had missing genotyping data. UK Biobank genotyping was conducted by Affymetrix (High Wycombe, UK) using the UK Biobank AxiomTM Array by Affymetrix (N = 49,950; UK BiLEVE; Santa Clara, CA) and the closely related UK Biobank AxiomTM Array (N = 438,427) and was already imputed to The 1000 Genomes and Haplotype Reference Consortium (HRC) panels by the data source. Four male controls were matched to each case based on age (birth year and month) and ten genetic principal components (principal components provided by UK Biobank, field 22009 [60]) using the methods below. Logistic regression analysis was used to evaluate the association between each SNP and TGCT in males <50 years of age and effect estimates were meta-analysed and used for MR. Assuming a Type-1 error rate (α) of 0.05 and a sample size of 1980 with 0.20 proportion of cases in the replication sample, we estimated between 6 and 84% power using the Codd et al. six SNP genetic predictor (variance explained 0.92%) and 7–94% power using the Li et al. 18 SNP genetic predictor (variance explained = 1.22%) to detect an OR = 1.2–3.0 for the association between LTL and TGCT. We estimated our statistical power in the replication analysis using the online tool mRnd (http://cnsgenomics.com/shiny/mRnd/) [61].
Matching algorithm
Cases were matched to four controls each based on demographics and metrics of genetic distance, with the goal of finding a set of controls with the most similar genetic background that was the same sex and had similar ages (age was only relevant for the UK Biobank data). This matching was performed using the software MatchSamples (https://github.com/PankratzLab/MatchSamples), which searches for the “n” nearest neighbour controls for a given case across multiple dimensions. The dimensions, in this case, were sex (0–1), age (for UK Biobank) and ten dimensions that captured genetic ancestry.
MatchSamples implements a k-d tree data structure [62] to efficiently perform the nearest neighbour search on numeric data (such as age and principal components). If the same control is selected for multiple cases, MatchSamples applies the Hungarian Algorithm [63] to optimally assign a unique set of controls to each case. MatchSamples automatically normalises all quantitative variables to have a mean of zero and a standard deviation of one prior to matching. MatchSamples also has a visualisation module that flags pairs where the distance metric is significantly larger than average, which would indicate a poor genetic match. This is often a problem when there are relatively few controls available for each control, but this was not an issue with either the UK Biobank or Geisinger data, where each case had hundreds or thousands of possible matches.
Results
A total of 1413 children and adolescents (610 COG case–parent trios, 74 COG case-only, 634 CBP and 95 MINNBB participants) with GCT were included in the analysis (Table 1). In our analysis of individual LTL-related SNPs and paediatric GCT risk, we observed an association between the LTL lengthening allele and decreased risk for GCT in all three TERT SNPs that reached statistical significance (P < 0.001; Table 2) after controlling for multiple comparisons (P = 0.007 for Codd et al. and P = 0.003 for Li et al.). A nominal association was observed for five SNPs including TERC/rs170974795 (P = 0.018), PARP1/rs3219104 (P = 0.024), TERC/rs1093600 (P = 0.021), POT1/rs59294613 (P = 0.049), and MPHOSPH6/rs7194734 (P = 0.026). No other associations were observed for the individual SNPs and paediatric GCT. In our analysis of UK Biobank adult TGCT, we observed a nominal association between four SNPs including all three TERT SNPs (P < 0.024) and TERF2/rs3785074 (P = 0.004) with decreased risk for TGCT after controlling for multiple comparisons.
Table 1.
Characteristics of the paediatric germ cell tumour cases overall and by study.
| Overall cases | TDT analysis cases | Case–control analysis cases | |
|---|---|---|---|
| (N = 1413) | (N = 610) | (N = 803) | |
| Sex | |||
| Male | 866 (61.3) | 311 (51.0) | 555 (69.1) |
| Female | 547 (38.7) | 299 (49.0) | 248 (30.9) |
| Location | |||
| Extragonadal | 260 (18.4) | 156 (25.6) | 105 (13.1) |
| Intracranial | 314 (22.2) | 177 (29.0) | 137 (17.1) |
| Ovary | 314 (22.2) | 150 (24.6) | 165 (20.5) |
| Testis | 525 (37.2) | 127 (20.8) | 396 (49.3) |
| Age group | |||
| <11 years | 439 (31.1) | 249 (40.8) | 192 (23.9) |
| 11–19 years | 974 (68.9) | 361 (59.2) | 611 (76.1) |
| Race/ethnicity | |||
| African American | 62 (4.4) | 29 (4.8) | 34 (4.2) |
| Asian | 87 (6.2) | 27 (4.4) | 60 (7.5) |
| Hispanic | 610 (43.2) | 176 (28.9) | 433 (53.9) |
| Non-Hispanic White | 654 (46.3) | 378 (62.0) | 276 (34.4) |
| Tumour histologya | |||
| Germinoma | 350 (24.8) | 146 (23.9) | 204 (25.4) |
| Teratomas | 338 (23.9) | 145 (23.8) | 193 (24.0) |
| Yolk sac tumours | 212 (15.0) | 134 (22.0) | 78 (9.7) |
| Mixed/other | 490 (34.7) | 162 (26.6) | 328 (40.8) |
aN = 23 cases were missing histology information.
Table 2.
Individual effect of leukocyte telomere length (LTL)-related variants on germ cell tumour risk (GCT).
| Paediatric GCT | UKB TGCT | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TDT (N = 610) | Case–control analysisa (N = 803) | Meta-analysis (N = 1413) | Case–Control (N = 396) | |||||||||
| SNP | Chr | Gene | Lb allele | Sc allele | ORd (95% CI) | P value | ORd (95% CI) | P value | ORd (95% CI) | P value | ORd (95% CI) | P value |
| Codd et al. [27] – 7 genetic variants predict LTL | ||||||||||||
| rs11125529 | 2 | ACYP2 | A | C | 1.02 (0.81, 1.30) | 0.855 | 1.02 (0.83, 1.26) | 0.853 | 1.02 (0.87, 1.19) | 0.793 | 0.92 (0.72, 1.17) | 0.480 |
| rs10936599 | 3 | TERC | C | T | 1.12 (0.93, 1.34) | 0.234 | 1.17 (1.01, 1.36) | 0.041 | 1.15 (1.02, 1.29) | 0.018 | 1.10 (0.92, 1.32) | 0.315 |
| rs7675998 | 4 | NAF1 | G | A | 1.24 (1.02, 1.52) | 0.033 | 0.94 (0.79, 1.12) | 0.485 | 1.06 (0.93, 1.20) | 0.399 | 1.17 (0.96, 1.42) | 0.118 |
| rs2736100e | 5 | TERT | C | A | 0.88 (0.75, 1.03) | 0.112 | 0.77 (0.67, 0.89) | 0.0004 | 0.82 (0.74, 0.91) | 0.0003 | 0.83 (0.71, 0.97) | 0.020 |
| rs9420907 | 10 | OBFC1 | C | A | 0.89 (0.72, 1.11) | 0.313 | 1.05 (0.85, 1.29) | 0.645 | 0.97 (0.83, 1.13) | 0.665 | 0.80 (0.63, 1.02) | 0.073 |
| rs8105767f | 19 | ZNF208 | G | A | 1.05 (0.89, 1.25) | 0.544 | 1.02 (0.88, 1.18) | 0.804 | 1.03 (0.92, 1.15) | 0.580 | 1.03 (0.87, 1.22) | 0.715 |
| rs755017 | 20 | RTEL1 | G | A | 1.12 (0.89, 1.42) | 0.337 | 1.06 (0.89, 1.26) | 0.508 | 1.08 (0.94, 1.25) | 0.270 | 0.93 (0.74, 1.18) | 0.560 |
| Li et al. [32] – 20 genetic variants predict LTL | ||||||||||||
| rs3219104 | 1 | PARP1 | C | A | 0.94 (0.77, 1.15) | 0.575 | 0.82 (0.70, 0.96) | 0.015 | 0.87 (0.76, 0.98) | 0.024 | 0.94 (0.76, 1.16) | 0.570 |
| rs55749605 | 3 | SENP7 | C | A | 1.10 (0.92, 1.30) | 0.294 | 1.03 (0.88, 1.20) | 0.716 | 1.06 (0.94, 1.19) | 0.331 | 0.90 (0.76, 1.05) | 0.183 |
| rs10936600 | 3 | TERC | A | T | 1.11 (0.92, 1.32) | 0.272 | 1.18 (1.01, 1.37) | 0.036 | 1.15 (1.02, 1.29) | 0.021 | 0.90 (0.75, 1.09) | 0.282 |
| rs13137667 | 4 | MOB1B | C | T | 1.18 (0.77, 1.82) | 0.442 | 0.75 (0.52, 1.09) | 0.131 | 0.91 (0.69, 1.21) | 0.515 | 0.90 (0.53, 1.52) | 0.683 |
| rs4691895 | 4 | NAF1 | C | G | 1.23 (1.01, 1.50) | 0.037 | 0.89 (0.75, 1.05) | 0.155 | 1.02 (0.90, 1.15) | 0.806 | 1.12 (0.92, 1.35) | 0.263 |
| rs7705526e | 5 | TERT | A | C | 0.81 (0.64, 1.03) | 0.086 | 0.86 (0.72, 1.02) | 0.077 | 0.84 (0.73, 0.97) | 0.015 | 0.82 (0.69, 0.97) | 0.024 |
| rs2853677e | 5 | TERT | G | A | 0.85 (0.70, 1.03) | 0.105 | 0.77 (0.66, 0.90) | 0.001 | 0.80 (0.71, 0.91) | <0.001 | 0.80 (0.68, 0.94) | 0.007 |
| rs34991172 | 6 | CARMIL1 | T | G | 0.66 (0.44, 0.99) | 0.041 | 0.95 (0.63, 1.44) | 0.826 | 0.79 (0.59, 1.06) | 0.111 | 1.16 (0.83, 1.61) | 0.384 |
| rs2736176 | 6 | PRRC2A | C | G | 0.89 (0.74, 1.06) | 0.179 | 0.92 (0.79, 1.07) | 0.256 | 0.90 (0.81, 1.01) | 0.082 | 1.08 (0.91, 1.28) | 0.386 |
| rs59294613 | 7 | POT1 | C | A | 1.00 (0.84, 1.21) | 0.963 | 1.20 (1.02, 1.42) | 0.028 | 1.11 (0.98, 1.25) | 0.096 | 0.93 (0.78, 1.11) | 0.411 |
| rs9419958 | 10 | STN1 (OBFC1) | T | C | 0.90 (0.72, 1.12) | 0.340 | 1.06 (0.86, 1.31) | 0.571 | 0.98 (0.84, 1.14) | 0.809 | 1.24 (0.97, 1.57) | 0.082 |
| rs228595 | 11 | ATM | G | A | 0.91 (0.77, 1.06) | 0.232 | 0.96 (0.83, 1.10) | 0.522 | 0.93 (0.84, 1.04) | 0.206 | 0.89 (0.76, 1.04) | 0.149 |
| rs2302588 | 14 | DCAF4 | C | G | 1.09 (0.83, 1.43) | 0.534 | 1.20 (0.94, 1.52) | 0.138 | 1.15 (0.96, 1.38) | 0.127 | 0.89 (0.69, 1.15) | 0.382 |
| rs3785074 | 16 | TERF2 | G | A | 0.92 (0.77, 1.11) | 0.400 | 0.93 (0.79, 1.10) | 0.402 | 0.93 (0.82, 1.05) | 0.235 | 0.76 (0.64, 0.92) | 0.004 |
| rs62053580 | 16 | RFWD3 | A | G | 0.97 (0.76, 1.26) | 0.846 | 1.14 (0.92, 1.41) | 0.243 | 1.07 (0.91, 1.26) | 0.442 | 0.93 (0.75, 1.16) | 0.530 |
| rs7194734 | 16 | MPHOSPH6 | C | T | 0.79 (0.65, 0.96) | 0.018 | 0.95 (0.79, 1.13) | 0.548 | 0.87 (0.76, 1.00) | 0.043 | 0.97 (0.81, 1.16) | 0.706 |
| rs8105767f | 19 | ZNF208 | G | A | 1.05 (0.89, 1.25) | 0.544 | 1.02 (0.88, 1.18) | 0.804 | 1.03 (0.93, 1.15) | 0.560 | 1.03 (0.87, 1.22) | 0.715 |
| rs75691080 | 20 | RTEL1/STMN3 | C | T | 1.04 (0.76, 1.41) | 0.814 | 1.07 (0.80, 1.44) | 0.639 | 1.06 (0.85, 1.30) | 0.616 | 1.31 (1.00, 1.70) | 0.046 |
| rs34978822 | 20 | RTEL1 | C | G | 2.00 (0.90, 4.45) | 0.083 | 0.92 (0.46, 1.84) | 0.825 | 1.28 (0.76, 2.16) | 0.348 | 1.30 (0.78, 2.16) | 0.315 |
| rs73624724 | 20 | RTEL1/ZBTB46 | C | T | 1.10 (0.87, 1.39) | 0.431 | 1.05 (0.88, 1.25) | 0.611 | 1.07 (0.92, 1.23) | 0.379 | 0.97 (0.77, 1.21) | 0.775 |
TDT transmission disequilibrium test.
aModel adjusted for ancestry-specific PCs (2 PCs for European Ancestry, 3 PCs for Hispanic and Asian Ancestry and 4 PCs for African Ancestry), sex (male and female) and study site (COG, Michigan or California).
bL allele--allele associated with longer leukocyte telomere length.
cS allele--allele associated with shorter telomere length.
dOR and 95% confidence intervals--the odds ratio for the association between lengthening allele and GCT risk.
eTERT SNP evaluated separately due to reported association with testicular GCT.
fSNP appears in both Codd et al. [27] and Li et al. [32] genetic instruments.
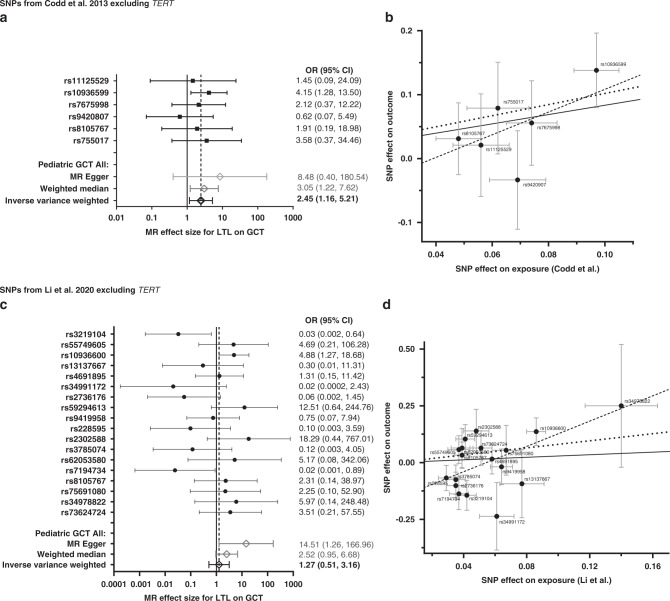
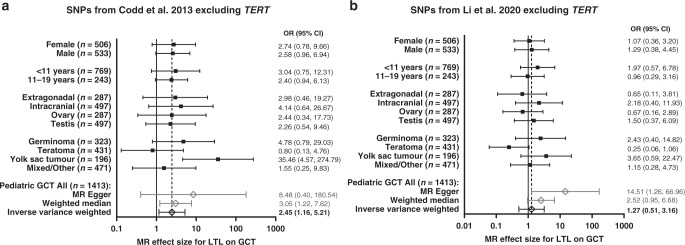
Our primary model did not include variants in TERT as this variant was previously identified as a risk variant in TGCT [22–25], the risk allele associated with telomere lengthening is the opposite allele associated with increased TGCT, and previous literature reporting associations with TGCT use a genetic predictor, not including TERT [39, 40]. In the overall paediatric GCT MR analysis using the Codd et al. SNPs excluding TERT/rs2736100, we observed a significant association between longer predicted LTL and increased risk of paediatric GCT in the IVW and the weighted median models (OR [95% CI] = 2.45 [1.16, 5.21] per kb of LTL; P = 0.019 and OR [95% CI] = 3.05 [1.22, 7.62]; P = 0.017, respectively; Fig. 1a) but not with the MR Egger model (OR [95% CI] = 8.48 [0.40, 180.54]; P = 0.243). The scatter plot in Fig. 1b depicts the relationship between the effect of each SNP associated with LTL and their effect on paediatric GCT overlaid with the three MR method slopes. The MR Egger intercept did not provide sufficient evidence of pleiotropy. Cochran’s Q-statistic indicated no evidence of instrumental heterogeneity in either the IVW or MR Egger models and there was no influence of outliers according to the MR-Radial models (Supplemental Table S2). In addition, the leave-one-out analysis, which excluded each SNP and re-estimated the causal effect using the IVW) method, identified ORs all similar in direction. Therefore, the sensitivity analysis suggested no evidence of heterogeneity or bias in the IVW estimate. Effect estimates for all variants are in the same direction except for OBFC1/rs9420807. In subgroup-specific analysis, effect estimates are all similar in direction and magnitude with the exception of teratomas (Fig. 2a). Individual MR results by paediatric GCT subgroup for the Codd et al. SNPs are presented in Supplemental Fig. S1.
Fig. 1. Association between individual single nucleotide polymorphisms (SNPs) that genetically predict leukocyte telomere length (LTL) and paediatric germ cell tumour (GCT) risk: results from the Mendelian Randomization (MR) analysis.
a, b Presents results using SNPs from the Codd et al. [27] genetic instrument excluding TERT and c, d presents results using SNPs from the Li et al. [32] genetic instrument excluding TERT. Forest plots (left) show the effect estimate per variant using MR Wald Ratio single SNP test, and overall MR estimates using MR Egger, weighted median, and inverse-variance-weighted (IVW) estimators (N = 1413). Odds ratio is for every 1 kb increase in LTL. Scatter plots (right) show the per-allele association with paediatric GCT risk (outcome) plotted against the per‐allele association with kb of LTL (exposure). The vertical and horizontal grey lines represent the standard error for each SNP. The slope of the scatter plot is overlaid with the Mendelian randomisation IVW estimate (solid black line), the MR Egger estimate (dotted black line) and the weighted median estimate (dashed black line) of the effect of LTL on GCT risk.
Fig. 2. Sub-group specific association between genetically predicted leukocyte telomere length (LTL) and paediatric germ cell tumour (GCT) risk: results from the Mendelian Randomization (MR) analysis.
a Presents results using single nucleotide polymorphisms (SNPs) from the Codd et al. [27] genetic instrument excluding TERT and b presents results using SNPs from Li et al. [32] genetic instrument excluding TERT. Forest plots (left) show the effect estimate per subgroup from the meta-analysis, and overall MR estimates using MR Egger, weighted median, and inverse-variance-weighted (IVW) estimators (N = 1413). Odds ratio is for every 1 kb increase in LTL. Footnote: Sample size indicated in parenthesis (n = #) represents the total number of cases which includes those in the complete trios and in the case–control analyses. Subgroup analysis included trios, Hispanic and White case–control only samples to get stable estimates. African American and Asian case–control samples were not included in subgroup analyses (only overall analyses) due to the small sample size.
In the overall MR analysis using the Li et al. SNPs excluding TERT/rs2853677 and TERT/rs7705526, we did not observe evidence of a significant association between longer predicted LTL and increased risk of paediatric GCT in our IVW (OR [95% CI] = 1.27 [0.51, 3.16]; P = 0.673) or weighted median models (OR [95% CI] = 2.52 [0.95, 6.68]; P = 0.064; Fig. 2c), but we do observe significance with the MR Egger model (OR [95% CI] = 14.51 [95% CI: 1.26, 166.96]; P = 0.047). Here the trend is less clear with about half the individual SNP MR effect estimates in the positive direction (increased risk) and the other half in the negative direction (decreased risk). However, Cochran’s Q-statistic indicated evidence of instrumental heterogeneity in the IVW model but not in the MR Egger model (Supplemental Table S2). This suggested the IVW estimate violated its assumption (all variants are valid instruments) and MR Egger was less likely to be biased as it takes into account directional pleiotropy. The scatter plot in Fig. 1d depicts the relationship between the effect of each SNP associated with LTL and its effect on paediatric GCT overlaid with the three MR method slopes. In subgroup-specific analysis, effect estimates are all similar in direction and magnitude with the exception of adolescents (11–19 years of age), extragonadal, ovary, and teratomas (Fig. 2b). Individual MR results by paediatric GCT subgroup for the Li et al. SNPs are presented in Supplemental Fig. S2.
Since TERT codes for an integral part of the telomerase enzyme that increases the length of telomeres, it is rational to expect that TERT’s association with GCT is mediated by changes in telomere length. Therefore, we also performed an analysis of the full LTL genetic predictors, including TERT. In the overall paediatric GCT MR analysis using the seven Codd et al. SNPs that genetically predict LTL, we did not observe evidence of a significant association between longer predicted LTL and increased risk of paediatric GCT in any of the three models (ORIVW [95% CI] = 1.10 [0.32, 3.80], P = 0.884; ORMR-Egger [95% CI] = 1.66 [0.004, 758.24], P = 0.878; ORweighted median [95% CI] = 2.08 [0.78, 5.53], P = 0.143; Supplemental Table S2). However, evidence of instrumental heterogeneity was observed within the seven Codd et al. SNP genetic instrument, as identified by Cochran’s Q-statistic, in both the IVW and MR Egger models, and MR-Radial models indicated TERT/rs2736100 as the outlier responsible for the heterogeneity (Supplemental Table S2). Similarly, in the overall MR analysis using the 20 Li et al. SNPs that genetically predict LTL, we did not observe a significant association between longer predicted LTL and increased risk of paediatric GCT in any of the three models (ORIVW [95% CI] = 0.68 [0.26, 1.75], P = 0.425; ORMR-Egger [95% CI] = 1.88 [0.012, 30.69], P = 0.664; ORweighted median [95% CI] = 1.34 [0.45, 3.99], P = 0.594; Supplemental Table S2). However, evidence of instrumental heterogeneity was observed by Cochran’s Q-statistic and MR-Radial models indicated TERT/rs2853677 and TERT/rs7705526 as the influential outliers responsible.
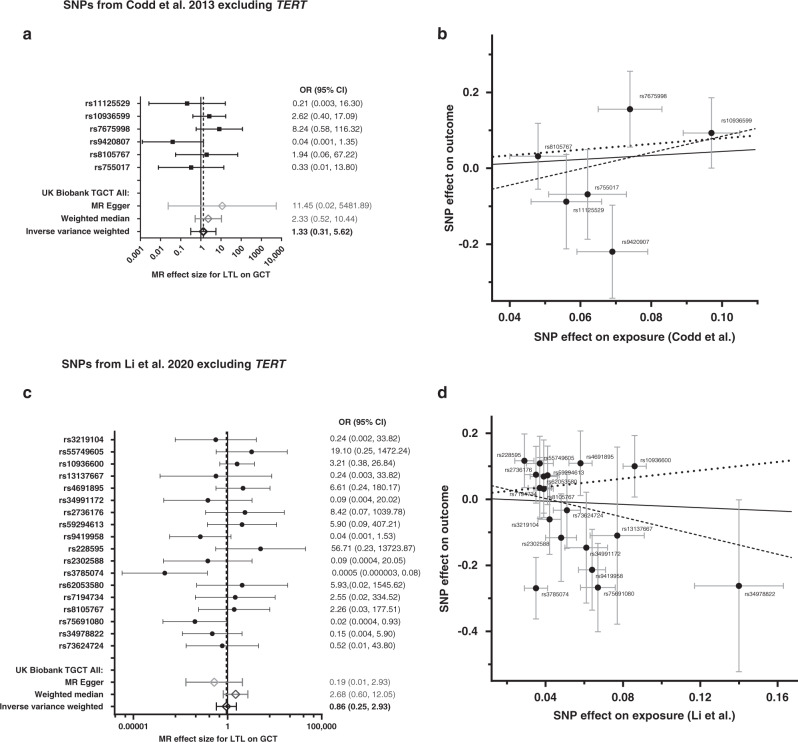
In our MR replication analysis of 396 UK Biobank adult TGCT cases and 1584 controls using the Codd et al. six SNPs (excluding TERT/rs2736100), we observed a positive trend between longer predicted LTL and overall increased risk for adult TGCT in all three MR models that was similar in magnitude to our discovery analyses but did not reach statistical significance (ORIVW [95% CI] = 1.33 [0.31, 5.62], P = 0.698; ORMR-Egger [95% CI] = 11.45 [0.02, 5481.89], P = 0.482; ORweighted median [95% CI] = 2.33 [0.52, 10.44], P = 0.270; Fig. 3a). The MR Egger intercept (−0.157; P value = 0.519; Supplemental Table S2) suggests no evidence of directional pleiotropy with a non-significant P value although the non-zero estimate may be indicative of possible directional pleiotropy. In addition, Cochran’s Q-statistic does not show evidence of heterogeneity in the analysis. Therefore, the sensitivity analysis suggested that there is no evidence the assumption of IVW was violated. Effect estimates for all variants are in the same direction except for ACYP2/rs11125529, OBFC1/rs9420807 and RTEL1/rs755017. In the MR analysis of the Li et al. 18 SNPs (excluding TERT/rs2853677 and TERT/rs7705526), we observed discordant results across the three MR models; for the IVW and MR Egger models we observed a trend in the opposite direction than we did for the Codd et al. genetic predictor (OR [95% CI] = 0.86 [0.25, 2.93] per kb of LTL; P = 0.814; and OR [95% CI] = 0.19 [0.006, 6.201]; P = 0.195, respectively; Fig. 3c) indicating longer predicted LTL and decreased risk for adult TGCT. Alternatively, the weighted median model was similar in direction and magnitude as the Codd et al. predictor (OR [95% CI] = 2.68 [0.60, 12.04] per kb of LTL; P = 0.198). Similar to our discovery analysis, half the individual SNP MR effect estimates were in the positive direction and the other half in the negative direction although Cochran’s Q-statistic did not demonstrate significant evidence of heterogeneity (P = 0.062). In addition, we found little evidence for the presence of pleiotropy, as indicated by the MR Egger intercept (Supplemental Table S2) for the Li et al. genetic predictor on adult TGCT. The scatter plots in Fig. 3b, d depicts the relationship between the effect of each SNP associated with LTL and its effect on adult TGCT overlaid with the three MR method slopes. Interestingly, comparing the individual SNP MR effect estimates between our adult and paediatric population, SNPs were in the opposite direction for seven SNPs including PRRC2A/rs2736176, ATM/rs228595, DCAF4/rs2302588, MPHOSPH6/rs7194734, RTEL1/STMN3/rs75691080, RTEL1/rs34978822 and RTEL1/ZBTB46/rs73624724.
Fig. 3. Association between individual single nucleotide polymorphisms (SNPs) that genetically predict leukocyte telomere length (LTL) and UKB testicular germ cell tumour (TGCT) risk: results from the Mendelian Randomization analysis.
a, b Presents results using SNPs from the Codd et al. [27] genetic instrument excluding TERT and c, d presents results using SNPs from the Li et al. [32] genetic instrument excluding TERT. Forest plots (left) show the effect estimate per variant using MR Wald Ratio single SNP test, and overall MR estimates using MR Egger, weighted median, and inverse-variance-weighted (IVW) estimators (N = 396). Odds ratio is for every 1 kb increase in LTL. Scatter plots (right) show the per-allele association with paediatric GCT risk (outcome) plotted against the per‐allele association with kb of LTL (exposure). The vertical and horizontal grey lines represent the standard error for each SNP. The slope of the scatter plot is overlaid with the Mendelian randomisation IVW estimate (solid black line), the MR Egger estimate (dotted black line) and the weighted median estimate (dashed black line) of the effect of LTL on GCT risk.
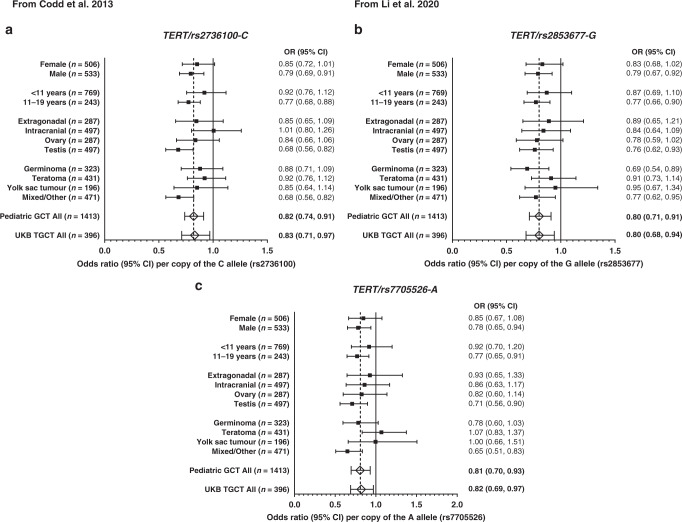
When we evaluated the three TERT SNPs removed from the LTL predictors, which are all in moderate linkage disequilibrium with one another (high D’ but lower R2; see Supplemental Table S3), we observed a significant association between the LTL lengthening allele and decreased risk for paediatric GCT for each SNP (P < 0.003 for all; Fig. 4). For all three TERT SNPs, the associations were strongest in males, among the adolescent age group (age at diagnosis 11–19 years), for tumours located in the testis, and for tumours with mixed/other histology. The direction of effect for point estimates overall and across the three TERT SNPs were consistent except for intracranial tumours for TERT/rs2736100-C (OR [95% CI] = 1.01 [0.80, 1.26]) and teratomas for TERT/rs7705526-A (OR [95% CI] = 1.07 [0.83, 1.37]). Similar in magnitude to our discovery sample, a significant association was observed between the LTL lengthening allele and decreased risk for adult testicular cancer in all three TERT SNPs (P < 0.024).
Fig. 4. Meta-analysis results for subgroup-specific associations between TERT single nucelotide polymorphisms (SNPs) and germ cell tumor (GCT) risk.
Odds ratios (OR and 95% CI) are presented as the risk allele associated with longer telomere length. a TERT SNP from Codd et al. 2013 and b, c TERT SNPs from Li et al. 2020. Footnote: Sample size indicated in parenthesis (n = #) represents the total number of cases which includes those in the complete trios and in the case–control analyses. Subgroup analysis included trios, Hispanic and White case–control only samples to get stable estimates. African American and Asian case–control samples were not included in subgroup analyses (only overall analyses) due to the small sample size.
Discussion
Using the largest collection of paediatric GCT cases available, we identified significant associations between GCTs and the TERT risk alleles previously found to be associated with adult TGCTs. In contrast, we did not observe strong evidence for an association between other variants associated with longer predicted LTL and increased risk for GCT in children or adults. For paediatric GCTs, we observed a weak but significant association with longer LTL using the more commonly used Codd et al. LTL genetic predictor excluding TERT in the primary IVW model (P = 0.019). In contrast, when we use the newer Li et al. genetic predictor of LTL that captures a slightly larger percent variation in LTL, we did not see a significant association with increased risk for GCT in the IVW model (P = 0.673) although the effect estimates were similar in direction. The inconsistent results between genetic predictors may be an indication of bias resulting from pleiotropy--where a SNP(s) within the genetic predictor is associated with both telomere length (exposure) and GCT (outcome), thus violating the assumptions necessary for valid causal inference [64]. We see some suggestion of this in the sensitivity analysis of the Li et al. genetic predictor, although when the TERT variants were removed from the genetic predictor, the MR Egger intercept differs from zero and is close to significance (−0.128; P = 0.054) thus suggesting directional pleiotropy within the instrument and therefore evidence of a biased IVW estimate. In the adult TGCT, we did not observe consistent associations or direction in effect estimates between LTL variants and TGCT risk using either LTL predictor.
Previous studies have identified associations between genetically predicted telomere length and adult TGCT using different variants in their genetic instruments and reported contradictory results [34, 39, 40]. These differences may be due to the inclusion of TERT, the previously identified susceptibility locus for adult TGCT [22–25], in the genetic instrument for predicted telomere length. When TERT is not included in the genetic predictor, increased telomere length is associated [39] or suggestively associated [40] with TGCT. On the contrary, when TERT is included in the genetic predictor, the direction of effect flips and there is a null association between shorter telomere length and increased risk for adult TGCT [34]. In the present study, we report associations both with and without variants at the TERT locus. Consistent with the literature, when TERT is included in the Li et al. genetic predictor, we observed a null association between shorter telomere length and increased risk for GCTs. When we remove TERT from the Li et al. genetic predictor, although still null, the effect estimate agrees with previously reported associations between longer telomere length and GCT risk. The direction of effect did not differ with the Codd et al. genetic predictor, although it was strengthened when TERT was removed. Furthermore, we observed inconsistent results across our two populations using the two predictors of LTL and GCT risk. We observed a trend for longer predicted telomere length being associated with increased risk for paediatric GCT, but if this is a true effect it appears to be weak, and statistical significance depends on the variants included. Similarly, in adult TGCT we observed a weak positive association that was not statistically significant with the Codd et al. genetic predictor and a weak inverse association with the Li et al. genetic predictor. The reason for the differences in directionality and associations with the two genetic predictors is not entirely clear but is likely related to the fact that the scores are made up of different variants. Although both genetic predictors were identified using a large GWAS meta-analysis, the Li et al. genetic predictor explains a larger percent variance in telomere length. However, this comes with some limitations as it also increases the possibility of heterogeneity within the genetic instrument that we observed in our sensitivity analysis. In addition, power was limited in the replication set due to the much smaller number of cases. Overall, these data suggest that telomere length variants other than TERT do not make a significant contribution to paediatric or adult GCT risk.
One driving principle of MR is that the genetic variants used in the instrument are associated with the exposure of interest (e.g., telomere length), independent of the outcome (e.g., GCTs), and not related to confounders in order to produce a non-biased causal estimate [53, 54]. Our sensitivity analysis demonstrated that for both genetic instruments, the TERT SNPs were responsible for substantial heterogeneity. This could indicate a variety of potential problems; most notably that TERT is exhibiting pleiotropic effects, where TERT affects more than one trait. In fact, several GWA studies have reported an association between TERT and adult TGCT [22–25]. However, we cannot distinguish whether the effects of TERT on TGCT are independent or mediated via LTL as we do not have enough data. Future studies controlling for prediagnostic measured telomere length could provide further insight.
It is important to note that the TERT risk allele associated with shorter telomere length is the risk allele associated with testicular cancer while most previous studies have reported associations between longer predicted telomere length and cancer [30, 37–39]. We show a reduction in risk because we have modelled this association using the risk allele associated with longer telomere length for consistency in our overall analysis. We were able to replicate these findings in the UK Biobank data using three individual TERT SNPs from the LTL predictor. In the paediatric GCTs, we also observed significant associations between these variants and GCT overall, although the association appears to be stronger in adolescent males with TGCT. This is likely due to biological differences in paediatric and adult TGCTs. All germ cell tumours arise from the PGC, however the precursor cells that give rise to GCTs in adolescents and adults arise from PGCs at a more advanced developmental stage than those that occur in children [65]. To our knowledge, the exact biological mechanism and functional impact by which TERT influences TGCT risk is complicated and remains less clear. Tissue-specific expression quantitative trait loci (eQTL) data from The Genotype-Tissue Expression (GTEx) project demonstrates different allele-specific expression levels associated with the TERT SNPs we evaluated, although expression differences were reported in the skin (not sun-exposed suprapubic) for rs2736100 and rs253677 and small intestine for rs7705526. No expression differences associated with these SNPs were reported in testis tissue, the most relevant tissue type for this analysis, or any other tissue types [66]. Furthermore, Litchfield et al. did not report eQTL associations for TERT/rs2736100 in normal testis tissue, TGCT tissue, or other tissue types [23]. Larger sample sizes in relevant tissues are necessary to draw conclusions on whether allele-specific expression of TERT is directly related to longer LTL.
The activity of telomerase differs between germ cells and somatic cells. Thus, the effect of telomere length and maintenance may be different in the germline versus somatic cells or cancer [67]. Telomerase is the ribonucleoprotein enzyme responsible for synthesising telomere repeats onto the ends of chromosomes to maintain telomere stability and length [68]. It is a multi-unit complex consisting of two core components: the catalytic subunit telomerase reverse transcriptase protein encoded by TERT and a telomerase RNA TERC. While telomerase is present during normal cell development, it is typically silenced in human somatic cells after birth which leads to age-related telomere shortening. In contrast, it has been reported that telomerase activity remains active and TERT is expressed in the highly proliferative cells of the germline and in cancer cells, including adult TGCT [69]. This evidence likely explains why we see an association between TERT and GCT but not between genetically predicted LTL and GCT and suggests that TERT plays a dual role in cancer risk and telomere biology but with opposite alleles. It is currently unknown how the exact mechanism of these particular variants in TERT affects the function of the resulting protein.
This is the first study to investigate the association between genetically predicted LTL and paediatric GCT. This study has many strengths including the use of an established racially and ethnically diverse cohort of paediatric GCT, including case–parent trios obtained through the COG, and cases and controls obtained from newborn screening biobanks in California and Michigan. The use of MR methods and validated genetic instruments to measure the association between LTL and GCT risk eliminates the potential of confounding or reverse causation associated with measuring LTL directly and provides more precise effect estimates. The results of this study should be interpreted with the following limitations in mind. First, although we had a large sample size overall, our power was limited to look at associations by subgroup. Second, MR is subject to important assumptions and limitations that may be violated, including a lack of pleiotropy, genetic heterogeneity, linkage disequilibrium, and population stratification [53]. We do not have enough data to show that TERT is pleiotropic, so we report results both with and without SNPs in TERT (rs2736100, rs2853677 and rs7705526), all of which demonstrated heterogeneity. Third, the discovery GWAS that identified the genetic instruments was conducted across a European population and thus could attenuate the effect estimates on a per SNP basis in other populations. In our population, we did not observe heterogeneity across ancestries for either the case–control meta-analysis or the combined case–control-TDT meta-analysis for any SNP used in the genetic instruments. Fourth, both the Codd et al. and Li et al. genetic predictor only explain a small percentage of telomere length variation and therefore the instruments used in this analysis are not fully accounting for telomere length. Further, we have not considered the role of telomere length in parental gametes (paternal sperm/maternal egg) although literature suggests there is not a strong maternal or paternal age effect on paediatric GCT [70–75].
In conclusion, the association with GCT and the variants in TERT is quite robust across sex, age and tumour type. However, the pattern is not as clear with the other variants associated with telomere length where we did not observe consistent associations in either children or adults. Since the TERT variants we found to be associated with decreased risk of GCT are also associated with longer LTL, our findings suggest that TERT is acting through a pathway other than the direct lengthening of telomeres. These results indicate further analyses of variants in genes relevant for telomere function and maintenance are warranted to better understand the potential role of telomere biology in GCTs.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Supplemental Table S1. Individual effect of leukocyte telomere length (LTL) related variants on germ cell tumor risk (GCT)
Supplemental Table S2. Sensitivity analysis for the MR analyses of genetically predicted telomere length on pediatric germ cell tumor
Supplemental Table S3 - Linkage relationship of TERT variants in previously reported loci.
Supplemental Figure S1. Mendelian Randomization (MR) analysis results for the association between individual SNPs that genetically predict leukocyte telomere length (LTL) and pediatric germ cell tumor
Supplemental Figure S2. Mendelian Randomization (MR) analysis results for the association between individual SNPs that genetically predict leukocyte telomere length (LTL) and pediatric germ cell tumor
Acknowledgements
The authors would like to acknowledge the University of Minnesota Genomics Center for performing the genotyping assays and the University of Minnesota Supercomputing Institute for providing hardware and support for the statistical analyses.
Author contributions
SSC analysed and interpreted the data, conducted the literature review, and drafted the article. TY assisted with the interpretation of the data and power calculations. NP, JJM, ACK, JAL and BRC conceptualised the study design, processed the genetic data and assisted with data analysis. EKL and AJH conducted laboratory work and supervised the data collection. MK and ALF contributed to the study design. JNP designed the study, conceptualised the analysis and interpreted the data. All authors contributed to the drafting, review and approval of the final article.
Funding
This work was supported by the National Institutes of Health (grant R01 CA151284 to Jenny N. Poynter, National Clinical Trials Network Operations Center grant U10CA180886, and National Clinical Trials Network Statistics and Data Management Center grant U10CA180899), and Epidemiology Award from Alex’s Lemonade Stand Foundation (Wynnewood, Pennsylvania), and the Children’s Cancer Research Fund (Minneapolis, Minnesota).
Data availability
The paediatric and adolescent datasets generated from the Children’s Oncology Group (COG) and used for analyses in the current study will be shared publicly on dbGaP. Data generated from the California and Michigan Biobank samples cannot be shared due to state-specific restrictions regarding the use of the data. The UK Biobank data is publicly available and can be accessed on the UK Biobank website (https://www.ukbiobank.ac.uk/).
Code availability
Representative code used to analyse the data in this study is provided in Supplemental Materials.
Competing interests
ALF has acted as a paid consultant for Decibel Therapeutics for work performed outside of the current study. The remaining authors declare no competing interests.
Ethics approval and consent to participate
All study procedures were approved by the University of Minnesota Institutional Review Board. The California Committee for the Protection of Human Subjects and the Michigan Department of Health and Human Services Institutional Review Board approved the use of the biobank samples.
Consent to publish
This manuscript does not contain any individual person’s data in any form.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01798-3.
References
- 1.Hug N, Lingner J. Telomere length homeostasis. Chromosoma. 2006;115:413–25. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- 2.Needham BL, Adler N, Gregorich S, Rehkopf D, Lin J, Blackburn EH, et al. Socioeconomic status, health behavior, and leukocyte telomere length in the National Health and Nutrition Examination Survey, 1999–2002. Soc Sci Med. 2013;85:1–8. doi: 10.1016/j.socscimed.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown L, Needham B, Ailshire J. Telomere length among older U.S. adults: differences by race/ethnicity, gender, and age. J Aging Health. 2017;29:1350–66. doi: 10.1177/0898264316661390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaziri H, Dragowska W, Allsopp RC, Thomas TE, Harley CB, Lansdorp PM. Evidence for a mitotic clock in human hematopoietic stem cells: loss of telomeric DNA with age. Proc Natl Acad Sci USA. 1994;91:9857–60. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith EM, Pendlebury DF, Nandakumar J. Structural biology of telomeres and telomerase. Cell Mol Life Sci. 2020;77:61–79. doi: 10.1007/s00018-019-03369-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shay JW. Role of telomeres and telomerase in aging and cancer. Cancer Discov. 2016;6:584–93. doi: 10.1158/2159-8290.CD-16-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wentzensen IM, Mirabello L, Pfeiffer RM, Savage SA. The association of telomere length and cancer: a meta-analysis. Cancer Epidemiol Biomark Prev. 2011;20:1238–50. doi: 10.1158/1055-9965.EPI-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamoto K, Seimiya H. Revisiting telomere shortening in cancer. Cells. 2019;8:107. doi: 10.3390/cells8020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma H, Zhou Z, Wei S, Liu Z, Pooley KA, Dunning AM, et al. Shortened telomere length is associated with increased risk of cancer: a meta-analysis. PLoS ONE. 2011;6:e20466. [DOI] [PMC free article] [PubMed]
- 10.Pooley KA, Sandhu MS, Tyrer J, Shah M, Driver KE, Luben RN, et al. Telomere length in prospective and retrospective cancer case-control studies. Cancer Res. 2010;70:3170–6. doi: 10.1158/0008-5472.CAN-09-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steenstrup T, Hjelmborg JVB, Kark JD, Christensen K, Aviv A. The telomere lengthening conundrum - Artifact or biology? Nucleic Acids Res. 2013;41:1–7. doi: 10.1093/nar/gkt370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baird DM. Mechanisms of telomeric instability. Cytogenetic Genome Res. 2009;122:308–14. doi: 10.1159/000167817. [DOI] [PubMed] [Google Scholar]
- 13.Kawanishi S, Oikawa S. Mechanism of telomere shortening by oxidative stress. Ann N. Y Acad Sci. 2004;1019:278–84. doi: 10.1196/annals.1297.047. [DOI] [PubMed] [Google Scholar]
- 14.Valdes AM, Andrew T, Gardner JP, Kimura M, Oelsner E, Cherkas LF, et al. Obesity, cigarette smoking, and telomere length in women. Lancet-. 2005;366:662–4. doi: 10.1016/S0140-6736(05)66630-5. [DOI] [PubMed] [Google Scholar]
- 15.Turner KJ. Telomere biology and human phenotype. Cells. 2019;8:73. [DOI] [PMC free article] [PubMed]
- 16.Factor-Litvak P, Susser E, Kezios K, McKeague I, Kark JD, Hoffman M, et al. Leukocyte telomere length in newborns: implications for the role of telomeres in human disease. Pediatrics. 2016;137:e20153927. doi: 10.1542/peds.2015-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oosterhuis JW, Looijenga LHJ. Human germ cell tumours from a developmental perspective. Nat Rev Cancer. 2019;19:522–37. doi: 10.1038/s41568-019-0178-9. [DOI] [PubMed] [Google Scholar]
- 18.Poynter JN, Amatruda JF, Ross JA. Trends in incidence and survival of pediatric and adolescent patients with germ cell tumors in the United States, 1975 to 2006. Cancer. 2010;116:4882–91. doi: 10.1002/cncr.25454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajpert-De Meyts E, McGlynn KA, Okamoto K, Jewett MAS, Bokemeyer C. Testicular germ cell tumours. Lancet. 2016;387:1762–74. doi: 10.1016/S0140-6736(15)00991-5. [DOI] [PubMed] [Google Scholar]
- 20.Litchfield K, Thomsen H, Mitchell JS, Sundquist J, Houlston RS, Hemminki K, et al. Quantifying the heritability of testicular germ cell tumour using both population-based and genomic approaches. Sci Rep. 2015;5:1–7. doi: 10.1038/srep13889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampson JN, Wheeler WA, Yeager M, Panagiotou O, Wang Z, Berndt SI, et al. Analysis of heritability and shared heritability based on genome-wide association studies for thirteen cancer types. J Natl Cancer Inst. 2015;107:djv279. doi: 10.1093/jnci/djv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nat Genet. 2010;42:604–7. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litchfield K, Levy M, Orlando G, Loveday C, Law PJ, Migliorini G, et al. Identification of 19 new risk loci and potential regulatory mechanisms influencing susceptibility to testicular germ cell tumor. Nat Genet. 2017;49:1133–40. doi: 10.1038/ng.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, McGlynn KA, Rajpert-De Meyts E, Bishop DT, Chung CC, Dalgaard MD, et al. Meta-analysis of five genome-wide association studies identifies multiple new loci associated with testicular germ cell tumor. Nat Genet. 2017;49:1141–6. doi: 10.1038/ng.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruark E, Seal S, McDonald H, Zhang F, Elliot A, Lau K, et al. Identification of nine new susceptibility loci for testicular cancer, including variants near DAZL and PRDM14. Nat Genet. 2013;45:686–9. doi: 10.1038/ng.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pooley KA, Bojesen SE, Weischer M, Nielsen SF, Thompson D, Amin Al Olama A, et al. A genome-wide association scan (GWAS) for mean telomere length within the COGS project: identified loci show little association with hormone-related cancer risk. Hum Mol Genet. 2013;22:5056–64. doi: 10.1093/hmg/ddt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Codd V, Nelson CP, Albrecht E, Mangino M, Deelen J, Buxton JL, et al. Identification of seven loci affecting mean telomere length and their association with disease. Nat Genet. 2013;45:422–7. doi: 10.1038/ng.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mangino M, Hwang SJ, Spector TD, Hunt SC, Kimura M, Fitzpatrick AL, et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum Mol Genet. 2012;21:5385–94. doi: 10.1093/hmg/dds382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Codd V, Mangino M, van der Harst P, Braund PS, Beveridge AJ, Rafelt S, et al. Variants near TERC are associated with mean telomere length. Nat Genet. 2010;42:197–9. doi: 10.1038/ng.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh KM, Whitehead TP, de Smith AJ, Smirnov IV, Park M, Endicott AA, et al. Common genetic variants associated with telomere length confer risk for neuroblastoma and other childhood cancers. Carcinogenesis. 2016;37:576–82. doi: 10.1093/carcin/bgw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy D, Neuhausen SL, Hunt SC, Kimura M, Hwang SJ, Chen W, et al. Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc Natl Acad Sci USA. 2010;107:9293–8. doi: 10.1073/pnas.0911494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, Stoma S, Lotta LA, Warner S, Albrecht E, Allione A, et al. Genome-wide association analysis in humans links nucleotide metabolism to leukocyte telomere length. Am J Hum Genet. 2020;106:389–404. doi: 10.1016/j.ajhg.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Rice SV, Chang TC, Liu Y, Liu Q, Qin N, et al. Molecular mechanism of telomere length dynamics and its prognostic value in pediatric cancers. Intergovernmental Panel on Climate Change, editor. JNCI: J Natl Cancer Inst. 2019;112:756–64. doi: 10.1093/jnci/djz210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao Y, Wei Y, Zhou X, Huang S, Zhao H, Zeng P. Assessing the relationship between leukocyte telomere length and cancer risk/mortality in UK biobank and TCGA datasets with the genetic risk score and Mendelian randomization approaches. Front Genet. 2020;11:1270. [DOI] [PMC free article] [PubMed]
- 35.Pierce BL, Kraft P, Zhang C. Mendelian randomization studies of cancer risk: a literature review. Curr Epidemiol Rep. 2018;5:184–96. doi: 10.1007/s40471-018-0144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C, Ostrom QT, Semmes EC, Ramaswamy V, Hansen HM, Morimoto L, et al. Genetic predisposition to longer telomere length and risk of childhood, adolescent and adult-onset ependymoma. Acta Neuropathologica Commun. 2020;8:173. doi: 10.1186/s40478-020-01038-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ojha J, Codd V, Nelson CP, Samani NJ, Smirnov IV, Madsen NR, et al. Genetic variation associated with longer telomere length increases risk of chronic lymphocytic leukemia. Cancer Epidemiol Biomark Prev. 2016;25:1043–9. doi: 10.1158/1055-9965.EPI-15-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walsh KM, Codd V, Rice T, Nelson CP, Smirnov IV, Mccoy LS, et al. Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget. 2015;6:42468. [DOI] [PMC free article] [PubMed]
- 39.Haycock PC, Burgess S, Nounu A, Zheng J, Okoli GN, Bowden J, et al. Association between telomere length and risk of cancer and non-neoplastic diseases a Mendelian randomization study. JAMA Oncol. 2017;3:636–51. doi: 10.1001/jamaoncol.2017.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown DW, Lan Q, Rothman N, Pluta J, Almstrup K, Dalgaard MD, et al. Genetically inferred telomere length and testicular germ cell tumor risk. Cancer Epidemiol Biomark Prevention. 2021;30:1275–8. [DOI] [PMC free article] [PubMed]
- 41.Musselman JRB, Spector LG, Krailo MD, Reaman GH, Linabery AM, Poynter JN, et al. The Children’s Oncology Group Childhood Cancer Research Network (CCRN): case catchment in the United States. Cancer. 2014;120:3007–15. doi: 10.1002/cncr.28813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poynter JN, Richardson M, Roesler M, Krailo M, Amatruda JF, Frazier AL. Family history of cancer in children and adolescents with germ cell tumours: a report from the Children’s Oncology Group. Br J Cancer. 2018;118:121–6. doi: 10.1038/bjc.2017.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 44.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–9. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuchsberger C, Abecasis GR, Hinds DA. Minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–4. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira M, Bender D, et al. PLINK: whole genome data analysis toolset. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spielman RS, Mcginnis RE, Ewenst WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet. 1993;52:506. [PMC free article] [PubMed]
- 50.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics Appl Note. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–29. doi: 10.2307/3001666. [DOI] [Google Scholar]
- 52.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-base platform supports systematic causal inference across the human phenome. eLife. 2018;7:e34408. [DOI] [PMC free article] [PubMed]
- 53.Didelez V, Sheehan N. Mendelian randomization as an instrumental variable approach to causal inference. Stat Methods Med Res. 2007;16:309–30. doi: 10.1177/0962280206077743. [DOI] [PubMed] [Google Scholar]
- 54.Smith GD, Ebrahim S. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 55.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377–89. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowden J, Davey, Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bowden J, Spiller W, Del Greco FM, Sheehan N, Thompson J, Minelli C, et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J Epidemiol. 2018;47:1264–78. doi: 10.1093/ije/dyy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–9. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.UK Biobank. UKB: Data-Field 22009 Genetic principal components [Internet]. [cited 2021 May 6]. Available from: https://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=22009.
- 61.Brion MJA, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42:1497–501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bentley JL. Multidimensional binary search trees used for associative searching. Commun ACM. 1975;18:509–17. doi: 10.1145/361002.361007. [DOI] [Google Scholar]
- 63.Kuhn HW. The Hungarian method for the assignment problem. Nav Res Logist Q. 1955;2:83–97. doi: 10.1002/nav.3800020109. [DOI] [Google Scholar]
- 64.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oosterhuis JW, Looijenga LHJ. Testicular germ-cell tumours in a broader perspective. Nat Rev Cancer. 2005;5:210–22. doi: 10.1038/nrc1568. [DOI] [PubMed] [Google Scholar]
- 66.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eisenberg DTA, Kuzawa CW. The paternal age at conception effect on offspring telomere length: mechanistic, comparative and adaptive perspectives. Philos Trans R Soc B: Biol Sci. 2018;373:20160442. doi: 10.1098/rstb.2016.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Developmental Genet. 1996;18:173–9. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 69.Schrader M, Burger AM, Müller M, Krause H, Straub B, Schostak M, et al. The differentiation status of primary gonadal germ cell tumors correlates inversely with telomerase activity and the expression level of the gene encoding the catalytic subunit of telomerase. BMC Cancer. 2002;2:32. doi: 10.1186/1471-2407-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnston HE, Mann JR, Williams J, Waterhouse JAH, Birch JM, Cartwright RA, et al. The Inter-Regional, Epidemiological Study of Childhood Cancer (IRESCC): case-control study in children with germ cell tumours. Carcinogenesis. 1986;7:717–22. doi: 10.1093/carcin/7.5.717. [DOI] [PubMed] [Google Scholar]
- 71.Chen Z, Robison L, Giller R, Krailo M, Davis M, Gardner K, et al. Risk of childhood germ cell tumors in association with parental smoking and drinking. Cancer. 2005;103:1064–71. doi: 10.1002/cncr.20894. [DOI] [PubMed] [Google Scholar]
- 72.Johnson KJ, Carozza SE, Chow EJ, Fox EE, Horel S, McLaughlin CC, et al. Parental age and risk of childhood cancer: a pooled analysis. Epidemiology. 2009;20:475–83. doi: 10.1097/EDE.0b013e3181a5a332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shu XO, Nesbit ME, Buckley JD, Krailo MD, Robison LL. An exploratory analysis of risk factors for childhood malignant germ-cell tumors: report from the Childrens Cancer Group (Canada, United States) Cancer Causes Control. 1995;6:187–98. doi: 10.1007/BF00051790. [DOI] [PubMed] [Google Scholar]
- 74.Stephansson O, Wahnström C, Pettersson A, Sørensen HT, Tretli S, Gissler M, et al. Perinatal risk factors for childhood testicular germ-cell cancer: a Nordic population-based study. Cancer Epidemiol. 2011;35:e100–4. doi: 10.1016/j.canep.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 75.Wanderas EH, Grotmol T, Fossa SD, Tretli S. Maternal health and pre- and perinatal characteristics in the etiology of testicular cancer: a prospective population- and register-based study on Norwegian males born between 1967 and 1995. Cancer Causes Control: CCC. 1998;9:475–86. doi: 10.1023/A:1008857702380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1. Individual effect of leukocyte telomere length (LTL) related variants on germ cell tumor risk (GCT)
Supplemental Table S2. Sensitivity analysis for the MR analyses of genetically predicted telomere length on pediatric germ cell tumor
Supplemental Table S3 - Linkage relationship of TERT variants in previously reported loci.
Supplemental Figure S1. Mendelian Randomization (MR) analysis results for the association between individual SNPs that genetically predict leukocyte telomere length (LTL) and pediatric germ cell tumor
Supplemental Figure S2. Mendelian Randomization (MR) analysis results for the association between individual SNPs that genetically predict leukocyte telomere length (LTL) and pediatric germ cell tumor
Data Availability Statement
The paediatric and adolescent datasets generated from the Children’s Oncology Group (COG) and used for analyses in the current study will be shared publicly on dbGaP. Data generated from the California and Michigan Biobank samples cannot be shared due to state-specific restrictions regarding the use of the data. The UK Biobank data is publicly available and can be accessed on the UK Biobank website (https://www.ukbiobank.ac.uk/).
Representative code used to analyse the data in this study is provided in Supplemental Materials.