Abstract
Paraspeckles are mammalian-specific nuclear bodies built on the long noncoding RNA NEAT1_2. The molecular mechanisms of paraspeckle formation have been mainly studied using human or mouse cells, and it is not known if the same molecular components are involved in the formation of paraspeckles in other mammalian species. We thus investigated the expression pattern of NEAT1_2 in naked mole-rats (nNEAT1_2), which exhibit extreme longevity and lower susceptibility to cancer. In the intestine, nNEAT1_2 is widely expressed along the entire intestinal epithelium, which is different from the expression of mNeat1_2 that is restricted to the cells of the distal tip in mice. Notably, the expression of FUS, a FET family RNA binding protein, essential for the formation of paraspeckles both in humans and mice, was absent in the distal part of the intestinal epithelium in naked mole-rats. Instead, mRNAs of other FET family proteins EWSR1 and TAF15 were expressed in the distal region. Exogenous expression of these proteins in Fus-deficient murine embryonic fibroblast cells rescued the formation of paraspeckles. These observations suggest that nNEAT1_2 recruits a different set of RNA binding proteins in a cell type-specific manner during the formation of paraspeckles in different organisms.
Keywords: FUS, NEAT1, naked mole-rat, paraspeckles
INTRODUCTION
A large number of noncoding RNAs are transcribed from the genomes of higher eukaryotes, constituting a significant portion of the transcriptional output (Clark et al. 2011; Djebali et al. 2012). Noncoding RNA transcripts longer than 200 nt are collectively called long noncoding RNAs (lncRNAs), and a series of studies revealed that they are functionally divided into several groups (for reviews, see Kopp and Mendell 2018; Ali and Grote 2020; Gil and Ulitsky 2020; Nakagawa et al. 2021; Statello et al. 2021). One group of lncRNAs are epigenetic regulators that control gene expression via association with chromatin modifying complexes in cis or trans (for reviews, see Ali and Grote 2020; Gil and Ulitsky 2020; Statello et al. 2021). Others function as molecular sponges that sequester associating RNA binding proteins or microRNAs and negatively regulate their function (for review, see Kopp and Mendell 2018). Architectural RNAs (arcRNAs) are an emerging group of lncRNAs that serve as a structural component of submicron-scale nonmembranous organelles, which are often referred to as molecular condensates (for review, see Nakagawa et al. 2021).
MALAT1 and NEAT1 are highly abundant lncRNAs, which are located at the same syntenic chromosomal regions in humans and mice (Hutchinson et al. 2007). MALAT1 is localized to nuclear speckles that are enriched in splicing regulators such as SR proteins and UsnRNPs. While MALAT1 is not required for the formation of nuclear speckles (Eissmann et al. 2012; Nakagawa et al. 2012; Zhang et al. 2012), it has been proposed to regulate pre-mRNA splicing via association with splicing regulators (Tripathi et al. 2010). NEAT1 is the most extensively studied arcRNA that plays an essential role to build mammalian-specific nuclear bodies called paraspeckles (for reviews, see Fox et al. 2018; Nakagawa et al. 2018), which were originally defined as nuclear bodies enriched in the RNA binding protein PSPC1 and 2 (paraspeckle protein 1 and 2) (Fox et al. 2002). Two isoforms of NEAT1, NEAT1_1 and NEAT1_2 are transcribed from the same promoter, and the longer isoform NEAT1_2 plays the architectural role, whereas NEAT1_1 alone cannot induce the formation of paraspeckles (Sasaki et al. 2009; Naganuma et al. 2012; Li et al. 2017; Isobe et al. 2020). Paraspeckles contain more than 30 RNA binding proteins (Naganuma et al. 2012; Fong et al. 2013), which are radially arranged to form spheres with characteristic core-shell structure (Souquere et al. 2010; West et al. 2016). A group of paraspeckle proteins including NONO and FUS are indispensable for the formation of paraspeckles (Naganuma et al. 2012; West et al. 2016). Many of these essential paraspeckle proteins contain distinct intrinsically disordered regions or prion-like domains, which undergo phase transition in vitro to form a hydrogel (Hennig et al. 2015). NEAT1 induces the formation of phase-separated condensates by recruiting paraspeckle proteins onto distinct regions of its transcript (Yamazaki et al. 2018). Paraspeckle spheres fuse to form sausage- or tube-like structures with distinct radial diameters, suggesting that they behave as flexible block copolymer micelles rather than rigid complexes with components strictly located at specific positions (Yamazaki et al. 2021).
Paraspeckles are ubiquitously observed in most cultured cell lines (for reviews, see Fox et al. 2018; Nakagawa et al. 2018), with a few exceptions such as ES cells that do not express enough NEAT1_2 (Modic et al. 2019). On the other hand, prominent expressions of Neat1_2 in mice (mNeat1_2) are restricted to a subset of particular cell types, including distal tip cells of intestinal epithelium and corpus luteal cells in female ovaries (Nakagawa et al. 2011; Isobe et al. 2020). mNeat1 knockout mice exhibit various phenotypes including decreased fertility and impaired mammary tract formation in female animals (Nakagawa et al. 2014; Standaert et al. 2014); however, the penetrance of the subfertile phenotype is around 50% and is variable even in the same individuals (Nakagawa et al. 2014). In addition, the lack of mNeat1 results in an increase or decrease in tumorigenesis in different tumor models, although the expression of mNeat1 is under the control of the p53 pathway in both cases (Adriaens et al. 2016; Mello et al. 2017). These observations suggest that mNeat1 plays a physiological role in a condition- or context-dependent manner in different cell types or experimental models (for review, see Nakagawa et al. 2018).
Given Neat1 expression and paraspeckle formation in tissues of living animals have been mostly studied in mice, it is possible that the molecular components of paraspeckles that associate with Neat1 could vary between different mammalian species. To investigate the molecular conservation of paraspeckle components between species, here we studied the expression pattern of NEAT1 in naked mole-rats (nNEAT1), an animal characterized by longevity and low susceptibility to cancer formation (for review, see Buffenstein et al. 2022). We found characteristic expression of nNEAT1_2 in the intestinal epithelium of naked mole-rats, however this was observed throughout the entire length of the intestinal epithelium, unlike the restricted expression in mice. In addition, nFUS, an essential RNA binding protein needed to form paraspeckles, was absent in the distal part of intestinal epithelium in naked mole-rats. Instead, mRNAs of nEWSR1 and nTAF15 were broadly expressed throughout the epithelium, suggesting that the FET family proteins substitute for the function of nFUS to form paraspeckles in the distal regions of intestinal epithelium in naked mole-rats. These observations suggest that paraspeckle components are variable between species, raising a possibility of species-specific function of paraspeckles in certain tissues or cell types.
RESULTS
Identification of MALAT1 and NEAT1 in naked mole-rats
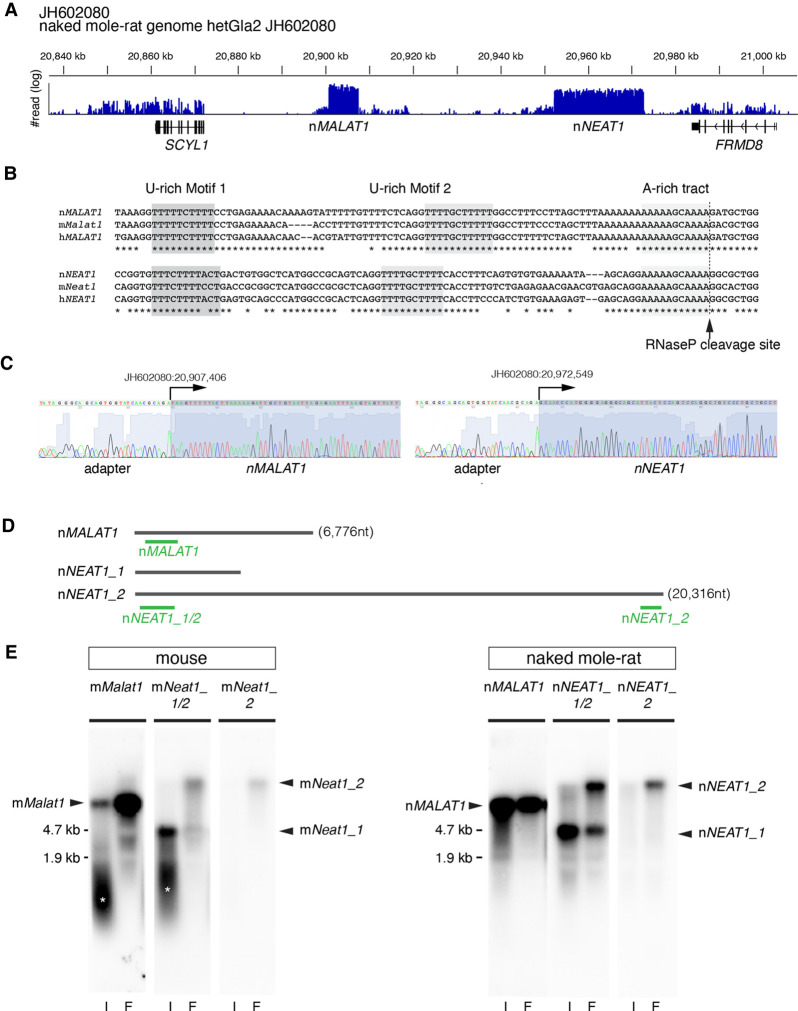
To gain insight into the role of lncRNA in naked mole-rats, we initially tried to identify MALAT1 and NEAT1, two of the most abundant lncRNAs, whose genes are adjacently located in the human and mouse genome (Hutchinson et al. 2007). We performed RNA-seq analyses using RNAs obtained from naked mole-rat skin fibroblast cells (NS-fibroblasts), and found that a large number of reads were mapped to two distinct regions in the assembly JH602080 of the naked mole-rat genome (hetGla2), flanked by SCYL1 and FRMD8 (Fig. 1A), which corresponds to a region syntenic to human chromosome 11 and mouse chromosome 19 containing the two lncRNAs and the same neighboring genes (Fig. 1A). Sequences at the 3′ regions of naked mole-rat MALAT1 and NEAT1 (hereafter nMALAT1 and nNEAT1) exhibited remarkable homology with mouse/human MALAT1/NEAT1 (hereafter mMalat1, mNeat1, hMALAT1, hNEAT1, respectively) (Fig. 1B), suggesting that nMALAT1 and nNEAT1 receive 3′ processing characteristics of these lncRNAs, which include the cleavage by RNaseP and the formation of triple-helix structures that stabilize the transcripts (Wilusz et al. 2008, 2012). To determine the transcription start sites of nMALAT1 and nNEAT1, we performed 5′ RACE (rapid amplification of cDNA ends) PCR. The PCR fragments were cloned into a plasmid vector and we sequenced three clones for each gene, which yielded the same sequences shown in Figure 1C. Probes designed against nMALAT1 and nNEAT1 detected bands of the same size as mNeat1 and mMalat1 by northern blot (Fig. 1D,E), where the sizes were consistent with the aforementioned prediction. Probes that detect the 5′ region of nNEAT1 detected two bands (Fig. 1E), suggesting that naked mole-rats also have a short (nNEAT1_1) and a long (nNEAT1_2) isoform of nNEAT1, like humans and mice (Sasaki et al. 2009; Sunwoo et al. 2009).
FIGURE 1.
Identification of MALAT1 and NEAT1 in naked mole-rats. (A) Distribution of RNA-seq reads mapped to the region of the naked mole-rat genome syntenic to mouse chromosome 19 containing mMalat1 (mouse Malat1) and mNeat1 (mouse Neat1), which are flanked by SCYL1 and FRMD8. Note that two regions were abundantly transcribed into RNA, which corresponded to nMALAT1 (naked mole-rat MALAT1) and nNEAT1 (naked mole-rat NEAT1), respectively. (B) Sequence alignment of the 3′ regions of NEAT1 and MALAT1 in naked mole-rats (n), mouse (m), and human (h). The tandem U-rich motifs followed by A-rich tract, which form triple helix structures, are highlighted in shadow boxes and are highly conserved between the three species. The arrow indicates the putative RNaseP cleavage site that generates the 3′ end of these transcripts. (C) Sequence analyses of PCR fragments obtained by the 5′ RACE analyses. Arrows indicate the positions of the transcription start sites of nMALAT1 and nNEAT1. (D) Schematic drawing indicating the positions of probes used for northern blot and in situ hybridization studies. (E) Northern blot analyses of nMALAT1 and nNEAT1 expression in embryonic fibroblast cells and intestine. (I) intestine, (F) fibroblasts. Asterisks show the signals derived from degraded RNAs.
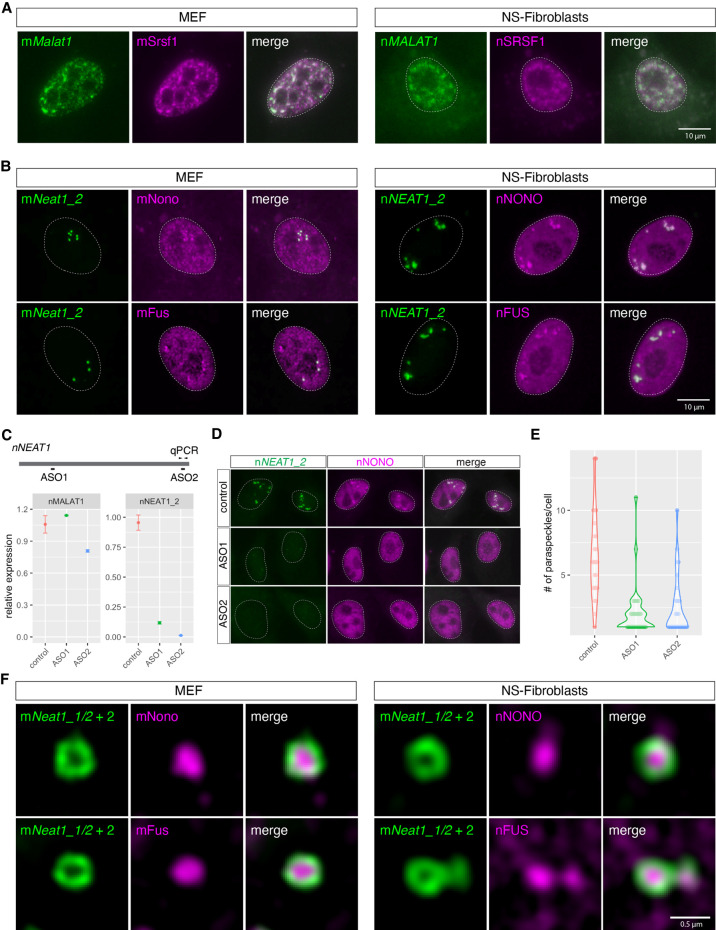
To further characterize nMALAT1 and nNEAT1, we examined the subcellular distribution of these transcripts in skin fibroblast cells (NS-fibroblasts) (Fig. 2). hMALAT1/mMalat1 is localized in nuclear speckles that are enriched in splicing regulators (Hutchinson et al. 2007; Tripathi et al. 2010). The localization of nMALAT1 coincided with nSRSF1, a marker for nuclear speckles in NS-fibroblasts, which was indistinguishable from the localization of mMalat1 in mouse embryonic fibroblast cells (MEF) (Fig. 2A; Supplemental Fig. S1). To test if nNEAT1 is localized in paraspeckles, we examined if a series of antibodies that recognize paraspeckle marker proteins in mice (West et al. 2016) also recognized antigens in naked mole-rats. However, only two of these antibodies, those that detect nNONO and nFUS, were effective in naked mole-rats (A Yamada and S Nakagawa, unpubl.), and both of these proteins are essential for the formation of paraspeckles in mice. nNEAT1_2, the architectural isoform of NEAT1, exhibited the same localization in paraspeckles visualized by marker proteins nNONO and nFUS in NS-fibroblasts, as was the case for the colocalization of mNeat1_2 and these marker proteins in MEF (Fig. 2B; Supplemental Fig. S1).
FIGURE 2.
The same subcellular distribution of mMalat1/nMALAT1 and mNeat1/nNEAT1 in MEF and NS-fibroblasts. (A) Simultaneous detection of mMalat1/nMALAT1 (green) and the nuclear speckle marker Srsf1/SRSF1 (magenta) in MEF and NS-fibroblasts. Note that both mMalat1 and nMALAT1 are enriched in nuclear speckles identified by mSrsf1/nSRSF1 expression. (B) Simultaneous detection of mNeat1_2/nNEAT1_2 (green) and the paraspeckle markers mNono/mFus and nNONO/nFUS in MEF and NS-fibroblasts, respectively. (C) Expression of nMALAT1 and nNEAT1_2 in NS-fibroblasts upon treatment of ASO. The positions of ASOs are shown in the schematic drawing. Arrowheads indicate the position of qPCR primers to detect the expression of nNEAT1_2. (D) Simultaneous detection of nNEAT1_2 (green) and nNONO (magenta) in NS-fibroblasts treated with ASOs. (E) Quantification of the number of paraspeckles in the NS-fibroblast upon knockdown of nNEAT1_2. Each dot represents the number of paraspeckles in each cell. (F) Simultaneous detection of mNeat1_2/nNEAT1_2 (green) and mNono/mFus and nNONO/nFUS (magenta) at high magnification using structural illumination microscopy. Scale bars, 10 µm in A and B and 0.5 µm in C.
To confirm if nNEAT1 is required for the formation of paraspeckles as has been shown in human or mouse cells (for reviews, see Fox et al. 2018; Nakagawa et al. 2018), we knocked down nNEAT1_2 using two different antisense oligonucleotides (ASO) designed against the 5′ and 3′ regions of this transcript (Fig. 2C). As expected, the expression of nNEAT1_2 was reduced by ∼90% compared to the control cells, resulting in a decrease of paraspeckle formation identified by the enriched localization of nNONO (Fig. 2C–E).
To test if the paraspeckles in naked mole-rats possess the characteristic core-shell structures found in human or mouse paraspeckles (Souquere et al. 2010; West et al. 2016), we observed them in NS-fibroblasts using structured illumination microscopy (Fig. 2F; Supplemental Fig. S1). Simultaneous detection of nNEAT1 and the paraspeckle markers nNONO and nFUS revealed that the 5′ and 3′ regions of nNEAT1 were located at the peripheral regions of paraspeckles, whereas nNONO and nFUS were located in the core (Fig. 2F; Supplemental Fig. S1), as was reported in previous studies in humans and mice (Souquere et al. 2010; West et al. 2016). Taken together, we concluded we have correctly identified homologs of MALAT1 and NEAT1 in naked mole-rats, which exhibited almost identical subcellular distributions compared to the mouse homologs.
nMALAT1 is ubiquitously expressed in various tissues with a variable subcellular localization
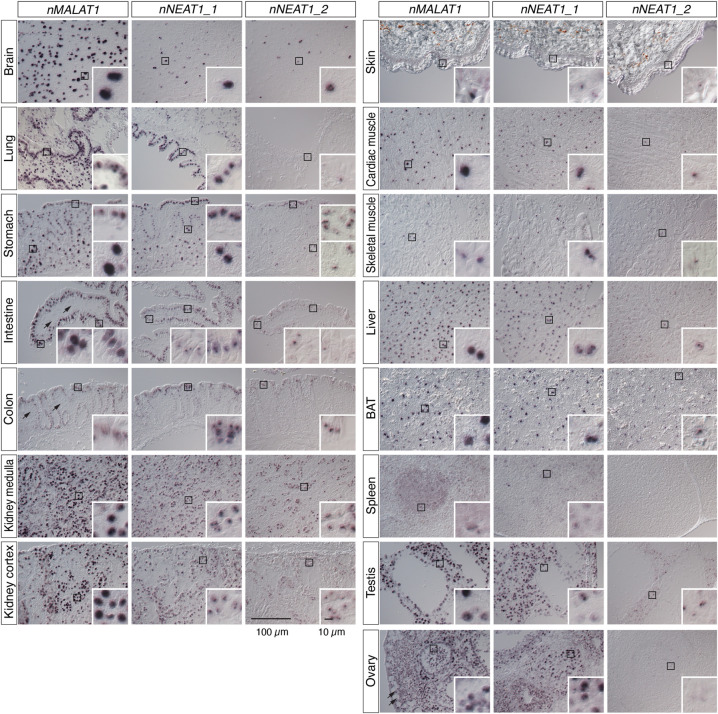
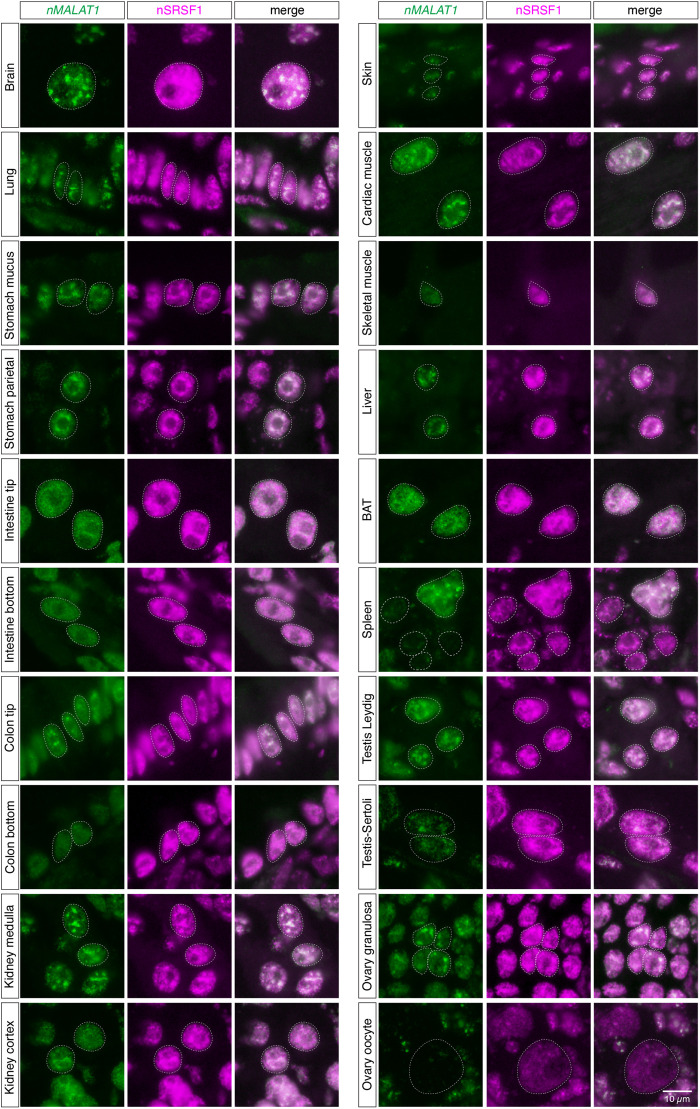
To study the expression pattern of nMALAT1 and nNEAT1 in tissues of naked mole-rats, we performed in situ hybridization using 14 different tissues (Fig. 3). Naked mole-rats live in eusocial colonies similar to those of ants and bees, consisting of one breeding queen, one to three breeding males, and many nonbreeding workers. For this experiment, we used one nonbreeder male and one nonbreeder female animal, and the same expression patterns were observed in all the tissues of these two animals except for reproductive tissues. As previously reported in mice (Nakagawa et al. 2012), intense expression of nMALAT1 was observed in a broad range of tissues and cell types including cortical neurons in the brain, mesenchymal and epithelial cells in lung, mucus epithelial cells in digestive system (stomach, intestine, and colon), parietal cells in stomach, renal tube cells in kidney, cardiac and skeletal muscles, hepatocytes in liver, brown adipose cells in brown adipose tissue (BAT), megakaryocytes in spleen, Sertoli and Leydig cells in testis, and granulosa cells in ovary (Fig. 3). On the other hand, certain cell types did not express nMALAT1, which included mesenchymal cells in intestine and colon, and oocytes in ovary (arrows in Fig. 3). To investigate if nMALAT1 was localized to nuclear speckles, we simultaneously detected nMALAT1 and nSRSF1 in these tissues (Fig. 4; Supplemental Fig. S2). The peaks of nMALAT1 signals generally coincided with the peaks of nSRSF1 signals, suggesting that nMALAT1 is mostly localized to nuclear speckles. However, nMALAT1 was diffusedly distributed in particular cell types despite the formation of nuclear speckles identified by the enrichment of nSRSF1, which included the bottom regions of intestinal and colon epithelium, and skeletal muscle cells (Fig. 4; Supplemental Fig. S2). On the other hand, nMALAT1 formed more condensed foci in certain cell types, which included cortical neurons, lung epithelial cells, renal tube cells in the medulla of kidney, cardiac muscle cells in heart, hepatocytes in liver, and a subset of granulosa cells in ovary (Fig. 4; Supplemental Fig. S2). These observations suggested that subnuclear localization of nMALAT1 is differentially regulated in each cell type.
FIGURE 3.
Expression of nMALAT1 and nNEAT1 in various tissues of naked mole-rats. Expression patterns of nMALAT1, nNEAT1_1, and nNEAT1_2 in brain, lung, stomach, intestine, colon, kidney, skin, cardiac muscle, skeletal muscle, liver, brown adipose tissue (BAT), spleen, testis, and ovary of naked mole-rats detected by in situ hybridization. All tissues are from a nonbreeder male animal (1 yr 8 mo) except for the ovary, which is derived from a non-queen female animal (1 yr 11 mo). Black squares represent areas shown in insets at a higher magnification. Arrows in the nMALAT1 panels indicate the cells that do not express nMALAT1. Scale bars are 100 µm for large panels and 10 µm for insets.
FIGURE 4.
Subnuclear localization of nMALAT1 exhibit various patterns in different cell types. Simultaneous detection of nMALAT1 and a nuclear speckle marker nSRSF1 in various tissues of naked mole-rats. Note that nMALAT1 was diffusedly distributed in the bottom regions of intestinal and colon epithelium and skeletal muscle cells, despite the speckled distribution of SRSF1. Scale bar, 10 µm.
The expression of nNEAT1_2 is observed in a broader range of tissues in naked mole-rats compared to the expression in mouse tissues
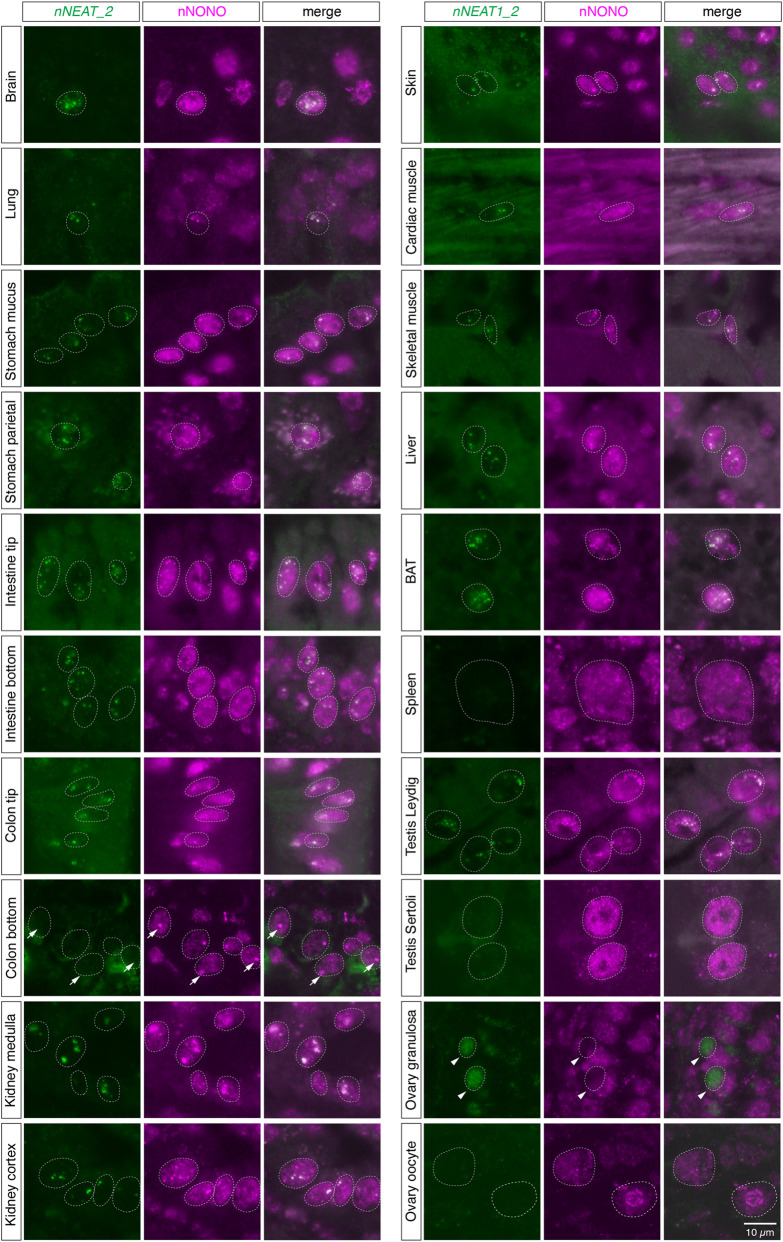
We then examined the expression pattern of nNEAT1_1 and nNEAT1_2 in the tissues of naked mole-rats. As has been reported in mouse tissues (Nakagawa et al. 2011), the expression of nNEAT1_1 was rather ubiquitously observed, whereas the expression of nNEAT1_2 was limited to particular cell types (Fig. 3). However, there are several marked differences between the reported expression of mNeat1_2 (Nakagawa et al. 2011) and the expression of nNEAT1_2 in terms of tissue distributions. For example, nNEAT1_2 was observed in a subpopulation of cortical neurons (Fig. 3), whereas mNeat1_2 expression is lacking in the same cell type in mouse brain. In addition, nNEAT1_2 expression was observed along the entire length of intestinal epithelium in naked mole-rats (Fig. 3), which was quite different to the restricted expression of mNeat1_2 in a small population of distal tip cells in the mouse intestinal epithelium (Nakagawa et al. 2011). Prominent expression of nNEAT1_2 was also observed in parietal cells in stomach, renal tube cells in kidney, and brown adipose cells (Fig. 3). Because paraspeckle formation is dependent on the expression of NEAT1_2 (Sasaki et al. 2009; Naganuma et al. 2012; Li et al. 2017; Isobe et al. 2020), these observations suggested that paraspeckle formation in naked mole-rats is more broadly observed in these cells than in mice. To confirm this hypothesis, we investigated the colocalization of nNEAT1_2 and the paraspeckle marker protein nNONO (Fig. 5; Supplemental Fig. S3). Simultaneous detection of nNEAT1_2 and nNONO indeed confirmed the formation of paraspeckles in these cells expressing nNEAT1_2 (Fig. 5; Supplemental Fig. S3). We noticed, however, there were certain cell types that highly express Neat1_2 in mice but not in naked mole-rats. For example, megakaryocytes in spleen did not express nNEAT1_2 and thus lacked the formation of paraspeckles (Figs. 3, 5; Supplemental Fig. S3). The expression of nNEAT1_2 was also absent in the ovary of naked mole-rats (Figs. 3, 5; Supplemental Fig. S3), where the strongest expression of mNeat1_2 is observed in ovaries in mice. Weak expression of nNEAT1_2 was observed in a subset of granulosa cells, but these cells lacked the expression of nNONO, and nNEAT1_2 transcripts were diffusedly localized in the nucleus without forming paraspeckles (arrowheads in Fig. 5; Supplemental Fig. S3). Along with these atypical expressions of nNEAT1_2, we found that the paraspeckle marker nNONO formed distinct large foci in the cells located at the bottom region of colon epithelium, even though they lacked the expression of nNEAT1_2 (arrows in Fig. 5; Supplemental Fig. S3).
FIGURE 5.
Formation of paraspeckles in various cell types in naked mole-rat tissues. Simultaneous detection of nNEAT1_2 and a paraspeckle marker nNONO in various tissues of naked mole-rats. Prominent paraspeckle formations identified by the colocalization of nNEAT1_2 and nNONO were observed in a wide range of cell types. Arrows indicate the large nNONO foci in the bottom regions of colon epithelium that lacked the expression of nNEAT1_2. Arrowheads indicate a subset of granulosa cells that lacked the expression of nNONO, where nNEAT1_2 was diffusedly localized in the nucleus. Scale bar, 10 µm.
The essential paraspeckle protein FUS is not expressed in the distal regions of intestinal epithelium despite the formation of paraspeckles in naked mole-rats
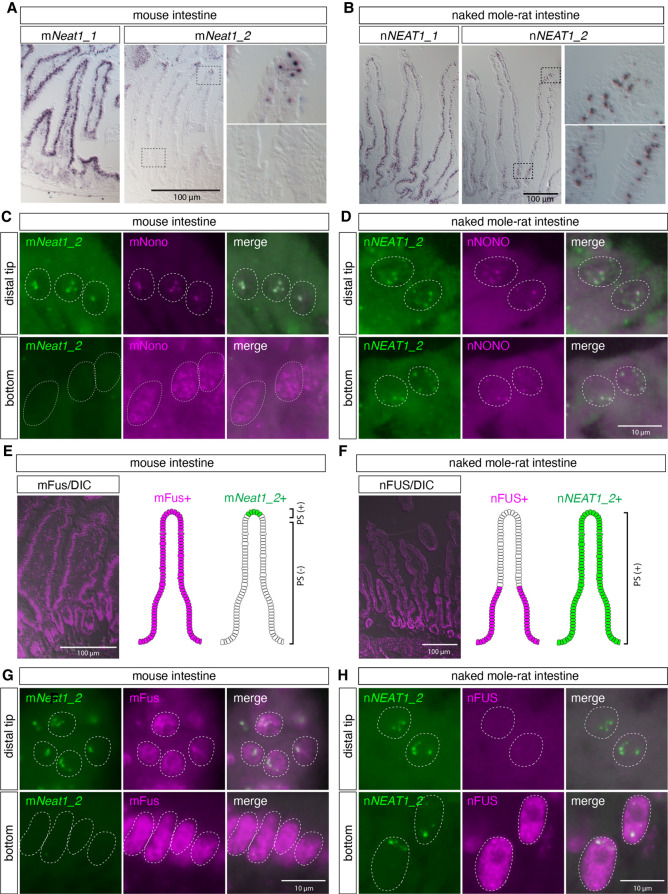
As described above, the expression of NEAT1_2 in intestinal epithelium of naked mole-rats was very different to that of mNeat1_2 in mice, which exhibited highly restricted expression, only in the distal tip cells of the intestinal epithelium (Fig. 6A,B). We thus decided to further investigate the expression of nNEAT1_2 and paraspeckle marker proteins in this tissue. As previously reported, colocalization of mNeat1_2 and mNono was found only in the tip of the intestinal epithelium in mice (Fig. 6C; Supplemental Fig. S1). On the other hand, nNEAT1_2 signals overlapped with nNONO, both in the distal tip, and the bottom region of the intestinal epithelium (Fig. 6D; Supplemental Fig. S1). These observations suggested that paraspeckle formation is differentially regulated by species-specific expression of NEAT1_2 in particular tissue or cell types in each species. To further confirm the formation of paraspeckles in the basal region of intestinal epithelium in naked mole-rats, we examined the localization of nFUS, an essential component of paraspeckles that assembles the NEAT1 ribonucleoprotein complex to build the nuclear bodies (Naganuma et al. 2012; Shelkovnikova et al. 2014; West et al. 2016). Unexpectedly, the expression of nFUS was not detected in the distal regions of intestinal epithelium in naked mole-rats (Fig. 6F,H), despite the distinct formation of paraspeckles confirmed by the colocalization of nNEAT1_2 and nNONO (Fig. 6D; Supplemental Fig. S1). On the other hand, mFus was ubiquitously expressed in the entire region of intestinal epithelium in mice (Fig. 6E,G). The lack of nFUS expression in the paraspeckle-forming cells was rather inconsistent with a previous observation showing that hFUS/mFus is essential for the formation of paraspeckles in human and mouse (Naganuma et al. 2012; Shelkovnikova et al. 2014; West et al. 2016).
FIGURE 6.
nNEAT1_2 is expressed in the entire regions of intestinal epithelium in naked mole-rats, whereas the essential paraspeckle component nFUS is not expressed in the distal region of intestinal epithelium in naked mole-rats. (A,B) In situ hybridization analyses of mNeat1/nNEAT1 in intestine of mice (A) and naked mole-rats (B). Note that the expression of mNeat1_2 is restricted to the distal tip cells in the intestinal epithelium of mice, whereas the expression of nNEAT1_2 is broadly observed throughout the epithelium in naked mole-rats. (C,D) Simultaneous detection of mNeat1_2/nNEAT1_2 (green) and the paraspeckle marker mNono/nNONO (magenta) in the top and the bottom regions of intestinal epithelium in mice (C) and naked mole-rats (D). Paraspeckle formation was observed in the bottom region of intestinal epithelium in naked mole-rats. (E,F) The expression of mFus/nFUS (magenta) in intestinal epithelium in mice (E) and naked mole-rats (F) and schematic drawing of the expression of mFus/nFUS and mNeat1_2/nNEAT1_2 (green) in the epithelium. DIC: differential interference contrast images. (G,H) Simultaneous detection mNeat1_2/nNEAT1_2 (green), and mFus/nFUS (magenta) in the top and the bottom regions of intestinal epithelium in mice (G) and naked mole-rats (H). Note the formation of paraspeckles in top region of the epithelium of naked mole-rats despite the lack of the expression of essential paraspeckle protein nFUS. Scale bars, 100 µm in A, B, E, F and 10 µm in C, D, G, H.
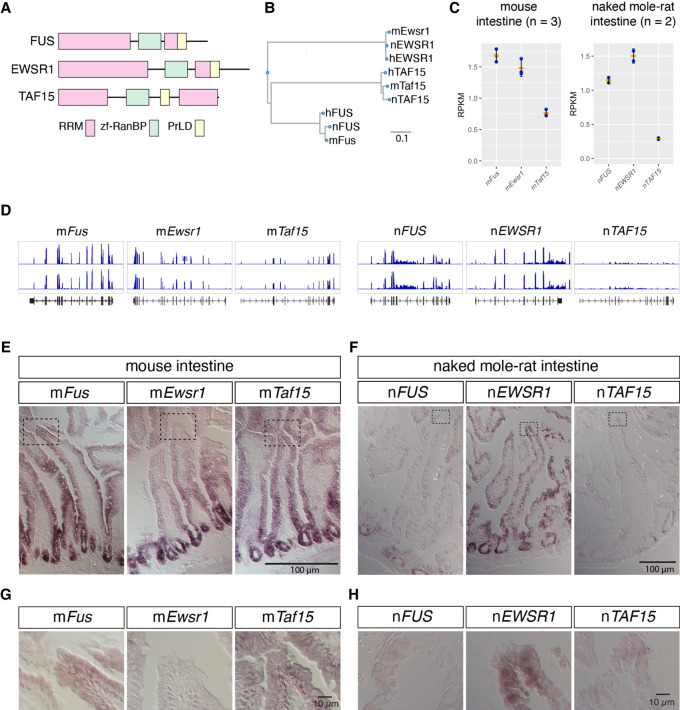
To investigate the molecular mechanisms that lead to the formation of paraspeckles in the absence of nFUS in the distal region of intestinal epithelium in naked mole-rats, we examined the expression of nEWSR1 and nTAF15, the two other members of the FET (FUS, EWS, and TAF15) protein family that share similar domain structures characterized by amino-terminally located long prion-like domains (PrLD) followed by RNA recognition motifs (RRM), zf-RanBP domain, and the second PrLD containing RGG repeats (Fig. 7A; Supplemental Fig. S4A; Schwartz et al. 2015). The putative amino acid sequences of naked mole-rat FET family proteins were highly homologous to human or mouse genes, showing >95% identity at the amino acid level (Fig. 7B; Supplemental Fig. S2B). Because antibodies that recognize nEWSR1 and nTAF15 in naked mole-rats were not available as far as we tested, we analyzed RNA-seq data and compared RPKM (reads per kilobase of transcript, per million mapped reads) for each FET family protein in the intestine of mice and naked mole-rats (Fig. 7C,D). We found that the RPKM of mFus was similar to that of mEwsr1 in mouse intestine, whereas RPKM of nEWSR1 was greater than nFUS in naked mole-rat intestine (Fig. 7C,D). The RPKM of TAF15 was smaller compared to the other two FET family proteins both in mice and naked mole-rats (Fig. 7C,D). To examine the spatial expression pattern of mRNAs of FET family proteins in the intestine of mice and naked mole-rats, we then performed in situ hybridization using RNA probes specific to these genes. In mice, mFus was widely expressed in the entire intestinal epithelium (Fig. 7E,G). On the other hand, the expression of nFUS was high in the crypt region of intestinal epithelium where proliferating stem cells reside, but only weak signals were detected in the other part of the epithelium (Fig. 7F,H). These observations were consistent with the gradual decrease of nFUS expression in the distally located cells that are displaced during the turnover of the epithelium (Fig. 6F). Almost complete lack of nFUS protein in the distal tip cells (Fig. 6H) might be due to translational repression or protein degradation, because we detected faint nFUS mRNA signals in this area (Fig. 7H). In contrast, nEWSR1 was highly expressed throughout the entire length of the intestinal epithelium, whereas only weak signals of nTAF15 were observed (Fig. 7F,H). These observations suggested that nEWSR1 functions redundantly in the distal regions of intestinal epithelium in naked mole-rats to compensate for the lack of nFUS expression during the formation of paraspeckles in this area.
FIGURE 7.
mRNAs of FET family proteins are redundantly expressed in intestine of naked mole-rats. (A) Domain structures of FET family proteins. PrLD: prion like domain, RRM: RNA recognition motif, zf-RanBP: zinc finger motif in Ran binding protein. The PrLD were predicted using PLAAC (Lancaster et al. 2014). (B) Phylogenetic tree of FET family proteins of human (h), mouse (m), and naked mole-rat (n). Scale bar indicates the amino acid substitution ratio per residue. (C) Quantification of gene expression of FET family proteins. The expression was estimated by RNA-seq analyses for mouse intestine and NS-fibroblasts. RPKM: reads per kilobase of transcript per million mapped reads. Note the higher expression of nEWSR1 compared to nFUS in naked mole-rats. Orange bars represent the mean value, blue dots represent the value of each sample, and black bars represent standard deviations. (D) Distribution of RNA-seq reads (blue) mapped to FET family genes in the genome of mice (mm10) and naked mole-rats (hetGla2). Gene structures are shown in black color in the bottom track. (E–H) In situ hybridization analyses of the expression of FET family proteins in the intestine of mice (E,G) and naked mole-rats (F,H). Dashed boxes in E and F indicate the regions shown at a higher magnification in G and H. Note that nEWSR1 was expressed in the distal tip cells of the intestinal epithelium of naked mole-rats. Scale bars, 100 µm in E and F, and 10 µm in G and H.
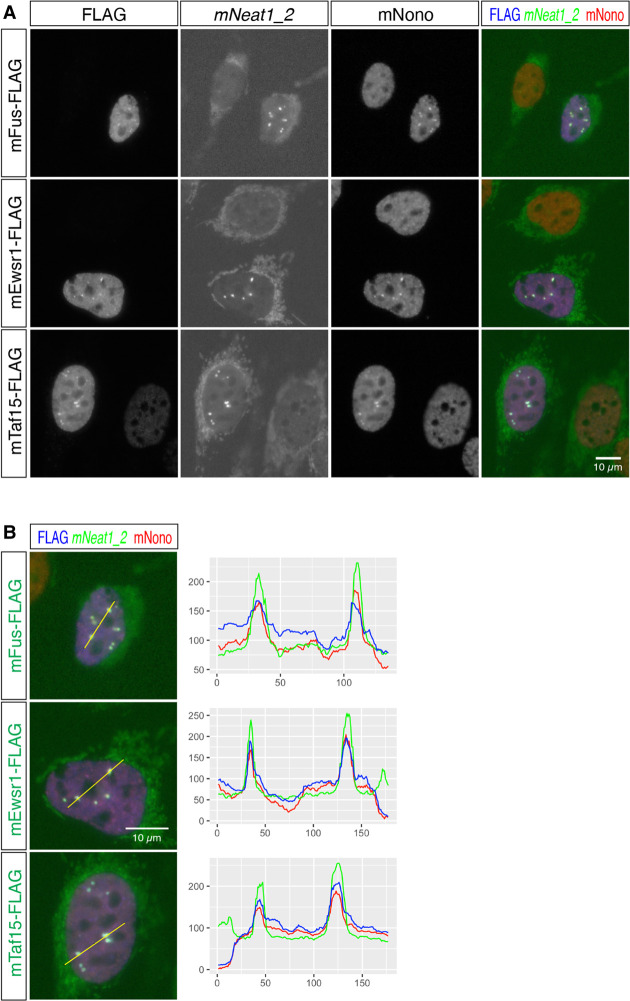
FET family proteins redundantly regulate the formation of paraspeckles in MEF derived from mFus knockout mice
Finally, we examined if mEwsr1 or mTaf15 can redundantly induce the formation of paraspeckles using MEF derived from mFus knockout (KO) mice (West et al. 2016). Because only putative protein sequences were available for naked mole-rats (i.e., RefSeq ID with suffix XP), we used FET family genes from mouse for the rescue experiments (Fig. 8A). As we have previously reported (West et al. 2016), the exogenous expression of mFus restored the formation of paraspeckles, which was confirmed by colocalization of mNeat1_2 and mNono (Fig. 8B). The formation of paraspeckles was also observed in the mFus KO MEF expressing mEwsr1 and mTaf15 (Fig. 8B), indicating that the FET family proteins redundantly function to regulate the formation of paraspeckles.
FIGURE 8.
FET family proteins redundantly rescue the formation of paraspeckles in MEF lacking the expression of mFus. (A) Simultaneous detection of mNeat1_2, exogenously expressed FLAG-tagged FET family proteins, and the paraspeckle marker mNono in MEF derived from mFus KO mice. Note the absence of paraspeckle formation in mFus KO MEF and the rescue of the paraspeckle formation in the cells expressing FET family proteins. The cytoplasmic signals in the green channel (mNeat1_2 channel) were derived from cross-reactions of streptavidin to endogenous biotinylated proteins in the mitochondria. (B) Merged images of exogenously expressed FLAG-tagged FET family proteins (blue), mNeat1_2 (green), and paraspeckle marker mNono (red), and the intensity profile plots. Yellow lines indicate the region analyzed in the intensity profile plots. X-axis represents the number of pixels and y-axis represents arbitrary units for the intensity of the signals of each channel. Scale bars, 10 µm.
DISCUSSION
Here we have revealed the expression patterns and subcellular localizations of the homologs of the two abundant lncRNAs, nMALAT1 and nNEAT1, in naked mole-rats. While the expression of nMALAT1 was almost the same as the expression of mMalat1, we observed striking differences between the expression of nNEAT1 and mNeat1 in intestinal epithelium. nNEAT1_2 was broadly expressed along the entire length of the epithelium, and paraspeckle formation was ubiquitously observed in the nNEAT1_2 expressing cells. The expression pattern of nNEAT1_2 is quite different from that of mNeat1_2, which is highly restricted to the distal tip cells of the intestinal epithelium in mice. On the other hand, the uniform expression of nNEAT1_1/mNeat1_1 was commonly observed in both mice and naked mole-rats. The production of hNEAT1_2 in HeLa cells is controlled by opposing activities of CPSF6 and HNRNPK that promote and inhibit the use of the alternative polyadenylation site required for hNEAT1_1 production, respectively (Naganuma et al. 2012). However, it remains unknown if a similar mechanism is operating in tissues of living animals, because the two RNA binding proteins are ubiquitously expressed in all of the intestinal epithelium in mice (S Nakagawa, unpubl.), despite the variable ratios of the expression of mNeat1_1 and mNeat1_2 in different tissues and cell types (Isobe et al. 2020).
It should be noted that the half-life of p53 is 10 times longer in embryonic fibroblast cells from naked mole-rats when compared with human or mouse embryonic fibroblast cells, resulting in constitutive accumulation of p53 in the nucleus in the absence of DNA damage (Deuker et al. 2020). Considering mNeat1 is regulated under the control of p53 (Adriaens et al. 2016; Mello et al. 2017), the increased amount of p53 possibly leads to hyper-activation of nNEAT1 promoter in naked mole-rats. The overproduction of nNEAT1 may induce preferential readthrough of 5′ located polyadenylation, resulting in the production of nNEAT1_2 in a broader range of cells in the intestinal epithelium. It is also possible that species-specific modification of HNRNPK and/or CPSF6 may modulate the activity of these proteins, resulting in the differential use of the polyadenylation signal in mice and naked mole-rats. It will be essential to investigate the expression pattern of paraspeckle proteins and other regulatory proteins in detail to further clarify the molecular mechanisms that lead to rather ubiquitous formation of paraspeckles in the intestine of naked mole-rats.
Although the distal region of the intestinal epithelium lacked the expression of nFUS in naked mole-rats, distinct formation of paraspeckles was nevertheless observed there. This observation contradicted the prior observations from mice and cell lines that hFUS/mFus plays an essential role to build paraspeckles (Naganuma et al. 2012; Shelkovnikova et al. 2014; West et al. 2016). We have shown that two FET-family RNA binding proteins, nEWSR1 and nTAF15, were expressed in the nFUS-negative distal regions, at least at the mRNA level, in naked mole-rats, and the expression of these proteins rescued the formation of paraspeckles in MEF lacking the expression of mFus. The formation of paraspeckles may thus be regulated by differential expression of paraspeckle proteins that have redundant functions in particular cell types in different species. While the domain structures of FET family proteins are conserved between the three proteins, the amino acid compositions of the PrLD are diverse. The amino-terminal PrLD is enriched in G, Q, S, Y residues in all FET family proteins; however, a much higher ratio of P and T is observed in EWSR1. The carboxy-terminal PrLDs commonly contain multiple RGG sequences, but an additional heptapeptide repeat of YGGDRGG is observed in TAF15. A recent study has shown that FET family proteins undergo a phase transition and form liquid droplets in vitro at different protein concentrations: FUS formed liquid droplets at the lowest concentration and EWSR1 at the highest concentration among the FET family proteins, and this property was regulated by the amino acid compositions of the amino-terminal PrLD (Wang et al. 2018). It would be intriguing to study if the dynamics and biophysical properties of paraspeckles are regulated by differential combinations of FET family proteins, which may result in cell type- or species-specific biological functions of paraspeckles.
We observed atypical expression of paraspeckle components nNEAT1_2 and nNONO in naked mole-rats. In the bottom regions of colon epithelium, nNONO formed distinct large foci despite the lack of nNEAT1_2 expression. These foci did not overlap with markers for the nucleolus (S Nakagawa, unpubl.), and may represent novel nuclear bodies. Considering that the formation of nuclear bodies are regulated by architectural noncoding RNAs (Nakagawa et al. 2021), it will be intriguing to investigate if the formation of these nNONO foci is sensitive to RNase treatment. Another atypical example for the localization of paraspeckle components was the expression of nNEAT1_2 in granulosa cells in naked mole-rat ovary. In mice, corpus luteal cells in the ovary express the highest level of mNeat1_2 among the tissues we examined (Nakagawa et al. 2011). Considering that only queen females can reproduce (Buffenstein et al. 2022), this cell type seems to be absent in the ovary of the nonbreeder female we examined, due to the lack of estrus cycle. Instead, we observed weak but distinct signals of nNEAT1_2 in a subset of granulosa cells that lacked the expression of nNONO. Because NONO is required for the stabilization of NEAT1_2 in human or mouse cells, the signals we detected with probes against nNEAT1_2 might be derived from partially degraded transcripts that failed to form paraspeckles. Alternatively, nNONO function as a type Ib paraspeckle protein (Naganuma et al. 2012), which is required for assembly of nNEAT1_2 ribonucleoprotein complex but is not involved in the stabilization of this transcript in this particular cell type. Whichever the case is, these observations raise the possibility that the formation and regulation of paraspeckles are differentially regulated in different cell types in the tissues of living animals.
MATERIALS AND METHODS
Animals
All experiments were approved by the safety divisions of Hokkaido University (#2015-079 and #14-0065) and Kumamoto University (A30-043 and A2020-042). C57BL6/N were used for all experiments for mouse studies. The naked mole-rat colonies were maintained in Hokkaido University and Kumamoto University. For anesthetization, medetomidine-midazolam-butorphanol (Kawai et al. 2011) was intraperitoneally injected at a volume of 10 µL/g of body weight. Animals used are as follows: BC16 (1 yr 8 mo, male) and GH42 (1 yr 11 mo, female) for expression studies shown in Figures 3–5; D21 (13 mo, male) and D22 (13 mo, male) for expression studies shown in Figure 6; R19 (14 mo, male) and R20 (14 mo, male) for expression studies shown in Figure 7; and L83 (12 mo, male) and CG14 (12 mo, male) for RNA sequencing and northern blot analyses shown in Figures 1 and 7.
Cell culture
NS-fibroblasts were prepared from adult skin as previously described (Miyawaki et al. 2016). NS-fibroblasts were cultured in DMEM (high glucose, Sigma #D5796) at 32°C under a humidified atmosphere containing 5% CO2 and 5% O2. MEF were cultured in DMEM/HamF12 (Wako #044-29765, Japan) at 37°C in 5% CO2 and 20% O2 conditions.
Deep sequencing and data analyses
Total RNAs were extracted from NS-fibroblasts and intestine using TRIzol (Thermo Fisher Scientific #15596018). The cellular lysate in TRIzol was incubated at 50°C for 5 min to enhance solubilization of semi-extractable lncRNAs as previously described (Chujo et al. 2017). RNA-seq libraries were made following a standard protocol using a Ribo-Zero Gold rRNA Removal Kit (Human/Mouse/Rat) (Illumina) and a TruSeq Stranded mRNA Library Prep (Illumina) for DRA013388; and a TruSeq Stranded Total RNA Library Prep Gold 20020598 for DRA014061. Deep sequencing was performed using HiSeq 4000 in QB3 Genomics at UC Berkeley for DRA013388 and at Macrogen for DRA014061. Sequence reads were mapped to the hetGl2 genome using HISAT2 (Kim et al. 2019). To identify nMALAT1 and nNEAT1, the syntenic region of the naked mole-rat genome containing SCYL1 and FRMD8 was identified by searching the gene names using the UCSC Genome Browser (https://genome.ucsc.edu/), and mapped reads in the corresponding genomic regions were visualized using the IGV Genome Browser (https://software.broadinstitute.org/software/igv/). To estimate the expression of FET family proteins, the number of reads mapped to each gene was counted using featureCounts (Liao et al. 2014) and normalized with the total number of reads and the length of the genes. For gene annotation, NCBI RefSeq GTF files were downloaded from the UCSC Table Browser. The sequencing data obtained in this study are available at DRA013388 and DRA014061. For the quantification of the expression of FET family protein mRNAs in mouse intestine, fastq files in a public database (SRR18508210, SRR18508211, SRR18508212) were used for the analyses.
5′ RACE
5′ RACE was performed using the SMARTer RACE 5′/3′ Kit (Takara #634858) following the manufacturer's instructions. cDNAs were synthesized using random primers and used for subsequent adapter ligation and nested PCR to obtain the 5′ end fragments. Obtained PCR products were cloned into a plasmid vector, and the sequences were determined following standard procedures. Primers used for the first round of PCR and the second nested PCR are listed in Supplemental Table S1.
Probe preparations
DNA was extracted from the finger of naked mole-rats using a standard protocol. cDNA was synthesized using ReverTra Ace (Toyobo #TRT-101) following manufacturer's instructions. To prepare templates for the probes, PCR was performed using Quick Taq HS DyeMix (Toyobo #DTM-101) and genomic DNAs or cDNAs as templates. Primers used to amplify the probe sequences are listed in Supplemental Table S1. PCR conditions used were predenatured at 94°C for 2 min, followed by 30 cycles of denature at 94°C for 30 sec, annealing at 58°C for 30 sec, and extension at 68°C for 1 min. Amplified PCR fragments were cloned into pCRII-TOPO using a TOPO TA Cloning Kit (Thermo Fisher #452640) following manufacturer's instructions. To prepare templates for RNA probe synthesis, PCR was performed using the M13 forward and reverse primers and the plasmid DNAs as templates, and the amplified DNA fragments were purified using the Wizard SV Gel and PCR Clean-Up System (Promega #A9282). DIG-labeled RNA probes were synthesized using T7 or SP6 RNA polymerase and DIG RNA Labeling Mix (Roche #11277073910), and free-nucleotides were removed using Centri-Sep Spin Columns (Princeton Separations #CS-901).
Northern blot analyses
Five micrograms of total RNAs were separated on 1% agarose gel containing 2% formaldehyde, and gels were treated with 0.05 N NaOH for 20 min to enhance transfer of high molecular weight RNAs, which step was essential to detect mNeat1_2/nNEAT1_2. RNAs were transferred on a positively charged Nylon membrane (Merck #11209299001) using standard protocol, and probe hybridization and detection were performed using DIG Easy Hyb (Merck #11603558001), anti-Dig AP (Merck #11093274910), and CSPD-Star (Merck #11685627001) following manufacturer's instructions. Chemiluminescence signals were detected with the ChemiDoc Touch Imaging System (Bio-Rad).
In situ hybridization and immunofluorescent staining
In situ hybridization was performed as previously described (Mito et al. 2016). Briefly, cultured cells were fixed in 4% paraformaldehyde in calcium-free Hepes-buffered saline HCMF (10 mM HEPES [pH 7.4], 137 mM NaCl, 5.4 mM KCl, 0.34 mM Na2HPO4, 5.6 mM glucose) for 10 min at room temperature, permeabilized with 0.1% TritonX-100 for 10 min at room temperature, and hybridized with DIG- or FITC-labeled probes overnight in 2× SSC containing 50% formamide at 55°C. After washing in 2× SSC containing 50% formamide, probes nonspecifically bound to the cells were removed by RNaseA treatment, and further washed in 2× SSC and 0.2× SSC for 30 min at 55°C. Hybridized probes and proteins were detected with primary and secondary antibodies listed in Supplemental Table S1. To detect mRNAs on tissue sections, tissues were embedded in OCT compounds (Sakura) and fresh-frozen in ethanol with dry ice. Cryosections at a thickness of 8 µm were collected on PLL-coated slide glasses, and fixed overnight in 4% paraformaldehyde in HCMF. The samples were then treated with 0.2N HCl, 3.3 µg/mL of proteinase K in a 10× TE buffer (100 mM Tris-Cl [pH 8.0] and 10 mM EDTA), and were acetylated with acetic anhydride in triethanolamine buffer before hybridization, as previously described (Mito et al. 2016). Probes were detected by chromogenic reaction using alkaline phosphatase-conjugated anti-DIG antibody and NBT/BCIP as substrates, or by antibodies against DIG or FITC followed by detection with fluorescently labeled secondary antibodies using standard immunofluorescent method. nFUS/mFUS and nNONO/mNono were detected using the post-hybridization tissue sections or cultured cells, because it gave stronger signals than normally fixed and permeabilized cells. The primary antibodies against these proteins were mixed with the primary antibody that detects FITC-labeled probes and detected with fluorescently labeled secondary antibodies. The fluorescent and DIC images were obtained using an epifluorescent microscope (BX51; Olympus) equipped with a CCD camera (DP74). Profiling of the signal intensity was measured by the “Color profiler” tool of Fiji (https://imagej.net/software/fiji/). The numbers of paraspeckles in the control and nNeat1_2 depleted cells were counted by processing the signals of nNONO with the “Find Maxima” function of Fiji, with the prominence parameter set as 40, which was optimized so that we could get the same number of paraspeckles with signals of nNONO and nNEAT1_2 in the control cells.
Depletion of nNEAT1_2 by ASO
ASO-mediated knockdown of nNEAT1_2 was performed following the procedures described previously (Ishida et al. 2015). Gapmer antisense oligonucleotides (ASOs) were synthesized by Integrated DNA Technologies. Briefly, ASOs were mixed with Lipofectamine 2000 (Thermo Fisher), and NS-fibroblast cells were reverse-transfected with the complex following the manufacturer's instructions. After 24 h, cells were fixed and processed for in situ hybridization or were lysed in TRIzol to purify RNAs. The sequences of ASOs are shown in Supplemental Table S1.
RT-qPCR analyses
cDNAs were synthesized using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo #FSQ-301) following manufacturer's instructions. An amount of 500 ng of total RNAs was used as templates and 1/50 volumes of synthesized cDNAs were used for subsequent qPCR analyses. qPCR reactions were performed using the THUNDERBIRD SYBR qPCR Mix (Toyobo #QPS-201) and the CFX Connect Real-Time System (Bio-Rad) with the following conditions: 95°C for 1 min followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec. The qPCR primers used in this study are listed in Supplemental Table S1. Relative expression for each gene was calculated by the ΔCt method using Gapdh as a standard.
Production and infection of lentivirus
Lentiviruses were prepared using the ViraPower Lentiviral Expression System (Thermo Fisher) according to the manufacturer's instructions. FLAG-tagged full-length cDNA sequences of mFus, mEwsr1, and mTaf15 were obtained by PCR using primers listed in Supplemental Table S1 and cloned into pLenti6/V5-DEST using the Gibson Assembly system, following the manufacturer's instruction. For lentiviral infection, mFus KO MEF cells were incubated overnight with culture supernatants obtained from virus-producing 293T cells transfected with packaging vectors, and were further incubated for 48 h in a fresh medium.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science/Ministry of Education, Culture, Sports, Science and Technology (JSPS/MEXT) granted to S.N. (21H05274, 21K19246, and 17H03604), K.M. (21H02392 and 21H05143), and Y.K. (19K06469); a Grant-in-Aid for Early-Career Scientists from JSPS granted to M.K. (21K15012); JST PRESTO Program (1159399) granted to M.K.; and JST FOREST Program (MJFR216C) granted to K.M.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079135.122.
REFERENCES
- Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W, et al. 2016. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med 22: 861–868. 10.1038/nm.4135 [DOI] [PubMed] [Google Scholar]
- Ali T, Grote P. 2020. Beyond the RNA-dependent function of LncRNA genes. Elife 9: e60583. 10.7554/eLife.60583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffenstein R, Amoroso V, Andziak B, Avdieiev S, Azpurua J, Barker AJ, Bennett NC, Brieno-Enriquez MA, Bronner GN, Coen C, et al. 2022. The naked truth: a comprehensive clarification and classification of current ‘myths’ in naked mole-rat biology. Biol Rev Camb Philos Soc 97: 115–140. 10.1111/brv.12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Yamazaki T, Kawaguchi T, Kurosaka S, Takumi T, Nakagawa S, Hirose T. 2017. Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. EMBO J 36: 1447–1462. 10.15252/embj.201695848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, et al. 2011. The reality of pervasive transcription. PLoS Biol 9: e1000625. discussion e1001102. 10.1371/journal.pbio.1000625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuker MM, Lewis KN, Ingaramo M, Kimmel J, Buffenstein R, Settleman J. 2020. Unprovoked stabilization and nuclear accumulation of the naked mole-rat p53 protein. Sci Rep 10: 6966. 10.1038/s41598-020-64009-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, et al. 2012. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol 9: 1076–1087. 10.4161/rna.21089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong KW, Li Y, Wang W, Ma W, Li K, Qi RZ, Liu D, Songyang Z, Chen J. 2013. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J Cell Biol 203: 149–164. 10.1083/jcb.201303145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. 2002. Paraspeckles: a novel nuclear domain. Curr Biol 12: 13–25. 10.1016/S0960-9822(01)00632-7 [DOI] [PubMed] [Google Scholar]
- Fox AH, Nakagawa S, Hirose T, Bond CS. 2018. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem Sci 43: 124–135. 10.1016/j.tibs.2017.12.001 [DOI] [PubMed] [Google Scholar]
- Gil N, Ulitsky I. 2020. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet 21: 102–117. 10.1038/s41576-019-0184-5 [DOI] [PubMed] [Google Scholar]
- Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, et al. 2015. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol 210: 529–539. 10.1083/jcb.201504117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. 2007. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39. 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K, Miyauchi K, Kimura Y, Mito M, Okada S, Suzuki T, Nakagawa S. 2015. Regulation of gene expression via retrotransposon insertions and the noncoding RNA 4.5S RNAH. Genes Cells 20: 887–901. 10.1111/gtc.12280 [DOI] [PubMed] [Google Scholar]
- Isobe M, Toya H, Mito M, Chiba T, Asahara H, Hirose T, Nakagawa S. 2020. Forced isoform switching of Neat1_1 to Neat1_2 leads to the loss of Neat1_1 and the hyperformation of paraspeckles but does not affect the development and growth of mice. RNA 26: 251–264. 10.1261/rna.072587.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S, Takagi Y, Kaneko S, Kurosawa T. 2011. Effect of three types of mixed anesthetic agents alternate to ketamine in mice. Exp Anim 60: 481–487. 10.1538/expanim.60.481 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F, Mendell JT. 2018. Functional classification and experimental dissection of long noncoding RNAs. Cell 172: 393–407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AK, Nutter-Upham A, Lindquist S, King OD. 2014. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 30: 2501–2502. 10.1093/bioinformatics/btu310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Harvey AR, Hodgetts SI, Fox AH. 2017. Functional dissection of NEAT1 using genome editing reveals substantial localization of the NEAT1_1 isoform outside paraspeckles. RNA 23: 872–881. 10.1261/rna.059477.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Mello SS, Sinow C, Raj N, Mazur PK, Bieging-Rolett K, Broz DK, Imam JFC, Vogel H, Wood LD, Sage J, et al. 2017. Neat1 is a p53-inducible lincRNA essential for transformation suppression. Genes Dev 31: 1095–1108. 10.1101/gad.284661.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito M, Kawaguchi T, Hirose T, Nakagawa S. 2016. Simultaneous multicolor detection of RNA and proteins using super-resolution microscopy. Methods 98: 158–165. 10.1016/j.ymeth.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Miyawaki S, Kawamura Y, Oiwa Y, Shimizu A, Hachiya T, Bono H, Koya I, Okada Y, Kimura T, Tsuchiya Y, et al. 2016. Tumour resistance in induced pluripotent stem cells derived from naked mole-rats. Nat Commun 7: 11471. 10.1038/ncomms11471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modic M, Grosch M, Rot G, Schirge S, Lepko T, Yamazaki T, Lee FCY, Rusha E, Shaposhnikov D, Palo M, et al. 2019. Cross-regulation between TDP-43 and paraspeckles promotes pluripotency-differentiation transition. Mol Cell 74: 951–965.e913. 10.1016/j.molcel.2019.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. 2012. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J 31: 4020–4034. 10.1038/emboj.2012.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, Hirose T. 2011. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol 193: 31–39. 10.1083/jcb.201011110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Ip JY, Shioi G, Tripathi V, Zong X, Hirose T, Prasanth KV. 2012. Malat1 is not an essential component of nuclear speckles in mice. RNA 18: 1487–1499. 10.1261/rna.033217.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Shimada M, Yanaka K, Mito M, Arai T, Takahashi E, Fujita Y, Fujimori T, Standaert L, Marine JC, et al. 2014. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 141: 4618–4627. 10.1242/dev.110544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Yamazaki T, Hirose T. 2018. Molecular dissection of nuclear paraspeckles: towards understanding the emerging world of the RNP milieu. Open Biol 8: 180150. 10.1098/rsob.180150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Yamazaki T, Mannen T, Hirose T. 2021. ArcRNAs and the formation of nuclear bodies. Mamm Genome 33: 382–401. 10.1007/s00335-021-09881-5 [DOI] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. 2009. MEN ε/β a noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci 106: 2525–2530. 10.1073/pnas.0807899106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Cech TR, Parker RR. 2015. Biochemical properties and biological functions of FET proteins. Annu Rev Biochem 84: 355–379. 10.1146/annurev-biochem-060614-034325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova TA, Robinson HK, Troakes C, Ninkina N, Buchman VL. 2014. Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum Mol Genet 23: 2298–2312. 10.1093/hmg/ddt622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souquere S, Beauclair G, Harper F, Fox A, Pierron G. 2010. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell 21: 4020–4027. 10.1091/mbc.e10-08-0690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standaert L, Adriaens C, Radaelli E, Van Keymeulen A, Blanpain C, Hirose T, Nakagawa S, Marine JC. 2014. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA 20: 1844–1849. 10.1261/rna.047332.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statello L, Guo CJ, Chen LL, Huarte M. 2021. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22: 96–118. 10.1038/s41580-020-00315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. 2009. MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359. 10.1101/gr.087775.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938. 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, et al. 2018. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174: 688–699.e616. 10.1016/j.cell.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Mito M, Kurosaka S, Takumi T, Tanegashima C, Chujo T, Yanaka K, Kingston RE, Hirose T, Bond C, et al. 2016. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J Cell Biol 214: 817–830. 10.1083/jcb.201601071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 2008. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919–932. 10.1016/j.cell.2008.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. 2012. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26: 2392–2407. 10.1101/gad.204438.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, Hirose T. 2018. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol Cell 70: 1038–1053.e1037. 10.1016/j.molcel.2018.05.019 [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Yamamoto T, Yoshino H, Souquere S, Nakagawa S, Pierron G, Hirose T. 2021. Paraspeckles are constructed as block copolymer micelles. EMBO J 40: e107270. 10.15252/embj.2020107270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al. 2012. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2: 111–123. 10.1016/j.celrep.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.