ABSTRACT
Next-generation sequencing (NGS) workflows applied to bronchoalveolar lavage (BAL) fluid specimens could enhance the detection of respiratory pathogens, although optimal approaches are not defined. This study evaluated the performance of the Respiratory Pathogen ID/AMR (RPIP) kit (Illumina, Inc.) with automated Explify bioinformatic analysis (IDbyDNA, Inc.), a targeted NGS workflow enriching specific pathogen sequences and antimicrobial resistance (AMR) markers, and a complementary untargeted metagenomic workflow with in-house bioinformatic analysis. Compared to a composite clinical standard consisting of provider-ordered microbiology testing, chart review, and orthogonal testing, both workflows demonstrated similar performances. The overall agreement for the RPIP targeted workflow was 65.6% (95% confidence interval, 59.2 to 71.5%), with a positive percent agreement (PPA) of 45.9% (36.8 to 55.2%) and a negative percent agreement (NPA) of 85.7% (78.1 to 91.5%). The overall accuracy for the metagenomic workflow was 67.1% (60.9 to 72.9%), with a PPA of 56.6% (47.3 to 65.5%) and an NPA of 77.2% (68.9 to 84.1%). The approaches revealed pathogens undetected by provider-ordered testing (Ureaplasma parvum, Tropheryma whipplei, severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2], rhinovirus, and cytomegalovirus [CMV]), although not all pathogens detected by provider-ordered testing were identified by the NGS workflows. The RPIP targeted workflow required more time and reagents for library preparation but streamlined bioinformatic analysis, whereas the metagenomic assay was less demanding technically but required complex bioinformatic analysis. The results from both workflows were interpreted utilizing standardized criteria, which is necessary to avoid reporting nonpathogenic organisms. The RPIP targeted workflow identified AMR markers associated with phenotypic resistance in some bacteria but incorrectly identified blaOXA genes in Pseudomonas aeruginosa as being associated with carbapenem resistance. These workflows could serve as adjunctive testing with, but not as a replacement for, standard microbiology techniques.
KEYWORDS: diagnostics, lower respiratory tract infection, next-generation sequencing
INTRODUCTION
Microbiological methods for bronchoalveolar lavage (BAL) fluid specimens are comprehensive, spanning disciplines of bacteriology, mycobacteriology, virology, mycology, and parasitology (1). However, these methods require extensive resources and may not identify fastidious pathogens, rare and atypical pathogens, or pathogens that are no longer viable due to antimicrobial therapy. Assays utilizing next-generation sequencing (NGS) technology have the potential to improve diagnostic testing by broadening detection, shortening the time to detection for certain microorganisms, and allowing the detection of unsuspected or treated pathogens (2–4). The utilization of NGS assays could directly impact patient care outcomes, including appropriate antibiotic use and a shortened duration of mechanical ventilation (5, 6). NGS approaches hold the possibility of consolidating some or all diagnostic approaches for pathogen identification and characterization into a single assay (2).
NGS workflows utilize simultaneous and parallel techniques to amplify and sequence large amounts of genetic material, with potential pathogens being identified via result comparison to databases by bioinformatic analysis (7). Metagenomic NGS (mNGS) workflows sequence as much DNA and/or RNA as possible in a sample, whereas targeted NGS (tNGS) workflows enrich specific genetic targets for sequencing. Metagenomic NGS has the advantage of potentially sequencing any genetic material present, increasing the ability to detect unforeseen pathogens, whereas targeted NGS has the advantage of enriching genetic targets for specific pathogens or pathogen groups as well as other genes of interest. This potentially increases the analytical sensitivity but could miss untargeted sequences. It is currently unknown if such trade-offs markedly impact the results generated by differing NGS workflows.
The Respiratory Pathogen ID/AMR enrichment (RPIP) kit (Illumina, Inc., San Diego, CA) is a targeted NGS assay developed to enrich the detection of pathogen and antimicrobial resistance (AMR) sequences from respiratory specimens. Following nucleic acid extraction and initial processing, RPIP reagents are designed to target specific bacterial, mycobacterial, viral, and fungal sequences, as well as select associated AMR sequences, using biotinylated capture probes. Sequences captured by probes are enriched through additional processing steps in an attempt to increase detection. Sequencing data generated in this process are analyzed using the IDbyDNA (Salt Lake City, UT) Explify bioinformatic pipeline, which is tailored to the targeted sequences and can detect nontargeted sequences as well.
The use of NGS technologies for pathogen identification is a burgeoning field, and optimal approaches are areas of active investigation. To date, no studies have directly evaluated the performances of metagenomic and targeted NGS approaches for infectious disease diagnostics. Accordingly, we sought to determine the performance characteristics of the RPIP targeted assay and a complementary metagenomic assay (utilizing a shotgun sequencing approach and in-house bioinformatic analysis) by comparing both workflows to a composite clinical standard consisting of provider-ordered microbiology testing, chart review, and orthogonal testing.
MATERIALS AND METHODS
Standard-of-care microbiology testing and sample enrollment.
BAL fluid specimens were enrolled from the Johns Hopkins Hospital (JHH) microbiology laboratory after provider-ordered testing was complete. Various testing methods were applied as part of provider-ordered testing, including Gram stain and quantitative aerobic bacterial culture (quantification range of 100 to >10,000 CFU/mL), Legionella culture, fungal stain and culture, Pneumocystis direct fluorescent-antibody stain or PCR (based on availability at the time of testing) (8), galactomannan antigen testing, mycobacterial stain and culture, Mycobacterium tuberculosis PCR (Xpert MTB/RIF; Cepheid, Sunnyvale, CA), respiratory virus multiplex PCR (Respiratory Pathogen panel; GenMark Diagnostics, Inc., Carlsbad, CA), cytomegalovirus (CMV) PCR (Quest Diagnostics, Chantilly, VA), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) PCR (RealStar; Altona Diagnostics, Hamburg, Germany). Testing was applied per ordering providers, and not all testing was performed on each specimen. Residual samples were required to meet minimal volume requirements for enrollment. Samples were not selected based on clinical syndrome or intention for testing. Enrolled samples were stored at 4°C until frozen at −80°C. Samples were collected from December 2019 through January 2021 from hospitals within the Johns Hopkins network served by the Johns Hopkins Hospital microbiology laboratory.
Clinical data collection.
Demographic and clinical patient data were collected through bulk query from the Johns Hopkins Hospital electronic medical record system. The Charlson comorbidity index was calculated based on ICD-10 codes without age adjustment (9). Results were confirmed by manually auditing 15% of records. Microbiology results from provider-ordered standard-of-care testing were manually extracted from the laboratory information system (LIS) for all samples.
Sample preparation for validation studies.
BAL fluid specimens negative by standard-of-care testing were selected for spike-in studies to establish the analytical sensitivity (limit of detection [LoD]) and precision of the targeted and metagenomic approaches. Specimens were spiked at concentrations 1 (1:10) dilution above, at, and 1 dilution below preliminary limits of detection as determined by pilot studies (data not shown). Samples containing pooled Streptococcus pneumoniae (ATCC 49619), Haemophilus influenzae (ATCC 10211), and Mycobacterium fortuitum (ATCC 6841) were spiked at 1 × 105 CFU/mL, 1 × 104 CFU/mL, and 1 × 103 CFU/mL. Samples containing pooled fungi were spiked at 1 × 106 CFU/mL, 1 × 105 CFU/mL, and 1 × 104 CFU/mL with Aspergillus fumigatus (clinical isolate) and Pneumocystis carinii (nuclei per milliliter) (ATCC PRA-159) as well as at 1 × 105 CFU/mL, 1 × 104 CFU/mL, and 1 × 103 CFU/mL with Cryptococcus neoformans (CBS 8710; Westerdijk Fungal Biodiversity Institute). Samples containing pooled viruses were spiked at 1 × 106 copies/mL, 1 × 105 copies/mL, and 1 × 104 copies/mL with influenza A and B viruses (clinical isolates) and at 1 × 105 copies/mL, 1 × 104 copies/mL, and 1 × 103 copies/mL with adenovirus C (ATCC VR-1516). Dilution concentrations were confirmed by plate counts for bacteria, mycobacteria, and yeasts. Spore counts determined by utilizing a hemocytometer were used to confirm the dilutions of A. fumigatus. Adenovirus and P. carinii concentrations established by the manufacturer were utilized. The concentration of influenza B virus was determined by droplet digital PCR. NGS testing was performed in triplicate for each pool at each dilution.
Sample processing.
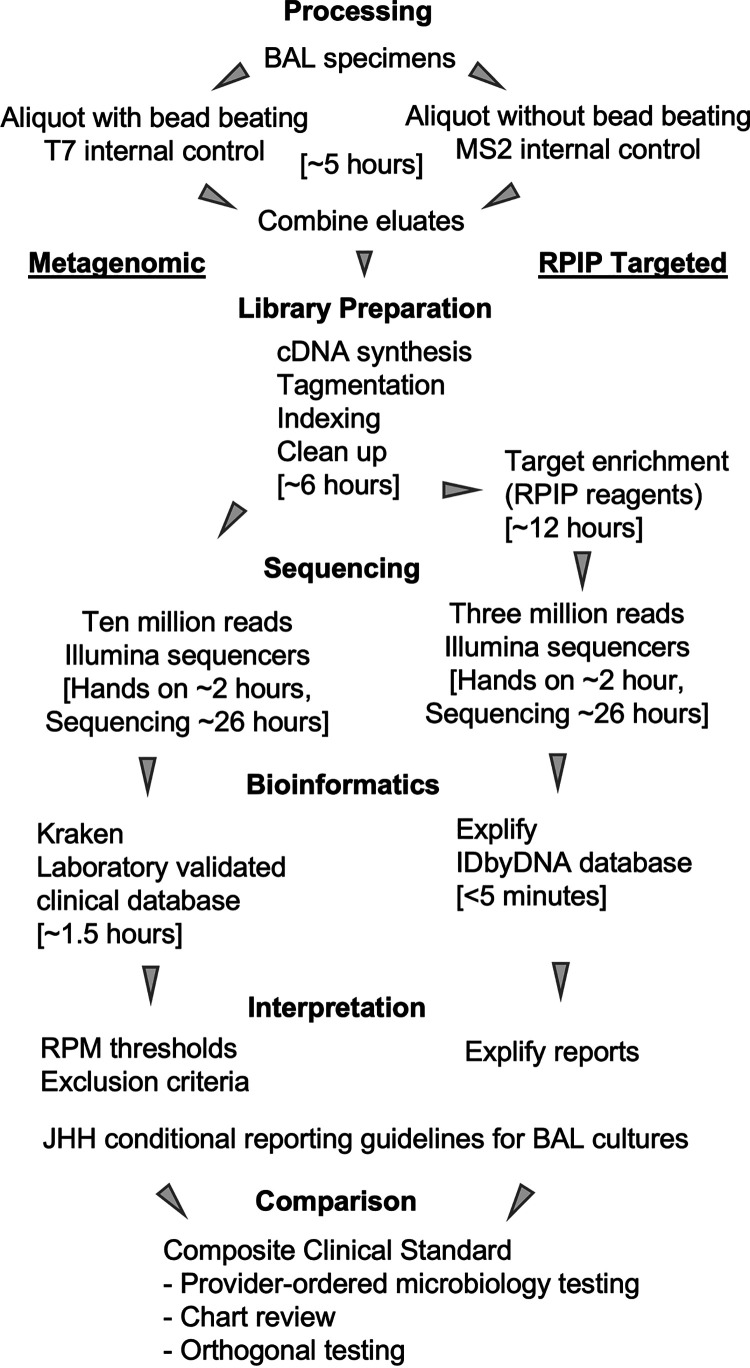
An overview of the methods is provided in Fig. 1. BAL fluid specimens were divided into aliquots that underwent bead beating (400 μL) and aliquots that did not (200 μL), as in a previous study, we found that bead beating increased the yield of reads for certain organism groups (e.g., Gram-positive bacteria and fungi) but detrimentally affected the recovery of others (e.g., nonenveloped viruses and Gram-negative organisms) (10). Samples too viscous to aliquot were diluted 1:1 with sterile saline. The 200-μL aliquots were spiked with an RNA internal control (bacteriophage MS2; Microbiologics, St. Cloud, MN) to a final concentration of 2.5 × 105 PFU/mL prior to nucleic acid extraction. The 400-μL aliquots were spiked with a DNA internal control (bacteriophage T7; Microbiologics, St. Cloud, MN) to a final concentration of 1.0 × 106 PFU/mL and underwent bead beating (lysing matrix D beads and FastPrep24 5G; MP Biomedicals, Santa Ana, CA) for 30 s. Samples rested at room temperature for approximately 20 min before 200 μL was removed for nucleic acid extraction.
FIG 1.
Overview of methods for performance studies. Time estimates included in brackets are based on runs containing 24 samples and reported by those performing the assay steps for this study. Each specimen underwent extraction with or without bead beating prior to the combination of eluates. Eluates from each specimen were processed with the metagenomic and RPIP targeted workflows. Data from each workflow were evaluated using the same conditional reporting guidelines and compared to composite clinical standard results obtained for the specimen. Identical processing and NGS workflows were utilized to establish analytical sensitivity using spiked samples, with comparisons being made to the organism pools rather than a composite clinical standard.
Nucleic acid extraction.
Automated extraction was separately performed for both aliquots of each sample (i.e., the aliquot that underwent bead beating and the aliquot that did not). Extraction was performed using the MagMAX pathogen RNA/DNA kit (Applied Biosystems, Waltham, MA) on the KingFisher Duo Prime system (Thermo Fisher Scientific, Waltham, MA) using the manufacturer-supplied high-volume program. This extraction method was optimized by the Johns Hopkins Hospital microbiology laboratory (10). Eluates were stored at −20°C until library preparation.
Library preparation and enrichment.
For each sample, eluates were thawed, and those that underwent bead beating were combined in equal volumes with those that had not. Nucleic acids were quantified using a Qubit 3.0 fluorometer with double-stranded DNA (dsDNA) and RNA high-sensitivity (HS) reagents. cDNA synthesis and library preparation were performed with the Respiratory Pathogen ID/AMR Enrichment (RPIP) kit according to the Illumina RNA prep with enrichment (L) tagmentation protocol. An aliquot was saved after the postindexing cleanup step as the product for metagenomic sequencing, with the remaining taken through library enrichment with RPIP reagents to generate the product for targeted sequencing. Target enrichment was performed by incubating RPIP probes with samples for approximately 12 h. The probe and bound products were captured, removed, and eluted and underwent a 14-cycle PCR for additional amplification. The final product for the targeted sequence was size selected using AMPure XP beads and eluted in Illumina resuspension (RSB) buffer.
Sequencing.
Products for metagenomic and targeted sequencing were quantified with a Qubit 3.0 instrument using dsDNA HS reagents. DNA fragment analysis was performed using the TapeStation 4200 system (Agilent Technologies, Santa Clara, CA) with a 5000 HS kit. Sequencing was performed on Illumina HiSeq2500 and NextSeq1000 instruments by the Johns Hopkins Hospital microbiology laboratory and using a NovaSeq or NextSeq2000 instrument by Illumina (University of Maryland BioPark, Baltimore, MD). Sequencing was performed with 200-bp paired-end read sequencing (2 × 100) to goal depths of 3 million reads for the targeted workflow and 10 million reads for the metagenomic workflow.
Bioinformatic analysis.
Analysis of sequencing data generated by the targeted RPIP workflow was accomplished using the automated Explify RPIP Data Analysis Solution (v1.0.1; IDbyDNA) accessed via Illumina BaseSpace. All results included in the Explify reports were taken forward for result interpretation. Analysis of sequencing data generated by the metagenomic workflow was performed in the Johns Hopkins Hospital microbiology laboratory with Kraken (11), using an in-house clinically validated database (2020v2). The highest 100 raw reads were evaluated for each sample. Raw read counts for viruses of ≥1 were taken forward for result interpretation. For bacteria, mycobacteria, and fungi, raw read counts for individual organisms were normalized per 1 million total sample reads to produce the reads per million (RPM) metric. Normalized RPM counts for each detected organism were compared to those of the corresponding organism in the negative extraction control (NEC) for each extraction run, and only sample read counts ≥10 times the values of the NEC were taken forward for result interpretation. Both bioinformatic approaches returned species-level identification for further analysis.
Result interpretation using conditional reporting criteria.
All organism identifications made by Explify quantification results or RPM counts passing the above-mentioned criteria were interpreted using conditional reporting criteria followed by the Johns Hopkins Hospital clinical microbiology laboratory for reporting culture-based results from BAL fluid (see Table S1 in the supplemental material). This document was adapted to account for organisms identified by NGS workflows but not routinely detected by standard aerobic culture, including anaerobic bacteria and resident microbiota members.
The potential pathogenicities of organisms identified by NGS workflows were interpreted on an organism-by-organism basis for each sample. If a bacterial organism with the highest Explify quantification or RPM counts was deemed to be a member of the resident microbiota by conditional reporting criteria, other bacteria with a lower Explify quantification or RPM counts were not recorded as pathogens unless they met predefined reporting criteria. If it was deemed a pathogen, the process was repeated with the bacteria having the next highest quantification or counts until the resident microbiota was documented. All results generated by the metagenomic workflow were assessed by a panel of three board-certified microbiologists (K. C. Carroll, P. J. Simner, and D. C. Gaston), and those not present in the conditional reporting criteria were included or excluded based on consensus. Reads for parasites were removed from the analysis given the low relative RPM counts and the consensus that these represented short host sequences aligning to parasitic eukaryotes. Torque teno viruses and bacterial phages were also excluded from the analysis. For a bacterial organism to be counted as a pathogen by the metagenomic workflow, the normalized RPM counts were required to be ≥5 times above the counts for resident microbiota, if it was not the only organism detected in a sample. All filamentous molds, mycobacteria, and viruses were included without a comparison of quantification methods to resident microbiota. Results of NGS testing were not released to providers.
Reporting criteria for Streptococcus species.
In reports where multiple Streptococcus species were identified, only the species with the highest normalized RPM count or Explify quantification that were not deemed members of resident microbiota were included as potential pathogens. In cases where Streptococcus pneumoniae was identified as the most abundant Streptococcus species but quantification was below that for the resident microbiota, this was indicated as the presence of resident microbiota including S. pneumoniae. S. pneumoniae was not identified as a pathogen for further analysis in these instances.
Comparison to standard-of-care microbiology results and orthogonal testing.
Results from NGS workflows meeting all criteria for inclusion as a potential pathogen were compared to standard-of-care microbiology results reported in the LIS. Comparisons were made separately for metagenomic and targeted workflows; NGS workflows were not directly compared due to differences between the workflows. Yeasts reported as members of the respiratory microbiota in culture were excluded from the analysis. Filamentous molds documented on <3 of 5 medium types or not deemed clinically significant by the treating providers were also excluded from the analysis. Individual organisms identified by testing methods were considered single analytes for performance calculations. Analytes detected by an NGS method but not tested for by standard-of-care microbiology were adjudicated by a composite clinical standard including the detection of the same organism from different samples or specimen types within 14 days of the BAL fluid specimen collection date. When unable to confirm results by the composite clinical standard, samples underwent orthogonal testing utilizing analyte-specific PCR. Orthogonal testing was performed as follows: Ureaplasma parvum testing was performed by the University of Alabama at Birmingham Diagnostic Mycoplasma Laboratory (Birmingham, AL); herpes simplex virus 1 (HSV-1), CMV, Epstein-Barr virus (EBV), human herpesvirus 6 (HHV-6), and SARS-CoV-2 testing was performed by the Johns Hopkins Hospital clinical microbiology laboratory; Tropheryma whipplei testing was performed by ARUP Laboratories (University of Utah, Salt Lake City, UT); and HHV-7 testing was performed by Eurofins Viracor (Lee’s Summit, MO).
Analytes detected by NGS workflows that were not present in provider-ordered testing, not adjudicated by the composite clinical standard, and not confirmed by orthogonal testing were counted as false positive (FP). Analytes detected by an NGS workflow and present in the composite clinical standard were counted as true positive (TP) for that workflow. Analytes present in the composite clinical standard but not detected by an NGS workflow were counted as false negative (FN) for that workflow. Analytes with an absence of detected analytes in the composite standard and an NGS workflow were counted as true negative (TN).
Statistics.
Statistical comparisons were performed using the Pandas package of Python (version 3.8.5), the epiR package of R (version 4.1.2), and GraphPad Prism (version 9.2.0). Error calculations represent 95% confidence intervals or standard deviations, where indicated. Statistical significance was set at an alpha value of <0.05.
IRB and ethics.
Sample collection, clinical data extraction, and storage of data containing protected health information were performed in compliance with the Health Insurance Portability and Accountability Act (HIPAA) and in accordance with the Johns Hopkins IRB-approved protocol 00264184.
RESULTS
Analytical sensitivity/limit-of-detection ranges, precision, and cross-reactivity.
Table 1 depicts the limit-of-detection (LoD) ranges for representative organism pools. Pools were utilized as the LoD cannot be established for all organism analytes identified by NGS assays given the breadth of potentially detectable analytes (12). Overall, the detection ranges for bacteria, fungi, and viruses were comparable between the metagenomic and targeted NGS workflows. Within the respective LoD ranges for each NGS workflow, the reproducibility of analyte detection was 100%. The metagenomic workflow detected all analytes within the type-specific limits of detection but identified Pneumocystis carinii as P. jirovecii due to taxonomic similarity and the absence of P. carinii in the Kraken database. The targeted workflow did not detect P. carinii at any tested dilution because RPIP reagents do not target this organism, and it is not present in the Explify database. Additionally, the targeted workflow reported Cryptococcus neoformans as Cryptococcus gattii.
TABLE 1.
Analytical sensitivity ranges and precisiona
| Spiked organism | No. of detected samples/no. of replicates |
|||||||
|---|---|---|---|---|---|---|---|---|
| Metagenomic NGS at dilution (CFU or copies/mL) of: |
RPIP targeted NGS at dilution (CFU or copies/mL) of: |
|||||||
| 106 | 105 | 104 | 103 | 106 | 105 | 104 | 103 | |
| Mycobacterium fortuitum | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 2/3 | ||
| Streptococcus pneumoniae | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | ||
| Haemophilus influenzae | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 2/3 | ||
| Aspergillus fumigatus | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 0/3 | ||
| Pneumocystis carinii | 3/3 | 3/3 | 0/3 | 0/3 | 0/3 | 0/3 | ||
| Cryptococcus neoformans | 3/3 | 1/3 | 0/3 | 3/3 | 3/3 | 1/3 | ||
| Adenovirus C | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | ||
| Influenza A virus | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | ||
| Influenza B virus | 3/3 | 3/3 | 1/3 | 3/3 | 3/3 | 3/3 | ||
Values in boldface type indicate the limit of detection (LoD) for the representative organisms; shaded values indicate LoD ranges for each organism type.
Cross-reactivity was assessed through detection by the NGS workflows of additional organisms not spiked into the samples. Detected organisms primarily represented members of the resident microbiota. Notable exceptions include reports of M. tuberculosis by both workflows in a single replicate spiked with M. fortuitum at the highest concentration of 1 × 105 CFU/mL, although read counts for M. fortuitum were markedly higher than those for M. tuberculosis in this replicate. The targeted workflow also reported Streptococcus mitis in 3/3 replicates spiked with S. pneumoniae at 1 × 105 CFU/mL and in a single replicate spiked with S. pneumoniae at 1 × 104 CFU/mL. The metagenomic workflow reported Lichtheimia corymbifera in 2/3 samples spiked with the fungal pathogen pool at the highest concentration range of 1 × 105 to 1 × 106 CFU/mL. Detection of these organisms did not occur at lower dilutions. Results were not investigated by orthogonal testing.
Evaluation of clinical specimens.
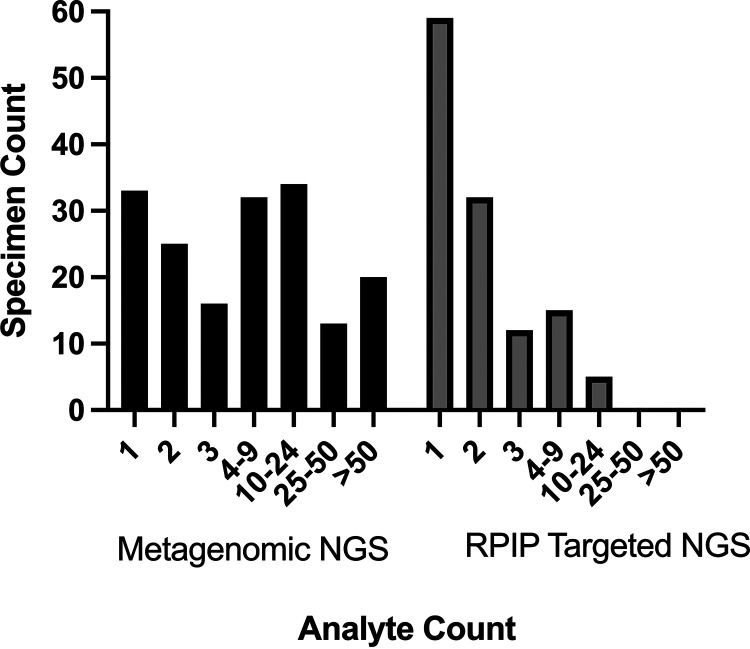
A total of 201 BAL fluid specimens from 177 patients underwent sequencing with the targeted and metagenomic workflows (Fig. 1). The demographics of patients providing BAL fluid specimens are included in Table S2 in the supplemental material. Prior to applying conditional reporting criteria, the metagenomic NGS workflow generated 2,995 hits (2,860 bacterial, 4 mycobacterial, 98 viral, 18 fungal, and 15 parasitic) from 173 specimens; 28 specimens were negative by this workflow. The targeted NGS workflow identified 294 potential pathogens (247 bacterial, 1 mycobacterial, 43 viral, and 3 fungal) from 123 BAL fluid specimens; 78 specimens were negative by this workflow. Ranges of the analytes detected per specimen for each workflow are presented in Fig. 2.
FIG 2.
Relative distribution of analytes detected by NGS workflows. Sample counts per number of analytes for the metagenomic NGS workflow are number of analytes (number of samples): 1 (33), 2 (25), 3 (16), 4 (32), 10 to 24 (34), 25 to 50 (13), and >50 (20); 98 analytes were detected in the sample containing the highest number for this workflow. Sample counts per number of analytes for the RPIP targeted NGS workflow are 1 (59), 2 (32), 3 (12), 4 to 9 (15), 10 (5), 25 to 50 (0), and >50 (0); 14 analytes were detected in the sample containing the highest number for this workflow.
Result interpretation.
Utilizing conditional reporting criteria to interpret NGS results allows comparison between provider-ordered standard-of-care testing and NGS workflows. NGS results were evaluated by a panel of three microbiologists and interpreted using criteria developed at the Johns Hopkins Hospital microbiology laboratory for BAL fluid cultures. Analytes passing the thresholds for inclusion as potential pathogens but not deemed pathogenic were excluded (Table S3), as were analytes deemed members of the resident microbiota (Table S4). The most frequently identified component of the resident microbiota by both workflows was Rothia mucilaginosa. Following R. mucilaginosa, the most frequently identified microbiota members for the targeted NGS workflow included Streptococcus mitis, Prevotella melaninogenica, Campylobacter concisus, and Veillonella parvula. The members of the resident microbiota most frequently identified by the metagenomic workflow included Schaalia odontolytica, Prevotella species (P. melaninogenica and P. jejuni), and Streptococcus species (S. mitis and genus-level calls for streptococci). A single genus-level call with multiple associated species was frequently encountered in data generated by the metagenomic workflow, particularly with streptococci. This analysis cannot determine if individual species were present or if the hits represented taxonomic “near-neighbor” effects and were accordingly counted as resident microbiota members according to interpretation criteria.
Following this analysis, 97 potential pathogens (29 bacterial, 4 mycobacterial, 56 viral, and 8 fungal) from 69 samples remained from the metagenomic NGS workflow, representing percent reductions of 96.8% for analytes and 60.1% for positive samples. Application of the same criteria to the analysis of results generated by the RPIP targeted NGS workflow revealed 73 potential pathogens (27 bacterial, 1 mycobacterial, 42 viral, and 3 fungal) from 55 samples, representing percent reductions of 75.2% for analytes and 55.3% for positive samples.
Performance.
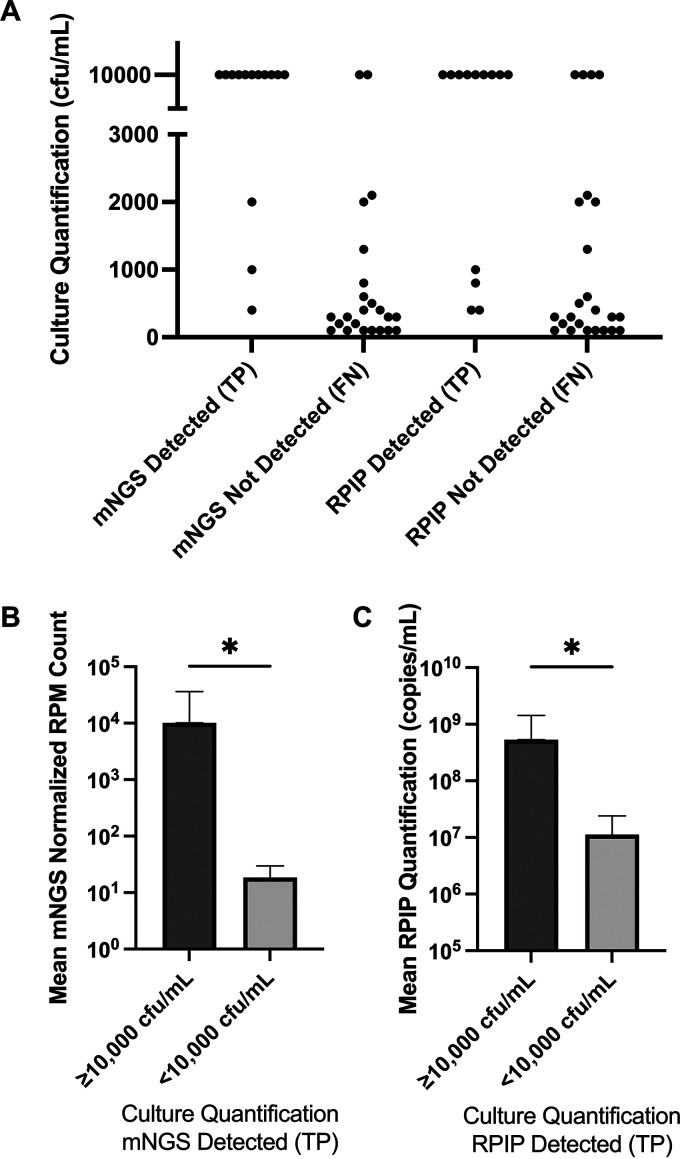
Results that remained after the application of interpretive criteria were compared to a composite clinical reference to determine performance characteristics. Potential pathogens for which directed testing was not ordered were confirmed by chart review or orthogonal testing and incorporated into standard-of-care results for comparisons (Table 2). The overall accuracy of the RPIP targeted NGS workflow was 65.6% (95% confidence interval, 59.2 to 71.5%), with a positive percent agreement (PPA) of 45.9% (36.8 to 55.2%) and a negative percent agreement (NPA) of 85.7% (78.1 to 91.5%). The overall accuracy of the metagenomic NGS workflow was 67.1% (60.9 to 72.9%), with a PPA of 56.6% (47.3 to 65.5%) and an NPA of 77.2% (68.9 to 84.1%). Table 3 shows performance characteristics per organism type, and data used to generate these characteristics are provided in the supplemental material. The majority of bacterial false-negative results were from samples with colony counts in standard aerobic cultures below 1 × 104 CFU/mL (Fig. 3). Bacterial analytes counted as false negative but for which reads were present at levels below those of the resident microbiota occurred in 2 of 24 analytes (8.3%) from the RPIP targeted workflow and 5 of 24 analytes (20.8%) from the metagenomic workflow. These analytes remained false negative according to the categorization method of using a higher abundance of resident microbiota as an exclusion criterion. The mean normalized read counts (mNGS workflow) and RPIP quantification (targeted workflow) of true-positive bacterial results quantified at <10,000 CFU/mL by standard culture were significantly lower than those at ≥10,000 CFU/mL (Fig. 3). No true positives for fungi were detected by either NGS workflow. Neither NGS workflow identified filamentous molds that were isolated in culture and deemed clinically significant by the treating providers (including Aspergillus, Rhizopus, Fusarium, and Cladosporium species), nor was a sample containing P. jirovecii identified by provider-ordered PCR detected by either NGS assay.
TABLE 2.
Analytes detected by NGS workflows but not by provider-ordered testinga
| Workflow, result, and organism type | Analyte | No. of samples with analyte detected |
|---|---|---|
| Metagenomic NGS workflow | ||
| True positive | ||
| Bacteria | Corynebacterium striatum b | 1 |
| Enterococcus faecium c | 1 | |
| Ureaplasma parvum | 1 | |
| Viruses | CMV | 5 |
| EBV | 16 | |
| HSV-1 | 4 | |
| HHV-6 | 5 | |
| HHV-7 | 4 | |
| Rhinovirus | 1 | |
| SARS-CoV-2 | 2 | |
| False positive | ||
| Bacteria | Achromobacter xylosoxidans | 2 |
| Acinetobacter haemolyticus | 1 | |
| Clostridium tetani | 1 | |
| Cupriavidus gilardii | 1 | |
| Listeria monocytogenes | 2 | |
| Orientia tsutsugamushi | 1 | |
| Rhodococcus sp. | 1 | |
| Stenotrophomonas maltophilia | 1 | |
| Mycobacteria | Mycobacterium avium complex | 1 |
| Fungi | Coccidioides posadasii | 2 |
| Cryptococcus neoformans | 1 | |
| Pneumocystis jirovecii | 1 | |
| Rhizophagus irregularis | 1 | |
| Scedosporium apiospermum | 1 | |
| Trichosporon asahii | 2 | |
| Viruses | CMV | 2 |
| EBV | 2 | |
| HSV-1 | 1 | |
| HHV-7 | 3 | |
| SARS-CoV-2 | 2 | |
| RPIP targeted NGS workflow | ||
| True positive | ||
| Bacteria | Corynebacterium striatum b | 1 |
| Enterococcus faecium c | 1 | |
| Tropheryma whipplei | 1 | |
| Ureaplasma parvum | 1 | |
| Viruses | EBV | 12 |
| HSV-1 | 4 | |
| HHV-6 | 4 | |
| Rhinovirus | 1 | |
| SARS-CoV-2 | 3 | |
| False positive | ||
| Bacteria | Burkholderia cepacia complex | 1 |
| Enterococcus faecalis | 2 | |
| Escherichia coli | 1 | |
| Fusobacterium nucleatum | 1 | |
| Ralstonia pickettii | 1 | |
| Stenotrophomonas maltophilia | 1 | |
| Fungi | Microascus cirrosus | 3 |
| Viruses | Human metapneumovirus | 1 |
| EBV | 1 | |
| SARS-CoV-2 | 5 | |
Counts represent the number of samples in which the analyte was detected per workflow.
Isolated from blood cultures.
Isolated from pleural fluid; vancomycin resistant by phenotypic testing and associated with a van gene by RPIP analysis.
TABLE 3.
Performance characteristics of targeted and metagenomic NGS workflows
| Performance category | No. of samples categorized as: |
Accuracy (%) (95% confidence interval) | PPA (%) (95% confidence interval) | NPA (%) (95% confidence interval) | |||
|---|---|---|---|---|---|---|---|
| TP | FP | TN | FN | ||||
| Metagenomic NGS workflow | |||||||
| Overall | 69 | 29 | 98 | 53 | 67.1 (60.9–72.9) | 56.6 (47.3–65.5) | 77.2 (68.9–84.1) |
| Bacterial | 20 | 10 | 153 | 24 | 83.6 (77.8–88.3) | 45.5 (30.4–61.2) | 93.9 (89.0–97.0) |
| Mycobacterial | 3 | 1 | 188 | 5 | 97.0 (93.5–98.9) | 37.5 (8.5–75.5) | 99.5 (97.1–100.0) |
| Fungal | 0 | 8 | 185 | 12 | 90.2 (85.3–93.9) | 0.0 (0.0–26.5) | 95.9 (92.0–98.2) |
| Viral | 46 | 10 | 116 | 12 | 88.0 (82.5–92.4) | 79.3 (66.6–88.8) | 92.1 (85.9–96.1) |
| RPIP targeted NGS workflow | |||||||
| Overall | 56 | 17 | 102 | 66 | 65.6 (59.2–71.5) | 45.9 (36.8–55.2) | 85.7 (78.1–91.5) |
| Bacterial | 20 | 7 | 158 | 24 | 85.2 (79.6–89.7) | 45.5 (30.4–61.2) | 95.8 (91.5–98.3) |
| Mycobacterial | 1 | 0 | 189 | 7 | 96.4 (92.8–98.6) | 12.5 (0.3–52.7) | 100 (98.1–100.0) |
| Fungal | 0 | 3 | 187 | 12 | 92.6 (88.0–95.8) | 0.0 (0.0–26.5) | 98.4 (95.5–99.7) |
| Viral | 35 | 7 | 115 | 23 | 83.3 (77.1–88.5) | 60.3 (46.6–73.0) | 94.3 (88.5–97.7) |
FIG 3.
Relationship of bacteria quantified by standard methods and those detected by NGS workflows. (A) True-positive (TP) and false-negative (FN) results per workflow. Each data point represents bacteria isolated and quantified from standard aerobic cultures (n = 37). Isolates reported as ≥10,000 CFU/mL were plotted at 10,000 CFU/mL. Bacteria detected by standard culture with semiquantification or without quantification were not included. The metagenomic NGS (mNGS) workflow detected 11 of 13 isolates (84.6%) quantified at >10,000 CFU/mL but did not detect 21 of 24 isolates (87.5%) quantified at <10,000 CFU/mL. Similarly, the RPIP targeted workflow detected 9 of 13 isolates (69.2%) quantified at >10,000 CFU/mL but did not detect 20 of 24 isolates (83.3%) quantified at <10,000 CFU/mL. (B and C) Relationship of NGS quantification methods to relative culture abundance for true-positive samples. Statistical comparisons were made using Mann-Whitney testing (P = 0.02 for mNGS, and P = 0.03 for RPIP targeted NGS). Error bars represent standard deviations. Note the difference in the y axes.
Analytes were detected by NGS workflows that were not detected by provider-ordered testing (Table 2). Notably, Ureaplasma parvum was detected by both NGS workflows from one specimen and confirmed by orthogonal testing. Corynebacterium striatum and Enterococcus faecium were detected by both workflows from specimens that were negative by routine aerobic culture of BAL fluid but positive by blood culture (C. striatum) or pleural fluid culture (E. faecium). Tropheryma whipplei was detected by the RPIP targeted workflow and confirmed by orthogonal testing. Additional analytes detected by NGS workflows were primarily viruses (44 of 46 positive orthogonal tests). Additional analytes with potential pathogenicity that did not have testing ordered by providers included SARS-CoV-2, CMV, and rhinovirus. All false-positive results for viruses were negative for the specific analyte by orthogonal testing. False-positive results for bacteria, mycobacteria, and fungi could not be directly assessed by orthogonal testing but lacked evidence of presence by chart review.
RPIP antimicrobial resistance markers.
The targeted workflow using RPIP reagents and Explify bioinformatic analysis enables the detection of bacterial genotypic AMR markers associated with antimicrobial susceptibility testing (AST) results for certain agents. AMR markers were detected in 136 of the 201 samples (67.7%) and associated with 16 potential pathogens in 15 samples. Of the 16 potential pathogens with associated AMR markers, standard-of-care methods detected 13 for which phenotypic resistance could be evaluated (Table 4). Full or partial agreement between the associated and tested resistance was found in 7 of 13 (53.8%) potential pathogens, including vancomycin-resistant E. faecium, methicillin-resistant Staphylococcus aureus, and M. tuberculosis with MICs for second-line agents at or near the proposed critical concentrations. Partial agreement occurred with one potential pathogen in which an extended-spectrum-beta-lactamase (ESBL)-producing Escherichia coli isolate was associated with blaCTX-M and blaOXA, although the blaOXA association was due to P. aeruginosa being present in the same sample. The E. coli isolate demonstrated susceptibility to meropenem by standard methods. Disagreement was found for 6 of 13 (46.2%) potential pathogens. Disagreement occurred because P. aeruginosa was associated with blaOXA genes identified as encoding carbapenemases, although the isolates tested susceptible to carbapenems. Additionally, a blaKPC gene harbored by a Klebsiella pneumoniae isolate detected by standard methods was not detected by the targeted workflow.
TABLE 4.
Antimicrobial resistance associations made by Explify analysis for the RPIP targeted workflowf
| Pathogen | Associated AMR marker(s)a | Susceptibility profile | Agreement |
|---|---|---|---|
| E. coli | ANT(3′), CTX-M, Dfr, MPH, OXA, Sul | ESBL; R-TMP-SMX | Agreeb (partial agreement) |
| E. faecalis | Erm | Pansusceptible (ampicillin, vancomycin, linezolid) | Agreec |
| E. faecium | APH(3′), Erm, Van | VRE; R-ampicillin, SDD-daptomycin, R-vancomycin | Agreed |
| M. tuberculosis | Qnr, RRS | Susceptible to first-line agents, prediction of resistance to second-line agentse | Agree |
| P. aeruginosa | CTX-M, Dfr, OXA | Pansusceptible | Disagree |
| P. aeruginosa | OXA | Pansusceptible | Disagree |
| P. aeruginosa | OXA | Pansusceptible | Disagree |
| P. aeruginosa | OXA, CrpP | Pansusceptible | Disagree |
| P. aeruginosa | OXA, CrpP | Pansusceptible | Disagree |
| P. aeruginosa | OXA, Dfr | S-meropenem, R-piperacillin-tazobactam, R-ceftazidime, R-cefepime, R-aztreonam | Disagree |
| S. aureus | ABC–F, APH(3′), MecA, MPH | R-oxacillin, R-erythromycin | Agree |
| S. aureus | Erm | R-clindamycin, R-erythromycin | Agree |
| S. aureus | MecA, Erm | PBP2a detected by LFA | Agree |
Associations are listed as reported by Explify analysis, which does not follow traditional gene-based reporting. Reported associations between AMR markers and drug classes are made by Explify as follows: ABC–F, macrolides; ANT(3′), aminoglycosides; APH(3′), aminoglycosides; CrpP, fluoroquinolones; CTX-M, cephalosporins and penicillins; Dfr, diaminopyrimidine; Erm, lincosamides and macrolides; MecA, beta-lactam/beta-lactamase inhibitors, carbapenems, cephalosporins, and penicillins; MPH, macrolides; OXA, carbapenems; Qnr, fluoroquinolones; RRS, aminoglycosides; Sul, sulfonamides; Van, glycopeptides. Associations were not made with all agents in each class.
Susceptible to meropenem, although OXA is associated. Macrolides were not tested for this isolate.
Macrolides were not tested for this isolate.
Aminoglycosides and macrolides were not tested for this isolate.
Second-line agents were moxifloxacin at an MIC of 0.25 μg/mL (at the proposed critical concentration), amikacin at an MIC of 0.5 μg/mL (below but within 1 dilution of the proposed critical concentration), and kanamycin at an MIC of 2.5 μg/mL (at the proposed critical concentration).
R, resistant; S, susceptible; TMP-SMX, trimethoprim-sulfamethoxazole; VRE, vancomycin-resistant Enterococcus; PBP2a, penicillin binding protein 2a; SDD = susceptible dose dependent; LFA = lateral flow assay.
DISCUSSION
NGS workflows for BAL fluid specimens are anticipated to aid in the identification of infectious pathogens. This study sought to evaluate the performance of a commercially manufactured targeted NGS workflow (RPIP with Explify analysis) and a complementary metagenomic NGS workflow. Using a composite clinical standard consisting of provider-ordered microbiology testing, clinical data, and orthogonal testing as the comparator, both NGS workflows demonstrated similar performances.
Organism types were detected by the NGS workflows with differing efficiencies. The detection of viruses demonstrated the highest PPA for both workflows. Similar to other reports, herpesviruses were frequently identified (13). The PPA for bacteria and mycobacteria was diminished by the LoD ranging between 103 and 104 CFU/mL, preventing the detection of organisms at lower abundances. Neither workflow reliably detected bacteria quantified in standard cultures near or below 103 CFU/mL from clinical samples. The enrichment of targeted sequences by the RPIP workflow did not overcome this challenge. The PPA for fungi was the lowest given the absence of true positives by either NGS assay. This is in contrast to other studies in which increased detection of fungal pathogens has been reported using metagenomic workflows (14, 15) but similar to studies noting decreased detection of fungi (16). Multiple reasons may underlie the differences in detection in this and other studies, such as extraction efficiency differences per organism type, library preparation techniques, sequencing depths, lower thresholds for detecting viral reads, and differences in sample selection (17).
The LoD ranges for both workflows were higher than anticipated based on previous studies. Our group previously reported LoD ranges from cerebrospinal fluid (CSF) of 101 to 103 U/mL depending on the pathogen and processing method (10). Whereas a comparable study utilizing a synthetic CSF matrix demonstrated lower LoD ranges of 10−1 to 102 U/mL (18), a study using BAL fluid specimens reported similarly wide LoD ranges (102 to 104 U/mL) overlapping those reported here (19). The analytical sensitivity of NGS assays is impacted by multiple variables, potentially involving all steps from sample collection to bioinformatic analysis (12). Our results suggest that using the same matrix for analytical sensitivity studies as the one for clinical testing may provide a more representative measure of performance.
Important considerations relating to bioinformatic analysis and database use are highlighted in the LoD studies. Neither workflow correctly identified P. carinii because this organism is not a human pathogen (20) and thus is absent from both databases, whereas the human pathogen P. jirovecii is present in both databases. The metagenomic workflow assigned an identity of P. jirovecii, the closest taxonomic assignment present in the Kraken database. The RPIP targeted workflow accurately made no association with P. jirovecii. Separately, C. neoformans was inaccurately identified as C. gattii by the RPIP targeted workflow. Both analytes are present in the Explify database, and this misidentification was due to the bioinformatic approach of Explify. This was resolved in a future version of the pipeline (IDbyDNA representatives Robert Schlaberg, personal communication). Thus, the bioinformatic approach and database used will directly impact the results, which must be considered when interpreting data generated by NGS workflows.
The multitude of organisms identified in individual samples by both NGS workflows demonstrates that interpretive criteria are necessary to determine potential pathological significance. Approximately 10-fold more results were generated by the metagenomic workflow than by the targeted workflow, and these results were reduced by 96.8% upon applying interpretive criteria. Although the targeted workflow involved two methods of enhancing the detection of potential pathogens, targeting with RPIP reagents and utilizing the paired Explify analysis, 75.2% of the reported analytes were deemed nonpathogenic members of the resident microbiota after applying interpretive criteria. The absence of predefined interpretive criteria could lead to the overreporting of microbiota members as pathogens, potentially leading to antimicrobial overuse and negative patient outcomes. Although the screening results using interpretive criteria masked the detection of bacterial pathogens by the NGS workflows in a small number of samples, utilizing such criteria is necessary given the abundance of organism reads that can be generated. Interpretive criteria are already in use in clinical microbiology laboratories to dictate the reporting of cultures containing mixed organisms that include members of the resident microbiota and should be adapted to interpret results generated by NGS assays as presented in this study. Such criteria include lists of organisms to conditionally report but could also incorporate surrogate methods of quantification such as read counts and relative abundances. These methods may prevent overcalling taxonomic near-neighbor effects, as in the misclassification of M. fortuitum reads as belonging to M. tuberculosis in one replicate of the analytical sensitivity studies.
The performance characteristics of the workflows in this study could allow their use as adjunctive tests with standard microbiology testing. The use of these assays independent of standard microbiology approaches would require comparable detection of all organism types at limits of detection afforded by culture and directed molecular testing and would be enhanced by the reduced detection of nonpathogenic members of the resident microbiota. Acceptable performance for clinical use has been demonstrated for particular pathogen groups (21–24), although the present study differs in sequencing methods, included higher numbers of samples, and represented ungoverned use through the lack of sample selection. Groups utilizing metagenomic sequencing methods similar to those studied here report performance characteristics below what is needed for independent use (25). Adjunctive use also involves diagnostic stewardship, and use with standard microbiology testing in select populations may be the most beneficial. The performance of NGS workflows is enhanced when samples from specific patient populations, such as those who are immunocompromised, are studied (26–29).
The use of these assays as adjunctive tests could detect pathogens that were not suspected or detected by standard-of-care methods. This was demonstrated with metagenomic workflows using the Explify platform for analysis, exhibiting the potential to improve pathogen identification from BAL fluid when utilized in conjunction with standard microbiology techniques (30). In the present work, Ureaplasma parvum was detected by NGS methods in a patient after lung transplantation with pneumonia with hyperuricemia, a clinical syndrome compatible with Ureaplasma infection (31). Additionally, T. whipplei, SARS-CoV-2, CMV, and rhinovirus were detected by NGS workflows that were not part of provider-ordered testing. Although limited, these cases illustrate how NGS testing can provide unexpected diagnostic information.
Antimicrobial resistance detection by the targeted RPIP workflow demonstrated concordance with results determined by phenotypic susceptibility testing in over half of the specimens with associated AMR genes and phenotypic AST results. These organisms were Gram positive (S. aureus and E. faecalis), Gram negative (E. coli), and mycobacterial (M. tuberculosis). Notably, the AMR detected for M. tuberculosis was for second-line agents, and the corresponding critical concentrations of these agents met proposed limits to define non-wild-type susceptibility (32). Associations were not made for all pathogens as demonstrated by the lack of identification of a blaKPC gene in a carbapenemase-producing carbapenem-resistant Klebsiella oxytoca isolate. AMR association failed for P. aeruginosa, wherein an endogenous, narrow-spectrum blaOXA-2 gene was associated with mediating carbapenem resistance. Additionally, the majority of AMR genes detected by the targeted workflow were not associated with detected pathogens. These were likely harbored by members of the resident microbiota that did not pass thresholds for inclusion in the reported data, further demonstrating difficulties encountered by testing AMR genes directly from specimens obtained from nonsterile sites. Issues of blaOXA-2 association with carbapenemase production in P. aeruginosa and reports of AMR markers unassociated with specific organisms have been addressed in a future version of the Explify bioinformatic pipeline (IDbyDNA representatives Robert Schlaberg, personal communication). These issues emphasize that AMR associations must be considered preliminary and not used for patient management decisions until associations generated by a given NGS workflow are demonstrated to be equivalent to those of standardized methods of AMR identification.
The primary functional differences between the NGS workflows evaluated in this study relate to library preparation, bioinformatic analysis, and the extent of result review and interpretation (Table 5). At the point in library preparation when the product is utilized for metagenomic sequencing, additional enrichment and cleanup steps with the RPIP reagents were performed for targeted sequencing. These steps were described as technically challenging by technologists performing the assay and require additional time. However, bioinformatic analysis performed with the Explify platform does not require in-depth knowledge of bioinformatic techniques and provides clear reports of potential pathogens detected in the specimens. This is in contrast to the metagenomic workflow, which is less technically demanding but requires a facility with bioinformatic analysis. The analysis pipeline used for the metagenomic workflow included data structure management, command-line interaction with Kraken, and manual analysis of taxonomically assigned read counts. Although this analysis allows more insight into microbial communities, it can be cumbersome and time-consuming. Additionally, computational infrastructure; data storage solutions; and database creation, curation, validation, and maintenance are needed to perform the in-house analysis. Without clear differences in performance, benefits may be realized in the technical or bioinformatic aspects at the level of individual users.
TABLE 5.
Practical considerations for mNGS and tNGS workflows
| Step | Consideration(s) for workflow |
|
|---|---|---|
| Metagenomic NGS | RPIP/Explify targeted NGS | |
| Processing and library preparation | Less extensive library prepn; lower cost for library prepn kits without targeted reagents | More extensive library prepn; additional time and higher reagent cost |
| Sequencing | Greater depth (goal of 10 million reads/sample); higher sequencing cost per sample | Shallower depth (goal of 3 million reads/sample); lower sequencing cost per sample |
| Bioinformatic analysis | Open-source bioinformatics; more involved and requires bioinformatic experience and database curation/maintenance; additional resource and time cost; ability to detect more organisms for other research questions | Less involved; does not require bioinformatic experience or database management |
| Result interpretation | Markedly more results requiring interpretation; possible missed pathogens with abundant resident microbiota | Easier interpretation, with potential pathogens listed on Explify report (but interpretation still required) |
| Detectable analytes | Broadest possible within processing and database limitations | Narrower; partially limited by bioinformatic analysis process |
| AMR prediction | Not studied but theoretically possiblea | Achievable but requires optimization |
See reference 33.
Workflow differences result in variabilities in cost. Library preparation reagents for the RPIP targeted assay are approximately $140 per sample. For a comparable metagenomic library preparation kit, costs are estimated to be approximately $60 per sample. Sequencing costs differ due to the numbers of reads utilized for each workflow. Using the NextSeq1000 platform as an example, the cost of sequencing 3 million reads (RPIP targeted workflow) is approximately $20 per sample, whereas the cost of sequencing 10 million reads (metagenomic workflow) is approximately $90 per sample. These costs are dependent upon the degree of multiplexing per run, and optimal multiplexing is assumed for this comparison. Although library preparation and sequencing costs are nearly equivalent at $150 to $160 per sample for each workflow, the additional technologist time required for the RPIP targeted workflow is not included in this estimation. Costs also differ for bioinformatic analyses. The Explify platform accessed via Illumina BaseSpace requires paid iCredits for use, but these costs must be considered in the context of those incurred for the metagenomic workflow (establishing computational infrastructure and performing bioinformatic analysis). This comparison is applicable only to the methods used in this study. Laboratories must consider diagnostic needs and available resources to determine the most valuable workflow for testing.
This study has multiple limitations. First, the patients included in this study represented a heavily pretreated population, with many treated with antimicrobial agents in the 14 days prior to the acquisition of the BAL fluid specimens. It is possible that false-positive bacterial and fungal results represented organisms that could not be recovered in culture due to antimicrobial use. Second, BAL fluid was not collected using protected techniques, potentially allowing contamination of samples by oropharyngeal flora. Third, the same provider-ordered tests were not applied to all samples. Although this is representative of provider ordering practices, all pathogens may not have been identified if missed by NGS workflows and the lack of standardized testing. Finally, reporting of BAL fluid cultures is standardized in the Johns Hopkins clinical microbiology laboratory, but variability by technologists in the extent of workup for small quantities of bacterial isolates may have occurred.
NGS assays have the potential to improve the clinical detection of pathogens from BAL fluid specimens. Additional optimization is required before the testing studied here can be used independently of standard microbiology techniques. Based on this study, NGS-based approaches for BAL fluid specimens should be considered an adjunct to standard methods, not a replacement. The value of these NGS workflows may be found in the detection of rare, atypical, or unsuspected pathogens. This study additionally highlights considerations for the clinical application of NGS workflows. Clinical laboratories must weigh technical and bioinformatic expertise before incorporating NGS workflows as laboratory-developed tests. Defining reporting criteria for respiratory NGS results is required for appropriate interpretation similarly to culture-based methods for respiratory specimens. Additionally, forming multidisciplinary teams to interpret results in the context of a specific syndrome as well as clinical microbiology and infectious diseases at large may benefit the use of NGS workflows for patient care. Studies evaluating NGS workflows in specific patient populations and how clinical outcomes are influenced by diagnostic NGS approaches are needed. The promise of these technologies for patient care remains, and optimal approaches require further investigation.
ACKNOWLEDGMENTS
We gratefully acknowledge Heba Mostafa and members of her clinical and research laboratories for performing orthogonal testing on selected specimens.
Study materials were provided by Illumina, Inc., and IDbyDNA, Inc., although these entities had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
This research was made possible by support from the Sherrilyn and Ken Fisher Center for Environmental Infectious Diseases, Division of Infectious Diseases, Johns Hopkins University School of Medicine. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the Fisher Center or Johns Hopkins University School of Medicine.
Footnotes
Supplemental material is available online only.
Contributor Information
David C. Gaston, Email: david.c.gaston@vumc.org.
Patricia J. Simner, Email: psimner1@jhmi.edu.
Yi-Wei Tang, Cepheid.
REFERENCES
- 1.Davidson KR, Ha DM, Schwarz MI, Chan ED. 2020. Bronchoalveolar lavage as a diagnostic procedure: a review of known cellular and molecular findings in various lung diseases. J Thorac Dis 12:4991–5019. doi: 10.21037/jtd-20-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simner PJ, Miller S, Carroll KC. 2018. Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin Infect Dis 66:778–788. doi: 10.1093/cid/cix881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filkins LM, Bryson AL, Miller SA, Mitchell SL. 2020. Navigating clinical utilization of direct-from-specimen metagenomic pathogen detection: clinical applications, limitations, and testing recommendations. Clin Chem 66:1381–1395. doi: 10.1093/clinchem/hvaa183. [DOI] [PubMed] [Google Scholar]
- 4.Miao Q, Ma Y, Wang Q, Pan J, Zhang Y, Jin W, Yao Y, Su Y, Huang Y, Wang M, Li B, Li H, Zhou C, Li C, Ye M, Xu X, Li Y, Hu B. 2018. Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin Infect Dis 67:S231–S240. doi: 10.1093/cid/ciy693. [DOI] [PubMed] [Google Scholar]
- 5.Zhou H, Larkin PMK, Zhao D, Ma Q, Yao Y, Wu X, Wang J, Zhou X, Li Y, Wang G, Feng M, Wu L, Chen J, Zhou C, Hua X, Zhou J, Yang S, Yu Y. 2021. Clinical impact of metagenomic next-generation sequencing of bronchoalveolar lavage in the diagnosis and management of pneumonia: a multicenter prospective observational study. J Mol Diagn 23:1259–1268. doi: 10.1016/j.jmoldx.2021.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Xie F, Duan Z, Zeng W, Xie S, Xie M, Fu H, Ye Q, Xu T, Xie L. 2021. Clinical metagenomics assessments improve diagnosis and outcomes in community-acquired pneumonia. BMC Infect Dis 21:352. doi: 10.1186/s12879-021-06039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu CY, Miller SA. 2019. Clinical metagenomics. Nat Rev Genet 20:341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu B, Totten M, Nematollahi S, Datta K, Memon W, Marimuthu S, Wolf LA, Carroll KC, Zhang SX. 2020. Development and evaluation of a fully automated molecular assay targeting the mitochondrial small subunit rRNA gene for the detection of Pneumocystis jirovecii in bronchoalveolar lavage fluid specimens. J Mol Diagn 22:1482–1493. doi: 10.1016/j.jmoldx.2020.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Glasheen WP, Cordier T, Gumpina R, Haugh G, Davis J, Renda A. 2019. Charlson comorbidity index: ICD-9 update and ICD-10 translation. Am Health Drug Benefits 12:188–197. [PMC free article] [PubMed] [Google Scholar]
- 10.Simner PJ, Miller HB, Breitwieser FP, Pinilla Monsalve G, Pardo CA, Salzberg SL, Sears CL, Thomas DL, Eberhart CG, Carroll KC. 2018. Development and optimization of metagenomic next-generation sequencing methods for cerebrospinal fluid diagnostics. J Clin Microbiol 56:e00472-18. doi: 10.1128/JCM.00472-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlaberg R, Chiu CY, Miller S, Procop GW, Weinstock G, Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology, Microbiology Resource Committee of the College of American Pathologists . 2017. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med 141:776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 13.Lewandowska DW, Schreiber PW, Schuurmans MM, Ruehe B, Zagordi O, Bayard C, Greiner M, Geissberger FD, Capaul R, Zbinden A, Boni J, Benden C, Mueller NJ, Trkola A, Huber M. 2017. Metagenomic sequencing complements routine diagnostics in identifying viral pathogens in lung transplant recipients with unknown etiology of respiratory infection. PLoS One 12:e0177340. doi: 10.1371/journal.pone.0177340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian Y-Y, Wang H-Y, Zhou Y, Zhang H-C, Zhu Y-M, Zhou X, Ying Y, Cui P, Wu H-L, Zhang W-H, Jin J-L, Ai J-W. 2021. Improving pulmonary infection diagnosis with metagenomic next generation sequencing. Front Cell Infect Microbiol 10:567615. doi: 10.3389/fcimb.2020.567615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Han Y, Feng J. 2019. Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med 19:252. doi: 10.1186/s12890-019-1022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng JM, Du B, Qin HY, Wang Q, Shi Y. 2021. Metagenomic next-generation sequencing for the diagnosis of suspected pneumonia in immunocompromised patients. J Infect 82:22–27. doi: 10.1016/j.jinf.2021.01.029. [DOI] [PubMed] [Google Scholar]
- 17.Leo S, Gaia N, Ruppe E, Emonet S, Girard M, Lazarevic V, Schrenzel J. 2017. Detection of bacterial pathogens from broncho-alveolar lavage by next-generation sequencing. Int J Mol Sci 18:2011. doi: 10.3390/ijms18092011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller S, Naccache SN, Samayoa E, Messacar K, Arevalo S, Federman S, Stryke D, Pham E, Fung B, Bolosky WJ, Ingebrigtsen D, Lorizio W, Paff SM, Leake JA, Pesano R, DeBiasi R, Dominguez S, Chiu CY. 2019. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res 29:831–842. doi: 10.1101/gr.238170.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W, Yin C, Wang G, Rosenblum J, Krishnan S, Dimitrova N, Fallon JT. 2019. Optimizing a metatranscriptomic next-generation sequencing protocol for bronchoalveolar lavage diagnostics. J Mol Diagn 21:251–261. doi: 10.1016/j.jmoldx.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Redhead SA, Cushion MT, Frenkel JK, Stringer JR. 2006. Pneumocystis and Trypanosoma cruzi: nomenclature and typifications. J Eukaryot Microbiol 53:2–11. doi: 10.1111/j.1550-7408.2005.00072.x. [DOI] [PubMed] [Google Scholar]
- 21.Huang J, Jiang E, Yang D, Wei J, Zhao M, Feng J, Cao J. 2020. Metagenomic next-generation sequencing versus traditional pathogen detection in the diagnosis of peripheral pulmonary infectious lesions. Infect Drug Resist 13:567–576. doi: 10.2147/IDR.S235182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Wu B, Yang D, Yang C, Jin Z, Cao J, Feng J. 2020. Optimal specimen type for accurate diagnosis of infectious peripheral pulmonary lesions by mNGS. BMC Pulm Med 20:268. doi: 10.1186/s12890-020-01298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan H, Li X, Mei A, Li P, Liu Y, Li X, Li W, Wang C, Xie S. 2021. The diagnostic value of metagenomic next-generation sequencing in infectious diseases. BMC Infect Dis 21:62. doi: 10.1186/s12879-020-05746-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Lu Z, Bao Y, Yang Y, de Groot R, Dai W, de Jonge MI, Zheng Y. 2020. Clinical diagnostic application of metagenomic next-generation sequencing in children with severe nonresponding pneumonia. PLoS One 15:e0232610. doi: 10.1371/journal.pone.0232610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Feng W, Ye K, Guo L, Xia H, Guan Y, Chai L, Shi W, Zhai C, Wang J, Yan X, Wang Q, Zhang Q, Li C, Liu P, Li M. 2021. Application of metagenomic next-generation sequencing in the diagnosis of pulmonary infectious pathogens from bronchoalveolar lavage samples. Front Cell Infect Microbiol 11:541092. doi: 10.3389/fcimb.2021.541092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Sun B, Tang X, Liu YL, He HY, Li XY, Wang R, Guo F, Tong ZH. 2020. Application of metagenomic next-generation sequencing for bronchoalveolar lavage diagnostics in critically ill patients. Eur J Clin Microbiol Infect Dis 39:369–374. doi: 10.1007/s10096-019-03734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun T, Wu X, Cai Y, Zhai T, Huang L, Zhang Y, Zhan Q. 2021. Metagenomic next-generation sequencing for pathogenic diagnosis and antibiotic management of severe community-acquired pneumonia in immunocompromised adults. Front Cell Infect Microbiol 11:661589. doi: 10.3389/fcimb.2021.661589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan T, Tan R, Qu H, Weng X, Liu Z, Li M, Liu J. 2019. Next-generation sequencing of the BALF in the diagnosis of community-acquired pneumonia in immunocompromised patients. J Infect 79:61–74. doi: 10.1016/j.jinf.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langelier C, Zinter MS, Kalantar K, Yanik GA, Christenson S, O’Donovan B, White C, Wilson M, Sapru A, Dvorak CC, Miller S, Chiu CY, DeRisi JL. 2018. Metagenomic sequencing detects respiratory pathogens in hematopoietic cellular transplant patients. Am J Respir Crit Care Med 197:524–528. doi: 10.1164/rccm.201706-1097LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azar MM, Schlaberg R, Malinis MF, Bermejo S, Schwarz T, Xie H, Dela Cruz CS. 2021. Added diagnostic utility of clinical metagenomics for the diagnosis of pneumonia in immunocompromised adults. Chest 159:1356–1371. doi: 10.1016/j.chest.2020.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Bharat A, Cunningham SA, Scott Budinger GR, Kreisel D, DeWet CJ, Gelman AE, Waites K, Crabb D, Xiao L, Bhorade S, Ambalavanan N, Dilling DF, Lowery EM, Astor T, Hachem R, Krupnick AS, DeCamp MM, Ison MG, Patel R. 2015. Disseminated Ureaplasma infection as a cause of fatal hyperammonemia in humans. Sci Transl Med 7:284re3. doi: 10.1126/scitranslmed.aaa8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clinical and Laboratory Standards Institute. 2018. Performance standards for susceptibility testing of mycobacteria, Nocardia spp., and other aerobic actinomycetes, 1st ed. CLSI guideline M62. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 33.Yee R, Breitwieser FP, Hao S, Opene BNA, Workman RE, Tamma PD, Dien-Bard J, Timp W, Simner PJ. 2021. Metagenomic next-generation sequencing of rectal swabs for the surveillance of antimicrobial-resistant organisms on the Illumina Miseq and Oxford MinION platforms. Eur J Clin Microbiol Infect Dis 40:95–102. doi: 10.1007/s10096-020-03996-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download jcm.00526-22-s0001.xlsx, XLSX file, 0.10 MB (102.8KB, xlsx)
Tables S1 to S4. Download jcm.00526-22-s0002.pdf, PDF file, 0.2 MB (253.8KB, pdf)