Abstract
Background
Fibromyalgia (FM) is characterized by chronic widespread pain. Its pathophysiological mechanisms remain poorly understood, and effective diagnosis and treatments are lacking. This study aimed to identify significantly changed biosignatures in FM and propose a novel classification for FM based on pain and soreness (sng) symptoms.
Methods
Urine and serum samples from 30 FM patients and 25 controls underwent metabolomic and proteomic profiling.
Results
Compared with controls, FM patients showed significant differential expression of three metabolites in urine and five metabolites and eight proteins in serum. Of them, DETP, 4‐guanidinobutanoic acid, SM(d18:1/18:0), PC(20:1(11Z)/18:0), S100A7, SERPINB3, galectin‐7 and LYVE1 were first reported as potential biomarkers for FM. Furthermore, lactate, 2‐methylmaleate and cotinine in urine and lactate, SM(d18:1/25:1), SM(d18:1/26:1) and prostaglandin D2 (PGD2) and PCYOX1, ITIH4, PFN1, LRG1, C8G, C8A, CP, CDH5 and DBH in serum could differentiate pain‐ (PG) and sng‐dominant groups (SG). Lactate, 2‐methylmaleate, cotinine, PCYOX1, ITIH4, PFN1 and DBH have a higher level in SG. SM(d18:1/25:1), SM(d18:1/26:1), PGD2, LRG1, C8G, C8A, CP and CDH5 in SG are lower than PG. The omics results indicated disordered free radical scavenging, and lipid and amino acid metabolism networks and resulting NF‐κB‐dependent cytokine generation in FM. Lactate level was altered simultaneously in urine and serum and significantly higher in sng‐dominant patients than others.
Conclusions
In this study, we identified potential biomarkers from FM patients. The selected biomarkers could discriminate sng and pain phenotypes in FM patients. These results could help elucidate the underlying pathological mechanisms for more effective diagnosis and therapy for FM.
1. INTRODUCTION
Fibromyalgia (FM) is a chronic, widespread pain disorder (Clauw et al., 2011). It is also considered to encompass a broad array of somatic and psychological symptoms as well as a ‘chronic pain amplification syndrome’ characterized by dysregulation of variable combinations of autonomic, neuroendocrine, immune and nociceptive processing functions (Smith et al., 2011). It is more prevalent in the female gender and is also usually accompanied by headaches, fatigue, sleep disturbances, cognitive dysfunctions and circadian rhythm disturbances (Jahan et al., 2012; Perrot, 2019; Wolfe et al., 1990, 2010). In addition to pain, soreness (also designated as ‘sng’, pronounced sә‐ng) is another common complaint amongst FM patients (Chang et al., 2020b; Lin et al., 2018). Sng represents the state of soreness whilst simultaneously imitating the natural vocalization of human beings feeling sore. Sngception (soreness sensation) can decrease maximal muscle strength and joint range of motion and limit activities of daily living (Mautner & Sussman, 2016). Until now, the mechanism through which sngception alters patients’ wellness and quality of life remains undetermined.
The clinical features of FM have been characterized, and FM is diagnosed solely on clinical grounds (Wolfe et al., 1990). FM diagnosis and therapy are challenging because of the lack of accurate diagnostic methods. FM has been intensively investigated, but the aetiology and pathogenesis remain elusive. Physicians currently rely on patient‐reported information about a multitude of symptoms to diagnose FM, which is frequently misdiagnosed or undiagnosed. Indeed, no reliable molecular biomarkers have been identified, and the diagnosis of FM still depends on in‐depth clinical evaluation. Some patients remain undiagnosed, leading to postponed care and poor management of symptoms (Arnold et al., 2011; Cohen, 2017).
Systems biology (e.g., metabolomics, an effective post‐genomic research tool) has been widely used to explore the mechanism, aetiology and/or biomarkers of complex disease (Zhang et al., 2015). The lipidome is a subset of the metabolome and plays multiple roles in cellular signalling, bioenergetics and membrane structure and function (Subramaniam et al., 2011). Proteomics is used for detecting diagnostic markers, understanding pathogenic mechanisms and interpreting functional protein pathways in various diseases by identifying and quantifying the ‘proteome’ of the cell, tissue or body fluids (Graves & Haystead, 2002). Integrating multi‐omics has been used to characterize and decipher the underlying pathomechanisms, explore potential pathogenic factors, and provide more effective diagnosis and treatment for the disease (Gross & Han, 2011).
In this study, we investigated the untargeted metabolomic, lipidomic and proteomic patterns in urine and serum from FM patients. We aimed to construct the possible pathogenic network of FM based on the correlation of levels of metabolites and clinical parameters. Furthermore, we aimed to explore the differences between pain and sng in FM.
2. MATERIALS AND METHODS
2.1. Participants, settings and clinical evaluations
Patients >20 years old who fulfilled the 2011 American College of Rheumatology criteria for FM were recruited from the outpatient clinics of the departments of Neurology and Physical Medicine and Rehabilitation in National Taiwan University Hospital from July 2017 to July 2018 (Wolfe et al., 2011). Briefly, patients had (1) a widespread pain index (WPI) ≧7 and symptom severity (SS) scale score ≧5 or WPI 3–6 and SS scale score ≧9; (2) symptoms lasting for at least 3 months and (3) no other disorders accounting for the pain. The WPI indicates whether patients have pain or tenderness in 19 regions, including the neck, chest and abdomen as well as bilateral temporomandibular joints, shoulders, arms, forearms, buttocks, thighs, calves and back. We excluded patients who were unable to express themselves clearly or who had an acute infection, malignancy or history of major surgery. To assess the impact of both pain and sng, each participant completed the Revised Fibromyalgia Impact Questionnaire with Integration of Soreness Assessment (FIQR‐S), including WPI, widespread sng index (WSI), pain visual analogue scale (P‐VAS), and sng visual analogue scale (S‐VAS) (Chang et al., 2020b; Lin et al., 2018). For omics studies, FM patients were further divided into three phenotypes: pain‐dominant group (PG), sng‐dominant group (SG) and no‐dominant group (NG) based on the difference between P‐VAS × WPI and S‐VAS × WSI and according to the following: S‐VAS × WSI − P‐VAS × WPI < −10; S‐VAS × WSI − P‐VAS × WPI > 10 and S‐VAS × WSI − P‐VAS × WPI = 10 to −10, respectively (Table 1). Age‐ and sex‐matched controls were recruited from adults receiving regular health check‐ups at the National Taiwan University Beihu Branch.
TABLE 1.
Clinical characteristics of patients with fibromyalgia
| Healthy controls | FM patients | ||||
|---|---|---|---|---|---|
| Total | SG | PG | NG | ||
| Age | 51.17 ± 1.76 | 52.07 ± 1.97 | 50.22 ± 3.54 | 51.88 ± 2.9 | 57.00 ± 1.41 |
| P‐VAS score | ND | 5.27 ± 0.35 | 4.33 ± 0.87 | 5.94 ± 0.31# | 4.50 ± 0.64 |
| WPI | ND | 7.97 ± 0.95 | 2.77 ± 0.79** | 10.88 ± 0.86* , ## | 8.00 ± 3.36 |
| S‐VAS score | ND | 4.03 ± 0.56 | 7.44 ± 0.68** | 2.17 ± 0.49* , ## | 4.50 ± 0.64 # , $ |
| WSI | ND | 6.33 ± 0.95 | 10.70 ± 1.15* | 3.64 ± 0.97 ## | 7.75 ± 3.47 |
Data are mean ± SD.
Abbreviations: ND, not detected; NG, no‐dominant sensation group; PG, pain‐dominant group; P‐VAS, pain visual analogue scale; SG, sng‐dominant group; S‐VAS, sng visual analogue scale; WPI, widespread pain index; WSI: widespread sng index.
p < 0.05
p < 0.01 compared with total.
p < 0.05 compared with SG.
p < 0.01 compared with SG.
p < 0.05 compared with PG.
2.2. Urine and serum sample collection
Each participant provided a 10‐ml sample of mid‐stream urine and 10 ml of peripheral blood collected from the antecubital vein upon recruitment. For FM patients, samples were collected before any treatment. Urine samples were centrifuged (1500 g for 10 min at 4°C), aliquoted, and stored at −80°C. Blood samples were collected in EDTA tubes on ice. Serum was separated immediately by centrifugation at 2000 rpm for 15 min and stored at −80°C until analysis.
2.3. Ethics
The study protocol was approved by the Institutional Review Board of National Taiwan University Hospital (IRB No. 201501081RINC). All participants provided written informed consent before entering the study. All clinical investigations were conducted according to the principles of the Declaration of Helsinki. The corresponding authors had full access to all data in the study and had final responsibility for the decision to submit the research for publication.
2.4. Metabolome and lipidome profiles for untargeted and lipid metabolites
Untargeted metabolomes in urine and serum were performed as described by Hsu et al. (Hsu et al., 2019, 2020). Briefly, liquid chromatography tandem mass spectrometry (LC‐MS) analysis involved using an Agilent 1290 UPLC system (ACQUITY UPLC HSS T3 column, 2.1 × 100 mm; 1.8 µm; Waters) coupled with the 6540‐Quadrupole‐Time‐of‐Flight (QTOF) mass system (Agilent Technologies). A mobile phase consisted of 0.1% formic acid in water (solvent A) and acetonitrile (ACN; solvent B) with a run program of 0–1.5 min, 2% B; linear gradient at 1.5–9 min, 2%–50% B and 9–14 min, 50%–95% B and isocratic at 14–15 min, 95% B. The injection volume was 2 μl with a flow rate of 0.3 ml/min in LC. A jet stream electrospray ionization (ESI) source was used for sample ionization. The following parameters were used throughout the study: curtain gas: gas temperature (325°C), gas flow (8 L/min), nebulizer pressure (40 psi), sheath gas temperature (325°C), sheath gas flow (10 L/min) and capillary voltage (40 kV for positive and 35 kV for negative). The mass scan range was set to 50–1700 m/z.
The LC‐MS lipidomic profiling in serum was analysed by using a ZORBAX Eclipse Plus C18 system (2.1 × 100 mm, 1.8 µm, Agilent Technologies) for QTOF with mobile phase A‐0.1% aqueous formic acid and 10 mM ammonium acetate, and mobile phase B‐0.1% formic acid and 10 mM ammonium acetate in ACN/isopropyl alcohol (50/50). The LC program was a linear gradient at 0–2.0 min, 35%–80% mobile phase B; 2.0–7 min, 80%–100% mobile phase B; isocratic from 7 to 14 min with 100% mobile phase B and column re‐equilibration with 100% mobile phase B for 2 min. The flow rate was 0.35 ml/min. The sample reservoir and column oven were maintained at 4°C and 55°C, respectively. The injection volume was 5 μl. MS processed with a positive electrospray ionization mode involved 300°C dry gas temperature, 5 L/min dry gas flow rate, 45 psi nebulizer pressure, 250°C sheath gas temperature, 11 L/min sheath gas flow rate, 3500 V capillary voltage and 500 V nozzle voltage. MS acquisition was executed in precursor ion scan mode. The autosampler and column oven were maintained at 4°C and 55°C, respectively. The injection volume was 5 μl. MS acquisition was performed in precursor ion scan mode and multiple reaction monitoring modes (Liao et al., 2020).
All MS raw data were converted to mzXML format by using Trapper (ISB) and normalized by TIPick, an in‐house package, as well as peak enhancement and peak chosen for the targeted metabolites. An in‐house database of sphingomyelin (SM), lysophosphatidylcholine, ceramides (Cer), phosphatidylcholines (PCs), phosphatidylinositol (PI), phosphatidylethanolamine (PE) and cerebroside (CB) was used for lipid screening. The analyst was blinded to patient group and disease classification.
2.5. Tandem mass tag (TMT)‐based quantitative proteomics
Serum samples were individually immunodepleted by using Proteome Purify 12 Human Serum Protein Immunodepletion Resin (R&D Systems) following the manufacturer's protocol. The immunodepleted serum samples of control participants were randomly pooled into three groups. A pooled sample, serving as an internal reference, consisted of aliquots of protein from all samples. The immunodepleted proteins were reduced, S‐alkylated, trypsin digested and desalted. The desalted peptides were TMT‐labelled by using the TMTsixplex™ Isobaric Label Reagent Set (Thermo Fisher Scientific). A total of eight batches of TMT‐labelled, mixed samples were prepared according to the channel arrangement. Each batch of TMT‐labelled samples was further fractionated by using the High pH Reversed‐Phase Peptide Fractionation Kit (Thermo Fisher Scientific).
The fractionated peptide samples were analysed with the use of Ultimate 3000 RSLCnano coupled with Thermo Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific) on a 75 μm × 25 cm Acclaim PepMapTM C18 column (Thermo Fisher Scientific) with a segmented gradient in 60 min from 5% to 45% solvent B (acetonitrile with 0.1% formic acid) at a flow rate of 300 nl/min. Solvent A was 0.1% formic acid in water. The mass spectrometer was operated in a data‐dependent mode. Survey scans of peptide precursors from m/z 400 to 1600 were performed at 120 K resolution with a 2 × 105 ion count target. The top 10 most intense precursor ions were selected for MS/MS by isolation window at 1.6 Da with the quadrupole, HCD fragmentation with a normalized collision energy of 30 and MS2 scan analysis at 30 K resolution in the orbitrap.
Raw data files from nanoLC‐MS/MS were searched against the Uniprot human database (August, 2020) using the Andromeda algorithm in MaxQuant software (v. 1.6.14.0). TMT quantitation was also performed in MaxQuant with the ‘matching‐between‐run’ function (Yu et al., 2020). Further data processing and statistics were performed using Perseus software (v. 1.6.14.0) as previously described (Yu et al., 2020). Signals from each channel were normalized with the internal reference channel, that is TMT131, in each TMT batch, and further normalized by the quantile normalization method. All nanoLC‐MS/MS raw files and MaxQuant search results were deposited at the ProteomeXchange Consortium (Deutsch et al., 2017) via the PRIDE partner repository data set identifier PXD022886. The analyst is blind to the patient group and disease categorization.
2.6. Statistics
For metabolomics and lipidomics, LC‐MS/MS spectrum data sets were exported to SIMCA‐P+ v12.0 (Umetrics) or MetaboAnalyst 5.0 (http://www.metaboanalyst.ca) for multivariate statistical analysis, principal component analysis (PCA), partial least squares discriminant analysis (PLS‐DA) and orthogonal PLS‐DA (OPLS‐DA) for determining the metabolites that most contributed in discriminating FM patients and controls. Using variable importance in projection (VIP) cut‐off value of 1, we determined whether or not metabolites were potential FM‐relevant signatures. Random forest (RF) classification and mean decrease accuracy (MDA) were used to further refine the features that could discriminate the metabolic changes, with out of bag (OOB) error of RF for serum and urine of 0.191 and 0.245, respectively. To increase the reliability of FM prediction, we calculated the receiver operating characteristic (ROC) curve on the basis of a logistic regression model to determine the area under the ROC curve (AUC). IBM SPSS 23.0 was used to analyse correlations between clinical parameters and targeted metabolites. Descriptive statistics are presented as mean ± SD, median (range) or number (percentage). Pearson correlation coefficients were used to estimate the correlation between the sng‐ and pain‐related indices and levels of metabolites. Pearson correlation analysis was used to evaluate the linear relation between the sng or pain score and metabolite levels after logarithmic transformation. p‐values were used to test the null hypothesis of the correlation between the level of an individual metabolite and clinical sng or pain score. Student t test was used to compare groups as appropriate. All calculated p‐values were two‐tailed. p < 0.05 was considered statistically significant. For proteomics, differences were evaluated with the Student t test, with p < 0.05 as the significance threshold.
2.7. Bioinformatics analyses
Ingenuity Pathway Analysis (IPA) Software (Ingenuity Systems), MetaboAnalyst 5.0 (http://www.metaboanalyst.ca) and ConsensusPathDB (CPDB) (http://cpdb.molgen.mpg.de/) were employed to analyse biological pathway and functional annotation of metabolomics or proteomics data.
To identify key biosignatures and correlation networks from proteomics, metabolomics and lipidomics data, integrative analyses were used with Data Integration Analysis for Biomarker discovery with the Latent cOmponents (DIABLO) program implemented in MixOmics R Bioconductor packages (Singh et al., 2019). DIABLO builds a classification framework with co‐expressed (or correlated) variables from multi‐omics data sets with the multivariate dimension reduction technique, which is a modification of the sGCCA algorithm (Tenenhaus et al., 2014). Candidate metabolites and proteins with at least 30% difference in level between SG and PG in FM patients were selected for DIABLO analyses, with the component number of two in the maximal distance, and three‐fold cross‐validation repeated 50 times.
3. RESULTS
3.1. Metabolomic and lipidomic profiling of urine and serum from FM patients
We recruited 55 participants, including 30 patients (male/female = 1/29) and 25 healthy controls (male/female = 1/24). The mean age was 52.07 ± 1.97 and 51.17 ± 1.76 years, respectively, with no significant difference between the two groups (Table 1). The PCA and OPLS‐DA score plots from multivariate analysis of untargeted metabolomics in urine and serum and lipidomics in serum are in Figure S1a (urine) and S1b (serum). A trend of inter‐group separation in the PCA score plots revealed the separation between controls and FM patients. OPLS‐DA plots showed two clusters clearly separated, thus suggesting that urine and serum metabolomes and lipidomes differed between FM patients and controls.
3.2. Identification of potential metabolites with discriminative features
Student t test analysis of patients and controls showed significantly different levels of 35 and 28 metabolites in serum and urine (Tables S1 and S2). Then, by combining VIP and ROC analyses, we selected six serum metabolites (isoleucine, l‐norleucine, diethylthiophosphate [DETP], tryptophan, PC(20:1(11Z)/18:0) and SM(d18:1/18:0)) and three urine metabolites (hypoxanthine, DETP and 4‐guanidinobutanoic acid) as the most contributing metabolites that simultaneously fulfilled the criteria of VIP >1, p < 0.05 and AUC > 0.75.
To complement the limitations of traditional VIP analysis, we performed RF analysis. The top 15 ranked differential metabolites in the respective models were selected according to MDA, which denoted the percentage decrease in accuracy when the trial was performed in the absence of the metabolites (Figure S2).
Finally, we integrated the metabolomic results (Figure S1) for serum‐ and urine‐derived metabolites for FM patients (Tables S1 and S2). Potential FM‐relevant biosignatures were isoleucine, DETP, tryptophan, PC(20:1(11Z)/18:0) and SM(d18:1/18:0) in serum and hypoxanthine, DETP, and 4‐guanidinobutanoic acid in the urine (Table 2). Notably, DETP was the common metabolite in urine and serum. Levels of hypoxanthine and SM(d18:1/18:0) were higher in FM patients than control and levels of 4‐guanidinobutanoic acid, isoleucine, DETP, tryptophan and PC(20:1(11Z)/18:0) were lower in FM patients than control. In addition, pairwise correlation analysis demonstrated that the level of DETP correlated with the level of tryptophan, isoleucine and SM(d18:1/18:0) and the level of isoleucine significantly correlated with levels of tryptophan and SM(d18:1/18:0) (Figure 1).
TABLE 2.
Potential metabolomic candidates in FM
| Metabolites | HMDB ID | FM/control | p value | VIP score | AUC value | |
|---|---|---|---|---|---|---|
| Urine | Hypoxanthine | HMDB0000157 | 1.782 ± 0.241 | 0.0104 | 1.9699 | 0.7509 |
| Diethylthiophosphate a | HMDB0001460 | 0.603 ± 0.115 | 0.0424 | 1.9671 | 0.7540 | |
| 4‐Guanidinobutanoic acid | HMDB0003464 | 0.587 ± 0.113 | 0.0411 | 4.5964 | 0.7668 | |
| Serum | SM(d18:1/18:0) | HMDB0001348 | 1.294 ± 0.093 | 0.0122 | 1.9189 | 0.7640 |
| Tryptophan | HMDB0000929 | 0.862 ± 0.035 | 0.0433 | 1.5268 | 0.7500 | |
| Isoleucine | HMDB0000172 | 0.804 ± 0.042 | 0.0088 | 1.0986 | 0.7506 | |
| PC(20:1(11Z)/18:0) | HMDB0008300 | 0.679 ± 0.069 | 0.0389 | 1.1639 | 0.7593 | |
| Diethylthiophosphate a | HMDB0001460 | 0.476 ± 0.117 | 0.0034 | 1.2454 | 0.8446 |
Data are mean ± SD unless indicated.
Abbreviations: AUC, area under the receiver‐operating characteristic curve; VIP, variable importance in projection.
Intersection of FM patient serum and urine.
FIGURE 1.
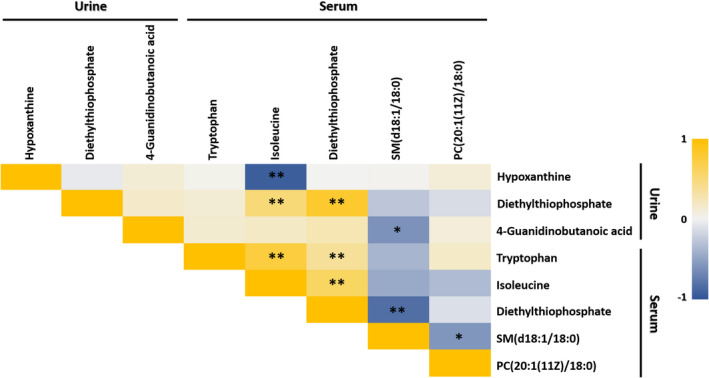
Heat map of correlations amongst all selected potential metabolomic biomarker candidates. Spearman's correlation heat map showing the correlation amongst all selected potential metabolomic and lipidomic biomarkers. Colour intensity represents the magnitude of correlation. Red represents positive correlations, and the green represents negative correlations. * p < 0.05; ** p < 0.01
3.3. Identification of potential proteins as biomarkers in FM patients
We selected eight proteins, including complement C1q C chain (C1qC), protein S100‐A7 (S100A7), serpin B3 (SERPINB3), galectin 7 (LGALS7), lymphatic vessel endothelial hyaluronan receptor 1 (LYVE1), fibrinogen alpha chain (FGA), fibrinogen beta chain (FGB) and fibrinogen gamma chain (FGG) with remarkable differential expression between FM patients and controls. Specifically, levels of S100A7, SERPINB3, LGALS7, FGA, FGB and FGG were lower in FM patients than controls and those of C1qC and LYVE1 were higher (Figure 2). S100A7, SERPINB3, galectin 7 and LYVE1 were first reported here as potential biomarkers in FM patients.
FIGURE 2.
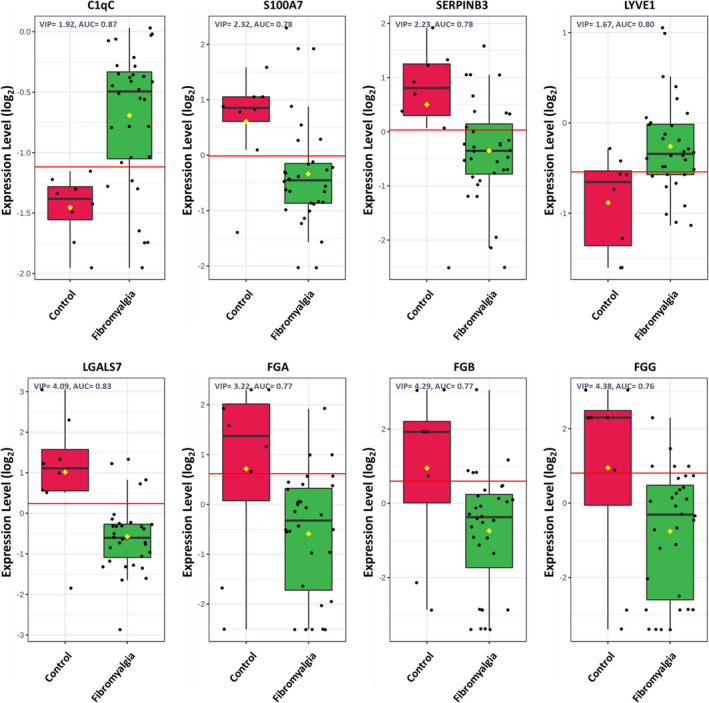
Potential proteomics biomarkers in fibromyalgia (FM). Graphs show serum proteins with a significant change in expression between FM patients and healthy controls for C1qC, S100A7, SERPINB3, LYVE1, LGALS7, FGA, FGB and FGG. The plot shows expression levels on the y‐axis and their group on the x‐axis. Values for all individual cases are shown as dots. Horizontal lines are median, box edges are interquartile range and whiskers are range
3.4. Integration of metabolic and proteomics network for FM
To correlate the FM‐related metabolite changes, we next used the MetaboAnalyst to elucidate the affected metabolic pathways between FM patients and controls. As shown in Figure 3a, several metabolic pathways were altered (impact >0.1, p < 0.05) in FM patients, including d‐glutamine and d‐glutamate metabolism, sphingolipid metabolism, aminoacyl‐tRNA biosynthesis, cysteine and methionine metabolism, glycine, serine and threonine metabolism, tryptophan metabolism and galactose metabolism.
FIGURE 3.
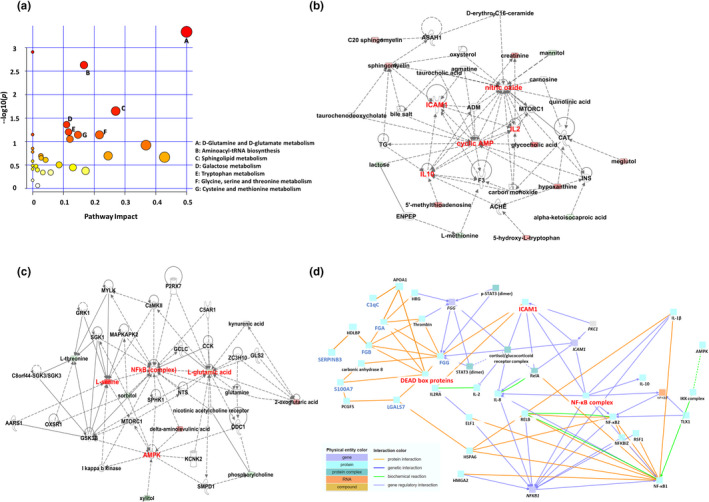
Summary of pathways related to FM and metabolomics–proteomics interaction network analysis. (a) Network pathways identified by using MetaboAnalyst. Metabolites were inferred in FM patients from changes in serum and urine levels of intermediates during substance metabolism. Network analysis of differentially expressed metabolites annotated in the Ingenuity database involved using ingenuity pathway tools (www.ingenuity.com). The plot shows logarithm p values on the y‐axis and their impact factors on the x‐axis. (b) Free radical scavenging and lipid metabolism networks. (c) Amino acid metabolism and molecular transport networks. (d) Use of ConsensusPathDB to analyse the interaction networks of proteomics and hub spots from ingenuity pathway analysis
Furthermore, we combined the differentially expressed metabolites in serum and urine (Tables S1 and S2) and determined the possible molecular mechanisms by using IPA network algorithm. The IPA network analysis exhibited significant perturbation, including in free radical scavenging and lipid metabolism networks (Figure 3b), as well as amino acid metabolism and molecular transport networks (Figure 3c). Nine hub spots, ICAM1, cyclic AMP, AMPK, L‐serine, L‐glutamic acid, nitric oxide, NF‐κB complex, IL‐2 and IL‐10 were identified in these two metabolomic networks. Subsequently, we used CPDB to integrate the proteomics results with hub spots of metabolic networks to construct relationship networks between proteomics and metabolomics (Figure 3d).
3.5. Distinguishing sng‐dominant and pain‐dominant in FM patients
In this study, we indicated that sngception can be evaluated accurately and reliably using a designed questionnaire and recorded in clinical FM diagnosis (Chang et al., 2020b). We analysed the parameters of the clinical questionnaire, P‐VAS, S‐VAS, WPI, WSI, P‐VAS × WPI and S‐VAS × WSI (Table S3), to identify important parameters contributing to the grouping of PG, SG and NG. Correlation analysis revealed that P‐VAS was significantly correlated with WPI and P‐VAS × WPI, whereas S‐VAS was significantly correlated with WSI and S‐VAS × WSI. However, P‐VAS showed no marked correlation with S‐VAS, WSI or S‐VAS × WSI. Likewise, S‐VAS showed no marked correlation with P‐VAS, WPI and P‐VAS × WPI (Figure 4a). These results suggest no correlation between pain and sng sensation. The data set from the clinical questionnaire was further processed by PCA and PLS analyses to generate an unbiased overview of the major clinical differences (Figure 4b). According to VIP (VIP > 1), the most significant discriminatory parameters between the three groups were S‐VAS × WSI (VIP = 2.13) and P‐VAS × WPI (VIP = 1.01) (Figure 4c). Consequently, we defined the value of S‐VAS × WSI as the ‘sng score’ and P‐VAS × WPI as the ‘pain score’. We then used S‐VAS × WSI and P‐VAS × WPI to build a scatter diagram for confirmation. The result substantially divided FM patients into three groups (Figure 4d). WPI was significantly lower, and S‐VAS and WSI were significantly higher for SG patients than all patients. WPI was significantly higher and S‐VAS was significantly lower for PG patients than all patients. PG and SG patients, both exhibited significant differences in WPI, S‐VAS and WSI, with much higher P‐VAS and WPI, as well as lower S‐VAS and WSI in the PG than SG group. S‐VAS was significantly lower in NG than SG group and significantly higher in the NG than SG group (Table 1). Besides, we found that recruited FM patients could be divided into three groups: (1) sng‐dominant (SG) patients, approximately one‐third of patients; (2) pain‐dominant (PG) patients, approximately two‐thirds of patients and (3) no‐dominant patients (NG, both sng and pain), the few remaining patients.
FIGURE 4.
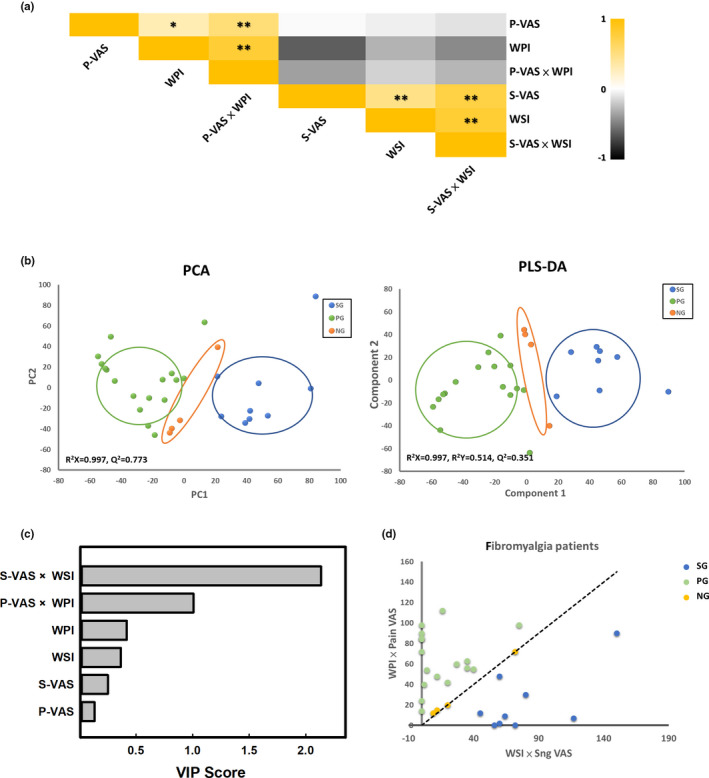
Distinction of different FM phenotypes. (a) Correlation heat map showing the correlation amongst all parameters from the clinical questionnaire. * p < 0.05; ** p < 0.01. (b) Principal component analysis (PCA) and partial least squares discriminant analysis (PLS‐DA) score plots were based on clinical questionnaire data for pain (green), sng (blue) and other (orange) groups. (c) Variable importance in projection analysis based on the weighted coefficients of the PLS‐DA model used to rank the contribution of parameters of the clinical questionnaire to the discrimination between the pain and sng groups in FM patients. (d) Scatter diagram of different phenotypes of FM patients
3.6. Metabolomics profiling analyses based on clinical manifestations
Following our grouping, we further examined the differences in differentially expressed metabolites amongst the PG, SG and NG groups. We found 27 and 20 metabolites in serum (Table S4) and urine (Table S5) with markedly differential expression amongst control, PG, SG and NG groups. Subsequently, we focused on PG and SG groups. In serum, 10 of 27 metabolites showed remarkable differences in levels between PG and SG groups. Levels of androstenedione, prostaglandin D2 (PGD2), SM(d18:1/25:1) and SM(d18:1/26:1) were higher in PG but lower in SG patients compared with controls. Levels of PC(18:2(9Z,12Z)/20:0), PC(18:1(9Z)/20:1(11Z)), PC(20:2(11Z,14Z)/18:0) and PC(20:2(11Z,14Z)/18:1(9Z)) were lower in SG patients than controls, with no significant change in PG patients. Levels of lactate and Cer(d18:1/22:1) were higher in SG but not PG patients than controls (Table S4). In urine, levels of cotinine, lactate and 2‐methylmaleate were increased in SG but decreased in PG patients, and the level of carnitine was decreased in SG but increased in PG patients (Table S5). Lactate was the common metabolite in serum and urine; its level was high in SG but low or with no change in PG patients, with a significant difference between PG and SG groups (p < 0.0105).
3.7. Correlation between differentially expressed metabolites and sng or pain scale
Levels of some metabolites showed a significant correlation with sng or pain scores (Table S6). We next integrated Tables S4–S6, and multiple correlation analysis showed a positive correlation (p < 0.05) between sng score and levels of lactate ( γ = 0.545) and 2‐methylmaleate ( γ = 0.505) as well as a negative correlation (p < 0.05) between pain score and level of cotinine ( γ = −0.441) in urine (Table 3). Moreover, sng score was negatively correlated (p < 0.05) with levels of SM(d18:1/25:1) ( γ = −0.594) and SM(d18:1/26:1) ( γ = −0.608) and positively (p < 0.05) with level of lactate ( γ = 0.612) in serum. Level of PGD2 showed a strong positive correlation ( γ = 0.499) with pain score (Table 3).
TABLE 3.
Potential metabolomic candidates for distinguishing FM subtypes
| Metabolites | HMDB ID | Sng‐dominant group (SG) | Pain‐dominant group (PG) | No‐dominant sensation group (NG) | VIP (SG vs PG) | AUC (SG vs PG) | Pearson correlation with sng VAS × WSI (γ1) | Pearson correlation with pain VAS × WPI (γ2) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FM/control | p value (vs control) | FM/control | p value (vs control) | p value (vs SG) | FM/control | p value (vs control) | |||||||
| Urine | Lactate a | HMDB0000190 | 1.579 ± 0.153 | 0.0915 | 0.753 ± 0.133 | 0.0811 | 0.0105 | 0.928 ± 0.222 | 0.8535 | 1.253 | 0.774 | 0.557** | −0.165 |
| 2‐Methylmaleate | HMDB0000634 | 1.372 ± 0.149 | 0.4245 | 0.782 ± 0.082 | 0.5232 | 0.0018 | 1.152 ± 0.079 | 0.8491 | 1.128 | 0.811 | 0.505** | −0.183 | |
| Cotinine | HMDB0001046 | 1.576 ± 0.272 | 0.0137 | 0.930 ± 0.043 | 0.6829 | 0.0470 | 2.737 ± 0.877 | 0.0001 | 1.128 | 0.754 | 0.176 | −0.441* | |
| Serum | Lactate a | HMDB0000190 | 1.399 ± 0.121 | 0.0441 | 0.958 ± 0.066 | 0.6860 | 0.0143 | 1.002 ± 0.218 | 0.9881 | 1.294 | 0.781 | 0.612* | −0.146 |
| SM(d18:1/25:1) | — | 0.625 ± 0.059 | 0.0426 | 1.683 ± 0.241 | 0.0071 | 0.0044 | 2.398 ± 0.718 | 0.0012 | 1.678 | 0.823 | −0.594* | −0.166 | |
| SM(d18:1/26:1) | HMDB0013461 | 0.744 ± 0.106 | 0.4176 | 1.321 ± 0.151 | 0.2103 | 0.0159 | 1.527 ± 0.503 | 0.2948 | 1.160 | 0.766 | −0.608* | 0.011 | |
| Prostaglandin D2 | HMDB0001403 | 0.558 ± 0.171 | 0.1194 | 1.643 ± 0.296 | 0.0475 | 0.0282 | 1.151 ± 0.353 | 0.6399 | 1.013 | 0.768 | −0.065 | 0.499* | |
Data are mean ± SD unless indicated.
Abbreviations: AUC, area under the receiver‐operating characteristic curve; VIP, variable importance in projection.
Intersection of FM patient serum and urine.
*p value < 0.05.
**p value < 0.01.
3.8. Changed protein levels in pain and sng clinical manifestations
In accordance with the above analysis, we investigated changes in protein levels between PG and SG groups. We found 18 proteins with significant differential expression in both groups (p < 0.05, VIP > 1, AUC > 0.75); levels of nine were correlated with sng or pain scores on Pearson correlation analysis. Sng score was positively correlated with levels of PFN1 ( γ = 0.3595), PCYOX1 ( γ = 0.4619) and ITIH4 ( γ = 0.4195) and negatively with level of LRG1 ( γ = −0.5444) (Table 4). In addition, pain score was positively correlated with levels of C8A ( γ = 0.5065), C8G ( γ = 0.5335), CDH5 ( γ = 0.3664), and CP ( γ = 0.4044) and negatively with level of DBH ( γ = −0.4836) (Table 4).
TABLE 4.
Potential proteomic candidates for distinguishing different FM subtypes
| Protein full name | Abbr. name | Sng‐dominant group (SG) | Pain‐dominant group (PG) | No‐dominant sensation group (NG) | VIP (SG vs PG) | AUC (SG vs PG) | Pearson correlation with sng VAS × WSI (γ1) | Pearson correlation with pain VAS × WPI (γ2) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Log2 (FM/control) | p value (vs control) | Log2 (FM/control) | p value (vs control) |
p value (vs SG) |
Log2 (FM/control) | p value (vs control) | ||||||
| Prenylcysteine oxidase 1 | PCYOX1 | 0.48 | 0.0028 | 0.13 | 0.3458 | 0.0178 | 0.50 | 0.6678 | 2.34 | 0.80 | 0.4619** | −0.2449 |
| Inter‐α‐trypsin inhibitor heavy chain H4 | ITIH4 | 0.71 | 0.0466 | 0.02 | 0.9491 | 0.0180 | 0.09 | 0.5553 | 1.35 | 0.78 | 0.4195* | −0.1658 |
| Profilin‐1 | PFN1 | 0.67 | 0.0456 | −0.16 | 0.4406 | 0.0024 | 0.52 | 0.0327 | 1.36 | 0.81 | 0.3595* | −0.2504 |
| Leucine‐rich alpha‐2‐glycoprotein | LRG1 | −0.48 | 0.0728 | 0.18 | 0.3661 | 0.0027 | −0.12 | 0.9681 | 1.91 | 0.86 | −0.5444** | 0.1765 |
| Complement C8 gamma chain | C8G | −0.62 | 0.1093 | 0.16 | 0.1653 | 0.0038 | 0.05 | 0.7766 | 2.24 | 0.77 | −0.2678 | 0.5335** |
| Complement C8 alpha chain | C8A | −0.18 | 0.433 | 0.30 | 0.020 | 0.0020 | 0.03 | 0.8966 | 1.95 | 0.77 | −0.2750 | 0.5065** |
| Ceruloplasmin | CP | −0.24 | 0.148 | 0.26 | 0.078 | 0.0020 | −0.19 | 0.2934 | 1.41 | 0.84 | −0.3181 | 0.4044* |
| Cadherin 5 | CDH5 | −0.30 | 0.002 | 0.16 | 0.381 | 0.0110 | −0.06 | 0.7195 | 1.27 | 0.86 | −0.2240 | 0.3664* |
| Dopamine β‐hydroxylase | DBH | 0.44 | 0.1342 | −0.27 | 0.3482 | 0.0103 | −0.05 | 0.5959 | 2.03 | 0.82 | 0.2661 | −0.4836** |
p < 0.05
p < 0.01.
3.9. Correlation between differentially expressed metabolites and proteins levels in PG and SG
Finally, we explored possible key features discriminating PG and SG in FM with the metabolomics, lipidomics and proteomics data sets. Here we used the DIABLO program, an integrative method for searching multi‐omics molecular features for phenotype discrimination (Singh et al., 2019) and found 17 key features (five proteins, seven lipids and five metabolites). Except for patient 13, these key features could divide FM patients into PG and SG phenotypes with unsupervised hierarchical clustering (Figure 5a). We also explored the correlation network between these key features (Figure 5b). Levels of PGD2, SM(d18:1/26:1) and SM(d18:1/25:1) were positively correlated with CP level and those of SM(d18:1/26:1) and SM(d18:1/25:1) positively correlated with C8A level. Lactate level was negatively correlated with CP level.
FIGURE 5.

Multi‐omics analyses of key features for classifying different FM phenotypes. (a) A heat map of unsupervised hierarchical clustering of multi‐omics signatures, selected by using the DIABLO program, showing that FM patients can be divided into pain‐dominant (PG) and sng‐dominant (SG) phenotypes. (b) Network visualization of the key features from DIABLO (absolute Pearson's correlation >0.5 or < −0.5). Rectangular and hexagonal boxes represent metabolites/lipids and proteins, respectively. The red‐lined boxes denote significant changes (p < 0.05) between PG and SG groups. Coloured lines between boxes represent Pearson's correlation. Cer, ceramide; LPC, lysophosphatidylcholine; PC, phosphatidylcholine; PI, phosphatidylinositol; SM, sphingomyelin
4. DISCUSSION
FM is a complex disease with unknown pathogenesis and diverse somatic complaints existed amongst FM patients. Muscle sng is an especially common complaint and is a distinguishable symptom from pain (Kawashita et al., 2020; Lin et al., 2018). In this study, we applied the Revised Fibromyalgia Impact Questionnaire with an Integration of Soreness Assessment (FIQR‐S), which was developed for delineating clinical conditions of sng sensation amongst FM patients (Chang et al., 2020a, 2020b), to divide the FM patients into three subtype groups (PG, SG and NG). We found that amongst FM patients, about one‐third were sng‐dominant (SG), two‐thirds pain‐dominant (PG) and the few remaining no‐dominant (NG, both sng and pain). These observations were similar to previous research (Lin et al., 2018). In addition, we performed integrated multi‐omics approaches to identify the potential metabolic and proteomic signatures associated with the FM patients as well as the FM subtypes. To our knowledge, this is the first multi‐omics study to differentiate FM based on the symptoms such as sng and pain. We anticipate that our study might provide valuable insights for identifying FM and might be used as potential disease‐relevant targets for developing subtype‐specific treatments.
According to our untargeted metabolomic and lipidomic results, potential biomarkers for FM were hypoxanthine, DETP and 4‐guanidinobutanoic acid in urine, and isoleucine, tryptophan, DETP, SM(d18:1/18:0), and PC(20:1(11Z)/18:0) in serum. Several researchers have reported an increased level of kynurenine, an intermediate in the major pathway for tryptophan degradation (Hackshaw et al., 2013; Nemeth et al., 2005), and decreased levels of serotonin (5‐hydroxytryptamine, 5‐HT) and tryptophan, in serum of FM patients (Heils et al., 1996; Hrycaj et al., 1993; Schwarz et al., 1999; Wolfe et al., 1997). Previous studies have also demonstrated that gut microbiota (such as Bifidobacterium, Eubacterium, Blautia, Faecalibacterium, Bacteroides, etc.) disorder and deterioration result in low tryptophan absorption, which leads to low serotonin synthesis in FM patients (Clos‐Garcia et al., 2019; Lattanzio, 2017; Minerbi et al., 2019). Some gut microbiota (such as Bifidobacterium, Blautia, Streptococcus, Lactobacillus, and Akkermansia) also could affect serum branch‐chain amino acids (BCAAs, including isoleucine, leucine and valine) levels (Clos‐Garcia et al., 2019; Hsu et al., 2021; Malatji et al., 2019). Furthermore, patients with FM had significantly lower serum levels of isoleucine than normal controls (Maes et al., 2000). These results are in accordance with our data. Our former study also revealed lower serum levels of isoleucine in the intermittent cold stress (ICS)‐induced FM mice (Hsu et al., 2019). Moreover, here we first identified DETP, 4‐guanidinobutanoic acid, SM(d18:1/18:0) and PC(20:1(11Z)/18:0) as potential biomarkers for FM. DETP levels are correlated with organophosphate exposure. Urinary levels of DETP, dimethylthiophosphate, dialkylphosphates and free 3‐phenoxybenzoic acid were found lower in organic than conventional food consumers (Baudry et al., 2019). Also, reports showed that DETP levels might be related to organophosphate exposure. People exposed to increased organophosphate levels showed a higher level of DETP or other organophosphate metabolites in urine than others (Hernandez et al., 2019; Whyatt & Barr, 2001). However, we found lower DETP levels in FM patients than controls. We still need more evidence to support it. Furthermore, 4‐Guanidinobutanoic acid is a common urinary metabolite and an arginine metabolite involved in the metabolism of arginine and proline (creatinine pathway) (Hong et al., 2013; Romagnoli et al., 2014). A lower level of 4‐guanidinobutanoic acid in FM patients than controls was detected in this study. Previous studies have shown that a low level of arginine is associated with pain severity in both adults and children (Atzler et al., 2016; Bakshi & Morris, 2016; Shell et al., 2016).
IL‐6 and fibrinolysis proteins (F2, GP5, FGA, FGB, FGG, GP1BA, THBS1 and THBS2) were previously found significantly lower in FM patients than controls (Han et al., 2020). Levels of complementary proteins (C4A, C1S, CFAH, CO7, CO2, C1qC and CO9), IL‐1 receptor accessory protein and immunoglobulin gamma Fc region receptor III‐A and B, involved in coagulation and inflammation, were significantly increased, mainly in FM patients (Garcia Rodriguez & Abud, 2020; Han et al., 2020; Ramirez‐Tejero et al., 2018; Wahlen et al., 2020). These results were similar to our findings. Furthermore, we also found that galectin 7, SERPINB3, S100A7 and LYVE1 could be novel biomarkers for FM. Amongst them, LYVE1, also known as cell‐surface retention sequence binding protein‐1 (CRSBP‐1), is one of the most specific lymphoedema and lymphatic vessel markers (Liu et al., 2017). Patients with lymphoedema frequently experience FM, arthritis, carpel tunnel syndrome and neck and shoulder dysfunction (Ridner & Dietrich, 2008), which might explain the higher LYVE1 level in FM patients than controls in our data. Our network analysis findings agree with prior studies reporting a high level of NF‐κB, inducing NF‐κB‐dependent pro‐inflammatory cytokine generation, in FM patients (Cordero et al., 2013; Ruster et al., 2005). These results also agree well with a recent investigation indicating altered energy, lipid and amino acid metabolism in FM patients (Menzies et al., 2020). Indeed, oxidative stress with lipid peroxidation induced by reactive oxygen species may be a relevant event in the pathogenesis of FM (Cordero et al., 2010, 2011; Hung et al., 2020).
We found no significant difference in lactate levels in serum and urine between FM patients and controls. However, lactate level was significantly higher in the SG group than in controls. Accumulating evidence has demonstrated a notable correlation between blood lactate level and post‐exertional muscle soreness and fatigue (Blohm et al., 2020; Gleeson et al., 1998). In addition, blood lactate level was significantly correlated with muscle damage after exercise (Manojlovic & Erculj, 2019). The increased lactate level in SG patients is intriguing because it could offer a molecular diagnosis of the sng phenotype of FM and might suggest a unique disease status of FM required for different therapeutic strategies. Previous studies have demonstrated increased ITIH4 levels in serum after functional over‐reaching, which was correlated with muscle damage and fatigue (Merritt et al., 2019; Nieman et al., 2018). We found that urine cotinine levels could distinguish SG and PG patients. Cotinine is one of the routinely used biomarkers for detecting tobacco smoke exposure and green tobacco sickness (GTS) (Benowitz et al., 2017; Cezar‐Vaz & Cargnin, 2019), including nausea, vomiting, weakness, dizziness, headache, insomnia, abdominal pain and muscle soreness and loss of appetite (Fotedar & Fotedar, 2017). However, whether these patients were smokers or their occupation was related to tobacco production is unknown. Thus, cotinine levels in urine may have nothing to do with their FM status but may indicate that SG patients are more likely to be smokers, or the cohort was too small. This needs further proof.
Long‐term exercise (≥ 60 min) reduces leptin level in plasma (Kraemer et al., 2002), and a lower level of leptin decreases Sphingomyelin (SM) and ceramide levels (Boini et al., 2017). Besides, SM level was found decreased during recovery after exertion compared with at rest, and SM level reduction may be associated with muscle soreness (Bergman et al., 2015). SM is also a major lipid component of low‐density lipoprotein (LDL) and, together with PC, forms the polar surface of the lipoproteins (Craig et al., 1995; Deevska et al., 2012). Moreover, PCYOX1, which is a pro‐oxidant enzyme of LDL, hydrolyzes prenylcysteines to cysteine and a C‐1 aldehyde of the isoprenoid moiety and lead to some SMs reduction (Herrera‐Marcos et al., 2018). These findings may explain the high level of PCYOX1 and low levels of SM(d18:1/25:1) and SM(d18:1/26:1) in the SG group and a significant negative correlation with the sng score. Further studies are needed to determine how SM may regulate sngception and how it is associated with PCYOX1 expression. Interestingly, serum levels of PGD2 were higher in the PG group, but lower in the SG group, compared with controls. When tissues are injured, prostaglandin H2 (PGH2) is produced by invading neutrophils and macrophages and metabolized into PGE2, PGD2, PGI2 or TXA2 by means of specific synthases, then these prostaglandins promote neuronal pain signals (Jang et al., 2020). It is also worth noting that PGD2 signalling via the PGD2 receptor 2 (DP2) signalling pathway from microglia to neurons is a triggering factor for mechanical allodynia in neuropathic pain (Kanda et al., 2013). Therefore, PGD2 levels showed a significant positive correlation with pain scores. We also found that CP, cadherin 5, C8A, C8B and C8G exhibited a significant positive correlation with pain scores. A previous investigation showed that complementary proteins and CP were strongly correlated with pain intensity in chronic widespread pain (Wahlen et al., 2018). Based on our knowledge, PGD2 could induce pain signals (Jang et al., 2020; Kanda et al., 2013; Kawabata, 2011), hence PGD2 level was positively correlated with CP level. Overall, levels of lactate, 2‐methylmaleate, PGD2, C8G and DBH may have substantial potential to discriminate amongst PG, SG or NG groups in FM.
The results and interpretation of this study have the following limitations: (1) Selection bias: because all participants were recruited from clinics of physical medicine and rehabilitation or neurology, patients with minor symptoms might not be included in this research. (2) From the clinical criteria used for selecting FM patients, we consider the group representative of FM, in general; however, the control group was from a health check‐up clinic, and we did not attempt to match participants based on characteristics, lifestyle, conditions and treatments (e.g., sex, smoking, diabetes, hypertension, chronic metal poisoning, hypercholesteremia or other diseases), which may have an impact on an individual's metabolome and lipidome. Further, we did not administer the ACR 2011 criteria on the healthy controls, so certain undiagnosed FM in the Control group is possible. (3) Although large numbers of patients and control groups are advocated in studies, from our experience with untargeted metabolomic and lipidomic studies, groups of selected cases of 20 would suffice in a pilot study; in the future, more FM patients need to be recruited for validation. (4) In this study, we did not control for medications in the FM or control group, which could be addressed in the future investigations. (5) Also, we did not control for recent exercise (especially important for lactate and Sphingomyelin results), dietary intakes, fasting status, etc., which may have significant effects on short‐term metabolites. (6) Diethylthiophosphate (DETP) may be associated with organophosphate exposure. However, lower DETP was found in FM patients, which need to be clarified.
5. CONCLUSION
Combined clinical diagnosis, questionnaire and analysis of selected biomarkers in a first screening could achieve a more accurate diagnosis of FM and its subtypes. The identified biomarkers could be used to determine FM classification: PG, SG or NG. We also provide a novel perspective that sng and pain are distinct sensations. Moreover, sng and pain might share certain common mechanisms, whereas other mechanisms may be dissimilar. These metabolites and proteins might provide valuable insights for identifying FM and might be used as potential disease‐relevant targets for developing subtype‐specific treatments. These insights and applications merit future validation with larger FM populations and discrimination between sng and pain.
CONFLICT OF INTERESTS
The authors have disclosed no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Study concept and design: Y.L.L., W.H.H. and D.S.H.; acquired data and conducted analyses: W.H.H., D.S.H., W.C.K. and Y.M.C.; drafted the manuscript: W.H.H. and D.S.H.; reviewed and edited the manuscript: Y.L.L. and C.C.C.
CONSENT FOR PUBLICATION
All authors consent to publication.
Supporting information
Fig S1‐S2
Table S1‐S6
ACKNOWLEDGEMENTS
This work was supported by grants from the Ministry of Science and Technology of Taiwan (MOST 107‐2320‐B‐039‐001, MOST 107‐2811‐039‐519, MOST 108‐2320‐B‐039‐001 and MOST 108‐2811‐039‐526 to Y.L.L.). The funders had no role in study design, data collection and analysis, decision to publish or the preparation of the manuscript. The authors also thanked the Metabolomics Core Lab of Genomic and Precision Medicine, National Taiwan University, Taipei, Taiwan, for the QTOFMS analyses, and the Mass Spectrometry Laboratory of Tzong Jwo Jang, College of Medicine, Fu Jen Catholic University, New Taipei, Taiwan as well as the Medicinal Chemistry and Analytical Core Facilities, Biomedical Translation Research Center, Academia Sinica, Taipei, Taiwan, for the instrumental assistance to nanoLC‐MS/MS proteomics analyses.
Hsu, W.‐H. , Han, D.‐S. , Ku, W.‐C. , Chao, Y.‐M. , Chen, C.‐C. , & Lin, Y.‐L. (2022). Metabolomic and proteomic characterization of sng and pain phenotypes in fibromyalgia. European Journal of Pain, 26, 445–462. 10.1002/ejp.1871
Wei‐Hsiang Hsu, Der‐Sheng Han and Wei‐Chi Ku contributed equally.
Funding information
This work was supported by grants from the Ministry of Science and Technology of Taiwan (MOST 107‐2321‐B‐001‐020, MOST 108‐2321‐B‐001‐005, MOST 108‐2321‐B‐001‐028‐MY2 and MOST 110‐2321‐B‐001‐010 to Y.L.L. and C.C.C.).
Contributor Information
Chih‐Cheng Chen, Email: chih@ibms.sinica.edu.tw.
Yun‐Lian Lin, Email: yllin5212@gmail.com, Email: yllin@mail.cmu.edu.tw.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding authors on request.
REFERENCES
- Arnold, L. M. , Clauw, D. J. , McCarberg, B. H. , & FibroCollaborative . (2011). Improving the recognition and diagnosis of fibromyalgia. Mayo Clinic Proceedings, 86, 457–464. 10.4065/mcp.2010.0738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atzler, D. , Baum, C. , Ojeda, F. , Keller, T. , Cordts, K. , Schnabel, R. B. , Choe, C. U. , Lackner, K. J. , Munzel, T. , Boger, R. H. , Blankenberg, S. , Schwedhelm, E. , & Zeller, T. (2016). Low homoarginine levels in the prognosis of patients with acute chest pain. Journal of the American Heart Association, 5, e002565. 10.1161/JAHA.115.002565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi, N. , & Morris, C. R. (2016). The role of the arginine metabolome in pain: Implications for sickle cell disease. Journal of Pain Research, 9, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry, J. , Debrauwer, L. , Durand, G. , Limon, G. , Delcambre, A. , Vidal, R. , Taupier‐Letage, B. , Druesne‐Pecollo, N. , Galan, P. , Hercberg, S. , Lairon, D. , Cravedi, J. P. , & Kesse‐Guyot, E. (2019). Urinary pesticide concentrations in French adults with low and high organic food consumption: Results from the general population‐based NutriNet‐Sante. Journal of Exposure Science & Environmental Epidemiology, 29, 366–378. [DOI] [PubMed] [Google Scholar]
- Benowitz, N. L. , Jain, S. , Dempsey, D. A. , Nardone, N. , Helen, G. S. , & Jacob, P. 3rd . (2017). Urine cotinine screening detects nearly ubiquitous tobacco smoke exposure in urban adolescents. Nicotine & Tobacco Research, 19, 1048–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman, B. C. , Brozinick, J. T. , Strauss, A. , Bacon, S. , Kerege, A. , Bui, H. H. , Sanders, P. , Siddall, P. , Kuo, M. S. , & Perreault, L. (2015). Serum sphingolipids: Relationships to insulin sensitivity and changes with exercise in humans. American Journal of Physiology‐Endocrinology and Metabolism, 309, E398–E408. 10.1152/ajpendo.00134.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blohm, K. , Beidler, J. , Rosen, P. , Kressler, J. , & Hong, M. Y. (2020). Effect of acute watermelon juice supplementation on post‐submaximal exercise heart rate recovery, blood lactate, blood pressure, blood glucose and muscle soreness in healthy non‐athletic men and women. International Journal of Food Sciences and Nutrition, 71, 482–489. 10.1080/09637486.2019.1675604 [DOI] [PubMed] [Google Scholar]
- Boini, K. M. , Xia, M. , Koka, S. , Gehr, T. W. , & Li, P. L. (2017). Sphingolipids in obesity and related complications. Frontiers in Bioscience (Landmark Ed), 22, 96–116. 10.2741/4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cezar‐Vaz, M. R. , & Cargnin, M. (2019). Use of cotinine biomarker in workers to detect green tobacco sickness. Revista Latino‐Americana De Enfermagem, 27, e3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, K. V. , Hung, C. H. , Sun, W. Z. , Wu, W. T. , Lai, C. L. , Han, D. S. , & Chen, C. C. (2020a). Authors’ response to the letter to the editor on “clinical consideration in evaluating soreness symptoms of fibromyalgia”. Journal of the Formosan Medical Association, 119, 889–890. 10.1016/j.jfma.2019.11.024 [DOI] [PubMed] [Google Scholar]
- Chang, K. V. , Hung, C. H. , Sun, W. Z. , Wu, W. T. , Lai, C. L. , Han, D. S. , & Chen, C. C. (2020b). Evaluating soreness symptoms of fibromyalgia: Establishment and validation of the revised fibromyalgia impact questionnaire with integration of soreness assessment. Journal of the Formosan Medical Association, 119, 1211–1218. 10.1016/j.jfma.2019.10.018 [DOI] [PubMed] [Google Scholar]
- Clauw, D. J. , Arnold, L. M. , McCarberg, B. H. , & FibroCollaborative . (2011). The science of fibromyalgia. Mayo Clinic Proceedings, 86, 907–911. 10.4065/mcp.2011.0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clos‐Garcia, M. , Andres‐Marin, N. , Fernandez‐Eulate, G. , Abecia, L. , Lavin, J. L. , van Liempd, S. , Cabrera, D. , Royo, F. , Valero, A. , Errazquin, N. , Vega, M. C. G. , Govillard, L. , Tackett, M. R. , Tejada, G. , Gonzalez, E. , Anguita, J. , Bujanda, L. , Orcasitas, A. M. C. , Aransay, A. M. , … Falcon‐Perez, J. M. (2019). Gut microbiome and serum metabolome analyses identify molecular biomarkers and altered glutamate metabolism in fibromyalgia. EBioMedicine, 46, 499–511. 10.1016/j.ebiom.2019.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, H. (2017). Controversies and challenges in fibromyalgia: A review and a proposal. Therapeutic Advances in Musculoskeletal Disease, 9, 115–127. 10.1177/1759720X17699199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero, M. D. , Alcocer‐Gomez, E. , Cano‐Garcia, F. J. , De Miguel, M. , Carrion, A. M. , Navas, P. , & Sanchez Alcazar, J. A. (2011). Clinical symptoms in fibromyalgia are better associated to lipid peroxidation levels in blood mononuclear cells rather than in plasma. PLoS One, 6, e26915. 10.1371/journal.pone.0026915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero, M. D. , de Miguel, M. , Carmona‐Lopez, I. , Bonal, P. , Campa, F. , & Moreno‐Fernandez, A. M. (2010). Oxidative stress and mitochondrial dysfunction in fibromyalgia. Neuro Endocrinology Letters, 31, 169–173. https://pubmed.ncbi.nlm.nih.gov/20424583/ [PubMed] [Google Scholar]
- Cordero, M. D. , Diaz‐Parrado, E. , Carrion, A. M. , Alfonsi, S. , Sanchez‐Alcazar, J. A. , Bullon, P. , Battino, M. , & de Miguel, M. (2013). Is inflammation a mitochondrial dysfunction‐dependent event in fibromyalgia? Antioxidants & Redox Signaling, 18, 800–807. 10.1089/ars.2012.4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, W. Y. , Poulin, S. E. , Palomaki, G. E. , Neveux, L. M. , Ritchie, R. F. , & Ledue, T. B. (1995). Oxidation‐related analytes and lipid and lipoprotein concentrations in healthy subjects. Arteriosclerosis, Thrombosis, and Vascular Biology, 15, 733–739. 10.1161/01.ATV.15.6.733 [DOI] [PubMed] [Google Scholar]
- Deevska, G. M. , Sunkara, M. , Morris, A. J. , & Nikolova‐Karakashian, M. N. (2012). Characterization of secretory sphingomyelinase activity, lipoprotein sphingolipid content and LDL aggregation in ldlr−/− mice fed on a high‐fat diet. Bioscience Reports, 32, 479–490. 10.1042/BSR20120036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch, E. W. , Csordas, A. , Sun, Z. , Jarnuczak, A. , Perez‐Riverol, Y. , Ternent, T. , Campbell, D. S. , Bernal‐Llinares, M. , Okuda, S. , Kawano, S. , Moritz, R. L. , Carver, J. J. , Wang, M. , Ishihama, Y. , Bandeira, N. , Hermjakob, H. , & Vizcaino, J. A. (2017). The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Research, 45, D1100–D1106. 10.1093/nar/gkw936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotedar, S. , & Fotedar, V. (2017). Green tobacco sickness: A brief review. Indian Journal of Occupational and Environmental Medicine, 21, 101–104. 10.4103/ijoem.IJOEM_160_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Rodríguez D. F., & Abud Mendoza C. (2020). Physiopathology of fibromyalgia. Reumatología Clínica, (English Edition), 16(3), 191–194. 10.1016/j.reumae.2020.02.004 [DOI] [PubMed] [Google Scholar]
- Gleeson, M. , Blannin, A. K. , Walsh, N. P. , Field, C. N. , & Pritchard, J. C. (1998). Effect of exercise‐induced muscle damage on the blood lactate response to incremental exercise in humans. European Journal of Applied Physiology, 77, 292–295. 10.1007/s004210050336 [DOI] [PubMed] [Google Scholar]
- Graves, P. R. , & Haystead, T. A. (2002). Molecular biologist's guide to proteomics. Microbiology and Molecular Biology Reviews, 66, 39–63; table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, R. W. , & Han, X. (2011). Lipidomics at the interface of structure and function in systems biology. Chemistry & Biology, 18, 284–291. 10.1016/j.chembiol.2011.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackshaw, K. V. , Rodriguez‐Saona, L. , Plans, M. , Bell, L. N. , & Buffington, C. A. (2013). A bloodspot‐based diagnostic test for fibromyalgia syndrome and related disorders. Analyst, 138, 4453–4462. 10.1039/c3an36615d [DOI] [PubMed] [Google Scholar]
- Han, C. L. , Sheng, Y. C. , Wang, S. Y. , Chen, Y. H. , & Kang, J. H. (2020). Serum proteome profiles revealed dysregulated proteins and mechanisms associated with fibromyalgia syndrome in women. Scientific Reports, 10, 12347. 10.1038/s41598-020-69271-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heils, A. , Teufel, A. , Petri, S. , Stober, G. , Riederer, P. , Bengel, D. , & Lesch, K. P. (1996). Allelic variation of human serotonin transporter gene expression. Journal of Neurochemistry, 66, 2621–2624. 10.1046/j.1471-4159.1996.66062621.x [DOI] [PubMed] [Google Scholar]
- Hernandez, A. F. , Lozano‐Paniagua, D. , Gonzalez‐Alzaga, B. , Kavvalakis, M. P. , Tzatzarakis, M. N. , Lopez‐Flores, I. , Aguilar‐Garduno, C. , Caparros‐Gonzalez, R. A. , Tsatsakis, A. M. , & Lacasana, M. (2019). Biomonitoring of common organophosphate metabolites in hair and urine of children from an agricultural community. Environment International, 131, 104997. 10.1016/j.envint.2019.104997 [DOI] [PubMed] [Google Scholar]
- Herrera‐Marcos, L. V. , Lou‐Bonafonte, J. M. , Martinez‐Gracia, M. V. , Arnal, C. , Navarro, M. A. , & Osada, J. (2018). Prenylcysteine oxidase 1, a pro‐oxidant enzyme of low density lipoproteins. Frontiers in Bioscience (Landmark Ed), 23, 1020–1037. 10.2741/4631 [DOI] [PubMed] [Google Scholar]
- Hong, H. , Fill, T. , & Leadlay, P. F. (2013). A common origin for guanidinobutanoate starter units in antifungal natural products. Angewandte Chemie (International Ed. in English), 52, 13096–13099. 10.1002/anie.201308136 [DOI] [PubMed] [Google Scholar]
- Hrycaj, P. , Stratz, T. , & Muller, W. (1993). Platelet 3H‐imipramine uptake receptor density and serum serotonin levels in patients with fibromyalgia/fibrositis syndrome. Journal of Rheumatology, 20, 1986–1988. [PubMed] [Google Scholar]
- Hsu, W. H. , Lee, C. H. , Chao, Y. M. , Kuo, C. H. , Ku, W. C. , Chen, C. C. , & Lin, Y. L. (2019). ASIC3‐dependent metabolomics profiling of serum and urine in a mouse model of fibromyalgia. Scientific Reports, 9, 12123. 10.1038/s41598-019-48315-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, W. H. , Lin, L. J. , Lu, C. K. , Kao, S. T. , & Lin, Y. L. (2021). Effect of you‐gui‐wan on house dust mite‐induced mouse allergic asthma via regulating amino acid metabolic disorder and gut dysbiosis. Biomolecules, 11, 812. 10.3390/biom11060812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, W. H. , Wang, S. J. , Chao, Y. M. , Chen, C. J. , Wang, Y. F. , Fuh, J. L. , Chen, S. P. , & Lin, Y. L. (2020). Urine metabolomics signatures in reversible cerebral vasoconstriction syndrome. Cephalalgia, 40, 735–747. 10.1177/0333102419897621 [DOI] [PubMed] [Google Scholar]
- Hung, C. H. , Lee, C. H. , Tsai, M. H. , Chen, C. H. , Lin, H. F. , Hsu, C. Y. , Lai, C. L. , & Chen, C. C. (2020). Activation of acid‐sensing ion channel 3 by lysophosphatidylcholine 16:0 mediates psychological stress‐induced fibromyalgia‐like pain. Annals of the Rheumatic Diseases, 79, 1644–1656. 10.1136/annrheumdis-2020-218329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan, F. , Nanji, K. , Qidwai, W. , & Qasim, R. (2012). Fibromyalgia syndrome: An overview of pathophysiology, diagnosis and management. Oman Medical Journal, 27, 192–195. 10.5001/omj.2012.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, Y. , Kim, M. , & Hwang, S. W. (2020). Molecular mechanisms underlying the actions of arachidonic acid‐derived prostaglandins on peripheral nociception. Journal of Neuroinflammation, 17, 30. 10.1186/s12974-020-1703-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda, H. , Kobayashi, K. , Yamanaka, H. , & Noguchi, K. (2013). COX‐1‐dependent prostaglandin D2 in microglia contributes to neuropathic pain via DP2 receptor in spinal neurons. Glia, 61, 943–956. 10.1002/glia.22487 [DOI] [PubMed] [Google Scholar]
- Kawabata, A. (2011). Prostaglandin E2 and pain—An update. Biological and Pharmaceutical Bulletin, 34, 1170–1173. 10.1248/bpb.34.1170 [DOI] [PubMed] [Google Scholar]
- Kawashita, T. , Dunnsiri, T. , Shu, S. , & Woo, B. K. P. (2020). Clinical consideration in evaluating soreness symptoms of fibromyalgia. Journal of the Formosan Medical Association, 119, 888. 10.1016/j.jfma.2019.11.023 [DOI] [PubMed] [Google Scholar]
- Kraemer, R. R. , Chu, H. , & Castracane, V. D. (2002). Leptin and exercise. Experimental Biology and Medicine, 227, 701–708. 10.1177/153537020222700903 [DOI] [PubMed] [Google Scholar]
- Lattanzio, S. M. (2017). Fibromyalgia syndrome: A metabolic approach grounded in biochemistry for the remission of symptoms. Frontiers in Medicine (Lausanne), 4, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, H. W. , Kuo, C. H. , Chao, H. C. , & Chen, G. Y. (2020). Post‐column infused internal standard assisted lipidomics profiling strategy and its application on phosphatidylcholine research. Journal of Pharmaceutical and Biomedical Analysis, 178, 112956. 10.1016/j.jpba.2019.112956 [DOI] [PubMed] [Google Scholar]
- Lin, J. H. , Hung, C. H. , Han, D. S. , Chen, S. T. , Lee, C. H. , Sun, W. Z. , & Chen, C. C. (2018). Sensing acidosis: Nociception or sngception? Journal of Biomedical Science, 25, 85. 10.1186/s12929-018-0486-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, N. F. , Yu, Z. , Luo, Y. , & Sun, D. (2017). A LYVE‐1/CRSBP‐1 mutation in inherited primary lymphedema. Lymphology, 50, 9–15. [PubMed] [Google Scholar]
- Maes, M. , Verkerk, R. , Delmeire, L. , Van Gastel, A. , van Hunsel, F. , & Scharpe, S. (2000). Serotonergic markers and lowered plasma branched‐chain‐amino acid concentrations in fibromyalgia. Psychiatry Research, 97, 11–20. 10.1016/S0165-1781(00)00204-3 [DOI] [PubMed] [Google Scholar]
- Malatji, B. G. , Mason, S. , Mienie, L. J. , Wevers, R. A. , Meyer, H. , van Reenen, M. , & Reinecke, C. J. (2019). The GC‐MS metabolomics signature in patients with fibromyalgia syndrome directs to dysbiosis as an aspect contributing factor of FMS pathophysiology. Metabolomics, 15, 54. 10.1007/s11306-019-1513-6 [DOI] [PubMed] [Google Scholar]
- Manojlovic, V. , & Erculj, F. (2019). Using blood lactate concentration to predict muscle damage and jump performance response to maximal stretch‐shortening cycle exercise. Journal of Sports Medicine and Physical Fitness, 59, 581–586. 10.23736/S0022-4707.18.08346-9 [DOI] [PubMed] [Google Scholar]
- Mautner, K. , & Sussman, W. I. (2016). Delayed onset muscle soreness: Illustrative case with sonographic findings. Current Sports Medicine Reports, 15, 168–170. 10.1249/JSR.0000000000000258 [DOI] [PubMed] [Google Scholar]
- Menzies, V. , Starkweather, A. , Yao, Y. , Thacker, L. R., 2nd , Garrett, T. J. , Swift‐Scanlan, T. , Kelly, D. L. , Patel, P. , & Lyon, D. E. (2020). Metabolomic differentials in women with and without fibromyalgia. Clinical and Translational Science, 13, 67–77. 10.1111/cts.12679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, E. K. , Nieman, D. C. , Toone, B. R. , Groen, A. , & Pugachev, A. (2019). Proteomic markers of non‐functional overreaching during the race across america (RAAM): A case study. Frontiers in Physiology, 10, 1410. 10.3389/fphys.2019.01410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minerbi, A. , Gonzalez, E. , Brereton, N. J. B. , Anjarkouchian, A. , Dewar, K. , Fitzcharles, M. A. , Chevalier, S. , & Shir, Y. (2019). Altered microbiome composition in individuals with fibromyalgia. Pain, 160, 2589–2602. 10.1097/j.pain.0000000000001640 [DOI] [PubMed] [Google Scholar]
- Nemeth, H. , Toldi, J. , & Vecsei, L. (2005). Role of kynurenines in the central and peripheral nervous systems. Current Neurovascular Research, 2, 249–260. [DOI] [PubMed] [Google Scholar]
- Nieman, D. C. , Groen, A. J. , Pugachev, A. , & Vacca, G. (2018). Detection of functional overreaching in endurance athletes using proteomics. Proteomes, 6, 33. 10.3390/proteomes6030033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot, S. (2019). Fibromyalgia: A misconnection in a multiconnected world? European Journal of Pain, 23, 866–873. 10.1002/ejp.1367 [DOI] [PubMed] [Google Scholar]
- Ramirez‐Tejero, J. A. , Martinez‐Lara, E. , Rus, A. , Camacho, M. V. , Del Moral, M. L. , & Siles, E. (2018). Insight into the biological pathways underlying fibromyalgia by a proteomic approach. Journal of Proteomics, 186, 47–55. 10.1016/j.jprot.2018.07.009 [DOI] [PubMed] [Google Scholar]
- Ridner, S. H. , & Dietrich, M. S. (2008). Self‐reported comorbid conditions and medication usage in breast cancer survivors with and without lymphedema. Oncology Nursing Forum, 35, 57–63. 10.1188/08.ONF.57-63 [DOI] [PubMed] [Google Scholar]
- Romagnoli, G. , Verhoeven, M. D. , Mans, R. , Fleury Rey, Y. , Bel‐Rhlid, R. , van den Broek, M. , Seifar, R. M. , Ten Pierick, A. , Thompson, M. , Muller, V. , Wahl, S. A. , Pronk, J. T. , & Daran, J. M. (2014). An alternative, arginase‐independent pathway for arginine metabolism in Kluyveromyces lactis involves guanidinobutyrase as a key enzyme. Molecular Microbiology, 93, 369–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruster, M. , Franke, S. , Spath, M. , Pongratz, D. E. , Stein, G. , & Hein, G. E. (2005). Detection of elevated N epsilon‐carboxymethyllysine levels in muscular tissue and in serum of patients with fibromyalgia. Scandinavian Journal of Rheumatology, 34, 460–463. [DOI] [PubMed] [Google Scholar]
- Schwarz, M. J. , Spath, M. , Muller‐Bardorff, H. , Pongratz, D. E. , Bondy, B. , & Ackenheil, M. (1999). Relationship of substance P, 5‐hydroxyindole acetic acid and tryptophan in serum of fibromyalgia patients. Neuroscience Letters, 259, 196–198. 10.1016/S0304-3940(98)00937-9 [DOI] [PubMed] [Google Scholar]
- Shell, W. E. , Pavlik, S. , Roth, B. , Silver, M. , Breitstein, M. L. , May, L. , & Silver, D. (2016). Reduction in pain and inflammation associated with chronic low back pain with the use of the medical food theramine. American Journal of Therapeutics, 23, e1353–e1362. 10.1097/MJT.0000000000000068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. , Shannon, C. P. , Gautier, B. , Rohart, F. , Vacher, M. , Tebbutt, S. J. , & Le Cao, K. A. (2019). DIABLO: An integrative approach for identifying key molecular drivers from multi‐omics assays. Bioinformatics, 35, 3055–3062. 10.1093/bioinformatics/bty1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H. S. , Harris, R. , & Clauw, D. (2011). Fibromyalgia: An afferent processing disorder leading to a complex pain generalized syndrome. Pain Physician, 14, E217–245. [PubMed] [Google Scholar]
- Subramaniam, S. , Fahy, E. , Gupta, S. , Sud, M. , Byrnes, R. W. , Cotter, D. , Dinasarapu, A. R. , & Maurya, M. R. (2011). Bioinformatics and systems biology of the lipidome. Chemical Reviews, 111, 6452–6490. 10.1021/cr200295k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenhaus, A. , Philippe, C. , Guillemot, V. , Le Cao, K. A. , Grill, J. , & Frouin, V. (2014). Variable selection for generalized canonical correlation analysis. Biostatistics, 15, 569–583. 10.1093/biostatistics/kxu001 [DOI] [PubMed] [Google Scholar]
- Wahlen, K. , Ernberg, M. , Kosek, E. , Mannerkorpi, K. , Gerdle, B. , & Ghafouri, B. (2020). Significant correlation between plasma proteome profile and pain intensity, sensitivity, and psychological distress in women with fibromyalgia. Scientific Reports, 10, 12508. 10.1038/s41598-020-69422-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlen, K. , Ghafouri, B. , Ghafouri, N. , & Gerdle, B. (2018). Plasma protein pattern correlates with pain intensity and psychological distress in women with chronic widespread pain. Frontiers in Psychology, 9, 2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt, R. M. , & Barr, D. B. (2001). Measurement of organophosphate metabolites in postpartum meconium as a potential biomarker of prenatal exposure: A validation study. Environmental Health Perspectives, 109, 417–420. 10.1289/ehp.01109417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, F. , Clauw, D. J. , Fitzcharles, M. A. , Goldenberg, D. L. , Hauser, W. , Katz, R. S. , Mease, P. , Russell, A. S. , Russell, I. J. , & Winfield, J. B. (2011). Fibromyalgia criteria and severity scales for clinical and epidemiological studies: A modification of the ACR preliminary diagnostic criteria for fibromyalgia. Journal of Rheumatology, 38, 1113–1122. 10.3899/jrheum.100594 [DOI] [PubMed] [Google Scholar]
- Wolfe, F. , Clauw, D. J. , Fitzcharles, M. A. , Goldenberg, D. L. , Katz, R. S. , Mease, P. , Russell, A. S. , Russell, I. J. , Winfield, J. B. , & Yunus, M. B. (2010). The American College of Rheumatology preliminary diagnostic criteria for fibromyalgia and measurement of symptom severity. Arthritis Care & Research, 62, 600–610. 10.1002/acr.20140 [DOI] [PubMed] [Google Scholar]
- Wolfe, F. , Russell, I. J. , Vipraio, G. , Ross, K. , & Anderson, J. (1997). Serotonin levels, pain threshold, and fibromyalgia symptoms in the general population. Journal of Rheumatology, 24, 555–559. [PubMed] [Google Scholar]
- Wolfe, F. , Smythe, H. A. , Yunus, M. B. , Bennett, R. M. , Bombardier, C. , Goldenberg, D. L. , Tugwell, P. , Campbell, S. M. , Abeles, M. , Clark, P. , Fam, A. G. , Farber, S. J. , Fiechtner, J. J. , Michael Franklin, C. , Gatter, R. A. , Hamaty, D. , Lessard, J. , Lichtbroun, A. S. , Masi, A. T. , … Sheon, R. P. (1990). The American College of Rheumatology 1990 criteria for the classification of fibromyalgia. Report of the multicenter criteria committee. Arthritis and Rheumatism, 33, 160–172. 10.1002/art.1780330203 [DOI] [PubMed] [Google Scholar]
- Yu, S. H. , Kyriakidou, P. , & Cox, J. (2020). Isobaric matching between runs and novel PSM‐level normalization in MaxQuant strongly improve reporter ion‐based quantification. Journal of Proteome Research, 19, 3945–3954. 10.1021/acs.jproteome.0c00209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, A. , Sun, H. , Yan, G. , Wang, P. , & Wang, X. (2015). Metabolomics for biomarker discovery: Moving to the clinic. BioMed Research International, 2015, 354671. 10.1155/2015/354671 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S2
Table S1‐S6
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors on request.


