Abstract
Background
Lysine acetyltransferase 6 A (KAT6A) is a MYST-type histone acetyltransferase (HAT) enzyme, which contributes to histone modification and cancer development. However, its biological functions and molecular mechanisms, which respect to hepatocellular carcinoma (HCC), are still largely unknown.
Methods
Immunohistochemical, western blot and qRT-PCR analysis of KAT6A were performed. A series of in vitro and in vivo experiments were conducted to reveal the role of KAT6A in the progression of HCC.
Results
We demonstrated that KAT6A expression was upregulated in HCC tissues and cell lines. Clinical analysis showed that increased KAT6A was significantly associated with malignant prognostic features and shorter survival. Gain- and loss-of-function experiments indicated that KAT6A promoted cell viability, proliferation and colony formation of HCC cells in vitro and in vivo. We confirmed that KAT6A acetylates lysine 23 of histone H3 (H3K23), and then enhances the association of the nuclear receptor binding protein TRIM24 and H3K23ac. Consequently, TRIM24 functions as a transcriptional activator to activate SOX2 transcription and expression, leading to HCC tumorigenesis. Restoration of SOX2 at least partially abolished the biological effects of KAT6A on HCC cells. Overexpression of KAT6A acetyltransferase activity-deficient mutants or TRIM24 mutants lacking H3K23ac binding sites did not affect SOX2 expression and HCC biological function. Moreover, matrix stiffness can upregulate the expression of KAT6A in HCC cells.
Conclusions
Our data support the first evidence that KAT6A plays an oncogenic role in HCC through H3K23ac/TRIM24-SOX2 pathway, and represents a promising therapeutic strategy for HCC patients.
Subject terms: Hepatocellular carcinoma, Hepatocellular carcinoma
Background
Hepatocellular carcinoma (HCC) is the fifth most common malignancy and the third leading cause of cancer-related death worldwide [1]. Despite the great advancement in new effective systemic therapies, the survival of HCC patients remains dismal [2]. Recent global genomic and transcriptome analyses reveal that chromosomal aberrations and epigenetic changes are common mechanisms for pathway deregulation in HCC, including histone modifications and mutations [3]. However, the mechanisms by which histone modifiers regulate HCC remain poorly understood.
Lysine acetyltransferase 6 A (KAT6A, also known as MYST3 and MOZ) was found in the 8p11-p12 amplicon and first identified in 1996 as part of a chromosomal translocation t(8; 16)(p11; p13) with CREB-binding protein in a subtype of acute myeloid leukemia [4, 5]. Aberrant KAT6A expression or its histone acetyltransferase activity was involved in cancers [5, 6]. KAT6A suppresses cellular senescence through the regulation of suppressors of the CDKN2A locus and inhibitors of KAT6A induce senescence and arrest tumour growth [7]. KAT6A was involved in histone acetylation and activates Estrogen receptor α (ERα) expression by altering chromatin structure [8]. KAT6A inhibits senescence via the INK4A-ARF pathway [9]. KAT6A expression was associated with glioblastoma (GBM) patient survival and promoted cell proliferation, migration, colony formation and tumour development in GBM. KAT6A regulates stemness of aging bone-marrow-derived mesenchymal stem cells through NRF2/ARE signaling pathway [10]. KAT6A acetylates lysine 23 of histone H3 (H3K23) to activate PIK3CA transcription and enhance PI3K/AKT signaling and tumorigenesis [11]. Nevertheless, the functional importance of KAT6A and its molecular mechanisms in HCC remain largely unknown.
In this study, we aimed to investigate the expression level, clinical importance, functions and potential mechanisms of KAT6A in HCC. Gain- and loss-of-function experiment revealed that the biological function of KAT6A in vitro and in vivo on HCC progression. Mechanistic investigations demonstrated that KAT6A acetylates lysine 23 of histone H3 (H3K23), which recruits the nuclear receptor binding protein TRIM24 to activate SOX2 transcription. Finally, we examined that high matrix stiffness could increase KAT6A expression in HCC cells and KAT6A knockdown could inhibit the promoting effects of matrix stiffness on the HCC cells. Therefore, we demonstrated that KAT6A exerts an oncogenic role in HCC progression and represents a potential target for HCC diagnosis and treatment.
Methods
Patients’ tissues and cell culture
HCC tissues and corresponding paired adjacent non-tumour tissues were obtained from our hospital after the informed consent were signed from all patients. All patients did not receive any therapy before surgery. This study was approved by the Ethics Committee of this hospital. The normal immortalised human hepatocyte THLE-3 and a group of HCC cells (Hep3B, HepG2, Huh7, MHCC-97H and HCCLM3) (Chinese Academy of Sciences, Shanghai, China) were maintained in DMEM (Invitrogen, Carlsbad, USA) containing 10% FBS (Gibco, GrandIsland, USA) and 100 U/mL penicillin-streptomycin mixture (Beyotime Institute of Biotechnology, Haimen, China) in 37 °C with 5% CO2.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissues or cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. Quantitative real-time PCR was performed using SYBR Premix Ex Taq II (TaKaRa). The amount of each target gene was quantitated by the comparative C (T) method using GAPDH as the normalisation control. qPCR primers were purchased from Genecopoeia (Guangzhou, China).
Western blot
Cell lysates were prepared using RIPA lysis Buffer (Pierce, Rockford, IL). Then protease inhibitors were added into lysates. A bicinchoninic acid (BCA) kit (Beyotime Institute of Biotechnology, Shanghai, China) was used to determine protein concentration. Subsequently, protein was separated in 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) membrane and then transferred onto a polyvinylidene difluoride (PVDF) membrane (Mippore, Merck KGaA, Germany). Tris-buffered saline (TBS) containing 5% nonfat milk was used to block the membranes for an hour at room temperature. Thereafter, the membranes were incubated with specific primary antibodies at 4 °C overnight. Subsequently, HRP-conjugated secondary antibody goat anti-rabbit IgG (1:2000, Abcam) was added and incubated for 2 h at room temperate. GAPDH was considered as the internal control. Bands signal were detected using the enhanced chemiluminescence (ECL; Millipore, Germany).
Immunohistochemistry (IHC) staining and scoring
Immunohistochemical analysis status was performed as our previous publication [12].
Polyacrylamide hydrogels with incremental stiffness
Polyacrylamide hydrogels with stiffness were prepared as reported previously [13].
Cell viability, proliferation and colony formation assays
Cell Counting Kit-8 (CCK8) reagents (Dojindo, Kumamoto, Japan), Edu assay and clone formation assay were conducted according to the protocols described in our previous studies [14]. Briefly, with respect to plate clone formation assay, 1 × 103 cells were seeded in six-well plates. The cells were mixed and then cultured for 2 weeks in culture medium with 10% FBS. Clusters containing ≥30 cells were counted as a single colony. With respect to Edu assay, Cell-Light™ EdU Apollo®488 In Vitro Imaging Kit (#C10310-3, RiboBio Co., LTD, Guangzhou, China) was used. All of the above experiments were performed according to the manufacturer’s protocols.
Chromatin immunoprecipitation (ChIP) and ChIP-qPCR
ChIP was performed using an EZ-Magna-ChIP HiSens kit (Millipore, 17–10461) as we previously described [13]. In brief, cells fixed with 1% formaldehyde and scraped from culture plate were subjected to nuclear lysis to release crosslinked protein/DNAs. The nuclear extract was then subjected to sonication and immunoprecipitation with anti-TRIM24 or H3K23ac antibody or control rabbit IgG (Santa Cruz Technology, sc-2027). Precipitated DNA fragments containing SOX2 promotor were quantitated by qPCR.
In vivo experiments
Four-to-six-week-old male BALB/c nude mice (Centre of Laboratory Animals, The Medical College of Xi’an Jiaotong University, Xi’an, China) were used to establish the nude mouse xenograft model. Hep3B (5 × 106) cells that were transfected with KAT6A or control vectors were mixed in 150 µl of Matrigel and were inoculated subcutaneously into the flank of nude mice. The tumour volume for each mouse was determined by measuring two of its dimensions and then calculated as tumour volume = length × width × width/2. After 3 weeks, the mice were sacrificed by cervical dislocation under anesthesia with ether and the xenograft tumour tissue was explanted for examination. Animal protocols were approved by the Institutional Animal Care and Use Committee of Xi’an Jiaotong University.
Statistical analysis
Results are managed as the mean ± SD and analysed by SPSS software, 16.0 (SPSS, Chicago, USA). The statistical approaches mainly included a two-tailed Student’s t-test, a Kaplan–Meier plot, Pearson chi-squared test and so on. Difference with P < 0.05 was regarded to be significant. Graphs were mainly made by GraphPad Prism 6 (GraphPad, San Diego, USA).
Results
KAT6A is upregulated in HCC and correlates with poor prognosis
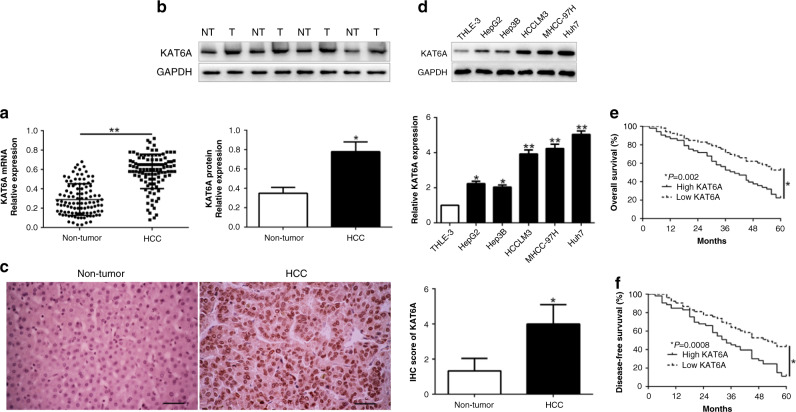
To explore the expression of KAT6A in HCC, we performed qRT-PCR and western blot to show that KAT6A mRNA and protein were significantly upregulated in HCC tissues compared to adjacent non-tumour tissues (P < 0.05, Fig. 1a, b). IHC staining assays validate the similar results (P < 0.05, Fig. 1c). Moreover, we demonstrated that KAT6A was increased in a panel of HCC cells compared with normal hepatic cells THLE-3 (P < 0.05, Fig. 1d). These results implied that KAT6A serves as a tumour promoter in HCC progression. To investigate the clinical role of KAT6A, we divided HCC patients into two subgroups with the median level of KAT6A as a cut-off value. As shown in Table 1, high KAT6A expression was significantly associated with large tumour sized (P = 0.002), high Edmondson-Steiner grade (P = 0.020) and advanced TNM stage (P = 0.034). More importantly, Kaplan–Meier survival cure showed that HCC patients with high KAT6A expression had a shorter overall survival (OS) (P = 0.002) and disease-free survival (DFS) (P = 0.0008, Fig. 1e, f). Thus, these data suggest a promising prognostic importance in HCC.
Fig. 1. KAT6A is frequently overexpressed in HCC tissues and cell lines.
a Relative KAT6A mRNA expression levels in HCC tissues and matched adjacent non-tumour tissues were determined by qRT-PCR. b Representative western blot analysis of KAT6A expression in the HCC (T) and non-tumour tissues (NT) was shown. c Representative images of IHC staining of KAT6A in HCC and adjacent non-tumour tissues. d Comparing differences in the expression level of KAT6A protein between HCC cell lines with the immortalised normal hepatic cell THLE-3. HCC patients with higher expression of KAT6A had worse overall survival e and disease-free survival f. *P < 0.05.
Table 1.
Clinical correlation of KAT6A expression in HCC (n = 106).
| Clinical parameters | Cases | Expression level | P-value | |
|---|---|---|---|---|
| (n) | KAT6Ahigh (n = 53) | KAT6Alow (n = 53) | (*p < 0.05) | |
| Age(years) | ||||
| <60 years | 64 | 29 | 35 | 0.233 |
| ≥60 years | 42 | 24 | 18 | |
| Gender | ||||
| Male | 81 | 42 | 39 | 0.492 |
| Female | 25 | 11 | 14 | |
| Tumour size (cm) | 0.002* | |||
| <5 cm | 58 | 21 | 37 | |
| ≥5 cm | 48 | 32 | 16 | |
| Tumour number | 0.403 | |||
| Solitary | 91 | 44 | 47 | |
| Multiple | 15 | 9 | 6 | |
| Edmondson | 0.020* | |||
| I + II | 24 | 7 | 17 | |
| III + IV | 82 | 46 | 36 | |
| TNM stage | 0.034* | |||
| I + II | 83 | 37 | 46 | |
| III + IV | 23 | 16 | 7 | |
| Venous infiltration | 0.296 | |||
| Present | 9 | 6 | 3 | |
| Absent | 97 | 47 | 50 | |
| AFP | 0.353 | |||
| <400 ng/ml | 24 | 14 | 10 | |
| ≥400 ng/ml | 82 | 39 | 43 | |
| HBsAg | 0.727 | |||
| Positive | 97 | 48 | 49 | |
| Negative | 9 | 5 | 4 | |
HCC hepatocellular carcinoma, AFP alpha-fetoprotein, TNM tumour-node-metastasis.
*Bold indicates statistically significant.
KAT6A promotes cell proliferation, colony formation and tumour growth
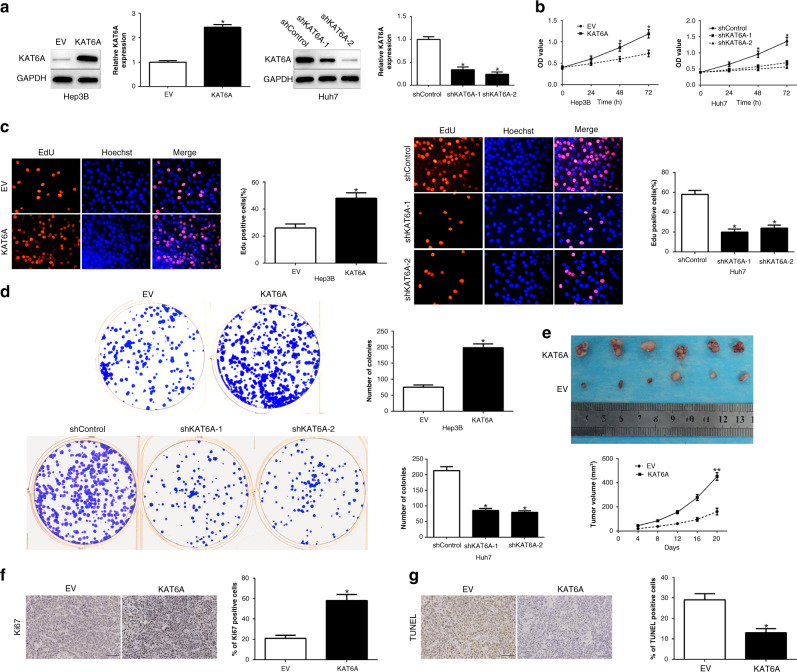
To confirm the biological function of KAT6A in HCC tumorigenesis, we overexpressed KAT6A in Hep3B cells and deleted KAT6A in Huh7 cells, which contained different endogenous KAT6A levels (P < 0.05, Fig. 2a). Functionally, CCK8, EdU and colony formation assays demonstrated that KAT6A overexpression significantly promoted cell viability, proliferation and colonies in Hep3B cells, whereas KAT6A knockdown showed the opposite effects on Huh7 cells (P < 0.05, Fig. 2b–d). To determine the effect of KAT6A in HCC tumorigenesis in vivo, we performed a subcutaneous tumour model and the tumour growth curves revealed that KAT6A overexpression significantly enhanced tumour growth (P < 0.05, Fig. 2e). Moreover, we used Ki-67 and TUNEL staining to evaluate the proliferative and apoptotic rate in the xenografted tissues. These stainings suggest that KAT6A regulated proliferation and impaired apoptosis in vivo (P < 0.05, Fig. 2f, g). Taken together, these data support that KAT6A was critical for HCC growth in vitro and in vivo.
Fig. 2. KAT6A promotes cell viability, proliferation and colony formation of HCC cells in vitro and in vivo.
a Hep3B and Huh7 cells that were transfected with corresponding KAT6A vectors were subjected to WB for KAT6A expression. Overexpression of KAT6A promoted cell viability (b), proliferation (c) and colony formation (d) in Hep3B cells, while downregulation of KAT6A inhibited cell viability (b), proliferation (c) and colony formation (d) in Huh7 cells. e Tumour growth curve revealed that KAT6A overexpression significantly promoted tumour growth in vivo. Tumour nodules were subjected to immunohistochemical staining for Ki-67 (e) and TUNEL (f) assays and quantitative analysis. Representative immunostaining and TUNEL assays revealed that KAT6A overexpression significantly increased the number of Ki-67-positive cells and inhibited the number of apoptotic cells. *P < 0.05.
KAT6A regulates SOX2 expression in HCC cells
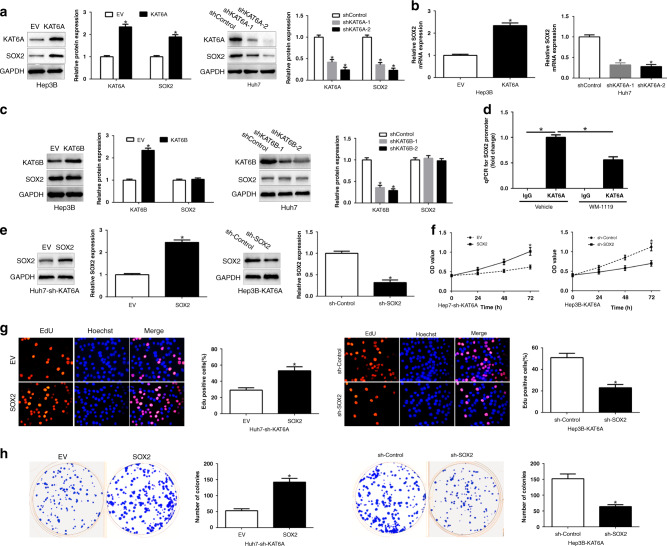
As SOX2 plays a critical role in driving tumour initiation and progression and we investigated the effects of KAT6A on SOX2 expression in HCC cells [15]. As shown in Fig. 3a and 3b, KAT6A obviously regulated SOX2 protein and mRNA expression in HCC cells. The KAT6A homologous KAT6B alternation did not impair SOX2 protein (Fig. 3c). Moreover, ChIP-qPCR suggest that KAT6A binds to the SOX2 promoter, additionally, KAT6A inhibitor WM-1119 reduced the binding (P < 0.05, Fig. 3d). To validate that SOX2 mediated the biological effects of KAT6A in HCC cells, we restored SOX2 in Huh7-sh-KAT6A cells or knockdown SOX2 in Hep3B-KAT6A cells (P < 0.05, Fig. 3e). SOX2 knockdown or inhibitor markedly abrogated the function of exogenous KAT6A overexpression, causing a significant decrease in the cell viability, proliferation and colonies (P < 0.05, Fig. 3f–h, Supplementary Fig. 1E-H). Similarly, SOX2 overexpression promoted HCC cell viability, proliferation and colonies in KAT6A-knockdown Huh7 cells (P < 0.05, Fig. 3f–h, Supplementary Fig. 1A–D). These data demonstrated that KAT6A upregulated SOX2 expression to enhance HCC tumorigenesis.
Fig. 3. KAT6A upregulates SOX2 expression in HCC cells.
a Effects of KAT6A overexpression or knockdown on SOX2 protein expression in Hep3B and Huh7 cells. b qRT-PCR analysis of effects of KAT6A overexpression or knockdown on SOX2 mRNA expression. c WB assays of KAT6B overexpression or knockdown on SOX2 expression. d ChIP-qPCR assays of the binding of KAT6A with the SOX2 promoter. After ChIP using antibodies against KAT6A or control IgG. Modulation of SOX2 partially abolishes KAT6A-mediated cellular processes in HCC. e KAT6A-suppressive Huh7 cells that were transfected with empty vector (EV) or SOX2 overexpression plasmid were subjected to western blot for SOX2. KAT6A-overexpressing Hep3B cells that were transfected with scrambled shRNA or SOX2 shRNA were subjected to western blot for SOX2. SOX2 restoration abrogated the effects of KAT6A knockdown on cell viability (f), proliferation (g) and colony formation (h) of Huh7 cells. SOX2 knockdown reversed the promotive effects of KAT6A overexpression in Hep3B cells (f–h). *P < 0.05.
KAT6A acetyltransferase activity is critical for SOX2 expression
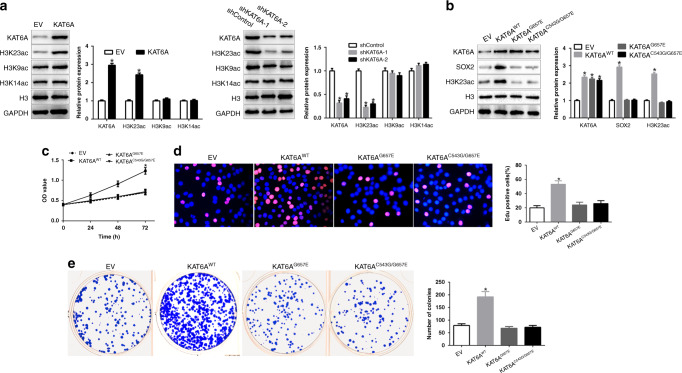
To investigate the potential mechanisms by which KAT6A regulates SOX2 expression, we measured the histone H3 acetylation including H3K9, H3K14 and H3K23, which were the targets of KAT6A [5]. As shown in Fig. 4a, KAT6A overexpression promoted the acetylation levels of H3K23; however, H3K9 and H3K14 did not change. KAT6A knockdown showed the opposite effects on H3K23. To determine H3K23ac is critical for KAT6A-mediated HCC tumour growth, we constructed two KAT6A acetyltransferase activity-deficient mutants, KAT6AG657E and KAT6AC543G/G657E. Overexpression of KAT6A wild-type (WT) in Hep3B cells promoted SOX2 expression, H3K23 acetylation, cell viability, proliferation and colony formation (P < 0.05, Fig. 4b–e), However, KAT6AG657E and KAT6AC543G/G657E overexpression showed no effects on SOX2 expression, H3K23 acetylation, cell viability, proliferation and colony formation (Fig. 4b–e). These results confirmed that KAT6A acetyltransferase activity in critical for SOX2 expression.
Fig. 4. KAT6A acetyltransferase activity is critical for SOX2 expression.
a WB measure the effects of KAT6A overexpression or knockdown on the acetylation of H3K23, H3K9 and H3K14. Histone H3 and GAPDH were used as controls. b Overexpression of KAT6A wild-type but not acetyltransferase activity-deficient mutant, G657E or C543G/G657E, restored KAT6A-knockdown-inhibited activation of SOX2 expression. c–e Effects of overexpression of KAT6A wild-type, acetyltransferase activity-deficient mutant, G657E or C543G/G657E, on cell viability (c), proliferation (d) and colony formation (e) in Hep3B cells. *P < 0.05.
H3K23 acetylation binds with TRIM24 to mediate SOX2 expression
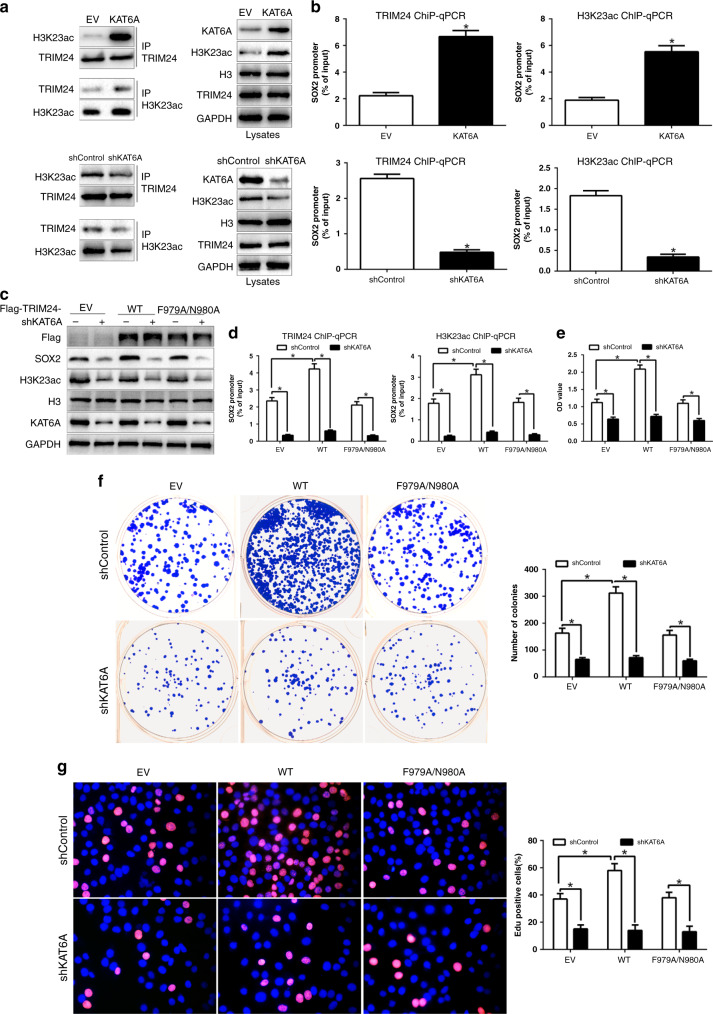
Previous studies reported that TRIM24 binds with H3K23ac to exert its effects in breast cancer and prostate cancer [16, 17]. To confirm it, we performed IP analysis in HCC cells with KAT6A vectors. TRIM24 interacted with H3K23ac in HCC cells and KAT6A overexpression promoted the association of TRIM24 and H3K23ac. However, KAT6A knockdown markedly inhibited the binding (Fig. 5a). To determine whether the association complex binds to SOX2 promoter in HCC cells, we used ChIP-qPCR assays to demonstrate that both TRIM24 and H3K23ac could bind to SOX2 promoter (Fig. 5b). Moreover, KAT6A could regulate the binding of TRIM24 and H3K23ac with SOX2 promoter. To investigate that TRIM24 was essential for KAT6A-mediated SOX2 expression, we restored Flag-tagged TRIM24 WT or F979A/N980A mutant in HCC cells with KAT6A shRNA. As shown in Fig. 5c, compared with the EV control, overexpression of Flag-TRIM24 WT promoted SOX2 expression in Huh7/shControl cells; however, Flag-TRIM24 WT overexpression did not affect SOX2 expression in Huh7/shKAT6A cells. Moreover, overexpression of Flag-TRIM24-F979A/N980A mutant had no effects on SOX2 expression in Huh7 cells with KAT6A shRNA or control shRNA (Fig. 5c). In addition, ChIP-qPCR analyses confirmed that overexpression of Flag-TRIM24 WT increased the binding of TRIM24 and H2K23ac with SOX2 promoter in Huh7/shControl cells but not in Huh7/shKAT6A cells (Fig. 5d). However, overexpression of Flag-TRIM24-F979A/N980A mutant showed no effects on the binding of TRIM24 and H2K23ac with SOX2 promoter. Moreover, Flag-TRIM24 WT overexpression enhanced the cell viability, proliferation and colony formation in Huh7/shControl cells but not in Huh7/shKAT6A cells (P < 0.05, Fig. 5e–g). However, Flag-TRIM24-F979A/N980A mutant did not exert its effects. These data revealed that KAT6A regulated SOX2 expression via the TRIM24/H3K23ac complex.
Fig. 5. H3K23 acetylation binds with TRIM24 to mediate SOX2 expression.
a Immunoprecipitation (IP) measure the effects of KAT6A overexpression or knockdown on H3K23ac association with TRIM24. b ChIP-qPCR assays of the binding of TRIM24 and H3K23ac with the SOX2 promoter. c Overexpression of Flag-TRIM24 wild-type but not the binding mutant of TRIM24 with H3K23ac, F979A/N980A, promoted SOX2 expression. d Effects of overexpression of Flag-TRIM24 wild-type or F979A/N980A mutant on the binding of TRIM24 and H3K23ac with SOX2 promoter. Effects of overexpression of Flag-TRIM24 wild-type or F979A/N980A mutant on cell viability e, proliferation f and colony formation g in Hep3B cells. *P < 0.05.
KAT6A is upregulated by high matrix stiffness and mediates its promoting effects
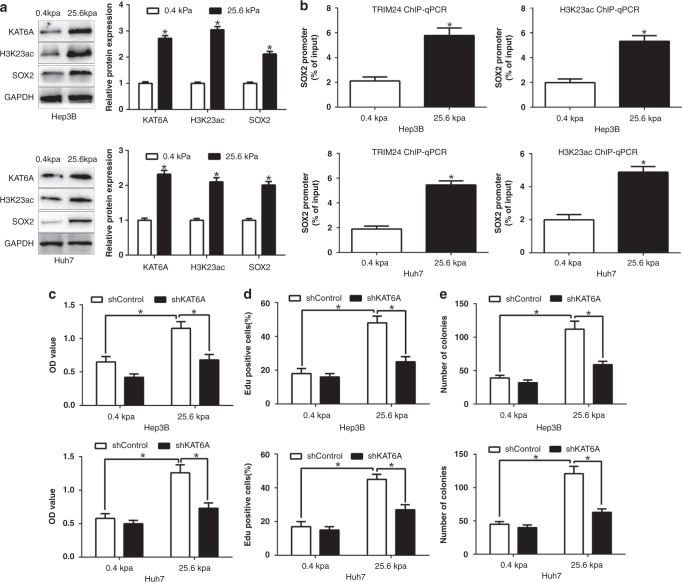
After confirming the functional effects and clinical importance of KAT6A upregulation in HCC, we further explored the reason responsible for the increased expression of KAT6A in HCC. Our previous studies demonstrated that matrix stiffness was an important feature of tumour microenvironments and a critical regulator of HCC progression [18]. Moreover, the expression of SOX2, which was confirmed as a downstream of KAT6A in this study, could be regulated by matrix stiffness. Therefore, we investigated whether the expression of KAT6A could be affected by matrix stiffness. Firstly, we confirmed that high matrix stiffness (25.6 kpa) obviously promoted KAT6A, H3K23ac and SOX2 expression in HCC cells by different matrix stiffness culture system (P < 0.05, Fig. 6a, Supplementary Fig. 2). Moreover, high matrix stiffness significantly promoted the binding of TRIM24 and H3K23ac with SOX2 promoter (Fig. 6b). Furthermore, KAT6A knockdown could abrogate the promoting effects of matrix stiffness on the cell viability, proliferation and colony formation of HCC cells (P < 0.05, Fig. 6c–e). Taken together, these data suggest that the increased expression of KAT6A in HCC is mediated by matrix stiffness and is critical for the promoting effects of matrix stiffness on the progression of HCC cells.
Fig. 6. KAT6A is upregulated by high matrix stiffness and mediates its promoting effects.
a The expression of KAT6A, H3K23ac and SOX2 in low or high matrix stiffness condition. b ChIP-qPCR assays of the binding of TRIM24 and H3K23ac with the SOX2 promoter in low or high matrix stiffness condition. KAT6A knockdown reversed the promotive effects of high matrix stiffness on cell viability c, proliferation d and colony formation e. *P < 0.05.
Discussion
Lysine acetyltransferases (KATs) catalyse lysine acetylation and presented a reversible protein modification in tumour initiation, development and progression [19]. KAT6A belongs to the MYST family of histone acetyltransferases and exert a critical role in the regulation of chromatin organisation and function [20]. Previous studies confirmed that KAT6A expression was upregulated in ovarian cancer and promoted tumorigenicity and chemoresistance in ovarian cancer by acetylating COP1 at K294 and led to the accumulation and enhanced activity of β-catenin [21]. KAT6A amplifications were associated with shorter progression-free survival and overall survival in patients with endometrial serous carcinoma [22]. Here, in present study, we described a novel mechanism that KAT6A is important in HCC growth and progression. The matrix stiffness tumour microenvironment promoted KAT6A expression. KAT6A promoted H3K23 acetylation and enhanced the association of TRIM24 and H3K23ac. TRIM24 functions as a transcriptional activator to upregulate SOX2 expression and promote HCC tumorigenesis.
Hepatocellular carcinoma develops in an altered biomechanical environment and the increased stiffness was an important predictor of HCC development [23]. Liver fibrosis was pathologically associated with increased rigidity of the extracellular matrix, involving multiple signal transduction pathways that regulate HCC development [24, 25]. Our data confirmed that CXCR4 was identified as a critical intra-cellular signal transducer that relays matrix stiffness signals to control mechano-sensitive cellular activities through URTD1-mediated YAP signaling pathway [18]. Matrix stiffness regulates α-TAT1-mediated acetylation of α-tubulin and promotes silica-induced epithelial-mesenchymal transition via DNA damage [26]. Here, we confirmed that high matrix stiffness could promote KAT6A expression and exerted its histone acetyltransferase activity to enhance H3K23 acetylation. We demonstrated that KAT6A was upregulated in HCC tissues and cell lines. Moreover, increased KAT6A level was obviously correlated with malignant clinicopathological features, including large tumour size, high histological grade and advanced TNM stage. In addition, KAT6A was a prognostic factor in predicting survival of HCC patients. These data demonstrated that KAT6A is involved in HCC progression and represents as a promising biomarker for HCC therapy.
In this study, our results showed that KAT6A promoted cell viability, proliferation and colony formation by gain- and loss-of function experiment in vitro and in vivo. KAT6A upregulates SOX2 transcription in HCC cells. Alteration of SOX2 expression could reverse the biological function of KAT6A in HCC cells. Moreover, KAT6A-regulated H3K23 acetylation and its acetyltransferase activity was important for SOX2 expression. KAT6A activated gene transcription by acetylating histones of target gene promoters through interacting with transcriptional factors, including p53 [27]. KAT6A interacts with and enhances p53 acetylation, which increased p53 activity and induce p21 expression [28, 29]. Here, we found that KAT6A obviously promoted H3K23 acetylation and enhances the association of TRIM24 with chromatin. Moreover, KAT6A or TRIM24 overexpression upregulates SOX2 transcription in HCC cells. However, KAT6A acetyltransferase-deficient mutant or TRIM24 binding mutant showed no effects. TRIM24 promotes stemness and invasiveness of glioblastoma cells via activating SOX2 expression [30]. In colorectal cancer, TRIM24 was activated by the DANCR/KAT6A complex and binds with H3K23ac to induce TRIM24-mediated recruitment of YAP to chromatin, which promoted the proliferation of colorectal cancer cells [31]. In conclusion, we demonstrated that KAT6A-regulated SOX2 expression depends on its acetyltransferase activity and H3K23/TRIM24 complex.
In conclusion, we reported for the first time that KAT6A was upregulated in HCC tissues and cells. Its overexpression was associated with malignant clinical features and unfavourable prognosis. We confirmed that KAT6A promotes cell viability, proliferation and colony formation through H3K23ac/TRIM24-SOX2 signaling pathway. Moreover, matrix stiffness can upregulate the expression of KAT6A in HCC cells. Taken together, this study demonstrates that KAT6A may potentially act as the novel biomarker and the attractive therapeutic target in HCC.
Supplementary information
Author contributions
Study concept and design: ZKL and NY. Acquisition of the data: WZ, HYM, RKL, TXC and NY. Management of data acquisition: WZ, HYM, RKL, TXC, ZKL and NY. Analysis of the present data: WZ, HYM and ZKL. Statistical analysis: WZ and NY. Critical interpretation of the present data: WZ, HYM, RKL, TXC and NY. Drafting of the paper: WZ, ZKL and NY. Critical revision of the paper for important intellectual content: WZ, HYM, RKL, TXC, ZKL and NY. Obtained funding: ZKL and NY. Technical or material support: WZ, ZKL and NY. Study supervision: ZKL and NY.
Funding information
This study was supported by grants from the National Natural Science Foundation of China (82103428), Natural Science Basic Research Program of Shaanxi (No.2020JQ-496, No.2020JQ-498); the Institutional Foundation of the First Affiliated Hospital of Xi’an Jiaotong University (2019QN-24, 2020QN-10); the Fundamental Research Funds for the Central Universities (Program No. xzy012020041).
Data availability
The datasets generated and/or analysed during the current study are not publicly available but are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All animal experiments in our study were carried out in accordance with the Helsinki Declaration, and approved by the Ethics Committee the First Affiliated Hospital of Xi’an Jiaotong University (XJTU-2021-668). Patients were informed that the resected specimens were stored by the hospital and potentially used for scientific research, and that their privacy would be maintained. All patients provided informed consent prior to undergoing screening procedures. Our study protocol was approved by the Ethics Committee of Xi’an Jiaotong University (Xi’an, China).
Consent to publish
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nan Yang, Email: nan_yang@xjtufh.edu.cn.
Zhikui Liu, Email: liuzk0319@xjtufh.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-022-01784-9.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Bruix J, Sherman M. American Association for the Study of Liver D. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–2. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura M, Chiba T, Kanayama K, Kanzaki H, Saito T, Kusakabe Y, et al. Epigenetic dysregulation in hepatocellular carcinoma: an up-to-date review. Hepatol Res. 2019;49:3–13. doi: 10.1111/hepr.13250. [DOI] [PubMed] [Google Scholar]
- 4.Borrow J, Stanton VP, Jr, Andresen JM, Becher R, Behm FG, Chaganti RS, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 5.Huang F, Abmayr SM, Workman JL. Regulation of KAT6 acetyltransferases and their roles in cell cycle progression, stem cell maintenance, and human disease. Mol Cell Biol. 2016;36:1900–7. doi: 10.1128/MCB.00055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lian J, Zhang H, Wei F, Li Q, Lu Y, Yu B, et al. Long non-coding RNA DANCR promotes colorectal tumor growth by binding to lysine acetyltransferase 6A. Cell Signal. 2020;67:109502. doi: 10.1016/j.cellsig.2019.109502. [DOI] [PubMed] [Google Scholar]
- 7.Baell JB, Leaver DJ, Hermans SJ, Kelly GL, Brennan MS, Downer NL, et al. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature. 2018;560:253–7. doi: 10.1038/s41586-018-0387-5. [DOI] [PubMed] [Google Scholar]
- 8.Yu L, Liang Y, Cao X, Wang X, Gao H, Lin SY, et al. Identification of MYST3 as a novel epigenetic activator of ERalpha frequently amplified in breast cancer. Oncogene. 2017;36:2910–8. doi: 10.1038/onc.2016.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheikh BN, Phipson B, El-Saafin F, Vanyai HK, Downer NL, Bird MJ, et al. MOZ (MYST3, KAT6A) inhibits senescence via the INK4A-ARF pathway. Oncogene. 2015;34:5807–20. doi: 10.1038/onc.2015.33. [DOI] [PubMed] [Google Scholar]
- 10.Fei D, Wang Y, Zhai Q, Zhang X, Zhang Y, Wang Y, et al. KAT6A regulates stemness of aging bone marrow-derived mesenchymal stem cells through Nrf2/ARE signaling pathway. Stem Cell Res Ther. 2021;12:104. doi: 10.1186/s13287-021-02164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lv D, Jia F, Hou Y, Sang Y, Alvarez AA, Zhang W, et al. Histone acetyltransferase KAT6A upregulates PI3K/AKT signaling through TRIM24 binding. Cancer Res. 2017;77:6190–201. doi: 10.1158/0008-5472.CAN-17-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z, Wang Y, Dou C, Xu M, Sun L, Wang L, et al. Hypoxia-induced up-regulation of VASP promotes invasiveness and metastasis of hepatocellular carcinoma. Theranostics. 2018;8:4649–63. doi: 10.7150/thno.26789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dou C, Liu Z, Tu K, Zhang H, Chen C, Yaqoob U, et al. P300 acetyltransferase mediates stiffness-induced activation of hepatic stellate cells into tumor-promoting myofibroblasts. Gastroenterology. 2018;154:2209–21. doi: 10.1053/j.gastro.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Wang Y, Dou C, Sun L, Li Q, Wang L, et al. MicroRNA-1468 promotes tumor progression by activating PPAR-gamma-mediated AKT signaling in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:49. doi: 10.1186/s13046-018-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Sun C, Sun L, Li Y, Kang X, Zhang S, Liu Y. Sox2 expression predicts poor survival of hepatocellular carcinoma patients and it promotes liver cancer cell invasion by activating Slug. Med Oncol. 2013;30:503. doi: 10.1007/s12032-013-0503-1. [DOI] [PubMed] [Google Scholar]
- 16.Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–32. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groner AC, Cato L, de Tribolet-Hardy J, Bernasocchi T, Janouskova H, Melchers D, et al. TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer Cell. 2016;29:846–58. doi: 10.1016/j.ccell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang N, Chen T, Wang L, Liu R, Niu Y, Sun L, et al. CXCR4 mediates matrix stiffness-induced downregulation of UBTD1 driving hepatocellular carcinoma progression via YAP signaling pathway. Theranostics. 2020;10:5790–801. doi: 10.7150/thno.44789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang F. New KAT6 inhibitors induce senescence and arrest cancer growth. Synth Syst Biotechnol. 2018;3:244–5. doi: 10.1016/j.synbio.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner-Ivey B, Guest ST, Irish JC, Kappler CS, Garrett-Mayer E, Wilson RC, et al. KAT6A, a chromatin modifier from the 8p11-p12 amplicon is a candidate oncogene in luminal breast cancer. Neoplasia. 2014;16:644–55. doi: 10.1016/j.neo.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W, Zhan Z, Zhang M, Sun B, Shi Q, Luo F, et al. KAT6A, a novel regulator of beta-catenin, promotes tumorigenicity and chemoresistance in ovarian cancer by acetylating COP1. Theranostics. 2021;11:6278–92. doi: 10.7150/thno.57455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saglam O, Tang Z, Tang G, Medeiros LJ, Toruner GA. KAT6A amplifications are associated with shorter progression-free survival and overall survival in patients with endometrial serous carcinoma. PloS ONE. 2020;15:e0238477. doi: 10.1371/journal.pone.0238477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albhaisi S, Sanyal AJ. Applying non-invasive fibrosis measurements in NAFLD/NASH: progress to date. Pharmaceut Med. 2019;33:451–63. doi: 10.1007/s40290-019-00305-z. [DOI] [PubMed] [Google Scholar]
- 24.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schrader J, Gordon-Walker TT, Aucott RL, van Deemter M, Quaas A, Walsh S, et al. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology. 2011;53:1192–205. doi: 10.1002/hep.24108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Chen S, Zhang Y, Xu H, Xu D, Wei Z, et al. Matrix stiffness regulates alpha-TAT1-mediated acetylation of alpha-tubulin and promotes silica-induced epithelial-mesenchymal transition via DNA damage. J Cell Sci. 2021;134:jcs243394. doi: 10.1242/jcs.243394. [DOI] [PubMed] [Google Scholar]
- 27.Rokudai S, Laptenko O, Arnal SM, Taya Y, Kitabayashi I, Prives C. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc Natl Acad Sci USA. 2013;110:3895–900. doi: 10.1073/pnas.1300490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang XJ. MOZ and MORF acetyltransferases: molecular interaction, animal development and human disease. Biochim Biophys Acta. 2015;1853:1818–26. doi: 10.1016/j.bbamcr.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Rokudai S, Aikawa Y, Tagata Y, Tsuchida N, Taya Y, Kitabayashi I. Monocytic leukemia zinc finger (MOZ) interacts with p53 to induce p21 expression and cell-cycle arrest. J Biol Chem. 2009;284:237–44. doi: 10.1074/jbc.M805101200. [DOI] [PubMed] [Google Scholar]
- 30.Zhang LH, Yin YH, Chen HZ, Feng SY, Liu JL, Chen L, et al. TRIM24 promotes stemness and invasiveness of glioblastoma cells via activating Sox2 expression. Neuro Oncol. 2020;22:1797–808. doi: 10.1093/neuonc/noaa138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie W, Zhang Y, Wang B, Hu Y, Zhan B, Wei F, et al. Tripartite motif containing 24 regulates cell proliferation in colorectal cancer through YAP signaling. Cancer Med. 2020;9:6367–76. doi: 10.1002/cam4.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are not publicly available but are available from the corresponding author on reasonable request.