Abstract
Inherited information is transmitted to progeny primarily by the genome through the gametes. However, in recent years, epigenetic inheritance has been demonstrated in several organisms, including animals. Although it is clear that certain post‐translational histone modifications, DNA methylation, and noncoding RNAs regulate epigenetic inheritance, the molecular mechanisms responsible for epigenetic inheritance are incompletely understood. This review focuses on the role of small RNAs in transmitting epigenetic information across generations in animals. Examples of documented cases of transgenerational epigenetic inheritance are discussed, from the silencing of transgenes to the inheritance of complex traits, such as fertility, stress responses, infections, and behavior. Experimental evidence supporting the idea that small RNAs are epigenetic molecules capable of transmitting traits across generations is highlighted, focusing on the mechanisms by which small RNAs achieve such a function. Just as the role of small RNAs in epigenetic processes is redefining the concept of inheritance, so too our understanding of the molecular pathways and mechanisms that govern epigenetic inheritance in animals is radically changing.
Keywords: epigenetics, gene regulation, gene silencing, miRNAs, piRNAs, RNA interference, small RNAs, transgenerational epigenetic inheritance
Small RNAs contribute to epigenetic inheritance in different animal model organisms. Small RNAs can store and transmit the memory of infections, environmental stresses, and nutritional changes. The mechanisms by which small RNAs achieve such functions are still not fully elucidated and will help advance our understanding of epigenetic inheritance.

Abbreviations
3’UTRs, 3’ untranslated regions
dsRNAs, double‐stranded RNAs
endo‐siRNAs, endogenous small interfering RNAs
EVs, extracellular vesicles
FHV, Flock House virus
H3K23me3, trimethylation of the lysine 23 of the histone H3
H3K27me3, trimethylation of the lysine 27 of the histone H3
H3K9me3, trimethylation of the lysine 9 of the histone H3
HPA, hypothalamic–pituitary–adrenal
miRNAs, microRNAs
Mrt, mortal germline
MSUS, unpredictable maternal separation combined with unpredictable maternal stress
ncRNAs, noncoding RNAs
NMD, nonsense‐mediated mRNA decay
nt, nucleotide
OV, Orsay virus
piRNAs, PIWI‐interacting small RNAs
Pol II, RNA polymerase II
PTMs, post‐translational modifications
RdRPs, RNA‐dependent RNA polymerases
risiRNAs, antisense ribosomal siRNAs
RNAi, RNA interference
siRNAs, small interfering RNAs
tRFs, tRNA fragments
viRNAs, virus‐derived small RNAs
WAGOs, worm‐specific Argonaute proteins
The inheritance of traits across generations largely depends on genetic information. However, the nongenetic inheritance of traits has been documented in several organisms, including animals [1]. The mechanisms responsible for such a type of inheritance are still incompletely understood. Epigenetic mechanisms, including DNA methylation, histone post‐translational modifications (PTMs), and small noncoding RNA pathways, have been proposed to transmit heritable information along with the genome [2].
Decades of research have elucidated the multiple functions of epigenetic mechanisms in gene regulation during animal development, cell division, and differentiation [3, 4, 5]. Histone PTMs and DNA methylation are dynamically added or removed by specific chromatin modifier enzymes [6]. These modifications can be specifically recognized and bound by proteins that can influence the genome's activity as well as the activation or repression of genes [6]. Small noncoding RNAs are also bound by a specific family of proteins, called Argonaute, which regulate the expression of genes by RNA base complementarity [7]. Some of these Argonaute proteins possess an endonucleolytic activity capable of slicing complementary targets [8, 9]. They interact with nascent transcripts during RNA polymerase II (Pol II) transcription and influence the composition of histone modifications and DNA methylation on specific loci or regulate target transcripts at the post‐transcriptional level [7]. Therefore, crosstalk between different epigenetic mechanisms might also occur. The abundance and the activity of these small RNA molecules, DNA, and histone modifications can also change in response to environmental stimuli [10]. This critical feature allows epigenetic mechanisms to propagate information acquired during the life of an organism, such as the encounter with a pathogen, traumatic experiences, or nutritional changes, as well as environmental changes such as heat shocks or the exposure to toxic agents [11]. Although such epigenetically inherited information can be detrimental when it contributes to propagating the inheritance of diseases [1], it may also have an adaptative function and contribute to the evolution of acquired traits [1].
DNA methylation, histone PTMs, and small RNA populations must be transmitted upon fertilization through the gametes so as to propagate epigenetic information. Once transmitted to the zygote, maintaining these epigenetic modifications and molecules in the following generations is also fundamental. For example, some levels of DNA methylation can be transmitted and maintained upon fertilization [12]. Similarly, the transmission of histone modifications associated with gene activation or repression into the zygote has been documented [13, 14, 15]. Research in animal model organisms, including nematodes (Caenorhabditis elegans), insects (Drosophila melanogaster), and mice (Mus musculus), has been fundamental to demonstrate the inheritance of such epigenetic modifications [2]. In this regard, C. elegans is an ideal animal model system to study epigenetic inheritance. Its short generation time of 3 days allows the collection of multiple generations in few weeks. Moreover, the ability to grow a large population of isogenic individuals helps minimize the effect of genetic variations. Indeed, several cases of transgenerational epigenetic inheritance have been documented in C. elegans, including the inheritance of complex traits such as longevity, fertility, stress responses, infections, and learned behavior [11]. DNA methylation, histone PTMs, and small RNAs also transmit heritable phenotypes in plants and play an essential role in heritable phenotypic variations and adaptation to environmental changes [16, 17].
The inheritance of small RNAs can also last for multiple generations. The heritable functions of small RNAs became evident with the discovery of the RNA interference (RNAi) phenomenon in C. elegans [18]. Several features make small RNAs ideal candidate molecules to transmit epigenetic information across generations. First, the movement of small RNAs across tissues and organs allows the transmission of the information acquired from soma to the germline and thereby to the next generation. Second, the amplification of small RNAs by specific small RNA machineries that use target mRNA templates ensures small RNAs' production across generations. Finally, small RNAs trigger the activity of other epigenetic mechanisms, such as histone PTMs and DNA methylation, which might help the propagation of epigenetic information in a small RNA‐independent manner.
This review attempts to cover the role of small RNAs in epigenetic inheritance in animals. The inheritance of small RNAs can occur from one generation to another or across multiple generations. I dedicate a large portion of the review to the phenomenon of RNAi in C. elegans, from the initiation of the RNA silencing to the propagation of the silencing signal across multiple generations. The inheritance of RNAi is one of the best‐characterized cases of small RNA‐mediated epigenetic inheritance in animals and also serves as a model for understanding other phenomena of small RNA inheritance. I detail the mechanisms by which small RNAs trigger the silencing of complementary targets and how small RNAs move across tissues and organs, and are transmitted from one generation to another. In addition to the RNAi, I review the classes of endogenous small RNAs found in different animal model organisms and their heritable epigenetic functions. Finally, I critically discuss well‐documented cases of small RNA‐mediated epigenetic inheritance of traits such as infection, stress responses, fertility, and acquired behavior in several animal model organisms.
RNA interference is an epigenetic phenomenon
The capacity of RNA to alter trait inheritance became evident with the discovery of RNAi over 20 years ago. Pioneering experiments by Mello and Fire showed that the nematode C. elegans injected with double‐stranded RNAs (dsRNAs) displayed gene silencing that persisted in descendants not exposed initially to the dsRNAs [18]. The inheritance of the phenotype induced by the targeted dsRNA could last for more than one generation leading to the hypothesis that an active mechanism was responsible for propagating the silencing signal through the gametes across multiple generations [19]. Indeed, some of the upstream factors involved in the initiation of silencing and the downstream effectors involved in the propagation of silencing across generations were readily discovered [19]. The RNAi phenomenon in C. elegans is still one of the most studied cases of epigenetic inheritance and perhaps the best understood.
From dsRNAs to the final effector molecules
The initial step of the RNA silencing process involves the production of primary small interfering RNAs (siRNAs). The endoribonucleases Dicer and its cofactor RDE‐4 generate siRNAs from the cleavage of injected dsRNA trigger [20] (see Fig. 1). The primary siRNAs derive from both strands and carry a 5' monophosphate terminus [21]. The Argonaute RDE‐1 loads these primary siRNAs [22]. The catalytic activity of RDE‐1 is not required to degrade the target mRNAs but for generating a single‐stranded small RNA [22]. The interaction between the primary siRNAs loaded into RDE‐1 and the complementary target mRNA leads to the recruitment of the endonuclease RDE‐8, which cleaves the mRNA target [23]. The ribonucleotidyltransferase RDE‐3 adds stretches of poly(UG) at the 3′end of the cleaved target mRNA [24]. Mutation in RDE‐8 abolishes pUGylated mRNAs production during RNAi, suggesting that the mRNA cleavage by RDE‐8 is essential for pUGylation [24]. Whether RDE‐8 directly recruits RDE‐3 on cleaved mRNA fragments or other intermediate steps are required remains to be elucidated. The direct injection of pUGylated mRNAs can bypass the requirement of RDE‐8 and the upstream components of the RNAi silencing machinery involved in the production of primary siRNAs [24]. The pUGylated mRNA fragments with at least 18 UG repeats interact with the RNA‐dependent RNA polymerase (RdRP) RRF‐1 and are sufficient to induce the production of secondary small RNAs, called 22G‐RNAs [24]. These 22G‐RNAs produced by the RdRP are single‐stranded small RNAs antisense to the mRNA target [25, 26]. They have specific features such as a 5' triphosphate terminus, a bias for guanosine starting at the first nucleotide position, and an average size of 22 nucleotides (nt) [27]. The secondary RdRP‐dependent small RNA production represents an amplification of the initial silencing signal [25, 26]. The 22G‐RNAs are then loaded into 12 downstream Worm‐specific Argonaute proteins (WAGOs) [21, 27], which are the effectors of the RNA silencing response and lead to the post‐transcriptional and the transcriptional silencing of the targeted mRNA. The production of secondary siRNAs continues to occur even in the absence of the initial dsRNA triggers [28]. Thus, progenies from worms exposed to dsRNA maintain the silencing for multiple generations even in the absence of the dsRNA. F1 worms not directly exposed to dsRNAs inherit secondary 22G‐RNAs [28] as well as pUGylated mRNAs [24]. These inherited molecules initiate de novo production of pUGylated mRNAs and 22G‐RNAs to maintain the heritable silencing response [24]. Mutations in all the 12 downstream WAGOs do not suppress the production of pUGylated mRNAs in worms exposed to dsRNAs but abolish pUGylated mRNAs in subsequent generations [24]. Moreover, only newly synthesized pUGylated RNAs and 22G‐RNAs are detected in progenies of RNAi‐treated worms [24]. Thus, inherited 22G‐RNAs are required to maintain the transgenerational silencing response; how the WAGOs and the inherited 22G‐RNAs trigger de novo production of small RNAs across generations are still unclear.
Fig. 1.
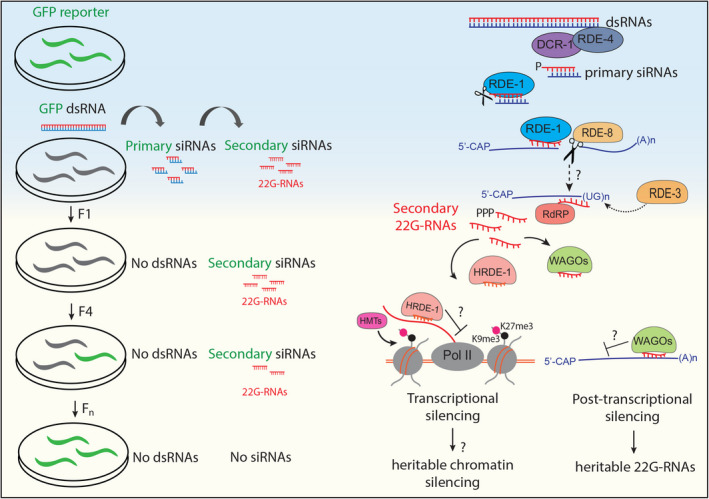
Schematic of the inheritance of C. elegans RNAi. The exposure to dsRNAs targeting a fluorescent GFP sequence leads to the inactivation of the GFP reporter (green and gray worms). The GFP reporter remains silenced even in subsequent generations that are not exposed to dsRNAs. The production of primary siRNAs generated by Dicer (DCR‐1) and its cofactor RDE‐4 are loaded into the Argonaute RDE‐1. The catalytic activity of RDE‐1 serves to remove one of the strands of the double‐stranded siRNA. The targeting of the complementary sequence by RDE‐1 leads to the recruitment of the endonuclease RDE‐8 and subsequently to the pUGylation of the cleaved mRNAs. pUGylated mRNAs become the substrate for the RdRP, which generates secondary single‐stranded 5′ triphosphorylated small RNAs called 22G‐RNAs. The 22G‐RNAs are loaded into downstream WAGO proteins, the effector Argonautes that trigger post‐transcriptional and transcriptional silencing. Such silencing persists for multiple generations and correlates with the presence of inherited 22G‐RNAs and pUGylated mRNAs. The nuclear Argonaute HRDE‐1 also triggers heritable histone PTMs.
Transcriptional and post‐transcriptional RNA‐mediated silencing
The loading of 22G‐RNAs into the twelve WAGOs responsible for the silencing responses generates a post‐transcriptional silencing in the cytosol and a co‐transcriptional silencing in the nucleus [27, 28, 29] (Fig. 1). In the cytosol, the post‐transcriptional degradation of the targeted mRNAs does not result from the Argonaute slicer activity. Even if most WAGOs, including the Argonaute WAGO‐1, trigger a post‐transcriptional downregulation of the mRNA target [27], they lack a catalytically active domain [30]. Thus, the degradation of the targeted mRNAs might result from other still unknown nucleases that are part of the WAGO RNAi silencing complex. Alternatively, the WAGOs might interact with different pathways involved in RNA metabolism. For example, some components of the nonsense‐mediated mRNA decay (NMD) participate in the production of 22G‐RNA from some WAGO endogenous targets [27]. Therefore, the WAGOs can recruit NMD components to trigger mRNA decay during RNAi.
Additionally, the RNAi pathway can directly or indirectly affect the translation of the mRNA target and the stability of the protein derived from the targeted mRNA. In the nucleus, the nuclear WAGO protein HRDE‐1 promotes the co‐transcriptional silencing in all germline nuclei [29]. The nuclear Argonaute NRDE‐3 plays a similar role in somatic nuclei [31, 32]. RNAi in animals lacking the nuclear Argonaute HRDE‐1 is functional, suggesting that the post‐transcriptional silencing is sufficient to repress the expression of the target genes in animals exposed to dsRNAs [29]. However, hrde‐1 mutant animals fail to inherit RNAi response, indicating that HRDE‐1 is required for heritable RNAi responses [29]. Thus, primary siRNAs and 22G‐RNAs loaded into WAGOs can initiate a post‐transcriptional silencing response, which is then maintained by the nuclear co‐transcriptional silencing across generations. Given its nuclear localization and the fact that RNAi induces the downregulation of the mature and nascent unspliced RNA target [29], it is believed that HRDE‐1 represses the transcription of target loci by interacting with nascent RNAs. HRDE‐1 also promotes the deposition of some repressive histone PTMs on RNAi‐targeted loci, such as the trimethylation of the lysine 9 of the histone H3 (H3K9me3) [28, 29], the trimethylation of the lysine 27 of the histone H3 (H3K27me3) [33], and the trimethylation of the lysine 23 of the histone H3 (H3K23me3) [34]. The transcriptional repression of the targeted locus by HRDE‐1 can occur by the direct recruitment of histone methyltransferases of repressive histone modifications. However, mass spec experiments aimed to detect the HRDE‐1 complex failed to reveal such interactions [35]. Moreover, the repressive histone modifications, such as H3K9me3, do not directly suppress Pol II transcription, even if the deposition of repressive histone modifications correlates with gene silencing [36]. Therefore, HRDE‐1 can also repress pol II transcription by other mechanisms uncoupled from the deposition of repressive histone modifications on the targeted locus. Nonetheless, progenies from RNAi‐treated parents inherit along with the 22G‐RNAs the repressive histone modifications on the RNAi‐targeted locus [28]. The Argonaute protein NRDE‐3 localizes to the nucleus upon RNAi treatment in somatic cells and co‐transcriptionally represses the targeted locus [31, 32]. NRDE‐3 is required to transmit heritable RNAi silencing in the somatic cells of F1 animals not exposed to dsRNAs like HRDE‐1 in the germline [37].
Propagation of the RNAi silencing response across tissues, organs, and generations
The RNAi phenomenon is not only triggered by the direct injection of dsRNAs into the body of the worms but also by feeding bacteria expressing dsRNAs [38]. Furthermore, the induction of RNAi by feeding can also last multiple generations [38]. Therefore, the bacteria's dsRNA delivered into the intestine and the small RNAs derived from it can travel across somatic tissues and organs to the germline and through the gametes to the next generations. I will discuss in this session how this is possibly achieved.
Soma to germline transfer of small RNAs
The dsRNAs released from ingested bacteria into the acidic C. elegans intestinal lumen are transported inside the cells by the apical intestinal membrane protein SID‐2 through endocytosis [39, 40]. SID‐2 is selective for dsRNAs and allows the uptake of dsRNA from the intestinal lumen [39, 40]. The release of dsRNAs into the cytosol of intestinal cells and the spreading of dsRNAs to other tissues or organs is mediated by the ubiquitously expressed transmembrane protein SID‐1, which is also selective for dsRNAs [40, 41]. Therefore, SID‐1 participates in both dsRNA release and uptake in donor and recipient cells. The C. elegans endosome‐associated protein SID‐5 is another factor required for systemic RNAi. SID‐5 localizes to endocytic vesicles implicated in exosome biogenesis and plays a role in dsRNA transport [42]. The injection of dsRNAs into the body cavity at the interface of the intestine and the germline can be transported into oocytes by SID‐1 and SID‐5 and extracellular vesicles (EVs) carrying yolk proteins from the intestine to mature oocytes [43, 44]. Consequently, it would be possible that dsRNAs can also be transported through extracellular vesicles across tissues and organs to the germline. The dsRNAs loaded into the oocytes are then released in the embryo by SID‐1 and SID‐5 [44].
Germ granules in heritable RNAi responses
How are germline small RNAs or somatic small RNAs delivered to the germline transmitted to the next generation? Recent studies suggest that perinuclear phase‐separated condensates called germ granules or nuage might serve such function [45, 46, 47] (Fig. 2). These germ granules reside near the nuclear pore [48] until the oocyte formation and are maternally transmitted through the cytosol to the zygote [49]. One of the key features of the components of the RNAi pathway is that they localize to germ granules. Most enzymes producing secondary 22G‐RNAs, like RdRPs and their cofactors, and cytoplasmic WAGOs also localize in germ granules [50, 51, 52]. For this reason, the synthesis and loading of 22G‐RNAs into WAGOs occur primarily on mature mRNAs entering germ granules from the nuclear pore. In support of this model, the removal of germ granules severely depletes the 22G‐RNAs loaded into WAGOs [53]. Interestingly, even the nuclear Argonaute HRDE‐1 is loaded with 22G‐RNAs derived from mature mRNAs [29], suggesting that HRDE‐1 transiently resides into the germ granules to load its 22G‐RNA cargo. The different activities of the RNAi pathway are also spatially separated in germ granules [52]. The germ granules form three distinct condensates: the P granule, Z granule, and mutator foci [52]. The endonuclease RDE‐8, the ribonucleotidyltransferase RDE‐3, and the RdRP RRF‐1 reside in the mutator foci separated from the P granules by the Z granules. However, most of the WAGOs localize to P granules, which are close to the nuclear pore. Thus, it is still unclear how the P granule localized WAGOs and the nuclear Argonaute HRDE‐1 load 22G‐RNAs in mutator foci. One possibility is that the membraneless features of these condensates allow the rapid diffusion of these factors from one condensate to another. Alternatively, other still unknown intermediate factors might be involved in connecting the synthesis and loading of 22G‐RNAs. Similarly puzzling is how the mRNA target exiting from the nuclear pore moves from the P granule to the mutator foci, where it is cleaved and pUGylated. Mutation in the structural Z granule component ZNFX‐1 and its co‐localized Argonaute WAGO‐4 specifically impair heritable RNAi response [52]. Structural components of P granules also participate in heritable RNAi responses [54]. Given that the germ granules are maternally inherited and segregate with the germline precursor [49], they also carry 22G‐RNAs and pUGylated RNAs into the zygote. Indeed, mutants that impair the assembly of germ granules into the zygote fail to transmit heritable RNAi even though germ granules are formed later in larvae [45, 46, 47]. In addition, the Z and P granules are fused in the zygotes during early embryogenesis and start to be separated only after zygotic transcription in the germline precursor [52]. It is not clear why the Z and P granules fuse in embryos before the zygotic genome activation. Perhaps this fusion facilitates the initiation of de novo 22G‐RNAs and pUGylated mRNA fragments. Alternatively, the inherited germ granules might serve to carry and protect 22G‐RNAs and pUGylated mRNAs from other degradation pathways operating in early embryogenesis before the zygotic activation of the genome. In this case, heritable 22G‐RNAs and pUGylated mRNA fragments might initiate de novo synthesis of 22G‐RNAs only after zygotic transcription.
Fig. 2.
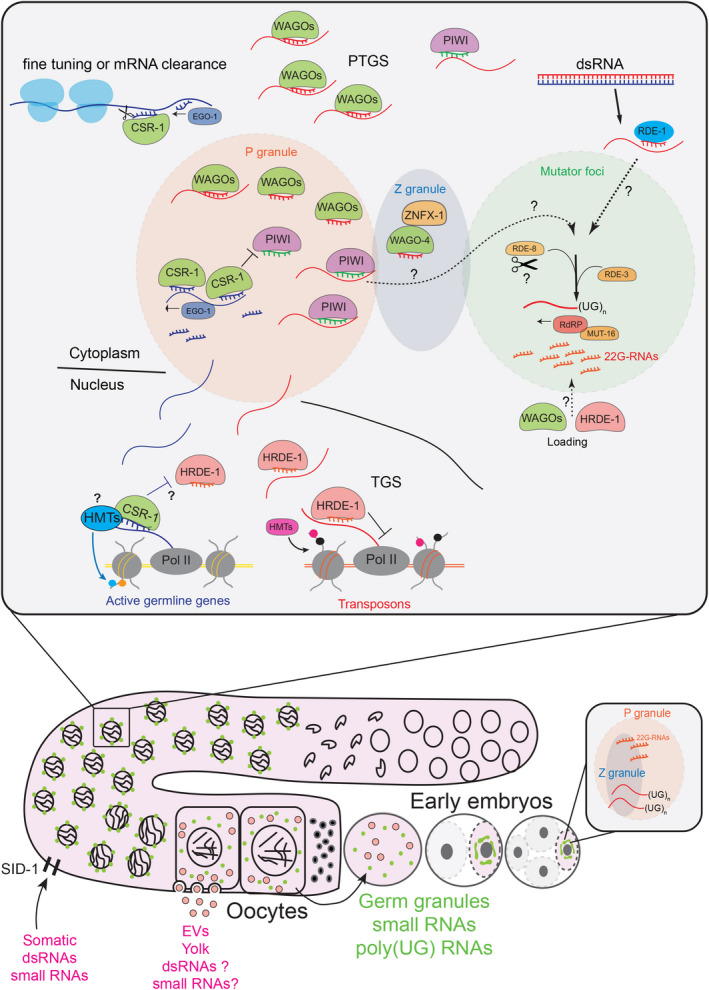
Schematic of the different endogenous germline small RNA pathways and their inheritance in C. elegans. Most of the Argonaute proteins localize to germ granules, phase‐separated condensates surrounding the nuclear membrane in the germline of C. elegans. Three different condensates characterize germ granules (P granules, Z granules, and mutator foci). The P granules contain most of the WAGOs, PIWI, and CSR‐1 proteins. The Z granules contain factors involved in heritable RNAi. The mutator foci include most of the enzymes required for the biogenesis of 22G‐RNAs triggered by dsRNAs or piRNAs. The 22G‐RNAs derived from active germline genes are produced by the RdRP EGO‐1 and are loaded into the Argonaute CSR‐1. These CSR‐1 22G‐RNAs can protect germline mRNAs from piRNA silencing and promote the degradation of the complementary mRNAs in the cytosol through the CSR‐1 catalytic activity. CSR‐1 can also bind nascent transcripts and prevent HRDE‐1 nuclear silencing. Most of the germline 22G‐RNAs and pUGylated mRNAs are transmitted to the embryos inside the germ granules. Somatic 22G‐RNAs and dsRNAs are transmitted to the germline through the dsRNA receptor SID‐1 or possibly through EVs carrying yolk protein from the intestine to the oocytes.
Heritability of the RNAi phenomenon
The RNAi inheritance is not permanent, and the silenced gene targeted by RNAi restores its expression after some generations [19, 28]. Multiple factors can influence the transgenerational duration of the RNAi response. First, technical variations affect the length of transgenerational silencing. For instance, the induction of RNAi is achieved by direct injection of dsRNA into the germline, the intestine, or the body cavity of the worms and by feeding bacteria expressing dsRNAs. Both the methods can generate some variation in the amount of dsRNAs that each worm receives, which can indirectly influence the penetrance of the RNAi response. Moreover, the inheritance of the silencing of a germline gene upon feeding bacteria expressing dsRNAs only efficiently works in adult worms and not in earlier larval stages [44]. Therefore, variation in the developmental stage of the worms, especially if working with mutants with developmental timing defects, affects the efficiency of heritable RNAi [44]. In addition to technical variations, active mechanisms might operate to prevent permanent silencing. For instance, mutations in the histone methyltransferase MET‐2, which is responsible for the deposition of H3K9me1/2, and in the chromodomain containing protein HERI‐1 increase the transgenerational length of RNAi [55, 56]. Conversely, the mutation of the histone methyltransferase SET‐25, which is responsible for the deposition of H3K9me3, almost immediately suppresses the inheritance of RNAi [55]. Thus, variation in the activity of histone modifier enzymes can modulate the length of RNAi inheritance. More recently, the mutation of another component of the endogenous small RNA pathway increases the duration of silencing for hundreds of generations [57]. These mutant worms continuously produce pUGylated RNAs and 22G‐RNAs, even hundreds of generations after the initial dsRNA exposure [57]. Consequently, the balance between the activities of different small RNA pathways might regulate the length of RNAi responses and avoid permanent heritable silencing. In addition, interindividual variation among each worm can directly influence the inheritance of RNAi. Differences in the expression of the heat‐shock transcriptional factor HSF‐1 among individual worms directly influence RNAi inheritance [58]. Using a different type of transgene as a target of RNAi experiments also causes variations in the duration of heritable RNAi. Most of the RNAi studies in C. elegans have used a multicopy germline‐expressed GFP transgene. The copy number and the chromatin environment of these multicopy transgenes influence the germline expression or the silencing of the reporter gene [59]. Due to the unstable nature of repetitive sequences, the variation in gene copy number and the number of active and inactive copies can indirectly affect the study of RNAi inheritance. Transgene copy number variation is also difficult to evaluate in each experiment and each worm, which can mislead the interpretation of RNAi inheritance experiments. Finally, the length of transgenerational silencing is variable even when targeting the same germline‐expressed sequence located in different genomic loci [60]. Single‐copy GFP transgenes or CRISPR‐Cas9 GFP tagged strains show different lengths of heritable silencing, suggesting that DNA regulatory elements might also influence the heritability of RNAi [60]. Thus, even if the use of RNAi in transgenic animals has dramatically advanced our understanding of RNAi inheritance, they might not necessarily reflect how an individual locus is susceptible to RNA silencing. Similarly, the inheritance of endogenous small RNAs might follow different rules from the inheritance of the RNAi phenomenon.
Endogenous small RNAs in epigenetic inheritance
The inheritance of RNAi upon exposure to exogenous dsRNAs is undoubtedly a powerful model system to study small RNA‐mediated epigenetic inheritance. But, given the known endogenous small RNA pathways, it is also essential to investigate whether these endogenous small RNAs provide similar heritable information and serve to transmit traits across generations. There are three major classes of small RNAs in animals: PIWI‐interacting small RNAs (piRNAs) produced from piRNA clusters, microRNAs (miRNAs) derived from hairpin‐shaped precursors and endogenous small interfering RNAs (endo‐siRNAs) generated from long dsRNAs or RdRPs. Other reviews have widely discussed the great potential of these endogenous small RNA classes to regulate gene expression and their biological functions during animal development. For this reason, the following session will be focused on their possible role in epigenetic inheritance and on how they can achieve such function.
The epigenetic functions of piRNAs
The class of piRNAs was first described in Drosophila testis [61]. Still, most of the studies have investigated the function of piRNAs in repressing the expression of repetitive elements, such as transposons, in the female germline (reviewed in ref. [62]). This small RNA class is associated with PIWI‐clade Argonautes, the expression of which is mostly in gonads of insects, mammals, nematodes, and fish. Large parts of the piRNA sequences in fly and mice match transposable elements, even though individual piRNA sequences are not conserved across species. Their biogenesis mechanisms are also different in insects, mammals, and nematodes (reviewed in ref. [62]). In general, piRNAs do not require dsRNAs and Dicer for their biogenesis. In Drosophila, long piRNA precursors derive from large genomic clusters containing the remnants of transposable elements [63]. In mouse, long noncoding piRNA precursors are generated from sparse genomic loci during the pachytene stage of spermatogenesis, and their sequences are largely devoided of transposable elements [64, 65]. In contrast, most mouse prepachytene piRNAs, which start to be expressed during embryogenesis, derive from clusters of transposable elements and 3′ untranslated regions (3′UTRs) of protein‐coding genes [65, 66]. In C. elegans, each piRNA derives from an individual locus that generates a short piRNA precursor molecule [67, 68, 69]. Most of the studies so far have identified a prominent role of piRNAs in transposon repression to preserve genome integrity in the germline of animals. Here, piRNAs trigger the post‐transcriptional silencing of these repetitive elements by cleaving complementary transcripts but also initiate an epigenetic transcriptional silencing through the deposition of repressive histone PTMs [70, 71, 72, 73] or DNA methylation [66]. Their role in transposon repression is predominant in the female germline in Drosophila and is essential for both male and female fertility. However, male piRNAs target different transposable elements than female piRNAs and regulate protein‐coding genes [74]. In mouse male germline, pachytene piRNAs are essential for spermatogenesis, where they might regulate protein‐coding genes important for male fertility [75, 76]. In contrast, female piRNAs in mouse oocytes are not essential, and they regulate transposons together with the endo‐siRNA pathway [77]. Male and female piRNAs can therefore regulate different targets and might differ in their impact on gametogenesis. In C. elegans, animals lacking piRNAs do not show global derepression of transposable elements [78, 79, 80, 81, 82]. C. elegans piRNAs are only required to initiate and not to maintain the transcriptional silencing of targeted sequences [73, 83]. Consequently, downstream chromatin silencing pathways and histone PTMs might help maintain the transposon silencing initiated by piRNAs. C. elegans piRNAs can also potentially regulate protein‐coding genes [84, 85]. One such example is the repression of a gene essential for sex determination [86]. The regulation of sex determination by a single piRNA was also previously found in the silkworm [87], suggesting another potentially conserved function of piRNAs.
The role of maternally inherited piRNAs
Studies in C. elegans and Drosophila suggest that maternally inherited piRNAs are required to initiate de novo establishment of chromatin silencing during early embryogenesis [88, 89]. This signature of chromatin silencing in embryos can be maintained in adult animals in a piRNA‐independent manner [88, 89]. Thus, inherited piRNAs are important to re‐initiate the memory of silencing at the beginning of each life cycle. piRNAs repress the expression of transposons by inducing chromatin changes and interfere with the splicing of the transposon‐derived mRNAs, causing the lack of translation of the transposase protein [90]. Maternally inherited piRNAs can also trigger the silencing of newly integrated transposable elements derived from the paternal genome after fertilization [91]. Flies that fail to inherit such a maternal pool of piRNAs become sterile, an epigenetic phenomenon called hybrid dysgenesis (reviewed in [92]). In natural Drosophila strains, the degree of variations to transposon invasions correlates with the abundance of germline piRNAs [93]. Thus, inherited piRNAs can provide a sort of innate immunity against transposable elements. Maternal inherited PIWI proteins also localize in somatic cells during early embryogenesis, where they appear to attenuate zygotic expression of transposable elements in somatic nuclei [94]. In addition to their role in repressing repetitive elements, maternally inherited piRNAs play a role in maternal mRNA clearance during zygotic genome activation in insects [95, 96]. Consequently, piRNAs could have a crucial function in initiating and maintaining their repressive functions across generations.
How do the piRNAs exert their transgenerational function?
The transgenerational function of piRNAs relies on an amplification system that generates and maintains an abundant level of small RNA population. In Drosophila and mouse, the generation of primary piRNAs triggers a secondary piRNA production through an amplification system that involves other Argonaute proteins. This amplification mechanism, called the ping pong cycle (reviewed in ref. [97]), is driven by the catalytic activity of other PIWI proteins, which generates secondary piRNAs from the cleavage of the target transcript [63, 65, 98]. In C. elegans, PIWI does not directly cleave its piRNA complementary targets but instead triggers the biogenesis of RdRP‐dependent secondary 22G‐RNAs [79, 84]. Thus, the C. elegans piRNAs silence their target through a mechanism similar to that regarding the RNAi (Fig. 2). Indeed, the same downstream effectors of the RNAi pathway are operating in the piRNA‐induced silencing pathway. Moreover, most of their sequences do not match transposons, yet they can trigger the silencing of repetitive elements. This is because the C. elegans piRNAs can initiate the biogenesis of 22G‐RNAs even by targeting transcripts with imperfect complementarity [85, 99]. A still unknown endonuclease generates cleaved fragments pUGylated by RDE‐3 [57], which act as a template suitable for an RdRP‐dependent 22G‐RNA amplification. The 22G‐RNA synthesis triggered by piRNAs is the prerequisite for loading WAGOs to activate the post‐transcriptional and transcriptional silencing of the targeted transcript [79, 84]. Thus, in C. elegans, the piRNAs are required to initiate the silencing performed by endo‐siRNAs. The piRNA‐induced silencing also generates a third wave of endo‐siRNAs in the F1 animals [100]. The nuclear WAGO protein HRDE‐1 is crucial for the generation of these tertiary siRNAs [100]. Thus, the nuclear silencing pathway may contribute to reinforcing the transgenerational production of siRNAs in germ granules. How the nuclear RNAi pathway influences the small RNA production in germ granules is still unknown. The repressive activity of piRNAs across multiple generations relies on the availability of the target templates and the amplification machinery. Therefore, the inheritance of piRNAs and the reinitiation of piRNA amplification signal is an essential feature of the piRNA function in epigenetic inheritance.
miRNAs in epigenetic inheritance
miRNAs are endogenous small regulatory RNAs produced from a longer stem‐loop RNA precursor (for a comprehensive review on miRNA biogenesis and functions, see ref. [101]). Their discovery occurred earlier than the RNAi phenomenon [102]. Nonetheless, it took some years to identify that the same molecular pathway involved in the RNAi phenomenon also participates in miRNA production and regulation [103]. The role of miRNAs in regulating endogenous genes by base complementarity was already proposed by Victor Ambros and colleagues when the miRNAs were first discovered [102]. The capacity of miRNAs to act as global post‐transcriptional regulators of gene expression programs is an essential function for animal development and is a well‐established function [101]. Whether miRNAs play a role in epigenetic inheritance is still largely uncharacterized. The evidence that both mature sperm and oocyte possess miRNA cargoes suggests that they can be inherited into the zygote and play a role in epigenetic inheritance [104]. However, whereas the role of zygotically expressed miRNAs in maternal mRNA decay is well established in the fruit fly, frog, and fish, the function of maternally inherited miRNAs is unclear [105]. In mouse oocytes and early embryos, miRNA function is globally suppressed [106, 107]. Moreover, maternally deposited miRNAs in Drosophila are adenylated and rapidly degraded during the maternal to zygotic transition [108]. These studies indicate that the role of maternally inherited miRNAs in embryos might be limited. Several studies have suggested that paternal miRNAs might instead contribute to epigenetic inheritance in the zygote through sperm [109, 110, 111, 112, 113]. The acquisition of the miRNA cargo in mature sperm occurs during the process of post‐testicular maturation, and they appear to play an essential role during early embryogenesis [109]. It is important to note that the crucial role of sperm miRNAs has only been observed under specific laboratory conditions and methodological preparation, raising the question of whether sperm miRNAs might be essential during natural mating conditions [114, 115]. The sperm miRNA payload changes during post‐testicular maturation in the epididymis, suggesting the transfer of miRNAs from epididymis epithelial cells to maturing sperm through EVs [116]. Thus, even somatic cells can contribute to the transfer of small RNAs to the next generation through sperm. The miRNA cargo in mature sperm also changes in response to environmental stress [111, 112, 113]. Whether the epididymis epithelial cells transmit the stress‐induced miRNAs to mature sperm remains unknown. The injection of sperm miRNAs from stressed mice appears to recapitulate stress‐induced phenotypes in F1 animals [111, 112, 113]. Nonetheless, the observed tiny contribution of sperm miRNAs to the pool of zygotic miRNAs during fertilization [117] challenges the possibility that the natural transfer of sperm miRNAs during fertilization has any biological effects. The mechanism by which inherited miRNAs can exert such prolonged epigenetic functions across multiple generations is also unclear, especially considering the absence of any amplification mechanisms in contrast to piRNAs. One possibility would be that miRNAs regulate the expression level of genes involved in epigenetic histone PTMs or other chromatin pathways and alter a specific pattern of chromatin marks inherited across multiple generations. In addition, miRNAs can also function in the nucleus. In human cell lines, a terminal hexanucleotide sequence allows a specific miRNA to localize in the nucleus [118]. Other miRNAs also localize in the nucleus [119], and hypoxic stress changes the distribution of cytoplasmic and nuclear miRNAs [120]. The human Argonaute protein Ago2 loads some nuclear miRNAs [121], suggesting they actively function in the nucleus. Thus, specific miRNAs can also exert some nuclear functions in normal conditions or during stress responses [122]. They can potentially promote epigenetic modifications at the chromatin level like piRNAs and some endo‐siRNAs [122]. Lastly, miRNA‐directed endo‐siRNA biogenesis has been documented in C. elegans [123], suggesting another potential mechanism for miRNAs to exert a transgenerational epigenetic function.
Inherited functions of endo‐siRNAs
Most endo‐siRNAs in Drosophila and mice derive from dsRNA intermediates processed by Dicer, like the siRNAs generated during the phenomenon of RNAi upon dsRNA injection. In C. elegans, however, the most abundant class of endo‐siRNAs are generated by RdRPs in a Dicer‐independent manner. I discuss in the following sections these two different types of endo‐siRNAs.
Endo‐siRNAs derived from endogenous dsRNA source
The production of siRNAs from injected dsRNA suggests that the RNAi pathway can encounter dsRNA molecules during animal life cycles. Viral infection is one of the natural sources of dsRNAs to which animals can be exposed. During viral infection, dsRNA replication intermediates can activate the production of virus‐derived small RNAs (viRNAs) by Dicer [124]. One of the proposed ancestral functions of the RNAi pathway is to protect cells from a viral infection. This function seems to be minor in vertebrates due to the presence of the interferon pathway [125]. Besides dsRNA derived from viruses, animals can also produce dsRNA molecules from their genome. A series of works in Drosophila have shown the existence of endo‐siRNAs in the soma and germline [126, 127, 128, 129, 130]. These endo‐siRNAs can be originated by heterochromatin‐derive sense and antisense transcripts, implying that the majority of the endo‐siRNAs map to transposons. Another source of endo‐siRNAs is dsRNAs originating from the convergent transcription of protein‐coding genes. These endo‐siRNAs can regulate in cis the levels of the mRNAs from which they come. Finally, endo‐siRNAs can originate from genes that generate long hairpin RNAs. Endo‐siRNAs from those genes can act in trans to regulate the expression of complementary transcripts. The expression of endo‐siRNAs in the Drosophila germline might allow their inheritance in embryos to exert some transgenerational epigenetic functions. For example, they can repress transposable elements like piRNAs or regulate the expression of protein‐coding genes [131, 132]. The production of dsRNAs from convergent transcripts might also be subjected to environmental changes and influence the biogenesis of siRNAs and their targeted transcripts. However, the absence of an amplification system that propagates Drosophila endo‐siRNAs might limit their possible heritable epigenetic functions. Also, it is still unclear whether they can act in the nucleus to promote histone modifications like the piRNAs. Thus, future studies will address whether Drosophila endo‐siRNAs might have heritable functions.
The discovery of endo‐siRNAs in Drosophila was concurrent with endo‐siRNAs in mouse oocytes and embryonic stem cells [133, 134, 135]. Like Drosophila endo‐siRNAs, the mouse endo‐siRNAs originate from dsRNAs derived from inverted repeat structures, bidirectional transcription, and antisense transcripts from various loci. Another source of endo‐siRNAs in mice is pseudogene RNAs that pair and form dsRNA intermediates with the complementary mRNA from the corresponding protein‐coding gene. These endo‐siRNAs can target transposons and protein‐coding genes to regulate their expression in oocytes and embryonic stem cells. In this regard, maternal inherited Ago2 and its interacting endo‐siRNAs have been proposed to clear maternal mRNAs during the oocyte to embryo transition [136]. Also, experiments in mouse embryonic stem cells have shown that in the absence of DNA methylation, endo‐siRNAs control the reactivation of transposable elements [137]. Thus, inherited mouse endo‐siRNAs can repress transposons and clear maternal mRNA in early embryogenesis. These functions might be essential given that components of the endo‐siRNA pathway are required for oocyte and early embryonic development. Furthermore, the production of endo‐siRNAs has been documented in mature sperm, where they can potentially target endogenous transcripts [138]. However, it is unknown whether they regulate those transcripts or complementary transcripts in embryos after fertilization. Even if endo‐siRNAs have the potential to be inherited from oocytes and sperm, the absence of a siRNA amplification mechanism, such as the presence of an RdRP, might prevent these molecules from having a potential impact across generations. Also, endo‐siRNAs appear to act through post‐transcriptional regulation of their target transcripts mainly. Therefore, in the absence of a link between the endo‐siRNA pathway and nuclear signaling through histone PTMs or DNA methylation, it is unlikely that endo‐siRNAs might have a long‐term transgenerational impact during animal development.
In C. elegans, endogenously produced dsRNAs can potentially trigger primary siRNAs like the ones generated during RNAi. However, because of the very low abundance of primary siRNAs compared to secondary siRNAs, the identification of endogenous primary siRNAs is difficult. Mutation in the catalytic activity of Dicer has revealed a subset of secondary 22G‐RNAs that depends on Dicer, suggesting a dsRNA source for possible primary siRNAs [139]. In contrast to Drosophila or mice, these endo‐siRNAs are not originating from a region producing dsRNAs [139]. DCR‐1 is instead required for the biogenesis of primary endo‐siRNAs called 26G‐RNAs, which are 26nt with a bias for a monophosphorylated 5′ guanosine [67, 78]. These monophosphate 26G‐RNAs are originated from a dsRNA precursor generated by the RdRP, which is then cleaved by Dicer and converted in monophosphate by a pyrophosphatase [140, 141]. The 26G‐RNAs are loaded by specific Argonaute proteins and trigger the production of secondary 22G‐RNAs. The 26G‐RNAs are mainly produced during spermatogenesis and the oocyte to embryo transition like the endo‐siRNAs produced in mice [142, 143]. The spermatogenic 26G‐RNAs have been shown to promote male fertility through the epigenetic transmission of male‐specific gene expression programs across generations [144]. Instead, no epigenetic functions have been associated with the 26G‐RNAs produced during the oocyte to embryo transition.
Dicer‐independent endo‐siRNAs
Most endogenously produced 22G‐RNAs in C. elegans germline do not require Dicer for their biogenesis. The RdRP directly synthesizes these 22G‐RNAs as single‐stranded small RNAs without needing a dsRNA intermediate or Dicer protein [26]. Two major dicer‐independent 22G‐RNA pathways act in C. elegans germline: (a) 22G‐RNAs triggered by the piRNAs and the PIWI protein PRG‐1 [84] and (b) the 22G‐RNAs that are associated with the Argonaute protein CSR‐1 [50, 84] (Fig. 2). Numerous studies have revealed the complex interplay between these two main germline 22G‐RNA pathways, essential for germline development and functions. Most, if not all, CSR‐1 22G‐RNAs are antisense to germline‐expressed genes [50, 53]. Given the capacity of piRNAs to target and silence mRNAs even by imperfect complementarity, they can potentially target the whole germline transcriptome [85, 99]. Thus, CSR‐1 22G‐RNAs might function in binding and protecting germline mRNAs from piRNAs silencing. The presence of CSR‐1 22G‐RNAs complementary to a transgenic reporter can license and activate in trans a piRNA silenced reporter with homologous sequence [146]. Moreover, the tethering of CSR‐1 on a transgenic reporter mRNA can counteract the transgenerational piRNA silencing effect [145]. The results from these transgenic models lead to the proposition that CSR‐1 protects endogenous ‘self’ germline‐expressed mRNAs from piRNA silencing and that the silencing capacity of piRNAs is only effective on ‘non‐self’ mRNAs [145]. More recent work has challenged this model by expressing a piRNA complementary to an endogenous germline‐expressed gene. The germline gene has resulted in being protected by piRNA silencing [99]. However, the targeted gene remains protected even in the absence of CSR‐1 [99]. Thus, CSR‐1 might not be the only factor preventing the production of 22G‐RNAs on endogenous ‘self’ mRNAs by piRNA targeting. Indeed, sequences encoded into exons [147] or introns [99] seem to protect germline mRNAs from piRNA silencing.
The protective function of CSR‐1 contrasts with another proposed role of CSR‐1 in mRNA degradation. CSR‐1 is responsible for most of the Argonaute‐mediated cleavage activity in C. elegans extracts, and CSR‐1 22G‐RNAs cleave complementary mRNAs in vitro [148]. The catalytic activity of CSR‐1 is essential for fertility and embryonic development [149, 150] and fine tunes the levels of complementary mRNA targets in the animal germline [150]. Whether CSR‐1 plays a role in preventing piRNA‐dependent silencing of germline mRNA targets or it cleaves and fine tunes the level of its mRNA targets remains debated. Recent work has shown that CSR‐1 catalytic activity is compatible with mRNA protection and the mRNA cleavage functions [53]. The cleavage activity of CSR‐1 has an impact on the expression level of a small subset of its mRNA targets, which are the ones with the most abundant antisense CSR‐1 22G‐RNAs [53]. Most CSR‐1 targets are instead unaffected in CSR‐1 catalytic mutant. But, in the absence of CSR‐1 protein, some of these unaffected targets show reduced transcription and antisense 22G‐RNA loading into the nuclear Argonaute HRDE‐1 [53]. Therefore, it is possible that CSR‐1 actively participates in protecting endogenous ‘self’ germline mRNAs from piRNA silencing. The protective function of CSR‐1 is consistent with other reports showing that CSR‐1 promotes the transcription of its targets [144, 151] and can antagonize the deposition of repressive histone PTMs at the chromatin level [152]. Thus, CSR‐1 can prevent the binding of piRNAs on mature mRNAs in germ granules or also regulate co‐transcriptionally the activity of the nuclear Argonaute protein HRDE‐1 and prevent the deposition of repressive histone PTMs [53, 152]. It is important to note that the CSR‐1‐protected genes do not show complete silencing in the absence of CSR‐1 [53], suggesting the existence of other mechanisms of protection at the chromatin level for this specific set of genes.
The biogenesis of CSR‐1 22G‐RNAs also differs from the piRNA‐dependent 22G‐RNAs. Whereas the synthesis of most piRNA‐dependent 22G‐RNAs occurs in germ granules, the production of most CSR‐1 22G‐RNAs by the RdRP EGO‐1 occurs in the cytosol on translating mRNAs [53]. What triggers the biogenesis of EGO‐1 22G‐RNAs is still unknown. Furthermore, the presence of two isoforms of CSR‐1 further complicates the understanding of CSR‐1 functions [153, 154]. These two CSR‐1 isoforms are differentially expressed during gametogenesis and appear to bind 22G‐RNAs targeting a different set of genes [153, 154]. A short isoform of CSR‐1 is expressed during the whole germline development in female and male germlines [153, 154]. The long isoform is instead specifically expressed during spermatogenesis and in some somatic tissues [153, 154]. Given its specific expression during spermatogenesis, CSR‐1 long isoform might promote the expression of spermatogenesis genes [153, 154]. However, spermatogenic genes are only mildly downregulated in the absence of CSR‐1 long isoform [153, 154]. Moreover, only the removal of the short isoform has a severe impact on gametogenesis [153, 154].
In addition to the epigenetic function of CSR‐1 in the germline, maternal inherited CSR‐1 22G‐RNAs are also essential for embryonic viability [149]. In contrast to most germline WAGOs and RNAi factors that segregate exclusively with the germline blastomeres inside the germ granules, the maternal CSR‐1 is also temporarily localized to the cytoplasm of somatic blastomeres [149]. Here, CSR‐1 catalytic activity contributes to promoting the degradation of maternal mRNAs in somatic blastomeres during the activation of the genome. This mechanism relies on a maternal pool of CSR‐1 22G‐RNAs generated on the mRNAs deposited in oocytes, which degrades complementary mRNA targets in embryos [149]. Furthermore, the CSR‐1 catalytic activity preferentially cleaves the complementary maternal mRNAs when not engaged in translation [149]. Thus, the loading of maternal mRNAs and CSR‐1 22G‐RNAs in embryos facilitates the removal of the maternal mRNAs only after their translation. Interestingly, the mouse endo‐siRNAs and the catalytic activity of maternal inherited Ago2 appear to be essential for early embryonic development [132]. Moreover, Ago2 and its endo‐siRNAs might play a role in maternal mRNA clearance during the early phase of embryonic development [105, 136]. Therefore, inherited endo‐siRNAs seem to play a conserved function in clearing complementary maternal mRNAs during early embryogenesis in animals.
Small RNA‐mediated inheritance of traits in animal model organisms
The transmission of small RNAs to the progeny and the amplification mechanism that allows the propagation of small RNAs for a certain number of generations raises the question of whether they can serve as molecules to transmit traits across generations. Given that small RNAs can be generated at any time during animal development upon an initial trigger and can move from different tissues or organs to the gametes, they constitute an ideal molecule to transmit acquired traits. For instance, changes in nutrition, environmental stresses, and pathogen or viral infections can trigger the production of small RNAs. The transmission of these small RNAs to the next generation can propagate the memory of such changes. This type of small RNA‐mediated inheritance is usually not permanent. Still, it can last for just one or multiple generations based on the perpetuation of the small RNA signal in the descendants. Moreover, small RNAs can also trigger chromatin changes through the deposition of histone modifications. Thus, chromatin pathways can continue to propagate heritable changes in the absence of small RNA inheritance. I focus the following paragraphs on well‐characterized examples of inherited traits by small RNAs documented in the worm, fruit fly, and mouse.
Lessons from worms
The nematode C. elegans is a fertile hermaphrodite; this feature allows producing a large population of isogenic individuals, which minimizes the effect of genetic variations. Furthermore, C. elegans’ short generation time of 3 days facilitates the study of epigenetic inheritance across multiple generations. For these reasons, C. elegans has been extensively used as a model organism to study epigenetic inheritance. Indeed, several cases of transgenerational epigenetic inheritance have been documented, including the inheritance of traits such as infections, stress responses, fertility, and learned behavior.
Small RNAs in heritable responses to viral infection
The heritable response to viral infection is one of the first studied traits linked to small RNAs inheritance [155]. Transgenic worms which express a modified version of a Flock House virus (FHV) fused to a fluorescent reporter under an inducible heat‐shock promoter have been used to stimulate antiviral RNAi responses [155]. The silencing of the fluorescent reporter fused to the FHV occurs across multiple generations in an RNAi‐dependent manner [155]. Inherited primary small RNAs derived from the FHV transgene were also detected, suggesting the possible inheritance of dsRNA‐derived small RNAs [155]. The FHV transgene is present in every cell of the animal, including the germline, where RNAi generally silences every transgene. Thus, it is not possible to untangle virus‐specific heritable RNAi responses from the well‐documented heritable transgene silencing effects [73, 156]. The discovery of the first natural C. elegans virus [157], the Orsay virus (OV), allowed researchers to test whether the resistance to viral infection was heritable through small RNAs [158]. This study shows that the small RNA pathway can interfere with viral replication during OV infection in the intestine. Furthermore, the generation of primary siRNAs and secondary 22G‐RNAs occurs in response to viral infection in the intestine [157]. Nevertheless, neither the primary siRNAs nor the secondary 22G‐RNAs are inherited in the following generation [158]. Moreover, progenies from worms previously exposed to the virus do not show inherited resistance to viral infection [158]. The lack of heritable responses might be the consequence of the fact that the OV exclusively replicates in intestinal cells [157]. Therefore, in the absence of an RNA template in the germline, the small RNAs produced in the intestine cannot be amplified and transmitted to the next generation. To overcome this issue, a fluorescent reporter strain of the vesicular stomatitis virus (VSV) [159], which is known to replicate in C. elegans cell cultures [160], has been microinjected in C. elegans. RNAi deficient mutant worms showed vertical transmission of the virus, and progenies from animals previously exposed to VSV showed reduced infection to VSV in an RNAi‐dependent manner [159]. Thus, RNAi prevents the vertical transmission of VSV, and inherited small RNAs can help to protect progenies from future infections. Nonetheless, small RNAs were not detected in progeny from animals previously exposed to the VSV [159]. Therefore, it remains unclear whether the inherited protection against future infections depends on heritable small RNAs. Since all the system used to investigate the heritable response to viral infection does not represent natural infection, discovering more natural C. elegans viruses will help elucidate whether small RNAs participate in heritable responses to viral infection.
Small RNAs in heritable stress responses
The first study that showed the heritability of small RNAs upon stress responses used starvation during the first larval stage (L1) and small RNA sequencing profiles to detect small RNAs inherited in progenies derived from stressed parents [161]. Among the many loci showing differential gene expression and small RNA changes in adult worms after starvation, a significant percentage showed a similar trend after three generations of growing under normal conditions [161]. Some of these small RNAs are antisense to metabolic genes, suggesting that the inheritance of these starvation‐induced small RNAs might have some physiological functions [161]. The F3 progenies from L1 starved parents displayed an increased life span than the control [161]. However, whether starvation in the early life stage led to the small RNA‐mediated inheritance of stress resistance phenotype has not been evaluated [161]. A careful evaluation of the transgenerational inheritance of stress resistance phenotype in progenies derived from L1 starved parents did not confirm these findings on a population level [162]. Starvation in early life has a detrimental fitness cost in F1 progenies, and only F2 and F3 progenies derived from selected severely affected individuals show increased stress resistance and life span [162]. Thus, only some individuals among the population can inherit stress resistance phenotypes. The fact that the epigenetic inheritance of stress responses does not occur in all individuals renders the experiments based on the whole population difficult to interpret. In addition to starvation, heat‐shock and osmotic stress can also alter the composition of heritable small RNAs and affect the inheritance of RNAi silencing [163]. However, the role of these small RNAs in transmitting heritable stress responses is unclear. The inheritance of stress resistance phenotype in progenies derived from stressed parents has been recently linked to small RNAs [164, 165]. Progenies from adult worms that have experienced a short period of stress, such as starvation, heavy metal, or hyperosmosis, inherit resistance to oxidative stress up to two generations [164]. The inheritance of the stress resistance phenotype depends on the heat shock‐induced transcriptional factor HSF‐1 [164] and RNAi factors, such as germline WAGOs, miRNA biogenesis factors expressed in neurons, and a small RNA transport machinery in the intestine [165]. The link between the transcriptional factor HSF‐1 and the small RNA pathways in transmitting heritable responses has also been observed during RNAi inheritance [58]. Thus, a general mechanism linking stress response pathways and small RNA production might operate in worms to regulate small RNA inheritance in different environmental conditions. Because these heritable stress resistance phenotypes might occur in few animals among the population of worms, the molecular and biochemical understanding of this phenomenon is currently tricky. Strategies to overcome this limitation could be undoubtedly helpful in future studies to elucidate the molecular mechanism of heritable stress responses.
Transgenerational loss of fertility in animals lacking piRNAs
In many animals, a mutation in components of the piRNA pathway causes sterility [166]. But in C. elegans, animals lacking piRNAs are initially fertile and gradually lose their fertility across generations until they become sterile after many generations [80, 167]. This phenomenon, also referred to as mortal germline (Mrt) phenotype [168], may occur upon mutation of components of germ granules, downstream components of the piRNA pathway, histone modifier enzymes, chromatin factors, and even in several C. elegans wild isolates [29, 54, 169, 170]. However, in these cases, the progressive loss of fertility is only induced in animals grown in laboratory conditions at the restrictive temperature of 25 °C. Instead, the sterility of animals lacking piRNAs occurs even at the standard temperature of growth of 20 °C [167]. Given the role of piRNAs in transposon silencing, the sterility observed in animals lacking piRNAs is commonly associated with the derepression of transposons and, consequently, with DNA damage [171]. Microarray analysis in piwi mutant worms has suggested that the progressive desilencing of some transposons and tandem repeats might be the underlying cause of sterility [167]. However, recent transcriptomic and small RNA analyses across multiple generations upon piwi mutation have revealed no correlation between small RNAs and transposon desilencing [80, 82]. In addition, worms with a mutation in the nuclear Argonaute HRDE‐1, the final effector of piRNA‐induced silencing, do not show the same phenotype at 20 °C, even though transposons and other repetitive elements are desilenced [80]. The reversibility of the transgenerational loss of fertility suggests that an epigenetic mechanism rather than DNA mutations might cause the inheritance of this phenotype [54]. One of the proposed causes of sterility is the epigenetic silencing of histone mRNAs which gradually occurs across generations without piRNAs [80]. The silencing of histone mRNAs is post‐transcriptional and mediated by histone 22G‐RNAs loaded in WAGO proteins, such as WAGO‐1, which normally acts downstream of the piRNA pathway [80]. Proteomic analysis has revealed that in the absence of PIWI, the downstream WAGO proteins interacting with PIWI lose their interaction with germ granule components and localize to the cytosol [80]. The loss of WAGOs germ granule localization does not occur immediately upon PIWI mutation, possibly because the interaction between WAGOs and P granule proteins still occurs in the absence of PIWI for a certain number of generations [80]. The removal of factors involved in the biogenesis of 22G‐RNAs is sufficient to remove histone 22G‐RNAs, increase histone mRNA levels, and restore the fertility of piwi mutant almost at wild‐type levels [80]. Similarly, removing some WAGOs downstream of the piRNA pathway contributes to restoring the fertility of piwi mutant worms [80]. More recent studies have revealed another class of genes that also accumulate 22G‐RNAs in the absence of piRNA, the ribosomal RNA genes [81, 172]. However, in the case of the inheritance of 22G‐RNAs antisense to the ribosomal RNAs, called antisense ribosomal siRNAs (risiRNAs), it is not clear whether they interfere with the transcription or the processing of ribosomal RNAs and how they can contribute to the sterility phenotype [173, 174, 175]. The biogenesis of the histone 22G‐RNAs and risiRNAs in the piwi mutant is also different. The histone 22G‐RNAs are triggered by the catalytic activity of the Argonaute CSR‐1 in the cytosol [80], whereas the risiRNAs require Dicer and the nuclear Argonaute HRDE‐1 [81]. Whether the inheritance of two classes of small RNAs causes the transgenerational loss of fertility phenotype remains unclear. Similarly, the transgenerational loss of fertility phenotype revealed at restrictive temperature in other mutants might not be caused by the same accumulation of 22G‐RNAs. In this regard, a recent study has shown that sterile mutant animals that grow at restrictive temperatures have reduced or abnormal germ granules [176]. Thus, germ granules dysfunction might also contribute to the sterility phenotype. This is consistent with the loss of WAGO‐1 germ granule localization in piwi mutant [80]. However, structural germ granule components are still perinuclear localized in the sterile piwi mutant, implying that defects in the protein composition of germ granules might contribute to the sterility phenotype rather than the absence of germ granules [80].
Transgenerational transmission of learned behavior
Behavioral adaptation to odors in the neurons appears to be mediated by endogenous small RNAs [177]. Antisense endo‐siRNAs loaded into the somatic nuclear Argonaute NRDE‐3 in neurons repress odr‐1 mRNAs, a gene whose downregulation is required for adaptation [177]. Components of the somatic nuclear RNAi pathway and the homologue of the H3K9me3 binding protein HP1 are required for olfactory adaptation, suggesting the role of nuclear RNAi and chromatin silencing in behaviors [177]. Small RNAs can also contribute to the inheritance of acquired behavioral traits in neurons [178, 179]. The exposure of worms to the pathogenic bacteria Pseudomonas aeruginosa PA14 induces aversion from PA14 in few hours [180]. Strikingly, the inherited learned aversive behavior occurs for up to four generations [179]. This observation suggests that the acquired behavior is epigenetically transmitted from the neuron to the progenies through the germline. Interestingly, piRNA mutants display a lack of inherited behavior, implying that the exposure of worms to PA14 induces the piRNA‐dependent transcriptional silencing of crucial genes essential for avoidance behavior [179]. Given that the piRNA pathway operates in the germline, some other factors should participate in transmitting some signals from the neurons to the germline or vice versa. In a follow‐up study, the same team discovered how PA14 triggered the pathogen avoidance behavior and showed the small RNA and piRNA pathways requirement in this process [181]. They found a small noncoding RNA (ncRNA) produced by PA14 that is sufficient to trigger the transgenerational pathogen avoidance phenotype [181]. Dicer process the bacteria's small ncRNAs in the intestine, and small RNAs can target an endogenous gene, maco‐1, usually downregulated in the neuron upon PA14 exposure [181]. The germline piRNA pathway is somehow crucial for this regulation [181]. However, it is still unclear how the bacterial small RNAs in the intestine trigger the piRNA silencing in the germline and how the piRNAs signal such regulation to neurons. Another study has shown how the role of endo‐siRNAs in neurons and piRNAs in the germline is involved in the inheritance of chemotaxis behavior [178]. A mutant of the double‐stranded RNA binding protein RDE‐4, which acts together with Dicer to generate siRNAs from dsRNA [182], complemented with a neuronal‐specific expression of RDE‐4, has been used to identify RDE‐4‐dependent neuronal 22G‐RNAs as well as germline 22G‐RNAs that depends on neuronal RDE‐4 activity [178]. The neuronal expression of RDE‐4 appears to be responsible for more than a thousand genes with altered 22G‐RNAs in the germline [178]. Some of these genes continue to produce altered 22G‐RNAs three generations after, suggesting that the neuronal RDE‐4 activity might trigger transgenerational gene expression changes through germline 22G‐RNAs [178]. The repression of one of these genes appears to be partially required to restore chemotaxis defects displayed by the rde‐4 mutant [178]. Moreover, rde‐4 mutant worms derived from parents with neuronal expression of RDE‐4 show ameliorated chemotaxis defects up to three generations [178]. Thus, the neuronal activity of RDE‐4 might participate in heritable neuronal phenotype through the RNA silencing of germline genes. Whether neuronal small RNAs are transferred to the germline or some other neuronal signals, elicit the biogenesis of germline 22G‐RNAs remains to be determined. Despite the lack of detailed mechanistic understanding, these studies have established that some sort of intertissues or organs communication of endogenous small RNAs can trigger transgenerational inheritance of acquired traits through the regulation of gene expression.
Cases of epigenetic inheritance in the fruit fly
Transgenerational epigenetic inheritance of stress response and nutrition also occurs in D. melanogaster [183]. In most cases, heterochromatin factors and polycomb group proteins change the chromatin states of different loci involved in the transmission of traits [183]. However, it is still unclear whether the small RNA pathways may contribute to the inheritance of such environmental‐induced heritable traits. Progenies from Drosophila exposed to RNA virus show multigenerational protection from viral infection [184]. The protection against novel infections persists for up to five generations. However, the RNAi pathway is not involved in this process [184]. Moreover, small RNAs are also undetectable in progenies derived from parents infected with RNA viruses. Thus, in contrast to C. elegans, Drosophila might rely on chromatin pathways rather than on small RNAs to transmit such traits [2]. Yet, small RNAs might be involved in initiating a signal in the parental generation, which is then maintained and propagated by chromatin pathways in following unstimulated generations. The only well‐documented case of small RNA‐mediated inheritance of traits in Drosophila is the protection from transposon silencing mediated by inherited piRNAs [91, 98]. As mentioned above, the silencing of transposons can be inherited through maternally provided piRNAs in a chromatin‐independent manner to propagate a robust silencing response in the progeny and protect them from potential deleterious invasive genomic elements [98].
Paternal inheritance of traits in mouse
Paternal inheritance, possibly via RNA transfer from sperm, has been documented in mice for traits such as diet‐induced metabolic disorders [185, 186], trauma symptoms [111], stress [110, 112], and behavior disorders [113]. Even if small RNAs derived from tRNA fragments (tRFs) appear to participate in the inheritance of diet‐induced metabolic disorders, whether Argonautes load these tRFs to promote gene repression remains unclear [185, 186]. The majority of tRFs are produced in a dicer‐independent manner, and only some of them appear to be loaded into Argonaute proteins [187]. Thus, whether the paternal inheritance of tRFs requires the canonical RNAi pathway to regulate gene expression in progenies should be determined. The mouse model of unpredictable maternal separation combined with unpredictable maternal stress (MSUS) revealed the role of small RNAs in early traumatic stress inheritance [111]. The inheritance of behavioral response to MSUS correlates with increased expression of some miRNAs in sperm of F1 animals [111]. Some of these miRNAs were also increased in the hippocampus of F1 and F2 animals, suggesting that they might directly regulate genes necessary for the inherited behavior [111]. Despite the inheritance of MSUS‐induced behavior up to F3 animals [188], these miRNAs were not upregulated in the sperm of F2 animals [111]. Thus, how miRNAs in sperm can influence the expression of the same miRNAs in the hippocampus remains unclear. Moreover, it remains undetermined whether the sperm transmit upregulated miRNAs to the fertilized oocyte. Some other molecules might trigger the expression of these miRNAs in the progenies. The inheritance of MSUS‐induced behavior in F3 animals occurs even if sperm miRNAs are unchanged in F2 animals, suggesting that different mechanisms, such as the inheritance of DNA methylation, might also play a role in such heritable traumatic‐induced behaviors [188].
The exposure of male mice to chronic stress before breeding also leads to the inheritance of hypothalamic–pituitary–adrenal (HPA) axis dysregulation in adult F1 mice [110]. In this study, nine miRNAs have resulted in being upregulated in sperm upon chronic stress, suggesting they can transmit the inheritance of stress responses [110]. The injection of the nine miRNAs into single‐cell zygotes recapitulates the paternal stress phenotype in adult mice [112]. Gene expression analysis 24 h postinjection has revealed the downregulation of some maternal mRNAs with known developmental functions and putatively targeting by the nine miRNAs [112]. However, the maternal mRNA targets remained unchanged in adult mice brains, indicating that the regulation of these mRNA targets occurs in the early zygote [112]. Moreover, some of these maternal mRNA targets belong to chromatin regulators [112]. Thus, the proposed model for this case of paternal transmission of stress is that the sperm transmit an altered miRNA cargo to the embryo, where they can silence maternal mRNAs essential for developing the stress‐related phenotype. Furthermore, some putative mRNA targets encode for chromatin regulators, so the paternal‐inherited miRNAs in embryos can indirectly affect chromatin changes. Consequently, epigenetic chromatin changes might impact adult brains' development and transmit the phenotype in the following generation.
Behavioral disorders, such as depression‐like phenotype, can also be transmitted to F1 progenies from stressed male sperm [113]. The injection of purified small RNAs from stressed male sperm recapitulates the depression‐like phenotype. The sequencing analysis has revealed the upregulation of miRNAs among other types of small RNAs [113]. The microinjection in normal zygotes of the 16 more abundant miRNAs in sperm upon stress is sufficient to induce the depression‐like phenotype [113]. However, contrary to the other inherited phenotypes described in the other studies, the depression‐like phenotype is not transmitted to the F2.
All these studies suggest that environmental stresses can alter the composition of small RNAs, particularly miRNAs, in mature sperm and that the transfer of these miRNAs in progenies contributes to the transmission of phenotypes associated with the stress response (Fig. 3). However, given that these studies rely on the microinjection of small RNAs, it would be necessary for the future to establish whether the cargo of small RNAs delivered by sperm during normal mating is sufficient to transmit such phenotypes.
Fig. 3.
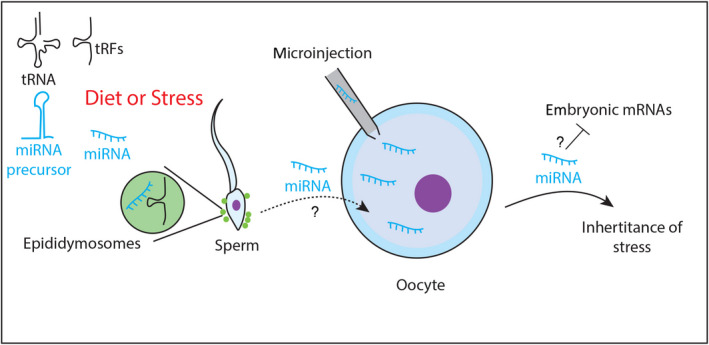
Paternal inheritance of miRNAs in the mouse. Metabolic changes or different types of stress alter the small RNA cargoes loaded into mature mouse sperm. Diet‐induced tRNA fragmentation results in high production of tRFs loaded into mature sperm through epididymosomes vesicles. These vesicles might also transport miRNAs induced by stress. The microinjection of miRNAs extracted from mature sperm produced by animals subjected to stresses recapitulates the stress‐induced phenotypes in subsequent generations. It is unclear whether the miRNA cargoes loaded into mature sperm can effectively regulate embryonic mRNAs after fertilization.
Conclusions and perspectives
The elucidation of the transgenerational phenomenon of RNAi in C. elegans has been instrumental in understanding small RNA‐based heritable traits in animal model organisms. However, many steps in the mechanisms of RNAi remain unclear. For instance, how the inherited small RNAs induce the de novo production of small RNAs in embryos, the role of germ granules in this process, and how nuclear small RNA pathways trigger the transcriptional silencing of their target genes. In addition, while the crosstalk between nuclear small RNAs and histone PTMs is well‐known, future work will need to explore the requirement of histone PTMs in heritable RNAi responses and whether histone PTMs can maintain the initiation of heritable RNAi by small RNAs in a small RNA‐independent manner. Moreover, it is still puzzling how chromatin factors and histone PTMs influence the duration of heritable RNAi.
Endogenous germline small RNA pathways in C. elegans are fundamental to incorporate somatic signals that acquire heritable phenotypes. The mechanisms underlying this soma to germline RNA communication are still unclear. A key feature of C. elegans endo‐siRNAs is the ability to be replicated and amplified by RdRP. Indeed, long transgenerational effects of small RNAs are only clearly reported for C. elegans endo‐siRNAs. However, it is unclear how animals lacking RdRP enzymes, such as Drosophila and the mouse, can replicate and amplify endo‐siRNAs across multiple generations. Endo‐siRNAs can potentially act directly in embryos, where they regulate essential genes that can influence animal development. But, it is difficult to predict how such endo‐siRNAs can propagate epigenetic traits across multiple generations without the support of an amplification mechanism. One possibility is the direct involvement of endo‐siRNAs in nuclear processes and histone PTMs, a role that is still largely uncharacterized. Heritable miRNAs also lack an amplification mechanism. Nonetheless, their epigenetic effects, such as the transmission of stresses, have been documented for more than one generation. How the low quantity of miRNAs delivered from sperm can exert gene regulatory functions in embryos, and whether the paternally transmitted miRNAs persist in adult mice are still unclear. Paternally transmitted miRNAs might have a positive feedback regulation on their own expression or regulate genes in the embryo, which in turn are essential for tissue‐specific expression of paternally transmitted miRNAs in adults. In addition to endo‐siRNAs, piRNAs can also play a transgenerational function in repressing repetitive elements, such as transposons, in the nucleus. Moreover, since a piRNA amplification mechanism is known, it can propagate piRNAs across multiple generations. The crosstalk between piRNAs and histone PTMs or DNA methylation can also propagate the piRNA silencing signal in a piRNA‐independent manner. Whether their primary function in repressing repetitive elements is coupled with other developmental processes remains to be determined. Given that not all the piRNAs map to transposable elements [62], they could exert similar epigenetic functions to control the expression of protein‐coding genes.
The potential capacity of small RNAs to acquire information from different tissues and transmit it across generations would revolutionize biological research and medicine. Therefore, identifying the endogenous RNAs and the mechanisms involved in the exchange of information from the soma to the germline, and ultimately to the next generation, could radically change our notion of inheritance. Research on laboratory model organisms remains crucial to elucidate the molecular mechanisms underlying transgenerational phenomena. However, given the importance of environmental cues in epigenetic inheritance, it would be necessary to expand our understanding of epigenetics from laboratory animal models toward organisms in their natural environment. Novel approaches to studying the wild population of individuals in different environmental settings and their interaction with other organisms will significantly contribute to understanding epigenetic inheritance and its role in animal evolution and traits heritability.
Acknowledgements
I would like to thank Dr. Meetali Singh and Dr. Nicola Iovino for their comments on the manuscript. GC has received funding from the Institut Pasteur, the CNRS, and the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement No. 679243 and No. 101002999).
Edited by Tamas Dalmay
References
- 1. Miska EA and Ferguson‐Smith AC (2016) Transgenerational inheritance: models and mechanisms of non–DNA sequence–based inheritance. Science 354, 59–63. [DOI] [PubMed] [Google Scholar]
- 2. Skvortsova K, Iovino N and Bogdanović O (2018) Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol 19, 774–790. [DOI] [PubMed] [Google Scholar]
- 3. Chen T and Dent SYR (2014) Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet 15, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parry A, Rulands S and Reik W (2021) Active turnover of DNA methylation during cell fate decisions. Nat Rev Genet 22, 59–66. [DOI] [PubMed] [Google Scholar]
- 5. Atlasi Y and Stunnenberg HG (2017) The interplay of epigenetic marks during stem cell differentiation and development. Nat Rev Genet 18, 643–658. [DOI] [PubMed] [Google Scholar]
- 6. Talbert PB, Meers MP and Henikoff S (2019) Old cogs, new tricks: the evolution of gene expression in a chromatin context. Nat Rev Genet 20, 283–297. [DOI] [PubMed] [Google Scholar]
- 7. Holoch D and Moazed D (2015) RNA‐mediated epigenetic regulation of gene expression. Nat Rev Genet 16, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song J‐J, Hammond SM, Joshua‐Tor L and Hannon GJ (2004) Argonaute 2 is the catalytic engine of mammalian RNAi. Science 305(5689), 1437–1441. [DOI] [PubMed] [Google Scholar]
- 9. Song J‐J, Smith SK, Hannon GJ and Joshua‐Tor L (2004) Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437. [DOI] [PubMed] [Google Scholar]
- 10. Feil R and Fraga MF (2012) Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet 13, 97–109. [DOI] [PubMed] [Google Scholar]
- 11. Perez MF and Lehner B (2019) Intergenerational and transgenerational epigenetic inheritance in animals. Nat Cell Biol 21, 143–151. [DOI] [PubMed] [Google Scholar]
- 12. Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA and Surani MA (2013) Germline DNA demethylation dynamics and imprint erasure through 5‐hydroxymethylcytosine. Science 339, 448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaydos LJ, Wang W and Strome S (2014) H3K27me and PRC2 transmit a memory of repression across generations and during development. Science 345(6203), 1515–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Samata M, Alexiadis A, Richard G, Georgiev P, Nuebler J, Kulkarni T, Renschler G, Basilicata MF, Zenk FL, Shvedunova M et al. (2020) Intergenerationally maintained histone H4 lysine 16 acetylation is instructive for future gene activation. Cell 182, 127–144.e23. [DOI] [PubMed] [Google Scholar]
- 15. Zenk F, Loeser E, Schiavo R, Kilpert F, Bogdanović O and Iovino N (2017) Germ line–inherited H3K27me3 restricts enhancer function during maternal‐to‐zygotic transition. Science 357, 212–216. [DOI] [PubMed] [Google Scholar]
- 16. Bond DM and Baulcombe DC (2014) Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol 24, 100–107. [DOI] [PubMed] [Google Scholar]
- 17. Heard E and Martienssen RA (2014) Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE and Mello CC (1998) Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans . Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 19. Grishok A, Tabara H and Mello CC (2000) Genetic requirements for inheritance of RNAi in C. elegans . Science 287, 2494–2497. [DOI] [PubMed] [Google Scholar]
- 20. Tabara H, Yigit E, Siomi H and Mello CC (2002) The dsRNA binding protein RDE‐4 interacts with RDE‐1, DCR‐1, and a DExH‐Box‐helicase to direct RNAi in C. elegans . Cell 109(7), 861–871. [DOI] [PubMed] [Google Scholar]
- 21. Yigit E, Batista PJ, Bei Y, Pang KM, Chen C‐CG, Tolia NH, Joshua‐Tor L, Mitani S, Simard MJ and Mello CC (2006) Analysis of the C. elegans argonaute family reveals that distinct argonautes act sequentially during RNAi. Cell 127, 747–757. [DOI] [PubMed] [Google Scholar]
- 22. Steiner FA, Okihara KL, Hoogstrate SW, Sijen T and Ketting RF (2009) RDE‐1 slicer activity is required only for passenger‐strand cleavage during RNAi in Caenorhabditis elegans . Nat Struct Mol Biol 16, 207–211. [DOI] [PubMed] [Google Scholar]
- 23. Tsai H‐Y, Chen C‐CG, Conte D, Moresco JJ, Chaves DA, Mitani S, Yates JR, Tsai M‐D and Mello CC (2015) A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell 160, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pak J and Fire A (2007) Distinct populations of primary and secondary effectors during RNAi in C. elegans . Science 315, 241–244. [DOI] [PubMed] [Google Scholar]
- 25. Sijen T, Steiner FA, Thijssen KL and Plasterk RHA (2007) Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315, 244–247. [DOI] [PubMed] [Google Scholar]
- 26. Gu W, Shirayama M, Conte D, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ et al. (2009) Distinct argonaute‐mediated 22G‐RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36, 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shukla A, Yan J, Pagano DJ, Dodson AE, Fei Y, Gorham J, Seidman JG, Wickens M and Kennedy S (2020) poly(UG)‐tailed RNAs in genome protection and epigenetic inheritance. Nature 582, 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gu SG, Pak J, Guang S, Maniar JM, Kennedy S and Fire A (2012) Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence‐targeted histone H3 lysine 9 methylation footprint. Nat Genet 44, 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A and Kennedy S (2012) A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489, 447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Youngman EM and Claycomb JM (2014) From early lessons to new frontiers: the worm as a treasure trove of small RNA biology. Front Genet 5, 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM and Kennedy S (2010) Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465(7301), 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J and Kennedy S (2008) An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science 321, 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mao H, Zhu C, Zong D, Weng C, Yang X, Huang H, Liu D, Feng X and Guang S (2015) The Nrde pathway mediates small‐RNA‐directed histone H3 lysine 27 trimethylation in Caenorhabditis elegans . Curr Biol 25, 2398–2403. [DOI] [PubMed] [Google Scholar]
- 34. Schwartz‐Orbach L, Zhang C, Sidoli S, Amin R, Kaur D, Zhebrun A, Ni J and Gu SG (2020) Caenorhabditis elegans nuclear RNAi factor SET‐32 deposits the transgenerational histone modification, H3K23me3. eLife 9, e54309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Akay A, Domenico TD, Suen KM, Nabih A, Parada GE, Larance M, Medhi R, Berkyurek AC, Zhang X, Wedeles CJ et al. (2017) The helicase aquarius/EMB‐4 is required to overcome intronic barriers to allow nuclear RNAi pathways to heritably silence transcription. Dev Cell 42, 241–255.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duempelmann L, Mohn F, Shimada Y, Oberti D, Andriollo A, Lochs S and Bühler M (2019) Inheritance of a phenotypically neutral epimutation evokes gene silencing in later generations. Mol Cell 74, 534–541.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burton NO, Burkhart KB and Kennedy S (2011) Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans . Proc Natl Acad Sci USA 108, 19683–19688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vastenhouw NL, Brunschwig K, Okihara KL, Müller F, Tijsterman M and Plasterk RHA (2006) Long‐term gene silencing by RNAi. Nature 442, 882. [DOI] [PubMed] [Google Scholar]
- 39. Winston WM, Sutherlin M, Wright AJ, Feinberg EH and Hunter CP (2007) Caenorhabditis elegans SID‐2 is required for environmental RNA interference. Proc Natl Acad Sci USA 104, 10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McEwan DL, Weisman AS and Hunter CP (2012) Uptake of extracellular double‐stranded RNA by SID‐2. Mol Cell 47(5), 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Feinberg EH and Hunter CP (2003) Transport of dsRNA into cells by the transmembrane protein SID‐1. Science 301, 1545–1547. [DOI] [PubMed] [Google Scholar]
- 42. Hinas A, Wright AJ and Hunter CP (2012) SID‐5 is an endosome‐associated protein required for efficient systemic RNAi in C. elegans . Curr Biol 22, 1938–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marré J, Traver EC and Jose AM (2016) Extracellular RNA is transported from one generation to the next in Caenorhabditis elegans . Proc Natl Acad Sci USA 113, 12496–12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang E and Hunter CP (2017) SID‐1 functions in multiple roles to support parental RNAi in Caenorhabditis elegans . Genetics 207, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dodson AE and Kennedy S (2019) Germ granules coordinate RNA‐based epigenetic inheritance pathways. Dev Cell 50, 704–715.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ouyang JPT, Folkmann A, Bernard L, Lee C‐Y, Seroussi U, Charlesworth AG, Claycomb JM and Seydoux G (2019) P granules protect RNA interference genes from silencing by piRNAs. Dev Cell 50, 716–728.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lev I, Toker IA, Mor Y, Nitzan A, Weintraub G, Antonova O, Bhonkar O, Shushan IB, Seroussi U, Claycomb JM et al. (2019) Germ granules govern small RNA inheritance. Curr Biol 29, 2880–2891.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pitt JN, Schisa JA and Priess JR (2000) P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol 219, 315–333. [DOI] [PubMed] [Google Scholar]
- 49. Strome S and Wood WB (1982) Immunofluorescence visualization of germ‐line‐specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans . Proc Natl Acad Sci USA 79, 1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF et al. (2009) The Argonaute CSR‐1 and its 22G‐RNA cofactors are required for holocentric chromosome segregation. Cell 139, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Phillips CM, Montgomery TA, Breen PC and Ruvkun G (2012) MUT‐16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Gene Dev 26, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wan G, Fields BD, Spracklin G, Shukla A, Phillips CM and Kennedy S (2018) Spatiotemporal regulation of liquid‐like condensates in epigenetic inheritance. Nature 557, 679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Singh M, Cornes E, Li B, Quarato P, Bourdon L, Dingli F, Loew D, Proccacia S and Cecere G (2021) Translation and codon usage regulate Argonaute slicer activity to trigger small RNA biogenesis. Nat Commun 12, 3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Spracklin G, Fields B, Wan G, Becker D, Wallig A, Shukla A and Kennedy S (2017) The RNAi inheritance machinery of Caenorhabditis elegans . Genetics 206, 1403–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lev I, Seroussi U, Gingold H, Bril R, Anava S and Rechavi O (2017) MET‐2‐dependent H3K9 methylation suppresses transgenerational small RNA inheritance. Curr Biol 27, 1138–1147. [DOI] [PubMed] [Google Scholar]
- 56. Perales R, Pagano D, Wan G, Fields BD, Saltzman AL and Kennedy SG (2018) Transgenerational epigenetic inheritance is negatively regulated by the HERI‐1 chromodomain protein. Genetics 210, 1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shukla A, Perales R and Kennedy S (2021) piRNAs coordinate poly(UG) tailing to prevent aberrant and perpetual gene silencing. Curr Biol 31(20), 4473–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Houri‐Zeevi L, Kohanim YK, Antonova O and Rechavi O (2020) Three rules explain transgenerational small RNA inheritance in C. elegans . Cell 182, 1186–1197.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aljohani MD, Mouridi SE, Priyadarshini M, Vargas‐Velazquez AM and Frøkjær‐Jensen C (2020) Engineering rules that minimize germline silencing of transgenes in simple extrachromosomal arrays in C. elegans . Nat Commun 11, 6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Devanapally S, Raman P, Chey M, Allgood S, Ettefa F, Diop M, Lin Y, Cho YE and Jose AM (2021) Mating can initiate stable RNA silencing that overcomes epigenetic recovery. Nat Commun 12, 4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM and Gvozdev VA (2001) Double‐stranded RNA‐mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11, 1017–1027. [DOI] [PubMed] [Google Scholar]
- 62. Ozata DM, Gainetdinov I, Zoch A, O’Carroll D and Zamore PD (2019) PIWI‐interacting RNAs: small RNAs with big functions. Nat Rev Genet 20, 89–108. [DOI] [PubMed] [Google Scholar]
- 63. Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R and Hannon GJ (2007) Discrete small RNA‐generating loci as master regulators of transposon activity in Drosophila . Cell 128, 1089–1103. [DOI] [PubMed] [Google Scholar]
- 64. Girard A, Sachidanandam R, Hannon GJ and Carmell MA (2006) A germline‐specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202. [DOI] [PubMed] [Google Scholar]
- 65. Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T and Hannon GJ (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31, 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Aravin AA, Sachidanandam R, Girard A, Fejes‐Toth K and Hannon GJ (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747. [DOI] [PubMed] [Google Scholar]
- 67. Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H and Bartel DP (2006) Large‐scale sequencing reveals 21U‐RNAs and additional microRNAs and endogenous siRNAs in C. elegans . Cell 127, 1193–1207. [DOI] [PubMed] [Google Scholar]
- 68. Cecere G, Zheng GXY, Mansisidor AR, Klymko KE and Grishok A (2012) Promoters recognized by forkhead proteins exist for individual 21U‐RNAs. Mol Cell 47, 734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gu W, Lee H‐C, Chaves D, Youngman EM, Pazour GJ, Conte D and Mello CC (2012) CapSeq and CIP‐TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell 151, 1488–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sienski G, Dönertas D and Brennecke J (2012) Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151, 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Thomas AL, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA and Tóth KF (2013) Piwi induces piRNA‐guided transcriptional silencing and establishment of a repressive chromatin state. Gene Dev 27(4), 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rozhkov NV, Hammell M and Hannon GJ (2013) Multiple roles for Piwi in silencing Drosophila transposons. Gene Dev 27, 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shirayama M, Seth M, Lee H‐C, Gu W, Ishidate T, Conte D and Mello CC (2012) piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chen P, Kotov AA, Godneeva BK, Bazylev SS, Olenina LV and Aravin AA (2021) piRNA‐mediated gene regulation and adaptation to sex‐specific transposon expression in D.melanogaster male germline. Gene Dev 35(11‐12), 914–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Goh WSS, Falciatori I, Tam OH, Burgess R, Meikar O, Kotaja N, Hammell M and Hannon GJ (2015) piRNA‐directed cleavage of meiotic transcripts regulates spermatogenesis. Gene Dev 29, 1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu P‐H, Fu Y, Cecchini K, Özata DM, Arif A, Yu T, Colpan C, Gainetdinov I, Weng Z and Zamore PD (2020) The evolutionarily conserved piRNA‐producing locus pi6 is required for male mouse fertility. Nat Genet 52, 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Taborska E, Pasulka J, Malik R, Horvat F, Jenickova I, Matošević ZJ and Svoboda P (2019) Restricted and non‐essential redundancy of RNAi and piRNA pathways in mouse oocytes. Plos Genet 15, e1008261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S et al. (2008) PRG‐1 and 21U‐RNAs interact to form the piRNA complex required for fertility in C. elegans . Mol Cell 31, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bagijn MP, Goldstein LD, Sapetschnig A, Weick E‐M, Bouasker S, Lehrbach NJ, Simard MJ and Miska EA (2012) Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Barucci G, Cornes E, Singh M, Li B, Ugolini M, Samolygo A, Didier C, Dingli F, Loew D, Quarato P et al. (2020) Small‐RNA‐mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat Cell Biol 22, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wahba L, Hansen L and Fire AZ (2021) An essential role for the piRNA pathway in regulating the ribosomal RNA pool in C. elegans . Dev Cell 56, 2295–2312.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Reed KJ, Svendsen JM, Brown KC, Montgomery BE, Marks TN, Vijayasarathy T, Parker DM, Nishimura EO, Updike DL and Montgomery TA (2019) Widespread roles for piRNAs and WAGO‐class siRNAs in shaping the germline transcriptome of Caenorhabditis elegans . Nucleic Acids Res 48(4), 1811–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ashe A, Sapetschnig A, Weick E‐M, Mitchell J, Bagijn MP, Cording AC, Doebley A‐L, Goldstein LD, Lehrbach NJ, Le Pen J et al. (2012) piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans . Cell 150, 88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lee H‐C, Gu W, Shirayama M, Youngman E, Conte D and Mello CC (2012) C. elegans piRNAs mediate the genome‐wide surveillance of germline transcripts. Cell 150, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shen E‐Z, Chen H, Ozturk AR, Tu S, Shirayama M, Tang W, Ding Y‐H, Dai S‐Y, Weng Z and Mello CC (2018) Identification of piRNA binding sites reveals the argonaute regulatory landscape of the C. elegans germline. Cell 172, 937–951.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tang W, Seth M, Tu S, Shen E‐Z, Li Q, Shirayama M, Weng Z and Mello CC (2018) A sex chromosome piRNA promotes robust dosage compensation and sex determination in C. elegans . Dev Cell 44, 762–770.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, Ishihara G, Kawaoka S, Sugano S, Shimada T et al. (2014) A single female‐specific piRNA is the primary determiner of sex in the silkworm. Nature 509, 633–636. [DOI] [PubMed] [Google Scholar]
- 88. Akkouche A, Mugat B, Barckmann B, Varela‐Chavez C, Li B, Raffel R, Pélisson A and Chambeyron S (2017) Piwi is required during Drosophila embryogenesis to license dual‐strand piRNA clusters for transposon repression in adult ovaries. Mol Cell 66, 411–419.e4. [DOI] [PubMed] [Google Scholar]
- 89. de Albuquerque BFM, Placentino M and Ketting RF (2015) Maternal piRNAs are essential for germline development following de novo establishment of endo‐siRNAs in Caenorhabditis elegans . Dev Cell 34, 448–456. [DOI] [PubMed] [Google Scholar]
- 90. Teixeira FK, Okuniewska M, Malone CD, Coux R‐X, Rio DC and Lehmann R (2017) piRNA‐mediated regulation of transposon alternative splicing in the soma and germ line. Nature 552, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A and Hannon GJ (2008) An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322, 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Malone CD, Lehmann R and Teixeira FK (2015) The cellular basis of hybrid dysgenesis and Stellate regulation in Drosophila . Curr Opin Genet Dev 34, 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ryazansky S, Radion E, Mironova A, Akulenko N, Abramov Y, Morgunova V, Kordyukova MY, Olovnikov I and Kalmykova A (2017) Natural variation of piRNA expression affects immunity to transposable elements. PLoS Genet 13, e1006731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fabry MH, Falconio FA, Joud F, Lythgoe EK, Czech B and Hannon GJ (2021) Maternally inherited piRNAs direct transient heterochromatin formation at active transposons during early Drosophila embryogenesis. eLife 10, e68573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Rouget C, Papin C, Boureux A, Meunier A‐C, Franco B, Robine N, Lai EC, Pelisson A and Simonelig M (2010) Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467, 1128–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Halbach R, Miesen P, Joosten J, Taşköprü E, Rondeel I, Pennings B, Vogels CBF, Merkling SH, Koenraadt CJ, Lambrechts L et al. (2020) A satellite repeat‐derived piRNA controls embryonic development of Aedes. Nature 580, 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Czech B and Hannon GJ (2016) One loop to rule them all: the ping‐pong cycle and piRNA‐guided silencing. Trends Biochem Sci 41, 324–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Grentzinger T, Armenise C, Brun C, Mugat B, Serrano V, Pelisson A and Chambeyron S (2012) piRNA‐mediated transgenerational inheritance of an acquired trait. Genome Res 22, 1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang D, Tu S, Stubna M, Wu W‐S, Huang W‐C, Weng Z and Lee H‐C (2018) The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 359, 587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sapetschnig A, Sarkies P, Lehrbach NJ and Miska EA (2015) Tertiary siRNAs mediate paramutation in C. elegans . PLoS Genet 11, e1005078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bartel DP (2018) Metazoan microRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lee RC, Feinbaum RL and Ambros V (1993) The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- 103. Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G and Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34. [DOI] [PubMed] [Google Scholar]
- 104. Sharma U (2019) Paternal contributions to offspring health: role of sperm small RNAs in intergenerational transmission of epigenetic information. Front Cell Dev Biol 7, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Svoboda P and Flemr M (2010) The role of miRNAs and endogenous siRNAs in maternal‐to‐zygotic reprogramming and the establishment of pluripotency. EMBO Rep 11, 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P and Schultz RM (2010) microRNA activity is suppressed in mouse oocytes. Curr Biol 20, 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J and Blelloch R (2010) microRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol 20, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lee M, Choi Y, Kim K, Jin H, Lim J, Nguyen TA, Yang J, Jeong M, Giraldez AJ, Yang H et al. (2014) Adenylation of maternally inherited microRNAs by Wispy. Mol Cell 56, 696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Conine CC, Sun F, Song L, Rivera‐Pérez JA and Rando OJ (2018) Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev Cell 46, 470–480.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rodgers AB, Morgan CP, Bronson SL, Revello S and Bale TL (2013) Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation. J Neurosci 33, 9003–9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E and Mansuy IM (2014) Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 17, 667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Rodgers AB, Morgan CP, Leu NA and Bale TL (2015) Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc Natl Acad Sci USA 112, 13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang Y, Chen Z‐P, Hu H, Lei J, Zhou Z, Yao B, Chen L, Liang G, Zhan S, Zhu X et al. (2021) Sperm microRNAs confer depression susceptibility to offspring. Sci Adv 7, eabd7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wang Y, Yamauchi Y, Wang Z, Zheng H, Yanagimachi R, Ward MA and Yan W (2020) Both cauda and caput epididymal sperm are capable of supporting full‐term development in FVB and CD‐1 mice. Dev Cell 55, 675–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Conine CC, Sun F, Song L, Rivera‐Pérez JA and Rando OJ (2020) Sperm head preparation and genetic background affect caput sperm ICSI embryo viability: cauda‐enriched miRNAs only essential in specific conditions. Dev Cell 55, 677–678. [DOI] [PubMed] [Google Scholar]
- 116. Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL and Rando OJ (2018) Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev Cell 46, 481–494.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yang Q, Lin J, Liu M, Li R, Tian B, Zhang X, Xu B, Liu M, Zhang X, Li Y et al. (2016) Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci Adv 2, e1501482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Hwang H‐W, Wentzel EA and Mendell JT (2007) A Hexanucleotide element directs microRNA nuclear import. Science 315, 97–100. [DOI] [PubMed] [Google Scholar]
- 119. Liao J‐Y, Ma L‐M, Guo Y‐H, Zhang Y‐C, Zhou H, Shao P, Chen Y‐Q and Qu L‐H (2010) Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One 5, e10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Turunen TA, Roberts TC, Laitinen P, Väänänen M‐A, Korhonen P, Malm T, Ylä‐Herttuala S and Turunen MP (2019) Changes in nuclear and cytoplasmic microRNA distribution in response to hypoxic stress. Sci Rep 9, 10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Gagnon KT, Li L, Chu Y, Janowski BA and Corey DR (2014) RNAi factors are present and active in human cell nuclei. Cell Rep 6, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Roberts TC (2014) The microRNA biology of the mammalian nucleus. Mol Ther Nucleic Acids 3, e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Corrêa RL, Steiner FA, Berezikov E and Ketting RF (2010) microRNA–directed siRNA biogenesis in Caenorhabditis elegans . PLoS Genet 6, e1000903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ding S‐W and Voinnet O (2007) Antiviral Immunity Directed by Small RNAs. Cell 130, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Maillard PV, van der Veen AG , Poirier EZ and Reis e Sousa C (2019) Slicing and dicing viruses: antiviral RNA interference in mammals. EMBO J 38, e100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC and Siomi H (2008) Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453, 793–797. [DOI] [PubMed] [Google Scholar]
- 127. Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler ELW, Zapp ML, Weng Z et al. (2008) Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320, 1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R et al. (2008) An endogenous small interfering RNA pathway in Drosophila . Nature 453, 798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Okamura K, Chung W‐J, Ruby JG, Guo H, Bartel DP and Lai EC (2008) The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453, 803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Okamura K, Balla S, Martin R, Liu N and Lai EC (2008) Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster . Nat Struct Mol Biol 15, 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Stein P, Rozhkov NV, Li F, Cárdenas FL, Davydenko O, Davydenk O, Vandivier LE, Gregory BD, Hannon GJ and Schultz RM (2015) Essential role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet 11, e1005013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Lykke‐Andersen K, Gilchrist MJ, Grabarek JB, Das P, Miska E and Zernicka‐Goetz M (2008) Maternal Argonaute 2 is essential for early mouse development at the maternal‐zygotic transition. Mol Biol Cell 19, 4383–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi‐Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T et al. (2008) Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543. [DOI] [PubMed] [Google Scholar]
- 134. Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM et al. (2008) Pseudogene‐derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453, 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Babiarz JE, Ruby JG, Wang Y, Bartel DP and Blelloch R (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor‐independent, dicer‐dependent small RNAs. Gene Dev 22, 2773–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Zhang J‐M, Hou W‐B, Du J‐W, Zong M, Zheng K‐L, Wang W‐J, Wang J‐Q, Zhang H, Mu Y‐S, Yin Z et al. (2020) Argonaute 2 is a key regulator of maternal mRNA degradation in mouse early embryos. Cell Death Discov 6, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Berrens RV, Andrews S, Spensberger D, Santos F, Dean W, Gould P, Sharif J, Olova N, Chandra T, Koseki H et al. (2017) An endosiRNA‐based repression mechanism counteracts transposon activation during global DNA demethylation in embryonic stem cells. Cell Stem Cell 21, 694–703.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Song R, Hennig GW, Wu Q, Jose C, Zheng H and Yan W (2011) Male germ cells express abundant endogenous siRNAs. Proc Natl Acad Sci USA 108, 13159–13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Welker NC, Pavelec DM, Nix DA, Duchaine TF, Kennedy S and Bass BL (2010) Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 16, 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Thivierge C, Makil N, Flamand M, Vasale JJ, Mello CC, Wohlschlegel J, Conte D and Duchaine TF (2012) Tudor domain ERI‐5 tethers an RNA‐dependent RNA polymerase to DCR‐1 to potentiate endo‐RNAi. Nat Struct Mol Biol 19, 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Chaves DA, Dai H, Li L, Moresco JJ, Oh ME, Conte D, Yates JR, Mello CC and Gu W (2021) The RNA phosphatase PIR‐1 regulates endogenous small RNA pathways in C. elegans . Mol Cell 81, 546–557.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, Duchaine TF, Mello CC and Conte D (2010) Sequential rounds of RNA‐dependent RNA transcription drive endogenous small‐RNA biogenesis in the ERGO‐1/Argonaute pathway. Proc Natl Acad Sci USA 107, 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M and Mello CC (2010) Argonautes ALG‐3 and ALG‐4 are required for spermatogenesis‐specific 26G‐RNAs and thermotolerant sperm in Caenorhabditis elegans . Proc Natl Acad Sci USA 107, 3588–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Conine CC, Moresco JJ, Gu W, Shirayama M, Conte D, Yates JR and Mello CC (2013) Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans . Cell 155, 1532–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wedeles CJ, Wu MZ and Claycomb JM (2013) Protection of germline gene expression by the C. elegans argonaute CSR‐1. Dev Cell 27, 664–671. [DOI] [PubMed] [Google Scholar]
- 146. Seth M, Shirayama M, Gu W, Ishidate T, Conte D and Mello CC (2013) The C. elegans CSR‐1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev Cell 27, 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Seth M, Shirayama M, Tang W, Shen E‐Z, Tu S, Lee H‐C, Weng Z and Mello CC (2018) The coding regions of germline mRNAs confer sensitivity to Argonaute regulation in C. elegans . Cell Rep 22, 2254–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Aoki K, Moriguchi H, Yoshioka T, Okawa K and Tabara H (2007) In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans . EMBO J 26, 5007–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Quarato P, Singh M, Cornes E, Li B, Bourdon L, Mueller F, Didier C and Cecere G (2021) Germline inherited small RNAs facilitate the clearance of untranslated maternal mRNAs in C. elegans embryos. Nat Commun 12, 1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Gerson‐Gurwitz A, Wang S, Sathe S, Green R, Yeo GW, Oegema K and Desai A (2016) A small RNA‐catalytic argonaute pathway tunes germline transcript levels to ensure embryonic divisions. Cell 165, 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Cecere G, Hoersch S, O’Keeffe S, Sachidanandam R and Grishok A (2014) Global effects of the CSR‐1 RNA interference pathway on the transcriptional landscape. Nat Struct Mol Biol 21, 358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Gushchanskaia ES, Esse R, Ma Q, Lau NC and Grishok A (2019) Interplay between small RNA pathways shapes chromatin landscapes in C. elegans . Nucleic Acids Res 47, 5603–5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Nguyen DAH and Phillips CM (2021) Arginine methylation promotes siRNA‐binding specificity for a spermatogenesis‐specific isoform of the Argonaute protein CSR‐1. Nat Commun 12, 4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Charlesworth AG, Seroussi U, Lehrbach NJ, Renaud MS, Sundby AE, Molnar RI, Lao RX, Willis AR, Woock JR, Aber MJ et al. (2021) Two isoforms of the essential C. elegans Argonaute CSR‐1 differentially regulate sperm and oocyte fertility. Nucleic Acids Res 49, 8836–8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Rechavi O, Minevich G and Hobert O (2011) Transgenerational inheritance of an acquired small RNA‐based antiviral response in C. elegans . Cell 147, 1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Minkina O and Hunter CP (2017) Stable heritable germline silencing directs somatic silencing at an endogenous locus. Mol Cell 65, 659–670.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Félix M‐A, Ashe A, Piffaretti J, Wu G, Nuez I, Bélicard T, Jiang Y, Zhao G, Franz CJ, Goldstein LD et al. (2011) Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol 9, e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Ashe A, Sarkies P, Pen JL, Tanguy M and Miska EA (2015) Antiviral RNA interference against Orsay virus is neither systemic nor transgenerational in Caenorhabditis elegans . J Virol 89, 12035–12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Gammon DB, Ishidate T, Li L, Gu W, Silverman N and Mello CC (2017) The antiviral RNA interference response provides resistance to lethal arbovirus infection and vertical transmission in Caenorhabditis elegans . Curr Biol 27, 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Wilkins C, Dishongh R, Moore SC, Whitt MA, Chow M and Machaca K (2005) RNA interference is an antiviral defence mechanism in Caenorhabditis elegans . Nature 436, 1044–1047. [DOI] [PubMed] [Google Scholar]
- 161. Rechavi O, Houri‐Ze’evi L, Anava S, Goh WSS, Kerk SY, Hannon GJ and Hobert O (2014) Starvation‐induced transgenerational inheritance of small RNAs in C. elegans . Cell 158, 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Jobson MA, Jordan JM, Sandrof MA, Hibshman JD, Lennox AL and Baugh LR (2015) Transgenerational effects of early life starvation on growth, reproduction, and stress resistance in Caenorhabditis elegans . Genetics 201, 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Zeevi LH, Teichman G, Gingold H and Rechavi O (2021) Stress resets ancestral heritable small RNA responses. eLife 10, e65797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kishimoto S, Uno M, Okabe E, Nono M and Nishida E (2017) Environmental stresses induce transgenerationally inheritable survival advantages via germline‐to‐soma communication in Caenorhabditis elegans . Nat Commun 8, 14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Okabe E, Uno M, Kishimoto S and Nishida E (2021) Intertissue small RNA communication mediates the acquisition and inheritance of hormesis in Caenorhabditis elegans . Commun Biol 4, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Thomson T and Lin H (2009) The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 25, 355–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Simon M, Sarkies P, Ikegami K, Doebley A‐L, Goldstein LD, Mitchell J, Sakaguchi A, Miska EA and Ahmed S (2014) Reduced insulin/IGF‐1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep 7, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Ahmed S and Hodgkin J (2000) MRT‐2 checkpoint protein is required for germline immortality and telomere replication in C. elegans . Nature 403, 159–164. [DOI] [PubMed] [Google Scholar]
- 169. Frézal L, Demoinet E, Braendle C, Miska E and Félix M‐A (2018) Natural genetic variation in a multigenerational phenotype in C. elegans . Curr Biol 28, 2588–2596.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Katz DJ, Edwards TM, Reinke V and Kelly WG (2009) A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137, 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Tóth KF, Pezic D, Stuwe E and Webster A (2015) Non‐coding RNA and the reproductive system. Adv Exp Med Biol 886(v–vi), 51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Montgomery BE, Vijayasarathy T, Marks TN, Reed KJ and Montgomery TA. (2020) piRNAs prevent runaway amplification of siRNAs from ribosomal RNAs and histone mRNAs. Biorxiv 2020.06.15.153023. [Google Scholar]
- 173. Zhou X, Feng X, Mao H, Li M, Xu F, Hu K and Guang S (2017) RdRP‐synthesized antisense ribosomal siRNAs silence pre‐rRNA via the nuclear RNAi pathway. Nat Struct Mol Biol 24, 258–269. [DOI] [PubMed] [Google Scholar]
- 174. Zhu C, Yan Q, Weng C, Hou X, Mao H, Liu D, Feng X and Guang S (2018) Erroneous ribosomal RNAs promote the generation of antisense ribosomal siRNA. Proc Natl Acad Sci USA 115, 201800974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Liao S, Chen X, Xu T, Jin Q, Xu Z, Xu D, Zhou X, Zhu C, Guang S and Feng X (2021) Antisense ribosomal siRNAs inhibit RNA polymerase I‐directed transcription in C. elegans . Nucleic Acids Res 49, 9194–9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Spichal M, Heestand B, Billmyre KK, Frenk S, Mello CC and Ahmed S (2021) Germ granule dysfunction is a hallmark and mirror of Piwi mutant sterility. Nat Commun 12, 1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Juang B‐T, Gu C, Starnes L, Palladino F, Goga A, Kennedy S and L’Etoile ND (2013) Endogenous nuclear RNAi mediates behavioral adaptation to odor. Cell 154, 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Posner R, Toker IA, Antonova O, Star E, Anava S, Azmon E, Hendricks M, Bracha S, Gingold H and Rechavi O (2019) Neuronal small RNAs control behavior transgenerationally. Cell 177, 1814–1826.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Moore RS, Kaletsky R and Murphy CT (2019) Piwi/PRG‐1 Argonaute and TGF‐β mediate transgenerational learned pathogenic avoidance. Cell 177, 1827–1841.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Zhang Y, Lu H and Bargmann CI (2005) Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans . Nature 438, 179–184. [DOI] [PubMed] [Google Scholar]
- 181. Kaletsky R, Moore RS, Vrla GD, Parsons LR, Gitai Z and Murphy CT (2020) C. elegans interprets bacterial non‐coding RNAs to learn pathogenic avoidance. Nature 586, 445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Parker GS, Eckert DM and Bass BL (2006) RDE‐4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA 12, 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Cavalli G and Heard E (2019) Advances in epigenetics link genetics to the environment and disease. Nature 571, 489–499. [DOI] [PubMed] [Google Scholar]
- 184. Mondotte JA, Gausson V, Frangeul L, Suzuki Y, Vazeille M, Mongelli V, Blanc H, Failloux A‐B and Saleh M‐C (2020) Evidence for long‐lasting transgenerational antiviral immunity in insects. Cell Rep 33, 108506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F et al. (2016) Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng G, Peng H, Zhang X, Zhang Y et al. (2016) Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400. [DOI] [PubMed] [Google Scholar]
- 187. Kumar P, Kuscu C and Dutta A (2016) Biogenesis and function of transfer RNA‐Related Fragments (tRFs). Trends Biochem Sci 41, 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Franklin TB, Russig H, Weiss IC, Gräff J, Linder N, Michalon A, Vizi S and Mansuy IM (2010) Epigenetic transmission of the impact of early stress across generations. Biol Psychiat 68, 408–415. [DOI] [PubMed] [Google Scholar]


