Abstract
Over the past decade, cancer diagnosis has expanded to include liquid biopsies in addition to tissue biopsies. Liquid biopsies can result in earlier and more accurate diagnosis and more effective monitoring of disease progression than tissue biopsies as samples can be collected frequently. Because of these advantages, liquid biopsies are now used extensively in clinical care. Liquid biopsy samples are analysed for circulating tumour cells (CTCs), cell-free DNA, RNA, proteins and exosomes. CTCs originate from the tumour, play crucial roles in metastasis and carry information on tumour heterogeneity. Multiple single-cell omics approaches allow the characterisation of the molecular makeup of CTCs. It has become evident that CTCs are robust biomarkers for predicting therapy response, clinical development of metastasis and disease progression. This review describes CTC biology, molecular heterogeneity within CTCs and the involvement of EMT in CTC dynamics. In addition, we describe the single-cell multi-omics technologies that have provided insights into the molecular features within therapy-resistant and metastasis-prone CTC populations. Functional studies coupled with integrated multi-omics analyses have the potential to identify therapies that can intervene the functions of CTCs.
Subject terms: Cancer, Cell biology
Introduction
Liquid biopsies are minimally invasive and have the potential to provide diagnostic and prognostic information that can aid in the treatment of patients with various solid tumours [1–3]. Circulating tumour cells (CTCs) and circulating tumour DNA (ctDNA), two prime biomarker candidates detected through liquid biopsy, can provide insight into tumour evolution, tumour biology, cancer progression and therapy resistance [4–6]. CTCs and ctDNA have great translational potential, and a plethora of clinical trials are currently utilising CTCs and ctDNA as biomarkers. These markers reflect different biological aspects of the disease, with CTCs crucial for evaluating prognosis in early and advanced disease (colorectal cancer, breast cancer and prostate cancer) [7] and ctDNA vital for monitoring treatment response and relapse [8, 9]. Despite active research in the ctDNA field, the lack of standardised methods for ctDNA extraction and analysis makes its utility controversial.
Research over the past decade has resulted in immense improvements in methods for CTC detection, separation, microfluidic isolation and genomic analysis. Whereas ctDNA can only be analysed at a genomic level, CTCs can be dissected at transcriptomic, genomic and proteomic levels either in bulk or as single cells. The precision medicine era has led to an in-depth investigation of CTCs. In this review, we discuss what is known about CTC biology and explore how single-cell multi-omics has contributed to our current understanding of these rare cancer cells and their potential in personalised medicine.
CTCs are shed from the primary tumour and are directly involved in the metastatic cascade based on Paget’s “seed and soil” hypothesis. CTCs are extraordinary cancer cells disseminated from the primary tumour that survive in the systemic circulation and initiate tumour formation at a site distant from the primary tumour. During the metastatic process, cancer cells undergo the epithelial-mesenchymal transition (EMT), through which they acquire the migratory and invasive ability and invade the surrounding stroma. They then intravasate and survive in the vasculature as CTCs, eventually extravasating at distant organs resulting in metastatic tumours [10, 11]. Although this cascade is very complex, it is inefficient as only a small fraction of cells progresses through all these stages. Not every CTC in the vascular system, and even fewer have the ability to establish metastases [12, 13].
The mesenchymal-epithelial transition (MET), which is the reverse of EMT, plays a crucial role in the colonisation and formation of macro-metastases. CTCs can exist in different states across the EMT spectrum [14, 15]. However, little is known regarding the influence of EMT and MET on CTC biology. CTCs can migrate as single cells or as clusters [16], and both single cells and clusters have been shown to have metastatic and invasive potential [17, 18]. CTCs are heterogeneous both within an individual patient and across cohorts. Properties, including cell size, cell-surface markers and tumour-initiating potentials, differ [19, 20]. In addition, accumulating evidence suggests that components of the tumour microenvironment bolster the metastatic potential of CTCs [21–25].
CTCs serve as a privileged gateway to study mechanisms of metastasis and have prognostic and predictive value in breast and other cancers [26–31]. The ability to non-invasively draw blood frequently allows for longitudinal sampling, and thus liquid biopsy is a better strategic approach for a personalised and precise treatment strategy for cancer patients than is tissue biopsy [32, 33]. Advancements in single-cell technologies have allowed the identification of rare subpopulations of CTCs and the study of their dynamics. This review summarises relevant clinical and biological properties of CTCs, technical aspects of CTC analysis, the use of CTC analysis as a powerful tool in clinical diagnosis and prognosis and the potential for CTC-targeting therapies.
CTC heterogeneity—through the EMT lens
The activation of EMT occurs at the early stage of metastatic progression. During EMT, carcinoma cells lose their ability to adhere to other cells and the matrix, lose apicobasal polarity and gain mesenchymal characteristics. The gain of mesenchymal-like features enables cells to migrate from the primary tumour [34] and promotes their entry into the circulation [35, 36]. During EMT, expression of epithelial markers such as E-cadherin, EpCAM and cytokeratins is lost and there is a gain in mesenchymal markers such as N-cadherin, vimentin and fibronectin [37, 38]. EMT is a reversible process that is driven by tumour-intrinsic as well as tumour-extrinsic mechanisms mediated by growth factors, including transforming growth factor beta (TGF-β) and epidermal growth factor (EGF), by EMT-inducing transcription factors such as Snail, Twist, Slug, ZEB1 and FOXC2, and by the suppression of microRNAs (miRNAs) such as miR-200 and miR155 [39–45]. The EMT program enhances tumorigenicity, metastatic ability, radio-resistance and chemoresistance [46–50]. The EMT program also imparts stem cell-like properties, including self-renewal ability, to cancer cells [51–53].
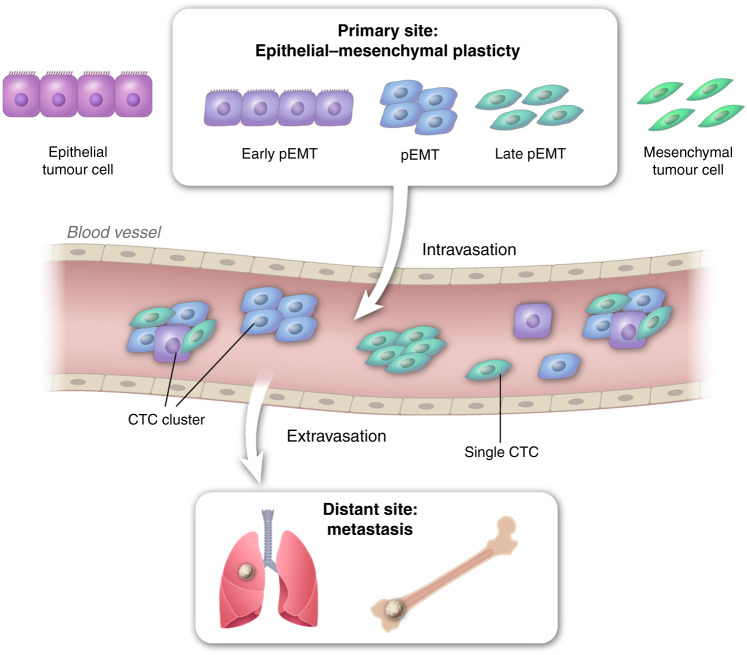
EMT is a spectrum rather than a process with binary states [54–56]. Three types of CTCs have been described: epithelial-like, mesenchymal-like and hybrid epithelial/mesenchymal (E/M) phenotypes [57]. CTCs that display a hybrid E/M phenotype have the plasticity to interconvert between epithelial and mesenchymal states enabling adaptation to the changing tumour microenvironment (Fig. 1) [58]. Invasion of the basement membrane by tumour cells is essential for metastatic cascade [40]. Following intravasation, stem cell-like features of CTCs help resist fluid shear stress [59–61]. To establish a metastasis, CTCs have to regain their epithelial phenotype [62]. Thus, CTCs undergo EMT when they start their journey from the primary tumour, and MET to form a metastatic lesion (Fig. 1) [63–65]. Cellular plasticity is linked with tumour progression, enhanced migration, metastasis and therapy resistance [66–70] and CTCs with E/M hybrid phenotypes and stem cell-like signatures are correlated with significantly reduced progression-free survival [71] and more aggressive and metastatic disease [72, 73]. CTC clusters with E/M hybrid plasticity display stemness, indicating that EMT contributes to the self-renewal of CTCs [74].
Fig. 1. Impact of CTC E/M plasticity on metastasis.
A small proportion of carcinoma cells exhibit E/M plasticity and display an increased capacity for intravasation and survival in the vasculature. Such CTC clusters extravasate and form metastatic lesions.
The detection and isolation of CTCs from the blood is routinely performed using the epithelial marker EpCAM [75], but levels of this marker are significantly reduced or completely lost on CTCs that have undergone EMT. In breast cancer patients, it has been reported that CTCs are enriched in mesenchymal markers (N-cadherin, TWIST and Vimentin) and have decreased epithelial marker expression (E-cadherin, EpCAM and CK8/18/19) [14]. In a study involving patients with progressive colon cancer, CTCs expressed transcription factors that initiate EMT (FOXC2, SNAIL and TWIST1) and vimentin but have low levels of epithelial markers E-cadherin and EpCAM [76]. Lecharpentier et al. demonstrated that single CTCs as well as clusters of CTCs from patients with non-small-cell lung cancer co-expressed the mesenchymal marker vimentin and the epithelial marker keratin [64]. These findings underscore the need for the discovery of markers that can capture CTCs independently of EMT state. Vimentin might be a suitable candidate [77].
CTCs with hybrid E/M properties express aldehyde dehydrogenase 1 (ALDH1), which is associated with stemness, and is expressed in hybrid E/M CTCs together with cytokeratins and nuclear TWIST1 [78] these CTCs are chemo-resistant [79]. CTCs isolated from patients with endometrial cancer display E/M plasticity and stemness resulting in enhanced survival and chemoresistance [80]. Collectively, these studies suggest that hybrid E/M CTCs are more metastatic than epithelial-like or mesenchymal-like CTCs and targeting hybrid E/M CTCs could improve patient survival. However, additional phenotypic and molecular characterisation of hybrid E/M CTCs is necessary.
CTC capture techniques
CTC detection could play an important role in the early detection of cancer and recurrence. Phenotype identification and molecular analysis of CTCs are expected to yield a significant amount of information about metastatic disease progression. However, CTCs are rare cells in the blood, are present at very low and undetectable levels. This poses a major challenge for detection, isolation and further downstream applications. Due to their rarity, a preceding CTC enrichment step is essential to separate CTCs based on the physical and biological properties of the tumour cells.
Several CTC detection platforms have emerged during the past decade. These platforms exploit a specific CTC feature and/or function for isolation, enumeration and functional characterisation. CellSearch (Janssen Diagnostics) was the first FDA-approved technology for CTC capture. It is based on the selection of EpCAM+ CTCs [81]. The Adna Test (Adnagen) uses captures CTCs with antibody-coated magnetic beads and has been shown to detect CTCs in blood of prostate and metastatic breast cancer patients [82, 83]. The MACS system employs a cytokeratin-based CTC counting approach [84], and geometrically enhanced differential immunocapture (GEDI), which has high specificity, captures CTCs on microfluidics with immobilised antibodies to proteins such as HER2 and PSMA [85]. Other microfluidics-based technologies utilise dual-modality approaches to capture CTCs using flow and immunomagnetic beads (IsoFlux) [86], viscous flow stress and magnetic force (the Quadrupole Magnetic Separator) [87], or a combination of microfluidics with sequential positive and negative CTC enrichment on a microchip (CTC-iChip) [88]. Such technologies enable robust molecular characterisation of rare cells like CTCs and have the potential to identify patients with nascent-stage disease.
Apart from label-dependent techniques, CTCs can be isolated using a plethora of marker-independent approaches based on physical properties, such as size [89, 90], charge [91, 92], density [93, 94] and elasticity [95, 96]. Moreover, CTCs can be isolated based on their biological properties, such as their invasive capability [97] or the presence of specific surface antigens that distinguish CTCs from other immune cells in the blood (negative selection of immune cells to enrich CTCs) [88, 98]. Isolation by size of tumour cells (ISET) is a filter-based isolation and enrichment technique that has higher efficiency than CellSearch [99, 100]. Similar approaches include the MetaCell system [101], ScreenCell Cyto [102] and CellSieve [103]. Although readily automated, filter-based exclusion is often prone to pore-clogging and requires large amounts of blood.
Additionally, the viability of CTCs in blood and purity of blood samples are major challenges [104]. The Cyttel system combines anti-CD45 immunohistochemistry and fluorescence in situ hybridisation (FISH) and involves the application of density-based centrifugation; it has a high detection rate in non-small cell lung cancer samples [105]. Other platforms, including OncoQuick [106], Percoll and Ficoll-Hypaque [107], are used to segregate whole blood components for CTC isolation; however, the formation of CTC aggregates interferes with these methods. Direct imaging offers an advantage over size-dependent isolation as it enables CTC detection and localisation using markers, such as cytokeratins, DAPI and CD45 and excludes the need for enrichment. FASTcell, CytoTrack and ImageStream are some of the platforms used in clinical studies to detect individual CTCs and clusters of CTCs as biomarkers [108–110]. Overcoming limitations of low sensitivity and resolution will make these powerful platforms for CTC detection and enumeration.
The CTC armour
CTCs shield themselves with protective armour to withstand the pressures of the microenvironment. Survival of CTCs is influenced by hypoxia, autophagy, inflammation and immune suppression, which all favour cancer progression (Fig. 2). CTCs interact with various components in the blood that nurture their survival, and they receive cues from the blood microenvironment that contribute to resistance to immune effector cells, anoikis and chemotherapy [111, 112]. CTCs express various cancer stem cell markers associated with decreased sensitivity to chemotherapies and cytotoxic immune effector cells [22, 113]. Further, high-resolution copy-number analysis revealed genetic alterations in CTCs in genes related to cancer cell dormancy, such as AKT2, PTEN and CADM2, genes associated with invasion and metastasis, such as ANGPTL4, BSG, miR-373 and LTBP4 and anti-apoptosis genes, such as miR-24, LTBP4, TFF3, NUMBL and miR-181 [114]. This suggests that CTCs can exist in subpopulations with different metastatic potentials.
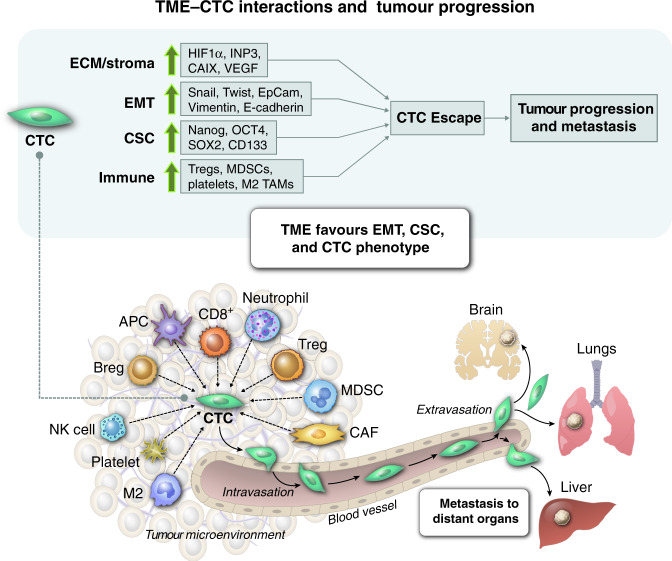
Fig. 2. The impact of tumour cell-intrinsic and extrinsic factors on establishing CTCs with E/M plasticity.
Upregulation of genes related to hypoxia (HIF-1a, INP3, CAIX and VEGF), EMT (SNAIL, TWIST, EpCAM, Vimentin and E-cadherin), cancer stem cells (Nanog, OCT4, SOX2 and CD133) and immune cell function in the tumour microenvironment remodels CTCs and primes these cells for escape from primary tumour site.
Additionally, survivin mRNA was detected in peripheral blood of patients with breast cancer, metastatic colorectal cancer, esophageal squamous cell carcinoma and non-small-cell lung cancer [115–118]. Moreover, CTCs with high survivin expression is correlated with poor overall survival in patients with metastatic colorectal cancer [117]. Survivin inhibits apoptosis of cancer cells and assists in immune evasion via blockade of cytotoxic anti-tumour activity of natural killer (NK) cells [119]. Extracellular survivin has been shown to decrease NKG2D receptor levels and affect the functionality of NK cells through alteration of activation markers, such as perforin, granzyme B, TNF-α and IFN-γ [120]. HER2 also plays a role in cell survival and proliferation in CTCs by activating the PI3K/Akt and Ras/Raf/MAPK signaling pathways [121, 122].
The presence of upregulated immune-suppressive elements in the tumour microenvironment may also facilitate CTC survival. A study of hormone receptor-positive breast cancer patients showed that the immune checkpoint regulator PD-L1 is upregulated on CTCs [123]. PD-L1 mediates the activity of regulatory T cells, which exert immunosuppressive action and induce apoptosis of activated cytotoxic T cells [124]. CTCs can induce apoptosis of peripheral T-helper cells by upregulation of CD95L/FasL on their surface. Sustained activation of Toll-like receptor signaling may also allow CTCs to evade the immune system. This activation generally attracts host immune cells but can also escalate inflammation and have deleterious effects [125]. Cytokines like IL-6 and IL-8, which are secreted by osteosarcoma cells, play a role in recruitment and clonal expansion of CTCs in distant organs to promote osteosarcoma outgrowth and act as tumour-derived attractants of CTCs; this phenomenon is called ‘tumour self-seeding of CTCs’ and can accelerate tumour growth [126, 127]. Thus, CTCs are efficient at modulating the immune microenvironment and favor self-survival in the circulation.
Platelets also play essential roles in the survival of CTCs by rendering them resistant to shear stress and NK cell cytotoxicity [128, 129]. Proteins like P-selectin and integrins expressed on the platelet cell surface contribute to tumorigenesis by promoting adhesion between CTCs and platelets [130, 131]. Several mechanisms mediated by platelet-derived TGF-β enable the escape of CTCs from immune clearance. TGF-β-mediated downregulation of anti-apoptotic Bcl-2 influences apoptosis, differentiation and cell proliferation [132] and downregulates NKG2D expression, thereby impairing NK cell cytotoxicity [133]. Platelet aggregation serves as a shield for CTCs, as CTC-platelet complexes escape immune attacks [134]. Blockade of CTC-platelet interactions by modifying platelets to express TRAIL resulted in the eradication of tumour cells in a model of prostate cancer metastasis [135].
In summary, CTCs expressing survivin, HER2 or PD-L1 or with mutations mentioned above could be valuable indicators of the survival capacity of the CTCs and could be used for the prediction of recurrences and metastasis. Further, abrogation of CTC survival mechanisms could create an environment hostile for CTCs and reduce their potential for survival and metastasis, especially in breast and gastric cancers where HER2 is overexpressed. A better understanding of CTC heterogeneity and the phenotypic and genotypic characterisation of CTCs could identify the ‘culprit’ CTC populations responsible for cancer progression, which could be targeted therapeutically. Moreover, immunosuppression-dependent CTC survival mechanisms could be targeted to abrogate the metastatic cascade.
Dissecting CTCs using a multi-omics microscope
Singularity matters
With the advent of various CTC capture technologies, research in the CTC field was no longer limited to detection and enumeration. Specifically, genomes, transcriptomes and proteomes of CTCs have been assessed. Single-cell multi-omics methods can dissect intra- and inter-tumour heterogeneity and provide insight into the roles of rare cells, such as CTCs, cancer stem cells and cells enriched for EMT [25, 136–140]. One single CTC can now be analysed to generate proteomic, transcriptomic and epigenomic data, yielding valuable information on molecular mechanisms and transcriptional programs active in these cells [141]. Since the analysis of CTCs gives insight into primary tumour characteristics, a minimally invasive yet maximally informative CTC multi-omics analysis would be an excellent approach to relate acquired genomic variation with changes in cellular function and phenotype in patients with different stages of cancer progression (Fig. 3). Therefore, CTC analysis should facilitate the personalisation of therapeutic options.
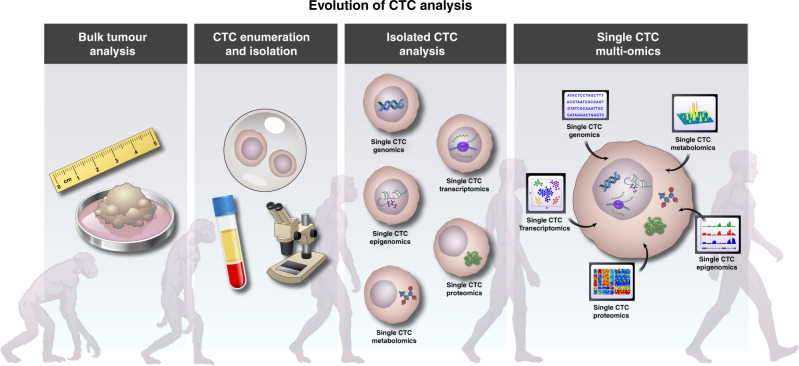
Fig. 3. Evolution of CTC analysis.
Development of sophisticated single-cell analysis technologies has allowed the dissection of heterogeneity within CTCs. Integration of multi-omics technologies will provide comprehensive knowledge of the landscape of CTC biology.
Single-CTC genomics
Understanding the genome at single-cell resolution across various human tissues has changed the way we look at human anatomy and has led to the discovery of new cell phenotypes. The genomic analysis of CTCs has similarly increased our understanding of tumour metastasis, intra-tumour heterogeneity and genetic mutations [142–145]. Whole-genome amplification (WGA), performed using methods such as multiple displacement amplification (MDA), degenerate oligonucleotide-primed polymerase chain reaction, multiple annealing and looping based amplification cycles (MALBAC) or linear amplification via transposon insertion (LIANTI), yields accurate genomic analysis of CTCs if performed with precision [146, 147]. MDA-based methods are well suited for amplifying point mutations because of the higher fidelity than PCR-based methods. MDA has been used to analyse the genomes of patient-derived CTCs [143, 148]. MALBAC combines MDA- and PCR-based methods and generates, using a specific oligonucleotide design, looped DNA molecules during multiple initial rounds of displacement preamplification. These looped DNAs are then further amplified using PCR [149]. MALBAC has been successfully used to analyse single-nucleotide variant (SNVs) profiles in CTCs [150]. LIANTI amplifies T7 promoter-tagged DNA fragments into thousands of RNA copies; this method covers 97% of the genome with a reduced false-negative rate compared to other WGA methods and could be applied to single-CTC profiling [151].
Lu et al. systematically evaluated four different WGA methods for copy-number variation (CNV) profiling of single CTCs from non-small cell lung cancer patients. The group showed that MALBAC and MDA-based Repli-g methods yield significantly higher genomic coverage than PCR-based methods (GenomPlex and Ampli1) [152]. Another study by Malihi and colleagues used the Epic Sciences CTC platform for genomic characterisation of CTCs based on WGA analysis of copy-number alterations (CNA) to evaluated associations between single-CTC genomics and clinical features, progression-free survival and overall survival in prostate cancer [153]. It currently remains challenging to achieve high genome coverage, low allele dropout and low amplification errors; however, single-CTC genomic analysis has the potential to become a powerful non-invasive diagnostic tool to study gene expression changes in cancer patients with localised, metastatic and recurrent diseases.
Single-CTC epigenomics
The cancer cell epigenome harbors myriad abnormalities that are potential cancer biomarkers and that could be targeted using therapeutic strategies [154]. Epigenetic modifiers are central components in regulating gene expression, and it is of prime importance to study epigenomes on the single-cell level to identify rare populations and characterise intra-tumour heterogeneity. During the past 20 years, mechanisms of epigenetic regulation have been thoroughly mapped and studied, and, with the emergence of single-cell technologies over the last 6 years, it has become possible to identify aberrations in major mechanisms like DNA methylation, chromosome conformation, histone modifications on a single-cell level [155–157].
Genes encoding tumour suppressors are silenced in CTCs of breast cancer patients [158, 159]; DNA methylation patterns and miRNA expression are altered in these CTCs [159–161]. DNA methylation and miRNA expression patterns provide crucial insights into molecular mechanisms of EMT and metastasis with important therapeutic implications [162]. Whole-Genome Bisulfite Sequencing (WGBS) offers a comprehensive DNA methylation profile in a broad range of biological systems. It can be used to analyse single-cell methylomes, whereas ChIP-seq allows the study of histone modifications on the single-cell level [163, 164].
DNA methylation signatures have been studied in CTCs of breast and prostate cancers [159, 165]. A CTC line was established using a mouse hepatocellular carcinoma model and overexpression of HGF and c-MET was linked with decreased DNA methylation at the promoters of genes that contributed to EMT [166]. Hypomethylation of stem cell genes and increased expression of pluripotency networks were also detected, suggesting increased metastatic potential due to the acquisition of a stem cell-like phenotype [18]. In prostate CTCs, increased methylation of promoters of known EMT-repressors (miR-200c/141, miR-200b/a/429 and CDH1) and epigenetic regulation of EMT-associated genes during bloodborne dissemination were detected [167]. Thus, analysis of the methylation status of CTCs can inform about hybrid E/M states associated with malignant phenotypes.
miRNA dysregulation is associated with different pathologies including cancer [168]. miR-21 expression was elevated in keratin-positive CTCs in 11 out of 25 patients with metastatic breast cancer, as shown by single-CTC analyses using in situ hybridisation [169]. In these breast cancer patients, miR-21 is a marker of CTCs with an E/M plasticity phenotype as demonstrated by comparing miR-21 expression in CTCs that co-expressed miR-21 and keratin and by analysis of MCF-7 cells induced to undergo EMT that had lost keratin expression but retained miR-21. Another study showed elevated miR-10b expression in CTCs isolated from metastatic breast, colorectal and prostate cancer patients [170]. miR-10b is linked with tumorigenesis and metastasis and is a promising target for miRNA-based therapy [171]. Analysis for the presence of miRNAs in single CTCs is evolving and has the potential to identify miRNAs that are essential for the maintenance of EMT and metastatic progression, which could be potential targets for treatment. The development of analyses of the epigenome that can be conducted on liquid biopsy samples is critical due to their non-invasive accessibility. We expect that single-cell epigenomics will enable us to map the changes in epigenetic marks throughout cancer evolution and identify the critical and highly dynamic epigenetic marks, paving the way for personalised therapies.
Single-CTC transcriptomics
Single-cell RNA sequencing (scRNA-seq) provides high-resolution information on cellular differences that bulk RNA-seq does not reveal. Several methods have been developed for whole-transcriptome analysis (WTA), such as Smart-seq [172], Smart-seq2, Quartz-seq [173], CEL-seq [174] and STRT-seq [175]. The Smart-seq technologies utilise a unique reverse transcriptase that anchors to both cDNA ends, thereby increasing the number of full-length RNAs sequenced; this raises the efficiency of detection of alternatively spliced isoforms and genetic variants [176]. CEL-seq and STRT-seq target the 5’ ends and the 3’ ends of the mRNA, respectively, and are not suitable for detecting variants in the coding region or alternatively spliced transcripts [174, 177]. Each of these methods suffer from certain biases, and it is rare to achieve a full-length sequence from a single cell, and low-abundance transcripts are usually not detected [178].
Despite these current limitations, single-cell analysis at the transcriptomic level provides a novel tool for the identification of cell phenotypes and biomarkers. For instance, a combination of flow cytometry and high-density scRNA-seq identified molecular changes in CTCs from hepatocellular carcinoma (HCC) patients and oncogenic drivers such as IGF2 [179, 180]. This integrated approach could provide a novel tool for biomarker development in various cancers. scRNA-seq analysis of EMT phenotypes within CTCs across different vascular compartments revealed that CTCs predominantly have an epithelial phenotype at release but show EMT activation via Smad2 and β-catenin signaling during hematogenous transit [180]. A comprehensive scRNA-seq analysis of CTCs from mouse and human pancreatic ductal adenocarcinoma revealed enrichment of SPARC, which encodes an extracellular matrix-derived protein that enhances migration and invasion properties of cancer cells [25]. Through scRNA-seq on CTCs from 13 patients with drug-resistant prostate cancer, a novel drug resistance mechanism involving non-canonical Wnt signaling was identified [181]. Another discovery made using scRNA-seq is that high plakoglobin levels are associated with CTC cluster formation and can contribute to the metastatic spread of breast cancer [16]. These findings could aid in the customisation of existing therapeutic interventions. The recent development of Hydro-seq, a scalable hydrodynamic scRNA-seq barcoding technique, shows promising efficiency for high throughput whole-transcriptome CTC analysis, which will allow analyses of higher numbers of CTCs more efficiently and will likely increase the discovery of rare clones that are critical for cancer progression and metastasis [182].
Single-CTC proteomics
Single-cell proteomic techniques are indispensable tools for exploring cellular mechanisms and processes. Proteins carry out functions like signal transduction and regulation of transcription and cytokine/chemokine secretion and mediate cell migration and invasion. Moreover, despite having the same genomic sequences, CTC subpopulations can differ in function and phenotype; hence, proteomics research holds great promise for the characterisation of the functions of CTCs. Current proteome profiling technologies can be broadly divided into cytometry-based and microfluidics-based platforms.
Microfluidics-based immunofluorescence techniques combine spatial and spectral encoding approaches and can detect up to 42 secreted proteins from a single cell [183]. Microfluidic western blotting integrates electrophoretic separations and multiple-cycle immunoassays, thereby allowing protein measurement in single rare cells such as CTCs. Sinkala and colleagues utilised single-cell resolution western blot [184] to evaluate a panel of eight surface and intracellular proteins in single CTCs isolated from patients with primary ER+ breast cancer and identified differences in expression patterns [185]. Single-cell western blot data are archivable and can be expanded to characterise subpopulations like CTCs undergoing EMT/immune escape and stem-like CTCs. Wang et al. developed a microchip-assisted single-cell proteomic method based on antibody and cellular DNA barcoding strategy for profiling CTC surface proteins [186]. However, these immunoassay-based approaches have shortcomings, including detecting a limited number of proteins and the need for antibodies [187].
Use of cytometry by time-of-flight (CyTOF) mass spectrometry for protein analysis has rapidly moved from the research laboratory to the clinical settings. CyTOF methodologies allow for the highly multiplexed and quantitative measurement of 35 or more proteins in a single cell [188]. Imaging mass cytometry employs mass spectroscopy-based analysis of tissue sections to yield information on the spatial expression of tens of proteins simultaneously [189],. IMC allows for simultaneous detection of multiple markers in a single cell coupled with greater protein quantification and co-localisation information compared to other methods [190]. Gerdtsson et al. combined high-definition single-cell analysis (HD-SCA) and imaging mass cytometry to evaluate subcellular localisation of markers of prostate cancer in a clonally related population of CTCs and disseminated tumour cells [176]. HD-SCA enables simultaneous classification and monitoring of cancer [191] [192].
Proteomics studies are far more challenging than nucleic acid analyses mainly due to the complexity of the proteome and the need for highly specific probes and amplification tools. Moreover, highly sensitive detection methods are urgently needed to allow the study of low-abundance proteins and post-translational modifications. The use of proteomics to study rare CTCs using panels that include established therapeutic markers will help discover novel markers and mechanisms behind CTC immune escape. Targeted single-CTC proteomics has the potential to crucial insights into the process of metastatic progression [193].
Personalised precision medicine targeting CTCs
Liquid biopsies to analyse cell-free DNA, exosomes and CTCs present in the blood can provide information on the tumour in a far less invasive way than a tumour biopsy. Currently, the enumeration of CTCs in a blood sample is used to guide treatment decisions. Although many cancer patients have very low or undetectable levels of CTCs, the presence of CTCs at the time of cancer diagnosis or recurrence is correlated with prognosis, and sequential liquid biopsy collection is used to evaluate disease progression. Several studies determined that between 60 and 80% of breast early breast cancer patients have detectable CTCs [194, 195]. The risk of recurrence increases and overall survival is reduced when CTCs are present in the blood sample in these patients. The recurrence risk correlates with the number of CTCs detected: The more CTCs, the higher the risk of recurrence [194, 195]. However, although at higher risk when CTCs were present, over 88% of women with CTCs had no cancer recurrence after 3 years of follow-up and over 87% had no metastatic progression [195]. Patients with CTCs are usually treated with chemotherapy; many of these patients are overtreated and may unnecessarily suffer chemotherapy’s side effects and complications. The other side of the coin is that many patients have very low or undetectable levels of CTCs but progress and develop metastatic disease [195]. This stresses the point that a higher resolution characterisation of CTCs is necessary to establish a treatment plan for patients with CTCs.
Clinically useful single-cell ‘omics approaches that can reveal prognostic phenotypic information about CTCs are needed. Epithelial markers, such as cytokeratin and EpCAM, are often used to detect CTCs [196–198]. Currently, the only FDA-approved test for CTC detection is CellSearch by Janssen Diagnostics based on the selection of EpCAM+ CTCs [81]. CTCs that have progressed toward the mesenchymal state have low expression of these epithelial markers (Fig. 1), and these dangerous CTCs potentially go undetected with the consequence that a patient does not receive the appropriate treatment needed to prevent recurrence and progression. Better assays must be developed that detected multiple biomarkers that are indicative of whether CTCs will cause cancer progression and metastasis. Single-cell multi-omics analysis together with long-term follow-up data will be able to identify biomarkers associated with progression.
All single-cell technologies give molecular insight into the makeup of CTCs, which we can correlate with clinical predictors and outcomes to identify CTC features that are detrimental for patients with solid cancers. Targeting these CTCs is challenging, not only because of our limited knowledge of which CTCs cause metastasis but also due to their heterogeneity. In-depth analyses of CTCs might provide insight into the cells that drive metastatic progression since the subclones that can become CTCs are subclones within the tumour. The true drivers of cancer progression might go undetected by performing multi-omics on the tumour bulk because these are rare clones with stem cell properties. The fitness to metastasize is represented in the CTCs, which can be observed in early disease before the existence of metastases. Likewise, drug resistance predictions could be determined based on the molecular makeup of CTCs. Over 90% of patients with cancer die of their metastases [199]. CTCs are the cells that cause metastasis and recurrences and thus should be eliminated to prevent metastatic progression and increase survival. CTC profiling should become an added tool together with current tools and approaches for an oncologist to determine patient diagnosis and prognosis. Like tumours, CTCs can have a very different molecular makeup between patients; hence CTC characterisation and application of personalised precision medicine go hand in hand. Until the true identity of CTCs responsible for cancer progression is identified, integration as part of the routine clinical analysis will be impossible. For CTC profiling to become an integrated part of routine clinical practice, a small biomarker set identifying and characterising CTCs is needed.
Targeting alterations in CTCs identified from the tumour with established compounds is an attractive approach to treat cancer. For instance, HER2 is frequently amplified and overexpressed in breast cancer, and the detection of changes in HER2 gene expression in CTCs might contribute to the effective stratification of breast cancer patients and personalised treatment strategies targeting HER2. Mutations in the ESR1, which encodes an estrogen receptor, can have large consequences on the efficacy of hormone therapy [200, 201]. These mutations can be detected in tumours, and two recent studies were able to identify activating mutations in ESR1 in CTCs [202, 203]. CTCs isolated from colorectal cancer (CRC) patients harbored KRAS mutations, which are a strong predictor for resistance to EGFR inhibitors [204]. These studies demonstrate that applying current knowledge from tumours to CTCs can be efficacious when sensitive detection methods are used.
Conclusion
Isolation and analysis of CTCs from liquid biopsies can offer non-invasive diagnostic and therapeutic information in a wide variety of cancers. Emerging research suggests the existence of different CTC subpopulations with differing phenotypic compositions and metastatic capabilities in individual patients. This underscores the importance of targeted studies to identify the phenotypes and genotypes of CTCs with specific emphasis on characterising their metastatic potential. Although the detection of CTCs remains a challenge due to low numbers of CTCs in the blood, recent advancements in isolation methods and analysis technologies suggest that CTC phenotyping will soon be used clinically.
Characterisation of single CTCs via the integration of genomic, epigenomic, transcriptomic and proteomic approaches has shed much light on intra-tumour heterogeneity and therapeutic resistance mechanisms (Table 1) [141, 205–209]. Single-cell multi-omics approaches have the potential to map the evolution of CTCs and thereby build an atlas of tumour evolution that includes a plethora of therapeutically viable targets. Profiling of CTC-derived biomarkers holds greater clinical significance than the current diagnostic and prognostic tools alone and their circulating characteristic is a considerable advantage. With the current pace of CTC research and the potential for the discovery of biomarkers for CTCs likely to cause cancer progression, liquid biopsies have the potential to become a routine for screening and monitoring cancer patients, paving the way toward more personalised therapies.
Table 1.
Multi-omics strategies for analyses of single CTCs.
 |
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Author contributions
Conception and design have been done by SAM and TV. Writing, reviewing and revising the manuscript have been done by TV, PDH, MC and SAM. Figures—design and concept by TV and PDH. Fig 1—concept by Abhijeet Deshmukh. Figures generated by Jordan Pietz, MA, CMI of MD Anderson Cancer Center. All authors have seen the manuscript and have agreed to the submission.
Funding
This research was awarded to SAM and supported by CPRIT (RP160710/RP170172), NSF (PHY-1935762), NIH/NCI (R01CA200970, 2R01CA155243, the Bowes Foundation, and in part by the MD Anderson Cancer Center Support Grant CA016672 (MRP to SAM).
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Since this review article is based on the literature and does not include patients or experimental animals, the ethics approval and consent do not apply.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01768-9.
References
- 1.Hench IB, Hench J, Tolnay M. Liquid biopsy in clinical management of breast, lung, and colorectal cancer. Front Med. 2018;5:9. doi: 10.3389/fmed.2018.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anker P, Mulcahy H, Chen XQ, Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev. 1999;18:65–73. doi: 10.1023/A:1006260319913. [DOI] [PubMed] [Google Scholar]
- 3.Murtaza M, Dawson S-J, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 4.Lim M, Kim C-J, Sunkara V, Kim M-H, Cho Y-K. Liquid biopsy in lung cancer: clinical applications of circulating biomarkers (CTCs and ctDNA) Micromachines. 2018;9:100. doi: 10.3390/mi9030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidess E, Jeffrey SS. Circulating tumor cells versus tumor-derived cell-free DNA: rivals or partners in cancer care in the era of single-cell analysis? Genome Med. 2013;5:1–4. doi: 10.1186/gm474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castro-Giner F, Gkountela S, Donato C, Alborelli I, Quagliata L, Ng CK, et al. Cancer diagnosis using a liquid biopsy: challenges and expectations. Diagnostics. 2018;8:31. doi: 10.3390/diagnostics8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das A, Kunkel M, Joudeh J, Dicker DT, Scicchitano A, Allen JE, et al. Clinico-pathological correlation of serial measurement of circulating tumor cells in 24 metastatic colorectal cancer patients receiving chemotherapy reveals interpatient heterogeneity correlated with CEA levels but independent of KRAS and BRAF mutation. Cancer Biol Ther. 2015;16:709–13. doi: 10.1080/15384047.2015.1030555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frattini M, Gallino G, Signoroni S, Balestra D, Battaglia L, Sozzi G, et al. Quantitative analysis of plasma DNA in colorectal cancer patients: a novel prognostic tool. Ann N Y Acad Sci. 2006;1075:185–90. doi: 10.1196/annals.1368.025. [DOI] [PubMed] [Google Scholar]
- 9.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–92. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–91. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameron MD, Schmidt EE, Kerkvliet N, Nadkarni KV, Morris VL, Groom AC, et al. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res. 2000;60:2541–6. [PubMed] [Google Scholar]
- 13.Morris VL, Schmidt EE, MacDonald IC, Groom AC, Chambers AF. Sequential steps in hematogenous metastasis of cancer cells studied by in vivo videomicroscopy. Invasion Metastasis. 1997;17:281–96. [PubMed] [Google Scholar]
- 14.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang Y, Pantel K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell. 2013;23:573–81. doi: 10.1016/j.ccr.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158:1110–22. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Micalizzi DS, Maheswaran S, Haber DA. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 2017;31:1827–40. doi: 10.1101/gad.305805.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176:98–112.e14. doi: 10.1016/j.cell.2018.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Bode AM, Dong Z. Circulating tumor cells: moving biological insights into detection. Theranostics. 2017;7:2606. doi: 10.7150/thno.18588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gkountela S, Szczerba B, Donato C, Aceto N. Recent advances in the biology of human circulating tumour cells and metastasis. ESMO Open. 2016;1:e000078. doi: 10.1136/esmoopen-2016-000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mlecnik B, Bindea G, Kirilovsky A, Angell HK, Obenauf AC, Tosolini M, et al. The tumor microenvironment and Immunoscore are critical determinants of dissemination to distant metastasis. Sci Transl Med. 2016;8:327ra26–ra26. doi: 10.1126/scitranslmed.aad6352. [DOI] [PubMed] [Google Scholar]
- 22.Noman MZ, Messai Y, Muret J, Hasmim M, Chouaib S. Crosstalk between CTC, immune system and hypoxic tumor microenvironment. Cancer Microenviron. 2014;7:153–60. doi: 10.1007/s12307-014-0157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goubran HA, Kotb RR, Stakiw J, Emara ME, Burnouf T. Regulation of tumor growth and metastasis: the role of tumor microenvironment. Cancer Growth Metastasis. 2014;7:S11285. doi: 10.4137/CGM.S11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinert G, Schölch S, Niemietz T, Iwata N, García SA, Behrens B, et al. Immune escape and survival mechanisms in circulating tumor cells of colorectal cancer. Cancer Res. 2014;74:1694–704. doi: 10.1158/0008-5472.CAN-13-1885. [DOI] [PubMed] [Google Scholar]
- 25.Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM, Miyamoto DT, et al. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 2014;8:1905–18. doi: 10.1016/j.celrep.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang WL, Pleskow HM, Miyamoto DT. Molecular analysis of circulating tumors cells: biomarkers beyond enumeration. Adv Drug Deliv Rev. 2018;125:122–31. doi: 10.1016/j.addr.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Pierga JY, Bidard FC, Cropet C, Tresca P, Dalenc F, Romieu G, et al. Circulating tumor cells and brain metastasis outcome in patients with HER2-positive breast cancer: the LANDSCAPE trial. Ann Oncol. 2013;24:2999–3004. doi: 10.1093/annonc/mdt348. [DOI] [PubMed] [Google Scholar]
- 28.Banys-Paluchowski M, Schneck H, Blassl C, Schultz S, Meier-Stiegen F, Niederacher D, et al. Prognostic relevance of circulating tumor cells in molecular subtypes of breast cancer. Geburtshilfe Frauenheilkd. 2015;75:232–7. doi: 10.1055/s-0035-1545788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang FB, Yang XQ, Yang S, Wang BC, Feng MH, Tu JC. A higher number of circulating tumor cells (CTC) in peripheral blood indicates poor prognosis in prostate cancer patients-a meta-analysis. Asian Pac J Cancer Prev. 2011;12:2629–35. [PubMed] [Google Scholar]
- 30.Nesteruk D, Rutkowski A, Fabisiewicz S, Pawlak J, Siedlecki JA, Fabisiewicz A. Evaluation of prognostic significance of circulating tumor cells detection in rectal cancer patients treated with preoperative radiotherapy: prospectively collected material data. BioMed Res Int. 2014;2014:712827. doi: 10.1155/2014/712827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shishido SN, Carlsson A, Nieva J, Bethel K, Hicks JB, Bazhenova L, et al. Circulating tumor cells as a response monitor in stage IV non-small cell lung cancer. J Transl Med. 2019;17:294. doi: 10.1186/s12967-019-2035-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16:398–406. doi: 10.1016/j.molmed.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Toss A, Mu Z, Fernandez S, Cristofanilli M. CTC enumeration and characterization: moving toward personalized medicine. Ann Transl Med. 2014;2:108. doi: 10.3978/j.issn.2305-5839.2014.09.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J, Antin P, Berx G, Blanpain C, Brabletz T, Bronner M, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2020;21:341–52. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang SP, Cabrera RM, Segall JE. Tumor cell intravasation. Am J Physiol Cell Physiol. 2016;311:C1–c14. doi: 10.1152/ajpcell.00238.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wells A, Yates C, Shepard CR. E-cadherin as an indicator of mesenchymal to epithelial reverting transitions during the metastatic seeding of disseminated carcinomas. Clin Exp Metastasis. 2008;25:621–8. doi: 10.1007/s10585-008-9167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15:117–34. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 40.Yokoyama K, Kamata N, Fujimoto R, Tsutsumi S, Tomonari M, Taki M, et al. Increased invasion and matrix metalloproteinase-2 expression by Snail-induced mesenchymal transition in squamous cell carcinomas. Int J Oncol. 2003;22:891–8. [PubMed] [Google Scholar]
- 41.Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, et al. MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28:6773–84. doi: 10.1128/MCB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA. 2007;104:10069–74. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–73. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 44.Sleeman JP, Thiery JP. SnapShot: The epithelial-mesenchymal transition. Cell. 2011;145:162–e1. doi: 10.1016/j.cell.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 45.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–37. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nieto MA, Huang RY-J, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 47.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–59. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 50.Balic M, Lin H, Young L, Hawes D, Giuliano A, McNamara G, et al. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res. 2006;12:5615–21. doi: 10.1158/1078-0432.CCR-06-0169. [DOI] [PubMed] [Google Scholar]
- 51.Mani SA, Guo W, Liao M-J, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morel A-P, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Eaton EN, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–60. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jolly MK, Tripathi SC, Jia D, Mooney SM, Celiktas M, Hanash SM, et al. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7:27067–84. doi: 10.18632/oncotarget.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boareto M, Jolly MK, Goldman A, Pietilä M, Mani SA, Sengupta S, et al. Notch-Jagged signalling can give rise to clusters of cells exhibiting a hybrid epithelial/mesenchymal phenotype. J R Soc Interface. 2016;13:2015110. 10.1098/rsif.2015.1106. [DOI] [PMC free article] [PubMed]
- 56.Jia D, Jolly MK, Tripathi SC, Den Hollander P, Huang B, Lu M, et al. Distinguishing mechanisms underlying EMT tristability. Cancer Converg. 2017;1:2. doi: 10.1186/s41236-017-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qi LN, Xiang BD, Wu FX, Ye JZ, Zhong JH, Wang YY, et al. Circulating tumor cells undergoing EMT provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Res. 2018;78:4731–44. doi: 10.1158/0008-5472.CAN-17-2459. [DOI] [PubMed] [Google Scholar]
- 58.Barriere G, Fici P, Gallerani G, Fabbri F, Zoli W, Rigaud M. Circulating tumor cells and epithelial, mesenchymal and stemness markers: characterization of cell subpopulations. Ann. Transl Med. 2014;2:109. 10.3978/j.issn.2305-5839.2014.10.04. [DOI] [PMC free article] [PubMed]
- 59.Barnes JM, Nauseef JT, Henry MD. Resistance to fluid shear stress is a conserved biophysical property of malignant cells. PLoS ONE. 2012;7:e50973. doi: 10.1371/journal.pone.0050973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:1–9. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ip CK, Li S-S, Tang MY, Sy SK, Ren Y, Shum HC, et al. Stemness and chemoresistance in epithelial ovarian carcinoma cells under shear stress. Sci Rep. 2016;6:1–11. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao D, Dai C, Peng S. Mechanism of the mesenchymal-epithelial transition and its relationship with metastatic tumor formation. Mol Cancer Res. 2011;9:1608–20. doi: 10.1158/1541-7786.MCR-10-0568. [DOI] [PubMed] [Google Scholar]
- 63.Jolly MK, Boareto M, Huang B, Jia D, Lu M, Ben-Jacob E, et al. Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol. 2015;5:155. doi: 10.3389/fonc.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lecharpentier A, Vielh P, Perez-Moreno P, Planchard D, Soria JC, Farace F. Detection of circulating tumour cells with a hybrid (epithelial/mesenchymal) phenotype in patients with metastatic non-small cell lung cancer. Br J Cancer. 2011;105:1338–41. doi: 10.1038/bjc.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.George JT, Jolly MK, Xu S, Somarelli JA, Levine H. Survival outcomes in cancer patients predicted by a partial EMT gene expression scoring metric. Cancer Res. 2017;77:6415–28. doi: 10.1158/0008-5472.CAN-16-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck. Cancer Cell. 2017;171:1611–24.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hendrix MJ, Seftor EA, Seftor RE, Trevor KT. Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am J Pathol. 1997;150:483–95. [PMC free article] [PubMed] [Google Scholar]
- 69.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–26. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 70.Bhatia S, Monkman J, Toh AKL, Nagaraj SH, Thompson EW. Targeting epithelial–mesenchymal plasticity in cancer: clinical and preclinical advances in therapy and monitoring. Biochem J. 2017;474:3269–306. doi: 10.1042/BCJ20160782. [DOI] [PubMed] [Google Scholar]
- 71.Ning Y, Zhang W, Hanna DL, Yang D, Okazaki S, Berger MD, et al. Clinical relevance of EMT and stem-like gene expression in circulating tumor cells of metastatic colorectal cancer patients. Pharmacogenomics J. 2018;18:29–34. doi: 10.1038/tpj.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jie X-X, Zhang X-Y, Xu C-J. Epithelial-to-mesenchymal transition, circulating tumor cells and cancer metastasis: mechanisms and clinical applications. Oncotarget. 2017;8:81558–71. [DOI] [PMC free article] [PubMed]
- 73.Bednarz-Knoll N, Alix-Panabières C, Pantel K. Plasticity of disseminating cancer cells in patients with epithelial malignancies. Cancer Metastasis Rev. 2012;31:673–87. doi: 10.1007/s10555-012-9370-z. [DOI] [PubMed] [Google Scholar]
- 74.Grosse-Wilde A, Fouquier d’Hérouël A, McIntosh E, Ertaylan G, Skupin A, Kuestner RE, et al. Stemness of the hybrid epithelial/mesenchymal state in breast cancer and its association with poor survival. PLoS ONE. 2015;10:e0126522. doi: 10.1371/journal.pone.0126522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ring A, Zabaglo L, Ormerod M, Smith I, Dowsett M. Detection of circulating epithelial cells in the blood of patients with breast cancer: comparison of three techniques. Br J Cancer. 2005;92:906–12. doi: 10.1038/sj.bjc.6602418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Satelli A, Mitra A, Brownlee Z, Xia X, Bellister S, Overman MJ, et al. Epithelial–mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin Cancer Res. 2014;21:899–906. [DOI] [PMC free article] [PubMed]
- 77.Gorges TM, Tinhofer I, Drosch M, Röse L, Zollner TM, Krahn T, et al. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papadaki MA, Kallergi G, Zafeiriou Z, Manouras L, Theodoropoulos PA, Mavroudis D, et al. Co-expression of putative stemness and epithelial-to-mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer. 2014;14:1–10. doi: 10.1186/1471-2407-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Papadaki MA, Stoupis G, Theodoropoulos PA, Mavroudis D, Georgoulias V, Agelaki S. Circulating tumor cells with stemness and epithelial-to-mesenchymal transition features are chemoresistant and predictive of poor outcome in metastatic breast cancer. Mol Cancer Ther. 2019;18:437–47. doi: 10.1158/1535-7163.MCT-18-0584. [DOI] [PubMed] [Google Scholar]
- 80.Alonso-Alconada L, Muinelo-Romay L, Madissoo K, Diaz-Lopez A, Krakstad C, Trovik J, et al. Molecular profiling of circulating tumor cells links plasticity to the metastatic process in endometrial cancer. Mol Cancer. 2014;13:1–10. doi: 10.1186/1476-4598-13-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang L, Balasubramanian P, Chen AP, Kummar S, Evrard YA, Kinders RJ, editors. Promise and limits of the CellSearch platform for evaluating pharmacodynamics in circulating tumor cells. Seminars in oncology. Elsevier; 2016. [DOI] [PMC free article] [PubMed]
- 82.Danila DC, Samoila A, Patel C, Schreiber N, Herkal A, Anand A, et al. Clinical validity of detecting circulating tumor cells by AdnaTest assay compared to direct detection of tumor mRNA in stabilized whole blood, as a biomarker predicting overall survival for metastatic castration-resistant prostate cancer patients. Cancer J (Sudbury, Mass) 2016;22:315. doi: 10.1097/PPO.0000000000000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Müller V, Riethdorf S, Rack B, Janni W, Fasching PA, Solomayer E, et al. Prognostic impact of circulating tumor cells assessed with the CellSearch System™ and AdnaTest Breast™ in metastatic breast cancer patients: the DETECT study. Breast Cancer Res. 2012;14:1–8. doi: 10.1186/bcr3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chinen LTD, De Carvalho FM, Rocha BMM, Aguiar CM, Abdallah EA, Campanha D, et al. Cytokeratin-based CTC counting unrelated to clinical follow up. J Thorac Dis. 2013;5:593. doi: 10.3978/j.issn.2072-1439.2013.09.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, et al. Capture of circulating tumor cells from whole blood of prostate cancer patients using geometrically enhanced differential immunocapture (GEDI) and a prostate-specific antibody. Lab Chip. 2010;10:27–9. doi: 10.1039/B917959C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harb W, Fan A, Tran T, Danila DC, Keys D, Schwartz M, et al. Mutational analysis of circulating tumor cells using a novel microfluidic collection device and qPCR assay. Transl Oncol. 2013;6:528–IN1. doi: 10.1593/tlo.13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu Y, Deighan CJ, Miller BL, Balasubramanian P, Lustberg MB, Zborowski M, et al. Isolation and analysis of rare cells in the blood of cancer patients using a negative depletion methodology. Methods. 2013;64:169–82. doi: 10.1016/j.ymeth.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Karabacak NM, Spuhler PS, Fachin F, Lim EJ, Pai V, Ozkumur E, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9:694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vona G, Sabile A, Louha M, Sitruk V, Romana S, Schütze K, et al. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am J Pathol. 2000;156:57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marrinucci D, Bethel K, Bruce RH, Curry DN, Hsieh B, Humphrey M, et al. Case study of the morphologic variation of circulating tumor cells. Hum Pathol. 2007;38:514–9. doi: 10.1016/j.humpath.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 91.Gascoyne PR, Noshari J, Anderson TJ, Becker FF. Isolation of rare cells from cell mixtures by dielectrophoresis. Electrophoresis. 2009;30:1388–98. doi: 10.1002/elps.200800373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moon HS, Kwon K, Kim SI, Han H, Sohn J, Lee S, et al. Continuous separation of breast cancer cells from blood samples using multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP) Lab Chip. 2011;11:1118–25. doi: 10.1039/c0lc00345j. [DOI] [PubMed] [Google Scholar]
- 93.Gertler R, Rosenberg R, Fuehrer K, Dahm M, Nekarda H, Siewert JR. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Results Cancer Res. 2003;162:149–55. doi: 10.1007/978-3-642-59349-9_13. [DOI] [PubMed] [Google Scholar]
- 94.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS, et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mohamed H, Murray M, Turner JN, Caggana M. Isolation of tumor cells using size and deformation. J Chromatogr A. 2009;1216:8289–95. doi: 10.1016/j.chroma.2009.05.036. [DOI] [PubMed] [Google Scholar]
- 96.Liu Z, Huang F, Du J, Shu W, Feng H, Xu X, et al. Rapid isolation of cancer cells using microfluidic deterministic lateral displacement structure. Biomicrofluidics. 2013;7:11801. doi: 10.1063/1.4774308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fan T, Zhao Q, Chen JJ, Chen WT, Pearl ML. Clinical significance of circulating tumor cells detected by an invasion assay in peripheral blood of patients with ovarian cancer. Gynecol Oncol. 2009;112:185–91. doi: 10.1016/j.ygyno.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lara O, Tong X, Zborowski M, Chalmers JJ. Enrichment of rare cancer cells through depletion of normal cells using density and flow-through, immunomagnetic cell separation. Exp Hematol. 2004;32:891–904. doi: 10.1016/j.exphem.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 99.Krebs MG, Hou J-M, Sloane R, Lancashire L, Priest L, Nonaka D, et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and-independent approaches. J Thorac Oncol. 2012;7:306–15. doi: 10.1097/JTO.0b013e31823c5c16. [DOI] [PubMed] [Google Scholar]
- 100.Morris KL, Tugwood JD, Khoja L, Lancashire M, Sloane R, Burt D, et al. Circulating biomarkers in hepatocellular carcinoma. Cancer Chemother Pharmacol. 2014;74:323–32. doi: 10.1007/s00280-014-2508-7. [DOI] [PubMed] [Google Scholar]
- 101.Bobek V, Gurlich R, Eliasova P, Kolostova K. Circulating tumor cells in pancreatic cancer patients: enrichment and cultivation. World J Gastroenterol. 2014;20:17163–70. doi: 10.3748/wjg.v20.i45.17163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schuur E, editor. Rapid and Simple Isolation of Circulating Tumor Cells for Clinical and Research Applications Using ScreenCell ®2012. https://www.semanticscholar.org/paper/Rapid-and-Simple-Isolation-of-Circulating-Tumor-for-Schuur/c8ce414bed0455d52ecacb3977708dd7260495d7.
- 103.Adams DL, Stefansson S, Haudenschild C, Martin SS, Charpentier M, Chumsri S, et al. Cytometric characterization of circulating tumor cells captured by microfiltration and their correlation to the cellsearch® CTC test. Cytom Part A. 2015;87:137–44. doi: 10.1002/cyto.a.22613. [DOI] [PubMed] [Google Scholar]
- 104.Coumans FA, van Dalum G, Beck M, Terstappen LW. Filtration parameters influencing circulating tumor cell enrichment from whole blood. PLoS ONE. 2013;8:e61774. doi: 10.1371/journal.pone.0061774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tong B, Xu Y, Zhao J, Chen M, Xing J, Zhong W, et al. Prognostic significance of circulating tumor cells in non-small cell lung cancer patients undergoing chemotherapy. Oncotarget. 2017;8:86615. doi: 10.18632/oncotarget.21255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Königsberg R, Obermayr E, Bises G, Pfeiler G, Gneist M, Wrba F, et al. Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta Oncol. 2011;50:700–10. doi: 10.3109/0284186X.2010.549151. [DOI] [PubMed] [Google Scholar]
- 107.Rosenberg R, Gertler R, Friederichs J, Fuehrer K, Dahm M, Phelps R, et al. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry J Int Soc Anal Cytol. 2002;49:150–8. doi: 10.1002/cyto.10161. [DOI] [PubMed] [Google Scholar]
- 108.Somlo G, Lau SK, Frankel P, Hsieh HB, Liu X, Yang L, et al. Multiple biomarker expression on circulating tumor cells in comparison to tumor tissues from primary and metastatic sites in patients with locally advanced/inflammatory, and stage IV breast cancer, using a novel detection technology. Breast Cancer Res Treat. 2011;128:155–63. doi: 10.1007/s10549-011-1508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hillig T, Horn P, Nygaard A-B, Haugaard AS, Nejlund S, Brandslund I, et al. In vitro detection of circulating tumor cells compared by the CytoTrack and CellSearch methods. Tumor Biol. 2015;36:4597–601. doi: 10.1007/s13277-015-3105-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ogle LF, Orr JG, Willoughby CE, Hutton C, McPherson S, Plummer R, et al. Imagestream detection and characterisation of circulating tumour cells—a liquid biopsy for hepatocellular carcinoma? J Hepatol. 2016;65:305–13. doi: 10.1016/j.jhep.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 111.Po JW, Roohullah A, Lynch D, DeFazio A, Harrison M, Harnett PR, et al. Improved ovarian cancer EMT-CTC isolation by immunomagnetic targeting of epithelial EpCAM and mesenchymal N-cadherin. J Circ Biomark. 2018;7:1849454418782617. doi: 10.1177/1849454418782617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Woodward WA, Sulman EP. Cancer stem cells: markers or biomarkers? Cancer Metastasis Rev. 2008;27:459–70. doi: 10.1007/s10555-008-9130-2. [DOI] [PubMed] [Google Scholar]
- 113.Yang MH, Imrali A, Heeschen C. Circulating cancer stem cells: the importance to select. Chin J Cancer Res. 2015;27:437–49. doi: 10.3978/j.issn.1000-9604.2015.04.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kanwar N, Hu P, Bedard P, Clemons M, McCready D, Done SJ. Identification of genomic signatures in circulating tumor cells from breast cancer. Int J Cancer. 2015;137:332–44. doi: 10.1002/ijc.29399. [DOI] [PubMed] [Google Scholar]
- 115.Rosato A, Pivetta M, Parenti A, Iaderosa GA, Zoso A, Milan G, et al. Survivin in esophageal cancer: an accurate prognostic marker for squamous cell carcinoma but not adenocarcinoma. Int J Cancer. 2006;119:1717–22. doi: 10.1002/ijc.21923. [DOI] [PubMed] [Google Scholar]
- 116.Yie S-M, Luo B, Ye N-Y, Xie K, Ye S-R. Detection of Survivin-expressing circulating cancer cells in the peripheral blood of breast cancer patients by a RT-PCR ELISA. Clin Exp Metastasis. 2006;23:279–89. doi: 10.1007/s10585-006-9037-7. [DOI] [PubMed] [Google Scholar]
- 117.Ning Y, Hanna DL, Zhang W, Mendez A, Yang D, El-Khoueiry R, et al. Cytokeratin-20 and survivin-expressing circulating tumor cells predict survival in metastatic colorectal cancer patients by a combined immunomagnetic qRT-PCR approach. Mol Cancer Ther. 2015;14:2401–8. doi: 10.1158/1535-7163.MCT-15-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yie S-m, Lou B, Ye S-r, He X, Cao M, Xie K, et al. Clinical significance of detecting survivin-expressing circulating cancer cells in patients with non-small cell lung cancer. Lung Cancer. 2009;63:284–90. doi: 10.1016/j.lungcan.2008.05.024. [DOI] [PubMed] [Google Scholar]
- 119.Végran F, Boidot R. Survivin-3B promotes chemoresistance and immune escape by inhibiting caspase-8 and -6 in cancer cells. Oncoimmunology. 2013;2:e26328. doi: 10.4161/onci.26328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ferguson Bennit HR, Gonda A, Kabagwira J, Oppegard L, Chi D, Licero Campbell J, et al. Natural killer cell phenotype and functionality affected by exposure to extracellular survivin and lymphoma-derived exosomes. Int J Mol Sci. 2021;22:1255. 10.3390/ijms22031255. [DOI] [PMC free article] [PubMed]
- 121.Jiang H, Li Q, Yu S, Yu Y, Wang Y, Li W, et al. Impact of HER2 expression on outcome in gastric cancer patients with liver metastasis. Clin Transl Oncol. 2017;19:197–203. doi: 10.1007/s12094-016-1523-z. [DOI] [PubMed] [Google Scholar]
- 122.Elster N, Collins DM, Toomey S, Crown J, Eustace AJ, Hennessy BT. HER2-family signalling mechanisms, clinical implications and targeting in breast cancer. Breast Cancer Res Treat. 2015;149:5–15. doi: 10.1007/s10549-014-3250-x. [DOI] [PubMed] [Google Scholar]
- 123.Mazel M, Jacot W, Pantel K, Bartkowiak K, Topart D, Cayrefourcq L, et al. Frequent expression of PD-L1 on circulating breast cancer cells. Mol Oncol. 2015;9:1773–82. doi: 10.1016/j.molonc.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.DiDomenico J, Lamano JB, Oyon D, Li Y, Veliceasa D, Kaur G, et al. The immune checkpoint protein PD-L1 induces and maintains regulatory T cells in glioblastoma. Oncoimmunology. 2018;7:e1448329. doi: 10.1080/2162402X.2018.1448329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Green TL, Santos MF, Ejaeidi AA, Craft BS, Lewis RE, Cruse JM. Toll-like receptor (TLR) expression of immune system cells from metastatic breast cancer patients with circulating tumor cells. Exp Mol Pathol. 2014;97:44–8. doi: 10.1016/j.yexmp.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 126.Kim MY, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH, Norton L, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–26. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Y, Ma Q, Liu T, Guan G, Zhang K, Chen J, et al. Interleukin-6 suppression reduces tumour self-seeding by circulating tumour cells in a human osteosarcoma nude mouse model. Oncotarget. 2016;7:446–58. doi: 10.18632/oncotarget.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cravioto-Villanueva A, Luna-Perez P, Gutierrez-de la Barrera M, Martinez-Gómez H, Maffuz A, Rojas-Garcia P, et al. Thrombocytosis as a predictor of distant recurrence in patients with rectal cancer. Arch Med Res. 2012;43:305–11. doi: 10.1016/j.arcmed.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 129.Leblanc R, Peyruchaud O. Metastasis: new functional implications of platelets and megakaryocytes. Blood. 2016;128:24–31. doi: 10.1182/blood-2016-01-636399. [DOI] [PubMed] [Google Scholar]
- 130.Qi C, Li B, Guo S, Wei B, Shao C, Li J, et al. P-Selectin-mediated adhesion between platelets and tumor cells promotes intestinal tumorigenesis in ApcMin/+ mice. Int J Biol Sci. 2015;11:679. doi: 10.7150/ijbs.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Weber C, Springer TA. Neutrophil accumulation on activated, surface-adherent platelets in flow is mediated by interaction of Mac-1 with fibrinogen bound to alphaIIbbeta3 and stimulated by platelet-activating factor. J Clin Invest. 1997;100:2085–93. doi: 10.1172/JCI119742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim ES, Kim MS, Moon A. Transforming growth factor (TGF)-beta in conjunction with H-ras activation promotes malignant progression of MCF10A breast epithelial cells. Cytokine. 2005;29:84–91. doi: 10.1016/j.cyto.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 133.Guo S-W, Du Y, Liu X. Platelet-derived TGF-β1 mediates the down-modulation of NKG2D expression and may be responsible for impaired natural killer (NK) cytotoxicity in women with endometriosis. Hum Reprod. 2016;31:1462–74. doi: 10.1093/humrep/dew057. [DOI] [PubMed] [Google Scholar]
- 134.Palumbo JS, Talmage KE, Massari JV, La Jeunesse CM, Flick MJ, Kombrinck KW, et al. Platelets and fibrin(ogen) increase metastatic potential by impeding natural killer cell-mediated elimination of tumor cells. Blood. 2005;105:178–85. doi: 10.1182/blood-2004-06-2272. [DOI] [PubMed] [Google Scholar]
- 135.Li J, Sharkey CC, Wun B, Liesveld JL, King MR. Genetic engineering of platelets to neutralize circulating tumor cells. J Control Release. 2016;228:38–47. doi: 10.1016/j.jconrel.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ortega MA, Poirion O, Zhu X, Huang S, Wolfgruber TK, Sebra R, et al. Using single-cell multiple omics approaches to resolve tumor heterogeneity. Clin Transl Med. 2017;6:1–16. doi: 10.1186/s40169-017-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhao L, Lu YT, Li F, Wu K, Hou S, Yu J, et al. High‐purity prostate circulating tumor cell isolation by a polymer nanofiber‐embedded microchip for whole exome sequencing. Adv Mater. 2013;25:2897–902. doi: 10.1002/adma.201205237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–96. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Macaulay IC, Teng MJ, Haerty W, Kumar P, Ponting CP, Voet T. Separation and parallel sequencing of the genomes and transcriptomes of single cells using G&T-seq. Nat Protoc. 2016;11:2081–103. doi: 10.1038/nprot.2016.138. [DOI] [PubMed] [Google Scholar]
- 140.Navin NE. The first five years of single-cell cancer genomics and beyond. Genome Res. 2015;25:1499–507. doi: 10.1101/gr.191098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35:936–9. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 142.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–60. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Polzer B, Medoro G, Pasch S, Fontana F, Zorzino L, Pestka A, et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol Med. 2014;6:1371–86. doi: 10.15252/emmm.201404033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Alderton GK. One cell at a time. Nat Rev Cancer. 2011;11:312. [DOI] [PubMed]
- 146.Deleye L, Tilleman L, Vander Plaetsen A-S, Cornelis S, Deforce D, Van, Nieuwerburgh F. Performance of four modern whole genome amplification methods for copy number variant detection in single cells. Sci Rep. 2017;7:3422. doi: 10.1038/s41598-017-03711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–88. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 148.Leung ML, Wang Y, Waters J, Navin NE. SNES: single nucleus exome sequencing. Genome Biol. 2015;16:1–10. doi: 10.1186/s13059-015-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Huang L, Ma F, Chapman A, Lu S, Xie XS. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
- 150.Wang Y, Guo L, Feng L, Zhang W, Xiao T, Di X, et al. Single nucleotide variant profiles of viable single circulating tumour cells reveal CTC behaviours in breast cancer. Oncol Rep. 2018;39:2147–59. doi: 10.3892/or.2018.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen C, Xing D, Tan L, Li H, Zhou G, Huang L, et al. Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI) Science. 2017;356:189–94. doi: 10.1126/science.aak9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lu S, Chang C-J, Guan Y, Szafer-Glusman E, Punnoose E, Do A, et al. Genomic analysis of circulating tumor cells at the single-cell level. J Mol Diagnostics. 2020;22:770–81. doi: 10.1016/j.jmoldx.2020.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Malihi PD, Graf RP, Rodriguez A, Ramesh N, Lee J, Sutton R, et al. Single-cell circulating tumor cell analysis reveals genomic instability as a distinctive feature of aggressive prostate cancer. Clin Cancer Res. 2020;26:4143–53. doi: 10.1158/1078-0432.CCR-19-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11:726–34. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Clark SJ, Lee HJ, Smallwood SA, Kelsey G, Reik W. Single-cell epigenomics: powerful new methods for understanding gene regulation and cell identity. Genome Biol. 2016;17:72. doi: 10.1186/s13059-016-0944-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kelsey G, Stegle O, Reik W. Single-cell epigenomics: recording the past and predicting the future. Science. 2017;358:69–75. doi: 10.1126/science.aan6826. [DOI] [PubMed] [Google Scholar]
- 157.Schwartzman O, Tanay A. Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet. 2015;16:716–26. doi: 10.1038/nrg3980. [DOI] [PubMed] [Google Scholar]
- 158.Lianidou ES, Markou A, Strati A. Molecular characterization of circulating tumor cells in breast cancer: challenges and promises for individualized cancer treatment. Cancer Metastasis Rev. 2012;31:663–71. doi: 10.1007/s10555-012-9366-8. [DOI] [PubMed] [Google Scholar]
- 159.Pixberg CF, Schulz WA, Stoecklein NH, Neves RP. Characterization of DNA methylation in circulating tumor cells. Genes (Basel) 2015;6:1053–75. doi: 10.3390/genes6041053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chimonidou M, Strati A, Tzitzira A, Sotiropoulou G, Malamos N, Georgoulias V, et al. DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clin Chem. 2011;57:1169–77. doi: 10.1373/clinchem.2011.165902. [DOI] [PubMed] [Google Scholar]
- 161.Paolillo C, Londin E, Fortina P, Single-Cell Genomics. Clin Chem. 2019;65:972–85. doi: 10.1373/clinchem.2017.283895. [DOI] [PubMed] [Google Scholar]
- 162.Park JW, Han J-W. Targeting epigenetics for cancer therapy. Arch Pharm Res. 2019;42:159–70. doi: 10.1007/s12272-019-01126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lang JE, Scott JH, Wolf DM, Novak P, Punj V, Magbanua MJ, et al. Expression profiling of circulating tumor cells in metastatic breast cancer. Breast Cancer Res Treat. 2015;149:121–31. doi: 10.1007/s10549-014-3215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Farlik M, Sheffield NC, Nuzzo A, Datlinger P, Schönegger A, Klughammer J, et al. Single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386–97. doi: 10.1016/j.celrep.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pixberg CF, Raba K, Müller F, Behrens B, Honisch E, Niederacher D, et al. Analysis of DNA methylation in single circulating tumor cells. Oncogene. 2017;36:3223–31. doi: 10.1038/onc.2016.480. [DOI] [PubMed] [Google Scholar]
- 166.Ogunwobi OO, Puszyk W, Dong H-J, Liu C. Epigenetic upregulation of HGF and c-Met drives metastasis in hepatocellular carcinoma. PLoS ONE. 2013;8:e63765. doi: 10.1371/journal.pone.0063765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang L, Vertes A. Single-cell mass spectrometry approaches to explore cellular heterogeneity. Angew Chem Int Ed Engl. 2018;57:4466–77. doi: 10.1002/anie.201709719. [DOI] [PubMed] [Google Scholar]
- 168.Godfrey AC, Xu Z, Weinberg CR, Getts RC, Wade PA, DeRoo LA, et al. Serum microRNA expression as an early marker for breast cancer risk in prospectively collected samples from the Sister Study cohort. Breast Cancer Res. 2013;15:1–10. doi: 10.1186/bcr3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ortega FG, Lorente JA, Garcia Puche JL, Ruiz MP, Sanchez-Martin RM, de Miguel-Pérez D, et al. miRNA in situ hybridization in circulating tumor cells-MishCTC. Sci Rep. 2015;5:9207. doi: 10.1038/srep09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gasch C, Plummer PN, Jovanovic L, McInnes LM, Wescott D, Saunders CM, et al. Heterogeneity of miR-10b expression in circulating tumor cells. Sci Rep. 2015;5:15980. doi: 10.1038/srep15980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yoo B, Kavishwar A, Wang P, Ross A, Pantazopoulos P, Dudley M, et al. Therapy targeted to the metastatic niche is effective in a model of stage IV breast cancer. Sci Rep. 2017;7:45060. doi: 10.1038/srep45060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–8. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 173.Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, et al. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol. 2013;14:3097. doi: 10.1186/gb-2013-14-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hashimshony T, Wagner F, Sher N, Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2:666–73. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 175.Islam S, Kjällquist U, Moliner A, Zajac P, Fan JB, Lönnerberg P, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–7. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ellsworth DL, Blackburn HL, Shriver CD, Rabizadeh S, Soon-Shiong P, Ellsworth RE. Single-cell sequencing and tumorigenesis: improved understanding of tumor evolution and metastasis. Clin Transl Med. 2017;6:15. doi: 10.1186/s40169-017-0145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Islam S, Kjällquist U, Moliner A, Zajac P, Fan J-B, Lönnerberg P, et al. Highly multiplexed and strand-specific single-cell RNA 5′ end sequencing. Nat Protoc. 2012;7:813. doi: 10.1038/nprot.2012.022. [DOI] [PubMed] [Google Scholar]
- 178.Marinov GK, Williams BA, McCue K, Schroth GP, Gertz J, Myers RM, et al. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res. 2014;24:496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.D’Avola D, Villacorta-Martin C, Martins-Filho SN, Craig A, Labgaa I, von Felden J, et al. High-density single cell mRNA sequencing to characterize circulating tumor cells in hepatocellular carcinoma. Sci Rep. 2018;8:1–7. doi: 10.1038/s41598-018-30047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sun Y-F, Guo W, Xu Y, Shi Y-H, Gong Z-J, Ji Y, et al. Circulating tumor cells from different vascular sites exhibit spatial heterogeneity in epithelial and mesenchymal composition and distinct clinical significance in hepatocellular carcinoma. Clin Cancer Res. 2018;24:547–59. doi: 10.1158/1078-0432.CCR-17-1063. [DOI] [PubMed] [Google Scholar]
- 181.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–6. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cheng Y-H, Chen Y-C, Lin E, Brien R, Jung S, Chen Y-T, et al. Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells. Nat. Commun. 2019;10:2163. doi: 10.1038/s41467-019-10122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lu Y, Xue Q, Eisele MR, Sulistijo ES, Brower K, Han L, et al. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc Natl Acad Sci USA. 2015;112:E607–E15. doi: 10.1073/pnas.1413483112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hughes AJ, Spelke DP, Xu Z, Kang C-C, Schaffer DV, Herr AE. Single-cell western blotting. Nat Methods. 2014;11:749–55. doi: 10.1038/nmeth.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sinkala E, Sollier-Christen E, Renier C, Rosas-Canyelles E, Che J, Heirich K, et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat Commun. 2017;8:1–12. doi: 10.1038/ncomms14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang C, Yang L, Wang Z, He J, Shi Q. Highly multiplexed profiling of cell surface proteins on single circulating tumor cells based on antibody and cellular barcoding. Anal Bioanal Chem. 2019;411:5373–82. doi: 10.1007/s00216-019-01666-9. [DOI] [PubMed] [Google Scholar]
- 187.Sinkala E, Sollier-Christen E, Renier C, Rosàs-Canyelles E, Che J, Heirich K, et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat Commun. 2017;8:14622. doi: 10.1038/ncomms14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165:780–91. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gerdtsson E, Pore M, Thiele JA, Gerdtsson AS, Malihi PD, Nevarez R, et al. Multiplex protein detection on circulating tumor cells from liquid biopsies using imaging mass cytometry. Converg Sci Phys Oncol. 2018;4:2057–1739 [DOI] [PMC free article] [PubMed]
- 190.Baharlou H, Canete NP, Cunningham AL, Harman AN, Patrick E. Mass cytometry imaging for the study of human diseases—applications and data analysis strategies. Front Immunol. 2019;10:2657. [DOI] [PMC free article] [PubMed]
- 191.Wistuba-Hamprecht K, Martens A, Weide B, Teng KW, Zelba H, Guffart E, et al. Establishing high dimensional immune signatures from peripheral blood via mass cytometry in a discovery cohort of stage IV melanoma patients. J Immunol. 2017;198:927–36. doi: 10.4049/jimmunol.1600875. [DOI] [PubMed] [Google Scholar]
- 192.Gruber I, Landenberger N, Staebler A, Hahn M, Wallwiener D, Fehm T. Relationship between circulating tumor cells and peripheral T-cells in patients with primary breast cancer. Anticancer Res. 2013;33:2233–8. [PubMed] [Google Scholar]
- 193.Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells—mechanisms of immune surveillance and escape. Nat Rev Clin Oncol. 2017;14:155–67. doi: 10.1038/nrclinonc.2016.144. [DOI] [PubMed] [Google Scholar]
- 194.Pierga JY, Hajage D, Bachelot T, Delaloge S, Brain E, Campone M, et al. High independent prognostic and predictive value of circulating tumor cells compared with serum tumor markers in a large prospective trial in first-line chemotherapy for metastatic breast cancer patients. Ann Oncol. 2012;23:618–24. doi: 10.1093/annonc/mdr263. [DOI] [PubMed] [Google Scholar]
- 195.Rack B, Schindlbeck C, Juckstock J, Andergassen U, Hepp P, Zwingers T, et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J Natl Cancer Inst. 2014;106:dju273. [DOI] [PMC free article] [PubMed]
- 196.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 198.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, et al. Circulating tumor cells: a novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol. 2005;23:1420–30. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 199.Seyfried TN, Huysentruyt LC. On the origin of cancer metastasis. Crit Rev Oncog. 2013;18:43–73. doi: 10.1615/CritRevOncog.v18.i1-2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jordan VC, Curpan R, Maximov PY. Estrogen receptor mutations found in breast cancer metastases integrated with the molecular pharmacology of selective ER modulators. J Natl Cancer Inst. 2015;107:djv075. doi: 10.1093/jnci/djv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013;45:1446–51. doi: 10.1038/ng.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Paolillo C, Mu Z, Rossi G, Schiewer MJ, Nguyen T, Austin L, et al. Detection of activating estrogen receptor gene (ESR1) mutations in single circulating tumor cells. Clin Cancer Res. 2017;23:6086–93. doi: 10.1158/1078-0432.CCR-17-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Beije N, Sieuwerts AM, Kraan J, Van NM, Onstenk W, Vitale SR, et al. Estrogen receptor mutations and splice variants determined in liquid biopsies from metastatic breast cancer patients. Mol Oncol. 2018;12:48–57. doi: 10.1002/1878-0261.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Denis JA, Patroni A, Guillerm E, Pepin D, Benali-Furet N, Wechsler J, et al. Droplet digital PCR of circulating tumor cells from colorectal cancer patients can predict KRAS mutations before surgery. Mol Oncol. 2016;10:1221–31. doi: 10.1016/j.molonc.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Macaulay IC, Haerty W, Kumar P, Li YI, Hu TX, Teng MJ, et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12:519–22. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 206.Dey SS, Kester L, Spanjaard B, Bienko M, van Oudenaarden A. Integrated genome and transcriptome sequencing of the same cell. Nat Biotechnol. 2015;33:285–9. doi: 10.1038/nbt.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hu Y, Huang K, An Q, Du G, Hu G, Xue J, et al. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 2016;17:1–11. doi: 10.1186/s13059-016-0950-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–19. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–8. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.