Abstract
Background
Butyrate is a major subgingival microbial metabolite that is closely related to periodontal disease. It affects the proliferation and differentiation of mesenchymal stem cells. However, the mechanisms by which butyrate affects the osteogenic differentiation of periodontal ligament stem cells (PDLSCs) remain unclear. Here, we investigated the effect of sodium butyrate (NaB) on the osteogenic differentiation of human PDLSCs.
Methods
PDLSCs were isolated from human periodontal ligaments and treated with various concentrations of NaB in vitro. The cell counting kit-8 assay and flow cytometric analysis were used to assess cell viability. The osteogenic differentiation capabilities of PDLSCs were evaluated using the alkaline phosphatase activity assay, alizarin red staining, RT-PCR, western blotting and in vivo transplantation.
Results
NaB decreased PDLSC proliferation and induced apoptosis in a dose- and time-depend manner. Additionally, 1 mM NaB reduced alkaline phosphatase activity, mineralization ability, and the expression of osteogenic differentiation-related genes and proteins. Treatment with a free fatty acids receptor 2 (FFAR2) antagonist and agonist indicated that NaB inhibited the osteogenic differentiation capacity of PDLSCs by affecting the expression of Smad1.
Conclusion
Our findings suggest that NaB inhibits the osteogenic differentiation of PDLSCs by activating FFAR2 and decreasing the expression of Smad1.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12903-022-02255-6.
Keywords: Sodium butyrate, Periodontal ligament stem cells, Proliferation, Osteogenic differentiation, Smad1, Free fatty acids receptor 2
Background
Periodontitis is a microbially associated chronic inflammatory disease that is characterized by loss of periodontal tissue integrity [1]. In the Global Burden of Disease 2010 study, severe periodontitis was reported as the sixth most prevalent health condition worldwide [2]. During the inflammation period of periodontitis, bacterial cytotoxins and metabolites can promote the host defence response, which increases the secretion of protein-rich gingival crevicular fluid and facilitates the growth of pathogenic periodontal bacteria [3, 4].
Short chain fatty acids (SCFAs), including butyrate, are the major metabolic by-products produced by subgingival pathogenic bacteria, such as Porphyromonas gingivalis and Fusobacterium nucleatum [4–6]. Higher concentration of butyrate can be detected in the gingival crevicular fluid of patients with periodontitis [7, 8]. In the oral cavity, butyrate decreases cytokine-induced intercellular adhesion molecule-1 expression in epithelial cells, and destroys gingival epithelial cell homeostasis by reducing the intercellular junctions [9, 10]. In addition, butyrate causes gingival epithelial cell autophagy via the AMP-activated protein kinase signalling pathway and results in subsequent cell death [11]. Butyrate exposure promotes gingival fibroblast apoptosis via extrinsic and intrinsic pathways and increases pro-inflammatory cytokine production [12].
Mesenchymal stem cells (MSCs) have self-renewal and multi-lineage differentiation abilities, and play major roles in periodontal tissue regeneration [13]. Recent studies have shown that 0.5 μM sodium butyrate (NaB) promotes the osteogenic and dentinogenic differentiation of mouse dental pulp stem cells by inhibiting histone deacetylation [14]. However, 0.5 mM NaB decreases mineralised nodule formation and the expression of runt-related transcription factor 2 (Runx2), osteocalcin (OCN), and type I collagen (Col I) in mouse bone marrow MSCs [15]. Periodontal ligament stem cells (PDLSCs) possess multiple differentiation capabilities, including differentiation into osteogenic and adipogenic cells [16]. Human PDLSCs have potential for use in cell-based treatments for mediating periodontal regeneration [17]. However, the effect of butyrate on PDLSCs remains unclear.
Butyrate can act as a signalling molecule that binds to free fatty acid receptors (FFAR), which are expressed in various tissues [18–20]. FFAR2 plays a major role in orthodontic tooth movement in C57BL/6 mice, and adipogenic differentiation of human adipose-derived MSCs [21, 22]. Smad proteins function as major signalling mediators during the osteogenic differentiation of MSCs. When receptor-associated Smad1/5/8 is phosphorylated by the activin receptor-like kinase 1, it forms a complex with Smad4 and translocates into the nucleus, where it regulates the expression of osteogenic genes, such as Runx2 and osterix (OSX) [23–25]. Here, we investigate the effects of NaB on human PDLSC osteogenesis and the signalling pathways that influence the biological behaviour of PDLSCs.
Methods
Cell culture of human PDLSCs
All experiments were performed according to the ISSCR Guidelines for Stem Cell Research and Clinical Translation, and approved by the Ethics Committee of Capital Medical University School of Stomatology (Beijing, China) (CMUSH-IRB-KJ-PJ-2020-11). Human premolars and impacted third molars were collected from healthy patients at the Department of Oral and Maxillofacial Surgery in Affiliated Stomatological Hospital of Capital Medical University. Informed consents were obtained from all donors. PDLSCs were isolated and cultured according to previously reported protocols [16]. The periodontal ligament tissues were gently separated from the middle third of the tooth roots and digested with 3 mg/ml collagenase type I (Sigma-Aldrich, USA) and 4 mg/ml dispase (Sigma-Aldrich, USA) for 1 h at 37 °C. After digestion, the tissues were cultured in α-modified Eagle's medium (Gibco, USA) containing 20% fetal bovine serum (Gibco, USA), 1% penicillin/streptomycin (Gibco, USA) and 1% glutamine (Gibco, USA) in 5% CO2 at 37 °C. PDLSCs from passages 3–5 were used in the following experiments.
Immunofluorescence staining
PDLSCs were cultured on 24-well plates with glass coverslips at a density of 105 cells/well overnight. Then, PDLSCs were fixed with 4% paraformaldehyde and incubated with anti-CD146 (1:100; Abcam, USA), anti-CD105 (1:100; Abcam, USA), and anti-CD45 (1:200; Abcam, USA) primary antibodies. The samples were then treated with rhodamine/FITC-conjugated secondary antibodies (1:1000; Sigma-Aldrich, USA) and stained with 4,6-diamidino-2-phenylindole (Sigma-Aldrich, USA). Jurkat cells were stained with anti-CD45 antibody as a positive control. The images were captured with a fluorescence microscopy (OLYMPUS, Japan).
Flow cytometric analysis
For stem cell identification, 106 PDLSCs were collected and fixed with 70% ethanol. Antibodies, including PE-labeled anti-CD146 (Biolegend, USA), FITC-labeled anti-CD105 (Biolegend, USA) and APC-labeled anti-CD45 (Biolegend, USA), were used to incubate the cells for 30 min in darkness. Jurkat cells were stained with anti-CD45 antibody as a positive control. Flow cytometry (FACS Calibur, BD Bioscience, USA) was used to test the samples.
PDLSCs were cultured on 6-well plates at a density of 106 cells/well overnight and treated with 0, 1 and 5 mM NaB for 24 and 72 h. The cells were stained according to the manufacturer’s protocol (Annexin V-FITC Apoptosis Detection Kit, BD Bioscience, USA). The percentage of apoptosis rate was then detected by flow cytometry.
Cell counting kit-8 (CCK-8) assay
Cell proliferation was measured using the Cell Counting Kit-8 (Dojindo, Japan) according to the manufacturer’s instruction. PDLSCs were cultivated on 96-well plates at a density of 5 × 103 cells/well and treated with 0, 0.625, 1.25, 2.5, 5 mM NaB for 24, 48 and 72 h. The absorbance (OD) value was read at a wavelength of 450 nm.
Alkaline phosphatase (ALP) activity assay and alizarin red staining
PDLSCs were cultivated on 6-well plates at a density of 2 × 105 cells/well. When reaching 80–90% confluence, the culture medium was replaced by osteogenesis-inducing media containing 100 mM ascorbic acid, 2 mM β-glycerophosphate, and 10 nM dexamethasone. The cells were treated with 0, 0.1 and 1 mM NaB at the same time. Based on the preliminary data, we used 10 μM FFAR2-selective antagonist (GLPG0974, Sigma-Aldrich, USA) and 10 μM agonist (4-CMTB, MedChemExpresss, USA) to repress and activate FFAR2, respectively during PDLSC osteogenesis (Additional files 1, 2: Figs. S1, S2).
After 5 days of osteogenic induction, PDLSCs were stained with ALP capacity kit (Sigma-Aldrich, USA). The OD value was read at a wavelength of 495 nm. The results were standardized on the basis of protein concentration. After 2 weeks of induction, PDLSCs were fixed with 70% ethanol for 1 h, and 1% alizarin red solution was utilized to stain the mineralized nodules (Solarbio, China). The relative concentration of calcium was measured after solubilizing in 10% cetylpyridinium chloride (Sigma-Aldrich, USA) for 30 min. The OD value was read at a wavelength of 562 nm.
Adipogenic differentiation assay
PDLSCs were cultivated on 6-well plates at a density of 2 × 105 cells/well. When reaching 80–90% confluence, the culture medium was replaced by adipogenesis-inducing medium (Cyagen Biosciences Inc, China). After 21 days of adipogenic induction, PDLSCs were fixed with 4% paraformaldehyde for 30 min. Cells were stained with Oil red O solution at room temperature for 15 min. The images were captured with an inverted microscopy (OLYMPUS, Japan).
Real-time reverse transcriptase-polymerase chain reaction (real-time RT-PCR)
Following 7 and 14 days of induction, total RNA was isolated from PDLSCs by using Trizol reagents (Cwbio, China). Reverse transcription was completed according to the manufacturer’s protocol (Takara, China). Then, RT-PCR reactions were performed using the SYBR Premix Ex Taq™ (Takara, China) and an icycler iQ Multi-color Real-time PCR Detection System. The primers for specific genes are listed in Table 1.
Table 1.
Primers sequences used in the real-time RT-PCR
| Gene symbol | Primer sequences (5′-3′) |
|---|---|
| GAPDH-F | CGGACCAATACGACCAAATCCG |
| GAPDH-R | AGCCACATCGCTCAGACACC |
| RunX2-F | GACTGTGGTTACCGTCATGGC |
| RunX2-R | ACTTGGTTTTTCATAACAGCGGA |
| OSX-F | CCTCCTCAGCTCACCTTCTC |
| OSX-R | GTTGGGAGCCCAAATAGAAA |
| OCN-F | CAGACAAGTCCCACACAGCA |
| OCN-R | CTTGGCATCTGTGAGGTCAG |
| OPN-F | AGCCACATCGCTCAGACACC |
| OPN-R | TGAAATTCATGGCTGTGGAA |
Western blotting analysis
Following 7 and 14 days of osteogenic induction, total protein was obtained from PDLSCs. The protocol has been described previously.14 The following primary antibodies were used: anti-Runx2 (1:1000; Cell Signaling Technology, USA), anti-Smad1 (1:1000; Cell Signaling Technology, USA), anti-p-Smad1/5/8 (1:1000; Cell Signaling Technology, USA), anti-OCN (1:1000; Abcam, USA), anti-osteopontin (anti-OPN) (1:1000; Abcam, USA), and anti-OSX (1:1000; Abcam, USA). GAPDH (1:2000; Abclonal, China) was used as a control.
Transplantation in nude mice
Six 10-week-old immunocompromised beige mice (nu/nu nude mice) were purchased from the Institute of Animal Science of the Vital River (Beijing, China). The experiments were performed according to ARRIVE guidelines, and institutionally set guidelines for animal research approved by the Animal Care and Use Committee of the Beijing Stomatological Hospital, Capital Medical University, Beijing, China (KQYY-201907–003). Approximately 2 × 106 human PDLSCs treated with or without 1 mM NaB were mixed with 20 mg hydroxyapatite/tricalcium phosphate (HA/TCP) ceramic particles (Engineering Research Center for Biomaterials, Sichuan University, China). Then, the mixtures were transplanted separately into the dorsal surface of the mice. After 3 months, the implants were harvested, fixed with 4% paraformaldehyde, and decalcified with 10% EDTA in PBS (pH 7.4) at 4 °C. The decalcified samples were then dehydrated and embedded in paraffin. The samples were sectioned at 5 μm and stained with hematoxylin and eosin (H&E). Six sections were evaluated per sample, and the blind evaluation was performed. The bone-like areas were measured using the Image-Pro Plus 6.0 program.
Statistical analysis
Statistical analysis was performed using SPSS 26.0 software. The Student’s t test or one-way ANOVA was used to assess statistical significance, with a p ≤ 0.05 was regarded as significant.
Results
Human PDLSCs culture and identification
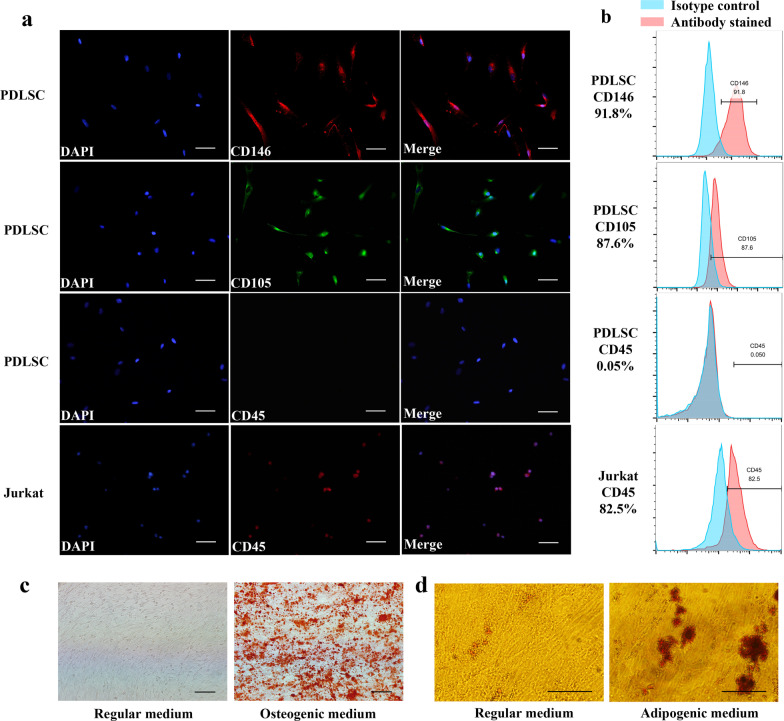
Human PDLSCs displayed a long spindle shape after passage. Flow cytometry and immunofluorescence revealed that human PDLSCs expressed MSC markers, including CD146 and CD105. Different from Jurkat cells, PDLSCs were negative for CD45 (Fig. 1a, b). The formation of calcium deposits and Oil red O-positive lipid clusters proved that PDLSCs had osteogenic and adipogenic differentiation abilities (Fig. 1c, d). These results demonstrated that human PDLSCs had characteristics similar to those of MSCs.
Fig. 1.
Identification of human periodontal ligament stem cells (PDLSCs). a Immunofluorescence showed that PDLSCs used in this study express CD146 and CD105, but negative for CD45. Jurkat cells were positive for CD45. Scale bars, 100 μM. b Flow cytometry showed that human PDLSCs exhibited the expression of cell surface markers, including CD146 (91.8%), CD105 (87.6%) and CD45 (0.05%). Isotype control was used in this experiment. Jurkat cells exhibited the expression of CD45 (82.5%). c Alizarin red staining assay was performed to prove the mineralized nodules formation after 14-days osteogenic induction. Scale bars, 100 μm. d The cellular Oil red O-positive lipid clusters were stained after 21-days adipogenic induction. Scale bars, 50 μm
Effects of NaB on the proliferation and apoptosis of PDLSCs
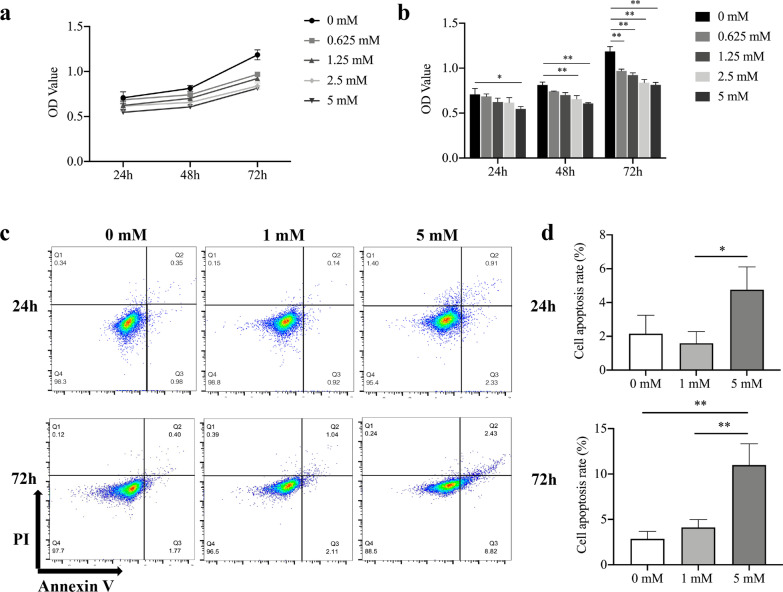
To explore the effects of NaB on cell proliferation, PDLSCs were treated with different concentrations of NaB. The results indicated that the cell proliferation decreased in a dose- and time-dependent manner (Fig. 2a, b). The cell proliferation was significantly decreased by 5 mM NaB after 24 h of treatment. After 48 h of treatment, the cell proliferation was markedly inhibited by 2.5 mM NaB and 5 mM NaB. A significant decrease was found in each concentration of NaB after 72 h of treatment. Additionally, flow cytometry demonstrated that 5 mM NaB significantly induced higher apoptosis rates at 24 and 72 h, while 1 mM NaB did not have a significant effect on PDLSC apoptosis (Fig. 2c, d).
Fig. 2.
Sodium butyrate (NaB) reduced the viability of periodontal ligament stem cells (PDLSCs). a, The cell proliferation curve of PDLSCs was depicted after the treatment of NaB for 24, 48, and 72 h. b, The absorbance values showed that NaB inhibited PDLSC proliferation in a dose- and time-dependent manner. c, d Flow cytometry showed that 5 mM NaB induced human PDLSC apoptosis in 24 and 72 h. Error bars represent the standard deviation (n = 3). *p < 0.05, **p < 0.01, using Student’s t test and one-way ANOVA
NaB inhibited the osteogenic differentiation of PDLSCs
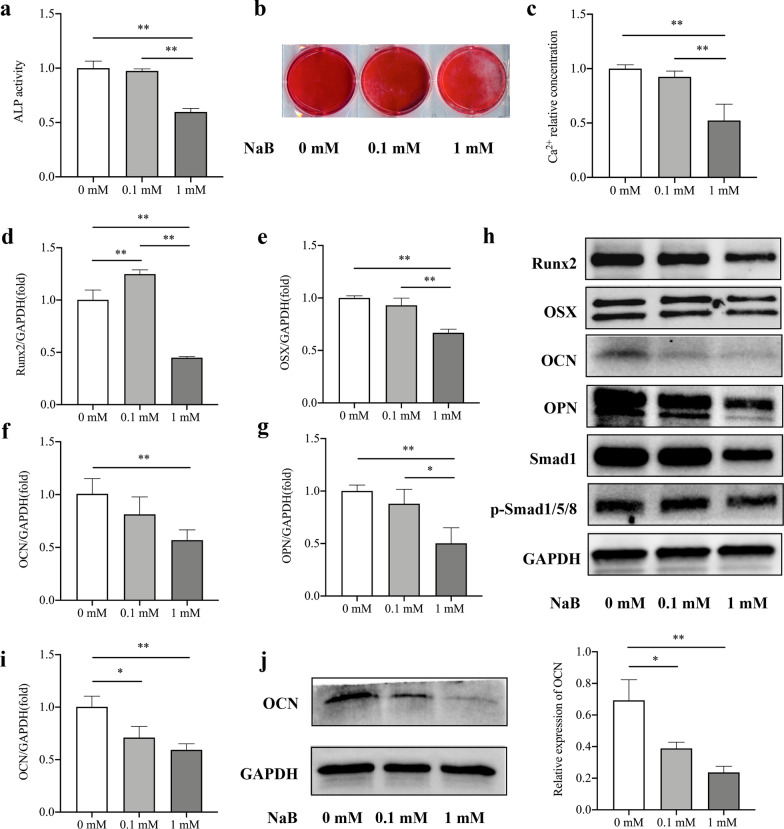
PDLSCs were cultured in osteogenesis-inducing media. After 5 days of osteogenesis induction, 1 mM NaB decreased ALP activity by 50–60% (compared to 0 mM and 0.1 mM NaB) (Fig. 3a). After 2 weeks of induction, the formation of calcium deposits decreased in the 1 mM NaB group (compared to the 0 mM and 0.1 mM groups) (Fig. 3b, c). The effects of NaB on the osteogenic differentiation of PDLSCs were also evaluated by RT-PCR and western blotting. The results showed that after 7 days of induction, the expression of Runx2, OSX, OCN, OPN, Smad1 and p-Smad1/5/8 in the 1 mM NaB group was markedly lower than that in the 0 mM and 0.1 mM groups (Fig. 3d–h). After 14 days of induction, the expression of OCN in the 0 mM and 0.1 mM groups was significantly decreased compared to the 0 mM group (Fig. 3i, j).
Fig. 3.
Sodium butyrate (NaB) decreased the osteogenic capacity of periodontal ligament stem cells (PDLSCs). a Evaluation of alkaline phosphatase (ALP) activity 5 days after osteogenic induction indicated that 1 mM NaB decreased ALP activity in PDLSCs. b, c Alizarin red staining assay and cetylpyridinium chloride assay performed 14 days after induction showed that 1 mM NaB decreased the formation of mineralized nodules in PDLSCs. d–g Following 7 days of osteogenic induction, RT-PCR showed that 1 mM NaB downregulated Runx2, OSX, OCN, and OPN. h Western blotting showed that 1 mM NaB reduced the protein expression of Runx2, OSX, OCN, OPN, Smad1 and p-Smad1/5/8. i Following 14 days of osteogenic induction, RT-PCR showed that 0.1 mM and 1 mM NaB downregulated OCN. j Western blotting and densitometric analysis showed that 0.1 mM and 1 mM NaB reduced the protein expression of OCN. The length of the blots was appropriate in the manuscript. Error bars represent the standard deviation (n = 3). *p < 0.05, **p < 0.01, using one-way ANOVA
NaB downregulated Smad1 by activating FFAR2 in PDLSCs
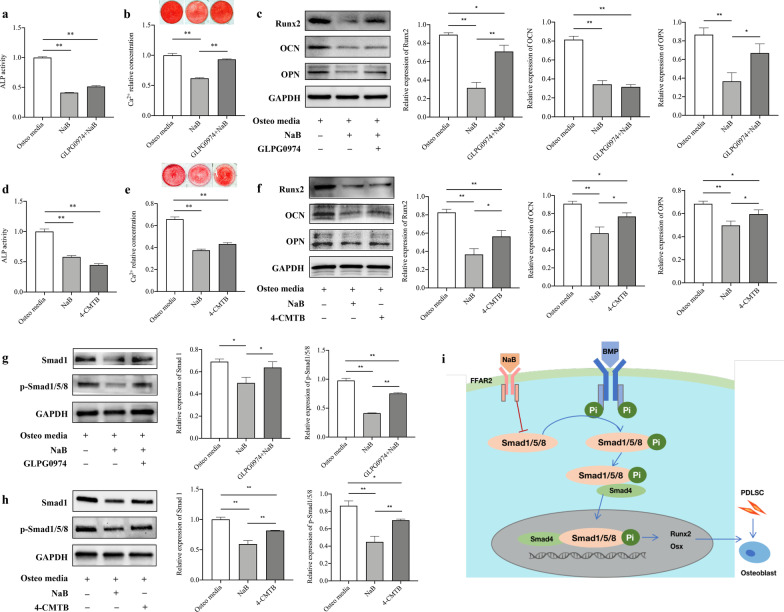
GLPG0974 treatment in the background of NaB stimulation slightly increased ALP activity (compared to that in the NaB group), but this was still lower than that detected in the control group (Fig. 4a). GLPG0974 significantly reversed the effect of NaB with respect to mineralised nodule formation, and expression of Runx2 and OPN (but not OCN) (Fig. 4b, c). Similar to that in the NaB group, 4-CMTB decreased ALP activity, mineralised nodule formation, and the expression of Runx2, OCN, and OPN (Fig. 4d–f). Western blotting showed that GLPG0974 treatment in the background of NaB stimulation reversed the reduction in Smad1 and p-Smad1/5/8 expression (compared to the NaB group), and the expression of Smad1 and p-Smad1/5/8 was reduced in response to 4-CMTB treatment (Fig. 4g, h). Figure 4i shows the mechanism proposed to explain these results, whereby NaB decreases the expression of Smad1 by activating FFAR2, thereby reducing p-Smad1/5/8-Smad4 translocation into the nucleus and downregulating Runx2 and OSX. This ultimately results in the inhibition of PDLSC osteogenic differentiation.
Fig. 4.
Sodium butyrate (NaB) inhibited periodontal ligament stem cell (PDLSC) osteogenesis by downregulating Smad1. a Free fatty acid receptor 2 (FFAR2) antagonist (GLPG0974) slightly increased alkaline phosphatase (ALP) activity in NaB-treated cells (compared to the NaB group). b, c GLPG0974 significantly reversed the NaB-mediated inhibition of mineralized nodule formation and the expression of Runx2 and OPN. d–f FFAR2 agonist (4-CMTB) decreased ALP activity, mineralized nodule formation and the expression of Runx2, OCN and OPN in NaB-treated cells. g, h Western blotting and densitometric analysis showed that GLPG0974 reversed the NaB-mediated reduction of Smad1 and p-Smad1/5/8 (compared to the NaB group), while 4-CMTB reduced the the expression of Smad1 and p-Smad1/5/8. The length of the blots was appropriate in the manuscript. i NaB decreased the expression of Smad1 by activating FFAR2, thus reducing the translocation of the p-Smad1/5/8-Smad4 complex into the nucleus, downregulating the expression of Runx2 and OSX, and inhibiting the osteogenic differentiation of PDLSCs. Error bars represent the standard deviation (n = 3). *p < 0.05, **p < 0.01, using one-way ANOVA
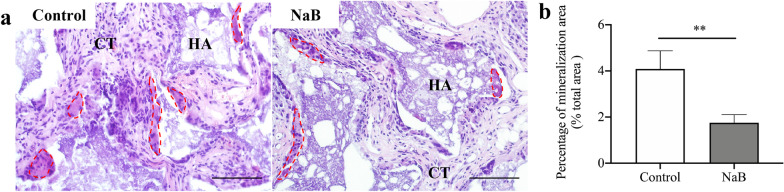
NaB decreased human PDLSC-mediated bone formation in nude mice
To explore the effect of NaB on the osteogenic differentiation of PDLSCs in vivo, human PDLSCs were treated with or without NaB and transplanted into the dorsal surfaces of nude mice. NaB-treated PDLSCs generated less bone-like mineralised tissue than untreated PDLSCs (Fig. 5a). Quantitative measurement of the mineralisation area showed less bone-like tissue formation in the NaB-treated group than in the control group (Fig. 5b).
Fig. 5.
Sodium butyrate (NaB) decreased human periodontal ligament stem cell-mediated bone formation in nude mice. a H&E staining revealed less bone-like tissue formation around HA/TCP in the NaB-treated group compared to the control. b Quantitative measurement of the mineralization area revealed less bone-like tissue formation in the NaB-treated group than in the control. Red lines represent bone-like tissue; HA = HA/TCP; CT = connective tissue. Scale bars, 100 μM. Error bars represent the standard deviation (n = 3). *p < 0.05, **p < 0.01, using Student’s t test
Discussion
In this study, we demonstrated that NaB decreased PDLSC proliferation and induced apoptosis in a dose- and time-depend manner. Additionally, 1 mM NaB reduced the osteogenesis ability in PDLSC. Treatment with a FFAR2 antagonist and agonist indicated that NaB inhibited the osteogenic differentiation capacity of PDLSCs by affecting the expression of Smad1.
Gut and oral bacteria are the major metabolic sources of SCFAs. SCFAs produced in the gut can be passively absorbed or actively transported by epithelial cells. Although butyrate is present at high concentrations in the colon, it does not inhibit the proliferation of normal epithelial cells because it is used as the main energy source for colonocytes. In contrast, because of the different metabolic patterns, butyrate accumulates three-fold in cancerous epithelial cells and affects cell growth as a histone deacetylase (HDAC) inhibitor [18]. A study proposed that the specific structure and metabolism of the crypt protected stem/progenitor cells that are located near the bottom of the crypt from butyrate-induced growth impairment [26]. In conclusion, the effect of butyrate is not only concentration dependent, but is also related to tissue structure, transportation, and metabolic patterns.
In the periodontal tissue, the gingival epithelium comprises squamous epithelial cells and has a thinner mucosal layer than the gut mucosa, which could allow butyrate to easily penetrate and remain within the gingiva [27]. A study has shown that butyrate downregulates intercellular junction‒related genes in human oral squamous cells [10]. It is speculated that unlike in the gut, lower concentrations of butyrate could penetrate the periodontal epithelial structure and impair the functions of human PDLSCs. Here, we verified that NaB inhibited human PDLSC proliferation and induced apoptosis in a dose- and time-dependent manner. The cell proliferation results indicated that 2.5 mM NaB reduced PDLSCs proliferation at 48 h and 72 h, and 5 mM NaB reduced cell proliferation at all time points. The cell apoptosis results demonstrated that 5 mM NaB induced PDLSCs apoptosis at 24 h and 72 h, while 1 mM NaB did not affect PDLSCs apoptosis. Thus, we used 1 mM NaB as the maximum concentration for cell osteogenic differentiation. We used 0.1 mM NaB to explore whether lower concentration of NaB inducing the osteogenic differentiation in PDLSCs.
To date, the effect of butyrate on osteogenesis remains unclear. Tomoko et al. demonstrated that NaB (< 1 mM) could induce mineralised nodule formation in human osteoblasts [28]. Butyrate-fed C57BL/6 J mice exhibited significantly higher tibial bone mass than the control group and a reduction in osteoclast number, while osteoblast number and OCN expression remained unchanged [29]. In contrast, Chang et al. found that NaB decreased Col I expression in MG-63 osteoblasts (between 43 and 77% at concentrations ranging between 2 and 16 mM, respectively) [30]. Here, 0.1 mM NaB only decreased the expression of OCN after 14 days osteogenic induction, while 1 mM NaB significantly decreased PDLSC osteogenesis both in vitro and in vivo.
In addition to being an HDAC inhibitor, butyrate can bind to G-protein receptors, including GPR41/FFAR3 and GPR43/FFAR2, which are expressed in many tissue types and play various roles in physiological functions. FFAR2−/− mice exhibited a significant increase in orthodontic tooth movement and a reduction in osteoblast counts compared to those in wild-type mice, while ALP was upregulated in the alveolar bone of FFAR2−/− mice [21]. In human adipose-derived mesenchymal stem cells, FFAR2 upregulation was observed under conditions of adipogenic differentiation, while FFAR3 expression was below the detection limit [22]. Therefore, butyrate may exhibit converse functions during the adipogenic and osteogenic differentiation of MSCs by acting as an FFAR agonist. Here, we used an FFAR2 antagonist and agonist to confirm that NaB inhibited the osteogenic differentiation of PDLSCs by downregulating Smad1 via FFAR2 and reducing ALP activity, mineralised nodule formation, and Runx2, OCN, and OPN expression. However, in the antagonist group, ALP activity and the expression of OCN significantly reduced (compared to the control group). Thus, NaB may affect PDLSC osteogenesis by activating FFAR3.
This study demonstrates the inhibitory effect of NaB on human PDLSC viability and osteogenic differentiation capability. By activating FFAR2, NaB inhibits the expression of Smad1, thereby downregulating osteogenesis-related genes and proteins. Our findings indicated that butyrate—a major subgingival bacterial metabolite—exhibited an important inhibitory effect on PDLSC osteogenic differentiation and periodontal tissue regeneration. It has been confirmed that many signalling pathways, such as the Smad, MAPK, and NF-κB pathways, are related to MSC osteogenic differentiation [31–33]. Here, we only explored the effect of NaB on the Smad pathway. Thus, further studies are required to explore the effects of NaB on signalling crosstalk.
At present, the prevalence of periodontal disease in adults is still at a high level. The improvement of periodontal disease treatment is the key factor to improve periodontal condition. The application of MSCs to the regeneration of periodontal tissue in patients with periodontal disease is the most advanced treatment method at present. This study found that NaB inhibits the viability and inhibits the osteogenic differentiation in PDLSCs. The results suggest that NaB may affect the therapeutic effect of MSCs. Therefore, exploring the pathogenic mechanism of butyrate and finding ways to block the production and combination of butyrate are urgent problems to be solved.
Conclusions
In summary, our results indicated that butyrate inhibits human PDLSC osteogenesis by activating FFAR2 to suppress Smad1 activation. Our findings contribute to the understanding of mechanisms underlying the following: (1) effects of butyrate on PDLSC functions and (2) effects of butyrate on periodontal tissue regeneration.
Supplementary Information
Additional file 1: Fig. S1. The dose optimization assay of free fatty acid receptor 2 (FFAR2) antagonist (GLPG0974). a The cell proliferation curve of PDLSCs was depicted after the treatment of GLPG0974 for 24, 48, and 72 h. b The absorbance values showed that 20 μM GLPG0974 inhibited PDLSC proliferation at 24 h and 72 h, and 40 μM GLPG0974 inhibited PDLSC proliferation at all time points. c Western blotting and densitometric analysis showed that 10 μM and 20 μM GLPG0974 reversed the NaB-mediated reduction of p-Smad1/5/8 (compared to the NaB group) at 24 h. The length of the blots was appropriate in the manuscript. Error bars represent the standard deviation (n = 3). *p < 0.05, **p < 0.01, using one-way ANOVA.
Additional file 2: Fig. S2. The dose optimization assay of free fatty acid receptor 2 (FFAR2) agonist (4-CMTB). a The cell proliferation curve of PDLSCs was depicted after the treatment of 4-CMTB for 24, 48, and 72 h. b The absorbance values showed that 20 μM 4-CMTB inhibited PDLSC proliferation at 72 h. c Western blotting and densitometric analysis showed that 5, 10, 20 and 40 μM 4-CMTB decreased the expression of p-Smad1/5/8 at 24 h (compared to the control group). The length of the blots was appropriate in the manuscript. Error bars represent the standard deviation (n = 3). *p < 0.05, **p < 0.01, using one-way ANOVA.
Acknowledgements
We would like to thank Editage (www.editage.cn) for English language editing.
Abbreviations
- SCFAs
Short chain fatty acids
- MSCs
Mesenchymal stem cells
- NaB
Sodium butyrate
- Runx2
Runt-related transcription factor 2
- OCN
Osteocalcin
- Col
Type I collagen
- OSX
Osterix
- PDLSCs
Periodontal ligament stem cells
- FFAR2
Free fatty acids receptor 2
- CCK-8
Cell counting kit-8
- OD
Absorbance
- ALP
Alkaline phosphatase
- RT-PCR
Reverse transcriptase-polymerase chain reaction
- OPN
Osteopontin
- HA/TCP
Hydroxyapatite/tricalcium phosphate
- H&E
Hematoxylin and eosin
- HDAC
Histone deacetylase
Author contributions
JH carried out the phenomenon research, experiments process and participated in drafting the manuscript. JX and HZ carried out the signaling pathway study. SW and YJ participated in collecting periodontal tissue. YL participated in the statistical analysis. LG and SL conceived of the study, and participated in its design. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Nature Science Foundation of China (81991504 and 81974149 to Y.L, 81600891 to L.G, 81570998 to S.L), the Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (ZYLX202121 to Y.L), the Beijing Baiqianwan Talents Project (2017A17 to Y.L), Beijing Municipal Administration of Hospitals’ Ascent Plan (DFL20181501 to Y.L), Beijing Municipal Science & Technology Commission (Z16100000516203 to L.G), Beijing Municipal Administration of Hospitals’ Youth Programme (QML20181501), Beijing Dongcheng Excellent Talent (to L.G). Innovation Foundation of Beijing Stomatological Hospital, Capital Medical University (21-09-18 to L.G).
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
The study was performed according to an informed protocol for handling human tissue approved by the Research Ethical Committee of Capital Medical University, Beijing, China (CMUSH-IRB-KJ-PJ-2020-11). All participants informed consent volunteered to participate in the study. The animal experiments were performed according to ARRIVE guidelines, and institutionally set guidelines approved by the Animal Care and Use Committee of the Beijing Stomatological Hospital, Capital Medical University, Beijing, China (KQYY-201907-003).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lijia Guo, Email: orthoest@163.com.
Song Li, Email: dentistli@263.net.
References
- 1.Hajishengallis G, Chavakis T. Local and systemic mechanisms linking periodontal disease and inflammatory comorbidities. Nat Rev Immunol. 2021;21:426–440. doi: 10.1038/s41577-020-00488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kassebaum NJ, Bernabé E, Dahiya M, et al. Global burden of severe periodontitis in 1990–2010: a systematic review and meta-regression. J Dent Res. 2014;93:1045–1053. doi: 10.1177/0022034514552491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16:745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi N. Oral microbiome metabolism: From“who are they?” to“What Are They Doing?”. J Dent Res. 2015;94:1628–1637. doi: 10.1177/0022034515606045. [DOI] [PubMed] [Google Scholar]
- 5.Abe K. Butyric acid induces apoptosis in both human monocytes and lymphocytes equivalently. J Oral Sci. 2012;54:7–14. doi: 10.2334/josnusd.54.7. [DOI] [PubMed] [Google Scholar]
- 6.Szafrański SP, Deng ZL, Tomasch J, et al. Functional biomarkers for chronic periodontitis and insights into the roles of Prevotella nigrescens and Fusobacterium nucleatum; a metatranscriptome analysis. NPJ Biofilms Microbiomes. 2015;1:15017. doi: 10.1038/npjbiofilms.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niederman R, Buyle-Bodin Y, Lu BY, Robinson P, Naleway C. Short-chain carboxylic acid concentration in human gingival crevicular fluid. J Dent Res. 1997;76:575–579. doi: 10.1177/00220345970760010801. [DOI] [PubMed] [Google Scholar]
- 8.Li QQ, Meng HX, Gao XJ. Longitudinal study of volatile fatty acids in the gingival crevicular fluid of patients with periodontitis before and after nonsurgical therapy. J Periodont Res. 2012;47:740–749. doi: 10.1111/j.1600-0765.2012.01489.x. [DOI] [PubMed] [Google Scholar]
- 9.Magrin GL, Di Summa F, Strauss FJ, et al. Butyrate decreases ICAM-1 expression in human oral squamous cell carcinoma cells. Int J Mol Sci. 2020;21:1679. doi: 10.3390/ijms21051679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Wang YX, Meng HX. Butyrate rather than LPS subverts gingival epithelial homeostasis by downregulation of intercellular junctions and triggering pyroptosis. J Clin Periodontol. 2019;46:894–907. doi: 10.1111/jcpe.13162. [DOI] [PubMed] [Google Scholar]
- 11.Evans M, Murofushi T, Tsuda H, et al. Combined effects of starvation and butyrate on autophagy-dependent gingival epithelial cell death. J Periodontal Res. 2017;52:522–531. doi: 10.1111/jre.12418. [DOI] [PubMed] [Google Scholar]
- 12.Shirasugi M, Nishioka K, Yamamoto T, Nakaya T, Kanamura N. Normal human gingival fibroblasts undergo cytostasis and apoptosis after long-term exposure to butyric acid. Biochem Biophys Res Commun. 2017;482:1122–1128. doi: 10.1016/j.bbrc.2016.11.168. [DOI] [PubMed] [Google Scholar]
- 13.Monsarrat P, Vergnes JN, Nabet C, et al. Concise review: mesenchymal stromal cells used for periodontal regeneration: a systematic review. Stem Cells Transl Med. 2014;3:768–774. doi: 10.5966/sctm.2013-0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren Y, Su SP, Liu XY, et al. Microbiota-derived short-chain fatty acids promote BMP signaling by inhibiting histone deacetylation and contribute to dentinogenic differentiation in murine incisor regeneration. Stem Cells Dev. 2020;29:1201–1214. doi: 10.1089/scd.2020.0057. [DOI] [PubMed] [Google Scholar]
- 15.Xie JY, Lou Q, Zeng YX, et al. Single-cell atlas reveals fatty acid metabolites regulate the functional heterogeneity of mesenchymal stem cells. Front Cell Dev Biol. 2021;9:653308. doi: 10.3389/fcell.2021.653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seo BM, Miura M, Gronthos S, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–155. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 17.Hu L, Liu Y, Wang SL. Stem cell-based tooth and periodontal regeneration. Oral Dis. 2017;24:696–705. doi: 10.1111/odi.12703. [DOI] [PubMed] [Google Scholar]
- 18.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 19.Kimura I, Ichimura A, Ohue-Kitano R, Igarashi M. Free fatty acid receptors in health and disease. Physiol Rev. 2020;100:171–210. doi: 10.1152/physrev.00041.2018. [DOI] [PubMed] [Google Scholar]
- 20.Van der Hee B, Wells JM. Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol. 2021;29:700–712. doi: 10.1016/j.tim.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Montalvany-Antonucci CC, Duffles LF, De Arruda JAA, et al. Short-chain fatty acids and FFAR2 as suppressors of bone resorption. Bone. 2019;125:112–121. doi: 10.1016/j.bone.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Iván J, Major E, Sipos A, et al. The short-chain fatty acid propionate inhibits adipogenic differentiation of human chorion-derived mesenchymal stem cells through the free fatty acid receptor 2. Stem Cells Dev. 2017;26:1724–1733. doi: 10.1089/scd.2017.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez-Puerto MC, Iyengar PV, de Vinuesa AG, et al. Bone morphogenetic protein receptor signal transduction in human diseases. J Pathol. 2018;247:9–20. doi: 10.1002/path.5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu MR, Chen GQ, Li P. TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4:16009. doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo K. Signaling cross talk between TGF-beta/Smad and other signaling pathways. Cold Spring Harb Perspect Biol. 2016;9:a022137. doi: 10.1101/cshperspect.a022137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiko GE, Ryu SH, Koues OI, et al. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;165:1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cueno ME, Ochiai K. Re-discovering periodontal butyric acid: New insights on an old Metabolite. Microb Pathog. 2016;94:48–53. doi: 10.1016/j.micpath.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Katono T, Kawato T, Tanabe N, et al. Sodium butyrate stimulates mineralized nodule formation and osteoprotegerin expression by human osteoblasts. Arch Oral Biol. 2008;53:903–909. doi: 10.1016/j.archoralbio.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Lucas S, Omata Y, Hofmann J, et al. Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat Commun. 2018;9:55. doi: 10.1038/s41467-017-02490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang MC, Tsai YL, Liou JW, et al. Effect of butyrate on collagen expression, cell viability, cell cycle progression and related proteins expression of MG-63 osteoblastic cells. PLoS ONE. 2016;11:e0165438. doi: 10.1371/journal.pone.0165438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li XY, Guo LJ, Liu Y, et al. MicroRNA-21 promotes osteogenesis of bone marrow mesenchymal stem cells via the Smad7-Smad1/5/8-Runx2 pathway. Biochem Biophys Res Commun. 2017;493:928–933. doi: 10.1016/j.bbrc.2017.09.119. [DOI] [PubMed] [Google Scholar]
- 32.Yang S, Guo LJ, Su YY, et al. Nitric oxide balances osteoblast and adipocyte lineage differentiation via the JNK/MAPK signaling pathway in periodontal ligament stem cells. Stem Cell Res Ther. 2018;9:118. doi: 10.1186/s13287-018-0869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li N, Li ZH, Wang YQ, et al. CTP-CM enhances osteogenic differentiation of hPDLSCs via NF-κB pathway. Oral Dis. 2021;27:577–588. doi: 10.1111/odi.13567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. The dose optimization assay of free fatty acid receptor 2 (FFAR2) antagonist (GLPG0974). a The cell proliferation curve of PDLSCs was depicted after the treatment of GLPG0974 for 24, 48, and 72 h. b The absorbance values showed that 20 μM GLPG0974 inhibited PDLSC proliferation at 24 h and 72 h, and 40 μM GLPG0974 inhibited PDLSC proliferation at all time points. c Western blotting and densitometric analysis showed that 10 μM and 20 μM GLPG0974 reversed the NaB-mediated reduction of p-Smad1/5/8 (compared to the NaB group) at 24 h. The length of the blots was appropriate in the manuscript. Error bars represent the standard deviation (n = 3). *p < 0.05, **p < 0.01, using one-way ANOVA.
Additional file 2: Fig. S2. The dose optimization assay of free fatty acid receptor 2 (FFAR2) agonist (4-CMTB). a The cell proliferation curve of PDLSCs was depicted after the treatment of 4-CMTB for 24, 48, and 72 h. b The absorbance values showed that 20 μM 4-CMTB inhibited PDLSC proliferation at 72 h. c Western blotting and densitometric analysis showed that 5, 10, 20 and 40 μM 4-CMTB decreased the expression of p-Smad1/5/8 at 24 h (compared to the control group). The length of the blots was appropriate in the manuscript. Error bars represent the standard deviation (n = 3). *p < 0.05, **p < 0.01, using one-way ANOVA.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.