Abstract
The brine-seawater interface of the Kebrit Deep, northern Red Sea, was investigated for the presence of microorganisms using phylogenetic analysis combined with cultivation methods. Under strictly anaerobic culture conditions, novel halophiles were isolated. The new rod-shaped isolates belong to the halophilic genus Halanaerobium and are the first representatives of the genus obtained from deep-sea, anaerobic brine pools. Within the genus Halanaerobium, they represent new species which grow chemoorganotrophically at NaCl concentrations ranging from 5 to 34%. The cellular fatty acid compositions are consistent with those of other Halanaerobium representatives, showing unusually large amounts of Δ7 and Δ11 16:1 fatty acids. Phylogenetic analysis of the brine-seawater interface sample revealed the presence of various bacterial 16S rRNA gene sequences dominated by cultivated members of the bacterial domain, with the majority affiliated with the genus Halanaerobium. The new Halanaerobium 16S rRNA clone sequences showed the highest similarity (99.9%) to the sequence of isolate KT-8-13 from the Kebrit Deep brine. In this initial survey, our polyphasic approach demonstrates that novel halophiles thrive in the anaerobic, deep-sea brine pool of the Kebrit Deep, Red Sea. They may contribute significantly to the anaerobic degradation of organic matter enriched at the brine-seawater interface.
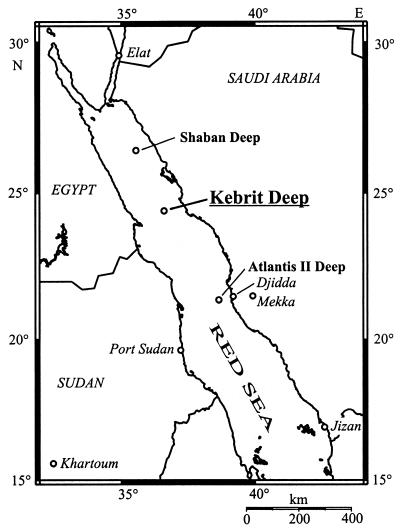
Hypersaline ecosystems are one of the most unusual and extreme environments on earth (15, 25, 36). Anaerobic, deep-sea brine pools, which are located along various tectonic rift systems, represent a special type of hypersaline environment. During the last 50 years about 25 deep-sea brine pools (Fig. 1) with highly saline waters were identified in the Red Sea, an ocean in statu nascendi within the East African Rift Valley system (1, 6, 11, 13, 14, 18, 46, 53, 60, 65). The brines of the Red Sea are typical athalassohaline waters which are in the main a reflection of the geology, geography, and topography of the areas where they develop (18, 25, 27). The high salinity is formed when seawater circulates through subbottom Miocene evaporite deposits, obtaining geothermal heat and dissolved solids before surfacing in the depression of the deeps (1, 12, 62, 72). Characteristic of the brine pools is the formation of gradients along the brine-seawater interface, e.g., salinity, pH, temperature, and oxygen gradients. Brine pools of different origin are also found in the Gulf of Mexico (e.g., the Orca Basin) and in the Mediterranean Sea (e.g., the Tyro, Bannock, or Urania Basin) (19, 37, 44, 61, 63).
FIG. 1.
Outline of the Red Sea, showing representative brine pools. The topographical map was generated with the Online Map Creation program (Geomar, Kiel, Germany [http://www.aquarius.geomar.de]).
The Kebrit Deep (in Arabic, kebrit means sulfur) in the northern Red Sea was first explored during a Valdivia cruise in 1971 and consists of a basin of approximately 1 by 2.5 km in size (27, 52, 59). The deep is filled with a brine of 84 m in thickness at a maximum depth of 1,549 m (27). At the brine-seawater interface of the Kebrit Deep there is a steep increase in salinity from 4 to 26% (wt/vol) NaCl (within only 3 m), an increase in temperature from 21.6 to 23.4°C (within about 7 m), an increase of the CH4 concentration from 50 nl/liter to 22 ml/liter, and a measurable brine pool H2S content of up to 12 to 14 mg of S/liter. Over the same interface the pH drops from 8.1 to 5.5 and the O2 concentration decreases from 3.2 ml of O2/liter to zero (23, 27, 64). At the same time, the density gradient created at the brine-seawater interface acts as an in situ particle trap for organic and inorganic materials from the Red Sea water (27–29, 40, 57, 60, 66).
During the last 30 years, detailed geological and geochemical investigations were carried out in the Kebrit Deep. In contrast, information about the microbial communities of this deep is very rare. Recently, novel bacterial and archaeal 16S rRNA gene sequences have been retrieved from brine sediments (21, 48, 50). These investigations showed that novel groups of Archaea and Bacteria (KB1 sequence group) thrive in the extreme environment of the Kebrit Deep (21). The presence of archaeal methanogenesis is also suggested by the biochemical characterization of C40 isoprenoids, an archaeal biomarker, in sedimentary organic matter (45), and an apparent biotic methane oxidation at the brine-seawater interface (23). Biochemical investigations in similar brine pools (Orca and Bannock Basins) indicated a high microbial potential at the brine-seawater interface and suggest the presence of halophilic microorganisms within the brine (17, 22, 39, 40, 68).
A great diversity of microorganisms have been isolated from high-salinity environments, including aerobic and anaerobic organisms of the bacterial and archaeal domains (25). These halophilic Bacteria include sulfate reducers (reference 4 and references therein) and anaerobic phototrophs, gram-positive heterotrophs, and cyanobacteria (references 8 and 25 and references therein). Some isolates, like Flexistipes sinusarabici, represent separate lineages (24). In addition, the Halanaerobiaceae, which represent a monophyletic lineage within the Bacteria, are specifically adapted to their high-salt milieu (49, 55, 71). The halophilic Archaea known to date comprise aerobic halophiles of the family Halobacteriaceae and anaerobic methanogens of the family Methanosarcinaceae (16, 26, 47).
The goal of this research was to assess, for the first time, the bacterial diversity of the brine-seawater interface of the Kebrit Deep, Red Sea. In this initial survey, which is preliminary, a twofold approach was used. This includes (i) phylogenetic analysis of 16S rRNA gene sequences as indicators of prokaryotic diversity and (ii) isolation and cultivation of halophilic representatives to establish physiological and function potential within the ecosystem community.
MATERIALS AND METHODS
Sampling.
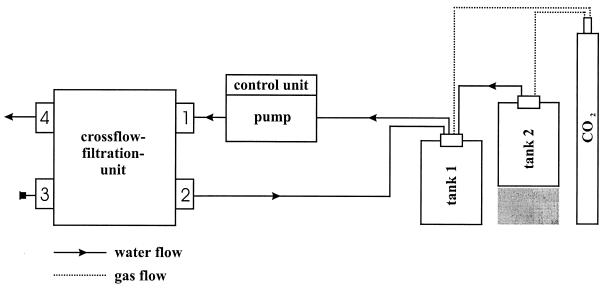
Brine from the brine-seawater interface of the Kebrit Deep, Red Sea (Fig. 1), was sampled during RV Sonne cruise SO 121 in 1997 using a rosette sampler equipped with 24 niskin bottles (10 liters) and a conductivity-temperature-depths (CTD) unit for monitoring salinity, temperature, transmission, and pressure (Sea-Bird Electronic, Bellevue, Wash.). The salinity of brine samples was measured with a hand refractometer (Atago, Tokyo, Japan). Microorganisms were concentrated by pumping anaerobic brine across a crossflow tangential filtration unit (Pellicon Kasettensystem; Millipore, Eschborn, Germany) (Fig. 2) under a CO2 protective atmosphere (cell concentration factor, 400-fold). Concentrated brine sample KT-2 was reduced with sodium dithionite (about 0.1 μM), and KT-3 was used without further treatment. In addition, sample KT-8 was taken from surface sediment of the Kebrit Deep by a chain-sack dredge (station no. 17034-2). The sample located near the brine-seawater interface consisted of oily ore rocks and brine (salinity, 15.6%). The samples were transported to the laboratory by air at ambient temperature and were stored at 4°C.
FIG. 2.
Schematic drawing of the anaerobic filtration system. The crossflow filtration unit was equipped with five Durapore membranes (pore size, 0.2 μm; filter surface, 0.46 m2) (Millipore). The cells were kept in suspension and were pumped under pressure over the filter surface. Before filtration, the complete system was flushed for 10 min with CO2 to remove oxygen. During the filtration procedure, a protective atmosphere of CO2 was maintained to prevent cellular damage due to oxygen sensitivity, to prevent precipitation of inorganic compounds, and to keep the pH of the sample constant. The concentrated brine was collected in tank 1; tank 2 was for storage. Brine flowed into the filtration unit at valve 1, concentrated cells exited at valve 2, and the filtrate outflow was at valve 4. Valve 3 is a closed valve.
Strains.
Halanaerobium praevalens DSM 2228, Halothermothrix orenii DSM 9562, and Halanaerobacter lacunaris DSM 6640 were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ), Braunschweig, Germany, and were cultivated by using the indicated DSMZ media.
Culture conditions.
Enrichment cultures from samples KT-2 and KT-3 (KT-2/3) and KT-8 were established in 28-ml serum tubes on board the RV Sonne. Brine used for enrichments was supplemented with various sterile organic and inorganic compounds (e.g., yeast extract, peptone, NaNO3, and Na2SO4) and chemically reduced by the addition of 1 ml of 50% (vol/vol) H2S per 10 ml of brine. In the laboratory, positive enrichments were transferred several times in supplemented brine and then grown in a synthetic medium whose composition was based on a chemical analysis of a Red Sea brine-seawater interface (brine interface medium [BI medium]) (D. Garbe-Schönberg [Institut für Geowissenschaften, Universität Kiel, Germany], personal communication). BI medium contained (per liter) 100 g of NaCl, 5.11 g of MgSO4 · 7H2O, 2.0 g of CaCl2 · 2H2O, 107 mg of MgCl2 · 6H2O, 19 mg of MnSO4 · H2O, 10 mg of SrCl2 · 6H2O, 2.7 mg of FeCl2 · 4H2O, 0.075 mg of NaNO3, 0.7 mg of ZnSO4 · 7H2O, 0.02 mg of Na2MoO4 · 2H2O, 100 mg of KH2PO4, and 1.0 g of NaHCO3. The pH was adjusted to 6.5 with HCl. The gas phase over the medium in incubation tubes was replaced with N2, and residual O2 was chemically reduced by the addition of 1 ml of 50% (vol/vol) H2S per 10 ml of medium. Growth was determined by direct cell counting with a Thoma chamber (depth, 0.02 mm).
Cell masses of Halanaerobium praevalens, Halothermothrix orenii, Halanaerobacter lacunaris, and the novel isolates were obtained by growth at 30°C (60°C for Halothermothrix orenii) with stirring (100 rpm) in an 80-liter enamel-protected fermentor (Bioengineering, Wald, Switzerland) pressurized with 300 kPa of N2 (N2-CO2 [80:20, vol/vol] for Halanaerobacter lacunaris and Halothermothrix orenii).
Light and electron microscopy.
Light microscopy, electron microscopy, and photography were carried out as described elsewhere (31).
Lipid analyses.
Freeze-dried cells (about 1 g) were extracted twice using chloroform-methanol (2:1, vol/vol) under reflux for 1 h. Fatty acid methyl esters (FAME) were prepared from a portion of the total lipid extract by a modification of the mild alkaline methanolysis procedure of White et al. (67), which involves heating at 37°C for 1 h and extraction with hexane-chloroform (4:1, vol/vol). FAME were separated by thin-layer chromatography on Silica Gel G plates (Merck, Darmstadt, Germany) using methylene chloride as the solvent. Gas chromatographic analysis was done using a Perkin-Elmer model Sigma 3B equipped with a flame ionization detector and a DB-5 megabore column (J&W Scientific, Folsom, Calif.), with methyl tricosanoate as an internal standard. Double-bond positions, cis-trans configurations, and confirmations of cyclopropane rings were determined with dimethyl disulfide adducts (69) using gas chromatography-mass spectrometry (35).
DNA extraction, PCR, and cloning.
Nucleic acids were extracted from 120 ml of the brine-seawater interface sample KT-2 with the IsoQuick Nucleic Acid Extraction kit (ORCA Research, Bothell, Wash.) according to the manufacturer's instructions, followed by RNase treatment for 30 min and precipitation of the nucleic acids with 1 volume of isopropanol. The nucleic acids of the halophilic isolates were extracted as described elsewhere (7). PCR amplifications of the rRNA genes between Escherichia coli positions (5) 9 and 1406 or 9 and 1512 (9bF, bacterial primer; 1406uR and 1512uR, universal primers) were carried out as described previously (21). PCR products of sample KT-2 were purified (Microcon 100; Amicon, Witten, Germany), and the 16S rRNA gene fragments were cloned into the pCR2.1 vector (Invitrogen, Leek, The Netherlands) according to the manufacturer's instructions. The resulting ligation products were used to transform E. coli TOP10F′ cells. The presence of inserts of the appropriate size in the transformants was identified by direct PCR screening; amplified ribosomal DNA (rDNA) restriction analysis was performed as described previously (21). Representative transformants were selected based on the fingerprinting pattern of the rRNA gene clones, and the corresponding plasmid DNAs were obtained using a QIAprep Spin Miniprep kit (Qiagen, Hilden, Germany).
Sequencing of rRNA genes.
16S rRNA gene sequences of the halophilic isolates and 16S rDNA clone sequences of sample KT-2 were sequenced with an ABI Prism 310 capillary DNA sequencer (PE Applied Biosystems, Foster City, Calif.), using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer), at the Institute for Genetics, University of Regensburg, Regensburg, Germany. The bacterial sequences were determined using a set of specific and universal primers (21).
Phylogenetic analyses.
For the analyses, an alignment of about 11,000 homologous full and partial primary sequences available in public databases (ARB project [41; W. Ludwig and O. Strunk, http://www.mikro.biologie.tu-muenchen.de/pub/ARB/documentation/arb.ps]) was used. The new bacterial 16S rRNA gene sequences (1,365 to 1,473 nucleotides) were fitted in the 16S rRNA tree by using the automated tools of the ARB software package (http://www.mikro.biologie.tu-muenchen.de/pub/ARB/documentation/arb.ps). Distance matrix (Jukes-Cantor correction), maximum-parsimony, and maximum-likelihood (fastDNAml) methods were applied as implemented in the ARB software package (42, 51). Insignificant branching points were shown by multifurication. Phylogenetic distances were determined using distance matrix analysis without applying a correction factor. Each sequence alignment was checked manually, and the sequences were analyzed with the CHECK_CHIMERA program of the Ribosomal Database Project (43) to detect the presence of possible chimeric artifacts.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences were submitted to EMBL and have been assigned accession numbers AJ309519 to AJ309527.
RESULTS
Transmission-temperature-depth profile of the brine-seawater interface.
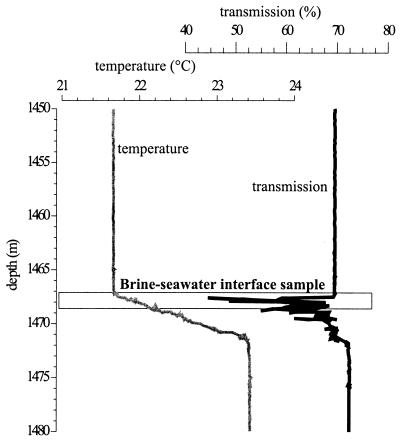
The data were collected during RV Sonne cruise SO 121. Figure 3 shows the transmission-versus-depth profile of the Kebrit Deep brine-seawater interface, including data for temperature. At a depth of 1,468 ± 3 m, 240 liters of brine was obtained (station no. 17029-8; 24°43.16′N, 36°16.42′E). The original sample temperature was 22°C, with a pH of 6.5. O2 was not detected, and the water smelled strongly of H2S. Over the 1-m length of the niskin bottles the salinity varied from 4.6% at the top to 9.6% at the bottom. Samples KT-2/3 were retrieved from the upper part of this brine-seawater interface, indicated by the beginning of an increase in temperature and salinity (4.6 to 9.6%) and an decrease in transmission (Fig. 3).
FIG. 3.
Transmission-temperature-depth profile of the brine-seawater interface of Kebrit Deep, Red Sea. The salinities at water depths of 1,465, 1,467, 1,469, 1,470, and 1,471 m (corrected depths) are 4.0, 4.2, 17.2, 24.5, and 26.0% (wt/vol) NaCl, respectively.
Enrichment and isolation.
In order to isolate representative microorganisms from the Kebrit brine, enrichment cultures were established in 28-ml serum tubes on board ship by adding different sterile organic and inorganic nutrients directly to the brine. Two successful enrichments designated KT-2/3-3 (from the brine-seawater interface, 1,468 ± 3 m) and KT-8-13 (from a brine surface sediment near the brine-seawater interface) were grown anaerobically in original brine supplemented with 0.1% (wt/vol) thiosulfate and 0.01% (wt/vol) of a mixture of equal parts of yeast extract, meat extract, peptone, and brain heart infusion (C-Org). In the laboratory, the enrichment cultures were transferred several times in the original brine and then into synthetic BI medium. From each enrichment culture, a single cell, which was optically trapped by using a strongly focused infrared laser beam (“optical tweezers”), was separated under visual control from the mixed culture and was grown as a pure culture (selected cell cultivation [2, 30, 32]). The pure cultures KT-2/3-3 and KT-8-13 (for simplicity, designated the enrichment cultures) were chosen as representatives for the experiments described below. Unless indicated otherwise, the novel isolates were cultivated in BI medium supplemented with 0.01% C-Org. Several further enrichment cultures resulted in single clones after selected cell cultivation, but they showed 16S rRNA gene sequences identical to that of either KT-2/3-3 or KT-8-13 and therefore were not further characterized (data not shown).
Morphology.
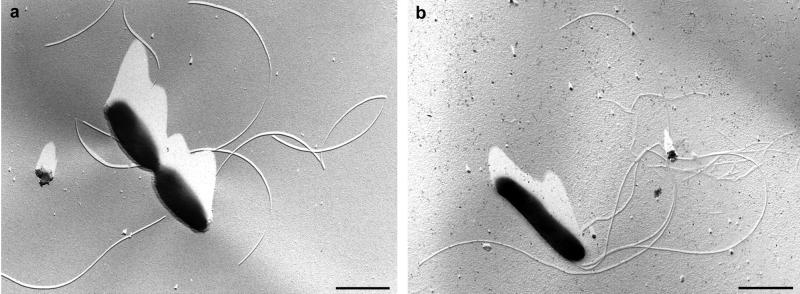
Cells of isolates KT-2/3-3 and KT-8-13 were gram-negative, motile rods with rounded ends; the average cell length was 1 to 2 μm, and the average cell width was 0.3 to 0.4 μm. Addition of glucose (0.1%, wt/vol) resulted in an increase of the cell length up to 6.5 μm (for KT-8-13). During growth, cells of the two isolates appeared singly or in pairs. No evidence of spore formation was observed. In transmission electron micrographs, the new isolates exhibited monopolar, polytrichous flagellation with up to three flagella per cell (Fig. 4).
FIG. 4.
Transmission electron micrographs of a dividing cell of Halanaerobium sp. strain KT-2/3-3 showing flagella (a) and of a single flagellated cell of Halanaerobium sp. strain KT-8-13 (b). The cells were air dried and platinum shadowed. Bars, 1 μm.
Growth and physiological characterization.
Isolates KT-2/3-3 and KT-8-13 grew chemoorganoheterotrophically under strictly anaerobic culture conditions. Both isolates were obligate halophiles, with an optimal NaCl requirement of between 10 and 20% (wt/vol). They grew at NaCl concentrations of between 5 and 34% at pH 6.5. In the presence of H2 (5%, vol/vol), thiosulfate or S0 (but not sulfate) was reduced to H2S without stimulating the growth rate.
Isolate KT-8-13 grew at temperatures of between 18 and 48°C, with an optimum of between 30 and 45°C. KT-8-13 was able to grow heterotrophically on C-Org (0.001 and 0.1%) and brain heart infusion (0.01%). No growth was observed on glucose (0.1%), fructose (0.1%), saccharose (0.1%), maltose (0.1%), lactose (0.1%), xylose (0.1%), betaine (0.1%), cholesterol (0.1%), acetate (0.01%), yeast extract (0.01%), peptone (0.01%) (Merck), Trypticase-peptone (0.01%) (BBL), meat extract (0.01%), or Casamino Acids (0.01%). H2S concentrations of 0.5 to 50% (vol/vol) were tolerated by isolate KT-8-13.
Cellular fatty acid composition.
The cellular fatty acid compositions of isolates KT-2/3-3 and KT-8-13 in comparison with those of representatives of three different halophilic genera are given in Table 1. The novel bacterial halophiles Halanaerobium praevalens, Halothermothrix orenii, and Halanaerobacter lacunaris were cultured under optimal growth conditions (see Materials and Methods) and were harvested in the exponential growth phase. The FAME analysis of the Kebrit Halanaerobium isolates, KT-2/3-3 and KT-8-13, showed large amounts of 16:0 and 16:1 fatty acids and minor amounts of 18:0 and 18:1 fatty acids, consistent with the data for Halanaerobium praevalens (Table 1) (71). The two Kebrit isolates contained a relatively large proportion of monounsaturated 16:1 with unusual bond positions at Δ7 and Δ11, closely matching the 16:1 isomer composition of Halanaerobium praevalens. Halothermothrix orenii contained little unsaturated FAME, while Halanaerobacter lacunaris had 16:1 with the commonly encountered Δ9 position but an unusual 18:1 for a bacterium, with the bond at Δ9. Hopanoids were not detected in the organisms analyzed (data not shown).
TABLE 1.
Comparison of the cellular fatty acid compositions of isolates KT-2/3-3 and KT-8-13 with those of Halanaerobium praevalens, Halothermothrix orenii, and Halanaerobacter lacunaris
| Fatty acida | % in:
|
||||
|---|---|---|---|---|---|
| KT-2/3-3 | KT-8-13 | Halanaerobium praevalens | Halothermothrix orenii | Halanaerobacter lacunaris | |
| 14 | 0.8 | 1.2 | 9.6 | 3.5 | 3.7 |
| 14:1, Δ7 | 0.2 | 0.2 | 2.9 | ND | 0.5 |
| 14:1, Δ9 | 0.1 | 0.0 | 0.9 | ND | ND |
| i15 | 0.1 | <0.1 | 0.1 | 0.1 | 0.2 |
| a15 | 0.1 | <0.1 | <0.1 | 0.1 | 0.2 |
| 15 | 0.2 | 0.4 | 0.3 | 0.2 | 0.6 |
| 15:1, Δ7 | 0.1 | 0.1 | 0.2 | ND | ND |
| 15:1, Δ9 | 0.1 | 0.1 | 0.4 | ND | ND |
| i16 | 0.2 | 0.1 | <0.1 | 0.4 | 0.2 |
| 16 | 17.3 | 24.8 | 17.9 | 54.9 | 20.9 |
| 16:1, Δ7cis | 7.3 | 10.1 | 12.3 | ND | 1.2 |
| 16:1, Δ9cis | 34.2 | 35.0 | 28.3 | 1.4 | 34.2 |
| 16:1, Δ9trans | 8.5 | 2.2 | 4.8 | 0.5 | 4.9 |
| 16:1, Δ11cis | 20.4 | 13.3 | 17.5 | ND | 1.1 |
| i17 | 0.2 | <0.1 | 0.1 | 0.2 | 0.1 |
| a17 | 0.2 | 0.2 | 0.1 | 0.3 | ND |
| 17 | 0.2 | 0.2 | <0.1 | 0.6 | 0.5 |
| cy17 | 1.7 | 1.6 | 0.2 | 0.2 | 0.6 |
| 17:1 | 0.2 | 0.6 | 0.1 | ND | 2.6 |
| i18 | NDb | <0.1 | 0.1 | 0.1 | ND |
| a18 | ND | ND | <0.1 | <0.1 | ND |
| 18 | 2.5 | 1.9 | 2.7 | 29.4 | 5.3 |
| 18:1, Δ9cis | 0.5 | 1.8 | 0.9 | 3.3 | 10.3 |
| 18:1, Δ9trans | 0.5 | 0.3 | 0.1 | 2.0 | 2.2 |
| 18:1, Δ11cis | 2.0 | 5.0 | 0.4 | 1.0 | 1.0 |
| 18:1, Δ11trans | 0.6 | ND | ND | 0.1 | 0.4 |
| 18:1, Δ13cis | 1.4 | 0.4 | ND | ND | ND |
| i19 | ND | ND | ND | 0.3 | 1.4 |
| a19 | 0.5 | 0.4 | 0.1 | 1.2 | 7.5 |
| 19 | ND | ND | <0.1 | 0.2 | 0.4 |
Fatty acids are designated by total number of carbon atoms:number of double bonds. The number after Δ indicates the position of the double bond relative to the carboxylic (Δ) end of the molecule, with cis and trans geometry indicated. cy, cyclopropyl ring; i and a, iso- and anteiso-branched fatty acids, respectively. The concentrations of total fatty acids of KT-2/3-3, KT-8-13, Halanaerobium praevalens, Halothermothrix orenii, and Halanaerobacter lacunaris were 160.9, 228.2, 1,017.4, 187.3, and 45.6 μg g (dry weight)-1, respectively.
ND, none detected.
16S rRNA phylogeny. (i) Halophilic isolates.
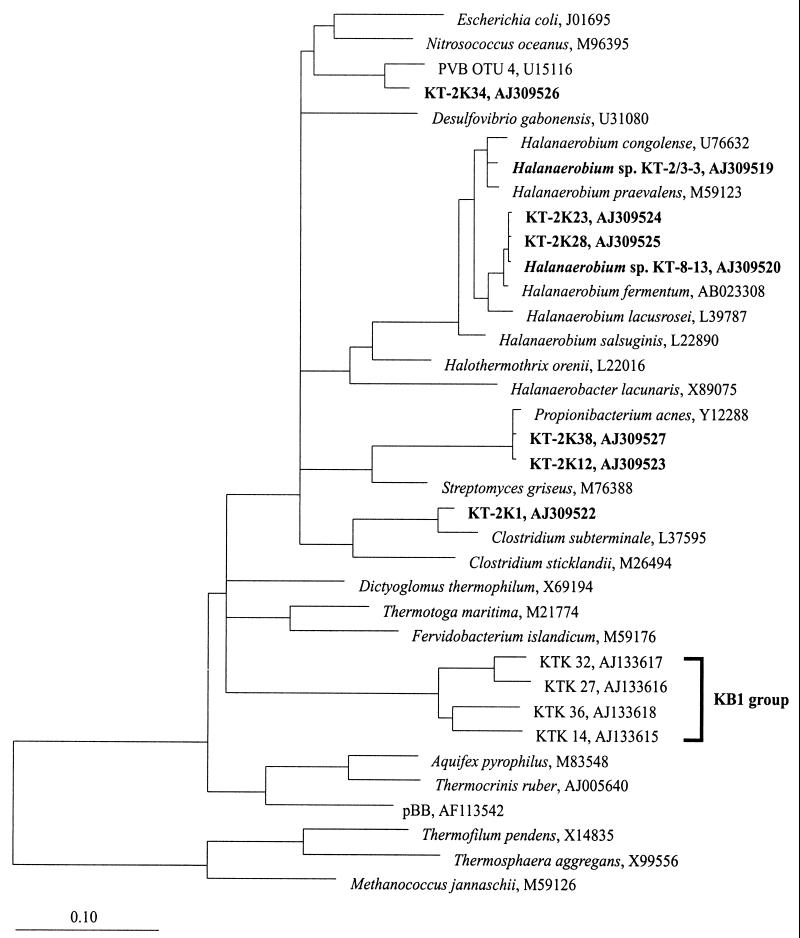
A comparative analysis of the 16S rRNA gene sequences revealed that isolates KT-2/3-3 and KT-8-13 belong to the Halanaerobiaceae within the monophyletic lineage of the Halanaerobiales (49, 55). Further sequence alignments and phylogenetic analyses showed the taxonomic and phylogenetic positions of the new isolates to be among those of the members of the genus Halanaerobium (Fig. 5). The sequence similarity of isolates KT-2/3-3 and KT-8-13 is 97.1%. The sequence similarities of KT-2/3-3 and KT-8-13 to other Halanaerobium species ranged from 95.7 to 99% and from 95.5 to 99.4%, respectively.
FIG. 5.
16S rRNA gene-based phylogenetic tree of the bacterial domain, including the 16S rDNA sequences from brine-seawater interface sample KT-2 and the 16S rRNA gene sequences of the new halophilic isolates from Kebrit Deep, Red Sea. The KB1 group marks a cluster of closely related environmental sequences, which were obtained from a sediment sample (depth, 1,515 m) of the Kebrit Deep (21). The topology of the tree is based on results of a maximum-parsimony analysis. Reference sequences were chosen to represent the broadest diversity of Bacteria. Only sequence positions that share 50% or more residues identical to the 16S rRNA sequences of a corresponding group were included for tree reconstruction. Accession numbers for the sequences are indicated. The scale bar represents 0.10 fixed mutation per nucleotide position.
(ii) Brine-seawater interface.
Specific (9bF) and universal (1406uR) 16S rRNA primers were used to amplify bacterial sequences from bulk DNA derived from the brine-seawater interface sample KT-2 (salinity, 4.6 to 9.6% NaCl). About 40 clones were obtained from the extracted nucleic acids. After cloning, the 16S rRNA gene fragments were further characterized by restriction endonuclease digestion. Based on a comparison of the restriction patterns on agarose gels, eight different bacterial groups were identified. From a representative of each restriction pattern group, the 16S rRNA gene sequence was determined and aligned with 16S rRNA sequences derived from the ARB database. Clone sequences KT-2K20 and KT-2K29 (representing 26% of the clones) were chimeras: KT-2K20 of KT-2K1 and KT-2K23/KT-2K28, and KT-2K29 of KT-2K1 and KT-2K38/KT-2K12.
The analysis of the clone sequences KT-2K1, KT-2K12, KT-2K23, KT-2K28, KT-2K34, and KT-2K38 showed a high phylogenetic diversity within the bacterial domain. The phylogenetic positions of the derived clone sequences were supported by the different tree reconstruction methods (see Materials and Methods). KT-2K23 and KT-2K28 (together, 41% of the derived clones) were affiliated with cultivated members of the genus Halanaerobium (Fig. 5). They showed highest sequence similarity to Halanaerobium sp. strain KT-8-13 (99.9%), Halanaerobium fermentans (99.3%), and Halanaerobium sp. strain KT-2/3-3 (97%). Sequence clones KT-2K1, KT-2K12, and KT-2K38 (representing 26% of the clones) showed the closest relationship to Clostridium subterminale or Propionibacterium acnes, whereas KT-2K34 (7% of the clones) is related to sequence clone PVB OTU 4 (U15116) within the γ-proteobacteria. The G+C contents of the rRNA gene sequences range from 51 to 57%.
DISCUSSION
In brine-filled Red Sea deeps, the brine-seawater interface represents a unique microbial ecosystem mainly determined by a steep salt gradient (4 to 26% NaCl) within only a few meters. The combination of salt, oxygen, and pH gradients may be responsible for organisms specifically adapted to this environment. To date, nothing is known about the morphological and physiological features of these organisms. The presence of microorganisms in hypersaline brines (e.g., the Gulf of Mexico and the Mediterranean Sea) was suggested by biochemical investigations which included measurements of ATP or CH4, lipid analysis, and epifluorescence microscopy (17, 20, 40, 54, 68). The novel halophilic isolates from the Kebrit Deep (KT-2/3-3 and KT-8-13) are the first organisms to have been cultivated and isolated from a brine-seawater interface or the deeper anaerobic, brine pool.
Based on 16S rRNA gene sequence comparisons, these Kebrit isolates were identified as members of the Halanaerobiales, a group of anaerobic, halophilic, fermentative bacteria. The Halanaerobiales represent a separate lineage in the bacterial domain (49, 55). Within this order, the isolates KT-2/3-3 and KT-8-13 showed the closest relationship to cultivated members of the genus Halanaerobium and most likely comprise new species in the genus (3, 9, 38, 49, 55, 56, 71). Prior to our investigations, members of the genus Halanaerobium have been shown to occur only in sediments from salt lakes and offshore oil fields; this study thus increases our knowledge of the ecological distribution of these organisms (3, 9, 55, 56). The results of the FAME analysis for the new Halanaerobium species are consistent with the 16S rRNA data in that the new species exhibit fatty acid patterns similar to those of Halanaerobium praevalens (71). The FAME are dominated by 16:0 and 16:1 and minor amounts of 18:0 and 18:1. In contrast to Halothermothrix orenii, the new isolates show the presence of large amounts of unsaturated fatty acids (Table 1) (10). The presence of relatively large amounts of Δ7 and Δ11 16:1 is very unusual and may serve as a biomarker for Halanaerobium. Small amounts of Δ11 16:1 are also present in type I methanotrophs; however, these bacteria also contain Δ8, Δ9, and Δ10 16:1 (33, 34). Like most other Halanaerobium representatives, Halanaerobium sp. strains KT-2/3-3 and KT-8-13 grew over a wide NaCl range (5 to 34% NaCl). This physiological flexibility allows the organisms to grow within the salt gradient of the brine-seawater interface and in the highly saline lower brine pool. The brine-seawater interface also exhibits a density gradient which acts as an in situ particle trap for organic material (Fig. 3) (29, 40, 57, 58, 66), providing an appropriate environmental niche for heterotrophic bacteria. Indeed, these chemoorganotrophic halophiles may contribute significantly to the anaerobic degradation of suspended organic material enriched at the brine-seawater interface of the Kebrit Deep. However, the occurrence of Halanaerobium representatives is not restricted to the Kebrit Deep. Recently, a new member of the genus Halanaerobium (isolate S5L4, AJ309521) was obtained from the brine-seawater interface (salinity, 24.2%) of the Shaban Deep, Red Sea (W. Eder and R. Huber, unpublished results).
For phylogenetic analysis, the brine sample was concentrated anaerobically on board ship using a crossflow tangential filtration unit operated under a protective CO2 atmosphere (Fig. 2). Two hundred forty liters of water of the brine-seawater interface was concentrated about 400-fold. Anaerobic sampling of the brine is essential to circumvent the precipitation of reduced inorganic compounds such as Fe(II) in the presence of O2 or the toxic effect of O2 on strict anaerobes like methanogens (60, 70). From the concentrated brine sample KT-2, the nucleic acids were extracted, and the 16S rRNA gene sequences were PCR amplified and cloned. The 16S rRNA gene fragments were further characterized by amplified rDNA restriction analysis, and different restriction pattern groups were identified. The phylogenetic analysis of a representative of each restriction group showed that the majority of the sequences had high sequence similarity to cultivated microorganisms within the bacterial domain. From the same bulk DNA, archaeal PCR products were obtained, but these were not further investigated for this study (Eder and Huber, unpublished).
Using different tree reconstruction methods (see Materials and Methods), the majority of the bacterial clone sequences represented by KT-2K23 and KT-2K28 showed the closest relationship with members of the genus Halanaerobium (55) and exhibited the highest sequence similarity to the isolated Halanaerobium sp. strain KT-8-13. Although KT-8-13 was enriched from the sedimentary portion of the brine pool, this sequence was not detected in a previous in situ analysis of the brine-sediment interface (21). That phylogenetic analysis identified a deep-branching KB1 sequence group (Fig. 5) (21). This KB1 group could not be retrieved from the brine-seawater interface sample, suggesting that representatives of the KB1 group are specifically adapted to the higher salinities within the lower brine body (Fig. 3) and the sediments (the salinity of the pore water is up to 26%), while KT-8-13, like KT-2/3-3, is more naturally adapted to the interface environment.
ACKNOWLEDGMENTS
We are grateful to Karl O. Stetter for stimulating and critical discussions. For helpful discussions we thank Reiner Botz, Roger Summons, and Gerald Maroske. Furthermore, we thank Reinhard Rachel and Peter Hummel for electron microscopy and Thomas Hader, Konrad Eichinger, and Marcus Koch for technical assistance. We are grateful for the valuable help of the crew on board the RV Sonne (SO 121 cruise) and for the valuable support of cruise leader Peter Stoffers.
This work was financially supported by the BMBF (grant no. 03G0121B to Karl O. Stetter and grant no. 03G0121A to Peter Stoffers) and by the Fonds der Chemischen Industrie (to Karl O. Stetter). The work of L.L.J. was supported by a grant from NASA's Exobiology program.
REFERENCES
- 1.Bäcker H, Schoell M. New deeps with brines and metalliferous sediments in the Red Sea. Nat Phys Sci. 1972;240:153–158. [Google Scholar]
- 2.Beck P, Huber R. Detection of cell viability in cultures of hyperthermophiles. FEMS Microbiol Lett. 1997;147:11–14. [Google Scholar]
- 3.Bhupathiraju V K, McInerney M J, Woese C R, Tanner R S. Haloanaerobium kushneri sp. nov., an obligately halophilic, anaerobic bacterium from an oil brine. Int J Syst Bacteriol. 1999;49:953–960. doi: 10.1099/00207713-49-3-953. [DOI] [PubMed] [Google Scholar]
- 4.Brandt K K, Patel B K C, Ingvorsen K. Desulfocella halophila gen. nov., sp. nov., a halophilic, fatty-acid-oxidizing, sulfate-reducing bacterium isolated from sediments of the Great Salt Lake. Int J Syst Bacteriol. 1999;49:193–200. doi: 10.1099/00207713-49-1-193. [DOI] [PubMed] [Google Scholar]
- 5.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 6.Bruneau L, Jerlov N G, Koczy F F. Physical and chemical methods (+ Appendix), p. 99–112. Reports of the Swedish deep-sea expedition, vol. III. Physics and chemistry. 1953. , no. 4. [Google Scholar]
- 7.Burggraf S, Huber H, Stetter K O. Reclassification of the crenarchaeal orders and families in accordance with 16S ribosomal RNA sequence data. Int J Syst Bacteriol. 1997;47:657–660. doi: 10.1099/00207713-47-3-657. [DOI] [PubMed] [Google Scholar]
- 8.Caumette P. Ecology and physiology of phototrophic bacteria and sulfate-reducing bacteria in marine salterns. Experientia. 1993;49:473–481. [Google Scholar]
- 9.Cayol J-L, Ollivier B, Patel B K C, Ageron E, Grimont P A D, Prensier G, Garcia J L. Haloanaerobium lacusroseus sp. nov., an extremely halophilic fermentative bacterium from the sediments of a hypersaline lake. Int J Syst Bacteriol. 1995;45:790–797. doi: 10.1099/00207713-45-4-790. [DOI] [PubMed] [Google Scholar]
- 10.Cayol J-L, Ollivier B, Patel B K C, Prensier G, Guezennec J, Garcia J L. Isolation and characterization of Halothermothrix orenii gen. nov., sp. nov., a halophilic, thermophilic, fermentative, strictly anaerobic bacterium. Int J Syst Bacteriol. 1994;44:534–540. doi: 10.1099/00207713-44-3-534. [DOI] [PubMed] [Google Scholar]
- 11.Charnock H. Anomalous bottom water in the Red Sea. Nature. 1964;203:591. [Google Scholar]
- 12.Craig H. Geochemistry and origin of the Red Sea brines. In: Degens E T, Ross D A, editors. Hot brines and recent heavy metal deposits in the Red Sea. New York, N.Y: Springer-Verlag; 1969. pp. 208–242. [Google Scholar]
- 13.Cruise report Menor 1. Preussag Meerestechnik, confidential report PREE-CR-03-4 (1403 AH). Jeddah, Saudi-Arabia: Ministry of Petroleum and Mineral Resources, Deputy Ministry for Mineral Resources; 1983. [Google Scholar]
- 14.Cruise report Menor 2. Preussag Meerestechnik, confidential report PREE-CR-04-10 and PREE-CR-05-4 (1404AH und 1405AH). Jeddah, Saudi-Arabia: Ministry of Petroleum and Mineral Resources, Deputy Ministry for Mineral Resources; 1984. [Google Scholar]
- 15.DasSarma S, Arora P. Embryonic encyclopedia of life sciences. [Online.] Nature Publishing Group, London, England. http://www.els.net. 1999. Halophiles. [Google Scholar]
- 16.Davidova I A, Harmsen H J M, Stams A J M, Belyaev S S, Zehnder A J B. Taxonomic description of Methanococcoides euhalobius and its transfer to the Methanohalophilus genus. Antonie Leeuwenhoek. 1997;71:313–318. doi: 10.1023/a:1000103618451. [DOI] [PubMed] [Google Scholar]
- 17.De Domenico M, De Domenico E. Bannock Basin: first approach to water microbiology. In: Cita M B, Camerlenghi A, Corselli C, editors. Anoxic basins of the eastern Mediterranean: results of the third conference, Bergamo (Italy), December 14–16, 1988. Mailand, Italy: Ricerca Scientifica Educazione Permanente; 1989. pp. 100–102. [Google Scholar]
- 18.Degens E, Ross D A. Hot brines and recent heavy metal deposits in the Red Sea. New York, N.Y: Springer-Verlag; 1969. [Google Scholar]
- 19.de Lange G J, ten Haven H L. Recent sapropel formation in the eastern Mediterranean. Nature. 1983;305:797–798. [Google Scholar]
- 20.Dickins H D, van Vleet E S. Archaebacterial activity in the Orca Basin determined by the isolation of characteristic isopranyl ether-linked lipids. Deep-Sea Res. 1992;39:521–536. [Google Scholar]
- 21.Eder W, Ludwig W, Huber R. Novel 16S rRNA gene sequences retrieved from highly saline brine sediments of Kebrit Deep, Red Sea. Arch Microbiol. 1999;172:213–218. doi: 10.1007/s002030050762. [DOI] [PubMed] [Google Scholar]
- 22.Erba E. Deep mid-water bacterial mats from anoxic basins of the Eastern Mediterranean. Mar Geol. 1991;100:83–101. [Google Scholar]
- 23.Faber E, Botz R, Poggenburg J, Schmidt M, Stoffers P, Hartmann M. Methane in Red Sea brines. Org Geochem. 1998;29:363–379. [Google Scholar]
- 24.Fiala G, Woese C R, Langworthy T A, Stetter K O. Flexistipes sinusarabici, a novel genus and species of eubacteria occurring in the Atlantis II Deep brines of the Red Sea. Arch Microbiol. 1990;154:120–126. [Google Scholar]
- 25.Grant W D, Gemmell R T, McGenity T J. Halophiles. In: Horikoshi K, Grant W D, editors. Extremophiles: microbial life in extreme environments. New York, N.Y: Wiley-Liss; 1998. pp. 93–132. [Google Scholar]
- 26.Grant, W. D., M. Kamekura, T. J. McGenity, and A. Ventosa. Family Halobacteriaceae. In: Garrity G. (ed.), Bergey's manual of systematic bacteriology, vol. 1. The Archaea, cyanobacteria, phototrophs and deeply branching genera, in press. Springer Verlag, New York, N.Y.
- 27.Hartmann M, Scholten J C, Stoffers P, Wehner F. Hydrographic structure of brine-filled deeps in the Red Sea—new results from the Shaban, Kebrit, Atlantis II, and Discovery Deep. Mar Geol. 1998;144:311–330. [Google Scholar]
- 28.Hemleben C, Roether W, Stoffers P. Meteor-Berichte 96–4; Östliches Mittelmeer, Rotes Meer, Arabisches Meer, Cruise No. 31. Meteor-Berichte. Hamburg, Germany: Institut für Meereskunde der Universität Hamburg; 1996. [Google Scholar]
- 29.Henneke E, de Lange G J. The distribution of DOC and POC in the water column and brines of the Tyro and Bannock Basins. Mar Chem. 1990;31:113–122. [Google Scholar]
- 30.Huber R, Burggraf S, Mayer T, Barns S M, Rossnagel P, Stetter K O. Isolation of a hyperthermophilic archaeum predicted by in situ RNA analysis. Nature. 1995;376:57–58. doi: 10.1038/376057a0. [DOI] [PubMed] [Google Scholar]
- 31.Huber R, Eder W, Heldwein S, Wanner G, Huber H, Rachel R, Stetter K O. Thermocrinis ruber gen. nov., sp. nov., a pink-filament-forming hyperthermophilic bacterium isolated from Yellowstone National Park. Appl Environ Microbiol. 1998;64:3576–3583. doi: 10.1128/aem.64.10.3576-3583.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber R, Huber H, Stetter K O. Towards the ecology of hyperthermophiles: biotopes, new isolation strategies and novel metabolic properties. FEMS Microbiol Rev. 2000;24:615–623. doi: 10.1111/j.1574-6976.2000.tb00562.x. [DOI] [PubMed] [Google Scholar]
- 33.Jahnke L L, Nichols P D. Methyl sterol and cyclopropane fatty acid composition of Methylococcus capsulatus grown at low oxygen tensions. J Bacteriol. 1986;167:238–242. doi: 10.1128/jb.167.1.238-242.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jahnke L L. The effects of growth temperature on the methyl sterol and phospholipid fatty acid composition of Methylococcus capsulatus (Bath) FEMS Microbiol Lett. 1992;93:209–212. doi: 10.1111/j.1574-6968.1992.tb05099.x. [DOI] [PubMed] [Google Scholar]
- 35.Jahnke L L, Summons R E, Dowling L M, Zahiralis K D. Identification of methanotrophic lipid biomarkers in cold-seep mussel gills: chemical and isotopic analysis. Appl Environ Microbiol. 1995;61:576–582. doi: 10.1128/aem.61.2.576-582.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Javor B. Hypersaline environments: microbiology and biochemistry. Berlin, Germany: Springer-Verlag; 1989. [Google Scholar]
- 37.Jongsma D, Fortuin A R, Huson W, Troelstra S R, Klaver G T, Peters J M, van Harten D, de Lange G J, ten Haven L. Discovery of an anoxic basin within the Strabo Trench, eastern Mediterranean. Nature. 1983;305:795–797. [Google Scholar]
- 38.Kobayashi T, Kimura B, Fujii T. Haloanaerobium fermentans sp. nov., a strictly anaerobic, fermentative halophile isolated from fermented puffer fish ovaries. Int J Syst Evol Bacteriol. 2000;50:1621–1627. doi: 10.1099/00207713-50-4-1621. [DOI] [PubMed] [Google Scholar]
- 39.La Ferla R, Crisafi E. Preliminary study on vertical distribution of microorganisms in the Bannock Basin waters (Eastern Mediterranean Sea) Mar Ecol Prog Ser. 1991;75:309–311. [Google Scholar]
- 40.LaRock P A, Lauer R D, Schwarz J R, Watanabe K K, Wiesenburg D A. Microbial biomass and activity distribution in an anoxic, hypersaline basin. Appl Environ Microbiol. 1979;37:466–470. doi: 10.1128/aem.37.3.466-470.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ludwig W. Sequence databases (3.3.5) In: Akkermans A D L, Van Elsas J D, De Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–22. [Google Scholar]
- 42.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K-H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 43.Maidak B L, Cole J R, Lilburn T G, Parker C T J, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MEDRIFF Consortium. Three brine lakes discovered in the seafloor of the eastern Mediterranean. EOS. 1995;76:313–318. [Google Scholar]
- 45.Michaelis W, Jenisch A, Richnow H H. Hydrothermal petroleum generation in Red Sea sediments from the Kebrit and Shaban Deeps. Appl Geochem. 1990;5:103–114. [Google Scholar]
- 46.Miller A R. Highest salinity in the world ocean? Nature. 1964;203:590–591. [Google Scholar]
- 47.Ollivier B, Caumette P, Garcia J-L, Mah R A. Anaerobic bacteria from hypersaline environments. Microbiol Rev. 1994;58:27–38. doi: 10.1128/mr.58.1.27-38.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olsen G J, Lane D J, Giovannoni S J, Pace N R, Stahl D A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 49.Oren A. Change of the names Haloanaerobiales, Haloanaerobiaceae and Haloanaerobium to Halanaerobiales, Halanaerobiaceae and Halanaerobium, respectively, and further nomenclatural changes within the order Halanaerobiales. Int J Syst Evol Bacteriol. 2000;50:2229–2230. doi: 10.1099/00207713-50-6-2229. [DOI] [PubMed] [Google Scholar]
- 50.Pace N R, Stahl D A, Lane D J, Olsen G J. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 51.Page R D M, Holmes E C. Molecular evolution: a phylogenetic approach. Oxford, England: Blackwell Science; 1998. [Google Scholar]
- 52.Pautot G. Les fosses de la Mer Rouge: approche géomorphologique d'un stade initial d'ouverture océanique réalisée à l'aide du Seabeam. Oceanol Acta. 1983;6:235–244. [Google Scholar]
- 53.Pautot G, Guennoc P, Coutelle A, Lyberis N. Discovery of a large brine deep in the northern Red Sea. Nature. 1984;310:133–136. [Google Scholar]
- 54.Pease T K, van Vleet E S, Barre J S. Diphytanyl glycerol ether distributions in sediments of the Orca Basin. Geochim Cosmochim Acta. 1992;56:3469–3479. doi: 10.1016/0016-7037(92)90391-u. [DOI] [PubMed] [Google Scholar]
- 55.Rainey F A, Zhilina T N, Boulygina E S, Stackebrandt E, Tourova T P, Zavarzin G A. The taxonomic status of the fermentative halophilic anaerobic bacteria: description of Haloanaerobiales ord. nov., Halobacteroidaceae fam. nov., Orenia gen. nov. and further taxonomic rearrangements at the genus and species level. Anaerobe. 1995;1:185–199. doi: 10.1006/anae.1995.1018. [DOI] [PubMed] [Google Scholar]
- 56.Ravot G, Magot M, Ollivier B, Patel B K C, Ageron E, Grimont P A D, Thomas P, Garcia J L. Haloanaerobium congolense sp. nov., an anaerobic, moderately halophilic, thiosulfate- and sulfur-reducing bacterium from an African oil field. FEMS Microbiol Lett. 1997;147:81–88. doi: 10.1111/j.1574-6968.1997.tb10224.x. [DOI] [PubMed] [Google Scholar]
- 57.Ryan W B F, Thorndike E M, Ewing M, Ross D A. Suspended matter in the Red Sea brines and its detection by light scattering. In: Degens E, Ross D A, editors. Hot brines and recent heavy metal deposits in the Red Sea. New York, N.Y: Springer-Verlag; 1969. pp. 153–157. [Google Scholar]
- 58.Sackett W M, Brooks J M, Bernard B B, Schwab C R, Chung H, Parker R A. A carbon inventory for Orca Basin brines and sediments. Earth Planet Sci Lett. 1979;44:73–81. [Google Scholar]
- 59.Schoell M. Valdivia VA 01/03, Hydrographie II und III. Hannover, Germany: Bundesanstalt für Bodenforschung; 1974. [Google Scholar]
- 60.Scholten J C, Stoffers P, Garbe-Schönberg D, Moammar M. Hydrothermal mineralization in the Red Sea. In: Cronan D S, editor. Handbook of marine mineral deposits. Boca Raton, Fla: CRC Press; 1999. pp. 369–395. [Google Scholar]
- 61.Scientific staff of Cruise Bannock 1984–12. Gypsum precipitation from cold brines in an anoxic basin in the eastern Mediterranean. Nature. 1985;314:152–154. [Google Scholar]
- 62.Shanks W C, Bischoff J L. Ore transport and deposition in the Red Sea geothermal system: a geochemical model. Geochim Cosmochim Acta. 1977;41:1507–1519. [Google Scholar]
- 63.Shokes R F, Trabant P K, Presley B, Reid D F. Anoxic, hypersaline basin in the northern Gulf of Mexico. Nature. 1977;196:1443–1446. doi: 10.1126/science.196.4297.1443. [DOI] [PubMed] [Google Scholar]
- 64.Stoffers P, Moammar M, Abu-Ouf M, Ackermand D, Alassif O, Al-Hazim Y, Boldt S, Botz R, Eder W, El-Garafi A, El-Mamoney M, Fleitmann D, Garbe-Schönberg D, Geiselhart S, Goedecke D, Hartmann M, Klauke S, Moussa K, Mülhan N, Mühlstrasser T, Poggenburg J, Rehder W, Schmidt M, Schmitt M, Schoeps D, Scholten J, Shbalaby M, Wismann A, Yohannes E. Cruise report SONNE 121, Red Sea. Hydrography, hydrothermalism and paleoceanography in the Red Sea. No. 88. Kiel, Germany: Geologisch-Paläontologisches Institut der Universität Kiel; 1998. [Google Scholar]
- 65.Swallow J C, Crease J. Hot salty water at the bottom of the Red Sea. Nature. 1965;205:165–166. [Google Scholar]
- 66.Trefry J H, Presley B J, Keeney-Kennicutt W L, Trocine R P. Distribution and chemistry of manganese, iron and suspended particles in Orca Basin brine. Geomar Lett. 1984;3:125–130. [Google Scholar]
- 67.White D C, Davis W M, Nickels J S, King J D, Bobbie R J. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia. 1979;40:51–62. doi: 10.1007/BF00388810. [DOI] [PubMed] [Google Scholar]
- 68.Wiesenburg D A, Brooks J M, Bernard B B. Biogenic hydrocarbon gases and sulfate reduction in the Orca basin brine. Geochim Cosmochim Acta. 1985;49:2069–2080. [Google Scholar]
- 69.Yamamoto K, Shibahara A, Nakayama T, Kajimoto G. Determination of double-bond positions in methylene-interrupted dienoic fatty acids by GC-MS as their dimethyl disulfide adducts. Chem Phys Lipids. 1991;60:39–50. [Google Scholar]
- 70.Zeikus J G. The biology of methanogenic bacteria. Bacteriol Rev. 1977;41:514–541. doi: 10.1128/br.41.2.514-541.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zeikus J G, Hegge P W, Thompson T E, Phelps T J, Langworthy T A. Isolation and description of Haloanaerobium praevalens gen. nov. and sp. nov., an obligately anaerobic halophile common to Great Salt Lake sediments. Curr Microbiol. 1983;9:225–234. [Google Scholar]
- 72.Zierenberg R A, Shanks W C I. Isotopic constraints on the origin of the Atlantis II, Suakin and Valdivia brines, Red Sea. Geochim Cosmochim Acta. 1986;50:2205–2214. [Google Scholar]