Abstract
PLAG1 rearrangements have been described as a molecular hallmark of salivary gland pleomorphic adenoma (PA), carcinoma ex pleomorphic adenoma (CEPA), and myoepithelial carcinoma (MECA). Several fusion partners have been described, however, commonly no further assignment to the aforementioned entities or a morphological prediction can be made based on the knowledge of the fusion partner alone. In contrast, TGFBR3‐PLAG1 fusion has been specifically described and characterized as an oncogenic driver in MECA, and less common in MECA ex PA. Here, we describe the clinicopathological features of three TGFBR3‐PLAG1 fusion‐positive salivary gland neoplasms, all of which arose in the deep lobe of the parotid gland. Histopathology showed high morphological similarities, encompassing encapsulation, a polylobular growth pattern, bland basaloid and oncocytoid cells with myoepithelial differentiation, and a distinct sclerotic background. All cases showed at least limited, unusual foci of minimal invasion into adjacent salivary gland tissue, including one case with ERBB2 (Her2/neu) amplified, TP53 mutated high‐grade transformation, and lymph node metastases. Of note, all cases illustrated focal ductal differentiation. Classification remains difficult, as morphological overlaps between myoepithelial‐rich cellular PA, myoepithelioma, and MECA were observed. However, evidence of minimal invasion advocates classification as low‐grade MECA. This case series further characterizes the spectrum of uncommon cellular myoepithelial neoplasms harboring TGFBR3‐PLAG1 fusion, which show recurrent minimal invasion of the adjacent salivary gland tissue, a predilection to the deep lobe of the parotid gland, and potential high‐grade transformation.
Keywords: adenoma, parotid gland, pleomorphic, salivary gland neoplasms, transforming growth factor beta
1. INTRODUCTION
PLAG1 rearrangements have been described as molecular hallmark of salivary gland pleomorphic adenoma (PA), 1 its dermal and soft tissue counterpart mixed tumor, 2 lipoblastoma, 3 and rare uterine myxoid leiomyosarcoma. 4 , 5 In salivary gland neoplasms, carcinoma ex pleomorphic adenoma (CEPA) 6 , 7 , 8 and myoepithelial carcinoma (MECA) 9 , 10 have also been reported to harbor PLAG1 rearrangements. In CEPA, the underlying PLAG1 fusions are most likely the initial clonal event of PA. 8 In addition, several second hits, such as ERBB2 (Her2/neu) amplification and/or TP53 mutation, are common in salivary duct carcinoma ex pleomorphic adenoma. 11 In contrast, recurrent activating HRAS mutations have been described as a very common feature of epithelial‐myoepithelial carcinoma. 12 , 13 However, HRAS mutations are rare in PLAG1 or HMGA2 fusion‐positive CEPA, indicating that development of (low‐grade) CEPA is driven by distinct molecular mechanisms. 14 Recently, TGFBR3‐PLAG1 fusions have been described as a distinctive finding of MECA and myoepithelial carcinoma ex PA (MECEPA), whereas only six cases have been reported yet. 9 In our current study, we thoroughly characterize the clinicopathological features of three additional cases of TGFBR3‐PLAG1 rearranged neoplasms of the salivary gland.
2. MATERIALS AND METHODS
2.1. Case selection
All cases were identified by the distinct gene fusion and retrieved from the archives of the Department of Pathology and Molecular Pathology, University Hospital Zurich in the period from January 2018 to December 2020.
2.2. Immunohistochemistry
The following primary antibodies were used in pairing with the Leica Bond automated staining system: SOX10 (BC34, 1:150, Biocare Medical), SMA (asm‐1, 1:50, Leica). For the following primary antibodies: S100 (polyclonal, 1:2000, DAKO A/S), CK7 (OV‐TL 12/30, 1:100, DAKO A/S), CK5/6 (D5/16B4, prediluted, Ventana‐Roche), calponin (CALP, 1:50, DAKO A/S), p40 (BC28, 1:100, Zytomed Systems), p63 (4A4, prediluted, Ventana‐Roche), Ki‐67 (30‐9, prediluted, Ventana‐Roche), p53 (DO‐7, 1:80, DAKO A/S), Her2/neu (4B5, prediluted, Ventana‐Roche), the Ventana Benchmark automated staining system was applied. Development was performed as described previously. 15
2.3. Next generation sequencing
Small punch biopsies were taken from the tumor areas of interest from the FFPE specimens. DNA and RNA were isolated using the Maxwell® 16 FFPE Tissue LEV DNA Purification Kit16 or LEV RNA FFPE Purification Kit, respectively (Promega, Madison, WI, USA). Library preparation for fusion analysis was performed using the Archer FusionPlex technology (ArcherDX, Boulder, CO, USA) and a custom primer set for salivary gland neoplasms (“SalvGlandDx panel,” described previously 16 ). Libraries were sequenced paired end on a NextSeq™550 (Illumina, San Diego, CA, USA) and data analysis was performed using the Archer analysis software (ArcherDX, Boulder, CO, USA). To assess copy number variations and possible mutations, the DNA part of the Oncomine™ Comprehensive v3 Assay (ThermoFisher Scientific, Waltham, MA, USA) was performed. Libraries were prepared according to the manufacturer's manual, templating was done using the IonChef system, and sequencing was conducted on the Ion S5 system (ThermoFisher Scientific, Waltham, MA, USA). Data were analyzed using the Ion Reporter v5.12 software (ThermoFisher Scientific, Waltham, MA, USA).
2.4. Ethical statement
All patients gave written informed consent and the study was approved by the local ethics committee (Kantonale Ethikkommission Zurich; BASEC No 2020‐1663).
3. RESULTS
3.1. Case 1
3.1.1. Clinical and imaging findings
This 30‐year‐old patient presented to our ENT outpatient department due to a painful swelling of his right parotid gland. An ultrasound examination revealed a lobulated hypoechogenic mass in the deep lobe (Table 1). Subsequent fine needle aspiration (FNA) yielded a diagnosis of a cellular salivary gland neoplasm. In synopsis with the following magnetic resonance imaging, which revealed a lobulated, centrally hypointense, peripherally contrast‐enhancing, and infiltrative mass (Figure 1A), high suspicion of malignancy was raised. 18F‐Fluorodeoxyglucose positron emission tomography (18F‐FDG‐PET) showed high FDG avidity of the lesion (maximum standardized uptake value [SUVmax] 12.9; Figure 1B). The patient underwent uneventful subtotal parotidectomy. Facial nerve function was intact post‐surgery.
TABLE 1.
Overview of the clinicopathological data
| Case | Age | Sex | Localization | Size (cm) | History | Morphology | Molecular alteration | Perineural invasion | Metastases | Therapy | Follow‐up |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 30 | M | Parotid gland, deep lobe | 2.1 | None | Encapsulated, basaloid, polylobular, low‐grade, minimal invasive | TGFBR3‐PLAG1 | No | No | Resection + ND, adjuvant RT | 27 months, NED |
| 2 | 39 | M | Parotid gland, deep lobe/parapharyngeal space | 3.5 | None | Encapsulated, basaloid, oncocytoid, low‐grade, minimal invasive | TGFBR3‐PLAG1 | No | No | Resection | 1 month, NED |
| 3 | 79 | M | Parotid gland, deep lobe, superficial part | 4.2 | Resection of PA, 48 and 29 years ago | Encapsulated, basaloid, oncocytoid, polylobular, low‐grade, minimal invasive |
TGFBR3‐PLAG1 ERBB2 non‐amplified TP53 wildtype |
No | No | Resection + ND, adjuvant RT | 16 months, NED |
| Parotid gland, deep lobe, deeper part |
Hemangiopericytoma‐like, polylobular, low‐grade, minimal invasive; in addition pleomorphic, high‐grade, brisk mitoses, broadly invasive, necrosis, undifferentiated |
TGFBR3‐PLAG1 ERBB2 non‐amplified, TP53 wildtype in low‐grade part; in undifferentiated part ERBB2 amplified, TP53 mutated |
Yes | Yes | Resection + ND, adjuvant RT | 16 months, NED |
Abbreviations: ND, neck dissection; NED, no evidence of disease; RT, radiotherapy.
FIGURE 1.
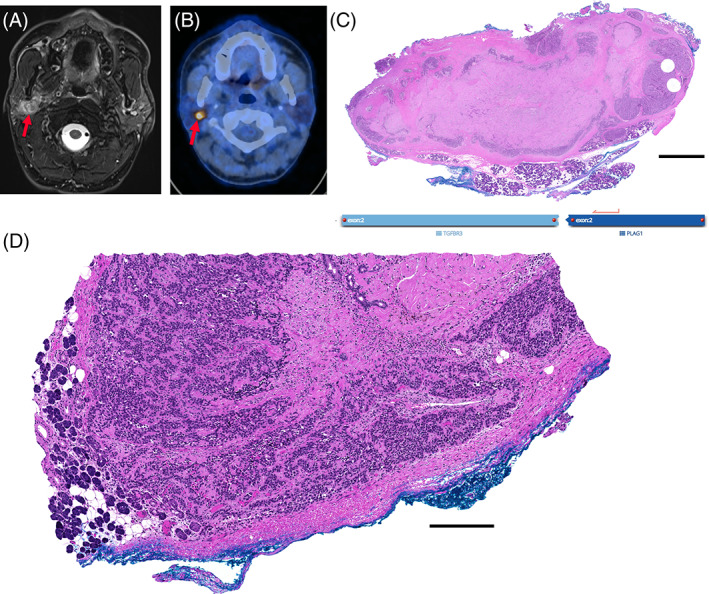
Imaging and pathological overview of case 1. The tumor (arrow) in the right‐sided parotid gland shows a central hypointensity after fine needle aspiration (FNA) and appears to diffusely infiltrate normal surrounding parotid tissue on T2‐weighted fat‐suppressed magnetic resonance (MR) image (A), the tumor shows high FDG‐avidity (SUVmax 12.9; arrow) on fused positron emission tomography/computed tomography (PET/CT) image (B). Histopathological H&E overview in (C) shows polylobulated and centrally sclerosed, cellular neoplasm with fibrous pseudocapsule. Magnification of different section (D) shows focal infiltration of the neoplastic cells through the encapsulation and in direct contact with adjacent salivary gland tissue. Scale bar: 2.5 mm (C), 250 μm (D)
3.1.2. Histopathological features
The resection specimen showed a polylobulated cellular neoplasm with central largely acellular fibrosis and focal squamous metaplasia, most likely due to the previous FNA, or alternatively, spontaneous infarction. The neoplasm showed a quite well‐preserved circumscription including a fibrous pseudocapsule and consisted mainly of bland, myoepithelial differentiated cells (Figure 1C). Focal limited areas could be seen, in which the bland, myoepithelial‐like cells were leaving the circumscription, infiltrating into the adjacent benign salivary gland tissue (Figures 1D and 2A,B). Overall growth pattern included a predominantly trabecular morphology (Figure 2C); however, focal ductal structures could also be noted (Figure 2D). No chondro‐myxoid stroma was evident. On immunohistochemistry, the tumor cells were diffusely positive for SOX10, S100, p40, p63, and CK5/6, whereas SMA and calponin were negative (Figure 2E; Table 2). Proliferation fraction (Ki‐67) encompassed 10%–20% of the tumor cells.
FIGURE 2.
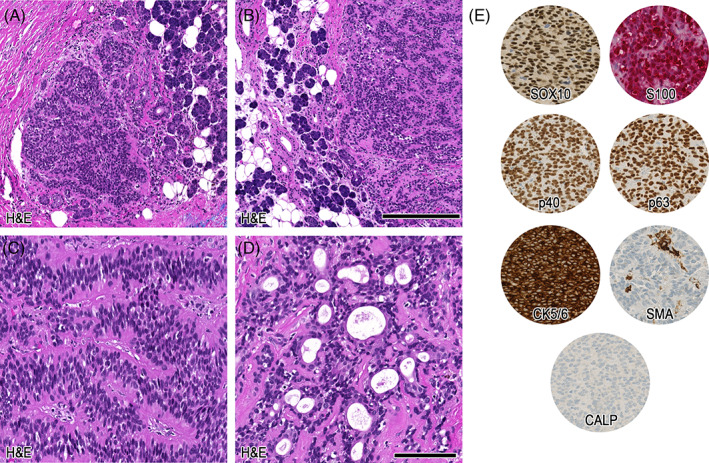
Histopathological details of case 1. In the magnifications (A + B) infiltration of bland, myoepithelial differentiated cells into adjacent salivary gland tissue. Further, in (C) trabecular growth pattern and in (D) obvious ductal differentiation can be appreciated. In (E) the relevant immunohistochemical patterns are depicted. Scale bars: 250 μm (A + B), 100 μm (C–E)
TABLE 2.
Overview of the immunohistochemical profile
| Case | SOX10 | S100 | p40 | p63 | CK5/6 | SMA | Calponin | Ki‐67 (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | pos | pos | pos | pos | pos | neg | neg | 10–20 |
| 2 | pos | pos | pos (partial) | pos (partial) | pos | pos (partial) | pos (partial) | 5–10 |
| 3 (low‐grade part) | pos | pos | pos | pos | pos | neg | pos (partial) | 10–20 |
| 3 (high‐grade part) | neg | Single cells | neg | neg | neg | neg | Single cells | 70–80 |
Abbreviations: neg, negative; pos, positive.
3.1.3. Molecular features
The SalvGlandDx panel detected a TGFBR3 (Exon 2)–PLAG1 (Exon 2) fusion (chr1:92327028, chr8:57092072) with 64.7% reads spanning the breakpoint.
3.1.4. Clinical follow‐up
Diagnosis of minimal invasive low‐grade MECA ex cellular pleomorphic adenoma was rendered. Due to the polylobular growth pattern and deep localization, tumor cells were focally detectable in the surgical margin. Our multidisciplinary tumor board recommended adjuvant radiotherapy, which was performed. After 27 months of follow‐up, there is no clinical evidence of tumor recurrence.
3.2. Case 2
3.2.1. Clinical and imaging findings
This 39‐year‐old patient presented to his general practitioner with head and neck pain and paresthesia of the distant upper extremity. Subsequent magnetic resonance (MR) imaging showed a large well‐defined tumor in the left‐sided parapharyngeal space, abutting the deep lobe of the parotid gland and displacing the medial pterygoid muscle (Figure 3A; Table 1). Based on MR, a schwannoma was suspected.
FIGURE 3.
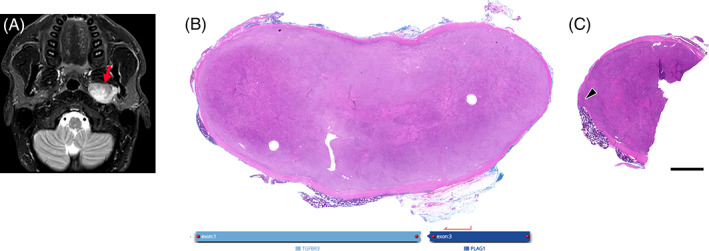
Imaging and pathological overview of case 2. Axial T2‐weighted fat‐suppressed magnetic resonance (MR) image (A) shows a large well‐defined, centrally inhomogeneous lesion (arrow) in the left‐sided parapharyngeal space, abutting the deep lobe of the parotid gland and displacing the medial pterygoid muscle. In (B) overview of the well‐circumscribed, encapsulated cellular neoplasm is shown, whereas (C) illustrates a different section with focal minimal infiltration through the encapsulation (arrowhead). Scale bar: 2.5 mm
3.2.2. Histopathological features
The mass was resected and showed a lobulated, prominent cellular, encapsulated neoplasm on histology (Figure 3B). Similar to case 1, focally monomorphic, myoepithelial‐like tumor cells were perforating through the fibrous pseudocapsule and were in direct contact with the adjacent serous salivary gland tissue in terms of a minimal infiltration (Figure 3C, magnification Figure 4A,B). Overall, cells showed a medium‐sized mixed morphology, including a largely oncocytoid aspect, compatible with a myoepithelial differentiation (Figure 4C). However, also sections depicting focal ductal differentiation were evident. Furthermore, focal lipomatous differentiation, and intracytoplasmic globules could be noted (Figure 4C, insets). Stromal background was strikingly sclerotic (Figure 4D). Strong immunolabeling for SOX10, S100, and CK5/6 was observed, whereas p40, p63, SMA, and calponin were partially expressed (Figure 4E; Table 2). Ki‐67 proliferation index was 5%–10%.
FIGURE 4.
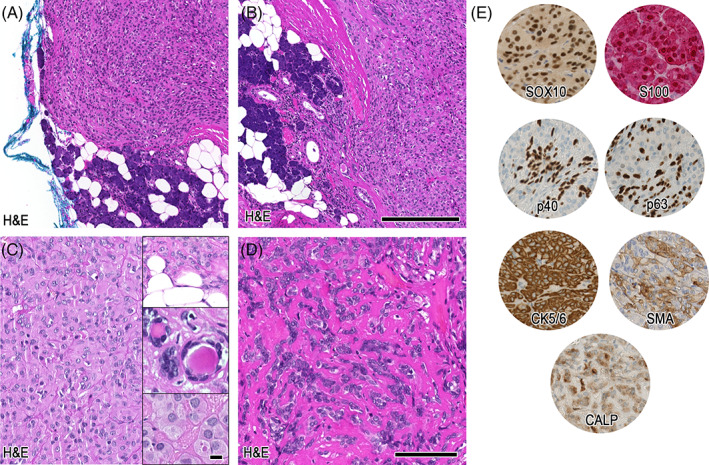
Histopathological details of case 2. In (A + B) the magnifications show two foci of infiltrating bland myoepithelial cells perforating through the encapsulation and depicting direct contact with adjacent salivary gland tissue. Further magnification in (C) shows oncocytoid differentiated cells with evidence of lipomatous differentiation, focal ductal differentiation, and intracellular globules (insets). (D) Illustrates sclerotic background dissecting the myoepithelial cells, whereas in (E) the relevant immunohistochemical stainings are shown. Scale bars: 250 μm (A + B), 100 μm (C–E). Insets: 10 μm
3.2.3. Molecular features
A TGFBR3 (Exon 1)–PLAG1 (Exon 3) (chr1:92351435, chr8:57083748) fusion with 80.2% reads spanning the breakpoint was detected by the SalvGlandDx panel.
3.2.4. Clinical follow‐up
A diagnosis of minimal invasive low‐grade carcinoma with myoepithelial phenotype, presumably ex cellular pleomorphic adenoma, and TGFBR3‐PLAG1 fusion was made. The multidisciplinary tumor board recommended follow‐up, as the tumor was resected without positive margins. Since this is a very recent case, the follow‐up period is currently limited to 1 month and is unremarkable.
3.3. Case 3
3.3.1. Clinical and imaging findings
This 79‐year‐old patient noticed a left preauricular swelling in the parotid region, where he underwent surgery for a mixed tumor 29 years and 48 years ago. No paraffin material or slides from these previous specimens were available for further review. On ultrasound examination, several hypoechogenic nodules could be noted, including a large mass in the (deep) parotid compartment/parapharyngeal space (Table 1). FNA of a cervical lymph node revealed cells of an undifferentiated malignant neoplasm. On T2‐weighted fat‐suppressed MR image, two distinct nodules in the left parotid compartment were seen (Figure 5A). Both nodules were extremely FDG‐avid on PET/CT (Figure 5B; arrow SUVmax 102.2, arrowhead SUVmax 37.6).
FIGURE 5.
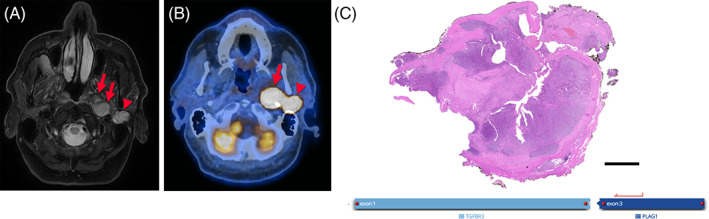
Imaging and pathological overview of case 3, low‐grade morphology. On T2‐weighted fat‐suppressed magnetic resonance (MR) image (A), two distinct nodules (arrows, arrowhead) are seen as hyperintense, well‐defined, lobulated lesions, the medial one consisting of two parts (arrows) in the left‐sided parotid gland region. Both nodules are extremely FDG‐avid as seen on fused positron emission tomography/computed tomography (PET/CT) image (B; arrow SUVmax 102.2, arrowhead SUVmax 37.6). (C) Overview of the superficial node, showing a quite well‐circumscribed and encapsulated cellular neoplasia. Scale bar: 2.5 mm
3.3.2. Histopathological features
The resection specimen of the more superficial nodule showed a polylobulated, variously cellular neoplasm with extensive acellular fibrosis (Figure 5C). A fibrous pseudocapsule was evident (Figure 6A), as well as focal minimal invasion of myoepithelial‐like monomorphic cells into adjacent fat and acinary salivary gland tissue (Figure 6B, including inset). A largely oncocytoid differentiation similar to case 2 was observed, including distinct ductal formation (Figure 6C, including inset). Further, a similar sclerotic background could be noted (Figure 6D). Cells were diffusely positive for SOX10, S100, p40, p63, CK5/6, and in 10%–20% for Ki‐67. SMA was negative, whereas calponin showed partial strong expression. Her2 was completely negative in this part of the tumor (Figure 6E, Table 2). In the sections from the deeper nodule, two further distinct components were visible (Figure 7A). Cellular regions encompassing small monomorphic cells compatible with myoepithelial differentiation were evident, including a distinct hemangiopericytoma‐like growth pattern (Figure 7B). Besides, in the acellular fibrosis, focal dissolute highly atypical, large cells displaying atypical mitotic figures were visible. In addition, the immunohistochemical myoepithelial markers were almost completely lost in this undifferentiated high‐grade fraction including a significantly increased Ki‐67 proliferation index (Table 2), consistent with a high‐grade transformation/dedifferentiation (Figure 7C, including inset). Metastases from this component were corresponding to the initial FNA and resected lymph nodes (not shown). Within the morphological bland, hemangiopericytoma‐like pattern, Her2 immunohistochemistry showed an increased expression (2+) in comparison to the Her2 negative superficial nodule, and a p53 staining pattern consistent with wildtype (Figure 7D). In the high‐grade transformed areas, Her2 expression was even stronger (3+), and p53 was diffusely positive, consistent with a mutant phenotype (Figure 7E, Table 2). Androgen receptor was negative, and pancytokeratin expression was lost in the metastasized high‐grade component (not shown).
FIGURE 6.
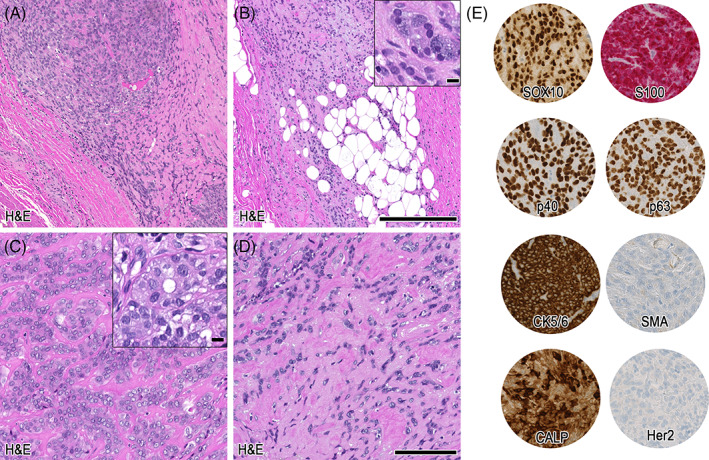
Histopathological details of case 3, low‐grade morphology. The magnification in (A) shows the encapsulation and bland myoepithelial‐differentiated cells, which show invasion into adjacent fatty and acinar salivary gland tissue (B, inset). In (C) a more oncocytoid differentiation with evidence of ductal structures (inset) is depicted. (D) Shows sclerotic background with dissection of the myoepithelial cells. (E) Illustrates immunohistochemical profile. Scale bars: 250 μm (A + B), 100 μm (C–E). Inset: 10 μm
FIGURE 7.
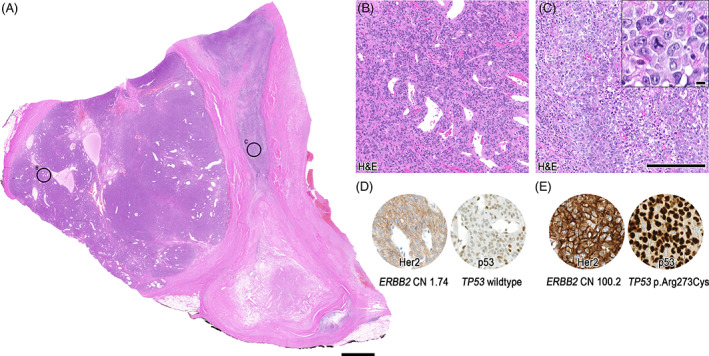
Pathological overview of case 3, low‐grade and high‐grade morphology. Overview of the deeper nodule showing further parts of the cellular lesion (A), including high‐grade transformation/dedifferentiation in the right part. (B) Shows magnification of the bland, cellular myoepithelial differentiated part with hemangiopericytoma‐like growth pattern, and (C) highly atypical cells including necrosis and atypical mitoses (inset). In (D) the equivocal Her2/neu expression, and wild‐type staining pattern of p53 is depicted, corresponding to no ERBB2 copy number (CN) gain, and TP53 wildtype. Immunohistochemical pattern in (E) illustrates strong membranous Her2/neu expression, and strong, diffuse p53 expression, fitting to significant ERBB2 copy number gain (CN: 100.2), and TP53 p.Arg273Cys missense mutation. Scale bars: 2.5 mm (A), 250 μm (B–E), Inset: 10 μm
3.3.3. Molecular features
The SalvGlandDx panel was performed and displayed a TGFBR3 (Exon 1)–PLAG1 (Exon 3) fusion with the identical breakpoint as case 2 (chr1:92351435, chr8:57083748), and 54.7% of reads covering the breakpoint. To investigate the Her2 (ERBB2) and p53 (TP53) status, the DNA part of the Oncomine Comprehensive v3 assay was performed. While the Her2/neu‐negative and the Her2/neu‐equivocal (2+) areas showed no copy number gain of ERBB2, the Her2/neu‐positive (3+) area revealed a clear copy number (CN) gain (CN 100.2). Moreover, in concordance with the p53 positivity, indicative for a mutant phenotype, an underlying pathogenic TP53 (p.Arg273Cys) missense mutation with a frequency of 46.9% was detected (Figure 7D,E).
3.3.4. Clinical follow‐up
Histological features were summarized to high‐grade transformed/dedifferentiated MECA ex cellular pleomorphic adenoma. Due to the advanced stage and high‐grade transformation, adjuvant radiotherapy was performed based on the decision of the multidisciplinary tumor board. After a follow‐up of 16 months, there is no evidence of tumor recurrence.
4. DISCUSSION
Here, we describe three cases of TGFBR3‐PLAG1 rearranged salivary gland neoplasms, whereas only six cases have been reported to date in one previous study. 9 The published cases encompassed mainly MECA; however, also two cases of MECA ex PA have been included. 9 Diagnosis of MECA/MECA ex PA is often challenging, and misinterpretation with benign lesions is common, in particular as cells might be deceptively bland and Ki‐67 proliferation index very low. 17 Our cases showed high morphological similarities, mainly consisting of encapsulated bland basaloid myoepithelial‐like cells and oncocytoid differentiation. A striking sclerotic background dissecting the individual tumor cells was also observed. Furthermore, focal ductal differentiation was evident in all cases. In addition, all cases showed an unusual minimal invasion composed of small tumor foci protruding through the fibrous pseudocapsule. A direct contact of neoplastic myoepithelial cells with adjacent salivary gland tissue was evident, rendering the diagnosis of an infiltrating/low‐grade malignant neoplasm in this anatomical field. Classification remains difficult, as no chondro‐myxoid background was visible for a confident diagnosis of an underlying classical pleomorphic adenoma. However, at least focal mixed differentiation with ductal structures was observed, as well as focal lipomatous cells amid the encapsulated tumor, in general, compatible with a mixed tumor, such as (myoepithelial‐rich) cellular PA. 18 Certainly, also myoepithelioma might show lipomatous foci 18 and occasional ducts. 19 According to the current WHO classification, MECA are typically unencapsulated and do classically show a more diffuse infiltration pattern. 19 In addition, hypocellular centers are described as a typical feature, 20 which were not evident in our cases. Furthermore, Her2/neu positive salivary duct CEPA is commonly encountered, 11 whereas high‐grade transformation has been only described in one case of MECA, 21 , 22 without information about the Her2/neu status. An initial diagnosis of minimal invasive low‐grade MECA of probable origin ex cellular pleomorphic adenoma was considered. However, due to a recurrent finding of minimal invasion, it is difficult to support this diagnosis, as no change in cell type or profile was observed between the invasive and non‐invasive parts. Androgen receptor was negative in our case 3. Hence, the overall immunophenotype of the high‐grade component points to a high‐grade transformed or dedifferentiated MECA. This phenomenon is not well‐described in MECA; however, our case is in concordance with the single published case, 22 reporting a similar high‐grade undifferentiated carcinoma with loss of myoepithelial markers. Pseudoinvasion can sometimes be observed in PA, such as “pseudopodia” and capsular perforation; however, PA is typically not encountered in direct contact with adjacent salivary gland tissue. 23 Furthermore, these phenomena are more commonly observed in relapsing tumors with less cellular chondro‐myxoid nodules, 20 including reports on intravascular tumor complexes after FNA. 23 Two of our reported cases represent de novo tumors, thereof one was not investigated by FNA or biopsy prior to resection.
In summary, the features of our cases show encapsulated, bland cellular myoepithelial neoplasms defined by a rare TGFBR3‐PLAG1 fusion and recurrently depict minimal invasion. Interestingly, no typical/benign PA harboring TGFBR3‐PLAG1 fusion has been described yet, whereas in the previously mentioned study 442 PA were investigated. 9 Furthermore, in vitro studies did show a significantly increased oncogenic potential when overexpressing PLAG1 and TGFBR3. 9 This finding underpins the importance of this distinct oncogenic fusion and the probability that at least minimal invasion seems to be innate to this type of neoplasm. High‐grade transformation can occur, as described in case 3. Minor ductal differentiation in MECA is accepted by some authors, 20 , 24 although a potential cutoff is difficult to determine.
The genomic breakpoints found in our cases 2 and 3 are identical to the breakpoints of one of the previously reported cases, classified as MECA de novo. 9 Interestingly, case MECA 03 9 in the large previous study (shown in the supplementary material of the referenced study) depicts a similar oncocytoid morphology to our cases 2 and 3. One other case in their study (MECA 07) 9 shares the same breakpoint of TGFBR3 with our case 1, however, depicting distinct morphology. Interestingly, cases harboring the TGFBR3‐PLAG1 fusion were prognostically favorable in the study by Dalin et al., 9 which is in concordance with our cases. Of note, also the case with high‐grade transformation shows no evidence of disease after 16 months of follow‐up. Unfortunately, due to the long period of > 25 years, the initial resection specimens of case 3 were not available for review.
To date, there is no report stating the existence of a (purely) benign TGFBR3‐PLAG1 fusion‐positive salivary gland neoplasm. This may impact the interpretation of sequencing results in FNA or on biopsies with such result. Classification remains tough, as encapsulation and ductal structures are typically absent in ordinary MECA. 19 Conversely, (low‐grade) MECA ex cellular pleomorphic adenoma might be an alternate diagnosis and was initially considered, although a significant separation of different tumor types with change of cell type or growth pattern was not observable in our cases. Furthermore, a typical chondro‐myxoid matrix was not evident in any case, and no benign counterpart of TGFBR3‐PLAG1 fusion‐positive tumor has been described despite a large cohort of investigated cases. 9 Moreover, the observed high‐grade transformation including Her2/neu overexpression in our case 3 is usually not seen in classical MECA, and presents rather a feature observed in CEPA. 11 However, in the same case, a history of longstanding recurrent mixed tumor was documented. Therefore, the presence of acellular fibrosis could also lead to the differential diagnosis of a MECA arising ex pleomorphic adenoma which slowly developed over the course of time. In conclusion, it cannot be ruled out that TGFBR3‐PLAG1 fusion could also occur in benign pleomorphic adenoma. Still, we believe the recurrent minimal infiltration is due to a genuine (low‐grade) malignant behavior derived from the oncogenic potential of the underlying distinct fusion. 9 The observed direct infiltration into adjacent salivary gland tissue is generally not compatible with a benign salivary gland neoplasm. However, further cases have to address the alternate consideration of a physiological phenomenon in a narrow anatomic location. Nevertheless, the unusual morphological features and current knowledge about the oncogenic potential of TGFBR3‐PLAG1 fusion should lead to a diagnosis of an at least atypical myoepithelial neoplasm to ensure clinical follow‐up in our view.
In this study, we further characterize three cases of a rare TGFBR3‐PLAG1 fusion‐positive salivary gland tumor. These neoplasms showed (i) a predisposition to the deep lobe of the parotid gland, (ii) predominant myoepithelial differentiation with focal ductal features including encapsulation, (iii) at least minimal invasion of the adjacent salivary gland tissue, and (iv) high‐grade transformation in one case.
Classification of our cases remains difficult; however, accepting focal ductal differentiation as part of the spectrum—diagnosis of a distinct type of minimal invasive low‐grade MECA with morphological similarities to myoepithelial‐rich cellular PA or myoepithelioma is warranted. Finally, our limited follow‐up data corroborate the published data indicating a rather less aggressive biological behavior, although longer follow‐up periods are necessary for a more confident classification.
CONFLICT OF INTEREST
NJR discloses an advisory board function and receipt of honoraria from F. Hoffmann‐La Roche AG. The custom SalvGlandDx panel has been developed with support from a grant by the Iten‐Kohaut Foundation/University Hospital Zurich Foundation to NJR. SH has received consultation fees from Bayer AG, outside of the submitted work. MWH is a recipient of grants from GE Healthcare, grants for translational and clinical cardiac and oncological research from the Alfred and Annemarie von Sick legacy, and grants from the Artificial Intelligence in oncological Imaging Network by the University of Zurich. SNF received honoraria from Oncobit AG. MB, DV, GBM, and MAB have no potential conflicts of interest to declare.
AUTHOR CONTRIBUTIONS
Conceptualization: Niels J. Rupp, Sandra N. Freiberger. Data curation: Muriel Brada, Sandra N. Freiberger. Formal analysis: Sandra N. Freiberger. Investigation: Niels J. Rupp, Sylvia Höller. Resources: Sylvia Höller, Domenic Vital, Grégoire B. Morand, Martina A. Broglie, Martin W. Huellner. Visualization: Niels J. Rupp, Martin W. Huellner. Writing – original draft preparation: Niels J. Rupp, Sandra N. Freiberger. Writing – review & editing: Niels J. Rupp, Sylvia Höller, Muriel Brada, Domenic Vital, Grégoire B. Morand, Martina A. Broglie, Martin W. Huellner, Sandra N. Freiberger.
ACKNOWLEDGMENTS
We thank Norbert Wey, Marion Bawohl, and Annette Bohnert for excellent technical support. Open access funding provided by Universitat Zurich.
Rupp NJ, Höller S, Brada M, et al. Expanding the clinicopathological spectrum of TGFBR3‐PLAG1 rearranged salivary gland neoplasms with myoepithelial differentiation including evidence of high‐grade transformation. Genes Chromosomes Cancer. 2022;61(2):94‐104. doi: 10.1002/gcc.23009
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1. Matsuyama A, Hisaoka M, Nagao Y, Hashimoto H. Aberrant PLAG1 expression in pleomorphic adenomas of the salivary gland: a molecular genetic and immunohistochemical study. Virchows Arch. 2011;458(5):583‐592. doi: 10.1007/s00428-011-1063-4 [DOI] [PubMed] [Google Scholar]
- 2. Bahrami A, Dalton JD, Krane JF, Fletcher CDM. A subset of cutaneous and soft tissue mixed tumors are genetically linked to their salivary gland counterpart. Genes Chromosomes Cancer. 2012;51(2):140‐148. doi: 10.1002/gcc.20938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lopez‐Nunez O, Alaggio R, Ranganathan S, et al. New molecular insights into the pathogenesis of lipoblastomas: clinicopathologic, immunohistochemical, and molecular analysis in pediatric cases. Hum Pathol. 2020;104:30‐41. doi: 10.1016/j.humpath.2020.07.016 [DOI] [PubMed] [Google Scholar]
- 4. Arias‐Stella JA, Benayed R, Oliva E, et al. Novel PLAG1 gene rearrangement distinguishes a subset of uterine myxoid leiomyosarcoma from other uterine myxoid mesenchymal tumors. Am J Surg Pathol. 2019;43(3):382‐388. doi: 10.1097/PAS.0000000000001196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thiryayi SA, Turashvili G, Latta EK, et al. PLAG1‐rearrangement in a uterine leiomyosarcoma with myxoid stroma and heterologous differentiation. Genes Chromosomes Cancer. 2021;60:713‐717. doi: 10.1002/gcc.22980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Katabi N, Ghossein R, Ho A, et al. Consistent PLAG1 and HMGA2 abnormalities distinguish carcinoma ex‐pleomorphic adenoma from its de novo counterparts. Hum Pathol. 2015;46(1):26‐33. doi: 10.1016/j.humpath.2014.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiosea SI, Thompson LDR, Weinreb I, et al. Subsets of salivary duct carcinoma defined by morphologic evidence of pleomorphic adenoma, PLAG1 or HMGA2 rearrangements, and common genetic alterations. Cancer. 2016;122(20):3136‐3144. doi: 10.1002/cncr.30179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Asahina M, Saito T, Hayashi T, Fukumura Y, Mitani K, Yao T. Clinicopathological effect of PLAG1 fusion genes in pleomorphic adenoma and carcinoma ex pleomorphic adenoma with special emphasis on histological features. Histopathology. 2019;74(3):514‐525. doi: 10.1111/his.13759 [DOI] [PubMed] [Google Scholar]
- 9. Dalin MG, Katabi N, Persson M, et al. Multi‐dimensional genomic analysis of myoepithelial carcinoma identifies prevalent oncogenic gene fusions. Nat Commun. 2017;8:1197. doi: 10.1038/s41467-017-01178-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Skálová A, Agaimy A, Vanecek T, et al. Molecular profiling of clear cell myoepithelial carcinoma of salivary glands with EWSR1 rearrangement identifies frequent PLAG1 gene fusions but no EWSR1 fusion transcripts. Am J Surg Pathol. 2021;45(1):1‐13. doi: 10.1097/PAS.0000000000001591 [DOI] [PubMed] [Google Scholar]
- 11. Shimura T, Tada Y, Hirai H, et al. Prognostic and histogenetic roles of gene alteration and the expression of key potentially actionable targets in salivary duct carcinomas. Oncotarget. 2017;9(2):1852‐1867. doi: 10.18632/oncotarget.22927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Urano M, Nakaguro M, Yamamoto Y, et al. Diagnostic significance of HRAS mutations in epithelial‐myoepithelial carcinomas exhibiting a broad histopathologic spectrum. Am J Surg Pathol. 2019;43(7):984‐994. doi: 10.1097/PAS.0000000000001258 [DOI] [PubMed] [Google Scholar]
- 13. Rupp NJ, Brada M, Skálová A, et al. New insights into tumor heterogeneity: a case of solid‐oncocytic epithelial‐myoepithelial carcinoma of the parotid gland harboring a HRAS and heterogeneous terminating ARID1A mutation. Head Neck Pathol. 2020;14(2):554‐558. doi: 10.1007/s12105-019-01055-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. El Hallani S, Udager AM, Bell D, et al. Epithelial‐myoepithelial carcinoma: frequent morphologic and molecular evidence of preexisting pleomorphic adenoma, common HRAS mutations in PLAG1‐intact and HMGA2‐intact cases, and occasional TP53, FBXW7, and SMARCB1 alterations in high‐grade cases. Am J Surg Pathol. 2018;42(1):18‐27. doi: 10.1097/PAS.0000000000000933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rupp NJ, Rechsteiner M, Freiberger SN, et al. New observations in tumor cell plasticity: mutational profiling in a case of metastatic melanoma with biphasic sarcomatoid transdifferentiation. Virchows Arch. 2018;473(4):517‐521. doi: 10.1007/s00428-018-2376-3 [DOI] [PubMed] [Google Scholar]
- 16. Freiberger SN, Brada M, Fritz C, et al. SalvGlandDx – a comprehensive salivary gland neoplasm specific next generation sequencing panel to facilitate diagnosis and identify therapeutic targets. Neoplasia. 2021;23(5):473‐487. doi: 10.1016/j.neo.2021.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu B, Mneimneh W, Torrence DE, et al. Misinterpreted myoepithelial carcinoma of salivary gland: a challenging and potentially significant pitfall. Am J Surg Pathol. 2019;43(5):601‐609. doi: 10.1097/PAS.0000000000001218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Agaimy A. Fat‐containing salivary gland tumors: a review. Head Neck Pathol. 2013;7(Suppl 1):90‐96. doi: 10.1007/s12105-013-0459-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. El‐Naggar A, Chan J, Grandis J, Takata T, Slootweg P. WHO Classification of Head and Neck Tumours (4th ed.). IARC; 2017. Accessed November 29, 2020. https://publications.iarc.fr/Book-And-Report-Series/Who-Classification-Of-Tumours/WHO-Classification-Of-Head-And-Neck-Tumours-2017 [Google Scholar]
- 20. Xu B, Katabi N. Myoepithelial carcinoma. Surg Pathol Clin. 2021;14(1):67‐73. doi: 10.1016/j.path.2020.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skalova A, Leivo I, Hellquist H, et al. High‐grade transformation/dedifferentiation in salivary gland carcinomas: occurrence across subtypes and clinical significance. Adv Anat Pathol. 2021;28(3):107‐118. doi: 10.1097/PAP.0000000000000298 [DOI] [PubMed] [Google Scholar]
- 22. Ogawa I, Nishida T, Miyauchi M, Sato S, Takata T. Dedifferentiated malignant myoepithelioma of the parotid gland. Pathol Int. 2003;53(10):704‐709. doi: 10.1046/j.1440-1827.2003.01536.x [DOI] [PubMed] [Google Scholar]
- 23. Hernandez‐Prera JC, Skálová A, Franchi A, et al. Pleomorphic adenoma: the great mimicker of malignancy. Histopathology. 2021;79:279‐290. doi: 10.1111/his.14322 [DOI] [PubMed] [Google Scholar]
- 24. Bishop JA, Thompson LDR, Wakely PE Jr., Weinreb I. Tumors of the Salivary Glands. AFIP Atlas of Tumor Pathology, Series 5, Fascicle 5. ARP Press; 2021. Accessed July 17, 2021. https://www.arppress.org/product-p/5f05.htm [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


