Abstract
Aim
To assess the microbial effects of mechanical debridement in conjunction with a mouthrinse on sites with peri‐implant mucositis and gingivitis.
Materials and methods
Eighty‐nine patients with peri‐implant mucositis were included in a double‐blinded, randomized, placebo‐controlled trial with mechanical debridement and 1‐month use of either delmopinol, chlorhexidine (CHX), or a placebo mouthrinse. Submucosal and subgingival plaque samples of implants and teeth were collected at baseline and after 1 and 3 months, processed for 16S V4 rRNA gene amplicon sequencing, and analysed bioinformatically.
Results
The sites with peri‐implant mucositis presented with a less diverse and less anaerobic microbiome. Exposure to delmopinol or CHX, but not to the placebo mouthrinse resulted in microbial changes after 1 month. The healthy sites around the teeth harboured a more diverse and more anaerobe‐rich microbiome than the healthy sites around the implants.
Conclusions
Peri‐implant sites with mucositis harbour ecologically less complex and less anaerobic biofilms with lower biomass than patient‐matched dental sites with gingivitis while eliciting an equal inflammatory response. Adjunctive antimicrobial therapy in addition to mechanical debridement does affect both dental and peri‐implant biofilm composition in the short term, resulting in a less dysbiotic subgingival biofilm.
Keywords: chlorhexidine, delmopinol, dental implant, microbiome, peri‐implant mucositis, subgingival plaque, submucosal plaque
Clinical Relevance.
Scientific rationale of the study: Current therapy of peri‐implant diseases is similar to that of periodontal diseases but results in a lower success rate. Additionally, microbiological knowledge on peri‐implant mucositis and its treatment is limited.
Principal findings: Antimicrobial adjuvant therapy of peri‐implant mucositis affects the microbiome in the short term without any clinically detectable differences. The submucosal microbiome of both peri‐implant mucositis and healthy peri‐implant sites is ecologically less complex and more aerobic than the subgingival microbiome with equal pro‐inflammatory potential.
Practical implications: Submucosal and subgingival microbiomes are distinct, and peri‐implant diseases require tailored therapeutic approaches that consider the local implant‐related environmental factors.
1. INTRODUCTION
Implant‐supported dental restorations have gained in popularity and have become a feasible solution for the replacement of missing teeth. However, the prevalence of peri‐implant diseases, such as mucositis (soft‐tissue inflammation alone) and peri‐implantitis (inflammation with bone loss), is increasing over time and may become a major burden on oral health care (Lee et al., 2017; Kumar et al., 2018).
The traditional assumptions that periodontal and peri‐implant diseases share their aetiological factors and therefore require identical therapy, are being put in doubt. There is increasing evidence that peri‐implant diseases are a specific group of diseases with a distinct aetiology and pathogenesis different from periodontal diseases (Robitaille et al., 2016; Belibasakis & Manoil, 2021; Kotsakis & Olmedo, 2021). Although several studies have addressed the microbiological differences between dental and peri‐implant plaque (Kumar et al., 2012; Dabdoub et al., 2013; Daubert et al., 2018), there are no longitudinal studies available that simultaneously compare the response to mechanical debridement with or without antimicrobial adjuvants on dental and peri‐implant sites with inflammation.
Here we report the microbiological findings from a randomized clinical study where the effects of three mouth rinses, namely delmopinol, chlorhexidine, and a placebo, were assessed in patients with peri‐implant mucositis on subgingival biofilms of peri‐implant sites in comparison with the patient‐matched dental sites.
2. MATERIALS AND METHODS
2.1. Study design and clinical procedures
This was a 3‐month, double‐blinded, randomized clinical trial (The Netherlands Trial Register NL5159 [NTR5299], Ethical Approval of VU METC no. 2015.370), with three parallel groups, as described in detail elsewhere (Philip et al., 2020). In brief, 89 patients with at least one implant diagnosed with peri‐implant mucositis (pocket depth <5 mm and bleeding on gentle probing) were randomly assigned to one of the three study groups: delmopinol (0.2% delmopinol hydrochloride containing mouthwash decapinol; Sinclair Pharmaceutical Ltd, UK), chlorhexidine (containing 0.2% chlorhexidine digluconate), or a placebo. Subjects were instructed to rinse the mouth twice a day, after brushing, with 10 ml of the mouth rinse for at least 1 min during 1 month.
At the screening examination, a single implant, diagnosed with peri‐implant mucositis, and a single tooth without periodontal disease were selected for microbiological sample collection throughout the study based on the highest number of bleeding sites per tooth or implant, respectively. At the baseline visit, first, the samples were collected as described below. Then, the clinical examination (plaque index [PI], pocket depth [PPD], and bleeding on probing [BOP]) was performed as described previously (Philip et al., 2020). To compare the clinical results with the microbiological composition, we calculated the BOP of the sampled sites and PI of the sampled sites from full‐mouth clinical data. For this, six individual values around the sampled tooth/implant were averaged. Thereafter, the participants received treatment including full‐mouth supra‐gingival scaling and polishing according to their periodontal conditions; the implants were debrided using an ultrasonic scaler. Supra‐gingival maintenance care was provided after 1 month (follow‐up 1, FU1) and 3 months (follow‐up 2, FU2). At each visit, the dentition was debrided and polished, and oral hygiene instructions were provided.
2.2. Subgingival plaque sample collection
The selected implant and tooth were isolated using cotton rolls, air‐dried, and the supra‐gingival biofilm was carefully removed using a scaler. The subgingival plaque sample around the selected implant or tooth was obtained with a sterile implant deplaquer (KerrHawe SA, Bioggio, Switzerland) with a vertical stroke at a moderate pressure against the root surface and wiped off on the inside edge of the lid of an Eppendorf tube containing 50 μl reduced transfer fluid (RTF) (Syed & Loesche, 1972). Plaque was collected from four sites around individual tooth/implant and pooled in a single tube, centrifuged, and stored at −80°C until analysis.
2.3. Sample processing and 16S rRNA gene amplicon sequencing
Tubes containing the samples were thawed and centrifuged for 10 s at 15,871g. The pellet was resuspended in the supernatant fluid and transferred to a 1.1‐ml deep‐well plate (Axygen Scientific Inc., CA). The tubes were washed with 150 μl Tris‐EDTA buffer and the fluid was transferred to the same well. After a 2‐min bead‐beating step, DNA was isolated and bacterial DNA concentration was determined using 16S rDNA qPCR, followed by amplicon generation (Kahharova et al., 2020). In addition to the clinical samples, duplicate sample blanks from sterile curettes and extraction blanks were included. The V4 hypervariable region of the 16S rRNA gene was amplified using 1 ng DNA, and samples and controls were pooled and sequenced on the Illumina MiSeq platform as described previously (Kahharova et al., 2020).
2.4. Sequencing data processing
The paired‐end reads were merged, quality‐filtered, and clustered into operational taxonomic units (OTUs) at 97% similarity, as described previously (Koopman et al., 2016). However, a maximum of 25 mismatches (10%, as reads were 2 × 251 nt) were allowed during read‐merging, before quality filtering at 0.5 maximum expected error. The most abundant sequence of each OTU was classified using the RDP classifier (Wang et al., 2007) (min. confidence 0.8) and the SILVA rRNA database v 128 (Quast et al., 2013), as described previously (Koopman et al., 2016).
2.5. Statistical analyses
The distribution of the univariate variables was tested using the Kolmogorov–Smirnov test for normality. The univariate data among the three groups were compared using the Kruskal–Wallis test followed by the Mann–Whitney test. The differences between the categorical variables were assessed using the Chi‐square test. Pair‐wise comparisons were performed using the Friedman test, followed by Wilcoxon signed ranks tests. The BOP and PI from the sampled sites were correlated with the microbial alpha diversity using Spearman correlation. When relative abundances of major genera were compared among the groups or between the sample types, the p‐values were not corrected for multiple comparisons; p < .05 was considered statistically significant. The statistical analyses above were performed with SPSS 25.0 (IBM Corporation, Armonk, NY).
The OTU table was randomly subsampled at 3400 reads/sample. The Shannon diversity index and species richness (number of OTUs/sample) were calculated on the sub‐sampled dataset, followed by principal component analysis (PCA) on the log 2‐transformed sub‐sampled dataset in PAST v 3.05 (Hammer et al., 2001).
To compare the microbial profiles between independent sample groups, one‐way permutational multivariate analysis of variance (PERMANOVA) with Bray–Curtis similarity and 9999 permutations, was used, while for comparisons of related samples PERMANOVA with restricted permutations was used. Both methods were applied using adonis2 (R v 3.6.1 [R Core Team, 2019]; vegan v. 2.4‐6 [Oksanen et al., 2019]).
To identify discriminatory OTUs, the linear discriminant analysis effect size (LEfSe) biomarker discovery tool (Segata et al., 2011) was used with default settings.
3. RESULTS
3.1. Clinical outcomes
The full description of the demographic data and the clinical results of the study have been reported previously (Philip et al., 2020). In brief, 89 patients diagnosed with peri‐implant mucositis underwent a professional debridement and were randomly assigned to use one of the three mouth rinses, namely delmopinol, CHX, or placebo, twice daily for 1 month. The study comprised a baseline visit, a follow‐up at 1 month (FU1), and a follow‐up 3 months (FU2) later. At baseline, no significant differences between the three groups were found for sex, age, ASA score, smoking habit, the mean number of implants, implant brand, type of suprastructure, bone level and augmentation, periodontal treatment history, keratinized tissue width, location of the implant, or the clinical indices (PI, BI, BOP, PPD) at the implant level. After 3 months, at FU2, all three groups showed significant improvement in the clinical parameters and this improvement was comparable among the groups (Philip et al., 2020).
3.2. Overall sequencing results and bacterial DNA concentration in the samples
In total, 532 samples were sequenced (N = 266 dental and N = 266 peri‐implant plaque), which resulted in an OTU table with 600 OTUs and 6,850,791 sequencing reads, with on average 12,877 reads per sample (min. 2, max. 27,062 reads, SD 5211) (Figure S1A). Of these, 52 samples did not pass the sub‐sampling cut‐off (3400 reads/sample) and were excluded from the analyses (Table S1, Figure S1B). The quality controls (n = 31) had on average 32 reads (SD 74, median 5, range 2–316) and were clustered together (Figure S1B). The blank isolation controls (n = 16) had on average 0.0045 ng/μl of bacterial DNA (SD 0.001), the RTF controls (n = 9) 0.0057 ng/μl (SD 0.004). The samples with less than 3400 reads and therefore excluded from the analyses (n = 52) had 0.008 ng/μl (SD 0.011, median 0.005) of bacterial DNA, while the successfully sequenced samples (n = 480) had 4.4 ng/μl (SD 5.9, median 2.1) of bacterial DNA per sample.
Peri‐implant plaque samples had significantly lower bacterial DNA concentration than dental plaque samples in all groups and timepoints except at FU1 after CHX use (Figure S2). For dental plaque, all excluded samples (n = 8) belonged to the CHX group (six samples were collected after FU1 and two after the FU2). For peri‐implant plaque, no difference among the groups in the numbers of the discarded samples (n = 44) was found (p > .05, Chi‐square test). Regarding the timepoints, significantly more peri‐implant samples were lost at FU1 (n = 19) and FU2 (n = 21) than at baseline (n = 4) (p = .001, Chi‐square test).
The final OTU table contained 258 dental and 222 peri‐implant plaque samples and 568 OTUs that were assigned to 151 genera or higher level taxa.
3.3. Comparison between dental and peri‐implant sites at baseline
At baseline, all 89 sampled peri‐implant sites were diagnosed with peri‐implant mucositis, while only 54 of the sampled dental sites were diagnosed with gingivitis, 6 with a point bleeding, and the remaining 29 healthy. For the comparison of the two conditions, namely gingivitis and peri‐implant mucositis, at the baseline, we included only those paired samples where both mucositis and gingivitis were diagnosed (n = 54).
Although no difference in BOP was observed between the sampled dental and implant sites, the peri‐implant sites had a significantly lower plaque index (p < .0001, Wilcoxon signed ranks test) (Figure 1A) and a higher PPD (median 3.33 mm, range 1.8–4.7 mm) than the sampled dental sites (median 3.0 mm, range 1.8–4.0) of the same individual (p = .005).
FIGURE 1.
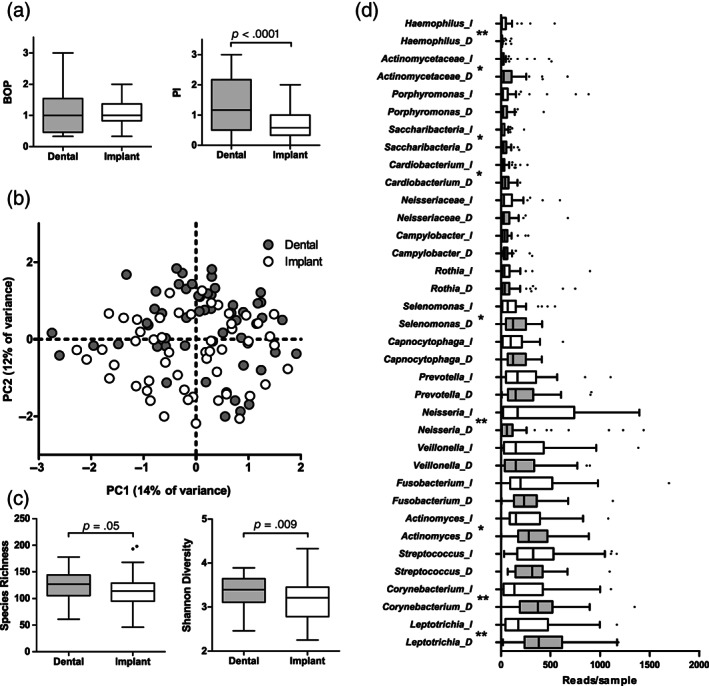
Comparison of baseline subgingival plaque samples collected from paired dental sites with gingivitis and implant surfaces with mucositis. (a) Bleeding on probing (BOP) and plaque index (PI) around the sampled teeth and implants (p‐value based on Wilcoxon signed ranks test). (b) Principal component analysis (PCA) plot of the bacterial profile data (p = .0001, F = 2.84; restricted PERMANOVA). (c) Species richness (number of OTUs/sample) and Shannon diversity index (p‐values based on Wilcoxon signed ranks test) and (d) top 18 most abundant genera or higher taxa by sample type. D, samples from dental surfaces; I, samples from implant surfaces. Boxplots are based on Tukey. Statistical significance between the paired samples (Wilcoxon signed ranks test) at p < .05 is indicated with *, while p < .005 with **. p‐Values were not corrected for multiple comparisons. N = 54 dental and implant site pairs
Microbial profiles of the subgingival plaque collected around the teeth with gingivitis differed significantly from subgingival plaque collected around the implants with mucositis from the same patients (p = .0001, F = 2.84, restricted PERMANOVA) (Figure 1b). The dental plaque had a significantly higher diversity (Figure 1c) and a significantly higher proportion of the genera Leptotrichia, Corynebacterium, Actinomyces, Selenomonas, Cardiobacterium, Saccharibacteria, unclassified Actinomycetaceae, Lachnoanaerobaculum, Tannerella, and Aggregatibacter than the respective samples around the implants, while peri‐implant subgingival plaque had a significantly higher proportion of the genera Neisseria and Haemophilus (Figure 1d).
At the individual OTU level, 99 OTUs discriminated between two sample types, of which 75 were associated with dental plaque and only 24 with peri‐implant plaque (Table S2A).
The Shannon diversity index of dental plaque correlated positively with PI (r = .285, p = .037) at the sampled sites with gingivitis, while no such relation was observed between peri‐implant plaque at the sampled sites with mucositis (r = .188, p = .178).
3.4. Effects of the therapy on the subgingival microbiome
Within each group, significant microbial shifts were observed in both dental and peri‐implant subgingival plaque after the use of delmopinol (Figure 2a) and CHX (Figure 2b) but not after the use of the placebo mouth rinse (Figure 2c). Within the delmopinol and CHX groups, the microbial shift was significant between the baseline and FU1 visit (delmopinol, dental: F = 4.14, p = .0004, peri‐implant: F = 4.07, p = .0001; CHX, dental: F = 6.04, p = .0001, peri‐implant: F = 3.54, p = .0002). At FU2, only within the CHX group the samples still differed significantly from the baseline samples (dental: F = 1.03, p = .04; peri‐implant: F = 1.46, p = .006).
FIGURE 2.
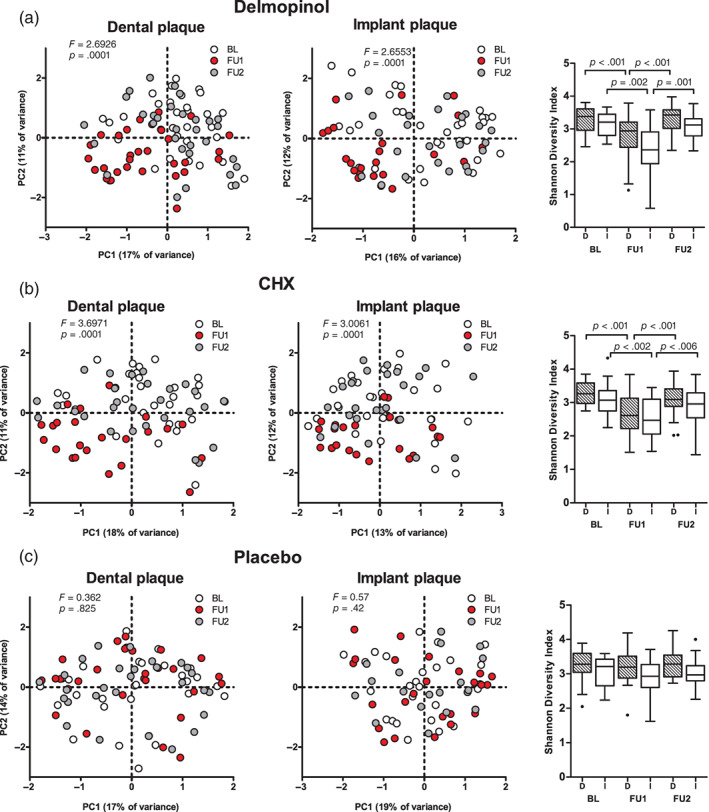
Effects of 1‐month use of delmopinol (a), chlorhexidine (CHX); b), and placebo (c) mouthrinses on microbial profiles and diversity of subgingival plaque from dental surfaces and implant surfaces in time. BL, baseline visit; FU1, first follow‐up (1 month); FU2, second follow‐up (3 months). F‐ and p‐values are based on restricted PERMANOVA. Boxplots are based on Tukey. D, dental plaque; I, implant plaque. Connectors indicate a significant difference in diversity between timepoints (Wilcoxon signed ranks test) [Colour figure can be viewed at wileyonlinelibrary.com]
Across the groups, no differences among them were observed at baseline or FU2, while at FU1 there was a significant difference among the three groups in both dental (F = 3.92; p = .0001, PERMANOVA) and peri‐implant plaque microbiomes (F = 2.43; p = .0006) (Figure S3). The largest difference in dental plaque was between CHX and the other two groups (p = .0003, for both comparisons), followed by the comparison between delmopinol and placebo (p = .008). For peri‐implant plaque, CHX differed most from placebo (p = .01) and delmopinol (p = .012), while delmopinol and placebo differed from each other at the border of significance (p = .039).
Bacterial diversity was significantly reduced at FU1, both on dental and implant surfaces, in delmopinol and CHX groups (Figure 2a,b). At FU1, delmopinol and CHX use led to a significant reduction in the Shannon diversity index in dental plaque and both in Shannon diversity (Figure S3B) and species richness (data not shown) in peri‐implant plaque compared to placebo. No difference in diversity was observed between the delmopinol and CHX groups. At FU2, microbial diversity of the delmopinol group had recovered both on the dental and the implant surfaces, while implant surfaces exposed to CHX at FU2 still had significantly lower species richness (median 94, range 43–188 taxa) compared to the baseline visit (median 115, range 75–198) (p = .005).
At the genus level, the proportion of the genus Streptococcus was significantly higher at FU1 in dental plaque exposed to CHX and in peri‐implant plaque exposed to both delmopinol and CHX at FU1, while the genera Leptotrichia, Actinomyces, and Corynebacterium showed a significantly lower proportion (Figure 3, Table S3). Exposure to delmopinol resulted in a significantly higher proportion of the genus Rothia both in dental and peri‐implant plaque at FU1. Notably, several genera in the peri‐implant plaque samples showed significant decrease or increase at FU1 also in the placebo group but not in the dental plaque (Table S3).
FIGURE 3.
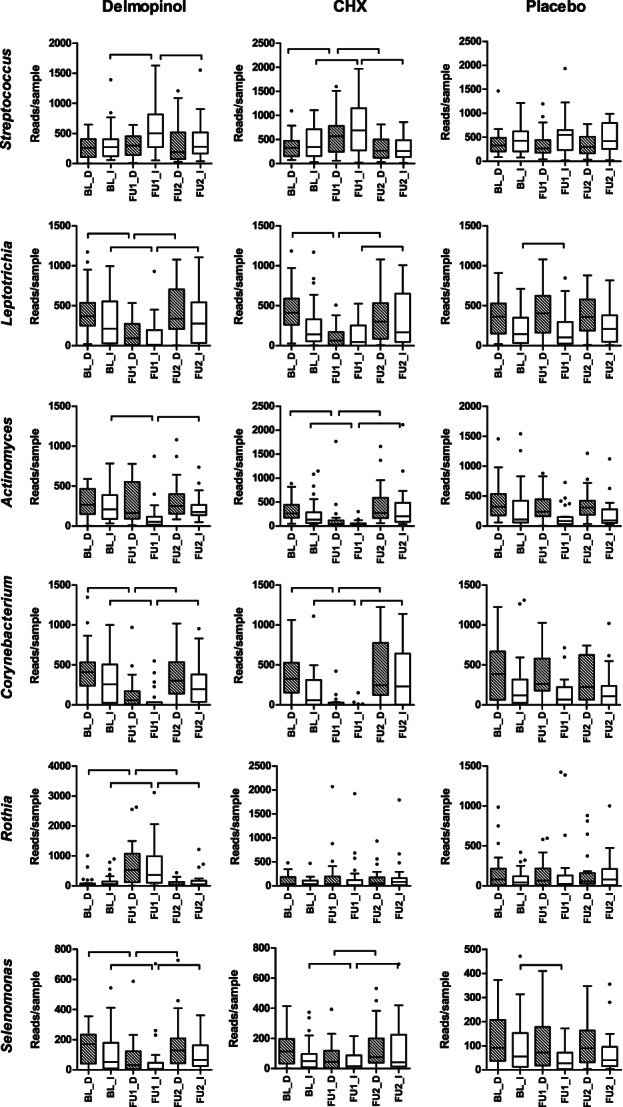
Examples of bacterial genera that were affected most significantly by the therapy. Hatched boxes indicate dental plaque and white boxes implant plaque. Connectors indicate a statistically significant difference (p < .05, Wilcoxon signed ranks test, p‐values not corrected for multiple comparisons). Full results on the 30 most abundant genera are shown in Table S3
At the individual OTU level, there were 29 OTUs in the dental plaque samples and 16 OTUs in the implant plaque samples, which discriminated between the delmopinol and the placebo groups at FU1 (Table S2B). Between the CHX and the placebo exposed samples, there were 45 OTUs in dental plaque and 23 OTUs in peri‐implant plaque, which discriminated the two groups, while no OTUs differed between the delmopinol and the CHX groups at FU1.
3.5. Subgingival plaque composition in relation to the clinical diagnosis after the therapy
In all groups, 3 months since the start of the therapy (FU2), except in the peri‐implant sites in the CHX group, proportionally more sampled sites were healthy (no bleeding or a point bleeding) than unhealthy (gingivitis or mucositis) (Figure 4a,b). In dental plaque, significantly more healthy sites were observed in the placebo group than in the delmopinol (p = .007) or the CHX (p = .021) group, while around the implants there was a significantly higher frequency of healthy sites in the delmopinol group compared to the CHX group (p = .013).
FIGURE 4.
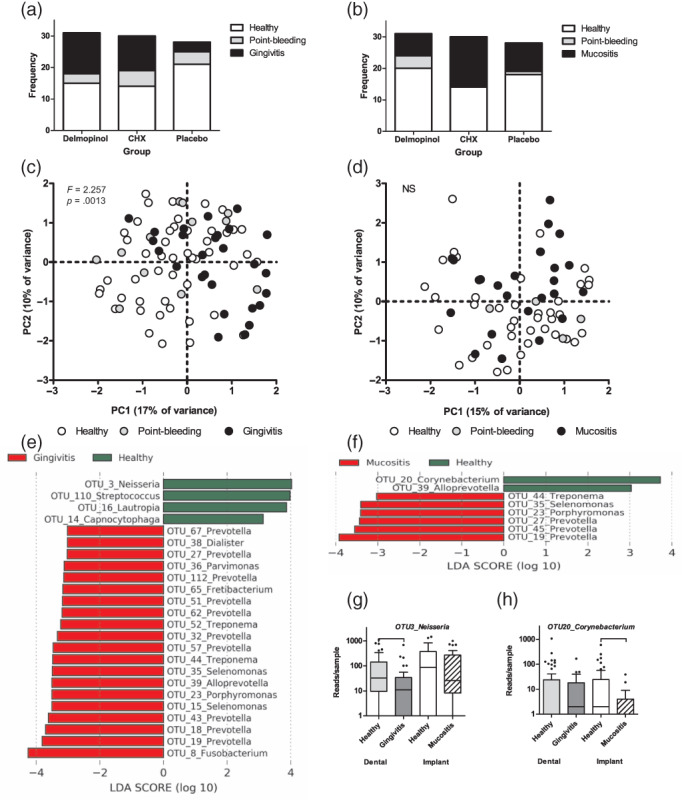
Comparison of sampled sites 2 months after the therapy by diagnosis: frequency distribution by diagnosis per group at the dental sites (a) and the peri‐implant sites (b). Principal component analysis (PCA) plot of the bacterial profiles data of dental plaque (c) and peri‐implant plaque samples (d) by clinical diagnosis (F‐ and p‐values based on PERMANOVA, NS ‐ p > .05); the most significantly discriminating OTUs between healthy and gingivitis cases (e) and between healthy and mucositis cases (f) (here, LDA > 3; LEfSe; full list of discriminatory OTUs at LDA > 2 is presented in Table S2C). (g) Boxplots of OTU3_Neisseria, discriminating healthy and gingivitis samples. (h) Boxplots of OTU20_Corynebacterium, discriminating the healthy and mucositis samples. The two examples were the most discriminatory features from LEfSe output associated with soft tissue health [Colour figure can be viewed at wileyonlinelibrary.com]
Next, we compared the microbial profiles based on the clinical diagnosis at the sampled site at FU2, irrespective of the treatment group. In dental plaque, there was a significant difference in microbiome composition by the diagnosis (p = .0013, F = 2.257, PERMANOVA). Although the PCA plot (Figure 4c,d) suggested grouping by diagnosis also among the peri‐implant biofilms, no statistical difference by diagnosis was observed. This lack of significance could be due to a smaller number of peri‐implant samples (n = 68) than dental plaque samples (n = 87) and unequal distribution of samples per diagnosis (n = 40 healthy, n = 4 point‐bleeding, n = 24 mucositis). Dental plaque samples with gingivitis (n = 27) differed from those with point bleeding (n = 12) (p = .02, F = 2.5, PERMANOVA, after Bonferroni correction) and those without bleeding (n = 48) (p = .0012, F = 3.517), while no difference in microbial profiles was found between the samples with a point bleeding and samples without any bleeding. Therefore, for further analyses, these two diagnoses were combined into a diagnosis “healthy”.
When the cases diagnosed with gingivitis or mucositis at baseline and as healthy at FU2 were assessed (n = 38 for teeth, n = 43 for implants), there was a significant reduction in bacterial diversity (species richness: p = .005; Shannon diversity index: p = .038) in the transition from mucositis to health in the peri‐implant samples, but not from gingivitis to health in the dental plaque samples.
At the genus level, healthy sites around teeth had a significantly higher proportion of Rothia (p = .043) and a lower proportion of Fusobacterium (p = .006) and Prevotella (p < .0001) than plaque from sites with gingivitis. Sites with peri‐implant mucositis had a significantly higher proportion of the genera Prevotella (p = .003), Porphyromonas (p = .045), Treponema (p = .021), and Alloprevotella (p = .017) than the healthy sites.
At the individual OTU level, 11 OTUs were significantly associated with healthy gingiva, while 79 OTUs were associated with gingivitis (Table S2C, Figure 4e). In peri‐implant sites, only 3 OTUs were associated with health and 20 with peri‐implant mucositis (Table S2C, Figure 4f). The main taxa associated with mucositis overlapped with those of gingivitis, while health was discriminated by different OTUs. For instance, in dental plaque, OTU3 (Neisseria) was the most discriminatory for health, while in the peri‐implant biofilms this OTU was relatively abundant irrespective of the diagnosis (Figure 4g). Healthy peri‐implant sites had a significantly higher proportion of OTU20 (Corynebacterium) (Figure 4h).
To assess the differences between the dental and peri‐implant sites at health, we compared the pairs of samples that were diagnosed as healthy (no bleeding or a point bleeding) (n = 28) at FU2 (Figure 5). A weak, though significant, difference in microbial profiles was observed (p = .033, F = 1.264, restricted PERMANOVA) (Figure 5a), with dental plaque samples having significantly higher species richness and Shannon diversity index than the respective peri‐implant samples (Figure 5b). At the genus level, healthy dental plaque had a significantly lower proportion of Streptococcus (p = .02) and Neisseria (p = .02) and a higher proportion of Leptotrichia (p = .012), Corynebacterium (p = .006), Selenomonas (p = .006) (Figure 5c), Tannerella (p = .045), and unclassified Actinomycetaceae (p = .004) than healthy peri‐implant biofilms. At the OTU level, 25 OTUs were discriminatory for healthy sites around teeth and 9 OTUs around healthy implants (Table S2D). The latter included non‐typical oral taxa such Peptoniphilus (OTU162), which were not associated with healthy dental sites.
FIGURE 5.
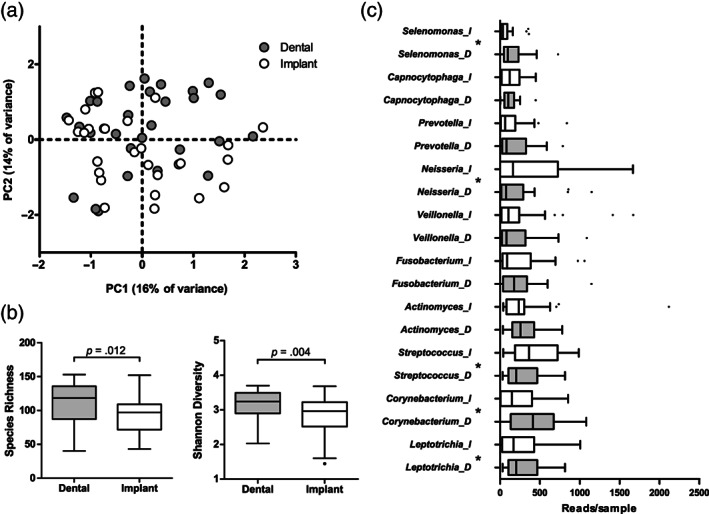
Microbial profiles of healthy sites 2 months after the therapy (FU2) by sample type: (a) Principal component analysis (PCA) plot of the bacterial profile data of dental plaque and peri‐implant plaque (p = .033, F = 1.264, restricted PERMANOVA). (b) Species richness (number of OTUs/sample) and Shannon diversity index (p‐values based on Wilcoxon signed ranks test) and (c) top 10 most abundant genera by sample type. D, dental and I, peri‐implant plaque samples. Boxplots are based on Tukey. *Statistical significance between the paired samples at p < .05 (Wilcoxon signed ranks test). p‐Values were not corrected for multiple comparisons. N = 28 dental and implant sample pairs
4. DISCUSSION
In this study, we addressed the differences between the subgingival dental plaque and peri‐implant plaque by the soft‐tissue diagnosis (gingivitis/mucositis and healthy) and mechanical debridement with the adjuvant antimicrobial therapy in time.
Before the therapy, the mucositis‐associated peri‐implant plaque community was distinct from that of gingivitis and had more aerobic taxa such as Neisseria and Haemophilus, while gingivitis was associated with higher plaque index, higher alpha diversity, and a higher proportion of strict anaerobes. Higher diversity at the dental sites compared to the implant sites is in line with previous reports (Kumar et al., 2012; Dabdoub et al., 2013; Payne et al., 2017). Kumar and colleagues also reported distinct microbial communities between the dental and peri‐implant sites (Kumar et al., 2012; Dabdoub et al., 2013). In contrast, a study of limited sample size (N = 15) failed to find any difference in bacterial composition or alpha diversity in subject‐matched teeth and implants (Schincaglia et al., 2017).
To our knowledge, there are no other randomized clinical trials on humans that have assessed the effects of delmopinol on peri‐implant mucositis. At the end of the one‐month treatment (FU1), both delmopinol and CHX had resulted in a significant microbial shift both in the dental and in the peri‐implant plaque. Two months after the end of the treatment (FU2), only CHX‐exposed samples still differed from the baseline samples in their composition. Nevertheless, all groups, including placebo, presented with comparable clinical improvement (Philip et al., 2020). This is in line with previous reports that have reported the effectiveness of mechanical debridement alone (Máximo et al., 2009), and similar results after combining mechanical debridement with antimicrobials (Thöne‐Mühling et al., 2010; Heitz‐Mayfield et al., 2011), antibiotics (Hallström et al., 2012), probiotics (Galofré et al., 2018) or essential oil‐containing mouthrinse (Pulcini et al., 2019). The potential Hawthorne effect (McCambridge et al., 2014), when all participants of a clinical study may have improved their normal oral hygiene habits just because they are participating in a study, cannot be excluded.
The comparison of the microbial profiles by the disease status at the end of the study revealed that the subgingival microbiome of the dental plaque, but not the peri‐implant plaque, differed by diagnosis and that, similar to the differences during disease, the healthy sites around the teeth harboured more diverse and more anaerobe‐rich microbiome than the healthy sites around the implants. Additionally, the inflamed implant sites had a lower plaque index than the dental sites with gingivitis. In other words, inflammation in the peri‐implant tissues seems to be triggered by a thinner and healthier biofilm than in the gingival tissues. These findings support the notion that the inflammatory response around the implants is (co)triggered by the presence of the implant (both its structure and material) and not the oral biofilm per se (Belibasakis, 2014).
Several factors such as friction against the bone during implant placement, wear due to mechanical load as well as wear generated by debridement, exposure to bacteria, oral environment, and chemical agents have been linked to the damage of the titanium oxide layer and corrosion of implant materials (Rodrigues et al., 2013; Delgado‐Ruiz & Romanos, 2018; Kotsakis & Olmedo, 2021). This corrosion may result in the release of titanium and other metal ions, which in turn are shown to relate with clinical outcomes and survival of the implants (Safioti et al., 2017; Delgado‐Ruiz & Romanos, 2018; Noumbissi et al., 2019). A recent study assessed the effects of titanium release in plaque on clinical and microbiological outcomes and found that titanium concentration in plaque correlated negatively with microbiome diversity and positively with certain bacterial taxa (Daubert et al., 2018). Titanium ion particles are shown to stimulate inflammasomes in macrophages and IL‐1ß release, resulting in pro‐inflammatory reaction (Pettersson et al., 2017). Taken together, future research should focus on tailored peri‐implant disease therapies that consider implant‐related environmental factors.
A serious drawback of this study was the loss of nearly 10% of the samples after the sequencing. These samples had low bacterial DNA concentration and faint to no band on the agarose gel used to assess the quality of amplification, already before the sequencing. Our experience shows that in some cases the absence of a clear band on the gel does not preclude a successful sequencing output. Therefore, all samples were included in the equimolar mix, after the adjustment for the bacterial DNA concentration. Most (85%) of the discarded samples originated from the peri‐implant subgingival biofilm, of which 91% were collected after the therapy (FU1 and FU2), irrespective of the mouthrinse group. This loss of the samples for the microbiome data has lowered the power of the study regarding the follow‐up peri‐implant samples and might have influenced the results. Additionally, since this study did not address the role of implant type and surface characteristics on the microbiological composition, the influence of these characteristics on the obtained results cannot be excluded and should be addressed in future studies.
5. CONCLUSION
Peri‐implant sites with mucositis harbour ecologically less complex and less anaerobic biofilms with lower biomass than patient‐matched dental sites with gingivitis while eliciting an equal inflammatory response, suggesting the responsiveness of the host tissue to the implant itself. Adjunctive antimicrobial therapy in addition to mechanical debridement does affect both dental and peri‐implant biofilm composition in the short term, resulting in a less dysbiotic subgingival biofilm. However, these microbiological effects are not reflected in the clinical response.
CONFLICT OF INTEREST
The authors declare no conflict of interests.
ETHICS STATEMENT
Ethical Approval of VU METC no. 2015.370.
Supporting information
Figure S1. Sequencing quality assessment: (A) Rarefaction plots showing OTUs per sample (observed species) and Shannon diversity index before sub‐sampling to 3400 reads/sample. (B) Left plot: PCA plot of all samples and controls coloured by the 3400 reads/sample cut‐off of the sub‐sampling depth. Right plot: PCA plot by the type of sample: bl.isol, blank DNA isolation controls (n = 16); implant, peri‐implant subgingival biofilm (n = 266); neg. PCR, negative controls of the PCR (n = 6); plaque, subgingival dental plaque samples (n = 266); RTF, reduced transport fluid controls (n = 9). As can be seen from this plot, all but a single blank isolation control (red dot) cluster tightly together with the negative PCR and RTF controls. This particular blank isolation sample had the most (316) reads and was a single outlier.
Figure S2. Bacterial DNA (16S rDNA) concentration (ng/μl sample) per group, sample type, and timepoint. Horizontal lines connect the subgroups that were statistically significantly different (p < .005, Wilcoxon signed ranks test) either by sample type or by timepoint.
Figure S3. Comparisons of the bacterial profiles (PCA, PERMANOVA) and bacterial diversity (Shannon diversity index) among the three groups (delmopinol, CHX, and placebo) per timepoint: (A) baseline visit; (B) follow‐up after 1 month of mouthrinse use (FU1); (C) follow‐up 3 months since the start of the therapy and 2 months after the end of the mouthrinse use (FU2).
Table S1. Number of samples that were either excluded due to low sequencing reads or included in the analyses (passed sub‐sampling cut‐off of 3400 reads/sample) per group, sample type (dental or peri‐implant), and timepoint (BL, baseline; FU1, 1 month follow‐up; FU2, 3‐months follow‐up).
Table S2. Significantly discriminatory OTUs between the groups: (A) Baseline unhealthy (gingivitis/mucositis) by sample type. (B) Effects of mouthrinses. (C) Diagnosis at FU2. (D) Healthy sites at FU2 by sample type. LDA scores and p‐values (output from LEfSe).
Table S3. Effects of the mouthrinses on the top 30 most abundant bacterial genera or higher taxa in dental and peri‐implant plaque samples in time. BL–FU1, comparison between the baseline and follow‐up 1 (1 month after the start of the therapy and at the end of the mouthrinse use); FU1–FU2, comparison between FU1 and FU2 (3 months after the start of the therapy and 2 months after the end of the mouthrinse use). NS, no statistically significant changes. ****p < .00005; ***p < .0005; **p < .005; *p < .05 (Wilcoxon signed ranks test). p‐Values are not corrected for multiple comparisons. Cells in red indicate significant decrease, and cells in green indicate significant increase in the proportion of the respective taxa in the respective comparison.
ACKNOWLEDGEMENTS
We thank the participants of the clinical study, the AUMC Cancer Center Amsterdam (Amsterdam, the Netherlands) for sequencing the amplicon on the Illumina Miseq system, and Elly van Deutekom and Wendy de Wit for their contribution in sample preparation for sequencing.
Philip, J. , Buijs, M. J. , Pappalardo, V. Y. , Crielaard, W. , Brandt, B. W. , & Zaura, E. (2022). The microbiome of dental and peri‐implant subgingival plaque during peri‐implant mucositis therapy: A randomized clinical trial. Journal of Clinical Periodontology, 49(1), 28–38. 10.1111/jcpe.13566
Funding informationThe work presented here was funded by the Department of Preventive Dentistry, Academic Center for Dentistry Amsterdam, The Netherlands.
DATA AVAILABILITY STATEMENT
Sequencing data is available upon request.
REFERENCES
- Belibasakis, G. N. (2014). Microbiological and immuno‐pathological aspects of peri‐implant diseases. Archives of Oral Biology, 59, 66–72. [DOI] [PubMed] [Google Scholar]
- Belibasakis, G. N. , & Manoil, D. (2021). Microbial community‐driven etiopathogenesis of peri‐implantitis. Journal of Dental Research, 100, 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabdoub, S. M. , Tsigarida, A. A. , & Kumar, P. S. (2013). Patient‐specific analysis of periodontal and peri‐implant microbiomes. Journal of Dental Research, 92, 168S–175S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubert, D. , Pozhitkov, A. , McLean, J. , & Kotsakis, G. (2018). Titanium as a modifier of the peri‐implant microbiome structure. Clinical Implant Dentistry and Related Research, 20, 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado‐Ruiz, R. , & Romanos, G. (2018). Potential causes of titanium particle and ion release in implant dentistry: A Systematic review. International Journal of Molecular Sciences, 19, 3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galofré, M. , Palao, D. , Vicario, M. , Nart, J. , & Violant, D. (2018). Clinical and microbiological evaluation of the effect of Lactobacillus reuteri in the treatment of mucositis and peri‐implantitis: A triple‐blind randomized clinical trial. Journal of Periodontal Research, 53, 378–390. [DOI] [PubMed] [Google Scholar]
- Hallström, H. , Persson, G. R. , Lindgren, S. , Olofsson, M. , & Renvert, S. (2012). Systemic antibiotics and debridement of peri‐implant mucositis. A randomized clinical trial. Journal of Clinical Periodontology, 39, 574–581. [DOI] [PubMed] [Google Scholar]
- Hammer, O. , Harper, D. A. T. , & Ryan, P. D. (2001). PAST: Paleontological statistics software package for education and data analysis. Palaeontologia Electronica, 4, 1–9. [Google Scholar]
- Heitz‐Mayfield, L. J. A. , Salvi, G. E. , Botticelli, D. , Mombelli, A. , Faddy, M. , Lang, N. P. , & the Implant Complication Research . (2011). Anti‐infective treatment of peri‐implant mucositis: A randomised controlled clinical trial. Clinical Oral Implants Research, 22, 237–241. [DOI] [PubMed] [Google Scholar]
- Kahharova, D. , Brandt, B. W. , Buijs, M. J. , Peters, M. , Jackson, R. , Eckert, G. , Katz, B. , Keels, M. A. , Levy, S. M. , Fontana, M. , & Zaura, E. (2020). Maturation of the oral microbiome in caries‐free toddlers: A longitudinal study. Journal of Dental Research, 99, 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman, J. E. , Buijs, M. J. , Brandt, B. W. , Keijser, B. J. F. , Crielaard, W. , & Zaura, E. (2016). Nitrate and the origin of saliva influence composition and short chain fatty acid production of oral microcosms. Microbial Ecology, 72, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsakis, G. A. , & Olmedo, D. G. (2021). Peri‐implantitis is not periodontitis: Scientific discoveries shed light on microbiome‐biomaterial interactions that may determine disease phenotype. Periodontology 2000, 86, 231–240. [DOI] [PubMed] [Google Scholar]
- Kumar, P. S. , Dabdoub, S. M. , Hegde, R. , Ranganathan, N. , & Mariotti, A. (2018). Site‐level risk predictors of peri‐implantitis: A retrospective analysis. Journal of Clinical Periodontology, 45, 597–604. [DOI] [PubMed] [Google Scholar]
- Kumar, P. S. , Mason, M. R. , Brooker, M. R. , & O'Brien, K. (2012). Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. Journal of Clinical Periodontology, 5, 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.‐T. , Huang, Y.‐W. , Zhu, L. , & Weltman, R. (2017). Prevalences of peri‐implantitis and peri‐implant mucositis: Systematic review and meta‐analysis. Journal of Dentistry, 62, 1–12. [DOI] [PubMed] [Google Scholar]
- Máximo, M. B. , De Mendonça, A. C. , Renata Santos, V. , Figueiredo, L. C. , Feres, M. , & Duarte, P. M. (2009). Short‐term clinical and microbiological evaluations of peri‐implant diseases before and after mechanical anti‐infective therapies. Clinical Oral Implants Research, 20, 99–108. [DOI] [PubMed] [Google Scholar]
- McCambridge, J. , Witton, J. , & Elbourne, D. R. (2014). Systematic review of the Hawthorne effect: New concepts are needed to study research participation effects. Journal of Clinical Epidemiology, 67, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noumbissi, S. , Scarano, A. , & Gupta, S. (2019). A literature review study on atomic ions dissolution of titanium and its alloys in implant dentistry. Materials, 12, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J. , Guillaume Blanchet, F. , Friendly, M. , Kindt, R. , Legendre, P. , McGlinn, D. , Minchin, P. , O'Hara, R. , Simpson, G. , Solymos, P. , Stevens, M. , Szoecs, E. , & Wagner, H. (2019). Vegan: Community Ecology Package. R package version 2.4‐6. https://CRAN.R-project.org/package=vegan
- Payne, J. B. , Johnson, P. G. , Kok, C. R. , Gomes‐Neto, J. C. , Ramer‐Tait, A. E. , Schmid, M. J. , & Hutkins, R. W. (2017). Subgingival microbiome colonization and cytokine production during early dental implant healing. mSphere, 2, e00527‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson, M. , Kelk, P. , Belibasakis, G. N. , Bylund, D. , Molin Thorén, M. , & Johansson, A. (2017). Titanium ions form particles that activate and execute interleukin‐1β release from lipopolysaccharide‐primed macrophages. Journal of Periodontal Research, 52, 21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip, J. , Laine, M. L. , & Wismeijer, D. (2020). Adjunctive effect of mouthrinse on treatment of peri‐implant mucositis using mechanical debridement: A randomized clinical trial. Journal of Clinical Periodontology, 47, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulcini, A. , Bollaín, J. , Sanz‐Sánchez, I. , Figuero, E. , Alonso, B. , Sanz, M. , & Herrera, D. (2019). Clinical effects of the adjunctive use of a 0.03% chlorhexidine and 0.05% cetylpyridinium chloride mouth rinse in the management of peri‐implant diseases: A randomized clinical trial. Journal of Clinical Periodontology, 46, 342–353. [DOI] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. , Peplies, J. , & Glöckner, F. O. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web‐based tools. Nucleic Acids Research, 41, D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . (2019) R: A Language and Environment for Statistical Computing. Foundation for Statistical Computing, Vienna, Austria.
- Robitaille, N. , Reed, D. N. , Walters, J. D. , & Kumar, P. S. (2016). Periodontal and peri‐implant diseases: Identical or fraternal infections? Molecular Oral Microbiology, 31, 285–301. [DOI] [PubMed] [Google Scholar]
- Rodrigues, D. C. , Valderrama, P. , Wilson, T. G. , Palmer, K. , Thomas, A. , Sridhar, S. , Adapalli, A. , Burbano, M. , & Wadhwani, C. (2013). Titanium corrosion mechanisms in the oral environment: A retrieval study. Materials, 6, 5258–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safioti, L. M. , Kotsakis, G. A. , Pozhitkov, A. E. , Chung, W. O. , & Daubert, D. M. (2017). Increased levels of dissolved titanium are associated with peri‐implantitis – A cross‐sectional study. Journal of Periodontology, 88, 436–442. [DOI] [PubMed] [Google Scholar]
- Schincaglia, G. P. , Hong, B. Y. , Rosania, A. , Barasz, J. , Thompson, A. , Sobue, T. , Panagakos, F. , Burleson, J. A. , Dongari‐Bagtzoglou, A. , & Diaz, P. I. (2017). Clinical, immune, and microbiome traits of gingivitis and peri‐implant mucositis. Journal of Dental Research, 96, 47–55. [DOI] [PubMed] [Google Scholar]
- Segata, N. , Izard, J. , Waldron, L. , Gevers, D. , Miropolsky, L. , Garrett, W. , & Huttenhower, C. (2011). Metagenomic biomarker discovery and explanation. Genome Biology, 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed, S. A. , & Loesche, W. J. (1972). Survival of human dental plaque flora in various transport media. Applied Microbiology, 24, 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöne‐Mühling, M. , Swierkot, K. , Nonnenmacher, C. , Mutters, R. , Flores‐de‐Jacoby, L. , & Mengel, R. (2010). Comparison of two full‐mouth approaches in the treatment of peri‐implant mucositis: A pilot study. Clinical Oral Implants Research, 21, 504–512. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Garrity, G. M. , Tiedje, J. M. , & Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73, 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequencing quality assessment: (A) Rarefaction plots showing OTUs per sample (observed species) and Shannon diversity index before sub‐sampling to 3400 reads/sample. (B) Left plot: PCA plot of all samples and controls coloured by the 3400 reads/sample cut‐off of the sub‐sampling depth. Right plot: PCA plot by the type of sample: bl.isol, blank DNA isolation controls (n = 16); implant, peri‐implant subgingival biofilm (n = 266); neg. PCR, negative controls of the PCR (n = 6); plaque, subgingival dental plaque samples (n = 266); RTF, reduced transport fluid controls (n = 9). As can be seen from this plot, all but a single blank isolation control (red dot) cluster tightly together with the negative PCR and RTF controls. This particular blank isolation sample had the most (316) reads and was a single outlier.
Figure S2. Bacterial DNA (16S rDNA) concentration (ng/μl sample) per group, sample type, and timepoint. Horizontal lines connect the subgroups that were statistically significantly different (p < .005, Wilcoxon signed ranks test) either by sample type or by timepoint.
Figure S3. Comparisons of the bacterial profiles (PCA, PERMANOVA) and bacterial diversity (Shannon diversity index) among the three groups (delmopinol, CHX, and placebo) per timepoint: (A) baseline visit; (B) follow‐up after 1 month of mouthrinse use (FU1); (C) follow‐up 3 months since the start of the therapy and 2 months after the end of the mouthrinse use (FU2).
Table S1. Number of samples that were either excluded due to low sequencing reads or included in the analyses (passed sub‐sampling cut‐off of 3400 reads/sample) per group, sample type (dental or peri‐implant), and timepoint (BL, baseline; FU1, 1 month follow‐up; FU2, 3‐months follow‐up).
Table S2. Significantly discriminatory OTUs between the groups: (A) Baseline unhealthy (gingivitis/mucositis) by sample type. (B) Effects of mouthrinses. (C) Diagnosis at FU2. (D) Healthy sites at FU2 by sample type. LDA scores and p‐values (output from LEfSe).
Table S3. Effects of the mouthrinses on the top 30 most abundant bacterial genera or higher taxa in dental and peri‐implant plaque samples in time. BL–FU1, comparison between the baseline and follow‐up 1 (1 month after the start of the therapy and at the end of the mouthrinse use); FU1–FU2, comparison between FU1 and FU2 (3 months after the start of the therapy and 2 months after the end of the mouthrinse use). NS, no statistically significant changes. ****p < .00005; ***p < .0005; **p < .005; *p < .05 (Wilcoxon signed ranks test). p‐Values are not corrected for multiple comparisons. Cells in red indicate significant decrease, and cells in green indicate significant increase in the proportion of the respective taxa in the respective comparison.
Data Availability Statement
Sequencing data is available upon request.


