Abstract
Hypertension is a common bevacizumab-induced toxicity. No markers are available to predict patients at risk of developing hypertension. We hypothesized that genetic risk of essential hypertension, as measured by a blood pressure polygenic risk score (PRS), would be associated with risk of severe bevacizumab-induced hypertension. PRS were calculated for 1,027 bevacizumab-treated cancer patients of European descent from four clinical trials (Alliance/CALGB 80303, 40503, 90401, 40502) using summary systolic (SBP) and diastolic (DBP) genome-wide association results obtained from 757,601 European descent individuals. The association between PRS and grade 3 bevacizumab-induced hypertension (CTCAE v3) in each trial was performed by multivariable logistic regression. Fixed-effect meta-analyses odds ratios (OR) per standard deviation (SD) of the association of PRS (quantitative) and hypertension across trials were estimated by inverse-variance weighting. PRS were additionally stratified into quintiles, with the bottom quintile as the referent group. OR of the association between hypertension and each quintile versus the referent group was determined by logistic regression. The most significant PRS (quantitative)-hypertension association included up to 67 SNPs associated with SBP (p=0.0077, OR per SD=1.31, 95% CI: 1.07–1.60), and up to 53 SNPs associated with DBP (p=0.0209, OR per SD=1.27, 95% CI: 1.04–1.56). Patients in the top quintile had a higher risk of developing bevacizumab-induced hypertension compared to patients in the bottom quintile using SNPs associated with SBP (p=4.75×10−4, OR=3.72, 95% CI: 1.84–8.16) and DBP (p=0.076, OR=1.83, 95% CI: 0.95–3.64). Genetic variants associated with essential hypertension, mainly SBP, increase the risk of severe bevacizumab-induced hypertension.
Keywords: bevacizumab, hypertension, polygenic risk score, blood pressure
Introduction
Bevacizumab is a VEGF-pathway inhibitor approved in the US and Europe to treat numerous cancers, including metastatic colorectal cancer, metastatic breast cancer, unresectable advanced, metastatic or recurrent non-small cell lung cancer, advanced or metastatic renal cell cancer, recurrent glioblastoma, advanced ovarian and cervical cancers, and hepatocellular carcinoma 1,2. The antitumor efficacy of bevacizumab relies on direct neutralization of VEGF and hence inhibition of the VEGF-signaling pathway, which is involved in endothelial survival, vascular permeability, and therefore tumor angiogenesis 3.
Hypertension, which can be life-threatening, is one of the most frequent toxicities induced by bevacizumab and other VEGF-pathway inhibitors, with severe cases (grades 3–4) reported in up to 27.6% of patients treated with bevacizumab 4. A meta-analysis showed that bevacizumab use is associated with a higher risk of severe hypertension with a relative risk of 5.28 (95% confidence interval, CI of 4.15–6.71) 5. The primary proposed mechanism of hypertension induced by VEGF-pathway inhibitors involves blockage of the nitric oxide (NO) pathway 6. The VEGF signaling pathway augments the transcription of endothelial NO synthase. The blockage of NO production leads to vasoconstriction, increased vascular tone, and hence hypertension.
There are no markers applied in the clinic to inform decisions for risk assessment of patients before treatment with bevacizumab or any other VEGF-pathway inhibitors. A few studies have reported associations of single nucleotide polymorphisms (SNPs) with hypertension induced by these drugs 7–9, and we have previously discovered and validated a SNP in KCNAB1 associated with the risk of grade ≥2 bevacizumab-induced hypertension in the largest genome-wide association study (GWAS) meta-analysis of bevacizumab-induced hypertension patients to date 4. However, the SNPs previously identified had a relatively small effect size. Polygenic risk scores (PRS) have emerged in recent years as a way of quantifying the combined effect of SNPs on phenotype risk, where PRS are estimated values of an individual genetic liability to a phenotype based on the effect size of a group of genetic variants. This approach is particularly relevant for polygenic traits, such as drug-induced hypertension, for which it is much more likely that a group of genetic variants explains the variance between phenotypes than a single genetic variant.
The calculation of PRS to predict patients at risk of drug-induced toxicities has been essentially unexplored, with the exception of a few studies for drug-induced liver injury 10. Essential hypertension is one of the major modifiable risks of cardiovascular diseases 11 and is likely to share genetic factors that are modifiers of the risk of drug-induced hypertension. From this perspective, essential hypertension and drug-induced hypertension might be considered similar phenotypes (proxy traits). The effect of PRS calculated based on proxy traits has been demonstrated in several studies 12–14. Therefore, herein we report results of the generation of PRS for severe bevacizumab-induced hypertension using the largest cohort of genome-wide data available for bevacizumab treated patients 4 and based on data from the largest GWAS meta-analyses conducted for blood pressure (BP).
Methods
Study population
This study included patients with advanced or metastatic cancer treated with bevacizumab from four phase III clinical trials from the Cancer and Leukemia Group B (CALGB, now part of the Alliance for Clinical Trials in Oncology, Alliance), CALGB 80303, 40503, 90401, and 40502. Only genetically determined European ancestry patients were included, according to the principal component analysis for clustering by Eigenstrat 15.
CALGB 80303, 40503, and 90401 were randomized studies placebo-controlled versus bevacizumab, while CALGB 40502 was randomized to bevacizumab combined with different chemotherapies. CALGB 80303 included patients with advanced pancreatic cancer treated with gemcitabine and 10 mg/kg bevacizumab. CALGB 40503 included patients with hormone receptor-positive advanced-stage breast cancer treated with letrozole and 15 mg/kg bevacizumab. CALGB 90401 included patients with metastatic castration-resistant prostate cancer treated with docetaxel in combination with prednisone and 15 mg/kg bevacizumab. CALGB 40502 included patients with recurrent or metastatic breast cancer treated with paclitaxel, or nab-paclitaxel, or ixabepilone with 10 mg/kg bevacizumab. Additional details on the design of the clinical trials, patient eligibility, clinical baseline characteristics, and treatments are reported in previous publications 4,16–19 and in Table 1.
Table 1. Characteristics of cancer patients treated with bevacizumab included in each clinical trial of the study population.
No grade 4 toxicity was reported. CALGB: Cancer and Leukemia Group B; SD: standard deviation.
| Clinical trial | CALGB 80303 (n=154) |
CALGB 40503 (n=105) |
CALGB 90401 (n=312) |
CALGB 40502 (n=456) |
|---|---|---|---|---|
| Age (mean ± SD) | 64.4 ± 10.5 | 56.9 ± 11.7 | 68.4 ± 8.3 | 57.1 ± 10.6 |
| Male | 90 | 0 | 312 | 0 |
| Female | 64 | 105 | 0 | 456 |
| Cancer type | Advanced pancreatic cancer | Hormone receptor-positive, advanced-stage breast cancer | Metastatic castration-resistant prostate cancer | Recurrent or metastatic breast cancer |
| Drug treatment combined with bevacizumab | Gemcitabine | Letrozole | Docetaxel | Paclitaxel or nab-paclitaxel or ixabepilone |
| Hypertension grade 3 (CTCAE v 3.0) | 19 (12.3%) | 29 (27.6%) | 21 (6.7%) | 49 (10.7%) |
Bevacizumab-induced hypertension
Bevacizumab-induced hypertension was recorded by the CALGB (Alliance) Statistics and Data Center according to pre-specified criteria described in the protocol for each trial 16–19. In the four clinical trials, blood pressure was measured prior to bevacizumab treatment on day 1 of the first cycle. Hypertension was recorded according to the Common Toxicity Criteria for Adverse Events (CTCAE) version 3.0. Grade 3 hypertension (severe cases) was used as the cut-off for the analysis and it is defined as recurrent or persistent (≥24 h) or symptomatic blood pressure increase by >20 mm Hg (diastolic) or to >150/100 mm Hg, requiring more than one drug or more intensive therapy than previously. Only hypertension with attribution of “possibly related” or higher, according to the National Cancer Institute (NCI) Guidelines 20, was included. Only patients with no hypertension before receiving bevacizumab were enrolled in the four clinical trials, and more details on the inclusion criteria for the cardiovascular status prior to treatment have been reported previously 4 and are also described in the Supplementary File.
Genotyping, quality control, and imputation
Germline DNA was obtained from peripheral blood and genotyped using the Illumina HumanHap550-Quad platform (CALGB 80303 and 90401) and the Illumina Human OmniExpressExome-8 platform (CALGB 40503 and 40502). Details on quality control (QC) and number of SNPs excluded due to call rate less than 95%, minor allele frequency (MAF) less than 5%, and Hardy-Weinberg equilibrium with p-value <1.00×10−8 can be found in previous publications 4,21–24. This study represents the first time that imputation for unobserved and missing genotypes has been performed in CALGB 80303, 40503, 90401, and 40502. Imputation was performed using the TOPMed Imputation Server (TIS, https://imputation.biodatacatalyst.nhlbi.nih.gov/). Following the practices used for eMERGE3 25, all eligible samples were uploaded together for imputation. Genotype data were pre-processed for imputation using the latest version of the Rayner imputation data preparation utilities (https://www.well.ox.ac.uk/~wrayner/tools/), modified to work with the TOPMed Freeze5 GRCh38 reference panel. Pre-phasing was performed on the TIS using the reference-based phasing algorithm Eagle 26 v2.4. Post-imputation quality control included filtering out all variants with an imputation quality r2 < 0.3 and examining overall allele frequency concordances for respective genetic ancestry groups.
PRS calculation
A summarized scheme of the methods for PRS analysis is shown in Figure 1. PRS were calculated using two input datasets, target and base datasets. PRS combine the effects of the risk alleles at pre-specified p-value thresholds from a base GWAS summary statistics into a single risk score. The number of risk alleles that each subject in the target dataset possesses for a specific SNP is weighted by the effect size of that SNP in the base dataset. The base dataset included two GWAS meta-analyses for BP, one of systolic BP (SBP) and one of diastolic BP (DBP), of 757,601 individuals of European descent from the UK Biobank and the International Consortium of Blood Pressure Genome-Wide Association Studies (ICBP) 27. The GWAS summary statistics of the base dataset were downloaded from the NHGRI-EBI Catalog of human GWAS 28. LiftOver 29 was used for standardizing genome builds across the target (hg38) and base (hg19) datasets. PRS were calculated in each study of the target dataset (CALGB 80303, 40503, 90401, and 40502) using PRSice-2 30. PRSice-2 uses the classic “clumping and thresholding” method for PRS calculation, which combines linkage disequilibrium (LD) clumping with p-value thresholding. As default in PRSice-2, LD clumping was performed with any SNP within 250 kb and in LD (r2 > 0.1) with the leading SNP (lowest p-value) in a given region. PRS for grade 3 bevacizumab-induced hypertension were calculated at forty different p-value thresholds (1×10−40, 1×10−39, 1×10−38… 0.01), using standardized effect size and including age, sex, and the first three principal components as covariates. Because the optimal p-value threshold is unknown a prior, the use of a range of thresholds is preferred instead of using a single arbitrary p-value threshold to calculate PRS in order to avoid underperformance of the PRS prediction 31. A liberal range of forty p-value thresholds was chosen because drug-induced hypertension is a highly polygenic phenotype with many loci with very small effects, and a large GWAS was used as the base dataset. For guarding against generation overfit prediction models and to correct for multiple testing, permutation testing with 10,000 permutations of the model was performed. The distributions of pseudo explained variance (Nagelkerke’s R2) and optimal p-value thresholds were calculated. The proportion of variance explained by PRS was estimated as the difference in Nagelkerke’s R2 between the full model (including PRS plus covariates) and the null model (only covariates). Receiver operating characteristic curves (ROC) were used to estimate the area under the curve (AUC).
Figure 1. Scheme of the method for polygenic risk scores (PRS) analysis for bevacizumab-induced hypertension based on GWAS for increased systolic blood pressure (SBP) and diastolic blood pressure (DBP).
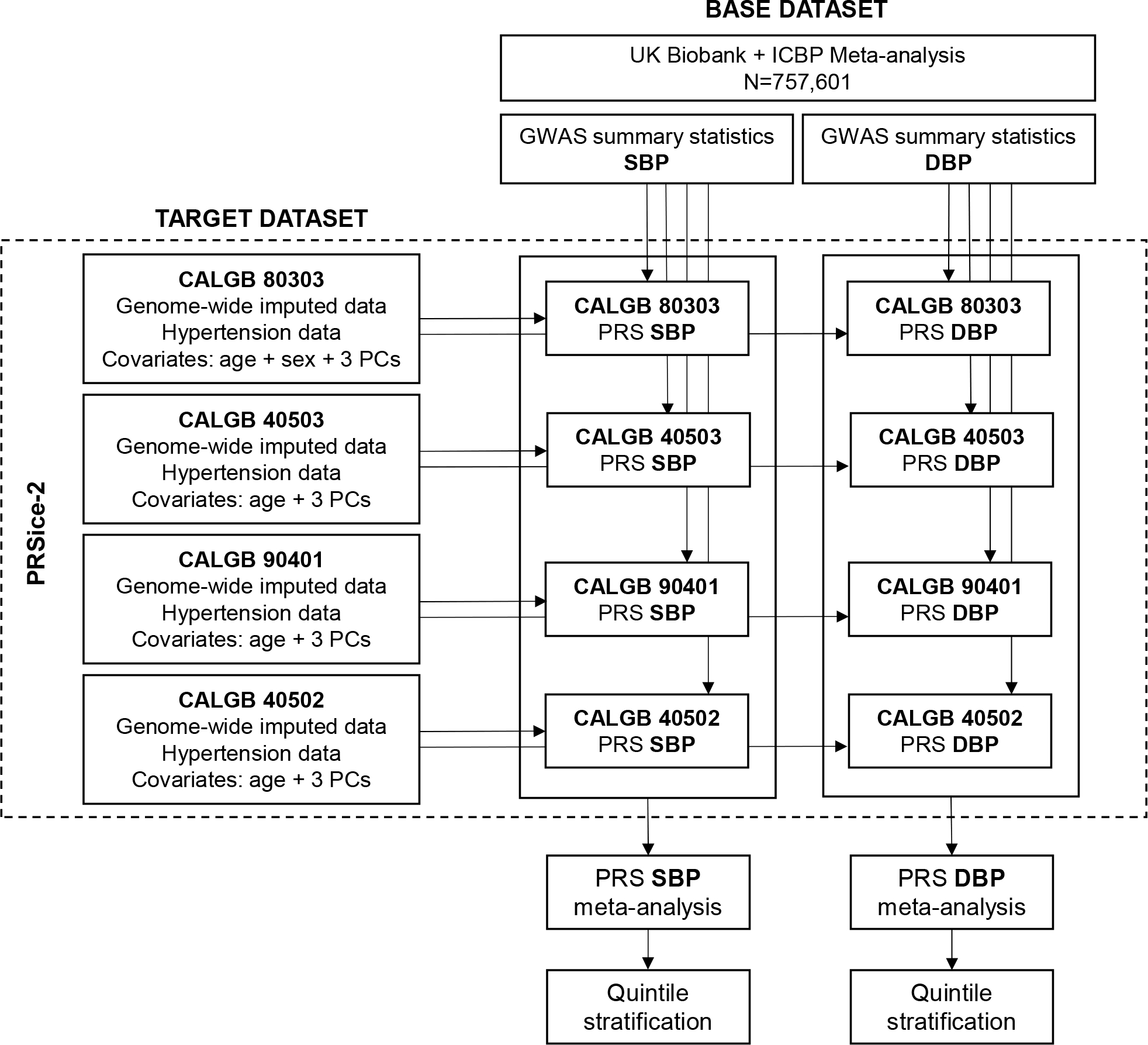
CALGB: Cancer and Leukemia Group B; ICBP: International Consortium of Blood Pressure; PCs: principal components.
Statistical analysis
In each trial, the association between PRS (quantitative) and grade 3 bevacizumab-induced hypertension was evaluated by multivariable logistic regression by PRSice-2 as described above. Fixed-effect meta-analyses odds ratio (OR) per standard deviation (SD) of the association of PRS (quantitative) and hypertension across trials was obtained by inverse variance for PRS generated in each p-value threshold for SBP and DBP, and a meta-analysis p-value <0.05 was considered the cut-off for statistical significance. For the p-value threshold corresponding to the most statistically significant association between a PRS (quantitative) and grade 3 bevacizumab-induced hypertension, PRS were stratified into quintiles for additional interpretation, with the bottom quintile modeled as the referent group. The OR of the association between hypertension and each quintile versus the referent group was estimated by logistic regression and a p-value <0.05 was considered the cut-off for statistical significance. All analyses were performed using the statistical software R v4.0.2 (R Core Team, Vienna, Austria) and its extension packages, including PRSice-2 30 and meta 32. Data quality was ensured by review of data by the Alliance Statistics and Data Management Center and by the study chairperson following Alliance policies.
Gene set enrichment analysis
An unsupervised gene set enrichment analysis using the Ingenuity Pathway Analysis (IPA®, Qiagen) software was performed to identify the main signaling pathways involving genes located in or near (upstream or downstream) SNPs used to calculate PRS for SBP and PRS for DBP.
Sex-based analysis
Because of the higher prevalence of bevacizumab-induced hypertension in female than male patients, we have also performed meta-analyses for the association between the PRS and grade 3 bevacizumab-hypertension only in either female or male patients.
Results
A total of 1,027 patients with cancer treated with bevacizumab were included in this study. Details on the demographics and characteristics of patients included in each trial are described in Table 1. The majority of patients was female (60.9%) with a mean age of 61.7 ± 11.3 years. Grade 3 hypertension was reported in 6.7–27.6% between the trials, with a higher prevalence observed in the clinical trials that included only female patients (breast cancer studies, CALGB 40503 and 40502) as discussed in the previous publication 4. CALGB 80303 was the only study that included both female and male patients and grade 3 hypertension was recorded in 14.1% of females (9 patients) and in 11.1% of males (10 patients). No grade 4 hypertension was reported.
PRS-hypertension association
The numbers of SNPs overlapping between the target and base datasets were 2,070,648 (CALGB 80303), 2,073,393 (CALGB 40503), 2,070,154 (CALGB 90401), 2,082,251 (CALGB 40502).
SNPs associated with SBP
The most statistically significant meta-analysis association between PRS (calculated based on SNPs associated with SBP) and grade 3 bevacizumab-induced hypertension included up to 67 SNPs within a p-value threshold = 1.0×10−29 (p=0.0077, OR per SD =1.31, 95% CI: 1.07–1.60) and were used for the downstream analysis. No covariates were statistically significant associated with grade 3 bevacizumab-induced hypertension (p-value >0.05). The results of each study association analysis using the p-value threshold of 1.0×10−29 are described in Table 2. The list of SNPs associated with SBP and included in the PRS calculation and their weight in the base dataset are shown in Table S1. The results of the association between PRS and grade 3 bevacizumab-induced hypertension for all p-value thresholds tested are reported in Table S2.
Table 2. Association between polygenic risk score (PRS quantitative) and grade 3 bevacizumab-induced hypertension in each study for the most statistically significant meta-analysis p-value using SNPs associated with systolic blood pressure (SBP) and diastolic BP (DBP).
OR per SD: odds ratio per standard deviation; CI: confidence interval; AUC: area under the curve.
| p-value | OR per SD (95% CI) | PRS R2 (adjusted) | AUC (95% CI) | Number of SNPs | |
|---|---|---|---|---|---|
| SBP (p-value threshold <10×10−29) | |||||
| Meta-analysis | 0.0077 | 1.31 (1.07–1.60) | |||
| CALGB 80303 | 0.3359 | 0.85 (0.52–1.39) | 0.012 | 0.62 (0.48–0.76) | 67 |
| CALGB 40503 | 0.1511 | 1.40 (0.89–2.22) | 0.028 | 0.64 (0.52–0.75) | 66 |
| CALGB 90401 | 0.0012 | 1.97 (1.19–3.26) | 0.086 | 0.73 (0.62–0.83) | 66 |
| CALGB 40502 | 0.0956 | 1.22 (0.90–1.65) | 0.012 | 0.56 (0.50–0.66) | 65 |
| DBP (p-value threshold <10×10−39) | |||||
| Meta-analysis | 0.0209 | 1.27 (1.04–1.56) | |||
| CALGB 80303 | 0.5245 | 0.79 (0.49–1.27) | 0.005 | 0.60 (0.46–0.73) | 53 |
| CALGB 40503 | 0.1572 | 1.39 (0.89–2.18) | 0.029 | 0.57 (0.44–0.71) | 53 |
| CALGB 90401 | 0.0087 | 2.15 (1.33–3.47) | 0.061 | 0.69 (0.59–0.80) | 53 |
| CALGB 40502 | 0.2093 | 1.29 (0.96–1.74) | 0.007 | 0.56 (0.48–0.64) | 49 |
Three out of the four studies showed a concordant direction of effect and patients with higher PRS are at higher risk of developing grade 3 bevacizumab-induced hypertension (Table 2). The PRS explained between 1.2% to 8.6% (Nagelkerke’s R2 = 0.012–0.086) of the variance of grade 3 bevacizumab-induced hypertension, with an AUC between 0.56 and 0.73 (Table 2). In addition, we observed a 3.72-fold higher odds of grade 3 bevacizumab-induced hypertension between the top and the referent quintiles of the PRS (OR=3.72, 95% CI: 1.84–8.16, p=4.75×10−4, Figure 2).
Figure 2.
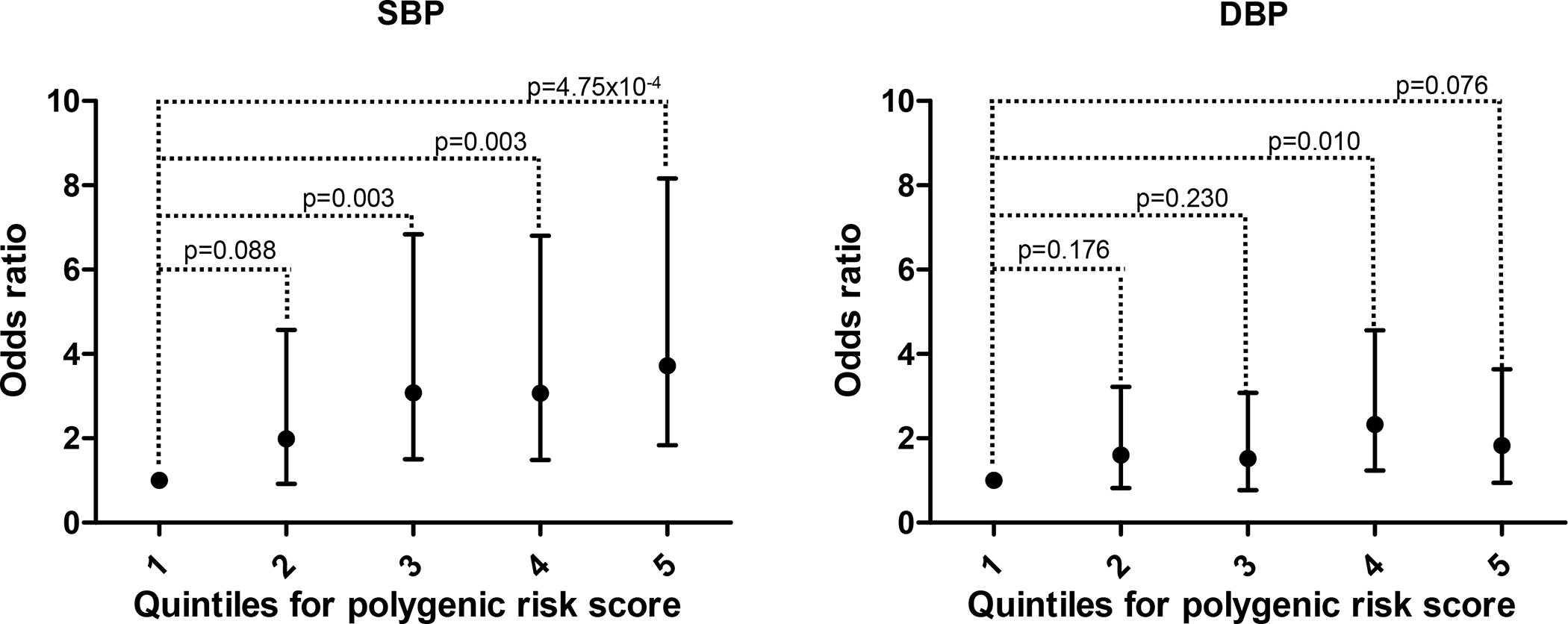
Association between quintiles of the polygenic risk score (PRS) based on SNPs associated with systolic blood pressure (SBP) on the left and SNPs associated with diastolic blood pressure (DBP) on the right.
SNPs associated with DBP
The most statistically significant meta-analysis association between PRS (calculated based on SNPs associated with DBP) and grade 3 bevacizumab-induced hypertension included up to 67 SNPs within a p-value threshold = 10×10−39 (p=0.021, OR per SD=1.27, 95% CI: 1.04–1.56) and were used for the downstream analysis. No covariates were statistically significant associated with grade 3 bevacizumab-induced hypertension (p-value >0.05). The results of each study association analysis using the p-value threshold of 10×10−39 are described in Table 2. The list of SNPs associated with DBP and included in the PRS calculation and their weight in the base dataset are shown in Table S3. The results of the association between PRS and grade 3 bevacizumab-induced hypertension for all p-value thresholds tested are reported in Table S2.
Three out of the four studies showed a concordant direction of effect and patients with higher PRS are at higher risk of developing grade 3 bevacizumab-induced hypertension (Table 2). The PRS explained between 0.7% to 6.1% (Nagelkerke’s R2 = 0.007–0.061) of the variance of grade 3 bevacizumab-induced hypertension across studies, with an AUC between 0.56 and 0.69 (Table 2). In addition, we observed a 1.83-fold higher odds of grade 3 bevacizumab-induced hypertension between the top and the referent quintiles of the PRS, but without reaching statistical significance (OR=1.83, 95% CI: 0.95–3.64, p=0.076, Figure 2).
Gene set enrichment analysis
Gene set enrichment analysis was performed with 73 and 62 genes located in or near (upstream or downstream) SNPs used to calculate PRS for SBP and PRS for DBP, respectively (Tables S1 and S3), and the top 50 signaling pathways are shown in Figure S1. Supplementary File 2 contains the details on all signaling pathways identified in this analysis with their respective p-values and genes.
Sex-based analysis
The meta-analyses for the association between PRS and grade 3 bevacizumab-induced hypertension in either female (n=625) and male patients (n=402) showed a statistically significant association between PRS and hypertension in female patients for PRS calculated based on SNPs associated with SBP (p=0.0238, OR per SD=1.30, 95% CI: 1.03–1.65) and DBP (p=0.0485, OR per SD=1.27, 95% CI: 0.84–1.86) (Table S4). The association between PRS and hypertension in male patients did not reach statistical significance (Table S4).
Discussion
Calculation of PRS for target traits based on GWAS of a proxy trait is a powerful strategy to optimize the identification of causal genetic variants when there are no large GWAS on the target trait available or feasible 31. The present study represents the first time that such an approach was applied to assess the risk of drug-induced toxicity. Using the largest GWAS meta-analyses for BP traits ever conducted as a base dataset, we have calculated PRS for severe bevacizumab-induced hypertension in 1,027 cancer patients. We found that genetic variants associated with essential hypertension, mainly SBP, increase the risk of severe bevacizumab-induced hypertension.
We have provided evidence for the shared genetic effects between increased SBP and DBP and severe bevacizumab-induced hypertension. The PRS calculated based on SNPs associated with increased SBP explained up to 8.6% variance of severe bevacizumab-induced hypertension with an AUC up to 0.73 (95% CI: 0.62–0.83), while PRS calculated based on SNPs associated with increased DBP explained up to 6.1% variance of severe bevacizumab-induced hypertension with an AUC up to 0.69 (95% CI: 0.59–0.80). The AUC observed are similar to previous reported PRS for drug-induced toxicity, more specifically liver injury 10.
SBP is more predictive of cardiovascular outcomes when compared to DBP 33, and an increase in SBP frequently occurs without an increase in DBP 34, known as isolated systolic hypertension. In the present study, patients with a moderate to high PRS for increased SBP (quintiles 3–5) were more than 3-fold more likely to develop severe hypertension when treated with bevacizumab when compared to patients with lower PRS (quintiles 1 and 2). Patients with a high PRS for increased DBP (quintiles 4 and 5) were only 1.8-fold more likely to develop severe hypertension compared to patients with lower PRS (quintiles 1–3) for increased DBP. Therefore, our findings also indicate that the genetics of SBP is more associated with drug-induced hypertension than DBP.
Endothelial dysfunction is one of the major determinants for isolated systolic hypertension due to increased arterial stiffness 35. Thus, a possible explanation for the effect of SBP on bevacizumab-induced hypertension is that endothelium dysfunction and reduced NO due to bevacizumab treatment increase arterial stiffness, and associated with an increased genetic risk for increased SBP, lead to an increased risk of severe bevacizumab-induced hypertension when compared with patients with a low PRS for increased SBP. Isolated systolic hypertension comprises 80% of the hypertension cases of individuals 60 years older 36, and a significant proportion of cancer patients treated with bevacizumab might have genetic factors associated with increased SBP. The calculation of PRS before treatment with bevacizumab and other VEGF-pathway inhibitors can be used to identify these patients prior to treatment.
The gene set enrichment analysis revealed an enrichment of critical cardiovascular signaling pathways for genes in or near SNPs used to calculate both PRS for SBP and PRS for DBP (Figure S1). The cardiac hypertrophy signaling pathway was the most statistically significant pathway in the enrichment analysis for both PRS for SBP and PRS for DBP, which was expected because hypertension is the primary cause of left ventricular hypertrophy 37. Interestingly, the NO signaling pathway in the cardiovascular system was also statistically significant associated with genes in or near SNPs in both PRS for SBP and PRS for DBP, showing that SNPs used for PRS calculation are directly associated with the function of the endothelium and possibly increasing the risk of endothelial dysfunction with bevacizumab therapy. Other cardiovascular related signaling pathways associated with either PRS for SBP or PRS for DBP include dilated cardiomyopathy, cardiac rhythm, apelin cardiomyocyte, and renin-angiotensin signaling pathways. The role of these pathways in bevacizumab-induced hypertension should be further investigated.
A higher prevalence of bevacizumab-induced hypertension was observed in female patients compared to male patients, as previously reported 4, potentially because of a lower clearance of bevacizumab in females compared to males 2. CALGB 80303 was the only study that included both female and male patients and showed an opposite direction of effect in both PRS calculated based on SNPs associated with SBP and DBP. However, in the sex-based analysis (Table S4), both PRS for SBP and PRS for DBP in only female patients of CALGB 80303 showed the same direction of effect increasing the risk of grade 3 bevacizumab-induced hypertension. In addition, when meta-analyzed with studies that included only female patients (CALGB 40502 and 40503), the association reached statistical significance. The opposite direction of effect observed in the analysis of all patients in CALGB 80303 was driven by the association between PRS and male patients (Table S4).
The emerging area of cardio-oncology has been focused on understanding the risk of toxicities induced by cancer treatment. A scientific statement published by the American Heart Association 38 highlights the future research directions in vascular cardio-oncology, including the better identification of patients at risk for cardiovascular toxicities during treatment with VEGF-pathway inhibitors and genetic inquiries for risk of toxicity. In addition, the very novel concept of onco-hypertension has been proposed to disseminate the need for further studies to uncover the complex pathophysiology of drug-induced hypertension in cancer patients 39. This present study is the first to investigate the influence of genetic factors associated with blood pressure on the risk of drug-induced hypertension during cancer treatment, leading to future insights and applications. Our findings might have important implications for identifying patients at higher risk of developing severe hypertension during cancer therapy and for understanding the complex polygenic relationship underlying this toxicity.
This study has some limitations. The relatively small sample size of the target dataset when compared to the base dataset and the inclusion of patients from different clinical trials can introduce some heterogeneity in the sample. All four clinical trials were conducted in a single cancer network, and the standardized collection of the phenotypic and genotypic minimizes the effects of possible confounding due to these differences. Moreover, we have used data from the largest cohort comprising genome-wide and hypertension data of patients treated with bevacizumab, performing the analyses in the largest sample size available to us. The associations between PRS and bevacizumab-induced hypertension were concordant for three out of the four studies and were driven mainly by CALGB 90401. The fact that patients in CALBG 90401 had a higher age when compared to the other studies and received concomitant docetaxel, which can also induce cardiovascular toxicities 38, should be considered when interpreting the results. Still, docetaxel-induced cardiovascular toxicity is characterized by capillary leak rather than hypertension 38. We have only calculated PRS for patients of European descent, as we do not have access to genomic and hypertension data of a large number of patients of other ethnicities treated with bevacizumab, and more efforts in this regard need to be applied to extend the application of PRS to other ethnicities as well. Finally, the evaluation of PRS in additional cohorts of patients treated with VEGF-pathway inhibitors needs to be further investigated to validate our findings.
In conclusion, we have generated PRS for the risk assessment of severe bevacizumab-induced hypertension in cancer patients. Patients with moderate to high PRS for increased SBP are at high risk of developing severe hypertension when treated with bevacizumab and possibly with other VEGF-pathway inhibitors. To the best of our knowledge, this is the first study showing the contribution of genetics for increased SBP for drug-induced hypertension in cancer patients. The availability of the list of genetic variants used to calculate PRS in this study allows for testing for replication of our findings in future studies, and, if validated in additional cohorts, the SNPs selected can be genotyped for PRS calculation before treatment with bevacizumab for a better risk assessment of cancer therapy.
Supplementary Material
Study Highlights.
What is the current knowledge on the topic?
Hypertension is a common toxicity experienced by patients treated with bevacizumab and other VEGF-pathway inhibitors, compromising the duration and efficacy of the therapy with these agents. Although few studies have reported associations of SNPs with hypertension induced by these drugs, there are no biomarkers applied in the clinic to inform decisions for risk assessment of patients before treatment with bevacizumab or any other VEGF-pathway inhibitors.
What question did this study address?
We hypothesized that genetic risk of essential hypertension, as measured by a blood pressure polygenic risk scores (PRS), would be associated with risk of severe bevacizumab-induced hypertension.
What does this study add to our knowledge?
This study provides evidence that genetic variants associated with blood pressure, mainly systolic blood pressure, can be used to predict severe drug-induced hypertension in cancer patients treated with bevacizumab.
How might this change clinical pharmacology or translational science?
In addition to the contribution for understanding the mechanism of bevacizumab-induced hypertension, if validated in additional cohorts, the SNPs selected by this study can be genotyped for PRS calculation before treatment with bevacizumab for a better risk assessment of cancer therapy.
Acknowledgments:
We acknowledge RIKEN for the genotyping of samples of CALGB 80303.
Funding:
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Numbers U10CA180821, U10CA180882 and U24CA196171 (to the Alliance for Clinical Trials in Oncology), UG1CA232760 and UG1CA233373. https://acknowledgments.alliancefound.org. JCFQ was supported by the American Heart Association (AHA Award Number 826128). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest: JCFQ and FI are coinventors of a patent application, serial number 16/932,002: Methods of identifying risk of bevacizumab-induced proteinuria and hypertension. FI is an AbbVie employee and receives stocks from AbbVie; this work was conceived when FI was a faculty at the University of North Carolina at Chapel Hill, and this work does not represent a potential conflict of interest. All other authors declared no competing interests for this work.
Ethics approval: The clinical studies were conducted in accordance with recognized ethical guidelines. The studies were performed in accordance with the Declaration of Helsinki and were approved by the local IRB.
SUPPORTING INFORMATION
Supplementary information accompanies this paper on the Clinical Pharmacology & Therapeutics website (www.cpt-journal.com).
References
- 1.EMA Summary of Product Characteristics. Available online: https://www.ema.europa.eu/en/documents/product-information/avastinepar-product-information_en.pdf. Accessed November 9, 2021.
- 2.FDA AVASTIN®Prescribing Information. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/125085s332lbl.pdf. Accessed November 9, 2021.
- 3.Ferrara N, Hillan KJ, Gerber HP, Novotny W Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat. Rev. Drug. Discov. 3, 391–400 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Quintanilha JCF et al. Bevacizumab-induced hypertension and proteinuria: a genome-wide study of more than 1000 patients. Br. J. Cancer 26, 265–274 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranpura V, Pulipati B, Chu D, Zhu X, Wu S Increased risk of high-grade hypertension with bevacizumab in cancer patients: a meta-analysis. Am. J. Hypertens. 23, 460–468 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Maitland ML et al. Ambulatory monitoring detects sorafenib-induced blood pressure elevations on the first day of treatment. Clin. Cancer Res. 15, 6250–6257 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin C et al. The influence of genetic variants of sorafenib on clinical outcomes and toxic effects in patients with advanced renal cell carcinoma. Sci. Rep. 6, 20089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eechoute K et al. Polymorphisms in endothelial nitric oxide synthase (eNOS) and vascular endothelial growth factor (VEGF) predict sunitinib-induced hypertension. Clin. Pharmacol. Ther. 92, 503–510 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Schneider BP et al. Genetic variant predicts bevacizumab-induced hypertension in ECOG-5103 and ECOG-2100. Br. J. Cancer. 111, 1241–1248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koido M et al. Polygenic architecture informs potential vulnerability to drug-induced liver injury. Nat Med. 26, 1541–1548 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.GBD 2017 Risk Factor Collaborators. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390, 1345–1422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.International Schizophrenia Consortium, Purcell SM et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selzam S Predicting educational achievement from DNA. Mol. Psychiatry 22, 267–272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JJ et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 50, 1112–1121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price AL, Patterson NJ, Plenge RM, Weinblatt ME., Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Kindler HL et al. Gemcitabine plus bevacizumab compared with gemcitabine plus placebo in patients with advanced pancreatic cancer: phase III trial of the Cancer and Leukemia Group B (CALGB 80303). J. Clin. Oncol. 28, 3617–3622 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly WK et al. Randomized, double-blind, placebo-controlled phase III trial comparing docetaxel and prednisone with or without bevacizumab in men with metastatic castration-resistant prostate cancer: CALGB 90401. J. Clin. Oncol. 30, 1534–1540 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickler MN et al. Phase III Trial Evaluating Letrozole As First-Line Endocrine Therapy With or Without Bevacizumab for the Treatment of Postmenopausal Women With Hormone Receptor-Positive Advanced-Stage Breast Cancer: CALGB 40503 (Alliance). J. Clin. Oncol. 34, 2602–2609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rugo HS et al. Randomized Phase III Trial of Paclitaxel Once Per Week Compared With Nanoparticle Albumin-Bound Nab-Paclitaxel Once Per Week or Ixabepilone With Bevacizumab As First-Line Chemotherapy for Locally Recurrent or Metastatic Breast Cancer: CALGB 40502/NCCTG N063H (Alliance). J. Clin. Oncol. 33, 2361–2369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Institute of Cancer (NCI) Guidelines for Investigators. 2021. https://ctep.cancer.gov/protocoldevelopment/electronic_applications/docs/aeguidelines.pdf. Accessed on 08 Jan 2021.
- 21.Quintanilha JCF et al. Genome-wide association studies of survival in 1520 cancer patients treated with bevacizumab-containing regimens. Int. J. Cancer 150, 279–289 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Innocenti F et al. Genomic Analysis of Germline Variation Associated with Survival of Patients with Colorectal Cancer Treated with Chemotherapy Plus Biologics in CALGB/SWOG 80405 (Alliance). Clin. Cancer Res. 27, 267–275 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rashkin SR et al. A Pharmacogenetic Prediction Model of Progression-Free Survival in Breast Cancer using Genome-Wide Genotyping Data from CALGB 40502 (Alliance). Clin. Pharmacol. Ther. 105, 738–745 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hertz DL et al. Pharmacogenetic Discovery in CALGB (Alliance) 90401 and Mechanistic Validation of a VAC14 Polymorphism that Increases Risk of Docetaxel-Induced Neuropathy. Clin. Cancer Res. 22, 4890–4900 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanaway IB et al. The eMERGE genotype set of 83,717 subjects imputed to ~40 million variants genome wide and association with the herpes zoster medical record phenotype. Genet. Epidemiol. 43, 63–81 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh PR et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evangelou E et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 50, 1412–1425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buniello A et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinrichs AS et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 34, D590–598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi SW, O’Reilly PF PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience 8, giz082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi SW, Mak TS, O’Reilly PF Tutorial: a guide to performing polygenic risk score analyses. Nat. Protoc. 15, 2759–2772 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarzer G, Carpenter JR, Gerta R Meta-Analysis with R. 2015.
- 33.Sever P Abandoning diastole. BMJ 318, 1773 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chobanian AV et al. Seventh report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Hypertension 42, 1206–1252 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Wallace SM et al. Isolated systolic hypertension is characterized by increased aortic stiffness and endothelial dysfunction. Hypertension 50, 228–233 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Franklin SS et al. Does the relation of blood pressure to coronary heart disease risk change with aging? The Framingham Heart Study. Circulation 103, 1245–1249 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Yildiz M, Oktay AA, Stewart MH, Milani RV, Ventura HO, Lavie CJ Left ventricular hypertrophy and hypertension. Prog Cardiovasc Dis. 2020, 63, 10–21. [DOI] [PubMed] [Google Scholar]
- 38.Campia U et al. Cardio-Oncology: Vascular and Metabolic Perspectives: A Scientific Statement From the American Heart Association. Circulation 139, e579–e602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kidoguchi S et al. New Concept of Onco-Hypertension and Future Perspectives. Hypertension 77, 16–27 (2021). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


