Figure 3.
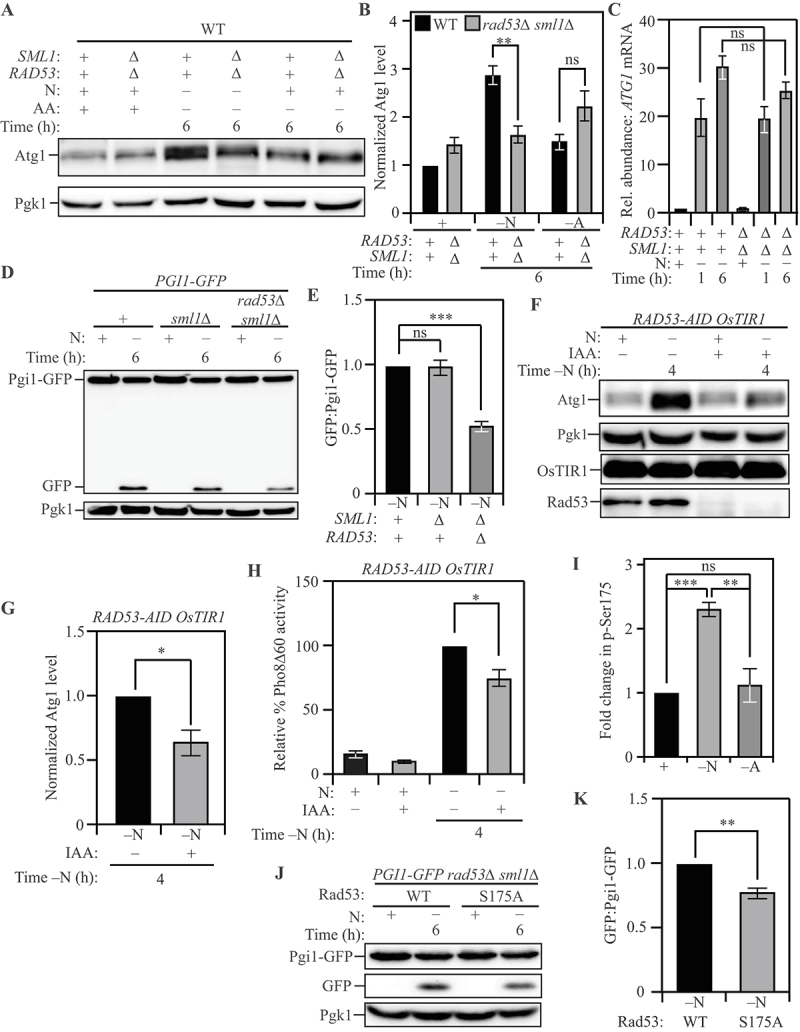
Post-transcriptional regulation of ATG1 expression by Rad53 facilitates nitrogen starvation-induced autophagy. (A) Atg1 levels exhibit a significantly greater increase in WT (SEY6210) cells relative to rad53∆ sml1∆ cells upon nitrogen starvation but not upon amino acid starvation: Cells of the indicated genotypes were harvested during nutrient-replete conditions or after nitrogen or amino acid starvation for the indicated times and protein level examined by western blot. Pgk1 was used as a loading control. (B) Densitometric analyses for (A) from three independent biological replicates. (C) A similar abundance of ATG1 transcript was detected in WT (SEY6210) and rad53∆ sml1∆ cells after nitrogen starvation: Cells of the indicated genotypes were harvested during nutrient-replete conditions or after starvation. qRT-PCR was used to determine ATG1 transcript abundance using ALG9 as the reference gene for normalization. Data from three independent biological replicates. (D) Autophagy flux during nitrogen starvation, assessed by the Pgi1-GFP processing assay, is reduced in rad53∆ sml1∆ cells compared to WT (SEY6210) and sml1∆ cells: Cells of the indicated genotypes, expressing chromosomally tagged Pgi1-GFP were harvested and examined as in (A). The appearance of free GFP indicates autophagy flux. (E) Densitometric analysis of (D) from three independent biological replicates. (F) The acute loss of Rad53 leads to a reduction in Atg1 expression during nitrogen starvation: WT (WLY176) RAD53-AID cells expressing the OsTIR1 ubiquitin ligase were treated with either IAA or DMSO and harvested during nutrient-replete conditions or after nitrogen starvation for the indicated time. IAA treatment activates the ligase activity and targets Rad53-AID for proteasomal degradation. Pgk1 was used as a loading control. (G) Densitometric analysis of (F) from three independent biological replicates. (H) The Pho8Δ60 assay reveals a reduction in autophagy flux during nitrogen starvation caused by the acute loss of Rad53: WT (WLY176) RAD53-AID OsTIR1 cells were harvested during nutrient-replete conditions or after starvation for the indicated time with or without IAA treatment, and Pho8Δ60 enzyme activity was measured by colorimetry. An increase in Pho8Δ60 activity indicates increased autophagic flux. Data are from three independent biological replicates. (I) Rad53 S175 phosphorylation levels are significantly higher in nitrogen-starvation compared to amino-acid starvation or nutrient-replete conditions: Phosphoproteome analysis of SEY6210 arg4∆ cells comparing nitrogen and amino acid starvation using triplex-SILAC labeling and LC-MS/MS analysis. The plot represents data from four independent biological replicates. (J) A non-phosphorylatable mutation of Rad53 S175 (Rad53S175A) reduces autophagy flux during nitrogen starvation, as examined by the Pgi1-GFP processing assay: WT (SEY6210) rad53∆ sml1∆ PGI1-GFP cells expressing either Rad53 or Rad53S175A were harvested during nutrient-replete conditions or after starvation for the indicated time. The appearance of free GFP indicates autophagy flux. Pgk1 used as loading control (K) Densitometric analysis of (J) from three independent biological replicates. Data in (B), (C), (E), (G-I) and (K) represent the mean ± SEM from the indicated number of replicates. Statistical analysis for (B), (C), (E) and (H) was carried out using one-way analysis of variance (ANOVA). (G) and (K) were analyzed using unpaired Student’s t-test whereas (I) was analyzed using paired Student’s t-test. Multiple comparisons were carried out using Tukey’s multiple comparisons test. *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001 ns: not significant.
