Abstract
Since Pseudomonas aeruginosa is capable of biosynthesis of polyhydroxyalkanoic acid (PHA) and rhamnolipids, which contain lipid moieties that are derived from fatty acid biosynthesis, we investigated various fab mutants from P. aeruginosa with respect to biosynthesis of PHAs and rhamnolipids. All isogenic fabA, fabB, fabI, rhlG, and phaG mutants from P. aeruginosa showed decreased PHA accumulation and rhamnolipid production. In the phaG (encoding transacylase) mutant rhamnolipid production was only slightly decreased. Expression of phaG from Pseudomonas putida and expression of the β-ketoacyl reductase gene rhlG from P. aeruginosa in these mutants indicated that PhaG catalyzes diversion of intermediates of fatty acid de novo biosynthesis towards PHA biosynthesis, whereas RhlG catalyzes diversion towards rhamnolipid biosynthesis. These data suggested that both biosynthesis pathways are competitive. In order to investigate whether PhaG is the only linking enzyme between fatty acid de novo biosynthesis and PHA biosynthesis, we generated five Tn5 mutants of P. putida strongly impaired in PHA production from gluconate. All mutants were complemented by the phaG gene from P. putida, indicating that the transacylase-mediated PHA biosynthesis route represents the only metabolic link between fatty acid de novo biosynthesis and PHA biosynthesis in this bacterium. The transacylase-mediated PHA biosynthesis route from gluconate was established in recombinant E. coli, coexpressing the class II PHA synthase gene phaC1 together with the phaG gene from P. putida, only when fatty acid de novo biosynthesis was partially inhibited by triclosan. The accumulated PHA contributed to 2 to 3% of cellular dry weight.
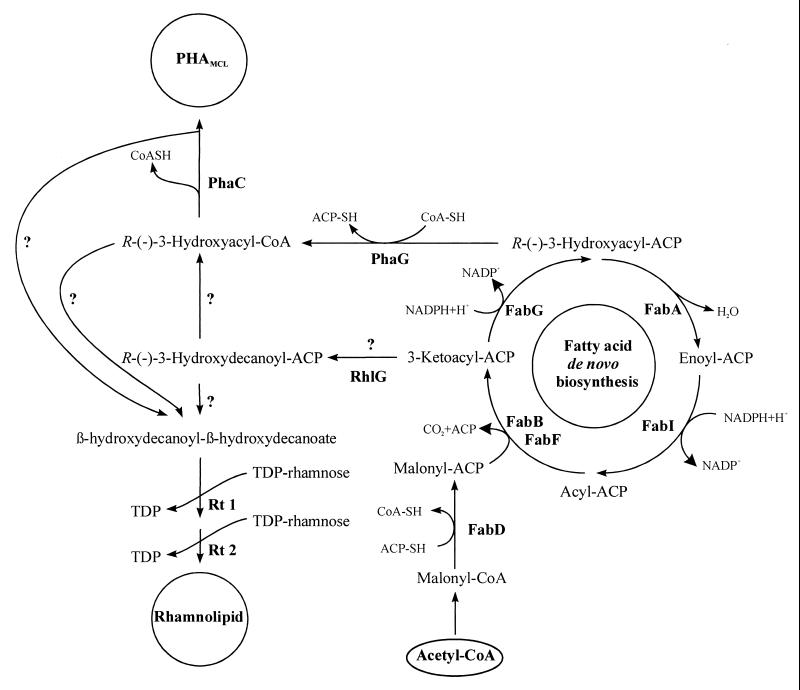
A wide variety of microorganisms accumulate polyhydroxyalkanoic acids (PHAs), mostly polyhydroxybutyrate, as metabolic storage materials, which are deposited as intracellular water-insoluble inclusions (1, 22). Meanwhile, more than 150 constituents of PHAs have been found (38). Recently, it was shown that provision of 3-mercaptopropionic acid as a carbon source resulted in biosynthesis of a novel sulfur-containing polyester with thioester linkages by Ralstonia eutropha (21). Most fluorescent pseudomonads belonging to rRNA homology group I, e.g., Pseudomonas aeruginosa and Pseudomonas putida, are able to synthesize and accumulate large amounts of PHAs consisting of various 3-hydroxy fatty acids with carbon chain lengths ranging from 6 to 14 carbon atoms (medium chain length [MCL] PHAs [PHAMCL]) as carbon and energy storage compounds from cheap carbon sources, e.g., low-rank coal liquefaction products or waste oil from biotechnological rhamnose production (1, 9, 10, 22, 39, 40). The composition of PHA depends on the PHA synthases, the carbon source, and the metabolic routes involved (29, 31, 32). β-Oxidation is the main pathway when fatty acids are used as a carbon source, and fatty acid de novo biosynthesis is the main route during growth on carbon sources which are metabolized to acetyl coenzyme A (acetyl-CoA), like gluconate, acetate, or ethanol (16, 28, 32). Recently, recombinant PHAMCL synthesis was also obtained in β-oxidation mutants of Escherichia coli LS1298 (fadB) or RS3097 (fadR) expressing PHA synthase genes from P. aeruginosa (20, 24, 25), indicating that the β-oxidation pathway in E. coli provides precursors for PHA synthesis. It has also been recently shown that coexpression of the thioesterase genes with a PHA synthase gene in E. coli fad mutants causes synthesis of PHAMCL from the carbon source gluconate (18, 30). These data suggested that the fatty acid de novo synthesis as well as the β-oxidation pathways were involved. It was recently confirmed that the purified PHAMCL synthases from P. aeruginosa exhibit in vitro enzyme activity with (R)-3-hydroxydecanoyl-CoA as the substrate (26). Thus, to serve as a substrate for the PHA synthase, (R)-3-hydroxyacyl-acyl carrier protein [(R)-3-hydroxyacyl-ACP], which is an intermediate of fatty acid de novo synthesis, must be converted to the corresponding CoA-derivative. Recently, the transacylase PhaGPp from P. putida, which catalyzes the transfer of the (R)-3-hydroxydecanoyl moiety from the ACP thioester to CoA, has been identified and characterized (28). Thus, PhaG directly links fatty acid de novo biosynthesis with PHA biosynthesis (Fig. 1). Meanwhile, phaG genes were isolated and characterized from Pseudomonas oleovorans and P. aeruginosa and evidence was obtained that this transacylase-mediated pathway is widespread among pseudomonads (14, 15). Interestingly, in P. aeruginosa about 40% of the accumulated PHA is provided via alternative pathways from gluconate as the carbon source independent of the transacylase PhaG (14). In non-PHA-accumulating Pseudomonas fragi the coexpression of phaGPp with phaC1Pa established a new pathway for PHAMCL biosynthesis from fatty acid de novo biosynthesis using nonrelated carbon sources, e.g., gluconate or acetate (8).
FIG. 1.
Proposed pathways for PHAMCL and rhamnolipid biosynthesis. PhaC, PHA synthase; PhaG, 3-hydroxydecanoyl-ACP–CoA transacylase; RhlG, β-ketoacyl reductase; FabG, β-ketoacyl-ACP reductase; FabA, 3-hydroxydecanoyl-ACP dehydrase; FabB, β-ketoacyl-ACP synthase I; FabF, β-ketoacyl-ACP synthase II; FabI, enoyl-ACP reductase; FabD, malonyl-CoA–ACP transacylase. The question marks indicate hitherto-unconfirmed metabolic routes.
P. aeruginosa is capable of producing various exoproducts, such as exoenzymes, pyocyanine, the exopolysaccharide alginate, and rhamnolipids. Rhamnolipids are glycolipids, which reduce water surface tension and emulsify oil. These rhamnolipids produced by P. aeruginosa in liquid cultures are mainly rhamnosyl-β-hydroxydecanoyl-β-hydroxydecanoate (monorhamnolipid) and rhamnosyl-rhamnosyl-β-hydroxydecanoyl-β-hydroxydecanoate (dirhamnolipids). Rhamnolipid biosynthesis proceeds through transfer of two rhamnose moieties from TDP-l-rhamnose (3). For the synthesis of monorhamnolipid, the enzyme rhamnosyltransferase 1 (Rt 1) catalyzes the rhamnose transfer to β-hydroxydecanoyl-β-hydroxydecanoate, while Rt 2 synthesizes dirhamnolipid from TDP-l-rhamnose and monorhamnolipid. Genes coding for biosynthesis, regulation, and induction of the Rt 1 enzyme are organized in tandem in the rhlABRI gene cluster (23). The gene rhlC, which encodes the Rt 2 enzyme, has been very recently described (27). This enzyme is homologous to rhamnosyltransferases involved in lipopolysaccharide biosynthesis. Recently, Campos-Garcia et al. (4) identified the rhlG gene encoding a β-ketoacyl reductase, which is presumably involved in the biosynthesis of rhamnolipids. RhlG is supposed to catalyze the NADPH-dependent reduction of β-ketodecanoyl-ACP, which is an intermediate of fatty acid de novo biosynthesis, resulting in β-hydroxydecanoyl-ACP, a putative precursor for rhamnolipid biosynthesis (Fig. 1).
Since both PHA and rhamnolipid contain lipid moieties which are derived from fatty acid biosynthesis, we investigated various fab mutants from P. aeruginosa with respect to the biosynthesis of PHA and rhamnolipid. Furthermore, the influence of the transacylase PhaG and the β-ketoacyl reductase RhlG on the synthesis of PHA and rhamnolipid, respectively, was studied. Moreover, we generated Tn5 mutants of P. putida deficient in PHAMCL biosynthesis from gluconate in order to investigate whether other genes are required for this specific pathway. Finally, the transacylase-mediated pathway for PHAMCL synthesis from gluconate was established in recombinant E. coli.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth of bacteria.
Pseudomonads and E. coli strains as well as the plasmids used in this study are listed in Table 1. E. coli was grown at 37°C in complex Luria-Bertani (LB) medium. Pseudomonads were grown at 30°C in 300-ml baffled flasks containing 50 ml of either LB medium, PPGAS medium (NH4Cl, 0.02 M; KCl, 0.02 M; Tris-HCl, 0.12 M; MgSO4, 0.0016 M; glucose, 0.5% [wt/vol]; peptone, 1% [wt/vol] [44]), or mineral salts medium (MM) containing 0.05% (wt/vol) ammonium chloride and a carbon source as indicated (34), and if required antibiotics were added to appropriate concentrations. The amounts of antibiotics used for P. aeruginosa were as follows (per milliliter): 300 μg of carbenicillin, 250 μg of gentamicin, 150 μg of tetracycline, and 300 μg of kanamycin. The amounts of antibiotics used for P. putida were as follows (per milliliter): 10 μg of gentamicin and 50 μg of kanamycin. The amounts of antibiotics used for E. coli were as follows (per milliliter): 50 μg of kanamycin and 100 μg of ampicillin.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| P. putida | ||
| KT2440 | Wild type | 43 |
| GPp104 | PHA synthase negative mutant of P. putida KT2442; mt-2 hsdR1 (r− m+) without TOL plasmid | 17 |
| PHAGN-21 | PhaG-negative mutant of P. putida KT2440 | 28 |
| B349/Tn5-1 | Tn5 mutant, PhaG negative | This study |
| B349/Tn5-2 | Tn5 mutant, PhaG negative | This study |
| B349/Tn5-3 | Tn5 mutant, PhaG negative | This study |
| B349/Tn5-4 | Tn5 mutant, PhaG negative | This study |
| B349/Tn5-5 | Tn5 mutant, PhaG negative | This study |
| P. aeruginosa | ||
| PAO1 | Wild type | ATCC 15692 |
| KO2 | PAO1 with phaG::Gmr | 14 |
| ACP5 | PAO1 with rhlG::Tcr | 4 |
| PAO191 | PAO1 with fabA::Gmr | 12 |
| PAO192 | PAO1 with fabB::Gmr | 12 |
| PAO235 | PAO1 with fabI::FRT | 13 |
| E. coli | ||
| S17-1 | recA; harbors the tra genes of plasmid RP4 in the chromosome; proA thi-1 | 3 |
| JM109 | recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 λ− lac [F′ proAB lacIqZΔM15] | 33 |
| RS3097 | el4- fadR(Ts) zcg-101::Tn10 fabA fadR tyrT mel-1 | 36 |
| DC170 | 6 | |
| IP1111 | Hfr galE45 fabI(Ts) relA1 spoT1 | 42 |
| Plasmids | ||
| pBBR1MCS-2 | Kmr; broad host range; lacPOZ′ | 19 |
| pBHR81 | pBBR1MCS-2 containing coding region of phaG gene from P. putida downstream of lac promoter | 28 |
| pUCP20 | Cbr; broad host range; lacPOZ′ | 35 |
| pJC3 | pUCP20 containing rhlG gene from P. aeruginosa PAO1 | 4 |
| pBHR71 | pBluescript SK(−) containing phaC1 gene from P. aeruginosa PAO1 | 20 |
| pBHR75 | pUCP27 containing phaG gene from P. putida KT2440 | 28 |
| pBHR86 | pBBR1MCS-2 containing coding region of phaC1 gene from P. aeruginosa downstream of lac promoter and the coding region of phaG from P. putida downstream of phaC1 including the native promoter | 8 |
| pBHR87 | pBBR1MCS-2 containing coding region of phaC1 gene from P. aeruginosa downstream of lac promoter and the coding region of phaG from P. putida downstream of phaC1 | This study |
Isolation, analysis, and manipulation of DNA.
DNA sequences of new plasmid constructs were confirmed by DNA sequencing according to the chain termination method using the model 4000L automatic sequencer LI-COR (MWG-Biotech, Ebersberg, Germany). All other genetic techniques were performed as described by Sambrook et al. (33).
Tn5 mutagenesis.
In order to generate mutants of P. putida, which are defective in PHAMCL biosynthesis from nonrelated carbon sources, such as gluconate, we performed Tn5 mutagenesis. The suicide plasmid pMON5302 (Monsanto, St. Louis, Mo.) was constructed by insertion of the Tn5 IS50L and IS50R regions comprising a gentamicin resistance cassette [AAC(3)-I gene] into plasmid pACYC (Monsanto). The plasmid was transferred into P. putida KT2440 by conjugation as previously described (27), and Tn5 mutants were screened on MM agar plates containing 1.5% (wt/vol) gluconate as the sole carbon source. Colonies which appeared nonopaque were isolated, and after cultivation in the same medium the cells were analyzed with respect to PHA accumulation.
Plasmid construction.
The coding region of the P. putida phaGPp gene was amplified by tailored PCR using primers with noncomplementary 5′ ends, introducing BamHI and XbaI restriction sites at either end of the PCR product by using plasmid pBHR75 as the template (28). The coding region of the PHA synthase gene phaClPa from P. aeruginosa was amplified by tailored PCR using plasmid pBHR71 as the template (20), introducing the EcoRI restriction site and the ribosome binding site at the 5′ end and the BamHI restriction site at the 3′ end. The following oligonucleotides were applied for the PCRs: 5′-CCCGAATTCAATAAGGAGATATACATATGAGTCAG-3′ (5′ end) and 5′-TGCTCTAGAGGGCCCCCCCTCGAGGTC-3′ (3′ end) (phaClPa); and 5′-CGCGGATCCAAGGAGTCGATGACATG-3′ (5′ end) and 5′-GCGTCTAGACTACAAGGCGCCGAGCCG-3′ (3′ end) (phaGPp). Both PCR products were simultaneously subcloned into restriction sites EcoRI and XbaI of the vector pBBR1MCS-2, resulting in the insertion of the PHA synthase gene and the transacylase gene collinear to the lac promoter. The resulting plasmid, pBHR87, enabled functional coexpression of phaClPa and phaGPp.
Functional expression of PHAMCL synthase gene.
PHA synthase activity was confirmed by expression of the respective PHA synthase gene in various metabolic backgrounds favoring PHAMCL synthesis, e.g., E. coli RS3097 and P. putida GPp104 (25, 26). Recombinant bacteria harboring the respective plasmid were cultivated in the presence of 0.25% (wt/vol) decanoate. PHA accumulation was determined by gas chromatography (GC) analysis of lyophilized cells and indicated in vivo PHA synthase activity.
Functional expression of PhaG (transacylase) gene.
Functional expression of phaG [encoding the (R)-3-hydroxydecanoyl-CoA–ACP transacylase] based on pBHR86 or pBHR87 was confirmed by complementation of phaG mutants P. aeruginosa KO2 and P. putida PhaGN-21 and establishment of the PhaG-mediated pathway in P. oleovorans (8, 14, 28). Recombinant cells were cultivated in MM plus 1.5% (wt/vol) sodium gluconate, and after 48 h of incubation at 30°C the PHA content of lyophilized cells was determined by GC analysis. PHA accumulation from gluconate indicated in vivo activity of PhaG.
Functional expression of Rh1G (β-ketoacyl reductase) gene.
Functional expression of rhlG (encoding the β-ketoacyl reductase) based on pJC3 was confirmed by complementation of the rhlG mutant P. aeruginosa ACP5 (4). Recombinant cells were cultivated on PPGAS medium, and after 24 h of incubation at 37°C the rhamnolipid concentration in the cell supernatant was determined and indicated in vivo activity of β-ketoacyl reductase.
GC analysis of polyester and fatty acids in cells.
PHAs and fatty acids were qualitatively and quantitatively analyzed by GC. Liquid cultures were centrifuged at 10,000 × g for 15 min, and then the cells were washed twice in saline and lyophilized overnight. Lyophilized cell material (8 to 10 mg) was subjected to methanolysis in the presence of 15% (vol/vol) sulfuric acid. The resulting methyl esters of the constituent 3-hydroxyalkanoic acids were assayed by GC according to the method of Brandl et al. (2) and as described in detail recently (40). GC analysis was performed by injecting 3 μl of sample into a Perkin-Elmer (Überlingen, Germany) 8420 gas chromatograph using a 0.5-μm-diameter Permphase PEG 25 Mx capillary column 60 m in length.
Analysis of rhamnolipids.
The orcinol assay (5) was used to directly assess the amount of rhamnolipids in the sample: 333 μl of the culture supernatant was extracted twice with 1 ml of diethyl ether. The ether fractions were pooled and evaporated to dryness, and 0.5 ml of H2O was added. To 100 μl of each sample 900 μl of a solution containing 0.19% orcinol (in 53% [vol/vol] H2SO4) was added; after being heated for 30 min at 80°C, the samples were cooled for 15 min at room temperature, and the A421 was measured. The concentration of rhamnolipids was calculated by comparing the data with those obtained with rhamnose standards between 0 and 50 μg/ml.
RESULTS
Analysis of various isogenic P. aeruginosa mutants with respect to PHA and rhamnolipid synthesis.
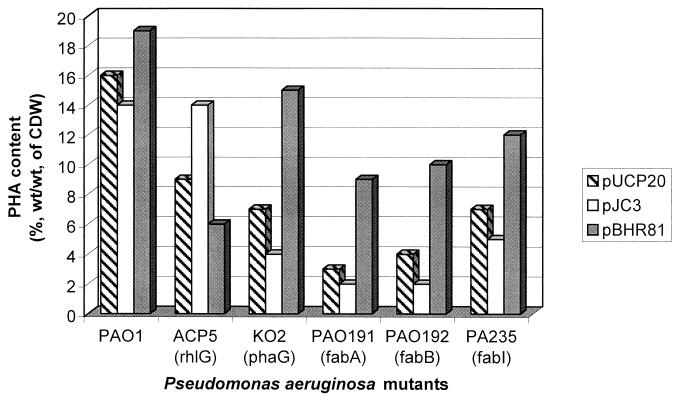
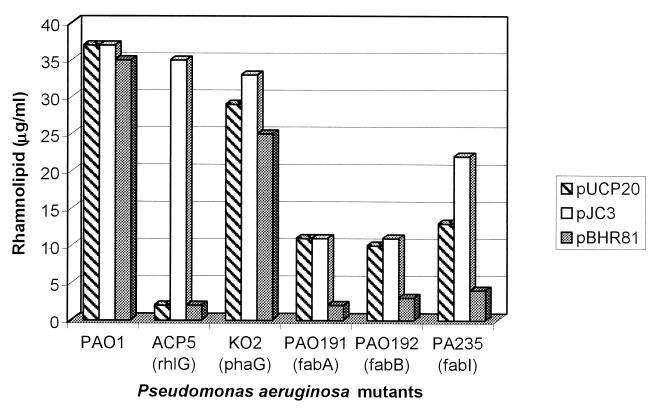
Since precursors for PHA and rhamnolipid biosynthesis are derived from fatty acid de novo biosynthesis, various fab mutants of P. aeruginosa were analyzed. For PHA biosynthesis analysis cells were cultivated under PHA-accumulating conditions on MM containing 1.5% (wt/vol) sodium gluconate, 0.01% (vol/vol) oleate, and 0.05% (wt/vol) ammonium chloride. For rhamnolipid biosynthesis analyses cells were cultivated in PPGAS medium containing 0.5% (wt/vol) glucose as the carbon source. The fabA mutant PAO191 carries a Gmr cassette in the fabA gene, which encodes β-hydroxyacyl-ACP dehydratase. The fabB mutant PAO192 was constructed by insertion of the Gmr cassette into the fabB gene, which encodes β-ketoacyl-ACP synthase I (12). Moreover, the isogenic fabI mutant PAO235 was employed, which carries an insertionally inactivated fabI gene that encodes enoyl-ACP reductase (13). In addition to the various fab mutants, we used the isogenic phaG mutant KO2, which is impaired in PHAMCL accumulation from nonrelated carbon sources (14), and the isogenic rhlG mutant ACP5, which is almost deficient of rhamnolipid production, i.e., it produced less than 1.3% of the rhamnolipid produced by the wild type (4). These mutants were studied with respect to their capability to produce PHAMCL and rhamnolipids. The fab mutants were strongly impaired in PHAMCL accumulation and rhamnolipid production, whereas the the phaG mutant showed only 40% of wild-type PHAMCL accumulation and only a slightly decreased rhamnolipid production (Fig. 2 and 3). The rhlG mutant showed a decreased PHAMCL accumulation and rhamnolipid production. Transfer of the phaG gene into the fab mutants showed a strong increase in PHAMCL accumulation, whereas rhamnolipid biosynthesis was almost abolished (Fig. 2 and 3). In the rhlG mutant PHAMCL accumulation was slightly decreased and no effect on rhamnolipid synthesis was observed when phaG was expressed (Fig. 2 and 3). Transfer of the rhlG gene into these mutants mediated an increase of rhamnolipid production in mutants ACP5 (complementation of mutation), KO2, and PA235, whereas PHAMCL accumulation was decreased in all mutants except in ACP5 (Fig. 2 and 3). In order to obtain evidence for the potential precursor of PHAMCL synthesis, the PHAMCL composition of all mutants was analyzed. GC analysis of the accumulated PHA showed that in the phaG knockout mutant KO2 the molar fraction of the 3-hydroxydecanoate (3HD) in PHAMCL was decreased by approximately 20%, whereas in the rhlG mutant ACP5 this molar fraction was slightly increased (Table 2).
FIG. 2.
PHAMCL accumulation by mutants of P. aeruginosa from gluconate. Cultivations were performed under PHA-accumulating conditions on MM containing 1.5% (wt/vol) sodium gluconate, 0.01% (vol/vol) oleate, and 0.05% (wt/vol) ammonium chloride. Cells were grown for 48 h at 37°C. PHA content and composition of comonomers were analyzed by GC. The gene affected by each mutation is given in parentheses. CDW, cellular dry weight.
FIG. 3.
Rhamnolipid production by mutants of P. aeruginosa. Rhamnolipid concentration is expressed as micrograms of rhamnose in rhamnolipids per milliliter of culture supernatant. The PPGAS medium contained 0.5% (wt/vol) glucose as a carbon source. The gene affected by each mutation is given in parentheses.
TABLE 2.
Accumulation of PHAs in various P. aeruginosa mutants from gluconatea
| Strain | PHA content (% [wt/wt]b) | Composition of PHA (mol%)c
|
||||
|---|---|---|---|---|---|---|
| 3HHx | 3HO | 3HD | 3HDD | 3HDD:1 | ||
| PAO1 | 18.5 | 5 | 20 | 63 | 12 | NDd |
| KO2 (phaG)e | 7 | 4 | 32 | 50 | 14 | ND |
| ACP5 (rhlG)e | 9 | 6 | 20 | 70 | 4 | ND |
| PAO191 (fabA)e | 3 | 5 | 13 | 65 | 17 | ND |
| PAO192 (fabB)e | 3.5 | 2 | 18 | 66 | 16 | ND |
| PA235 (fabI)e | 7 | 5 | 24 | 63 | 8 | ND |
Cultivations were performed under PHA-accumulating conditions on MM containing 1.5% (wt/vol) sodium gluconate and 0.05% (wt/vol) ammonium chloride. Cells were grown for 48 h at 30°C. PHA content and composition of comonomers were analyzed by GC.
Dry weight.
Abbreviations: 3HHx, 3-hydroxy-hexanoate; 3HO, 3-hydroxyoctanoate; 3HDD:1, 3-hydroxydodecenoate.
ND, not detectable.
Gene affected by insertional inactivation.
Generation of independent Tn5 mutants of P. putida deficient in PHAMCL accumulation.
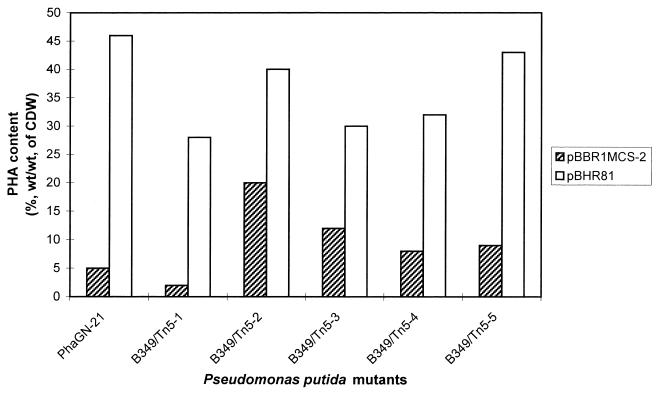
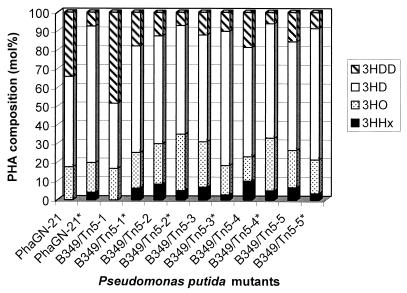
Since the transacylase PHA biosynthesis pathway was described in Pseudomonas, the only gene involved that has been identified so far is phaG, capable of restoring PHAMCL biosynthesis in the N-methyl-N′-nitro-N-nitrosoguanidine mutant PhaGN-21 of P. putida, which was strongly impaired in PHAMCL synthesis from nonrelated carbon sources. The establishment of this pathway in non-Pseudomonas species has not been achieved yet. In order to further analyze this possibility, we generated independent Tn5 mutants of P. putida. For this, we constructed the suicide plasmid pMON5302, which enabled Tn5-mediated random insertion of a Gmr cassette into the P. putida chromosome. After transfer of plasmid pMON5302 into P. putida, cells were screened on MM containing gluconate as the sole carbon source. Five nonopaque mutants (B349/Tn5-1 to -5) were isolated that were strongly impaired in PHAMCL biosynthesis from nonrelated carbon sources but accumulated significantly higher levels of PHAMCL from decanoate as the carbon source (data not shown). Transfer of plasmid pBHR81, which carries the phaGPp gene under the lac promoter's control, into these Tn5 mutants showed restoration of PHAMCL accumulation from nonrelated carbon sources comparable to the level of PHAMCL accumulation from decanoate as the carbon source (Fig. 4). Analysis of PHAMCL composition by GC-mass spectrometry showed that the molar fraction of 3HD was decreased, whereas the molar fraction of 3-hydroxydodecanoate (3HDD) was increased in the Tn5 mutants when compared with that in wild-type P. putida (Fig. 5). Functional expression of phaGPp in these mutants again enhanced the molar fraction of 3HD and decreased the molar fraction of 3HDD of the accumulated PHAMCL.
FIG. 4.
PHAMCL accumulation by Tn5 mutants of P. putida. Cultivations were performed under PHA-accumulating conditions on MM containing 1.5% (wt/vol) sodium gluconate and 0.05% (wt/vol) ammonium chloride. Cells were grown for 48 h at 30°C. PHA content and composition of comonomers were analyzed by GC. Plasmid pBHR81 enables functional expression of the phaGPp gene. CDW, cellular dry weight.
FIG. 5.
Composition of PHAMCL accumulated by Tn5 mutants of P. putida. Cultivations were performed under PHA-accumulating conditions on MM containing 1.5% (wt/vol) sodium gluconate and 0.05% (wt/vol) ammonium chloride. Cells were grown for 48 h at 30°C. PHA content and composition of comonomers were analyzed by GC. 3HHx, 3-hydroxy-hexanoate; 3HO, 3-hydroxyoctanoate. An asterisk indicates that cells harbor plasmid pBHR81. Plasmid pBHR81 enables functional expression of the phaGPp gene.
Establishment of transacylase-mediated PHAMCL biosynthesis in recombinant E. coli.
We recently reported the establishment of the transacylase-mediated PHAMCL biosynthesis pathway in the non-PHA accumulating P. fragi by functional expression of phaGPp plus phaC1Pa using plasmid pBHR86 (8). These data clearly demonstrated that diversion of intermediates from fatty acid β-oxidation is not required to establish the transacylase-mediated pathway. Therefore, we introduced plasmid pBHR86, which contains phaClPa and phaGPp collinear to the lac promoter with phaGPp still preceded by its native promoter, into E. coli JM109. Cells were cultivated either in LB medium or M9 medium containing gluconate as the sole carbon source. No PHAMCL accumulation was observed. To avoid transcriptional deficiency of phaGPp in E. coli due to its native promoter, we constructed plasmid pBHR87, containing the phaGPp gene without its native promoter. However, plasmid pBHR87 did not mediate PHAMCL biosynthesis in E. coli JM109. Since (R)-3-hydroxyacyl-ACP, an intermediate of fatty acid de novo biosynthesis and substrate for the transacylase PhaG, has to be available for PHAMCL biosynthesis from nonrelated carbon sources, we employed two E. coli fab mutants, which might contain higher levels of (R)-3-hydroxyacyl-ACP. The fabA mutant E. coli DC170 and the fabI mutant E. coli IP1111 were used to establish the transacylase-mediated pathway. However, transfer of plasmids pBHR86 and pBHR87 into each of the mutants, respectively, alone did not mediate PHAMCL accumulation. Therefore, we used inhibitors of fatty acid de novo biosynthesis in order to generate an intermediate pool, which might favor provision of the substrate for the transacylase. We employed cerulenin, which specifically inhibits FabB (β-ketoacyl-ACP synthase I) and FabF (β-ketoacyl-ACP synthase II), which catalyze the condensation of malonyl-ACP with acyl-ACP (7). The application of the inhibitor cerulenin did not show any effect on PHAMCL synthesis from nonrelated carbon sources in recombinant E. coli S17-1. However, application of triclosan, which specifically inhibits the enoyl-ACP reductase (11), led to PHAMCL accumulation contributing to about 2 to 3% of cellular dry weight in recombinant E. coli harboring either pBHR86 or pBHR87, when grown on LB medium plus gluconate as the carbon source (Table 3).
TABLE 3.
Analysis of recombinant E. coli S17-1 harboring plasmids with respect to PHA accumulationa
| E. coli-borne plasmid | Composition of PHA (% dry wt)b
|
|||
|---|---|---|---|---|
| 3HB | 3HO | 3HD | 3HDD | |
| pBBR1MCS-1 | ND | ND | ND | ND |
| pBHR86 | ND | ND | 3.3 | ND |
| pBHR87 | ND | ND | 1.5 | ND |
Recombinant E. coli was cultivated in LB medium containing 1.5% (wt/vol) gluconate, 1 mM isopropyl-β-d-thiogalactopyranoside and kanamycin (50 μg/ml). After 24 h of cultivation at 37°C, triclosan was added to the medium at a final concentration of 0.1 μg/ml, and cells were harvested after a further incubation of 24 h.
Abbreviations: 3HB, 3-hydroxybutyrate; 3HDD, 3-hydroxydodecanoate.
DISCUSSION
In this study, we evaluated the role of fatty acid de novo biosynthesis for PHAMCL synthesis and rhamnolipid production, particularly considering the role of the linking enzymes PhaG, transacylase, and Rh1G, β-ketoacyl reductase. Analysis of PHAMCL synthesis and rhamnolipid production in various isogenic fab mutants of P. aeruginosa, impaired in fatty acid de novo biosynthesis, strongly suggested that precursors for PHAMCL biosynthesis and rhamnolipid biosynthesis are provided via fatty acid de novo biosynthesis. This is consistent with the observation that the PHAMCL biosynthesis transacylase PhaG and the rhamnolipid biosynthesis β-ketoacyl reductase Rh1G use intermediates of fatty acid de novo biosynthesis as the substrate, which are converted by the respective enzyme to a direct precursor of PHAMCL or rhamnolipid biosynthesis, respectively (4, 28). It is still not clear why the rhlG mutant ACP5 is strongly impaired in PHAMCL biosynthesis and why the phaG mutant KO2 showed a slightly decreased rhamnolipid production. Functional expression of phaGPp in the fab mutants enhanced carbon flux towards PHAMCL biosynthesis, whereas rhamnolipid production was almost abolished, which indicated that PhaG catalyzes conversion of a molecule that plays a role in rhamnolipid biosynthesis. Interestingly, expression of phaGPp in P. aeruginosa KO2 (phaG mutant) did not abolish rhamnolipid biosynthesis, suggesting that the original genomic phaG gene is required in addition to phaGPp gene copies, provided by plasmid pBHR81, for efficient diversion of intermediates towards PHA biosynthesis. The finding that expression of rhlGPa decreased PHAMCL accumulation in all mutants, except the ACP5 mutants, indicated that PHAMCL biosynthesis and rhamnolipid biosynthesis interfere with each other, presumably by competing for intermediates. Mutant ACP5 (rhlG) harboring plasmid pJC3 (rhlGPa) might not exhibit the same phenotype, because of the missing genomic rhlGPa gene.
Five independent Tn5 mutants of P. putida which are deficient in PHAMCL accumulation from nonrelated carbon sources were all at least to some extent complemented by constitutive expression of phaG, which indicated that PhaG is the only key enzyme linking fatty acid de novo biosynthesis with PHAMCL biosynthesis. The compositional analysis of the PHAMCL accumulated by the Tn5 mutants suggested that PhaG contributed to PHAMCL biosynthesis by strong provision of 3-hydroxydecanoyl–CoA, presumably reflecting the substrate specificity of PhaG. Since the transacylase PhaG seems to be the only required enzyme for PHAMCL biosynthesis from nonrelated carbon sources, and since fatty acid de novo biosynthesis plays a crucial role in the provision of precursors for PHAMCL biosynthesis from nonrelated carbon sources, we employed fab mutants of E. coli as well as specific inhibitors of fatty acid de novo biosynthesis in order to establish the transacylase-mediated route in recombinant E. coli. However, only the application of triclosan, a specific inhibitor of the enoyl-ACP reductase FabI, enabled weak PHAMCL accumulation from nonrelated carbon sources in E. coli when phaGPp and phaClPa were functionally expressed. Overall, this study indicates that carbon flux through the fatty acid de novo biosynthesis, i.e., the pool of intermediates, is crucial for PHAMCL biosynthesis as well as rhamnolipid production and that fatty acid de novo biosynthesis in pseudomonads is different from that in E. coli.
ACKNOWLEDGMENTS
This study was supported by research grant Re1097/4-1 from the Deutsche Forschungsgemeinschaft.
We acknowledge the provision of triclosan as a gift by Ciba Spezialitätenchemie AG (Basel, Switzerland). We also thank H. P. Schweizer for provision of the P. aeruginosa fab mutants and G. Soberon-Chavez for provision of plasmid pJC3 as well as the P. aeruginosa rhlG mutant.
REFERENCES
- 1.Anderson A J, Haywood G W, Dawes E A. Biosynthesis and composition of bacterial poly(hydroxyalkanoates) Int J Biol Macromol. 1990;12:102–105. doi: 10.1016/0141-8130(90)90060-n. [DOI] [PubMed] [Google Scholar]
- 2.Brandl H, Gross R A, Lenz R W, Fuller R C. Pseudomonas oleovorans as a source of poly(3-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl Environ Microbiol. 1988;54:1977–1982. doi: 10.1128/aem.54.8.1977-1982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burger M M, Glaser L, Burton R M. The enzymatic synthesis of rhamnose-containing glycolipids by extracts of Pseudomonas aeruginosa. J Biol Chem. 1963;238:2595–2602. [PubMed] [Google Scholar]
- 4.Campos-Garcia J, Caro A D, Najera R, Miller-Maier R M, Al-Tahhan R A, Soberon-Chavez G. The Pseudomonas aeruginosa rhlG gene encodes an NADPH-dependent β-ketoacyl reductase which is specifically involved in rhamnolipid synthesis. J Bacteriol. 1998;180:4442–4451. doi: 10.1128/jb.180.17.4442-4451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandrasekan E V, Bemiller J N. Constituent analyses of glycosaminoglycans. In: Whistler R L, editor. Methods in carbohydrate chemistry. New York, N.Y: Academic Press, Inc.; 1980. pp. 89–96. [Google Scholar]
- 6.Clark D P, DeMendoza D, Polacco M, Cronan J E. Beta-hydroxydecanoyl thioester dehydrase does not catalyze a rate-limiting step in Escherichia coli unsaturated fatty acid synthesis. Biochemistry. 1983;22:5897–5900. doi: 10.1021/bi00294a032. [DOI] [PubMed] [Google Scholar]
- 7.D'Agnolo G, Rosenfeld I S, Awaya J, Omura S, Vagelos P R. Inhibition of fatty acid synthesis by the antibiotic cerulenin. Specific inactivation of beta-ketoacyl-acyl carrier protein synthetase. Biochim Biophys Acta. 1973;326:155–166. doi: 10.1016/0005-2760(73)90241-5. [DOI] [PubMed] [Google Scholar]
- 8.Fiedler S, Steinbüchel A, Rehm B H A. PhaG-mediated synthesis of poly(3-hydroxyalkanoates) consisting of medium-chain-length constituents from nonrelated carbon sources in recombinant Pseudomonas fragi. Appl Environ Microbiol. 2000;66:2117–2124. doi: 10.1128/aem.66.5.2117-2124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Füchtenbusch B, Steinbüchel A. Biosynthesis of polyhydroxyalkanoates from low-rank coal liquefaction products by Pseudomonas oleovorans and Rhodococcus ruber. Appl Microbiol Biotechnol. 1999;52:91–95. doi: 10.1007/s002530051492. [DOI] [PubMed] [Google Scholar]
- 10.Füchtenbusch B, Wullbrandt D, Steinbüchel A. Production of polyhydroxyalkanoic acids by Ralstonia eutropha and Pseudomonas oleovorans from an oil remaining from biotechnological rhamnose production. Appl Microbiol Biotechnol. 2000;53:167–172. doi: 10.1007/s002530050004. [DOI] [PubMed] [Google Scholar]
- 11.Heath R J, Yu Y T, Shapiro M A, Olson E, Rock C O. Broad spectrum antimicrobial biocides target the FabI component of fatty acid synthesis. J Biol Chem. 1998;273:30316–30320. doi: 10.1074/jbc.273.46.30316. [DOI] [PubMed] [Google Scholar]
- 12.Hoang T T, Schweizer H P. Fatty acid biosynthesis in Pseudomonas aeruginosa: cloning and characterization of the fabAB operon encoding beta-hydroxyacyl-acyl carrier protein dehydratase (FabA) and beta-ketoacyl-acyl carrier protein synthase I (FabB) J Bacteriol. 1997;179:5326–5332. doi: 10.1128/jb.179.17.5326-5332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang T T, Schweizer H P. Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (FabI): a target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J Bacteriol. 1999;181:5489–5497. doi: 10.1128/jb.181.17.5489-5497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann N, Steinbüchel A, Rehm B H A. The Pseudomonas aeruginosa phaG gene product is involved in the synthesis of polyhydroxyalkanoic acid consisting of medium-chain-length constituents from non-related carbon sources. FEMS Microbiol Lett. 2000;184:253–259. doi: 10.1111/j.1574-6968.2000.tb09023.x. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann N, Steinbüchel A, Rehm B H A. Polyhydroxyalkanoate synthesis in Pseudomonas oleovorans from simple carbon sources: homologous functional expression of cryptic phaG establishes a transacylase-mediated pathway. Appl Microbiol Biotechnol. 2000;54:665–670. doi: 10.1007/s002530000441. [DOI] [PubMed] [Google Scholar]
- 16.Huijberts G N M, de Rijk T C, de Waard P, Eggink G. 13C-nuclear magnetic resonance studies of Pseudomonas putida fatty acid metabolic routes involved in poly(3-hydroxyalkanoate) synthesis. J Bacteriol. 1994;176:1661–1666. doi: 10.1128/jb.176.6.1661-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huisman G W, Wonink E, Meima R, Kazemier B, Terpstra P, Witholt B. Metabolism of poly(3-hydroxyalkanoates) by Pseudomonas oleovorans: identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J Biol Chem. 1991;266:2191–2198. [PubMed] [Google Scholar]
- 18.Klinke S, Ren Q, Witholt B, Kessler B. Production of medium-chain length poly(3-hydroxyalkanoates) from gluconate by recombinant Escherichia coli. Appl Environ Microbiol. 1999;65:540–548. doi: 10.1128/aem.65.2.540-548.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop R M, 2nd, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBRIMCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 20.Langenbach S, Rehm B H A, Steinbüchel A. Functional expression of the PHA synthase gene phaC1 from Pseudomonas aeruginosa in Escherichia coli results in poly(3-hydroxyalkanoate) synthesis. FEMS Microbiol Lett. 1997;150:303–309. doi: 10.1016/s0378-1097(97)00142-0. [DOI] [PubMed] [Google Scholar]
- 21.Lütke-Eversloh T, Bergander K, Luftmann H, Steinbüchel A. Identification of a new class of biopolymer: bacterial synthesis of a sulfur-containing polymer with thioester linkages. Microbiology. 2001;147:11–19. doi: 10.1099/00221287-147-1-11. [DOI] [PubMed] [Google Scholar]
- 22.Madison L L, Huisman G W. Metabolic engineering of poly(3-hydroxy-alkanoates): from DNA to plastic. Microbiol Mol Biol Rev. 1999;63:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochser U, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi Q, Rehm B H A, Steinbüchel A. Synthesis of poly(3-hydroxyalkanoates) in Escherichia coli expressing the PHA synthase gene phaC2 from Pseudomonas aeruginosa: comparison of PhaC1 and PhaC2. FEMS Microbiol Lett. 1997;157:155–162. doi: 10.1111/j.1574-6968.1997.tb12767.x. [DOI] [PubMed] [Google Scholar]
- 25.Qi Q, Steinbüchel A, Rehm B H A. Metabolic routing towards polyhydroxyalkanoic acid synthesis in recombinant Escherichia coli (fadR): inhibition of fatty acid β-oxidation by acrylic acid. FEMS Microbiol Lett. 1998;167:89–94. doi: 10.1111/j.1574-6968.1998.tb13212.x. [DOI] [PubMed] [Google Scholar]
- 26.Qi Q, Steinbüchel A, Rehm B H A. In vitro synthesis of poly(3-hydroxydecanoate): purification and enzymatic characterization of type II polyhydroxyalkanoate synthases PhaC1 and PhaC2 from Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2000;54:37–43. doi: 10.1007/s002530000357. [DOI] [PubMed] [Google Scholar]
- 27.Rahim, R., C. Olivera, M. Graninger, P. Messner, U. A. Ochsner, J. S. Lam, and G. Soberon-Chavez. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for dirhamnolipid biosynthesis. Mol. Microbiol., in press. [DOI] [PubMed]
- 28.Rehm B H A, Krüger N, Steinbüchel A. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. J Biol Chem. 1998;273:24044–24051. doi: 10.1074/jbc.273.37.24044. [DOI] [PubMed] [Google Scholar]
- 29.Rehm B H A, Steinbüchel A. Biochemical and genetic analysis of PHA synthases and other proteins required for PHA synthesis. Int J Biol Macromol. 1999;13:83–88. doi: 10.1016/s0141-8130(99)00010-0. [DOI] [PubMed] [Google Scholar]
- 30.Rehm B H A, Steinbüchel A. Heterologous expression of the acyl-acyl carrier protein thioesterase gene from the plant Umbellularia californica mediates polyhydroxyalkanoate biosynthesis in recombinant Escherichia coli. Appl Microbiol Biotechnol. 2001;55:205–209. doi: 10.1007/s002530000541. [DOI] [PubMed] [Google Scholar]
- 31.Rehm, B. H. A., and A. Steinbüchel. PHA synthases—the key enzyme of PHA synthesis. In A. Steinbüchel and Y. Doi (ed.), Biopolymers, in press. Wiley-VCH, Heidelberg, Germany.
- 32.Rehm, B. H. A., N. Hoffmann, Q. Qi, S. Fiedler, and A. Steinbüchel. Biosynthesis of latex-like polyhydroxyalkanoates. In Proceedings of the International Symposium “More Quality of Life by Means of Biotechnology: the Bioconversion of Renewable Raw Materials,” in press. GBF, Braunschweig, Germany.
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Schlegel H G, Kaltwasser H, Gottschalk G. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- 35.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 36.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 37.Simons R W, Egan P A, Chute H T, Nunn W D. Regulation of fatty acid degradation in Escherichia coli: isolation and characterization of strains bearing insertion and temperature-sensitive mutations in gene fadR. J Bacteriol. 1980;142:621–632. doi: 10.1128/jb.142.2.621-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinbüchel A, Valentin H E. Diversity of microbial polyhydroxyalkanoic acids. FEMS Microbiol Lett. 1995;128:219–228. [Google Scholar]
- 39.Steinbüchel A, Füchtenbusch B, Gorenflo V, Hein S, Jossek R, Langenbach S, Rehm B H A. Biosynthesis of polyester in bacteria and recombinant organism. Polym Degrad Stabl. 1997;59:177–182. [Google Scholar]
- 40.Steinbüchel A, Füchtenbusch B. Bacterial and other biological systems for polyester production. Trends Biotechnol. 1998;16:419–427. doi: 10.1016/s0167-7799(98)01194-9. [DOI] [PubMed] [Google Scholar]
- 41.Timm A, Steinbüchel A. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl Environ Microbiol. 1990;56:3360–3367. doi: 10.1128/aem.56.11.3360-3367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turnowsky F, Fuchs K, Jeschek C, Hoegenauer G. envM genes of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1989;171:6555–6565. doi: 10.1128/jb.171.12.6555-6565.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Worsey M J, Williams P A. Metabolism of toluene and the xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Miller R M. Enhanced octadecane dispersion and biodegradation by a Pseudomonas rhamnolipid surfactant (biosurfactant) Appl Environ Microbiol. 1992;58:3276–3282. doi: 10.1128/aem.58.10.3276-3282.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]