Abstract
Background
Ferroptosis has attracted increasing interest in cancer therapy. Emerging evidences suggest that naturally occurring naphthoquinones exhibit potent anti-glioma effects via various mechanisms.
Methods
The anti-glioma effects of plumbagin were evaluated by in vitro and in vivo experiments. Anti-glioma mechanism of plumbagin was studied by proteomics, flow cytometry, MDA assay, western blot, and RT-PCR. Gene knockdown/overexpression, molecular docking, PharmMappper database, and coimmunoprecipitation were used to study the targets of plumbagin.
Results
Plumbagin showed higher blood–brain barrier penetration ability than that of lapachol and shikonin and elicited significant growth inhibitory effects in vitro and in vivo. Ferroptosis was the main mechanism of plumbagin-induced cell death. Mechanistically, plumbagin significantly downregulated the protein and mRNA levels of xCT and decreased GPX4 protein levels. NAD(P)H quinone dehydrogenase 1 (NQO1) was revealed as a plumbagin predictive target using PharmMappper database and molecular docking. Plumbagin enhanced NQO1 activity and decreased xCT expression, resulting in NQO1-dependent cell death. It also induced GPX4 degradation via the lysosome pathway and caused GPX4-dependent cell death.
Conclusions
Plumbagin inhibited in vitro and in vivo glioma growth via targeting NQO1/GPX4-mediated ferroptosis, which might be developed as a novel ferroptosis inducer or anti-glioma candidate.
Subject terms: Cancer therapy, Drug screening
Background
Gliomas are the most common malignant primary brain tumours with poor prognosis. Patients with newly diagnosed glioma are currently treated with maximum surgical resection followed by chemotherapy and radiotherapy as the standard [1]. The most commonly used chemotherapeutic agent for gliomas is a DNA-alkylating agent called temozolomide (TMZ). However, most glioma patients are prone to relapsing and developing resistance to TMZ, ultimately leading to treatment failure. Ferroptosis, a new type of cell death discovered in recent years, has been reported to be strongly associated with TMZ resistance [2]. Tumour cells can maintain sensitivity to ferroptosis while escaping other forms of cell death [3]. Moreover, brain tumours are reported to be more sensitive to ferroptosis induction than other cancer types [4, 5]. Thus, ferroptosis induction might be a promising strategy to improve the chemotherapeutic outcomes of gliomas.
Naphthoquinones are an important class of naturally occurring ingredients with multiple pharmacological effects, such as anti-cancer, anti-bacterial, and anti-malarial properties [6, 7]. Quinone moieties are present in many clinically used anti-cancer drugs, such as doxorubicin, mitomycin, mitoxantrone, daunorubicin, and saintopin. Many naturally derived naphthoquinones (e.g. lapachol, shikonin, and naphthazarin) show potential efficacy for further development as anti-cancer drugs [8].
Our previous study showed that lapachol (4-hydroxy-3-(3-methylbut-2-enyl) naphthalene-1,2-dione, Fig. 1a), a natural naphthoquinone isolated from many species of Bignoniaceae, exhibited potential anti-glioma effects on rat C6 glioma cells in vitro and in vivo [9]. Plumbagin (PLB; 5-hydroxy-2-methyl-1,4-naphthoquinone, Fig. 1a), isolated from the root of Plumabago zeylanica L., is a structural analogue of lapachol [10]. Numerous studies have provided compelling evidences that PLB can inhibit tumour growth and progression in a variety of tumours in vitro and in vivo [11]. It can inhibit tumour growth by apoptosis induction, cell cycle arrest, anti-angiogenesis, and generation of reactive oxygen species (ROS) [12, 13]. However, the anti-glioma effects and mechanism of PLB has not been clearly revealed.
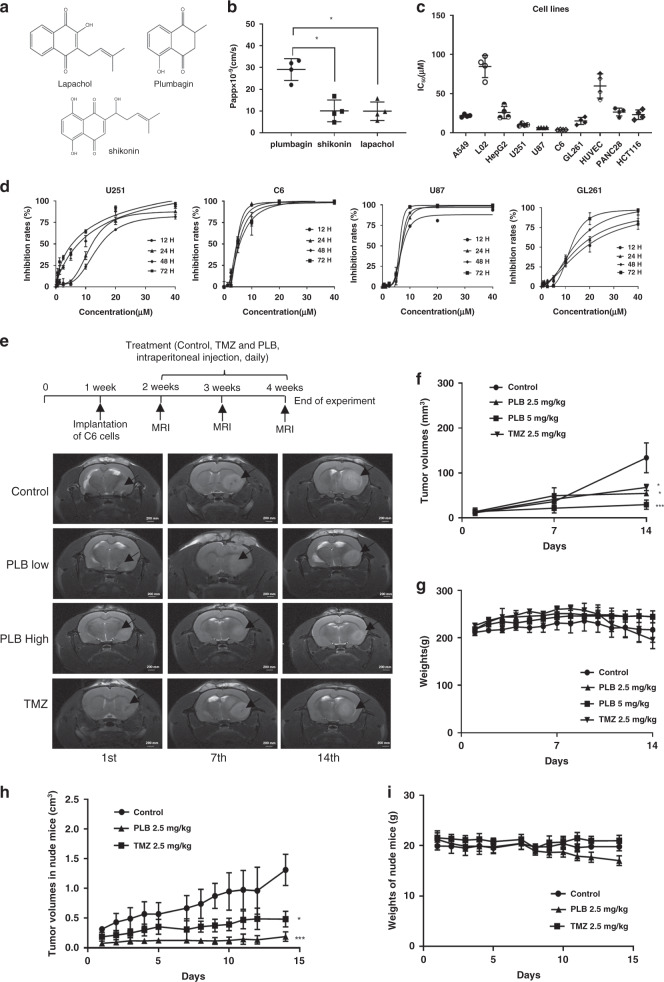
Fig. 1. PLB significantly inhibited glioma growth in vitro and in vivo.
a Chemical structures of PLB, lapachol, and shikonin; b calculated Papp × 10−9 of PLB, lapachol, and shikonin in the in vitro BBB cell model (n = 4, *P < 0.05). c IC50 of PLB in different cancer cells and normal cells via the MTS assay (n = 4). d Dose–response curves of PLB in U251, C6, U87, and GL261 glioma cells at 12, 24, 48, and 72 h (n = 3). e Schematic chart of rat orthotopic transplantation model and MRI images of C6 glioma in the brains of the rats indifferent groups on the 1st, 7th, and 14th days. f Tumour volumes of the rats in the control, TMZ, and PLB groups (n = 6, *P < 0.05, ***P < 0.005). g Body weights of the rats in the control, TMZ, and PLB groups (n = 6). h Tumour volumes of the nude mice in the control, TMZ, and PLB groups recorded during the experiment (n = 6, *P < 0.05, ***P < 0.005). i Body weights of the mice in the control, TMZ, and PLB groups recorded during the experiment (n = 6). The results were all expressed as mean ± S.D.
In the present study, the blood–brain barrier (BBB) penetration abilities of several natural naphthoquinones (lapachol, PLB, and shikonin) were compared in vitro, and the results showed that the BBB penetration ability of PLB was relatively higher than that of the other naphthoquinones. Thus, in vitro and in vivo anti-glioma effects and the underlying mechanisms of PLB were further studied.
Methods
Materials and reagents
The materials and reagents used in this study are shown in Table S1.
Cell lines and cell culture conditions
Human glioma cell lines (U251 and U87), rat glioma cell line (C6), mouse glioma cell line (GL261), human non-small cell lung cancer cell line (A549), human hepatocellular carcinoma cell line (HepG2), human pancreatic cancer cell line (PANC28), human colon cancer cell line (HCT116), human normal liver cell line (L02), and human venous endothelial cell line (HUVEC) were obtained from the Cell Resource Center of Peking Union Medical College, Beijing, China. All cells were cultured at 37 °C in a humidified incubator containing 5% CO2. C6, U87, and U251 cells were authenticated by STR profiling and tested for mycoplasma contamination. Cell viabilities were evaluated with Trypan blue staining before each experiment. Only cell suspensions with >95% cell viability (passage numbers: 3–8) were acceptable for the following experiments.
Anti-proliferation assays
Cell proliferations were detected with 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay. After drug exposure, premixed MTS and phenazinemethosulfate at a ratio of 20:1 was immediately added to each well. After 2 h, formazan production was determined at 490 nm by using a microplate reader (Thermo Scientific Multiskan GO, USA). The inhibitory rates and half inhibitory concentrations (IC50) were then calculated with GraphPad Prism 6.0 (GraphPad Software, Inc., San Diego, USA). The morphology changes of these glioma cells with or without PLB treatment were observed under an inverted microscope (Olympus, Japan).
BBB penetration assay in vitro
Rat brain microvascular endothelial cells (BMECs) were separated from the cerebral cortex of 3-week-old Sprague–Dawley rats, and primary astrocytes were separated from the cerebral cortex of newborn Sprague–Dawley rats as previously described methods [14]. The co-culture system of rat BMECs and astrocytes in Transwell inserts was used to establish the in vitro BBB model as shown in Fig. S1A. Transepithelial electrical resistance (TEER) value was measured every day by using Millicell-ERS 2 (Millipore, USA) until the TEER increased to 300 Ω/cm2. Only the inserts with TEER of >300 Ω/cm2 were used for BBB penetration assay (Fig. S1B).
The effects of lapachol, shikonin, and PLB on the proliferations of BMECs were detected via MTS assay before the experiment. Hank’s balanced salt solution (HBSS) containing 75 μM lapachol, 2.0 μM shikonin, or 2.0 μM PLB was added to the apical side of the inserts, while 600 μL of HBSS was added to the basolateral side. Drug concentrations in the samples at the time points of 15, 30, 60, 90, 120, and 180 min were determined by high-performance liquid chromatography (HPLC) under the following conditions: Agilent XDB-C18 column (150 × 4.6 mm, i.d. 5 μm); mobile phase, methanol: 1% ethanoic acid (from 10:90 to 90:10, v/v); flow rate, 0.8 mL/min; detection wavelength, 260 nm. Apparent permeability coefficients (Papp) were calculated as previously reported (n = 4) [14].
Establishment of rat orthotopic transplantation model and PLB treatment
A rat orthotopic transplantation model was established in accordance with our previously published protocol [9]. Wistar rats (male, weighing 180–200 g) were anaesthetised with isoflurane, and C6 cells were injected as previously described. Tumorigenesis was detected with a Bruker 7.0 T Micro-MRI system (Bruker BioSpec 70/20 USR, Germany) 7 days after implantation. The rats with a maximum intracerebral tumour diameter of >5.0 ± 0.5 mm were considered to have gliomas. These rats were double-blindly and randomly divided into four groups with minimisation: control group (20% hydroxyl-β-cyclodextrin, n = 6), TMZ group (2.5 mg/kg, dissolved in 20% hydroxyl-β-cyclodextrin, n = 6), and PLB groups (2.5 mg/kg, 5.0 mg/kg, dissolved in 20% hydroxyl-β-cyclodextrin, n = 6 each). The doses of PLB were set based on our pre-experiment. The drugs were intraperitoneally injected daily for 2 weeks. The general status and body weights were recorded each day. Tumours in the brains of the rats were detected with Bruker 7.0 T Micro-MRI on the 1st, 7th, and 14th days after administration.
Establishment of tumour-bearing nude mouse model and PLB treatment
U251 cells were subcutaneously inoculated into the right flank of BABL/c-nu/nu nude mice (female, 4–6 weeks, 18–20 g) [15]. When the tumour nodules grew to approximately 100 mm3, the mice were double-blindly and randomly divided into three groups with minimisation: control group (20% hydroxyl-β-cyclodextrin, n = 6), TMZ group (2.5 mg/kg, dissolved in 20% hydroxyl-β-cyclodextrin, n = 6), and PLB group (2.5 mg/kg, dissolved in 20% hydroxyl-β-cyclodextrin, n = 6). The drugs were administered intragastrically daily for 2 weeks. One mouse in the control group was killed before the end of the experiment because of excessively large tumour volumes. Tumour sizes and body weights were measured every day for 2 weeks. Then, all the mice were sacrificed, and tumour tissues were collected.
Immunohistochemistry
In the rat C6 orthotopic transplantation experiment, the rats were sacrificed after detection and their brain tissues were removed and fixed with 4% formalin. Then tissues were embedded in paraffin, cut into 5 μm-thick sections, and subjected to haematoxylin–eosin (HE) staining and immunochemical staining for glial fibrillary acidic protein (GFAP) and proliferation marker protein Ki-67 (Ki67) (n = 5 each group). Tumour tissues were collected from tumour-bearing nude mice with or without drug treatment and subjected to HE staining and immunochemical staining (GFAP, Ki67, xCT, and GPX4). All the staining images were recorded with a high-capacity digital slide scanner system (3DHISTECH Ltd., Budapest, Hungary).
Label-free quantitative proteomic and bioinformatic analysis
U251 cells with or without PLB treatment (4 μM) for 48 h were collected and analysed via label-free quantitative proteomics at Jingjie PTM BioLab, Hangzhou, China [16]. The main process is shown in Fig. S2A. In brief, cells were lysed, and protein solutions were reduced with dithiothreitol, alkylated with iodoacetamide, and digested with trypsin. The obtained peptides were fractionated using an EASY-nLC 1000 UPLC system with a reversed-phase C18 analytical column (15 cm long, 75 μm i.d.), and then analysed through tandem mass spectrometry (MS/MS) in Q ExactiveTM Plus (Thermo Fisher Scientific, Waltham, MA, USA). The parameters were as follows: electrospray voltage, 2.0 kV; m/z scan range, 350–1800; fixed first mass, 100 m/z; and automatic gain control, 5E4. Peptides were selected for MS/MS with an NCE of 28. Intact peptides were detected in Orbitrap at a resolution of 70,000, and fragments were detected in Orbitrap at a resolution of 17,500. The mass tolerance for precursor and fragment ions were set to 20 p.p.m. and 0.02 Da, respectively. The false discovery rate for peptides was adjusted to <1%, and the minimum score for the modified peptides was set at >40.
Gene Ontology (GO) annotation was performed using the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). InterProScan software was used for the identified proteins that were not annotated by the UniProt-GOA database. The threshold of differential expression change was set at 1.5 times, and P < 0.05 was set as the significance threshold. Then, the proteins were classified into three categories: cellular component, biological process, and molecular function. The Kyoto Encyclopaedia of Genes and Genomes (KEGG) database was used to annotate protein pathways. Two-tailed Fisher’s exact test was performed to examine the enrichment of differentially expressed proteins against all the identified proteins, and corrected P < 0.05 was considered statistically significant.
Transmission electron microscopy
U251 and C6 cells with or without PLB treatment (4 μM) for 6 h were collected, fixed in ice-cold 2.5% glutaraldehyde in phosphate-buffered saline (PBS; pH 7.3), washed, post-fixed in 1% osmium tetroxide with 0.1% potassium ferricyanide, dehydrated with a graded series of ethanol, embedded in Epon, and sectioned. Ultrathin sections (65 nm) were stained with 1% uranyl acetate and 0.1% lead citrate and examined on a HT7700 transmission electron microscope (HITACHI, Japan).
Measurement of malondialdehyde (MDA)
After exposure to different concentrations of PLB, erastin, PLB + Ferrostatin-1 (F1), or PLB + desferrioxamine mesylate (DFM) for 48 h, the cells were lysed, and protein concentrations in the supernatants were quantified via a BCA assay. Each sample (100 μL) was incubated with 100 μL of the test solution for 15 min at 100 °C. Then optical densities (ODs) were measured at 530 nm by using a microplate reader.
Measurement of intracellular glutathione (GSH) levels
After exposure to 2, 4, and 6 μM PLB for 48 h, the cells were collected and lysed. GSH levels were measured using a commercially available kit (BC1170, Solarbio, Beijing, China) in accordance with the manufacturer’s instruction.
Determination of intracellular ROS levels
U251 and C6 cells were seeded into 96-well plates overnight and then exposed to 2, 4, and 6 μM PLB for 48 h. After washing with PBS, the cells were incubated with DCFH-DA probe (20 µM) in the dark for 30 min at 37 °C. Fluorescence intensities were measured using a fluorometric plate reader (SpectraMax M5, Molecular Devices, CA, USA).
Lipid peroxidation detection by flow cytometry
After exposure to 2, 4, and 6 μM PLB for 48 h, the cells were harvested and resuspended in Hank’s solution containing C11-BODIPY581/591 (2 μM) for 30 min. Flow cytometry was carried out on a Guava easyCyte flow cytometer system (Merck Millipore, Billerica, MA, USA). The results were analysed with FlowJo V10.
Western blot
The cells were exposed to different drugs, collected, and lysed. The proteins in the supernatants were quantified, and the samples were separated on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. The membranes were blocked with TBST containing 5% albumin bovine V for 60 min and incubated with primary antibodies at 4 °C for 1 h. After incubation with secondary antibodies, immunoblots were imaged on a molecular imager (Bio-Rad Laboratories, Inc., Hercules, CA, USA). ImageJ was used to quantify the intensity of the bands.
Quantitative reverse transcription–polymerase chain reaction (RT-PCR)
After exposure to 2, 4, and 6 μM PLB for 48 h, total RNA was isolated and reverse-transcribed into cDNA by using a cDNA transcription kit. Quantitative RT-PCR was carried out with a real-time fluorescence quantitative PCR detection system IQ5 (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The primers are shown in Table S2. The relative expression levels of target genes were normalised to GAPDH via the 2−ΔΔCt method.
Prediction and screening of PLB targets
The PharmMappper database (http://www.lilab-ecust.cn/pharmmapper/) was used by uploading PLB’s mol2 format to determine PLB-related predictive targets [17]. The following parameters were used: generate conformers, yes; maximum generated conformations, 300; select target set, human protein target only; number of reserved matched targets, 300. PLB-related predictive targets were inputted into the UniProtKB database (http://www.uniprot.org/) to collect the official symbols by eliminating duplicate and nonstandard targets. Then, a protein–protein interaction (PPI) network was obtained from STRING database 11.0 (http://string-db.org/) by uploading these predictive targets and the experimentally confirmed ferroptosis targets (xCT and GPX4). The species was limited to “Homo sapiens” and the minimum required interaction score was 0.400.
Molecular docking studies
PLB and NAD(P)H quinone dehydrogenase 1 (NQO1) were subjected to molecular docking by using AutoDockTools-1.5.6. The crystal structure of NQO1 was obtained from Protein Data Bank (http://www1.rcsb.org/, PDB Entry: 2F1O, Resolution: 2.75 Å), and water molecules were removed with PyMOLWin. The three-dimensional structure of PLB and β-Lapachone was prepared and modified to its optimum conformation by using the Chemdraw software. The receptor and ligand molecules were then imported into AutoDockTools-1.5.6. The binding active pocket of Dicoumarol in NQO1 protein crystal structure was set to GridBox. Then, the standard docking procedure for a rigid protein and a flexible ligand was performed using “autogrid4” and “autodock” model. β-Lapachone was set as the positive control.
NQO1 activity assay
The effects of PLB on NQO1 activities were determined with an NQO1 activity assay kit. After exposure to 2, 4, and 6 μM PLB for 6 h, cells were collected, lysed, and quantified. After adding 50 μL samples to each well, 50 μL reaction buffer + inhibitor or reaction buffer was rapidly added to the appropriate paired sample wells. The OD values were immediately recorded at 440 nm with the elapsed time (interval 20 sec, up to 5 min) in the microplate reader.
Transient NQO1 small interfering RNA (siRNA) transfection
Non-targeting control siRNA (siNC) or siRNA targeting NQO1 (siNQO1, sense, 5′-GCCUCUCCGACCUAAACUU-3′; antisense, 5′-AAGUUUAGGUCAAAGAGGC-3′) were synthesised by Shanghai GenePharma Co., Ltd. (Shanghai, China). The cells were transfected with siNC or siNQO1 by using Lipofectamine 3000, and NQO1 expression levels were determined via western blot.
Construction of GPX4-overexpressing cells
U251 and U87 cells in the logarithmic growth phase were transfected with an overexpression plasmid (pcDNA3.1-GPX4) by using Lipofectamine 3000. Subsequently, the cells were collected and subjected to western blot to confirm GPX4 expression in the transfected cells.
Coimmunoprecipitation
A coimmunoprecipitation assay was performed to verify the interaction between GPX4 and HSC70 or Lamp-2a. GPX4-overexpressing U251 cells were treated with PLB (6 μM), BafA1 (100 nM), and PLB (6 μM) + Baf-A1 (100 nM) for 48 h and then lysed. The cell lysates were collected and incubated with anti-GPX4 antibody for 1 h. Then, protein A/G agarose beads were added to each tube and incubated with cell lysates at 4 °C for 2 h. After the beads were washed, an immunocomplex was eluted from the beads by using a loading buffer. Then, western blot was performed to detect the expression levels of GPX4, HSC70, and Lamp-2a in the immunocomplex and input samples. The interaction between NQO1 and HSC70 or P53 in U251 and U87 cells with or without PLB treatment was detected using similar method.
Statistical analysis
Results were expressed as mean ± S.D. of at least three independent experiments and technical replicates per experiment (n values in the corresponding figure legends). One-way analysis of variance was performed to analyse the differences between groups by using GraphPad Prism 6.0. Student’s t test was conducted to compare the two groups. Differences were considered statistically significant when *P < 0.05, **P < 0.01, ***P < 0.005, or ****P < 0.001.
Results
PLB showed higher BBB penetration ability in vitro
The co-culture system of rat BMECs and astrocytes in Transwell inserts was established (Fig. S1A). Only the inserts with TEER values of >300 Ω/cm2 were used for the BBB penetration assay (Fig. S1B). The nontoxic concentrations of PLB, lapachol, and shikonin were chosen for the BBB penetration assay based on the MTS results shown in Supplemental Fig. S1C–E. The HPLC chromatogram of PLB, lapachol, and shikonin is presented in Fig. S1F. The standard curves for calculating Papp of PLB, lapachol, and shikonin in the in vitro BBB cell model are shown in Table S3. The BBB penetration ability of PLB was higher than that of lapachol and shikonin (**P < 0.01). In particular, PLB, lapachol, and shikonin had Papp of 24.56 × 10−9, 7.42 × 10−9, and 9.53 × 10−9 cm/s, respectively (Fig. 1b and Table S3).
PLB significantly inhibited the proliferation of glioma cells
IC50 values of PLB in different cell lines are shown in Fig. 1c. The inhibitory effects of PLB on glioma cell lines (U251, U87, C6, and GL261) were stronger than those on other cancer cell lines (HCT116, A549, Panc28, and HepG2) and normal cell lines (L02 and HUVEC). Further evaluation showed that PLB dose dependently and time dependently inhibited the proliferations of U251, U87, C6, and GL261 glioma cells (Fig. 1d). The calculated IC50 values of PLB in these cell lines at the time points of 12, 24, 48, and 72 h are shown in Table S4. As shown in Fig. S1G, the cells in the control group grew normally in an adherent way. After the PLB treatment, the cell shapes were obviously changed, and the number of cells decreased dose dependently.
PLB significantly inhibited glioma growth in vivo
Schematic chart of establishment of rat C6 glioma model and drug treatment is shown in Fig. 1e. The tumour volumes of the rats in PLB (2.5 and 5.0 mg/kg)-treated groups and TMZ (2.5 mg/kg)-treated group significantly decreased compared with those in the control group (*P < 0.05, ***P < 0.005; Fig. 1f). The body weights had no significant differences among the groups (P > 0.05; Fig. 1g). HE staining result showed that the glioma cells in the tumour areas of the control group grew diffusely without an obvious boundary, whereas agglomerate growth with an obvious boundary could be observed in the PLB- or TMZ-treated groups (Fig. S2A). Immunohistochemical staining results revealed a positive GFAP expression in the tumour areas of all the groups, confirming that the tumour tissues had an astrocytic origin (Fig. S2B). The Ki67 expression significantly decreased in the PLB- or TMZ-treated groups compared with that in the control group (***P < 0.005; Fig. S2B, C).
The anti-glioma effects of PLB were also evaluated using a xenograft glioma in a nude mouse model. In Figs. 2h and S2D, the inhibitory effects of PLB on tumour growth were stronger than those of TMZ (*P < 0.05) and the control group (***P < 0.005). The body weights did not differ among the three groups (P > 0.05; Fig. 2i). HE staining and immunohistochemical staining for GFAP confirmed the astrocytic origin of the tumours (Fig. S2E, F). In Fig. S2E, the tumour tissues in the PLB- and TMZ-treated groups showed a decreased cancer cell density, an obvious pyknotic nucleus, and cell rupture compared with those in the control group. Immunohistochemical staining result revealed that the positive staining rates of Ki67 in the tumour tissues of PLB- and TMZ-treated groups significantly decreased compared with those in the control group (***P < 0.005; Fig. S2F, G). These results suggested that PLB elicited strong anti-glioma effects in vivo without affecting the body weights of the animals.
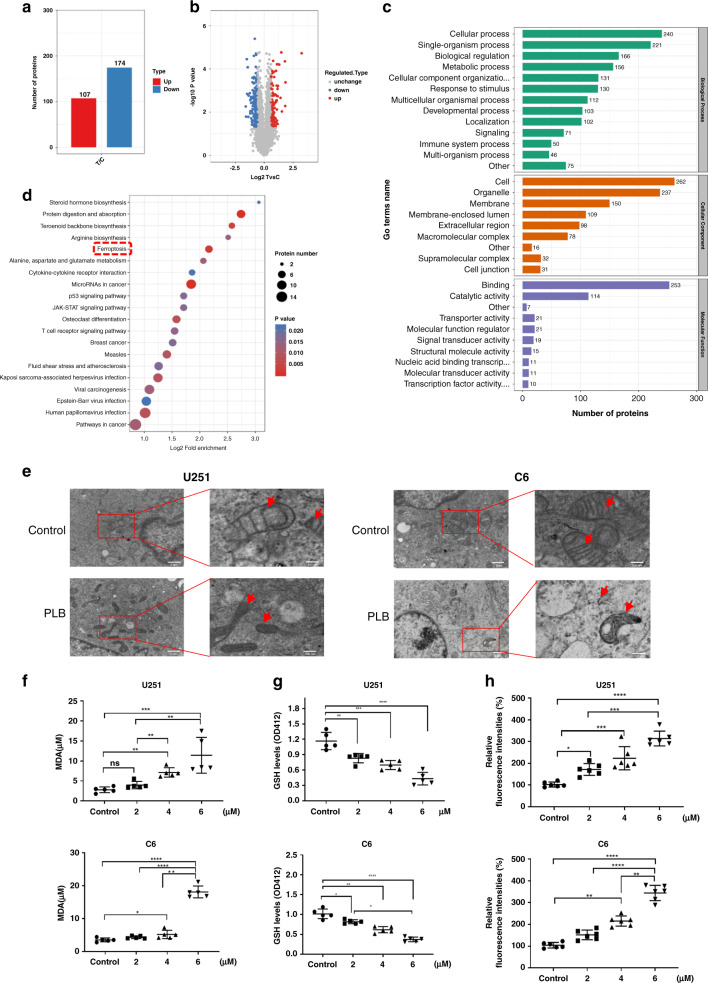
Fig. 2. PLB induced ferroptosis in glioma cells.
a Label-free quantitative proteomic analysis of the number of differentially expressed proteins in the control and PLB-treated cells (n = 3). b Volcano plot of differentially expressed proteins. The threshold of the differential expression change was set at 1.5 times, and P < 0.05 was set as the significance threshold. c Differentially expressed proteins were classified into three main categories (cellular component, biological process and molecular function) via GO annotation. d Bubble diagram of KEGG enrichment results of differentially expressed proteins. e Transmission electronic microscopy images of U251 and C6 cells with or without 4 μM PLB treatment (n = 3). f Malondialdehyde (MDA) levels in U251 and C6 cells with or without 2, 4, and 6 μM PLB treatment (n = 5). g Intracellular GSH levels in U251 and C6 cells treated with 0, 2, 4, and 6 μM PLB (n = 5). h Relative fluorescence intensities of DCFH-DA in U251 and C6 cells treated with 0, 2, 4, and 6 μM PLB (n = 6). All results are expressed as mean ± S.D. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001.
PLB induced ferroptosis-dependent cell death
Label-free quantitative proteomics was performed to identify the differentially expressed proteins in the control and PLB treated cells (Fig. S3A). A total of 6161 proteins were identified, and 5385 of these proteins contained quantitative information (Fig. S3B). Among them, 107 upregulated proteins and 174 downregulated proteins were detected in PLB-treated cells compared with those in the control cells (Fig. 2a). The volcano plot of differentially expressed proteins is illustrated in Fig. 2b. The proteins were categorised into three main categories (cellular component, biological process, and molecular function) via GO annotation (Fig. 2c).
The top 20 pathways most significantly changed were displayed on a bubble map based on KEGG pathway enrichment analysis (Fig. 2d). Ferroptosis was found to be one of the most significantly changed pathways. The heatmap of the differentially expressed proteins based on KEGG pathway analysis also revealed that ferroptosis was one of the most significantly changed pathways (top fourth; Fig. S3C). Since ferroptosis was closely related to cancer therapy, it might act as the main mechanism of PLB-induced cell death.
Transmission electronic microscopy was used to confirm PLB-induced ferroptosis, and the results showed that PLB treatment resulted in obvious morphological features of ferroptosis in U251 and C6 cells. PLB-treated cells exhibited vesicle formation, shrinking mitochondria, or even the disappearance of the mitochondrial ridge (Fig. 2e). Subsequently, the lipid peroxidation levels in the PLB-treated cells were examined by measuring MDA, an important indicator of lipid peroxidation. The results showed that PLB treatment significantly increased the MDA contents in dose-dependent manners in U251 and C6 cells (*P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001; Fig. 2f). The intracellular GSH detection result showed that PLB dose dependently decreased the intracellular GSH levels in U251 and C6 cells (*P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001; Fig. 2g). Also, PLB dose dependently decreased the relative fluorescence intensities of DCFH-DA (*P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001; Fig. 2h), indicating that the intracellular ROS levels were significantly increased in U251 and C6 cells.
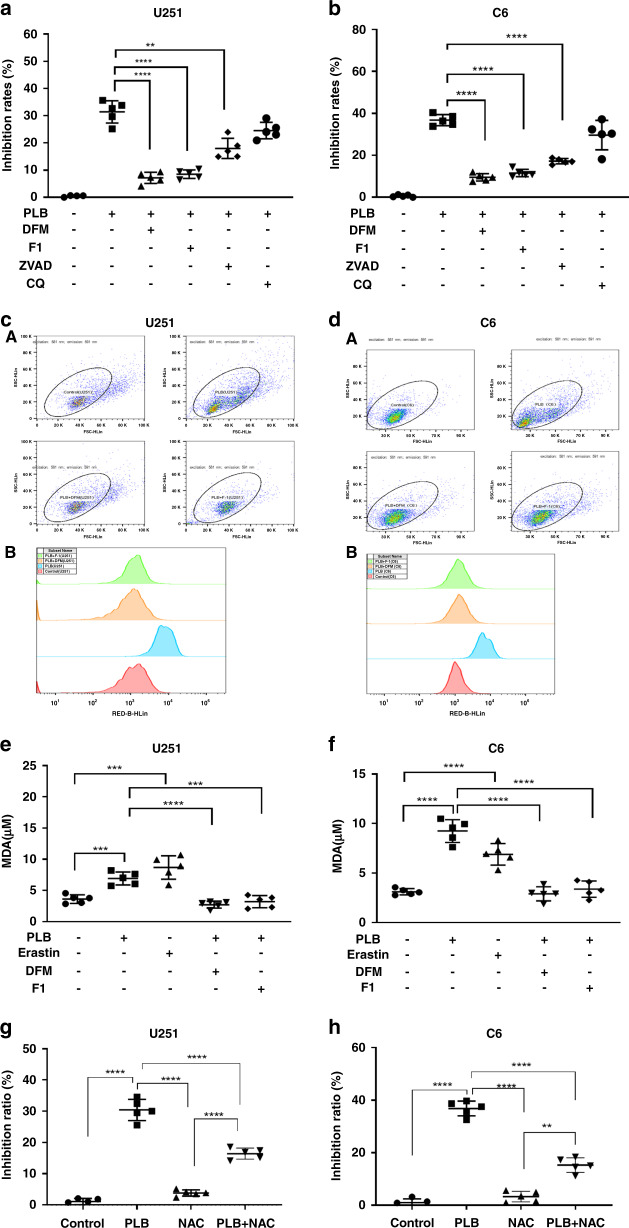
Apoptosis inhibitor (Z-VAD-FMK, ZVAD), autophagy inhibitor (chloroquine (CQ)), and two ferroptosis inhibitors (DFM and F1) were used to verify whether PLB-induced cell death was dependent on ferroptosis. Of note, only nontoxic concentrations of ZVAD, CQ, F1, and DFM were used (Fig. S4). The results showed that ZVAD, DFM, and F1 significantly rescued the growth inhibitory effects of PLB on C6 and U251 cells (**P < 0.01 and ****P < 0.001, Fig. 3a, b). Also, the rescue effects of ZVAD were weaker than that of DFM and F1.
Fig. 3. PLB induced ferroptosis-dependent cell death.
a, b Inhibitory effects of PLB on U251 and C6 cells in the presence of ferroptosis inhibitors (10 μM DFM and 1.25 μM F-1), apoptosis inhibitor (2 μM ZVAD), and autophagy inhibitor (10 μM CQ) via the MTS assay (n = 5). cA, dA Fluorescence intensities of C11-BODIPY581/591 in PLB treated U251 and C6 cells in the presence of ferroptosis inhibitors (10 μM DFM and 1.25 μM F-1) detected via flow cytometry. cB, dB Quantification of the fluorescence intensities detected with flow cytometry in cA, dA, respectively (n = 3). e, f MDA levels observed through the TBARS assay in U251 and C6 cells treated with 4 μM PLB in the presence of ferroptosis inhibitors (10 μM DFM and 1.25 μM F-1) (n = 5). g, h Inhibitory effects of PLB on U251 and C6 cells in the presence of NAC (10 μM) (n = 5). Results are expressed as mean value ± S.D. **P < 0.01, ***P < 0.005, and ****P < 0.001 versus the control group.
Lipid peroxidation levels and MDA accumulation were also determined in the cells treated with PLB in the presence of DFM or F1. In Fig. 3c, d, PLB treatment induced the significant enhancement of fluorescence intensities of C11-BODIPY581/591 in C6 and U251 cells, and the enhanced fluorescence intensities significantly decreased in the presence of DMF or F1. Furthermore, DFM and F1 significantly rescued the increased MDA levels induced by PLB (***P < 0.005 and ****P < 0.001, Fig. 3e, f). These results suggested that PLB induced ferroptosis-dependent cell death in C6 and U251 glioma cells.
To verify whether the cell death induced by PLB was dependent on ROS, we determined the effects of PLB on proliferations of C6 and U251 cells in the presence of NAC, a ROS Scavenger. The nontoxic concentration of NAC (10 μM) was used (Supplemental Fig. S5A, B). As shown in Fig. 3g, h, NAC (10 μM) significantly rescued the growth inhibitory effects of PLB on U251 and C6 cells (****P < 0.001). The result indicated that PLB-induced cell death was dependent on ROS levels.
PLB inhibited cystine-glutamate antiporter (xCT) and glutathione peroxidase 4 (GPX4) expression
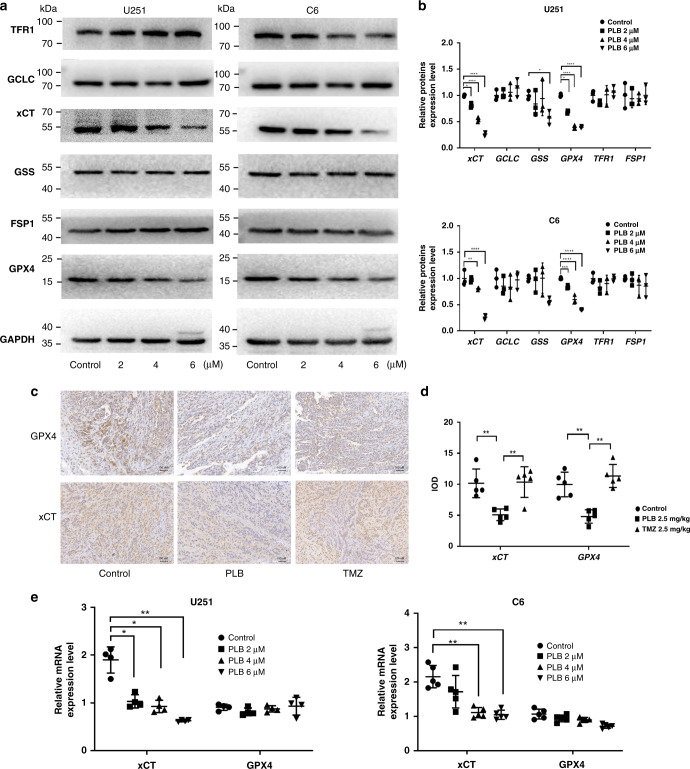
Ferroptosis can be triggered via glutathione biosynthesis, Fe3+ transport, and glutathione-independent pathway, among others [4]. Thus, the expression levels of glutamate cysteine ligase catalytic subunit (GCLC), glutathione synthetase (GSS), xCT, and GPX4 in the glutathione biosynthesis pathway, transferrin receptor protein 1 (TFR1) in the Fe3+ transport pathway, and ferroptosis suppressor protein 1 (FSP1) in the glutathione-independent pathway were determined using western blot. As shown in Fig. 4a, b, the expression levels of xCT and GPX4 were significantly downregulated in the PLB-treated cells compared with that in the control cells (*P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.0001). Significant changes in the protein levels of GCLC, GSS, TFR1, and FSP1 among the groups were not found.
Fig. 4. xCT and GPX4 expression levels decreased in PLB-induced ferroptosis.
a Representative western blot results of ferroptosis-related protein expression levels in U251 and C6 cells treated with PLB. b Quantitation of western blot results of ferroptosis-related proteins. GAPDH served as the loading control (n = 4). c Representative immunohistochemical staining results of xCT and GPX4 in xenograft glioma tissues in nude mice treated with PLB or TMZ. Scale bar, 100 μm. d Statistical analysis of immunohistochemical staining results of xCT and GPX4 (n = 5). e mRNA expression levels of xCT and GPX4 in U251 (n = 4) and C6 cells (n = 5) treated with PLB. The results are expressed as mean value ± S.D. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001 versus the control group.
Immunohistochemical staining was performed to check the GPX4 and xCT expression levels in the tumour tissues of PLB-treated tumour-bearing nude mice. The results showed that the expression levels of xCT and GPX4 decreased significantly in the tumour tissues of mice in the PLB-treated group compared with those in the control group (**P < 0.01, Fig. 4c, d). RT-PCR results demonstrated that PLB dose dependently decreased the mRNA levels of xCT in C6 and U251 cells (*P < 0.05 and **P < 0.01), but no significant changes in the mRNA levels of GPX4 among the groups were found (P > 0.05, Fig. 4e).
PLB induced GPX4-dependent cell death by causing GPX4 degradation
GPX4-overexpressing cells (U251 and U87 cells) were constructed to examine the effect of GPX4 in PLB-induced cell death (Fig. 5a, b). In Fig. 5c, d, GPX4 overexpression rescued the growth inhibitory effects of PLB on U251 and U87 cells (*P < 0.05 and **P < 0.01). Similar results were found in U87 cells. These results suggested that PLB induced cell death via a GPX4-dependent pathway.
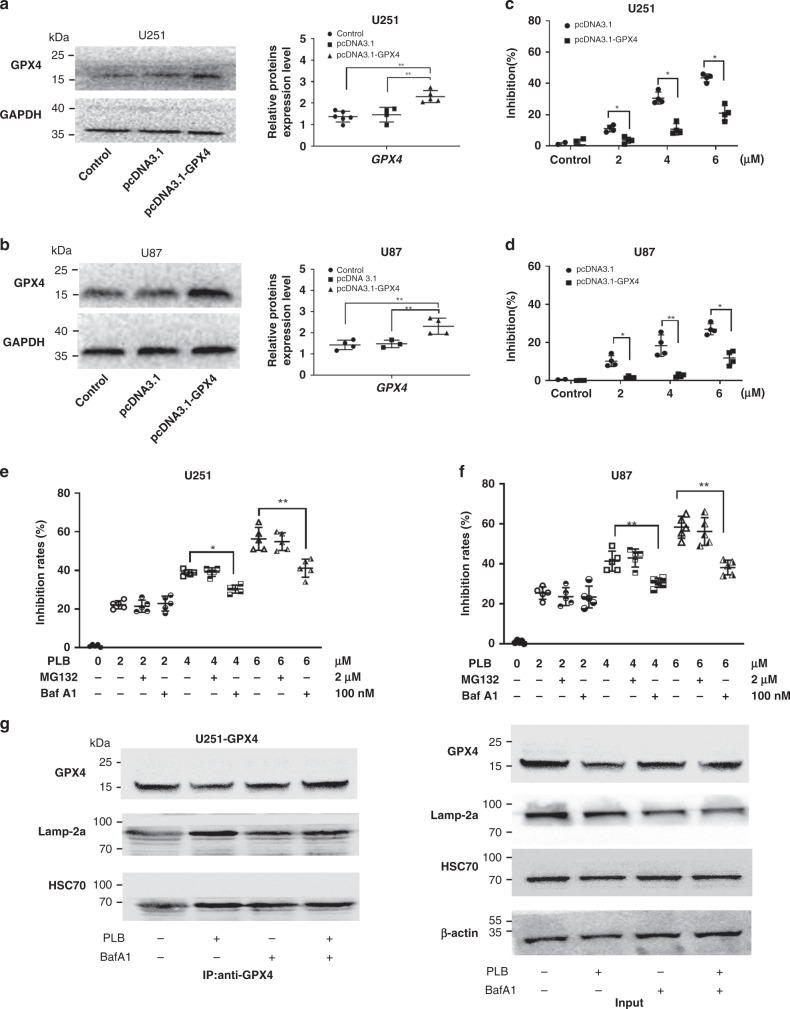
Fig. 5. PLB induced GPX4-dependent cell death by causing GPX4 degradation.
a, b Western blot and quantitation results of the GPX4 expression in GPX4 plasmid-transfected U87 (n = 4) and U251 cells (n = 5). c, d Inhibitory effects of PLB on GPX4 overexpressed and vector control U87 and U251 cells via the MTS assay (n = 4). e, f Cell viabilities of U251 and U87 cells treated with PLB in the presence of MG132 (2 μM) or Baf-A1 (100 nM) for 48 h (n = 5). g Coimmunoprecipitation results of GPX4 and HSC70 or Lamp-2a in GPX4-overexpressing U251 cells treated with 6 μM PLB in the presence of Baf-A1 (100 nM). Cell lysates were immunoprecipitated with anti-GPX4 antibody and subjected to western blot with the indicated antibodies. *P < 0.05 and **P < 0.01, versus the control group.
PLB treatment resulted in the downregulation of GPX4 protein levels without affecting its mRNA levels (Fig. 4a–e), as such, PLB might induce the protein degradation of GPX4. The effects of proteasome inhibitor (MG-132) and lysosome inhibitor (Bafilomycin A1 (Baf-A1)) on PLB-induced cell death were detected. The results showed that only Baf-A1 could rescue PLB-induced cell death in U251 and U87 cells (Fig. 5e, f), indicating that PLB might cause GPX4 degradation via the lysosome pathway. Lamp-2a and HSC70 are required for GPX4 protein degradation through the lysosome pathway [18], so a coimmunoprecipitation assay was performed in this study to detect the effect of PLB on the interaction between GPX4 and Lamp-2a or HSC70. In Fig. 5g, the levels of HSC70 and Lamp-2a binding to GPX4 were increased by PLB, whereas the increased binding decreased in the presence of Baf-A1 in GPX4-overexpressing U251 cells. Western blot results of the input proteins demonstrated that PLB led to a decreased GPX4 expression, and the changed levels of GPX4 could be rescued by the addition of Baf-A1 (Fig. 5g).
PLB induced cell death by targeting NQO1
A total of 153 PLB-related predictive targets were identified from the PharmMapper database (Table S5). Then, STRING database 11.0 was used to construct the PPI network of the experimentally confirmed ferroptosis targets (xCT and GPX4) and PLB-related predictive targets. In Fig. 6a, 15 PLB-related predictive targets were found to be functionally related to xCT or GPX4. Among them, NQO1 interacted with xCT and GPX4 and played a significant role in PLB-induced ferroptosis. Molecular docking study was then performed to verify the binding ability of PLB to NQO1. PLB mainly formed hydrogen bonds with GLU-117 and TRP-105 in NQO1 (Fig. 6b). The binding energy calculation indicated that the binding energy of PLB-NQO1 (binding energy of −6.14 kcal/mol) was similar to that of PLB-β-lapachone (binding energy of −6.75 kcal/mol).
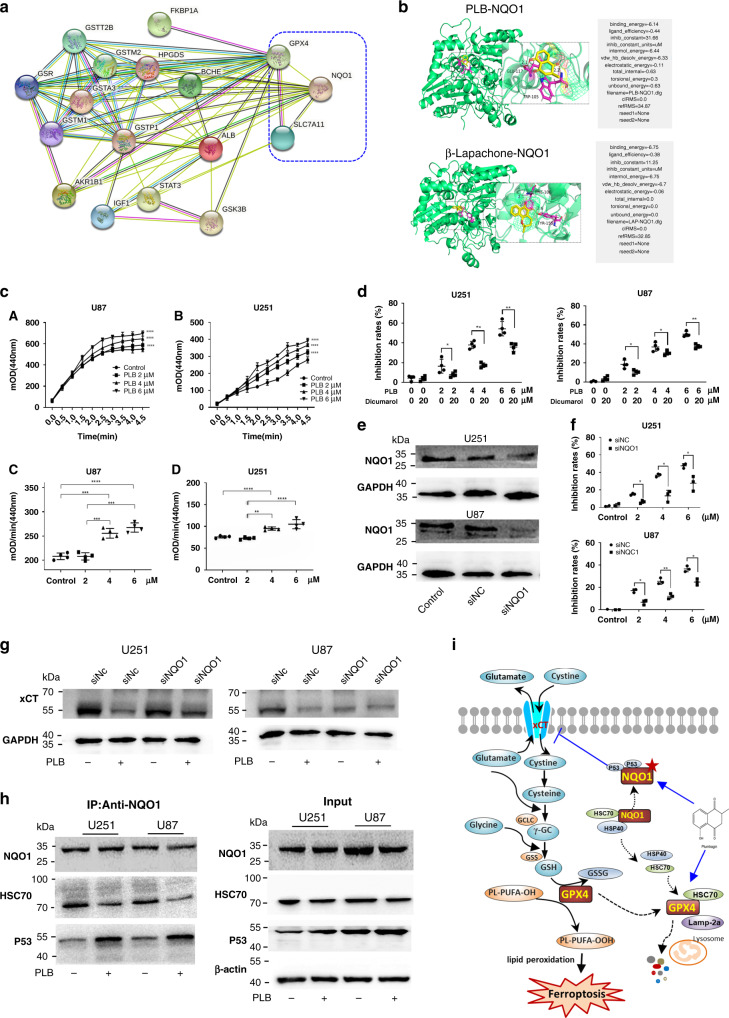
Fig. 6. PLB induced ferroptosis by targeting NQO1.
a Functional protein association network of GPX4, xCT, and PLB-related predictive targets by using the STRING database 11.0 (http://string-db.org/). PLB-related predictive targets were identified from the PharmMapper database (http://www.lilab-ecust.cn/pharmmapper/). b Molecular docking result for the binding of PLB to NQO1. β-Lapachone was used as the positive control. cA, cB The OD dynamic change curve of U251 and U87 cells with or without PLB treatment (n = 4). cC, cD Quantitation of NQO1 activity in U251 and U87 cells with or without PLB treatment (n = 4). d Cell viabilities of U251 and U87 cells treated with PLB in the presence of dicumarol (20 μM) for 48 h (n = 4). e Western blot results of NQO1 expression in siNQO1- or siNC-transfected U87 and U251 cells (n = 4). f Inhibitory effects of PLB on siNQO1- or siNC-transfected U87 and U251 cells via the MTS assay (n = 3). g Western blot results of xCT expression levels in siNQO1- or siNC-transfected U87 and U251 cells with or without 6 μM PLB treatment (n = 3). h Coimmunoprecipitation results of NQO1 and p53 or HSC70 in U251 and U87 cells treated with or without 6 μM PLB (n = 3). i Schematic of PLB-induced ferroptosis in glioma cells. Results are expressed as mean ± S.D. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001.
The effects of PLB on NQO1 activities were examined by using an NQO1 activity assay kit. The results showed that PLB treatment significantly increased NQO1 activities in U251 and U87 cells (**P < 0.01, ***P < 0.005, and ****P < 0.001; Fig. 6c), suggesting that PLB might serve as a substrate for NQO1. The growth inhibitory effects of PLB on U251 and U87 cells significantly decreased in the presence of dicumarol, a known inhibitor of NQO1 (Fig. 6d, *P < 0.05 and **P < 0.01). NQO1 was downregulated using the siRNA approach in U251 and U87 cells to further analyse the effect of NQO1 on PLB-induced cell death (Fig. 6e and Supplemental Fig. S6A, B, *P < 0.05 and ***P < 0.005). In Fig. 6f, the knockdown of NQO1 rescued the growth inhibitory effects of PLB on U251 and U87 cells (*P < 0.05 and **P < 0.01). Similar results were found in U87 cells. These results suggested that PLB induced cell death via an NQO1-dependent pathway.
NQO1 could interact with p53 and protect p53 from degradation, which sensitised cells to ferroptosis by reducing the xCT expression [19]. Thus, the effects of PLB on xCT expression in NQO1 knocked down cells and vector control cells were detected. In Fig. 6g and Supplemental Fig. S6C, D, PLB significantly decreased the xCT expression in vector control cell lines, whereas the knockdown of NQO1 significantly rescued the decreased xCT expression by PLB (**P < 0.01 and ***P < 0.005). This result indicated that PLB decreased the xCT expression via an NQO1-dependent manner.
A coimmunoprecipitation assay was performed to detect the effect of PLB on the interaction between NQO1 and p53. As shown in Fig. 6h, the amount of p53 binding to NQO1 was significantly increased by PLB in U251 and U87 cells. These results suggested that PLB promoted the interaction between NQO1 and p53 to protect p53 from degradation, which reduced the expression of xCT. In normal conditions, NQO1 can form stable complex with HSC70. Coimmunoprecipitation assay showed that the amount of HSC70 binding to NQO1 was obviously decreased by PLB in U251 and U87 cells (Fig. 6h). Taken together, PLB markedly decreased the binding of NQO1 to HSC70, which then interacted with GPX4 to promote the degradation of GPX4 through the lysosome pathway.
Discussion
Natural products have long been used to treat central nervous system diseases, including glioma. Many naturally derived naphthoquinones have great potentials to be developed as anti-glioma drugs [8, 9]. Our study focussed on the anti-glioma effects of PLB, a natural naphthoquinone isolated from the root of the plant P. zeylanicas L.[10]. The BBB penetration ability of PLB in vitro was relatively higher than that of its structural analogues, namely, lapachol and shikonin. PLB showed strong growth inhibitory effects on glioma cell lines, xenograft glioma in nude mice, and orthotopic implanted glioma in Wistar rats. These results suggested that PLB had the potential to be developed as a promising anti-glioma drug.
With the recent progress in proteomic methodologies, the application of proteomics in the drug discovery process has widely increased [20]. In our study, label-free quantitative proteomics was first applied to explore the anti-glioma mechanism of PLB, and the results showed that ferroptosis was the main mechanism of PLB-induced cell death. Furthermore, a variety of experiments confirmed that PLB induced ferroptosis-dependent cell death. Also, apoptosis inhibitor (ZVAD) showed some rescue effects after PLB treatment (Fig. 3a), while autophagy inhibitor (CQ) showed no significant rescue effects. In fact, it was reported that PLB induced apoptosis of many kinds of tumour cells [11]. Our result showed that the rescue effects of two ferroptosis inhibitors (DFM and F1) on PLB-induced cell death were stronger than that of apoptosis inhibitor ZVAD, indicating that ferroptosis might play more important role in PLB-induced cell death.
Ferroptosis has emerged as an iron-dependent oxidative form of cell death that differs from apoptosis, necrosis, and autophagy [21, 22]. Treatments based on triggering ferroptosis show potential application in cancer therapy [23]. The central nervous system contains the highest levels of polyunsaturated fatty acids (PUFAs) in the body, and the peroxidation of PUFAs drives ferroptosis; as such, brain tumours are more sensitive to ferroptosis induction [24]. Therefore, triggering ferroptosis is a promising strategy of glioma treatment.
Advancements have been achieved in studies on the molecular mechanism of ferroptosis in recent years [21, 22]. Cystine-glutamate antiporter xCT exports intracellular glutamate with extracellular cystine, a precursor of cysteine for GSH synthesis with the help of GCL and GSS. Ferroptosis requires the depletion of GSH or the inactivation of GPX4, triggering the accumulation of lipid ROS and the peroxidation of PUFAs. Disruption in iron homoeostasis is also essential for toxic lipid peroxidation to cause ferroptosis [22]. Iron is regulated by proteins such as ferritin and TFR1, which are also implicated in ferroptosis regulation. Additionally, FSP1 is a newly found glutathione-independent ferroptosis suppressor [25]. Our results showed that PLB treatment significantly decreased the protein and mRNA levels of xCT and the protein levels of GPX4.
PharmMappper database was applied to predict PLB-binding targets in the ferroptosis pathway. A total of 153 PLB-related predictive targets were identified from the PharmMapper database. Among them, NQO1 likely participated in PLB-induced ferroptosis. Natural naphthoquinone β-lapachone has been shown to be a substrate for NQO1, generating a futile redox cycle, wherein quinone and hydroquinone form a redox cycle with a net concomitant loss of reduced NAD(P)H [26]. Our results demonstrated that PLB treatment significantly increased NQO1 activities in U251 and U87 cells. The growth inhibitory effects of PLB were influenced by NQO1, and suppressing (with dicumarol) or eliminating (NQO1 knockdown) NQO1 dramatically reduced the growth inhibitory effects of PLB (Fig. 6). Thus, as a structural analogue of β-lapachone, PLB was possibly an NQO1 bioactivatable drug.
The roles of NQO1 in cancer are well established and documented [27, 28]. NQO1 has been proposed as a potential therapeutic target for many different cancer types. NQO1 likely plays an important role in the brain because of the high PUFA levels, high oxygen consumption, and low antioxidant enzyme levels in the brain [28]. NQO1 binds to and stabilises p53, which sensitise cells to ferroptosis by reducing xCT expression [19]. Our results indicated that PLB decreased the xCT expression via promoting the interaction between NQO1 and p53 to protect p53 from degradation.
GPX4 is regarded as a central regulator of ferroptosis-induced cell death [29]. Our results showed that GPX4 overexpression significantly rescued PLB-induced cell death, indicating that GPX4 participated in PLB-induced ferroptosis. Since PLB treatment resulted in the downregulation of GPX4 protein levels without affecting its mRNA levels, the decreased GPX4 protein levels induced by PLB were mediated by post-transcriptional events. Ubiquitin proteasome system and autophagy lysosome pathway are key mechanisms of protein degradation. Further studies involving a proteasome inhibitor (MG-132) and a lysosome inhibitor (Baf-A1) have showed that PLB may cause GPX4 degradation via the lysosome pathway. Lamp-2a acts as a receptor to transport substrates into the lysosome matrix for degradation, and HSC70 is responsible to recognise and bind to substrates to deliver them to the lysosomal membrane [30, 31]. Our immunoprecipitation result revealed that PLB treatment increased the interactions between GPX4 and HSC70 or Lamp-2a to promote the degradation of GPX4 through lysosome pathway (Fig. 5). NQO1 can form stable complex with HSC70 in normal conditions. In the presence of PLB, the amount of HSC70 binding to NQO1 was obviously decreased in U251 and U87 cells (Fig. 6h). Taken together, we concluded that PLB markedly decreased the binding of NQO1 with HSC70, which then interacted with GPX4 to promote the degradation of GPX4 through the lysosome pathway.
Conclusion
As a novel form of regulated cell death, ferroptosis has become an attractive target for cancer therapy. Ferroptosis inducers are generally divided into two classes: xCT inhibitors (e.g. Erastin, sorafenib, sulfasalazine) and GPX4 inhibitors (RSL3 and FIN56) [21, 29]. NQO1 is a promising target for cancer treatment. Our study was the first to report that PLB induced ferroptosis by dual targeting of NQO1 and GPX4. PLB can interact with NQO1, increase NQO1 activity, and decrease xCT expression levels. It can also cause the GPX4 degradation via the lysosome pathway (Fig. 6i). These results suggested that PLB had the potential to be developed as a novel ferroptosis inducer for cancer treatment.
Supplementary information
Acknowledgements
We thank Mrs. Yang Hui in core facility centre of Capital Medical University for their technique assistance in transmission electron microscopy analysis.
Author contributions
Conceptualisation, H-lX, X-kL, MX; methodology, SZ, LL, R-rM, X-qW, S-sP; formal analysis, SZ, H-lX; writing—original draft preparation, SZ, LL; writing—review and editing, H-lX, X-kL, MX; funding acquisition, H-lX, X-kL. All authors have approved the final article.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Nos. 81774191 and 82174265), the International Collaborative of Ministry of Science and Technology (No. 2017YEE0915000), and National Science Foundation of Shandong Province in China (Nos. ZR201911020118 and ZR201911110299).
Data availability
Data are also available as Supplementary Information.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All animal experiments were approved by the Committee of Animal Experiments and Experimental Animal Welfare of Capital Medical University in Beijing, China (Nos. AEEI-2019078 and 37363) and performed in accordance with the National Institutes of Health guide for the care and use of laboratory animals (NIH Publications No. 8023, revised 1978).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01800-y.
References
- 1.Herrlinger U, Tzaridis T, Mack F, Steinbach JP, Schlegel U, Sabel M, et al. Lomustine-temozolomide combination therapy versus standard temozolomide therapy in patients with newly diagnosed glioblastoma with methylated MGMT promoter (CeTeG/NOA-09): a randomised, open-label, phase 3 trial. Lancet. 2019;393:678–88. doi: 10.1016/S0140-6736(18)31791-4. [DOI] [PubMed] [Google Scholar]
- 2.Hu Z, Mi Y, Qian H, Guo N, Yan A, Zhang Y, et al. A potential mechanism of temozolomide resistance in glioma–ferroptosis. Front Oncol. 2020;10:897. doi: 10.3389/fonc.2020.00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang WS, Kim KJ, Gaschler MM, Patel M, Shchepinov MS, Stockwell BR. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci USA. 2016;113:E4966–75. doi: 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiland A, Wang Y, Wu W, Lan X, Han X, Li Q, et al. Ferroptosis and its role in diverse brain diseases. Mol Neurobiol. 2019;56:4880–93. doi: 10.1007/s12035-018-1403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu Y, Wang J, Ray PS, Guo H, Huang J, Shin-Sim M, et al. Thioredoxin-like 2 regulates human cancer cell growth and metastasis via redox homeostasis and NF-κB signaling. J Clin Investig. 2011;121:212–25. doi: 10.1172/JCI43144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuzbasioglu Baran M, Guvenalp Z, Saracoglu I, Kazaz C, Salih B, Demirezer LO, et al. Cytotoxic naphthoquinones from Arnebia densiflora (Nordm.) Ledeb and determining new apoptosis inducers. Nat Prod Res. 2020;34:1669–77. doi: 10.1080/14786419.2018.1525714. [DOI] [PubMed] [Google Scholar]
- 7.Aminin D, Polonik S. 1,4-Naphthoquinones: some biological properties and application. Chem Pharm Bull. 2020;68:46–57. doi: 10.1248/cpb.c19-00911. [DOI] [PubMed] [Google Scholar]
- 8.Redaelli M, Mucignat-Caretta C, Isse AA, Gennaro A, Pezzani R, Pasquale R, et al. New naphthoquinone derivatives against glioma cells. Eur J Med Chem. 2015;96:458–66. doi: 10.1016/j.ejmech.2015.04.039. [DOI] [PubMed] [Google Scholar]
- 9.Xu H, Chen Q, Wang H, Xu P, Yuan R, Li X, et al. Inhibitory effects of lapachol on rat C6 glioma in vitro and in vivo by targeting DNA topoisomerase I and topoisomerase II. J Exp Clin Cancer Res. 2016;35:178. doi: 10.1186/s13046-016-0455-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin Z, Zhang J, Chen L, Guo Q, Yang B, Zhang W, et al. Anticancer effects and mechanisms of action of plumbagin: review of research advances. Biomed Res Int. 2020;2020:6940953. doi: 10.1155/2020/6940953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao YY, Yu J, Liu TT, Yang K, Yang L, Chen Q, et al. Plumbagin inhibits the proliferation and survival of esophageal cancer cells by blocking STAT3-PLK1-AKT signaling. Cell Death Dis. 2018;9:17. doi: 10.1038/s41419-017-0068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang Y, Zhou R, Liang X, Kong X, Yang B. Pharmacological targets and molecular mechanisms of plumbagin to treat colorectal cancer: a systematic pharmacology study. Eur J Pharmacol. 2020;881:173227. doi: 10.1016/j.ejphar.2020.173227. [DOI] [PubMed] [Google Scholar]
- 13.Li N, Ou J, Bao N, Chen C, Shi Z, Chen L, et al. Design, synthesis and biological evaluation of novel plumbagin derivatives as potent antitumor agents with STAT3 inhibition. Bioorg Chem. 2020;2020:104208. doi: 10.1016/j.bioorg.2020.104208. [DOI] [PubMed] [Google Scholar]
- 14.Yang T, Zang D, Shan W, Guo A, Wu J, Wang YJ, et al. Synthesis and evaluations of novel apocynin derivatives as anti-glioma agents. Front Pharmacol. 2019;10:951. doi: 10.3389/fphar.2019.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu L, Zhan S, Liu X, Zhao X, Lin X, Xu H. Antitumor effects and the compatibility mechanisms of herb pair Scleromitrion diffusum (Willd.) R. J. Wang–Sculellaria barbata D. Don. Front Pharmacol. 2020;11:292. doi: 10.3389/fphar.2020.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li FD, Yang YC, Li Y, Yang H, Wang H. Quantitative analysis of the global proteome in peripheral blood mononuclear cells from patients with new-onset psoriasis. Proteomics. 2018;18:e1800003. doi: 10.1002/pmic.201800003. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Shen Y, Wang S, Li S, Zhang W, Liu X, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45:W356–360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Z, Geng Y, Lu X, Shi Y, Wu G, Zhang M, et al. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc Natl Acad Sci USA. 2019;116:2996–3005. doi: 10.1073/pnas.1819728116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song X, Long D. Nrf2 and ferroptosis: a new research direction for neurodegenerative diseases. Front Neurosci. 2020;14:267. doi: 10.3389/fnins.2020.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu K, Wang Z, Wu Z, Tan H, Mishra A, Peng J. High-throughput profiling of proteome and posttranslational modifications by 16-Plex TMT labeling and mass spectrometry. Methods Mol Biol. 2021;2228:205–24. doi: 10.1007/978-1-0716-1024-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battaglia AM, Chirillo R, Aversa I, Sacco A, Costanzo F, Biamonte F. Ferroptosis and cancer: mitochondria meet the “iron maiden” cell death. Cells. 2020;9:1505. doi: 10.3390/cells9061505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conrad M, Pratt DA. The chemical basis of ferroptosis. Nat Chem Biol. 2019;15:1137–47. doi: 10.1038/s41589-019-0408-1. [DOI] [PubMed] [Google Scholar]
- 23.Zou Y, Palte MJ, Deik AA, Li H, Eaton K, Wang W, et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun. 2019;10:1617. doi: 10.1038/s41467-019-09277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexiou GA, Gerogianni P, Vartholomatos E, Kyritsis AP. Deferiprone enhances temozolomide cytotoxicity in glioma cells. Cancer Invest. 2016;34:489–95. doi: 10.1080/07357907.2016.1233424. [DOI] [PubMed] [Google Scholar]
- 25.Doll S, Freitas FP, Shah R, Aldrovandi M, da Silva MC, Ingold I, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–8. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 26.Gong Q, Hu J, Wang P, Li X, Zhang X. A comprehensive review on beta-lapachone: mechanisms, structural modifications, and therapeutic potentials. Eur J Med Chem. 2021;210:112962. doi: 10.1016/j.ejmech.2020.112962. [DOI] [PubMed] [Google Scholar]
- 27.Li X, Liu Z, Zhang A, Han C, Shen A, Jiang L, et al. NQO1 targeting prodrug triggers innate sensing to overcome checkpoint blockade resistance. Nat Commun. 2019;10:3251. doi: 10.1038/s41467-019-11238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang K, Chen D, Ma K, Wu X, Hao H, Jiang S. NAD(P)H:Quinone oxidoreductase 1 (NQO1) as a therapeutic and diagnostic target in cancer. J Med Chem. 2018;61:6983–7003. doi: 10.1021/acs.jmedchem.8b00124. [DOI] [PubMed] [Google Scholar]
- 29.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–5. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 31.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–3. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are also available as Supplementary Information.