Abstract
The prominence of the α-subclass of Proteobacteria in the marine bacterioplankton community and their role in dimethylsulfide (DMS) production has prompted a detailed examination of dimethylsulfoniopropionate (DMSP) metabolism in a representative isolate of this phylotype, strain LFR. [1-13C]DMSP was synthesized, and its metabolism and that of its cleavage product, [1-13C]acrylate, were studied using nuclear magnetic resonance (NMR) spectroscopy. [1-13C]DMSP additions resulted in the intracellular accumulation and then disappearance of both [1-13C]DMSP and [1-13C]β-hydroxypropionate ([1-13C]β-HP), a degradation product. Acrylate, the immediate product of DMSP cleavage, apparently did not accumulate to high enough levels to be detected, suggesting that it was rapidly β-hydroxylated upon formation. When [1-13C]acrylate was added to cell suspensions of strain LFR it was metabolized to [1-13C]β-HP extracellularly, where it first accumulated and was then taken up in the cytosol where it subsequently disappeared, indicating that it was directly decarboxylated. These results were interpreted to mean that DMSP was taken up and metabolized by an intracellular DMSP lyase and acrylase, while added acrylate was β-hydroxylated on (or near) the cell surface to β-HP, which accumulated briefly and was then taken up by cells. Growth on acrylate (versus that on glucose) stimulated the rate of acrylate metabolism eightfold, indicating that it acted as an inducer of acrylase activity. DMSP, acrylate, and β-HP all induced DMSP lyase activity. A putative model is presented that best fits the experimental data regarding the pathway of DMSP and acrylate metabolism in the α-proteobacterium, strain LFR.
Dimethylsulfoniopropionate (DMSP) is an abundant sulfonium compound in marine environments (1, 30, 33), where it appears to function as a compatible solute for osmoregulation in marine algae and phytoplankton (10, 11, 34). DMSP is released into the water column or sediment pore water by autolytic processes related to algal senescence or zooplankton predation and by leaching from zooplankton fecal pellets (8, 23, 27, 31). The enzymatic cleavage of DMSP by DMSP lyase in some microbial species results in the production of dimethylsulfide (DMS) and acrylate (5, 7, 9, 18, 22), while other species demethylate it to methyl-3-mercaptopropionate and mercaptopropionate (21, 22, 32). The positive correlation between chlorophyll a and bacterial numbers in DMSP-producing phytoplankton blooms (4, 13, 29, 35), where intracellular DMSP can range from 0.01 to >100 mM (17), suggests that DMSP and acrylate could be an important carbon source for bacterioplankton. The accumulation of 1 to 7 mM acrylate in the colony mucus of Phaeocystis spp. (28) is another source that would be readily available at the time of senescence to microbes in the vicinity. The products of DMSP metabolism are all substrates for various marine bacteria (3, 16, 19, 20, 28a, 32, 37).
Understanding the biochemistry of DMSP metabolism has resulted from work with anoxic sediments, anaerobic isolates (20, 36, 38) and, more recently, aerobic marine (3, 12, 24, 40) and freshwater (D. C. Yoch, R. N. Hardee, R. Friedman, and N. Kulkarni, submitted for publication) isolates. The most detailed analysis of DMSP metabolism has been on the salt marsh isolate, Alcaligenes faecalis strain M3A. Its metabolism of DMSP to acrylate and DMS and of acrylate to β-hydroxypropionate (β-HP) (HOCH2CH2CO2−) all occurs on the cell surface, with only the β-HP being transported into the cytoplasm, where it serves as an energy source and inducer of DMSP lyase and acrylase (3). While there is no evidence that the β-subclass of Proteobacteria (β-Proteobacteria; like A. faecalis) are major players in DMS production (2), this process is common among Roseobacter isolates, an abundant subclass of the α-Proteobacteria in marine surface waters (12, 13, 14). Since DMSP metabolism is not well understood in this prominent group of DMS producers, it was the goal of this work to analyze this process in an isolate from this group; strain LFR, which has a cytosolic DMSP lyase (26), was chosen.
MATERIALS AND METHODS
Growth and preparation of cell suspensions.
Strain LFR was grown in 50-ml batch cultures of seawater-based f/2 medium (15) supplemented with either glucose or acrylate (5 mM) as the sole carbon and energy source. Cultures were incubated on a rotary shaker (140 rpm) for 24 h at 30°C, harvested by centrifugation (11,000 × g for 10 min), and resuspended in half the volume of filter-sterilized seawater, and 10-ml aliquots were placed in 36-ml glass serum bottles. Cultures were allowed to equilibrate for 30 min at 100 rpm prior to the addition of DMSP or acrylate. To monitor the metabolism of DMSP to acrylate and the disappearance of the latter from the medium, cells were removed at various time intervals and centrifuged in an Eppendorf Microfuge for 1 min, and the supernatant was analyzed by high-performance liquid chromatography as described previously (3). DMSP, acrylate, and β-HP were tested as inducers of DMSP lyase in strain LFR by adding various concentrations of these putative inducers to glucose-grown cell suspensions. Aliquots were removed at 2-h intervals, microcentrifuged for 30 s, and resuspended in an equal volume of seawater to which the putative inducer was added. DMSP metabolism was monitored by gas chromatography, and the rate of DMS production by the cells removed at each time point was assumed to be proportional to the extent of DMSP lyase induction. Data presented below in Results are representative of at least two experiments.
NMR analysis of [1-13C]DMSP and [1-13C]acrylate metabolites.
[1-13C]DMSP was synthesized as described previously (6) using [1-13C]acrylate in place of unlabeled acrylate. Nuclear magnetic resonance (NMR) analysis determined the [1-13C]DMSP preparation to be >95% pure, and the concentrations of the reactants (DMS and acrylate) in the product were below detection limits. To have a basis for identifying labeled DMSP metabolites in the cells, the gradient-enhanced 1H 13C heteronuclear multiplequantum coherence (1H{13C} HMQC) spectrum of the [1-13C]DMSP standard was analyzed. It showed one 13C resonance at 177 ppm which had two 1H correlations at 3.38 and 2.65 ppm, which were indicative of two methylene groups adjacent to the 1-13C-labeled carboxyl group of DMSP (data not shown).
To determine the pathway of [1-13C]DMSP and [1-13C]acrylate metabolism by using NMR analysis, cells were grown for 24 h in 250 ml of f/2 medium supplemented with acrylate (final A600 ≈ 0.32), harvested by centrifugation, resuspended in 12 ml of filter-sterilized seawater, and placed in a 64-ml glass serum bottle. The cells were incubated at 140 rpm for 30 min prior to the addition of 5 mM [1-13C]DMSP or [1-13C]acrylate. After the addition of 13C-labeled DMSP or acrylate, the metabolites were identified by removing 1-ml aliquots of the cell suspensions at 0.5-h (for DMSP) or 5-min (for acrylate) time intervals and centrifuging them in an Eppendorf Microfuge for 2 min. The supernatant (the extracellular medium) was separated from the cell pellet and immediately frozen until needed for NMR analysis. The cell pellet was washed once with seawater, and the intracellular constituents were extracted in 1 ml of 100% ethanol by gentle agitation overnight at room temperature on a rotary shaker. The mixture was then centrifuged for 2 min to remove all nonsoluble debris. The ethanol extract was dried in a rotary evaporator for 2 h and resuspended in 1 ml of deuterium oxide for NMR analysis. NMR data on [1-13C]DMSP, [1-13C]acrylate, and their metabolic product(s) were collected on a Varian Unity Inova 500 NMR spectrometer as described previously (3).
RESULTS
DMSP and acrylate metabolism.
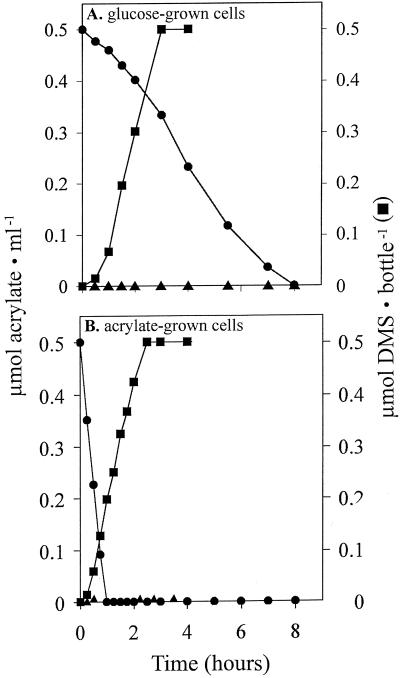
The metabolism of DMSP and acrylate by cell suspensions of strain LFR grown in f/2 medium supplemented with either glucose or acrylate is presented in Fig. 1A and B, respectively. DMSP metabolism was determined by measuring DMS production, while acrylate metabolism was measured by following its disappearance from the extracellular medium. The addition of DMSP (500 μM) to suspensions of either glucose- or acrylate-grown cell suspensions resulted in a low level of DMS production followed by a rapid increase in this rate. The DMSP added at time zero served as the substrate for DMSP lyase, as evidenced by DMS released in the gas phase; however, acrylate could not be detected in the extracellular medium of cells grown in either glucose or acrylate, suggesting that acrylate was sequestered inside the cell.
FIG. 1.
DMSP and acrylate metabolism in cell suspensions of strain LFR cultured on either glucose (A) or acrylate (B). Substrates and products were measured in either the extracellular medium or gas phase following the addition of DMSP or acrylate (500 μM) to the cell suspension. The cells had an A600 of ca. 0.5 and protein concentration of 0.12 mg · ml−1. Symbols: ■, DMS production from added DMSP; ▴, extracellular acrylate resulting from DMSP cleavage; ●, uptake (or metabolism) of added acrylate.
When acrylate was added to a parallel set of glucose- and acrylate-grown cell suspensions it was consumed without a lag; however, the rate of disappearance differed dramatically. In glucose-grown cells (Fig. 1A), the low initial rate of acrylate disappearance from the extracellular medium during the first several hours was followed by a threefold increase in this rate. In the presence of the protein synthesis inhibitor gentamicin, the rate of acrylate disappearance in glucose-grown cells remained low (data not shown), indicating that this organism had a low constitutive level of the acrylate-degrading (or transporting) enzyme. In suspensions of acrylate-grown cells, the rate of disappearance of added acrylate was ca. eight times faster than that for glucose-grown cells, resulting in a depletion of the acrylate from the extracellular medium within 1 h. These results confirm that acrylate metabolism, like that of DMSP, is an inducible process in strain LFR.
[1-13C]DMSP metabolism.
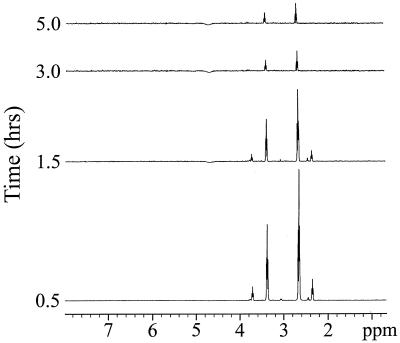
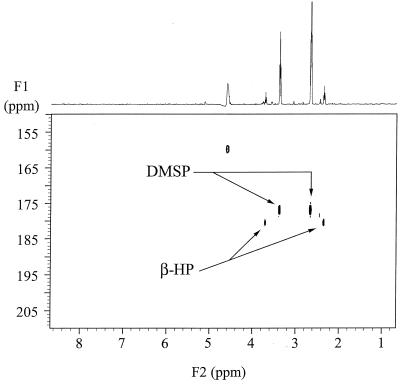
1H and 13C NMR analyses were used to determine the fate of [1-13C]DMSP added to concentrated cell suspensions of strain LFR. The NMR characteristics of [1-13C]DMSP are given in Materials and Methods. To identify the metabolite(s) produced as a result of DMSP catalysis, [1-13C]DMSP was added to a concentrated suspension of acrylate-grown cells. The intracellular and extracellular metabolites were both analyzed by NMR at half-hour intervals. 1H spectra from gradient-enhanced 1H{13C} HMQC analyses of the cytosolic extracts prepared at various time points following the addition of [1-13C]DMSP are presented in Fig. 2. Four resonances, centered at 3.71, 3.38, 2.65, and 2.35 ppm, were seen in the cytosol at the first time point. To confirm the identity of the molecules represented by these signals, gradient-enhanced 1H{13C} HMQC spectrum of the 0.5-h sample was collected (Fig. 3). The proton chemical shifts at 3.38 and 2.65 ppm correlated to a 13C chemical shift of 177 ppm, identical to that of [1-13C]DMSP. The second molecule in the cytosol had proton chemical shifts at 3.71 and 2.35 ppm, indicative of the two methylene groups adjacent to a 13C-labeled carboxyl group of [1-13C]β-HP (3). Both of the 13C-labeled molecules began to decrease in concentration after the first half hour, and by 3 h, only a small amount of the [1-13C]DMSP could be detected in the cytosol. Conspicuously absent in the cytosol was the appearance of an acrylate signal following the addition of labeled-DMSP.
FIG. 2.
1H NMR analysis of intracellular [1-13C]DMSP and its metabolite(s) following the addition of 5 mM [1-13C]DMSP to a cell suspension of strain LFR (acrylate-grown). Samples were taken at the indicated time intervals and prepared as described in Materials and Methods. The NMR spectra were acquired by using the same conditions and number of scans for each sample so that the peak intensity would represent an approximate concentration of each metabolite(s) relative to each sample. 1H resonances centered at 3.38 and 2.65 ppm and 3.71 and 2.35 ppm were identified as those belonging to [1-13C]DMSP and [1-13C]β-HP, respectively.
FIG. 3.
The gradient-enhanced 1H{13C} HMQC spectrum of the cell extract taken at 0.5 h after the addition of 5 mM [1-13C]DMSP to a concentrated cell suspension of strain LFR. NMR parameters were exactly the same as those reported elsewhere (3).
The extracellular medium analyzed at the first time point (0.5 h) after labeled DMSP was added showed two sets of resonances. One set, centered at 3.38 and 2.65 ppm, was from [1-13C]DMSP, and it disappeared with time (data not shown). The second set of resonances, having proton chemical shifts centered at 3.71 and 2.35 ppm, was recognized as β-HP. The β-HP was observed only at this first time point, after which it disappeared from the extracellular medium (data not shown).
[1-13C]acrylate metabolism.
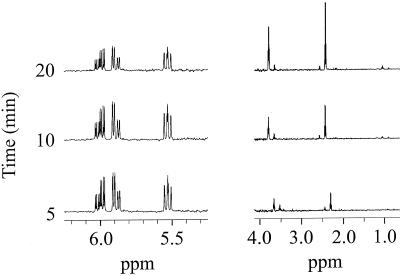
To determine why [1-13C]acrylate was not detected as a result of [1-13C]DMSP cleavage in strain LFR, [1-13C]acrylate was added to cell suspensions and both the extracellular medium and the ethanol-extracted cell pellets (cytosol) were analyzed by NMR spectroscopy. Both the extracellular (Fig. 4) and cytosolic (Fig. 5) metabolites were examined at 5-min intervals over a 30-min time course. The extracellular sample indicated that at the first time point (5 min), proton resonances due to both acrylate (6.05, 5.90, and 5.55 ppm, which correlate with a 13C resonance at 175 ppm) and β-HP (resonances at 3.64 and 2.35 ppm) were observed (Fig. 4). The decrease in the intensity of the acrylate proton resonances over time corresponded to an increase in the intensity of the β-HP resonances. After the β-HP concentration reached a maximum in the extracellular medium, it decreased, suggesting that it was taken up by the cells (data not shown).
FIG. 4.
1H NMR analysis of the extracellular medium following the addition of 5 mM [1-13C]acrylate to a concentrated cell suspension. Samples were taken at the various time intervals indicated; the cells were removed by centrifugation and the supernatant was assayed directly. The NMR spectra were acquired using the same conditions and number of scans for each sample so that the peak intensity would represent an approximate concentration of each metabolite relative to each sample. 1H resonances at 5.55, 5.90, and 6.05 ppm and at 2.35 and 3.64 ppm were identified as those belonging to that of acrylate and β-HP, respectively.
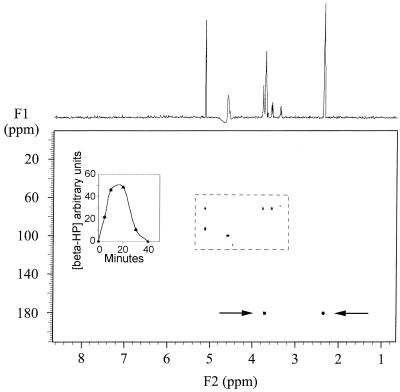
FIG. 5.
Gradient-enhanced 1H{13C} HMQC spectrum of the cell extract (cytosol) taken at 10 min after the exposure of cell suspension to [1-13C]acrylate. NMR parameters are as reported previously (3). (Inset) Cytosolic concentration of β-HP following the addition of [1-13C]acrylate, as measured by the intensity of the α-C protons (3.71 ppm) of the 180-ppm 13C-labeled component from HMQC data (i.e., β-HP).
Next, it was determined if any of the [1-13C]acrylate added to cell suspensions, or products derived from its metabolism, could be found in the cytosol of strain LFR. The 1HNMR spectrum of the ethanol extracts showed many overlapping peaks which could not be interpreted; however, the gradient-enhanced 1H{13C} HMQC spectrum showed one resonance at 180 ppm (Fig. 5). This resonance was assumed to be an acrylate metabolite due to its high intensity, and it resembled β-HP. Cytosol samples of strain LFR prepared over the time course showed an increase in this 180-ppm signal, which reached its maximum at 20 min and thereafter declined (Fig. 5, inset). The intracellular intermediate was confirmed as being β-HP by adding this compound to the cytosolic sample and showing that the intensity of the resonance centered at 180 ppm increased dramatically. Because this cytosolic signal disappeared with time and no other resonances appeared, it indicated the 13C-labeled carboxyl group of β-HP was subsequently decarboxylated.
In addition to the resonances attributed to β-HP, several other peaks were observed in the gradient-enhanced 1H{13C} HMQC spectrum between 65 and 100 ppm in the 13C dimension and between 5.2 and 3.2 ppm in the 1H dimension as seen in Fig. 5. Since these peaks did not increase in intensity before or after the addition of [1-13C]acrylate, they are not products of either [1-13C]acrylate or [1-13C]β-HP metabolism. This indicated that strain LFR produced a metabolite(s) that was stored at high enough concentrations within the cell cytoplasm to be detected by NMR spectroscopy, suggesting that it may function as a compatible solute. No attempt was made to identify this molecule(s); however, there was a resemblance to [13C]sucrose chemical shifts observed in extracts of Chromatium salexigens (39). It is noteworthy that none of the [1-14C]acrylate added to the cell suspensions could be detected in the cytosol.
DMSP lyase induction.
Both DMSP and acrylate are known to induce DMSP lyase activity in strain LFR (24). With the discovery that β-HP was the product of both DMSP and acrylate metabolism by strain LFR, we reexamined the inducer profile for DMSP lyase and found that β-HP was an even better inducer than DMSP and acrylate. The effectiveness of DMSP and its metabolites as putative inducers of DMSP lyase is proportional to the initial rate of DMS production from cells preincubated with the putative inducer and washed before DMSP is added. Induction rates for β-HP, DMSP, acrylate, and the constitutive activity were 0.35, 0.23, 0.2, and 0.09 μmol · ml of culture−1, respectively. The inducers were tested over a range of 100 to 1,000 μM and each was maximally effective at approximately 500 μM.
DISCUSSION
Studies of DMSP and acrylate metabolism in natural marine samples yield results that are the sum of the activities of the diverse population of microbes carrying out these processes (1, 18, 23, 25). Such results can be difficult to interpret, and they therefore provide a rationale for our focus on the pathway of DMSP cleavage of individual β- and γ-Proteobacteria isolates (3, 40). As those two isolates, A. faecalis and Pseudomonas doudoroffii, showed considerable variability in this process, there was no way to know a priori how the α-Proteobacteria, now recognized as probably the most prominent group of DMS producers in the marine environment (12–14), would metabolize DMSP. This report describes the mechanism of DMSP and acrylate metabolism in one culturable isolate of α-Proteobacteria, strain LFR.
While strain LFR was similar to A. faecalis in metabolizing DMSP to acrylate and then to β-HP, the details relating to location of the relevant enzymes and uptake of intermediates differed. The facts that DMSP was not taken up by A. faecalis and that the product of DMSP lyase was seen only outside the cell both suggested that the lyase was an extracellular enzyme in that organism. Conversely, strain LFR took up DMSP before it was cleaved, suggesting an intracellular lyase (24), which was confirmed here with 13C-labeled DMSP (Fig. 2). Acrylase (the acrylate-hydroxylating enzyme) in A. faecalis was extracellular (3), while the NMR results provided convincing evidence for both an intracellular and extracellular acrylase in strain LFR. Specifically, the intracellular 13C-labeled DMSP and β-HP detected following [1-13C]DMSP addition indicated an intracellular acrylase, even though the acrylate intermediate could not be detected. The 13C-labeled β-HP detected extracellularly following [1-13C]acrylate addition indicated an external location. The term “extracellular enzyme” is loosely interpreted here to mean an enzyme whose product(s) was released to the solution outside the cell. No claim can be made as to whether the enzyme is periplasmic or bound to cytoplasmic or outer membranes. Our observations, in fact, have not ruled out the possibility of a single transmembrane acrylase that could hydroxylate both extracellular and cytosolic acrylate.
A couple of apparent anomalies in the results need to be discussed. The first is the absence of detectable acrylate in the cytosol following the addition and cleavage of DMSP. This observation is rationalized by suggesting that strain LFR has a very active acrylase such that the acrylate pool does not accumulate to levels that can be detected by NMR. The second anomaly is the observation of extracellular β-HP following the addition of DMSP to cell suspensions. This might be explained if strain LFR had an extracellular DMSP lyase, but this is contradicted by the kinetic data of Ledyard and Dacey (24). They clearly showed that DMSP uptake into the cytosol preceded its cleavage to form DMS, thus proving that the DMSP lyase resides in the cytosol. A likely, though unproven, explanation for the brief appearance of β-HP outside the cells is that leakage occurred during the cell processing procedure. The fact that extracellular β-HP was not detected at later time points coincided with its decrease in the cytosol (Fig. 2), suggesting that β-HP leakage stopped when the cytosolic levels decreased.
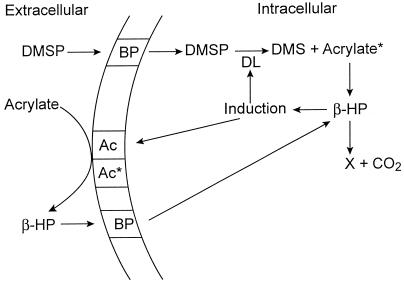
The progressive disappearance of the [1-13C]β-HP signal from the cytosol following addition of [1-13C]acrylate (Fig. 5, inset) indicates that β-HP is decarboxylated to some unknown compound X by strain LFR. If a metabolite of β-HP was decarboxylated, a new set of NMR signals would have appeared, and they did not, meaning that β-HP itself was decarboxylated. The model for DMSP metabolism of this α-proteobacterial isolate is putative but best fits the experimental data presented here (Fig. 6). It remains to be determined if other members of the α-Proteobacteria metabolize DMSP and acrylate by a similar pathway.
FIG. 6.
A model comparing the metabolism of DMSP and acrylate in strain LFR and A. faecalis M3A. Abbreviations: DL, DMSP lyase; Ac*, constitutive levels of acrylase; Ac, inducible acrylase; BP, uptake/binding proteins; acrylate*, intracellular acrylate (assumed to be present, but not actually observed, presumably due to low pool size).
ACKNOWLEDGMENTS
This work was supported in part by grants from the S.C. Sea Grant Consortium (R-MX-8) and from DOE/SCUREF.
REFERENCES
- 1.Andreae M O. The emission of sulfur to the remote atmosphere. In: Galoway J N, Charlson R J, Andreae M O, Rode H, editors. The biogeochemical cycling of sulfur and nitrogen in the remote atmosphere. D. Boston, Mass: Redial Publishing Co.; 1985. pp. 5–25. [Google Scholar]
- 2.Ansede J H, Friedman R, Yoch D C. Phylogenetic analysis of culturable dimethylsulfide-producing bacteria from a Spartina-dominated salt marsh and estuarine water. Appl Environ Microbiol. 2001;67:1210–1217. doi: 10.1128/AEM.67.3.1210-1217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansede J H, Pellechia P J, Yoch D C. Metabolism of acrylate to β-hydroxypropionate and its role in dimethylsulfoniopropionate lyase induction by a salt marsh sediment bacterium, Alcaligenes faecalis M3A. Appl Environ Microbiol. 1999;65:5075–5081. doi: 10.1128/aem.65.11.5075-5081.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billin G, Joiris C, Myer-Reil L, Lindeboom H. Role of bacteria in the North Sea ecosystem. Neth J Sea Res. 1990;26:265–293. [Google Scholar]
- 5.Cantoni G L, Anderson D G. Enzymatic cleavage of dimethylthetin by Polysiphonia lanosa. J Biol Chem. 1956;222:171–177. [PubMed] [Google Scholar]
- 6.Chambers S T, Kunin C M, Miller D, Hamada A. Dimethylthetin can substitute for glycine betaine as an osmoprotectant molecule for Escherichia coli. J Bacteriol. 1987;169:4845–4847. doi: 10.1128/jb.169.10.4845-4847.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dacey J W H, Blough N V. Hydroxide decomposition of DMSP to form DMS. Geophys Res Lett. 1987;14:1246–1249. [Google Scholar]
- 8.Dacey J W H, Wakeham S G. Oceanic dimethylsulfide: production during zooplankton grazing on phytoplankton. Science. 1986;233:1314–1316. doi: 10.1126/science.233.4770.1314. [DOI] [PubMed] [Google Scholar]
- 9.de Souza M P, Yoch D C. Purification and characterization of dimethylsulfoniopropionate lyase from an Alcaligenes-like dimethyl sulfide-producing marine isolate. Appl Environ Microbiol. 1995;61:21–26. doi: 10.1128/aem.61.1.21-26.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickson D M J, Kirst G O. The role of dimethylsulfoniopropionate, glycine betaine and homarine in the osmoacclimation of Platymonas subcordiformis. Planta. 1986;167:536–543. doi: 10.1007/BF00391230. [DOI] [PubMed] [Google Scholar]
- 11.Dickson D M J, WynJones R G, Davenport J. Steady state osmotic adaptation in Ulva lactuca. Planta. 1980;150:158–165. doi: 10.1007/BF00582360. [DOI] [PubMed] [Google Scholar]
- 12.González J M, Kiene R P, Moran M A. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria. Appl Environ Microbial. 1999;65:3810–3819. doi: 10.1128/aem.65.9.3810-3819.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González J M, Simó R, Massana R, Covert J S, Casamayor E O, Pedrós-Alió C, Moran M A. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol. 2000;66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González J M, Moran M A. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillard R R L. Culture of phytoplankton for feeding marine invertebrates. In: Smith W L, Chanley M H, editors. Culture of marine invertebrate animals. New York, N.Y: Plenum Press; 1975. pp. 29–60. [Google Scholar]
- 16.Kanagara T, Kelly D P. Breakdown of dimethyl sulfide by mixed cultures and by Thiobacillus thioparus. FEMS Microbiol Lett. 1986;34:13–19. [Google Scholar]
- 17.Keller M D, Bellows W K, Guillard R R L. Dimethyl sulfide production in marine phytoplankton. In: Salzman E S, Cooper W J, editors. Biogenic sulfur in the environment. American Chemical Society Symposium Series No. 393. Washington, D.C.: American Chemical Society; 1989. pp. 167–182. [Google Scholar]
- 18.Kiene R P. Dimethyl sulfide production from dimethylsulfoniopropionate in coastal seawater samples and bacterial cultures. Appl Environ Microbiol. 1990;56:3292–3297. doi: 10.1128/aem.56.11.3292-3297.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiene R P, Oremland R S, Catena A, Miller L G, Capone D G. Metabolism of reduced methylated sulfur compounds in anaerobic sediments and by a pure culture of an estuarine methanogen. Appl Environ Microbiol. 1986;52:1037–1045. doi: 10.1128/aem.52.5.1037-1045.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiene R P, Taylor B F. Metabolism of acrylate and 3-mercaptopropionate. In: Salzman E S, Cooper W J, editors. Biogenic sulfur in the environment. American Chemical Society Symposium Series No. 393. Washington, D.C.: American Chemical Society; 1989. pp. 222–230. [Google Scholar]
- 21.Kiene R P, Taylor B F. Demethylation of dimethylsulfoniopropionate and production of thiols in anoxic marine sediment. Appl Environ Microbiol. 1988;54:2208–2212. doi: 10.1128/aem.54.9.2208-2212.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiene R P, Visscher P T. Production and fate of methylated sulfur compounds from methionine and dimethylsulfoniopropionate in anoxic salt marsh sediments. Appl Environ Microbiol. 1987;53:2426–2434. doi: 10.1128/aem.53.10.2426-2434.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwint R L J, Irigoien X, Kramer K J M. Kinetics of DMSP-lyase activity in coastal seawater. In: Kiene R P, Visscher P T, Keller M D, Kirst G O, editors. Environmental and biological chemistry on dimethylsulfoniopropionate and related sulfonium compounds. New York, N.Y: Plenum Press; 1996. pp. 239–252. [Google Scholar]
- 24.Ledyard K M, Dacey J W H. Dimethylsulfide production from dimethylsulfoniopropionate by a marine bacterium. Mar Ecol Prog Ser. 1994;110:95–103. [Google Scholar]
- 25.Ledyard K M, Dacey J W H. Kinetics of DMSP-lyase activity in coastal seawater. In: Kiene R P, Visscher P T, Keller M D, Kirst G O, editors. Environmental and biological chemistry on dimethylsulfoniopropionate and related sulfonium compounds. New York, N.Y: Plenum Press; 1996. pp. 325–336. [Google Scholar]
- 26.Ledyard K M, DeLong E F, Dacey J W H. Characterization of a DMSP-degrading bacterial isolate from the Sargasso Sea. Arch Microbiol. 1993;160:312–318. [Google Scholar]
- 27.Matrai P, Keller M D. DMS in a large scale coccolithophore bloom in the Gulf of Maine. Cont Shelf Res. 1993;13:831–843. [Google Scholar]
- 28.Noordkamp D J B, Gieskes W W C, Gottschal J C, Forney L J, van Rijssel M. Acrylate in Phaeocystis colonies does not affect the surrounding bacteria. J Sea Res. 2000;43:287–296. [Google Scholar]
- 28a.Oremland R S, Kiene R P, Mathrani I, Whiticar M J, Boone D R. Description of an estuarine methylotrophic methanogen which grows on dimethyl sulfide. Appl Environ Microbiol. 1989;55:994–1002. doi: 10.1128/aem.55.4.994-1002.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Putt M, Miceli G, Stoecker D K. Association of bacteria with Phaeocystis sp. in McMurdo Sound, Antarctica. Mar Ecol Prog Ser. 1994;105:179–189. [Google Scholar]
- 30.Salzman E S, Cooper W J, editors. Biogenic sulfur in the environment. American Chemical Society Symposium, Series No. 393. Washington, D. C.: American Chemical Society; 1989. [Google Scholar]
- 31.Stefels J, van Boekel W H M. Production of DMS from dissolved DMSP in axenic cultures of the marine phytoplankton species Phaeocystis sp. Mar Ecol Prog Ser. 1993;97:11–18. [Google Scholar]
- 32.Taylor B T, Gilchrist D C. New routes for aerobic biodegradation of dimethylsulfoniopropionate. Appl Environ Microbiol. 1991;57:3581–3584. doi: 10.1128/aem.57.12.3581-3584.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner S M, Malin G, Liss P S, Harbor D S, Holligan P M. The seasonal variation of dimethyl sulfide and dimethylsulfoniopropionate concentrations in nearshore waters. Limnol Oceanogr. 1988;33:364–375. [Google Scholar]
- 34.Vairavamurthy A, Andreae M O, Iversen R L. Biosynthesis of dimethyl sulfide and dimethyl propiothetin by Hymenomonas carterae in relation to sulfur source and salinity variations. Limnol Oceanogr. 1985;30:59–70. [Google Scholar]
- 35.van Boekel W H M, Hansen F C, Riegman R, Bak R P M. Lysis-induced decline of a Phaeocystis spring bloom and coupling with the microbial foodweb. Mar Ecol Prog Ser. 1992;81:269–276. [Google Scholar]
- 36.van der Maarel M J E C, van Bergeijk S, van Werkhoven A F, Laverman A M, Meijer W G, Stam W T, Hansen T A. Cleavage of dimethylsulfoniopropionate and reduction of acrylate by Desulfovibrio acrylicus sp. nov. Arch Microbiol. 1996;166:109–115. [Google Scholar]
- 37.Visscher P T, Diaz M R, Taylor B F. Enumeration of bacteria which cleave or demethylate dimethylsulfoniopropionate in the Caribbean Sea. Mar Ecol Progr Ser. 1992;89:293–296. [Google Scholar]
- 38.Wagner C, Stadtman E R. Bacterial fermentation of dimethyl-β-propiothetin. Arch Biochem Biophys. 1962;98:331–336. doi: 10.1016/0003-9861(62)90191-1. [DOI] [PubMed] [Google Scholar]
- 39.Welch D T, Herbert R A. Identification of organic solutes accumulated by purple and green sulfur bacteria during osmotic stress using natural abundance 13C nuclear magnetic resonance spectroscopy. FEMS Microbiol Ecol. 1993;13:145–150. [Google Scholar]
- 40.Yoch D C, Ansede J H, Rabinowitz K S. Evidence for intracellular and extracellular dimethylsulfoniopropionate (DMSP) lyases and DMSP uptake sites in two species of marine bacteria. Appl Environ Microbiol. 1997;63:3182–3188. doi: 10.1128/aem.63.8.3182-3188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]