Abstract
The candidate phyla radiation (CPR) is a large bacterial group consisting mainly of uncultured lineages. They have small cells and small genomes, and they often lack ribosomal proteins uL1, bL9, and/or uL30, which are basically ubiquitous in non-CPR bacteria. Here, we comprehensively analyzed the genomic information on CPR bacteria and identified their unique properties. The distribution of protein lengths in CPR bacteria peaks at around 100–150 amino acids, whereas the position of the peak varies in the range of 100–300 amino acids in free-living non-CPR bacteria, and at around 100–200 amino acids in most symbiotic non-CPR bacteria. These results show that the proteins of CPR bacteria are smaller, on average, than those of free-living non-CPR bacteria, like those of symbiotic non-CPR bacteria. We found that ribosomal proteins bL28, uL29, bL32, and bL33 have been lost in CPR bacteria in a taxonomic lineage-specific manner. Moreover, the sequences of approximately half of all ribosomal proteins of CPR differ, in part, from those of non-CPR bacteria, with missing regions or specifically added regions. We also found that several regions in the 16S, 23S, and 5S rRNAs of CPR bacteria are lacking, which presumably caused the total predicted lengths of the three rRNAs of CPR bacteria to be smaller than those of non-CPR bacteria. The regions missing in the CPR ribosomal proteins and rRNAs are located near the surface of the ribosome, and some are close to one another. These observations suggest that ribosomes are smaller in CPR bacteria than those in free-living non-CPR bacteria, with simplified surface structures.
Keywords: candidate phyla radiation, ribosome, rRNA, ribosomal protein, bioinformatics
INTRODUCTION
Studies of microbial communities based on sequence analyses of DNA extracted directly from the environment without culturing the microorganisms, such as metagenomic analyses or 16S rRNA gene sequencing, have revealed large numbers of microbial lineages that do not belong to known classification groups (Castelle and Banfield 2018). The candidate phyla radiation (CPR) is a monophyletic group in the bacterial domain that is mainly composed of uncultured lineages (Brown et al. 2015; Hug et al. 2016). At least 74 candidate phyla belonging to the CPR have been reported, and these bacteria are widely distributed in various environments, including soil, sediments, groundwater, fresh water, and the human oral cavity (Wrighton et al. 2012; Kantor et al. 2013; Rinke et al. 2013; Brown et al. 2015; Luef et al. 2015; Anantharaman et al. 2016). None of these bacteria have been cultured, except Saccharibacteria, derived from the human oral cavity. When prokaryotic genomes are clustered according to the presence or absence of 921 widely distributed protein families, CPR bacteria are clearly seen to have evolved separately from other bacteria (Meheust et al. 2019). On a phylogenetic tree of the three domains of life (Bacteria, Archaea, and Eukaryota) constructed by Hug et al. (2016) based on ribosomal proteins, the CPR diverge at the base of the bacterial domain, forming a clade separated from all other bacteria. However, the reliability of the branches at deep positions is considered poor on that phylogenetic tree. In contrast, in a bacterial phylogenetic analysis by Coleman et al., CPR is a sister group of the phylum Chloroflexi in the Terrabacteria group, which consists of multiple phyla (Coleman et al. 2021). Therefore, no conclusions have been reached regarding the phylogenetic position of the CPR bacteria. The scale of the diversity among the CPR bacteria is also unclear. The phylogenetic analysis of Hug et al. (2016) suggested that CPR bacteria account for about half of all bacterial diversity. In contrast, Parks et al. (2017) predicted that CPR bacteria account for 26.3% of all bacterial diversity. In either case, the presence of CPR cannot be ignored in any discussion of bacterial diversity and evolution.
Regardless of the extent of their phylogenetic diversity, the CPR bacteria have some common characteristics. First, they have small cells, reflected in the fact that they are abundant in samples that have been passed through filters with a pore size of 0.2 µm (Miyoshi et al. 2005; Brown et al. 2015; Luef et al. 2015) and in electron microscopic observations (He et al. 2015; Luef et al. 2015). Their genomes are also small, and most genomic sequences determined with metagenomics or single-cell genomics indicate sizes of ≤1.5 Mb, which is close to those of symbiotic bacterial genomes. These genomes lack genes of important metabolic pathways, such as the tricarboxylic acid cycle and amino acid and nucleotide biosynthesis pathways (Kantor et al. 2013; Nelson and Stegen 2015; Danczak et al. 2017; Suzuki et al. 2017; Castelle et al. 2018). The characteristics of CPR bacteria, such as their small genomes and reduced biosynthetic capacity, are similar to those of symbiotic organisms. However, the lifestyles of most CPR bacteria have not been clarified, except for Saccharibacteria, which attaches to a bacterium of the genus Actinomyces (He et al. 2015), and a member of the Parcubacteria, “Candidatus Sonnebornia yantaiensis,” which lives in the cytoplasm of Paramecium bursaria (Gong et al. 2014).
The distinctiveness of CPR bacteria also extends to their ribosome-related genes. The bacterial ribosome is composed of the 30S small (SSU) and 50S large (LSU) subunits, which consist of multiple ribosomal proteins and three rRNAs (16S rRNA in the SSU, and 23S and 5S rRNAs in the LSU) (Schuwirth et al. 2005). In Escherichia coli, the SSU contains 21 proteins and the LSU contains 33 proteins (Kaczanowska and Ryden-Aulin 2007). Some CPR bacteria contain one or more introns in their 16S rRNA and 23S rRNA genes, although these are very rare in other bacteria (Brown et al. 2015). All CPR bacteria lack the ribosomal protein uL30; uL1 is absent in some species of Parcubacteria (a CPR subgroup); and bL9 is absent in most Microgenomates (a CPR subgroup) and other candidate phyla, including Dojkabacteria, WWE3, and Saccharibacteria (Brown et al. 2015). Although these ribosomal proteins are not considered essential for bacterial survival, they are known to occur widely in bacteria, although not in some symbionts (Yutin et al. 2012; Nikolaeva et al. 2021). Because ribosomes have fundamental functions associated with basic life processes, their structures are considered highly conserved (Bernier et al. 2018). However, in recent years, large-scale studies of the distribution of ribosomal proteins have shown that bacteria with small genomes often lack certain ribosomal proteins. It has been proposed that the structures near the surface of the ribosome vary in regions that were acquired in the “late phase” of the molecular evolution of the ribosome (Yutin et al. 2012; Nikolaeva et al. 2021). In general, reduced genomes are found at the level of a single phylum or genus, whereas in CPR bacteria, small genomes occur throughout a large clade containing multiple phyla (Brown et al. 2015; Castelle et al. 2018). Therefore, understanding the structure of the CPR ribosome will not only clarify its origin but will also be important in any discussion of the diversity and evolution of bacterial ribosomes in general. However, although missing ribosomal proteins have been identified over a wide range of CPR bacterial lineages, little is known about the sizes and structures of individual proteins.
In recent years, the number of prokaryote genomes registered in public databases has increased steadily. Those of CPR bacteria are no exception, and approximately 70 complete genomes and many partial or draft genomes have been registered. In this study, we attempted to characterize the ribosomes of CPR bacteria based on a comparison of 69 complete and 828 draft genomes of CPR bacteria with known non-CPR bacterial genomes. The size distribution of all bacterial proteins predicted from each individual genome showed that CPR bacteria, like some parasites, have smaller proteins on average. Moreover, some CPR bacteria lack several more ribosomal proteins than have been noted previously. A comparison of the amino acid sequences of ribosomal proteins revealed regions that are absent only in CPR bacteria and regions that are present only in CPR bacteria. A comparison of the ribonucleotide sequences of each rRNA also revealed RNA regions that are only absent in CPR bacteria or only present in CPR bacteria. In three-dimensional ribosomal structures, these missing regions in rRNAs and ribosomal proteins are unevenly distributed on the ribosomal surface. These results suggest that the ribosomes of CPR bacteria are small, with relatively simple surface structures.
RESULTS AND DISCUSSION
Smaller proteins in CPR bacteria
To characterize the CPR bacterial genomes using as many examples as possible, we comprehensively compared the gene lengths of CPR bacteria with those of other well-known (non-CPR) bacteria. For this purpose, 69 complete and 828 draft genomes of CPR bacteria and 1661 complete genomes of non-CPR bacteria were collected from the National Center for Biotechnology Information (NCBI) database (Supplemental Tables S1–S4; Supplemental Fig. S1). The non-CPR bacteria included 167 endosymbiotic or parasitic lineages (non-CPR symbiotic) and 1494 other lineages (non-CPR free-living). Because symbiotic bacteria often have small genomes and/or reduced biosynthetic capacities, in common with CPR bacteria (Castelle et al. 2018), comparing CPR bacterial genomes with symbiotic bacterial genomes should highlight the similarities and differences among their reduced genomes. Based on 43 single-copy genes, the genomes of CPR bacteria were estimated to range from approximately 0.3 to 1.7 Mb. In contrast, the genome sizes of free-living and symbiotic non-CPR bacteria were calculated to range in size from approximately 1.3 to 13.0 Mb and from 0.3 to 8.8 Mb, respectively. This confirms that CPR bacterial genomes are as small as those of symbiotic bacteria, as reported previously (Castelle et al. 2018).
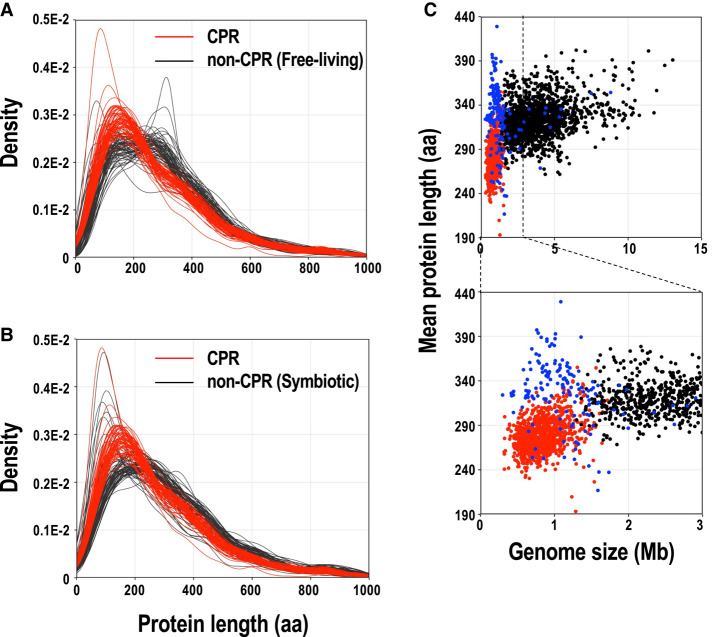
A total of 721,344 proteins (191–1723 proteins per genome) of CPR bacteria were identified with the gene-finding program Prodigal (Hyatt et al. 2010), and 5,388,587 proteins (237–10,234 proteins per genome) of the non-CPR bacteria were identified according to the genome annotations in NCBI RefSeq. Figure 1A,B show the distribution of protein lengths in the 69 complete genomes of CPR bacteria and in representative genomes of non-CPR bacteria (70 free-living and 70 symbiotic species), respectively, together with density curves (see Supplemental Fig. S2 for the distribution of protein lengths in all genomes used in this study). In the distribution of protein lengths of CPR bacteria, the peak was at around 100–150 amino acids, whereas the position of the peak varied in the range of 100–300 amino acids in free-living non-CPR bacteria (Fig. 1A). Among the free-living non-CPR bacteria, some genomes show multimodal distributions of protein lengths, with one peak at approximately 150 amino acids and another at approximately 300 amino acids (e.g., Bacteroides fragilis and Bacillus subtilis). Compared with the distribution of protein lengths in free-living non-CPR bacteria, CPR bacteria tended to have a higher proportion of proteins of ≤250 amino acids. These results show that CPR bacteria not only have smaller genomes, they also have smaller proteins, on average, than free-living non-CPR bacteria. Among the non-CPRs, the peak in the protein length distribution was around 100–200 amino acids in most symbiotic bacteria, which is closer to that of CPR bacteria than to that of other (free-living) non-CPR bacteria (Fig. 1B). For example, the protein length distributions of TC1 (an endosymbiont of Trimyema compressum), Spiroplasma mirum, Coxiella burnetii, and Wolbachia sp., which are intracellular endosymbionts or pathogens with genomes of <1.6 Mb, were highly skewed to the left, with sharp peaks at around 100 amino acids.
FIGURE 1.
Comparison of the lengths of proteins encoded in complete CPR and non-CPR genomes. (A) Comparison of the lengths of proteins from CPR and non-CPR (free-living species) bacteria. (B) Comparison of the lengths of proteins from CPR and non-CPR (symbiotic species) bacteria. Distributions of lengths of proteins are shown as density curves using the deduced amino acid sequences of the protein genes for 69 CPR bacteria (red line) and 140 representative non-CPR bacteria (70 free-living lineages and 70 symbiotic lineages; each black line). (C) CPR bacteria have smaller genomes with shorter proteins. We used 897 CPR bacterial genomes (69 complete and 828 draft genomes; red dots), 167 symbiotic non-CPR bacterial genomes (blue dots), and 1494 free-living non-CPR bacterial genomes (black dots) in the analysis. The lower panel is an enlarged view of part of the upper panel.
Plots of the mean protein length per genome (Fig. 1C) showed similar results. The mean protein length per CPR bacterial genome (193–355 amino acids; mean, 279 amino acids) was smaller than that of free-living non-CPR bacteria (262–402 amino acids; mean, 325 amino acids; P < 0.01, Mann–Whitney U-test). Although most symbiotic non-CPR bacteria have small genomes like CPR bacteria, the mean protein length in their genomes (217–429 amino acids; mean, 329 amino acids) was larger than that in CPR genomes (P < 0.01, Mann–Whitney U-test). As an exception, some non-CPR bacteria, such as TC1 (217 amino acids, on average), Coxiella sp. (237 amino acids long, on average), and Spiroplasma citri (237 amino acids long, on average), had particularly small mean protein lengths. Interestingly, the mean protein length correlated weakly with genome size in the CPR bacteria (R = 0.36, P < 0.01) and free-living non-CPR bacteria (R = 0.30, P < 0.01), but no correlation was observed in symbiotic non-CPR bacteria.
Lack of certain ribosomal proteins in CPR bacterial lineages
The CPR genomes examined in this study were classified into 65 candidate phyla with reference to the NCBI taxonomy (Schoch et al. 2020) and a previously reported phylogenetic tree (Hug et al. 2016). Some CPR phyla were classified into subgroups according to the recent phylogenetic tree by Jaffe et al. (2020), including Microgenomates, Parcubacteria 1–4, and other Parcubacteria (Supplemental Table S1A; Supplemental Fig. S1). To comprehensively extract the 54 ribosomal proteins that are widely distributed in bacteria from the genomes of the CPR bacteria, we obtained known ribosomal protein sequences registered in the NCBI Clusters of Orthologous Groups of proteins (COG) database (Supplemental Table S5; Galperin et al. 2021b) and domain data for the ribosomal proteins registered in the Pfam database (Supplemental Table S6; Mistry et al. 2021). Using these data, the ribosomal proteins were extracted from the open reading frames (ORFs) in the CPR genomes, and 443–890 sequences for each ribosomal protein were obtained. The ribosomal protein sequences of non-CPR bacteria were extracted according to the annotations in RefSeq, and 1454–2613 sequences were obtained for each ribosomal protein.
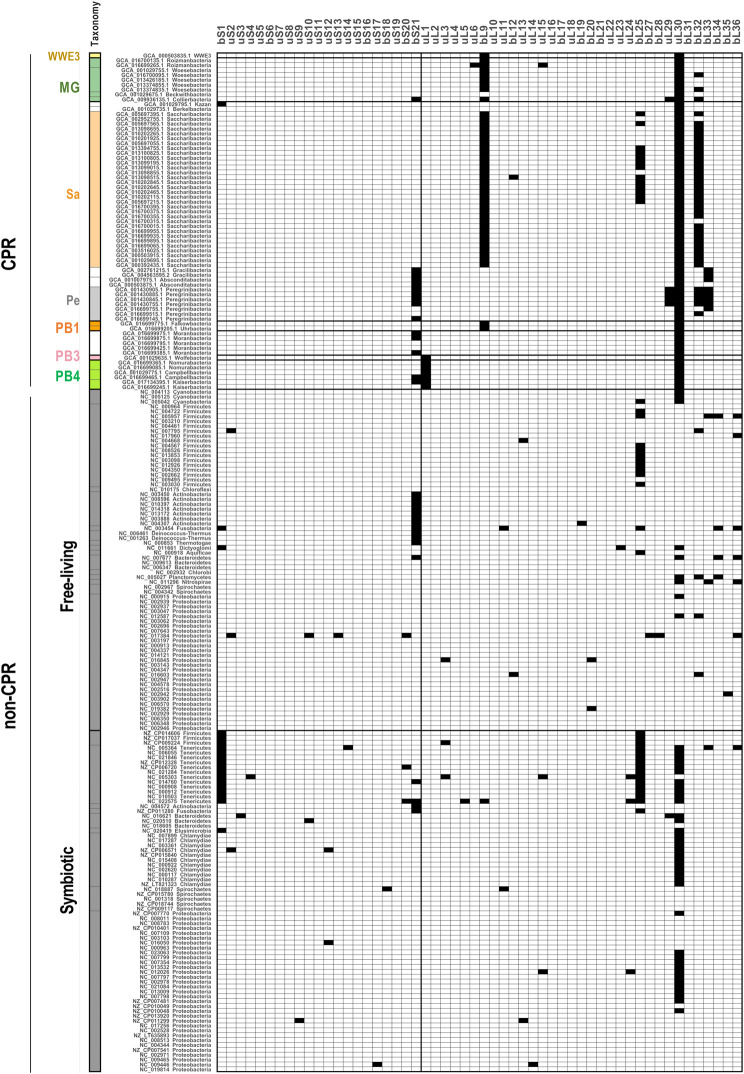
Figure 2 shows the presence or absence of each ribosomal protein in the 69 complete genomes of CPR bacteria and 140 representative non-CPR bacteria (70 free-living lineages and 70 symbiotic lineages). Consistent with a previous report (Brown et al. 2015), no uL30 was detected in the CPR bacteria. uL30 is reportedly encoded in a gene cluster and is located between uS5 and uL15 in many bacteria (Cerretti et al. 1983; Roberts et al. 2008), but uS5 and uL15 are located close to each other in CPR bacterial genomes. We also confirmed that uL1 is lacking in a subgroup of Parcubacteria, and that bL9 is lacking in Microgenomate (excluding Beckwithbacteria), Saccharibacteria, WWE3, and Dojkabacteria.
FIGURE 2.
Presence and absence of ribosomal proteins encoded in complete CPR and non-CPR genomes. The distributions of 54 ribosomal proteins (columns) across representative bacterial genomes (rows) are shown. The presence (white) or absence (black) of each ribosomal protein is indicated for 69 complete genomes of CPR bacteria and 140 complete genomes of representative non-CPR bacteria (70 free-living lineages and 70 symbiotic lineages; see Supplemental Tables S2–S4). Ribosomal protein names follow the nomenclature proposed by Ban et al. (2014). The rows were sorted based on a phylogenetic tree (Jaffe et al. 2020), and the labels provide NCBI accession numbers and the taxonomy of each genome. The left panel shows the phylum-level classification and is colored according to taxonomic group (see Supplemental Fig. S1). MG: Microgenomates; Sa: Saccharibacteria; Pe: Peregrinibacteria; PB: Parcubacteria.
Recently reported complete CPR genomes also lack other ribosomal proteins. Clades containing Peregrinibacteria, Gracilibacteria, and Absconditabacteria frequently lacked bS21 and bL33, and some Peregrinibacteria also lacked uL29 and bL32. Saccharibacteria usually lacked bL32, except for one complete genome. Approximately half of the phyla also lacked bL25. Although the use of complete genomes is appropriate for assessing the loss of specific genes encoding ribosomal proteins, the number of CPR bacterial lineages for which complete genomes are available is currently limited, and 57% of the complete genomes used in the present study were concentrated in Saccharibacteria and Peregrinibacteria (Supplemental Table S1A). Therefore, to investigate the distribution of ribosomal proteins throughout the CPR bacteria, the frequency of loss of each ribosomal protein in each phylum was also calculated using all of the available data, including both the complete and draft genomes (Supplemental Fig. S3). Here, we targeted all CPR phyla for which three or more genomes could be obtained (either complete or draft). The ribosomal proteins that were not detected in >80% of the genomes from any of these phyla were then identified. The results showed that bS21 was also frequently absent from Parcubacteria, Collierbacteria in Microgenomates, as in Peregrinibacteria, Gracilibacteria, and Absconditabacteria (as mentioned above). bL28 was lacking in all of the draft genomes of Daviesbacteria of Microgenomates. In contrast, most ribosomal proteins were widely conserved in the non-CPR bacterial genomes, although ribosomal proteins bS1, bS21, bL25, and uL30 were absent across at least two non-CPR phyla (Fig. 2; Supplemental Fig. S3). These ribosomal proteins have already been reported being as readily lost in non-CPR bacteria (Yutin et al. 2012; Grosjean et al. 2014). In particular, Mollicutes, a phylum of symbiotic bacteria, always lacked at least two (and up to eight) ribosomal proteins. In summary, bS21, bL25, and uL30 were frequently absent from both CPR and non-CPR bacteria, and uL1, bL9, bL28, uL29, bL32, and bL33 were also preferentially absent from CPR. It seems that LSU proteins are more likely to be lacking in CPR than SSU proteins. Although these proteins are widely distributed in non-CPR, any ribosomal protein can be lost in a specific non-CPR bacterial genome, particularly in small genomes (Lecompte et al. 2002; Nikolaeva et al. 2021).
The pattern of gene lacking in the CPR bacteria is similar to that in the mitochondrial ribosomes. For example, bS21, bL9, bL25, bL28, uL29, uL30, bL32, and bL33 are often lacking in mitochondrial genomes (Maier et al. 2013; Petrov et al. 2019). Furthermore, in mitochondrial ribosomes, LSU proteins are more frequently lacking than SSU proteins (Maier et al. 2013). The core ribosomal proteins in the mitochondrial ribosome are also conserved in the CPR bacteria. Based on assembly maps, no ribosomal proteins are strongly dependent on the ribosomal proteins that are not encoded in some CPR genomes, such as bS21, uL1, bL9, bL25, uL30, bL32, and bL33 (Herold and Nierhaus 1987; Grondek and Culver 2004). In E. coli, bL25 is involved in the assembly of uL6, but this dependence is weak (Herold and Nierhaus 1987). Although bL28 is involved in binding bL33, together with uL3 and bL25 (Herold and Nierhaus 1987), Daviesbacteria (NCBI: txid 1752718) in the CPR bacteria does not encode bL28 but does encode bL33.
Many ribosomal proteins in CPR bacteria differ from those in non-CPR bacteria
We analyzed the presence or absence of ribosomal proteins in CPR bacteria and clarified the lineage-specific absence of several ribosomal proteins. Because the molecules that make up the ribosome interact in a complex manner, it is possible that the lack of individual specific ribosomal proteins causes changes in other proteins. Therefore, we examined the sizes and amino acid sequences of individual ribosomal proteins in CPR bacteria and compared them with those in non-CPR bacteria. The summary statistics for each ribosomal protein length are available in Supplemental Table S7, and the sequence alignment data are available in Supplemental Table S8. Figure 3 shows the size distributions and amino acid sequence alignments of three ribosomal proteins—uL13, uS19, and uL1—as examples of sequences that differ significantly between CPR and non-CPR bacteria. Although the uL13, uS19, and uL1 proteins each show constant size distributions in most species of non-CPR bacteria, the size distributions of these proteins in CPR bacteria have two peaks, and the proteins in one group are significantly smaller (uL13) or larger (uS19 and uL1) than the corresponding proteins in non-CPR bacteria (Fig. 3A–C). When these amino acid sequences were aligned, regions missing in particular lineages of CPR bacteria and regions present only in some CPR bacteria were detected. Because genes encoding ribosomal protein of abnormal lengths were sometimes found in CPR bacterial genomes, presumably as a result of sequencing or assembly errors, we focused only on features that were shared among closely related genomes. The ribosomal protein uL13 tends to lack its amino-terminal (approximately 12 amino acids) and carboxy-terminal (approximately 16 amino acids) regions in most Parcubacteria (Fig. 3D). In the carboxy-terminal region of the ribosomal protein uS19, an alanine- and lysine-rich region (approximately 25 amino acids) is specifically present in Parcubacteria and a paraphyletic group of Parcubacteria (Fig. 3E). As mentioned above, the ribosomal protein uL1 is lacking in a group of Parcubacteria (Fig. 2; Supplemental Fig. S3; Brown et al. 2015), and the amino acid sequences of the uL1 protein in lineages other than Parcubacteria also differ in some characteristics from those of non-CPR bacteria (Fig. 3F). In Berkelbacteria and in Microgenomates other than Woykebacteria, a few internal regions of the uL1 protein (approximately 20–40 amino acids, in total) are absent, and a specific region of about 40–130 amino acids is inserted at the amino terminus. This amino-terminal region is also present in Saccharibacteria, Woykebacteria, and some members of WWE3 in which the internal region is not missing. The carboxy-terminal region of the uL13 protein and the internal region of the uL1 protein that are missing in CPR bacteria correspond to regions containing highly conserved amino acid residues in non-CPR bacteria. The CPR-specific regions inserted at the carboxyl terminus of the uS19 protein and the amino terminus of the uL1 protein do not match any known functional domain registered in the Pfam database (Mistry et al. 2021).
FIGURE 3.
Multiple amino acid sequence alignments of three selected ribosomal proteins that differ in length between CPR and non-CPR bacteria. (A–C) Distributions of the lengths of three ribosomal proteins (A: uL13; B: uS19; and C: uL1) with distinctly different lengths in CPR and non-CPR bacteria. Density curves for each protein length were calculated based on all sequences in the data set (CPR: light red; non-CPR: light blue). The total number of sequences is indicated in each panel. The length of each ribosomal protein in E. coli is indicated with a black inverted triangle. (D–F) Multiple amino acid sequence alignments of three ribosomal proteins corresponding to panels A–C (D: uL13; E: uS19; and F: uL1) are shown in the order based on a phylogenetic tree (Hug et al. 2016). Representative sequences were selected from each CPR and non-CPR group, and poorly aligned columns were removed (see Materials and Methods). The consensus residues in each column are highlighted in a blue gradient, according to the percentage identity calculated when gaps were ignored. The magnification and aspect ratio of each alignment panel have been adjusted for clarity. The scale bars of the alignment columns (amino acid lengths) are shown at the top right. The number of sequences included in the alignment is shown at the bottom. Panels on the left side of the alignment are colored according to the taxonomic group of each sequence (see Supplemental Fig. S1). MG: Microgenomates; Sa: Saccharibacteria; Pe: Peregrinibacteria; PB: Parcubacteria.
Supplemental Figure S4 shows the length distributions and amino acid sequence alignments of 53 ribosomal proteins, excluding the uL30 protein, which is completely absent from CPR bacterial genomes. Surprisingly, in about half the ribosomal proteins, including the abovementioned three ribosomal proteins (uL13, uL1, and uS19), the positions and numbers of peaks in the size distributions differ significantly between CPR and non-CPR bacteria. Ribosomal proteins bS1, uS3, uS14, uL2, uL3, uL5, uL15, and bL20 tend to be smaller in CPR bacteria than in non-CPR bacteria. In particular, like the uL13 protein (Fig. 3A,D), uL2, uL3, uL5, and uL15 proteins are significantly smaller in some CPR bacteria than in non-CPR bacteria, and these small proteins lack an amino-terminal (uL2 and uL5 proteins), carboxy-terminal (uL15 protein), or internal region (uL3 protein). The bS1, uS3, uS14, and bL20 proteins have various patterns of length in non-CPR bacteria, but in most CPR bacteria they are similar in size to the smallest group of proteins in the non-CPR bacteria. Of these proteins, bS1 is known to consist of a different number of S1 domains, depending on the species (Machulin et al. 2019). In non-CPR bacteria, bS1 proteins with six domains account for around 60% of the total known bS1 proteins, whereas most bS1 proteins in CPR bacteria are composed of 3–4 S1 domains (Supplemental Fig. S4). The ribosomal proteins with regions that are specifically missing in CPR bacteria are uL1, uL2, uL3, uL5, uL13, and uL15. These proteins occur preferentially in the LSU, as do those proteins completely absent from CPR bacteria (Fig. 2). In addition to uS19 (Fig. 3B,E) and uL1 (Fig. 3C,F) mentioned above, several ribosomal proteins are large in the CPR bacteria; i.e., bS6, uS10, uS11, uS12, uS13, uS15, bS21, bL12, bL19, uL22, uL23, bL25, bL27, bL31, and bL32. In particular, regions specifically present in only a proportion of CPR bacteria were detected in the carboxy-terminal regions of the uS13 and uS15 proteins, in the amino-terminal region of the uL23 protein, and in the internal region of the bL27 protein. The internal region of uS12, which only occurs in Tenericutes, Chloroflexus, and some Firmicutes in non-CPR bacteria, is present in almost all uS12 sequences of CPR bacteria. Similar to the uL1 protein (Fig. 3C,F), the bS20 protein amino acid sequence has both a missing region (center) and a specific extra (carboxy-terminal) region. However, it should be noted that in the proteins with an additional amino-terminal region, the start codon of the ORF was predicted based on the location of the first start codon and is not necessarily the actual start codon. Although the folding structures and functions of the CPR-specific protein regions have not been characterized, in some examples, the carboxy-terminal extensions of the ribosomal proteins appear to improve the stability of rRNA folding and also contribute to the environmental adaptation of Thermus thermophilus (Melnikov et al. 2018).
Smaller rRNAs and insertion-sequence-containing rRNAs in CPR bacteria
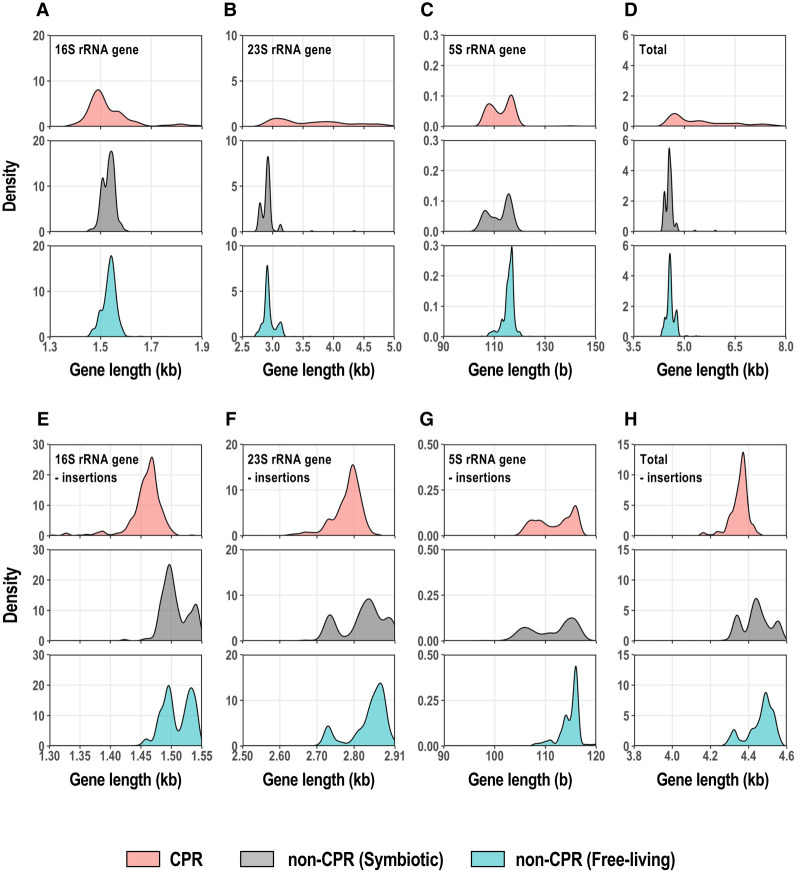
Ribosomal RNAs form the basis of the ribosome and play a major role in translation (Nissen et al. 2000). By comparing the sizes of the rRNA genes in CPR and non-CPR bacteria, we found that the rRNAs of CPR bacteria are rather small. With an Infernal search of rRNA secondary structure models, 375 full-length genes for 16S rRNA, 347 full-length genes for 23S rRNA, and 630 full-length genes for 5S rRNA were obtained from the complete and draft genomes of CPR bacteria. The CPR bacteria basically have one copy of each rRNA gene per genome. Figure 4A–D shows the distribution of rRNA gene lengths in CPR, symbiotic non-CPR, and free-living non-CPR bacteria. Some CPR bacteria have long 16S and 23S rRNA genes, containing an insertion sequence (or sequences; Fig. 4A,B). Most insertion sequences in the rRNA genes of CPR bacteria were predicted to be introns based on a comparison of the genes and their transcripts (Brown et al. 2015). In particular, we found that most 23S rRNAs in CPR bacteria contain insertion sequences, ranging from 0.5 kb to several kilobases in total length. However, because we have not compared all rRNA gene sequences with their transcripts, we refer to them as insertion sequences in this paper. When the insertion sequences were extracted, based on alignments with the rRNA genes of E. coli K-12, 42% of the 16S rRNA genes and 77% of the 23S rRNA genes of the CPR bacteria contained long insertions of ≥100 bases. The average length of insertions per gene was 623 bases (maximum 5.5 kb) for the 16S rRNA gene and 1232 bases (maximum 5.7 kb) for the 23S rRNA gene. Because the inserted regions were estimated by comparison with E. coli rRNAs, they do not exactly reflect the regions excluded after RNA splicing, but it is obvious that the 16S and 23S rRNA genes of CPR bacteria frequently contain insertions. The length profile of the 5S rRNA in CPR bacteria closely resembles that of parasitic non-CPR bacteria, with two peaks at 105 and 120 bases, whereas the peak in the 5S rRNA length distribution of free-living non-CPR bacteria is predominantly at 120 bases (Fig. 4C). Therefore, about half the 5S rRNAs in CPR bacteria are much smaller than those in free-living non-CPR bacteria.
FIGURE 4.
Length distributions of rRNA genes encoded in CPR and non-CPR genomes. (A–D) Density distributions of the whole-length 16S, 23S, and 5S rRNA genes and the total combined lengths of these three genes. Each horizontal axis is limited to the range in which the main peak occurs, and a few genes beyond this limit (A: 5.7%; B: 3.0%; C: 0.3%; and D: 1.5% of the total data) were omitted from the figures. (E–H) Density distributions of the 5S, 16S, and 23S rRNA gene lengths, excluding the insertion sequences, and their combined length for each genome. The numbers of data were: 16S rRNA genes (CPR bacteria, n = 375; symbiotic non-CPR bacteria, n = 167; free-living non-CPR bacteria, n = 1494), 23S rRNA genes (CPR bacteria, n = 347; symbiotic non-CPR bacteria, n = 167; free-living non-CPR bacteria, n = 1494), 5S rRNA genes (CPR bacteria, n = 630; symbiotic non-CPR bacteria, n = 167; free-living non-CPR bacteria, n = 1494), and the three genes combined (CPR bacteria, n = 240; symbiotic non-CPR bacteria, n = 167; free-living non-CPR bacteria, n = 1494). Density curves are colored according to group (CPR, light red; symbiotic non-CPR, light gray; free-living non-CPR, light blue).
We removed the insertion sequences from each CPR rRNA gene based on a comparison with the corresponding E. coli rRNA genes to roughly estimate the length distributions of the mature rRNAs (after RNA processing) in CPR bacteria (Fig. 4E–G). The distributions of rRNA lengths in non-CPR bacteria showed 2–3 peaks for all rRNA types, and the proportion of shorter RNAs was greater in symbiotic bacteria than in free-living bacteria. In contrast, the size distributions of the 16S and 23S rRNA genes (without insertion sequences) in the CPR bacteria usually had one large peak. The standard size (∼1.47 kb) of 16S rRNA in CPR bacteria, estimated from the location of the peak, was about 30 bases smaller than the smaller 16S rRNA gene group in non-CPR bacteria (Fig. 4E). However, most of the 23S rRNAs in CPR bacteria had an intermediate size (∼2.80 kb) between the two peaks found in the 23S rRNA size distribution of the non-CPR bacteria. This was about 50 bases smaller than the larger 23S genes in the non-CPR bacteria (peak on the right in Fig. 4F), which accounted for the majority of non-CPR bacteria, although non-CPR also included smaller 23S rRNA genes, at ∼2.72 kb (peak on the left in Fig. 4F). Among the non-CPR bacteria, these very small 23S rRNAs were abundant in the Proteobacteria, whereas among the CPR bacteria, Magasanikbacteria of Parcubacteria also had small 23S rRNAs. The size distributions of the 5S rRNAs of CPR bacteria displayed two peaks, similar to the distribution in parasitic bacteria, and the smaller gene group, which accounted for about half the total genomes, had 105–110 bases and occurred mainly in Parcubacteria (Fig. 4G). In free-living non-CPR bacteria, the proportion of 5S rRNA genes with ≤110 bases was only 8%. Summing the lengths of the three rRNA genes and excluding each insertion sequence, the most frequent relative lengths were in the following order: CPR < symbiotic non-CPR < free-living non-CPR (P < 0.01 for each pair, Bonferroni's test; Fig. 4H). The most frequent value for the total rRNA length in CPR bacteria (4.37 kb) was 118 bases shorter than that in non-CPR bacteria. The total rRNA length of some non-CPR bacteria was the same as that of the CPR bacteria, but more often in symbiotic bacteria than in free-living bacteria. These results suggest that the core regions of rRNAs are smaller in CPR bacteria than in typical non-CPR bacteria.
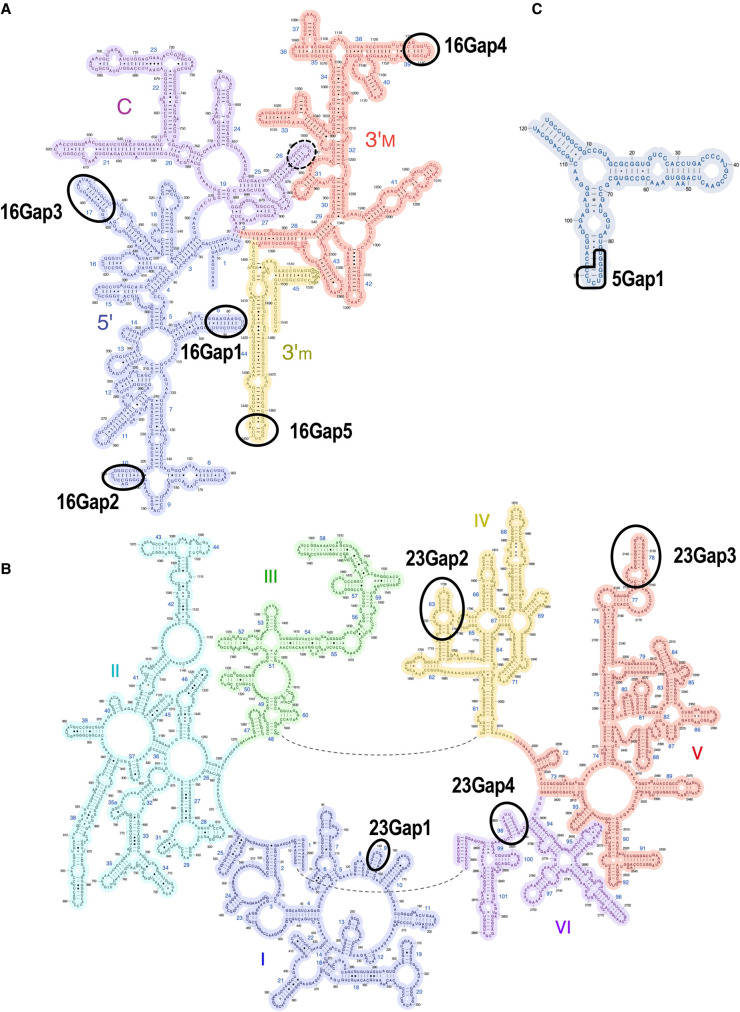
To investigate the smaller rRNA genes of the CPR bacteria at the nucleotide sequence level, a multiple alignment analysis of CPR and non-CPR rRNA genes was performed. Using the E. coli rRNA genes as the reference sequences, the insertion sequences were removed from each rRNA gene of the CPR bacteria, and the resulting nucleotide sequences of the CPR and non-CPR rRNA genes were compared. The results showed that all three types of rRNA genes in the CPR bacteria frequently lacked one region (in 5S rRNA) or several regions (in 16S and 23S rRNAs; Supplemental Fig. S5, alignment data are available in Supplemental Table S9). The 16S rRNA genes of the CPR bacteria had five gap regions (regions with higher gap rates in CPR bacteria than in non-CPR bacteria were designated 16Gap1–16Gap5), and the number of non-CPR bacterial lineages lacking regions 16Gap4 and 16Gap5 was extremely small. The 16Gap4 region was missing in the phylum Chloroflexi and in part of the class Tenericutes, which contains symbiotic bacteria, and the 16Gap5 region was missing in the phyla Bacteroidetes and Chlorobi. Although the anti-SD sequence in the 16S rRNA is rarely lost (Lim et al. 2012; Amin et al. 2018), the anti-SD motif (CCTCCT) (Nikolaeva et al. 2021) at the 3′ end of the 16S rRNA was detected less frequently in a subgroup within Parcubacteria (detection rate in Parcubacteria = 48%) in our data. The lineages without the anti-SD motif overlapped with most of the uL1-lacking group (Fig. 2; Supplemental Fig. S3), but the functional association between the anti-SD sequence and uL1 is unclear. In the 23S rRNA genes, four regions (designated 23Gap1–23Gap4) were lost throughout the CPR bacteria or in a lineage-specific manner. The 23Gap3 region was lost in some Parcubacteria 1 (e.g., Magasanikbacteria, Uhbacteria, and Falkowbacteria), Microgenomates, WWE3, and Berkelbacteria in the CPR bacteria, but in only two species of the non-CPR bacteria. The 5S rRNA genes of the CPR bacteria had one gap region (designated 5Gap1), which was missing in most Parcubacteria and some non-CPR bacteria. Unexpectedly, although there were missing regions in all three rRNA genes of CPR bacteria, a 16S rRNA gene region corresponding to nucleotides 837–850 of the E. coli gene was highly conserved in the CPR bacteria, especially in Parcubacteria and the paraphyletic group of Parcubacteria, although it was weakly conserved in the non-CPR bacterial data set and is reported to be commonly lost in small genomes (Nikolaeva et al. 2021).
Mapping the missing regions in each rRNA gene of the CPR bacteria against the known E. coli rRNA secondary structures revealed that all gap regions correspond to whole or the tips of particular stem–loop structures in each type of rRNA (Fig. 5). For example, the lack of 16Gap5 in the CPR bacteria indicates that the tip of helix 44 is lost and the helix is slightly shortened. Although helix 44 contributes to the accuracy of translation initiation (Qin et al. 2012), the position of 16Gap5 does not affect known functional sites. 5Gap1 in the 5S rRNA corresponds to helix IV and loop D, a region that shows large structural variation in bacteria, and can be lost (Szymanski et al. 2016; Stepanov and Fox 2021). Some rRNA helices are known to be deleted in small non-CPR genomes (Nikolaeva et al. 2021), but no case of 23SGap3 (the loss of 23S rRNA helix 78) has yet been reported. Therefore, the rRNAs of CPR bacteria have structures in which multiple stem–loops are lacking or shortened relative to the corresponding E. coli rRNA structures. In contrast, although all CPR bacteria lack uL30, they retain the sequence encoding the region in the vicinity of loop E, which contains the uL30 binding site for 5S rRNA (Sun and Caetano-Anolles 2009). The 16S rRNA region highly conserved in Parcubacteria and the paraphyletic group of Parcubacteria (surrounded by dashed lines in Fig. 5A) corresponds to helix h26, which interacts with the SD helix (base pairs with the SD sequence in mRNAs and the anti-SD sequence of 16S rRNA) and is considered to contribute to the start of translation (Korostelev et al. 2007). It is thought that the SD helices do not form in some Parcubacteria because they lack the anti-SD motif, but the role of conserved helix h26 is unknown.
FIGURE 5.
rRNAs of CPR bacteria lack terminal stem–loop domain(s) in their RNA secondary structures. RNA regions that are lacking in CPR bacteria are indicated by ellipses in the secondary structures of (A) 16S, (B) 23S, and (C) 5S rRNAs of E. coli. The dashed line indicates a region highly conserved in CPR bacteria. See Supplemental Figure S5 for details. These RNA secondary structures were obtained from the Center for Molecular Biology of RNA (http://rna.ucsc.edu/rnacenter/ribosome_images.html).
Ribosomes of CPR bacteria lack RNA and protein regions present on the ribosomal surface
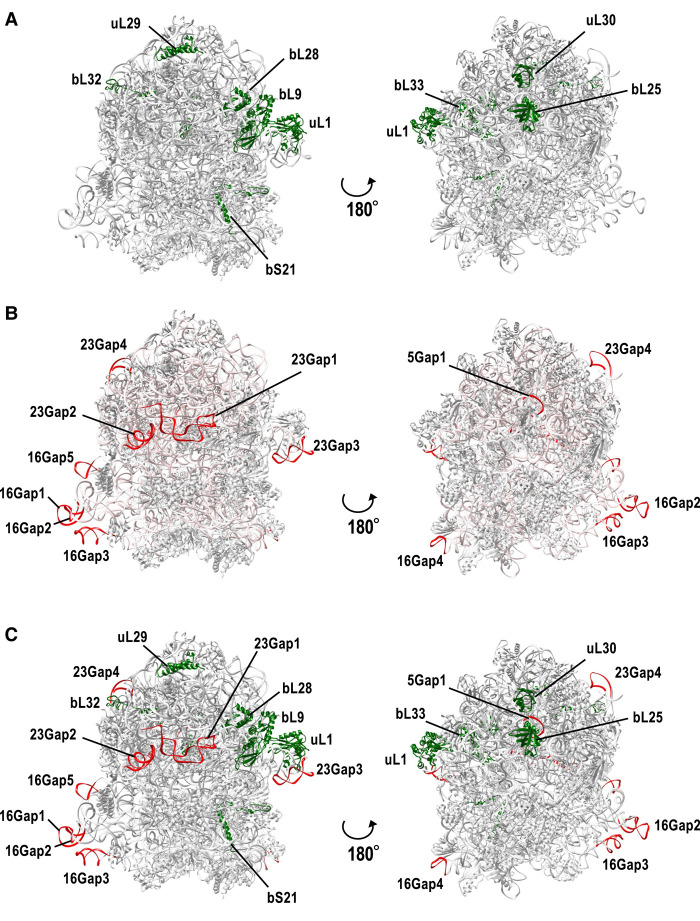
To roughly estimate the shape of the CPR bacterial ribosomes, the ribosomal proteins and rRNA regions missing in all or some CPR bacteria were mapped onto a well-studied ribosomal structural model of E. coli strain K-12 (Fig. 6A,B). Ribosomal proteins bind around the rRNA backbone to form the outer part of the ribosome. As mentioned above, most of the ribosomal proteins lacking in the CPR bacteria occur in the large subunit, and all but the bL33 protein are exposed on the ribosomal surface. The uL1, bL9, and bL28 proteins are located close to each other on the ribosome surface (Fig. 6A). Although rRNAs form the core of the ribosome, the regions in the 16S and 23S rRNAs that are lacking in CPR bacteria (Supplemental Fig. S5) are all exposed on the surface of the ribosome (Fig. 6B). Moreover, the regions lacking in the 16S rRNA (16Gap1, −2, and −3, corresponding to helices h6, h10, and h17, respectively) and the regions lacking in the 23S rRNA (23Gap1 and −2, corresponding to helices h9 and h63, respectively) are located close to each other in the ribosome tertiary structure, suggesting that the local structures formed by these helices are lost on the surface of the ribosome in CPR bacteria. When we compared the missing parts of the rRNAs and ribosomal proteins, uL1 and 23Gap3 (helix 78), bL32 and 23Gap4 (helix 98), and bL25 and 5Gap1 were located close to each other (Fig. 6C). These positional relationships were confirmed with a 2D structural analysis using RiboVision2 (Supplemental Fig. S6), except when related to uL1. It is noteworthy that the structural data used as the template in RiboVision2 did not contain the ribosomal protein uL1.
FIGURE 6.
Missing rRNA regions and missing ribosomal proteins in CPR bacteria map to the surface of the 3D E coli ribosomal structure. A three-dimensional structural model of the E. coli K-12 ribosome (PDB ID: 5U9G) was used for the analysis. (A) Ribosomal proteins missing in some or all CPR bacteria (see Fig. 2; Supplemental Fig. S3) are shown in green. (B) 16S, 23S, and 5S rRNA regions missing in CPR bacteria (see Fig. 5) are colored red. Entire rRNA regions are colored light pink. (C) Mixed view of A and B.
The uL1 protein and 23S rRNA helices 76–78 are known to form a mobile structure called the “L1-stalk,” which contributes to translation efficiency (Trabuco et al. 2010; Reblova et al. 2012), but both uL1 and 23Gap3 (corresponding to 23S rRNA helix 78) are specifically absent in some CPR bacteria (Supplemental Fig. S5). Although uL1 and 23Gap3 are not always lacking in the same lineages, some CPR bacteria lacking the 23Gap3 region (all Microgenomates, WWE3, Berkelbacteria) have a unique uL1 sequence (amino-terminal insertion and/or deletion of the internal region; see Fig. 3F). Moreover, bL9 is known to contact the base of the L1-stalk and may interact with helix 78 (Tishchenko et al. 2012). Here, we also found that the loss of bL9 in CPR bacteria is often found in lineages lacking the 23Gap3 region and containing a unique uL1 sequence. This suggests that in at least some CPR bacterial ribosomes, the L1-stalk is absent or incomplete.
Mapping the contact sites for the ribosomal proteins to the rRNA secondary structure using RiboVision2 (Bernier et al. 2014) showed that none of the lost regions in CPR ribosomal RNAs other than 23Gap3 are involved in their binding to ribosomal proteins. That is, the lost regions (other than 23Gap3) we identified in CPR bacterial rRNAs are unlikely to affect their binding to ribosomal proteins. Similarly, the lack of certain ribosomal proteins in CPR does not result in the loss of their binding sites on rRNAs (Supplemental Fig. S6). This has also been reported in non-CPR bacteria (Nikolaeva et al. 2021). In particular, for uL1, bL9, and uL30, which are frequently lost in the ribosomes of CPR bacteria, we confirmed that the sequences of their binding sites on the 23S rRNA are conserved, and that there are no significant changes in these sites compared with those in non-CPR bacteria. We also analyzed the base–base interactions between rRNAs using RiboVision2 and confirmed that 16Gap2 and 16Gap3, which are lost in all CPR bacteria, interact with each other in the E. coli ribosome.
Some ribosomal proteins have regions that are specifically present in only some CPR bacterial lineages (Fig. 3; Supplemental Fig. S4). However, these do not necessarily occur in the same vicinity in the ribosome's three-dimensional structure, so it is unclear whether these regions compensate for the missing regions found in these bacteria. The simplified structure of the CPR bacterial ribosome may provide another evolutionary option (shape) for constructing the ribosome. To clarify this, a structural analysis of the ribosomes of CPR bacteria will be essential in future research.
Simplified ribosome structure in CPR bacteria supports the theory of ribosome evolution
In this study, we detected the lack of several ribosomal regions in all or some lineages of CPR bacteria. These missing regions occur preferentially on the outside surface of the ribosomal complex and are thought to simplify the surface structure of the ribosomes of CPR bacteria. It has been reported that the evolution of ribosomes (i.e., the expansion of rRNA molecules and the acquisition of new ribosomal proteins) progressed from the center of the ribosome to the outer surface. Previous studies have proposed that the evolution of the ribosome progressed in six phases, based on predictions of the order of acquisition of each segment of the prokaryotic rRNA (Petrov et al. 2014, 2015). According to that theory, the rRNAs in the SSU and LSU evolved independently from phase 1 to phase 3, and that the interaction between the subunits formed in phases 3 and 4. In phase 5, the acquisition of functional ribosomal proteins began, with the integration of the 5S rRNA. The ribosomal proteins strengthened the binding between the subunits and formed the binding sites for translation factors. In the final phase 6, the rRNA regions located on the ribosome surface were acquired, and the surface was covered with proteins that bound to regions of the rRNAs (proteinizing). In this evolutionary model, all the regions missing in the CPR rRNAs (Fig. 5; Supplemental Fig. S5) are considered to have been acquired in phase 5 or 6. The 16S rRNA region (nucleotides 837–850 in E. coli 16S rRNA) that shows much greater conservation in CPR than in non-CPR was also acquired in phase 6. The detailed order of acquisition of the ribosomal proteins has not yet been predicted completely. However, the ribosomal proteins absent in CPR, except for bL33, are exposed on the ribosome surface when they are present in other bacteria (Fig. 6A), so it is thought that they were acquired in the latest phase of ribosome evolution. The central regions of the ribosome, which formed in the early stage of molecular evolution, are conserved throughout the three domains of life (Bacteria, Archaea, and Eukaryota) and play a central role in translation (Melnikov et al. 2012; Bernier et al. 2018). However, although the ribosomal proteins in surface regions, acquired in the late stage of evolution, contribute to the efficiency of translation and the stability of the ribosome, they are not essential for organismal survival in many cases (Galperin et al. 2021a). In the rRNAs, regions corresponding to the peptidyl transfer center and exit tunnel, which are important for mRNA translation, are well conserved, whereas the rRNA regions on the ribosome surface are often lost. The regions lost in the 16S and 23S rRNAs of CPR bacteria are also lost in the truncated rRNAs of mitochondria (Sharma et al. 2003, 2009; Petrov et al. 2019). The 5S rRNA is also often lost in mitochondrial genomes (Petrov et al. 2019). Furthermore, mapping the contact sites of ribosomal proteins to the secondary structures of rRNAs using RiboVision2 (Bernier et al. 2014) showed that none of the lost regions, other than 23Gap3, are involved in the binding of the rRNAs to proteins. Because the central regions of the ribosome are strongly conserved, even in CPR bacteria, the lack of certain molecules (described in this paper) is thought to simplify the structure of the ribosome because their absence does not basically affect the regions essential for translation. Our study provides concrete examples that support the theory of ribosome evolution.
MATERIALS AND METHODS
Data sources
We downloaded 897 publicly available genomes of CPR bacteria (69 complete and 828 draft) from GenBank at the NCBI site (https://ftp.ncbi.nlm.nih.gov/genomes/genbank/) (Supplemental Tables S1A, S2; Sayers et al. 2019). This data set consisted of 837 phylogenetically characterized genomes (nine complete and 828 draft), estimated to be >70% complete by Hug et al. (2016), and an additional 60 recently sequenced complete genomes. The classification of these genomes at the phylum level was assigned based on the NCBI Taxonomy (Schoch et al. 2020) or the taxonomic assignments by Hug et al. (2016). Some CPR phyla were classified into subgroups according to Jaffe et al. (2020): Microgenomates, Parcubacteria 1–4, and other Parcubacteria. Three representative phyla outside the Parcubacteria and Microgenomates were also considered: Katanobacteria (formerly known as WWE3), Saccharibacteria (Sa), and Peregrinibacteria (Pe; Supplemental Table S1A; Supplemental Fig. S1). As the control, we downloaded 1661 complete genomes of non-CPR bacteria, described as “Reference” or “Representative” from the NCBI Reference Sequence Database (RefSeq; https://ftp.ncbi.nlm.nih.gov/genomes/refseq/; accessed on November 1, 2018) (O'Leary et al. 2016). These non-CPR bacterial genomes were classified into endosymbiotic or parasitic groups (symbiotic non-CPR bacteria, n = 167) and others (free-living non-CPR bacteria, n = 1494; Supplemental Table S1B). Seventy genomes from each of these two non-CPR groups were selected as representative (Supplemental Tables S3, S4). Protein-coding genes in the CPR genomes were predicted with Prodigal v2.6.3 (Hyatt et al. 2010) and used to analyze the protein length distributions (Fig. 1; Supplemental Fig. S2). The genes of Absconditabacteria and Gracilibacteria were predicted with genetic code 25 (Campbell et al. 2013), in which the UGA stop codon is translated as glycine. The genes of the other genomes were predicted with the standard bacterial genetic code.
Estimation of genome sizes in CPR bacteria
The genome sizes of the CPR draft genomes were estimated by assessing their completeness using 43 universal single-copy genes (SCGs) for CPR bacteria (Brown et al. 2015). Each CPR genome was translated in six frames with the getORF program in EMBOSS 6.6.0 (Rice et al. 2000). Genetic code 25 was used for Absconditabacteria and Gracilibacteria (Campbell et al. 2013), and code 11 was used for the other taxa. All possible ORFs with a minimum length of 10 amino acids were extracted for subsequent analysis. Using SCG proteins from the NCBI COG database (https://www.ncbi.nlm.nih.gov/COG/) (Galperin et al. 2021b) as queries, BLASTP searches (blast+ version 2.9.0) (Camacho et al. 2009) were performed against the ORFs of the CPR bacteria (E-value threshold: 1 × 10−5) to identify SCG candidates. These candidate SCG proteins were then subjected to a reverse BLASTP search against the same SCG protein set from the COG database, and the top hits were used to confirm the assignments. The ratio of SCGs detected in each genome was defined as the genome restoration rate, and the estimated genome size was obtained by dividing the total length of the scaffold by the genome restoration rate.
Search for ribosomal protein genes and rRNA genes
The ribosomal protein genes and rRNA genes in the CPR bacterial genomes were detected with sequence similarity searches. Ribosomal protein sequences were compared with the ORFs in the CPR bacterial genomes (see the previous section for the detection of ORFs), using a combination of BLASTP and hmmscan. Using the amino acid sequence sets of 54 bacterial ribosomal proteins from the NCBI COG database as the queries (Supplemental Table S5), BLASTP searches (blast+ version 2.9.0) (Camacho et al. 2009) were performed against the ORFs in the CPR bacterial genomes (E-value threshold: 1 × 10−5; query cover threshold: 50%) to identify candidate ribosomal proteins. These candidate proteins were then subjected to a reverse BLASTP search against the same set of 54 bacterial ribosomal proteins from the COG database. The top hits obtained confirmed the assignments. The ORFs from the CPR bacterial genomes were then compared with the Pfam ribosomal protein HMM profiles (Mistry et al. 2021) using hmmscan from the HMMER 3.3.1 package (Eddy 2011). A set of ribosomal protein sequences was generated by combining the sequences found with these two methods and was inspected to remove false positive hits, particularly those observed in targets containing ubiquitous RNA-binding domains.
To detect the 16S, 23S, and 5S rRNA sequences in the CPR genomes, the cmsearch program from the Infernal package (version 1.1.3) was used (E-value threshold: 1 × 10−4) (Nawrocki and Eddy 2013). Here, we used RNA secondary structure models for 5S rRNA (RF00001), 16S rRNA (RF00177), and 23S rRNA (RF02541) obtained from the Rfam database (http://rfam.xfam.org/) (Kalvari et al. 2018). Because CPR bacteria often have long insertions within their 16S and 23S rRNA genes, several partial hits were identified. If the partial hits were adjacent on the same scaffold (i.e., the gaps between hits were ≤5000 bases and no hits for other rRNAs were identified in the gap), those hits were considered to be single genes. Ribosomal protein and rRNA genes in non-CPR bacteria were identified according to the RefSeq annotations, and one representative sequence per genome was extracted for each protein and rRNA. Because one non-CPR genome (Streptococcus pyogenes, accession: NC_002737) lacked annotation of the 5S rRNA gene, it was evaluated with cmsearch. The partial gene sequences truncated at the end of the scaffold were removed from the subsequent analysis. The sequences of the CPR and non-CPR rRNA genes obtained were aligned with MAFFT L-INS-i (v7.407) (Katoh and Standley 2013), and the insertion sequences were identified based on a comparison with the well-studied E. coli rRNA genes (16S, 1542 bases; 23S, 2904 bases; 5S, 120 bases), which were included in the non-CPR data set.
Comparative sequence analysis and structural mapping
Representative sequences of ribosomal proteins and rRNA genes were selected for alignment and visualization. After sequences containing unknown amino acids (“X”) or unknown nucleotides (“N”) were removed, the genes were clustered based on their sequence identity using the UCLUST algorithm (cluster_fast command) in USEARCH v11 (Edgar 2010), and the cluster centroids (typical sequences) were selected as representative. The identity thresholds were 80% for the ribosomal proteins, 5S rRNA genes, and 23S rRNA genes, and 85% for the 16S rRNA genes, based on the number of clusters generated. The representative sequences were aligned with MAFFT L-INS-i (v7.407) (Katoh and Standley 2013). To visualize the alignment of ribosomal protein sequences, columns with gap frequency of >90% in both the CPR and non-CPR groups were removed. To visualize the rRNA gene sequence alignment, insertions with respect to the E. coli K-12 genes were removed. The alignments were visualized with Jalview 2.11 (Waterhouse et al. 2009). The locations of the missing regions in the CPR bacterial ribosomal proteins and rRNAs were estimated by mapping the genes and rRNAs onto the tertiary structure of the E. coli K-12 ribosome (PDB ID: 5U9G) (Demo et al. 2017). Three-dimensional mapping was performed with UCSF Chimera (Pettersen et al. 2004). The ribosomal protein contacts and internucleotide interactions in the secondary structures of the rRNAs were mapped with RiboVision2 (Bernier et al. 2014). The structural data (PDB ID: 4V9D) used as the template in RiboVision2 did not contain the ribosomal protein uL1.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Shigenori Maruyama for his critical suggestions. We also thank all members of the RNA Group at the Institute for Advanced Biosciences of Keio University, Japan, for their insightful discussions. This work was supported, in part, by a KAKENHI Grant-in-Aid for Japan for the Society for the Promotion of Science (JSPS) Fellows (21J12231) and research funds from the Yamagata Prefectural Government and Tsuruoka City, Japan. The funding bodies played no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079103.122.
Freely available online through the RNA Open Access option.
MEET THE FIRST AUTHOR
Megumi Tsurumaki.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Megumi Tsurumaki is the first author of this paper, “Features of smaller ribosomes in candidate phyla radiation (CPR) bacteria revealed with a molecular evolutionary analysis.” Megumi is a Ph.D. student in Dr. Akio Kanai's laboratory in the Systems Biology Program at Keio University, Japan. Her research interests include genomic microbiology, evolutionary biology, and bioinformatics.
What are the major results described in your paper and how do they impact this branch of the field?
The candidate phyla radiation (CPR) is a monophyletic supergroup mainly composed of uncultured bacterial lineages that is important in the discussion of bacterial diversity and evolution. Here we performed a bioinformatic analysis focusing on the ribosomes of CPR bacteria, and suggested that CPR bacterial ribosomes are smaller than those in other bacteria and are characterized by a simplified surface structure. We believe that our results provide a new characterization of CPR which may provide another evolutionary option for constructing the ribosome.
What led you to study RNA or this aspect of RNA science?
When I entered graduate school, I got informed about the topic of CPR and became interested in the features and evolutionary background of this novel bacterial supergroup. I learned that CPR bacteria lack some ribosomal proteins from previous research and wanted to know the overall shape of their ribosome, including rRNAs. While working on our current study about simplified ribosomes of CPR, I developed an interest in the structure and evolution of the translation system.
If you were able to give one piece of advice to your younger self, what would that be?
My professor often says to students, “Work gradually but steadily. Never stop even if each step is small.” I would like to pass this message to my teenage and undergraduate self. I want her to cherish her interests, build up knowledge little by little, and not be afraid to discuss with others.
What are your subsequent near- or long-term career plans?
I will continue to work in my current laboratory, focusing on CPR bacteria and their ribosomes, while aiming to submit my Ph.D. thesis. I would like to be able to study life sciences through both informatics and experimental approaches, as well as pursue a career in the field of molecular biology in either academia or industry.
REFERENCES
- Amin MR, Yurovsky A, Chen Y, Skiena S, Futcher B. 2018. Re-annotation of 12,495 prokaryotic 16S rRNA 3′ ends and analysis of Shine-Dalgarno and anti-Shine-Dalgarno sequences. PLoS One 13: e0202767. 10.1371/journal.pone.0202767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, Thomas BC, Singh A, Wilkins MJ, Karaoz U, et al. 2016. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun 7: 13219. 10.1038/ncomms13219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol 24: 165–169. 10.1016/j.sbi.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier CR, Petrov AS, Waterbury CC, Jett J, Li F, Freil LE, Xiong X, Wang L, Migliozzi BL, Hershkovits E, et al. 2014. RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss 169: 195–207. 10.1039/C3FD00126A [DOI] [PubMed] [Google Scholar]
- Bernier CR, Petrov AS, Kovacs NA, Penev PI, Williams LD. 2018. Translation: the universal structural core of life. Mol Biol Evol 35: 2065–2076. 10.1093/molbev/msy101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523: 208–211. 10.1038/nature14486 [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JH, O'Donoghue P, Campbell AG, Schwientek P, Sczyrba A, Woyke T, Soll D, Podar M. 2013. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci 110: 5540–5545. 10.1073/pnas.1303090110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelle CJ, Banfield JF. 2018. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell 172: 1181–1197. 10.1016/j.cell.2018.02.016 [DOI] [PubMed] [Google Scholar]
- Castelle CJ, Brown CT, Anantharaman K, Probst AJ, Huang RH, Banfield JF. 2018. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat Rev Microbiol 16: 629–645. 10.1038/s41579-018-0076-2 [DOI] [PubMed] [Google Scholar]
- Cerretti DP, Dean D, Davis GR, Bedwell DM, Nomura M. 1983. The spc ribosomal protein operon of Escherichia coli: sequence and cotranscription of the ribosomal protein genes and a protein export gene. Nucleic Acids Res 11: 2599–2616. 10.1093/nar/11.9.2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman GA, Davin AA, Mahendrarajah TA, Szantho LL, Spang A, Hugenholtz P, Szollosi GJ, Williams TA. 2021. A rooted phylogeny resolves early bacterial evolution. Science 372: eabe0511. 10.1126/science.abe0511 [DOI] [PubMed] [Google Scholar]
- Danczak RE, Johnston MD, Kenah C, Slattery M, Wrighton KC, Wilkins MJ. 2017. Members of the Candidate Phyla Radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome 5: 112. 10.1186/s40168-017-0331-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demo G, Svidritskiy E, Madireddy R, Diaz-Avalos R, Grant T, Grigorieff N, Sousa D, Korostelev AA. 2017. Mechanism of ribosome rescue by ArfA and RF2. Elife 6: e23687. 10.7554/eLife.23687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7: e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Galperin MY, Wolf YI, Garushyants SK, Alvarez RV, Koonin EV. 2021a. Nonessential ribosomal proteins in bacteria and archaea identified using clusters of orthologous genes. J Bacteriol 203: e0005–e8-21. 10.1128/JB.00058-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Wolf YI, Makarova KS, Vera Alvarez R, Landsman D, Koonin EV. 2021b. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res 49: D274–D281. 10.1093/nar/gkaa1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Qing Y, Guo X, Warren A. 2014. “Candidatus Sonnebornia yantaiensis”, a member of candidate division OD1, as intracellular bacteria of the ciliated protist Paramecium bursaria (Ciliophora, Oligohymenophorea). Syst Appl Microbiol 37: 35–41. 10.1016/j.syapm.2013.08.007 [DOI] [PubMed] [Google Scholar]
- Grondek JF, Culver GM. 2004. Assembly of the 30S ribosomal subunit: positioning ribosomal protein S13 in the S7 assembly branch. RNA 10: 1861–1866. 10.1261/rna.7130504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Breton M, Sirand-Pugnet P, Tardy F, Thiaucourt F, Citti C, Barre A, Yoshizawa S, Fourmy D, de Crecy-Lagard V, et al. 2014. Predicting the minimal translation apparatus: lessons from the reductive evolution of mollicutes. PLoS Genet 10: e1004363. 10.1371/journal.pgen.1004363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, et al. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci 112: 244–249. 10.1073/pnas.1419038112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold M, Nierhaus KH. 1987. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J Biol Chem 262: 8826–8833. 10.1016/S0021-9258(18)47489-3 [DOI] [PubMed] [Google Scholar]
- Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, et al. 2016. A new view of the tree of life. Nat Microbiol 1: 16048. 10.1038/nmicrobiol.2016.48 [DOI] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11: 119. 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AL, Castelle CJ, Matheus Carnevali PB, Gribaldo S, Banfield JF. 2020. The rise of diversity in metabolic platforms across the Candidate Phyla Radiation. BMC Biol 18: 69. 10.1186/s12915-020-00804-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowska M, Ryden-Aulin M. 2007. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev 71: 477–494. 10.1128/MMBR.00013-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI. 2018. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46: D335–D342. 10.1093/nar/gkx1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, Thomas BC, Banfield JF. 2013. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio 4: e00708–e00713. 10.1128/mBio.00708-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Asahara H, Laurberg M, Lancaster L, Noller HF. 2007. Interactions and dynamics of the Shine Dalgarno helix in the 70S ribosome. Proc Natl Acad Sci 104: 16840–16843. 10.1073/pnas.0707850104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecompte O, Ripp R, Thierry JC, Moras D, Poch O. 2002. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res 30: 5382–5390. 10.1093/nar/gkf693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K, Furuta Y, Kobayashi I. 2012. Large variations in bacterial ribosomal RNA genes. Mol Biol Evol 29: 2937–2948. 10.1093/molbev/mss101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luef B, Frischkorn KR, Wrighton KC, Holman HY, Birarda G, Thomas BC, Singh A, Williams KH, Siegerist CE, Tringe SG, et al. 2015. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun 6: 6372. 10.1038/ncomms7372 [DOI] [PubMed] [Google Scholar]
- Machulin AV, Deryusheva EI, Selivanova OM, Galzitskaya OV. 2019. The number of domains in the ribosomal protein S1 as a hallmark of the phylogenetic grouping of bacteria. PLoS One 14: e0221370. 10.1371/journal.pone.0221370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier UG, Zauner S, Woehle C, Bolte K, Hempel F, Allen JF, Martin WF. 2013. Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol Evol 5: 2318–2329. 10.1093/gbe/evt181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meheust R, Burstein D, Castelle CJ, Banfield JF. 2019. The distinction of CPR bacteria from other bacteria based on protein family content. Nat Commun 10: 4173. 10.1038/s41467-019-12171-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat Struct Mol Biol 19: 560–567. 10.1038/nsmb.2313 [DOI] [PubMed] [Google Scholar]
- Melnikov S, Manakongtreecheep K, Soll D. 2018. Revising the structural diversity of ribosomal proteins across the three domains of life. Mol Biol Evol 35: 1588–1598. 10.1093/molbev/msy021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, et al. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res 49: D412–D419. 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Iwatsuki T, Naganuma T. 2005. Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters. Appl Environ Microbiol 71: 1084–1088. 10.1128/AEM.71.2.1084-1088.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29: 2933–2935. 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WC, Stegen JC. 2015. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front Microbiol 6: 713. 10.3389/fmicb.2015.00713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaeva DD, Gelfand MS, Garushyants SK. 2021. Simplification of ribosomes in bacteria with tiny genomes. Mol Biol Evol 38: 58–66. 10.1093/molbev/msaa184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. 10.1126/science.289.5481.920 [DOI] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Rinke C, Chuvochina M, Chaumeil PA, Woodcroft BJ, Evans PN, Hugenholtz P, Tyson GW. 2017. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol 2: 1533–1542. 10.1038/s41564-017-0012-7 [DOI] [PubMed] [Google Scholar]
- Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM, et al. 2014. Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci 111: 10251–10256. 10.1073/pnas.1407205111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, et al. 2015. History of the ribosome and the origin of translation. Proc Natl Acad Sci 112: 15396–15401. 10.1073/pnas.1509761112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Wood EC, Bernier CR, Norris AM, Brown A, Amunts A. 2019. Structural patching fosters divergence of mitochondrial ribosomes. Mol Biol Evol 36: 207–219. 10.1093/molbev/msy221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Qin D, Liu Q, Devaraj A, Fredrick K. 2012. Role of helix 44 of 16S rRNA in the fidelity of translation initiation. RNA 18: 485–495. 10.1261/rna.031203.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reblova K, Sponer J, Lankas F. 2012. Structure and mechanical properties of the ribosomal L1 stalk three-way junction. Nucleic Acids Res 40: 6290–6303. 10.1093/nar/gks258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16: 276–277. 10.1016/S0168-9525(00)02024-2 [DOI] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, et al. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. 10.1038/nature12352 [DOI] [PubMed] [Google Scholar]
- Roberts E, Sethi A, Montoya J, Woese CR, Luthey-Schulten Z. 2008. Molecular signatures of ribosomal evolution. Proc Natl Acad Sci 105: 13953–13958. 10.1073/pnas.0804861105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, Karsch-Mizrachi I. 2019. GenBank. Nucleic Acids Res 47: D94–D99. 10.1093/nar/gky989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, McVeigh R, O'Neill K, Robbertse B, et al. 2020. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford) 2020: baaa062. 10.1093/database/baaa062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskay MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834. 10.1126/science.1117230 [DOI] [PubMed] [Google Scholar]
- Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. 2003. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115: 97–108. 10.1016/S0092-8674(03)00762-1 [DOI] [PubMed] [Google Scholar]
- Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK. 2009. Structure of a mitochondrial ribosome with minimal RNA. Proc Natl Acad Sci 106: 9637–9642. 10.1073/pnas.0901631106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanov VG, Fox GE. 2021. Expansion segments in bacterial and archaeal 5S ribosomal RNAs. RNA 27: 133–150. 10.1261/rna.077123.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun FJ, Caetano-Anolles G. 2009. The evolutionary history of the structure of 5S ribosomal RNA. J Mol Evol 69: 430–443. 10.1007/s00239-009-9264-z [DOI] [PubMed] [Google Scholar]
- Suzuki S, Ishii S, Hoshino T, Rietze A, Tenney A, Morrill PL, Inagaki F, Kuenen JG, Nealson KH. 2017. Unusual metabolic diversity of hyperalkaliphilic microbial communities associated with subterranean serpentinization at The Cedars. ISME J 11: 2584–2598. 10.1038/ismej.2017.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski M, Zielezinski A, Barciszewski J, Erdmann VA, Karlowski WM. 2016. 5SRNAdb: an information resource for 5S ribosomal RNAs. Nucleic Acids Res 44: D180–D183. 10.1093/nar/gkv1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishchenko S, Gabdulkhakov A, Nevskaya N, Sarskikh A, Kostareva O, Nikonova E, Sycheva A, Moshkovskii S, Garber M, Nikonov S. 2012. High-resolution crystal structure of the isolated ribosomal L1 stalk. Acta Crystallogr D Biol Crystallogr 68: 1051–1057. 10.1107/S0907444912020136 [DOI] [PubMed] [Google Scholar]
- Trabuco LG, Schreiner E, Eargle J, Cornish P, Ha T, Luthey-Schulten Z, Schulten K. 2010. The role of L1 stalk-tRNA interaction in the ribosome elongation cycle. J Mol Biol 402: 741–760. 10.1016/j.jmb.2010.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC, Wilkins MJ, Hettich RL, Lipton MS, Williams KH, et al. 2012. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337: 1661–1665. 10.1126/science.1224041 [DOI] [PubMed] [Google Scholar]
- Yutin N, Puigbo P, Koonin EV, Wolf YI. 2012. Phylogenomics of prokaryotic ribosomal proteins. PLoS One 7: e36972. 10.1371/journal.pone.0036972 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.