Abstract
Hybridization can serve as an evolutionary stimulus, but we have little understanding of introgression at early stages of hybrid zone formation. We analyze reproductive isolation and introgression between a range‐limited and a widespread species. Reproductive barriers are estimated based on differences in flowering time, ecogeographic distributions, and seed set from crosses. We find an asymmetrical mating barrier due to cytonuclear incompatibility that is consistent with observed clusters of coincident and concordant tension zone clines (barrier loci) for mtDNA haplotypes and nuclear SNPs. These groups of concordant clines are spread across the hybrid zone, resulting in weak coupling among barrier loci and extensive introgression. Neutral clines had nearly equal introgression into both species’ ranges, whereas putative cases of adaptive introgression had exceptionally wide clines with centers shifted toward one species. Analyses of cline shape indicate that secondary contact was initiated within the last 800 generations with the per‐generation dispersal between 200 and 400 m, and provide some of the first estimates of the strength of selection required to account for observed levels of adaptive introgression. The weak species boundary between these species appears to be in early stages of dissolution, and ultimately will precipitate genetic swamping of the range‐limited species.
Keywords: Adaptive introgression, coupling of barrier loci, cytonuclear incompatibility, peripatric speciation, Ranunculus, reproductive isolation, tension zone
The potential for hybridization to contribute to adaptation and diversification has been recognized since the early 1900s (Lotsy 1916; Anderson 1949; Abbott et al. 2013; Gompert et al. 2017), and it is becoming increasingly apparent that introgression during divergence has affected the evolution of a wide range of species (Payseur and Rieseberg 2016). The potential for interspecific genetic exchange occurs as diverging lineages come into contact following a period of isolation. Depending on the strength of reproductive isolation, there are a number of possible outcomes from secondary contact, including a complete melding of the two lineages at one extreme, or the maintenance of sharp clines and little or no introgression at the other (Arnold 1997; Abbott et al. 2016; Payseur and Rieseberg 2016; Butlin and Smadja 2018). Sharp clines can be maintained as “tension zones” when there is selection against hybrids (Key 1968; Moore 1977; Barton 1979; Barton and Hewitt 1985), and these regions of ongoing hybridization can persist over long periods of time (Szymura and Barton 1986; Harrison 1990; Rieseberg et al. 1999). Within hybrid zones, independent genomic regions may display heterogeneous cline shapes and degrees of introgression that may depend on neutral processes or the effects of selection (Payseur et al. 2004; Fitzpatrick 2013; Abbott et al. 2016; Gompert et al. 2017). The highest rates of introgression are expected for genomic regions under positive selection in the range of one species (Anderson and Stebbins 1954; Barton 2001; Fitzpatrick et al. 2010; Hedrick 2013; Suarez‐Gonzalez et al. 2018), but these events are difficult to detect because the large majority of hybrid zones appear to be relatively old, such that any adaptive alleles have already attained high frequency in the ranges of both species (Arnold 1997; Abbott et al. 2016; Payseur and Rieseberg 2016). Analyses of recent secondary contact are much less common (e.g., Fitzpatrick et al. 2010; Lamer et al. 2015; Lehnert et al. 2018), and our understanding of the dynamics of genetic exchange during early stages of secondary contact is incomplete.
Analyses of clines separating divergent lineages have been conducted primarily under the assumption that their characteristics reflect equilibrium conditions (Barton and Hewitt 1985; Arnold 1997; Abbott et al. 2016; Payseur and Rieseberg 2016), but this assumption may not apply during early stages of hybrid zone formation. After initial secondary contact, all loci with strong allele frequency differences between lineages will display sharp step‐clines (Fig. 1), but depending on rates of dispersal and the effects of selection, clines are likely to become wider over time (Barton and Hewitt 1985; Fitzpatrick 2013). In the absence of selection, cline centers will remain close to the region of initial contact, whereas the cline becomes wider as introgression progresses into the ranges of both lineages at a rate that is dependent on dispersal (Fisher 1937; Piálek and Barton 1997). Alternatively, the rate of introgression can be accelerated if an allele from one lineage is subject to positive selection in the genomic background of the second lineage (Fisher 1937; Barton 2001). In this case, we expect that introgression will be unidirectional, and the cline will shift away from the point of secondary contact at a velocity that depends on the rate of dispersal and the strength of selection (Fisher 1937; Kolmogorov et al. 1937; Fig. 1). Under equilibrium conditions, adaptive introgression is expected to occur as an advancing wave where the width of the cline stays constant. The third possible outcome is expected in cases where interspecific allelic combinations have lower fitness due to underdominance or epistasis (Barton and Hewitt 1985; Abbott et al. 2016). Under these conditions, a tension zone forms as the widening of the cline ceases and a balance between the per‐generation dispersal rate and the strength of selection is achieved (Fig. 1). When equilibrium conditions are attained, cline width will be proportional to the strength of selection acting against specific genetic combinations at each genomic region (Barton and Gale 1993).
Figure 1.
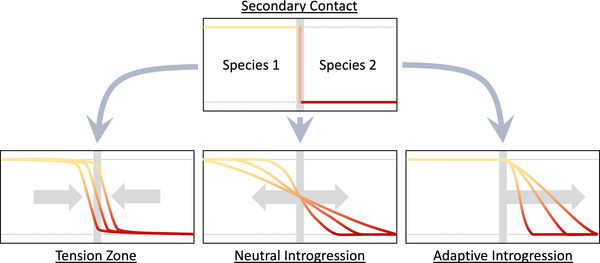
Three scenarios for changes in cline width and center following secondary contact. Sharp clines characteristic of tension zones are predicted if there is selection against hybrids due to underdominance or epistasis between interspecific genomic elements. In the absence of selection, neutral introgression will result in wider clines and cline centers will remain close to region of secondary contact. Adaptive introgression into one species is predicted to result in wide clines and displacement of the cline center. In each figure, the horizontal axis is geographic distance and the vertical axis is marker frequency.
The scenarios described above assume that population density and fitness of both lineages are equivalent, and that mating barriers between them are equal. Under these conditions, neutral clines will expand symmetrically, and the centers of tension zone clines are expected to coincide (Barton and Gale 1993; Abbott et al. 2016). When mating barriers, dispersal rates, or fitness are not equivalent, cline centers are expected to shift position away from the original location of secondary contact. Hybrid zone movement appears to be common and may occur for a variety of reasons (Cruzan 2005; Buggs 2007; Lehnert et al. 2018; van Riemsdijk et al. 2019; Wielstra 2019). For example, higher rates of dispersal for one lineage will shift cline centers toward the lineage with more limited dispersal (Barton and Hewitt 1985). With equal rates of dispersal, gene flow will be more prevalent toward the lineage with lower fitness due to higher numbers of emigrating individuals and spores (seeds and pollen in plants) from the high‐fitness lineage, resulting in a shift in all of the cline centers (the entire hybrid zone) and genetic swamping of the low‐fitness lineage (van Riemsdijk et al. 2019). Asymmetrical mating barriers have a similar effect as cline centers are expected to shift toward the lineage with weaker reproductive isolation (Payseur et al. 2004). As the hybrid zone moves, the centers of tension zone clines become staggered and spread out, and the direction of introgression toward one lineage is evident as higher levels of linkage disequilibria at the leading edge of hybrid zone movement (Barton and Gale 1993; Cruzan 2005; van Riemsdijk et al. 2019).
An appreciation of the processes contributing to the stability or movement of clines and the potential for hybrid zones to prevent introgression between divergent lineages depends on an evaluation of parental lineage and hybrid fitness, and the strength of reproductive barriers (e.g., Dasmahapatra et al. 2002; Payseur et al. 2004; Cruzan 2005; Buggs 2007; Wellenreuther et al. 2018). Isolation is most effective when lineages are ecologically and geographically separated (Ramsey et al. 2003; Christie and Strauss 2018; Cruzan 2018). When lineages are in close contact, post‐mating mechanisms, such as genetic incompatibility (i.e., Bateson‐Dobzhansky‐Muller [BDM] incompatibilities; Orr 1996), can present strong barriers (Hendry et al. 2007; Baack et al. 2015; Christie and Strauss 2018). In particular, BDM incompatibilities occurring between nuclear and organelle genomes (cytonuclear incompatibilities) generate asymmetrical mating barriers, and have been observed to arise early during divergence (Levin 2003; Greiner et al. 2011; Burton et al. 2013; Barnard‐Kubow et al. 2016). The effectiveness of BDM incompatibilities (barrier loci) to maintain hybrid zones that limit introgression depends on their number and geographic coincidence of their clines (Barton 1983; Ravinet et al. 2017; Butlin and Smadja 2018). With larger numbers of barrier loci, clines that are strongly coincident (having the same center) and concordant (having the same shape), there is a “coupling effect” that is thought to generate a strong barrier preventing neutral introgression and slowing the progress of adaptive introgression between hybridizing lineages. Conversely, the barrier would be weaker with fewer barrier loci and low coincidence, resulting in extensive introgression and eventually the dissolution of the hybrid zone.
In this study, we examine the strength of reproductive isolation and evaluate nuclear, cytoplasmic, and phenotypic trait clines associated with a hybrid zone between the range‐limited species, Ranunculus austro‐oreganus L.D. Benson (Southern Oregon buttercup), and its widespread congener, R. occidentalis Nutt. (Western buttercup; Ranunculaceae). These two species appear to be interbreeding in a region where their ranges come into contact, in a narrow hybrid zone in the Rogue River Valley of southern Oregon. Ranunculus occidentalis occurs across an extensive region, stretching from southern California to Alaska, whereas R. austro‐oreganus occupies a small area in Jackson County and is a candidate threatened species (Kagan et al. 2016). We first assess the strength of reproductive isolation between these species using niche modeling, evaluation of flowering times, and seed set from greenhouse crosses to test the strength and symmetry of mating barriers. We then conduct a SNP survey to identify the loci displaying the strongest allele frequency differences, and evaluate their cline characteristics across the region of secondary contact using geographic and genomic cline approaches (Gompert et al. 2017). We identify a range of nuclear and cytoplasmic clines that display large differences in their widths and displacement from the hybrid zone center. Analyses of cline width and displacement indicate that secondary contact was initiated only recently, and clines representing ongoing neutral and adaptive introgression are present.
Methods
MORPHOLOGICAL DIFFERENTIATION
Ranunculus austro‐oreganus and R. occidentalis display differences in their ventral petal color and in the density of leaf trichomes. We quantified these two morphological traits for plants across populations to characterize their geographic distribution across the hybrid zone. Putative hybrid populations were identified by the presence of plants representing both petal color morphs. Three petals from three separate flowers and two leaves (one basal and one cauline) were collected in the field from each plant in two flowering seasons. Petals and leaves were collected from 32 populations (eight R. austro‐oreganus, 11 R. occidentalis, and 13 putative hybrids) in 2017 and 2019 (Fig. S1).
Morphological characters were quantified using a Leica MZ16 stereomicroscope and ImageJ software (version 1.52c). Ventral petal color was quantified for 532 individuals by capturing images at 40× magnification. ImageJ was used to convert each petal image to 8‐bit gray scale using weighted RGB conversions to maintain luminosity (Rx0.30, Gx0.59, Bx0.11), where the yellow color is then perceived as lightness (high gray value) and the red/brown character is darkness (low gray value). A 500px2 selection was made over the center of the image and the ImageJ measurement tool was used to capture the mean grayscale value of the selection, where every pixel had a value from 0 to 255, black to white. Variation in leaf hair coverage was quantified in the same manner as petals, for one basal and one cauline leaf from 28 populations, resulting in 1184 leaf samples. Leaf and petal data were analyzed using linear models with SAS (2007) to test for differences between the two species and the putative hybrid populations. Residuals from these models displayed approximately normal distributions.
GREENHOUSE CROSSES
Seeds were collected from three R. occidentalis and three R. austro‐oreganus populations that appeared to lack hybrid individuals and were well displaced from the apparent region of secondary contact. Seeds were cold‐stratified in moist seedling potting mix (2°C) for 6 weeks. Germinated seedlings were grown for 2 months and were vernalized by half burying pots in soil outside before transfer into the greenhouse. We emasculated newly opened flowers with indehiscent anthers and hand‐pollinated emasculated flowers when stigmas became receptive (3 days after emasculation). Pollinations were made with dehisced anthers from flowers on the same plant (selfing), from different populations of the same species (intraspecific outcross), or from populations of the other species (interspecific crosses). In total, we performed 29 interspecific crosses, 31 intraspecific crosses, and 19 self‐pollinations with R. austro‐oreganus as the seed parent, and 23 interspecific, 17 intraspecific crosses, and nine self‐pollinations with R. occidentalis as the seed parent. We made final seed collections approximately 4 weeks after the emasculation date when filled seeds first appeared light brown.
Seeds were examined using light microscopy and characterized as filled, aborted, or unfertilized based on their color and size. Filled seeds were opaque and brown, often with spots; aborted seeds were small to large, green, and translucent; and unfertilized seeds were very small, yellow, and undeveloped. The data displayed high frequencies of zeros so we assumed a Poisson distribution to compare cross treatments with maternal collection site entered as a random variable and total number of ovules plus seeds entered as a covariate in a general linear model using the GENMOD procedure of SAS (SAS 2007).
The filled seeds from intra‐, inter‐, and self‐crosses were cold‐stratified, and germination success was measured over a 3‐week period from when they were first removed from cold stratification. Germination success was compared among cross treatments in a general linear model in SAS (SAS 2007) where maternal site was specified as a random effect and the total number of seeds planted was considered a covariate. Individual plants were assessed for the presence/absence of flower production. We analyzed flowering data across treatment types using categorical models in the CATMOD procedure of SAS (SAS 2007).
SEED SET UNDER FIELD CONDITIONS
To assess levels of seed set under field conditions, we collected fruiting heads (clusters of achenes) from three R. occidentalis, five R. austro‐oreganus, and two putative hybrid populations. In each population, up to three achene clusters from a maximum of 20 individuals were collected for a total of 146 seed clusters from 60 plants of R. occidentalis, 358 clusters from 164 plants of R. austro‐oreganus, and 59 clusters from 26 putative hybrid plants. Seed cluster constituents were sorted as filled, aborted, or unfertilized achenes using a stereomicroscope as described above. Data were analyzed to test for seed set (filled achenes) differences among populations identified as hybrid or as one of the two species assuming a negative binomial distribution. We compared the number of seeds per flower between species with population site as a random variable in a general linear mixed model using the lme4 R package (version 3.6.0). The model included number of achenes per flower as a covariate as it was inconsistent among individuals.
REPRODUCTIVE ISOLATION
We followed the methods of Ramsey et al. (2003) to calculate the absolute contribution (AC) of each component and total reproductive isolation (T) between R. occidentalis and R. austro‐oreganus. The combined effects of ecogeographic and ecological isolation were determined from ENM analyses (Appendix S1), and the contribution to reproductive isolation (RI) was calculated as ACEcoGeo = 1 – (co‐occurrences)/(total). To estimate isolation due to differences in flowering time between species, we used the collection dates of herbarium records (Appendix S1) with Julian Day corrected by elevation, year sampled, whether the record was an observation or a collection, and latitude. Corrected Julian Days were ranked in ascending order for both species combined and reproductive isolation was calculated as ACFlower = 1 – (days co‐flowering)/(total days). Total days was the earliest to the latest day when either species was flowering, and days co‐flowering was the first to the last day when both species were predicted to flower together.
Reproductive isolation components for seed set, seed germination, and offspring flowering were calculated based on both the combined data and data with each species separately acting as the seed parent to identify components contributing to asymmetrical isolation. For all three components, interspecific crosses were compared to crosses between populations of the same species (intraspecific) to control for the effects of drift load (Rhode and Cruzan 2005; Charlesworth and Willis 2009). Reproductive isolation at the seed set stage was calculated from the mean seeds per flower after inter‐ and intraspecific crosses as ACSeeds = 1 – (mean interspecific seed number)/(mean intraspecific seed number). For seed germination, we used mean proportion of seeds germinating as ACGerm = 1 – (mean interspecific germination)/(mean intraspecific germination). All seedlings were scored as either 0 for the plants that did not flower because they did not survive or were not observed to flower or 1 for the plants that survived and flowered before the end of the season. These scores were used to calculate average flowering frequency as ACFlowering = 1 – (interspecific flowering)/(intraspecific flowering). The total reproductive isolation for the combined data and for each species was calculated as T = ∑AC i , and the relative contribution of each stage as RC i = AC i /T.
GENETIC SAMPLING
Leaf tissue was collected from eight individuals in each of 48 populations from R. austro‐oreganus, R. occidentalis, and their putative hybrids through the 2015–2018 field seasons (Table S1). All plant materials were stored in coin envelopes and were dried and kept in silica gel until DNA extraction. Total genomic DNA was extracted following the Qiagen DNeasy 96 Plant Kit protocol (Qiagen, Germantown, MD). Isolated DNA was quantified using a Qubit 4 fluorometer (Invitrogen, ThermoFisher Scientific, Waltham, MA). Individuals with a DNA concentration greater than 20 ng/μL were prioritized and selected for Genotyping‐by‐Sequencing (GBS; Elshire et al. 2011). Samples were sent to the Biotechnology Center at University of Wisconsin‐Madison for enzyme optimization, library construction, and sequencing. Raw sequences were processed and SNPs were identified and filtered for quality following the protocol specified by the GBS‐SNP‐CROP pipeline (Melo et al. 2016; Appendix S2). Once filtered, a subset of 2196 SNPs remained and were used in subsequent analyses.
IDENTIFICATION OF OUTLIER LOCI
Loci displaying strong allele frequency differences between populations of each species due to selection or drift can be used for analyses of clines (Nosil et al. 2009; Gompert and Buerkle 2011; Stankowski et al. 2017), and for assessing patterns of introgression when rates of gene flow are high (Guichoux et al. 2013). Although F ST is often used to identify outlier loci (Beaumont 2005), this approach does not work well when loci are nearly homozygous in allopatric populations (Harrison and Larson 2016), so we identified outlier loci based on the absolute differences in allele frequencies (Larson et al. 2014). Outlier loci in this study were identified as SNPs displaying the largest allelic frequency differences (>0.3) between the two populations of each species having the lowest variance in ventral petal color (Fig. S2). These criteria identified a total of 61 outlier SNPs out of the original 2196 detected.
GEOGRAPHIC CLINE ANALYSIS
We estimated cline centers and widths to evaluate the extent of admixture and patterns of introgression across the region of hybridization using a maximum likelihood approach for both of the morphological traits and each of the 61 outlier SNP loci. The position of populations along a latitudinal line is based on the position of a geographic marker placed ∼50 m west of the west‐most population. Cline models were estimated with the R package HZAR (Derryberry et al. 2014) for both the phenotypic traits (ventral petal color and leaf trichome density) and allele frequencies at each of the outlier loci. A null model, which tests the hypothesis that there is no cline in the sampled region, along with 15 alternative cline models was analyzed for each locus or trait. Models differ based on combinations of trait frequency [pMin, pMax] (fixed to 0 and 1; observed values; estimated values) and combinations of fitting cline tails (none fitted; left only; right only; mirror tails; both tails estimated separately; Derryberry et al., 2014). Each model was run on a separate seed and with an MCMC length set to 100,000 with a burn‐in of 10,000. All cline models were checked for convergence and the model with the lowest AIC score was selected to extract summary statistics and the maximum‐likelihood clines for each variant. Based on the criteria described above, cline centers and widths were used to classify each marker as fitting a neutral (low displacement from the hybrid zone center and introgression toward both species’ ranges), adaptive (high displacement and introgression in one direction), or tension zone model (low displacement and narrow cline width; Fig. 1).
GENOMIC CLINE ANALYSIS
Genomic clines for the 50 SNP loci that had significant fits to one of the cline models in HZAR (see below) were analyzed using Bayesian estimation of genomic clines (bgc; Gompert and Buerkle 2012) and ClinePlotR (Martin et al. 2020). All populations were aggregated into three categories: Ranunculus occidentalis and R. austro‐oreganus parental samples (pooling two populations of each species with the lowest variance in ventral petal color; Fig. S2) and admixed samples. Five runs were completed with 170,000 MCMC steps and a burn‐in period of 70,000 samples. In each run, data were thinned by recording every 10th sample. Because the loci were identified using next‐generation sequencing, the genotype‐uncertainty model was used in conjunction with a sequence error probability of 0.0001. Cline parameters α and β were calculated.
Estpost software (http://repec.sowi.unibe.ch/stata/estout/) was used to summarize the results from each run, and runs were checked for convergence using the plot_traces function in ClinePlotR. Outlier loci were identified with the get_bgc_outliers function using the overlap.zero method, which identifies loci with cline parameters that exclude zero for α, β, or both parameters. If a locus was identified as an outlier, the direction of displacement (positive or negative) was found through comparison to the linear model for the relationship between the hybrid index and the ancestry coefficient (Φ).
LINKAGE DISEQUILIBRIA AMONG OUTLIER LOCI
Levels of linkage (gametic) disequilibria (D) provide evidence of recent gene flow and can be used to infer the direction of hybrid zone movement (Barton and Gale 1993; Cruzan 2005; van Riemsdijk et al. 2019). We used methods to estimate the average level of disequilibria () developed by Barton and Gale (1993), which is based on the variance in the hybrid index (H) among individuals within populations (VH ). This approach assumes that VH is due to variance in allele frequencies across n loci, and is inflated if alleles at different loci are in disequilibria with each other. The average disequilibrium in each population can be calculated as
where is the mean hybrid index and Vp is the variance in allele frequencies for each population, calculated as 1/n∑(pi – ) across n loci (Barton and Gale 1993). We estimated the hybrid index (H) for each individual based on the 50 F ST outlier loci found to have significant clines (see below) using the R package introgress (Gompert and Buerkle 2010). Standard deviations for and tests for significant differences from zero within each population were estimated using 100 bootstrap replicates with replacement.
We evaluated whether different groups of SNPs were tending toward introgression together into the range of each species by examining patterns of disequilibria among the 50 outlier loci. We pooled individuals from the five populations with the highest estimates of for each species and tallied instances of significant pairwise disequilibria (P < 0.05) with Arlequin (Excoffier and Lischer 2010) using 10,000 permutations with haplotype phase unknown. Separate tallies were made for significant estimates of disequilibria among SNPs with clines displaced toward the range of R. austro‐oreganus, R. occidentalis, or both.
Results
MORPHOLOGICAL VARIATION
We found strong differences between the two Ranunculus species and the hybrid populations for both ventral petal color and leaf trichome density. Ventral petals of R. austro‐oreganus were much darker compared to R. occidentalis, and hybrids were intermediate (F 2/25 = 61.36, P < 0.0001 tested over maternal site; Fig. 2A). Leaves of R. austro‐oreganus were much brighter than R. occidentalis, and the hybrids were intermediate between the two species (F 2/25 = 5.03, P = 0.0146 tested over maternal site; Fig. 2B). There were significant differences among sampling locations for petals (F 25/501 = 14.79, P < 0.001) and leaf hair gray values (F 25/28 = 21.52, P < 0.001), but the difference for gray value between basal and cauline leaves was not significant (F 28/1005 = 0.78, P = 0.793). There was a significant correspondence between the mean ventral petal color and the variance in color, with populations having intermediate means displaying the highest variance in color (Fig. S2; quadratic model adjusted R 2 = 0.66, F 2/27 = 29.2, P < 0.0001).
Figure 2.
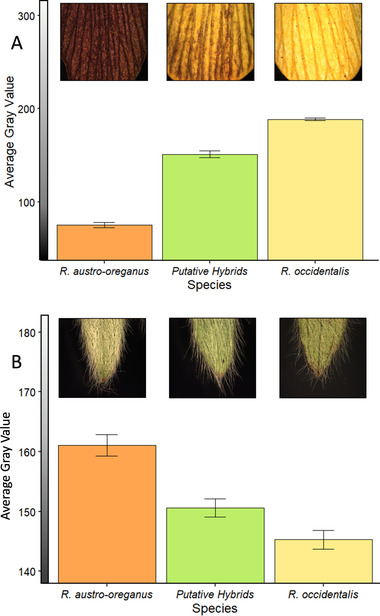
Morphological differentiation measured in the average gray values for petal color (A) and trichome density (B) in Ranunculus austro‐oreganus, R. occidentalis, and their putative hybrids.
GREENHOUSE CROSSES
Significant differences were found in the number of filled seeds among the types of crosses (x 2 5 = 88.84, P < 0.001) and the maternal sampling location nested within cross type (x 2 11 = 136.17, P < 0.001). The covariate of total number of ovules was also significant (x 2 1 = 9.89, P = 0.002). For each type of cross (interspecific, intraspecific, and self), R. occidentalis had significantly higher seed set than R. austro‐oreganus (Table 1). Ranunculus occidentalis intraspecific crosses also had a significantly higher seed set than their self‐pollinations (Table 1).
Table 1.
Numbers of seeds produced, seeds germinated, and seedlings surviving to flowering after self, intraspecific (outcrossing between populations), and interspecific pollinations in Ranunculus occidentalis (Ranocc) and R. austro‐oreganus (Ranaus). Model effects for each analysis included treatment (species/pollination treatment combined) and maternal site (not included in the categorical model for flowering). For each analysis, contrast statements were used to compare self versus intraspecific and intraspecific versus interspecific pollinations within each species. Between species comparisons included self, intraspecific, and interspecific pollinations. Significant probabilities are in bold
| Seeds | Germination | Flowering | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DF | χ 2 | Prob. | DF | F | Prob. | DF | χ 2 | Prob | ||
| Treatment | 5 | 88.84 | <0.0001 | 5 | 2.27 | 0.0568 | 5 | 12.17 | 0.0325 | |
| Maternal site | 11 | 136.17 | <0.0001 | 11 | 1.36 | 0.2109 | ||||
| total | 1 | 9.89 | 0.0017 | 1 | 123.25 | <0.0001 | ||||
| Ranocc | Self vs. Intra | 1 | 4.50 | 0.0338 | 1 | 11.13 | 0.0014 | 1 | 0.80 | 0.3717 |
| Intra vs. Inter | 1 | 0.01 | 0.9183 | 1 | 1.30 | 0.2582 | 1 | 0.33 | 0.5652 | |
| Ranaus | Self vs. Intra | 1 | 0.09 | 0.7658 | 1 | 0.02 | 0.8755 | |||
| Intra vs. Inter | 1 | 0.03 | 0.8716 | 1 | 0.02 | 0.8823 | 1 | 10.07 | 0.0015 | |
| Ranocc vs. Ranaus | Self | 1 | 11.74 | 0.0006 | 1 | 3.49 | 0.0659 | |||
| Intra | 1 | 37.01 | <0.0001 | 1 | 1.34 | 0.2513 | 1 | 2.74 | 0.098 | |
| Inter | 1 | 36.27 | <0.0001 | 1 | 0.07 | 0.791 | 1 | 5.85 | 0.0156 | |
From the 617 seeds planted, 353 germinated. Germination success was not significantly different between types of crosses, although there appeared to be a trend toward a treatment effect (F 5/70 = 2.27, P = 0.0568); the contrasts revealed R. occidentalis self‐pollinations had significantly lower germination rates than the intraspecific crosses in this species, and all other contrasts were not significant (Table 1). The maternal sampling location nested within treatments did not have a significant effect on germination (F 11/70 = 1.36, P = 0.2109), whereas the covariate of total number of seeds planted was significant (F 1/70 = 123.25, P < 0.001).
Of the 353 germinated seeds, 232 grew to adult size. We found significant differences between crosses on flowering success (x 2 = 12.17, P = 0.0325). This was driven largely by seedlings from interspecific crosses to R. austro‐oreganus as the seed parent having a lower flowering frequency, as compared to intraspecific seedlings in the same species, and interspecific seedlings from R. occidentalis as the seed parent, which both had higher flowering frequencies (Tables 1 and S2).
SEED SET UNDER FIELD CONDITIONS
Levels of seed set and ovule abortion under field conditions were similar to the pattern found between species in the greenhouse. Seed set was highest in R. occidentalis (4.72) followed by putative hybrid and R. austro‐oreganus populations (4.44 and 4.02, respectively; P > 0.05 for all comparisons). Embryo abortion was highest in R. austro‐oreganus populations (9.07 seeds) followed by putative hybrid and R. occidentalis (7.67 and 4.96, respectively; P = 0.004 for the contrast between R. austro‐oreganus and R. occidentalis; P > 0.05 for both contrasts with plants in putative hybrid populations).
REPRODUCTIVE ISOLATION
Reproductive isolation between these species of Ranunculus appears to be fairly strong overall, but this is mostly due to ecological niche models, which predicted large geographic regions to be suitable habitat for only R. occidentalis (Table S2). In contrast, there was only a few small regions that were predicted to be suitable exclusively for R. austro‐oreganus. The contribution of flowering time to isolation, on the other hand, was small and was driven by the flowering period of R. occidentalis being extended by about 20 days compared to R. austro‐oreganus. Fecundity (seed set), seed germination, and the flowering frequencies of each offspring group did not contribute much to overall isolation, but seed set and flowering frequencies presented strong barriers for introgression into R. austro‐oreganus, and generated asymmetrical isolation when each species was considered separately (Table S2). Postzygotic barriers for R. austro‐oreganus as the maternal parent were primarily due to low seed set and flowering frequencies for offspring from interspecific crosses. In contrast, for interspecific crosses to R. occidentalis as the maternal parent, seed and flowering frequencies were higher than for intraspecific crosses (Table S2). The low negative value of reproductive isolation for R. occidentalis indicates that pollen‐mediated introgression from R. austro‐oreganus is strongly favored when these two species are in sympatry.
GENETIC DIFFERENTIATION AND CLINE CLASSIFICATION
Results from the intraspecific crosses and reproductive barrier analyses provide opposite predictions for the direction of introgression across the hybrid zone. On the one hand, the lower average seed set observed in the greenhouse and in the field for R. austro‐oreganus would generate a fitness disadvantage resulting in higher rates of introgression into its range (Buggs 2007; van Riemsdijk et al. 2019). On the other hand, strong asymmetrical mating barriers are expected to favor introgression from R. austro‐oreganus into the range of R. occidentalis (Crochet et al. 2003; Devitt et al. 2011; Johnson et al. 2015). We further expect that clines representing different genomic regions may display heterogenous shapes and degrees of displacement from the original region of secondary contact, which will depend on the strength of selection, dispersal rates, and interactions among genomic regions (Hu and Li 2002; Johnson et al. 2015).
Structure analysis (Pritchard et al. 2000) indicates that genetic differentiation among populations and species based on all 2196 SNPs was relatively weak (Fig. S4), but stronger differences in allele frequencies between two species were found for the 50 outlier loci displaying significant clines (Fig. S5). Eight of these SNPs displayed strong departures from Hardy‐Weinberg equilibrium due to a complete lack of observed heterozygotes (blue boxes in Fig. S5). By comparing the genomic region surrounding these SNPs (based on the mock reference genome), we were able to determine that five of them had strong sequence similarity (89.53–100%) to plant mitochondrial genomes (Table S3). One group of four SNPs and one group of two displayed strong correlations in frequencies among populations (r ≥ 0.99), but weak correlations with other SNPs (r < 0.6), and were assumed to represent two haplotypes (shaded blue and green, respectively, in Fig. S5 and Table S4). Based on the lack of observed heterozygotes and patterns of correlations, these eight SNPs were assumed to represent four different cytoplasmic haplotypes.
Clines for the 42 nuclear SNPs and four cytoplasmic haplotypes displayed a wide range of variation in cline centers and widths (Figs. 3 and S5; Table S4). Nine of the clines ranged in width between 30,000 m and close to 140,000 m, with large displacements from the hybrid zone center (see below) that were proportional to their widths (i.e., displacement/width ratios ranged from 0.17 to 0.39; Fig. 3B). Six of these nine clines had centers displaced toward R. austro‐oreganus, and three had centers displaced toward R. occidentalis (including one cytoplasmic haplotype; Fig. S5). Given the extreme widths and patterns of displacement toward one species or the other, these nine clines best fit the expectations for adaptive introgression (Fig. 1).
Figure 3.
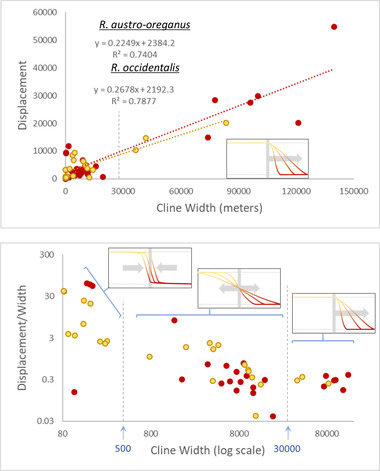
Classification of the clines for 50 outlier SNP loci based on the characteristics presented in Figure 1. Separate regression analyses for the relationship between cline width and displacement from the hybrid zone center (A) were conducted for clines displaced toward the Ranunculus occidentalis allopatric region (yellow markers and yellow dotted line) and for clines displaced toward R. austro‐oreganus (red markers and red dotted line). A subset of the widest clines (w > 30 km) had centers displaced from the hybrid zone proportional to their width (A and B) and were classified as putative cases of adaptive introgression. A second subset of clines were exceptionally narrow (w < 500 m) and were classified as tension zones (B). Neutral introgression clines displayed introgression into both species ranges with centers having moderate levels of displacement from the hybrid zone center (B).
To estimate the hybrid zone center, we excluded the nine clines appearing to be spreading by adaptive introgression, and took the average of the centers of the remaining clines for 37 SNP markers and haplotypes (hybrid zone center at 20,128 m; Figs. 4 and 5). Ten of these clines (seven with centers displaced toward R. occidentalis, and three with centers toward R. austro‐oreganus) had extremely narrow widths (<500 m) and were assumed to represent tension zone clines. Two of these tension zone clines were cytoplasmic haplotypes: one displaced in the direction of R. occidentalis and represented by four SNPs and the other displaced in the direction of R. austro‐oreganus and represented by two SNPs (Figs. 5 and S5). The centers of tension zone clines occurred in three groups that were strongly coincidental: one group of three (two nuclear and one cytoplasmic) displaced toward the R. austro‐oreganus range, and two groups of seven and four (three nuclear and one cytoplamic) each displaced toward R. occidentalis (Fig. 5). The remaining 27 clines had widths falling between 700 and 30,000 m with their centers relatively close to the hybrid zone center, and were assumed to represent neutral introgression, or perhaps weak selection (Figs. 3 and S5). Twelve of these clines (including one cytotype) had centers displaced toward R. occidentalis, and 15 had centers displaced toward R. austro‐oreganus. For the two phenotypic traits, the ventral petal color best fit the tension zone model, and trichome density best fit a model of neutral introgression (Fig. 4).
Figure 4.
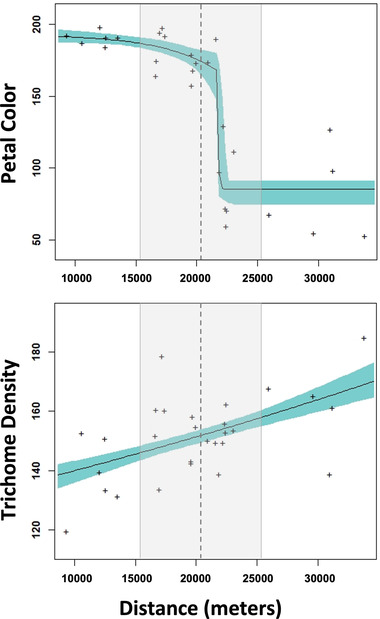
Cline models for two phenotypic traits across the hybrid zone between Ranunculus occidentalis and R. austro‐oreganus. The cline for ventral petal color displays a pattern consistent with a tension zone, whereas the cline for leaf trichome density is most consistent with neutral introgression. The gray shaded area represents the 95% confidence intervals of centers for the neutral introgression SNP clines, and the vertical dashed line is the hybrid zone center.
Figure 5.
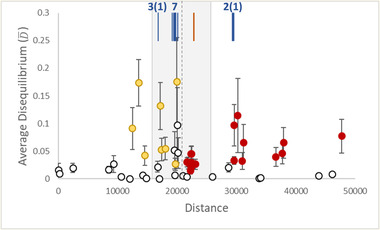
Levels of average linkage (gametic) disequilibria () for populations sampled across the hybrid zone between Ranunculus occidentalis (yellow) and R. austro‐oreganus (red). Filled markers represent estimates that are significantly different from zero. Error bars represent the standard deviation. The gray shaded area indicates the 95% confidence intervals of centers for the neutral introgression SNP clines, and the vertical dashed line is the hybrid zone center. The blue vertical lines indicate the centers of clines classified as tension zones, with the number in each cluster that are nuclear SNPs and cytoplasmic (in parentheses) haplotypes (each represented by multiple SNPs; Fig. S5). The orange vertical line represents the center of the ventral petal color cline.
GENOMIC CLINE ANALYSIS
Out of the 50 clines analyzed, 26 of them displayed levels of R. occidentalis ancestry that were higher or lower than expected (positive or negative α outliers, respectively), had higher or lower rates of introgression (negative or positive β outliers, respectively), or both (Figs. S7 and S8; Table S4). Consistent with the geographic cline analysis, 19 of the cline centers were displaced in the direction of R. austro‐oreganus, and only seven were displaced in the direction of R. occidentalis. One cytoplasmic haplotype represented by two clines had a lower rate of introgression indicating a sharp cline (positive β outlier) and one nuclear cline had a higher rate of introgression (negative β outlier) and displacement in the direction of R. austro‐oreganus (Table S4; Fig. S8).
LINKAGE DISEQUILIBRIA
Variation in the hybrid index (VH ) exceeded the variance in allele frequencies (Vp ) across the 50 outlier loci with significant clines for all populations (Fig. 5), and resulted in significant estimates of average linkage disequilibria () for 24 of the 48 populations. Fifteen of the significant disequilibria estimates were for populations within the range of R. austro‐oreganus, whereas nine were for R. occidentalis populations (Fig. 5).
There were large numbers of significant disequilibria estimates among the 50 outlier loci for the two groups of populations with the highest estimates of within each species’ range (Figs. 5 and S6). The majority of significant estimates were associated with SNP markers having clines categorized as tension zones, and a good proportion of those were due to the SNPs representing cytoplasmic haplotypes. The highest frequencies of significant disequilibria were for tension zone SNPs in R. austro‐oreganus populations, or for tension zone SNPs in populations with the highest estimates for both species (Fig. S6).
CLINE ANALYSES
We analyzed the clines fitting expectations for neutral introgression to obtain estimates for per‐generation dispersal (δ; per‐generation dispersal) and the generations since initial secondary contact (t) using the relationship w = 2.51δ√t for cline width (w) under neutral introgression (Barton and Gale 1993). Some of these clines may be weakly affected by selection, which may limit the rate of introgression, so we chose a range of w between 10,000 and 20,000 m as representative of the seven widest neutral clines (Table S3; Fig. 3B). Within this range, predicted values of δ fell between 200 and 800 m per‐generation and t ranged from 25 to 1600 generations (Fig. 6). These plants require vernalization after the first season of growth, and most flower in the second season. Assuming a generation time of 2 years, this analysis predicts that the Ranunculus hybrid zone was first formed between 50 and 3200 years in the past.
Figure 6.
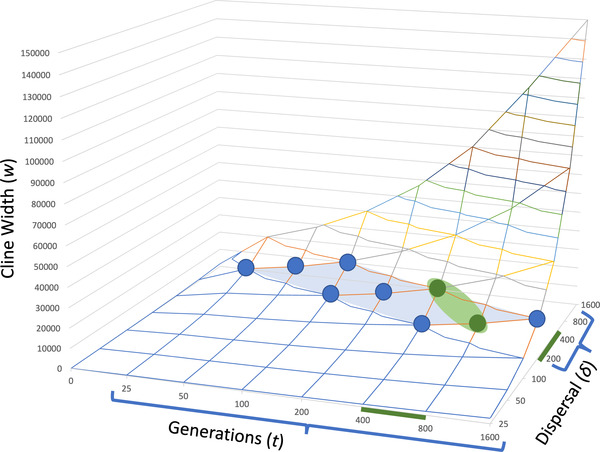
The range of cline widths (w) predicted from the per‐generation dispersal (δ) and the number of generations since secondary contact (t) under neutral introgression. The blue markers and shaded area encompass the combination of values that are consistent with observed neutral cline widths in the Ranunculus occidentalis and R. austro‐oreganus hybrid zone (blue brackets on the X and Y axes). The smaller green region and markers represent the subset of values producing reasonable estimates of s for clines displaying adaptive introgression (green bars on the X and Y axes).
For cases of advantageous introgression, we estimated the selective advantage of the beneficial allele (s) driving introgression using the relationship ν = δ√(2s), where δ is the per‐generation dispersal and ν is the velocity of introgression (Fisher 1937; Kolmogorov et al. 1937; Ralph and Coop 2010). We estimated the velocity based on the degree to which cline width exceeded the rate of neutral introgression as ν = νA – N where νA = wA /t and N is the average of the seven widest neutral clines divided by generations (t). After testing the range of values for δ and t predicted from the analysis of neutral introgression, we found a smaller subset of values provided reasonable estimates of s (i.e., s < 1.0) for all of the clines displaying adaptive introgression (green‐shaded region in Fig. 6). From values of δ between 200 and 400 m per‐generation and t between 400 and 800 generations, estimates of s ranged from 0.01 to 0.31 (Table 2). Qualitatively similar results are obtained if we do not adjust for neutral introgression (s ranges from 0.026 to 0.381). Based on these refined estimates of t, secondary contact between these Ranunculus species was initiated between 800 and 1600 years in the past.
Table 2.
Estimates of selection for beneficial alleles (s) from the per‐generation dispersal (δ) and the widths of clines (w) displaying adaptive introgression in the region of admixture between Ranunculus occidentalis (Ranocc) and R. austro‐oreganus (Ranaus). Introgression velocities (v) were based on the number of generations since secondary contact (t), and were adjusted for the rate of neutral introgression (average of the seven widest neutral clines)
| Cline | Neutral | t = 400/δ = 400 | t = 600/δ = 300 | t = 800/δ = 200 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Locus | w | w | v | s | v | s | v | s |
| Ranocc | 141124 | 41671.84 | 13776.29 | 69.74 | 0.015 | 46.49 | 0.012 | 34.87 | 0.015 |
| Ranocc | 186072 | 36485.87 | 13776.29 | 56.77 | 0.010 | 37.85 | 0.008 | 28.39 | 0.010 |
| Ranocc | 801503 | 83682.22 | 13776.29 | 174.76 | 0.095 | 116.51 | 0.075 | 87.38 | 0.095 |
| Ranaus | 237266 | 74044.15 | 13776.29 | 150.67 | 0.071 | 100.45 | 0.056 | 75.33 | 0.071 |
| Ranaus | 319835 | 100197.30 | 13776.29 | 216.05 | 0.146 | 144.04 | 0.115 | 108.03 | 0.146 |
| Ranaus | 695100 | 77793.23 | 13776.29 | 160.04 | 0.080 | 106.69 | 0.063 | 80.02 | 0.080 |
| Ranaus | 1202109 | 96143.65 | 13776.29 | 205.92 | 0.133 | 137.28 | 0.105 | 102.96 | 0.133 |
| Ranaus | 1274729 | 139669.30 | 13776.29 | 314.73 | 0.310 | 209.82 | 0.245 | 157.37 | 0.310 |
| Ranaus | 4657214 | 121159.00 | 13776.29 | 268.46 | 0.225 | 178.97 | 0.178 | 134.23 | 0.225 |
Discussion
The hybrid zone separating the widespread R. occidentalis species from the range‐limited R. austro‐oreganus species appears to be a rare example of recent secondary contact. With the exception of introduced species (e.g., Rosenthal et al. 2008; Fitzpatrick et al. 2010; Ward et al. 2012; Lamer et al. 2015; Lehnert et al. 2018), it is difficult to identify cases where admixture has initiated very recently. In fact, the large majority of hybrid zones that have been characterized represent stable species boundaries that may shift geographic position and display different degrees of permeability to introgression (Buggs 2007; Abbott et al. 2016; Harrison and Larson 2016; Gompert et al. 2017; McEntee et al. 2020). In the case of the Ranunculus hybrid zone, the clines that were generated have not yet attained shapes that represent a balance between dispersal and selection. This is evident by the number of clines that appear to be undergoing bidirectional neutral introgression, the shape and displacement of the clines representing adaptive introgression, and the narrow widths of the clines representing tension zones relative to the per‐generation dispersal. The unique characteristics of these clines have allowed us to make estimates of time since the initiation of secondary contact. Moreover, the early age of this hybrid zone has allowed us to estimate the strength of selection driving the movement of adaptive clines. The observed levels of neutral and adaptive introgression are consistent with a weak barrier between these species, and it is likely that this hybrid zone is at early stages of dissolution, ultimately resulting in the genetic swamping of the range‐limited species.
The limited range, weak reproductive isolation, and low degree of genetic and morphological divergence of R. austro‐oreganus are consistent with its recent origin through the process of peripatric (budding) speciation (Mayr 1942; Frey 1993; Patiño et al. 2014; Cruzan 2018). Cases for peripatric speciation are typically built on the observation of contemporary geographic distributions, with one species having a limited distribution at the periphery of the other (Grossenbacher et al. 2014; Christie and Strauss 2018). On the other hand, many of these pairs of species display substantial phylogenetic divergence, suggesting that their contemporary ranges may not accurately reflect historical distributions (Warren et al. 2014; Christie and Strauss 2018). Ranunculus austro‐oreganus may be an exceptional example of peripatric divergence because it appears to be recently derived. The close relationship between these species is supported by weak reproductive barriers, low levels of phenotypic and nuclear genetic divergence, weak ecological differentiation, and a high degree of cpDNA haplotype sharing (unpubl. data). Moreover, the low seed set after interpopulation crosses, which is similar to seed set after selfing in R. austro‐oreganus, indicates that these populations are fixed for similar complements of deleterious alleles (i.e., they share substantial amounts of drift load; Charlesworth and Willis 2009). The high level of drift load shared among populations of R. austro‐oreganus suggests this lineage recently passed through a genetic bottleneck, which is consistent with its recent peripatric origin.
Under most circumstances, it is expected that a recently derived inbred lineage with low fecundity would be short lived, but during divergence R. austro‐oreganus appears to have acquired cytoplasmic factors that may inhibit genetic swamping from R. occidentalis. The presence of groups of coincident and concordant abrupt clines for both cytoplasmic haplotypes and nuclear SNPs, and the asymmetrical mating barriers due to low seed set for crosses to R. austro‐oreganus as the seed parent, is consistent with the effects of cytonuclear epistasis (Asmussen et al. 1989; Arnold 1993; Burton et al. 2013; McKenzie et al. 2016). Such cytonuclear incompatibilities, constituting Bateson‐Muller‐Dobzhansky incompatibilities (BDMI; Orr 1996), appear to be common and have been observed to arise during early stages of divergence (Levin 2003; Greiner et al. 2011; Burton et al. 2013; Barnard‐Kubow et al. 2016). Asymmetrical mating is evident because seed set and seedling fitness were higher with R. occidentalis acting as the maternal parent compared to the reciprocal cross, perhaps because of the effects of heterosis (Rhode and Cruzan 2005). However, we would normally expect a similar level of heterosis after interspecific crosses with R. austro‐oreganus as the maternal parent, so the low seed numbers and offspring fitness indicate the effects of cytonuclear incompatibilities. Cytonuclear incompatibility would be expected to result in asymmetrical introgression (Tiffin et al. 2001; Lowry et al. 2008), but in this case, it appears that the barrier is not strong enough to prevent substantial levels of gene flow from R. occidentalis into R. austro‐oreganus populations.
Ranunculus austro‐oreganus may represent an exception among cases of peripatric derivation because its current distribution may correspond closely to its geographic origin. The two phenotypic traits differentiating R. austro‐oreganus are consistent with its origin in a nearby high‐elevation location; higher leaf trichome density provides UV protection at high elevation, and anthocyanin pigments provide better heat absorption in cold climates for more rapid floral development (Landi et al. 2015; Buckley et al. 2019). The only taxonomic character that clearly delineates these species is the ventral petal color, but in its current range, it is difficult to find occurrences of R. austro‐oreganus where all individuals have consistently dark red ventral petal coloration (personal observation). Although it may be debatable whether R. austro‐oreganus is a separate species, such unique peripheral populations are valuable targets for conservation efforts (Lesica and Allendorf 1995).
CLINE ANALYSES
The recent initiation of secondary contact between R. occidentalis and R. austro‐oreganus has allowed us to estimate parameters for time since establishment of the hybrid zone, the per‐generation dispersal rate, and the selection coefficients for SNPs representing adaptive introgression. This region of secondary contact is also unusual because it appears to be undergoing bidirectional expansion, not only for (nearly) neutral clines, but also for the movement of clines representing tension zones and adaptive introgression. The 95% confidence intervals for neutral and tension zone cline centers covered a 10‐km hybrid zone region bounding the presumed location of secondary contact. The position of clusters of tension zone cline centers and high levels of linkage disequilibria in populations on both sides of the hybrid zone indicate ongoing and extensive bidirectional introgression, with some degree of asymmetry biased toward the range of R. austro‐oreganus. The observation of extensive neutral and adaptive introgression along with the lack of coincidence among tension zone clines (barrier loci) indicates a low degree of coupling (Ravinet et al. 2017; Butlin and Smadja 2018), and consequently a weak boundary between these recently diverged species.
The results of the genomic cline analyses were consistent with the geographic cline analysis in several respects. Compared to previous studies (e.g., Gompert and Buerkle 2012; Gompert et al. 2017; Jahner et al. 2021; Wen and Fu 2021), the range of the hybrid index was limited (i.e., from ∼0.5 to ∼0.85), which is consistent with a low level of divergence between these two Ranunculus species. Moreover, a high proportion of the clines analyzed displayed excess ancestry, and most of these had centers displaced toward R. austro‐oreganus. The correspondence between classifications of individual clines based on the geographic and genomic analyses was low. Most previous studies have also found that correlations between genomic and geographic cline parameters were weak or absent (e.g., Teeter et al. 2010; Grossen et al. 2016; Souissi et al. 2018), but one study found high correspondence for an abrupt hybrid zone between strongly differentiated species (Larson et al. 2014). In the case of this Ranunculus hybrid zone, the inclusion of geographic information in the analysis of clines appears to be critical for understanding the strength of reproductive isolation and the potential for selection to affect rates of introgression.
There are many examples of the genomic and fitness effects of adaptive introgression after hybridization (e.g., Martin et al. 2005; Martin et al. 2006; Whitney et al. 2010; Pardo‐Diaz et al. 2012; Hedrick 2013; Martin and Jiggins 2017; Suarez‐Gonzalez et al. 2018; Grant and Grant 2019), but few previous studies have characterized the waves of advance for clines of putatively advantageous alleles following secondary contact (e.g., Menon et al. 2020). By comparing the widths of putatively neutral and adaptive clines, we estimated the velocity of introgression and the selection coefficients required to account for the observed rates of cline expansion. Although the mathematical treatment conducted by Fisher (1937) predicted that cline width would be inversely proportional to the selection coefficient, we found the opposite, as clines that have introgressed further were also wider. However, Fisher's analyses did not account for the effects genetic drift within populations, which would slow the rate of fixation within populations. Fixation rates within populations would also be slowed by the longevity of individual plants, whereas the spread of alleles at the leading edge of the wave could occur on an annual basis through pollen and seed dispersal, and could be accelerated by long‐distance dispersal (Piálek and Barton 1997). Landscape genetic studies on seed‐mediated gene flow for other species in the Rogue River Valley indicate that large mammals may be important dispersal vectors for plants in this geographic region (Cruzan and Hendrickson 2020). Based on the average ratio of displacement of clines to cline width for advantageous alleles (∼0.3), the migration rate is about three times the rate of increase in allele frequencies within populations. Given the early stages of hybrid zone development, there may not have been enough time for cline shape to reflect a balance among the processes of dispersal, drift, and selection.
We do not have information on the phenotypic effects of the expressed genomic regions associated with the advantageous SNPs, but they are unlikely to be linked to environmental (extrinsic) effects of selection because there was little evidence of niche differentiation between these species. On the other hand, six of the nine SNPs displaying putatively adaptive clines, and most of those with the highest selection coefficients, displayed introgression toward R. austro‐oreganus. Given the high level of drift load in this species, these SNPs could be linked to regions representing fixed deleterious alleles in R. austro‐oreganus. The three SNPs representing adaptive clines introgressing into the range of R. occidentalis had the smallest selection coefficients. Two of these represent cytoplasmic haplotypes, and all three are apparently associated with novel mutations acquired during divergence of the R. austro‐oreganus lineage. The presence of novel advantageous mutations in this lineage is unexpected because the overall level of genetic divergence is low, and beneficial mutations are generally considered to be rare (Orr 2010). Further characterization of the genomic regions associated with these SNPs and their phenotypic effects would be required to determine the nature of their associated fitness advantages.
The hybrid zone separating these two lineages of Ranunculus is unique because it is only around 10‐km wide and the clines representing tension zones are narrower than the per‐generation dispersal rate. In contrast, almost all other hybrid zones extend over tens to hundreds of kilometers and are characterized by large numbers of coincident and concordant clines representing stable species boundaries (Toews and Brelsford 2012; De La Torre et al. 2015; Gompert et al. 2017; Stankowski et al. 2017; McEntee et al. 2020; Slager et al. 2020). Such abrupt clines are unusual, but similarly narrow clines have been recorded for a field cricket hybrid zone (Larson et al. 2014). The cricket clines differed from the abrupt clines in the Ranunculus hybrid zone because they were coincident and appeared to be maintained by prezygotic mating barriers. The noncoincidental clusters of tension zone clines separating the two species of Ranunculus are more likely to represent groups of interacting genomic regions contributing to postzygotic barriers. Prezygotic barriers are unlikely because the bowl‐shaped flowers of these plants are visited by a variety of insects, and based on levels of seed set, there does not appear to be selection against intermediate flower morphologies (unpubl. data), which would be required for the maintenance of sharp clines.
The majority of clines observed between these two species of Ranunculus were characterized as neutral, or nearly neutral to selection; they are expanding into the ranges of both species with their centers remaining close to the original location of secondary contact. The bidirectional expansion of these clines, and the hybrid zone in general, is supported by the observation of high levels of linkage disequilibria in populations on both sides of the region defined by the majority of neutral and tension zone cline centers. High levels of linkage disequilibria are typical of hybrid zone movement (Barton and Hewitt 1985; Cruzan 2005; van Riemsdijk et al. 2019; Wielstra 2019), but the Ranunculus hybrid zone is unique because there are high levels of disequilibria on both sides, indicating that introgression is progressing into the ranges of both species. This bidirectional expansion appears to include the movement of clusters of tension zone clines, waves of adaptive introgression, and the widening of putatively neutral clines. For the neutral clines, there is a range of cline widths, so it is possible that some of the genomic regions represented by these SNPs could be affected by selection. Selection estimates for the narrowest adaptive clines were minimal, so it is more likely that the narrower neutral clines are weakly affected by selection against interspecific genomic combinations. Although the taxonomic distinction between these species may persist by the presence of differences in ventral petal color, their genetic distinction will continue to erode with the progress of introgression across this weak species boundary.
The clines for the two phenotypic traits differentiating these two species of Ranunculus appear to be similar to the neutral (leaf trichome density) or tension zone (ventral petal color) SNP clines. The trichome density cline was relatively wide with its center close to the region of secondary contact suggesting that this trait is neutral or only weakly affected by selection. In contrast, the petal color cline was abrupt and similar in shape to the SNP tension zone clines. Given that intermediate petal color morphs did not have lower seed set, this cline is more likely to be maintained by epistasis among interspecific genomic regions, but the nature of this interaction and the causes of low fitness are unknown. Although the petal color cline was not coincident with any of the other tension zone clines, the SNPs analyzed represent only a sample of all of the genomic regions that might be under selection. Although the petal color difference is striking, this trait may be effectively neutral across the current ranges of these species, and the appearance of strong selection may be only due to genomic interactions that have yet to be characterized.
Conclusions
The distribution of phenotypic traits and the weak ecological and genetic differentiation between these species are consistent with the hypothesis that the range‐limited Ranunculus austro‐oreganus originated through peripatric divergence. Unlike other putative examples of peripatric speciation, R. austro‐oreganus displays characteristics of a lineage that has recently recovered from a severe population bottleneck, which is consistent with divergence through selection and genetic drift in a peripherally isolated population. These two species are separated by a narrow region of secondary contact. The structure of this hybrid zone is affected to some degree by cytonuclear incompatibility, which is supported by asymmetrical cross‐compatibility and the presence of clusters of coincident and concordant tension zone clines that include nuclear SNPs and cytoplasmic haplotypes. Although cytonuclear epistasis is predicted to favor introgression from the range‐limited species into the range of the widespread species, the low fitness of R. austro‐oreganus populations would favor introgression in the opposite direction. The clusters of tension zone clines are not coincidental, resulting in weak coupling and high levels of introgression across the species boundary. The observed high levels of linkage disequilibria on both sides of the hybrid zone indicate that introgression is bidirectional and ongoing. This region of secondary contact is unique in many respects, including its recent origin, bidirectional expansion, and the presence of clines representing neutral and adaptive introgression. The hybrid zone between these species of Ranunculus appears to be at early stages of dissolution, and ongoing introgression is predicted to ultimately result in genetic swamping of the range‐limited species.
AUTHOR CONTRIBUTIONS
PGT managed field sampling, helped design the greenhouse experiments, and contributed to data analysis and writing of the manuscript. KAK conducted the flowering phenology and flower morphology analyses, and assisted with field work as an REU student. KRG conducted the ecological niche modeling and assisted with field work as part of her Senior Honors Thesis. HMM conducted the field seed set surveys and image analyses of phenotypic traits as an REU student. NAD conducted niche modeling, helped generate geographic maps, and contributed to writing of the manuscript. ECH conducted the genomic cline analyses and wrote a portion of the manuscript. JMP conducted lab and bioinformatic analyses to identify SNPs, helped design the greenhouse experiments, assisted with field work, and contributed to writing of the manuscript as part of her MS thesis. MBC helped with experimental design, field sampling, conducted cline analyses, linkage disequilibria estimates, and hybrid zone age estimates, and wrote the majority of the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA ARCHIVING
All data presented in this study are publicly available through deposits in Data DRYAD (https://doi.org/10.5061/dryad.66t1g1k2z).
Associate Editor: T. Flatt
Handling Editor: T. Chapman
Supporting information
Fig.S1. Sampling locations for the Ranunculus occidentalis (yellow), R. austro‐oreganus (orange), and their putative hybrids (green) in Jackson County, Oregon. The inset shows the geographic range of R. occidentalis in green and of R. austro‐oreganus (small orange rectangle).
Fig. S2. The relationship between the mean and the variance in ventral petal color for populations of Ranunculus occidentalis (yellow markers), R. austro‐oreganus (red markers), and hybrids (green markers) in Jackson County, Oregon. (quadratic model adjusted R 2 = 0.66, F 2/27 = 29.2, P < 0.0001). Marker colors correspond to the sampling locations shown in Figure 1. The two populations of each species with the lowest variance in ventral petal color (blue boxes) were used to identify SNP outlier loci.
Fig. S3. Predicted geographic distributions for Ranunculus austro‐oreganus (red) and R. occidentalis (yellow), and areas where both species are predicted to occur (orange) in the Rogue River Valley region (Jackson County) of southern Oregon (inset map) based on ecological niche modeling. Sample locations are indicated by black markers.
Fig. S4. Genetic cluster analysis based on 2196 SNPs for K = 2. Populations are ordered by location from west to east. Populations are categorized as representing Ranunculus occidentalis, R. austro‐oreganus, or putative hybrids based on the presence of ventral petal color morphs. The populations with green stars within the R. austro‐oreganus range had some plants with yellow ventral petals present.
Fig. S5. The significant cline models from HZAR for 50 SNP outlier loci from the hybrid zone between Ranunculus occidentalis (yellow outlines) and R. austro‐oreganus (red outlines). Cines are grouped according to their classification as tension zones, or cases of neutral or adaptive introgression. Nine of the SNPs only occurred as homozygotes or their adjacent regions matched plant mtDNA (Table S3) and were assumed to represent cytoplasmic genomes (blue outlines). Cytoplasmic SNPs within two groups (four shaded blue and two shaded green) appeared to be segregating together and were assumed to represent two different haplotypes.
Fig. S6. A summary of the numbers of pairs of loci having significant linkage disequilibria for populations with the highest estimates of in the range of Ranunculus occidentalis (yellow squares), R. austro‐oreganus (red squares), or both for the three groups of clines (tension zones, and neutral and adaptive introgression) in Fig. S6.
Fig. S7. Top: The histogram describes the distribution of hybrid index values across all 50 loci fall between 0.5 and 0.85. Bottom: Parameter outliers from Bayesian genomic cline analysis (bgc; Gompert and Buerkle 2012) for the 50 clines between Ranunculus occidentalis and R. austro‐oreganus. The dashed line indicates the expected relationship when R. occidentalis ancestry (Φ) is equivalent to the hybrid index. Cline with greater than expected R. occidentalis ancestry (yellow; α 95% credible interval > 0) or rate (red; β 95% credible interval > 0) or both (green) are above the dashed line (Φ > hybrid index), those with lower than expected ancestry are below the dashed line (Φ < hybrid index).
Fig. S8. Scatterplot of the cline center (α) and rate (β) parameters from Bayesian genomic cline analysis (bgc; Gompert and Buerkle 2012) for the 50 clines between Ranunculus occidentalis and R. austro‐oreganus. Yellow markers and polygons for α < 0 indicate clines with excess R. austro‐oreganus ancestry, and for α > 0 indicate clines with excess R. occidentalis ancestry. Gray markers were not outliers for either parameter. The darker blue shades indicate larger concentrations of loci. The red markers indicate clines with lower than expected rates of introgression (β > 0), and the green marker indicates a cline with excess ancestry and a higher than expected rate of introgression.
Appendix 1. Reproductive isolation estimates.
Appendix 2. GBS and bioinformatics processing.
Appendix 3. Supplemental Tables
ACKNOWLEDGMENTS
The authors are grateful for assistance in the field and the lab from former REU students, A. Hamilton and A. Pheil, and graduate students, T. Arredondo, M. Grasty, B. Kohrn, and J. Schwoch. The authors thank N. Barton, K. de Lima Berg, and M. Streisfeld for comments on the manuscript. The authors thank M. Sultany, high school science teacher, for volunteering to take microscopy images of petals. The authors are also grateful to M. Morrison at The Nature Conservancy and K. Mergenthaler with the Southern Oregon Land Conservancy for assistance in locating and accessing Ranunculus populations. Specimen data were provided by L. Hardison from the Oregon Flora Project, D. Giblin from the Consortium of Pacific Northwest Herbaria, and M. Link‐Perez from the OSU Herbarium. This research was supported by National Science Foundation Macrosystems Biology award NSF‐MSB‐1340746 to MBC.
LITERATURE CITED
- Abbott, R. , Albach D., Ansell S., Arntzen J. W., Baird S. J. E., Bierne N., Boughman J. W., Brelsford A., Buerkle C. A., Buggs R., et al. 2013. Hybridization and speciation. J. Evol. Biol. 26:229–246. [DOI] [PubMed] [Google Scholar]
- Abbott, R. J. , Barton N. H., and Good J. M.. 2016. Genomics of hybridization and its evolutionary consequences. Mol. Ecol. 25:2325–2332. [DOI] [PubMed] [Google Scholar]
- Anderson, E. 1949. Introgressive hybridization. John, Wiley & Sons, New York. [Google Scholar]
- Anderson, E. , and Stebbins G. L. Jr. 1954. Hybridization as an evolutionary stimulus. Evolution 8:378–388. [Google Scholar]
- Arnold, J. 1993. Cytonuclear disequilibria in hybrid zones. Annu. Rev. Ecol. Syst. 24:521–554. [Google Scholar]
- Arnold, M. L. 1997. Natural hybridization and evolution. Oxford Univ. Press, New York. [Google Scholar]
- Asmussen, M. A. , Arnold J., and Avise J. C.. 1989. The effects of assortative mating and migration on cytonuclear associations in hybrid zones. Genetics 122:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baack, E. , Melo M. C., Rieseberg L. H., and Ortiz‐Barrientos D.. 2015. The origins of reproductive isolation in plants. New Phytol. 207:968–984. [DOI] [PubMed] [Google Scholar]
- Barnard‐Kubow, K. B. , So N., and Galloway L. F.. 2016. Cytonuclear incompatibility contributes to the early stages of speciation. Evolution 70:2752–2766. [DOI] [PubMed] [Google Scholar]
- Barton, N. H. 1979. The dynamics of hybrid zones. Heredity 43:341–359. [Google Scholar]
- ———. 1983. Multilocus clines. Evolution 37:454–471. [DOI] [PubMed] [Google Scholar]
- ———. 2001. The role of hybridization in evolution. Mol. Ecol. 10:551–568. [DOI] [PubMed] [Google Scholar]
- Barton, N. H. , and Hewitt G. M.. 1985. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 16:113–148. [Google Scholar]
- Barton, N. H. , and Gale K. S.. 1993. Genetic analysis of hybrid zones. Pp. 13–45 in Harrison R. G., ed. Hybrid zones and the evolutionary process. Oxford Univ. Press, New York. [Google Scholar]
- Beaumont, M. A. 2005. Adaptation and speciation: what can Fst tell us? Trends Ecol. Evol. 20:435–440. [DOI] [PubMed] [Google Scholar]
- Buckley, J. , Widmer A., Mescher M. C., and De Moraes C. M.. 2019. Variation in growth and defence traits among plant populations at different elevations: implications for adaptation to climate change. J. Ecol. 107:2478–2492. [Google Scholar]
- Buggs, R. J. A. 2007. Empirical study of hybrid zone movement. Heredity 99:301–312. [DOI] [PubMed] [Google Scholar]
- Burton, R. S. , Pereira R. J., and Barreto F. S.. 2013. Cytonuclear genomic interactions and hybrid breakdown. Annu. Rev. Ecol. Evol. Syst. 44:281–302. [Google Scholar]
- Butlin, R. K. , and Smadja C. M.. 2018. Coupling, reinforcement, and speciation. Am. Nat. 191:155–172. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D. , and Willis J. H.. 2009. The genetics of inbreeding depression. Nat. Rev. Genet. 10:783–796. [DOI] [PubMed] [Google Scholar]
- Christie, K. , and Strauss S. Y.. 2018. Along the speciation continuum: quantifying intrinsic and extrinsic isolating barriers across five million years of evolutionary divergence in California jewelflowers. Evolution 72:1063–1079. [DOI] [PubMed] [Google Scholar]
- Crochet, P. A. , Chen J. Z., Pons J.‐M., Lebreton J. D., Hebert P. D. N., and Bonhomme F.. 2003. Genetic differentiation at nuclear and mitochondrial loci among large white‐headed gulls: sex‐biased interspecific gene flow? Evolution 57:2865–2878 [DOI] [PubMed] [Google Scholar]
- Cruzan, M. B. 2005. Patterns of introgression across an expanding hybrid zone: analyzing historical patterns of gene flow using non‐equilibrium approaches. New Phytol. 167:267–278. [DOI] [PubMed] [Google Scholar]
- ———. 2018. Evolutionary biology ‐ a plant perspective. Oxford Univ. Press, New York. [Google Scholar]
- Cruzan, M. B. , and Hendrickson E. C.. 2020. Landscape genetics of plants – challenges and opportunities. Plant Commun. 1:100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmahapatra, K. K. , Blum M. J., Aiello A., Hackwel S., Davies N., Bermingham E. P., and Mallet J.. 2002. Inferences from a rapidly moving hybrid zone. Evolution 56:741–753. [DOI] [PubMed] [Google Scholar]
- De La Torre, A. , Ingvarsson P. K., and Aitken S. N.. 2015. Genetic architecture and genomic patterns of gene flow between hybridizing species of Picea . Heredity 115:153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derryberry, E. P. , Derryberry G. E., Maley J. M., and Brumfield R. T.. 2014. hzar: hybrid zone analysis using an R software package. Mol. Ecol. Resour. 14:652–663. 10.1111/1755-0998.12209 [DOI] [PubMed] [Google Scholar]
- Devitt, T. J. , Baird S. J. E., and Moritz C.. 2011. Asymmetric reproductive isolation between terminal forms of the salamander ring species Ensatina eschscholtzii revealed by fine‐scale genetic analysis of a hybrid zone. BMC Evol. Biol. 11:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire, R. J. , Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K., Buckler E. S., and Mitchell S. E.. 2011. A robust, simple genotyping‐by‐sequencing (GBS) approach for high diversity species. PLoS ONE 6:e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier, L. , and Lischer H. E. L.. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10:564–567. [DOI] [PubMed] [Google Scholar]
- Fisher, R. A. 1937. The wave of advance of advantageous genes. Ann. Eugen. 7:355–369. [Google Scholar]
- Fitzpatrick, B. M. 2013. Alternative forms for genomic clines. Ecol. Evol. 3:1951–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, B. M. , Johnson J. R., Kump D. K., Smith J. J., Voss S. R., and Shaffer H. B.. 2010. Rapid spread of invasive genes into a threatened native species. Proc. Natl. Acad. Sci. USA 107:3606–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, J. K. 1993. Modes of peripheral isolate formation and speciation. Syst. Biol. 42:373–381. [Google Scholar]
- Gompert, Z. , and Buerkle A. C.. 2010. introgress: a software package for mapping components of isolation in hybrids. Mol. Ecol. Resour. 10:378–384. [DOI] [PubMed] [Google Scholar]
- ———. 2011. Bayesian estimation of genomic clines. Mol. Ecol. 20:2111–2127. [DOI] [PubMed] [Google Scholar]
- ———. 2012. bgc: software for Bayesian estimation of genomic clines. Mol. Ecol. Resour. 12:1168–1176. [DOI] [PubMed] [Google Scholar]
- Gompert, Z. , Mandeville E. G., and Buerkle C. A.. 2017. Analysis of population genomic data from hybrid zones. Annu. Rev. Ecol. Evol. Syst. 48:207–229. [Google Scholar]
- Grant, P. R. , and Grant B. R.. 2019. Hybridization increases population variation during adaptive radiation. Proc. Natl. Acad. Sci. USA 116:23216–23224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner, S. , Rauwolf U., Meurer J., and Herrmann R. G.. 2011. The role of plastids in plant speciation. Mol. Ecol. 20:671–691. [DOI] [PubMed] [Google Scholar]
- Grossen, C. , Seneviratne S. S., Croll D., and Irwin D. E.. 2016. Strong reproductive isolation and narrow genomic tracts of differentiation among three woodpecker species in secondary contact. Mol. Ecol. 25:4247–4266. [DOI] [PubMed] [Google Scholar]
- Grossenbacher, D. L. , Veloz S. D., and Sexton J. P.. 2014. Niche and range size patterns suggest that speciation begins in small, ecologically diverged populations in North American monkeyflowers (Mimulus spp.). Evolution 68:1270–1280. [DOI] [PubMed] [Google Scholar]
- Guichoux, E. , Garnier‐Géré P., Lagache L., Lang T., Boury C., and Petit R. J.. 2013. Outlier loci highlight the direction of introgression in oaks. Mol. Ecol. 22:450–462. [DOI] [PubMed] [Google Scholar]
- Harrison, R. G. 1990. Hybrid zones: windows on evolutionary process. Oxford Surv. Evol. Biol. 7:69–128. [Google Scholar]
- Harrison, R. G. , and Larson E. L.. 2016. Heterogeneous genome divergence, differential introgression, and the origin and structure of hybrid zones. Mol. Ecol. 25:2454–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick, P. W. 2013. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 22:4606–4618. [DOI] [PubMed] [Google Scholar]
- Hendry, A. P. , Nosil P., and Rieseberg L. H.. 2007. The speed of ecological speciation. Funct. Ecol. 21:455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X. S. , and Li B.. 2002. Seed and pollen flow and cline discordance among genes with different modes of inheritance. Heredity 88:212–217. [DOI] [PubMed] [Google Scholar]
- Jahner, J. P. , Parchman T. L., and Matocq M. D.. 2021. Multigenerational backcrossing and introgression between two woodrat species at an abrupt ecological transition. Mol. Ecol. 30:4245–4258. [DOI] [PubMed] [Google Scholar]
- Johnson, B. B. , White T. A., Phillips C. A., and Zamudio K. R.. 2015. Asymmetric introgression in a spotted salamander hybrid zone. J. Hered. 106:608–617. [DOI] [PubMed] [Google Scholar]
- Kagan, J. , Vrilakas S., Christy J., Gaines E., Wise L., Pahl C., and Howell K.. 2016. Rare, threatened and endangered species of Oregon. Oregon Biodiversity Information Center, Institute for Natural Resources, Portland State University, Portland, OR. [Google Scholar]
- Key, K. H. L. 1968. The concept of stasipatric speciation. Syst. Zool. 17:14–22. [Google Scholar]
- Kolmogorov, A. N. , Petrovskii I., and Piscunov N.. 1937. A study of the equation of diffusion with increase in the quantity of matter, and its application to a biological problem. Mosc. Univ. Bull. Math. 1:1–25. [Google Scholar]
- Lamer, J. T. , Ruebush B. C., Arbieva Z. H., McClelland M. A., Epifanio J. M., and Sass G. G.. 2015. Diagnostic SNPs reveal widespread introgressive hybridization between introduced bighead and silver carp in the Mississippi River Basin. Mol. Ecol. 24:3931–3943. [DOI] [PubMed] [Google Scholar]
- Landi, M. , Tattini M., and Gould K. S.. 2015. Multiple functional roles of anthocyanins in plant‐environment interactions. Environ. Exp. Bot. 119:4–17. [Google Scholar]
- Larson, E. L. , White T. A., Ross C. L., and Harrison R. G.. 2014. Gene flow and the maintenance of species boundaries. Mol. Ecol. 23:1668–1678. [DOI] [PubMed] [Google Scholar]
- Lehnert, S. J. , DiBacco C., Jeffery N. W., Blakeslee A. M. H., Isaksson J., Roman J., Wringe B. F., Stanley R. R. E., Matheson K., McKenzie C. H., Hamilton L. C., and Bradbury I. R.. 2018. Temporal dynamics of genetic clines of invasive European green crab (Carcinus maenas) in eastern North America. Evol. Appl. 11:1656–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesica, P. , and Allendorf F. W.. 1995. When are peripheral populations valuable for conservation. Conserv. Biol. 9:753–760. [Google Scholar]
- Levin, D. A. 2003. The cytoplasmic factor in plant speciation. Syst. Bot. 28:5–11. [Google Scholar]
- Lotsy, J. P. 1916. Evolution by means of hybridization. Martinus Nijhoff, The Hague, The Netherlands. [Google Scholar]
- Lowry, D. B. , Modliszewski J. L., Wright K. M., Wu C. A., and Willis J. H.. 2008. The strength and genetic basis of reproductive isolating barriers in flowering plants. Philos. Trans. R. Soc. B Biol. Sci. 363:3009–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, B. T. , Chafin T. K., Douglas M. R., and Douglas M. E.. 2020. ClinePlotR: visualizing genomic clines and detecting outliers in R. bioRxiv 10.1101/2020.09.05.284109. [DOI] [Google Scholar]
- Martin, N. H. , Bouck A. C., and Arnold M. L.. 2005. Loci affecting long‐term hybrid survivorship in Louisiana irises: implications for reproductive isolation and introgression. Evolution 59:2116–2124, 2119. [PubMed] [Google Scholar]
- ———. 2006. Detecting adaptive trait introgression between Iris fulva and I. brevicaulis in highly selective field conditions. Genetics 172:2481–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. H. , and Jiggins C. D.. 2017. Interpreting the genomic landscape of introgression. Curr. Opin. Genet. Dev. 47:69–74. [DOI] [PubMed] [Google Scholar]
- Mayr, E. 1942. Systematics and the origin of species. Columbia Univ. Press, New York. [Google Scholar]
- McEntee, J. P. , Burleigh J. G., and Singhal S.. 2020. Dispersal predicts hybrid zone widths across animal diversity: implications for species borders under incomplete reproductive isolation. Am. Nat. 196:9–28. [DOI] [PubMed] [Google Scholar]
- McKenzie, J. L. , Dhillon R. S., and Schulte P. M.. 2016. Steep, coincident, and concordant clines in mitochondrial and nuclear‐encoded genes in a hybrid zone between subspecies of Atlantic killifish, Fundulus heteroclitus . Ecol. Evol. 6:5771–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, A. T. O. , Bartaula R., and Hale I.. 2016. GBS‐SNP‐CROP: a reference‐optional pipeline for SNP discovery and plant germplasm characterization using variable length, paired‐end genotyping‐by‐sequencing data. BMC Bioinformatics 17:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon, M. , Landguth E., Leal‐Saenz A., Bagley J. C., Schoettle A. W., Wehenkel C., Flores‐Renteria L., Cushman S. A., Waring K. M., and Eckert A. J.. 2020. Tracing the footprints of a moving hybrid zone under a demographic history of speciation with gene flow. Evol. Appl. 13:195–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, W. S. 1977. An evaluation of narrow hybrid zones in vertebrates. Q. Rev. Biol. 52:263–277. [Google Scholar]
- Nosil, P. , Funk D. J., and Ortiz‐Barrientos D.. 2009. Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 18:375–402. [DOI] [PubMed] [Google Scholar]
- Orr, H. A. 1996. Dobzhansky, Bateson, and the genetics of speciation. Genetics 144:1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2010. The population genetics of beneficial mutations. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 365:1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo‐Diaz, C. , Salazar C., Baxter S. W., Merot C., Figueiredo‐Ready W., Joron M., McMillan W. O., and Jiggins C. D.. 2012. Adaptive introgression across species boundaries in heliconius butterflies. PLoS Genet. 8:e1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patiño, J. , Carine M., Fernández‐Palacios J. M., Otto R., Schaefer H., and Vanderpoorten A.. 2014. The anagenetic world of spore‐producing land plants. New Phytol. 201:305–311. [DOI] [PubMed] [Google Scholar]
- Payseur, B. A. , and Rieseberg L. H.. 2016. A genomic perspective on hybridization and speciation. Mol. Ecol. 25:2337–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur, B. A. , Krenz J. G., and Nachman M. W.. 2004. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution 58:2064–2078. [DOI] [PubMed] [Google Scholar]
- Piálek, J. , and Barton N. H.. 1997. The spread of an advantageous allele across a barrier: the effects of random drift and selection against heterozygotes. Genetics 145:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens M., and Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph, P. , and Coop G.. 2010. Parallel adaptation: one or many waves of advance of an advantageous allele? Genetics 186:647–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, J. , Bradshaw H. D., and Schemske D. W.. 2003. Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phrymaceae). Evolution 57:1520–1534. [DOI] [PubMed] [Google Scholar]
- Ravinet, M. , Faria R., Butlin R. K., Galindo J., Bierne N., Rafajlović M., Noor M. A. F., Mehlig B., and Westram A. M.. 2017. Interpreting the genomic landscape of speciation: a road map for finding barriers to gene flow. J. Evol. Biol. 30:1450–1477. [DOI] [PubMed] [Google Scholar]
- Rhode, J. M. , and Cruzan M. B.. 2005. Contributions of heterosis and epistasis to hybrid fitness. Am. Nat. 166:E124–E139. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H. , Whitton J., and Gardner K.. 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152:713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal, D. M. , Ramakrishnan A. P., and Cruzan M. B.. 2008. Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Mol. Ecol. 17:4657–4669. [DOI] [PubMed] [Google Scholar]
- SAS . 2007. SAS/STAT user's guide version 9.02. SAS institute, Carey, NC. [Google Scholar]
- Slager, D. L. , Epperly K. L., Ha R. R., Rohwer S., Wood C., Van Hemert C., and Klicka J.. 2020. Cryptic and extensive hybridization between ancient lineages of American crows. Mol. Ecol. 29:956–969. [DOI] [PubMed] [Google Scholar]
- Souissi, A. , Bonhomme F., Manchado M., Bahri‐Sfar L., and Gagnaire P.‐A.. 2018. Genomic and geographic footprints of differential introgression between two divergent fish species (Solea spp.). Heredity 121:579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankowski, S. , Sobel J. M., and Streisfeld M. A.. 2017. Geographic cline analysis as a tool for studying genome‐wide variation: a case study of pollinator‐mediated divergence in a monkeyflower. Mol. Ecol. 26:107–122. [DOI] [PubMed] [Google Scholar]
- Suarez‐Gonzalez, A. , Lexer C., and Cronk Q. C. B.. 2018. Adaptive introgression: a plant perspective. Biol. Lett. 14:20170688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymura, J. M. , and Barton N. H.. 1986. Genetic analysis of a hybrid zone between the fire‐bellied toads, Bombina bombina and Bombina variegata, near Cracow in Southern Poland. Evolution 40:1141–1159. [DOI] [PubMed] [Google Scholar]
- Teeter, K. C. , Thibodeau L. M., Gompert Z., Buerkle C. A., Nachman M. W., and Tucker P. K.. 2010. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution 64:472–485. [DOI] [PubMed] [Google Scholar]
- Tiffin, P. , Olson M. S., and Moyle L. C.. 2001. Asymmetrical crossing barriers in angiosperms. Proc. R. Soc. B Biol. Sci. 268:861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toews, D. P. L. , and Brelsford A.. 2012. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 21:3907–3930. [DOI] [PubMed] [Google Scholar]
- van Riemsdijk, I. , Butlin R. K., Wielstra B., and Arntzen J. W.. 2019. Testing an hypothesis of hybrid zone movement for toads in France. Mol. Ecol. 28:1070–1083. [DOI] [PubMed] [Google Scholar]
- Ward, J. L. , Blum M. J., Walters D. M., Porter B. A., Burkhead N., and Freeman B.. 2012. Discordant introgression in a rapidly expanding hybrid swarm. Evol. Appl. 5:380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, D. L. , Cardillo M., Rosauer D. F., and Bolnick D. I.. 2014. Mistaking geography for biology: inferring processes from species distributions. Trends Ecol. Evol. 29:572–580. [DOI] [PubMed] [Google Scholar]
- Wellenreuther, M. , Muñoz J., Chávez‐Ríos J. R., Hansson B., Cordero‐Rivera A., and Sánchez‐Guillén R. A.. 2018. Molecular and ecological signatures of an expanding hybrid zone. Ecol. Evol. 8:4793–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, G. , and Fu J.. 2021. Isolation and reconnection: demographic history and multiple contact zones of the green odorous frog (Odorrana margaretae) around the Sichuan Basin. Mol. Ecol. 30:4103–4117. [DOI] [PubMed] [Google Scholar]
- Whitney, K. D. , Randell R. A., and Rieseberg L. H.. 2010. Adaptive introgression of abiotic tolerance traits in the sunflower Helianthus annuus. New Phytol. 187:230–239. [DOI] [PubMed] [Google Scholar]
- Wielstra, B. 2019. Historical hybrid zone movement: more pervasive than appreciated. J. Biogeogr. 46:1300–1305. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig.S1. Sampling locations for the Ranunculus occidentalis (yellow), R. austro‐oreganus (orange), and their putative hybrids (green) in Jackson County, Oregon. The inset shows the geographic range of R. occidentalis in green and of R. austro‐oreganus (small orange rectangle).
Fig. S2. The relationship between the mean and the variance in ventral petal color for populations of Ranunculus occidentalis (yellow markers), R. austro‐oreganus (red markers), and hybrids (green markers) in Jackson County, Oregon. (quadratic model adjusted R 2 = 0.66, F 2/27 = 29.2, P < 0.0001). Marker colors correspond to the sampling locations shown in Figure 1. The two populations of each species with the lowest variance in ventral petal color (blue boxes) were used to identify SNP outlier loci.
Fig. S3. Predicted geographic distributions for Ranunculus austro‐oreganus (red) and R. occidentalis (yellow), and areas where both species are predicted to occur (orange) in the Rogue River Valley region (Jackson County) of southern Oregon (inset map) based on ecological niche modeling. Sample locations are indicated by black markers.
Fig. S4. Genetic cluster analysis based on 2196 SNPs for K = 2. Populations are ordered by location from west to east. Populations are categorized as representing Ranunculus occidentalis, R. austro‐oreganus, or putative hybrids based on the presence of ventral petal color morphs. The populations with green stars within the R. austro‐oreganus range had some plants with yellow ventral petals present.
Fig. S5. The significant cline models from HZAR for 50 SNP outlier loci from the hybrid zone between Ranunculus occidentalis (yellow outlines) and R. austro‐oreganus (red outlines). Cines are grouped according to their classification as tension zones, or cases of neutral or adaptive introgression. Nine of the SNPs only occurred as homozygotes or their adjacent regions matched plant mtDNA (Table S3) and were assumed to represent cytoplasmic genomes (blue outlines). Cytoplasmic SNPs within two groups (four shaded blue and two shaded green) appeared to be segregating together and were assumed to represent two different haplotypes.
Fig. S6. A summary of the numbers of pairs of loci having significant linkage disequilibria for populations with the highest estimates of in the range of Ranunculus occidentalis (yellow squares), R. austro‐oreganus (red squares), or both for the three groups of clines (tension zones, and neutral and adaptive introgression) in Fig. S6.
Fig. S7. Top: The histogram describes the distribution of hybrid index values across all 50 loci fall between 0.5 and 0.85. Bottom: Parameter outliers from Bayesian genomic cline analysis (bgc; Gompert and Buerkle 2012) for the 50 clines between Ranunculus occidentalis and R. austro‐oreganus. The dashed line indicates the expected relationship when R. occidentalis ancestry (Φ) is equivalent to the hybrid index. Cline with greater than expected R. occidentalis ancestry (yellow; α 95% credible interval > 0) or rate (red; β 95% credible interval > 0) or both (green) are above the dashed line (Φ > hybrid index), those with lower than expected ancestry are below the dashed line (Φ < hybrid index).
Fig. S8. Scatterplot of the cline center (α) and rate (β) parameters from Bayesian genomic cline analysis (bgc; Gompert and Buerkle 2012) for the 50 clines between Ranunculus occidentalis and R. austro‐oreganus. Yellow markers and polygons for α < 0 indicate clines with excess R. austro‐oreganus ancestry, and for α > 0 indicate clines with excess R. occidentalis ancestry. Gray markers were not outliers for either parameter. The darker blue shades indicate larger concentrations of loci. The red markers indicate clines with lower than expected rates of introgression (β > 0), and the green marker indicates a cline with excess ancestry and a higher than expected rate of introgression.
Appendix 1. Reproductive isolation estimates.
Appendix 2. GBS and bioinformatics processing.
Appendix 3. Supplemental Tables
Data Availability Statement
All data presented in this study are publicly available through deposits in Data DRYAD (https://doi.org/10.5061/dryad.66t1g1k2z).


