Abstract
Synechococcus sp. strain SH-94-5 is a nitrate assimilation-deficient cyanobacterium which was isolated from an ammonium-replete hot spring in central Oregon. While this clone could grow on ammonium and some forms of organic nitrogen as sole nitrogen sources, it could not grow on either nitrate or nitrite, even under conditions favoring passive diffusion. It was determined that this clone does not express functional nitrate reductase or nitrite reductase and that the lack of activity of either enzyme is not due to inactivation of the cyanobacterial nitrogen control protein NtcA. A few other naturally occurring cyanobacterial strains are also nitrate assimilation deficient, and phylogenetic analyses indicated that the ability to utilize nitrate has been independently lost at least four times during the evolutionary history of the cyanobacteria. This phenotype is associated with the presence of environmental ammonium, a negative regulator of nitrate assimilation gene expression, which may indicate that natural selection to maintain functional copies of nitrate assimilation genes has been relaxed in these habitats. These results suggest how the evolutionary fates of conditionally expressed genes might differ between environments and thereby effect ecological divergence and biogeographical structure in the microbial world.
In the cyanobacteria, nitrogen (N) comprises approximately 10% of cell dry weight (9). These oxygen-evolving, photoautotrophic bacteria primarily use inorganic N sources to meet their nutritional requirements, of which nitrate is generally the most available form of combined N (19). Nitrate is actively transported across the cell membrane by either an ATP-binding cassette transporter or a nitrate permease (45), and once inside the cell, it is sequentially reduced to nitrite and ammonium through the activities of nitrate reductase (Nar) and nitrite reductase (Nir), respectively (9, 35). The eight electrons required to reduce nitrate to ammonium are donated by photosynthetically reduced ferredoxin (30), and ammonium is subsequently incorporated into amino acids, primarily via the glutamine synthetase (GS)-glutamate synthase cycle (31).
The genes encoding the ATP-binding cassette transporter (nrtABCD), Nar (narB), and Nir (nirA) cluster as a similarly organized, cotranscribed unit (the nirA operon) in Synechococcus sp. strain PCC 7942 (35) and Anabaena sp. strain PCC 7120 (12). However, differences in the genetic organization of nitrate assimilation genes have been described for Synechocystis sp. strain PCC 6803 (22) and Phormidium laminosum (32). Despite these differences in genetic architecture, cyanobacteria regulate the expression of nitrate assimilation genes in a similar manner. These organisms preferentially use ammonium if it is available, presumably because it can be directly incorporated into organic matter without the associated costs of reducing the power required for the utilization of other forms of inorganic N (9). While ammonium has been shown to posttranslationally inhibit nitrate transport in Anacystis nidulans PCC 6301 (26), the principal way ammonium regulates nitrate utilization in cyanobacteria is by negative transcriptional control of nitrate assimilation gene expression (29). Transcription is derepressed in the absence of ammonium but must be activated by the cyanobacterial N control protein NtcA, a member of the Crp family of DNA-binding proteins (44).
Genes which are conditionally expressed may evolve with different dynamics in different environmental contexts and therefore provide the raw material for ecological diversification. Here we report the isolation in laboratory culture of a nitrate assimilation-deficient clone of the thermophilic cyanobacterium Synechococcus from an ammonium-rich hot spring in central Oregon. We characterized the N sources which support its growth and investigated possible mechanisms by which the ability to use nitrate may have been lost in this clone. While this phenotype is rare among cyanobacterial isolates, it is exhibited by a few other strains from diverse habitats. We first inferred the phylogenetic distribution of this phenotype among cyanobacteria to evaluate whether the loss of the ability to assimilate nitrate has a single or multiple evolutionary origins. We further identified an apparent association between the evolution of this trait and the presence of environmental ammonium and discussed the potential biogeographical implications of this association.
MATERIALS AND METHODS
Isolation and culture maintenance.
The inoculum was a microbial mat sample collected from an area with a temperature of 50°C in the main channel of South Harney Hot Springs, a slightly alkaline (pH 7.4) spring located near the southern end of Harney Lake, Harney County, Oreg. The cyanobacterial cell density of a homogenized suspension was quantified with a hemocytometer, and aliquots containing an estimated five cyanobacterial cells were delivered to flasks of BG11, D, and NT media (5) and incubated under low cool white fluorescent light at 45°C. An axenic isolate (Synechococcus sp. strain SH-94-5) was obtained from an NT enrichment flask after repeated streakings on 1.5% agar plates and incubating as described above. SH-94-5 was maintained under the above conditions both on plates and in flasks of NDA medium [N-free ND medium (5) supplemented with 0.38 mM (NH4)2S2O4 and 0.4 mM Na2S2O3 · 5H2O], which contains ammonium as the sole utilizable N source. The addition of sodium thiosulfate has been found to improve the survival and growth of many cyanobacteria isolated from hot spring environments replete with reduced sulfur and nitrogen (5). This strain has been deposited in the Culture Collection of Microorganisms from Extreme Environments at the University of Oregon (strain CCMEE-5231).
N sources supporting growth.
SH-94-5 was screened for growth on a number of N sources. For all treatments, duplicate flasks were incubated at 45°C with 100 microeinsteins of cool white fluorescent light m−2 s−1 in ND medium supplemented with 100 mg of Na2S2O3 · 5H2O liter−1 either without or with one of the following N sources: (NH4)2S2O4 (0.5, 2, or 5 mM), NaNO3 (5, 15, 30, 60, or 120 mM), NaNO2 (0.1, 0.5, 1, 5, 15, 30, 60, or 120 mM), glutamine (0.5, 2, or 5 mM), glutamate (0.5, 2, or 5 mM), urate (0.5, 2, or 5 mM), allantoin (0.5, 2, or 5 mM), and urea (0.5, 2, or 5 mM). Organic N sources in ND medium were added by sterile filtration following autoclaving, and the pH of all media was 7.6. Each flask was inoculated with N-starved SH-94-5 cells to a final density of 105 cells ml−1, and growth was monitored as absorbance at 750 nm (A750). The turbidity at this wavelength was found to be linearly related to cell density over the range of values used in the experiments (A750 < 0.45 [data not shown]).
Reversion assay.
Ammonium-grown SH-94-5 cells starved for N for 60 h were washed in ND medium, centrifuged, and resuspended in ND medium to a final density of 109 cells ml−1. Subsamples (500 μl) of the suspension were spread plated onto ND plates supplemented with 0.9 mM NaNO3, 0.4 mM Na2S2O3 · 5H2O, and 6.67 mM NaHCO3. A total of 1.3 × 1010 cells were plated. To estimate plating efficiency on NDA medium, triplicate spread plates of 2 × 104 cells each were made. Plates were incubated as described above and monitored for colonies over 3 weeks.
Nar and Nir assays.
Nar activity was assayed as nitrate reduction (with sodium dithionite-reduced methyl viologen as the electron donor) by cells permeabilized with mixed alkyl trimethylammonium bromide at a final concentration of 75 μg ml−1 (19). NDA medium-grown, mid-exponential-phase cells were harvested by filtration and rinsed with and resuspended in D medium, which contains nitrate as the sole N source. After incubation for 5 h under standard maintenance conditions, cells were concentrated by filtration, rinsed, and resuspended in double-distilled H2O. Aliquots of cells containing ∼200 μg of protein, as determined with the Sigma bicinchoninic acid assay (40), were added to the reaction mixture, incubated for 20 min under maintenance conditions, and assayed every 5 min for nitrite production using the colorimetric assay of Snell and Snell (41). Activity was expressed as nanomoles of NO2− produced per minute per milligram of protein. No activity was ever observed in reaction mixtures which lacked cells, contained cells boiled for 15 min prior to assaying, lacked methyl viologen, or lacked sodium dithionite.
Nir was assayed according to a modification of the method of Rigano (39). Cells grown as described above were filtered, rinsed with 0.3 M phosphate buffer (pH 7.5) containing 1 mM cysteine, and resuspended in 10 ml of buffer. Crude extracts were produced by sonication, and cell debris was removed by centrifugation at 15,000 × g for 30 min at 4°C. Aliquots of extract containing 300 μg of protein were added to a reaction mixture containing 400 nmol of KNO2, 100 μmol of phosphate buffer (pH 7.5), 100 nmol of methyl viologen, and 4.6 μmol of Na2S2O4 in a final volume of 2 ml. Assay reactions were terminated periodically as described by in Rigano (39) over a 30-min duration for nitrite determination, and Nir activity was expressed as nanomoles of NO2− removed per minute per milligram of protein. No detectable activity was observed in extracts boiled for 15 min prior to assaying.
GS assay.
Subcultures of mid-exponential-phase, NDA medium-grown SH-94-5 cells were rinsed with either NDA medium or N-NDA medium [NDA without (NH4)2S2O4] and subsequently incubated for 48 h under standard maintenance conditions in that medium (NDA), after which time the A750 of both subcultures was ∼0.25. A portion (260 ml) of each subculture was filtered, rinsed with 0.3 M phosphate buffer (pH 7.5) containing 1 mM cysteine, and resuspended in 10 ml of buffer. Crude extracts were produced as described above. The GS activities of five replicate aliquots of cell extracts containing 30 to 50 μg of protein were estimated using transferase assays (7). The assay duration was 20 min, and activity was expressed as nanomoles of γ-glutamyl hydroxamate produced per minute per milligram of protein. γ-Glutamyl hydroxamate production in control assays containing enzyme extracts boiled for 15 min was less than 5% of reported GS activities.
DNA isolation, amplification, and sequencing.
Genomic DNA was isolated by the method of Pitcher et al. (37). Fragments of the 16S rRNA gene spanning Escherichia coli nucleotide positions 360 to 1335 were amplified in 50-μl reaction mixtures containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, a 200 μM concentration of each deoxynucleoside triphosphate, 1.12 mM MgCl2, 1.25 U of Taq polymerase (Perkin-Elmer), ∼10 ng of genomic DNA, and 0.2 μM (each) primer CYA359F (34) and primer PLG2.3 [5′CTTCA(C/T)G(C/T)AGGCGAGTTGCAGC3′], a modification of PLG2.1 (43). Reaction conditions were 40 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 2 min. Amplification products were purified with the QIAquick-spin PCR purification kit (Qiagen), and cleaned amplification products were directly sequenced with an ABI Prism 377 at the University of Oregon's DNA sequencing facility.
Sequence alignment and phylogenetic analyses.
Gene sequences were aligned using Malign 2.77 (47) as described by Miller and Castenholz (33). Briefly, this was a pairwise alignment which accounted for secondary structure of the mature 16S rRNA molecule with a gap cost to substitution cost of three.
Phylogenetic trees were constructed according to three optimality criteria. A neighbor-joining tree was built with MEGA (version 1.01; S. Kumar, K. Tamura, and M. Nei, The Pennsylvania State University, University Park). For the calculation of a distance matrix, Kimura's two-parameter model was assumed (24), and nucleotide positions containing gaps or missing data were deleted in a pairwise fashion. The tree was inferred with pairwise deletion of gaps and with 1,000 bootstrap pseudoreplicates. A maximum-parsimony phylogeny was constructed using PAUP (version 3.1.1; D. L. Swofford, Sinauer Associates, Sunderland, Mass.) by a heuristic search using the tree-bisection-reconnection branch-swapping algorithm. Starting trees were obtained by stepwise addition of sequences and 10 replications of random sequence addition. The analysis was bootstrap pseudoreplicated 100 times. A maximum-likelihood tree was created with the “dnaml” program in PHYLIP (version 3.5c; J. Felsenstein, Department of Genetics, University of Washington, Seattle) with a transition-to-transversion ratio of two, sequential addition of sequences, and 100 bootstrap pseudoreplicates. All trees were rooted with Aquifex pyrophilus.
The ability of cyanobacterial ancestors (the internodes in the phylogenies described above) to use nitrate was reconstructed for each of the three phylogenies described above according to the maximum-parsimony algorithm of Maddison and Maddison (MacClade, version 3.01; W. P. Maddison and D. R. Maddison, Sinauer Associates, Sunderland, Mass.). This analysis estimated the minimum number of evolutionary steps (i.e., losses of the ability to assimilate nitrate) required to produce the observed distribution of the nitrate assimilation phenotype among extant cyanobacteria.
GenBank nucleotide accession numbers for organisms used in phylogenetic analyses are as follows: Aquifex pyrophilus, M83548; E. coli, J01859; Bacillus subtilis, X60646; Synechococcus sp. strain PCC 6307, AF001477; Synechococcus sp. strain PCC 6301, X03538; Leptolyngbya sp. strain PCC 7375, AF132786; Leptolyngbya sp. strain PCC 73110, X84810; Leptolyngbya foveolarum, X84808; Microcoleus sp. strain PCC 7420, X70770; “Mastigocladus” sp. strain PCC 7518, X68780; Cylindrospermum sp. strain PCC 7417, AJ133163; Nostoc sp. strain PCC 7120, X59559; Nostoc sp. strain PCC 73102, AF027655; Prochloron sp., X63141; Pleurocapsa sp. strain PCC 7516, X78681; Synechococcus sp. strain PCC 7002, D88289; Spirulina sp. strain PCC 6313, X75044; Microcystis sp. strain PCC 7941, U40340; Synechocystis sp. strain PCC 6803, D64000; Synechococcus sp. strain C9, L35481 to 83; and Synechococcus sp. strain SH-94-5, AF285260. Sequences for Chamaesiphon sp. strain PCC 7430, Chroococcidiopsis sp. strain PCC 7203, Gloeobacter sp. strain PCC 7421, Myxosarcina sp. strain PCC 7312, and Oscillatoria amphigranulata NZ-Concert-Oa were obtained from Ribosomal Database Project II (http://www.cme.msu.edu/RDP).
Nucleotide sequence accession numbers.
The 16S rRNA gene sequence data obtained for the strains used in this study have been submitted to GenBank and can be found under the accession numbers AF317503 to AF317510.
RESULTS
Enrichment and isolation.
The three media used for the enrichment of the South Harney Hot Spring sample differ both in quality and in quantity of combined N. D medium contains 9.2 mM nitrate but no ammonium, BG11 medium has 5.9 mM nitrate and 25 μM ammonium, and NT medium contains 9.2 mM nitrate and 0.76 mM ammonium. The most successful enrichment was in NT medium, in which a dense, chlorophyll-rich suspension of the unicellular cyanobacterium Synechococcus developed. Small pellets of yellow Synechococcus cells formed in six BG11 enrichments, while no growth was observed in D medium. Transfer of pellets formed in BG11 medium to NT medium resulted in greening of the cells and exponential growth, while transfer of cells from NT medium to either BG11 or D medium led to rapid yellowing of Synechococcus cells (not shown). Based on these observations and the ammonium levels of the respective media, it was suspected that the enriched Synechococcus could not grow on inorganic N sources other than ammonium. An axenic isolate, Synechococcus sp. strain SH-94-5 (CCMEE-5231), was obtained from the NT enrichment for characterization of N sources supporting growth.
N sources supporting SH-94-5 growth.
Consistent with predictions based on observations from the enrichments, ammonium was the only inorganic N source tested which supported SH-94-5 growth, although decreases in the growth rate occurred with increasing concentrations of (NH4)2S2O4 (Table 1). SH-94-5 could not grow with either nitrate or nitrite as the sole N source (Table 1), even at concentrations great enough for the substrate to be available by passive diffusion (35, 36). These results suggest that the failure of SH-94-5 to grow with these N sources is not simply due to the lack of active transport of these substrates. Cells bleached within ∼72 h of exposure to NaNO2 at a concentration of ≥5 mM. SH-94-5 also did not grow with N2, indicating that this clone cannot fix N under aerobic conditions.
TABLE 1.
Growth of Synechococcus sp. strain SH-94-5 on various nitrogen sources
| N source | Concn (mM) | Mean doubling time (h) ± SE |
|---|---|---|
| None | No growth | |
| Ammonium sulfate | 0.5 | 14.2 ± 1.42 |
| 2 | 23.4 ± 2.52 | |
| 5 | 51.5 ± 0.44 | |
| Sodium nitrate | 5 | No growth |
| 15 | No growth | |
| 30 | No growth | |
| 60 | No growth | |
| 120 | No growth | |
| Sodium nitrite | 0.1 | No growth |
| 0.5 | No growth | |
| 1 | No growth | |
| 5 | No growth | |
| 15 | No growth | |
| 30 | No growth | |
| 60 | No growth | |
| 120 | No growth | |
| Glutamine | 0.5 | 16.4 ± 1.15 |
| 2 | 25.3 ± 1.06 | |
| 5 | 13.2 ± 0.20 | |
| Glutamate | 0.5 | No growth |
| 2 | No growth | |
| 5 | 121.1 ± 46.88 | |
| Urate | 0.5 | 154.5 ± 3.96 |
| 2 | No growth | |
| 5 | No growth | |
| Allantoin | 0.5 | 196.0 ± 18.97 |
| 2 | 141.9 ± 16.50 | |
| 5 | 128.1 ± 2.73 | |
| Urea | 0.5 | 89.9 ± 21.54 |
| 2 | 64.9 ± 26.31 | |
| 5 | 25.8 ± 6.89 |
SH-94-5 growth on a number of organic N sources was also assessed. Glutamine supported growth at rates comparable to those on ammonium (Table 1), but SH-94-5 grew at only the highest concentration of glutamate tested. Growth with urea and two intermediates of the purine degradation pathway (allantoin and urate) was poor (Table 1), and cells exhibited the yellow pigmentation characteristic of N starvation.
Reversion assay.
No revertants capable of growth on nitrate were obtained upon the plating of 1.3 × 1010 N-starved SH-94-5 cells on medium containing 0.9 mM NaNO3. The mean plating efficiency ± the standard error on NDA plates was 16.6% ± 1.62%.
Lack of Nar and Nir activities in SH-94-5.
SH-94-5 cells lacked both Nar and Nir activities (Table 2). Enzyme activities were assayed after 5 h of preincubation in nitrate-containing medium, conditions under which Nar and Nir activities would be expected to be maximized (18, 20). The positive control, Synechococcus sp. strain C9 (8), previously determined to be the sister organism of SH-94-5 by phylogenetic analysis (33), exhibited activity for both enzymes (Table 2).
TABLE 2.
Activities of Nar, Nir, and GS in Synechococcus sp. strains SH-94-5 and C9a
| Clone | Mean Nar activitya ± SE | Mean Nir activityb ± SE | Mean GS activitya ± SE in:
|
|
|---|---|---|---|---|
| NH4+-replete cells | N-starved cells | |||
| SH-94-5 | 0c | 0c | 711.5 ± 37.96 | 1,272.4 ± 25.97 |
| C9 | 73.0 ± 5.18 | 18.8 ± 0.08 | NDd | ND |
Activities are expressed as nanomoles of product produced per milligram of protein per minute.
Activities are expressed as nanomoles of nitrite removed per milligram of protein per minute.
No activity was found for two separate experiments.
ND, not determined.
Presence of functional NtcA in SH-94-5.
The lack of Nar and Nir activities under nitrate-assimilating conditions can result from nonfunctional reductases and/or a nonfunctional regulator of nitrate assimilation. The protein NtcA is required for the transcription of nitrate assimilation genes. It also has a positive effect on the transcription of glnA, which encodes GS (29), and cyanobacteria have higher GS activity in the absence of ammonium as a consequence of NtcA activity (49). However, N-starved and ammonium-replete cells of laboratory mutants of Synechococcus sp. strain PCC 7942 lacking functional NtcA show no difference in GS activity, whereas the wild type exhibits approximately twofold-greater activity under N starvation (44). To determine whether SH-94-5 has functional NtcA, the presence of active protein was assessed indirectly by estimating the GS activities of crude extracts from ammonium-replete and N-starved SH-94-5 cells. The activity of ammonium-replete cell extracts was ∼56% of that of N-starved cell extracts (Table 2). The inability of SH-94-5 to assimilate nitrate and nitrite is therefore not due to the loss of NtcA activity.
Distribution and evolutionary origins of nitrate assimilation deficiencies in cyanobacteria.
In addition to Synechococcus sp. strain SH-94-5, several other cyanobacterial isolates representing three genera lack the ability to grow with nitrate (Table 3). We took a phylogenetic comparative approach to establish whether this phenotype had a single origin predating diversification of these organisms or whether it evolved multiple times during the evolutionary history of the cyanobacteria.
TABLE 3.
Clones from which 16S rRNA gene sequence data were obtained for phylogenetic analysis of the nitrate assimilation phenotype in cyanobacteria
| Clone | Origina | Ability to use nitrate | Reference |
|---|---|---|---|
| Synechococcus sp. strain SH-94-45 | South Harney Hot Springs, Oreg. | + | This study |
| O. amphigranulata 11-3 | Spte-2 Hot Spring, Rotorua, New Zealand | − | 13 |
| O. amphigranulata 13-1 | Cirque Hot Spring, Whakarewarewa, New Zealand | + | 13 |
| O. amphigranulata 14-1 | TB-86 Hot Spring, Rotorua, New Zealand | − | 13 |
| O. amphigranulata 19-2 | Cirque Hot Spring, Whakarewarewa, New Zealand | − | 13 |
| O. amphigranulata 23-3 | Par-8 Hot Spring, Rotoiti, New Zealand | + | 13 |
| Oscillatoria sp. strain OH25 | Hunter's Hot Springs, Oreg. | + | This study |
| Leptolyngbya sp. strain PCC 9207 | Lake Olalla, Doñana National Park, Spain | − | 41 |
| Leptolyngbya sp. strain PCC 9221 | Lake Chiuchiu, Chile | − | 41 |
| Geitlerinema sp. strain PCC 9222 | Loa River, Chile | − | 41 |
Spte-2, Sulphur Point-2; TB-86, Tudor Boil-86; Par-8, Parangarenga-8.
16S rRNA gene sequence data were collected for the clones listed in Table 3 for phylogenetic analysis. Some interesting patterns emerged from the sequence data themselves. Leptolyngbya sp. strains PCC 9207 and PCC 9221 have identical sequences over the entire ca. 1-kb amplified gene product. In addition, clones of the morphospecies Oscillatoria amphigranulata, known only from New Zealand hot springs (4, 13), fell into two groups based on sequence identity. The sequences of O. amphigranulata isolates 11-3, 19-2, and NZ-Concert-Oa (6, 14) were identical, while that of O. amphigranulata 14-1, for which only partial sequence data were obtained, was identical to the above sequences over 560 bp. These clones are similar phenotypically in that they are nitrate assimilation deficient and produce the light-harvesting protein phycoerythrin (PE). The sequence of O. amphigranulata strains 13-1 and 23-3, which lack PE and exhibit poor growth on nitrate, are identical to each other over the 560-bp partial sequence obtained for 13-1. These PE+ and nitrate− and PE− and nitrate+ O. amphigranulata sequences are 2.8% divergent over the entire fragment.
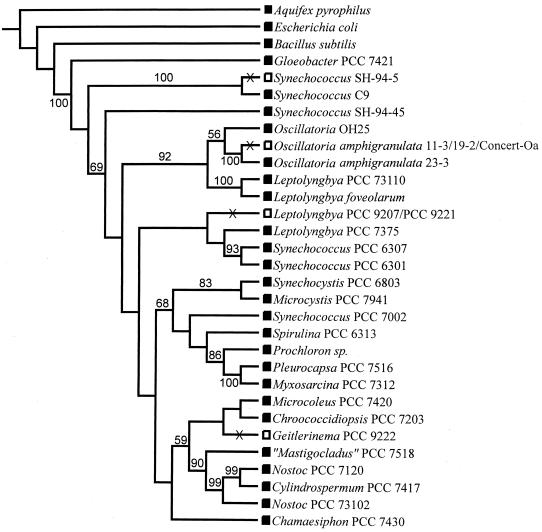
A neighbor-joining phylogeny indicates that none of the organisms which are unable to utilize nitrate are closely related (Fig. 1). This phylogeny exhibits many similarities with both a maximum-likelihood tree and the single most parsimonious tree obtained using maximum parsimony (not shown). Synechococcus sp. strain SH-94-5 was found to be the sister organism of Synechococcus sp. strain C9 in all trees with strong bootstrap support (Fig. 1) (33). Their sequences are identical at 99.0% of the nucleotide positions, excluding 27 bp of missing data in the C9 sequence and six ambiguous nucleotides. These Synechococcus strains did not group with Synechococcus sp. strain SH-94-45, a South Harney isolate identical to group I hot spring Synechococcus (33) which can grow on nitrate (Table 3). The two sequence groups of O. amphigranulata clustered together with strong bootstrap support in all phylogenies (Fig. 1). Similarly, in all three trees Leptolyngbya sp. strains PCC 9207 and PCC 9221 formed a clade with Leptolyngbya sp. strain PCC 7375 and Synechococcus sp. strains PCC 6301 and PCC 6307, while Geitlerinema sp. strain PCC 9222 was part of a clade consisting of Chroococcidiopsis sp. strain PCC 7203, Microcoleus sp. strain PCC 7420, and the heterocystous cyanobacteria, although the topology of this clade varies across trees.
FIG. 1.
Ancestral state reconstruction of the cyanobacterial nitrate assimilation phenotype along a neighbor-joining phylogeny inferred using ∼950 nucleotides of 16S rRNA gene sequence data. A nodal value represents the percentage of time that the organisms to the right of that node formed a clade for 1,000 bootstrap pseudoreplicates. Only bootstrap values greater than 50% are indicated. Extant organisms are coded to indicate whether they are either able to utilize nitrate (shaded rectangles) or nitrate assimilation deficient (open rectangles). X, inferred timing of losses of ability to assimilate nitrate during cyanobacterial evolution according to the maximum-parsimony method of Maddison and Maddison (MacClade, version 3.01).
For all three phylogenies, the most parsimonious ancestral state reconstruction of the ability to use nitrate strongly indicated that nitrate assimilation deficiencies have been independently derived at least four times during the evolutionary history of the cyanobacteria (Fig. 1).
DISCUSSION
A number of lines of evidence indicate that Synechococcus sp. strain SH-94-5 was derived from a population of nitrate assimilation-deficient cells comprising a numerically significant component of the South Harney Hot Spring microbiota at the time of collection. That these cells were abundant community members is supported by the enrichment results, since several enrichments like the one from which SH-94-5 was isolated were obtained from inocula which had been diluted to a total of 5 cells from a homogenate with an initial density of ∼107 cyanobacterial cells ml−1. The fact that SH-94-5-like cells came up in multiple enrichment flasks also makes it likely that cells with this phenotype were present in the collected sample rather than a product of mutation in laboratory culture. Results of the reversion assay also support the conclusion that this phenotype is not a product of recent mutation in the laboratory. The reversion rates of nitrate assimilation mutants of Synechococcus sp. strain PCC 7942 created by mutagenization with either UV radiation or nitrosoguanidine ranged from 10−7 to 10−9 (44), but we found no revertants to a nitrate-utilizing phenotype among > 1010 SH-94-5 cells.
SH-94-5 can grow on a number of N sources, with ammonium and glutamine being the best observed substrates for growth (Table 1). The doubling time of ammonium-grown SH-94-5 increased with increasing ammonium concentration over the tested range of 0.5 to 5 mM (Table 1). This pattern may result from the inhibitory effects of high ammonia concentrations on the water-splitting enzyme of photosystem II (21). Like many cyanobacteria, SH-94-5 can grow with urea as the sole N source, but only at rates much lower than those reported for other strains (see references 23 and 25 for examples). SH-94-5 showed slow growth on two intermediates of the purine degradation pathway, urate and allantoin (Table 1). These results differed from those obtained for another nitrate assimilation-deficient cyanobacterium, O. amphigranulata NZ-Concert-Oa, which exhibited higher growth rates with ∼1 mM urea, urate, and allantoin than with 4 mM ammonium (E. Jensen and R. W. Castenholz, unpublished data).
The inability of SH-94-5 to grow on either nitrate or nitrite over the wide range of concentrations tested (Table 1) suggests that the lack of growth is not simply due to a failure to actively take up these N sources. A transport mutant of Synechococcus sp. strain PCC 7942 with nrtA interrupted by the insertion of an aminoglycoside 3′-phosphotransferase gene did not grow in medium containing 2 mM KNO3, while the wild type doubled about twice a day at this concentration (36). The growth rate of this mutant, however, increased with increasing [NO3−] between ca. 5 and 40 mM, as more substrate became available for intracellular reduction by passive diffusion. Additionally, at the pH of the media used in this study, nitrite is expected to be available (as nitrous acid) by passive diffusion at concentrations greater than about 100 μM (35). SH-94-5 exhibited no growth at increasing concentrations of either N source, which is consistent with the interpretation that this clone cannot reduce intracellular nitrate or nitrite.
The enzyme assays provided further evidence that SH-94-5 does not have functional Nar or Nir (Table 2). O. amphigranulata NZ-Concert-Oa also lacks activity of these enzymes (Jensen and Castenholz, unpublished). The failure of SH-94-5 to express Nar and Nir is not due to nonfunctional NtcA (Table 2). Like nitrate assimilation genes, ntcA transcription is itself subject to ammonium repression, although weak constitutive transcription has been found for Synechococcus sp. strain PCC 7942 and Anabaena sp. strain PCC 7120 (29, 38). An ntcA mutant of Anabaena sp. strain PCC 7120 exhibits wild-type growth rates on ammonium but appears to have a poorer yield (46). In addition to failing to up-regulate GS activity and ammonium transport during N starvation, ntcA mutants of Synechococcus sp. strain PCC 7942 were estimated to perform worse than the wild type for both of these functions under ammonium-replete conditions (44). The above results suggest that there might be selection to maintain functional NtcA even under conditions of ammonium availability.
It should be considered whether other proteins involved in the regulation of N assimilation may affect the SH-94-5 phenotype. In the presence of nitrite, NtcB, a member of the LysR family of transcription factors, specifically promotes nirA operon transcription in Synechococcus sp. strain PCC 7942 (1). However, since an ntcB deletion mutant of this cyanobacterium can grow with nitrate at ∼70% of the wild-type rate (42), it is doubtful that the inability of SH-94-5 to use nitrate is due to the loss of NtcB activity.
PII, an NtcA-regulated phosphoprotein homologous to GlnB in enterobacteria, coordinates carbon and N metabolism in Synechococcus sp. strain PCC 7942 and may exist in one of four phosphorylation states (10, 27). PII is always dephosphorylated in the presence of ammonium and highly phosphorylated during N starvation, whereas the degree of phosphorylation in nitrate-grown cells depends upon factors such as CO2 availability (10, 11). PII does not appear to be involved in the regulation of N assimilation genes during ammonium repression, since the PII null mutant MP2 of Synechococcus sp. strain PCC 7942 displays the wild-type phenotype of increased Nar, Nir, and GS activities after transfer from ammonium- to nitrate-containing medium (11). The authors' observation that MP2 cannot grow in ammonium-containing medium at a pH greater than 8 suggests that a PII null mutant might actually be selected against in some ammonium-containing environments. On the other hand, MP2 is released from wild-type reduction in nitrate assimilation under conditions of nitrate repletion but low CO2 availability (11), and it has been suggested that dephosphorylated PII is involved in the posttranslational negative regulation of nitrate uptake under these conditions. If this is the case, it is expected that a hypothetical mutant with permanently dephosphorylated PII resulting from a nonfunctional kinase would not assimilate nitrate except under passive-diffusion conditions. It is unlikely that SH-94-5 is such a mutant since it cannot grow with a high concentration of nitrate (Table 2).
We conclude that the inability of SH-94-5 to use either nitrate or nitrite is probably due at least in part to mutational inactivation of narB and nirA, which encode Nar and Nir, respectively. However, we presently cannot rule out the possibility that the lack of expression of these enzymes is not simply due to changes in an upstream promoter sequence (or sequences) such that it can no longer be recognized by NtcA, although we would have expected to find detectable rates of reversion to a nitrate-utilizing phenotype for such a regulatory mutant.
Ancestral state reconstruction of discrete characters along a phylogeny is a useful method for estimating the timing of evolutionary events and for testing for shared versus independent origins of phenotypic traits, particularly when transitions between character states are rare (MacClade, version 3.01; Maddison and Maddison), as is the case with our data set. Because nitrate assimilation deficiencies appear to have evolved multiple times in distantly related cyanobacteria from diverse habitats in different geographical locations (Fig. 1; Table 3), we expect that the molecular genetic basis of inactivation of nitrate assimilation has differed in different lineages. Inactivation may have been the result of mutations of different classes (e.g., nonsense mutation versus deletion) at different sites or potentially different loci (e.g., narB versus nirA). Determining the actual mutational event which inactivated nitrate assimilation in a particular lineage, however, would likely be difficult unless it were of very recent evolutionary origin. This is because a gene would be expected to evolve at an accelerated (neutral) rate following inactivation. Nonetheless, we would expect the sequences of inactivated nitrate assimilation genes from different cyanobacteria to exhibit unique molecular signatures reflecting their independent histories of mutation accumulation, a pattern which would corroborate the hypothesis of independent evolutionary origins supported by the phylogenetic distribution of nitrate assimilation deficiencies (Fig. 1).
It is possible that these independent evolutionary events share an ecological correlate. Since nitrate assimilation is repressed by ammonium, populations occupying ammonium-replete habitats may exist for generations under conditions in which mutational inactivation of genes involved in the assimilation of nitrate and nitrite would not be deleterious. Synechococcus sp. strain SH-94-5 was isolated from an ammonium-rich hot spring (955 μg liter−1), (C. E. Wingard, J. R. Schiller, S. R. Miller, and R. W. Castenholz, unpublished data), while its closest known relative, Synechococcus sp. strain C9, which grows well on nitrate, was obtained from Octopus Spring, Yellowstone National Park, a spring with no detectable ammonium (3). O. amphigranulata (Table 3) is found in New Zealand hot springs with ammonium levels varying between 200 and 1,200 μg liter−1 (16). Ammonium is continuously supplied to the above habitats by their respective geothermal sources. The peridunar pond from which Leptolyngbya sp. strain PCC 9207 (Table 3) was isolated varied in ammonium concentration from ∼10 to 200 μg liter−1 during a 2-year limnological survey (28). We were not able to obtain water chemistry data for the collection sites from which Leptolyngbya sp. strain PCC 9221 and Geitlerinema sp. strain PCC 9222 were isolated (Table 3). However, all strains for which we have environmental data were isolated from ammonium-replete habitats, which is consistent with the hypothesis that nitrate assimilation deficiencies can attain high frequency or fixation only in populations in which individuals are released from selection to maintain functional nitrate assimilation genes by persistent environmental ammonium.
The observation of convergent phenotypes in unrelated organisms with shared ecologies is often considered to be evidence for adaptation (15). It is not clear, however, whether natural selection favored these losses of the ability to assimilate nitrate. Although no transcripts of nitrate assimilation genes can be detected during ammonium repression (9), cell extracts and permeabilized cells of a few cyanobacterial strains do exhibit very low activities of Nar and Nir (18, 20). Expression of these proteins would serve little purpose in vivo under these conditions, however, since ammonium very efficiently inhibits all nitrate and nitrite transport across the cell membrane (19). A mutant which diverted resources ordinarily allocated toward basal expression of the nitrate assimilation machinery toward other purposes might therefore have a selective advantage under ammonium repression. On the other hand, if the loss of nitrate assimilation genes were somehow favored during ammonium repression, we might not expect to find cyanobacteria capable of utilizing more oxidized forms of inorganic N in ammonium-replete habitats. This is not the case, for we have isolated a number of clones from South Harney Hot Spring which grow well with nitrate as the sole N source, including Synechococcus sp. strain SH-94-45 (Table 3; Fig. 1) and a filamentous cyanobacterium (unpublished results). The latter was cloned from an enrichment in D medium of 10 cells/trichome, which indicates that it was derived from an abundant natural population.
Alternatively, these independently derived losses of the ability to use nitrate may be examples of neutral, nonadaptive evolution in which one or more nitrate assimilation genes have degenerated by mutation into pseudogenes over generations of transcriptional repression by environmental ammonium. These neutral alleles could attain high frequency or fixation in a population by drift or, more likely, by being linked to favorable alleles at other loci during episodes of periodic directional selection (2). Evidence for genetic hitchhiking of neutral alleles during such selective sweeps in natural populations of E. coli has been provided by Guttman and Dykhuizen (17). Diversification of ecological requirements, therefore, is not necessarily always adaptive.
Regardless of the evolutionary basis of nitrate assimilation deficiencies in these cyanobacteria, the loss of the ability to utilize nitrate would be deleterious in some environments and consequently could act as a barrier to migration. That is, while the ability of nitrate assimilation-deficient cyanobacteria to disperse between ammonium-containing habitats is suggested by the demonstration of clones with identical 16S rRNA gene sequences from lakes in Europe and South America (Leptolyngbya sp. strains PCC 9207 and PCC 9221), a dispersing nitrate assimilation-deficient cell would not be expected to successfully colonize ammonium-depleted but otherwise suitable habitats. For example, a Synechococcus sp. strain SH-94-5 cell would be expected to starve for N in Octopus Spring, Yellowstone National Park, from which its nitrate-utilizing sister, Synechococcus sp. strain C9, was isolated. The derivation of nitrate assimilation-deficient cyanobacteria in ammonium-replete habitats therefore may illustrate how ecology and evolution can interact to shape biogeography in the microbial world.
ACKNOWLEDGMENTS
We thank Carrie Inman for her helpful assistance and two anonymous reviewers for their comments and suggestions.
This work was supported by U.S. National Science Foundation grants IBN-9219273 and IBN-9630674 to R.W.C. and by an NSF research training grant (DBI-9413223) to the Department of Biology at the University of Oregon.
REFERENCES
- 1.Aichi M, Omata T. Involvement of NtcB, a LysR family transcription factor, in nitrite activation of the nitrate assimilation operon in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1997;179:4671–4675. doi: 10.1128/jb.179.15.4671-4675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atwood K C, Schneider L K, Ryan F J. Periodic selection in Escherichia coli. Proc Natl Acad Sci USA. 1951;37:146–155. doi: 10.1073/pnas.37.3.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brock T D. Thermophilic microorganisms and life at high temperatures. New York, N.Y: Springer-Verlag; 1978. [Google Scholar]
- 4.Castenholz R W. The effect of sulfide on the blue-green algae of hot springs. I New Zealand and Iceland J Phycol. 1976;12:54–68. [Google Scholar]
- 5.Castenholz R W. Culturing methods for cyanobacteria. Methods Enzymol. 1988;167:68–93. [Google Scholar]
- 6.Castenholz R W, Utkilen H C. Physiology of sulfide tolerance in a thermophilic Oscillatoria. Arch Microbiol. 1984;136:299–305. [Google Scholar]
- 7.Dobrogosz W J. Enzymatic activity. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 365–392. [Google Scholar]
- 8.Ferris M J, Ruff-Roberts A L, Kopezynski E D, Bateson M M, Ward D M. Enrichment culture and microscopy conceal diverse thermophilic Synechococcus populations in a single hot spring microbial mat habitat. Appl Environ Microbiol. 1996;62:1045–1050. doi: 10.1128/aem.62.3.1045-1050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flores E, Herrero A. Assimilatory nitrogen metabolism and its regulation. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 487–517. [Google Scholar]
- 10.Forchhammer K, Tandeau de Marsac N. The PII protein in the cyanobacterium Synechococcus sp. strain PCC 7942 is modified by serine phosphorylation and signals the cellular N-status. J Bacteriol. 1994;176:84–91. doi: 10.1128/jb.176.1.84-91.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forchhammer K, Tandeau de Marsac N. Functional analysis of the phosphoprotein PII (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1995;177:2033–2040. doi: 10.1128/jb.177.8.2033-2040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frías J E, Flores E, Herrero A. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:477–486. doi: 10.1128/jb.179.2.477-486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Pichel F, Castenholz R W. Comparative anoxygenic photosynthetic capacity in 7 strains of a thermophilic cyanobacterium. Arch Microbiol. 1990;153:344–351. [Google Scholar]
- 14.Giovannoni S J, Turner S, Olsen G J, Barns S, Lane D J, Pace N R. Evolutionary relationships among cyanobacteria and green chloroplasts. J Bacteriol. 1988;170:3584–3592. doi: 10.1128/jb.170.8.3584-3592.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Givnish T J. Adaptive radiation and molecular systematics: issues and approaches. In: Givnish T J, Sytsma K J, editors. Molecular evolution and adaptive radiation. Cambridge, England: Cambridge University Press; 1997. pp. 1–54. [Google Scholar]
- 16.Glover R B. The chemistry of thermal waters at Rotorua. N Z J Sci. 1967;10:70–96. [Google Scholar]
- 17.Guttman D, Dykhuizen D. Detecting selective sweeps in naturally occurring Escherichia coli. Genetics. 1994;138:993–1003. doi: 10.1093/genetics/138.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrero A, Flores E, Guerrero M G. Regulation of nitrate reductase levels in the cyanobacteria Anacystis nidulans, Anabaena sp. strain 7119, and Nostoc sp. strain 6719. J Bacteriol. 1981;145:175–180. doi: 10.1128/jb.145.1.175-180.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrero A, Flores E, Guerrero M. Regulation of nitrate reductase cellular levels in the cyanobacteria Anabaena variabilis and Synechocystis sp. FEMS Microbiol Lett. 1985;26:21–25. [Google Scholar]
- 20.Herrero A, Guerrero M. Regulation of nitrite reductase in the cyanobacterium Anacystis nidulans. J Gen Microbiol. 1986;132:2463–2468. [Google Scholar]
- 21.Izawa S. Inhibitors of electron transport. In: Trebst A, Avron M, editors. Photosynthesis I: photosynthetic electron transport and photophosphorylation. Berlin, Germany: Springer-Verlag; 1977. pp. 266–282. [Google Scholar]
- 22.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions (supplement) DNA Res. 1996;3:185–209. doi: 10.1093/dnares/3.3.185. [DOI] [PubMed] [Google Scholar]
- 23.Kapp R, Stevens S E, Fox J L. A survey of available nitrogen sources for the growth of the blue-green alga, Agmenellum quadruplicatum. Arch Microbiol. 1975;104:135–138. doi: 10.1007/BF00447313. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 25.Kratz W A, Meyers J. Nutrition and growth of several blue-green algae. Am J Bot. 1955;42:282–287. [Google Scholar]
- 26.Lara C, Romero J M, Guerrero M G. Regulated nitrate transport in the cyanobacterium Anacystis nidulans. J Bacteriol. 1987;169:4376–4378. doi: 10.1128/jb.169.9.4376-4378.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H-M, Vázquez-Bermúdez M F, Tandeau de Marsac N. The global nitrogen regulator NtcA regulates transcription of the signal transducer PII (GlnB) and influences its phosphorylation level in response to nitrogen and carbon supplies in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1999;181:2697–2702. doi: 10.1128/jb.181.9.2697-2702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez T, Toja J, Gabellone N. Limnological comparison of two peridunar ponds in the Donana National Park (Spain) Arch Hydrobiol. 1991;120:357–378. [Google Scholar]
- 29.Luque I, Flores E, Herrero A. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 1994;13:2862–2869. doi: 10.1002/j.1460-2075.1994.tb06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manzano C, Candau P, Gomez-Moreno C, Relimpio A M, Losada M. Ferredoxin-dependent photosynthetic reduction of nitrate and nitrite by particles of Anacystis nidulans. Mol Cell Biochem. 1976;10:161–169. doi: 10.1007/BF01731687. [DOI] [PubMed] [Google Scholar]
- 31.Meeks J C, Wolk C P, Thomas J, Lockau W, Shaffer P W, Austin S M, Chien W-S, Galonsky A. The pathways of assimilation of 13NH4+ by the cyanobacterium, Anabaena cylindrica. J Biol Chem. 1977;252:7894–7900. [PubMed] [Google Scholar]
- 32.Merchan F, Kindle K L, Llama M J, Serra J L, Fernandez E. Cloning and sequencing of the nitrate transport system from the thermophilic, filamentous cyanobacterium Phormidium laminosum: comparative analysis with the homologous system from Synechococcus sp. PCC 7942. Plant Mol Biol. 1995;28:759–766. doi: 10.1007/BF00021199. [DOI] [PubMed] [Google Scholar]
- 33.Miller S R, Castenholz R W. Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl Environ Microbiol. 2000;66:4222–4229. doi: 10.1128/aem.66.10.4222-4229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omata T. Transcriptional and post-translational regulation of nitrate utilization in the cyanobacterium Synechococcus sp. strain PCC 7942. In: Satoh K, Murata N, editors. Stress responses of photosynthetic organisms. Amsterdam, The Netherlands: Elsevier; 1998. pp. 197–214. [Google Scholar]
- 36.Omata T, Ohmori M, Arai N, Ogawa T. Genetically engineered mutant of the cyanobacterium Synechococcus PCC 7942 defective in nitrate transport. Proc Natl Acad Sci USA. 1989;86:6612–6616. doi: 10.1073/pnas.86.17.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 38.Ramasubramanian T S, Wei T-F, Oldham A K, Golden J W. Transcription of the Anabaena sp. strain PCC 7120 ntcA gene: multiple transcripts and NtcA binding. J Bacteriol. 1996;178:922–926. doi: 10.1128/jb.178.3.922-926.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rigano C. Studies on nitrate reductase from Cyanidium caldarium. Arch Mikrobiol. 1971;76:265–276. doi: 10.1007/BF00409121. [DOI] [PubMed] [Google Scholar]
- 40.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke M N, Olson B J, Clenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 41.Snell F D, Snell C T. Colorimetric methods of analysis. Vol. 3. New York, N.Y: Van Nostrand; 1949. [Google Scholar]
- 42.Suzuki I, Horie N, Sugiyama T, Omata T. Identification and characterization of two nitrogen-regulated genes of the cyanobacterium Synechococcus sp. strain PCC7942 required for maximum efficiency of nitrogen assimilation. J Bacteriol. 1995;177:290–296. doi: 10.1128/jb.177.2.290-296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urbach E, Robertson D, Chisholm S W. Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation. Nature. 1992;355:267–269. doi: 10.1038/355267a0. [DOI] [PubMed] [Google Scholar]
- 44.Vega-Palas M A, Madueño F, Herrero A, Flores E. Identification and cloning of a regulatory gene for nitrogen assimilation in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1990;172:643–647. doi: 10.1128/jb.172.2.643-647.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Li H, Post A F. Nitrate assimilation genes of the marine diazotrophic, filamentous cyanobacterium Trichodesmium sp. strain WH9601. J Bacteriol. 2000;182:1764–1767. doi: 10.1128/jb.182.6.1764-1767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei T-F, Ramasubramanian T S, Golden J W. Anabaena sp. strain PCC 7120 ntcA gene required for growth on nitrate and heterocyst development. J Bacteriol. 1994;176:4473–4482. doi: 10.1128/jb.176.15.4473-4482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wheeler W, Gladstein D. MALIGN: a multiple sequence alignment program. J Hered. 1994;85:417–418. [Google Scholar]