Summary
Some plant traits may be legacies of coevolution with extinct megafauna. One example is the convergent evolution of ‘divaricate’ cage architectures in many New Zealand lineages, interpreted as a response to recently extinct flightless avian browsers whose ancestors arrived during the Paleogene period. Although experiments have confirmed that divaricate habit deters extant browsers, its abundance on frosty, droughty sites appears consistent with an earlier interpretation as a response to cold, dry Plio‐Pleistocene climates.
We used 45 protein‐coding sequences from plastid genomes to reconstruct the evolutionary history of the divaricate habit in extant New Zealand lineages. Our dated phylogeny of 215 species included 91% of New Zealand eudicot divaricate species.
We show that 86% of extant divaricate plants diverged from non‐divaricate sisters within the last 5 Ma, implicating Plio‐Pleistocene climates in the proliferation of cage architectures in New Zealand.
Our results, combined with other recent findings, are consistent with the synthetic hypothesis that the browser‐deterrent effect of cage architectures was strongly selected only when Plio‐Pleistocene climatic constraints prevented woody plants from growing quickly out of reach of browsers. This is consistent with the abundance of cage architectures in other regions where plant growth is restricted by aridity or short frost‐free periods.
Keywords: avian herbivory, cage architectures, convergent evolution, divaricating shrubs, moa, New Zealand, plant structural defences
Introduction
As early as the 19th century, Alfred Russell Wallace and other scientists recognised that ‘we live in a zoologically impoverished world from which all the hugest and fiercest and strangest forms have recently disappeared’ (Wallace, 1876). Although much research has focused on the causes of late Quaternary extinctions (Martin & Klein, 1989; Stuart, 2015; Saltré et al., 2016), biologists have increasingly also inquired into the ecological roles played by extinct megafauna, and the resulting evolutionary legacies. A range of puzzling features of plants has been interpreted as anachronistic legacies of herbivory past or of broken mutualistic interactions (Bond & Silander, 2007; Kistler et al., 2015; Galetti et al., 2018).
One such possible legacy is the convergent evolution of cage architectures in many plant lineages in New Zealand (Greenwood & Atkinson, 1977). Cage architectures occur there in as many as 80 eudicot and one conifer species from 20 families, representing c. 13% of the indigenous woody flora (Maurin & Lusk, 2021). Although spinescence is the best‐known type of structural defence against vertebrate browsers (Hanley et al., 2007; Charles‐Dominique et al., 2016), densely branched ‘cage’ architectures with small widely separated leaves have also been shown to deter browsing (Charles‐Dominique et al., 2017). Cage architectures in other regions are often accompanied by spinescence (Cavagnaro & Golluscio, 2017; Charles‐Dominique et al., 2017), but most New Zealand cage‐like plants are markedly nonspinescent, and have historically been termed ‘divaricate’ or ‘divaricating’ plants (Greenwood & Atkinson, 1977; Fig. 1).
Fig. 1.

Architectural comparison of seedlings of two New Zealand congeners that frequently hybridise. The broadleaved Coprosma robusta is on the left, and the divaricate Coprosma propinqua on the right. Reproduced from Lusk et al. (2021), with permission from the publisher.
Nevertheless, the putative selective forces driving this celebrated case of convergent evolution in the New Zealand flora has been the subject of intense debate. Early botanists, noting its prominence in the rain shadow of New Zealand’s Southern Alps, interpreted the divaricate habit as an adaptation to cold, dry Plio‐Pleistocene climates (Diels, 1896; Cockayne, 1912), a hypothesis developed further by McGlone & Webb (1981). Greenwood & Atkinson (1977) proposed an alternative hypothesis, that the divaricate habit arose as a defence against now‐extinct flightless avian browsers, nine species of moa. Although experiments have since confirmed that the divaricate habit can deter browsing by both avian and ungulate herbivores (Bond et al., 2004; Pollock et al., 2007), there is also experimental evidence that the small leaves of divaricate plants help them cope with frost (Lusk et al., 2018), and small leaves in general reduce vulnerability to overheating during drought (Yates et al., 2010). The climatic and moa‐browsing hypotheses are not mutually exclusive (Cooper et al., 1993), and a recent synthetic hypothesis proposes that cage architectures did not become advantageous in New Zealand as an antibrowsing defence until climatic adversity restricted growth rates of young trees and shrubs, preventing them from growing quickly out of reach of ground‐dwelling herbivores (Lusk et al., 2016).
Cooper et al. (1993) suggested that phylogenetic dating of divergences between New Zealand divaricate plants and their non‐divaricate relatives would help to distinguish between these hypotheses. Fossils date moa presence in New Zealand to at least 16–19 million years ago (Ma) (Tennyson et al., 2010) and genetic evidence suggest their volant ancestors arrived c. 60 Ma (Phillips et al., 2010), therefore, under the moa‐browsing hypothesis, one might expect the divaricate habit to date at least as far back as the early Miocene age. The cold, dry climates purported to have favoured selection for the divaricate habit under the climate hypothesis arose from the Plio‐Pleistocene combination of global cooling (Hornibrook, 1992) and the uplift of the Southern Alps (Batt et al., 2000), therefore, under both the climate hypothesis and the synthetic browsing‐climate hypothesis, one might expect the divaricate habit to be at most c. 5 Ma old. Dated phylogenies including divaricate species indicate ages from <1 Ma to >20 Ma for such divergences (Maurin & Lusk, 2021), but these studies did not necessarily sample the most closely related pairs of extant non‐divaricate and divaricate species, as it was not their objective; the resulting phylogenies may therefore overestimate the age of some divergences.
Here, we present the first study with the objective of dating the origin of the divaricate habit in extant lineages of the New Zealand flora. We propose a dated phylogeny of 215 species, with an extensive sampling of divaricate New Zealand eudicots (91%). It is based on DNA sequences of protein‐coding plastid genes extracted from complete or near‐complete plastid genomes, built under a maximum likelihood framework and calibrated using 10 fossils and one secondary calibration.
Materials and Methods
Sampling plan
We gathered a dataset of 215 complete or near‐complete plastid DNA sequences of eudicot species. This included 73 divaricate species, representing 91% of the list of eudicot divaricate species of New Zealand compiled in Maurin & Lusk (2021). We managed to include 100% of the divaricate species from 21 out of the 23 eudicot genera with divaricate representatives of this list; 82% of divaricate Coprosma species were included (Table 1, see Results section). We aimed at including the most closely related non‐divaricate species as possible to each divaricate species we considered, based on morphological characters and previous phylogenetic studies. In total, 190 of the sequences we used were newly generated for this study from herbarium specimens or newly collected vouchered samples, while the others were sourced from GenBank. Supporting Information Table S1 presents the sampling plan with the relevant information regarding the specimens.
Table 1.
Maximum estimated ages of the divergences between divaricate and non‐divaricate species for each genus represented in our phylogeny.
| Genus | Proportion of divaricate species sampled | Older CI bound of divaricate/non‐divaricate MRCA (Ma) |
|---|---|---|
| Raukaua (Araliaceae) | 1/1 (100%) | 0.0 |
| Myrsine (Primulaceae) | 1/1 (100%) | 0.1 |
| Discaria (Rhamnaceae) | 1/1 (100%) | 0.2 |
| Hoheria (Malvaceae) | 2/2 (100%) | 0.3 |
| Corokia (Argophyllaceae) | 1/1 (100%) | 0.5 |
| Pennantia (Pennantiaceae) | 1/1 (100%) | 0.6 |
| Sophora (Fabaceae) | 2/2 (100%) | 0.6 |
| Streblus (Moraceae) | 1/1 (100%) | 0.7 |
| Aristotelia (Elaeocarpaceae) | 1/1 (100%) | 0.9 |
| Lophomyrtus (Myrtaceae) | 1/1 (100%) | 1.8 |
| Melicope (Rutaceae) | 1/1 (100%) | 1.8 |
| Plagianthus (Malvaceae) | 2/2 (100%) | 1.8 |
| Melicytus (Violaceae) | 6/6 (100%) | 2.0 |
| Pittosporum (Pittosporaceae) | 7/7 (100%) | 2.2 |
| Olearia (Asteraceae) | 9/9 (100%) | 2.4 |
| Elaeocarpus (Elaeocarpaceae) | 1/1 (100%) | 3.1 |
| Muehlenbeckia (Polygonaceae) | 3/3 (100%) | 4.1 |
| Carpodetus (Rousseaceae) | 1/1 (100%) | 4.6 |
| Coprosma (Rubiaceae) | 28/34 (82%) | 4.9 |
| Teucrium (Lamiaceae) | 1/1 (100%) | 7.8 |
| Neomyrtus (Myrtaceae) | 1/1 (100%) | 10.9 |
| Rhabdothamnus (Gesneriaceae) | 1/1 (100%) | 20.6 |
In a genus, this age is taken as the older confidence interval (CI) bound of the age of the most recent common ancestor (MRCA) of divaricate species or clades and their non‐divaricate sisters. Genera are ordered by increasing age. Also provided is the proportion of divaricate species of each genus (according to the list compiled by Maurin & Lusk, 2021) that we sampled. Ma, million years ago.
Gene flow between species that have only recently diverged can complicate the estimation of their relatedness by way of plastid DNA phylogenies, yet this issue can be circumvented for our research question. Plastid introgression (or ‘capture’; Soltis & Kuzoff, 1995) is likely to result in underestimates of divergence ages in species‐rich genera known for frequent interspecific hybridisation, such as Coprosma, Olearia and Sophora (Allan, 1961; Wichman et al., 2002; Shepherd & Heenan, 2021); however, our dependence on single accessions of each species precludes the identification of potential occurrences of this phenomenon. We circumvent this risk of underestimating the age of the divaricate habit with a combination of two methodological elements: (1) within these potentially problematic genera, we sampled allopatric species from different landmasses around the Southern Hemisphere, between which gene flow is very unlikely to have occurred; (2) we conservatively considered the age of the most recent common ancestor (MRCA) of each of these genera rather than the date of divergence between each divaricate species and its nearest non‐divaricate relative. However, our trees should not be considered reliable estimates of phylogeny at the species level in such groups.
Plastid DNA has important advantages in studies focused on dating of divergences, rather than on detailed phylogenetic relationships. Firstly, it can be modelled as a haploid single nonrecombining linkage (Nock et al., 2011) that is generally structurally conserved (Tonti‐Filippini et al., 2017) allowing the alignment of long stretches of DNA and increasing the precision of date estimates for relatively recent divergences through increased sampling of sites. Secondly, coding regions of plastid DNA can be unambiguously aligned across broad phylogenetic sampling such as ours and suffer less than nuclear makers from substitution saturation of variable regions.
DNA extraction
The DNA of the samples was extracted following one of two methods:
Using a CTAB‐based protocol (Doyle & Dickson, 1987) modified as in Smissen & Heenan (2007) to include a phenol : chloroform extraction and recovery using spin columns (Zymo IIC; Zymo Research, Orange County, CA, USA).
Following the DNA tissue protocol of the Maxwell 16 instrument (Promega, Madison, WI, USA) and further purified by phenol : chloroform extraction and recovery in spin columns.
Detailed step‐by‐step protocols are available upon request. The DNA concentration of the extracts was measured using the Qubit dsDNA high‐sensitivity assay (Thermo Fisher Scientific, Waltham, MA, USA).
Library preparation and sequencing
Genomic DNA libraries were prepared following one of two methods:
Using Illumina Nextera DNA Library Prep kits (Illumina, San Diego, CA, USA), following the manufacturer’s instructions (Reference Guide, #15027987 v01, January 2016) except that we halved the quantities of reagents and the target amount of input DNA.
Using Illumina TruSeq Nano DNA Library Prep kits, according to the manufacturer’s instructions (Reference Guide, #15041110 Rev. D, June 2015) again using halved reagent quantities and target input DNA; genomic DNA was fragmented using a Covaris ME220 Focused‐ultrasonicator (Covaris, Woburn, MA, USA; settings: 75 s duration – 40 W peak power – 25% duty factor – 50 cycles per burst).
The concentration and size range of libraries were measured using a LabChip GX Touch HT instrument (PerkinElmer, Hopkinton, MA, USA). Libraries were enriched for plastid DNA using a custom myBaits kit (Arbor Biosciences, Ann Arbor, MI, USA) modified from Stull et al. (2013; baits provided in Dataset S1) using the manufacturer’s instructions (v.3.02, July 2016 or v.4.01, April 2018). Illumina HiSeq sequencing was carried out by Otago Genomics Facility (Dunedin, New Zealand) using paired‐end 2 × 125 bp reads.
Plastid DNA assembly and annotation
Reads were first trimmed using Trimmomatic v.0.38 software (Bolger et al., 2014) with the following settings: ILLUMINACLIP:[path/to/NexteraPE‐PE.fa or TruSeq3‐PE‐2.fa according to the library preparation method]:1:30:10 SLIDINGWINDOW:10:20 MINLEN:40. For each family we included, a complete plastid genome as closely related as possible to our samples was selected from GenBank as a reference against which to map the reads of samples from that family. Then, in each family, the best quality (i.e. best compromise between highest HQ%, lowest percentage of ambiguous bases and highest coverage) resulting consensus sequence was selected and its reads mapped against itself to create an assembly of improved quality. Finally, the reads of the other members of the family were mapped against this new improved consensus. Mappings were performed with Bwa, using the Bwa‐Mem algorithm (Li, 2013).
Sequence annotation was carried out as follows: (1) the improved reference sequence for each family was aligned to the GenBank sequence initially used to map its reads using the Mafft algorithm v.7.388 (Katoh et al., 2002; Katoh & Standley, 2013) plugin in Geneious Prime 2019.2.1; (2) the annotations of the GenBank references were transferred to the improved references and manually checked; (3) the other sequences of each family were aligned to their corresponding improved references, again with Mafft within Geneious Prime, and annotations transferred across and manually checked.
After making test phylogenies of the resulting sequences, unexpected relationships within the genus Coprosma led us to investigate these sequences more deeply. Consensus sequences generated from mapped reads for many of these samples had a high level of ambiguous sites. It appeared that many of these ambiguous sites were the result of pseudogene sequences, in that they contained frame‐shift mutations and substitutions leading to inferred stop codons in coding regions. Nonfunctional nuclear or mitochondrial DNAs resulting from gene transfer from the plastid genome are both well documented (Cummings et al., 2003; Goremykin et al., 2009; Zhang et al., 2020), although we are not aware of other reports of them impacting assemblies of massively parallel sequence data. Because mitochondrial genomes occur in plant cells at much higher copy number than nuclear genomes, they are likely to be the source of the pseudogenes in our Coprosma data, but proliferation of a plastid‐derived sequence or sequences within the nuclear genome cannot be excluded. To minimise this issue, we mapped these reads against the GenBank sequence of Anthospermum spathulatum (accession no. KY378687), a plastid sequence missing one copy of the inverted repeat region, with custom mapping parameters in Geneious Prime (available upon request to the authors). Despite many trials with different sets of mapping parameters, we could not obtain genomes of good and consistent quality over their entire sequences; the compromise we found allowed us to obtain protein‐coding sequences of good quality, but the intergenic portions of genome were of very bad quality. As a result, the Rubiaceae sequences could not be deposited on GenBank: they are instead provided in a Geneious file as Dataset S6.
Data partitioning
We used 45 protein‐coding sequences from the long and short single‐copy regions. They were aligned in Geneious Prime using the Mafft plugin, and the alignment manually checked. We partitioned this alignment into 1st + 2nd codon position on the one hand and 3rd codon position on the other hand. Sites containing at least one unresolved base were removed before conducting the phylogenetic analyses described below, resulting in a final alignment of 31 248 sites.
Reconstruction and dating of the phylogeny
We reconstructed the phylogeny of our samples with RAxML v.8.2.12 (Stamatakis, 2014), run on CIPRES (Miller et al., 2010), following the guide by Maurin (2020). The search for the best ML tree was conducted with the following settings: 10 random alternative starting trees, 1000 bootstrap (BS) replicates, GTRGAMMA model, and fixing the sequence of Ranunculus sceleratus (Ranunculales) as an outgroup to the rest of the Eudicots. From this best ML tree, we generated 1000 BS replicates to produce a dated phylogeny with a 95% confidence interval (CI) on the age at the nodes using treePL (Smith & O’Meara, 2012), again following the guide by Maurin (2020). To calibrate the tree, we used 10 fossils assigned to internal nodes (all outside our study clades) and one secondary calibration for the root of the tree. Our calibration strategy was based on strategies designed for recently published dated phylogenies of angiosperms (Magallón et al., 2015; Li et al., 2019), and is explained in Methods S1. The Phylip alignment used in RAxML and its associated partitioning file are provided as Datasets S2 and S3, respectively, while the treePL configuration file built is provided as Dataset S4.
This treePL phylogeny was compared with a phylogeny built under Bayesian inference. We used Beast 2.6.2 software (Bouckaert et al., 2019) with the following settings: Birth‐Death model (Gernhard, 2008), bModelTest (Bouckaert & Drummond, 2017) and relaxed clock with rates drawn from a lognormal distribution (Drummond et al., 2006) for each partition. We combined the results of three chains started from different seeds, which we sampled once every 50k generations and ran until their combination resulted in effective sampling size > 200 for all parameters, which required chains of 520.6–597.8 M generations. We calibrated the same nodes as for the treePL‐built phylogeny, and provide details about the calibration strategy in Methods S1. The XML file used for in Beast2 is provided as Dataset S5.
Finally, the resulting trees were first formatted in FigTree v.1.4.4 (Rambaut, 2018) then refined in Inkscape v.0.92.3. The treePL and Beast2 files used for the phylogenetic analyses are provided, ‘see Data availability section’.
Results
Congruence between our treePL phylogeny and prior knowledge
The maximum likelihood and Bayesian inference approaches produced virtually the same result, in terms both of topology and dates (Figs S1, S2, respectively). Both these phylogenies led to the same conclusions regarding our research question; we chose to focus our discussion on the treePL results only.
The dated phylogeny we built with treePL is robust to the choice of parameters. Even though this analysis used the best optimisation and smoothing parameters suggested by treePL, we tested other sensible values: they resulted in trees that had similar dates as the tree built from the best parameters. The differences in values (10−1–100 order of magnitude) do not impinge on our conclusions, as the great majority of divergence dates between divaricate clades and their non‐divaricate sister groups remained < 5 Ma regardless of choice of parameters. The variation in dates observed can at least partly be attributed to stochasticity of the treePL dating process (Maurin, 2020a).
The topology of our treePL phylogeny from the root to the crown of the families is consistent with current ideas about the relationships among the corresponding clades (APG IV, 2016; Stevens, 2017; Fig. 2). This backbone has a BS support of ≥ 94%, with all nodes but two having BS support of 100%. Furthermore, the great majority of genera are monophyletic with BS support of 100%, the exceptions being a few genera of doubtful or contentious monophyly, as suggested by previous studies of those groups (e.g. Coprosma, Cantley et al., 2016 and Teucrium, Salmaki et al., 2016). Finally, the relationships among species within genera are also consistent with previous work (except for Muehlenbeckia, see next paragraph), some based on plastid DNA (Maurin, 2020b for Pennantia, Maurin & Smissen, 2021 for Corokia); others on nuclear ribosomal markers (e.g. Gardner et al., 2021 for Streblus; Mitchell et al., 2009 for Melicytus; Appelhans et al., 2014 for Melicope) or based on morphological characters (e.g. Aagesen, 1999 for Discaria).
Fig. 2.
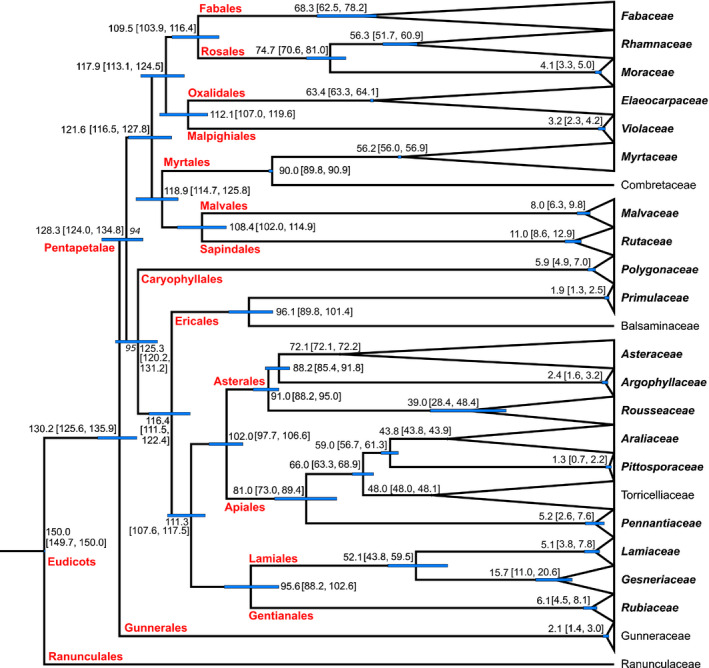
treePL phylogeny (Supporting Information Fig. S1) with nodes collapsed at the family level. In bold italics are the 19 eudicot families containing divaricate species. Mean node ages and 95% confidence intervals on node ages (also represented by blue bars) are displayed at the corresponding nodes. Names of orders and more inclusive clades are indicated at the crown node of said ranks.
Our estimated ages of orders and more inclusive clades are largely consistent with previous knowledge (Stevens, 2017). Similarly, our age estimates of family and genus crowns and/or stems are consistent with previous studies with comparable taxon sampling and dating methods but various sets of DNA markers (Heenan & McGlone, 2019). There are two notable exceptions however: the split between Coprosma moorei and the clade composed of Nertera and the rest of Coprosma, and the crown age of Muehlenbeckia. Our date is significantly younger than that of Cantley et al. (2016) for Coprosma, most probably because their calibration was conservatively old but also perhaps because of our much‐increased sampling of nucleotides. Schuster et al. (2013) provided an age range of 14.2–33.5 Ma for the crown age of Muehlenbeckia based on an analysis of plastid and nuclear ribosomal DNA, with their tree showing a relatively early divergence of M. astonii within Muehlenbeckia as well as a topology inconsistent with our tree. However, some of the Muehlenbeckia nrDNA sequences used in Schuster et al. (2013) were replaced without comment with more recent sequences in Schuster et al. (2015) and we note that in both cases cloning of PCR products was used to generate sequences. It is possible that a mixture including paralogous sequences or pseudogenes were analysed in Schuster et al. (2013).
Divergence time between divaricate species and their closest non‐divaricate relatives
The older CI bound of the age of the > 90% BS‐supported MRCA of a divaricate species (or clade) and its closest non‐divaricate sister is ≤ 5 Ma in all but three cases (Table 1). In cases of genera with numerous species whose relationships are not well supported or when there is reason to believe that plastid phylogenies may be poor reflections of species phylogenies (e.g. Coprosma, Olearia, Sophora), we conservatively considered the MRCA of the entire genus. For Coprosma, C. moorei was excluded because phylogenetic evidence suggests that it should be excluded from that genus (Cantley et al., 2016; this study). Figs S1 and S2 show which nodes we decided to examine under these rules. Because our discussion about the evolution of divaricate habit in New Zealand is centred around whether or not the divaricate habit proliferated within the last c. 5 Ma, we considered the older CI bounds of these ages instead of the mean estimated ages, to ensure a conservative treatment of the question. The only three cases for which older dates are suggested are Teucrium, Neomyrtus and Rhabdothamnus. Neomyrtus and Rhabdothamnus are both monospecific; even if we sampled their closest extant non‐divaricate relatives, it is likely that extinctions of closer non‐divaricate relatives have occurred. For Neomyrtus, phylogenies of nuclear internal transcribed spacer sequences suggest that the monotypic New Caledonian endemic genus Myrtastrum, for which limited plastid DNA sequence is available, is a closer relative than any taxon we sampled (Smissen et al., 2021). The difference for Teucrium may not be significant: the placement of T. parvifolium is only supported by a BS value of 73; a better resolved and more thoroughly sampled phylogeny based on similar genetic markers and dating approach to ours may find a younger MRCA of T. parvifolium and a non‐divaricate relative.
Discussion
The divergence dates we obtained indicate a proliferation of the divaricate habit in New Zealand within the last 5 Ma (Table 1; Fig. 3). The only three species associated with divergences older than 5 Ma (T. parvifolium, Neomyrtus pedunculata, Rhabdothamnus solandri) have only weakly developed cage architectures and were described by Greenwood & Atkinson (1977) as ‘semidivaricate’. By contrast, divergences that gave rise to highly developed cage architectures occurred exclusively after this date. This Plio‐Pleistocene concentration of divergences implicates climatic adversity as, at least partly, responsible for the evolution of the divaricate habit in many New Zealand lineages. The combined effects of the rise of the Southern Alps (Batt et al., 2000) and global cooling (Hornibrook, 1992) created new frosty and droughty environments in New Zealand during the last 5 Ma, especially in the eastern South Island (Fig. 3).
Fig. 3.
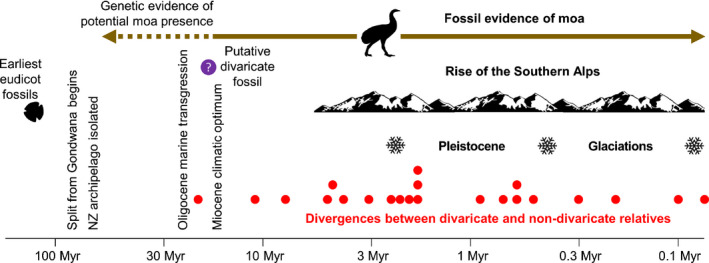
Log time‐scale plot of the maximum estimated ages of the divergences between divaricate and non‐divaricate species (Table 1). The selective pressures purported to have favoured the evolution of the divaricate habit in New Zealand (NZ) and landmarks in its geological history are also shown. Although the NZ biota has in the past been regarded as a Gondwanan legacy, molecular clock dating studies have shown that the vast majority of extant angiosperm lineages arrived by trans‐oceanic dispersal during the Cenozoic era (Heenan & McGlone, 2019). Similarly, moa are now believed to have evolved from volant ancestors that arrived by long‐distance dispersal (Phillips et al., 2010). Although the extent of the NZ landmass was greatly reduced during the Oligocene period (Mildenhall et al., 2014), fossil and genetic evidence suggests that this marine transgression has had little effect on the formation of the extant flora (Heenan & McGlone, 2019). Myr, million years.
Our results are difficult to reconcile with an explanation of divaricate evolution based solely on avian browsing (Greenwood & Atkinson, 1977). Fossil evidence indicates that moa were present in New Zealand at least 16 Ma (Tennyson et al., 2010), and the date of divergence of moa from their closest extant relatives (South American tinamous) has been estimated at c. 60 Ma (Phillips et al., 2010; Fig. 3). If moa browsing alone had driven the development of the divaricate habit, we might therefore expect to find highly developed cage architectures associated with many divergences before 5 Ma and possibly whole clades of species showing cage architectures, as seen in the Miocene proliferation of spinescence in Africa, coincident with the rise of bovids (Charles‐Dominique et al., 2016).
Although it is possible that Neogene plant extinctions have obscured evidence of earlier proliferation of cage architectures in New Zealand, overall the plant fossil record is not consistent with this scenario. The New Zealand flora in general has undergone massive turnover since the mid‐Miocene age, with an estimated 89% of the extant vascular plant species of New Zealand originating within the last 15 Ma (Heenan & McGlone, 2019; see also Wallis & Jorge, 2018). Large moa were present at least as early as 16 Ma (Tennyson et al., 2010; Fig. 3), and probably much earlier (Phillips et al., 2010). Earlier divaricate lineages might conceivably have been present during the Miocene, only to have died out and been replaced by new lineages that independently evolved the divaricate habit anew in response to moa browsing. According to this scenario, the cold and dry climates arising since 5 Ma would not have been critical for the local evolution of cage architectures. A macrofossil of a divaricate‐like plant with nanophyll leaves has been reported from the early Miocene age of New Zealand (Campbell et al., 2000), although the equivalence of this plant with contemporary divaricate plants is by no means certain. More conclusively, the typical leaf class sizes of extant divaricate species (nanophylls and leptophylls, sensu Wolfe, 1993) make up only 1.5% of New Zealand macrofossil assemblages from the warmer climates of the early to mid‐Miocene (Reichgelt et al., 2017). This palaeobotanical evidence seems telling, as small leaves are normally an integral part of contemporary cage architectures in other regions (McQueen, 2000; Bond & Silander, 2007; Charles‐Dominique et al., 2017). Cage architectures are therefore unlikely to have been widespread in early to mid‐Miocene New Zealand. Although Pole & Moore (2011) reported fossil leaves very similar to those of the extant New Zealand divaricate Myrsine divaricata from near the end of the Miocene (6.5–6.0 Ma), by that time global temperatures had cooled considerably.
Although our chronology of divergences (Table 1; Fig. 3) is compatible with an explanation based solely on climate (Diels, 1896; McGlone & Webb, 1981), other studies have implicated browsing in the evolution of cage architectures. Experiments have shown New Zealand divaricate plants to be less attractive to both avian and ungulate browsers than larger leaved, more sparsely‐branched relatives (Pollock et al., 2007; Lusk et al., 2021), as also seen with cage architectures in southern Africa (Charles‐Dominique et al., 2017). These studies indicated a selective advantage of the divaricate habit and of cage architecture, in general, in deterring browsers.
The sum of evidence from this and previous research is therefore best explained by the synthetic hypothesis that cage architectures were not strongly selected in New Zealand until cold, dry Plio‐Pleistocene climates prevented juvenile trees from growing quickly out of the browse zone (Lusk et al., 2016). This hypothesis is consistent with the abundance of cage architectures in other regions where plant growth is restricted by aridity or short frost‐free periods, such as Patagonian steppe (McQueen, 2000), African savannas (Charles‐Dominique et al., 2017), and Madagascan thickets (Bond & Silander, 2007). It is also consistent with the low incidence of spinescence (a more well known structural defence) in moist tropical forests (Charles‐Dominique et al., 2016; da Silva‐Luz et al., 2019), where juvenile pioneer trees can rapidly escape from ground‐based browsers by growing several metres in height per year beneath treefall gaps (Brokaw, 1987). As well as reconciling the two competing explanations of the divaricate habit in New Zealand, this study therefore adds to evidence that climate modulates the adaptive value of structural defences against browsing, worldwide. The especially high incidence of spinescence in fertile savannas (Scholes, 1990), and of cage architectures on alluvial soils in New Zealand (Lusk et al., 2020), suggest that a selection for structural defences is strongest when high nutrient availability coincides with strong climatic constraints on plant growth rates (Lusk et al., 2016).
Author contributions
CHL and RDS conceived the original idea; KJLM developed and designed the project, with input from CHL and RDS; KJLM and RDS generated the sequence data; KJLM performed the phylogenetic analyses; KJLM led the writing of the manuscript and produced the figures and tables with input from CHL and RDS All the authors read and approved the final manuscript.
Supporting information
Dataset S1 Set of baits used for the enrichment of the library for chloroplast DNA.
Dataset S2 Alignment of our 215 sequences in Phylip format for reconstructing an undated phylogeny with RAxML.
Dataset S3 Partitioning of the Phylip alignment.
Dataset S4 treePL configuration file used to date the RAxML‐generated trees.
Dataset S5 XML file used for the Beast2 analysis.
Dataset S6 Geneious file containing the Rubiaceae sequences, which could not be deposited on GenBank.
Fig. S1 Dated phylogeny of our 215 species, built with treePL from RAxML‐generated trees (in three figures: bootstrap support; 95% confidence intervals on node ages; mean node ages).
Fig. S2 Dated phylogeny of our 215 species, built with Beast2 (in three figures: posterior probabilities; 95% highest posterior densities on node ages; mean node ages).
Methods S1 Calibration strategy we designed for our phylogeny.
Table S1 DNA sequence information for the 215 species used for the phylogenetic analyses.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This research is supported by the Royal Society of New Zealand through Marsden contract 16‐UOW‐029 and the Faculty of Science and Engineering of the University of Waikato through FSEN Student Trust Fund (#P102218 SoS/PG Support). We thank Ana Podolyan, Carina Davis, Caroline Mitchell and Kat Trought for laboratory support; Ines Schönberger and Mary Korver for herbarium support at CHR; the herbaria BRI, CANB, CHR, HO, MEL, NOU, OTA, P, and Kerry Ford (CHR), Jason Cantley (San Francisco State University) and Patricio Saldivia Pérez (University of Otago) for samples; the Direction de l’Environnement of Province Sud of New Caledonia for local sampling permits; the Department of Conservation of New Zealand for local sampling permits, and the Sanctuary Mountain Maungatautari, the Waipa District Council, the Pukemokemoke Bush Trust, Otari Native Botanic Garden and the iwi Mōkai Pātea, Ngāti Apa, Ngāti Hauiti, Ngāti Rangi, Pirirakau Hapū, Taumutu, Waikato‐Tainui for local sampling agreements; Alison Devault (Arbor Biosciences) for myBaits design; and two anonymous referees for insightful comments. The authors declare no competing interests.
Data availability
All sequences used in this study except those from Rubiaceae species can be downloaded from the GenBank database; their accession numbers are provided in Table S1. The Rubiaceae sequences are available in Dataset S6. All files used to build the phylogenies are also provided: Datasets S2–S4 for the treePL phylogeny, Dataset S5 for the Beast2 phylogeny.
References
- Aagesen L. 1999. Phylogeny of the tribe Colletieae, Rhamnaceae. Botanical Journal of the Linnean Society 131: 1–43. [Google Scholar]
- Allan HH. 1961. Flora of New Zealand, vol. I. Wellington, New Zealand: Government Printer. [Google Scholar]
- Appelhans MS, Wen J, Wagner WL. 2014. A molecular phylogeny of Acronychia, Euodia, Melicope and relatives (Rutaceae) reveals polyphyletic genera and key innovations for species richness. Molecular Phylogenetics and Evolution 79: 54–68. [DOI] [PubMed] [Google Scholar]
- Batt GE, Braun J, Kohn BP, McDougall I. 2000. Thermochronological analysis of the dynamics of the Southern Alps, New Zealand. Geological Society of America Bulletin 112: 250–266. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond WJ, Lee WG, Craine JM. 2004. Plant structural defences against browsing birds: a legacy of New Zealand’s extinct moas. Oikos 104: 500–508. [Google Scholar]
- Bond WJ, Silander JA. 2007. Springs and wire plants: anachronistic defences against Madagascar’s extinct elephant birds. Proceedings of the Royal Society of London B: Biological Sciences 274: 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Vaughan TG, Barido‐Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N et al. 2019. Beast 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Computational Biology 15: e1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert RR, Drummond AJ. 2017. bModelTest: bayesian phylogenetic site model averaging and model comparison. BMC Evolutionary Biology 17: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw N. 1987. Gap‐phase regeneration of three pioneer tree species in a tropical forest. Journal of Ecology 75: 9–19. [Google Scholar]
- Campbell J, Lee D, Lee W. 2000. A woody shrub from the Miocene Nevis Oil Shale, Otago, New Zealand – a possible fossil divaricate? Journal of the Royal Society of New Zealand 30: 147–153. [Google Scholar]
- Cantley JT, Markey AS, Swenson NG, Keeley SC. 2016. Biogeography and evolutionary diversification in one of the most widely distributed and species rich genera of the Pacific. AoB Plants 8: plw043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavagnaro FP, Golluscio RA. 2017. Structural anti‐herbivore defense reduction of two Patagonian spiny shrubs in response to long time exclusion of large herbivores. Journal of Arid Environments 142: 36–40. [Google Scholar]
- Charles‐Dominique T, Barczi J, Le Roux E, Chamaillé‐Jammes S. 2017. The architectural design of trees protects them against large herbivores. Functional Ecology 31: 1710–1717. [Google Scholar]
- Charles‐Dominique T, Davies TJ, Hempson GP, Bezeng BS, Daru BH, Kabongo RM, Maurin O, Muasya AM, Van der Bank M, Bond WJ. 2016. Spiny plants, mammal browsers, and the origin of African savannas. Proceedings of the National Academy of Sciences, USA 113: E5572–E5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockayne L. 1912. Observations concerning evolution, derived from ecological studies in New Zealand. Transactions of the New Zealand Institute 44: 1–50. [Google Scholar]
- Cooper A, Atkinson I, Lee WG, Worthy T. 1993. Evolution of the moa and their effect on the New Zealand flora. Trends in Ecology & Evolution 8: 433–437. [DOI] [PubMed] [Google Scholar]
- Cummings MP, Nugent JM, Olmstead RG, Palmer JD. 2003. Phylogenetic analysis reveals five independent transfers of the chloroplast gene rbcL to the mitochondrial genome in angiosperms. Current Genetics 43: 131–138. [DOI] [PubMed] [Google Scholar]
- da Silva‐Luz CL, Pirani JR, Mitchell JD, Daly D, do Valle Capelli N, Demarco D, Pell SK, Plunkett GM. 2019. Phylogeny of Schinus L. (Anacardiaceae) with a new infrageneric classification and insights into evolution of spinescence and floral traits. Molecular Phylogenetics and Evolution 133: 302–351. [DOI] [PubMed] [Google Scholar]
- Diels L. 1896. Vegetations‐biologie von Neu‐Seeland. Leipzig, Germany: Wilhelm Engelmann. [Google Scholar]
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon 36: 715–722. [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biology 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galetti M, Moleón M, Jordano P, Pires MM, Guimarães PR Jr., Pape T, Nichols E, Hansen D, Olesen JM, Munk M et al. 2018. Ecological and evolutionary legacy of megafauna extinctions. Biological Reviews 93: 845–862. [DOI] [PubMed] [Google Scholar]
- Gardner EM, Garner M, Cowan R, Dodsworth S, Epitawalage N, Arifiani D, Sahromi, Baker WJ, Forest F, Maurin O et al. 2021. Repeated parallel losses of inflexed stamens in Moraceae: phylogenomics and generic revision of the tribe Moreae and the reinstatement of the tribe Olmedieae (Moraceae). Taxon. doi: 10.1002/tax.12526. [DOI] [Google Scholar]
- Gernhard T. 2008. The conditioned reconstructed process. Journal of Theoretical Biology 253: 769–778. [DOI] [PubMed] [Google Scholar]
- Goremykin VV, Salamini F, Velasco R, Viola R. 2009. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Molecular Biology and Evolution 26: 99–110. [DOI] [PubMed] [Google Scholar]
- Greenwood R, Atkinson I. 1977. Evolution of divaricating plants in New Zealand in relation to moa browsing. Proceedings of the New Zealand Ecological Society 24: 21–33. [Google Scholar]
- Hanley ME, Lamont BB, Fairbanks MM, Rafferty CM. 2007. Plant structural traits and their role in anti‐herbivore defence. Perspectives in Plant Ecology, Evolution and Systematics 8: 157–178. [Google Scholar]
- Heenan PB, McGlone MS. 2019. Cenozoic formation and colonisation history of the New Zealand vascular flora based on molecular clock dating of the plastid rbcL gene. New Zealand Journal of Botany 57: 204–226. [Google Scholar]
- Hornibrook NB. 1992. New Zealand Cenozoic marine paleoclimates: a review based on the distribution of some shallow water and terrestrial biota. In: Tsuchi R, Ingle JC Jr, eds. Pacific Neogene: environment, evolution, and events. Tokyo, Japan: University of Tokyo Press, 83–106. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. Mafft: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software v.7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler L, Newsom LA, Ryan TM, Clarke AC, Smith BD, Perry GH. 2015. Gourds and squashes (Cucurbita spp.) adapted to megafaunal extinction and ecological anachronism through domestication. Proceedings of the National Academy of Sciences, USA 112: 15107–15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with Bwa‐Mem . arXiv: 1303.3997. [Google Scholar]
- Li H‐T, Yi T‐S, Gao L‐M, Ma P‐F, Zhang T, Yang J‐B, Gitzendanner MA, Fritsch PW, Cai J, Luo Y et al. 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants 5: 461–470. [DOI] [PubMed] [Google Scholar]
- Lusk CH, Clearwater MJ, Laughlin DC, Harrison SP, Prentice IC, Nordenstahl M, Smith B. 2018. Frost and leaf‐size gradients in forests: global patterns and experimental evidence. New Phytologist 219: 565–573. [DOI] [PubMed] [Google Scholar]
- Lusk CH, McGlone MS, Overton JM. 2016. Climate predicts the proportion of divaricate plant species in New Zealand arborescent assemblages. Journal of Biogeography 43: 1881–1892. [Google Scholar]
- Lusk CH, Wiser SK, Laughlin DC. 2020. Macroclimate and topography interact to influence the abundance of divaricate plants in New Zealand. Frontiers in Plant Science 11: 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk CH, Wiser SK, Laughlin DC. 2021. Climate influences the value of a plant structural defence against browsing. Journal of Ecology 109: 1411–1423. [Google Scholar]
- Magallón S, Gómez‐Acevedo S, Sánchez‐Reyes LL, Hernández‐Hernández T. 2015. A metacalibrated time‐tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist 207: 437–453. [DOI] [PubMed] [Google Scholar]
- Martin PS, Klein RG. 1989. Quaternary extinctions: a prehistoric revolution. Tucson, AZ, USA: University of Arizona Press. [Google Scholar]
- Maurin KJL. 2020a. An empirical guide for producing a dated phylogeny with treePL in a maximum likelihood framework. arXiv: 2008.07054. [Google Scholar]
- Maurin KJL. 2020b. A dated phylogeny of the genus Pennantia (Pennantiaceae) based on whole chloroplast genome and nuclear ribosomal 18S–26S repeat region sequences. PhytoKeys 155: 15–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurin KJL, Lusk CH. 2021. 120 years of untangling the divaricate habit: a review. New Zealand Natural Sciences 46: 1–29. [Google Scholar]
- Maurin KJ, Smissen RD. 2021. A dated phylogeny of Argophyllaceae (Asterales) is consistent with spread by long‐distance dispersal. New Zealand Journal of Botany. doi: 10.1080/0028825X.2021.1905671. [DOI] [Google Scholar]
- McGlone MS, Webb C. 1981. Selective forces influencing the evolution of divaricating plants. New Zealand Journal of Ecology 4: 20–28. [Google Scholar]
- McQueen D. 2000. Divaricating shrubs in Patagonia and New Zealand. New Zealand Journal of Ecology 24: 69–80. [Google Scholar]
- Mildenhall DC, Mortimer N, Bassett K, Kennedy E. 2014. Oligocene paleogeography of New Zealand: maximum marine transgression. New Zealand Journal of Geology and Geophysics 57: 107–109. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments workshop (GCE). New Orleans, LA, USA: IEEE, doi: 10.1109/GCE.2010.5676129. [DOI] [Google Scholar]
- Mitchell A, Heenan PB, Murray B, Molloy B, de Lange P. 2009. Evolution of the south‐western Pacific genus Melicytus (Violaceae): evidence from DNA sequence data, cytology and sex expression. Australian Systematic Botany 22: 143–157. [Google Scholar]
- Nock CJ, Waters DL, Edwards MA, Bowen SG, Rice N, Cordeiro GM, Henry RJ. 2011. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnology Journal 9: 328–333. [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Gibb GC, Crimp EA, Penny D. 2010. Tinamous and moa flock together: mitochondrial genome sequence analysis reveals independent losses of flight among ratites. Systematic Biology 59: 90–107. [DOI] [PubMed] [Google Scholar]
- Pole M, Moore PR. 2011. A late Miocene leaf assemblage from Coromandel Peninsula, New Zealand, and its climatic implications. Alcheringa 35: 103–121. [Google Scholar]
- Pollock ML, Lee WG, Walker S, Forrester G. 2007. Ratite and ungulate preferences for woody New Zealand plants: influence of chemical and physical traits. New Zealand Journal of Ecology 31: 68–78. [Google Scholar]
- Rambaut A. 2018. FigTree v.1.4.4. Computer program distributed by the author. [WWW document] URL https://github.com/rambaut/figtree/releases [accessed 27 November 2018]. [Google Scholar]
- Reichgelt T, Lee WG, Lusk CH, Kennedy EM. 2017. Changes in leaf physiognomy of New Zealand woody assemblages in response to Neogene environmental cooling. Journal of Biogeography 44: 1160–1171. [Google Scholar]
- Salmaki Y, Kattari S, Heubl G, Bräuchler C. 2016. Phylogeny of non‐monophyletic Teucrium (Lamiaceae: ajugoideae): implications for character evolution and taxonomy. Taxon 65: 805–822. [Google Scholar]
- Saltré F, Rodríguez‐Rey M, Brook BW, Johnson CN, Turney CSM, Alroy J, Cooper A, Beeton N, Bird MI, Fordham DA et al. 2016. Climate change not to blame for late Quaternary megafauna extinctions in Australia. Nature Communications 7: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes R. 1990. The influence of soil fertility on the ecology of southern African dry savannas. Journal of Biogeography 17: 415–419. [Google Scholar]
- Schuster TM, Reveal JL, Bayly MJ, Kron KA. 2015. An updated molecular phylogeny of Polygonoideae (Polygonaceae): relationships of Oxygonum, Pteroxygonum, and Rumex, and a new circumscription of Koenigia. Taxon 64: 1188–1208. [Google Scholar]
- Schuster TM, Setaro SD, Kron KA. 2013. Age estimates for the buckwheat family Polygonaceae based on sequence data calibrated by fossils and with a focus on the Amphi‐Pacific Muehlenbeckia . PLoS ONE 8: e61261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd LD, Heenan PB. 2021. Phylogenomic analyses reveal a history of hybridisation and introgression between Sophora sect. Edwardsia (Fabaceae) species in New Zealand. New Zealand Journal of Botany. doi: 10.1080/0028825X.2021.1960567. [DOI] [Google Scholar]
- Smissen R, Heenan P. 2007. DNA fingerprinting supports hybridisation as a factor explaining complex natural variation in Phormium (Hemerocallidaceae). New Zealand Journal of Botany 45: 419–432. [Google Scholar]
- Smissen RD, Heenan PB, Maurin KJ. 2021. New Zealand endemic Neomyrtus is sister to New Caledonian endemic Myrtastrum (Myrtaceae, Myrteae). New Zealand Journal of Botany. doi: 10.1080/0028825X.2021.1965629. [DOI] [Google Scholar]
- Smith SA, O’Meara BC. 2012. treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28: 2689–2690. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Kuzoff RK. 1995. Discordance between nuclear and chloroplast phylogenies in the Heuchera group (Saxifragaceae). Evolution 49: 727–742. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML v.8: a tool for phylogenetic analysis and post‐analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens P. 2017. Angiosperm phylogeny website. v.14. [WWW document] URL http://www.mobot.org/MOBOT/research/APweb/ [accessed 22 July 2020]. [Google Scholar]
- Stuart AJ. 2015. Late Quaternary megafaunal extinctions on the continents: a short review. Geological Journal 50: 338–363. [Google Scholar]
- Stull GW, Moore MJ, Mandala VS, Douglas NA, Kates H‐R, Qi X, Brockington SF, Soltis PS, Soltis DE, Gitzendanner MA. 2013. A targeted enrichment strategy for massively parallel sequencing of angiosperm plastid genomes. Applications in Plant Sciences 1: 1200497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson AJ, Worthy TH, Jones CM, Scofield RP, Hand SJ. 2010. Moa’s Ark: miocene fossils reveal the great antiquity of moa (Aves: Dinornithiformes) in Zealandia. Records of the Australian Museum 62: 105–114. [Google Scholar]
- The Angiosperm Phylogeny Group , Chase MW, Christenhusz M, Fay M, Byng J, Judd WS, Soltis D, Mabberley D, Sennikov A, Soltis PS et al. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181: 1–20. [Google Scholar]
- Tonti‐Filippini J, Nevill PG, Dixon K, Small I. 2017. What can we do with 1000 plastid genomes? The Plant Journal 90: 808–818. [DOI] [PubMed] [Google Scholar]
- Wallace AR. 1876. The geographical distribution of animals: with a study of the relations of living and extinct faunas as elucidating the past changes of the earth’s surface. London, UK: MacMillan. [Google Scholar]
- Wallis GP, Jorge F. 2018. Going under down under? Lineage ages argue for extensive survival of the Oligocene marine transgression on Zealandia. Molecular Ecology 27: 4368–4396. [DOI] [PubMed] [Google Scholar]
- Wichman SR, Wright SD, Cameron EK, Keeling DJ, Gardner RC. 2002. Elevated genetic heterogeneity and Pleistocene climatic instability: inferences from nrDNA in New Zealand Coprosma (Rubiaceae). Journal of Biogeography 29: 943–954. [Google Scholar]
- Wolfe JA. 1993. A method of obtaining climatic parameters from leaf assemblages. Washington, DC, USA: US Government Printing Office. [Google Scholar]
- Yates MJ, Verboom GA, Rebelo AG, Cramer MD. 2010. Ecophysiological significance of leaf size variation in Proteaceae from the Cape Floristic Region. Functional Ecology 24: 485–492. [Google Scholar]
- Zhang G‐J, Dong R, Lan L‐N, Li S‐F, Gao W‐J, Niu H‐X. 2020. Nuclear integrants of organellar DNA contribute to genome structure and evolution in plants. International Journal of Molecular Sciences 21: 707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1 Set of baits used for the enrichment of the library for chloroplast DNA.
Dataset S2 Alignment of our 215 sequences in Phylip format for reconstructing an undated phylogeny with RAxML.
Dataset S3 Partitioning of the Phylip alignment.
Dataset S4 treePL configuration file used to date the RAxML‐generated trees.
Dataset S5 XML file used for the Beast2 analysis.
Dataset S6 Geneious file containing the Rubiaceae sequences, which could not be deposited on GenBank.
Fig. S1 Dated phylogeny of our 215 species, built with treePL from RAxML‐generated trees (in three figures: bootstrap support; 95% confidence intervals on node ages; mean node ages).
Fig. S2 Dated phylogeny of our 215 species, built with Beast2 (in three figures: posterior probabilities; 95% highest posterior densities on node ages; mean node ages).
Methods S1 Calibration strategy we designed for our phylogeny.
Table S1 DNA sequence information for the 215 species used for the phylogenetic analyses.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
All sequences used in this study except those from Rubiaceae species can be downloaded from the GenBank database; their accession numbers are provided in Table S1. The Rubiaceae sequences are available in Dataset S6. All files used to build the phylogenies are also provided: Datasets S2–S4 for the treePL phylogeny, Dataset S5 for the Beast2 phylogeny.


