Abstract
The continuous spread of African swine fever virus (ASFV) in Europe and Asia represents a major threat to livestock health, with billions of dollars of income losses and major perturbations of the global pig industry. One striking feature of African swine fever (ASF) is the existence of different forms of the disease, ranging from acute with mortality rates approaching 100% to chronic, with mild clinical manifestations. These differences in pathogenicity have been linked to genomic alterations present in attenuated ASFV strains (and absent in virulent ones) and differences in the immune response of infected animals. In this mini-review, we summarized current knowledge on the connection between ASFV pathogenicity and the innate immune response induced in infected hosts, with a particular focus on the pathways involved in ASFV detection. Indeed, recent studies have highlighted the key role of the DNA sensor cGAS in ASFV sensing. We discussed what other pathways may be involved in ASFV sensing and inflammasome activation and summarized recent findings on the viral ASFV genes involved in the modulation of the interferon (IFN) and nuclear factor kappa B (NF-κB) pathways.
Keywords: ASFV, CGAS, NFkapapB, innate immunity, STING
Introduction
Since its identification in 1921 in Kenya (Eustace Montgomery, 1921), African swine fever (ASF) has been detected in wild African suids (warthogs, bush pigs, and giant forest hogs), wild boars, and domestic pigs in over 60 countries (Gaudreault et al., 2020). African swine fever represents an increasing economic burden worldwide: following its emergence in China in 2018 (Ge et al., 2018), an estimated 300 million pigs have died from infection or preventive culling, with economic losses ranging in the range of 100 billion dollars. ASF is caused by African swine fever virus (ASFV), a large enveloped DNA virus belonging to the Asfarviridae family. Different strains of ASFV have been associated with varying levels of virulence. Highly virulent ASFV strains induce a severe disease, with clinical symptoms and pathological lesions reminiscent of hemorrhagic fevers, and mortality rates often close to 100%. In contrast, attenuated ASFV isolates only cause a moderate disease with few, and for the most part unspecific, symptoms. These different clinical outcomes may be the result of differences in the immune response to ASFV. Indeed, although infection of aortic endothelial cells has been reported (Vallée et al., 2001), the main target cells of ASFV are likely macrophages and dendritic cells (Franzoni et al., 2017, 2018a,b), which play pivotal roles in the host immune response. In these myeloid cells, virulent strains of ASFV induce high levels of inflammation, which may be deleterious to the host, while attenuated strains ASFV stimulate the secretion of type I interferon (IFN-I), which may help to control infection (Gil et al., 2008). Important genomic differences between ASFV strains have been reported: indeed, the size of ASFV genomes ranges from 170 to 194 kbp (Malogolovkin and Kolbasov, 2019). Most naturally or laboratory-attenuated ASFV strains carry large deletions and lack many so called “virulence factors” that have been reported to antagonize innate immune pathways (Dixon et al., 2019). However, basic knowledge on the cellular pathways involved in ASFV detection remains limited. In this mini-review, we summarized the current knowledge on the pattern recognition receptors (PRRs) involved in ASFV sensing, the innate immune pathways being activated following ASFV infection, and the mechanisms of viral innate immune escape.
Activation of the cGas/Sting Pathway by African Swine Fever Virus
While ASFV infection of porcine macrophages (the main target of the virus) triggers a potent innate immune response, the PRRs involved in ASFV detection remain only partially characterized. The cGAS/STING signaling pathway has emerged as a key mediator of immunity to infection, cellular stress, and tissue damage. It has a broad capacity to sense and regulate the cellular response toward microbial and host-derived DNAs, which serve as ubiquitous danger-associated molecules (Decout et al., 2021). Recent studies indicate that the cGAS/STING pathway is stimulated upon ASFV infection (Wang et al., 2018; García-Belmonte et al., 2019), as described for many other DNA viruses (Cai et al., 2014). Interestingly, comparative structural and functional analyses of porcine, murine, and human cGAS have confirmed that these orthologs share key features (Kato et al., 2013), which may allow to extrapolate some knowledge on human cGAS to the innate sensing of other less-studied viruses such as ASFV. In addition, several ASFV virulence factors inhibit its activation: both DP96R and MGF-360-11L inhibit cGAS activation and TBK1 phosphorylation (Wang et al., 2018; Yang et al., 2021c), while MGF-505-7R and MGF-505-11R degrade STING (Li et al., 2021a; Yang et al., 2021c). Other viral proteins not previously labeled as virulence factors also have the ability to impair cGAS activation: for instance, pS273R, a viral protease, prevents the interaction between STING and IKK-ε (Luo et al., 2021). Many of these studies, however, were conducted in human cell lines and relied on overexpression approaches. Studies using specific chemical inhibitors of cGAS and/or knock-down or knock-out approaches in porcine cells are necessary to formally confirm the involvement of cGAS in ASFV detection. With so many viral proteins targeting the cGAS/STING pathway (at least when expressed ectopically), it will be interesting to determine whether cGAS activation is indeed inhibited during the course of ASFV infection and to compare the ability of virulent and attenuated strains to do so.
While the mechanisms of regulation and inhibition of cGAS have been extensively studied in humans (Hertzog and Rehwinkel, 2020; Taffoni et al., 2021), less is known about these mechanisms in pigs and other livestock species. For instance, it is unknown whether cGAS binds directly to ASFV genomes in cytoplasmic viral factories; in the nucleus, where early genome replication occurs (Simões et al., 2019); or in other subcellular compartments, where cGAS can also be detected (de Oliveira Mann and Hopfner, 2021). It would also be interesting to determine whether cGAS activation by ASFV leads (as expected) to the production of the secondary messenger cGAMP and ultimately to its passive incorporation in viral particles, as described for other viruses (Bridgeman et al., 2015).
Potential Detection of African Swine Fever Virus by Other DNA Sensors
While the available data so far indicated a predominant role for the cGAS/STING pathway, other PRRs may also be involved in ASFV sensing. In particular, the potential roles of other DNA sensors, such as TLR9, AIM2, and IFI16, have not yet been interrogated.
TLR9 is a DNA sensor expressed in the endosomal compartment of numerous immune cell types, including porcine macrophages (Qin et al., 2016). It recognizes unmethylated CpG-rich DNA of bacterial and viral origins, as was demonstrated for example for porcine circovirus (PCV) DNA (Shi et al., 2021). TLR9 activation leads to the synthesis of proinflammatory cytokines such as IL-12, IL-6, and TNF-α, known to be induced by ASFV (Gil et al., 2008; Wang S. et al., 2021). Its involvement in ASFV detection has, however, never been directly assessed.
The family of AIM2-like receptors (ALRs) consists of four proteins in humans: MNDA, PYHIN1, IFI16, and AIM2. While the pig genome does include MNDA, PYHIN1, and IFI16, AIM2 seems to be missing (Dawson et al., 2017), suggesting that the porcine AIM2 inflammasome may not be functional (Brunette et al., 2012). However, comparison of porcine and murine AIM2 inflammasomes indicated that they are activated by the same triggers, which differ significantly from the other major inflammasome, the NLRP3-inflammasome (Kim et al., 2014; Ahn et al., 2018). This activation leads to a pathway of lytic cell death called pyroptosis, as a result of caspase-1 activation, maturation, and release of IL-1 and IL-18 and gasdermin D cleavage. ASFV inhibits this process through its viral protease pS273R, which has been reported to cleave gasdermin D in a non-canonical way that does not lead to pyroptosis (Zhao et al., 2022). ASFV also encodes many other inhibitors of apoptotic cell death, including the Bcl-2 like protein A179L; the C-type lectin AP153R; and the inhibitor of apoptosis protein (IAP) A224L (Dixon et al., 2017).
In the absence of AIM2, which other sensor may activate inflammasomes in response to ASFV infection? IFI16, which recognizes many DNA viruses (including Kaposi’s sarcoma-associated herpesvirus, Herpes simplex virus, and Epstein-Barr virus) is an interesting candidate because it is also able to detect genomic lesions following DNA damage (Taffoni et al., 2021). While ASFV replicates mostly in cytoplasmic viral factories (Netherton and Wileman, 2013), ASFV proteins p10 and p37 can accumulate in the nucleus, forming readily detectable foci (Eulálio et al., 2007; Nunes-Correia et al., 2008). Early ASFV genome replication events in the nucleus have been reported to induce the DNA damage response (DDR) pathway (Simões et al., 2013) and to alter the nuclear architecture and to induce changes in the epigenetic landscape (Ballester et al., 2011; Simões et al., 2015a,b, 2019). It is therefore an intruding thought that IFI16 may recognize ASFV, either directly (by binding to viral DNA) or indirectly (by recognizing ASFV-induced DNA damage). Similar to observations made in human macrophages (Jønsson et al., 2017, 16), porcine IFI16 has recently been described to regulate cGAS signaling (Zheng et al., 2020). Further studies are required to establish whether IFI16 (or other ALRs such as MNDA or PYHIN1) is involved in ASFV sensing, and whether this impacts ASFV cGAS/STING detection. While many studies in human cells suggest that cGAS is engaged in cross-talks with various sensing pathways, including some targeting RNA viruses (Anwar et al., 2021), this phenomenon remains poorly studied in pigs. If confirmed, it would then be interesting to study the effect of ASFV-encoded cGAS inhibitors on these other sensing pathways.
Recognition of African Swine Fever Virus by Other Sensors
There is preliminary evidence that ASFV may also be detected by other PRRs beyond DNA sensors. For instance, the viral protein CD2v, an attachment factor required for ASFV infection in the Ornithodoros soft tick vector (Rowlands et al., 2009) stimulates IFN production (Chaulagain et al., 2021) by an unknown mechanism. AT-rich regions in ASFV DNA may also be recognized by RNA Pol III, which in turn activates RIG-I (Ran et al., 2022), although the ASFV virulence factor I267L inhibits this process by impairing Riplet-mediated activation of RIG-I (Ran et al., 2022). Several studies also suggest a role for TLR3 in ASFV detection. Indeed, its expression (together with other TLRs) is upregulated following ASFV infection (Yang et al., 2021a), and ASFV encodes two inhibitors of TLR3, A276R, and I329L. A276R inhibits IFN production following stimulation of TLR3 by poly(I:C) (Correia et al., 2013) through an unknown mechanism. I329L is a functional homolog of TLR3 and blocks IFN signaling at the level of TRIF (de Oliveira et al., 2011; Correia et al., 2013). How would TLR3, a PRR detecting dsRNAs in endosomes, be involved in the detection of ASFV, a DNA virus mostly replicating in cytoplasmic viral factories (Netherton and Wileman, 2013)? ASFV enters cells through clathrin-mediated endocytosis (Galindo et al., 2015; Hernáez et al., 2016), and ASFV infection leads to the redistribution endosomal membranes to the site of viral replication (Cuesta-Geijo et al., 2022), so it is conceivable that TLR3 ends up in proximity of ASFV RNA transcripts. To our knowledge, the production of dsRNAs was not yet observed in the context of ASFV infection but was reported for other DNA viruses (Weber et al., 2006). Future studies are therefore required to formally demonstrate the involvement of RNA sensors in ASFV sensing and to characterize at which stage of the ASFV replication cycle this detection takes place. Interestingly, multiple single nucleotide polymorphisms (SNPs) in TLRs have been observed in wild boars (Bergman et al., 2010) and in some specific breeds of domestic pigs (Palermo et al., 2009; Chen et al., 2021), and hence it would therefore be interesting to assess their effect on ASFV innate immune sensing.
Nuclear Factor κB Activation and Stimulation of Inflammation by African Swine Fever Virus
Modulation of the inflammatory response has been suggested to be (at least in part) responsible for the observed differences in clinical outcomes following ASFV infection (Zhu et al., 2019). Indeed, in vivo studies showed that pigs infected with virulent ASFV exhibited a delayed onset of the inflammatory response, allowing the virus to replicate and to escape immune defenses, to later trigger a fatal cytokine storm (Wang S. et al., 2021). In this study, we discussed how ASFV modulates activation of the NF-κB signaling pathway.
The NF-κB transcription factor family is formed by five members, RelA (p65), RelB, c-Rel, p50/p105 (NF-κB1), and p52/p100 (NF-κB2) and plays a role as a master regulator of cell death and innate immune responses. It integrates the signals triggered by different PRRs to mount a suitable cellular response (Dorrington and Fraser, 2019). This makes this pathway a perfect target for viruses which have evolved diverse strategies to dampen or modulate the cellular innate immune response it in order to promote viral replication (Hayden and Ghosh, 2008; Schmitz et al., 2014). This is also the case for ASFV. However, NF-κB modulation by ASFV is a double-edged sword: on the one hand, the virus exploits the antiapoptotic functions of NF-κB to promote early viral replication; on the other hand, ASFV employs mechanisms to evade NF-κB-mediated antiviral cytokine responses. Primary alveolar macrophages (the most suitable cellular infection model) infected with highly virulent ASFV display a strong upregulation of genes belonging to this pathway (Cackett et al., 2022) and blocking its activation with the inhibitor BAY11-7082 limits viral replication in vitro (Gao et al., 2022). Upon PRR stimulation, signal transduction will activate the IKK complex (formed by IKK-α, IKK-β, and IKK-γ), which, in turn, will phosphorylate IκB proteins, leading to their degradation and the release of free NF-κB dimers that will translocate to the nucleus. There, NF-κB will trigger the expression of proinflammatory cytokines and negative feedback loops to ultimately shut down the inflammatory response (Ashall et al., 2009). Thus, rather than acting as a simple on/off switch for inflammatory responses, NF-κB activity is an intricate, tightly regulated and dynamic process. This inflammatory response needs to be strong enough to eliminate the infection, but at the same time, it must not cause too much damage to the host (Gottschalk et al., 2016). Single cell studies show that differences in NF-κB signaling dynamics will lead to differences in the subset of genes expressed by the cell (Tay et al., 2010; Kellogg et al., 2017; Lane et al., 2017). NF-κB activation dynamics seem to be cell- and stimulus-dependent for instance, macrophages show a gradual activation of NF-κB with a long-lasting nuclear translocation, while non-inflammatory cells seem to activate it in an “all or nothing” manner showing NF-κB oscillations (Dorrington and Fraser, 2019; López-Gálvez et al., 2021).
African swine fever virus disrupts NF-κB signaling at different levels. The ubiquitin-conjugating enzyme (UBCv1) and F317L are early viral proteins blocking NF-κB activation upstream or at the level of the IKK complexes (Barrado-Gil et al., 2021; Yang et al., 2021b). F317L interacts with IKK-β to block IκBα phosphorylation, and its overexpression promotes viral replication (Yang et al., 2021b). ASFV also blocks NF-κB signaling through the A238L protein, an IκBα homolog binding directly to NF-κB heterodimers and inhibiting their nuclear translocation (Powell et al., 1996; Silk et al., 2007). Furthermore, A238L has been suggested to interact with the CBP/P300 transcription coactivator proteins (Granja et al., 2006), which are involved in the regulation of NF-κB-dependent transcription (Mukherjee et al., 2013) to inhibit the synthesis of TNF-α. Interestingly, many ASFV genes do not only block NF-κB activation but also fulfill other functions. For instance, A224L is an inhibitor of apoptosis (IAP)-like protein, which also activates NF-κB signaling (Rodríguez et al., 2002). In addition, the K205R protein triggers endoplasmic reticulum stress and subsequently increases NF-κB activation (Wang Q. et al., 2022). Another multifunctional ASFV viral protein is MGF-505-7R (Huang et al., 2022), which, in addition to its ability to target the IFN pathway, suppresses TLR8-mediated phosphorylation of p65 phosphorylation (Liu X. et al., 2021) and inhibits IL-1 production by interfering with the formation of the NLRP3 inflammasome (Li et al., 2021c).
Altogether, ASFV infection results in a complex interactome of viral proteins that can activate or inhibit NF-κB. Further studies are needed to better understand how this mosaic of interactions modulate NF-κB and determine cell fate. A key question would be to understand the mechanisms through which virulent ASFV strains overcome the cellular regulatory “checkpoints” and shift the immune response from a low-degree inflammation to a sudden (and fatal) cytokine storm. For instance, assessing the kinetics of expression of virulence genes across different ASFV strains would allow to build a model explaining how virulent strains manipulate the early inflammatory response and trigger excessive cytokine production leading to endothelial dysfunction, coagulopathy and ultimately fatal hemorrhages. Future studies may also assess whether differences in NF-κB pathway activation can be responsible for the absence of symptoms in some ASFV-infected animals such as wild African suids. Although one report has demonstrated that warthog-specific signatures in RelA/p65 (Palgrave et al., 2011) do not confer ASFV resistance to domestic pigs (McCleary et al., 2020), it is still conceivable that other components in the NF-κB pathway may differ between warthogs (or other wild African suids) and pigs, either at the genetic level or in their activation/regulation dynamics.
Viral Antagonism of Interferon and Nuclear Factor κB Pathways and Virulence: a Complex Picture
As discussed earlier, several ASFV viral genes seem to be able to prevent the induction of the cGAS/STING pathway. In addition, many other ASFV genes have a more downstream effect on IFN signaling. Early genomic studies comparing ASFV genomes from virulent and attenuated ASFV have identified genes of the multi gene family (MGF) 360 and 505 as being deleted in attenuated ASFV (Neilan et al., 2002; Chapman et al., 2008; Portugal et al., 2015). Consistent with this, attenuated strains lacking genes from this family appear to be unable to shut down the IFN response (Portugal et al., 2018) and to be more sensitive to its antiviral effects (Golding et al., 2016). Deletion of MGF 360/505 genes from virulent ASFV strains led to attenuation in vivo (O’Donnell et al., 2015) confirming the relevance of this gene family. This prompted intense research efforts to uncover which virulence factors are responsible for the modulation of the innate immune response to ASFV, although the relative contributions of each individual gene remain elusive. In addition, the relationship between ASFV pathogenicity and viral replication is complex, as many attenuated ASFV strains are still able to sustain high levels of replication, at least in vitro (Lacasta et al., 2015; Reis et al., 2016; Franzoni et al., 2018b,2020). Since several good reviews have been published recently covering these topics (Dixon et al., 2019; Gladue and Borca, 2022; Wang Z. et al., 2022), we did not aim to make an exhaustive list of the ASFV genes targeting innate immune pathways but rather highlight recent discoveries and illustrate conserved antagonism strategies.
Recent study has suggested that MGF-505-1R and MGF-360-12L both inhibit the IFN response, albeit with different mechanisms, as only the latter impacted ASFV replication in macrophages (Rathakrishnan et al., 2022). Strikingly, some key proteins of the IFN pathway are the target of multiple ASFV inhibitors. For instance, TBK1 is inhibited (or degraded) by E120R (Liu H. et al., 2021), DP96R (Wang et al., 2018), MGF-360-11L (Yang et al., 2022), and A137R (Sun et al., 2022). Similarly, JAK/STAT are targeted by MGF-360-9L (Zhang et al., 2022) and MGF-505-7R (Li et al., 2021b). Some ASFV proteins, like I226R (Hong et al., 2022), I329L (de Oliveira et al., 2011), and MGF-360-12L (Zhuo et al., 2021) are able to target both the IFN and NF-κB pathways at the same time. Interestingly, multiple ASFV proteins, such as A137R (Sun et al., 2022), K205R (Wang Q. et al., 2022), and MGF-505-7R (Li et al., 2021a), induce the autophagy pathway, which may represent a strategy shared by multiple virulence genes to modulate the innate immune response to ASFV.
As summarized in Table 1, many ASFV viral genes modulate IFN and NF-κB signaling. Such a level of functional redundancy likely indicates that ASFV is under strong selective pressure to evade the antiviral effects of IFN and NF-κB. To date, however, only a very limited subset of IFN-stimulated genes (ISGs) have been studied in the context of ASFV. So far, only MxA (Netherton et al., 2009) and IFITM2/3 (Muñoz-Moreno et al., 2016) have been described to inhibit the replication of Ba71V, the Vero-adapted strain of ASFV. Whether ASFV evolved mechanisms of antagonism of antiviral ISGs (for instance through virulence factors) has not been studied either.
TABLE 1.
Examples of African swine fever virus (ASFV) viral genes modulating interferon (IFN) and nuclear factor kappa B (NF-κB) activation.
| Viral gene | Effect on virulence | Mechanism of action | References |
| S2736 | Unclear | Inhibition of the IKKε/STING interaction through IKKε binding. Involved in pyroptosis modulation | Luo et al., 2021; Zhao et al., 2022 |
| MGF-505-7R | Deletion leads to attenuation in the HLJ/18 strain | Upregulation of ULK1 and degradation of STING through autophagy. Interaction with IKKα inhibiting NF-κB activation | Li et al., 2021a,b |
| I267L | Deletion leads to attenuation in the CN/GS/201 strain | Inhibition of Riplet-mediated K63-polyubiquitination and activation of Rig-I | Ran et al., 2022 |
| MGF-360-9L | Deletion leads to attenuation in the CN/GS/201 strain | Degradation of STAT1 and STAT2 | Zhang et al., 2022 |
| MGF-360-11L | Unclear | Degradation of TBK1 and IRF7 | Yang et al., 2022 |
| MGF-360-14L | Unclear | Degradation of IRF3 through TRIM21-mediated K63-ubiquitination | Wang Y. et al., 2021 |
| MGF-505-11R | Unclear | Degradation of STING | Yang et al., 2021c |
| MGF-360-12L | Deletion leads to attenuation in Georgia strain | Blocks interaction between importins and NF-κB | Zhuo et al., 2021; Rathakrishnan et al., 2022 |
| K205R | Unclear | Stimulation of p65 nuclear translocation. Involved in ER stress and autophagy | Wang Q. et al., 2022 |
| E120R | Unclear | Interaction with IRF3 blocking its activation | Liu H. et al., 2021 |
| DP96R | Deletion leads to attenuation in some (but not all) strains | Inhibition of TBK1 phosphorylation | Wang et al., 2018; Ramirez-Medina et al., 2019 |
| A238L | Deletion in the E70 isolate has no effect | IκBα homolog binding inhibiting NF-κB translocation. Interaction with CBP | Powell et al., 1996; Granja et al., 2006; Silk et al., 2007; Salguero et al., 2008 |
For the indicated ASFV genes is indicated the effect on virulence (e.g., when deleted from virulent strains), the proposed mechanism of action and the references.
Attenuated and virulent strains of ASFV seem to differ in the innate immune response they induce, with different profiles of cytokines being produced after infection (Gil et al., 2008). However, the role of individual ASFV virulence genes remains unclear. Indeed, while deletion of some viral genes, such as I267L (Ran et al., 2022), MGF-505-5R (Li et al., 2021a, c; Huang et al., 2022), MGF-360-12L, and MGF-505-1R (Rathakrishnan et al., 2022) lead to ASFV attenuation in vivo, others, such as I329L (Reis et al., 2020) or A238L (Salguero et al., 2008), seem to have no effect. Moreover, these effects can be strain-dependent: for instance, deletion of DP96R lead to attenuation in the virulent strain E70 but not in the virulent Georgia strain that emerged in Europe in 2007 (Ramirez-Medina et al., 2019). In some cases, deletion of viral genes known to modulate the immune response had deleterious effects on the efficacy of live attenuated vaccines candidates (Reis et al., 2020; Lopez et al., 2021; Rathakrishnan et al., 2022). These studies highlight the complexity of the mechanisms underlying ASFV virulence and suggest that multiple viral proteins act in concert to shape the innate immune response to ASFV.
Conclusion
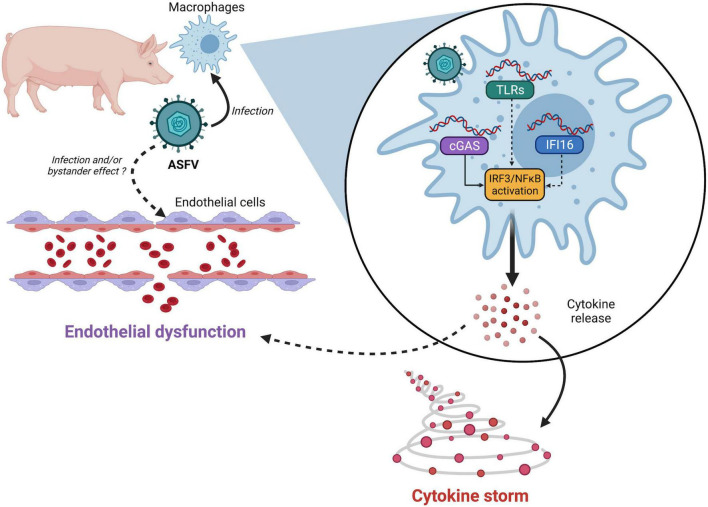
The mechanisms of innate immune detection of ASFV are just starting to be described. While most studies focused on the cGAS/STING pathway so far, ASFV may also be sensed by other DNA sensors such as TLR9 or IFI16 or by RNA sensors like TLR3 or RIG-I. Many ASFV genes (including virulence factors but also conserved genes) inhibit the activation of the IRF3 and NF-κB signaling pathways at different levels. Such functional redundancy strongly suggests that ASFV is under strong pressure to antagonize these pathways, although the roles of individual genes (and their mechanisms) remain to be fully characterized. It will be of particular interest to determine which viral genes shape the innate immune response to ASFV, and how this process leads to endothelial dysfunction and cytokine storm, the main hallmarks of virulent ASFV infection (Figure 1). While direct infection of endothelial cells has been reported to trigger apoptosis (Vallée et al., 2001), endothelial dysfunction may also be an indirect consequence of excessive proinflammatory cytokine production (Tatoyan et al., 2019). Further studies may also shed light on the mechanisms responsible for the lack of virulence of some naturally attenuated ASFV strains in domestic pigs, as well as for ASFV persistence in wild African suids, which do not develop symptoms upon infection (Oura et al., 1998). Ultimately, research efforts exploring the complex mechanisms of ASFV pathogenicity may guide the design of safe and robust live attenuated vaccines for this deadly pathogen.
FIGURE 1.
Links between African swine fever virus (ASFV) sensing and pathogenesis. ASFV nucleic acids—either incoming DNA or newly synthesized viral DNA/RNA—may be detected by several pattern recognition receptors (PRRs). While cGAS has been validated by multiple teams, the involvement of other PRRs like IFI16 or TLRs cannot be ruled out. ASFV detection then triggers IRF3 and NF-κB activation resulting in uncontrolled secretion of proinflammatory cytokines. Whether endothelial dysfunction is mediated by apoptosis of infected cells or through bystander effects remains to be determined.
Author Contributions
AA, ST, RG, IC, and FR wrote and proofread the manuscript and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
AA received a scholarship from the Erasmus+ programme of the European Union (EMJMD IDOH+, Infectious Diseases and One Health). Work on this study was supported by a grant from the EU Horizon 2020 programme (VetBioNet project, GA No. 731014) and intramural funding from the INRAE Animal Health Department (INRAE Département SA).
References
- Ahn H., Kim J., Kwon S., Kim P. H., Kwon H. M., Lee E., et al. (2018). Triggers of NLRC4 and AIM2 inflammasomes induce porcine IL-1β secretion. Vet. Res. Commun. 42 265–273. 10.1007/s11259-018-9729-x [DOI] [PubMed] [Google Scholar]
- Anwar S., Ul Islam K., Azmi M. I., Iqbal J. (2021). Cgas-sting-mediated sensing pathways in DNA and RNA virus infections: crosstalk with other sensing pathways. Arch. Virol. 166 3255–3268. 10.1007/s00705-021-05211-x [DOI] [PubMed] [Google Scholar]
- Ashall L., Horton C. A., Nelson D. E., Paszek P., Harper C. V., Sillitoe K., et al. (2009). Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science 324 242–246. 10.1126/science.1164860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester M., Rodríguez-Cariño C., Pérez M., Gallardo C., Rodríguez J. M., Salas M. L., et al. (2011). Disruption of nuclear organization during the initial phase of African swine fever virus infection. J. Virol. 85 8263–8269. 10.1128/JVI.00704-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrado-Gil L., Del Puerto A., Galindo I., Cuesta-Geijo M. Á., García-Dorival I., de Motes C. M., et al. (2021). African Swine Fever Virus Ubiquitin-Conjugating Enzyme Is an Immunomodulator Targeting NF-κB Activation. Viruses 13:1160. 10.3390/v13061160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman I. M., Rosengren J. K., Edman K., Edfors I. (2010). European wild boars and domestic pigs display different polymorphic patterns in the Toll-like receptor (TLR) 1, TLR2, and TLR6 genes. Immunogenetics 62 49–58. 10.1007/s00251-009-0409-4 [DOI] [PubMed] [Google Scholar]
- Bridgeman A., Maelfait J., Davenne T., Partridge T., Peng Y., Mayer A., et al. (2015). Viruses transfer the antiviral second messenger cGAMP between cells. Science 349 1228–1232. 10.1126/science.aab3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunette R. L., Young J. M., Whitley D. G., Brodsky I. E., Malik H. S., Stetson D. B. (2012). Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J. Exp. Med. 209 1969–1983. 10.1084/jem.20121960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cackett G., Portugal R., Matelska D., Dixon L., Werner F. (2022). African Swine Fever Virus and Host Response: transcriptome Profiling of the Georgia 2007/1 Strain and Porcine Macrophages. J. Virol. 96:e0193921. 10.1128/jvi.01939-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Chiu Y. H., Chen Z. J. (2014). The cGAS-cGAMP-STING Pathway of Cytosolic DNA Sensing and Signaling. Mol. Cell 54 289–296. 10.1016/j.molcel.2014.03.040 [DOI] [PubMed] [Google Scholar]
- Chapman D. A. G., Tcherepanov V., Upton C., Dixon L. K. (2008). Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J. Gen. Virol. 89 397–408. 10.1099/vir.0.83343-0 [DOI] [PubMed] [Google Scholar]
- Chaulagain S., Delhon G. A., Khatiwada S., Rock D. L. (2021). African Swine Fever Virus CD2v Protein Induces β-Interferon Expression and Apoptosis in Swine Peripheral Blood Mononuclear Cells. Viruses 13:1480. 10.3390/v13081480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. Y. A., Huang C. W., Liu S. H., Liu A. C., Chaung H. C. (2021). Single Nucleotide Polymorphisms of Immunity-Related Genes and Their Effects on Immunophenotypes in Different Pig Breeds. Genes 12:1377. 10.3390/genes12091377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia S., Ventura S., Parkhouse R. M. (2013). Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 173 87–100. 10.1016/j.virusres.2012.10.013 [DOI] [PubMed] [Google Scholar]
- Cuesta-Geijo M. Á., García-Dorival I., Del Puerto A., Urquiza J., Galindo I., Barrado-Gil L., et al. (2022). New insights into the role of endosomal proteins for African swine fever virus infection. PLoS Pathog. 18:e1009784. 10.1371/journal.ppat.1009784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson H. D., Smith A. D., Chen C., Urban J. F. (2017). An in-depth comparison of the porcine, murine and human inflammasomes; lessons from the porcine genome and transcriptome. Vet. Microbiol. 202 2–15. 10.1016/j.vetmic.2016.05.013 [DOI] [PubMed] [Google Scholar]
- de Oliveira V. L., Almeida S. C. P., Soares H. R., Crespo A., Marshall-Clarke S., Parkhouse R. M. E. (2011). A novel TLR3 inhibitor encoded by African swine fever virus (ASFV). Arch. Virol. 156 597–609. 10.1007/s00705-010-0894-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Mann C. C., Hopfner K. P. (2021). Nuclear cGAS: guard or prisoner?. EMBO J. 40:e108293. 10.15252/embj.2021108293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decout A., Katz J. D., Venkatraman S., Ablasser A. (2021). The cGAS–STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 21 548–569. 10.1038/s41577-021-00524-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L. K., Islam M., Nash R., Reis A. L. (2019). African swine fever virus evasion of host defences. Virus Res. 266 25–33. 10.1016/j.virusres.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L. K., Sánchez-Cordón P. J., Galindo I., Alonso C. (2017). Investigations of Pro- and Anti-Apoptotic Factors Affecting African Swine Fever Virus Replication and Pathogenesis. Viruses 9:241. 10.3390/v9090241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrington M. G., Fraser I. D. C. (2019). NF-κB Signaling in Macrophages: dynamics, Crosstalk, and Signal Integration. Front. Immunol. 10:705. 10.3389/fimmu.2019.00705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulálio A., Nunes-Correia I., Salas J., Salas M. L., Simões S., Pedroso de Lima M. C. (2007). African swine fever virus p37 structural protein is localized in nuclear foci containing the viral DNA at early post-infection times. Virus Res. 130 18–27. 10.1016/j.virusres.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Eustace Montgomery R. (1921). On A Form of Swine Fever Occurring in British East Africa (Kenya Colony). J. Comp. Pathol. Ther. 34 159–191. 10.1016/S0368-1742(21)80031-4 [DOI] [Google Scholar]
- Franzoni G., Dei Giudici S., Oggiano A. (2018a). Infection, modulation and responses of antigen-presenting cells to African swine fever viruses. Virus Res. 258 73–80. 10.1016/j.virusres.2018.10.007 [DOI] [PubMed] [Google Scholar]
- Franzoni G., Graham S. P., Sanna G., Angioi P., Fiori M. S., Anfossi A., et al. (2018b). Interaction of porcine monocyte-derived dendritic cells with African swine fever viruses of diverse virulence. Vet. Microbiol. 216 190–197. 10.1016/j.vetmic.2018.02.021 [DOI] [PubMed] [Google Scholar]
- Franzoni G., Graham S. P., Giudici S. D., Bonelli P., Pilo G., Anfossi A. G., et al. (2017). Characterization of the interaction of African swine fever virus with monocytes and derived macrophage subsets. Vet. Microbiol. 198 88–98. 10.1016/j.vetmic.2016.12.010 [DOI] [PubMed] [Google Scholar]
- Franzoni G., Razzuoli E., Dei Giudici S., Carta T., Galleri G., Zinellu S., et al. (2020). Comparison of Macrophage Responses to African Swine Fever Viruses Reveals that the NH/P68 Strain is Associated with Enhanced Sensitivity to Type I IFN and Cytokine Responses from Classically Activated Macrophages. Pathogens 9:209. 10.3390/pathogens9030209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo I., Cuesta-Geijo M. A., Hlavova K., Muñoz-Moreno R., Barrado-Gil L., Dominguez J., et al. (2015). African swine fever virus infects macrophages, the natural host cells, via clathrin- and cholesterol-dependent endocytosis. Virus Res. 200 45–55. 10.1016/j.virusres.2015.01.022 [DOI] [PubMed] [Google Scholar]
- Gao Q., Yang Y., Feng Y., Quan W., Luo Y., Wang H., et al. (2022). Effects of the NF-κB Signaling Pathway Inhibitor BAY11-7082 in the Replication of ASFV. Viruses 14:297. 10.3390/v14020297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Belmonte R., Pérez-Núñez D., Pittau M., Richt J. A., Revilla Y. (2019). African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the cGAS-STING Pathway. J. Virol. 93 e02298–18. 10.1128/JVI.02298-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreault N. N., Madden D. W., Wilson W. C., Trujillo J. D., Richt J. A. (2020). African Swine Fever Virus: an Emerging DNA Arbovirus. Front. Vet. Sci. 7:215. 10.3389/fvets.2020.00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S., Li J., Fan X., Liu F., Li L., Wang Q., et al. (2018). Molecular Characterization of African Swine Fever Virus. China, 2018. Emerg. Infect. Dis. 24 2131–2133. 10.3201/eid2411.181274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil S., Sepúlveda N., Albina E., Leitão A., Martins C. (2008). The low-virulent African swine fever virus (ASFV/NH/P68) induces enhanced expression and production of relevant regulatory cytokines (IFNα, TNFα and IL12p40) on porcine macrophages in comparison to the highly virulent ASFV/L60. Arch. Virol. 153 1845–1854. 10.1007/s00705-008-0196-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladue D. P., Borca M. V. (2022). Recombinant ASF Live Attenuated Virus Strains as Experimental Vaccine Candidates. Viruses 14:878. 10.3390/v14050878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding J. P., Goatley L., Goodbourn S., Dixon L. K., Taylor G., Netherton C. L. (2016). Sensitivity of African swine fever virus to type I interferon is linked to genes within multigene families 360 and 505. Virology 493 154–161. 10.1016/j.virol.2016.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk R. A., Martins A. J., Angermann B. R., Dutta B., Ng C. E., Uderhardt S., et al. (2016). Distinct NF-κB and MAPK Activation Thresholds Uncouple Steady-State Microbe Sensing from Anti-pathogen Inflammatory Responses. Cell Syst. 2 378–390. 10.1016/j.cels.2016.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granja A. G., Nogal M. L., Hurtado C., Del Aguila C., Carrascosa A. L., Salas M. L., et al. (2006). The viral protein A238L inhibits TNF-alpha expression through a CBP/p300 transcriptional coactivators pathway. J. Immunol. 176 451–462. 10.4049/jimmunol.176.1.451 [DOI] [PubMed] [Google Scholar]
- Hayden M. S., Ghosh S. (2008). Shared principles in NF-kappaB signaling. Cell 132 344–362. 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- Hernáez B., Guerra M., Salas M. L., Andrés G. (2016). African Swine Fever Virus Undergoes Outer Envelope Disruption, Capsid Disassembly and Inner Envelope Fusion before Core Release from Multivesicular Endosomes. PLoS Pathog. 12:e1005595. 10.1371/journal.ppat.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog J., Rehwinkel J. (2020). Regulation and inhibition of the DNA sensor cGAS. EMBO Rep. 21:e51345. 10.15252/embr.202051345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J., Chi X., Yuan X., Wen F., Rai K. R., Wu L., et al. (2022). I226R Protein of African Swine Fever Virus Is a Suppressor of Innate Antiviral Responses. Viruses 14:575. 10.3390/v14030575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Li J., Zheng J., Li D., Weng C. (2022). Multifunctional pMGF505-7R Is a Key Virulence-Related Factor of African Swine Fever Virus. Front. Microbiol. 13:852431. 10.3389/fmicb.2022.852431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jønsson K. L., Laustsen A., Krapp C., Skipper K. A., Thavachelvam K., Hotter D., et al. (2017). IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun. 8:14391. 10.1038/ncomms14391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K., Ishii R., Goto E., Ishitani R., Tokunaga F., Nureki O. (2013). Structural and Functional Analyses of DNA-Sensing and Immune Activation by Human cGAS. PLoS One 8:e76983. 10.1371/journal.pone.0076983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg R. A., Tian C., Etzrodt M., Tay S. (2017). Cellular Decision Making by Non-Integrative Processing of TLR Inputs. Cell Rep. 19 125–135. 10.1016/j.celrep.2017.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Ahn H., Woo H. M., Lee E., Lee G. S. (2014). Characterization of porcine NLRP3 inflammasome activation and its upstream mechanism. Vet. Res. Commun. 38 193–200. 10.1007/s11259-014-9602-5 [DOI] [PubMed] [Google Scholar]
- Lacasta A., Monteagudo P. L., Jiménez-Marín Á., Accensi F., Ballester M., Argilaguet J., et al. (2015). Live attenuated African swine fever viruses as ideal tools to dissect the mechanisms involved in viral pathogenesis and immune protection. Vet. Res. 46:135. 10.1186/s13567-015-0275-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane K., Van Valen D., DeFelice M. M., Macklin D. N., Kudo T., Jaimovich A., et al. (2017). Measuring Signaling and RNA-Seq in the Same Cell Links Gene Expression to Dynamic Patterns of NF-κB Activation. Cell Syst. 4 458–469.e5. 10.1016/j.cels.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Yang W., Li L., Li P., Ma Z., Zhang J., et al. (2021a). African Swine Fever Virus MGF-505-7R Negatively Regulates cGAS-STING-Mediated Signaling Pathway. J. Immunol. 206 1844–1857. 10.4049/jimmunol.2001110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Song J., Kang L., Huang L., Zhou S., Hu L., et al. (2021c). pMGF505-7R determines pathogenicity of African swine fever virus infection by inhibiting IL-1β and type I IFN production. PLoS Pathog. 17:e1009733. 10.1371/journal.ppat.1009733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Zhang J., Yang W., Li P., Ru Y., Kang W., et al. (2021b). African swine fever virus protein MGF-505-7R promotes virulence and pathogenesis by inhibiting JAK1- and JAK2-mediated signaling. J. Biol. Chem. 297:101190. 10.1016/j.jbc.2021.101190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhu Z., Feng T., Ma Z., Xue Q., Wu P., et al. (2021). African Swine Fever Virus E120R Protein Inhibits Interferon Beta Production by Interacting with IRF3 To Block Its Activation. J. Virol. 95:e0082421. 10.1128/JVI.00824-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ao D., Jiang S., Xia N., Xu Y., Shao Q., et al. (2021). African Swine Fever Virus A528R Inhibits TLR8 Mediated NF-κB Activity by Targeting p65 Activation and Nuclear Translocation. Viruses 13:2046. 10.3390/v13102046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez E., Bosch-Camós L., Ramirez-Medina E., Vuono E., Navas M. J., Muñoz M., et al. (2021). Deletion Mutants of the Attenuated Recombinant ASF Virus, BA71ΔCD2, Show Decreased Vaccine Efficacy. Viruses 13:1678. 10.3390/v13091678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Gálvez R., Fleurot I., Chamero P., Trapp S., Olivier M., Chevaleyre C., et al. (2021). Airway Administration of Flagellin Regulates the Inflammatory Response to Pseudomonas aeruginosa. Am. J. Respir. Cell Mol. Biol. 65 378–389. 10.1165/rcmb.2021-0125OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Ni J., Jiang S., Xia N., Guo Y., Shao Q., et al. (2021). The African swine fever virus protease pS273R inhibits DNA sensing cGAS-STING pathway by targeting IKKε. bioRxiv [Preprint]. 10.1101/2021.06.02.446855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malogolovkin A., Kolbasov D. (2019). Genetic and antigenic diversity of African swine fever virus. Virus Res. 271:197673. 10.1016/j.virusres.2019.197673 [DOI] [PubMed] [Google Scholar]
- McCleary S., Strong R., McCarthy R. R., Edwards J. C., Howes E. L., Stevens L. M., et al. (2020). Substitution of warthog NF-κB motifs into RELA of domestic pigs is not sufficient to confer resilience to African swine fever virus. Sci. Rep. 10:8951. 10.1038/s41598-020-65808-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S. P., Behar M., Birnbaum H. A., Hoffmann A., Wright P. E., Ghosh G. (2013). Analysis of the RelA:CBP/p300 Interaction Reveals Its Involvement in NF-κB-Driven Transcription. PLoS Biol. 11:e1001647. 10.1371/journal.pbio.1001647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Moreno R., Cuesta-Geijo M. Á, Martínez-Romero C., Barrado-Gil L., Galindo I., García-Sastre A., et al. (2016). Antiviral Role of IFITM Proteins in African Swine Fever Virus Infection. PLoS One 11:e0154366. 10.1371/journal.pone.0154366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilan J. G., Zsak L., Lu Z., Kutish G. F., Afonso C. L., Rock D. L. (2002). Novel swine virulence determinant in the left variable region of the African swine fever virus genome. J. Virol. 76 3095–3104. 10.1128/jvi.76.7.3095-3104.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netherton C. L., Simpson J., Haller O., Wileman T. E., Takamatsu H. H., Monaghan P., et al. (2009). Inhibition of a large double-stranded DNA virus by MxA protein. J. Virol. 83 2310–2320. 10.1128/JVI.00781-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netherton C. L., Wileman T. E. (2013). African swine fever virus organelle rearrangements. Virus Res. 173 76–86. 10.1016/j.virusres.2012.12.014 [DOI] [PubMed] [Google Scholar]
- Nunes-Correia I., Rodríguez J. M., Eulálio A., Carvalho A. L., Citovsky V., Simões S., et al. (2008). African swine fever virus p10 protein exhibits nuclear import capacity and accumulates in the nucleus during viral infection. Vet. Microbiol. 130 47–59. 10.1016/j.vetmic.2007.12.010 [DOI] [PubMed] [Google Scholar]
- O’Donnell V., Holinka L. G., Gladue D. P., Sanford B., Krug P. W., Lu X., et al. (2015). African Swine Fever Virus Georgia Isolate Harboring Deletions of MGF360 and MGF505 Genes Is Attenuated in Swine and Confers Protection against Challenge with Virulent Parental Virus. J. Virol. 89 6048–6056. 10.1128/JVI.00554-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oura C. A., Powell P. P., Anderson E., Parkhouse R. M. (1998). The pathogenesis of African swine fever in the resistant bushpig. J. Gen. Virol. 79 1439–1443. 10.1099/0022-1317-79-6-1439 [DOI] [PubMed] [Google Scholar]
- Palermo S., Capra E., Torremorell M., Dolzan M., Davoli R., Haley C. S., et al. (2009). Toll-like receptor 4 genetic diversity among pig populations. Anim. Genet. 40 289–299. 10.1111/j.1365-2052.2008.01833.x [DOI] [PubMed] [Google Scholar]
- Palgrave C. J., Gilmour L., Lowden C. S., Lillico S. G., Mellencamp M. A., Whitelaw C. B. A. (2011). Species-specific variation in RELA underlies differences in NF-κB activity: a potential role in African swine fever pathogenesis. J. Virol. 85 6008–6014. 10.1128/JVI.00331-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugal R., Coelho J., Höper D., Little N. S., Smithson C., Upton C., et al. (2015). Related strains of African swine fever virus with different virulence: genome comparison and analysis. J. Gen. Virol. 96 408–419. 10.1099/vir.0.070508-0 [DOI] [PubMed] [Google Scholar]
- Portugal R., Leitão A., Martins C. (2018). Modulation of type I interferon signaling by African swine fever virus (ASFV) of different virulence L60 and NHV in macrophage host cells. Vet. Microbiol. 216 132–141. 10.1016/j.vetmic.2018.02.008 [DOI] [PubMed] [Google Scholar]
- Powell P. P., Dixon L. K., Parkhouse R. M. (1996). An IkappaB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J. Virol. 70 8527–8533. 10.1128/jvi.70.12.8527-8533.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y., Li H., Qiao J. (2016). TLR2/MyD88/NF-κB signalling pathway regulates IL-8 production in porcine alveolar macrophages infected with porcine circovirus 2. J. Gen. Virol. 97 445–452. 10.1099/jgv.0.000345 [DOI] [PubMed] [Google Scholar]
- Ramirez-Medina E., Vuono E., O’Donnell V., Holinka L. G., Silva E., Rai A., et al. (2019). Differential Effect of the Deletion of African Swine Fever Virus Virulence-Associated Genes in the Induction of Attenuation of the Highly Virulent Georgia Strain. Viruses 11:599. 10.3390/v11070599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Y., Li D., Xiong M. G., Liu H. N., Feng T., Shi Z. W., et al. (2022). African swine fever virus I267L acts as an important virulence factor by inhibiting RNA polymerase III-RIG-I-mediated innate immunity. PLoS Pathog. 18:e1010270. 10.1371/journal.ppat.1010270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathakrishnan A., Connell S., Petrovan V., Moffat K., Goatley L. C., Jabbar T., et al. (2022). Differential Effect of Deleting Members of African Swine Fever Virus Multigene Families 360 and 505 from the Genotype II Georgia 2007/1 Isolate on Virus Replication, Virulence, and Induction of Protection. J. Virol. 96:e0189921. 10.1128/jvi.01899-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis A. L., Abrams C. C., Goatley L. C., Netherton C., Chapman D. G., Sanchez-Cordon P., et al. (2016). Deletion of African swine fever virus interferon inhibitors from the genome of a virulent isolate reduces virulence in domestic pigs and induces a protective response. Vaccine 34 4698–4705. 10.1016/j.vaccine.2016.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis A. L., Goatley L. C., Jabbar T., Lopez E., Rathakrishnan A., Dixon L. K. (2020). Deletion of the Gene for the Type I Interferon Inhibitor I329L from the Attenuated African Swine Fever Virus OURT88/3 Strain Reduces Protection Induced in Pigs. Vaccines 8:262. 10.3390/vaccines8020262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez C. I., Nogal M. L., Carrascosa A. L., Salas M. L., Fresno M., Revilla Y. (2002). African swine fever virus IAP-like protein induces the activation of nuclear factor kappa B. J. Virol. 76 3936–3942. 10.1128/jvi.76.8.3936-3942.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands R. J., Duarte M. M., Boinas F., Hutchings G., Dixon L. K. (2009). The CD2v protein enhances African swine fever virus replication in the tick vector. Ornithodoros erraticus. Virology 393 319–328. 10.1016/j.virol.2009.07.040 [DOI] [PubMed] [Google Scholar]
- Salguero F. J., Gil S., Revilla Y., Gallardo C., Arias M., Martins C. (2008). Cytokine mRNA expression and pathological findings in pigs inoculated with African swine fever virus (E-70) deleted on A238L. Vet. Immunol. Immunopathol. 124 107–119. 10.1016/j.vetimm.2008.02.012 [DOI] [PubMed] [Google Scholar]
- Schmitz M. L., Kracht M., Saul V. V. (2014). The intricate interplay between RNA viruses and NF-κB. Biochim. Biophys. Acta 1843 2754–2764. 10.1016/j.bbamcr.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Hou L., Liu J. (2021). Host immune response to infection with porcine circoviruses. Anim. Dis. 1:23. 10.1186/s44149-021-00027-3 [DOI] [Google Scholar]
- Silk R. N., Bowick G. C., Abrams C. C., Dixon L. K. (2007). African swine fever virus A238L inhibitor of NF-kappaB and of calcineurin phosphatase is imported actively into the nucleus and exported by a CRM1-mediated pathway. J. Gen. Virol. 88 411–419. 10.1099/vir.0.82358-0 [DOI] [PubMed] [Google Scholar]
- Simões M., Freitas F. B., Leitão A., Martins C., Ferreira F. (2019). African swine fever virus replication events and cell nucleus: new insights and perspectives. Virus Res. 270:197667. 10.1016/j.virusres.2019.197667 [DOI] [PubMed] [Google Scholar]
- Simões M., Martins C., Ferreira F. (2013). Host DNA damage response facilitates African swine fever virus infection. Vet. Microbiol. 165 140–147. 10.1016/j.vetmic.2013.01.007 [DOI] [PubMed] [Google Scholar]
- Simões M., Martins C., Ferreira F. (2015a). Early intranuclear replication of African swine fever virus genome modifies the landscape of the host cell nucleus. Virus Res. 210 1–7. 10.1016/j.virusres.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Simões M., Rino J., Pinheiro I., Martins C., Ferreira F. (2015b). Alterations of Nuclear Architecture and Epigenetic Signatures during African Swine Fever Virus Infection. Viruses 7 4978–4996. 10.3390/v7092858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Yu S., Ge H., Wang T., Li Y., Zhou P., et al. (2022). The A137R Protein of African Swine Fever Virus Inhibits Type I Interferon Production via the Autophagy-Mediated Lysosomal Degradation of TBK1. J. Virol. 96:e0195721. 10.1128/jvi.01957-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taffoni C., Steer A., Marines J., Chamma H., Vila I. K., Laguette N. (2021). Nucleic Acid Immunity and DNA Damage Response: new Friends and Old Foes. Front. Immunol. 12:660560. 10.3389/fimmu.2021.660560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatoyan M. R., Ter-Pogossyan Z. R., Semerjyan A. B., Gevorgyan V. S., Karalyan N. Y., Sahakyan C. T., et al. (2019). Serum Concentrations of Vascular Endothelial Growth Factor, Stromal Cell-Derived Factor, Nitric Oxide and Endothelial DNA Proliferation in Development of Microvascular Pathology in Acute African Swine Fever. J. Comp. Pathol. 167 50–59. 10.1016/j.jcpa.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Tay S., Hughey J. J., Lee T. K., Lipniacki T., Quake S. R., Covert M. W. (2010). Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature 466 267–271. 10.1038/nature09145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallée I., Tait S. W., Powell P. P. (2001). African swine fever virus infection of porcine aortic endothelial cells leads to inhibition of inflammatory responses, activation of the thrombotic state, and apoptosis. J. Virol. 75 10372–10382. 10.1128/JVI.75.21.10372-10382.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhou L., Wang J., Su D., Li D., Du Y., et al. (2022). African Swine Fever Virus K205R Induces ER Stress and Consequently Activates Autophagy and the NF-κB Signaling Pathway. Viruses 14:394. 10.3390/v14020394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Ai Q., Huang S., Ou Y., Gao Y., Tong T., et al. (2022). Immune Escape Mechanism and Vaccine Research Progress of African Swine Fever Virus. Vaccines 10:344. 10.3390/vaccines10030344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhang J., Zhang Y., Yang J., Wang L., Qi Y., et al. (2021). Cytokine Storm in Domestic Pigs Induced by Infection of Virulent African Swine Fever Virus. Front. Vet. Sci. 7:601641. 10.3389/fvets.2020.601641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cui S., Xin T., Wang X., Yu H., Chen S., et al. (2021). African Swine Fever Virus MGF360-14L Negatively Regulates Type I Interferon Signaling by Targeting IRF3. Front. Cell. Infect. Microbiol. 11:818969. 10.3389/fcimb.2021.818969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wu J., Wu Y., Chen H., Zhang S., Li J., et al. (2018). Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1. Biochem. Biophys. Res. Commun. 506 437–443. 10.1016/j.bbrc.2018.10.103 [DOI] [PubMed] [Google Scholar]
- Weber F., Wagner V., Rasmussen S. B., Hartmann R., Paludan S. R. (2006). Double-Stranded RNA Is Produced by Positive-Strand RNA Viruses and DNA Viruses but Not in Detectable Amounts by Negative-Strand RNA Viruses. J. Virol. 80 5059–5064. 10.1128/JVI.80.10.5059-5064.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Huang Q., Wang R., Zeng Y., Cheng M., Xue Y., et al. (2021c). African swine fever virus MGF505-11R inhibits type I interferon production by negatively regulating the cGAS-STING-mediated signaling pathway. Vet. Microbiol. 263:109265. 10.1016/j.vetmic.2021.109265 [DOI] [PubMed] [Google Scholar]
- Yang B., Shen C., Zhang D., Zhang T., Shi X., Yang J., et al. (2021a). Mechanism of interaction between virus and host is inferred from the changes of gene expression in macrophages infected with African swine fever virus CN/GS/2018 strain. Virol. J. 18:170. 10.1186/s12985-021-01637-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Li S., Feng T., Zhang X., Yang F., Cao W., et al. (2021b). African Swine Fever Virus F317L Protein Inhibits NF-κB Activation To Evade Host Immune Response and Promote Viral Replication. mSphere 6:e0065821. 10.1128/mSphere.00658-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Xue Y., Niu H., Shi C., Cheng M., Wang J., et al. (2022). African swine fever virus MGF360-11L negatively regulates cGAS-STING-mediated inhibition of type I interferon production. Vet. Res. 53:7. 10.1186/s13567-022-01025-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Yang B., Shen C., Zhang T., Hao Y., Zhang D., et al. (2022). MGF360-9L Is a Major Virulence Factor Associated with the African Swine Fever Virus by Antagonizing the JAK/STAT Signaling Pathway. mBio 13:e0233021. 10.1128/mbio.02330-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Li T., Liu X., Zhang T., Zhang Z., Kang L., et al. (2022). African swine fever virus cysteine protease pS273R inhibits pyroptosis by noncanonically cleaving gasdermin D. J. Biol. Chem. 298:101480. 10.1016/j.jbc.2021.101480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Zhou R., Li S., He S., Luo J., Zhu M., et al. (2020). Porcine IFI16 Negatively Regulates cGAS Signaling Through the Restriction of DNA Binding and Stimulation. Front. Immunol. 11:1669. 10.3389/fimmu.2020.01669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. J., Ramanathan P., Bishop E. A., O’Donnell V., Gladue D. P., Borca M. V. (2019). Mechanisms of African swine fever virus pathogenesis and immune evasion inferred from gene expression changes in infected swine macrophages. PLoS One 14:e0223955. 10.1371/journal.pone.0223955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo Y., Guo Z., Ba T., Zhang C., He L., Zeng C., et al. (2021). African Swine Fever Virus MGF360-12L Inhibits Type I Interferon Production by Blocking the Interaction of Importin α and NF-κB Signaling Pathway. Virol. Sin. 36 176–186. 10.1007/s12250-020-00304-4 [DOI] [PMC free article] [PubMed] [Google Scholar]