Abstract
Background
Prospective audit with feedback (PAF) is an impactful strategy for antimicrobial stewardship program (ASP) activities. However, because PAF requires reviewing large numbers of antimicrobial orders on a case-by-case basis, PAF programs are highly resource intensive. The current study aimed to identify predictors of ASP intervention (ie, feedback) and to build models to identify orders that can be safely bypassed from review, to make PAF programs more efficient.
Methods
We performed a retrospective cross-sectional study of inpatient antimicrobial orders reviewed by the University of Maryland Medical Center’s PAF program between 2017 and 2019. We evaluated the relationship between antimicrobial and patient characteristics with ASP intervention using multivariable logistic regression models. Separately, we built prediction models for ASP intervention using statistical and machine learning approaches and evaluated performance on held-out data.
Results
Across 17 503 PAF reviews, 4219 (24%) resulted in intervention. In adjusted analyses, a clinical pharmacist on the ordering unit or receipt of an infectious disease consult were associated with 17% and 56% lower intervention odds, respectively (adjusted odds ratios [aORs], 0.83 and 0.44; P ≤ .001 for both). Fluoroquinolones had the highest adjusted intervention odds (aOR, 3.22 [95% confidence interval, 2.63–3.96]). A machine learning classifier (C-statistic 0.76) reduced reviews by 49% while achieving 78% sensitivity. A “workflow simplified” regression model that restricted to antimicrobial class and clinical indication variables, 2 strong machine learning–identified predictors, reduced reviews by one-third while achieving 81% sensitivity.
Conclusions
Prediction models substantially reduced PAF review caseloads while maintaining high sensitivities. Our results and approach may offer a blueprint for other ASPs.
Keywords: ASP, antimicrobial stewardship, machine learning, prospective audit with feedback
Prospective audit with feedback (PAF) is an impactful, but resource-intensive, antimicrobial stewardship activity. Evaluating >17 000 PAF reviews, we built machine learning and statistical prediction models that substantially reduced review caseloads while maintaining high sensitivities, including with minimal or no automation.
Antimicrobial stewardship programs (ASPs) are tasked with improving antimicrobial use across many disciplines in and out of the hospital. A prospective audit with feedback (PAF) program is an impactful strategy for in-hospital ASP activities. Under this type of stewardship program, an antimicrobial stewardship team audits active antimicrobial orders within a specified timeframe. If the antimicrobial order is deemed noncompliant with recommended guidelines or needs optimization of dose, route, etc, the ASP team will intervene to provide therapeutic recommendations and feedback to treating providers. Compared to restrictive strategies, PAF is more effective at reducing antimicrobial utilization, and it is a favored ASP strategy in most inpatient settings [1–3].
In large medical centers with >200 new antimicrobial orders daily, however, this ASP task also becomes enormous. Understanding which clinical service and care locations, patient types, and antimicrobials have the highest or lowest likelihood of intervention may provide opportunities for streamlining PAF programs to reduce their resource-intensiveness. We were therefore interested in learning how PAF programs might perform activities more efficiently by identifying “hotspots” for ASP intervention in our medical center.
Motivated by the preceding considerations, the objectives of the current study were 2-fold: (1) to understand which patient and treatment characteristics are associated with either a higher or lower likelihood of intervention in a PAF program; and (2) to develop prediction models to identify antimicrobial orders that may be safely excluded from review due to a high probability that, even if they had been reviewed, they would not have triggered an intervention. To achieve these goals, we deployed both traditional regression analyses and newer machine learning techniques. We hope that our approach can provide a blueprint for other stewardship programs to undertake similar analyses on their own data.
METHODS
The University of Maryland Medical Center PAF Program
The University of Maryland Medical Center (UMMC) is a 750-bed tertiary care center in Baltimore, Maryland, with solid organ and hematopoietic stem cell transplant and oncology programs and 8 specialized intensive care units. At UMMC, there are approximately 100 new antimicrobial orders daily and 500 active antimicrobial orders at any one time. The UMMC ASP team consists of 2 infectious disease (ID) physicians (combined 0.3 full-time equivalent) and 3 ID pharmacists (combined 1.5 full-time equivalent). A PAF program was introduced in July 2017 and is the mainstay of the stewardship program [4]. Under this program, the ASP team aims to review all new inpatient antimicrobial orders within 3 days of order, to replicate the Centers for Disease Control and Prevention–endorsed 72-hour “antibiotic time-out” [5].
Each weekday, ASP staff obtain a list of antimicrobial orders from EPIC, the electronic medical record (EMR). Orders from patients who have not received ID consults are prioritized for review, as are intravenous antimicrobials and fluoroquinolones; however, our ASP policy is to review every new order that is not discontinued before day 3, and our program generally meets this goal. During review, prescriptions are evaluated for appropriateness against UMMC antimicrobial guidelines by an ASP team member; UMMC follows national guidelines, with guidance tailored to the UMMC formulary and antibiograms. If the ASP member deems the antibiotic order suboptimal or unnecessary, they will intervene to recommend a therapeutic change (eg, discontinuation, de-escalation, dosing modifications) or an ID consult. All reviews and interventions are documented electronically and maintained in an ASP PAF database.
Study Cohort and Collected Data
We conducted a 2.5-year cross-sectional study of inpatient antimicrobial orders that were reviewed by the PAF program between July 2017 and December 2019. All unique reviews were included, except for orders that resulted from previous ASP intervention (eg, if a patient was initiated on piperacillin-tazobactam and the ASP team recommended change to ceftriaxone, we excluded subsequent review[s] of ceftriaxone for this admission). For each included order, we classified it by antimicrobial class/spectrum of activity, route of administration and dose, and season. Antimicrobial classifications were selected based upon antimicrobial stewardship and clinical relevance, and decisions were completed prior to data analysis (Supplementary Table 1). We batch-extracted additional EMR data for each order: (1) patient encounter and demographic data (eg, age, sex); (2) clinical and provider data (eg, service, provider-entered clinical indication for antimicrobials, whether the patient received an ID consult); (3) location data (eg, prescribing unit, whether the unit staffs a clinical pharmacist); (4) and patient antimicrobial treatment and resistance history (eg, prior antimicrobial therapy, drug allergies, multidrug-resistant organism history). From our PAF database, we also extracted data capturing whether there was a positive culture at the time of review (as recorded by the ASP member contemporaneously with review). Because an important goal of the study was to develop a model for prospectively predicting antimicrobial orders to bypass from review, we restricted to information that was available at or before the review day. This study was determined to be exempt human subjects research by the University of Maryland School of Medicine Institutional Review Board.
Outcome and Statistical Methods
The primary study outcome was whether a review resulted in ASP intervention (ie, feedback to the treating provider). The relationship between each covariate and ASP intervention was evaluated using univariable and multivariable logistic regression models with general estimating equations to account for repeat observations by patient. Results were summarized by odds ratios (ORs) and corresponding 95% confidence intervals (CIs). Variables found to have a P value < .10 on univariable analysis were evaluated in a multivariable model.
To evaluate the discrimination of the multivariable model, we refit the model on an 80% random cohort sample (training set). We generated a receiver operating characteristic curve and calculated the C-statistic using the remaining, held-out 20% of the cohort (validation set). Given the intended real-world use of our models, we also evaluated “workflow simplified” models using the same process; these models used only statistically significant variables from the full multivariable model that would also be rapid to ascertain at review, without requiring extensive chart review. All tests were 2-tailed, and P values ≤ .05 were used for statistical significance testing. Analyses were performed using SAS version 9.4 (SAS Institute) and Stata 15.0 (StataCorp) software.
Sensitivity Analyses
To address clinical importance and generalizability considerations, we performed 3 prespecified sensitivity analyses. First, we evaluated an alternative outcome that restricted to de-escalation and escalation interventions, arguably the most “high-impact” interventions [6]; this outcome excluded interventions for dosing/route optimization, ID consult recommendations, and therapeutic duration/other modifications. Second and third, a relatively high percentage of UMMC patients receive ID consults, and some UMMC units also staff dedicated clinical pharmacists that round with the primary care team and often take on an antimicrobial stewardship role; these characteristics may not be widely generalizable. We therefore performed analyses restricting to patients who did not receive ID consults and to patients who received neither ID consults nor had a clinical pharmacist involved in their care.
Machine Learning–Based Predictive Modeling
We also developed a prediction model for ASP intervention using random forests, a machine learning algorithm [7–9]. Because random forest algorithms accommodate high predictor-to-outcome ratios, collinearities, and interaction effects by default [10, 11], we provided all variables to the algorithm during model-building, including permutations (eg, a composite multidrug-resistant organism [MDRO] history variable and MDRO history variables by organism, an alternative antimicrobial classification system developed by Moehring et al [12]) (see Supplementary Materials). We fit our model using 500 bootstrapped decision trees and calculated the sensitivity and specificity, the C-statistic, the out-of-bag error rate, and variable importance rankings. Machine learning analyses were performed using the randomForest package (version 4.6–14) in R version 4.1.2. To enable other institutions to perform similar analyses on their own data, we have provided statistical and machine learning programming code and implementation suggestions in the Supplementary Materials and Supplementary Appendix 1.
RESULTS
During the July 2017–December 2019 study period, the ASP team conducted 19 852 antimicrobial order reviews, 17 503 of which were included in the final study (Supplementary Figure 1). The 17 503 included reviews, conducted a mean of 3.7 days from antimicrobial order (standard deviation, 1.19), came from 9865 unique inpatient admissions and 8226 unique patients. Reviews of first-line antipseudomonal agents were most common (n = 4218 [24%]), followed by anti–methicillin-resistant Staphylococcus aureus (MRSA) agents (n = 2769 [16%]) and broad-spectrum agents (n = 2674 [15%]) (Table 1).
Table 1.
Patient Demographic, Clinical, and Antimicrobial Characteristics in a Cohort of Antimicrobial Stewardship Program–Reviewed Antimicrobial Orders at the University of Maryland Medical Center (2017–2019)
| Characteristic | Total (n = 17 503) |
No Intervention (n = 13 284) |
Resulted in Intervention (n = 4219) |
|---|---|---|---|
| Patient demographic and history characteristics | |||
| Male sex | 10 126 (58) | 7723 (58) | 2403 (57) |
| Age >55 ya | 9974 (57) | 7459 (56) | 2515 (60) |
| EMR-documented antibiotic allergy | 9015 (52) | 6862 (52) | 2153 (51) |
| Prior MDRO historyb | 5778 (33) | 6862 (52) | 2153 (51) |
| Patient clinical and treatment characteristics | |||
| Provider-entered clinical indication for antimicrobial orderc | |||
| Sepsis/bacteremia | 3375 (19) | 2633 (20) | 742 (18) |
| Bone/joint | 865 (5) | 718 (5) | 147 (4) |
| Central nervous system | 350 (2) | 302 (2) | 48 (1) |
| Cardiac/vascular | 578 (3) | 522 (4) | 56 (1) |
| Gastrointestinal | 1844 (11) | 1387 (10) | 457 (11) |
| Genitourinary | 1185 (7) | 738 (6) | 447 (11) |
| Respiratory | 2951 (17) | 2135 (16) | 816 (19) |
| Nonsurgical prophylaxis | 150 (1) | 118 (0.9) | 32 (0.8) |
| Skin and soft tissue infection | 2794 (16) | 2065 (16) | 729 (17) |
| Mycobacterial infection | 493 (3) | 391 (3) | 102 (2) |
| Neutropenia | 627 (4) | 556 (4) | 71 (2) |
| Surgical prophylaxis | 715 (4) | 386 (3) | 329 (8) |
| None provided | 1576 (9) | 1333 (10) | 243 (6) |
| Immunosuppressedd | 2607 (15) | 2151 (16) | 456 (11) |
| Received ID consulte | 12 830 (73) | 10 435 (79) | 2395 (57) |
| Clinical pharmacist–staffed unit | 10 056 (58) | 7877 (59) | 2179 (52) |
| Antimicrobial order characteristicsf | |||
| Antimicrobial class | |||
| Narrow-spectrum agents | 1252 (7) | 999 (8) | 253 (6) |
| Antiviral agents | 227 (1) | 197 (2) | 30 (1) |
| Broad-spectrum agents | 2674 (15) | 2066 (16) | 608 (14) |
| Antifungal agents | 1472 (8) | 1158 (8) | 314 (7) |
| Other | 1954 (11) | 1433 (11) | 521 (12) |
| First-line antipseudomonal agents | 4218 (24) | 3132 (24) | 1086 (26) |
| Protected agents | 1500 (9) | 1138 (9) | 362 (9) |
| Anti-MRSA agents | 2769 (16) | 2144 (16) | 625 (15) |
| Clostridioides difficile agents | 506 (3) | 453 (3) | 53 (1) |
| Fluoroquinolones | 931 (5) | 564 (4) | 367 (8) |
| Fall/winter season of order | 10 430 (60) | 8049 (61) | 2381 (56) |
| Positive culture by time of ASP review | 8289 (47) | 6642 (50) | 1647 (39) |
Data are presented as No. (%). Percentages may not sum to 100% due to rounding.
Abbreviations: ASP, antimicrobial stewardship program; EMR, electronic medical record; ID, infectious disease; MDRO, multidrug-resistant organism; MRSA, methicillin-resistant Staphylococcus aureus.
The variable for patient age was dichotomized at the mean age of the cohort, which was 55 years, for ease of implementation, internal validity, and generalizability considerations in the prediction models. Continuous variables such as age will require initial exploratory data analysis to confirm that regression assumptions are met in the data. Because age will not necessarily demonstrate log-linearity with the outcome of intervention at other hospitals, and not all ASPs are likely equipped to perform lengthy exploratory data analysis prior to model-building, we felt that dichotomization offered a preferred parameterization for this variable. Moreover, our dataset included unique antimicrobial orders, but not unique patients. We used general estimating equations in our logistic regression models to account for repeat observations by patient, and tested performance on held-out data, but highly granular variables like age could still pose some risk of overfitting in prediction models that include multiple observations per patient. Use of a less granular, dichotomized age variable helps to decrease this risk. An alternative option would be categorization into age brackets. The median age in our cohort was 57 (interquartile range, 44–67) years.
As defined by an infection control banner flag for MRSA, vancomycin-resistant enterococci, carbapenem-resistant Enterobacterales, an extended-spectrum β-lactamase–producing organism, multidrug-resistant Acinetobacter baumannii, or an otherwise-not-specific multidrug-resistant gram-negative organism.
We restricted to the provider-entered clinical indication, even when this indication was later corrected during review by the ASP team, to ensure that the prediction models only considered information that was available at or before review. Otherwise, allowing the model to consider the corrected indication would contaminate the model with information that only became available during or following order review.
Defined as patient presence on an oncology or solid-organ transplant unit at the time of antibiotic order.
By the time of ASP team review.
See Supplementary Table 1 for the list of agents included in each antimicrobial class.
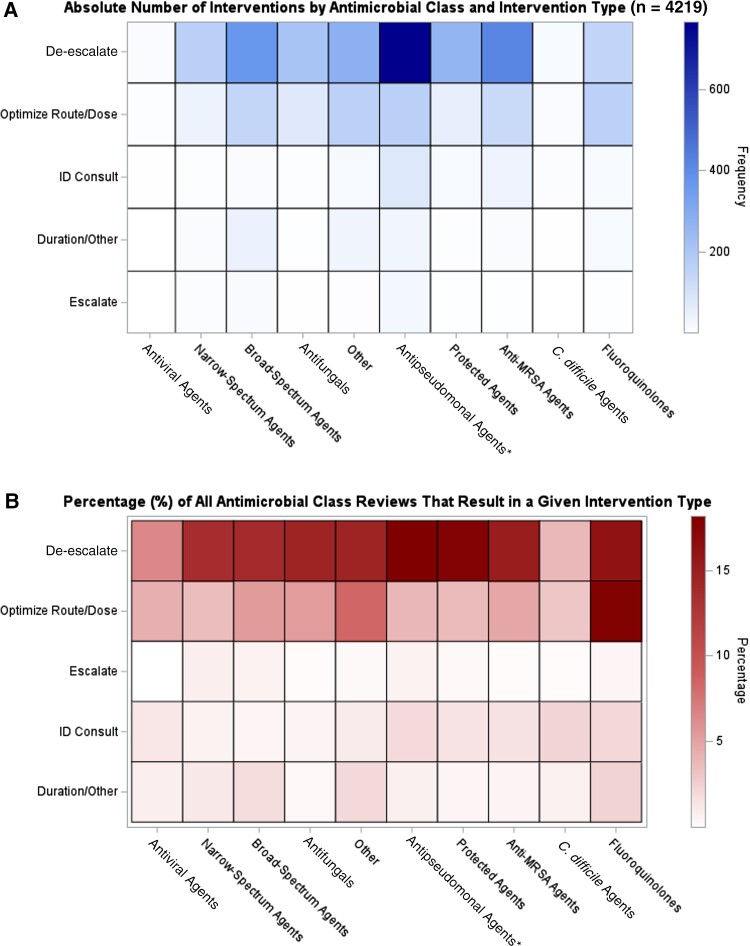
Twenty-four percent (4219) of reviews resulted in an intervention. Supplementary Figure 1 reflects the distribution of intervention types; recommendations to de-escalate were the most common (2690 [64%]), followed by recommendations to optimize route or dosing (995 [24%]). Figure 1 reflects a heatmap of ASP intervention by antimicrobial class and intervention type (see Supplementary Figures 2–4 for frequency counts by cell, and for heatmaps by provider-entered clinical indication). The ASP team intervened most frequently to recommend de-escalation of first-line antipseudomonal agents, corresponding to 766 interventions and 18% of all reviews of these agents. The ASP team also intervened a similarly high percentage of the time to de-escalate protected agents (18% of all protected agent reviews) and to de-escalate or to optimize dosing of fluoroquinolones. However, on an absolute basis these interventions were much fewer in number, due to less frequent use of protected agents and fluoroquinolones in the cohort. Intervention was generally rarest for antivirals and anti–Clostridioides difficile agents.
Figure 1.
Heatmaps of antimicrobial stewardship programs, by antimicrobial class and intervention type, on absolute (A) and relative (B) bases. Antimicrobial classifications are mutually exclusive. *“Antipseudomonal” refers to first-line antipseudomonal agents (see Supplementary Table 1 for all antimicrobial classifications). Abbreviations: C. difficile, Clostridioides difficile; ID, infectious disease; MRSA, methicillin-resistant Staphylococcus aureus.
Relationship Between Antimicrobial, Patient Demographic, and Clinical Characteristics and ASP Intervention
In univariable analyses, many antimicrobial, patient, and clinical characteristics were associated with ASP intervention at an α level of .10 (Table 2). Apart from age, all variables remained significantly associated with ASP intervention in a multivariable model (Table 2). Presence of a clinical pharmacist on the ordering unit or receipt of an ID consult were independently associated with 17% and 56% lower odds of intervention, respectively (adjusted ORs [aORs], 0.83 [95% CI, .76–.90] and 0.44 [95% CI, .40–.48]; P ≤ .001 for both). By antimicrobial class, fluoroquinolones and protected agents had the highest and second-highest relative adjusted odds of intervention, respectively, with intervention odds that were 3.22 (95% CI, 2.63–3.96) and 2.54 (95% CI, 2.08–3.10) times that of narrow-spectrum agents (Table 2). Relative to a provider-entered indication of sepsis or bacteremia, only genitourinary and surgical prophylaxis indications had higher adjusted odds of intervention (aORs, 1.68 [95% CI, 1.42–1.98] and 1.87 [95% CI, 1.53–2.29], respectively).
Table 2.
Association Between Patient Demographic, Clinical, and Antimicrobial Characteristics and Antimicrobial Stewardship Program Intervention in Univariable and Multivariable Models a
| Characteristic | OR (95% CI) (n = 17 503) |
P Value | Adjusted OR (95% CI) (n = 17 503) |
P Value |
|---|---|---|---|---|
| Patient demographic and history characteristics | ||||
| Male sex | 0.95 (.88–1.03) | 0.21 | … | |
| Age >55 yb | 1.16 (1.07–1.25) | <.001 | 1.06 (.98–1.15) | .15 |
| EMR-documented antibiotic allergy | 0.99 (.92–1.07) | .81 | … | |
| Prior MDRO historyc | 0.69 (.64–.75) | <.001 | 0.81 (.74–.89) | <.001 |
| Patient clinical and treatment characteristics | ||||
| Provider-entered clinical indication for antimicrobial orderd | ||||
| Sepsis/bacteremia | Ref | Ref | Ref | Ref |
| Bone/joint | 0.73 | .002 | 0.68 (.54–.84) | <.001 |
| Central nervous system | 0.55 | <.001 | 0.53 (.38–.75) | <.001 |
| Cardiac/vascular | 0.39 | <.001 | 0.43 (.31–.58) | <.001 |
| Gastrointestinal | 1.14 | .07 | 0.86 (.74–1.01) | .06 |
| Genitourinary | 2.10 | <.001 | 1.67 (1.42–1.98) | <.001 |
| Respiratory | 1.35 | <.001 | 1.07 (.94–1.22) | .28 |
| Nonsurgical prophylaxis | 0.97 | .89 | 0.56 (.36–.85) | .01 |
| Skin and soft tissue infection | 1.22 | .002 | 1.00 (.87–1.15) | 1.00 |
| Mycobacterial infection | 0.91 | .47 | 0.68 (.52–.90) | .01 |
| Neutropenia | 0.49 | <.001 | 0.50 (.38–.65) | <.001 |
| Surgical prophylaxis | 2.93 | <.001 | 1.87 (1.53–2.29) | <.001 |
| None provided | 0.65 | <.001 | 0.65 (.52–.81) | <.001 |
| Immunosuppressede | 0.64 (.57–.72) | <.001 | 0.78 (.68–.90) | <.001 |
| Received ID consultf | 0.37 (.34–.40) | <.001 | 0.44 (.40–.48) | <.001 |
| Clinical pharmacist-staffed unit | 0.76 (.70–.82) | <.001 | 0.83 (.76–.90) | <.001 |
| Antimicrobial order characteristics | ||||
| Antimicrobial classg | ||||
| Narrow-spectrum agents | Ref | Ref | Ref | Ref |
| Antiviral agents | 0.61 (.40–.93) | .02 | 1.38 (.85–2.25) | .19 |
| Broad-spectrum agents | 1.13 (.96–1.34) | .14 | 1.16 (.97–1.37) | .10 |
| Antifungal agents | 1.17 (.97–1.41) | .10 | 1.93 (1.58–2.36) | <.001 |
| Other | 1.45 (1.23–1.71) | <.001 | 1.90 (1.57–2.29) | <.001 |
| First-line antipseudomonal agents | 1.37 (1.17–1.60) | <.001 | 1.91 (1.62–2.27) | <.001 |
| Protected agents | 1.32 (1.10–1.59) | .003 | 2.54 (2.08–3.10) | <.001 |
| Anti-MRSA agents | 1.18 (1.00–1.39) | .05 | 1.94 (1.63–2.32) | <.001 |
| Clostridioides difficile agents | 0.48 (.36–.65) | <.001 | 1.11 (.76–1.63) | .57 |
| Fluoroquinolones | 2.61 (2.15–3.16) | <.001 | 3.22 (2.63–3.96) | <.001 |
| Fall/winter season of order | 0.85 (.79–.92) | <.001 | 0.84 (.78–.91) | <.001 |
| Positive culture by time of ASP review | 0.64 (.60–.70) | <.001 | 0.80 (.73–.88) | <.001 |
Abbreviations: ASP, antimicrobial stewardship program; CI, confidence interval; EMR, electronic medical record; ID, infectious disease; MDRO, multidrug-resistant organism; MRSA, methicillin-resistant Staphylococcus aureus; OR, odds ratio; Ref, reference group.
Associations were evaluated using logistic regression models with generalized estimating equations to account for repeat observations by patient. Variables with P values < .10 on univariable analysis were evaluated in the multivariable model. Using these criteria, variables excluded from the multivariable model are denoted in the table by “…”.
The variable for patient age was dichotomized at the mean age of the cohort, which was 55 years, for ease of implementation, internal validity, and generalizability considerations in the prediction models. Continuous variables such as age will require initial exploratory data analysis to confirm that regression assumptions are met in the data. Because age will not necessarily demonstrate log-linearity with the outcome of intervention at other hospitals, and not all ASPs are likely equipped to perform lengthy exploratory data analysis prior to model-building, we felt that dichotomization offered a preferred parameterization for this variable. Moreover, our dataset included unique antimicrobial orders, but not unique patients. We used general estimating equations in our logistic regression models to account for repeat observations by patient, and tested performance on held-out data, but highly granular variables like age could still pose some risk of overfitting in prediction models that include multiple observations per patient. Use of a less granular, dichotomized age variable helps to decrease this risk. An alternative option would be categorization into age brackets. The median age in our cohort was 57 (interquartile range, 44–67) years.
As defined by an infection control banner flag for MRSA, vancomycin-resistant enterococci, carbapenem-resistant Enterobacterales, an extended-spectrum β-lactamase–producing organism, multidrug-resistant Acinetobacter baumannii, or an otherwise-not-specific multidrug-resistant gram-negative organism.
We restricted to the provider-entered clinical indication, even when this indication was later corrected during review by the ASP team, to ensure that the prediction models only considered information that was available at or before review. Otherwise, allowing the model to consider the corrected indication would contaminate the model with information that only became available during or following order review, which might artificially inflate predictive performance and would pose threats to internal validity.
Defined as patient presence in an oncology or solid-organ transplant unit at the time of antibiotic order.
By the time of ASP team review.
See Supplementary Table 1 for a list of antibiotics included in each antimicrobial class.
Multivariable Logistic Regression Model Performance for Predicting ASP Intervention
The C-statistic (area under the curve [AUC]) for the full multivariable logistic regression model on the held-out validation set was 0.70 (95% CI, .68–.72). Using the intercept and coefficients from this model, a predicted probability of intervention was computed for each antimicrobial order in the validation set. Supplementary Table 2 shows model sensitivity, specificity, and the caseload reduction at each probability cutoff, by decile. At a review threshold of ≥20% probability (ie, review only those orders with a 20% or greater probability of intervention), the ASP team would reduce its review caseload by 44% while continuing to review three-fourths of all orders that result in intervention (model sensitivity: 76%). We selected this probability threshold because we believed that it represented the best balance between optimizing sensitivity and reducing review caseloads; however, other institutions could choose different thresholds.
Our “workflow simplified” logistic regression models only included variables that do not require extensive chart review; because variables’ ease-of-extraction likely varies across institutions, we evaluated 3 permutations: Each model included antimicrobial class, plus either provider-entered clinical indication (model A: validation set AUC [vAUC], 0.65 [95% CI, .63–.67]), whether the patient had an ID consult (model B: vAUC, 0.65 [95% CI, .63–.67]), or whether there was a positive culture at review (model C: vAUC, 0.59 [95% CI, .57–.61]). Focusing on the first 2 models, which had the highest discrimination, reviews could be reduced by approximately one-third (34% and 31%, respectively) while still achieving sensitivities ≥80%. The coefficients and sensitivities/specificities at various probability threshold cutoffs for these models are presented in Table 3.
Table 3.
Sensitivity, Specificity, and Caseload Reduction at Various Probability Cutoffs From the 2 Highest-Performing “Workflow Simplified” Models
| Probability Cutoff Threshold (ie, Review Only Those Orders With a Predicted Intervention Probability of the Below Value) | Sensitivity | Specificity | Total No. of Reviews Bypassed Using This Cutoffa (of a Possible 3435 Reviews) | % Reduction in PAF Review Caseloada | No. of “Missed” Interventionsa (of a Possible 849 Interventions) |
|---|---|---|---|---|---|
| Model A: Antimicrobial class + clinical indication variables | |||||
| ≥0% | 100.0% | 0.0% | 0 | 0% | 0 |
| ≥10% | 99.2% | 2.1% | 60 | 2% | 7 |
| ≥20% | 85.4% | 32.0% | 952 | 28% | 124 |
| ≥21%b | 81.0% | 38.8% | 1163 | 34% | 161 |
| ≥30% | 29.0% | 88.2% | 2884 | 84% | 603 |
| ≥40% | 12.7% | 94.9% | 3194 | 93% | 741 |
| ≥50% | 4.2% | 98.5% | 3360 | 98% | 813 |
| ≥60% | 0.1% | 99.9% | 3431 | 100% | 848 |
| >60% | 0.0% | 100.0% | 3435 | 100% | 849 |
| Model B: Antimicrobial class + ID consult variables | |||||
| ≥0% | 100.0% | 0.0% | 0 | 0% | 0 |
| ≥10% | 98.9% | 4.0% | 112 | 3% | 9 |
| ≥19%b | 79.9% | 34.1% | 1053 | 31% | 171 |
| ≥20% | 69.7% | 48.5% | 1511 | 44% | 257 |
| ≥30% | 45.1% | 79.3% | 2516 | 73% | 466 |
| ≥40% | 21.0% | 92.0% | 3049 | 89% | 671 |
| ≥50% | 5.3% | 98.5% | 3352 | 98% | 804 |
| >50% | 0.0% | 100.0% | 3435 | 100% | 849 |
Abbreviations: ID, infectious disease; PAF, prospective audit with feedback.
Data are from the held-out testing set (n = 3435).
Probability cutoffs were discretized by decile for initial evaluation. After identifying the upper and lower decile bands that would contain the acceptable sensitivity and caseload reduction values for our project (which we identified as approximately ≥70% sensitivity and ≥20% reduction in caseload), we evaluated single percentage-point cutoffs between these 2 deciles (results unshown, except for the selected cutoff in bold). For example, for model A we identified that the optimal probability cutoff would fall between 20% and 30%, and we evaluated cutoffs at 21%, 22%, etc, through to 29% (results unshown, except for the final selected cutoff in bold). For each model, the bold text represents the cutoff thresholds that we felt optimized the balance between sensitivity and caseload reduction, but other antimicrobial stewardship programs could choose different cutoffs depending upon their needs and preferences.
Finally, in a sensitivity analysis predicting only de-escalation and escalation interventions, model performance was similar to the primary model for predicting any intervention (vAUC, 0.69 [95% CI, .67–.72]). In sensitivity analyses predicting any intervention but restricting to patients who did not receive ID consults or who had neither ID consults nor clinical pharmacists involved in their care, AUCs were somewhat lower (0.64 and 0.65, respectively). In both instances, however, the number of reviews could be reduced by one-third (33%) while maintaining the same or higher sensitivities as the full cohort (80% and 76%, respectively).
Machine Learning–Based Modeling for Predicting ASP Intervention
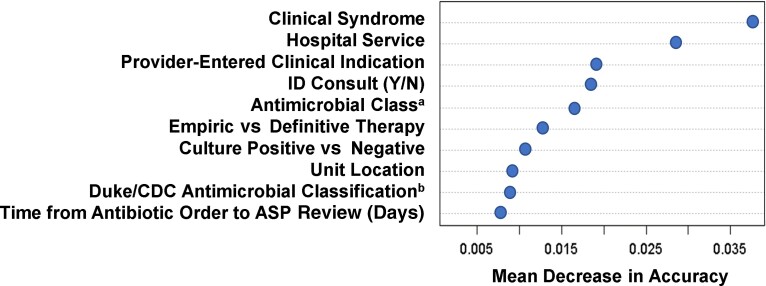
The random forest model had a C-statistic of 0.76 (95% CI, .75–.77), with a sensitivity and specificity of 78% and 58%, respectively (model parameters were weighted to prioritize sensitivity). Using this model would reduce review caseloads by 49%. The 2 most important predictors of ASP intervention were (1) clinical syndrome, which was a more granular parameterization of provider-entered clinical indication that also incorporated culture results (see Supplementary Appendix 2 for further description); and (2) primary hospital service at order (Figure 2). These variables were not evaluated in logistic regression models due to collinearity and high-dimensionality concerns (eg, hospital service had 47 unique values). The next 3 most important predictors were provider-entered clinical indication, antimicrobial class, and whether the patient had received an ID consult. Incidentally, these were the same variables included in the 2 best-performing “workflow simplified” models, suggesting that despite their simplicity, these simplified models included strong predictors.
Figure 2.
Variable importance plot from the random forest model, displaying the 10 most important variables for predicting antimicrobial stewardship program (ASP) intervention, in descending order of importance. Importance is measured by the mean decrease in model accuracy, which is roughly analogous to the loss in classifier accuracy when a given variable is excluded (ie, more important predictors will cause greater decreases in model predictive accuracy when they are removed from consideration during model-building). Some predictors may be collinear or represent similar concepts. For example, both our antimicrobial classification schemaa and an alternative Duke/Centers for Disease Control and Prevention antimicrobial classification schemab that we also provided to the model [12] both made it into the top 10 predictors list. This suggests that regardless of the exact classification schema used, antimicrobial class is an important variable for predicting which antimicrobial order reviews will result in ASP intervention. Abbreviations: ASP, antimicrobial stewardship program; CDC, Centers for Disease Control and Prevention; ID, infectious disease.
DISCUSSION
National guidelines recommend PAF as a core component of ASPs [2]. However, manually reviewing antimicrobial orders for appropriateness—the bedrock of PAF programs—is highly resource-intensive. These resource constraints create barriers to the successful implementation and sustainability of PAF programs in US hospitals. We hypothesized that by identifying intervention “hotspots,” PAF review caseloads could be safely reduced while maintaining high impact. Evaluating >17 000 PAF reviews across a 2.5-year period, we found that many clinical and antimicrobial characteristics were significantly associated with PAF intervention; these variables may help identity targets for increasing antibiotic order appropriateness. Moreover, prediction models built using these and other demographic, clinical, and antimicrobial characteristics substantially reduced PAF review caseloads while maintaining high sensitivities, including when using simplified models that are readily implementable with minimal to no automation.
Understanding which variables are independently associated with ASP intervention can highlight where we might intervene to reduce antimicrobial order inappropriateness. Holding other factors constant, presence of a clinical pharmacist on the ordering unit or receipt of an ID consult were associated with 17% and 56% lower odds of intervention, respectively. Our ID consult findings comport with previous research by our group linking ID consults to a higher likelihood of antibiotic appropriateness [4], as well as to research in pediatric populations [13] and in specific ID syndromes (eg, Staphylococcus aureus bacteremia) [14]. Similarly, clinical pharmacists are primed to engage in antimicrobial stewardship activities with a focus on optimization such as duration, dosing, and administration route adjustments, as well as de-escalation. Prior reports have identified positive impacts from non-ID-trained clinical pharmacists on antibiotic prescribing, including decreases in antibiotic utilization and antibiotic-associated costs [15, 16]. As such, clinical pharmacists provide a critical role in antimicrobial stewardship, and it is not surprising that ASP interventions were less likely in patients with clinical pharmacist oversight.
To predict which orders should be bypassed from ASP review, we evaluated multiple models using statistical and machine learning approaches. The highest-performing model (AUC, 0.76) was a machine learning–based classifier, which halved ASP review caseloads while achieving 78% sensitivity, that is, ensuring that nearly 8 of every 10 orders that result in intervention would still get reviewed by the ASP team. A full multivariable logistic regression model had somewhat lower discrimination (AUC, 0.70) but still reduced caseloads by 44% while maintaining 76% sensitivity. In a sensitivity analysis predicting only escalation or de-escalation interventions, model discrimination was very similar. Thus, there is no evidence that ASPs adopting this model would disproportionately miss the most high-impact interventions.
The previous work that is most similar to the current study was performed by Bystritsky et al at the University of California San Francisco Medical Center. Using a 2015–2017 cohort, they also developed a machine learning and a multivariable logistic regression model to predict ASP intervention [17]. Interestingly, although they used a different machine learning approach and their cohort restricted to patients receiving broad-spectrum antibiotics, our respective machine learning–based classifiers performed similarly (AUC, 0.75 vs 0.76 in our study). As the authors acknowledged, however, both of their models were highly complex: each included numerous time-varying variables, such as vitals and laboratory measurements, and implementation would almost certainly require sophisticated embedding within the electronic health record (EHR) operating environment [17]. Because our models included fewer and simpler variables, we expect that they would be more feasible to implement, including by ASPs at nonacademic medical centers. Nevertheless, they would still require sophisticated and automated implementation (the machine learning model) or somewhat lengthy manual calculations (the full multivariable logistic regression model).
To circumvent these limitations, we also built “workflow simplified” prediction models that included only 2, easily ascertainable variables: antimicrobial class, plus either provider-entered clinical indication (model A) or whether the patient received an ID consult (model B). When applied to held-out data, each model achieved sensitivities >80% while decreasing reviews by roughly one-third. Of all the models we developed, these may offer ASPs an optimal balance between predictive accuracy and practicality. To illustrate their real-world potential, provider-entered clinical indications appear alongside antimicrobial orders in our ASP database. It would be relatively straightforward and rapid for an ASP team member to manually calculate an order’s model A–predicted intervention probability using these 2 variables. Alternatively, automating model A within this database would only require minimal, relatively rudimentary programming (and could even be achieved using Excel formulas). We spend approximately 4 hours daily/19.5–20 hours weekly reviewing orders, which is similar to the ASPs nationally [18]. Therefore, an automated implementation of model A would save our ASP roughly 6.6 and 28 person-hours weekly and monthly, respectively (ie, a one-third reduction in reviews and, thus, review time). Time saved can be reallocated to other important ASP interventions and policies, such as education and quality improvement projects that offer sustainable impact and engage front-line providers [19]. Time-saving strategies became especially important during the pandemic, when shifting of clinical duties to coronavirus disease 2019 (COVID-19) therapeutics left ASPs deprioritizing stewardship activities [20]. Prediction models can keep ASP priorities durable despite new demands.
Alternative or adjunctive strategies for optimizing ASP workflows are EHR-automated scoring systems, which assign points to antimicrobial orders (ie, higher scores receive higher priority) and generate ASP alerts [21]. These decision support modules have been successfully implemented in other centers [22]. However, to perform well, they generally require substantial, front-end customization and build-times of 1–2 years, which is not practical for many institutions [22]; otherwise, assigned points can be somewhat arbitrary and not necessarily relevant to specific hospitals. Moreover, as “rule-based” systems, these scoring modules require ASP/programmer teams to specify review targets a priori (eg, patients with methicillin-susceptible S aureus on vancomycin). In contrast, our data-driven approach can uncover previously unknown patterns and intervention hotspots—for example, surgical prophylaxis and genitourinary indications in our institution. Therefore, even for ASPs that already use EHR scoring systems, incorporating data-driven approaches may further improve workflows.
This study is subject to several limitations. First, this was a single-center study at an academic medical center. To increase generalizability, we evaluated models on held-out, unseen data, and we performed multiple sensitivity analyses to ensure relevance to smaller and nonacademic medical centers, for example where ID consults may be less common. Nevertheless, these hospitals may differ in other, unmeasured ways that could affect model generalizability. For this reason, we encourage external validation of our models. To achieve maximal site-specific performance, we also encourage ASPs to develop models using their own data; to this end, we have provided programming code in the Supplementary Materials, along with implementation pointers and practical suggestions for replicating our approach. Second, we found that holding other factors constant, antimicrobial orders for patients who had received ID consults were significantly less likely to trigger intervention. However, we recognize that an ASP may be less likely to recommend second ID consults in patients who have already received initial ID consults; because ID consult recommendations were an intervention type, albeit a rare one (<6%), in our cohort, this could bias the ID consult effect estimate below the null. To assess this possibility, we refit the model on a subcohort that excluded recommendations for ID consults (results unshown), and receipt of an ID consult remained strongly protective against intervention (aOR, 0.58 [95% CI, .53–.64]). Third, our cohort preceded the COVID-19 pandemic, which precipitated important changes in inpatient antimicrobial prescribing [23–26]. It is unclear how our models would generalize to periods of high COVID-19 inpatient volumes, and whether optimal models would require COVID-19-specific predictors. With the approaches we have outlined and the programming tools that we provide, however, other ASPs can tailor models to COVID-19 populations and time periods as needed.
Overall, in this study of >17 000 antimicrobial orders, prediction models substantially reduced PAF caseloads while maintaining high sensitivities. By identifying orders that can be safely bypassed from ASP review, these models may help ensure that PAF programs are not curtailed or abandoned due to resource constraints. This workflow optimization can also enable ASPs to focus on other high-impact, but time-consuming, stewardship activities [19]. Our models should generalize well to other institutions with similar characteristics, but our approach and programming code also provide other ASPs a blueprint to undertake similar prediction model–building exercises on their own data.
Supplementary Material
Contributor Information
Katherine E Goodman, Department of Epidemiology and Public Health, University of Maryland School of Medicine, Baltimore, Maryland, USA.
Emily L Heil, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
Kimberly C Claeys, Department of Pharmacy Practice and Science, University of Maryland School of Pharmacy, Baltimore, Maryland, USA.
Mary Banoub, Department of Pharmacy, University of Maryland Medical Center, Baltimore, Maryland, USA.
Jacqueline T Bork, Department of Medicine, University of Maryland School of Medicine, Baltimore, Maryland, USA.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank and acknowledge Dr Michael Kleinberg, the University of Maryland Medical Center antimicrobial stewardship medical director from 2013 to 2021, and his dedicated work in maintaining the antimicrobial stewardship databases throughout his tenure.
Financial support. This work was supported by funding from the Agency for Healthcare Research and Quality (grant number K01-HS028363-01A1 to K. E. G.).
Potential conflicts of interest. E. L. H. reports consulting for Wolters-Kluwer (Lexi-Comp), outside the submitted work. K. C. C. is on the speaker’s bureau for BioFire Diagnostics and has investigator-initiated funding from Merck & Co, outside the submitted work. All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Tamma PD, Avdic E, Keenan JF, et al. What is the more effective antibiotic stewardship intervention: preprescription authorization or postprescription review with feedback? Clin Infect Dis 2017; 64:537–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barlam TF, Cosgrove SE, Abbo LM, et al. Implementing an antibiotic stewardship program: guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis 2016; 62:1197–202. [DOI] [PubMed] [Google Scholar]
- 3. Dorobisz MJ, Parente DM. Antimicrobial stewardship metrics: prospective audit with intervention and feedback. R I Med J (2013) 2018; 101:28. [PubMed] [Google Scholar]
- 4. Bork JT, Claeys KC, Heil EL, et al. A propensity score matched study of the positive impact of infectious diseases consultation on antimicrobial appropriateness in hospitalized patients with antimicrobial stewardship oversight. Antimicrob Agents Chemother 2020; 64:e00307–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Centers for Disease Control and Prevention . Core elements of hospital antibiotic stewardship programs. 2019. https://www.cdc.gov/antibiotic-use/core-elements/hospital.html. Accessed 17 June 2022.
- 6. Moehring RW, Anderson DJ, Cochran RL, et al. Expert consensus on metrics to assess the impact of patient-level antimicrobial stewardship interventions in acute-care settings. Clin Infect Dis 2017; 64:377–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Toth R, Schiffmann H, Hube-Magg C, et al. Random forest-based modelling to detect biomarkers for prostate cancer progression. Clin Epigenetics 2019; 11:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bi Q, Goodman KE, Kaminsky J, Lessler J. What is machine learning: a primer for the epidemiologist. Am J Epidemiol 2019; 188:2222–39. [DOI] [PubMed] [Google Scholar]
- 9. Strobl C, Malley J, Tutz G. An introduction to recursive partitioning: rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychol Methods 2009; 14:323–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and Regression Trees. 1st ed. New York: Routledge; 1984. [Google Scholar]
- 11. Roth JA, Battegay M, Juchler F, Vogt JE, Widmer AF. Introduction to machine learning in digital healthcare epidemiology. Infect Control Hosp Epidemiol 2018; 39:1457–62. [DOI] [PubMed] [Google Scholar]
- 12. Moehring RW, Ashley ESD, Davis AE, et al. Development of an electronic definition for de-escalation of antibiotics in hospitalized patients. Clin Infect Dis 2021; 73:e4507–14. [DOI] [PubMed] [Google Scholar]
- 13. Osowicki J, Gwee A, Noronha J, et al. The impact of an infectious diseases consultation on antimicrobial prescribing. Pediatr Infect Dis J 2014; 33:669–71. [DOI] [PubMed] [Google Scholar]
- 14. Buehrle K, Pisano J, Han Z, Pettit NN. Guideline compliance and clinical outcomes among patients with Staphylococcus aureus bacteremia with infectious diseases consultation in addition to antimicrobial stewardship-directed review. Am J Infect Control 2017; 45:713–6. [DOI] [PubMed] [Google Scholar]
- 15. Wang H, Wang H, Yu X, et al. Impact of antimicrobial stewardship managed by clinical pharmacists on antibiotic use and drug resistance in a Chinese hospital, 2010–2016: a retrospective observational study. BMJ Open 2019; 9:e026072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mas-Morey P, Valle M. A systematic review of inpatient antimicrobial stewardship programmes involving clinical pharmacists in small-to-medium-sized hospitals. Eur J Hosp Pharm 2018; 25:e69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bystritsky RJ, Beltran A, Young AT, Wong A, Hu X, Doernberg SB. Machine learning for the prediction of antimicrobial stewardship intervention in hospitalized patients receiving broad-spectrum agents. Infect Control Hosp Epidemiol 2020; 41:1022–7. [DOI] [PubMed] [Google Scholar]
- 18. Doernberg SB, Abbo LM, Burdette SD, et al. Essential resources and strategies for antibiotic stewardship programs in the acute care setting. Clin Infect Dis 2018; 67:1168–74. [DOI] [PubMed] [Google Scholar]
- 19. Jenkins TC, Tamma PD. Thinking beyond the “core” antibiotic stewardship interventions: shifting the onus for appropriate antibiotic use from stewardship teams to prescribing clinicians. Clin Infect Dis 2021; 72:1457–62. [DOI] [PubMed] [Google Scholar]
- 20. Ashiru-Oredope D, Kerr F, Hughes S, et al. Assessing the impact of COVID-19 on antimicrobial stewardship activities/programs in the United Kingdom. Antibiotics (Basel) 2021; 10:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kullar R, Goff DA, Schulz LT, Fox BC, Rose WE. The “epic” challenge of optimizing antimicrobial stewardship: the role of electronic medical records and technology. Clin Infect Dis 2013; 57:1005–13. [DOI] [PubMed] [Google Scholar]
- 22. Dzintars K, Fabre VM, Avdic E, et al. Development of an antimicrobial stewardship module in an electronic health record: options to enhance daily antimicrobial stewardship activities. Am J Health Syst Pharm 2021; 78:1968–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Evans TJ, Davidson HC, Low JM, Basarab M, Arnold A. Antibiotic usage and stewardship in patients with COVID-19: too much antibiotic in uncharted waters? J Infect Prev 2021; 22:119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pettit NN, Nguyen CT, Lew AK, et al. Reducing the use of empiric antibiotic therapy in COVID-19 on hospital admission. BMC Infect Dis 2021; 21:516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baghdadi JD, Coffey KC, Adediran T, et al. Antibiotic use and bacterial infection among inpatients in the first wave of COVID-19: a retrospective cohort study of 64,691 patients. Antimicrob Agents Chemother 2021; 65:e0134121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dieringer TD, Furukawa D, Graber CJ, et al. Inpatient antibiotic utilization in the Veterans’ Health Administration during the coronavirus disease 2019 (COVID-19) pandemic. Infect Control Hosp Epidemiol 2021; 42:751–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.