ABSTRACT
COVID-19 has brought unprecedented attention to the crucial role of diagnostics in pandemic control. We compared severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) test performance by sample type and modality in close contacts of SARS-CoV-2 cases. Close contacts of SARS-CoV-2-positive individuals were enrolled after informed consent. Clinician-collected nasopharyngeal (NP) swabs in viral transport media (VTM) were tested with a routine clinical reference nucleic acid test (NAT) and PerkinElmer real-time reverse transcription-PCR (RT-PCR) assay; positive samples were tested for infectivity using a VeroE6TMPRSS2 cell culture model. Self-collected passive drool was also tested using the PerkinElmer RT-PCR assay. For the first 4 months of study, midturbinate swabs were tested using the BD Veritor rapid antigen test. Between 17 November 2020 and 1 October 2021, 235 close contacts of SARS-CoV-2 cases were recruited, including 95 with symptoms (82% symptomatic for ≤5 days) and 140 asymptomatic individuals. Reference NATs were positive for 53 (22.6%) participants; 24/50 (48%) were culture positive. PerkinElmer testing of NP and saliva samples identified an additional 28 (11.9%) SARS-CoV-2 cases who tested negative by reference NAT. Antigen tests performed for 99 close contacts showed 83% positive percent agreement (PPA) with reference NAT among early symptomatic persons, but 18% PPA in others; antigen tests in 8 of 11 (72.7%) culture-positive participants were positive. Contacts of SARS-CoV-2 cases may be falsely negative early after contact, but more sensitive platforms may identify these cases. Repeat or serial SARS-CoV-2 testing with both antigen and molecular assays may be warranted for individuals with high pretest probability for infection.
KEYWORDS: COVID-19, SARS-CoV-2, rapid diagnostics, test performance
INTRODUCTION
As global COVID-19 cases exceed 250 million by December 2021 (1), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) diagnosis remains a critically important public health priority for individuals to know their status and, from a public health standpoint, to understand the amount of circulating virus and risk of infection. Sustained demand for SARS-CoV-2 testing has accelerated the development and deployment of multiple competing testing technologies, sample types, and approaches (2, 3). Rapid diagnostic tests that use a lateral flow assay (LFA) to detect SARS-CoV-2 antigen offer scale, convenience, and potential for home use but suffer from reduced sensitivity compared to nucleic acid tests (NAT) such as real-time reverse transcription-PCR (RT-PCR) (4).
In early symptomatic disease—within 5 days of symptom onset—infected persons are likely to have high viral loads such that both antigen and molecular tests are likely to perform well. For individuals who are close contacts (5), the viral burden of the index case has been associated with risk of transmission (6). However, in the presymptomatic phase when viral burdens are still low (7), false-negative antigen tests can occur. A recent study from the Netherlands in close contacts showed that only 64% of SARS-CoV-2 molecular-test-positive close contacts were identified by antigen testing; antigen test sensitivity was lower in asymptomatic individuals (58.7%) than in symptomatic individuals (84.2%) (8).
We sought to evaluate the performances of molecular and antigen testing in symptomatic and asymptomatic close contacts of SARS-CoV-2-confirmed individuals. We also sought to assess the sensitivity of both molecular and antigen tests in nasal swabs and molecular tests in saliva to diagnose close contacts with infectious virus in nasopharyngeal (NP) swab viral transport media (VTM) using the VeroE6TMPRS2 cell model.
MATERIALS AND METHODS
Patient population and study design.
Participants were recruited through posters, online advertisement, and social media written in English and Spanish. Direct outreach targeted close contacts of persons who tested positive for SARS-CoV-2. Under a partial waiver of HIPAA authorization, study staff called patients who tested positive for SARS-CoV-2 at any Johns Hopkins Medical System site to provide study information to their close contacts, defined as individuals who spent >15 min at <6 feet in the 5 days after symptom onset or test positivity (9). Interested close contacts were able to participate by contacting a dedicated study phone number. Study staff obtained informed consent from willing and eligible participants using a script administered either over the phone or in person. Participants were scheduled for same- or next-day testing at one of two outdoor outpatient testing sites. Two weeks after specimen collection, the study staff called the participant to administer a brief questionnaire, which assessed ongoing or new COVID-19 symptoms, hospitalization, and any other SARS-CoV-2 test results that the participant may have obtained after enrollment. The study was approved by the Johns Hopkins School of Medicine Institutional Review Board.
Sample collection.
NP swabs were collected by a trained nurse on all participants and immediately inoculated into VTM. Study staff additionally observed self-collection of a midturbinate swab for antigen testing and 2 mL of passive drool saliva in a sterile urine cup. All specimens were transported on ice to the clinical laboratory using a medical courier service. The transit time from sample collection sites to the laboratory was 15 to 20 min, and samples were processed immediately upon receipt.
Reference nucleic acid testing.
All molecular testing took place in the Johns Hopkins Molecular Virology Laboratory, which is Clinical Laboratory Improvement Amendments of 1988 (CLIA) certified. In order to provide results to participants without delay, NATs were performed as part of the routine clinical workflow on a range of available platforms. The NAT performed and resulted to patients in real-time was considered to be the reference NAT. For each of the following assays, testing was performed per manufacturer Food and Drug Administration emergency use authorization (EUA) instructions: NeuMoDx SARS-CoV-2 assay (NeuMoDx, Ann Arbor, MI) (10), Roche cobas 6800 (Roche Molecular Systems, Pleasanton, CA), RealStar SARS-CoV-2 RT-PCR (Altona Diagnostics, Hamburg, Germany), Hologic Aptima SARS-CoV-2 assay (Hologic, Bedford, MA) (11), PerkinElmer new coronavirus nucleic acid detection kit (PerkinElmer, Inc. Austin, TX) (12), and BD Veritor SARS-CoV-2 (Becton, Dickinson, Sparks, MD); results were interpreted using the Veritor Plus analyzer (13). The reference clinical assay performed on fresh NP swab VTM was the NeuMoDx, Roche, Aptima, Cepheid Xpert, or Altona assay. Remnant specimen was aliquoted and stored frozen at −80°C and then 300 μL thawed for extraction followed by RT-PCR on the PerkinElmer instrument. Similarly, fresh saliva was aliquoted, frozen, and then, in batches, 300 μL of saliva was processed using the same methods. BD Veritor testing occurred within 6 h of collection, with all midturbinate swabs stored on ice. At the time of test performance and interpretation, staff were blinded to results of other testing modalities and sample types. Antigen testing was performed in a separate laboratory space independent of molecular test facilities.
Virus culture from nasal swabs.
VeroE6TMPRSS2 cells were grown in complete medium (CM) consisting of Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) (Gibco), 1 mM glutamine (Invitrogen), 1 mM sodium pyruvate (Invitrogen), 100 U/mL of penicillin (Invitrogen), and 100 μg/mL of streptomycin (Invitrogen) (14). Viral infectivity was assessed on VeroE6TMPRSS2 cells as previously described using infection media (IM) (identical to CM except the FBS is reduced to 2.5%) (15). When a cytopathic effect was visible in >50% of cells in a given well, the supernatant was harvested. The presence of SARS-CoV-2 was confirmed through reverse transcription-quantitative PCR (RT-qPCR) as described previously by extracting RNA from the cell culture supernatant using the Qiagen viral RNA isolation kit and performing RT-qPCR using the N1 and N2 SARS-CoV-2-specific primers and probes in addition to primers and probes for the human RNaseP gene using synthetic RNA target sequences to establish a standard curve (16).
Statistical analysis.
Clinical characteristics of participants with positive clinical reference test results were compared to participants with negative results. Categorical variables were compared using the chi-square test or exact Fisher test for comparisons, including groups with <5 expected frequencies. Continuous variables were compared using the Kruskal-Wallis test. As SARS-CoV-2 molecular tests are highly specific, in cases of discordant molecular results by modality (standard clinical reference test or PerkinElmer RT-PCR) or sample type (NP or saliva), participants who tested positive by any molecular test were considered to have SARS-CoV-2. Simple frequencies were calculated for the proportion of infectious samples (as measured by cell culture) that were positive by antigen and molecular testing. Antigen test performance characteristics were reported as positive and negative percent agreement with the clinical reference test and calculated as simple proportions with two-sided 95% confidence intervals (CI).
RESULTS
Clinical characteristics among all participants by SARS-CoV-2 diagnosis.
Between 17 November 2020 and 1 October 2021, 327 participants were consented, among whom 301 presented for sample collection and provided information to evaluate their status as a close contact of a laboratory-confirmed SARS-CoV-2 case (Fig. 1; see also Fig. S1 in the supplemental material). There were 235 close contacts (upon presentation, n = 66 were deemed not to meet close contact criteria) (Fig. 1) who comprise the analyzed cohort, among whom 95 were symptomatic and 140 were asymptomatic. Of the symptomatic contacts, 82% (78/95) had symptoms for ≤5 days. Table 1 shows the baseline characteristics of the participants. Overall, the median age was 38.0 (interquartile range [IQR], 29.0 to 50.5), and 52.3% were male. There were 148 (63%) participants who reported having health care coverage. The standard clinical reference in NP samples was positive for 53 (23%) participants. Those who were SARS-CoV-2 test positive compared to those who were negative were more likely to be unvaccinated (83.0% versus 71.4%; P = 0.050), black (22.6% versus 15.9%), and of Hispanic ethnicity (54.7% versus 39.6%). Among the cohort of COVID-19 contacts, 32.1% of those who were test positive were asymptomatic, which may include contacts in the early, presymptomatic phase or asymptomatic infections. The most common symptoms among the NP swab SARS-CoV-2-positive patients were cough (41.5%), runny nose (41.5%), muscle aches (35.8%), fever or chills (32.1%), and scratchy throat (26.4%).
FIG 1.
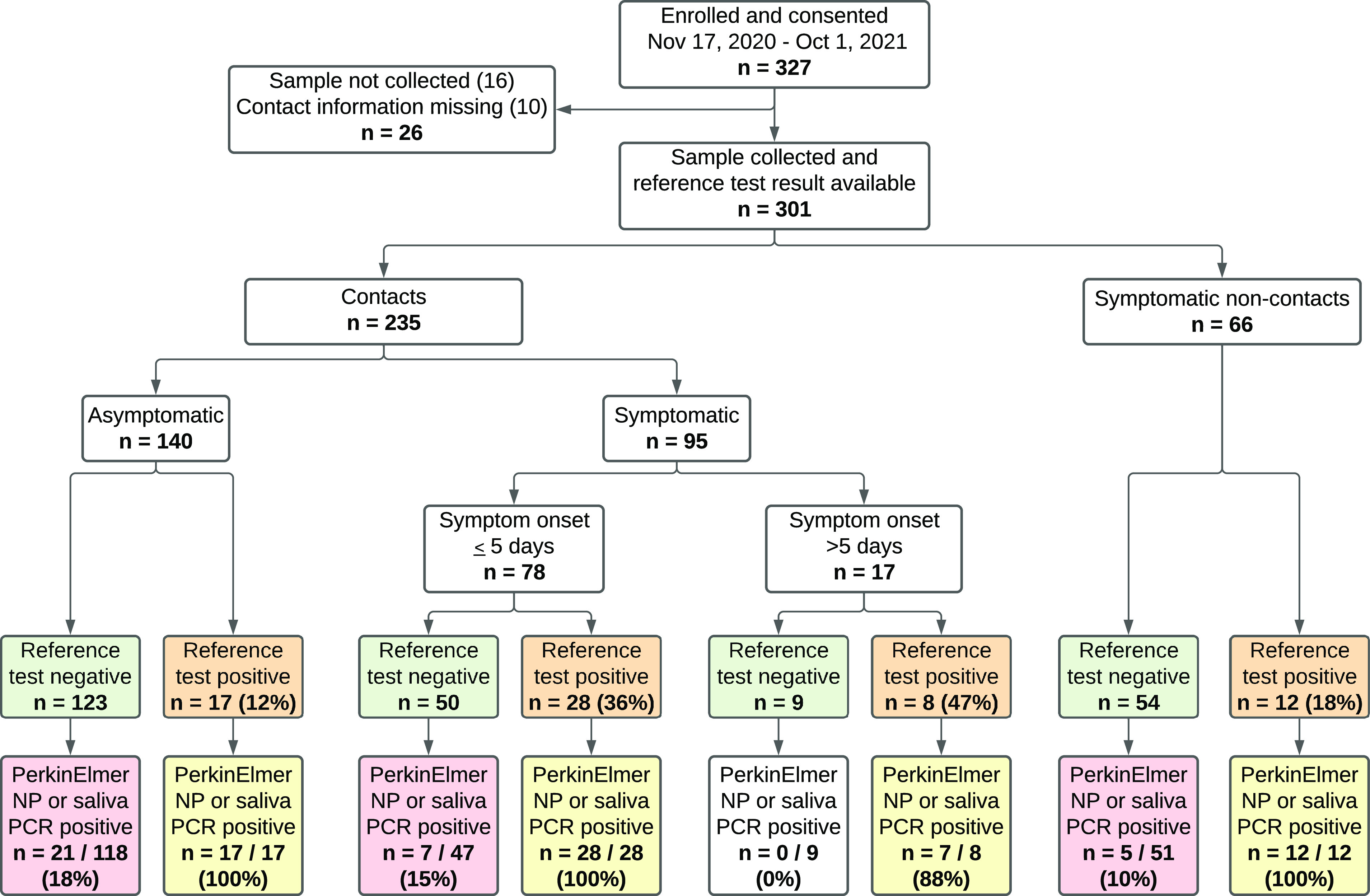
Participants enrolled and molecular test results.
TABLE 1.
Clinical characteristics among close contacts stratified by SARS-CoV-2 reference test result
| Clinical characteristica | Total no. (%) contacts | No. (%) reference test negative | No. (%) reference test positive | P value |
|---|---|---|---|---|
| n = 235 | n = 182 | n = 53 | ||
| Age, median (IQR) | 38.0 (29.0–50.5) | 37.0 (29.2–50.8) | 41.0 (27.0–49.0) | 0.495 |
| Male | 123 (52.3) | 95 (52.2) | 28 (52.8) | 1.000 |
| Race and ethnicity | 0.068 | |||
| Asian | 3 (1.3) | 3 (1.6) | 0 (0.0) | |
| Black or African American | 41 (17.4) | 29 (15.9) | 12 (22.6) | |
| Hispanic | 101 (43.0) | 72 (39.6) | 29 (54.7) | |
| Other | 2 (0.9) | 2 (1.1) | 0 (0.0) | |
| White | 88 (37.4) | 76 (41.8) | 12 (22.6) | |
| Vaccine status | 0.050 | |||
| Fully vaccinated | 43 (18.3)b | 39 (21.4) | 4 (7.5) | |
| Not vaccinated | 174 (74.0) | 130 (71.4) | 44 (83.0) | |
| Partially vaccinated | 18 (7.7) | 13 (7.1) | 5 (9.4) | |
| Tobacco use | 23 (9.8) | 17 (9.3) | 6 (11.3) | 0.870 |
| Health care coverage | 148 (63.0) | 117 (64.3) | 31 (58.5) | 0.544 |
| Diabetes | 15 (6.4) | 11 (6.0) | 4 (7.5) | 0.750 |
| Kidney failure | 2 (0.9) | 1 (0.5) | 1 (1.9) | 0.401 |
| Hypertension | 37 (15.7) | 26 (14.3) | 11 (20.8) | 0.356 |
| Cancer, under active treatment | 1 (0.4) | 1 (0.5) | 0 (0.0) | 1.000 |
| Asthma or COPD | 22 (9.4) | 17 (9.3) | 5 (9.4) | 1.000 |
| Symptom onset | <0.001 | |||
| Asymptomatic | 140 (59.6) | 123 (67.6) | 17 (32.1) | |
| Symptomatic, onset >5 days or unknown | 17 (7.2) | 9 (4.9) | 8 (15.1) | |
| Symptomatic, onset within 5 days | 78 (33.2) | 50 (27.5) | 28 (52.8) | |
| Scratchy throat | 32 (13.6) | 18 (9.9) | 14 (26.4) | 0.004 |
| Painful sore throat | 25 (10.6) | 14 (7.7) | 11 (20.8) | 0.014 |
| Cough worse than usual | 40 (17.0) | 18 (9.9) | 22 (41.5) | <0.001 |
| Runny nose | 45 (19.1) | 23 (12.6) | 22 (41.5) | <0.001 |
| Symptoms of fever or chills | 25 (10.6) | 8 (4.4) | 17 (32.1) | <0.001 |
| Temperature >38.0°C | 13 (5.5) | 4 (2.2) | 9 (17.0) | <0.001 |
| Muscle aches | 31 (13.2) | 12 (6.6) | 19 (35.8) | <0.001 |
| Nausea, vomiting, diarrhea | 17 (7.2) | 10 (5.5) | 7 (13.2) | 0.071 |
| Shortness of breath | 20 (8.5) | 9 (4.9) | 11 (20.8) | 0.001 |
| Unable to taste or smell | 11 (4.7) | 2 (1.1) | 9 (17.0) | <0.001 |
| Red or painful eyes | 13 (5.5) | 5 (2.7) | 8 (15.1) | 0.002 |
IQR, interquartile range; COPD, chronic obstructive pulmonary disease.
Among asymptomatic close contacts, 28 (20%) were fully vaccinated; among symptomatic close contacts, 15 (16%) were fully vaccinated.
Molecular test and viral culture results.
The NP swab reference NAT was positive for 22.6% (53/235) of participants who were close contacts of SARS-CoV-2 cases (Table 2; see Fig. S1). Among 50 of 53 reference-positive samples which underwent viral culture, 25 (50%) were culture positive. Remnant frozen NP swab VTM for 158 of the 182 SARS-CoV-2 close contacts who tested negative by the reference test were retested by PerkinElmer RT-PCR and positive for 13 (8%) samples with a median cycle threshold (CT) of 36.8 (IQR, 36.0 to 38.6) (see Table S1 in the supplemental material). Saliva testing using PerkinElmer RT-PCR was performed for 186 (79%) close contacts, including 143 of 182 (79%) reference test-negative participants. Saliva was positive for 37 of 43 (86%) reference-positive participants and 17 of 143 (12%) reference-negative participants. Combined, PerkinElmer RT-PCR of NP and saliva samples identified SARS-CoV-2 in an additional 21 asymptomatic close contacts and 7 symptomatic close contacts not identified by the NP reference test. The median CT value of symptomatic close contacts was lower than that of asymptomatic close contacts for both NP (22.0 with an IQR of 17.7 to 28.5 versus 34.8 with an IQR of 31.4 to 36.9) and saliva (25.8 with an IQR of 22.5 to 31.5 versus 33.4 with an IQR of 26.5 to 36.7) samples. Among the 67 participants that tested positive by any NAT and had a saliva NAT result, the sensitivity of saliva NAT was 80.6% and NP was 77.6%. Among symptomatic patients, patients with systemic symptoms of fever and muscle ache were more likely to test positive by both NP and saliva samples, and patients with runny nose were more likely to test positive by NP only.
TABLE 2.
SARS-CoV-2 testing among contacts by presence of symptoms
| Assay and result | Total no. (%) | No. (%) asymptomatic | No. (%) symptomatic | P value |
|---|---|---|---|---|
| Reference NAT, NP | 235 | 140 | 95 | <0.001 |
| Positive | 53 (22.6) | 17 (12.1) | 36 (37.9) | |
| Negative | 182 (77.4) | 123 (87.9) | 59 (62.1) | |
| Reference CT value (IQR), NP (n = 51)a | 24.7 (20.4–31.1) | 30.9 (25.3–32.2) | 21.8 (19.4–30.0) | 0.002 |
| Viral cultureb | 50 | 16 | 34 | 0.077 |
| Positive for COVID-19 | 25 (50.0) | 5 (31.2) | 20 (58.8) | |
| Negative for COVID-19 | 23 (46.0) | 11 (68.8) | 12 (35.3) | |
| Inconclusive | 2 (4.0) | 0 (0.0) | 2 (5.9) | |
| PerkinElmer, NPc | 209 | 129 | 80 | 0.004 |
| Positive | 61 (29.2) | 28 (21.7) | 33 (41.2) | |
| Negative | 148 (70.8) | 101 (78.3) | 47 (58.8) | |
| PerkinElmer CT value (IQR), NP | 30.0 (19.9–34.3) | 34.8 (31.4–36.9) | 22.0 (17.7–28.5) | <0.001 |
| PerkinElmer, Salivad | 186 | 108 | 78 | 0.010 |
| Positive | 54 (29.0) | 23 (21.3) | 31 (39.7) | |
| Negative | 132 (71.0) | 85 (78.7) | 47 (60.3) | |
| PerkinElmer CT value (IQR), Saliva | 28.9 (23.0–34.8) | 33.4 (26.5–36.7) | 25.8 (22.5–31.5) | 0.020 |
| BD Veritor (antigen), midturbinate | 99 | 55 | 44 | <0.001 |
| Positive | 14 (14.1) | 1 (1.8) | 13 (29.5) | |
| Negative | 85 (85.9) | 54 (98.2) | 31 (70.5) |
Reference NAT for 2 participants was performed on Panther; CT values not available.
Evaluable culture results were not available for 3 of the reference NAT-positive samples.
Twenty-six samples were insufficient for PerkinElmer testing.
dForty-nine participants either did not produce saliva or had insufficient quantities to test.
Antigen test results.
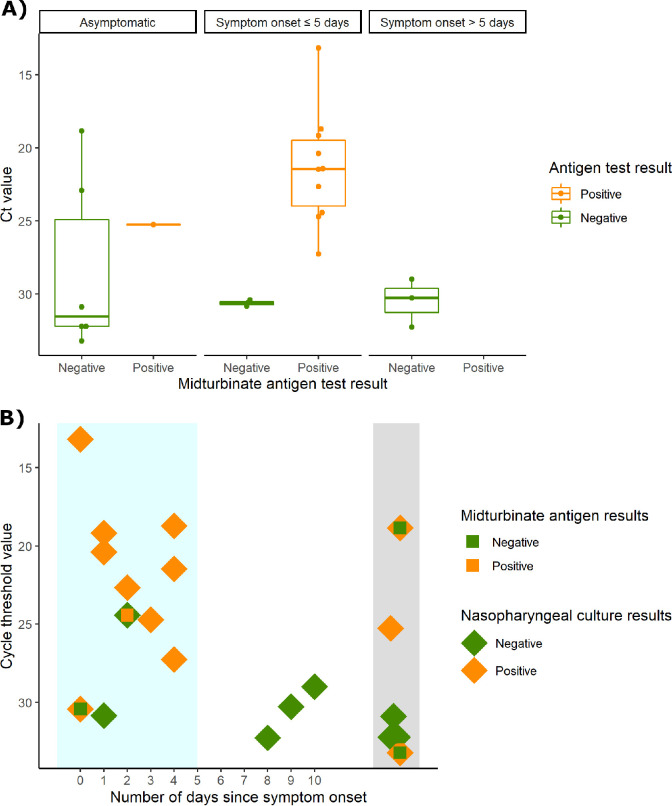
From 17 November 2020 to 11 March 2021, the initial 126 participants underwent antigen testing of a midturbinate swab using the BD Veritor lateral flow assay, and among these participants, 99 were close contacts. Among close contacts who underwent antigen testing, 55 (56%) were asymptomatic, 35 (35%) were symptomatic and within the first 5 days of symptom onset, and 9 (9%) were symptomatic but beyond 5 days of symptom onset. Among symptomatic close contacts in the first 5 days of symptom onset (intended use case for the BD Veritor test), the positive percent agreement (PPA) was 83.3% (95% CI, 50.9 to 97.1) and the negative percent agreement was 95.7% (95% CI, 76.0 to 99.8) compared to the reference molecular test. However, among asymptomatic persons or persons after 5 days of symptom onset, the PPA was 18.2% (95% CI, 3.2 to 52.2). The median reference test CT for participants who tested positive by reference test and negative by BD Veritor was 31.6 for asymptomatic participants, 30.8 for symptomatic in the first 5 days of symptom onset, and 30.3 for participants beyond 5 days of symptom onset (Fig. 2A). Among the 11 close contacts that were culture positive with BD Veritor results, 8 (72.7%) were positive by BD Veritor (Fig. 2B). Among the 3 that were missed, 2 were asymptomatic.
FIG 2.
Antigen test positivity of contacts. (A) Cycle threshold (CT) values stratified by asymptomatic, symptom onset of ≤5 days, and symptom onset of >5 days for the RT-PCR-positive reference assays, for which there were antigen lateral flow results (n = 99). (B) Culture, antigen result, and CT of reference assay for all RT-PCR-positive NP samples arrayed by early symptomatic (blue shaded), symptomatic (white), and asymptomatic (gray).
DISCUSSION
In this study, we report the relative performance of multiple molecular test approaches in a large number of known SARS-CoV-2 NAT-confirmed case contacts. As expected, a high proportion (22.6%) of these contacts were reference lab NP NAT positive. However, an additional 11.6% were positive when both NP VTM and saliva were tested using a more sensitive platform; more than half of asymptomatic close contacts tested falsely negative using a high-complexity reference lab test. Patients with symptomatic SARS-CoV-2 were less likely to be missed by standard NAT testing, and there were no missed cases among patients who were not close contacts. Although prior data has shown that approximately half of patients who eventually test positive for SARS-CoV-2 initially test negative and test performance is worse in the first few days after exposure and before symptom onset (17, 18), work describing the potential to detect additional cases using more sensitive assays and additional specimen types is limited.
The FDA requires that SARS-CoV-2 NATs demonstrate concordance with existing authorized tests to gain EUA (19), which gives the impression that the clinical performance of all EUA SARS-CoV-2 NATs are approximately equivalent. However, among SARS-CoV-2 NATs with FDA EUA, there is a 100-fold difference between the highest and lowest levels of detection (LOD) reported using comparable FDA reference materials (20). In this study, testing using the PerkinElmer NAT, which has the lowest reported LOD, identified many cases missed by standard reference tests. Another source of missed positives were samples that tested positive in saliva but not NP samples. Similar to prior work, saliva was not more or less sensitive than NP samples, but it did identify a population of SARS-CoV-2 cases that was only partially overlapping with those found using NP samples (21–23). If testing resources were not scarce, a strategy of performing high sensitivity NAT on both saliva and NP specimens may be considered to exclude infection in patients with the highest pretest probability for infection.
Although antigen testing identified the majority of culture-positive contacts, a few patients with culture-positive SARS-CoV-2 were missed. It is expected that antigen testing will miss a proportion of SARS-CoV-2 cases, particularly those with lower viral burden (identified by higher CT values) and particularly among asymptomatic individuals; a recent meta-analysis reported a sensitivity of 58.1% (95% CI, 40.2% to 74.1%) (24), and more focused studies including asymptomatic close contacts reported a sensitivity of 59 to 82% compared to NAT (8, 25). However, the hope has been that persons with potential to transmit SARS-CoV-2 may be excluded, at least in the moment, by a negative antigen test (15, 26, 27). Used as intended, in the first 5 days of symptom onset, only one patient with culture-positive SARS-CoV-2 tested negative by antigen testing, but two asymptomatic patients who tested negative by antigen tests had positive cultures. Recent data from our group and others have shown that serial testing may be a strategy to overcome the lack of sensitivity in presymptomatic patients (23, 28). Daily antigen testing is now being used by some school systems to forego quarantine after exposure (29, 30).
False negative SARS-CoV-2 test results for asymptomatic close contacts may provide false reassurance, leading to relaxation of isolation measures and greater onward transmission. Our findings suggest that for asymptomatic close contacts with a high pretest probability for infection, testing at a single time point with a standard reference NAT misses some cases, and negative antigen testing does not exclude transmissibility. SARS-CoV-2 exposures that are to household members, indoors, and prolonged pose a higher risk of transmission (31), which may warrant additional vigilance, including repeat testing for close contacts with high pretest probability for infection. Newer SARS-CoV-2 variants, including delta and omicron, have higher capacity for transmission than earlier circulating genotypes (32, 33). Therefore, the consequences for each false-negative test on community transmission may be greater now that currently circulating variants are more transmissible than earlier circulating SARS-CoV-2.
Limitations include the possibility that other commercially available antigen tests may outperform the one chosen for use in this study, although direct comparisons of the Veritor antigen test used in this study has shown similar performance characteristics to others (8, 34). Similarly, differences in the analytical sensitivity of SARS-CoV-2 test platforms may impact the results if this study were to be replicated using alternative SARS-CoV-2 NAT. The antigen tests were not performed as intended at the point of collection, which could reduce their sensitivity. Most of the positive cases were from unvaccinated persons prior to circulation of delta and omicron variants—as more vaccinated persons test positive for COVID, new data regarding the relationship between pretest probability of infection, test performance characteristics, SARS-CoV-2 variant, and culture positivity may be required. Data regarding timing of participant exposure to an individual with COVID were not available, which limits the ability to identify subgroups of close contacts with better or worse test performance.
Ongoing SARS-CoV-2 transmission with increasingly contagious variants highlights the continued importance of widespread and accurate testing even as the pandemic approaches its third year. Our findings in a large cohort of close contacts demonstrate the limitations of cross-sectional antigen testing to exclude transmissibility and standard NAT to diagnose COVID-19. Close contacts of individuals with COVID-19 should isolate and may need to test more than once, especially those with highest risk exposures such as household contacts. Future work is necessary to improve test performance characteristics or implement serial testing to identify patients with SARS-CoV-2 when they are most likely to have transmissible disease.
ACKNOWLEDGMENTS
We acknowledge Madison Conte and Elena Konstant for their contributions to the processing and storage of specimens used in this study.
Becton Dickenson provided Veritor test kits for use in this study but did not contribute to the writing of this manuscript nor critically review its content.
This study was funded by the NIH RADx-Tech program under 3U54HL143541-02S2 and U54EB007958-12S1.
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Institute of Biomedical Imaging and Bioengineering; the National Heart, Lung, and Blood Institute; the National Institutes of Health, or the U.S. Department of Health and Human Services.
Salary support came from the National Institutes of Health U54EB007958-13 (Y.C.M., M.L.R., J.H.), AI272201400007C (A.P., Y.C.M.), UM1AI068613 (Y.C.M.), U54HL143541, R01HL141434, R01HL137784, R01HL155343, R61HL158541, R01HL137734 (D.D.M.).
D.D.M. reports consulting and research grants from Bristol Myers Squibb and Pfizer, consulting and research support from Fitbit, consulting and research support from Flexcon, a research grant from Boehringer Ingelheim, a consulting grant from Avania, nonfinancial research support from Apple Computer, and consulting/other support from Heart Rhythm Society. L.G.J. is on a scientific advisory board for Moderna on projects unrelated to SARS-CoV-2. H.H.M. reports receipt of research contracts from Bio-Rad, DiaSorin, and Hologic. Y.C.M. has received tests from Quanterix, Becton, Dickinson, Ceres, and Hologic for research-related purposes, consults for Abbott on subjects unrelated to SARS-CoV-2, and receives funding support to Johns Hopkins University from miDiagnostics.
Footnotes
Supplemental material is available online only.
Contributor Information
Matthew L. Robinson, Email: mrobin85@jhmi.edu.
Yi-Wei Tang, Cepheid.
REFERENCES
- 1.Dong E, Du H, Gardner L. 2020. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20:533–534. 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mackay MJ, Hooker AC, Afshinnekoo E, Salit M, Kelly J, Feldstein JV, Haft N, Schenkel D, Nambi S, Cai Y, Zhang F, Church G, Dai J, Wang CL, Levy S, Huber J, Ji HP, Kriegel A, Wyllie AL, Mason CE. 2020. The COVID-19 XPRIZE and the need for scalable, fast, and widespread testing. Nat Biotechnol 38:1021–1024. 10.1038/s41587-020-0655-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.U.S. Food and Drug Administration. 2020. Coronavirus (COVID-19) update: FDA publishes comparative performance data for COVID-19 molecular diagnostic tests. U.S. Food and Drug Administration, Washington, DC. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-publishes-comparative-performance-data-covid-19-molecular-diagnostic. [Google Scholar]
- 4.Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S, Emperador D, Takwoingi Y, Cunningham J, Beese S, Dretzke J, Ferrante di Ruffano L, Harris IM, Price MJ, Taylor-Phillips S, Hooft L, Leeflang MM, Spijker R, Van den Bruel A, Cochrane COVID-19 Diagnostic Test Accuracy Group. 2020. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev 8:CD013705. 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2021. Appendix A – glossary of key terms, close contact. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/php/contact-tracing/contact-tracing-plan/appendix.html#contact. Accessed 15 July 2021. [Google Scholar]
- 6.Marks M, Millat-Martinez P, Ouchi D, Roberts CH, Alemany A, Corbacho-Monné M, Ubals M, Tobias A, Tebé C, Ballana E, Bassat Q, Baro B, Vall-Mayans M, G-Beiras C, Prat N, Ara J, Clotet B, Mitjà O. 2021. Transmission of COVID-19 in 282 clusters in Catalonia, Spain: a cohort study. Lancet Infect Dis 21:629–636. 10.1016/S1473-3099(20)30985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sethuraman N, Jeremiah SS, Ryo A. 2020. Interpreting diagnostic tests for SARS-CoV-2. JAMA 323:2249–2251. 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 8.Schuit E, Veldhuijzen IK, Venekamp RP, van den Bijllaardt W, Pas SD, Lodder EB, Molenkamp R, GeurtsvanKessel CH, Velzing J, Huisman RC, Brouwer L, Boelsums TL, Sips GJ, Benschop KSM, Hooft L, van de Wijgert JHHM, van den Hof S, Moons KGM. 2021. Diagnostic accuracy of rapid antigen tests in asymptomatic and presymptomatic close contacts of individuals with confirmed SARS-CoV-2 infection: cross sectional study. BMJ 374:n1676. 10.1136/bmj.n1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2021. Interim guidance on developing a COVID-19 investigation & contact tracing plan: appendices. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/php/contact-tracing/contact-tracing-plan/appendix.html#contact. Accessed 8 December 2021. [Google Scholar]
- 10.NeuMoDx Molecular. 2020. NeuMoDx™ SARS-CoV-2 assay. NeuMoDx Molecular, Ann Arbor, MI. https://www.fda.gov/media/136565/download. [Google Scholar]
- 11.Hologic. 2020. Aptima SARS-CoV-2 assay (Panther® system). Hologic, San Diego, CA. https://www.fda.gov/media/138096/download. [Google Scholar]
- 12.PerkinElmer. 2021. Instructions for PerkinElmer new coronavirus nucleic acid detection kit. PerkinElmer, Austin, TX. https://www.fda.gov/media/136410/download. [Google Scholar]
- 13.Becton, Dickinson and Company. 2021. Veritor system for rapid detection of SARS-CoV-2. Becton, Dickinson and Company, Sparks, MD. https://www.fda.gov/media/139755/download. [Google Scholar]
- 14.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, Nagata N, Sekizuka T, Katoh H, Kato F, Sakata M, Tahara M, Kutsuna S, Ohmagari N, Kuroda M, Suzuki T, Kageyama T, Takeda M. 2020. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci USA 117:7001–7003. 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pekosz A, Parvu V, Li M, Andrews JC, Manabe YC, Kodsi S, Gary DS, Roger-Dalbert C, Leitch J, Cooper CK. 2021. Antigen-based testing but not real-time polymerase chain reaction correlates with severe acute respiratory syndrome coronavirus 2 viral culture. Clin Infect Dis 73:e2861–e2866. 10.1093/cid/ciaa1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waggoner JJ, Stittleburg V, Pond R, Saklawi Y, Sahoo MK, Babiker A, Hussaini L, Kraft CS, Pinsky BA, Anderson EJ, Rouphael N. 2020. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1633–1635. 10.3201/eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arevalo-Rodriguez I, Buitrago-Garcia D, Simancas-Racines D, Zambrano-Achig P, Del Campo R, Ciapponi A, Sued O, Martinez-García L, Rutjes AW, Low N, Bossuyt PM, Perez-Molina JA, Zamora J. 2020. False-negative results of initial RT-PCR assays for COVID-19: a systematic review. PLoS One 15:e0242958. 10.1371/journal.pone.0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. 2020. Variation in false-negative rate of reverse transcriptase polymerase chain reaction–based SARS-CoV-2 tests by time since exposure. Ann Intern Med 173:262–267. 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Food and Drug Administration. 2021. In vitro diagnostics EUAs. Food and Drug Administration, Washington, DC. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas. Accessed 8 December 2021. [Google Scholar]
- 20.Food and Drug Administration. 2020. SARS-CoV-2 reference panel comparative data. Food and Drug Administration, Washington, DC. https://www.fda.gov/medical-devices/coronavirus-covid-19-and-medical-devices/sars-cov-2-reference-panel-comparative-data. Accessed 8 December 2021. [Google Scholar]
- 21.Bastos ML, Perlman-Arrow S, Menzies D, Campbell JR. 2021. The sensitivity and costs of testing for SARS-CoV-2 infection with saliva versus nasopharyngeal swabs. Ann Intern Med 174:501–510. 10.7326/M20-6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manabe YC, Reuland C, Yu T, Azamfirei R, Hardick JP, Church T, Brown DM, Sewell TT, Antar A, Blair PW, Heaney CD, Pekosz A, Thomas DL, Cox A, Keller S, Keruly J, Klein S, Mehta S, Mostafa H, Pisanic N, Sauer L, Tornheim J, Townsend J, Armstrong D, Bachu V, Barnaba B, Charles C, Dai W, Ganesan A, Holden J, Jang M, Johnstone JR, Kruczynski K, Kusemiju O, Lambrou A, Li L, Littlefield K, Park H-S, Tuchler A, Montana MP, Prizzi M, Ursin R, Ambulatory COVID Team. 2021. Self-collected oral fluid saliva is insensitive compared with nasal-oropharyngeal swabs in the detection of severe acute respiratory syndrome coronavirus 2 in outpatients. Open Forum Infect Dis 8:ofaa648. 10.1093/ofid/ofaa648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith RL, Gibson LL, Martinez PP, Ke R, Mirza A, Conte M, Gallagher N, Conte A, Wang L, Fredrickson R, Edmonson DC, Baughman ME, Chiu KK, Choi H, Jensen TW, Scardina KR, Bradley S, Gloss SL, Reinhart C, Yedetore J, Owens AN, Broach J, Barton B, Lazar P, Henness D, Young T, Dunnett A, Robinson ML, Mostafa HH, Pekosz A, Manabe YC, Heetderks WJ, McManus DD, Brooke CB. 2021. Longitudinal assessment of diagnostic test performance over the course of acute SARS-CoV-2 infection. J Infect Dis 224:976–982. 10.1093/infdis/jiab337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dinnes J, Deeks JJ, Berhane S, Taylor M, Adriano A, Davenport C, Dittrich S, Emperador D, Takwoingi Y, Cunningham J, Beese S, Domen J, Dretzke J, Ferrante Di Ruffano L, Harris IM, Price MJ, Taylor-Phillips S, Hooft L, Leeflang MM, McInnes MD, Spijker R, Van Den Bruel A, Cochrane COVID-19 Diagnostic Test Accuracy Group. 2021. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev 3:CD013705. 10.1002/14651858.CD013705.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siddiqui ZK, Chaudhary M, Robinson ML, McCall AB, Peralta R, Esteve R, Callahan CW, Manabe YC, Campbell JD, Johnson JK, Elhabashy M, Kantsiper M, Ficke JR, Mostafa HH. 2021. Implementation and accuracy of BinaxNOW rapid antigen COVID-19 test in asymptomatic and symptomatic populations in a high-volume self-referred testing site. Microbiol Spectr 9:e01008-21. 10.1128/Spectrum.01008-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mina MJ, Peto TE, García-Fiñana M, Semple MG, Buchan IE. 2021. Clarifying the evidence on SARS-CoV-2 antigen rapid tests in public health responses to COVID-19. Lancet 397:1425–1427. 10.1016/S0140-6736(21)00425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petersen I, Crozier A, Buchan I, Mina MJ, Bartlett JW. 2021. Recalibrating SARS-CoV-2 antigen rapid lateral flow test relative sensitivity from validation studies to absolute sensitivity for indicating individuals shedding transmissible virus. Clin Epidemiol 13:935–940. 10.2147/CLEP.S311977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young BC, Eyre DW, Kendrick S, White C, Smith S, Beveridge G, Nonnenmacher T, Ichofu F, Hillier J, Oakley S, Diamond I, Rourke E, Dawe F, Day I, Davies L, Staite P, Lacey A, McCrae J, Jones F, Kelly J, Bankiewicz U, Tunkel S, Ovens R, Chapman D, Bhalla V, Marks P, Hicks N, Fowler T, Hopkins S, Yardley L, Peto TEA. 2021. Daily testing for contacts of individuals with SARS-CoV-2 infection and attendance and SARS-CoV-2 transmission in English secondary schools and colleges: an open-label, cluster-randomised trial. Lancet 398:1217–1229. 10.1016/S0140-6736(21)01908-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.State of Vermont Agency of Education. 2021. COVID-19 testing - test to stay. Vermont Agency of Education, Montpelier, VT. https://education.vermont.gov/covid19/testing-test-to-stay. Accessed 8 December 2021. [Google Scholar]
- 30.New York Department of Health. 2021. Updated testing and quarantine supplemental information. New York Department of Health, Albany, NY. https://coronavirus.health.ny.gov/system/files/documents/2021/11/tts-and-toq-memo.pdf. Accessed 8 December 2021. [Google Scholar]
- 31.Andrejko KL, Pry J, Myers JF, Openshaw J, Watt J, Birkett N, DeGuzman JL, Barbaduomo CM, Dong ZN, Fang AT, Frost PM, Ho T, Javadi MH, Li SS, Tran VH, Wan C, Jain S, Lewnard JA, Samani H, Walas N, Xavier E, Poindexter DJ, Dabbagh N, Spinosa MM, Saretha S, Cornejo AF, Park H, Bermejo MI, Lam A, Kaur A, Dyke A, Felipe D, Spencer M, Corredor S, Abdulrahim Y, California COVID-19 Case-Control Study Team. 21 December 2021. Predictors of SARS-CoV-2 infection following high-risk exposure. Clin Infect Dis 10.1093/cid/ciab1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen H, Vusirikala A, Flannagan J, Twohig KA, Zaidi A, Chudasama D, Lamagni T, Groves N, Turner C, Rawlinson C, Lopez-Bernal J, Harris R, Charlett A, Dabrera G, Kall M. 2022. Household transmission of COVID-19 cases associated with SARS-CoV-2 delta variant (B.1.617.2): national case-control study. Lancet Region Health Europe 12:100252. 10.1016/j.lanepe.2021.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.UK Health Security Agency. 2021. SARS-CoV-2 variants of concern and variants under investigation in England. Technical briefing 32. UK Health Security Agency, London, UK. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1042688/RA_Technical_Briefing_32_DRAFT_17_December_2021_2021_12_17.pdf. Accessed 20 December 2021. [Google Scholar]
- 34.Young S, Taylor SN, Cammarata CL, Varnado KG, Roger-Dalbert C, Montano A, Griego-Fullbright C, Burgard C, Fernandez C, Eckert K, Andrews JC, Ren H, Allen J, Ackerman R, Cooper CK, Miller MB. 2020. Clinical evaluation of BD Veritor SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS antigen point-of-care test. J Clin Microbiol 59:e02338-20. 10.1128/JCM.02338-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and Table S1. Download jcm.00187-22-s0001.pdf, PDF file, 0.6 MB (576.6KB, pdf)