Abstract
Humans and pigs share a close contact relationship, similar biological traits, and one of the highest estimated number of viruses compared to other mammalian species. The contribution and directionality of viral exchange between humans and pigs remain unclear for some of these viruses, but their transmission routes are important to characterize in order to prevent outbreaks of disease in both host species. This review collects and assesses the evidence to determine the likely transmission route of 27 viruses between humans and pigs.
Keywords: Zoonosis, reverse zoonosis, viruses, pig, human
INTRODUCTION
Viruses circulating in wildlife reservoirs can spillover into susceptible human populations and contribute significantly to the global burden of human infectious diseases, which cause approximately 2.5 billion infections and 2.7 million deaths each year [1, 2]. Before emerging as zoonotic human pathogens, wildlife‐adapted viruses must first overcome a series of epidemiological barriers, such as behavioral barriers (level of human exposure to zoonotic viruses), interspecies barrier, and immunological barriers [3].
Livestock are able to facilitate viral spillover from wildlife to humans by acting as “epidemiological bridges” or intermediate hosts in the transmission chain [4, 5]. Unsurprisingly, through thousands of years of close contact animal husbandry and intensive farming in recent decades, domesticated animals harbor eight times more zoonotic viruses than predicted in other non‐domesticated mammalian species [6]. Opportunities for viral zoonosis accompany the expansion of human agricultural activities, which provoked over 50% of zoonotic emerging infectious disease (EID) events during the past 70 years [7]. Wildlife, however, is not the only threat to livestock; close contact humans can also be a source of viral zoonosis (hereafter referred to as reverse zoonosis and also known as zooanthroponosis and anthroponosis), which is somewhat understudied [8].
A recent study estimated that humans exchange the highest number of viruses with domesticated pigs (Sus scrofa domesticus) (n ≈ 31 viruses), cattle (n ≈ 31 viruses), horses (n ≈ 31 viruses), and dogs (n ≈ 27 viruses), surpassing both domestic cats (n ≈ 16 viruses) and goats (n ≈ 22 viruses) [6]. Pigs have served as intermediate, amplification, and “mixing” hosts in past human epidemics and pandemics (e.g., Japanese encephalitis [9], Nipah [10], and influenza A viruses [11]), and humans have spread viruses to pigs in return (e.g., influenza A virus [12]). Global demand for pork continues to rise and, although pig farming practices differ worldwide, the movement of swine and multiple contact points with humans, i.e., at farms, breeding facilities, slaughterhouses, wet markets, and trade shows, intensifies the opportunities for viral transmission [13, 14, 15]. Furthermore, pigs are increasingly used for xenotransplantation and as animal models for human diseases and conditions due to their physiological, genetic, and immunological similarities to humans [16, 17, 18, 19]. Therefore, understanding the viral exchange at the swine–human interface can help prevent zoonotic and reverse zoonotic viral outbreaks, leading to disease, deaths, culling of swine herds, and economic losses [20].
Predicting EIDs in humans and pigs is challenging. Viral zoonoses are considered rare in humans relative to the extensive viral diversity in the animal kingdom, and viral dynamics are strongly amenable to selection mechanisms resulting in rapid changes to viral landscapes [21, 22, 23, 24]. Spillover events can occur incidentally into “dead‐end” hosts, or viral outbreaks can ensue with sustained onward transmission within the novel host population, and can even become a persistent endemic threat [23, 25]. Determining the natural reservoir species and intermediate hosts of EIDs after a spillover event is also demanding when routine surveillance is not in place [26]. Furthermore, the novel host of an EID can become a newfound viral reservoir and spillover into the next susceptible species, e.g., SARS‐CoV‐2 transmission chain from horseshoe bats‐to‐unknown mammalian intermediate‐to‐humans‐to‐mink‐to‐humans [26, 27, 28].
In this review, we collect genetic‐, pathogenic‐, and immunological‐based evidence to determine the likely direction of viral transmission between humans and pigs with the purpose of identifying viral threats to human and pig health, and the roles humans and pigs play as direct viral reservoirs for each other.
MATERIALS AND METHODS
A framework of factors (Table S1) was designed and applied in scientific literature surveys to assess the infectivity and transmissibility of 27 viruses naturally found in humans and pigs within the past 70 years. The focus is largely on the detection of human or pig‐associated viruses in the secondary host, genetic variation between viral strains isolated from the two hosts, viral entry into target host cells, detection of viral shedding that indicates viral replication in the host and transmission potential, viral dissemination in the host, and the ability for the host’s immune system to suppress infection. This information is highlighted in Table S2 with distinctions drawn between humans and pigs where appropriate. The viruses were then determined to demonstrate zoonotic, reverse zoonotic, or bidirectional viral transmission according to the definitions in Box 1, and the results are summarized in Table 1.
Table 1.
Summary of transmission routes and sources of the 27 reviewed viruses.
| Virus and taxonomy |
Transmission route (→ denotes direction) |
Significant viral reservoir |
|---|---|---|
| Zoonotic viruses (1): Pigs as major sources of viruses | ||
| Eastern equine encephalitis (EEEV); Alphavirus; Togaviridae. |
Mosquito (Aedes, Coquillettidia, and Uranotaenia species) → human/pig [143]: vector‐borne. Pig → mosquito: vector‐borne [43]. Pig → pig/human: oronasal contact with infected oropharyngeal secretions or fecal‐oral [43]. |
Birds are natural hosts (e.g., wading birds, passerine songbirds, and starlings) [143]. Pigs are potential amplification hosts [43]. |
| Japanese encephalitis (JEV); Flavivirus; Flaviviridae. |
Mosquito (Culex and Aedes species) → human/pig: vector‐borne [143]. Pig → mosquito: viremia, vector‐borne [44, 45]. Pig → human: oronasal contact with infected oronasal secretions oronasal secretions [47]. Mosquito → mosquito: transovarial [9]. |
Aquatic birds are natural hosts. Pigs are amplification hosts [9]. |
| Menangle (MenPV); Rubulavirus; Paramyxoviridae. |
Fruit bat (Pteropus species) → pig: oronasal contact with environmental contamination [59, 62]. Pig → pig: fecal‐oral or urinary‐oral or transplacental [144, 145]. Pig → human: possibly infected bodily fluid in cuts [60]. |
Fruit bats (Pteropus species) are natural hosts [59, 62]. Pigs are possible intermediate hosts [60, 61]. |
| Nipah (NiV); Henipavirus; Paramyxoviridae. |
NiV‐Malaysia: Fruit bat (Pteropus species) → pig: oronasal contact with environmental contamination [146]. Pig → pig: airborne or oronasal contact with infected oronasal secretions [147]. Pig → human: airborne or oronasal contact with infected oronasal secretions [148]. NiV‐Bangladesh: Fruit bat (Pteropus species) → human: food‐borne consumption of contaminated date palm sap [149]. Human → human: oronasal contact with infected human bodily fluids, limited transmission chain but caused ˜50% of cases [149]. Pig → human: undocumented but possible [150]. |
Fruit bats (Pteropus species) [151, 152]. Pigs are amplifications hosts for NiV‐Malaysia and potentially for NiV‐Bangladesh [10, 150]. |
| Reston ebola (RESTV); Ebolavirus; Filoviridae. |
Fruit bat (likely Miniopterus species) → pig: oronasal contact with environmental contamination [153]. Pig → pig: oronasal contact with infected nasopharyngeal secretions [58]. Pig → human: oronasal contact with infected nasopharyngeal secretions [58, 154]. |
Fruit bats (likely Miniopterus species) are natural hosts [153]. Pigs are intermediate hosts [154]. |
| Tioman (TioV); Rubulavirus; Paramyxoviridae. |
Fruit bat (Pteropus species) → pig/humans: oronasal contact with environmental contamination [64]. Pig → pig/human: possible airborne or oronasal contact with oronasal secretions [31]. |
Fruit bats (Pteropus species) are natural hosts [31, 63]. Pigs are potentially intermediate hosts [64]. |
| Vesicular stomatitis (VSV); Vesiculovirus; Rhabdoviridae. |
Vertebrate reservoir → biting insect: vector (biological and mechanical [50, 155]). Biting insect → pig/human: vector. Pig → pig/human: possible vector [46, 50], airborne, oronasal contact with infected oronasal secretions, or contact with infected vesicular lesions [48, 49, 50]. |
Unknown vertebrate reservoir host but likely multiple livestock (including pigs) and wildlife species [156]. |
| Zoonotic viruses (2): Pigs as minor sources of viruses | ||
| Banna (BAV); Seadornavirus; Reoviridae. | Mosquito (Culex and Aedes species) → human/pig: vector‐borne [157, 158]. | Potentially mosquito as replication has been demonstrated in mosquito cell line (C6/36) and replication in mammalian cell lines is not possible (BHK‐21 and Vero) [159]. Although replication in mice has been demonstrated (develop viremia), re‐infection was not possible [160]. |
| Cache Valley (CVV); Orthobunyavirus; Bunyaviridae. |
Mosquito (Aedes, Coquillettidia, Culex, Culiseta, Orthopodomyia, Psorophora, and Uranotaenia species) → human/pig: vector‐borne [161, 162]. Mosquito → mosquito: transovarial demonstrated experimentally [163]. |
Deer [164, 165]. |
| Chandipura (CHPV); Vesiculovirus; Rhabdoviridae. |
Sandfly (Phlebotomine) → human/pig: vector‐borne (demonstrated in mice [166]). Sandfly → sandfly: transovarial and venereal [167]. |
Potentially sandfly (Phlebotomine) species as replication has been demonstrated in vector [166]. |
| Encephalomyocarditis (EMCV); Cardiovirus; Picornaviridae. |
Rodent → human/pig: fecal/urinal‐oral [168]. Pig → pig: fecal‐oral or oronasal contact with infected nasal secretions [169]. |
Rodents [169]. |
| Foot‐and‐mouth disease (FMDV); Aphthovirus; Picornaviridae. |
Pig → pig: airborne, oronasal contact with infected oronasal secretions, physical contact with secretions in cuts, environmental contamination (equipment, clothing, animal feed) [170]. Pig → human: potentially by direct contact with secretions through damaged skin [171, 172]. |
African Cape buffalo (Syncerus caffer) (serotypes SAT‐1, 2, and 3) [173]. |
| Getah (GETV); Alphavirus; Togaviridae. |
Mosquito (Culex, Anopheles, Aedes, Armigeres, and Mansonia species) → human/pig: vector‐borne [174]. Pig → pig: vertically to fetus during early stage of pregnancy [175]. |
Potentially cattle (strong serological evidence) [174]. |
| Louping ill (LIV); Flavivirus; Flaviviridae. |
Tick (Ixodes ricinus) → human/pig: vector‐borne [176, 177]. Sheep → human: contact with infected sheep, sheep tissues, or raw milk [176, 177, 178]. |
Ticks (Ixodes ricinus), sheep, and red grouse [176, 177]. |
| Rabies (RABV); Lyssavirus; Rhabdoviridae. |
Canine (Carnivora) or bat (Chiroptera) → pig/human: bite with infected saliva [71]. Pig → pig: uncommon unless infected with “furious” form and bite [73]. Pig → human: undocumented but possible [73]. Human → pig: unlikely due to behavioral factors. Human → human: only through organ/tissue transplant [72]. |
Canine (Carnivora) and bat (Chiroptera) species are natural hosts [71]. |
| Toscana (TOSV); Phlebovirus; Bunyaviridae. |
Vertebrate → sandfly (Phlebotomus): vector‐borne → pig/human [68, 179, 180]. |
Vector reservoir is sandfly (Phlebotomus species). Unknown vertebrate reservoir host but likely multiple livestock and wildlife species. Unclear contribution of pigs in epidemiology [179, 180]. |
| Venezuelan equine encephalitis (VEEV); Alphavirus; Togaviridae. |
Horse or rodent → mosquito (Ochlerotatus or Culex species): vector‐borne [70]. Mosquito → pig/human: vector‐borne [69, 70] Mosquito → human → mosquito: possible humans can develop sufficient viremia to infect mosquito [181]. Human → human: airborne or oronasal contact possible but unproven [182]. |
Horses are amplification host for epidemic subtypes, and rodents are reservoirs for endemic subtypes [70]. |
| Reverse zoonotic viruses | ||
| Norovirus (NoV); Norovirus; Caliciviridae. |
Human → human: depending on strain fecal‐oral, vomit‐oral, food‐/water‐borne (dependent on strain) (reviewed in 86). Human → pig: possibly fecal‐oral, but not directly detected [84, 183, 184]. Pig → pig: fecal‐oral [83]. |
Unknown source of novel strains emerging in human populations but immunocompromised patients in nosomical settings are significant reservoirs [86]. |
| Severe acute respiratory syndrome related‐coronavirus (SARSr‐CoV); Betacoronavirus; Coronaviridae. |
Horseshoe bat (Rhinolophus species) → (unknown mammalian intermediary, possible recombination with pangolin‐CoV) → human: oronasal contact with infected secretions or excretions [26, 75, 185, 186]. Human → human: airborne [187]. Human → pig: foodborne via contaminated animal feed (restaurant leftovers) [76], possibly airborne/oronasal contact [78]. |
Horseshoe bat (Rhinolophus species) are natural hosts [185]. Humans are reservoir hosts [75]. |
| Swine vesicular disease (SVDV); Enterovirus; Picornaviridae. |
Human → pig: possibly fecal‐oral or oronasal contact with infected oronasal secretions or contaminated environment containing recombinant coxsackievirus B (CV‐B) and CV‐A9 [79, 80, 81]. Pig → pig: oronasal contact with environmental contamination during transportation [188]. |
Humans are reservoir hosts for ancestral strain [80]. Virulence decreased through subsequent passages in pigs [81, 189]. |
| Bidirectionally transmitted viruses | ||
| Hepatitis E (HEV); Orthohepevirus; Hepeviridae. |
Pig → human: foodborne, consumption of raw or undercooked pig products, or direct contact [102, 103]. Human → human: fecal‐oral via consumption of feces‐contaminated water (type 1 and 2 in developing countries), or blood transfusion [102, 103]. Pig → pig: fecal‐oral [103]. |
Pigs [102]. |
| Influenza A (IAV); Alphainfluenzavirus; Orthomyxoviridae. |
Human ↔ pig: airborne or oronasal contact with infectious oronasal secretions [190]. Human → human: airborne or oronasal contact with infectious oronasal secretions [190]. Pig → pig: airborne or oronasal contact with infectious oronasal secretions [190]. |
Wild aquatic birds are natural hosts [191]. IAV subtypes circulate in human and pig populations [12]. |
| Influenza C (ICV); Gammainfluenzavirus; Orthomyxoviridae. |
Human ↔ pig: possible but unknown if ICV transmitted from pigs to humans or from humans to pigs [111, 192]. Human → human: airborne or oronasal contact with infectious oronasal secretions [192]. Pig → pig: airborne or oronasal contact with infectious oronasal secretions, demonstrated in contact pigs experimentally infected with human and pig‐derived ICV [113]. |
Humans [192]. |
| Picobirnavirus (PBV); Picobirnavirus; Picobirnaviridae. | Human ↔ pig: fecal‐oral or oronasal contact with infected respiratory secretions [193, 194]. | Prokaryotes in host microbiome are likely hosts [98]. |
| Ross River (RRV); Alphavirus; Togaviridae. |
Marsupial or horse → mosquito (Ades and Culex species): vector‐borne. Mosquito → human/pig: vector‐borne [195]. Human → mosquito → human: vector‐borne, occurs during urban epidemics [115, 117]. Human/pig → mosquito → human/pig: possibly vector‐borne [116, 117, 196]. |
Marsupials in Australia [197] or horses in South Pacific islands [196]. |
| Rotavirus genogroup A (RVA); Rotavirus; Reoviridae. | Human ↔ pig: fecal‐oral, respiratory, food/water‐borne [108, 198, 199, 200]. | Diverse animal reservoirs including humans, porcine, bovine, ovine, pteropine, rodent, avian, and insectivore species [198, 200]. |
| Torque teno (TTV); Alphatorquevirus (huTTV), Iotatorquevirus (TTSuV1), Kappatorquevirus (TTSuVK2); Anelloviridae. | Human ↔ pig: contact with environmental contamination, e.g., contamination of TTSuV detected in veterinary vaccines, human drugs and pork products [92, 93], and TTV found ubiquitously in the environment including water sources and hospitals [91, 94]. | Unknown sources of emergent strains. |
BOX 1. Definitions of viral transmission and reservoirs used in this review.
| Zoonotic viruses amplify in pigs and shed sufficient amounts to infect close contact humans, but viruses infecting humans are unable to infect pigs, thereby, pigs are viral reservoirs for humans (pig‐to‐human transmission), or zoonotic viruses infect humans directly from another reservoir species without significant involvement of pigs. |
| Reverse zoonotic viruses amplify in humans and transmit to pigs, but pigs are unable to infect humans in return, in which case, humans are viral reservoirs for pigs (human‐to‐pig transmission). |
| Bi‐directional zoonotic viruses are exchanged between humans and pigs, whereby, both hosts are reservoirs for the other (both zoonotic and reverse zoonotic). |
The list of viruses shared by humans and pigs was taken from a recent study by Johnson et al., 2020 [6]. However, we were unable to find documentation of natural infection (either detection of viral genetic material or serological evidence of an antibody response against viral infection) in pigs for Ilheus, Ljungan, Monkeypox (experimental inoculation in pig skin only [29]), and Wesselsbron viruses (one study indicated serological evidence of infection in pigs but was inaccessible [30]). Tioman virus was included, despite undetected natural infection in pigs, due to evidence from an in vivo experimental infection study [31].
RESULTS AND DISCUSSION
Pigs as reservoirs for zoonotic viruses
The majority of the reviewed zoonotic viruses originate from wildlife reservoirs (Table 1). Pigs are significant intermediate and amplification hosts for the transmission of at least seven wildlife viruses to humans: Nipah (NiV), Japanese encephalitis (JEV), Eastern equine encephalitis (EEEV), Vesicular stomatitis (VSV), Reston ebola (RESTV), Menangle (MenPV), and potentially Tioman (TioV) (Table 1). Transmission routes of these zoonotic viruses from pigs to humans are illustrated in Fig. 1, which are generally linked to occupational exposure.
Fig. 1.
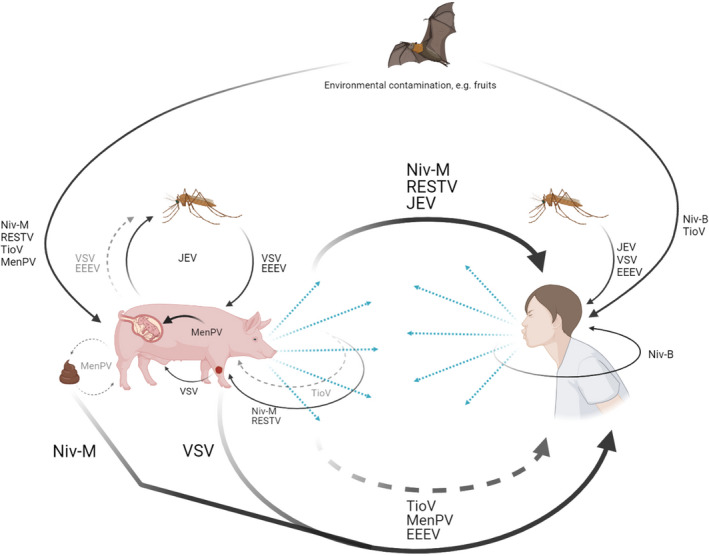
Transmission routes for seven zoonotic viruses. Solid arrows indicate transmission route, while dashed arrows indicate potential transmission route. The figure was created with BioRender.com.
Global livestock abundance and destruction of wildlife habitats have been associated with increased zoonotic spillover risk [6]. Following a rapid increase in the past few decades, approximately 800 million to 1 billion pigs are produced globally each year in often dense and genetically homogenous populations [32, 33], owing to 95% of genetic resources being exported from Europe and the USA to developing countries between 1990 and 2005 [34]. Although increased homogeneity in a swine herd is unlikely to increase their susceptibility to epidemics, the severity of epidemics is likely to be enhanced [35]. Furthermore, the frequency of animal turnover with immunologically naïve litters of piglets in swine herds can stunt the development of herd immunity against viral infections and enable viral persistence [36].
Deforestation and encroachment of pig farms into Pteropus fruit bat species habitats have been implicated in causing the zoonotic NiV epidemic in pigs and human pig farm workers in Malaysia and Singapore in 1999 [37]. The spillover of NiV‐Malaysia (NiV‐M) into pig herds was traced back to two introductions from fruit bats, with isolates from local bats, pigs, and humans sharing >99% nucleotide homology [10, 38, 39], indicating transmission between hosts required limited viral adaptation. However, humans developed more severe disease with 40% case fatality rate compared to 1‐5% in pigs [40]. This difference in disease severity could be linked to higher expression of the receptor ephrin‐B2 on human tracheal and bronchial airway epithelial cells than in pigs, leading to more efficient infection [41]. NiV‐M did not transmit between humans and viral RNA was isolated from 30% of infected throat swabs [42]; therefore, it seems unlikely that infected humans posed a risk to pigs.
Pigs contribute to the epidemiology of three zoonotic arthropod vector‐transmitted viruses: EEEV, JEV, and VSV. In addition to causing viremia in pigs [43, 44, 45], EEEV can be recovered from oropharyngeal, rectal, and tonsil swabs, JEV can shed in oronasal secretions, and VSV can exude from ruptured vesicular fluids, providing further transmission routes to close contact humans (Fig. 1) [43, 47, 48, 49, 50]. However, VSV has infrequently infected farm and laboratory workers [51], likely due to the capability of human myxovirus resistance protein dynamin‐like GTPase 1 (M×A) in reducing VSV replication by 90% compared to the porcine homolog Mx1, which inhibits only 25% of VSV replication [52, 53, 54].
Antibodies against RESTV were detected in 6.3% of exposed pig farm workers in the Philippines [55]. Unlike other ebolavirus species, which cause severe hemorrhagic fever in humans [56], RESTV is unable to suppress interferon (IFN) signaling immune response in humans [57]. However, pigs develop gross abnormalities in the lymphatic and respiratory systems after experimental infection and shed RESTV in nasopharyngeal secretions, which transmit RESTV to neighboring pigs [58].
An outbreak of MenPV occurred in an Australian piggery farm in 1997 with symptoms of reproductive disease in pigs, which included increased fetal death and abnormalities, and stillborn piglets [59]. Additionally, neutralizing antibodies were detected in adult pigs and two farm workers who developed an unexplained febrile illness [59, 60]. MenPV isolated from a stillborn piglet replicated in secondary lymphoid organs and intestines in experimentally infected pigs and shed in oronasal secretions, feces, and urine for under a week [61]. The source of MenPV was assumed to be local Pteropus fruit bat species based on serological evidence and later confirmed following the isolation of MenPV from fruit bat urine samples, which shared 94% nucleotide homology to the pig isolates [59, 62].
TioV was also discovered in Pteropus fruit bat species in Tioman Island, Malaysia [63]. Outbreaks of TioV have not been reported in either humans or pigs, but due to fruit bats harboring other zoonotic viruses, a serological survey of the Tioman Island population found 1.8% of islanders were seropositive for antibodies against TioV [64]. TioV is unable to inhibit IFN‐α/β signaling in human kidney cells, but can interfere with proinflammatory cytokine interleukin 6 (IL‐6) and IFN‐β promoter induction to cause infection [65]. Following experimental infection in pigs, TioV was isolated from oral swabs and neutralizing antibodies developed without inducing clinical signs [31]. This implicates pigs as potential amplification hosts if TioV spills over from bats.
Other reservoir host species for zoonotic viruses
Pigs appear to be minor, incidental hosts in the transmission chain for eleven zoonotic viruses. Although, more research is required to substantiate the insignificant contribution from pigs in the maintenance of many of these viruses. The majority are vector‐borne viruses: Toscana (TOSV), Venezuelan equine encephalitis (VEEV), Banna, Cache Valley, Chandipura, Getah, and Louping ill, and three are non‐vector‐borne viruses: rabies (RABV), encephalomyocarditis, and foot‐and‐mouth disease virus (Table 1).
Despite causing acute meningitis in humans [66, 67], the reservoir host species maintaining TOSV remains unknown, but likely involves a cyclic combination of arthropod, wildlife, and domesticated animals, akin to most other arbovirus maintenance cycles (Table 1). One serological survey detected IgG antibodies against TOSV in 22% of tested pigs in Spain [68], but further research efforts in pigs are lacking. Serological surveys for VEEV infection in pigs have also received limited attention since the last survey conducted in 1971 [69]. However, horses and rodents have been identified as the main amplifying hosts for epidemic and endemic strains of VEEV [70].
Other zoonotic viruses present a threat to the wider human population, beyond immediate farm and laboratory workers. Each year, RABV causes 59,000 deaths in humans usually bitten by rabid canines or bats [71]. Although RABV has been isolated from human secretions, the risk of human‐to‐human transmission is almost exclusively through organ transplantations [72]. RABV incidence in pigs is rare, and the “furious” form causing aggression with biting has only been recorded once in China [73]. As a generalist virus capable of infecting a wide range of species, RABV genetic diversity correlates with geographical origin rather than specialization in different host species, as RABV isolated from a pig shared 99.7% nucleotide homology in the partial N gene to a circulating “street” strain from a rabid canine isolated in the previous year [73].
Humans as reservoirs for reverse zoonotic viruses
Humans have spread three viruses: severe acute respiratory syndrome‐related coronaviruses (SARSr‐CoV), swine vesicular disease (SVDV), and noroviruses (NoV), to pigs through varied transmission routes (Table 1) illustrated in Fig. 2 together with bidirectionally transmitted viruses (addressed in the next section).
Fig. 2.
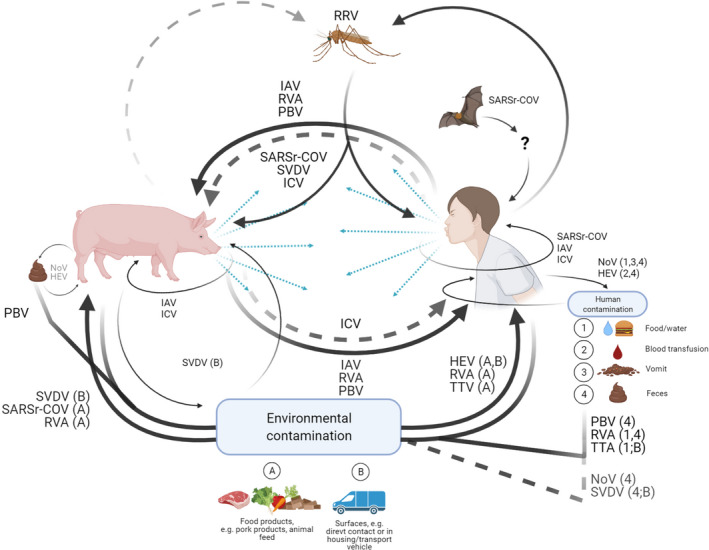
Transmission routes for three reverse zoonotic and seven bidirectionally transmitted viruses. Solid arrows indicate transmission route, while dashed arrows indicate potential transmission route. The figure was created with BioRender.com.
Although SARSr‐CoV originate from Rhinolophus horseshoe bat species and spilled over into humans through an intermediary species, humans rapidly became an effective transmitting host and viral reservoir for SARS‐CoV in 2003 and SARS‐CoV‐2 in 2019 [74, 75]. SARS‐CoV was transmitted to pigs in China presumably via contaminated feed from restaurant leftovers [76], but there has been no evidence of natural infection in swine with SARS‐CoV‐2. However, both SARSr‐CoV appear to replicate poorly in pigs [77, 78], possibly due to less efficient viral attachment to the porcine angiotensin‐converting enzyme 2 (ACE2) homolog receptor, which shares 81% nucleotide identity with the human ACE2 receptor [75, 78].
During human meningitis epidemics between 1948 and 1964, SVDV emerged in pigs as a genetic sublineage of human‐infecting coxsackievirus B (CV‐B) [79, 80, 81]. Periodic outbreaks in pigs arose in Europe and Asia until 2007 with SVDV becoming progressively adapted to swine as later SVDV isolates (post‐1990s) lost the ability to bind human decay‐accelerating factor as a co‐receptor and infect humans [82].
Highly genetically diverse NoV infect a broad range of species, but strains belonging to genogroup II (GII) exclusively infect humans and pigs [83]. Human‐associated NoV (huNoV) have been detected in pigs, but porcine‐associated NoV (porNoV) have never been detected in humans [84, 85, 86]. porNoV were unable to bind histo‐blood group antigens (HBGA) as co‐receptors on human cells, whereas huNoV‐GII.P4 was able to bind to duodenal and buccal tissues from either A+ or H+ phenotype HBGA pigs [84, 87].
Bidirectional viral transmission
Theoretically, a virus with the ability to infect and induce viral shedding in both humans and pigs can transmit between the two species. Non‐enveloped viruses are typically stable in the environment, which increases potential routes for transmission [88, 89, 90]. Seven viruses demonstrate bidirectional transmission by this principal (Table 1 and Fig. 2), four of which are non‐enveloped: Torque teno (TTV), picobirnavirus (PBV), hepatitis E (HEV), rotavirus A (RVA), and three are enveloped: influenza A (IAV), influenza C (ICV), and Ross River (RRV).
TTV and PBV are considered opportunistic pathogens due to their ubiquitous detection in both diseased and healthy human and pig populations and in various environments [91, 92, 93, 94, 95, 96]. Although specific TTV species of varying genome sizes are associated with human or pig infection, human‐associated Alphatorquevirus TTV species (huTTV) have been detected in 80% of pig sera samples and porcine‐associated Iotatorquevirus and Kappatorquevirus TTV species (TTSuV1 and TTSuVK2) have been detected in 92.5% of human sera samples [97], indicating viral exchange between the hosts. Growing evidence indicates PBV infects prokaryotes in the microbiome of humans and pigs [98]. Nevertheless, a genetic association between PBV isolated from humans and pigs has been suggested [99, 100, 101].
Humans are typically infected with HEV following the consumption of raw or undercooked pork products in developed countries and through the fecal‐oral transmission route in developing countries via consumption of water contaminated with human feces [102, 103]. Viremia peaks during the incubation period and the early symptomatic phase, with viral shedding in feces [102, 103]. While pigs are significant sources of HEV for humans, experimental infection in pigs with HEV isolated from humans has also been demonstrated [104, 105].
Similar to NoV, RVA attaches to HBGAs as co‐receptors to infect host cells, the phenotype of which depends on the VP8 domain of protease‐cleaved protein (P) types rather than the host species [106]. Unlike NoV, however, reassortant viruses with segments of human RVA origin have been found in pigs and vice versa [107, 108].
The exchange of IAV between humans and pigs is well known. Reassortant IAV generated with segments originating from human and swine IAV have been found in both host populations [12]. One high profile example was the novel genotype of H1N1 virus, which caused a human pandemic in 2009 after a quadruple reassortant IAV containing segments from avian IAV, human H3N2 subtype, Eurasian avian‐like swine IAV, and classical swine H1N1 subtype jumped from pigs into humans and back into pigs [109, 110].
Although humans were the only known natural host for ICV [111, 112], ICV has also been isolated from naturally infected pigs [109]. ICV strains isolated from humans during 1988‐1990 were highly related to the swine isolates obtained in China during 1981–1982 [111, 113], strongly suggesting interspecies transmission between humans and pigs; although, it is unknown whether the virus had transmitted from pigs to humans or from humans to pigs [111]. There is increasing evidence that other influenza species (influenza B and influenza D) are able to infect both humans and pigs and transmit between the two hosts [114].
Unlike all other zoonotic arboviruses in Table 1, RRV can potentially transmit between humans and pigs via mosquitoes. Human‐to‐mosquito‐to‐human transmission has been demonstrated during urban epidemics and pigs can also develop viremia, albeit at lower viral titers than humans [115, 116, 117].
Viral emergence, molecular evolution, and generation of diversity
To spill over into human or pig populations, either viruses possess intrinsic ability to pass through epidemiological barriers when the permitting factors align (without significant alteration to the viral genome) or viruses must first undergo substantial genetic changes to infect new host cells and evade host immune responses. Genetic divergence is driven by mutation, recombination, and reassortment and the resulting variants, haplotypes, or reassortants either propagate or diminish by various selective processes as the virus adapts to the new host [118, 119].
RNA viruses are exceedingly more likely to be zoonotic than DNA viruses [120], given their high nucleotide substitution rates of approximately 1 × 10 −3 nucleotide substitutions per site per year (ns/s/y) on average and rapid ability to adapt [121]. This is reflected in our review as all except one virus encode an RNA genome (Table S2). Nucleotide substitutions in most viruses with RNA genomes occur during replication by error‐prone, viral‐encoded RNA polymerases, while viruses with DNA genomes employ the host cell DNA polymerase with exonuclease activity to correct errors and are additionally subjected to post‐replication repair systems [119, 122]. However, TTV has a DNA genome with a comparable mutation rate to RNA viruses (0.53‐0.55 × 10‐3 ns/s/y [123]) and is highly genetically diverse, which could be attributed to the persistent nature of TTV infections in the host [124].
Nucleotide substitution rates and the number of susceptible host species are uncorrelated across the reviewed viruses (Table S2). Vector‐borne RNA viruses generally exhibit significantly lower mutation rates than non‐vector‐borne RNA viruses, with highly genetically similar strains infecting wide ranges of hosts (Table S2). For non‐vector‐borne RNA viruses, it is plausible that maintaining high mutation rates is necessary to adapt to a wide range of hosts. Encephalomyocarditis and foot‐and‐mouth disease viruses infect a broad range of hosts (30 and 72 documented hosts, respectively) and exhibit significantly higher mutation rates (1.61 and 1.45 × 10 −3 ns/s/y, respectively) than vector‐borne viruses [6, 121, 125]. However, the number of infected hosts is not a reliable proxy for mutation rate; Chandipura virus (CHPV) has a host range of 6 and the highest mutation rate at 6.577 × 10 −3 ns/s/y, RABV has the widest host range (126 known hosts) but a lower mutational rate (0.09 × 10 −3 ns/s/y), and SVDV rapidly adapted to swine after introduction from humans (3.84 × 10‐3 ns/s/y) (Table S2). Instead, mutation rates are more likely influenced by the efficiency of virus–host cell interactions, host immune evasion, and viral reproductive strategies, among many other biotic and abiotic factors.
Major genetic changes in viruses can occur by recombination and reassortment events when host cells are co‐infected with at least two viral strains (variants or distant relatives), which interact during replication to form progeny with genetic material from both strains [118, 119]. In general, recombination is prevalent in single‐stranded, positive‐sense RNA viruses with the exception of Flaviviruses where recombination is rarely observed [118]. Novel SVDV emerged in pigs because of a probable recombination event between human‐infecting coxsackievirus B (CV‐B) and CV‐A9; although, it is unknown whether the recombination event occurred in pigs or humans [80]. Polymerase (P) types of human‐ and pig‐associated NoV frequently recombine with common breakpoints between open reading frame junctions [126, 127, 128, 129, 130], but such recombinants have only been detected in pigs [85]. Even though single‐stranded, negative sense RNA viruses in general show lower rates of recombination, reassortment is frequently observed in Orthomyxoviridae, such as influenza A virus, which belong to the single‐stranded, negative sense RNA viruses. Reassortment is restricted to segmented RNA viruses and can result in rapid genetic change by formation of reassortants with novel genome combinations [118]. Twenty‐five percent of the assessed viruses in this review have a segmented genome, potentially making these viruses more disposed to fast adaptation to a new host/interspecies transmission.
Challenges in determining viral transmission
Our assessment of viral transmission is based on past strains of viruses. The viral landscape is under constant selective pressures, and the rapid and continuous generation of extensive genetic diversity is challenging to anticipate. Emergence of novel antigenic variants of viruses can undermine vaccination efforts, and vaccine availability against the majority of viruses is low (Table S2). Identifying the host factors a virus would need to adapt to is one modeling strategy to predict future variants, e.g., identifying viral–host protein interactions between the protein homologs in different hosts or the use of alternative host cell receptors.
RESTV is currently non‐pathogenic to humans, but substitutions of three amino acids in RESTV VP24 protein might enable binding to human karyopherin alpha5, which block innate immunity pathways in the same manner as other related pathogenic ebolaviruses [57, 131, 132]. In addition, a truncation in RESTV VP30 in a fraction of the RESTV isolates from pigs is characteristic of the Zaire ebolavirus adaptation to human cells during several months of human‐to‐human transmission in the 2013‐2016 ebolavirus disease outbreak [133].
Alternatively, wildlife viruses may attenuate as they passage through swine herds. NiV‐M, which was transmitted from bats‐to‐pigs‐to‐humans, caused a 40% case fatality rate in humans, while NiV‐Bangladesh genotype was transmitted directly from bats‐to‐humans via contaminated date palm sap causing over 70% case fatalities and has even transmitted onward to first contact humans [134]. The nucleotide difference between the two genotypes (8.2% [39]) is the most likely explanation for the difference in case fatality rates. Thus, viral attenuation through nucleotide changes in an intermediary host is a potential outcome.
Interactions between viruses and bacteria in the host microbiome may be another hidden factor facilitating viral transmission between humans and pigs. Certain bacteria express HBGAs to facilitate attachment of NoV to B cells, and CagA‐positive Helicobacter pylori induces HBGA expression in the mucosa of individuals without a functional FUT2 gene and HGBA phenotype [135, 136]. This can potentially increase the replication efficiency of particular NoV and RVA genotypes infecting humans and pigs.
Routine surveillance programs have been established for only some viruses in pigs (e.g., IAV [137]), and a few others are notifiable to international health bodies upon detection [138]. Many outbreaks lack real‐time monitoring and sampling in swine herds and humans, which can make retrospective analyses difficult and viral records incomplete (e.g., SARS‐CoV‐2 [26]). The choice of screening assays may also exclude some viruses. However, recent technical developments of next‐generation sequencing or probe‐based techniques with high‐throughput capabilities allow characterizing entire viromes of large populations a viable option. The overall aim of surveillance programs for emerging pathogens and zoonosis should be to act as early detection/warning systems because the success of limiting the spread of, e.g., a new zoonotic virus to a great extent relies on the possibility to contain it before it jumps to the first human. This in turn calls for more basic research into identification of reliable viral and host markers of species specificity for the different types of viruses combined with a One Health‐oriented design of the monitoring programs, i.e., by the inclusion of more targeted sampling of people in close contact with animals, e.g., swine.
Experimental studies involving human volunteers are rare. Only IAV, ICV, NoV, and RVA have been administered in challenge studies, usually with human‐derived isolates, common circulating genotypes in the population, or attenuated viral strains [139, 140, 141, 142]. Therefore, experiments with viruses to study human‐related dynamics rely on cell culture, explants, or animal models, which have some restrictions for application in a human population. Nevertheless, these experiments provide valuable data, particularly concerning specific virus–cell interactions.
CONCLUDING REMARKS
The list of 27 viruses shared by humans and pigs are generally regarded as zoonotic [6]. Reverse zoonosis or humans’ ability to transmit viruses to other animals is overlooked in some cases [8]. This review gathered evidence to assess the direction of viral transmission in the context of humans and pigs. Where direct detection was lacking, we theorized whether the virus could infect and transmit to the other host based on viral entry requirements, ability to establish infection, activation of immune responses, and shed in transmissible routes.
Transmission routes and viral sources are illustrated in Figs 1 and 2. Pigs are or have potential to be significant reservoirs, intermediaries, and amplifiers for at least seven zoonotic viruses; humans have been the source of three reverse zoonotic viruses in pigs; and humans and pigs possibly exchange seven viruses back and forth (Table 1).
The authors thank the FluZooMark group for contributing to discussions of the content of the review.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
Funding information
The work presented in this review is part of the FluZooMark project supported by Novo Nordisk Foundation (grant NNF19OC0056326).
Supporting information
Table S1. Framework of viral factors with associated relevance and assumptions considered in the review.
Table S2. Highlights of collected data based on the framework of factors in Table S1, which is used to inform viral transmission direction in Table 1. Distinctions are drawn between humans and pigs where appropriate.
Glud HA, George S, Skovgaard K, Larsen LE. Zoonotic and reverse zoonotic transmission of viruses between humans and pigs. APMIS. 2021; 129: 675–693.
REFERENCES
- 1. Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–3. 10.1038/nature06536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grace D, Mutua F, Ochungo P, Kruska R, Jones K, Brierley L, et al. Mapping of Poverty and Likely Zoonoses Hotspots. Zoonoses Project 4. Report to the UK Department for International Development. International Livestock Research Institute: Nairobi, Kenya. 2012.
- 3. Plowright RK, Parrish CR, McCallum H, Hudson PJ, Ko AI, Graham AL, et al. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 2017;15(8):502–10. 10.1038/nrmicro.2017.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones BA, Grace D, Kock R, Alonso S, Rushton J, Said MY, et al. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl Acad. Sci. 2013;110(21):8399–404. 10.1073/pnas.1208059110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kock R. Drivers of disease emergence and spread: Is wildlife to blame? Onderstepoort J Vet Res. 2014;81(2):1–4. [DOI] [PubMed] [Google Scholar]
- 6. Johnson CK, Hitchens PL, Pandit PS, Rushmore J, Evans TS, Young CCW, et al. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc Biol Sci. 2020;287(1924):20192736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rohr JR, Barrett CB, Civitello DJ, Craft ME, Delius B, DeLeo GA, et al. Emerging human infectious diseases and the links to global food production. Nature Sustainab. 2019;2(6):445–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Messenger AM, Barnes AN, Gray GC. Reverse zoonotic disease transmission (zooanthroponosis): a systematic review of seldom‐documented human biological threats to animals. PLoS One. 2014;9(2):e89055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Erlanger TE, Weiss S, Keiser J, Utzinger J, Wiedenmayer K. Past, present, and future of Japanese encephalitis. Emerg Infect Dis. 2009;15(1):1–7. 10.3201/eid1501.080311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chua KB, Bellini WJ, Rota PA, Harcourt BH, Tamin A, Lam SK, et al. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288(5470):1432–5. 10.1126/science.288.5470.1432 [DOI] [PubMed] [Google Scholar]
- 11. Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL, et al. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science. 2010;328(5985):1529. 10.1126/science.1189132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Anderson TK, Chang J, Arendsee ZW, Venkatesh D, Souza CK, Kimble JB, et al. Swine influenza a viruses and the tangled relationship with humans. Cold Spring Harb. Perspect Med. 2021;11(3):a038737– 10.1101/cshperspect.a038737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. United States Department of Agriculture (USDA) . Livestock and Poultry: World Markets and Trade. Foreign Agricultural Service. 2020 October 9. Retrieved from https://apps.fas.usda.gov/psdonline/circulars/livestock_poultry.pdf
- 14. Food and Agriculture Organisation of the United Nations (FAO) . Pigs and Management & Housing. 2014 Nov 23. Retrieved from: http://www.fao.org/ag/againfo/themes/en/pigs/AP_management.html
- 15. Bowman AS, Walia RR, Nolting JM, Vincent AL, Killian ML, Zentkovich MM, et al. Influenza A(H3N2) virus in swine at agricultural fairs and transmission to humans, Michigan and Ohio, USA, 2016. Emerg Infect Dis. 2017;23(9):1551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol. 2012;20(1):50–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dawson HD, Loveland JE, Pascal G, Gilbert JG, Uenishi H, Mann KM, et al. Structural and functional annotation of the porcine immunome. BMC Genom. 2013;15(14):332. 10.1186/1471-2164-14-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mair KH, Sedlak C, Käser T, Pasternak A, Levast B, Gerner W, et al. The porcine innate immune system: An update. Dev. Comp. Immunol. 2014;45(2):321–43. 10.1016/j.dci.2014.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Starbæk SMR, Brogaard L, Dawson HD, Smith AD, Heegaard PMH, Larsen LE, et al. Animal models for influenza a virus infection incorporating the involvement of innate host defenses: enhanced translational value of the porcine model. ILAR J. 2018;59(3):323–37. [DOI] [PubMed] [Google Scholar]
- 20. Uddin Khan S, Atanasova KR, Krueger WS, Ramirez A, Gray GC. Epidemiology, geographical distribution, and economic consequences of swine zoonoses: a narrative review. Emerg Microbes Infect. 2013;2(12):e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Warren CJ, Sawyer SL. How host genetics dictates successful viral zoonosis. PLoS Biol. 2019;17(4):e3000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carlson CJ, Zipfel CM, Garnier R, Bansal S. Global estimates of mammalian viral diversity accounting for host sharing. Nat Ecol Evol. 2019;3(7):1070–5. 10.1038/s41559-019-0910-6 [DOI] [PubMed] [Google Scholar]
- 23. Parrish CR, Holmes EC, Morens DM, Park EC, Burke DS, Calisher CH, et al. Cross‐species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72(3):457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park M, Loverdo C, Schreiber SJ, Lloyd‐Smith JO. Multiple scales of selection influence the evolutionary emergence of novel pathogens. Philos Trans R Soc Lond B Biol Sci. 2013;368(1614):20120333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Centers for Disease Control and Prevention (CDC) . Principles of Epidemiology in Public Health Practice. 2012 May. Retrieved from: https://www.cdc.gov/csels/dsepd/ss1978/SS1978.pdf
- 26. Friend T, Stebbing J. What is the intermediate host species of SARS‐CoV‐2? Future Virol. 2021;16(3):153–6. [Google Scholar]
- 27. Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, et al. Transmission of SARS‐CoV‐2 on mink farms between humans and mink and back to humans. Science. 2021;371(6525):172‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larsen HD, Fonager J, Lomholt FK, Dalby T, Benedetti G, Kristensen B, et al. Preliminary report of an outbreak of SARS‐CoV‐2 in mink and mink farmers associated with community spread, Denmark, June to November 2020. Euro Surveill. 2021;26(5):2100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soekawa M, Moriguchi R, Morita C, Kitamura T, Tanaka Y. Electron‐microscopical observations on the development of vaccinia, cowpox and monkeypox viruses in pig skin. Zentralbl Bakteriol Orig A. 1977;237(4):425–43. [PubMed] [Google Scholar]
- 30. Baba SS, Fagbami AH, Ojeh CK, Olaleye OD, Omilabu SA. Wesselsbron virus antibody in domestic animals in Nigeria: retrospective and prospective studies. New Microbiol. 1995;18(2):151–62. [PubMed] [Google Scholar]
- 31. Yaiw KC, Bingham J, Crameri G, Mungall B, Hyatt A, Yu M, et al. Tioman virus, a paramyxovirus of bat origin, causes mild disease in pigs and has a predilection for lymphoid tissues. J. Virol. 2008;82(1):565–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drew TW. The emergence and evolution of swine viral diseases: to what extent have husbandry systems and global trade contributed to their distribution and diversity? Rev Sci Tech. 2011;30(1):95–106. [DOI] [PubMed] [Google Scholar]
- 33. Food and Agriculture Organisation of the United Nations (FAO) . FAOSTAT Production of Pigs in. World 1980 ‐ 2019. Accessed: 2020 Mar 5. Available from: http://www.fao.org/faostat/en/#data/QA/visualize
- 34. Gollin D, Van Dusen E, Blackburn H. Animal genetic resource trade flows: Economic assessment. Livestock Sci. 2009;120(3):248–55. 10.1016/j.livsci.2008.07.017 [DOI] [Google Scholar]
- 35. Springbett AJ, MacKenzie K, Woolliams JA, Bishop SC. The contribution of genetic diversity to the spread of infectious diseases in livestock populations. Genetics. 2003;165(3):1465–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pitzer VE, Aguas R, Riley S, Loeffen WL, Wood JL, Grenfell BT. High turnover drives prolonged persistence of influenza in managed pig herds. J R Soc Interface. 2016;13(119):20160138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chua KB, Chua BH, Wang CW. Anthropogenic deforestation, El Niño and the emergence of Nipah virus in Malaysia. Malays J Pathol. 2002;24(1):15–21. [PubMed] [Google Scholar]
- 38. AbuBakar S, Chang LY, Ali AR, Sharifah SH, Yusoff K, Zamrod Z. Isolation and molecular identification of Nipah virus from pigs. Emerg Infect Dis. 2004;10(12):2228–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harcourt BH, Lowe L, Tamin A, Liu X, Bankamp B, Bowden N, et al. Genetic characterization of Nipah virus, Bangladesh, 2004. Emerg Infect Dis. 2005;11(10):1594–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Looi LM, Chua KB. Lessons from the Nipah virus outbreak in Malaysia. Malays J Pathol. 2007;29(2):63–7. [PubMed] [Google Scholar]
- 41. Sauerhering L, Zickler M, Elvert M, Behner L, Matrosovich T, Erbar S, et al. Species‐specific and individual differences in Nipah virus replication in porcine and human airway epithelial cells. J Gen Virol. 2016;97(7):1511–9. 10.1099/jgv.0.000483 [DOI] [PubMed] [Google Scholar]
- 42. Chua KB, Lam SK, Goh KJ, Hooi PS, Ksiazek TG, Kamarulzaman A, et al. The presence of Nipah virus in respiratory secretions and urine of patients during an outbreak of Nipah virus encephalitis in Malaysia. J Infect. 2001;42(1):40–3. 10.1053/jinf.2000.0782 [DOI] [PubMed] [Google Scholar]
- 43. Elvinger F, Baldwin CA, et al. Eastern equine encephalomyelitis virus. In: Straw BE, Zimmerman J, D’Allaire S, editors. Diseases of Swine, 9th edn. Blackwell Publishing Company; Ames, IA. 2006. p. 554–7. [Google Scholar]
- 44. Burke DS, Tingpalapong M, Ward GS, Andre R, Leake CJ. Intense transmission of Japanese encephalitis virus to pigs in a region free of epidemic encephalitis. Southeast Asian J Trop Med Public Health. 1985;16(2):199–206. [PubMed] [Google Scholar]
- 45. Ricklin ME, Garcìa‐Nicolàs O, Brechbühl D, Python S, Zumkehr B, Posthaus H, et al. Japanese encephalitis virus tropism in experimentally infected pigs. Vet Res. 2016;24(47):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Velazquez‐Salinas L, Pauszek SJ, Stenfeldt C, O'Hearn ES, Pacheco JM, Borca MV, et al. Increased virulence of an epidemic strain of vesicular stomatitis virus is associated with interference of the innate response in pigs. Front Microbiol. 2018;15(9):1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ricklin ME, García‐Nicolás O, Brechbühl D, Python S, Zumkehr B, Nougairede A, et al. Vector‐free transmission and persistence of Japanese encephalitis virus in pigs. Nat Commun. 2016;23(7):10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Howerth EW, Stallknecht DE, Dorminy M, Pisell T, Clarke GR. Experimental vesicular stomatitis in swine: effects of route of inoculation and steroid treatment. J Vet Diagn Invest. 1997;9(2):136–42. 10.1177/104063879700900205 [DOI] [PubMed] [Google Scholar]
- 49. Stallknecht DE, Perzak DE, Bauer LD, Murphy MD, Howerth EW. Contact transmission of vesicular stomatitis virus New Jersey in pigs. Am J Vet Res. 2001;62(4):516–20. 10.2460/ajvr.2001.62.516 [DOI] [PubMed] [Google Scholar]
- 50. Mead DG, Gray EW, Noblet R, Murphy MD, Howerth EW, Stallknecht DE. Biological transmission of vesicular stomatitis virus (New Jersey serotype) by Simulium vittatum (Diptera: Simuliidae) to domestic swine (Sus scrofa). J Med Entomol. 2004;41(1):78–82. 10.1603/0022-2585-41.1.78 [DOI] [PubMed] [Google Scholar]
- 51. Letchworth GJ, Rodriguez LL. Vesicular stomatitis. Vet J. 1999;157(3):239–60. 10.1053/tvjl.1998.0303 [DOI] [PubMed] [Google Scholar]
- 52. Asano A, Ko JH, Morozumi T, Hamashima N, Watanabe T. Polymorphisms and the antiviral property of porcine Mx1 protein. J Vet Med Sci. 2002;64(12):1085–9. 10.1292/jvms.64.1085 [DOI] [PubMed] [Google Scholar]
- 53. Sasaki K, Tungtrakoolsub P, Morozumi T, Uenishi H, Kawahara M, Watanabe T. A single nucleotide polymorphism of porcine MX2 gene provides antiviral activity against vesicular stomatitis virus. Immunogenetics. 2014;66(1):25–32. 10.1007/s00251-013-0745-2 [DOI] [PubMed] [Google Scholar]
- 54. Schwemmle M, Weining KC, Richter MF, Schumacher B, Staeheli P. Vesicular stomatitis virus transcription inhibited by purified MxA protein. Virology. 1995;206(1):545–54. 10.1016/s0042-6822(95)80071-9 [DOI] [PubMed] [Google Scholar]
- 55. Miranda ME, Miranda NL. Reston ebolavirus in humans and animals in the Philippines: a review. J Infect Dis. 2011;204(Suppl 3):S757–60. 10.1093/infdis/jir296 [DOI] [PubMed] [Google Scholar]
- 56. Kobinger GP, Leung A, Neufeld J, Richardson JS, Falzarano D, Smith G, et al. Replication, pathogenicity, shedding, and transmission of Zaire ebolavirus in pigs. J Infect Dis. 2011;204(2):200–8. 10.1093/infdis/jir077 [DOI] [PubMed] [Google Scholar]
- 57. Guito JC, Albariño CG, Chakrabarti AK, Towner JS. Novel activities by ebolavirus and marburgvirus interferon antagonists revealed using a standardized in vitro reporter system. Virology. 2017;15(501):147–65. 10.1016/j.virol.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Marsh GA, Haining J, Robinson R, Foord A, Yamada M, Barr JA, et al. Ebola Reston virus infection of pigs: clinical significance and transmission potential. J Infect Dis. 2011;204(Suppl 3):S804–9. 10.1093/infdis/jir300 [DOI] [PubMed] [Google Scholar]
- 59. Philbey AW, Kirkland PD, Ross AD, Davis RJ, Gleeson AB, Love RJ, et al. An apparently new virus (family paramyxoviridae) infectious for pigs, humans, and fruit bats. Emerg. Infect. Dis. 1998;4(2):269–71. 10.3201/eid0402.980214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chant K, Chan R, Smith M, Dwyer DE, Kirkland P. Probable human infection with a newly described virus in the family paramyxoviridae. Emerg. Infect. Dis. 1998;4(2):273–5. 10.3201/eid0402.980215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bowden TR, Bingham J, Harper JA, Boyle DB. Menangle virus, a pteropid bat paramyxovirus infectious for pigs and humans, exhibits tropism for secondary lymphoid organs and intestinal epithelium in weaned pigs. J Gen Virol. 2012;93(Pt 5):1007–16. 10.1099/vir.0.038448-0 [DOI] [PubMed] [Google Scholar]
- 62. Barr JA, Smith C, Marsh GA, Field H, Wang LF. Evidence of bat origin for Menangle virus, a zoonotic paramyxovirus first isolated from diseased pigs. J Gen Virol. 2012;93(Pt 12):2590–4. 10.1099/vir.0.045385-0 [DOI] [PubMed] [Google Scholar]
- 63. Chua KB, Wang LF, Lam SK, Crameri G, Yu M, Wise T, et al. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology. 2001;283(2):215–29. 10.1006/viro.2000.0882 [DOI] [PubMed] [Google Scholar]
- 64. Yaiw KC, Crameri G, Wang L, Chong HT, Chua KB, Tan CT, et al. Serological evidence of possible human infection with Tioman virus, a newly described paramyxovirus of bat origin. J Infect Dis. 2007;196(6):884–6. 10.1086/520817 [DOI] [PubMed] [Google Scholar]
- 65. Caignard G, Lucas‐Hourani M, Dhondt KP, Labernardière JL, Petit T, Jacob Y, et al. The V protein of tioman virus is incapable of blocking type I interferon signaling in human cells. PLoS One. 2013;8(1):e53881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Charrel RN, Gallian P, Navarro‐Mari JM, Nicoletti L, Papa A, Sánchez‐Seco MP, et al. Emergence of Toscana virus in Europe. Emerg Infect Dis. 2005;11(11):1657–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vilibic‐Cavlek T, Zidovec‐Lepej S, Ledina D, Knezevic S, Savic V, Tabain I, et al. Clinical, virological, and immunological findings in patients with toscana neuroinvasive disease in croatia: report of three cases. Trop Med Infect Dis. 2020;5(3):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Navarro‐Marí JM, Palop‐Borrás B, Pérez‐Ruiz M, Sanbonmatsu‐Gámez S. Serosurvey study of Toscana virus in domestic animals, Granada, Spain. Vector Borne Zoonotic Dis. 2011;11(5):583–7. 10.1089/vbz.2010.0065 [DOI] [PubMed] [Google Scholar]
- 69. Scherer WF, Dickerman RW, Campillo‐Sainz C, Zarate ML, Gonzales E. Ecologic studies of Venezuelan encephalitis virus in southeastern México. V. Infection of domestic animals other than equines. Am J Trop Med Hyg. 1971;20(6):989–93. 10.4269/ajtmh.1971.20.989 [DOI] [PubMed] [Google Scholar]
- 70. Weaver SC, Ferro C, Barrera R, Boshell J, Navarro JC. Venezuelan equine encephalitis. Annu Rev Entomol. 2004;49:141–74. 10.1146/annurev.ento.49.061802.123422 [DOI] [PubMed] [Google Scholar]
- 71. World Health Organization (WHO) . Rabies. 2020 Apr. Retrieved from: https://www.who.int/news‐room/fact‐sheets/detail/rabies
- 72. Manning SE, Rupprecht CE, Fishbein D, Hanlon CA, Lumlertdacha B, Guerra M, et al. Advisory Committee on Immunization Practices Centers for Disease Control and Prevention (CDC). Human rabies prevention‐‐United States, 2008: recommendations of the Advisory Committee on Immunization Practices. MMWR Recomm Rep. 2008;57(RR‐3):1‐28. [PubMed] [Google Scholar]
- 73. Jiang Y, Yu X, Wang L, Lu Z, Liu H, Xuan H, et al. An outbreak of pig rabies in Hunan province, China. Epidemiol. Infect. 2008;136(4):504–8. 10.1017/S0950268807008874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhong NS, Zheng BJ, Li YM, Poon XZH, Chan KH, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. The Lancet. 2003;362(9393):1353–8. 10.1016/S0140-6736(03)14630-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chen W, Yan M, Yang L, Ding B, He B, Wang Y, et al. SARS‐associated coronavirus transmitted from human to pig. Emerg Infect Dis. 2005;11(3):446–8. 10.3201/eid1103.040824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Weingartl HM, Copps J, Drebot MA, Marszal P, Smith G, Gren J, et al. Susceptibility of pigs and chickens to SARS coronavirus. Emerg Infect Dis. 2004;10(2):179–84. 10.3201/eid1002.030677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pickering BS, Smith G, Pinette MM, Embury‐Hyatt C, Moffat E, Marszal P, et al. Susceptibility of domestic swine to experimental infection with severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2021;27(1):104–12. 10.3201/eid2701.203399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang G, Haydon DT, Knowles NJ, McCauley JW. Molecular evolution of swine vesicular disease virus. J Gen Virol. 1999;80(Pt 3):639–51. 10.1099/0022-1317-80-3-639 [DOI] [PubMed] [Google Scholar]
- 80. Bruhn CA, Nielsen SC, Samaniego JA, Wadsworth J, Knowles NJ, Gilbert MT. Viral meningitis epidemics and a single, recent, recombinant and anthroponotic origin of swine vesicular disease virus. Evol Med Public Health. 2015;2015(1):289–303. 10.1093/emph/eov026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lomakina NF, Yu Shustova E, Strizhakova OM, Felix Drexler J, Lukashev AN. Epizootic of vesicular disease in pigs caused by coxsackievirus B4 in the Soviet Union in 1975. J Gen Virol. 2016;97(1):49–52. 10.1099/jgv.0.000318 [DOI] [PubMed] [Google Scholar]
- 82. Jimenez‐Clavero MA, Escribano‐Romero E, Ley V, Spiller OB. More recent swine vesicular disease virus isolates retain binding to coxsackie‐adenovirus receptor, but have lost the ability to bind human decay‐accelerating factor (CD55). J Gen Virol. 2005;86(Pt 5):1369–77. 10.1099/vir.0.80669-0 [DOI] [PubMed] [Google Scholar]
- 83. Vinjé J. Advances in Laboratory Methods for Detection and Typing of Norovirus. J. Clin. Microbiol. 2015;53(2):373–81. 10.1128/JCM.01535-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Farkas T, Nakajima S, Sugieda M, Deng X, Zhong W, Jiang X. Seroprevalence of noroviruses in swine. J Clin Microbiol. 2005;43(2):657–61. 10.1128/JCM.43.2.657-661.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Villabruna N, Koopmans MPG, de Graaf M. Animals as reservoir for human norovirus. Viruses. 2019;11(5):478. 10.3390/v11050478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. de Graaf M, van Beek J, Koopmans MP. Human norovirus transmission and evolution in a changing world. Nat Rev Microbiol. 2016;14(7):421–33. 10.1038/nrmicro.2016.48 [DOI] [PubMed] [Google Scholar]
- 87. Cheetham S, Souza M, McGregor R, Meulia T, Wang Q, Saif LJ. Patterns of human norovirus‐like particles to buccal and intestinal tissues of gnotobiotic pigs in relation to A/H histo‐blood group antigen expression. J. Virol. 2007;81(7):3535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Firquet S, Beaujard S, Lobert PE, Sané F, Caloone D, Izard D, et al. Survival of enveloped and non‐enveloped viruses on inanimate surfaces. Microbes Environ. 2015;30(2):140–4. 10.1264/jsme2.ME14145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Geoghegan JL, Senior AM, Di Giallonardo F , Holmes EC. Virological factors that increase the transmissibility of emerging human viruses. Proc. Natl Acad. Sci. 2016;113(15):4170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bushman FD, McCormick K, Sherrill‐Mix S. Virus structures constrain transmission modes. Nature Microbiol. 2019;4(11):1778–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Griffin JS, Plummer JD, Long SC. Torque teno virus: an improved indicator for viral pathogens in drinking waters. Virol J. 2008;3(5):112. 10.1186/1743-422X-5-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kekarainen T, Martínez‐Guinó L, Segalés J. Swine torque teno virus detection in pig commercial vaccines, enzymes for laboratory use and human drugs containing components of porcine origin. J Gen Virol. 2009;90(Pt 3):648–53. 10.1099/vir.0.006841-0 [DOI] [PubMed] [Google Scholar]
- 93. Jiménez‐Melsió A, Parés S, Segalés J, Kekarainen T. Detection of porcine anelloviruses in pork meat and human faeces. Virus Res. 2013;178(2):522–4. 10.1016/j.virusres.2013.09.035 [DOI] [PubMed] [Google Scholar]
- 94. D'Arcy N, Cloutman‐Green E, Klein N, Spratt DA. Environmental viral contamination in a pediatric hospital outpatient waiting area: implications for infection control. Am J Infect Control. 2014;42(8):856–60. 10.1016/j.ajic.2014.04.014 [DOI] [PubMed] [Google Scholar]
- 95. Martínez LC, Masachessi G, Carruyo G, Ferreyra LJ, Barril PA, Isa MB, et al. Picobirnavirus causes persistent infection in pigs. Infect Genet Evol. 2010;10(7):984–8. 10.1016/j.meegid.2010.06.004 [DOI] [PubMed] [Google Scholar]
- 96. Yinda CK, Vanhulle E, Conceição‐Neto N, Beller L, Deboutte W, Shi C, et al. Gut virome analysis of cameroonians reveals high diversity of enteric viruses. Including potential interspecies transmitted viruses. Msphere. 2019;4(1):e00585–18. 10.1128/mSphere.00585-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ssemadaali MA, Effertz K, Singh P, Kolyvushko O, Ramamoorthy S. Identification of heterologous Torque Teno Viruses in humans and swine. Sci Rep. 2016;25(6):26655. 10.1038/srep26655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Krishnamurthy SR, Wang D. Extensive conservation of prokaryotic ribosomal binding sites in known and novel picobirnaviruses. Virology. 2018;516:108–14. 10.1016/j.virol.2018.01.006 [DOI] [PubMed] [Google Scholar]
- 99. Bányai K, Martella V, Bogdán Á, Forgách P, Jakab F, Meleg E, et al. Genogroup I picobirnaviruses in pigs: evidence for genetic diversity and relatedness to human strains. J Gen Virol. 2008;89(Pt 2):534–9. 10.1099/vir.0.83134-0 [DOI] [PubMed] [Google Scholar]
- 100. Carruyo GM, Mateu G, Martínez LC, Pujol FH, Nates SV, Liprandi F, et al. Molecular characterization of porcine picobirnaviruses and development of a specific reverse transcription‐PCR Assay. J. Clin. Microbiol. 2008;46(7):2402–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Chen M, Sun H, Hua X. Segment 2 sequences analysis of genogroup II picobirnavirus in pig stool in China. Acta Virol. 2019;63(1):126–8. 10.4149/av_2019_108 [DOI] [PubMed] [Google Scholar]
- 102. Kamar N, Dalton HR, Abravanel F, Izopet J. Hepatitis E virus infection. Clin Microbiol Rev. 2014;27(1):116–38. 10.1128/CMR.00057-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Park WJ, Park BJ, Ahn HS, Lee JB, Park SY, Song CS, et al. Hepatitis E virus as an emerging zoonotic pathogen. J. Vet. Sci. 2016;17(1):1–11. 10.4142/jvs.2016.17.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Halbur PG, Kasorndorkbua C, Gilbert C, Guenette D, Potters MB, et al. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J Clin Microbiol. 2001;39(3):918–23. 10.1128/JCM.39.3.918-923.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Meng XJ, Halbur PG, Shapiro MS, Govindarajan S, Bruna JD, Mushahwar IK, et al. Genetic and experimental evidence for cross‐species infection by swine hepatitis E virus. J Virol. 1998;72(12):9714–21. 10.1128/JVI.72.12.9714-9721.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Guo Y, Candelero‐Rueda RA, Saif LJ, Vlasova AN. Infection of porcine small intestinal enteroids with human and pig rotavirus A strains reveals contrasting roles for histo‐blood group antigens and terminal sialic acids. PLoS Pathog. 2021;17(1):e1009237. 10.1371/journal.ppat.1009237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wu FT, Bányai K, Jiang B, Liu LT, Marton S, Huang YC, et al. Novel G9 rotavirus strains co‐circulate in children and pigs, Taiwan. Sci Rep. 2017;18(7):40731. 10.1038/srep40731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Abass G, Dubal ZB, Rajak KK, Kale BM, Raorane A, Dudhe N, et al. Molecular characterization of porcine rotavirus A from India revealing zooanthroponotic transmission. Animal Biotechnol. 2021;8:1–13. 10.1080/10495398.2020.1868486 [DOI] [PubMed] [Google Scholar]
- 109. Mostafa A, Abdelwhab EM, Mettenleiter TC, Pleschka S. Zoonotic Potential of influenza A viruses: a comprehensive overview. Viruses. 2018;10(9):497. 10.3390/v10090497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe. 2010;7(6):440–51. 10.1016/j.chom.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kimura H, Abiko C, Peng G, Muraki Y, Sugawara K, Hongo S, et al. Interspecies transmission of influenza C virus between humans and pigs. Virus Res. 1997;48(1):71–9. 10.1016/s0168-1702(96)01427-x [DOI] [PubMed] [Google Scholar]
- 112. Matsuzaki Y, Mizuta K, Sugawara K, Tsuchiya E, Muraki Y, Hongo S, et al. Frequent reassortment among influenza C viruses. J Virol. 2003;77(2):871–81. 10.1128/jvi.77.2.871-881.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Guo YJ, Jin FG, Wang P, Wang M, Zhu JM. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J Gen Virol. 1983;64(Pt 1):177–82. 10.1099/0022-1317-64-1-177 [DOI] [PubMed] [Google Scholar]
- 114. Lee J, Wang L, Palinski R, Walsh T, He D, Li Y, et al. Comparison of pathogenicity and transmissibility of influenza B and D viruses in pigs. Viruses. 2019;11(10):905. 10.3390/v11100905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Koolhof IS, Carver S. Epidemic host community contribution to mosquito‐borne disease transmission: Ross River virus. Epidemiol Infect. 2017;145(4):656–66. 10.1017/S0950268816002739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Spradbrow PB. Letter: Experimental infection of sheep and pigs with Ross River virus. Aust Vet J. 1973;49(8):403–4. [DOI] [PubMed] [Google Scholar]
- 117. Rosen L, Gubler DJ, Bennett PH. Epidemic polyarthritis (Ross River) virus infection in the Cook Islands. Am J Trop Med Hyg. 1981;30(6):1294–302. 10.4269/ajtmh.1981.30.1294 [DOI] [PubMed] [Google Scholar]
- 118. Pérez‐Losada M, Arenas M, Galán JC, Palero F, González‐Candelas F. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect Genet Evolut. 2015;30:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sanjuán R, Domingo‐Calap P. Genetic diversity and evolution of viral populations. Encyclopedia of Virology 4th ed. Academic Press; 2021. 53‐61 p. [Google Scholar]
- 120. Kreuder Johnson C, Hitchens PL, Smiley Evans T, Goldstein T, Thomas K, Clements A, et al. Spillover and pandemic properties of zoonotic viruses with high host plasticity. Sci Rep. 2015;7(5):14830. 10.1038/srep14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J Mol Evol. 2002;54(2):156–65. 10.1007/s00239-001-0064-3 [DOI] [PubMed] [Google Scholar]
- 122. Smith EC. The not‐so‐infinite malleability of RNA viruses: Viral and cellular determinants of RNA virus mutation rates. PLoS Pathog. 2017;13(4):e1006254. 10.1371/journal.ppat.1006254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Cadar D, Kiss T, Ádám D, Cságola A, Novosel D, Tuboly T. Phylogeny, spatio‐temporal phylodynamics and evolutionary scenario of Torque teno sus virus 1 (TTSuV1) and 2 (TTSuV2) in wild boars: fast dispersal and high genetic diversity. Vet Microbiol. 2013;166(1–2):200–13. 10.1016/j.vetmic.2013.06.010 [DOI] [PubMed] [Google Scholar]
- 124. Nieto D, Aramouni M, Grau‐Roma L, Segalés J, Kekarainen T. Dynamics of Torque teno sus virus 1 (TTSuV1) and 2 (TTSuV2) DNA loads in serum of healthy and postweaning multisystemic wasting syndrome (PMWS) affected pigs. Vet Microbiol. 2011;152(3–4):284–90. 10.1016/j.vetmic.2011.05.020 [DOI] [PubMed] [Google Scholar]
- 125. Hicks AL, Duffy S. Cell tropism predicts long‐term nucleotide substitution rates of mammalian RNA viruses. PLoS Pathog. 2014;10(1):e1003838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Bull RA, Hansman GS, Clancy LE, Tanaka MM, Rawlinson WD, White PA. Norovirus recombination in ORF1/ORF2 overlap. Emerg Infect Dis. 2005;11(7):1079–85. 10.3201/eid1107.041273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang QH, Han MG, Cheetham S, Souza M, Funk JA, Saif LJ. Porcine noroviruses related to human noroviruses. Emerg Infect Dis. 2005;11(12):1874–81. 10.3201/eid1112.050485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Shen Q, Zhang W, Yang S, Yang Z, Chen Y, Cui L, et al. Recombinant porcine norovirus identified from piglet with diarrhea. BMC Vet Res. 2012;3(8):155. 10.1186/1746-6148-8-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Chhabra P, de Graaf M, Parra GI, Chan MC, Green K, Martella V, et al. Updated classification of norovirus genogroups and genotypes. J Gen Virol. 2019;100(10):1393–406. 10.1099/jgv.0.001318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Cavicchio L, Tassoni L, Laconi A, Cunial G, Gagliazzo L, Milani A, et al. Unrevealed genetic diversity of GII Norovirus in the pig population of North East Italy. Sci Rep. 2020;10(1):9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wauquier N, Becquart P, Padilla C, Baize S, Leroy EM. Human fatal zaire ebola virus infection is associated with an aberrant innate immunity and with massive lymphocyte apoptosis. PLoS Negl Trop Dis. 2010;4(10):e837. 10.1371/journal.pntd.0000837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Pappalardo M, Juliá M, Howard MJ, Rossman JS, Michaelis M, Wass MN. Conserved differences in protein sequence determine the human pathogenicity of Ebolaviruses. Sci Rep. 2016;24(6):23743. 10.1038/srep23743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Albariño CG, Wiggleton Guerrero L, Jenks HM, Chakrabarti AK, Ksiazek TG, et al. Insights into Reston virus spillovers and adaption from virus whole genome sequences. PLoS One. 2017;12(5):e0178224. 10.1371/journal.pone.0178224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lo MK, Lowe L, Hummel KB, Sazzad HM, Gurley ES, Hossain MJ, et al. Characterization of Nipah virus from outbreaks in Bangladesh, 2008–2010. Emerg Infect Dis. 2012;18(2):248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 2014;346(6210):755–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Ruvoën‐Clouet N, Magalhaes A, Marcos‐Silva L, Breiman A, Figueiredo C, David L, et al. Increase in Genogroup II.4 norovirus host spectrum by CagA‐positive helicobacter pylori infection. J. Infect. Dis. 2014;210(2):183–91. [DOI] [PubMed] [Google Scholar]
- 137. Simon G, Larsen LE, Dürrwald R, Foni E, Harder T, Van Reeth K, et al. European surveillance network for influenza in pigs: surveillance programs, diagnostic tools and Swine influenza virus subtypes identified in 14 European countries from 2010 to 2013. PLoS One. 2014;9(12):e115815. 10.1371/journal.pone.0115815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. World Organisation for Animal Health (OIE) . OIE‐Listed diseases 2021. Retrieved from: https://www.oie.int/animal‐health‐in‐the‐world/oie‐listed‐diseases‐2021/
- 139. Sherman AC, Mehta A, Dickert NW, Anderson EJ, Rouphael N. The Future of flu: a review of the human challenge model and systems biology for advancement of influenza vaccinology. Front Cell Infect Microbiol. 2019;17(9):107. 10.3389/fcimb.2019.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Lindesmith L, Moe C, Lependu J, Frelinger JA, Treanor J, Baric RS. Cellular and humoral immunity following Snow Mountain virus challenge. J Virol. 2005;79(5):2900–9. 10.1128/JVI.79.5.2900-2909.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Frenck R, Bernstein DI, Xia M, Huang P, Zhong W, Parker S, et al. Predicting Susceptibility to Norovirus GII.4 by use of a challenge model involving humans. J. Infect. Dis. 2012;206(9):1386–93. 10.1093/infdis/jis514 [DOI] [PubMed] [Google Scholar]
- 142. Ward RL, Bernstein DI, Shukla R, McNeal MM, Sherwood JR, Young EC, et al. Protection of adults rechallenged with a human rotavirus. J Infect Dis. 1990;161(3):440–5. 10.1093/infdis/161.3.440 [DOI] [PubMed] [Google Scholar]
- 143. Go YY, Balasuriya UB, Lee CK. Zoonotic encephalitides caused by arboviruses: transmission and epidemiology of alphaviruses and flaviviruses. Clin Experi Vaccine Res. 2014;3(1):58–77. 10.7774/cevr.2014.3.1.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Kirkland PD, Love RJ, Philbey AW, Ross AD, Davis RJ, Hart KG. Epidemiology and control of Menangle virus in pigs. Aust Vet J. 2001;79(3):199–206. 10.1111/j.1751-0813.2001.tb14580.x [DOI] [PubMed] [Google Scholar]
- 145. Love RJ, Philbey AW, Kirkland PD, Ross AD, Davis RJ, Morrissey C, et al. Reproductive disease and congenital malformations caused by Menangle virus in pigs. Aust Vet J. 2001;79(3):192–8. 10.1111/j.1751-0813.2001.tb14578.x [DOI] [PubMed] [Google Scholar]
- 146. Halpin K, Hyatt AD, Fogarty R, Middleton D, Bingham J, Epstein JH, et al. Pteropid bats are confirmed as the reservoir hosts of henipaviruses: a comprehensive experimental study of virus transmission. Am J Trop Med Hyg. 2011;85(5):946–51. 10.4269/ajtmh.2011.10-0567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Middleton DJ, Westbury HA, Morrissy CJ, van der Heide BM, Russell GM, Braun MA, et al. Experimental Nipah virus infection in pigs and cats. J Comp Pathol. 2002;126(2‐3):124–36. 10.1053/jcpa.2001.0532 [DOI] [PubMed] [Google Scholar]
- 148. Goh KJ, Tan CT, Chew NK, Tan PS, Kamarulzaman A, Sarji SA, et al. Clinical features of Nipah virus encephalitis among pig farmers in Malaysia. N Engl J Med. 2000;342(17):1229–35. 10.1056/NEJM200004273421701 [DOI] [PubMed] [Google Scholar]
- 149. Luby SP, Hossain MJ, Gurley ES, Ahmed BN, Banu S, Khan SU, et al. Recurrent zoonotic transmission of nipah virus into humans, Bangladesh, 2001–2007. Emerg. Infect. Dis. 2009;15(8):1229–35. 10.3201/eid1508.081237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kasloff SB, Leung A, Pickering BS, Smith G, Moffat E, Collignon B, et al. Pathogenicity of Nipah henipavirus Bangladesh in a swine host. Sci Rep. 2019;9(1):5230. 10.1038/s41598-019-40476-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Yob JM, Field H, Rashdi AM, Morrissy C, van der Heide B, Rota P, et al. Nipah virus infection in bats (Order Chiroptera) in Peninsular Malaysia. Emerg. Infect. Dis. 2001;7(3):439–41. 10.3201/eid0703.010312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Chua KB, Koh CL, Hooi PS, Wee KF, Khong JH, Chua BH, et al. Isolation of Nipah virus from Malaysian Island flying‐foxes. Microbes Infect. 2002;4(2):145–51. 10.1016/s1286-4579(01)01522-2 [DOI] [PubMed] [Google Scholar]
- 153. Jayme SI, Field HE, de Jong C, Olival KJ, Marsh G, Tagtag AM, et al. Molecular evidence of Ebola Reston virus infection in Philippine bats. Virol J. 2015;17(12):107. 10.1186/s12985-015-0331-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Barrette RW, Metwally SA, Rowland JM, Xu L, Zaki SR, Nichol ST, et al. Discovery of swine as a host for the Reston ebolavirus. Science. 2009;325(5937):204–6. 10.1126/science.1172705 [DOI] [PubMed] [Google Scholar]
- 155. Smith PF, Howerth EW, Carter D, Gray EW, Noblet R, Mead DG. Mechanical transmission of vesicular stomatitis New Jersey virus by Simulium vittatum (Diptera: Simuliidae) to domestic swine (Sus scrofa). J Med Entomol. 2009;46(6):1537–40. 10.1603/033.046.0643 [DOI] [PubMed] [Google Scholar]
- 156. Rozo‐Lopez P, Drolet BS, Londoño‐Renteria B. Vesicular stomatitis virus transmission: a comparison of incriminated vectors. Insects. 2018;9(4):190. 10.3390/insects9040190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Attoui H, Mohd Jaafar F, de Micco P, de Lamballerie X. Coltiviruses and seadornaviruses in North America, Europe, and Asia. Emerg Infect Dis. 2005;11(11):1673–9. 10.3201/eid1111.050868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Jaafar FM, Attoui H, Mertens PPC, de Micco P, de Lamballerie X. Structural organization of an encephalitic human isolate of Banna virus (genus Seadornavirus, family Reoviridae). J Gen Virol. 2005;86(Pt 4):1147–57. 10.1099/vir.0.80578-0 [DOI] [PubMed] [Google Scholar]
- 159. Xia H, Liu H, Zhao L, Atoni E, Wang Y, Yuan Z. First Isolation and Characterization of a Group C Banna Virus (BAV) from Anopheles sinensis Mosquitoes in Hubei, China. Viruses. 2018;10(10):555. 10.3390/v10100555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Attoui H, Jaafar FM, Belhouchet M, Tao S, Chen B, Liang G, et al. Liao ning virus, a new Chinese seadornavirus that replicates in transformed and embryonic mammalian cells. J Gen Virol. 2006;87(Pt 1):199–208. 10.1099/vir.0.81294-0 [DOI] [PubMed] [Google Scholar]
- 161. Centers for Disease Control and Prevention (CDC) . Cache Valley virus. 2019 Nov. Retrieved from: https://www.cdc.gov/cache‐valley/index.html
- 162. Nguyen NL, Zhao G, Hull R, Shelly MA, Wong SJ, Wu G, et al. Cache valley virus in a patient diagnosed with aseptic meningitis. J. Clin. Microbiol. 2013;51(6):1966–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Hayles LB, Lversen JO. Cache Valley virus: experimental infection in Culiseta inornata. Can J Microbiol. 1980;26(3):287–90. [PubMed] [Google Scholar]
- 164. Armstrong PM, Andreadis TG, Anderson JF. Emergence of a new lineage of cache valley virus (Bunyaviridae: Orthobunyavirus) in the Northeastern United States. Am J Tropical Med Hygiene. 2015;93(1):11–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Andreadis TG, Armstrong PM, Anderson JF, Main AJ. Spatial‐temporal analysis of Cache Valley virus (Bunyaviridae: Orthobunyavirus) infection in anopheline and culicine mosquitoes (Diptera: Culicidae) in the northeastern United States, 1997–2012. Vector Borne Zoonotic Dis. 2014;14(10):763–73. 10.1089/vbz.2014.1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Mavale MS, Fulmali PV, Ghodke YS, Mishra AC, Kanojia P, Geevarghese G. Experimental transmission of Chandipura virus by Phlebotomus argentipes (diptera: psychodidae). Am J Trop Med Hyg. 2007;76(2):307–9. [PubMed] [Google Scholar]
- 167. Sudeep AB, Gurav YK, Bondre VP. Changing clinical scenario in Chandipura virus infection. Indian J Med Res. 2016;143(6):712–21. 10.4103/0971-5916.191929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Oberste MS, Gotuzzo E, Blair P, Nix WA, Ksiazek TG, Comer JA, et al. Human febrile illness caused by encephalomyocarditis virus infection, Peru. Emerg Infect Dis. 2009;15(4):640–6. 10.3201/eid1504.081428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Billinis C, Paschaleri‐Papadopoulou E, Anastasiadis G, Psychas V, Vlemmas J, Leontides S, et al. A comparative study of the pathogenic properties and transmissibility of a Greek and a Belgian encephalomyocarditis virus (EMCV) for piglets. Vet Microbiol. 1999;70(3–4):179–92. 10.1016/s0378-1135(99)00145-5 [DOI] [PubMed] [Google Scholar]
- 170. Paton DJ, Gubbins S, King DP. Understanding the transmission of foot‐and‐mouth disease virus at different scales. Curr Opin Virol. 2018;28:85–91. 10.1016/j.coviro.2017.11.013 [DOI] [PubMed] [Google Scholar]
- 171. Armstrong R, Davie J, Hedger RS. Foot‐and‐mouth disease in man. Br Med J. 1967;4(5578):529–30. 10.1136/bmj.4.5578.529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Prempeh H, Smith R, Müller B. Foot and mouth disease: the human consequences. BMJ. 2001;322(7286):565–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Hall MD, Knowles NJ, Wadsworth J, Rambaut A, Woolhouse ME. Reconstructing geographical movements and host species transitions of foot‐and‐mouth disease virus serotype SAT 2. Mbio. 2013;4(5):e00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Liu H, Zhang X, Li LX, Shi N, Sun XT, Liu Q, et al. First isolation and characterization of Getah virus from cattle in northeastern China. BMC Vet Res. 2019;15(1):320. 10.1186/s12917-019-2061-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Izumida A, Takuma H, Inagaki S, Kubota M, Hirahara T, Kodama K, et al. Experimental infection of Getah virus in swine. Nihon Juigaku Zasshi. 1988;50(3):679–84. [DOI] [PubMed] [Google Scholar]
- 176. Jeffries CL, Mansfield KL, Phipps LP, Wakeley PR, Mearns R, Schock A, et al. Louping ill virus: an endemic tick‐borne disease of Great Britain. J. Gen. Virol. 2014;95(5):1005–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Dobler G. Zoonotic tick‐borne flaviviruses. Vet Microbiol. 2010;140(3–4):221–8. 10.1016/j.vetmic.2009.08.024 [DOI] [PubMed] [Google Scholar]
- 178. Davidson MM, Williams H, Macleod JA. Louping ill in man: a forgotten disease. J Infect. 1991;23(3):241–9. 10.1016/0163-4453(91)92756-u [DOI] [PubMed] [Google Scholar]
- 179. Alkan C, Bichaud L, de Lamballerie X, Alten B, Gould EA, Charrel RN. Sandfly‐borne phleboviruses of Eurasia and Africa: epidemiology, genetic diversity, geographic range, control measures. Antiviral Res. 2013;100(1):54–74. 10.1016/j.antiviral.2013.07.005 [DOI] [PubMed] [Google Scholar]
- 180. Moriconi M, Rugna G, Calzolari M, Bellini R, Albieri A, Angelini P, et al. Phlebotomine sand fly‐borne pathogens in the Mediterranean Basin: Human leishmaniasis and phlebovirus infections. PLoS Negl Trop Dis. 2017;11(8):e0005660. 10.1371/journal.pntd.0005660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Wang E, Bowen RA, Medina G, Powers AM, Kang W, Chandler LM, et al. Virulence and viremia characteristics of 1992 epizootic subtype IC Venezuelan equine encephalitis viruses and closely related enzootic subtype ID strains. Am J Trop Med Hyg. 2001;65(1):64–9. 10.4269/ajtmh.2001.65.64 [DOI] [PubMed] [Google Scholar]
- 182. Beckham JD, Arbovirus TKL. Arbovirus Infections. Continuum: Life Learning Neurol. 2015;21:1599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Cheetham S, Souza M, Meulia T, Grimes S, Han MG, Saif LJ. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J Virol. 2006;80(21):10372–81. 10.1128/JVI.00809-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Souza M, Cheetham SM, Azevedo MS, Costantini V, Saif LJ. Cytokine and antibody responses in gnotobiotic pigs after infection with human norovirus genogroup II.4 (HS66 Strain). J. Virol. 2007;81(17):9183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Fan Y, Zhao K, Shi ZL, Zhou P. Bat Coronaviruses in China. Viruses. 2019;11(3):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res. 2018;100:163‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Hoffmann M, Kleine‐Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Dekker A, Moonen P, de Boer‐Luijtze EA, Terpstra C. Pathogenesis of swine vesicular disease after exposure of pigs to an infected environment. Vet Microbiol. 1995;45(2–3):243–50. 10.1016/0378-1135(95)00032-6 [DOI] [PubMed] [Google Scholar]
- 189. Bellini S, Alborali L, Zanardi G, Bonazza V, Brocchi E. Swine vesicular disease in northern Italy: diffusion through densely populated pig areas. Rev Sci Tech. 2010;29(3):639–48. 10.20506/rst.29.3.2006 [DOI] [PubMed] [Google Scholar]
- 190. Webster RG. Influenza virus: transmission between species and relevance to emergence of the next human pandemic. Arch Virol Suppl. 1997;13:105–13. 10.1007/978-3-7091-6534-8_11 [DOI] [PubMed] [Google Scholar]
- 191. Yoon SW, Webby RJ, Webster RG. Evolution and ecology of influenza A viruses. Curr Top Microbiol Immunol. 2014;385:359–75. 10.1007/82_2014_396 [DOI] [PubMed] [Google Scholar]
- 192. Sederdahl BK, Williams JV. Epidemiology and clinical characteristics of influenza C virus. Viruses. 2020;12(1):89. 10.3390/v12010089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Kylla H, Dutta TK, Roychoudhury P, Malik YS, Mandakini R, Subudhi PK. Prevalence and molecular characterization of porcine Picobirnavirus in piglets of North East Region of India. Trop. Anim. Health Prod. 2017;49(2):417–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Smits SL, Poon LL, van Leeuwen M, Lau PN, Perera HK, Peiris JS, et al. Genogroup I and II picobirnaviruses in respiratory tracts of pigs. Emerg Infect Dis. 2011;17(12):2328–30. 10.3201/eid1712.110934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Claflin SB, Webb CE. Ross river virus: many vectors and unusual hosts make for an unpredictable pathogen. PLoS Pathog. 2015;11(9):e1005070. 10.1371/journal.ppat.1005070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Togami E, Gyawali N, Ong O, Kama M, Cao‐Lormeau VM, Aubry M, et al. First evidence of concurrent enzootic and endemic transmission of Ross River virus in the absence of marsupial reservoirs in Fiji. Int J Infect Dis. 2020;96:94–6. 10.1016/j.ijid.2020.02.048 [DOI] [PubMed] [Google Scholar]
- 197. Harley D, Sleigh A, Ritchie S. Ross river virus transmission, infection, and disease: a cross‐disciplinary review. Clin. Microbiol. Rev. 2001;14(4):909–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. De Grazia S, Martella V, Rotolo V, Bonura F, Matthijnssens J, Bányai K, et al. Molecular characterization of genotype G6 human rotavirus strains detected in Italy from 1986 to 2009. Infect Genet Evol. 2011;11(6):1449–55. 10.1016/j.meegid.2011.05.015 [DOI] [PubMed] [Google Scholar]
- 199. Fu ZF, Hampson DJ. Natural transmission of group A rotavirus within a pig population. Res Vet Sci. 1989;46(3):312–7. [PubMed] [Google Scholar]
- 200. Vlasova AN, Amimo JO, Saif LJ. Porcine rotaviruses: epidemiology, immune responses and control strategies. Viruses. 2017;9(3):48. 10.3390/v9030048 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Framework of viral factors with associated relevance and assumptions considered in the review.
Table S2. Highlights of collected data based on the framework of factors in Table S1, which is used to inform viral transmission direction in Table 1. Distinctions are drawn between humans and pigs where appropriate.


