Abstract
Inflammation is an immune response to protect against various types of infections. When unchecked, acute inflammation can be life-threatening, as seen with the current coronavirus pandemic. Strong oxidants, such as peroxynitrite produced by immune cells, are major mediators of the inflammation-associated pathogenesis. Cellular thiols play important roles in mitigating inflammation-associated macromolecular damage including DNA. Herein, we have demonstrated a role of glutathione (GSH) and other thiols in neutralizing the effect of peroxynitrite-mediated DNA damage through stable GSH-DNA adduct formation. Our observation supports the use of thiol supplements as a potential therapeutic strategy against severe COVID-19 cases and a Phase II (NCT04374461) open-label clinical trial launched in early May 2020 by the Memorial Sloan Kettering Cancer Center.
Graphical Abstract
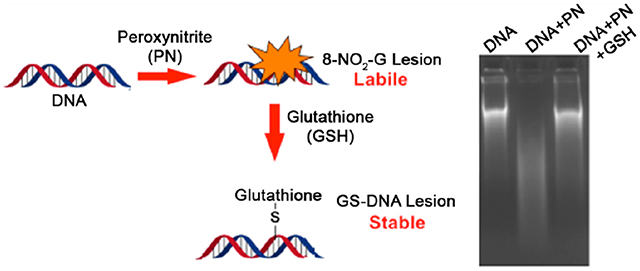
Peroxynitrite, a strong oxidant is produced at the site of acute inflammation causing macromolecular damages. Herein, we have shown that DNA-damage caused by peroxynitrite is mitigated by formation of stable adduct with cellular thiols like GSH.
INTRODUCTION
Inflammation is the body’s natural way of eradicating infectious agents including viruses, bacteria, fungi, parasites, and foreign substances.1–4 Acute inflammation triggers an influx of immune cells such as macrophages and neutrophils at the sites of infection.1,5 For respiratory pandemic viruses including influenza A, H5N1, 1918 influenza virus, SARS-CoV, and SARS-CoV-2, the surge of inflammatory cells causes severe lung injury, often contributing to patient mortality.6,7 Studies with H5N1 attributed much of the severe lung injury to excessive reactive oxygen (ROS) and nitrogen (RNS) species generated by the immune cells,8,9 which are indiscriminately cytotoxic to the infected cells and surrounding host tissue because of their abilities to cause DNA and protein damage and lipid peroxidation. Reactive-species-induced somatic mutations can also cause malignant transformation of the host cells.10
During inflammation, overproduction of nitric oxide (NO) and superoxide (O2•−) results in peroxynitrite (PN) formation from its combination.11 In vivo, PN reacts with CO2 to generate nitrosoperoxycarbonate species that rapidly decomposes to form CO3•− and NO2 radicals.12 The primary DNA lesions resulting from PN exposure is 8-nitroguanine (8-nitro-G), 8-oxo-7,8-dehydroguanine (8-oxo-G), and 5-guanidino-4-nitroimidazole (NIm). PN exposure also results in the formation of GT cross-links in DNA.12 8-NO2-G (half-life of ~1.6 and 2.4 h for single- and double-stranded DNA, respectively),13–15 being labile, undergoes spontaneous depurination to form abasic sites, ultimately resulting in DNA strand breaks (Scheme 1. In vitro studies with oligonucleotides containing 8-nitro-2′-methoxyguanosine at a defined site demonstrated it to be mutagenic.16
Scheme 1.
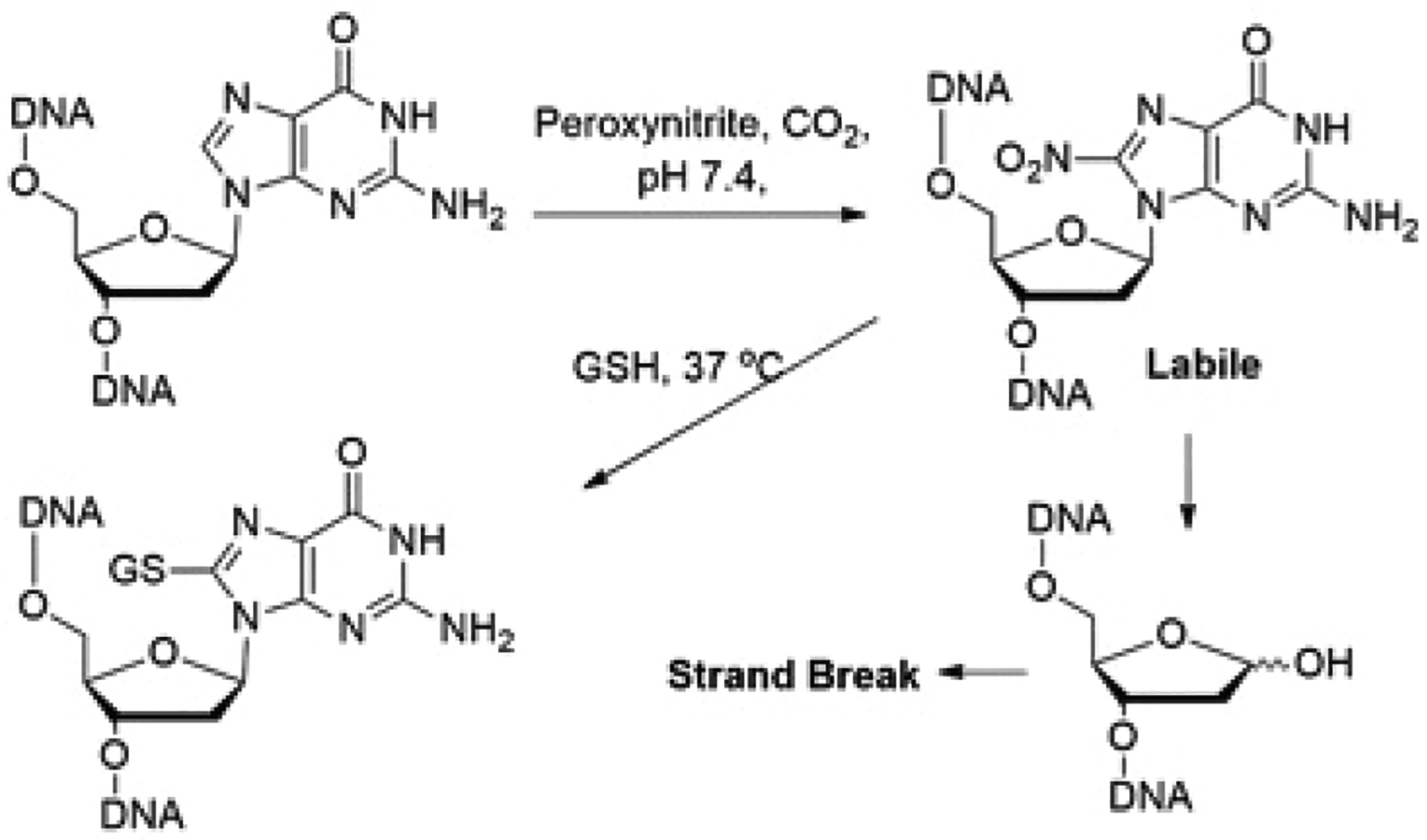
Cellular Fate of 8-Nitroguanine Formed in DNA
Glutathione (GSH), a tripeptide (l-c-glutamyl-l-cysteinylglycine) containing a thiol (SH) group, serves as an important cellular protective system against electrophiles and free radicals including those formed during inflammatory responses.17 GSH plays a critical role in lung defense mechanisms, particularly in protecting airspace epithelium (membrane integrity) from oxidative/free radical-mediated injury and inflammation.18 Various studies reported administration of extracellular GSH to reduce inflammation in lung diseases. Intravenous administration of N-acetylcysteine (NAC), a cell permeable precursor of cysteine (Cys), for 72 h improved systemic oxygenation and reduced the need for ventilatory support in patients with mild-to-moderate acute lung injury.19,20 The protective effect of GSH generally stems from its ability to quench free radicals and radical centers in DNA and other macromolecules.21 Herein, we report alteration of DNA damage inflicted by PN through formation of a stable GSDNA adduct, reducing its cytotoxicity. We hypothesized that the major PN-induced DNA lesion 8-NO2-G reacts with cellular thiols like GSH to form a nonlabile thiol adduct of dG in DNA and prevents DNA cleavage. Substitution of the nitro group in 8-NO2-G by thiols in nucleosides and Cys in oligonucleotides has been reported.22,23
EXPERIMENTAL PROCEDURE
Reagents and Cell Line.
All reagents were of the highest purity available and, unless otherwise mentioned, were obtained from Sigma-Aldrich (now Merck, St. Louis, MO, USA); 2′-deoxyguanosine, Agarose, ethidium bromide, ethanol, dimethyl sulfoxide (DMSO), NaCl, and ethylenediaminetetraacetic acid (EDTA) were obtained from HiMedia (Mumbai, India); N,N′-dimethylethylenediamine (DMEDA) from Tokyo Chemical Industry (Tokyo, Japan); acetonitrile from JT Baker (Center Valley, PA, USA); glutathione from Sisco Research Lab. Pvt Ltd. (Mumbai, India). HEK293T cell line was procured from NCCS (Pune, India) and authenticated by short tandem repeat profiling (Lifecode Technologies, New Delhi, India).
Synthesis of Peroxynitrite (PN).
PN was freshly prepared before each reaction using a published protocol.24 Briefly, the following three solutions were freshly prepared in distilled water: 0.7 M HCL + 0.6 M H2O2, 0.6 M sodium nitrite, and 3 M sodium hydroxide. The solutions 0.7 M HCl + 0.6 M H2O2 and 0.6 M sodium nitrite were loaded into individual syringes that were connected with the use of a “T” connector. Contents of the individual syringes were mixed with the help of a syringe pump running at a flow rate of 17 mL/min. After mixing, the reaction was quenched by collecting the eluent in a beaker containing 3 M NaOH on ice. The lengths of the tubing that determine the reaction time were optimized for the highest yields. Metals were avoided in all connections.
Transformation.
Twenty micrograms of pUC19 plasmid in 100 mM potassium phosphate (pH 7.5) buffer was treated with peroxynitrite (50 μM) on ice for 10 min followed by the addition of GSH (5 mM) and incubation at 37 °C for 2 h. After incubation, DNA was precipitated using 70% ethanol and 0.3 M sodium acetate. The precipitated plasmid DNA was pelleted by centrifugation at 10000g, washed with cold 70% ethanol, and air-dried. Competent ampicillin-sensitive E. coli cells were transformed following the standard protocol giving a heat shock at 42 °C and plated on Luria-Bertani (LB)-agar plates having ampicillin (100 μg/mL). The reactions were carried out in triplicate. The plates were incubated overnight at 37 °C and colonies counted.
Transfection.
Fifty micrograms of pEGFP-N1 plasmid was treated with PN (5 μM) in the presence of 1 mM carbonate and 100 mM potassium phosphate buffer (pH 7.5) on ice for 10 min. After the addition of GSH (5 mM), the reaction mixture was incubated at 37 °C for another 2 h. A control reaction was performed without the addition of GSH. After incubation, DNA was precipitated using 70% cold ethanol and 0.3 M sodium acetate. The precipitated plasmid DNA was pelleted by centrifugation at 10000g, washed with cold 70% ethanol, and air-dried. HEK-293T cells were transfected with precipitated plasmid (7.5 μg) using Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific) as per the manufacturer’s instruction. Imaging was done at 10× in Leica DMi1 (Wetzlar, Germany) after 24 h.
DNA Cleavage Assays.
In a typical DNA cleavage reaction, plasmid DNA (pUC19, 1 μg) in potassium phosphate buffer (100 mM, pH 7.4) was treated with PN (0–100 μM) on ice for 10 min followed by addition of 5 mM GSH and incubation at 37 °C for 2 h. For assays involving the effect of thiols on DNA cleavage by PN, plasmid DNA (pUC19, 1 μg) in potassium phosphate buffer (100 mM, pH 7.4) was treated with PN (50 μM) on ice for 10 min followed by the addition of 0–500 μM of various thiols (GSH, β-mercaptoethanol, or cysteine) and incubation at 37 °C for 2 h. The reactions were done in triplicate. Following incubations, reactions were subjected to N,N′-dimethylethylenediamine (DMEDA, 100 mM) workup for 2 h at 37 °C, quenched by the addition of 5 μL of glycerol loading buffer, and then electrophoresed for 45 min at 80 V in 1% agarose gel (w/v) containing 0.5 μg/mL ethidium bromide. DNA was visualized and quantified using a GE Image Quant LAS 500 gel documentation system (Massachusetts, USA). Strand breaks per plasmid DNA molecule (S) were calculated using the equation S = –ln fI, where fI is the fraction of plasmid present as form I.
UV–Vis and Fluorescence Spectroscopy.
Calf thymus DNA (20 μg) in potassium phosphate buffer (100 mM, pH 7.4) was treated with PN (1 mM) for 10 min on ice followed by the addition of GSH (5 mM) or synthesized dansylated GSH (5 mM) at 37 °C for 2 h.25 Following incubation, DNA was precipitated with 70% ethanol and 0.3 M sodium acetate. The precipitated DNA was pelleted by centrifugation at 10000g and washed three times with cold 70% ethanol to get rid of any unbound dansylated GSH. The DNA was air-dried and dissolved in water and the absorbance was analyzed at 340 nm by an Agilent Cary 8454 UV–vis diode array spectrophotometer and emission at 520 nm using an Horiba Fluorolog-3 spectrofluorometer.
Formation of DNA Adduct with Calf Thymus DNA.
In a typical reaction, 1 mg of calf thymus (sonicated) DNA and 500 μM PN in 100 mM potassium phosphate buffer (pH 7.5) was incubated for 10 min on a stirrer at 25 °C. Then GSH (5 mM) was added and the reaction mixture was incubated at 37 °C for another 2 h. Following incubation, the DNA was precipitated with 70% ethanol and 0.3 M sodium acetate. The precipitated DNA was pelleted by centrifugation at 10000g, washed three times with cold 70% ethanol, and air-dried. Precipitated DNA was redissolved in 50 mM potassium phosphate (pH 7.4) buffer containing MgCl2 (5 mM) and digested with DNase (250 units), phosphodiesterase (0.01 units), benzonase (0.1 unit), and alkaline phosphatase (2.5 units) for 6 h at 37 °C. After digestion, the reaction was quenched with cold acetonitrile (1:1 v/v) and denatured proteins were removed by centrifugation at 10000g for 20 min. The supernatant was dried under a stream of N2 and analyzed by LC-tandem MS.
Formation of GS-dG Adduct.
To form GS-dG adduct, we incubated peroxynitrite (1 mM) in an ice-cold solution of dG (500 μM) in 100 mM potassium phosphate (pH 7.5) for 10 min followed by the addition of GSH (5 mM) and incubation for another 2 h at 37 °C. The sample was analyzed with LC-tandem MS.
Stability of GS-dG Adduct.
In a vial, 2′-deoxyguanosine (500 μM) was dissolved in 100 mM potassium phosphate buffer, pH 7.4, and stirred on ice. To this ice-cold solution, peroxynitrite (1 mM) was added and stirred for another 5 min. After the addition of GSH (10 mM), the reaction mixture was incubated at 37 °C for 2 h. The GS-dG adduct formed was HPLC-purified to homogeneity. The purified GS-dG adduct was again incubated in 100 mM potassium phosphate buffer, pH 7.4, at 37 °C. At different time points aliquots were removed and analyzed using LCMS.
LC-MS Detection of GS-dG Adducts.
An Agilent 6540 UHD Accurate-Mass Q-TOF LC-MS System (Agilent Technologies, Santa Clara, CA, USA) with an Agilent UHPLC system was used for LC-tandem MS analysis. Data was analyzed using the MassHunter software (Agilent Technologies, Santa Clara, CA, USA). A Phenomenex (Torrance, CA, USA) Kinetex Polar C18 column (2.6 μm, 2.1 mm × 100 mm) was used for chromatography. GS-dG adducts were separated using solvent A (0.1% HCO2H and water, v/v) and solvent B (0.1% HCO2H and CH3CN, v/v) following a gradient program with a flow rate of 300 μL min−1: 0–2 min, 98% A (v/v); 2.0–12.5 min, linear gradient to 100% B; 12.5–15.5 min, hold at 100% B (v/v); 15.5–16.0 min, linear gradient to 98% A (v/v); 16–20 min, hold at 98% A (v/v). The temperature of the column was maintained at 30 °C and samples (20 μL) were infused with an autosampler. ESI conditions were as follows: gas temperature, 325 °C; drying gas flow rate, 8 L/min; nebulizer, 35 psi; sheath gas temperature, 300 °C; sheath gas flow rate, 10 L/min; capillary voltage, 3000 V; nozzle voltage, 1000 V; capillary current, 0.054 μA; chamber current, 4.23 μA; fragmentor voltage, 80 V; and skimmer voltage, 70 V. For MS/MS, a normalized collision energy of 20% was used.
RESULTS AND DISCUSSION
GSH Protects DNA from PN-Mediated Loss of Its Biological Effect.
To test our hypothesis, we first verified if the presence of GSH interferes with the biological and chemical properties of PN-treated DNA. Accordingly, we treated a pUC19 plasmid DNA bearing an ampicillin-resistance gene with PN (1 mM) in the presence of carbonate (1 mM). Under physiological conditions, PN reacts with CO2 to generate nitrosoperoxycarbonate anion.12 The reaction was allowed to proceed for 10 min on ice for complete disappearance of PN, followed by incubation with a physiological concentration of GSH (5 mM) at 37 °C for another 2 h. The pUC19 DNA was purified and transformed into E. coli. The cells were grown in the presence of ampicillin overnight and colonies were counted. We found significantly more colonies in cells transformed with pUC19 treated with PN + GSH than with only PN (Figure 1A). To confirm this observation, we performed parallel experiments in mammalian cells. A pEGFP-N1 plasmid bearing green fluorescence protein (GFP) cDNA was treated with PN alone or PN + GSH and transfected into the SV-40 T antigen-expressing immortal Human Embryonic Kidney (HEK)-293T cells. Twenty-four hours after transfection, cells were subjected to microscopic analysis for visualization of GFP expression. Consistent with the E. coli experiment, we found a comparable number of green cells in the PN + GSH treatment group to those of the control group (pEGFP-N1), with no detectable green cells in the PN group (Figure 1B). On the basis of these experiments, we conclude that GSH can reverse the effect of PN on DNA. The possibility of GSH directly neutralizing PN was avoided by adding GSH 10 min after PN treatment. Both PN and nitrosoperoxycarbonate anion are reactive species with half-lives of only a few seconds in aqueous neutral buffer.11,26
Figure 1.
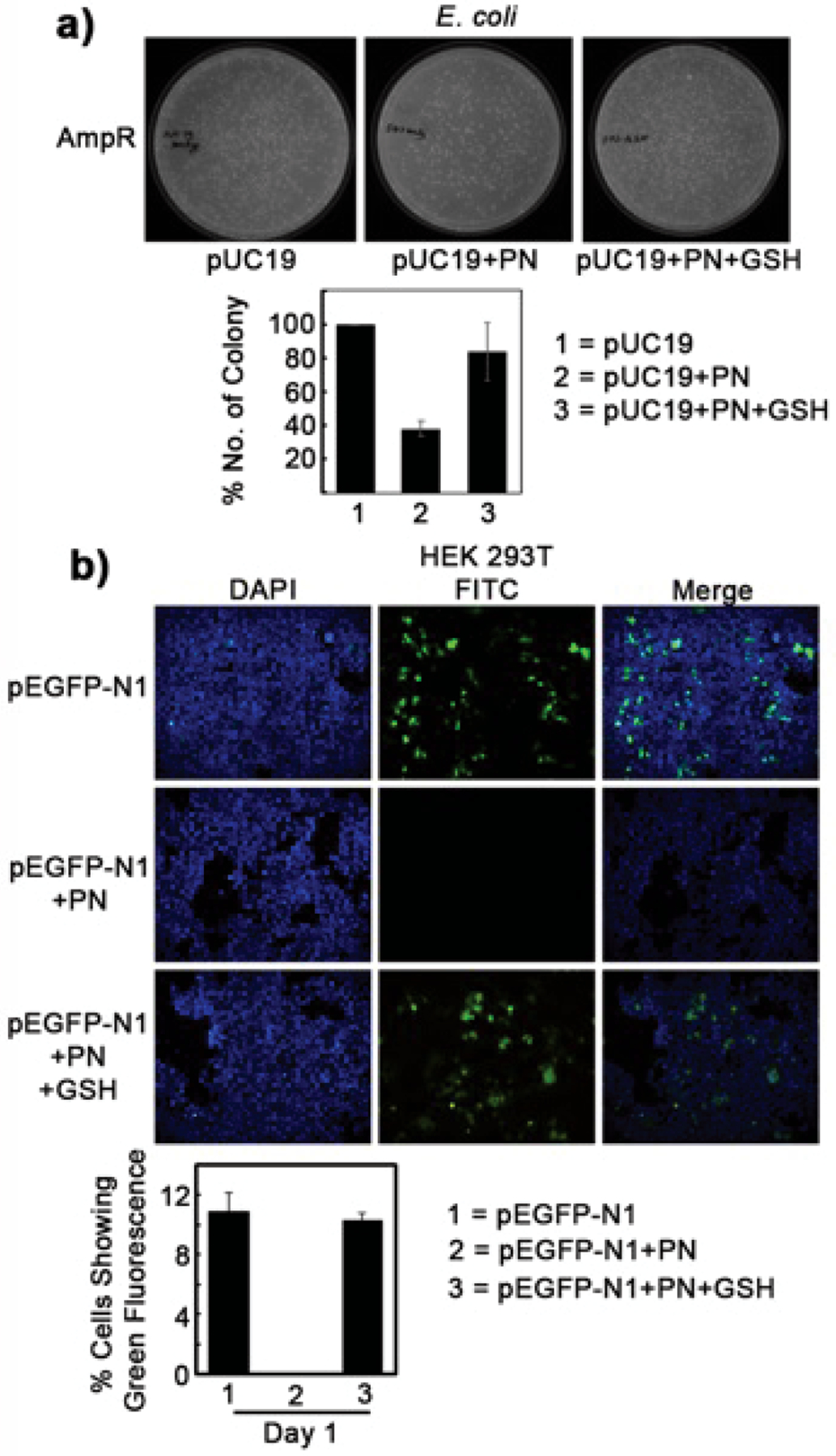
Protective effect of GSH on PN-mediated DNA damage. (A) Ampicillin-resistant colonies transformed with ampicillin-resistant pUC19 plasmid (20 μg) treated with PN (50 μM) or PN followed by GSH (5 mM). (B) Green fluorescence protein-expressing HEK-293T cells transiently transfected with green fluorescence plasmid pEGFPN1 (50 μg) treated with PN (5 μM) or PN followed by GSH (5 mM).
Effect of GSH and Thiols on PN-Mediated DNA Cleavage.
To further elaborate on this detoxifying effect of GSH against PN-mediated DNA damage, we treated calf thymus DNA with PN at 25 °C for 10 min followed by GSH addition. Because the half-life of 8-NO2-G in DNA is significantly higher, 25 °C was used. Labile sites including 8-NO2-G were converted into cleavage sites using DMEDA workup and analyzed by agarose gel electrophoresis.27 The extent of DNA cleavage was qualitatively detected by the length of DNA smear visible in an agarose gel. Typically, ctDNA produces a very tight smear consisting of comparable length DNA fragments. Treatment with PN resulted in a diffused smear, indicative of extensive DNA cleavage. Addition of GSH 10 min after PN treatment resulted in a smearing pattern similar to that observed for ctDNA alone, indicating the presence of larger fragments (Figure 2A, top panel). These results confirmed that GSH can prevent DNA cleavage caused by PN treatment.
Figure 2.
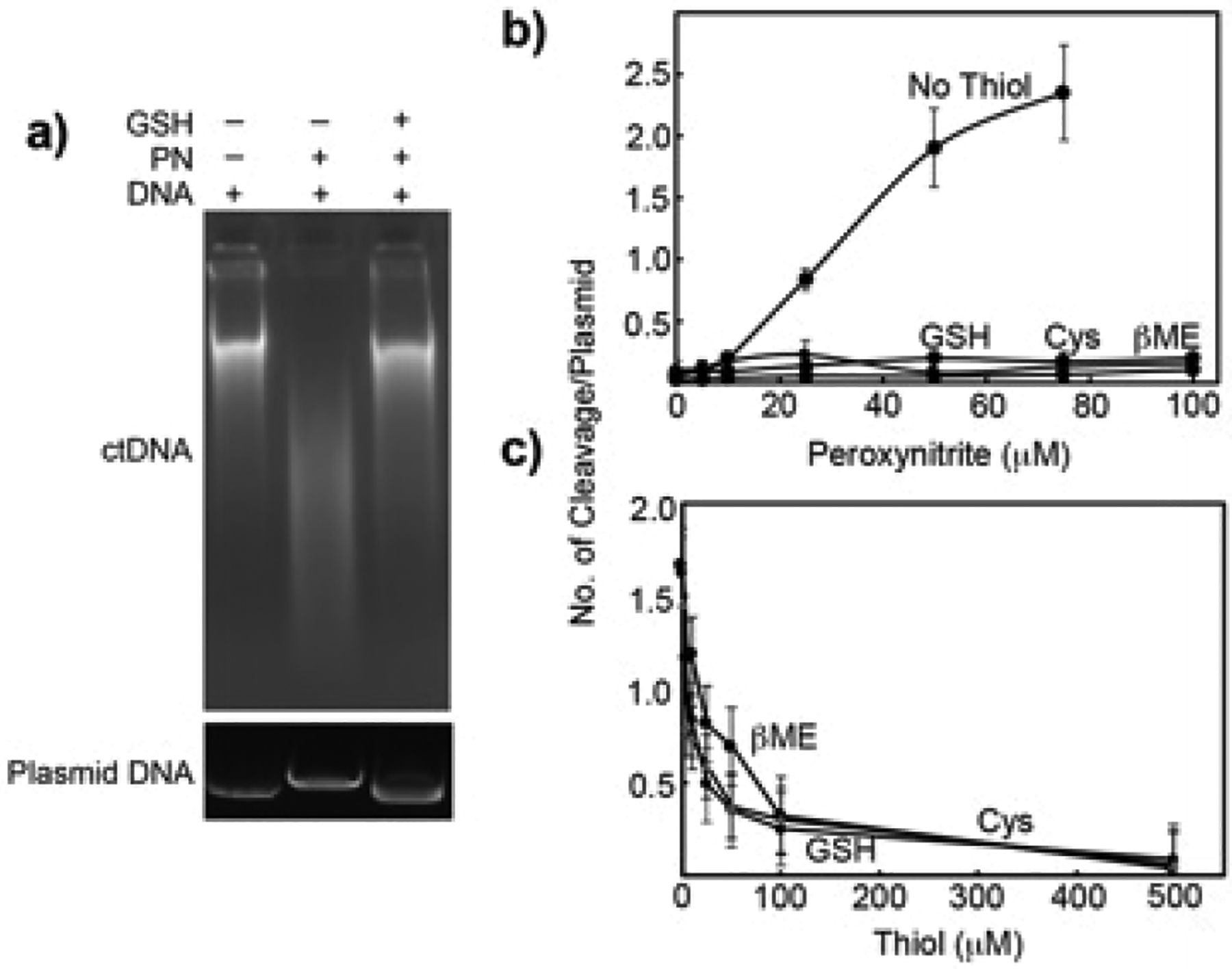
DNA cleavage assay. (A) Agarose gel images of ctDNA or plasmid following various treatments. ctDNA (1 mg) or pUC19 (1 μg) were incubated with PN (0.5 mM for ctDNA and 1 mM for pUC19) for 10 min at 25 °C (ctDNA) or on ice (pUC19) followed by the addition of GSH (5 mM, 37 °C for 2 h). (B) pUC19 (1 μg) DNA cleavage by PN in the presence and absence of thiols (5 mM). (C) pUC19 (1 μg) DNA cleavage by PN (50 μM) in the presence of varying concentrations of thiols.
We repeated the above experiment with the widely used supercoiled plasmid DNA cleavage assay for quantitative analysis.27,28 The results in Figure 2, parts A (bottom panel) and B, revealed significant DNA cleavage with an increasing concentration of PN that was almost completely rescued by the addition of 5 mM GSH, cysteine (Cys), and β-mercaptoethanol (βME). These data suggested that the addition of thiols converts labile sites in DNA to nonlabile sites. Repeating the experiment with 1 mM PN and increasing the concentration of thiols showed that 100 μM thiol was sufficient to inhibit DNA cleavage by PN (Figure 2C). Together, these data confirmed that thiols prevent the DMEDA-mediated conversion of the lesion formed as a result of PN treatment into DNA cleavage sites.
Detection and Identification of the GS-dG Adduct from 2′-Deoxyguanosine.
Although the DNA cleavage assay confirmed the protective roles of thiols on PN-exposed DNA, it did not provide any direct evidence for a stable adduct formation or any alternate mechanism. Therefore, instead of that with DNA, we performed a similar experiment with the nucleoside 2′-deoxyguanosine (dG) and analyzed the reaction products with LC-tandem MS (Figure 3). LC-MS revealed the presence of a peak eluting at 5.8 min having a mass of 573.1704 ([M + H]+, calculated 573.1722). LC-MS/MS of this m/z 573 peak generated major fragment ions at m/z 457 and 328 corresponding to the neutral loss of 116 (deoxyribose) and 116 + 129 (deoxyribose + glutamyl), respectively. Neutral loss of 116 amu is characteristic of nucleosides and 129 amu of GSH.29,30 Together, these data clearly indicated the formation of a GS-dG adduct, which was found to be significantly stable (Figure S1, Supporting Information).
Figure 3.
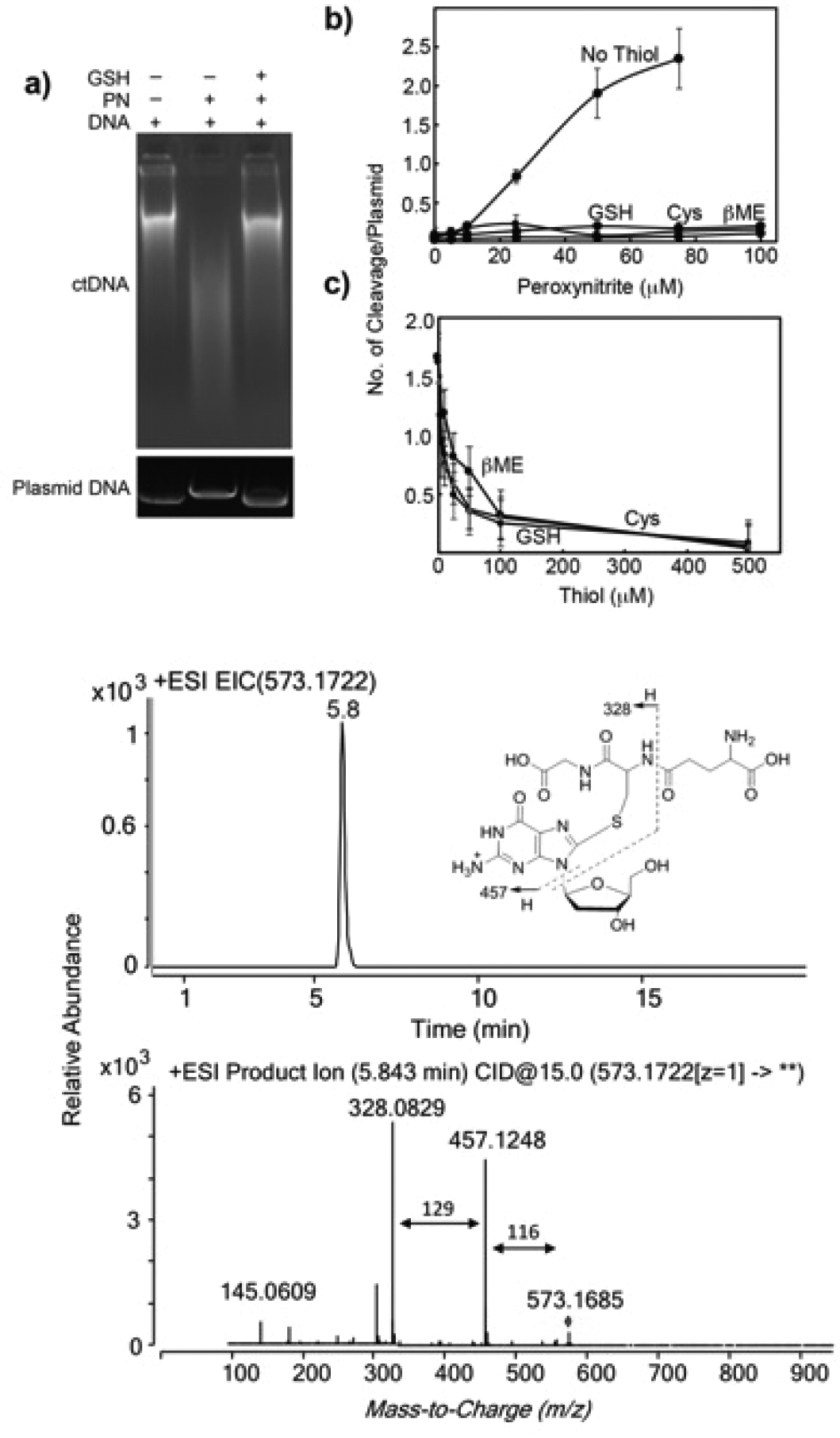
LC-MS extracted ion chromatogram and LC-MS/MS spectrum of m/z 573 peak eluting at 5.8 min (GS-dG adduct).
Spectroscopic Detection of GS-dG Adduct in Calf Thymus DNA.
To detect the presence of GS-dG adduct in PN-treated DNA, we repeated the above-mentioned experiment with calf thymus DNA and dansylated GSH (DGSH). Unlike GSH, DGSH has a specific UV–vis absorbance at 340, the specific UV–vis absorbance and fluorescence of DGSH to detect DGS-DNA adduct. Accordingly, DNA was treated with PN and DGSH following our standard protocol, precipitated, and washed to remove any unbound DGSH before UV–vis and fluorescence spectroscopic (Figure 4) analysis. As expected, peaks corresponding to DGSH were observed only in the DNA sample that was treated with PN and DGSH. This result clearly indicated the binding of DGSH to DNA.
Figure 4.
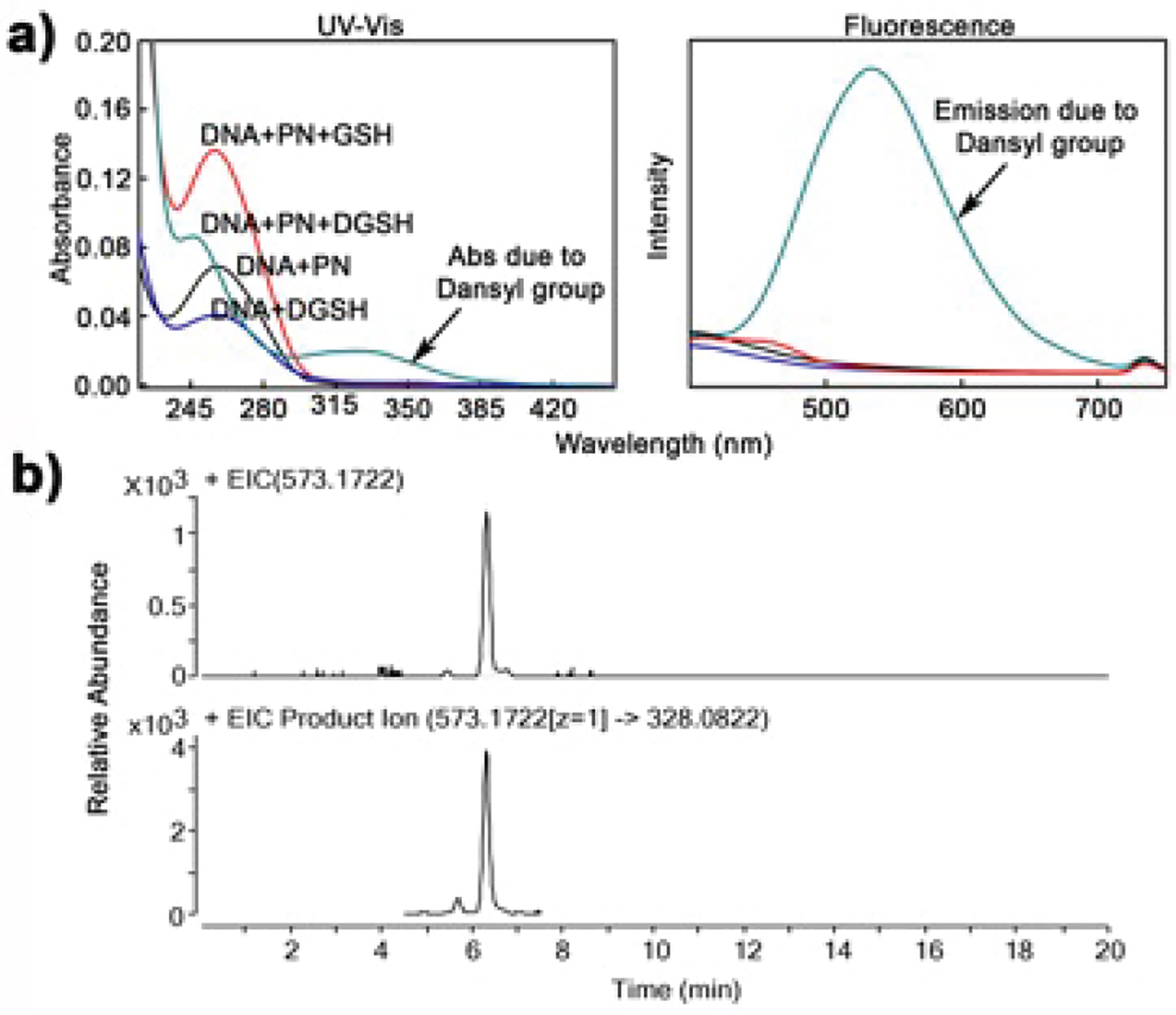
Detection of GS-DNA adduct. (a) UV–vis and fluorescence spectra of DNA after various treatments; (b) LC-MS and LC-MSMS chromatogram showing m/z 573.17 and 573 → 328 peak, respectively.
To characterize the GS-DNA adduct, we repeated the above experiment with GSH, enzymatically digested the adducted DNA to nucleosides, and analyzed them by LC-tandem MS.29 Consistent with the data obtained from dG, we found a peak at 6.2 min having m/z 573.1701 ([M + H]+, with the slight difference in retention time possibly being attributed to the age of the column). Monitoring of m/z 573 → 328 also produced a peak at 6.2 min. Dissociation of m/z 573 ion gave major fragments that are consistent with the GS-dG adduct (m/z 457 and 328). Together, the data confirmed the presence of a GS-dG adduct in DNA. To the best of our knowledge, this GS-dG adduct was not detected in PN and GSH-treated DNA previously.
Thus far, the results presented here clearly demonstrate that GSH mitigates the DNA cleaving property of PN through the formation of a stable GS-DNA adduct. This is consistent with previous reports that 8-NO2-G-containing nucleosides react with cellular thiols, including GSH and Cys residue of proteins to form adducts.22 A similar substitution of the nitro group by Cys was reported in an oligonucleotide containing the 8-NO2-G lesion.23 The protecting effect of GSH reported here may follow a similar mechanism involving the reaction of 8-NO2-G lesion in DNA with thiols (GSH) to form stable GS-DNA adducts.
GS-DNA Adduct Is Mutagenic.
Although GS-DNA adduct provides an immediate protection against the toxic effect of PN, it could be mutagenic, as observed with 1,2,3,4-diepoxybutane and ethylene dibromide.31,32 Accordingly, to test the mutagenic potential of the GS-DNA adduct reported here, we performed the Ames test in a TA100 histidine-dependent strain of Salmonella typhimurium bacteria. The number of revertants growing in the absence of histidine was counted and corrected with a degraded PN treatment control. Compared to GSH/Cys or PN, (PN + GSH)/Cys-treated cells produced more revertants than GSH/Cys and PN combined (additive effect), indicating the possibility of the GS-DNA adduct causing mutations (Figure 5). The results indicate that GS-dG adduct reported here may be mutagenic, consistent with other types of GS-DNA adducts.
Figure 5.
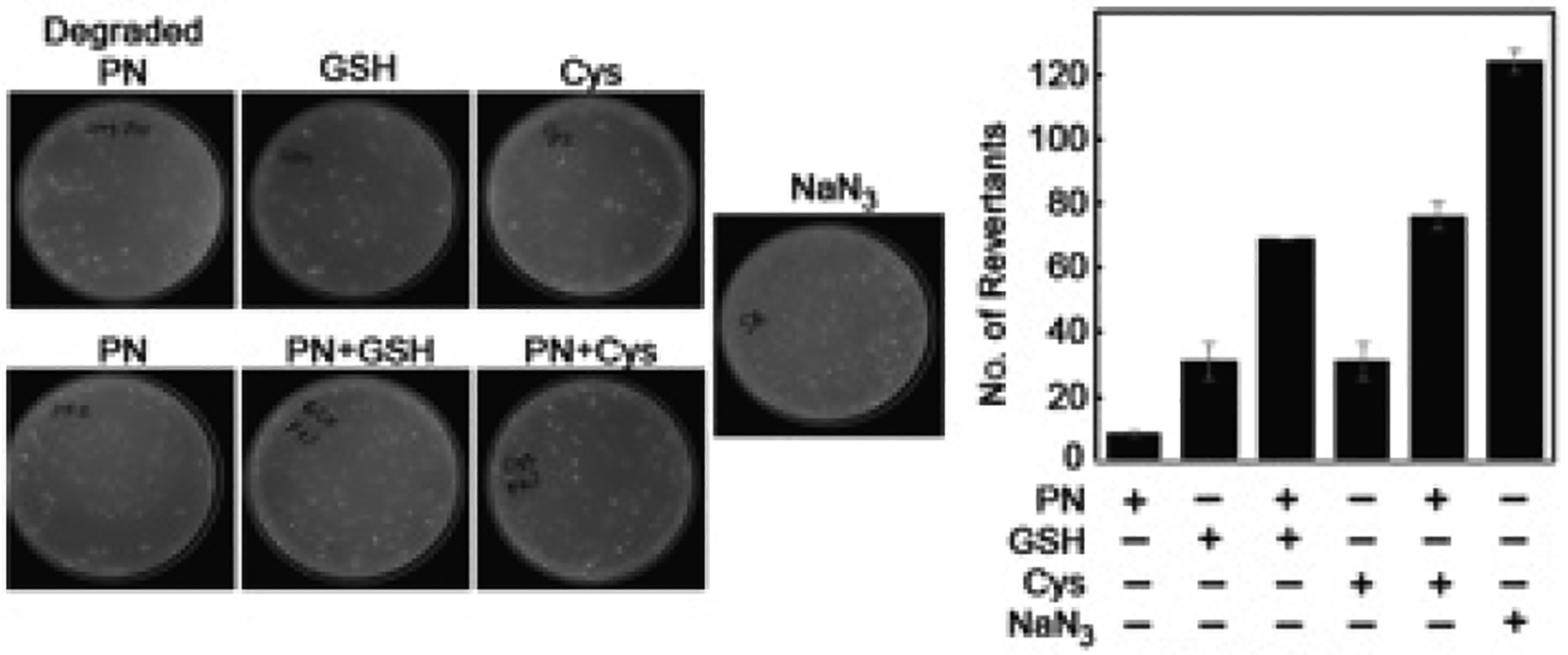
Ames mutagenicity test with PN and PN + GSH or PN + Cys. The figure shows pictures of colonies and its quantitation. Sodium azide (NaN3, 15 μM) and degraded PN were used as positive and negative controls, respectively. The concentration of PN was 10 μM and that of GSH/Cys was 5 mM.
CONCLUSION
In conclusion, we have presented strong evidence in support of the protective role of GSH against the PN-mediated toxic effect at the site of inflammation in vitro. We have shown that GSH is able to restore the property of DNA that is damaged by PN. Our observation is in agreement with the published data reporting the anti-inflammatory effect of NAC, a cellular precursor of GSH, and the beneficial role of long-term NAC treatment in attenuating influenza symptoms.17,20 NAC is approved for alleviating symptoms of lung inflammation in patients with cystic fibrosis and chronic obstructive pulmonary disease.17 A recent study reported endogenous GSH deficiency with poor clinical outcomes in SARS-CoV-2-infected patients.33 An ongoing Phase II (NCT04374461) open-label clinical trial at the Memorial Sloan Kettering Cancer Center aims to test the efficacy of NAC in severe or critically ill COVID-19 cases. The protective action of GSH/thiol compounds against inflammation-induced lung injury is widely accepted, although the underlying mechanisms are still areas of active investigation. In this study, we have uncovered a protective mechanism by which GSH/thiols mitigate PN-induced toxicity during inflammation. Interestingly, our preliminary data also indicates the possibility of such adduct to be mutagenic in bacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grants from DBT [BT/RLF/RE-ENTRY/18/2013: G.C. and BT/RLF/RE-ENTRY/35/2012: A.C.] and SERB [ECR/2015/000197: G.C. and ECR/2015/000198: A.C.] and the National Institutes of Health (R01 ES010546 (F.P.G.)). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also thank Shiv Nadar University, T. Wani, S. Siddiqui, and A. Jana.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.chemrestox.0c00299.
Time course of PN damage of DNA and stability of GS-dG adduct (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.chemrestox.0c00299
The authors declare no competing financial interest.
Contributor Information
Nabeel Ahmed, Department of Life Sciences, Shiv Nadar University, Greater Noida, Uttar Pradesh 201314, India.
Anindita Chakrabarty, Department of Life Sciences, Shiv Nadar University, Greater Noida, Uttar Pradesh 201314, India.
F. Peter Guengerich, Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, Tennessee 37232-0146, United States.
Goutam Chowdhury, Department of Chemistry, Shiv Nadar University, Greater Noida, Uttar Pradesh 201314, India.
REFERENCES
- (1).Fujiwara N, and Kobayashi K (2005) Macrophages in inflammation. Curr. Drug Targets: Inflammation Allergy 4, 281–286. [DOI] [PubMed] [Google Scholar]
- (2).Medzhitov R (2008) Origin and physiological roles of inflammation. Nature 454, 428–435. [DOI] [PubMed] [Google Scholar]
- (3).Medzhitov R (2010) Inflammation 2010: new adventures of an old flame. Cell 140, 771–776. [DOI] [PubMed] [Google Scholar]
- (4).Weiss U (2008) Inflammation. Nature 454, 427. [DOI] [PubMed] [Google Scholar]
- (5).Coussens LM, and Werb Z (2002) Inflammation and cancer. Nature 420, 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Channappanavar R, and Perlman S (2017) Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol 39, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Parohan M, Yaghoubi S, and Seraji A (2020) Liver injury is associated with severe coronavirus disease 2019 (COVID-19) infection: A systematic review and meta-analysis of retrospective studies. Hepatol. Res 50 (8), 924–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lin X, Wang R, Zou W, Sun X, Liu X, Zhao L, Wang S, and Jin M (2016) The influenza virus H5N1 infection can induce ROS production for viral replication and host cell death in A549 cells modulated by human Cu/Zn superoxide dismutase (SOD1) over-expression. Viruses 8, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Perrone LA, Belser JA, Wadford DA, Katz JM, and Tumpey TM (2013) Inducible nitric oxide contributes to viral pathogenesis following highly pathogenic influenza virus infection in mice. J. Infect. Dis 207, 1576–1584. [DOI] [PubMed] [Google Scholar]
- (10).Murata M (2018) Inflammation and cancer. Environ. Health Prev. Med 23, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Koppenol WH, Moreno JJ, Pryor WA, Ischiropoulos H, and Beckman JS (1992) Peroxynitrite, a cloaked oxidant formed by nitric oxide and superoxide. Chem. Res. Toxicol 5, 834–842. [DOI] [PubMed] [Google Scholar]
- (12).Yun BH, Geacintov NE, and Shafirovich V (2011) Generation of guanine-thymidine cross-links in DNA by peroxynitrite/carbon dioxide. Chem. Res. Toxicol 24, 1144–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys 98, 5648–5652. [Google Scholar]
- (14).Hu CW, Chang YJ, Chen JL, Hsu YW, and Chao MR (2018) Sensitive detection of 8-nitroguanine in DNA by chemical derivatization coupled with online solid-phase extraction LC-MS/MS. Molecules 23, 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Hu CW, Chang YJ, Hsu YW, Chen JL, Wang TS, and Chao MR (2016) Comprehensive analysis of the formation and stability of peroxynitrite-derived 8-nitroguanine by LC-MS/MS: Strategy for the quantitative analysis of cellular 8-nitroguanine. Free Radical Biol. Med 101, 348–355. [DOI] [PubMed] [Google Scholar]
- (16).Bhamra I, Compagnone-Post P, O’Neil IA, Iwanejko LA, Bates AD, and Cosstick R (2012) Base-pairing preferences, physicochemical properties and mutational behaviour of the DNA lesion 8-nitroguanine. Nucleic Acids Res. 40, 11126–11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Rahman I, and MacNee W (2000) Oxidative stress and regulation of glutathione in lung inflammation. Eur. Respir. J 16, 534–554. [DOI] [PubMed] [Google Scholar]
- (18).Li XY, Donaldson K, Rahman I, and MacNee W (1994) An investigation of the role of glutathione in increased epithelial permeability induced by cigarette smoke in vivo and in vitro. Am. J. Respir. Crit. Care Med 149, 1518–1525. [DOI] [PubMed] [Google Scholar]
- (19).Domenighetti G, Suter PM, Schaller MD, Ritz R, and Perret C (1997) Treatment with N-acetylcysteine during acute respiratory distress syndrome: a randomized, double-blind, placebo-controlled clinical study. J. Crit. Care 12, 177–182. [DOI] [PubMed] [Google Scholar]
- (20).Suter PM, Domenighetti G, Schaller MD, Laverrière MC, Ritz R, and Perret C (1994) N-acetylcysteine enhances recovery from acute lung injury in man. A randomized, double-blind, placebo-controlled clinical study. Chest 105, 190–194. [DOI] [PubMed] [Google Scholar]
- (21).Winterbourn CC (2016) Revisiting the reactions of superoxide with glutathione and other thiols. Arch. Biochem. Biophys 595, 68–71. [DOI] [PubMed] [Google Scholar]
- (22).Sawa T, Zaki MH, Okamoto T, Akuta T, Tokutomi Y, Kim-Mitsuyama S, Ihara H, Kobayashi A, Yamamoto M, Fujii S, Arimoto H, and Akaike T (2007) Protein S-guanylation by the biological signal 8-nitroguanosine 3′,5′-cyclic monophosphate. Nat. Chem. Biol 3, 727–735. [DOI] [PubMed] [Google Scholar]
- (23).Alexander KJ, McConville M, Williams KR, Luzyanin KV, O’Neil IA, and Cosstick R (2018) Chemistry of the 8-nitroguanine DNA lesion: Reactivity, labelling and repair. Chem. - Eur. J 24, 3013–3020. [DOI] [PubMed] [Google Scholar]
- (24).Robinson KM, and Beckman JS (2005) Synthesis of peroxynitrite from nitrite and hydrogen peroxide. Methods Enzymol. 396, 207–214. [DOI] [PubMed] [Google Scholar]
- (25).Gan J, Harper TW, Hsueh MM, Qu Q, and Humphreys WG (2005) Dansyl glutathione as a trapping agent for the quantitative estimation and identification of reactive metabolites. Chem. Res. Toxicol 18, 896–903. [DOI] [PubMed] [Google Scholar]
- (26).Augusto O, Goldstein S, Hurst JK, Lind J, Lymar SV, Merenyi G, and Radi R (2019) Carbon dioxide-catalyzed peroxynitrite reactivity - The resilience of the radical mechanism after two decades of research. Free Radical Biol. Med 135, 210–215. [DOI] [PubMed] [Google Scholar]
- (27).Wani TH, Chakrabarty A, Shibata N, Yamazaki H, Guengerich FP, and Chowdhury G (2017) The dihydroxy metabolite of the teratogen thalidomide causes oxidative DNA damage. Chem. Res. Toxicol 30, 1622–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Wani TH, Surendran S, Jana A, Chakrabarty A, and Chowdhury G (2018) Quinone-based antitumor agent sepantronium bromide (YM155) causes oxygen-independent redox-activated oxidative DNA damage. Chem. Res. Toxicol 31, 612–618. [DOI] [PubMed] [Google Scholar]
- (29).Chowdhury G, Cho SH, Pegg AE, and Guengerich FP (2013) Detection and characterization of 1,2-dibromoethane-derived DNA crosslinks formed with O(6) -alkylguanine-DNA alkyltransferase. Angew. Chem., Int. Ed 52, 12879–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Chowdhury G, Murayama N, Okada Y, Uno Y, Shimizu M, Shibata N, Guengerich FP, and Yamazaki H (2010) Human liver microsomal cytochrome P450 3A enzymes involved in thalidomide 5-hydroxylation and formulation of a glutathione conjugate. Chem. Res. Toxicol 23, 1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Cho SH, and Guengerich FP (2013) In vivo roles of conjugation with glutathione and O6-alkylguanine DNA-alkyltransferase in the mutagenicity of the bis-electrophiles 1,2-dibromoethane and 1,2,3,4-diepoxybutane in mice. Chem. Res. Toxicol 26, 1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sedgeman CA, Su Y, and Guengerich FP (2017) Formation of S-[2-(N(6)-deoxyadenosinyl)ethyl]glutathione in DNA and replication past the adduct by translesion DNA polymerases. Chem. Res. Toxicol 30, 1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Polonikov A (2020) Endogenous deficiency of glutathione as the most likely cause of serious manifestations and death in COVID-9 patients. ACS Infect. Dis 6, 1558–1562. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


