Summary
Existing global regionalization schemes for plants consider the compositional affinities among biotas, but these have not explicitly considered phylogenetic information. Here, we present for the first time, a phytogeographical delineation of the global vascular flora based on species‐level evolutionary relationships.
We analysed 8737 820 geographical occurrence records for vascular plants together with a time‐calibrated phylogeny including 67 269 species. We constructed a global phylogenetic regionalization by estimating species composition and phylogenetic beta diversity among 200 km × 200 km grid cells across the world.
We identified de novo 16 phytogeographical units that are deeply split into two clusters: Laurasian and Gondwanan. Our regionalization broadly matches previous schemes, but also highlights the separation of the Gondwanan biota into an Holotropical cluster and an Australian–Neozealandic–Patagonian cluster. In contrast, no clear split among Laurasian and Gondwanan biotas was retrieved when omitting phylogenetic information.
The integration of phylogenetic and geographical information provides new insights into the delineation of phytogeographical areas and their historical relationships, enabling the identification of three large, clearly differentiated biotas, here referred to as kingdoms: Holarctic, Holotropical, and Austral. Our results provide further evidence for delineating transition zones and show a clear latitudinal pattern of increasing evolutionary distinctiveness towards the poles.
Keywords: biogeographical regionalization, cluster analysis, evolutionary distinctiveness, phylogenetic beta diversity, vascular flora
Introduction
Each species has a unique geographic distribution, but many species show similar geographic ranges. Shared ranges between two (or more) species may be due to a common evolutionary history, physical barriers to dispersal, or ecological requirements that limit survival (Lomolino et al., 2004; Posadas et al., 2006). In many cases, this translates into closely related species being distributed within the same regions more often than expected by chance, which in turn results in distinct lineages having nonrandom and spatially clustered distribution ranges (Lomolino et al., 2004). As a result, this historical factor acts as a major driver of the current distribution patterns, so that different regions of the globe host different sets of living organisms (biotas).
These nonrandom geographic patterns are the prerequisite for dividing the Earth into distinct biogeographical units (de Candolle, 1820), which should also take into account the shared evolutionary history among taxa (Wallace, 1876). A biogeographical regionalization is typically seen as a hierarchical system that classifies geographic areas according to their shared biotic composition. These hierarchical relations implicitly represent a shared evolutionary history among areas (Daru et al., 2017). Thus, biogeographical regionalization is the underlying framework for many basic and applied issues in ecology, evolution, and conservation (Kreft & Jetz, 2010; Wen et al., 2013; Morrone, 2018). In principle, a biogeographical regionalization should be constructed based not only on the distribution of species, but also on their phylogenetic relationships. Whilst biogeographers have generally used data available for the contemporary species distributions (Good, 1947; Takhtajan, 1978; Cox, 2001), these analyses have been done without explicitly considering phylogenetic relationships among species, mostly due to the limited availability of large‐scale phylogenies. In recent decades, species‐level phylogenies based on molecular data have become increasingly available (e.g. Jetz et al., 2012; Jetz & Pyron, 2018; Smith & Brown, 2018; Upham et al., 2019), allowing a more robust delimitation of biogeographic units based on historical relationships (Ronquist & Sanmartín, 2011; Holt et al., 2013; Daru et al., 2016; Ye et al., 2019; Pataro et al., 2021; Qian et al., 2021).
Whilst global zoogeographical regions have been recently updated based on modern approaches (Holt et al., 2013), the most recent global proposals for plants heavily rely on the scheme advanced by Cox (2001), which in turn is a qualitative refining of the floristic kingdoms proposed by Takhtajan (1978, 1986). Several proposals of a global phytogeographical regionalization have been advanced to recognize between three and six floristic kingdoms (sensu Good, 1947; Table 1). However, all these proposals have relied on qualitative criteria for the recognition of floristic kingdoms, and none has used explicit phylogenetic information in their biogeographical schemes. Recently, Procheş & Ramdhani (2020) attempted a global phytogeographical regionalization based on the distribution and endemism of ancient plant lineages across the 35 phytogeographic units delimited by Good (1974). However, phylogenetic information (based on Harris & Davies, 2016) is only used for the identification of taxonomic units (lineages), but is not incorporated into the establishment of relationships among phytogeographic units.
Table 1.
Phytogeographic kingdoms (in bold) and their historical circumscriptions.
| Engler (1879, 1882) | Diels (1908) | Good (1947) | Takhtajan (1978) | Cox (2001) | Morrone (2015) | Our proposal | |
|---|---|---|---|---|---|---|---|
| Temperate and cold regions of the Northern Hemisphere | Arcto‐Tertiary | Arcto‐Tertiary | Holarktis | Holarctic | Holarctic | Holarctic | Holarctic |
| Holotropical | Holotropical | ||||||
| Old World tropics | Palaeotropical | Palaeotropical | Palaeotropis | Palaeotropical | African | Palaeotropical subkingdom | |
| Cape region here | Cape region here | ||||||
| New Zealand here | Indo‐Pacific | ||||||
| New World tropics | Neotropical | Neotropical | Neotropis | Neotropical | South American | Neotropical subkingdom | |
| Subantarctic islands here | |||||||
| Temperate and cold regions of the Southern Hemisphere | Ancient Ocean | Antarctic | Antarktis | Antarctic | Austral |
Austral Antarctic subkingdom |
|
| New Zealand here | Patagonia, New Zealand and southeast Australia here | ||||||
| Australian | Australis | Australian | Australian | Australian subkingdom | |||
| New Zealand here | New Zealand here | ||||||
| Cape region in South Africa | Cape | Capensis | Cape |
In this context, an integration of zoogeography and phytogeography into a single biogeographic scheme is needed (Morrone, 2015). However, the global biogeographic regionalization based on plant distribution has lagged behind those constructed based on animals. To fill this gap, we carry out a reassessment of the phytogeographic regionalization of the world that, for the first time, is based on a quantitative and phylogenetically informed big‐data approach. We take advantage of publicly available global occurrence databases and available species‐level molecular phylogenies for vascular plants, to integrate over eight million occurrence records with a dated phylogeny for more than sixty thousand vascular plants across the world. We applied pairwise phylogenetic (pβ) and taxonomic (β) beta‐diversity metrics to delimit geographical areas, discover their relationships, and identify the hierarchical organization of the regionalization. We then estimate the evolutionarily distinctiveness of the floras enclosed within phytogeographical units and discuss differences between our proposed framework with respect to previous regionalizations.
Materials and Methods
Geographic occurrence data
We retrieved geographic occurrence data for vascular plants across the globe from the Global Biodiversity Information Facility by querying all records under ‘Tracheophyta’ (we only considered ‘Preserved Specimens’ in our search); this database consisted of 68 570 538 occurrence records (GBIF.org, 2019). The occurrence records were taxonomically homogenized and cleaned following the procedures described in Edwards et al. (2015) and Ramírez‐Barahona et al. (2020), but using Kew’s Plants of the World database as the source of taxonomic information (POWO, 2019; last accessed June 2019). Basically, we discarded records without species identification, flagged records with missing or badly formatted global positioning system (GPS) coordinates, and identified records potentially associated with biodiversity institutions. We updated family and species names through cross‐validation with Kew’s taxonomic database, and used the coordinatecleaner package (Zizka et al., 2019) in R (R Core Team, 2020) to flag suspect occurrence records meeting one of the following criteria: (1) equal latitude and longitude; (2) zero latitude and longitude; (3) coordinates falling within a 5 km radius of a country’s political centroid; (4) coordinates falling within a 10 km radius of a country’s capital; (5) coordinates falling within a 2 km radius of biodiversity institutions; and (6) coordinates falling in the open ocean, using a reference landmass buffered by 1° from the coastline to avoid eliminating species living in the coast.
By this approach we retained 27 537 044 records (40% of the original records) for 277 597 species. However, after inspection of the resulting database, we still detected several problems with the distribution of species, mostly associated with possible nonnative records. Thus, we performed a complementary step to flag suspect records using Kew's Plants of the World database, which includes information on the native status for many species in countries across the world. We flagged an additional 11 783 185 records that potentially represent nonnative species distributions. The final geographic occurrence dataset used consisted of 15 753 859 records (23% of the original records) for 268 425 species of lycophytes, monilophytes, gymnosperms, and angiosperms across the world.
We incorporated point locality information into species distribution ranges (representing the maximum geographical extent of each species) by modelling point occurrences using hull geometries following the procedure described in Rabosky et al. (2016). Species with ≥ 6 unique occurrences (52 649 species) were modelled with alpha hulls (see Roll et al., 2017). Parameters for the delimitation of alpha hulls were adaptively selected using the function ‘getDynamicAlphaHull’ from the package rangebuilder (https://github.com/ptitle/rangeBuilder) in R (R Core Team, 2020). We started with initial alpha values of two and then adjusted for the distribution of records so that at least 95% of the records were included within the estimated range. Species with 3–5 unique occurrences (6767 species) were modelled using convex hulls. For species with 1–2 unique occurrences (7853 species), we used a 7584 km buffer based on the average spatial error reported in GBIF, to account for potential measurement and spatial errors. Species occurrences were extracted over an equal‐area grid (Mollweide projection) based on 200 km × 200 km grid cells. Finally, we also excluded grid cells containing fewer than two recorded species (Kreft & Jetz, 2010; Ramírez‐Barahona et al., 2020). Only a few records occurred in Antarctica (98% covered by ice), so this continent was not included in our analyses.
Phylogenetic data
Initially, we used the species‐level phylogenetic tree published by Jin & Qian (2019) (‘GBOTB.extended’), that includes 74 531 species of vascular plants, but with a poor representation of ferns (only 486 species out of c. 9000). To fill this gap, we combined the ‘GBOTB.extended’ tree with the fern phylogeny published by Testo & Sundue (2016) (‘TS.full’), which encompasses 4007 fern species. Prior to combining the trees, we performed a taxonomic homogenization of the names in the two trees using Kew’s Plants of the World database, resulting in: (1) 1720 synonyms flagged in the ‘GBOTB.extended’ tree; and (2) eight synonyms flagged in the ‘TS.full’ tree, with an additional 76 species that were not found in Kew’s database. After dropping these species from the trees, we merged the two trees by replacing the fern clade in the ‘GBOTB.extended’ tree with the ‘TS.full’ tree (without the outgroup); however, due to differing age estimates for the crown group of ferns between the two trees, prior to merging, we rescaled the ‘TS.full’ tree using the crown age for ferns provided in the ‘GBOTB.extended’. Thus, the final combined phylogenetic tree (‘GBOTB_TS’) keeps the original divergence age estimates of the ‘GBOTB.extended’ tree. The ‘GBOTB_TS’ tree was used in subsequent analyses and encompasses 76 226 species of lycophytes, monilophytes, gymnosperms, and angiosperms.
The taxonomic homogenization of the ‘GBOTB_TS’ tree allowed us to directly match species in the phylogeny to the geographic occurrence database. In sum, we obtained geographic data for 67 269 species of vascular plants included in the phylogeny, representing 8737 820 occurrence records across the world (Supporting Information Table S1; Fig. S1).
Computing phylogenetic and taxonomic beta diversity
Phylogenetic dissimilarity matrices among grid cells were calculated with Simpson's phylogenetic pairwise beta‐diversity metric (pβsim). Simpson index has the advantage of being independent of differences in species richness observed among sites (Kreft & Jetz, 2010). Phylogenetic beta diversity quantifies the turnover in phylogenetic composition between adjacent grid cells; this metric is an extension of the taxonomic beta‐diversity metric (βsim), where the proportion of shared species is instead substituted for the proportion of phylogenetic branch lengths represented by species shared between grid‐cells (after discounting differences in phylogenetic diversity between them). We constructed a matrix of pairwise pβsim among grid‐cells and, for comparison, we also constructed a matrix for the taxonomic beta‐diversity metric (βsim) among grid‐cells. All analyses were conducted using the package phyloregion (Daru et al., 2020) in R (R Core Team, 2020).
Cluster algorithm selection
The optimal number of units was defined using the ‘elbow’ method, setting the maximum number of clusters to k = 30 (Daru et al., 2020). The ‘elbow’ method identifies the optimal number of units based on the range of explained variances (Daru et al., 2020). We additionally evaluated the sensitivity of the resulting regionalization scheme by setting the number of units to match the number of previously recognized floristic kingdoms: k = 3, 4, 5, and 6. The phytogeographical units identified were mapped and visualized using a multidimensional scaling colour space, indicating the degree of phylogenetic (or taxonomic) differentiation between units: phytogeographical units with similar colours have a greater proportion of shared clades (or taxa) that those with diverging colours. We then used the function ‘phyloregion’ to estimate the evolutionary distinctiveness (ED) of each phytogeographical unit by computing the mean value of phylogenetic (or taxonomic) beta diversity between a focal unit and all other units (Holt et al., 2013; Daru et al., 2020). All the analyses were conducted using the pβsim and βsim separately, in order to delineate a taxonomic and a phylogenetic regionalization.
Nonmetric multidimensional scaling (NMDS) ordination plots and hierarchical clustering were used to evaluate the relationships among biogeographical units. Unit nomenclature mostly follows Takhtajan (1978). To identify spatial clusters of units across the world, we used the function ‘select_linkage’ from the phyloregion package. This function assesses the degree of data distortion with the cophenetic correlation coefficient, which is a measure of how a dendrogram preserves the pairwise distances between the original distance matrix, and has a value between 0 (poor correlation) and 1 (strong correlation). We tested eight, commonly used (Kreft & Jetz, 2010; Daru et al., 2020) hierarchical clustering algorithms on the pβsim and βsim matrices: single linkage, complete linkage, unweighted pair‐group method using arithmetic averages (UPGMA), unweighted pair‐group method using centroids (UPGMC), weighted pair‐group method using arithmetic averages (WPGMA), weighted pair‐group method using centroids (WPGMC), and Ward's minimum variance. UPGMA was identified as the best clustering algorithm for both matrices (cophenetic correlation r = 0.66 for pβsim and r = 0.807 for βsim; Table S2).
Results
We identified 16 phylogenetically distinct phytogeographical units across the world, which according to the hierarchical dendrogram of phylogenetic relationships are deeply split into two principal clusters that broadly match the separation between Gondwana and Laurasia (Fig. 1). The separation into these two clusters accounts for almost 50% of the total explained variance (0.31 out of 0.67; Fig. 1c). The hierarchical dendrogram also reveals that the Gondwanan cluster is divided into two subordinate clusters, albeit separated by lower pβsim values (Fig. 1d): one well defined Holotropical cluster (#2–9, 16 units), and a second Austral cluster composed of Australia (#7, 10 units) and a Neozealandic–Patagonian unit (#11). Indeed, the NMDS ordination (Fig. 1b) suggests that the Neozealandic–Patagonian and the Eremaean (#10) units are clearly isolated, but that northern Australia unit (#7) has a closer affinity to some tropical units, namely the Malesian (#8), the Indian–Indochinese (#9) and, to a lesser extent, the Madagascan (#6) units.
Fig. 1.
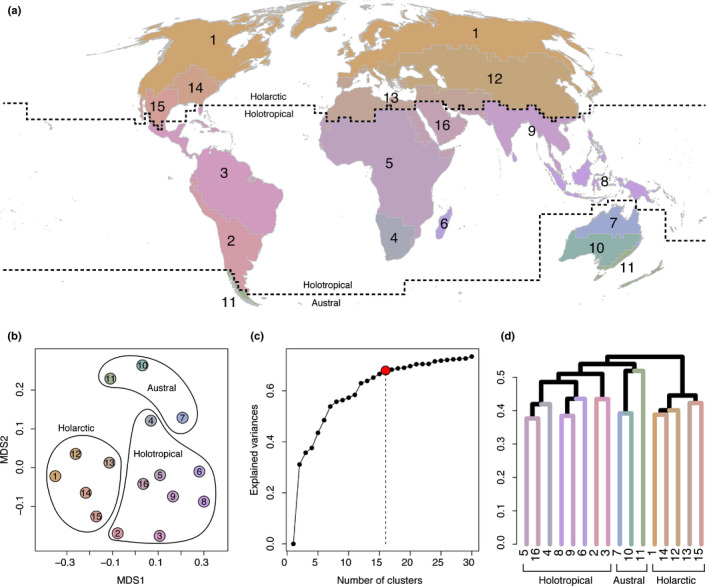
Map of the terrestrial phylogenetically distinct phytogeographic regions of the world (a), and their relationship presented as a nonmetric multidimensional scaling (NMDS) ordination plot (b) and as a unweighted pair‐group method using arithmetic averages (UPGMA) dendrogram (d) of beta diversity (pβsim) estimated across 200 km × 200 km grid cells. Sixteen units were defined based on the ‘elbow’ method, considering the range of explained variance (c). Colours differentiating between units in the NMDS plot, dendrogram and map are identical. Dashed lines highlight the borders of the three kingdoms: Holarctic, Holotropical, and Austral, corresponding to the three major clusters in (d). NMDS stress = 0.156.
The Holotropical cluster is further split into two clusters separating the Neotropics and the Paleotropics: one includes most of South America (excluding most of Patagonia), Mesoamerica and the Antilles (#2, 3 units), and the second includes most of Africa (excluding North Africa), southeast Asia, India, Malesia, and the Arabian peninsula (#4, 5, 6, 8, 9, 16 units). The Madagascan (#6) unit, which is the smallest (Table 2), is phylogenetically more similar to the Malesian (#8) and the Indian–Indochinese (#9) units than to continental Africa (Fig. 1b,c). Lastly, the Laurasian cluster exhibits a higher internal homogeneity and harbours the largest phytogeographical unit, the Circumboreal unit (#1, Table 2). Here, the Madrean unit (#15) and the Mediterranean–Iranian unit (#13) cluster together.
Table 2.
Summary of phytogeographic regions within each kingdom and subkingdom based on unweighted pair‐group method using arithmetic averages (UPGMA) clustering of phylogenetic beta diversity (pβsim) for assemblages of vascular plant species within 200 km × 200 km grid cells across the world.
| Kingdom | Subkingdom | Region | Unit No. | Area in Mkm2 | Total PD | SR | ED |
|---|---|---|---|---|---|---|---|
| Holarctic | Circumboreal | 1 | 50 920 | 113 364 | 11 058 | 0.561 | |
| Eurasiatic | 12 | 22 520 | 146 542 | 14 870 | 0.534 | ||
| Mediterranean–Iranian | 13 | 10 760 | 72 027 | 6867 | 0.498 | ||
| North American–Atlantic | 14 | 3520 | 56 317 | 3292 | 0.518 | ||
| Madrean | 15 | 2840 | 78 633 | 5907 | 0.517 | ||
| Holotropical | Neotropical | Andean–Argentinian | 2 | 7240 | 112 298 | 9367 | 0.518 |
| Neotropical | 3 | 20 480 | 168 800 | 17 878 | 0.527 | ||
| Palaeotropical | Southern African | 4 | 3480 | 70 906 | 6424 | 0.534 | |
| African | 5 | 22 920 | 89 369 | 5849 | 0.489 | ||
| Madagascan | 6 | 1640 | 53 441 | 2727 | 0.538 | ||
| Malesian | 8 | 12 680 | 103 389 | 5942 | 0.546 | ||
| Indian–Indochinese | 9 | 9200 | 138 093 | 10 010 | 0.513 | ||
| Arabian | 16 | 6200 | 35 171 | 1544 | 0.469 | ||
| Austral | Australian | Northern Australian | 7 | 4480 | 65 267 | 3166 | 0.526 |
| Eremaean | 10 | 4640 | 61 400 | 3785 | 0.551 | ||
| Antarctic | Neozealandic–Patagonian | 11 | 3680 | 65 911 | 3774 | 0.549 |
ED, evolutionary distinctiveness; PD, total phylogenetic diversity; SR, species richness.
Even when reducing the number of units to match the number of currently recognized floristic kingdoms (k = 3, 4, 5, and 6), the separation between the Gondwanan and Laurasian clusters is always maintained, as well as the independency of the Austral cluster, which includes Australia and the Neozealandic–Patagonian unit (Figs S2–S5). For instance, for k = 6, the three main clusters (each with two units) are identifiable: Holotropical, Austral, and Holarctic (Fig. S5). In this case, the Holarctic cluster is composed of a large Circumboreal unit and of a Trans‐Atlantic unit roughly corresponding to the transition zones between Laurasia and Gondwana. In turn, with k = 5 (Fig. S4), the Holotropical cluster is composed of a Indo‐Malesian unit and a large amphi‐Atlantic unit composed by Africa and the Neotropics, whereas the Austral cluster is divided into the Australian and Neozealandic–Patagonian units.
As expected, the pβsim and the βsim values are strongly correlated (Mantel test: r = 0.68, P = 0.001, 999 permutations). However, the best taxonomic regionalization is composed of 23 units (Fig. S6) and, despite the higher number of units, the total explained variance (0.5) is lower than that explained by the phylogenetic regionalization presented earlier (0.67). In addition, when omitting phylogenetic information, we found a less clear phytogeographic distinction between the Gondwanan and Laurasian clusters (Figs S6, S7). More specifically, in the taxonomic regionalization, the Mediterranean–Iranian unit (which is here split into two separate units) groups with the Paleotropical units, whereas the Madrean unit, together with an Appalachian and a Californian unit, is recovered within a mostly Neotropical cluster (Fig. S6). In addition, the Patagonian and Neozealandic units are not grouped together, with the former being placed within a Neotropical cluster and the latter placed within a Paleotropical cluster.
Overall, based on our phylogenetic regionalization, we show that the northernmost and southernmost units, together with the Malesian and Neotropical units, exhibit the highest evolutionary distinctiveness (Fig. 2a). Also, regions (#2, 7, 13, 15, 16) at the contact between main clusters generally show lower ED values. Since lycophytes, monilophytes, and gymnosperms could potentially have a strong impact on ED estimates because they represent less diverse, evolutionarily distinct clades, we re‐ran our analysis considering angiosperms only. This resulted in similar ED patterns (Fig. 2b), but with overall higher ED estimates for angiosperms (Fig. 2b). ED patterns for lycophytes and ferns are highest in extra‐tropical or arid regions (Fig. S8), where species richness concerning these clades is lowest. However, given the moderate sampling size of these lineages in our dataset (Table S1), these results should be taken with caution. Concerning gymnosperms, it seems that the regions showing highest ED (Fig. S8) correspond to those areas where the highest gymnosperm diversity has been documented, with co‐occurrence of different lineages with deep divergences within particular units (e.g. Cycadaceae (Cycadales) and Podocarpaceae (Cupressales) across Malesia (Stevens, 2001); Araucariaceae (Cupressales) and Ephedraceae (Gnetales) in South America (Stevens, 2001); Taxaceae (Cupressales) and Ephedraceae (Gnetales) in the Mediterranean region; Zamiaceae (Cycadales) and Cupressaceae (Cupressales) in southeast Australia and New Zealand (Stevens, 2001). Given the low number of species of living gymnosperms, this is likely sufficient to cause the pronounced differences observed in the map.
Fig. 2.
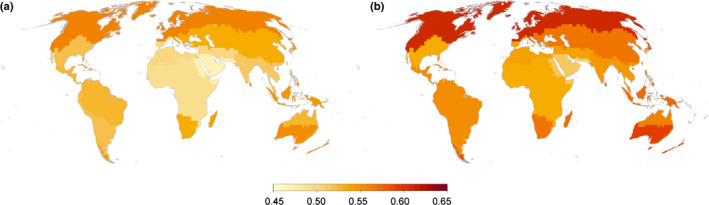
Evolutionary distinctiveness within the 16 phytogeographical units, considering all vascular plant species (a) and angiosperms only (b), quantified as the mean of pairwise pβsim values between each unit, contrasted with all other units. Darker regions indicate regions of higher evolutionary distinctiveness.
Discussion
Here we built a comprehensive phytogeographic regionalization scheme of the world based on the integration of large‐scale geographic and phylogenetic data for vascular plants. This allowed us to quantitively test, for the first time, century‐old regionalizations of the world based on plant distributions (Engler, 1882; Diels, 1908; Good, 1947; Takhtajan, 1978; Cox, 2001), using a species‐level phylogeny rather than a family or major clades approach (Procheş & Ramdhani, 2020). This is an important step forward in building a comprehensive, phylogenetically informed regionalization of the world (Morrone, 2015). Integrating phylogenetic and occurrence records on a global scale, we were able to retrieve three major clusters, which we identify here as kingdoms, and their cladistic relationships following the sequence: Holarctic, Holotropical, and Austral. Both Holotropical and Austral kingdoms are split into two main clusters, which we identify here as subkingdoms, while the 16 main phytogeographical units which best explain the variance observed in our dataset are here referred as regions (Table 2). Because our goal is to identify major clusters of phytogeographical units and their relationships at a global scale, the hierarchical organization of our regionalization is described using only higher rank categories (kingdom, subkingdom, region), whereas other lower‐rank categories (e.g. dominion, province, district) (Morrone, 2018) were not considered.
Our phylogenetic regionalization of the world’s vascular flora into distinct phytogeographical kingdoms, subkingdoms, and regions show substantial congruence to long‐recognized regionalization systems (Engler, 1882; Diels, 1908; Good, 1947; Takhtajan, 1978; Cox, 2001; Morrone, 2015; Fig. 3), including a deep separation of biotas broadly corresponding to the Laurasian–Gondwanan divergence (Raven & Axelrod, 1974; Morrone, 2015). This split is likely deeply rooted and related to the break up of Pangea into the two supercontinents Laurasia and Gondwana (Mao et al., 2012). Other phenomena, such as long‐distance intercontinental dispersal (Davis et al., 2002), niche conservatism, and local extinctions may have attenuated the signal of this deep split, thus generating transition zones; these factors are probably affecting the shallow definition of many phytogeographical regions. However, these factors, together with global climate change, may have also played a key role in further reinforcing the Laurasian/Gondawanan split in more recent times.
Fig. 3.
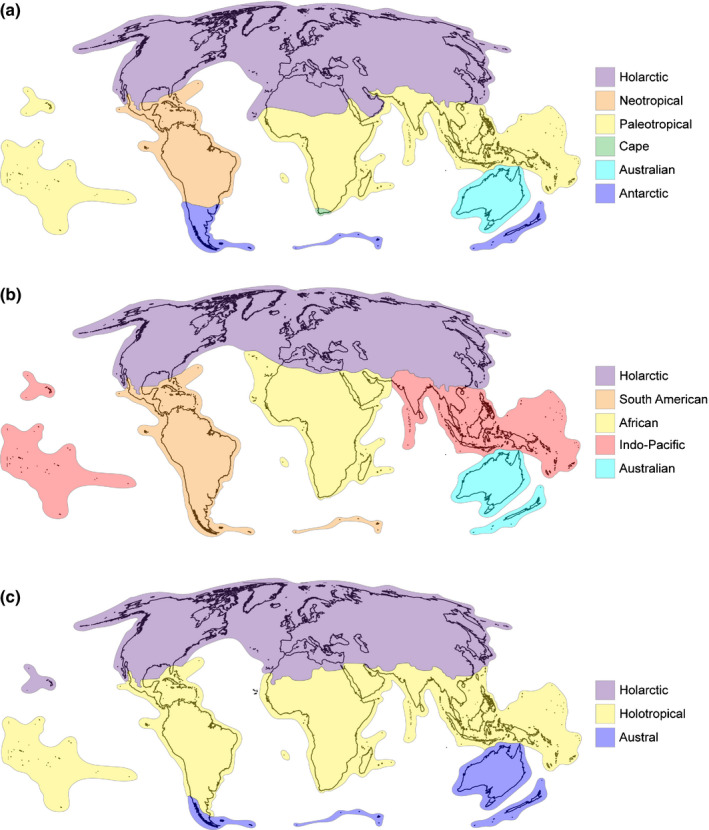
Maps of the floristic kingdoms of the world as for: (a) Takhtajan (1986); (b) Cox (2001); (c) the present study.
Irrespective of the origin of the current distribution patterns uncovered for the world’s vascular flora, our results are in agreement with the recent ‘integrative’ biogeographical proposal advanced by Morrone (2015). More specifically, this proposal recognizes just three major biogeographically unique areas of the world: Holarctic, Holotropical, and Austral. Our results recovered the general features of the regionalization proposed by Morrone (2015), but with some exceptions. The Holarctic kingdom herein recognized is not segregated into Nearctic and Palearctic components. Instead, this kingdom is split into two clusters: (1) Circumboreal and (2) temperate Asia and the Mediterranean. Repeated dispersal events between Eurasia and North America (Donoghue & Smith, 2004; Graham, 2018) likely explain the homogeneity in phylogenetic composition of the Holarctic cluster, while keeping a high evolutionary distinctiveness. The Austral kingdom coincides with that recognized by Morrone (2015) and by Procheş & Ramdhani (2020), except for the southern African region (linked to the Holotropical kingdom here) and the Antarctic territories (not assessed here). In turn, the Holotropical kingdom mostly corresponds to that recognized by Morrone (2015), but with a main split between the Neotropical and Palaeotropical subkingdoms, in agreement with most of classical phytogeographical proposals (Table 1). The split between Neotropics and Paleotropics has also been supported in a recent assessment of area relationships across the tropics (Slik et al., 2018).
The phytogeographical units identified here were defined de novo, without relying on previous schemes (as done by Procheş & Ramdhani, 2020), that is to say, our analyses are agnostic to previous regionalizations. Nevertheless, our results broadly match traditional regionalization schemes, with some discrepancies. For example, the phytogeographical relationships of New Zealand have long been debated (Moreira‐Muñoz, 2007), being alternatively placed within either the Palaeotropical, Australian, or Antarctic kingdoms (Table 1). Here, we clearly found a phylogenetically homogeneous unit, composed of New Zealand and Patagonia clustering with Australia, highlighting a common evolutionary history and isolation of these territories (Procheş & Ramdhani, 2020). As proposed by Gonzalez‐Orozco et al. (2014), Australia is separated into two regions (Morrone, 2002) that broadly correspond with tropical (north) and extratropical (south) Australia, the former showing a closer affinity with the Malesian region (Fig. 1). Although the South African region is clearly distinct from the rest of Africa (Fig. 1), this region is not recognized as a major unit (kingdom) as advocated by Diels (1908); Good (1947), and Takhtajan (1978), but instead it is nested within a broader African group; this is in agreement with Cox (2001), who did not recognize a Cape kingdom. Interestingly, the southern African region is more clearly differentiated from the rest of Africa when only taxonomic turnover is considered (Fig. S5). In line with Takhtajan (1978), we also recognize a well distinct Arabian region (Brenan, 1978). Despite the close geographical proximity of Madagascar to Africa, and their taxonomic affinity (Fig. S5), the former exhibits a remarkably higher phylogenetic affinity with the Indo‐Chinese, Malesian, and Australian regions, as already highlighted by Schatz (1996). The discrepancies between our results and traditional regionalization schemes highlight the importance of explicitly incorporating phylogenetic information to establish phytogeographical schemes (Escalante, 2017). This is further supported by the observed differences between the phylogenetically informed regionalization and that based only on taxonomic turnover.
Our results also support an intermediate position of some regions between kingdoms (Fig. 1b). More specifically, we found an intermediate position between Holarctic and Holotropical kingdoms of the Mediterranean and Madrean (= Mexican Transition Zone) regions, which also exhibit an overall lower evolutionary distinctiveness (Fig. 2; Table 2). Based on our results, we support a view of these regions as transition zones, which result from the intergrading of independent evolutionary histories (Morrone, 2015). Usually, clustering algorithms are unable to differentiate between core and transition zones, because the former are typically merged with core areas with which they show the greatest affinity (Kreft & Jetz, 2010). Despite this, however, we found additional evidence for these transition zones from the hierarchical relationships: the Madrean and the Mediterranean–Iranian regions (#15, 13) were retrieved as clustering separately from the rest of the Holarctic kingdom. These results support the hypothesis that taxa from these transitions zones are, in general, more closely related to Holarctic taxa than to taxa from tropical regions (Morrone, 2015).
The contrast between the phylogenetically‐based regionalization and that based only on taxonomic affinities, demonstrates the advantages of incorporating phylogenetic information for the delineation of phytogeographical regions on a global scale. The phylogenetically‐based regionalization mostly supports the proposed schematic diagram of historical area relationships proposed by Morrone (2015), which is mostly based on Engler (1882). Furthermore, the taxonomic‐only approach is less parsimonious, and explains a lower amount of variance in phytogeographic affinities among units, resulting in more shallow differences among major clusters (Figs S6, S7). In terms of the inferred relationships among units, one of the main differences between the phylogenetically informed and the taxonomic‐only approaches concerns the northern limit of the Holotropical kingdom. Using taxonomic data only, the Holotropical kingdom also includes the Mediterranean–Iranian and the Madrean regions, further confirming the view of these units as transition zones. Another important difference between the two approaches is that the taxonomic‐only regionalization recognizes three alternative main clusters: Neotropical, Paleotropical, and Laurasian. Here the Laurasian cluster mostly corresponds to the Holarctic kingdom. However, in this case, the units corresponding to the Austral kingdom (otherwise recognized as a distinct cluster, Fig. 1) are split up into two separate areas: the first (Patagonian unit) is identified within the Neotropical cluster, and the second (Neozealandic unit) within the Paleotropical cluster. This regionalization is less parsimonious than recognizing a single Austral kingdom, partly due to the high phylogenetic distinctiveness of its flora (Fig. 2). More specifically, the higher phylogenetic uniqueness of the Neozealandic–Patagonian region is consistent with the isolation of these areas and their connection through Antarctica, resulting in the presence of plant lineages (e.g. Berberidopsidales, Araucariaceae) with disjunct distributions (Winkworth et al., 2015; Procheş & Ramdhani, 2020).
The observed latitudinal patterns of evolutionary distinctiveness are suggestive of phylogenetic niche conservatism (Wiens, 2004) likely leading to the accumulation of phylogenetic diversity within tropical regions, particularly for angiosperms and ferns (Schuettpelz & Pryer, 2009; Ramírez‐Barahona et al., 2020). However, the latitudinal patterns of evolutionary distinctiveness are steeper for angiosperms than for gymnosperms and seed‐free vascular plants, and this can be interpreted in light of the presumed tropical origin of the group (Feild et al., 2009; Coiro et al., 2019; Condamine et al., 2020) and of the delayed, but accelerated, rise of flowering plant lineages in extratropical regions (Igea & Tanentzap, 2020; Ramírez‐Barahona et al., 2020). Ferns and lycophytes exhibit a similar, albeit shallower, latitudinal pattern of increasing ED towards the poles (Fig. S8). However, gymnosperms, and to some extent ferns and lycophytes, show a pattern of increasing ED in arid and semi‐arid regions across the world (e.g. Madrean, Mediterranean–Iranian units), which might explain the more shallow latitudinal pattern observed for the entire vascular flora (Fig. 2a). In this context, phylogenetic dissimilarity between the three kingdoms is not high, and instead is higher among phytogeographical regions. One possible biological interpretation for this pattern is that the world’s flora shares a recent evolutionary origin, especially for the overwhelmingly larger angiosperm component, and that early dispersal of this group across the world (Coiro et al., 2019) has played a key role in determining the distribution of plant lineages. This does not allow for stronger divergences among the three kingdoms, albeit these differences are sufficiently clear to recognize them as distinct biogeographical units.
The inclusion of phylogenetic information allowed a robust regionalization of vascular plant distributions at a global level and provided new insights into the historical relationships among phytogeographical regions. The match between our regionalization and earlier schemes is remarkable. Thus, we propose only some minor changes to the existing classification schemes, mainly at higher ranks. Our results could be a launch pad for further detailed studies, specifically devoted identifying and circumscribing lower rank units (i.e. below region). In addition, our regionalization scheme, as opposed to others, can be improved: not only by adding new information, but also by providing all codes and data. As such, the phytogeographic regionalization herein presented provides a baseline for future ecological, evolutionary, and conservation studies of vascular plants at global scale (see Mucina, 2020, and references cited therein) and represents, together with the zoogeographical scheme advanced by Holt et al. (2013), a major step forward towards the integration of zoogeography and phytogeography into a single biogeographical scheme.
Data limitations and caveats
In any biogeographical analysis, acknowledging the limitations of the geographic and phylogenetic data is fundamental to properly interpret the resulting geographical patterns. Here, some limitations of the occurrence data include the long‐recognized problems associated with publicly available collection data (e.g. misidentification of species, erroneous geographic records, nonnative records). The series of taxonomical and geographical filters we applied to the data were aimed at minimizing these problems. However, a main limitation in the data still remains: the geographical bias in occurrence records, which is evident at first sight in regions such as the Indian subcontinent and Siberia, which are well‐known to be considerably underrepresented in the GBIF database, and might lead to biased results. Other sources of distribution data are available (e.g. Flora of the USSR), but these data are often in the form of broadly‐defined expert maps. In spite the limitations of our data, we believe that the recognition of the three main biogeographic kingdoms is robust enough to these sources of bias, but these could potentially have an impact on the delimitation (and especially relationships) among individual phytogeographical regions.
Phylogenetic data also come with some limitations due to (1) incompletely sampled trees, (2) sampling bias across lineages, (3) topological resolution, and (4) uncertainty in divergence time estimates. The first two limitations probably have a minor impact on the results, given that the sampling is skewed towards the larger and more widespread lineages, yet some major lineages are poorly sampled in the phylogenetic tree, such as the lycophytes and some monilophyte lineages (e.g. tree ferns). The third limitation represents a more serious challenge to any proposed regionalization, but it would only seriously affect the shallower relationships between species and genera, therefore being of relevance only for the delimitation of more nested areas within the kingdoms. The fourth is a major, but often neglected limitation in biogeographic analyses, where divergence time estimates are accepted at face value without consideration of uncertainties stemming from model assumptions and fossil calibration schemes (Parham et al., 2012; Sauquet et al., 2012; Magallón, 2021). In this context, the phylogenies used here (after rescaling of the fern phylogeny) proved to be useful to address the phytoregionalization of the world, but differences in divergence times and branch lengths will most likely have a strong impact in the estimate of phylogenetic dissimilarity across regions. These sources of uncertainty would play a major role when attempting to interpret any proposed regionalization in terms of the evolutionary history of species.
Author contributions
AC and LP conceived the idea; AC designed the research; SR‐B compiled and curated the occurrence and phylogenetic data; AC performed the analysis; AC wrote the manuscript with contributions from LP and SR‐B.
Supporting information
Fig. S1 Species richness and number of occurrences across 200 km × 200 km grid cells.
Figs S2–S5 Map of the terrestrial phylogenetically distinct phytogeographic units of the world constraining the number of clusters to k = 3, 4, 5, and 6.
Fig. S6 Map of the terrestrial taxonomically distinct phytogeographic units of the world.
Fig. S7 Maps of the terrestrial phylogenetic and taxonomic regionalization of the world side by side.
Fig. S8 Evolutionary distinctiveness within the 16 phytogeographical units, considering only lycophytes, ferns and gymnosperms.
Table S1 Total number of species of vascular plants used in the delimitation of phytogeographical units across the globe.
Table S2 Performance of clustering algorithms for phylogenetic beta diversity (pβsim) and beta diversity (βsim) of global vascular species data.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgement
University of Pisa is gratefully acknowledged.
Data availability
Code and data supporting the results are available at https://github.com/spiritu‐santi/Floristic‐Kingdoms and at Zenodo with the identifier https://doi.org/10.5281/zenodo.4711219. The geographic occurrence data for vascular plants is available from the Global Biodiversity Information Facility with the identifier https://doi.org/10.15468/dl.bdxzkw.
References
- Brenan JPM. 1978. Some aspects of the phytogeography of tropical Africa. Annals of the Missouri Botanical Garden 65: 437–478. [Google Scholar]
- de Candolle AP. 1820. Essai élémentaire de géographie botanique. In: Dictionnaire des Sciences Naturelles, vol. 18. Paris, France: F. Levrault. [Google Scholar]
- Coiro M, Doyle JA, Hilton J. 2019. How deep is the conflict between molecular and fossil evidence on the age of angiosperms? New Phytologist 223: 83–99. [DOI] [PubMed] [Google Scholar]
- Condamine FL, Silvestro D, Koppelhus EB, Antonelli A. 2020. The rise of angiosperms pushed conifers to decline during global cooling. Proceedings of the National Academy of Sciences, USA 117: 28867–28875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox CB. 2001. The biogeographic regions considered. Journal of Biogeography 28: 511–523. [Google Scholar]
- Daru BH, Elliott TL, Park DS, Davies TJ. 2017. Understanding the processes underpinning patterns of phylogenetic regionalization. Trends in Ecology & Evolution 32: 845–860. [DOI] [PubMed] [Google Scholar]
- Daru BH, Karunarathne P, Schliep K. 2020. phyloregion: R package for biogeographical regionalization and macroecology. Methods in Ecology and Evolution 11: 1483–1491. [Google Scholar]
- Daru BH, Van der Bank M, Maurin O, Yessoufou K, Schaefer H, Slingsby JA, Davies TJ. 2016. A novel phylogenetic regionalization of phytogeographical zones of southern Africa reveals their hidden evolutionary affinities. Journal of Biogeography 43: 155–166. [Google Scholar]
- Davis CC, Bell CD, Mathews S, Donoghue MJ. 2002. Laurasian migration explains Gondwanan disjunctions: evidence from Malpighiaceae. Proceedings of the National Academy of Sciences, USA 99: 6833–6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diels L. 1908. Pflanzengeographie. Leipzig, Germany: Göschen. [Google Scholar]
- Donoghue MJ, Smith SA. 2004. Patterns in the assembly of temperate forests around the Northern Hemisphere. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 359: 1633–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ, de Vos JM, Donoghue MJ. 2015. Doubtful pathways to cold tolerance in plants. Nature 521: E5–E6. [DOI] [PubMed] [Google Scholar]
- Engler A. 1879. Versuch Einer Entwicklungsgeschichte der Pflanzenwelt, vol. 1. Leipzig, Germany: Verlag von Wilhelm Engelmann. [Google Scholar]
- Engler A. 1882. Versuch Einer Entwicklungsgeschichte der Pflanzenwelt, vol. 2. Leipzig, Germany: Verlag von Wilhelm Engelmann. [Google Scholar]
- Escalante T. 2017. A natural regionalization of the world based on primary biogeographic homology of terrestrial mammals. Biological Journal of the Linnean Society 120: 349–362. [Google Scholar]
- Feild TS, Chatelet DS, Brodribb TJ. 2009. Ancestral xerophobia: a hypothesis on the whole plant ecophysiology of early angiosperms. Geobiology 7: 237–264. [DOI] [PubMed] [Google Scholar]
- GBIF.org. 2019. GBIF occurrence download. doi: 10.15468/dl.bdxzkw [accessed 3 September 2019]. [DOI] [Google Scholar]
- Gonzalez‐Orozco CE, Ebach MC, Laffan S, Thornhill AH, Knerr NJ, Schmidt‐Lebuhn AN, Cargill CC, Clements M, Nagalingum NS, Mishler BD et al. 2014. Quantifying phytogeographical regions of Australia using geospatial turnover in species composition. PLoS ONE 9: e92558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good R. 1947. The geography of the flowering plants, 1 st edn. London, UK: Longman. [Google Scholar]
- Good R. 1974. The geography of the flowering plants, 4 th edn. London, UK: Longman. [Google Scholar]
- Graham A. 2018. The role of land bridges, ancient environments, and migrations in the assembly of the North American flora. Journal of Systematics and Evolution 56: 405–429. [Google Scholar]
- Harris LW, Davies TJ. 2016. A complete fossil‐calibrated phylogeny of seed plant families as a tool for comparative analyses: testing the ‘time for speciation’ hypothesis. PLoS ONE 11: e0162907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt BG, Lessard J‐P, Borregaard MK, Fritz SA, Araújo MB, Dimitrov D, Fabre P‐H, Graham CH, Graves GR, Jønsson KA et al. 2013. An update of Wallace's zoogeographic regions of the world. Science 339: 74–78. [DOI] [PubMed] [Google Scholar]
- Igea J, Tanentzap AJ. 2020. Angiosperm speciation cools down in the tropics. Ecology Letters 23: 692–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetz W, Pyron RA. 2018. The interplay of past diversification and evolutionary isolation with present imperilment across the amphibian tree of life. Nature Ecology & Evolution 1: 850–858. [DOI] [PubMed] [Google Scholar]
- Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. 2012. The global diversity of birds in space and time. Nature 491: 444–448. [DOI] [PubMed] [Google Scholar]
- Jin Y, Qian H. 2019. v.phylomaker: an R package that can generate very large phylogenies for vascular plants. Ecography 42: 1353–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft H, Jetz W. 2010. A framework for delineating biogeographical regions based on species distributions. Journal of Biogeography 37: 2029–2053. [Google Scholar]
- Lomolino MV, Sax DF, Brown JH, eds. 2004. Foundations of biogeography: classic papers with commentaries. Chicago, IL, USA: University of Chicago Press. [Google Scholar]
- Magallón S. 2021. Principles of molecular dating. In: Ho S, ed. The molecular evolutionary clock. Cham, Switzerland: Springer, 67–81. [Google Scholar]
- Mao K, Milne RI, Zhang L, Peng Y, Liu J, Thomas P, Mill RP, Renner SS. 2012. Distribution of living Cupressaceae reflects the breakup of Pangea. Proceedings of the National Academy of Sciences, USA 109: 7793–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira‐Muñoz A. 2007. The Austral floristic realm revisited. Journal of Biogeography 34: 1649–1660. [Google Scholar]
- Morrone JJ. 2002. Biogeographic regions under track and cladistic scrutiny. Journal of Biogeography 29: 149–152. [Google Scholar]
- Morrone JJ. 2015. Biogeographic regionalisation of the world: a reappraisal. Australian Systematic Botany 28: 81–90. [Google Scholar]
- Morrone JJ. 2018. The spectre of biogeographical regionalization. Journal of Biogeography 45: 282–288. [Google Scholar]
- Mucina L. 2020. Biomes are everybody's kingdom: a platform where ecology and biogeography meet: response to Procheş (2020) ‘Biomes are nobody’s kingdom: on environmental and historical plant geography’. New Phytologist 228: 1463–1466. [DOI] [PubMed] [Google Scholar]
- Parham JF, Donoghue PCJ, Bell CJ, Calway TD, Head JJ, Holroyd PA, Inoue JG, Irmis RB, Joyce WG, Ksepka DT et al. 2012. Best practices for justifying fossil calibrations. Systematic Biology 61: 346–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pataro L, Molina‐Venegas R, Calatayud J, Moreno‐Saiz JC, Rodríguez MÁ. 2021. An updated phylogenetic bioregionalization for the European fern flora. Biodiversity and Conservation 30: 201–215. [Google Scholar]
- Posadas P, Crisci JV, Katinas L. 2006. Historical biogeography: a review of its basic concepts and critical issues. Journal of Arid Environments 66: 389–403. [Google Scholar]
- POWO . 2019. Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. [WWW document] URL http://www.plantsoftheworldonline.org/ [accessed June 2019]. [Google Scholar]
- Procheş Ş, Ramdhani S. 2020. A global regionalisation based on the present‐day distribution of broad plant lineages. Phytotaxa 442: 20–26. [Google Scholar]
- Qian H, Jin Y, Leprieur F, Wang X, Deng T. 2021. Patterns of phylogenetic beta diversity measured at deep evolutionary histories across geographical and ecological spaces for angiosperms in China. Journal of Biogeography 48: 773–784. [Google Scholar]
- R Core Team . 2020. R: a language and environment for statistical computing (v.4.0.3). Vienna, Austria: R Foundation for Statistical Computing. [WWW document] URL http://www.R‐project.org/. [Google Scholar]
- Rabosky ARD, Cox CL, Rabosky DL, Title PO, Holmes IA, Feldman A, McGuire JA. 2016. Coral snakes predict the evolution of mimicry across New World snakes. Nature Communications 7: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez‐Barahona S, Sauquet H, Magallón S. 2020. The delayed and geographically heterogeneous diversification of flowering plant families. Nature Ecology and Evolution 4: 1232–1238. [DOI] [PubMed] [Google Scholar]
- Raven PH, Axelrod DI. 1974. Angiosperm biogeography and past continental movements. Annals of the Missouri Botanical Garden 61: 539–673. [Google Scholar]
- Roll U, Feldman A, Novosolov M, Allison A, Bauer AM, Bernard R, Bohm M, Castro‐Herrera F, Chirio L, Collen B et al. 2017. The global distribution of tetrapods reveals a need for targeted reptile conservation. Nature Ecology & Evolution 1: 1677–1682. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Sanmartín I. 2011. Phylogenetic methods in biogeography. Annual Review of Ecology, Evolution, and Systematics 42: 441–464. [Google Scholar]
- Sauquet H, Ho SYW, Gandolfo MA, Jordan GJ, Wilf P, Cantrill DJ, Bayly MJ, Bromham L, Brown GK, Carpenter RJ et al. 2012. Testing the impact of calibration on molecular divergence times using a fossil‐rich group: the case of Nothofagus (Fagales). Systematic Biology 61: 289–313. [DOI] [PubMed] [Google Scholar]
- Schatz GE. 1996. Malagasy/Indo‐australo‐malesian phytogeographic connections. In: Lourenço WR, ed. Biogeography of Madagascar. Paris, France: Editions ORSTOM, 73–83. [Google Scholar]
- Schuettpelz E, Pryer KM. 2009. Evidence for a Cenozoic radiation of ferns in an angiosperm‐dominated canopy. Proceedings of the National Academy of Sciences, USA 106: 11200–11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slik JWF, Franklin J, Arroyo‐Rodríguez V, Field R, Aguilar S, Aguirre N, Ahumada J, Aiba S‐I, Alves LF, K A et al. 2018. Phylogenetic classification of the world’s tropical forests. Proceedings of the National Academy of Sciences, USA 115: 1837–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Brown JW. 2018. Constructing a broadly inclusive seed plant phylogeny. American Journal of Botany 105: 302–314. [DOI] [PubMed] [Google Scholar]
- Stevens PF. 2001. onwards. Angiosperm Phylogeny Website. v.14, July 2017 [and more or less continuously updated since] (MOBOT). [WWW document] URL http://www.mobot.org/MOBOT/Research/APweb/welcome.html [accessed August 2021]. [Google Scholar]
- Takhtajan A. 1978. The floristic regions of the world. Moscow, Russia: Soviet Sciences Press; (in Russian). [Google Scholar]
- Takhtajan A. 1986. The floristic regions of the world. Berkeley, CA, USA: UC Press. [Google Scholar]
- Testo W, Sundue M. 2016. A 4000‐species dataset provides new insight into the evolution of ferns. Molecular Phylogenetics and Evolution 105: 200–211. [DOI] [PubMed] [Google Scholar]
- Upham NS, Esselstyn JA, Jetz W. 2019. Inferring the mammal tree: species‐level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biology 17: e3000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AR. 1876. Die geographische Verbreitung der Tiere. Dresden, Germany: Verlag von R. v. Zahn. [Google Scholar]
- Wen J, Ree RH, Ickert‐Bondl SM, Nie Z, Funk V. 2013. Biogeography: where do we go from here? Taxon 62: 912–927. [Google Scholar]
- Wiens JJ. 2004. Speciation and ecology revisited: phylogenetic niche conservatism and the origin of species. Evolution 58: 193–197. [DOI] [PubMed] [Google Scholar]
- Winkworth RC, Hennion F, Prinzing A, Wagstaff SJ. 2015. Explaining the disjunct distributions of austral plants: the roles of Antarctic and direct dispersal routes. Journal of Biogeography 42: 1197–1209. [Google Scholar]
- Ye J, Lu L, Liu B, Yang T, Zhang J, Hu H, Li R, Lu A, Liu H, Mao L et al. 2019. Phylogenetic delineation of regional biota: a case study of the Chinese flora. Molecular Phylogenetics and Evolution 135: 222–229. [DOI] [PubMed] [Google Scholar]
- Zizka A, Silvestro D, Andermann T, Azevedo J, Duarte Ritter C, Edler D, Farooq H, Herdean A, Ariza M, Scharn R et al. 2019. coordinatecleaner: standardized cleaning of occurrence records from biological collection databases. Methods in Ecology and Evolution 10: 744–751. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Species richness and number of occurrences across 200 km × 200 km grid cells.
Figs S2–S5 Map of the terrestrial phylogenetically distinct phytogeographic units of the world constraining the number of clusters to k = 3, 4, 5, and 6.
Fig. S6 Map of the terrestrial taxonomically distinct phytogeographic units of the world.
Fig. S7 Maps of the terrestrial phylogenetic and taxonomic regionalization of the world side by side.
Fig. S8 Evolutionary distinctiveness within the 16 phytogeographical units, considering only lycophytes, ferns and gymnosperms.
Table S1 Total number of species of vascular plants used in the delimitation of phytogeographical units across the globe.
Table S2 Performance of clustering algorithms for phylogenetic beta diversity (pβsim) and beta diversity (βsim) of global vascular species data.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
Code and data supporting the results are available at https://github.com/spiritu‐santi/Floristic‐Kingdoms and at Zenodo with the identifier https://doi.org/10.5281/zenodo.4711219. The geographic occurrence data for vascular plants is available from the Global Biodiversity Information Facility with the identifier https://doi.org/10.15468/dl.bdxzkw.


