Abstract
Purpose
Adaptive proton therapy (APT) of lung cancer patients requires frequent volumetric imaging of diagnostic quality. Cone‐beam CT (CBCT) can provide these daily images, but x‐ray scattering limits CBCT‐image quality and hampers dose calculation accuracy. The purpose of this study was to generate CBCT‐based synthetic CTs using a deep convolutional neural network (DCNN) and investigate image quality and clinical suitability for proton dose calculations in lung cancer patients.
Methods
A dataset of 33 thoracic cancer patients, containing CBCTs, same‐day repeat CTs (rCT), planning‐CTs (pCTs), and clinical proton treatment plans, was used to train and evaluate a DCNN with and without a pCT‐based correction method. Mean absolute error (MAE), mean error (ME), peak signal‐to‐noise ratio, and structural similarity were used to quantify image quality. The evaluation of clinical suitability was based on recalculation of clinical proton treatment plans. Gamma pass ratios, mean dose to target volumes and organs at risk, and normal tissue complication probabilities (NTCP) were calculated. Furthermore, proton radiography simulations were performed to assess the HU‐accuracy of sCTs in terms of range errors.
Results
On average, sCTs without correction resulted in a MAE of 34 ± 6 HU and ME of 4 ± 8 HU. The correction reduced the MAE to 31 ± 4HU (ME to 2 ± 4HU). Average 3%/3 mm gamma pass ratios increased from 93.7% to 96.8%, when the correction was applied. The patient specific correction reduced mean proton range errors from 1.5 to 1.1 mm. Relative mean target dose differences between sCTs and rCT were below ± 0.5% for all patients and both synthetic CTs (with/without correction). NTCP values showed high agreement between sCTs and rCT (<2%).
Conclusion
CBCT‐based sCTs can enable accurate proton dose calculations for APT of lung cancer patients. The patient specific correction method increased the image quality and dosimetric accuracy but had only a limited influence on clinically relevant parameters.
Keywords: adaptive proton therapy, cone‐beam computed tomography, deep learning, lung cancer, synthetic CT
1. INTRODUCTION
Proton therapy can deliver highly conformal dose distributions, leading to lower normal tissue dose, sparing of organs at risk (OAR), and target dose escalation. The dosimetric advantages of proton therapy can be achieved by the characteristic depth‐dose profile as protons traverse matter and deposit most of their dose at an energy dependent depth (Bragg peak) after which protons rapidly stop. 1 Compared to conventional photon beam therapy, this behavior results in a lower entrance and exit dose.
However, the beneficial depth‐dose characteristic of proton beams leads to an increased sensitivity of proton dose distributions to density changes along the beam path. Density shifts can occur due to anatomical variations, patient alignment errors, and changes in tumor size (growth/shrinkage). Adaptive proton therapy (APT) aims at detecting such anatomical changes and re‐adjusting treatment plans according to the updated patient anatomy. 2 , 3 , 4 Frequent patient imaging is a pivotal part of APT and provides the foundation for adaptation decisions. For daily APT, cone‐beam computed tomography (CBCT) images have the potential to serve as an alternative to conventional computed tomography (CT). In proton therapy, CBCTs are often routinely acquired for daily pre‐treatment patient alignment. The CBCT acquisition protocols used in radiotherapy are optimized to deliver significantly lower imaging dose than conventional diagnostic CT scans, making CBCTs more suitable for repeated imaging. Although the image quality of CBCTs is sufficient for position verification, they suffer from severe image artifacts which prevent them from being suitable for accurate proton dose calculations.
To correct CBCT deficiencies and enable CBCT‐based adaptive radiotherapy workflows, several methods have been developed and investigated in the context of photon and proton dose calculations in various anatomical locations. For the thorax, this includes techniques based on CBCT calibration, 5 , 6 , 7 HU‐overrides, 8 , 9 , 10 , 11 deformable image registration, 11 , 12 , 13 , 14 , 15 and Monte Carlo simulations. 16 , 17 Recently, research activities focused heavily on developing and evaluating deep learning methods to correct CBCTs and generate the so‐called synthetic CTs (sCTs). 18 , 19 , 20 , 21 , 22 In previous studies, deep learning methods have shown the ability to generate sCTs suitable for proton dose calculations for head and neck, 23 , 24 pelvis, 25 and prostate cancer patients. 26 , 27 However, regarding lung cancer treatment adaptation, only results for photon dose calculations have been reported. 22 , 28
In this study, we investigated the generation of CBCT‐based sCTs for adaptive proton therapy of lung cancer patients. Synthetic CTs were generated using a deep convolutional neural network (DCNN) which was previously evaluated for H&N cancer patients. 23 , 24 Furthermore, we proposed an accompanying patient‐specific correction strategy to further improve image quality of the resulting sCTs. Synthetic CTs were evaluated in terms of image quality and proton range error. The clinical suitability was assessed by recalculating clinically used treatment plans on sCTs and same‐day repeat CTs. Based on these dose distributions, gamma pass ratios, dose statistics for target volumes (TV) and organs at risk (OAR), and normal tissue complication probabilities (NTCP) were calculated.
2. MATERIALS AND METHODS
2.1. Patient datasets
A dataset containing 33 thoracic cancer patients, treated with pencil beam scanning proton therapy (PBS‐PT) at the University Medical Center Groningen (UMCG), was used in this study to train and evaluate a DCNN with an accompanying patient‐specific correction technique. 27 patients were treated for lung cancer, while the remaining six patients were either diagnosed with thoracic thymoma or mediastinal lymphoma. All 33 patients were imaged with the same acquisition protocols. Only lung cancer patients were included in the dosimetric evaluation of the sCTs. The lung cancer patients (15 female, 12 male) were aged between 46 and 83 years, with a median age of 69 years. For eight patients, the tumor was located in the left lung; for 18 in the right lung; and in one patient, tumor tissue was present on both sides. The tumor position also varied between upper (13 patients) and middle/lower lobe (14 patients). A table with patient demographic information is available in the Supporting Information (Table S1).
2.2. Imaging data
For each patient CBCT, planning CT and repeat CT images were used. CBCT images were acquired with an IBA Proteus Plus (IBA, Belgium) and reconstructed with the clinically used protocol. Repeat 4D‐CT and planning 4D‐CT scans were acquired on a Siemens SOMATOM Confidence (Siemens Healthineers, Germany) and on a Siemens SOMATOM Definition AS scanner, respectively, using the same imaging protocol. For treatment planning and dose calculation, average 4DCTs were generated from the 10 breathing phases of pCTs and rCTs. More detailed imaging and reconstruction parameters for CBCT, rCT, and pCT are listed in the Supporting Information (Table S2).
Repeat CT scans were acquired on the same day as the CBCT, used for training of the DCNN and selected as reference for image quality and dosimetric evaluation. There was a time difference of a few weeks between the pCT acquisition, used within the patient specific correction workflow, and the rCT/CBCT acquisition. For all patients, the first available rCT‐CBCT pair (acquired in the first week of treatment) was chosen.
2.3. Image pre‐processing
Before training the DCNN with CBCT‐rCT image pairs, several pre‐processing steps were performed. First, CBCTs were rigidly registered to the same‐day rCT and the patient outline was automatically segmented on CBCT and rCT using Plastimatch 29 (www.plastimatch.org). Voxels outside the patient outline were set to −1000 HU on CBCT and rCT. To account for the limited CBCT field‐of‐view (FOV) in superior‐inferior direction, the rCT and the respective mask were cropped to cover the same FOV. To reduce residual errors between CBCT and rCT, a deformable image registration (DIR) algorithm, implemented in the open‐source MATLAB toolbox openREGGUI (www.openreggui.org), was used to deformably register the rCT to the CBCT. This DIR‐algorithm has been found to be suitable for CBCT/CT image registration in previous studies. 14 , 30 , 31 The resulting image pairs of CBCT and deformed rCT were used to train the neural network.
2.4. Neural network
To generate CBCT‐based sCTs, a deep convolutional neural network (DCNN), previously described by Spadea et al. 32 was utilized. Figure S3 in the Supporting Information depicts the network architecture. The DCNN is composed of an encoding and decoding path to extract features from the CBCT and reconstruct it with accurate CT‐numbers. Similar to Spadea et al., three individual networks were trained exclusively with axial, sagittal, or coronal slices. A final sCT was created by averaging the network outputs from each training. Mean absolute error together with L1‐regularization was used as loss function to train the network. Due to the limited dataset size, threefold cross validation was applied. This allowed utilizing all 33 patients for evaluation purposes. We randomly split the dataset into three subsets of 11 patients each. Two subsets were used for training, and the third subset was used for evaluation. The training was repeated so that each subset was used for evaluation once. Based on previous experience with this network architecture, a batch size of 1 was used, and the training process was stopped when no decrease in loss was observed for five consecutive epochs. 23 , 24 A NVIDIA GTX 1080 Ti with 11 GB of VRAM was used for training and inference of the neural network.
2.5. Planning CT‐based patient‐specific correction method
In addition to the DCNN, we introduced a patient‐specific correction method. The correction workflow utilizes each patient's pCT, which contains accurate lung CT‐numbers but was acquired several weeks before the CBCT acquisition. In a first step, the planning CT was deformably registered to the synthetic CT using openREGGUI. Afterward, the registered pCT was subtracted from the original sCT (sCTorig), generating a difference image. A threshold was applied to the difference image to exclude differences bigger than ±150 HUs. This thresholding makes the correction method insensitive to major anatomical changes (e.g., tumor growth, alignment errors) since it is excluding areas that significantly change between pCT and CBCT acquisition. The threshold value (150 HU) controls the impact the pCT has on the final sCT and was found empirically by calculating gamma pass ratios for sCTs with a variety of threshold values. Afterwards, three HU‐regions were segmented on the sCT: region 1, containing air volumes and lung tissue (−1000 HU to −300 HU); region 2, covering soft tissues (−300 to 200 HU); and region 3, containing bones (> 200 HU). Using the above‐described segmentation masks, each HU‐region of the difference image was smoothed individually using a 3D Gaussian kernel (6 voxel standard deviation) and combined into the final correction map. Smoothing each region individually ensures sharp edges between varying tissues (e.g., lung tissue—soft tissue, soft tissue—bone). In a final step, the correction map was subtracted from the original sCT to create the corrected sCT (sCTcor).
2.6. Image evaluation
The image similarity between the synthetic CTs (sCTorig/ sCTcor) and the deformed same day rCT was evaluated by calculating mean absolute error (MAE), mean error (ME), peak signal noise ratio (PSNR), and structural similarity index (SSIM), 33 , 34 defined in Equations (1) to (4):
| (1) |
| (2) |
| (3) |
| (4) |
where and are the respective HU values of sCT and rCT, is the total number of voxels within the patient outline, is the maximum HU value of sCT and rCT, and are the mean pixel values of sCT and rCT, and are the variances of sCT and rCT, is the covariance between sCT and rCT, and is the dynamic range of sCT and rCT. All image similarity metrics were only calculated for voxels within the patient outline. To analyze the MAE of various tissues, an MAE spectrum was generated for sCTorig and sCTcor by grouping voxels in bins of 20 HU and calculating MAE for each bin. Wilcox signed‐rank tests were used to check for statistical significance of differences between sCTorig and sCTcor.
2.7. Dosimetric evaluation
For the 27 lung cancer patients, clinical treatment plans were recalculated on both sCTs (sCTorig,/sCTcor) and compared to the same‐day rCTs using global gamma analysis (dose threshold of 10%, 2%/2 mm, and 3%/3 mm criteria). Dose calculations were performed in RayStation Research (Version 9A) using the clinical Monte Carlo dose engine with an uncertainty of 1% and a dose grid of 3 × 3 × 3 mm3. Clinical treatment plans consisted of three beam directions and were generated on the average 4D pCT using multi‐field and robust optimization, with a range uncertainty of ±3% and a setup error of 6 mm as defined in our clinical protocol. 35 Dose was prescribed to the ITV. All dosimetric evaluations were performed for the entire plan (all fields combined). The clinical suitability was evaluated by calculating mean dose differences in TVs (GTV, CTV) and selected OARs (heart, lung, esophagus, spinal cord). For the CTV, additional dosimetric parameters were also investigated (Dmax, D95, D98, V95, and V100). For the spinal cord, maximum dose instead of mean dose was reported. Delineations of TVs and OARs were transferred from the pCT, which for this purpose was deformably registered to the rCT.
2.8. Comparison between sCTcor and deformed pCT
Deformable image registration (DIR)‐based strategies for CBCT‐based proton dose calculations have been investigated in previous studies. 14 , 31 Within these approaches, the patient‐specific pCT, containing accurate HUs, is deformed to the CBCT to represent the daily patient geometry. Our proposed sCT correction method also relies on the deformed pCT for HU correction but is combined with additional smoothing and thresholding operations. Therefore, we performed an image quality and dosimetric comparison between a pure DIR‐based strategy (pCTdef) and our corrected sCT (sCTcor). pCTdef was generated with the same DIR‐algorithm and settings used in the patient‐specific correction described above. The comparison includes image quality metrics (MAE and ME) and gamma analysis. The same‐day rCT was used as reference for image quality and dosimetric evaluations.
2.9. NTCP
Based on the recalculation of clinical treatment plans, we also calculated NTCP, using NTCP‐models described in the Dutch National Indication Protocol for proton therapy of lung cancer (NIPP). 36 NTCP models are used to estimate the risk of developing certain side effects during radiation therapy. The indication protocol for lung cancer includes models for radiation pneumonitis, 37 acute dysphagia, 38 and 2‐year mortality. 39 Certain clinical parameters (e.g., age, smoking status, and tumor location) and mean dose values of OARs (heart, lung, and esophagus) are used as input parameters for the NTCP models. At Dutch proton therapy centers, NTCP models are used within the patient selection process for proton therapy. In our work, we used these models to evaluate the clinical similarity of rCTs and sCTs, by calculating the NTCP difference (ΔNTCP) between them.
2.10. Radiography simulations
To visualize and quantify the similarity between rCT and sCTs in terms of proton range, we performed proton radiography simulations (PR) on rCT, sCTorig, and sCTcor using a dedicated proton radiography module of openREGGUI (ww.openreggui.org). It employs a direct ray tracing algorithm to simulate PRs as acquired with a multi‐layer ionization chamber. 40 PRs were simulated from a gantry angle of 0 degrees (anterior–posterior direction), a pencil beam spacing similar to the rCT imaging grid (1 mm left‐to‐right, 2 mm inferior–superior), and an energy of 210 MeV. Range error maps were computed between rCT and sCTorig and between rCT and sCTcor for all patients. Range error was calculated similarly to previous studies. 41 , 42 , 43 The resulting range error maps were analyzed by calculating mean absolute range error (MARE) and mean range error (MRE). The influence of lung tissue on range errors was investigated by calculating MARE only including pencil beams traversing the lungs. To select these beams, the lungs were segmented on the rCT and the resulting lung mask was projected along the proton beam direction. This resulting 2D‐lung mask was applied to the range error maps.
3. RESULTS
3.1. Image evaluation
Figure 1 presents an overview of CBCT, sCTorig, sCTcor, and the reference rCT together with difference images between rCT and sCTorig/sCTcor. On average, sCTcor resulted in a significantly lower MAE than sCTorig with respective values of 30.7 ± 4.4 HU and 34.1 ± 5.5 HU (p‐value: < 1 × 10–6). Average ME changed from 4.3 ± 7.7 HU for sCTorig to 2.4 ± 3.9 HU for sCTcor, but the difference was not found to be statistically significant (p‐value > 0.05). Overall, ME showed a trend toward positive values indicating lower HU values on sCTs when compared to rCTs. SSIM remained virtually unchanged with values of 0.938 ± 0.019 for sCTorig and 0.941 ± 0.019 for sCTcor. The PSNR showed a slight improvement from sCTorig to sCTcor with values of 30.7 ± 3.3 dB to 31.2 ± 3.4 dB respectively but was not statistically significant (p‐value > 0.05).
FIGURE 1.

Axial and coronal slices of CBCT, sCTorig, sCTcor, and the reference rCT together with difference maps between sCTorig/cor and rCT. A HU‐window of 2000 (width)/ 0 (level) was used for sCTorig, sCTcor, and rCT
Detailed MAE and ME results for each patient individually are visualized in Figure 2. Individual results for the other metrics are reported in the Supporting Information (Figure S4).
FIGURE 2.
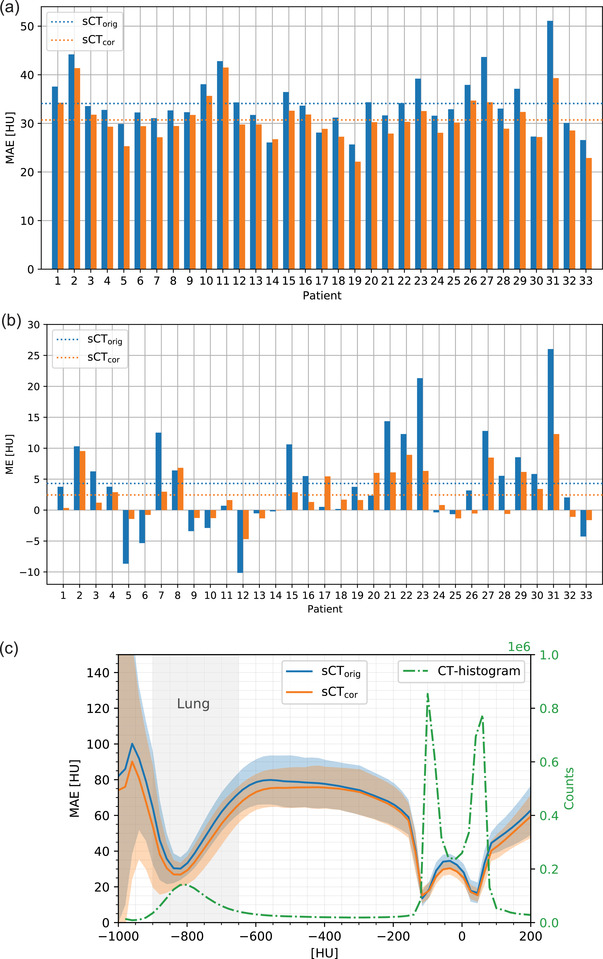
(a) MAE and (b) ME for sCTorig and sCTcor for each patient. The dashed lines indicate the average values. (c) Average MAE spectrum of sCTorig and sCTcor. The shaded area indicates one standard deviation. In green, an average image histogram is presented
Figure 2c shows the MAE spectrums for sCTorig and sCTcor. For voxels below 300 HU, sCTcor shows slightly lower MAE than sCTorig, indicating the effectiveness of the patient‐specific correction. Error regions however overlap for the entire HU‐range.
3.2. Dosimetric evaluation
Results from the gamma analysis of clinical treatment plans are shown in Figure 3a (3%/3 mm and 2%/2 mm criteria). The patient‐specific correction technique increased average 3%/3 mm gamma pass ratios from 93.7 ± 4.8% to 96.8 ± 2.4%. This difference was found to be statistically significant (p‐value: 4*10–4). Furthermore, the lowest observed 3%/3 mm pass ratio increased from 82.8% (sCTorig, patient 23) to 90.7% (sCTcor, patient 20). A similar trend was observed for 2%/2 mm pass ratios.
FIGURE 3.

(a) Gamma pass ratios (top: 3%/3 mm, bottom: 2%/2 mm) of sCTorig and sCTcor for each patient individually. The dotted line in the corresponding color indicates the mean value of sCTorig and sCTcor. This figure shows results for lung cancer patients only. (b) Relative dose differences between sCTs and rCT for target volumes and selected organs at risk. Mean dose was used for all structures, except the spinal cord (max dose)
Figure 3b presents boxplots of relative dose differences for TVs and OARs. For GTV and CTV, both sCTs showed very good agreement with mean doses calculated on the rCT. The mean dose differences were within ±0.5% for all patients. For the CTV, also Dmax, D95, D98, and V95 agreed well with the rCT for both sCTorig and sCTcor. Differences for all patients were within ±5% and mean differences close to 0%. Only V100 values showed larger discrepancies of up to −15% for sCTorig. Applying the correction reduced the maximum V100 difference to −7%. A figure containing CTV dose differences for these dosimetric parameters is presented in the Supporting Information (Figure S5). For OAR, mean doses varied greatly, with values between 0.3 and 37.9 GyRBE. Results for OAR with an absolute dose below 1 Gy were excluded. Overall, higher relative dose differences were observed for OAR. The largest differences occurred in the heart (mean dose) and the spinal cord (max dose), with values up to 10%. For lung and esophagus, values were within ±5% for all patients. Across all TVs and OARs, sCTcor resulted in a slightly lower variance than sCTorig.
In Figure 4, HU and dose profiles of rCT, sCTorig, and sCTcor are presented for patient 12. The selected profiles run parallel to the proton beam direction (gantry angle of 300 degrees) and the displayed dose values are only for the 300 degree beam direction instead of the entire plan. The HU‐ and dose‐profiles visualize the relationship of lung tissue inaccuracies of sCTorig and the resulting dose shift. Applying the patient‐based correction (sCTcor) restored accurate proton range, and good agreement with the rCT dose profile can be observed.
FIGURE 4.
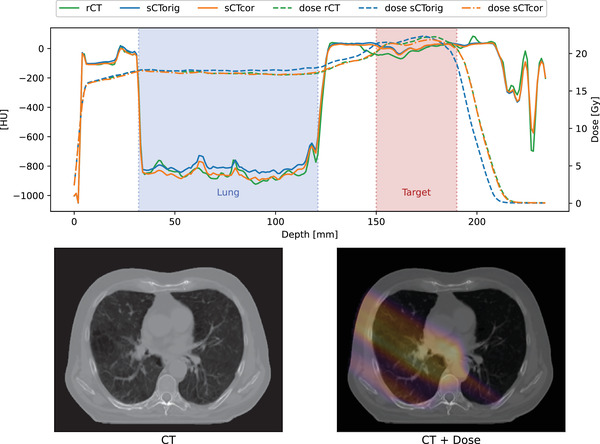
HU and dose profiles for rCT, sCTorig, and sCTcor. The selected profile is indicated with the blue arrow. Solid lines represent the HU‐profiles; dashed lines the corresponding dose profiles. The displayed dose is from the 330° beam direction only and does not represent the full plan dose
3.3. Comparison between pCTdef and sCTcor
Almost similar MAE and ME values were observed for pCTdef and sCTcor. Average MAE/ME values were 31.5 ± 6.6 HU/2.9 ± 4.7 HU for pCTdef and 30.7 ± 4.4 HU/2.4 ± 3.9 HU for sCTcor. The comparison of proton dose distributions resulted in higher gamma pass rates for sCTcor than for pCTdef (3%/3 mm: 96.8% vs. 95.6%, 2%/2 mm: 93.1% vs. 91.6%). For three patients, a 3%/3 mm pass rate below 85 % was observed for pCTdef, while for sCTcor, all patients achieved pass rates above 90%. These pCTdef outliers appeared when a significant anatomical change occurred between acquisition of pCT and CBCT. An example of such an anatomical change and figures showing evaluation results are presented in the Supporting Information (Figure S7).
3.4. NTCP
A boxplot of ΔNTCP values for sCTorig and sCTcor is presented in Figure 5. A high level of agreement between NTCP values, calculated on rCT and both sCTs, was observed for all patients and across all predicted toxicities. For radiation pneumonitis, the average ΔNTCP values were 0.0 ± 0.3% for sCTorig and 0.0 ± 0.2% for sCTcor (max. ΔNTCP values for pneumonitis: −1% for sCTorig, −0.9% for sCTcor). For dysphagia, average ΔNTCP values were −0.2 ± 0.4% for sCTorig and −0.1 ± 0.3% for sCTcor (max. values of −1.3% and −0.9%, respectively). The endpoint of 2‐year mortality resulted in average ΔNTCP values of 0.0 ± 0.3% for sCTorig and 0.0 ± 0.2% for sCTcor (max. values of −1.1% for sCTorig and −0.7% for sCTcor). Individual NTCP values for each patient and toxicity are presented in the Supporting Information (Figure S6).
FIGURE 5.
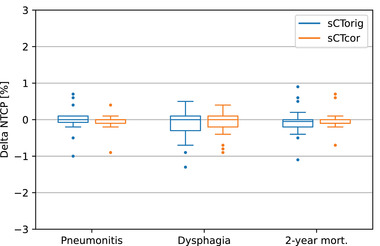
Delta NTCP values (NTCPrCT − NTCPsCT) for dysphagia, radiation pneumonitis, and 2‐year mortality, calculated on sCTorig and sCTcor
3.5. Proton radiography simulations
In Figure 6, range error maps between the reference rCT and the two synthetic CTs (Figure 6a: sCTorig; Figure 6b: sCTcor) are presented for patient 23, in which the largest relative MARE decrease was achieved by using the patient‐specific correction. Figure 6c shows an accompanying water‐equivalent thickness map. A reduction in range errors is clearly visible in the lungs, while the surrounding areas remain mainly unchanged. On average, MARE was reduced from 1.5 ± 0.5 mm on sCTorig to 1.1 ± 0.4 mm on sCTcor. This difference was found to be statistically significant (p‐value: < 10–5). MRE decreased from 0.6 ± 0.8 mm on sCTorig to 0.3 ± 0.6 mm on sCTcor (not significant, p‐value: 0.2).
FIGURE 6.

Range error maps for patient 23 between rCT and sCTorig (a) and between rCT and sCTcor (b). Panel (c) shows the corresponding water equivalent thickness map (calculated based on the rCT). Positive range errors indicate larger range on sCTs than rCTs; negative range errors lower range on sCTs than rCTs
Figure 7 shows MARE and MRE results for each patient individually. Overall, MRE is shifted toward positive values, indicating that an increased proton range was found on PR simulations based on sCTs. This is consistent with the observation of a positive shift in ME values. The MARE calculation considering only beams traversing lung tissue resulted in an average MARE of 2.1 ± 0.9 mm for sCTorig and 1.6 ± 0.6 for sCTcor. The MARE for the remaining beams is significantly lower on both sCTorig and sCTcor, with values of 0.9 ± 0.6 and 0.7 ± 0.5 mm, respectively. This indicates that the overall range error is mainly determined by the range error in lung tissue. Additional range error maps are presented in the Supporting Information.
FIGURE 7.
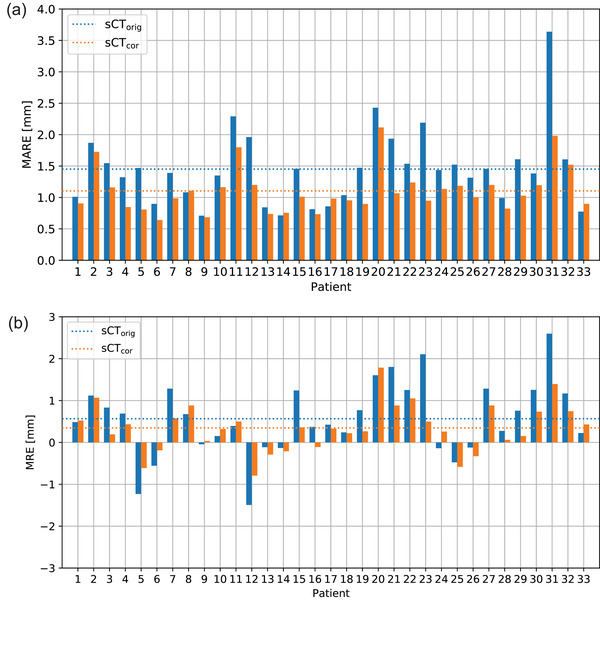
(a) Mean absolute range error for sCTorig and sCTcor. (b) Mean range error of sCTorig and sCTcor. The dashed lines indicate the mean values
4. DISCUSSION
Frequent imaging is a prerequisite for APT in lung cancer patients. Deep convolutional neural networks have previously shown their ability to correct HU deficiencies of routinely acquired CBCTs and thus enable CBCT‐based APT in other treatment sites. This study aimed at investigating image quality, dosimetric accuracy, and clinical suitability of deep learning based sCTs for lung cancer patients. We also proposed an accompanying patient‐specific correction technique, utilizing HU information from the pCT, which further improved the sCT in terms of dosimetric accuracy and image quality.
The patient‐specific correction method reduced the average MAE from 34.1 ± 5.5 HU to 30.7 ± 4.4 HU. This MAE is lower than previously reported results in literature. Maspero et al. achieved an MAE of 83 ± 10 HU. Their study used a single network for head and neck, lung, and breast cancer patients and a different network architecture (generative adversarial network, GAN). 22 Eckl et al. used a similar GAN architecture and reported a comparable MAE of 94 ± 32 HU. 28 However, image quality comparisons between different studies are challenging since sCT image quality depends on the specific CBCT acquisition protocols and image similarity metrics, such as MAE, are sensitive to the CBCT field‐of‐view and the used reference image (e.g., same‐ day, rigid‐ or deformable registration).
The studies by Maspero et al. and Eckl et al. only reported results for photon dose calculations. In the present study, for the first time, proton dose calculation accuracy of deep learning based synthetic CTs for APT of lung cancer patients was presented. We achieved average gamma pass ratios (3%/3 mm) of 93.7 ± 4.8% for the uncorrected and 96.8 ± 2.4% for the corrected sCTs. The lowest observed pass ratios increased from 82.8% to 90.7%. The results showed that sCTs with the lowest pass ratio benefited most from the patient‐specific correction strategy. This outcome is relevant for clinical implementation of sCTs, where outliers have to be avoided and consistent sCTs for a large patient cohort are desired. Besides investigating global dose differences using gamma analysis, we also performed a local dose evaluation for target volumes and organs at risk. The observed difference in mean dose to target volumes was below ± 0.5% for both sCTorig and sCTcor. Larger differences of up to 12% were measured for organs at risk (spinal cord and heart). However, the absolute dose in organs at risk varied greatly between patients and the largest relative deviations were seen for the lowest absolute doses. By calculating NTCP, we also showed that these dose differences are negligible for calculating the risk of developing certain side effects (dysphagia, radiation pneumonitis and 2‐year mortality). The average differences between NTCP calculated on the reference rCT and both sCTorig and sCTcor were below 0.2% and maximum ΔNTCP values did not exceed 2%.
The evaluation of several image quality and dose metrics shows the need for a broad evaluation of sCTs. Global image quality (MAE, ME, PSNR, SSIM) and dose (gamma analysis) metrics alone do not provide enough insights to assess the clinical suitability of sCTs. Local dose metrics (target and organ at risk doses, NTCP models) and proton radiography are valuable tools that provide evidence on the locality and clinical impact of sCT errors.
Proton radiography simulations served two purposes: 1) to visualize the similarity of HU values obtained in sCTs with respect to rCTs and thereby highlight anatomical areas with less accurate HUs and 2) to quantify the differences in proton range between rCTs and sCTs (with and without correction). Applying the patient‐specific correction reduced the overall MARE between rCT and sCT from 1.5 ± 0.5 mm to 1.1 ± 0.4 mm. This confirms the effectiveness of the correction strategy and is consistent with improvements seen in MAE and gamma pass ratios. Exclusively evaluating range errors for the lung region showed significantly higher MARE for both sCTorig (2.1 ± 0.9 mm) and sCTcor (1.6 ± 0.6 mm), which approximately doubles the range error observed in the remaining tissues. Based on these results, we conclude that it is more challenging for the DCNN to accurately reproduce HUs of lung tissues and that the patient‐specific correction technique is able to partially correct for it. The proton radiography simulations were performed for the entire patient, which were beneficial to assess the HU accuracy but do not represent a clinically feasible field size. Due to the detector size, in vivo proton radiography measurements using a multi‐layer ionization chamber are usually limited to small fields of a few square centimeters. 40 , 42 In vivo proton radiography measurements are envisioned to be utilized as a quality control tool to verify deep learning based sCTs within APT workflows. 44
The proposed patient‐specific correction technique was introduced to correct low frequency HU variations of the sCTs, which were most prevalent in lung tissue. Lung tissue is prone to scatter artifacts on the CBCT, which lead to a lack of detail and interferes with the fine structure. Correction maps and proton radiography simulations highlight the larger errors of lung tissue. The patient‐specific correction relies on accurate CT HU‐values. For the first few treatment fractions, the pCT is the only available CT image and was therefore chosen for our study. However, in a clinical workflow, pCTs can also be replaced with more recent rCT images if available. The results presented in our study show a worst‐case scenario, in which the correction is based on a CT image acquired several weeks before treatment (pCT). Although, by deforming the pCT to the sCT, thresholding the difference map and applying a smoothing filter, the influence of time and anatomical differences of the pCT are mitigated. Some patients (e.g., patients 8, 17, and 20) showed increased ME after applying the DIR‐based patient‐specific correction. However, no correlation between the deformation vector field properties (e.g., mean vector amplitude, Jacobian) and the ME was found. An increased ME after applying the correction does not necessarily indicate worse image quality. ME alone is not suitable as an image quality and similarity metric, since negative and positive HU errors can cancel each other out. ME should be considered in combination with a metric that also uses the absolute differences between two images (e.g., MAE).
The comparison of pure deformable image registration (pCTdef) and sCTcor highlighted the benefit of the combination of DCNN‐based sCT and patient‐specific HU‐correction. sCTcor resulted in an accurate representation of the daily anatomy and provided improved HU accuracy. For many patients, pCTdef resulted in comparable dosimetric accuracy as sCTcor, but for some patients, anatomical changes between acquisition of pCT and CBCT, which cannot always be modeled accurately by DIR, lead to outliers with significantly lower dose calculation performance (gamma pass ratio decrease of up to 17%). This makes clinical implementation of pure DIR‐based sCT generation challenging and favors DCNN‐based sCTs in combination with a pCT‐based correction method.
This study was performed with a limited dataset of 33 thorax cancer patients. To efficiently use the entire dataset for image and dose evaluation, a threefold cross validation procedure was used. Our dataset was limited since the treatment of lung cancer patients with proton therapy only recently started at our institution. The patient number will increase in the future and will enable studies to investigate the influence of the dataset size on image and dosimetric quality. The image quality of the initial CBCT has a major influence on sCT quality too. Higher image quality, especially of lung tissue, could further improve sCT accuracy. Currently, CBCT imaging parameters are chosen for patient alignment purposes. Better CBCT image quality would most likely be connected to an increased imaging dose, which would have a large impact on the imaging dose burden, particularly if CBCT images are acquired daily. Therefore, potential gains in image quality have to be carefully balanced with the dose exposure of patients. The influence of imaging parameters on lung sCTs should be investigated in further studies.
We used a similar neural network architecture in previous head and neck cancer patient studies, 23 , 24 which enables to establish a comparison of image quality and dosimetric accuracy between these anatomical locations. Lung sCTs (sCTorig) resulted in an average MAE of 34.1 ± 5.5 HU (sCTorig), which is lower than the MAE observed for H&N sCTs (40.2 ± 3.9 HU). This is contrary to the observed 3%/3 mm gamma pass ratios of clinical treatment plans, which were significantly higher for H&N cancer patients (98.8% vs. 93.7%). Even with the applied patient specific correction, lung sCTs resulted in a lower pass ratio (98.8% vs. 96.8%). This discrepancy is caused by the different tissue compositions, larger tissue heterogeneity, and the increased radiological depth in the lung region. The evaluation of NTCP values and target/OAR doses show similar accuracy for H&N and lung cancer patients, indicating similar clinical suitability of deep learning based sCTs for both treatment sites.
5. CONCLUSION
In this study, we proposed and evaluated a CBCT‐based sCT generation method for APT of lung cancer patients. We have shown that a DCNN in combination with a patient‐specific correction method can generate accurate sCTs for proton dose calculations. Clinically relevant dose statistics and NTCP values showed high agreement between sCTs and same‐day rCTs, indicating the potential suitability of the generated sCTs for application in APT workflows for lung cancer patients.
CONFLICT OF INTEREST
J. A. Langendijk is a consultant for International Scientific Advisory Committees of IBA and RaySearch. The Department of Radiation Oncology, University Medical Centre Groningen, has active research agreements with IBA, RaySearch, Siemens, Elekta, Leoni, and Mirada. A. Meijers is employed by Varian Medical Systems. Work in the context of this manuscript was conducted prior to the employment with Varian.
Supporting information
SUPPORTING INFORMATION
ACKNOWLEDGMENT
This study was financially supported by a grant from the Dutch Cancer Society (KWF research project 11518).
Thummerer A, Seller Oria C, Zaffino P, et al. Clinical suitability of deep learning based synthetic CTs for adaptive proton therapy of lung cancer. Med Phys. 2021;48:7673–7684. 10.1002/mp.15333
DATA AVAILABILITY STATEMENT
Research data cannot be shared.
REFERENCES
- 1. Newhauser WD, Zhang R. The physics of proton therapy. Phys Med Biol. 2015;60(8):R155–R209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sonke JJ, Belderbos J. Adaptive radiotherapy for lung cancer. Semin Radiat Oncol. 2010;20(2):94–106. [DOI] [PubMed] [Google Scholar]
- 3. Sonke JJ, Aznar M, Rasch C. Adaptive radiotherapy for anatomical changes. Semin Radiat Oncol. 2019;29(3):245–257. [DOI] [PubMed] [Google Scholar]
- 4. Albertini F, Matter M, Nenoff L, Zhang Y, Lomax A. Online daily adaptive proton therapy. Br J Radiol. 2020;93(1107):20190594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fotina I, Hopfgartner J, Stock M, Steininger T, Lütgendorf‐Caucig C, Georg D. Feasibility of CBCT‐based dose calculation: comparative analysis of HU adjustment techniques. Radiother Oncol. 2012;104(2):249–256. [DOI] [PubMed] [Google Scholar]
- 6. de Smet M, Schuring D, Nijsten S, Verhaegen F. Accuracy of dose calculations on kV cone beam CT images of lung cancer patients. Med Phys. 2016;43(11):5934–5941. [DOI] [PubMed] [Google Scholar]
- 7. Kaplan LP, Elstrøm UV, Møller DS, Hoffmann L. Cone beam CT based dose calculation in the thorax region. Phys Imaging Radiat Oncol. 2018;7:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Usui K, Ichimaru Y, Okumura Y, et al. Dose calculation with a cone beam CT image in image‐guided radiation therapy. Radiol Phys Technol. 2012;6(1):107–114. [DOI] [PubMed] [Google Scholar]
- 9. Dunlop A, McQuaid D, Nill S, et al. Comparison of CT number calibration techniques for CBCT‐based dose calculation. Strahlentherapie und Onkol. 2015;191(12):970–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen S, Le Q, Mutaf Y, et al. Feasibility of CBCT ‐based dose with a patient‐specific stepwise HU ‐to‐density curve to determine time of replanning. J Appl Clin Med Phys. 2017;18(5):64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giacometti V, King RB, Agnew CE, et al. An evaluation of techniques for dose calculation on cone beam computed tomography. Br J Radiol. 2019;92(1096):20180383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peroni M, Ciardo D, Spadea MF, et al. Automatic segmentation and online virtualCT in head‐and‐neck adaptive radiation therapy. Int J Radiat Oncol. 2012;84(3):e427–e433. [DOI] [PubMed] [Google Scholar]
- 13. Veiga C, McClelland J, Moinuddin S, et al. Toward adaptive radiotherapy for head and neck patients: feasibility study on using CT‐to‐CBCT deformable registration for “dose of the day” calculations. Med Phys. 2014;41(3):31703. [DOI] [PubMed] [Google Scholar]
- 14. Veiga C, Janssens G, Teng CL, et al. First clinical investigation of cone beam computed tomography and deformable registration for adaptive proton therapy for lung cancer. Int J Radiat Oncol Biol Phys. 2016;95(1):549–559. [DOI] [PubMed] [Google Scholar]
- 15. Yuan Z, Rong Y, Benedict SH, Daly ME, Qiu J, Yamamoto T. Dose of the day” based on cone beam computed tomography and deformable image registration for lung cancer radiotherapy. J Appl Clin Med Phys. 2019;21(1):88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thing RS, Bernchou U, Mainegra‐Hing E, Brink C. Patient‐specific scatter correction in clinical cone beam computed tomography imaging made possible by the combination of Monte Carlo simulations and a ray tracing algorithm. Acta Oncol (Madr). 2013;52(7):1477–1483. [DOI] [PubMed] [Google Scholar]
- 17. Thing RS, Bernchou U, Hansen O, Brink C. Accuracy of dose calculation based on artefact corrected cone beam CT images of lung cancer patients. Phys Imaging Radiat Oncol. 2017;1:6–11. [Google Scholar]
- 18. Kida S, Nakamoto T, Nakano M, et al. Cone beam computed tomography image quality improvement using a deep convolutional neural network. Cureus. 2018;10(4):e2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harms J, Lei Y, Wang T, et al. Paired cycle‐GAN‐based image correction for quantitative cone‐beam computed tomography. Med Phys. 2019;46(9):3998–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y, Lei Y, Wang T, et al. CBCT‐based synthetic CT generation using deep‐attention cycleGAN for pancreatic adaptive radiotherapy. Med Phys. 2020;47(6):2472–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liang X, Chen L, Nguyen D, et al. Generating synthesized computed tomography (CT) from cone‐beam computed tomography (CBCT) using CycleGAN for adaptive radiation therapy. Phys Med Biol. 2019;64(12):125002. [DOI] [PubMed] [Google Scholar]
- 22. Maspero M, Houweling AC, Savenije MH, et al. A single neural network for cone‐beam computed tomography‐based radiotherapy of head‐and‐neck, lung and breast cancer. Phys Imaging Radiat Oncol. 2020;14:24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thummerer A, Zaffino P, Meijers A, et al. Comparison of CBCT based synthetic CT methods suitable for proton dose calculations in adaptive proton therapy. Phys Med Biol. 2020;65(9):95002. [DOI] [PubMed] [Google Scholar]
- 24. Thummerer A, de Jong BA, Zaffino P, et al. Comparison of the suitability of CBCT‐ and MR‐based synthetic CTs for daily adaptive proton therapy in head and neck patients. Phys Med Biol. 2020;65(23):235036. [DOI] [PubMed] [Google Scholar]
- 25. Zhang Y, Yue N, Su M, et al. Improving CBCT quality to CT level using deep learning with generative adversarial network. Med Phys. 2021;48(6):2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Landry G, Hansen D, Kamp F, et al. Comparing Unet training with three different datasets to correct CBCT images for prostate radiotherapy dose calculations. Phys Med Biol. 2019;64(3):35011. [DOI] [PubMed] [Google Scholar]
- 27. Kurz C, Maspero M, Savenije MHF, et al. CBCT correction using a cycle‐consistent generative adversarial network and unpaired training to enable photon and proton dose calculation. Phys Med Biol. 2019;64(22):225004. [DOI] [PubMed] [Google Scholar]
- 28. Eckl M, Hoppen L, Sarria GR, et al. Evaluation of a cycle‐generative adversarial network‐based cone‐beam CT to synthetic CT conversion algorithm for adaptive radiation therapy. Phys Medica. 2020;80:308–316. [DOI] [PubMed] [Google Scholar]
- 29. Zaffino P, Raudaschl P, Fritscher K, Sharp GC, Spadea MF. Technical note: plastimatch mabs, an open source tool for automatic image segmentation. Med Phys. 2016;43(9):5155–5160. [DOI] [PubMed] [Google Scholar]
- 30. Veiga C, Alshaikhi J, Amos R, et al. Cone‐beam computed tomography and deformable registration‐based “dose of the day” calculations for adaptive proton therapy. Int J Part Ther. 2015;2(2):404–414. [Google Scholar]
- 31. Landry G, Nijhuis R, Dedes G, et al. Investigating CT to CBCT image registration for head and neck proton therapy as a tool for daily dose recalculation. Med Phys. 2015;42(3):1354–1366. [DOI] [PubMed] [Google Scholar]
- 32. Spadea MF, Pileggi G, Zaffino P, et al. Deep convolution neural network (DCNN) multiplane approach to synthetic CT generation from MR images—application in brain proton therapy. Int J Radiat Oncol Biol Phys. 2019;105(3):495–503. [DOI] [PubMed] [Google Scholar]
- 33. Wang Z, Simoncelli EP, Bovik AC, Multi‐scale structural similarity for image quality assessment. Conference Record of the Asilomar Conference on Signals, Systems and Computers. IEEE; 2003;2:1398–1402. 10.1109/acssc.2003.1292216 [DOI] [Google Scholar]
- 34. Renieblas GP, Nogués AT, González AM, Gómez‐Leon N, del Castillo EG. Structural similarity index family for image quality assessment in radiological images. J Med Imaging. 2017;4(3):035501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van der Laan HP, Anakotta RM, Korevaar EW, et al. Organ sparing potential and inter‐fraction robustness of adaptive intensity modulated proton therapy for lung cancer. Acta Oncol (Madr). 2019;58(12):1775–1782. [DOI] [PubMed] [Google Scholar]
- 36.Nederlandse vereniging voor Radiotherapie en Oncologie. Landelijk indicatieprotocol protonen thorax. https://nvro.nl/images/documenten/rapporten/LIPP_longen_final_01122019.pdf. Accessed November 9, 2021.
- 37. Appelt AL, Vogelius IR, Farr KP, Khalil AA, Bentzen SM. Towards individualized dose constraints: adjusting the QUANTEC radiation pneumonitis model for clinical risk factors. Acta Oncol (Madr). 2014;53(5):605–612. [DOI] [PubMed] [Google Scholar]
- 38. Dankers FJWM, Wijsman R, Troost EGC, et al. External validation of an NTCP model for acute esophageal toxicity in locally advanced NSCLC patients treated with intensity‐modulated (chemo‐)radiotherapy. Radiother Oncol. 2018;129(2):249–256. [DOI] [PubMed] [Google Scholar]
- 39. Defraene G, Dankers FJWM, Price G, et al. Multifactorial risk factors for mortality after chemotherapy and radiotherapy for non‐small cell lung cancer. Radiother Oncol. 2020;152:117–125. [DOI] [PubMed] [Google Scholar]
- 40. Farace P, Righetto R, Meijers A. Pencil beam proton radiography using a multilayer ionization chamber. Phys Med Biol. 2016;61(11):4078–4087. [DOI] [PubMed] [Google Scholar]
- 41. Farace P, Righetto R, Deffet S, Meijers A, Vander Stappen F. Technical note: a direct ray‐tracing method to compute integral depth dose in pencil beam proton radiography with a multilayer ionization chamber. Med Phys. 2016;43(12):6405–6412. [DOI] [PubMed] [Google Scholar]
- 42. Meijers A, Seller Oria C, Free J, Langendijk JA, Knopf AC, Both S. Technical note: first report on an in vivo range probing quality control procedure for scanned proton beam therapy in head and neck cancer patients. Med Phys. 2021;48(3):1372–1380. [DOI] [PubMed] [Google Scholar]
- 43. Seller Oria C, Marmitt GG, Both S, Langendijk JA, Knopf AC, Meijers A. Classification of various sources of error in range assessment using proton radiography and neural networks in head and neck cancer patients. Phys Med Biol. 2020;65(23):235009. [DOI] [PubMed] [Google Scholar]
- 44. Seller Oria C, Thummerer A, Free J, et al. Range probing as a quality control tool for CBCT‐based synthetic CTs: in vivo application for head and neck cancer patients. Med Phys. 2021;48(8):4498–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION
Data Availability Statement
Research data cannot be shared.


