Abstract
The enzyme glutamate decarboxylase (GAD) is prevalent in Escherichia coli but few strains in the various pathogenic E. coli groups have been tested for GAD. Using PCR primers that amplify a 670-bp segment from the gadA and gadB genes encoding GAD, we examined the distribution of the gadAB genes among enteric bacteria. Analysis of 173 pathogenic E. coli strains, including 125 enterohemorrhagic E. coli isolates of the O157:H7 serotype and its phenotypic variants and 48 isolates of enteropathogenic E. coli, enterotoxigenic E. coli, enteroinvasive E. coli, and other Shiga toxin-producing E. coli (STEC) serotypes, showed that gadAB genes were present in all these strains. Among the 22 non-E. coli isolates tested, only the 6 Shigella spp. carried gadAB. Analysis of naturally contaminated water and food samples using a gadAB-specific DNA probe that was labeled with digoxigenin showed that a gadAB-based assay is as reliable as standard methods that enumerate E. coli organisms on the basis of lactose fermentation. The presence of few E. coli cells initially seeded into produce rinsates could be detected by PCR to gadA/B genes after overnight enrichment. A multiplex PCR assay using the gadAB primers in combination with primers to Shiga toxin (Stx) genes stx1 and stx2 was effective in detecting STEC from the enrichment medium after seeding produce rinsate samples with as few as 2 CFU. The gadAB primers may be multiplexed with primers to other trait virulence markers to specifically identify other pathogenic E. coli groups.
At least six categories of enterovirulent Escherichia coli are recognized, with four responsible for illness transmitted by food or water (5, 8, 24). Development of widely applicable tests for enterovirulent E. coli is hampered by the physiological diversity of these groups and the loss of plasmids during some standard procedures (2, 3, 6, 9, 14, 23). Isolation of the most widely studied category, the enterohemorrhagic E. coli (EHEC), is routinely conducted on the basis of their sorbitol- and glucuronidase-negative traits; but numerous variants have now been reported, diminishing the utility of these tests (7, 10, 12, 13, 25).
There is currently no procedure for isolation of E. coli from food or water based on a characteristic unique to the species and found in all virulence subgroups. Existing methods for detection of pathogenic E. coli first require time-consuming biochemical procedures to determine if an isolate is E. coli, followed by molecular and cell assays to determine whether specific virulence markers are present (16). During outbreaks caused by food- or water-borne E. coli it is important that this process be conducted rapidly, and identification of a marker unique to the species but universal in all six subgroups would be valuable.
The enzyme glutamate decarboxylase (GAD) has been reported to be limited to E. coli and is encoded by two virtually identical genes, gadA and gadB (22, 27). In the present study we have expanded on these initial reports, emphasizing the question of the prevalence of gadAB in E. coli O157:H7 and other strains of greatest public health significance. In addition to using PCR primers (22) to determine the range of gadAB in Shiga toxin producing strains, we developed specific DNA probes to determine whether this marker could be used for primary isolation from food and water, as would be essential during an outbreak. Additionally, we combined gadAB primers and primers for specific virulence genes to determine whether they could be used in multiplex PCR to rapidly identify these genes once an E. coli strain was isolated from contaminated food or water.
(Portions of this study were presented at the 100th General Meeting of the American Society for Microbiology in Los Angeles, Calif., 2000.)
MATERIALS AND METHODS
Strains tested for gadAB genes.
As indicated in Table 1, 173 enterovirulent E. coli isolates and 22 other enterics were tested for the presence of gadAB, as described below. All 195 isolates were obtained from culture collections in two U.S. Food and Drug Administration facilities: Pacific Regional Laboratory Northwest (PRLNW) and the Center for Food Science and Applied Nutrition (CFSAN). The E. coli isolates had been collected over a period of 9 years during outbreaks of gastroenteritis caused by contaminated foods. These were provided to the Food and Drug Administration by the Washington State Department of Health, Oregon State Health Division, California State Department of Health, Canadian Food Inspection Agency, Phil Tarr, Children's Hospital and Regional Medical Center, Montana State Department of Health, and the Centers for Disease Control and Prevention (CDC). Serotyping was performed in these facilities, although the original designation of some isolates was kindly confirmed by the CDC or, in PRLNW or CFSAN, by using the RIM E. coli O157:H7 test kit (Remel, Lenexa, Kans.). Additional serological data on E. coli O157:NM and Rough strains was previously reported (7, 13). The presence of stx1 and stx2 genes was also confirmed in all putative Shiga toxin-containing isolates using digoxigenin-11-dUTP (DIG) probes specific for these genes (JIB9/4 and JIB5/6) as described below.
TABLE 1.
Bacteria strains tested for gadAB
| Enterovirulent E. coli serotype or other enteric species | No. of strainsa | No. positive by:
|
|
|---|---|---|---|
| PCR | Probe | ||
| Enterovirulent E. coli serotype | |||
| O157:H7 | 103 | 103 | 103 |
| O157:NM | 16 | 16 | 16 |
| O157:H | 5 | 5 | 5 |
| O157:H7R | 1 | 1 | 1 |
| O165:H25 | 4 | 4 | 4 |
| O146:H21 | 2 | 2 | 2 |
| O137:H41 | 2 | 2 | 2 |
| O126:H27 | 1 | 1 | 1 |
| O125:NM | 1 | 1 | 1 |
| O125AC:NM | 1 | 1 | 1 |
| O121:H19 | 1 | 1 | 1 |
| O111:NHT | 1 | 1 | 1 |
| O111:NM | 1 | 1 | 1 |
| O104:H21 | 2 | 2 | 2 |
| O103:H6 | 1 | 1 | 1 |
| O103:H2 | 1 | 1 | 1 |
| O85:NM | 1 | 1 | 1 |
| O55:H7 | 8 | 8 | 8 |
| O50:H7 | 1 | 1 | 1 |
| O48:H21 | 1 | 1 | 1 |
| O48:H? | 1 | 1 | 1 |
| O45:H2 | 1 | 1 | 1 |
| O28AC:H35 | 1 | 1 | 1 |
| O26:H11 | 5 | 5 | 5 |
| O26:NM | 1 | 1 | 1 |
| O26:H2 | 1 | 1 | 1 |
| O22:H8 | 1 | 1 | 1 |
| O15:H7 | 1 | 1 | 1 |
| O14:H21 | 1 | 1 | 1 |
| O14:H19 | 2 | 2 | 2 |
| O11:NHT | 1 | 1 | 1 |
| O4:NM | 1 | 1 | 1 |
| EIEC | 1 | 1 | 1 |
| ETEC | 1 | 1 | 1 |
| Other enteric species | |||
| Klebsiella pneumoniae | 3 | 0 | 0 |
| Klebsiella terrigena | 1 | 0 | 0 |
| Salmonella dublin | 1 | 0 | 0 |
| Salmonella enteritidis | 1 | 0 | 0 |
| Salmonella enterica serovar Typhimurium | 1 | 0 | 0 |
| Salmonella gaminara | 1 | 0 | 0 |
| Enterobacter amnigenus | 1 | 0 | 0 |
| Enterobacter sakizaki | 1 | 0 | 0 |
| Proteus mirabilis | 1 | 0 | 0 |
| Hafnia alvei | 2 | 0 | 0 |
| Shigella sonnei | 2 | 2 | 2 |
| Shigella flexneri | 4 | 4 | 4 |
| Serratia ficara | 1 | 0 | 0 |
| Schewanella putrificans | 1 | 0 | 0 |
| Yersinia enterocolitica | 1 | 0 | 0 |
Total number of enterovirulent E. coli strains, 173; total number of other enteric strains, 22.
Detection of gadAB genes.
The distribution of gadAB genes among the 195 isolates was examined by PCR. The primers, which specifically amplify a fragment of 670 bp from both gadA (GenBank accession number M84024) and gadB (GenBank accession number M84025) genes were described by McDaniels et al. (22). To prepare PCR templates, 1.0 ml of overnight cultures of bacterial strains were centrifuged and the pellets were rinsed in sterile physiological saline and then resuspended in 1 ml of sterile deionized water and heated for 10 min in a boiling water bath. PCR was performed as described previously (22) except 1.5 mM MgCl2 and 1 μl of template were used in a 50-μl reaction volume. Amplification was done using a GeneAmp 9700 thermal cycler (PE Applied Biosystems, Foster City, Calif.) programmed for 25 cycles of 1 min at 94, 58, and 72°C, followed by a final 7-min extension. After amplification, 10 μl of each PCR product was analyzed by agarose gel (1%) electrophoresis in Tris-borate-EDTA buffer (GibcoBRL, Gaithersburg, Md.) and examined by UV transillumination using the Gel Doc 2000 system (Bio-Rad, Hercules, Calif.).
Enumeration of E. coli using gadAB-specific DNA probe.
To determine whether the gadAB genes can serve as a reliable marker for detecting E. coli, the gadAB-specific PCR product was labeled with DIG and used as a DNA probe to enumerate E. coli in naturally contaminated samples. The gadAB probe was generated and labeled with DIG during PCR amplification using a described procedure (Boehringer Mannhein Biochemicals user's guide for filter hybridization, version 2.0, Boehringer Mannheim Corp. [Roche], Indianapolis, Ind.). DIG-labeled DNA was excised from agarose gel, purified with Gene Clean II (Bio101, La Jolla, Calif.), and stored at −20°C.
Samples of wastewater effluent, runoff water adjacent to cattle feed lots, and cheeses that are naturally contaminated with E. coli were tested by colony hybridization with a DIG-gadAB probe. For comparison, the same samples were tested by three standard E. coli enumeration methods that are based on lactose fermentation. These methods included direct plating on violet red bile agar with 4-methylumbelliferyl-β-d-glucuronide (VRBAM) (Difco, Detroit, Mich.) (16), the most probable number (MPN) assay (16), and membrane filtration using m-ColiBlue24 (CB) (Hach, Loveland, Colo.) (11).
Colony blots were prepared by spread plating serially diluted samples on tryptic soy agar with 0.6% yeast extract. After incubating at 45.5°C for 24 h, the plates were chilled for 1 h at 5°C; then Whatman 541 filters were placed on the agar surface for 5 min to pick up the colonies. Colonies adhering to the filters were lysed, denatured, fixed, and probed by DNA hybridization as previously described (28).
Data on enumeration of the natural E. coli populations by the four methods were collected on a Microsoft Excel spreadsheet and analyzed using paired two-tailed Student's t tests. Means were calculated on the basis of triplicate analysis for the colony hybridization, VRBAM, and CB methods and duplicate analysis for MPN.
Analysis of E. coli in foods by gadAB-specific PCR.
The detection sensitivity of the gadAB PCR assay was determined using produce rinsate that was spiked with E. coli strain JSW1, a culture previously isolated in PRLNW and identified according to the Bacteriological Analytical Manual (16) and lacking the stx1 and stx2 genes. The samples were prepared by placing 454 g of fresh produce (parsley, cilantro, or strawberry) in sterile plastic bags with 454 ml of Butterfield's diluent and rinsing by shaking at 100 rpm for 5 min at room temperature. The rinsate was then seeded with E. coli cells to yield final cell concentrations of 1,000, 99, 8, and 2 CFU/ml. In addition, 908-ml volumes of Butterfield's diluent (16) were spiked at the same levels to determine whether components of the rinsate exerted inhibitory effects on E. coli outgrowth. One milliliter portions of each spiked rinsate, or buffer, were then added to 9 ml of lauryl tryptose (LST) broth and incubated for 24 h at 35°C, and PCR templates were prepared from the resulting enrichments.
Multiplex PCR analyses of pathogenic E. coli in seeded produce samples.
To determine whether the gadAB gene target can be used in a multiplex PCR assay to detect pathogenic E. coli, the gadAB-specific primers were combined with primers for stx1 (JIB9/4) and stx2 (JIB5/6) to develop a multiplex PCR assay. The stx primer sequences and amplification conditions used are as previously described (15, 17, 28). Samples of rinsate from parsley, cilantro, and strawberry and inoculated buffer controls (positive controls) were prepared as described above and spiked with E. coli O157:H7 at levels of 1,000, 126, 15, and 2 CFU per 50 ml of rinsate or buffer. The strain used, SEA6341, produces both stx1 and stx2. The entire 50-ml volume of spiked rinsates was then combined with 50 ml of 2× EHEC enrichment broth (EEB) and incubated with shaking at 37°C for 24 h (16). Templates were prepared from the enrichment and used in PCR amplification of the gadAB, stx1, and stx2 genes.
RESULTS
Distribution of gadAB genes.
A total of 195 enteric strains were tested by PCR and probed for the presence of the gadAB genes. All 173 strains of E. coli examined had the 670-bp product and therefore carried gadAB genes (Table 1). The E. coli strains examined included 103 strains of O157:H7 serotype, 22 variant strains of O157 that were nonmotile, H−, or rough, and 48 strains of enteropathogenic E. coli, enteroinvasive E. coli, and other Shiga toxin E. coli that were not the O157 serotype. Of the 22 non-E. coli enteric strains tested, only the 6 Shigella spp. were positive for gadAB.
Enumeration of E. coli by gadAB probe.
One cheese and four water samples containing natural E. coli populations were analyzed by colony hybridization using a DIG-labeled gadAB DNA probe and compared to three standard enumeration methods that are based on lactose fermentation. Results in Table 2 show that the counts obtained by probe hybridization were higher than the MPN estimates in all samples except for cheese. Analysis of results by Student's t test indicated that the values obtained by the gadAB probe were significantly larger than those for VRBAM and MPN in both the primary effluent sample and runoff sample 2, significantly lower than CB in runoff sample 3, and not significantly different from other method means in any other samples.
TABLE 2.
Enumeration of natural E. coli populations using a gadAB hybridization probe and three standard methods
| Sample type and analysis methods |
E. coli population
|
|
|---|---|---|
| Mean (log10) | 95% CIa (log10) | |
| 1. Primary effluent, wastewater treatment plant | ||
| gadAB probe | 2.740 | 2.565–2.834 |
| VRBAM | 2.301 | 2.100–2.438 |
| m-ColiBlue24 | 2.643 | 2.429–2.786 |
| MPN | 0.556 | −0.769–1.253 |
| 2. Runoff water, cattle feedlot no. 1 | ||
| gadAB probe | 4.564 | 4.118–4.779 |
| VRBAM | 4.397 | 4.362–44.31 |
| m-ColiBlue24 | 4.819 | 4.472–4.885 |
| MPN | 2.544 | 1.819–3.176 |
| 3. Runoff water, cattle feedlot no. 2 | ||
| gadAB probe | 4.954 | 4.848–5.039 |
| VRBAM | 4.491 | 4.447–4.531 |
| m-ColiBlue24 | 5.066 | 5.002–5.121 |
| MPN | 4.041 | 3.255–4.613 |
| 4. Runoff water, cattle feedlot no. 3 | ||
| gadAB probe | 4.564 | 4.287–4.732 |
| VRBAM | 4.457 | 4.404–4.504 |
| m-ColiBlue24 | 4.888 | 4.854–4.919 |
| MPN | 3.381 | 2.623–4.000 |
| 5. Naturally contaminated cheese | ||
| gadAB probe | <1.519 | |
| VRBAM | <1.519 | |
| m-ColiBlue24 | 1.519 | −1.491–1.987 |
| MPN | 1.519 | 0.833–2.137 |
CI, confidence interval.
Detection of E. coli in spiked produce rinsates using gadAB PCR.
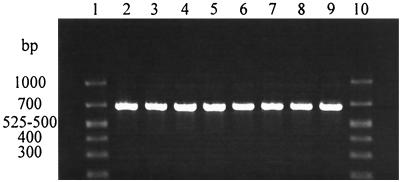
Rinsates of strawberries, parsley, and cilantro that were spiked with various levels of E. coli were tested by gadAB PCR after enrichment in LST for 24 h. Rinsates from all produce samples as well as inoculated medium controls (LST) that were spiked with as few as 8 CFU of E. coli cells/ml all yielded the 670-bp gadAB-specific PCR product (Fig. 1). The gadAB-specific amplicon was detected in rinsates of strawberry and parsley and inoculated medium controls that were spiked with as few as 2 CFU of E. coli/ml (data not shown).
FIG. 1.
Agarose gel of electrophoresis of products amplified by gadAB PCR from samples of produce rinsates spiked with E. coli at different levels. Lanes 1 and 10, BioMarker Low molecular size standards (BioVentures, Inc., Murfreesboro, Tenn.), with the sizes of certain fragments at the left; lanes 2 and 3, LST enrichment broth (positive medium control); lanes 4 and 5, parsley rinsate; lanes 6 and 7, strawberry rinsate; lanes 8 and 9, cilantro rinsate. Samples shown in lanes 2, 4, 6, and 8 were seeded with 99 CFU of E. coli cells, and those in lanes 3, 5, 7, and 9 were seeded with 8 CFU of E. coli cells.
Multiplex PCR for detection of pathogenic E. coli in produce rinsate.
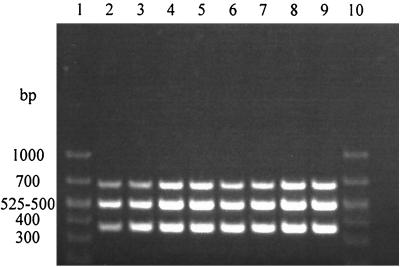
A multiplex PCR specific for the gadAB, stx1, and stx2 genes was used to detect the presence of EHEC in spiked produce rinsates. The specific products from this amplification reaction were 670, 513, and 364 bp for the gadAB, stx1, and stx2 genes, respectively. Rinsates of strawberries, parsley, cilantro, and inoculated medium controls (EEB) that were spiked with as few as 8 CFU of EHEC O157:H7 yielded the three specific PCR products after overnight enrichment in EEB (Fig. 2). All three PCR products were also observed in rinsates of strawberry, parsley, and inoculated controls (EEB) that were spiked with as few as 2 CFU of E. coli O157:H7 (data not shown).
FIG. 2.
Agarose gel of electrophoresis of products amplified by multiplex PCR from samples of produce rinsates spiked with E. coli O157:H7 at different levels. The products, with the expected base pair sizes in parenthesis, are: gadAB (670 bp), stx1 (513 bp), and stx2 (364 bp). Lanes 1 and 10, BioMarker Low molecular size standards (BioVentures, Inc., Murfreesboro, Tenn.), with the sizes of some of the fragments shown to the left of the gel; lanes 2 and 3, EEB enrichment broth (positive medium controls); lanes 4 and 5, parsley rinsate; lanes 6 and 7, strawberry rinsate; and lanes 8 and 9, cilantro rinsate. Samples shown in lanes 2, 4, 6, and 8 were seeded with 127 CFU of E. coli O157:H7 cells, and those in lanes 3, 5, 7, and 9 were seeded with 15 CFU of E. coli O157:H7 cells.
DISCUSSION
In the identification of pathogenic E. coli groups, isolates must first be identified as E. coli before they are tested for group-specific virulence traits. Because the groups are phenotypically diverse, existing methods for detecting E. coli are not always useful for pathogenic strains. Therefore we examined the potential of using gadAB genes as a prescreening marker for identifying pathogenic E. coli in foods. PCR analysis of various enteric species showed that the gadAB genes were present in all E. coli strains examined, including isolates of all pathogenic subgroups and phenotypic variants of EHEC of the O157:H7 serotype that were nonmotile (10), glucuronidase positive (13), and did not express the O157 antigen (7).
The only other enteric bacteria that carried the gadAB genes were the six strains of Shigella, a genus so closely related to E. coli that some investigators conclude they constitute a single species (4). An ideal diagnostic test might differentiate between the two but from a public health perspective, the detection of Shigella by a GAD-based assay would not detract from the utility of this test, as Shigella is also a human pathogen. Additionally, since GAD-positive isolates will be further tested for virulence genes, any Shigella isolates will be easily distinguished from pathogenic E. coli by these tests. One exception however, may be Shigella dysenteriae type I, which produces an stx1 that is virtually identical to that produced by EHEC strains (24). The results of our studies on GAD distribution are consistent with previous findings. Rice et al. (26) used a phenotypic GAD assay to show that this trait was 95% specific for E. coli strains. McDaniels et al. (22) used both phenotypic and genotypic assays and detected GAD in 62 of 64 E. coli isolates tested and gadAB in all 64, including the pathogenic groups that are implicated in food-borne illness. McDaniels et al. examined 32 enterovirulent E. coli strains. In the present study we extended the number of virulent strains known to contain gadAB by 173, including numerous atypical variants.
Standard methods to detect E. coli are based on this organism's ability to ferment lactose at elevated temperatures (16). To determine the reliability of gadAB as a detection marker for E. coli, we used a DIG-labeled DNA probe to gadAB to enumerate E. coli in naturally contaminated food and water samples. To our knowledge this is the first comparison of E. coli enumeration on the basis of gadAB versus growth in standard lactose-containing media. Comparative analysis with three standard enumeration methods showed that the E. coli counts based on gadAB were not significantly different from those obtained with m-ColiBlue24 in three of four samples and were not significantly different or significantly larger than those from MPN or VRBAM methods in all four environmental samples. Further testing is necessary, but this preliminary comparison of E. coli enumeration within these five samples indicates that the gadAB marker was as reliable as lactose fermentation-based standard tests. Furthermore, since gadAB genes are present in all E. coli tested to date, this marker has the advantage of being able to detect atypical, lactose-nonfermenting variants of E. coli, which may comprise as much as 10% of all isolates (5), and the strains of the pathogenic enteroinvasive E. coli group, which do not have the ability to ferment lactose (2).
In the analysis of E. coli using glucuronidase or the uidA gene that encodes the enzyme, Bej et al. (1) showed that a genotypic assay was more sensitive than phenotypic assays. Bacteria can sometimes carry genetic sequences that are not expressed, or perhaps the expression of the gene is repressed by physiological factors. In our study, a PCR assay specific for the gadAB gene was found to be very sensitive—capable of detecting, after overnight enrichment, as few as 2 CFU of E. coli initially seeded into various produce rinsate samples.
Law discussed the wide range of virulence among EHEC and even within the O157 serogroup. This variation may stem from the location of putative virulence factors on such mobile genetic elements as plasmids, bacteriophages, and pathogenicity islands, which may also be subject to numerous insertions and deletions (19). In contrast, gadAB genes are not only chromosomal but are found at two loci (27). Glutamate-dependent acid resistance is one of the two mechanisms by which E. coli survives exposure to low pH, such as passage through the stomach after ingestion (20, 21). Retention of this enzyme is therefore important for cellular survival, and GAD represents a stable marker for E. coli detection. GAD does not necessarily correlate with any specific virulence marker.
Since gadAB appeared to be a reliable and sensitive marker for E. coli, we examined whether these primers could be combined with primers to genes to simultaneously identify pathogenic E. coli. Primers to stx1 and stx2 were pooled with those for gadAB to form a multiplex PCR and used to test the presence of EHEC seeded into produce rinsates. PCR analysis of these samples after culture enrichment showed that as few as 2 CFU of E. coli O157:H7 seeded into various produce rinsates could be efficiently detected, with the positive samples showing all three specific PCR products.
In the PCR analysis of produce rinsates, we did not attempt to directly detect the presence of E. coli or E. coli O157:H7 in these samples without enrichment, because many foods are known to contain inhibitors that can affect PCR assays (18). Hence, the inclusion of a short culture enrichment was often beneficial in diluting out the inhibitory effects of these components and is standard practice in attempts to isolate potential pathogens from contaminated food. Furthermore, enterovirulent E. coli are thought to constitute a small percentage of total E. coli in most food or water samples and enrichment allows selective growth of the target population to levels detectible by molecular analysis. Current procedures mandate analysis for virulent E. coli only when specified levels of nonvirulent E. coli are found in food samples (16).
In conclusion, we have found that the gadAB gene was prevalent in E. coli, including in the pathogenic E. coli groups and its atypical phenotypic variants. DNA probe analysis showed that the gadAB marker was just as reliable as lactose fermentation in detecting E. coli in food and water samples. When gadAB was combined with primers to stx1 and stx2, the multiplex assay was effective in detecting the presence of Shiga toxin E. coli in seeded produce rinsates. The gadAB marker therefore appears to be a suitable prescreening marker for E. coli or may be combined with primers to other trait virulence genes to detect the presence of pathogenic E. coli in foods.
ACKNOWLEDGMENTS
We thank Karen Jinneman, Chuck Kaysner, and Walter Hill for helpful technical discussions, Stephanie Harris, USEPA, for providing runoff samples, and Joy Wells, CDC, for providing cultures.
REFERENCES
- 1.Bej A K, McCarty S C, Atlas R M. Detection of coliform bacteria and Escherichia coli by multiplex polymerase chain reaction: comparison with defined substrate and plating methods for water quality monitoring. Appl Environ Microbiol. 1991;57:2429–2432. doi: 10.1128/aem.57.8.2429-2432.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doyle M P, Padhye V V. Escherichia coli. In: Doyle M P, editor. Foodborne bacterial pathogens. New York, N.Y: Marcel Dekker, Inc; 1989. p. 235. [Google Scholar]
- 3.Doyle M P, Schoeni J L. Survival and growth characteristics of Escherichia coli associated with hemorrhagic colitis. Appl Environ Microbiol. 1984;48:855–856. doi: 10.1128/aem.48.4.855-856.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards B H. Salmonella and Shigella species. Clin Lab Med. 1999;19:469–487. [PubMed] [Google Scholar]
- 5.Ewing W H. Edward's and Ewing's identification of Enterobacteriaceae. 4th ed. New York, N.Y: Elsevier; 1986. [Google Scholar]
- 6.Feng P, Lampel K A. Genetic analysis of uidA expression in enterohaemorrhagic Escherichia coli serotype O157. Microbiology. 1994;140:2101–2107. doi: 10.1099/13500872-140-8-2101. [DOI] [PubMed] [Google Scholar]
- 7.Feng P, Sandlin R C, Clark C H, Wilson R A, Nishibuchi M. Identification of a rough strain of Escherichia coli O157:H7 that produces no detectable O157 antigen. J Clin Microbiol. 1998;36:2339–2341. doi: 10.1128/jcm.36.8.2339-2341.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, P.Escherichia coli. In R. G. Laffe and J. S. Garcia (ed.), Guide to foodborne pathogens, in press. Wiley and Sons, Inc., New York, N.Y.
- 9.Ferenc J, Oliver J, Witkowski R, McLandsborough L, Levin E E. Studies on the growth of Escherichia coli O157:H7 strains at 45.5°C. J Food Prot. 2000;63:1173–1178. doi: 10.4315/0362-028x-63.9.1173. [DOI] [PubMed] [Google Scholar]
- 10.Fields P I, Blom K, Hughes H J, Helsel L O, Feng P, Swaminathan B. Molecular characterization of gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for the identification of E. coli O157:H7 and O157:NM. J Clin Microbiol. 1997;35:1066–1070. doi: 10.1128/jcm.35.5.1066-1070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant M A. A new membrane filtration medium for simultaneous detection and enumeration of Escherichia coli and total coliforms. Appl Environ Microbiol. 1997;63:3526–3530. doi: 10.1128/aem.63.9.3526-3530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunzer F, Bohm H, Russman H, Bitzman M, Aleksic S, Karch H. Molecular detection of sorbitol-fermenting Escherichia coli O157 in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1046–1048. doi: 10.1128/jcm.30.7.1807-1810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes P S, Blom K, Feng P, Lewis J, Stockbine N A, Swaminathan B. Isolation and characterization of a β-d-glucuronidase-producing strain of Escherichia coli serotype O157:H7 in the United States. J Clin Microbiol. 1995;33:3347–3348. doi: 10.1128/jcm.33.12.3347-3348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill W E, Carlisle C L. Loss of plasmids during enrichment for Escherichia coli. Appl Environ Microbiol. 1981;41:1046–1048. doi: 10.1128/aem.41.4.1046-1048.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill W E, Jinneman K C, Trost P A, Bryant J L, Bond J, Wekell M M. Multiplex polymerase chain reaction detection of Shiga-like toxin genes in Escherichia coli. U.S. Food and Drug Administration Laboratory Information Bulletin No. 3811. U.S. Washington, D.C.: Food and Drug Administration; 1993. [Google Scholar]
- 16.Hitchins A D, Feng P, Watkins W D, Rippey S R, Chandler L A. FDA bacteriological analytical manual. 8th ed. 1999. Chapter 4. Escherichia coli and the coliform bacteria. , revision A. Association of Official Analytical Chemists International, Gaithersburg, Md. [Google Scholar]
- 17.Jinneman K C, Trost P A, Hill W E, Weagant S D, Bryant J L, Kaysner C A, Wekell M M. Comparison of template preparation methods from food for amplification of Escherichia coli O157 Shiga-like toxins type I and II DNA by multiplex polymerase chain reaction. J Food Prot. 1995;58:722–726. doi: 10.4315/0362-028X-58.7.722. [DOI] [PubMed] [Google Scholar]
- 18.Lantz P-G, Hahn-Hagerdal B, Radstrom P. Sample preparation methods in PCR-based detection of food pathogens. Trends Food Sci Technol. 1994;5:384–389. [Google Scholar]
- 19.Law D. Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli. J Appl Microbiol. 2000;88:729–745. doi: 10.1046/j.1365-2672.2000.01031.x. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, Lee I S, Frey J, Slonczewski J L, Foster J W. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Smith M P, Chapin K C, Baik H S, Bennett G N, Foster J W. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDaniels A E, Rice E W, Reyes A L, Johnson C H, Haugland R A, Stelma G N. Confirmational identification of Escherichia coli, a comparison of genotypic and phenotypic assays for glutamate decarboxylase and β-d-glucuronidase. Appl Environ Microbiol. 1996;62:3350–3354. doi: 10.1128/aem.62.9.3350-3354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng, J. H., P. Feng, and M. P. Doyle. Pathogenic Escherichia coli. In Compendium of methods for the microbiological examination of foods, in press. American Public Health Association, Washington, D.C.
- 24.Narato J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Padhye N V, Doyle M P. Escherichia coli O157:H7: epidemiology, pathogenesis, and methods for detection in food. J Food Prot. 1992;55:555–565. doi: 10.4315/0362-028X-55.7.555. [DOI] [PubMed] [Google Scholar]
- 26.Rice E W, Johnson C H, Dunnigan M E, Reasoner D J. Rapid glutamate decarboxylase assay for detection of Escherichia coli. Appl Environ Microbiol. 1993;59:4347–4349. doi: 10.1128/aem.59.12.4347-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith D K, Kassam T, Singh B, Elliot J F. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J Bacteriol. 1992;174:5820–5826. doi: 10.1128/jb.174.18.5820-5826.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weagant S D, Jagow J A, Omiecinski C J, Kaysner C A, Hill W E. Development of digoxigenin-labeled PCR amplicon probes for use in the detection and identification of enteropathogenic Yersinia and Shiga toxin-producing Escherichia coli from foods. J Food Prot. 1999;62:438–443. doi: 10.4315/0362-028x-62.5.438. [DOI] [PubMed] [Google Scholar]