Summary
The Pseudomonas syringae DC3000 type III effector HopAM1 suppresses plant immunity and contains a Toll/interleukin‐1 receptor (TIR) domain homologous to immunity‐related TIR domains of plant nucleotide‐binding leucine‐rich repeat receptors that hydrolyze nicotinamide adenine dinucleotide (NAD+) and activate immunity. In vitro and in vivo assays were conducted to determine if HopAM1 hydrolyzes NAD+ and if the activity is essential for HopAM1’s suppression of plant immunity and contribution to virulence.
HPLC and LC‐MS were utilized to analyze metabolites produced from NAD+ by HopAM1 in vitro and in both yeast and plants. Agrobacterium‐mediated transient expression and in planta inoculation assays were performed to determine HopAM1’s intrinsic enzymatic activity and virulence contribution.
HopAM1 is catalytically active and hydrolyzes NAD+ to produce nicotinamide and a novel cADPR variant (v2‐cADPR). Expression of HopAM1 triggers cell death in yeast and plants dependent on the putative catalytic residue glutamic acid 191 (E191) within the TIR domain. Furthermore, HopAM1’s E191 residue is required to suppress both pattern‐triggered immunity and effector‐triggered immunity and promote P. syringae virulence.
HopAM1 manipulates endogenous NAD+ to produce v2‐cADPR and promote pathogenesis. This work suggests that HopAM1’s TIR domain possesses different catalytic specificity than other TIR domain‐containing NAD+ hydrolases and that pathogens exploit this activity to sabotage NAD+ metabolism for immune suppression and virulence.
Keywords: cADPR variant, HopAM1, NAD+ , NAD+ hydrolase, Pseudomonas syringae, TIR domain, v2‐cADPR, v‐cADPR
Introduction
Toll/interleukin‐1 receptor (TIR) domain proteins are an ancient family of nicotinamide adenine dinucleotide (NAD+) hydrolases (Essuman et al., 2018). TIR domains are found across kingdoms including archaea, bacteria, animals and plants (Bayless & Nishimura, 2020). In animals, TIR domain‐containing Toll‐like receptors (TLRs) serve as pattern recognition receptors that recognize pathogen‐ or microbe‐associated molecular patterns (PAMPs or MAMPs) and activate innate immunity (O'Neill et al., 2013). One animal TIR domain‐containing protein, the highly conserved sterile alpha and TIR motif containing 1 (SARM1), serves as a TLR inhibitor and possesses NAD+ hydrolase activity that causes NAD+ depletion and neuronal cell death (Essuman et al., 2017; Carty & Bowie, 2019). In plants, TIR domain nucleotide‐binding leucine‐rich repeat receptors (TIR‐NLRs), together with coiled‐coil (CC)‐domain NLRs, make up the two major groups of intracellular receptors that activate immunity and serve as resistance proteins (R proteins) (Ma et al., 2020; Martin et al., 2020). Recently, it was found that direct or indirect recognition of pathogenic effectors in plants results in oligomerization of R‐gene NLRs mediated by the CC domain or TIR domain and this oligomerization of TIR‐NLRs leads to NAD+ cleavage that results in immune activation and cell death (Horsefield et al., 2019; Wan et al., 2019; Ma et al., 2020; Martin et al., 2020).
TIR domain‐containing proteins (Tcps) are widely distributed in prokaryotes but more sparsely in fungi and viruses and have been classified as a family of virulence factors, some of which disrupt TLR signaling in animal hosts (Bowie et al., 2000; Newman et al., 2006; Spear et al., 2009; Cirl & Miethke, 2010; Alaidarous et al., 2014; Patterson et al., 2014; Rosadini et al., 2015; Imbert et al., 2017). Prokaryotic TIR domain proteins can also hydrolyze NAD+, including TIR domain‐containing effectors BtpA and BtpB from the animal pathogen Brucella abortus that deplete host NAD+ while suppressing TLR signaling (Essuman et al., 2018; Coronas‐Serna et al., 2020). TIR domains present in a wide range of microbial genomes are probable components of prokaryotic antiviral immunity (Ma et al., 2020; Ofir et al., 2021). To date, no TIR domain‐containing effectors of phytopathogens have been reported to hydrolyze NAD+. A few non‐TIR domain effectors use NAD+ as a substrate for their immune‐suppressive activity: Pseudomonas syringae effectors HopU1 and HopF2 utilize NAD+ to ADP‐ribosylate host proteins and Xanthomonas effector AvrRxo1 phosphorylates NAD+ into NADP+ (Fu et al., 2007; Wang et al., 2010; Jeong et al., 2011; Shidore et al., 2017). Despite their similarity to eukaryotic TIR domains, bacterial TIR domains and their resulting cleavage products do not appear to activate plant immunity when expressed in planta (Duxbury et al., 2020). However, the influence of TIR domain effectors and NAD+ on the immune system remains unclear in plants.
Nicotinamide adenine dinucleotide is an essential dinucleotide molecule at the center of cellular metabolism and redox signaling (Noctor, 2006). In plants, NAD+ is either produced de novo using aspartate or via the salvage pathway using nicotinamide (Nam), nicotinamide mononucleotide or other intermediates (Gakière et al., 2018). NAD+ is functional across a range of diverse cellular activities including calcium signaling, ADP‐ribosylation and histone modification (Hunt et al., 2004). Although the role of NAD+ during pathogen infection is not fully understood, NAD+ is clearly implicated in a vital defense role in plant–pathogen interactions (Pétriacq et al., 2013, 2016). In barley, NAD+ content spiked in leaves infected with powdery mildew (Ryrie & Scott, 1968). Overproduction of endogenous NAD+ in Arabidopsis stimulated the salicylic acid pathway leading to enhanced resistance to the avirulent strain Pto‐AvrRpm1 (Pétriacq et al., 2012). Extracellular NAD+ is suggested to be an elicitor of plant immunity and a possible component of plant systemic acquired resistance (Wang et al., 2017). Phosphorylation of NAD+ by Xanthomonas effector AvrRxo1 mitigates the reactive oxygen species (ROS) burst component of the immune response in tobacco (Shidore et al., 2017). Recently, NAD+ hydrolysis and the derived products were associated with the cell death response to pathogens in animals and plants (Horsefield et al., 2019; Wan et al., 2019). Variable cleavage of NAD+ by TIR domain proteins produces nicotinamide and adenine diphosphate ribose (ADPR), cyclic adenine diphosphate ribose (cADPR) or a structurally unidentified cADPR variant (v‐cADPR) with equal mass theorized to be an alternative cyclic linkage (Essuman et al., 2017, 2018; Wan et al., 2019; Duxbury et al., 2020). cADPR is an adenine nucleotide cyclized at the N1 position of the adenine ring that acts as a second messenger of ABA and as a mediator of calcium (Ca2+) release (Lee, 1994; Wu et al., 1997). A clear picture has yet to emerge describing the function of NAD+, cADPR and v‐cADPR during pathogenic interactions.
To successfully colonize hosts, pathogens must circumvent immunity. Hosts are capable of recognizing PAMPs or MAMPs and activating pattern‐triggered immunity (PTI) to restrict pathogen growth (Zipfel, 2014). In plants, effector recognition by intracellular CC‐NLRs and TIR‐NLRs potentiates a more intense effector‐triggered immunity (ETI) resulting in a hypersensitive response (HR) and plant cell death (Wu et al., 2017; Wan et al., 2019). To combat immunity, many bacterial pathogens utilize secretion systems like the type III secretion system (T3SS) to deliver effectors that suppress PTI and/or ETI by interfering with host signaling or protein components of plant immunity (Guo et al., 2009; Buttner, 2012; Macho & Zipfel, 2015). Pathogenic effectors possess diverse enzymatic activity and target a broad range of host physiological processes as part of a concerted effort to disrupt the immune response (Toruno et al., 2016). Most described effectors target host proteinaceous components but a few are thought to indirectly alter specific plant metabolites: P. syringae effector HopZ1 suppresses the isoflavone biosynthesis enzyme GmHID1 (Zhou et al., 2011), whereas the maize pathogen Pantoea stewartii effector WtsE upregulates the shikimate and phenylpropanoid pathways (Asselin et al., 2015). The specific mechanisms of these effectors remain unclear and their effects appear to promote virulence only indirectly (Macho, 2016). Only a few effectors directly alter specific metabolites: Ralstonia solanacearum effector RipAY exhibits γ‐glutamyl cyclotransferase (GGCT) activity and degrades intracellular glutathione, another R. solanacearum effector RipTPS synthesizes the plant signaling metabolite trehalose‐6‐phosphate and Xanthomonas type III effector AvrRxo1 phosphorylates NAD+ into NADP+ (Poueymiro et al., 2014; Fujiwara et al., 2016; Shidore et al., 2017). The products of these effectors, however, are metabolites already endogenous to plant cells. Little is known about how effectors directly mediate conversion of host metabolites into novel compounds or about the consequences of this activity on plant immunity and virulence.
Pseudomonas syringae is a hemibiotrophic bacterial pathogen with pathovars that can infect many plant species, including crops of agronomic importance. P. syringae pv. tomato (Pto) DC3000 is pathogenic in the model plants tomato and Arabidopsis (Xin & He, 2013). Pto DC3000 injects > 30 type III effectors, of which key effectors have been identified as immunity suppressors and contribute to pathogenicity (Jamir et al., 2004; Guo et al., 2009; Wei et al., 2015; Wei & Collmer, 2018). While significant progress has been made towards understanding the virulence contribution and host targets of P. syringae effectors, the catalytic activity of the majority of effectors in plants is unknown (Block & Alfano, 2011). One such effector is HopAM1, which strongly suppresses plant immunity and manipulates responses to ABA (Jamir et al., 2004; Goel et al., 2008; Guo et al., 2009). Ectopic expression of HopAM1 is toxic to yeast (Munkvold et al., 2008). Unusually, resistance to Pto DC3000 expressing HopAM1 appears to be polygenic instead of monogenic, and the presence of HopAM1 restricts growth of Pto DC3000 on some Arabidopsis ecotypes (Iakovidis et al., 2016; Velásquez et al., 2017). HopAM1 is one of a minimal set of effectors sufficient to restore virulence and disease symptoms of the poly‐effector mutant DC3000 D28E (Cunnac et al., 2011). Here we show that HopAM1 contains a noncanonical TIR domain and has enzymatic activity that hydrolyzes the essential metabolite NAD+ in plants to promote virulence.
Materials and Methods
Bacterial cultures, DNA manipulation and purification of recombinant proteins
Bacterial cultures, growth conditions, plasmid assembly and protein purification are described in Supporting Information Methods S1. The plasmids and strains used in this study are listed in Table S1.
Generation of Pto DC3000 mutants
The Pto DC3000 mutants studied were generated by an unmarked mutagenesis strategy with modification (House et al., 2004) and are described in Methods S1.
Plant materials
Arabidopsis plants were grown at 24°C with a 10 h : 14 h, light : dark cycle in microclimate‐controlled growth chambers. Nicotiana benthamiana and Nicotiana tabacum cv Xanthi plants were grown under standard glasshouse conditions.
Agrobacterium‐mediated transient assays
Agrobacterium strains carrying binary vectors were infiltrated into N. benthamiana or N. tabacum leaves at an OD600 of 0.8. A detailed methodology is described in Methods S1.
HPLC
For samples extracted using perchloric acid, 90 µl of sample was mixed with 10 µl of 0.5 M phosphate buffer (pH 7.0). Metabolites were analyzed using an Agilent 1260 HPLC system (Agilent, Santa Clara, CA, USA) with an Agilent Poroshell 120 EC‐C18 column (4.6 mm × 150 mm, 2.7 µm) under isocratic conditions. The mobile phase consisted of 0.05 M potassium phosphate buffer (pH 7.0) and methanol (95 : 5, v/v) with a flow rate of 1.0 ml min−1. The eluent was monitored at 260 nm by a diode array detector. Alternatively, in vitro samples and standards were reconstituted in 0.05 M phosphate buffer (pH 7.0) and analyzed using an Agilent Nexera LC‐40 with a Kinetex C18 column (3 mm × 100 mm, 2.6 µM) under isocratic conditions. The mobile phase consisted of 0.05 M potassium phosphate buffer (pH 7.0).
For sample extracted using the methanol–chloroform method, lyophilized powder was resuspended in 100 µl of 0.05 M ammonium acetate buffer (pH 6.7). Metabolites were analyzed using an Agilent 1100 HPLC system with an Agilent Poroshell 120 EC‐C18 column (4.6 mm × 150 mm, 2.7 µm) under isocratic conditions. The mobile phase consisted of 0.05 M ammonium acetate buffer (pH 6.7) and methanol (95 : 5, v/v) with a flow rate of 1.0 ml min−1. The eluent was monitored at 260 nm by a variable‐wavelength detector. Standards NAD+ (Selleck Chemicals, Pittsburgh, PA, USA), Nam (Supelco, Bellefonte, PA, USA), and cADPR (Cayman Chemical, Ann Arbor, MI, USA) were run concurrently. HPLC chromatograms were prepared using OpenChrom (https://lablicate.com/platform/openchrom).
Mass spectrometry
Lyophilized powder was resuspended in 100 µl of 0.05 M ammonium acetate buffer. A sample of 100 µl was loaded and standards NAD+, Nam and cADPR were run concurrently. Metabolites were separated using a Waters NanoAcquity UPLC system with a Waters 1.0 × 50 mm column (1.8 μm packing HSS) under isocratic conditions. The mobile phase consisted of 0.05 M ammonium acetate buffer (pH 6.7) and methanol (95 : 5, v/v) with a flow rate of 40 µl min−1. Metabolites were analyzed using a Waters Xevo G2‐XS QTOF with electrospray ionization. MS and MS/MS chromatograms were prepared using MassLynx (https://www.waters.com/waters/en_US/MassLynx‐MS‐Software/nav.htm?cid=513662).
In vitro NAD hydrolase and enzyme kinetics assays
Protocols previously reported were followed (Essuman et al., 2017) and are described in Methods S1.
Yeast metabolite analysis
Yeast overnight cultures grown in liquid dropout medium Glucose‐His were centrifuged and washed with double distilled H2O and resuspended with Galactose‐His adjusted to an OD600 of 0.5. At the indicated time points, a specified volume of culture was removed and spun down for 30 s at 15 000 g . For HPLC analysis, 1 ml of culture was resuspended in 200 µl ice‐cold 0.5 M perchloric acid and incubated on ice for 10 min. Samples were then subjected to three cycles of freezing in liquid nitrogen and thawing on ice. Samples were then centrifuged at 15 000 g for 5 min, and 180 µl of supernatant was then mixed with 67 µl of 3 M K2CO3 and incubated for 10 min on ice. Samples were centrifuged again at 15 000 g for 5 min and 90 µl of supernatant was saved at −80°C until HPLC analysis, whereupon the samples were amended with 10 µl of 0.5 M phosphate buffer (26.2 g l−1 potassium phosphate monobasic, 53.5 g l−1 potassium phosphate dibasic).
For MS analysis, 1 ml culture was resuspended in 250 µl of 50% methanol at −40°C and vortexed, then mixed with 250 µl chloroform at −40°C. Samples were subjected to three freeze–thaw cycles alternating between liquid nitrogen and a −40°C ethanol bath. Samples were then centrifuged at 15 000 g for 5 min at −10°C, then the aqueous/methanol layer was removed, lyophilized and stored at −80°C until MS analysis.
Plant metabolite analysis
Nicotiana benthamiana leaf disks were excised 24 h after induction using a 16 mm diameter cork borer. Samples were frozen in liquid nitrogen and ground with a plastic pestle. The frozen powder was suspended in 500 µl of 50% methanol at −40°C and vortexed, then mixed with 500 µl chloroform at −40°C. Samples were subjected to two additional freeze–thaw cycles alternating between liquid nitrogen and a −40°C ethanol bath. Samples were then spun at 15 000 g for 5 min at −10°C, then the aqueous/methanol layer was removed, lyophilized and stored at −80°C until HPLC.
Overnight cultures of Pto DC3000 strains resuspended in 10 mM MgCl2 were infiltrated with a blunt syringe into leaves of 4‐wk‐old Arabidopsis plants. Pto DC3000 derivatives were infiltrated at a density of 1 × 105 colony‐forming units (CFU) ml−1 while Pto DC3000D28E derivatives were infiltrated at 1 × 108 CFU ml−1. Plants were kept in covered trays at room temperature. Infiltrated leaves were removed at the indicated times, weighed, frozen in liquid nitrogen and ground with a plastic pestle. The metabolites were extracted following the above protocol for N. benthamiana.
ROS assay
Production of ROS was determined following a previously described protocol (Asai et al., 2008). A detailed methodology is described in Methods S1.
Callose deposition assay
A previously described method was used for callose deposition assays (Adam & Somerville, 1996). A detailed methodology is described in Methods S1.
In planta bacterial growth assay
Pathogenicity assays were carried out as previously described (Guo et al., 2016). A detailed methodology is described in Methods S1.
Results
Conserved residues within HopAM1’s putative TIR domain are required to induce cell death in yeast and plants
Two identical hopAM1 alleles, each encoding a 276 amino acid protein, are present in the Pto DC3000 genome. hopAM1‐1 is located in the chromosome while hopAM1‐2 is found on the endogenous plasmid pDC3000A (Buell et al., 2003). Homologs of HopAM1 are sporadically distributed in multiple phytopathogenic strains, predominantly in Pseudomonas species but also in Xanthomonas spp. and Ralstonia spp. (Fig. S1a). HopAM1 is highly similar (> 95% identity in peptide sequence) among Pseudomonas species but less similar among Xanthomonas spp. (50% identity and 68% similarity) or Ralstonia spp. (54% identity and 71% similarity) (Fig. S1a). The primary peptide sequence of HopAM1 does not share significant similarity with proteins with known functional domains based on an iterative Psi‐Blast (Position‐Specific Iterated Blast) analysis. We then conducted a search for proteins with structural similarity to HopAM1 using the Phyre2 algorithm for structural modeling (Kelley et al., 2015). A short stretch of HopAM1 peptide sequence (amino acids 165–214) was predicted to share secondary structure with known TIR domains, including those found in animal bacterial pathogens, animal TLR receptors and resistance proteins of plants associated with immunity (Fig. 1a,b) (Cirl & Miethke, 2010). Within this region of similarity, we observed multiple highly conserved amino acid residues in HopAM1 homologs from phytobacterial pathogens, including I168, F177, I178, F187, E191, L195, E197, F207 and F210 (Fig. S1b). All known enzymatically active TIR domains contain a consensus glutamic acid residue essential for NAD+ hydrolase catalytic activity (Essuman et al., 2017, 2018). Among the conserved residues, E191 was invariant among all aligned proteins, implicating it as the putative catalytic residue. Phylogenetically, the homologs present in phytopathogenic bacteria form a distinctive clade from others, suggesting the existence of a lineage of noncanonical TIR domains (Fig. S1c).
Fig. 1.
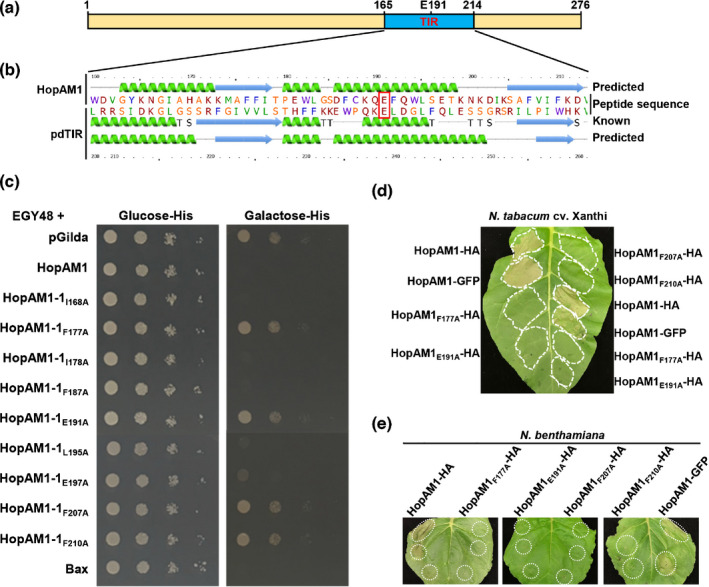
Conserved residues in HopAM1’s Toll/interleukin‐1 receptor (TIR) domain are required to trigger cell death in yeast and plants. (a) A schematic of the HopAM1 protein indicating its putative TIR domain shaded in blue and the putative catalytic glutamic acid 191 (E191). (b) Alignment of the Phyre2‐predicted secondary structure of HopAM1’s putative TIR domain with the known structure of pdTIR (PDB ID: 3H16) from Paracoccus denitrificans. The consensus putative catalytic glutamic acid residues are indicated in a red box. HopAM1 contains similar secondary structure to the pdTIR structure template with highest confidence in Phyre2 searches. (c) Yeast cell death assay with strains expressing HopAM1 wild‐type and mutants of conserved residues within the TIR domain. The conserved residues F177, E191, F207 and F210 were essential for HopAM1‐associated yeast cell death. Yeast EGY48 strains harboring plasmids containing hopAM1 and the mutants of conserved residues shown in Supporting Information Fig. S1(b), pGilda (vector) and Bax (positive control of cell death) were induced with galactose and assessed for survival. (d) HopAM1 triggers hypersensitive response‐like cell death in Nicotiana tabacum cv Xanthi and (e) Nicotiana benthamiana. In planta transient expression of HopAM1 and the indicated mutations except HopAM1‐GFP were estradiol‐inducible and mediated via Agrobacterium tumefaciens at an OD600 of 0.8. Images shown in (c–e) were taken 3 d after plating yeast cultures or inducing the expression with estradiol at 20 μM in plant leaves.
Previously, it was shown that overexpression of HopAM1 is lethal to yeast (Saccharomyces cerevisiae) (Jamir et al., 2004; Munkvold et al., 2008). To further examine HopAM1’s ability to induce lethality in yeast, site‐directed mutants were generated in the coding sequence of conserved residues within the putative TIR domain and cloned into pGilda. Expression of HopAM1 mutants was induced on galactose‐containing medium and all the HopAM1 mutants appeared to be stable (Fig. S1d). Strains expressing the mutants HopAM1F177A, HopAM1E191A, HopAM1F207A and HopAM1F210A no longer caused cell death (Fig. 1a), demonstrating that these residues are essential for the function of HopAM1’s putative TIR domain and cell death in yeast.
HopAM1 induces cell death in both N. tabacum and N. benthamiana (Choi et al., 2017). We next examined whether HopAM1‐associated cell death is also dependent on the same required conserved residues in yeast. HopAM1 and mutants HopAM1F177A, HopAM1E191A, HopAM1F207A and HopAM1F210A fused with either a hemagglutinin (HA) epitope tag or green fluorescence protein (GFP) were transiently expressed via Agrobacterium‐mediated transformation in N. tabacum cv Xanthi and N. benthamiana and assessed for their ability to induce cell death (Fig. S1e). Mutations of these essential TIR domain residues including the putative catalytic E191 abolished HopAM1‐induced cell death in both N. tabacum cv Xanthi and N. benthamiana. In line with cell death induction by HopAM1 in yeast, the results are consistent with the hypothesis that HopAM1’s putative TIR domain and its associated NAD+ hydrolase activity are required for HopAM1‐induced cell death (Fig. 1c–e).
HopAM1 is an active NAD+ hydrolyzing enzyme
TIR domains of prokaryotic and eukaryotic origins, including animal SARM1 and plant TIR‐containing NLRs, have been demonstrated to possess enzymatic activity that hydrolyzes NAD+ (Essuman et al., 2017, 2018; Horsefield et al., 2019; Wan et al., 2019). To determine whether HopAM1 hydrolyzes NAD+, we examined its NAD+ hydrolase activity in vitro. hopAM1 and hopAM1E191A were cloned into pET28a(+) and the recombinant His‐tagged proteins were purified. Purified HopAM1‐His and HopAM1E191A‐His proteins were incubated with 60 µM NAD+. The reaction products were analyzed with reversed‐phase HPLC. Only input NAD+ substrates were observed with all reactions at time 0 (Fig. S2a). Two peaks were detected in the products from the reaction with HopAM1‐His proteins with retention time of c. 6.5 and 6.8 min, respectively, while the substrate NAD+ peak was almost exhausted, indicative of enzymatic NAD+ hydrolase activity (Fig. 2a). By contrast, reactions with HopAM1E191A‐His and the vector control did not result in any detectable peaks corresponding to the products other than the input substrate NAD+ (Fig. 2a). The peak with a retention time at 6.5 min was distinct from NAD+, cADPR and Nam standards (Fig. 2a), suggesting it is a unique product.
Fig. 2.
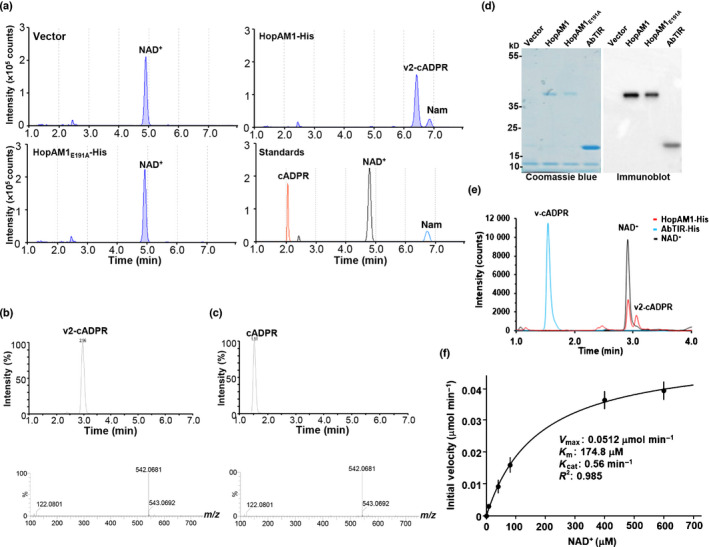
HopAM1 hydrolyzes nicotinamide adenine dinucleotide (NAD+) in vitro to produce a novel product. (a) In vitro assays with purified HopAM1‐His, HopAM1E191A‐His and mock preparation of empty vector control, and HPLC chromatographs with NAD+, cADPR and nicotinamide (Nam) standards. (b) Mass spectra of v2‐cADPR and (c) cADPR standard. v2‐cADPR and cADPR appeared to have the same mass but different retention time. (d) Coomassie staining and immunoblot of purified HopAM1, HopAM1E191A and AbTIR. (e) Overlaid HPLC chromatographs of NAD+ standard and in vitro products of HopAM1‐His and AbTIR‐His. (f) Enzymatic kinetics of HopAM1. Purified protein and substrate NAD+ at a series of concentration of 1–600 μM were incubated at room temperature for 60 min. Bars indicate standard error (SE). The reaction products were extracted, analyzed by HPLC and used to calculate the rate of initial NAD+ consumption. In (e), the unique product v2‐cADPR is present specifically in the HopAM1 partial hydrolysis at a retention time of c. 3.1 min while v‐cADPR is present at 1.6 min.
To identify this product, we performed LC‐MS time of flight (TOF) analysis on extracted metabolites from the in vitro reactions (Fig. S2b). The product was determined to have a mass of 542.0681 – identical to that of the cADPR standard. However, HopAM1’s product and the cADPR standard eluted at different times, indicating this product is a variant compound with different chemical properties from the canonical cADPR (Fig. 2b,c). The masses of Nam and remaining NAD+ from the in vitro reaction were also determined (Fig. S2c,d). Taken together, HPLC and LC‐MS analysis of the products of in vitro reactions confirmed HopAM1 is an active NAD+ hydrolyzing enzyme and the putative E191 residue within its TIR domain is required for catalytic activity.
A subset of prokaryotic TIR domains including AbTIR, a Tcp from the opportunistic bacterial pathogen Acinetobacter baumanni, generate a variant of cADPR hypothesized to be an alternative cyclization designated v‐cADPR (Essuman et al., 2018). To identify HopAM1’s unique in vitro product, we compared it with AbTIR’s known product via HPLC. NAD+ hydrolase reactions were conducted with purified HopAM1 and AbTIR (Fig. 2d). The HPLC chromatographs of their products revealed that HopAM1’s unique in vitro product eluted at a different time from v‐cADPR produced by AbTIR, indicating that HopAM1 hydrolyzes NAD+ into a product clearly distinct from v‐cADPR and designated and hereafter referred to as v2‐cADPR (Fig. 2e). HopAM1’s enzymatic properties were further determined with Michaelis–Menten kinetic assays and found to possess a Michaelis constant (K m) of 174.8 μM, a maximum velocity (V max) of 0.0512 μM min−1 and a catalyst rate constant (K cat) of 0.56 min−1 (Fig. 2f). HopAM1’s K m (174.8 μM) is comparable to that of TirS‐TIR (490 μM) and TcpC‐TIR (196 μM) but much higher than that of SARM1 (24 μM), which is close to the intracellular NAD+ concentrations of 200–2000 μM in plants (Essuman et al., 2017, 2018; Gakière et al., 2018). By contrast, HopAM1’s V max (0.0512 μM min−1) was low compared to that of TirS‐TIR (10 μM min−1), TcpC‐TIR (1.8 μM min−1) and SARM1 (3.57 μM min−1) (Essuman et al., 2017, 2018). Furthermore, HopAM1’s K cat (0.56 min−1) is much lower than that of SARM1 (10.3 min−1), suggesting that HopAM1 is a slow‐turnover enzyme, consistent with the time course enzymatic activity (Fig. S2e,f) (Essuman et al., 2017).
HopAM1 depletes NAD+ in yeast
To determine if HopAM1’s TIR domain is active in yeast, we extracted metabolites from yeast cultures induced to express HopAM1, HopAM1E191A or a vector control. A unique peak with a retention time of about 8 min was consistently seen in HPLC chromatograms from yeast cultures expressing HopAM1 but not HopAM1E191A or the vector control at 2 h postinduction (Fig. 3a). The appearance of this unique peak was concurrent with a marked depletion of NAD+ (Fig. 3b), while the other peaks were relatively unchanged among all samples, consistent with the product being v2‐cADPR directly derived from endogenous NAD+ in yeast cells. To rule out the possibility that depletion of NAD+ in yeast cultures was a byproduct of cell death, we compared the amount of NAD+ in yeast strains expressing HopAM1 or Bax (Lacomme & Santa Cruz, 1999), a well‐known inducer of yeast cell death, and HopA1 Psy 61, an effector from P. syringae pv. syringae 61 also known to be lethal to yeast. We found the amount of NAD+ in all strains except HopAM1 was similar (Fig. S3a). These data show HopAM1’s NAD+ hydrolase activity depletes NAD+ to induce yeast cell death. However, it is likely that the accumulation of v2‐cADPR is also responsible for such HopAM1‐induced cell death in combination with NAD+ depletion or independently.
Fig. 3.
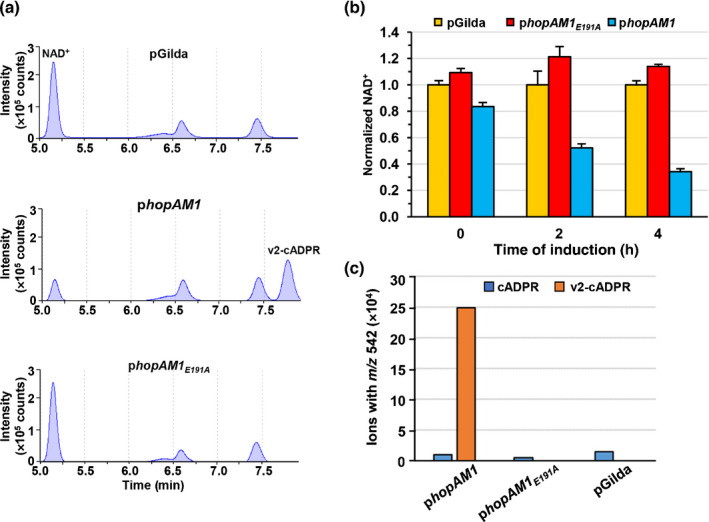
HopAM1 depletes nicotinamide adenine dinucleotide (NAD+) and produces v2‐cADPR in yeast. (a) A unique peak was detected with HPLC in metabolite extracts of yeast expressing HopAM1 but not HopAM1E191A or pGilda vector control. (b) Decrease of NAD+ during the induction of HopAM1. NAD+ levels were normalized to pGilda empty vector control at each time point. (c) Total ion counts of cADPR and the variant cADPR derived from MALDI‐TOF (matrix‐assisted laser desorption ionization time‐of‐flight) chromatograms of the samples in (a). EGY48 yeast carrying pGilda‐hopAM1, pGilda‐hopAM1E191A and empty vector were induced with galactose. Metabolite samples extracted from the respective yeast culture at time 0 and 3 h after induction were subjected to HPLC and MS/MS spectrum analysis. The error bars indicate SE.
The mass of the unique metabolites was determined with LC‐MS. Metabolites with the same mass at 542 m/z, the exact mass of cADPR, were detected at retention times of 3.8 and 4.5 min, respectively (Fig. S3b). The peaks detected at 3.8 min were invariant in all three strains, apparently representing the endogenous cADPR in yeast. The peak at 4.5 min was specific to HopAM1, consistent with v2‐cADPR derived from NAD+ detected in the in vitro assay (Figs 2, S3b). Surprisingly, the HopAM1‐specific metabolite was 56‐fold more abundant than endogenous cADPR (Figs 3c, S3b). These data indicate HopAM1 hydrolyzes NAD+ to produce and accumulate v2‐cADPR in yeast.
HopAM1 consumes NAD+ to produce v2‐cADPR in plants
To examine if HopAM1 is also active in plants, we transiently expressed HopAM1 and HopAM1E191A, both fused with an HA epitope tag and driven with estradiol inducible promoter, in N. benthamiana and subsequently extracted metabolites 48 h after estradiol induction. As in yeast, HPLC revealed that metabolic extracts from HopAM1‐expressing samples contained a unique peak with a retention time of the product v2‐cADPR concurrent with NAD+ depletion (Figs 2, 4a–c). In contrast, no metabolic changes were observed in samples expressing the TIR catalytic null mutant HopAM1E191A or the vector control (Fig. 4a). Overexpression of HopAM1 but not HopAM1E191A via estradiol induction further depleted endogenous NAD+ in the plants (Fig. 4b), validating that HopAM1 is active in consuming NAD+ in planta. Only estradiol‐induced HopAM1 overexpressions showed cell death, suggesting that that HopAM1‐associated cell death in N. benthamiana may be the result of either NAD+ depletion or the accumulation of v2‐cADPR or a combination of both via HopAM1’s endogenous activity (Figs 1d,e, 4b,c).
Fig. 4.
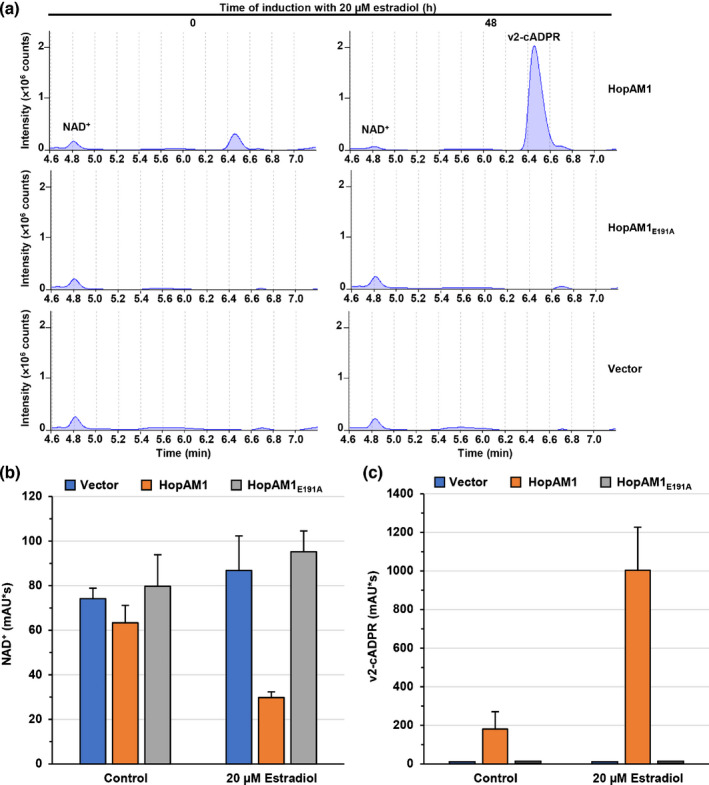
HopAM1‐dependent nicotinamide adenine dinucleotide (NAD+) depletion and production of variant v2‐cADPR in plants. (a) HPLC chromatograms of metabolite extracts from plants expressing HopAM1 and HopAM1E191A. A reduced NAD+ and a corresponding variant v2‐cADPR with peaks at retention times of 4.8 and 7 min in HPLC were present in samples expressing HopAM1 but not in vector control or HopAM1E191A. Metabolites from control plant leaves or those induced with 20 µM estradiol for 48 h were analyzed with HPLC. (b) Quantification of NAD+ and (c) variant v2‐cADPR production shown in (a). Nicotiana benthamiana leaves were infiltrated with Agrobacterium strains harboring the vector control, phopAM1 and phopAM1E191A and after 24 h were sprayed with 20 µM estradiol. Metabolites were extracted from induced plants and noninduced controls 48 h after estradiol application. The error bars indicate SE.
T3SS delivered HopAM1 is active in Arabidopsis plants
To determine whether HopAM1 activity is detectable in plant cells after type III secreted delivery by bacteria, Pto DC3000D28E was used to express HopAM1 and HopAM1E191A in trans via a broad host vector. Pto DC3000D28E is a polyeffector mutant of Pto DC3000 lacking 28 key effector genes, including both alleles of hopAM1, and is nearly nonpathogenic in Arabidopsis and N. benthamiana (Cunnac et al., 2011; Velásquez et al., 2017). HopAM1 triggers a hypersensitive‐like cell death response in resistant Arabidopsis accessions Xan‐2 and Xan‐5 (Velásquez et al., 2017). We then assessed metabolic changes over time in both Arabidopsis ecotypes Col‐0 and Xan‐2 infiltrated with Pto DC3000D28E(pML123‐hopAM1), Pto DC3000D28E(pML123‐hopAM1E191A ), Pto DC3000D28E(pML123), as well as the T3SS defective strain Pto DC3000D28E hrcU(pML123‐hopAM1). Expression of HA‐tagged HopAM1 and its mutants in DC3000D28E strains was detected with immunoblots and appeared to be stable (Fig. S6a). v2‐cADPR was detected within 24 h postinoculation with Pto DC3000D28E(pML123‐hopAM1) and the amount appeared to increase over the course of infection (Figs 5a,b, S4a). Nam was not distinguishable due to coelution with a large peak of another metabolite in common for all Arabidopsis samples. Neither Pto DC3000D28E(pML123‐hopAM1E191A ), the vector control, nor type III defective strain Pto DC3000D28E hrcU(pML123‐hopAM1) were able to produce v2‐cADPR in plants (Fig. 5a). Interestingly, despite the clear accumulation of v2‐cADPR, no significant changes were detected in the amount of NAD+ among plants infected with different strains, suggesting that DC3000 T3SS‐injected HopAM1 is sufficient to accumulate v2‐cADPR but insufficient to overcome NAD+ homeostasis (Figs 5a–c, S4a,b).
Fig. 5.
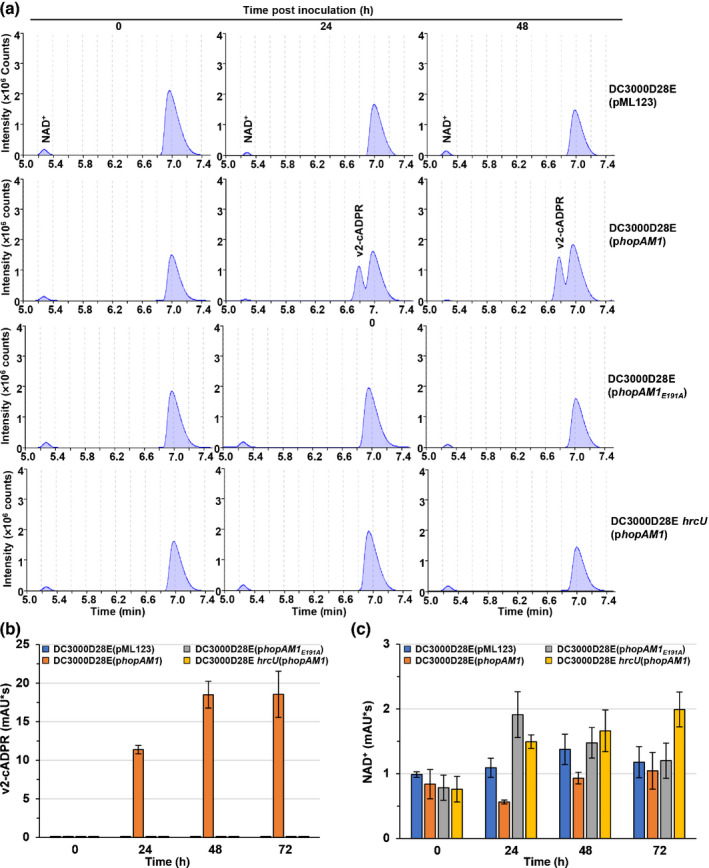
Type III secretion system (T3SS)‐delivered HopAM1 possesses in vivo activity in Arabidopsis. (a) HPLC analysis of metabolites in Arabidopsis Col‐0 leaves infected with DC3000D28E expressing HopAM1, HopAM1E191A, empty vector control or T3SS‐defective mutant DC3000D28E hrcU. (b) Quantification of v2‐cADPR in planta production and (c) nicotinamide adenine dinucleotide (NAD+) of the samples infected with the strains in (a). Values are means ± SE. The amount of v2‐cADPR is elevated by HopAM1 while NAD+ levels are not significantly altered in plants.
To rule out the possibility that other effectors in Pto DC3000 also possess the same NAD+ hydrolase activity, we analyzed metabolites in Arabidopsis plants infected with DC3000 mutants lacking one or both hopAM1‐1 and hopAM1‐2 alleles. The metabolic profile of Pto DC3000‐infected Arabidopsis Col‐0 was similar to that of Pto DC3000D28E(pML123‐hopAM1), whereas reduced amounts of v2‐cADPR were detected in leaves infected with the ΔhopAM1‐1 and ΔhopAM1‐2 single mutants (Figs 5a,b, S5a,b). No v2‐cADPR was detected in leaves infected with the ΔhopAM1‐1 ΔhopAM1‐2 (ΔhopAM1‐1/1‐2) double mutant lacking both hopAM1 alleles, but complementation with pML123‐hopAM1 was sufficient to restore v2‐cADPR production to wild‐type levels (Fig. S5a,b). Consistent with Pto DC3000D28E strains, the amount of NAD+ was not significantly altered during infection (Figs 5c, S5c). Together, these data show HopAM1 is the sole effector in Pto DC3000 and each allele encodes an active enzyme that displays NAD+ hydrolase activity in plants after delivery via the native Pto DC3000 T3SS.
HopAM1’s putative catalytic residue is required for ETI‐like responses in resistant Arabidopsis plants
To examine whether HopAM1’s putative catalytic residue is required for ETI‐like responses, Xan‐2 and Xan‐5 were inoculated with Pto DC3000D28E strains expressing HopAM1 or the catalytic null mutant HopAM1E191A, as well as T3SS‐defective strain DC3000D28E hrcU(pML123‐hopAM1). When infiltrated with Pto DC3000D28E(phopAM1) at 1 × 108 cells ml−1, leaves of both Xan‐2 and Xan‐5 displayed an ‘ETI‐like’ cell death hallmarked by severe chlorosis, consistent with the previous observation (Velásquez et al., 2017) (Figs 6a, S6b). By contrast, no chlorosis developed in leaves infiltrated with DC3000D28E(phopAM1E191A ), DC3000D28E hrcU(pML123‐hopAM1) or the vector control (Figs 6a, S6b). When the assay was repeated in susceptible Arabidopsis Col‐0 plants, a symptom‐like mild chlorosis or yellowing was observed in plants infiltrated with Pto DC3000D28E(phopAM1) often present with less severity than the chlorosis in Xan‐2 and Xan‐5, suggesting a virulent response in the susceptible Arabidopsis host (Figs 6a,b, S6b). Interestingly, ETI‐like responses were also abolished in Xan‐5 infiltrated with DC3000D28E(phopAM1F177A ) (Fig. S6b), suggesting that other conserved residues in the putative TIR domain may also affect HopAM1’s NAD+ hydrolase activity. It appears HopAM1’s activity has a prominent effect on symptom development. HopAM1’s putative catalytic residue is required for both ETI‐like responses in Xan‐2 and Xan‐5 plants and a symptom‐like chlorosis response in Col‐0, suggesting that there is a spectrum of genotype‐dependent responses to HopAM1’s NAD+ hydrolysis and v2‐cADPR production in planta (Figs 5, 6, S4, S5).
Fig. 6.
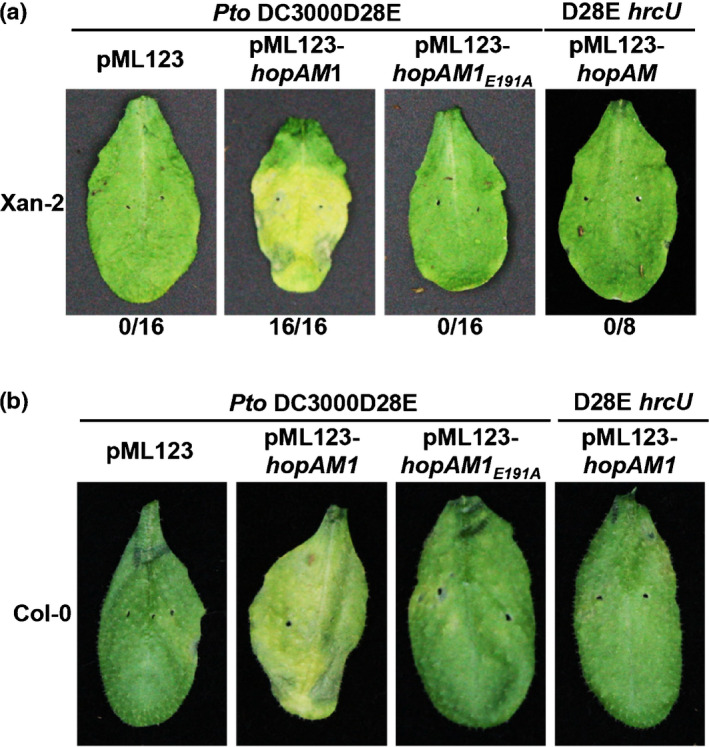
HopAM1‐induced responses in different Arabidopsis accessions. (a) Hypersensitive response‐like response in resistant Arabidopsis accession Xan‐2. (b) Virulence response in susceptible Arabidopsis Col‐0. Development of symptom‐like chlorosis was seen only in Col‐0 infiltrated with Pto DC3000D28E (pML123‐hopAM1), not vector control Pto DC3000D28E(pML123), Pto DC3000D28E(phopAM1E191A ) or Pto DC3000D28E hrcU(pML123‐hopAM1). Pto DC3000D28E or Pto DC3000D28E hrcU strains carrying the respective plasmids were infiltrated into Arabidopsis leaves at a cell density of 1 × 108 cells ml−1. Images were taken 4 d after inoculation.
Pto DC3000 HopAM1 virulence is dependent on its putative catalytic residue
HopAM1 suppresses both plant PTI and ETI (Jamir et al., 2004; Goel et al., 2008; Guo et al., 2009). To examine if HopAM1 PTI suppression is dependent on its TIR domain, we utilized the nonpathogenic Pseudomonas fluorescens (Pf)(pLN1965) strain, which expresses a functional T3SS and delivers heterologous type III effectors effectively for immunity suppression assays (Guo et al., 2009). Arabidopsis Col‐0 leaves were infiltrated with Pf(pLN1965) strains expressing HopAM1, putative catalytic mutant HopAM1E191A and a vector control and assayed for ROS production and callose deposition. Pf(pLN1965) expressing HopAM1 suppressed ROS production and callose deposition, while HopAM1E191A and the vector failed to suppress either response (Fig. 7a,b). HopAM1 was previously shown to suppress ETI in N. tabacum and Arabidopsis Ws‐0 induced by Pf(pHIR11), which encodes a functional T3SS and HopA1 Psy 61, an effector that triggers an ETI dependent on the TIR‐NLR RPS6 in Arabidopsis (Jamir et al., 2004; Kim et al., 2009). HopAM1 could suppress Pf(pHIR11)‐dependent ETI while HopAM1E191A and the vector control could not (Fig. 7c). These results clearly demonstrate that a putative catalytic site within the TIR domain is required for HopAM1’s suppression of plant immune responses.
Fig. 7.
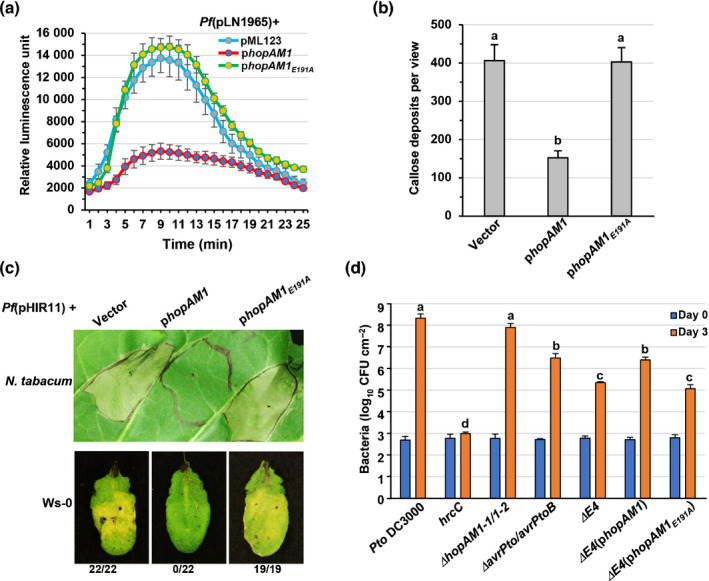
HopAM1 requires putative catalytic glutamic acid residue E191 to suppress pattern‐triggered immunity and effector‐triggered immunity and to contribute to virulence. (a) Reactive oxygen species (ROS) assays with Pf(pLN1965) strains carrying pML123 (Vector), pML123‐hopAM1 and pML123‐hopAM1E191A . Pf(pLN1965) strains were infiltrated into Arabidopsis Col‐0 leaves and ROS production was determined as relative luminescence. (b) Callose deposition assays of Pf(pLN1965) strains used in (a). (c) Hypersensitive response (HR) suppression assays in Nicotiana tabacum cv Xanthi (upper panel) and Arabidopsis Ws‐0 (lower panel) with Pf(pHIR11) strains carrying pML123 (vector), pML123‐hopAM1 or pML123‐hopAM1E191A . (d) In planta bacterial growth in Arabidopsis Col‐0. Pathogenicity assays were conducted with Pto DC3000, hrcC, a type III defective mutant, a double mutant ΔhopAM1‐1 ΔhopAM1‐2 (ΔhopAM1‐1/1‐2), a double mutant ΔavrPto ΔavrPtoB and a quadruple mutant ΔavrPto ΔavrPtoB ΔhopAM1‐1 ΔhopAM1‐2 (ΔE4), and the complementing strains expressing HopAM1 and HopAM1E191A. Pf(pHIR11) is a Pseudomonas fluorescens strain carrying a cosmid pHIR11 that encodes a full functional type III secretion system and effector HopA1 derived from Pseudomonas syringae pv. syringae 61, which enables delivery of HopA1 to trigger an HR in N. tabacum cv Xanthi and Arabidopsis Ws‐0 in a manner dependent on RPS6. Arabidopsis leaves were spray‐inoculated with the indicated strains. Bacterial growth in leaf tissue was quantified 0 and 3 d after inoculation. Bars denote SE. Letters denote statistical significance (P < 0.05). The bacterial growth assays with Col‐0 were repeated three times with similar results.
We further determined how HopAM1 contributes to Pto DC3000 virulence in Arabidopsis Col‐0 and tomato cv Money Maker plants. In assays with hopAM1 single mutants, neither ΔhopAM1‐1 nor ΔhopAM1‐2 was reduced in bacterial growth relative to the wild‐type, consistent with a previous report (Boch et al., 2002). The ΔhopAM1‐1/1‐2 double mutant displayed a slight but insignificant reduction in bacterial growth in Col‐0 and tomato plants (Figs 7d, S7). Due to redundancy of effectors in DC3000, single effector mutants often produce only subtle growth defects while polyeffector mutants more clearly demonstrate variations in virulence (Cunnac et al., 2011). To examine HopAM1’s contribution to Pto DC3000 virulence, HopAM1 was deleted in tandem with avrPto and avrPtoB, two well‐known key effectors in Pto DC3000. As expected, the ΔavrPto ΔavrPtoB double mutant displayed reduced growth in Col‐0 and tomato (Figs 7d, S7) (Lin & Martin, 2005). A quadruple effector mutant ΔhopAM1‐1 ΔhopAM1‐2 ΔavrPto ΔavrPtoB (ΔE4) was generated by further deletion of both hopAM1‐1 and hopAM1‐2 from the ΔavrPto ΔavrPtoB double mutant and assessed for its in planta bacterial growth. ΔE4 was further reduced relative to ΔavrPto ΔavrPtoB, and complementation with HopAM1 but not HopAM1E191A was able to restore the growth of ΔE4 to the level of ΔavrPto ΔavrPtoB (Figs 7d, S7). Bacterial growth assays indicate HopAM1 contributes to DC3000 virulence in plants in a manner dependent on the putative catalytic residue within its TIR domain concurrent with production of v2‐cADPR.
Discussion
We report that the Pto DC3000 effector HopAM1 contains a noncanonical TIR domain and possesses enzymatic activity that hydrolyzes NAD+ in vitro and produces nicotinamide and the novel metabolite v2‐cADPR, likely to be a unique cADPR isomer distinct from cADPR and v‐cADPR (Fig. 2). The existence of two cADPR variants suggests a family of alternative cyclic linkages for cADPR. The in vivo NAD+ hydrolase activity and production of v2‐cADPR is evidence that overexpression of HopAM1 in yeast and plants results in a drastic decrease of endogenous NAD+ (Figs 3, 4). Moreover, HopAM1’s enzymatic activity is detectable in planta after native T3SS delivery and is required for HopAM1’s suppression of plant immunity and contribution to Pto virulence (Figs 5, 6, 7). Although the majority of P. syringae type III effectors appear to suppress plant immunity and collectively contribute to virulence, the biochemical activity of most effectors is still elusive (Guo et al., 2009; Block & Alfano, 2011; Büttner, 2016). Most enzymatically active effectors of plant pathogens biochemically modify immunity‐associated host proteins to alter or suppress immunity and promote virulence (Block & Alfano, 2011). Only a few effectors have been demonstrated to interfere with plant metabolism or directly target essential metabolites of host plants (Poueymiro et al., 2014; Shidore et al., 2017). The study highlights a clearly novel biochemical activity for a phytopathogenic effector that hydrolyzes the essential metabolite NAD+ to generate a novel metabolite that probably increases virulence in host plants.
In plants, the NAD+ hydrolase activity of TIR domain‐containing NLRs during activation of immunity was recently revealed to be essential to the perception of cognate effectors and ETI (Horsefield et al., 2019; Wan et al., 2019; Ma et al., 2020; Martin et al., 2020). Though both HopAM1 and a subset of plant NLRs contain TIR domains, HopAM1 is distinguished by several key differences. While plant NLR NAD+ hydrolase activity activates immunity, HopAM1’s NAD+ hydrolase activity suppresses it. HopAM1 is active in different subcellular compartments than plant NLRs: HopAM1 localizes to the cytoplasm and probably acts in the cytosol (Fig. S8), whereas plant NLRs are plasma membrane‐bound (Dangl et al., 2013; Choi et al., 2017). More importantly, despite the same substrate NAD+, HopAM1’s product is different. HopAM1‐induced cell death is not EDS1‐dependent, suggesting it does not induce typical ETI‐associated cell death (Choi et al., 2017). It is most likely that TIR domain effectors such as HopAM1 evolved specific biochemical activity to generate a novel metabolite to subvert host immunity and promote virulence.
Toll/interleukin‐1 receptor domain‐containing proteins belong to a newly emerging class of virulence factors that are widely distributed among various animal bacterial pathogens, though they are also commonly seen in nonpathogenic organisms (Spear et al., 2009; Cirl & Miethke, 2010). Despite controversy, much attention has been given to the theory that bacteria have adapted TIR domain proteins to interfere with the host immune system via protein–protein interactions and molecular mimicry to promote virulence (Spear et al., 2009). However, recent findings that both eukaryotic and bacterial TIR domains belong to a large ancient family of NAD+ hydrolases have led to a better understanding of TIR domain‐containing virulence and effector proteins (Essuman et al., 2018; Essuman, 2020). Recently, the TIR domain‐containing type IV effectors BtpA and BtpB from B. abortus were found to interfere with NAD+ metabolism in human cells, suggesting that TIR domain enzymatic activity is important for virulence in animals (Coronas‐Serna et al., 2020). HopAM1 is an unprecedented phytopathogenic effector injected by the Pto T3SS that exerts NAD+ hydrolase activity in planta to promote virulence, reinforcing TIR domain‐associated NAD+ hydrolase activity as a component of pathogen virulence in diverse eukaryotic hosts.
ABA is a signaling target of plant bacterial pathogens. Elevated levels of ABA or increased sensitivity to ABA appear to be correlated with P. syringae virulence and enhancement of bacterial growth (Cao et al., 2011). Pseudomonas syringae type III effectors manipulate the ABA pathway or undermine the ABA network (de Torres‐Zabala et al., 2007; García‐Andrade et al., 2020). AvrPtoB promotes ABA accumulation for P. syringae virulence. Unlike AvrPtoB, HopAM1 and HopF2 act similarly to enhance ABA sensitivity and suppress immune responses for P. syringae virulence, but the underlying biochemical mechanism is not known (de Torres‐Zabala et al., 2007; Goel et al., 2008; Cao et al., 2019). However, recent studies have revealed an unexpected link between NAD+ and the ABA response pathway with NAD+ regulation probably upstream of ABA signaling (Feitosa‐Araujo et al., 2020; Hong et al., 2020). While endogenous NAD+ levels in plant leaves are depleted as a consequence of HopAM1 overexpression via Agrobacterium‐mediated transformation, NAD+ homeostasis is not changed significantly by T3SS‐delivered HopAM1. By contrast, HopAM1 mediates substantial accumulation of v2‐cADPR following both heterologous overexpression and native delivery. Thus, it is possible that previously observed effects of HopAM1 on ABA responses are indirect (Lee, 1994; Goel et al., 2008). Alternatively, given that cADPR acts as second messenger of ABA and v2‐cADPR is an isomer of cADPR, it is plausible that HopAM1‐mediated v2‐cADPR affects ABA responses by interfering with cADPR signaling (Wu et al., 1997).
HopAM1 induces HR‐like cell death in some plants while producing mild chlorosis in others. HopAM1 was originally identified in P. syringae pv. pisi as a plasmid‐borne avirulence gene avrPpiB based on the HR response in pea cultivars with specific R3 resistance locus (Cournoyer et al., 1995). HopAM1 induces cell death in Nicotiana species, but it is not clear if this recognition is specific to an R protein. HopAM1‐associated cell death is dependent on HopAM1’s catalytic activity and is observed both in experiments showing NAD+ depletion and those without significant NAD+ depletion. In mammalian neurons, SARM1‐mediated hydrolysis of NAD+ is the mechanism of execution of axon degeneration (Gerdts et al., 2016; Essuman et al., 2017). Activation of SARM1 results in NAD+ depletion and marked production of cADPR, but modulation of cADPR has no effect on axon degeneration (Sasaki et al., 2020). This model suggests that depletion of endogenous NAD+ may be one cause of HopAM1‐induced cell death seen in plant and yeast over‐expressions. Another cause of HopAM1‐induced cell death may be accumulation of v2‐cADPR. It is not clear whether HopAM1‐dependent cell death or severe chlorosis seen in Arabidopsis genotypes Xan‐2 and Xan‐5 is the same as cell death observed in genotype Bur‐0 and lesions seen in N. benthamiana (Cunnac et al., 2011; Iakovidis et al., 2016; Velásquez et al., 2017). Resistance of Xan‐2 and Xan‐5 to hopAM1‐carrying strains is atypical: instead of qualitative NLR (R)‐mediated resistance, two major quantitative trait loci are associated with HopAM1‐induced cell death and chlorosis (Iakovidis et al., 2016). It is possible that Xan‐2 and Xan‐5 do not exhibit canonical resistance but rather quantitative traits conferring sensitivity to HopAM1‐produced v2‐cADPR. HopAM1‐associated chlorosis in Col‐0 is less severe and resembles that seen in Ws‐0 upon bacterial expression of HopAM1 in trans or transgenic expression in planta (Goel et al., 2008). Unlike Xan‐2 and Xan‐5, both Col‐0 and Ws‐0 are susceptible to Pto DC3000, which suggests chlorosis results from virulence activity rather than an immune response (Goel et al., 2008). We confirmed that HopAM1‐associated chlorosis is reliant on HopAM1’s activity (Fig. 6), perhaps as a direct consequence of accumulation of the product v2‐cADPR. Interestingly, hopAM1 is absent in group II P. syringae strains that contain toxin (syringomycin, syringopeptin and syringolin) biosynthesis pathways (Baltrus et al., 2011). Nontoxin‐producing P. syringae strains may have acquired effector genes such as hopAM1 to take advantage of existing metabolites in hosts to produce de novo small molecules for symptom development and virulence during infection. The small molecule effector victorin of the necrotrophic fungus Cochliobolus victoriae behaves differently in the presence or absence of an NLR to induce host susceptibility (Lorang et al., 2012). Similarly, it is plausible that differences in sensitivity to v2‐cADPR lead to differential responses in the context of genotypes of different susceptibility. The identification of more effector‐mediated small molecules and determination of their role in pathogenesis deserve further investigation.
When delivered via DC3000, HopAM1 does not measurably deplete NAD+ but still results in v2‐cADPR accumulation. This suggests that, although NAD+ is an essential plant metabolite and coenzyme with involvement in immunity, HopAM1’s contribution to virulence may not be entirely due to depletion of NAD+ but instead accumulation of v2‐cADPR (Fig. 5). The chemical structures of v‐cADPR and v2‐cADPR remain to be resolved. The specific function of v2‐cADPR and its relationship to cADPR and v‐cADPR are still unknown in the context of either eukaryotic NLR‐associated immune activity or prokaryotic HopAM1‐associated virulence activity. However, the apparent similarity of v2‐cADPR to cADPR and v‐cADPR suggests that this novel isoform may interfere with cADPR and v‐cADPR signaling downstream of plant NLRs in order to suppress immunity.
Author contributions
MG conceived experiments. SE carried out most experiments and TS conducted HPLC analysis. MAZ and ND conducted kinetics experiments of HopAM1 and AbTIR. PK performed immunoblots to detect expression in yeast. SM conducted immunoblots to detect expression in bacterial strains. AD and JM provided key materials and insights for NAD+ hydrolase experiments. JRA provided intellectual insights. TEC oversaw the experiments. SE and MG contributed to manuscript writing.
Supporting information
Fig. S1 HopAM1 contains a putative Toll/interleukin‐1 receptor domain.
Fig. S2 HopAM1 hydrolyzes nicotinamide adenine dinucleotide in vitro.
Fig. S3 HopAM1’s nicotinamide adenine dinucleotide (NAD+) hydrolysis activity is associated with NAD+ depletion in yeast.
Fig. S4 HopAM1‐mediated metabolites in Arabidopsis Xan‐2.
Fig. S5 HopAM1 in Pto DC3000 is responsible for production of v2‐cADPR in Arabidopsis.
Fig. S6 HopAM1’s effector‐triggered immunity‐like response in Arabidopsis Xan‐5 is dependent on its conserved residues in the Toll/interleukin‐1 receptor domain.
Fig. S7 HopAM1 contributes to Pto DC3000 virulence in tomato plants in a manner dependent on its putative catalytic residue.
Fig. S8 Mutation of conserved residues in HopAM1’s Toll/interleukin‐1 receptor domain do not affect its subcellular localization.
Methods S1 Supporting Information for the Materials and Methods.
Table S1 Strains and plasmids used in this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This project was partially supported through funds provided by the Nebraska Soybean Board project no. 1734, and the Center for Plant Science Innovation, University of Nebraska‐Lincoln. We are indebted to Drs Rich Wilson, Lirong Zeng and Guangyong Li for their critical reviews of the manuscript. We thank Dr Kurt Wulser from UNL’s department of chemistry for performing LC‐MS analysis. We thank the UNL microscopy core facility and the Center for Biotechnology for the use of their facilities. We thank Drs Sheng Yang He and André C. Velásquez for sharing Arabidopsis accession Xan‐2 and Xan‐5 seeds.
Data availability
All vectors and strains are available by request to Ming Guo at mguo2@unl.edu.
References
- Adam L, Somerville SC. 1996. Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana . The Plant Journal 9: 341–356. [DOI] [PubMed] [Google Scholar]
- Alaidarous M, Ve T, Casey LW, Valkov E, Ericsson DJ, Ullah MO, Schembri MA, Mansell A, Sweet MJ, Kobe B. 2014. Mechanism of bacterial interference with TLR4 signaling by Brucella Toll/interleukin‐1 receptor domain‐containing protein TcpB. Journal of Biological Chemistry 289: 654–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai S, Ohta K, Yoshioka H. 2008. MAPK signaling regulates nitric oxide and NADPH oxidase‐dependent oxidative bursts in Nicotiana benthamiana . Plant Cell 20: 1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin JAE, Lin J, Perez‐Quintero AL, Gentzel I, Majerczak D, Opiyo SO, Zhao W, Paek S‐M, Kim MG, Coplin DL. 2015. Perturbation of maize phenylpropanoid metabolism by an AvrE family type III effector from Pantoea stewartii . Plant Physiology 167: 1117–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. 2011. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathogens 7: e1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless AM, Nishimura MT. 2020. Enzymatic functions for Toll/interleukin‐1 receptor domain proteins in the plant immune system. Frontiers in Genetics 11: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A, Alfano JR. 2011. Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Current Opinion in Microbiology 14: 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Joardar V, Gao L, Robertson TL, Lim M, Kunkel BN. 2002. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana . Molecular Microbiology 44: 73–88. [DOI] [PubMed] [Google Scholar]
- Bowie A, Kiss‐Toth E, Symons JA, Smith GL, Dower SK, O'Neill LA. 2000. A46R and A52R from vaccinia virus are antagonists of host IL‐1 and toll‐like receptor signaling. Proceedings of the National Academy of Sciences, USA 97: 10162–10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proceedings of the National Academy of Sciences, USA 100: 10181–10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner D. 2012. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant‐ and animal‐pathogenic bacteria. Microbiology and Molecular Biology Reviews 76: 262–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner D. 2016. Behind the lines–actions of bacterial type III effector proteins in plant cells. FEMS Microbiology Reviews 40: 894–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao FY, Khan M, Taniguchi M, Mirmiran A, Moeder W, Lumba S, Yoshioka K, Desveaux D. 2019. A host–pathogen interactome uncovers phytopathogenic strategies to manipulate plant ABA responses. The Plant Journal 100: 187–198. [DOI] [PubMed] [Google Scholar]
- Cao FY, Yoshioka K, Desveaux D. 2011. The roles of ABA in plant–pathogen interactions. Journal of Plant Research 124: 489–499. [DOI] [PubMed] [Google Scholar]
- Carty M, Bowie AG. 2019. SARM: from immune regulator to cell executioner. Biochemical Pharmacology 161: 52–62. [DOI] [PubMed] [Google Scholar]
- Choi S, Jayaraman J, Segonzac C, Park H‐J, Park H, Han S‐W, Sohn KH. 2017. Pseudomonas syringae pv. actinidiae type III effectors localized at multiple cellular compartments activate or suppress innate immune responses in Nicotiana benthamiana . Frontiers in Plant Science 8: 2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirl C, Miethke T. 2010. Microbial Toll/interleukin 1 receptor proteins: a new class of virulence factors. International Journal of Medical Microbiology 300: 396–401. [DOI] [PubMed] [Google Scholar]
- Coronas‐Serna JM, Louche A, Rodríguez‐Escudero M, Roussin M, Imbert PRC, Rodríguez‐Escudero I, Terradot L, Molina M, Gorvel J‐P, Cid VJ et al. 2020. The TIR‐domain containing effectors BtpA and BtpB from Brucella abortus impact NAD metabolism. PLoS Pathogens 16: e1007979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cournoyer B, Sharp JD, Astuto A, Gibbon MJ, Taylor JD, Vivian A. 1995. Molecular characterization of the Pseudomonas syringae pv. pisi plasmid‐borne avirulence gene avrPpiB which matches the R3 resistance locus in pea. Molecular Plant–Microbe Interactions 8: 700–708. [DOI] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A. 2011. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae . Proceedings of the National Academy of Sciences, USA 108: 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ. 2013. Pivoting the plant immune system from dissection to deployment. Science 341: 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres‐Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bogre L, Grant M. 2007. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO Journal 26: 1434–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxbury Z, Wang S, MacKenzie CI, Tenthorey JL, Zhang X, Huh SU, Hu L, Hill L, Ngou PM, Ding P et al. 2020. Induced proximity of a TIR signaling domain on a plant‐mammalian NLR chimera activates defense in plants. Proceedings of the National Academy of Sciences, USA 117: 18832–18839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essuman K, Summers DW, Sasaki Y, Mao X, DiAntonio A, Milbrandt J. 2017. The SARM1 Toll/interleukin‐1 receptor domain possesses intrinsic NAD(+) cleavage activity that promotes pathological axonal degeneration. Neuron 93: 1334–1343 e1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essuman K, Summers DW, Sasaki Y, Mao X, Yim AKY, DiAntonio A, Milbrandt J. 2018. TIR domain proteins are an ancient family of NAD(+)‐consuming enzymes. Current Biology 28: 421–430 e424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essuman KA. 2020. The enzymatic function of the TIR domain: from axon degeneration to innate immunity. PhD thesis, Washington University, St Louis, MO, USA. [Google Scholar]
- Feitosa‐Araujo E, Fonseca‐Pereira P, Pena MM, Medeiros DB, Perez de Souza L, Yoshida T, Weber APM, Araújo WL, Fernie AR, Schwarzländer M et al. 2020. Changes in intracellular NAD status affect stomatal development in an abscisic acid‐dependent manner. The Plant Journal 104: 1149–1168. [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. 2007. A type III effector ADP‐ribosylates RNA‐binding proteins and quells plant immunity. Nature 447: 284–289. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Kawazoe T, Ohnishi K, Kitagawa T, Popa C, Valls M, Genin S, Nakamura K, Kuramitsu Y, Tanaka N et al. 2016. RipAY, a plant pathogen effector protein, exhibits robust γ‐glutamyl cyclotransferase activity when stimulated by eukaryotic thioredoxins. Journal of Biological Chemistry 291: 6813–6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakière B, Hao J, de Bont L, Pétriacq P, Nunes‐Nesi A, Fernie AR. 2018. NAD+ biosynthesis and signaling in plants. Critical Reviews in Plant Sciences 37: 259–307. [Google Scholar]
- García‐Andrade J, González B, Gonzalez‐Guzman M, Rodriguez PL, Vera P. 2020. The role of ABA in plant immunity is mediated through the PYR1 receptor. International Journal of Molecular Sciences 21: 5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdts J, Summers Daniel W, Milbrandt J, DiAntonio A. 2016. Axon self‐destruction: new links among SARM1, MAPKs, and NAD+ metabolism. Neuron 89: 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel AK, Lundberg D, Torres MA, Matthews R, Akimoto‐Tomiyama C, Farmer L, Dangl JL, Grant SR. 2008. The Pseudomonas syringae type III effector HopAM1 enhances virulence on water‐stressed plants. Molecular Plant–Microbe Interactions 21: 361–370. [DOI] [PubMed] [Google Scholar]
- Guo M, Kim P, Li G, Elowsky CG, Alfano JR. 2016. A bacterial effector co‐opts calmodulin to target the plant microtubule network. Cell Host & Microbe 19: 67–78. [DOI] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, Alfano JR. 2009. The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Molecular Plant–Microbe Interactions 22: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Wang Z, Shi H, Yao J, Liu X, Wang F, Zeng L, Xie Z, Zhu J‐K. 2020. Reciprocal regulation between nicotinamide adenine dinucleotide metabolism and abscisic acid and stress response pathways in Arabidopsis. PLoS Genetics 16: e1008892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsefield S, Burdett H, Zhang X, Manik MK, Shi Y, Chen J, Qi T, Gilley J, Lai J‐S, Rank MX. 2019. NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365: 793–799. [DOI] [PubMed] [Google Scholar]
- House BL, Mortimer MW, Kahn ML. 2004. New recombination methods for Sinorhizobium meliloti genetics. Applied and Environmental Microbiology 70: 2806–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Lerner F, Ziegler M. 2004. NAD – new roles in signalling and gene regulation in plants. New Phytologist 163: 31–44. [DOI] [PubMed] [Google Scholar]
- Iakovidis M, Teixeira PJ, Exposito‐Alonso M, Cowper MG, Law TF, Liu Q, Vu MC, Dang TM, Corwin JA, Weigel D. 2016. Effector‐triggered immune response in Arabidopsis thaliana is a quantitative trait. Genetics 204: 337–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert PR, Louche A, Luizet JB, Grandjean T, Bigot S, Wood TE, Gagné S, Blanco A, Wunderley L, Terradot L. 2017. A Pseudomonas aeruginosa TIR effector mediates immune evasion by targeting UBAP 1 and TLR adaptors. EMBO Journal 36: 1869–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamir Y, Guo M, Oh H‐S, Petnicki‐Ocwieja T, Chen S, Tang X, Dickman MB, Collmer A, Alfano JR. 2004. Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. The Plant Journal 37: 554–565. [DOI] [PubMed] [Google Scholar]
- Jeong B‐R, Lin Y, Joe A, Guo M, Korneli C, Yang H, Wang P, Yu M, Cerny RL, Staiger D et al. 2011. Structure function analysis of an ADP‐ribosyltransferase type III effector and its RNA‐binding target in plant immunity. Journal of Biological Chemistry 286: 43272–43281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kwon SI, Saha D, Anyanwu NC, Gassmann W. 2009. Resistance to the Pseudomonas syringae effector HopA1 is governed by the TIR‐NBS‐LRR protein RPS6 and is enhanced by mutations in SRFR1. Plant Physiology 150: 1723–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme C, Santa Cruz S. 1999. Bax‐induced cell death in tobacco is similar to the hypersensitive response. Proceedings of the National Academy of Sciences, USA 96: 7956–7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC. 1994. Cyclic ADP‐ribose: a new member of a super family of signalling cyclic nucleotides. Cellular Signalling 6: 591–600. [DOI] [PubMed] [Google Scholar]
- Lin N‐C, Martin GB. 2005. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto‐mediated resistance and is less virulent on tomato. Molecular Plant–Microbe Interactions 18: 43–51. [DOI] [PubMed] [Google Scholar]
- Lorang J, Kidarsa T, Bradford C, Gilbert B, Curtis M, Tzeng S‐C, Maier C, Wolpert T. 2012. Tricking the guard: exploiting plant defense for disease susceptibility. Science 338: 659–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Lapin D, Liu LI, Sun Y, Song W, Zhang X, Logemann E, Yu D, Wang J, Jirschitzka J et al. 2020. Direct pathogen‐induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370: eabe3069. [DOI] [PubMed] [Google Scholar]
- Macho AP. 2016. Subversion of plant cellular functions by bacterial type‐III effectors: beyond suppression of immunity. New Phytologist 210: 51–57. [DOI] [PubMed] [Google Scholar]
- Macho AP, Zipfel C. 2015. Targeting of plant pattern recognition receptor‐triggered immunity by bacterial type‐III secretion system effectors. Current Opinion in Microbiology 23: 14–22. [DOI] [PubMed] [Google Scholar]
- Martin R, Qi T, Zhang H, Liu F, King M, Toth C, Nogales E, Staskawicz BJ. 2020. Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science 370: eabd9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold KR, Martin ME, Bronstein PA, Collmer A. 2008. A survey of the Pseudomonas syringae pv. tomato DC3000 type III secretion system effector repertoire reveals several effectors that are deleterious when expressed in Saccharomyces cerevisiae . Molecular Plant–Microbe Interactions 21: 490–502. [DOI] [PubMed] [Google Scholar]
- Newman RM, Salunkhe P, Godzik A, Reed JC. 2006. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin‐1 receptor family proteins. Infection and Immunity 74: 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G. 2006. Metabolic signalling in defence and stress: the central roles of soluble redox couples. Plant, Cell & Environment 29: 409–425. [DOI] [PubMed] [Google Scholar]
- Ofir G, Herbst E, Baroz M, Cohen D, Millman A, Doron S, Tal N, Malheiro DBA, Malitsky S, Amitai G et al. 2021. Antiviral activity of bacterial TIR domains via signaling molecules that trigger cell death. bioRxiv. doi: 10.1101/2021.01.06.425286. [DOI] [PubMed] [Google Scholar]
- O'Neill LA, Golenbock D, Bowie AG. 2013. The history of Toll‐like receptors – redefining innate immunity. Nature Reviews Immunology 13: 453–460. [DOI] [PubMed] [Google Scholar]
- Patterson NJ, Gunther J, Gibson AJ, Offord V, Coffey TJ, Splitter G, Monk I, Seyfert HM, Werling D. 2014. Two TIR‐like domain containing proteins in a newly emerging zoonotic Staphylococcus aureus strain sequence type 398 are potential virulence factors by impacting on the host innate immune response. Frontiers in Microbiology 5: 662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pétriacq P, de Bont L, Hager J, Didierlaurent L, Mauve C, Guérard F, Noctor G, Pelletier S, Renou J‐P, Tcherkez G et al. 2012. Inducible NAD overproduction in Arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst‐AvrRpm1. The Plant Journal 70: 650–665. [DOI] [PubMed] [Google Scholar]
- Pétriacq P, de Bont L, Tcherkez G, Gakière B. 2013. NAD: not just a pawn on the board of plant‐pathogen interactions. Plant Signaling & Behavior 8: e22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pétriacq P, Ton J, Patrit O, Tcherkez G, Gakière B. 2016. NAD acts as an integral regulator of multiple defense layers. Plant Physiology 172: 1465–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro M, Cazalé A‐C, François J‐M, Parrou J‐L, Peeters N, Genin S. 2014. A Ralstonia solanacearum type III effector directs the production of the plant signal metabolite trehalose‐6‐phosphate. mBio 5: e02065‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosadini CV, Zanoni I, Odendall C, Green Erin R, Paczosa Michelle K, Philip Naomi H, Brodsky Igor E, Mecsas J, Kagan JC. 2015. A single bacterial immune evasion strategy dismantles both MyD88 and TRIF signaling pathways downstream of TLR4. Cell Host & Microbe 18: 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryrie I, Scott K. 1968. Metabolic regulation in diseased leaves II. Changes in nicotinamide nucleotide coenzymes in barley leaves infected with powdery mildew. Plant Physiology 43: 687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Engber TM, Hughes RO, Figley MD, Wu T, Bosanac T, Devraj R, Milbrandt J, Krauss R, DiAntonio A. 2020. cADPR is a gene dosage‐sensitive biomarker of SARM1 activity in healthy, compromised, and degenerating axons. Experimental Neurology 329: 113252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shidore T, Broeckling CD, Kirkwood JS, Long JJ, Miao J, Zhao B, Leach JE, Triplett LR. 2017. The effector AvrRxo1 phosphorylates NAD in planta . PLoS Pathogens 13: e1006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear AM, Loman NJ, Atkins HS, Pallen MJ. 2009. Microbial TIR domains: not necessarily agents of subversion? Trends in Microbiology 17: 393–398. [DOI] [PubMed] [Google Scholar]
- Toruno TY, Stergiopoulos I, Coaker G. 2016. Plant‐pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annual Review of Phytopathology 54: 419–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velásquez AC, Oney M, Huot B, Xu S, He SY. 2017. Diverse mechanisms of resistance to Pseudomonas syringae in a thousand natural accessions of Arabidopsis thaliana. New Phytologist 214: 1673–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Essuman K, Anderson RG, Sasaki Y, Monteiro F, Chung E‐H, Nishimura EO, DiAntonio A, Milbrandt J, Dangl JL. 2019. TIR domains of plant immune receptors are NAD+‐cleaving enzymes that promote cell death. Science 365: 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhou M, Zhang X, Yao J, Zhang Y, Mou Z. 2017. A lectin receptor kinase as a potential sensor for extracellular nicotinamide adenine dinucleotide in Arabidopsis thaliana . eLife 6: e25474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li J, Hou S, Wang X, Li Y, Ren D, Chen S, Tang X, Zhou J‐M. 2010. A Pseudomonas syringae ADP‐ribosyltransferase inhibits Arabidopsis mitogen‐activated protein kinase kinases. Plant Cell 22: 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H‐L, Chakravarthy S, Mathieu J, Helmann TC, Stodghill P, Swingle B, Martin GB, Collmer A. 2015. Pseudomonas syringae pv. tomato DC3000 type III secretion effector polymutants reveal an interplay between HopAD1 and AvrPtoB. Cell Host & Microbe 17: 752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H‐L, Collmer A. 2018. Defining essential processes in plant pathogenesis with Pseudomonas syringae pv. tomato DC3000 disarmed polymutants and a subset of key type III effectors. Molecular Plant Pathology 19: 1779–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C‐H, Abd‐El‐Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, Kamoun S. 2017. NLR network mediates immunity to diverse plant pathogens. Proceedings of the National Academy of Sciences, USA 114: 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Kuzma J, Maréchal E, Graeff R, Lee HC, Foster R, Chua N‐H. 1997. Abscisic acid signaling through cyclic ADP‐ribose in plants. Science 278: 2126–2130. [DOI] [PubMed] [Google Scholar]
- Xin X‐F, He SY. 2013. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annual Review of Phytopathology 51: 473–498. [DOI] [PubMed] [Google Scholar]
- Zhou H, Lin J, Johnson A, Morgan RL, Zhong W, Ma W. 2011. Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host & Microbe 9: 177–186. [DOI] [PubMed] [Google Scholar]
- Zipfel C. 2014. Plant pattern‐recognition receptors. Trends in Immunology 35: 345–351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 HopAM1 contains a putative Toll/interleukin‐1 receptor domain.
Fig. S2 HopAM1 hydrolyzes nicotinamide adenine dinucleotide in vitro.
Fig. S3 HopAM1’s nicotinamide adenine dinucleotide (NAD+) hydrolysis activity is associated with NAD+ depletion in yeast.
Fig. S4 HopAM1‐mediated metabolites in Arabidopsis Xan‐2.
Fig. S5 HopAM1 in Pto DC3000 is responsible for production of v2‐cADPR in Arabidopsis.
Fig. S6 HopAM1’s effector‐triggered immunity‐like response in Arabidopsis Xan‐5 is dependent on its conserved residues in the Toll/interleukin‐1 receptor domain.
Fig. S7 HopAM1 contributes to Pto DC3000 virulence in tomato plants in a manner dependent on its putative catalytic residue.
Fig. S8 Mutation of conserved residues in HopAM1’s Toll/interleukin‐1 receptor domain do not affect its subcellular localization.
Methods S1 Supporting Information for the Materials and Methods.
Table S1 Strains and plasmids used in this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
All vectors and strains are available by request to Ming Guo at mguo2@unl.edu.


