Abstract
The mesophile Acinetobacter sp. strain BD413 and the extreme thermophile Thermus thermophilus HB27 display high frequencies of natural transformation. In this study we identified and characterized a novel competence gene in Acinetobacter sp. strain BD413, comA, whose product displays significant similarities to the competence proteins ComA and ComEC in Neisseria and Bacillus species. Transcription of comA correlated with growth phase-dependent transcriptional regulation of the recently identified pilin-like factors of the transformation machinery. This finding strongly suggests that comA is part of a competence regulon. Examination of the genome sequence of T. thermophilus HB27 led to detection of a comA/comEC-like open reading frame (ORF) which is flanked by an ORF whose product shows significant similarities to the Bacillus subtilis competence protein ComEA. To examine whether these two ORFs, designated comEC and comEA, are implicated in natural transformation of T. thermophilus HB27, both were disrupted by using a thermostable kanamycin resistance marker. Natural transformation in comEC mutants was reduced 1,000-fold, whereas in comEA mutants the natural transformation phenotype was completely eliminated. These results strongly suggest that both genes, comEC and comEA, are required for natural transformation in T. thermophilus HB27. Several transmembrane α-helices are predicted based on the amino acid sequences of ComA in Acinetobacter sp. strain BD413 and ComEC in T. thermophilus HB27, which suggests that ComA and ComEC are located in the inner membrane and function in DNA transport through the cytoplasmic membrane.
Increasingly, data obtained in studies of molecular microbial ecology and genome analyses indicate that horizontal gene flow between different species and genera plays a major role in acquisition of novel metabolic capabilities, of the different mechanisms of gene transfer, natural transformation is perhaps the most versatile and provides an important mechanism for increasing the genetic variability of microorganisms. The ability to take up free DNA via natural transformation is widely distributed among representatives of different phylogenetic and trophic groups inhabiting distinct natural ecosystems (35). To gain insight into the structure of the DNA uptake machinery and the mechanism of natural transformation of gram-negative soil bacteria, we used Acinetobacter sp. strain BD413 as a mesophilic model organism. This strain is known for its high competence for natural transformation (25). Acinetobacter sp. strain BD413 has been shown to induce maximal competence immediately after the transition from the lag phase to the exponential growth phase. During prolonged exponential growth competence is gradually lost (43, 48). Prerequisites for induction of competence for natural transformation in Acinetobacter sp. strain BD413 have been investigated thoroughly and are well understood (44). Since Acinetobacter sp. strain BD413 does not discriminate between heterologous and homologous DNA, this strain has been broadly used as a model strain to elucidate the significance of intra- and interspecies gene transfer in natural soil habitats (15, 29, 35, 38, 39).
Although many thermophilic bacteria are known to exhibit high levels of competence for natural transformation, information on natural transformation systems of extremely thermophilic microorganisms is very scarce, in contrast to the abundant data on the transformation mechanisms of and the potential for DNA transfer between mesophilic bacteria (31). One extreme thermophile, Thermus thermophilus HB27, an aerobic, rod-shaped, gram-negative bacterium that grows at temperatures between 50 and 82°C, is known to exhibit high frequencies of natural transformation (22, 31). Transformation frequencies ranging from 10−2 to 10−1 were found when proliferating cells incubated at pH 6 to 9 and 70°C were exposed to chromosomal DNA (31). As observed in Acinetobacter sp. strain BD413, the highest transformation frequencies were obtained in the presence of divalent cations (Ca2+ or Mg2+).
We recently identified five genes, comC, comP, comB, comE, and comF, that are required for DNA uptake in Acinetobacter sp. strain BD413 (1, 21, 34, 47, 48). Primary sequence comparisons, mutant studies, and biochemical analyses have suggested that ComP is part of a DNA binding and/or transport structure that is anchored in the cytoplasmic membrane and spans the periplasm (48). In contrast to our increasing knowledge concerning the structure and function of transformation mechanisms in mesophilic bacteria (8), nothing is known about the components of transformation systems of inhabitants of extreme environments, such as extremely thermophilic bacteria. The differences in physiology and environmental conditions between the genera Acinetobacter and Thermus led us to ask whether the transformation mechanisms of these phylogenetically distantly related bacteria are completely different or include conserved components.
In this study we identified a novel competence gene essential for natural transformation in Acinetobacter sp. strain BD413. The deduced protein encoded by this competence gene shows significant similarities to the competence proteins ComA of Neisseria gonorrhoeae, ComEC of Bacillus subtilis, Rec-2 of Haemophilus influenzae, and CelB of Streptococcus pneumoniae (2, 4, 10, 17). Due to the similarities, the novel competence gene in Acinetobacter sp. strain BD413 was designated comA. Examination of the genomic DNA sequence of T. thermophilus HB27 resulted in identification of a comA/comEC-like open reading frame (ORF) in Thermus, which is flanked by an ORF encoding a putative protein with significant similarities to the competence proteins ComEA of B. subtilis and CelA of S. pneumoniae. We obtained clear evidence that the comEA-comEC competence locus is essential for natural transformation in T. thermophilus HB27. Primary sequence comparisons and secondary structure predictions suggested that ComEC of Thermus and ComA of Acinetobacter are transmembrane proteins which may be involved in DNA transport through the cytoplasmic membrane. Our data strongly suggested that ComA- and ComEC-like proteins are ubiquitous, essential components of bacterial transformation mechanisms independent of the phylogenetic relationships and natural environments of the transformable bacteria.
MATERIALS AND METHODS
Strains, plasmids, and DNA manipulation.
Acinetobacter wild-type and mutant strains were grown in Luria-Bertani (LB) medium (50) or mineral medium (41). Growth conditions described previously were used (47). A p-hydroxybenzoate hydroxylase (pobA) mutant strain of BD413, designated Acinetobacter sp. strain ADP239 (19), was used in all studies. Escherichia coli DH5α was used as a host strain for cloning of genomic DNA (18). E. coli S17-1(λpir) was used as a donor strain for conjugative transposition of mini-Tn10pLOF/Km (Apr, Tn10 delivery plasmid with Kmr) (20). The E. coli strains were cultured at 37°C in LB medium. T. thermophilus HB27 wild-type and mutant strains were grown in a 1:1 mixture of TM broth (31) and LB medium at 60 to 70°C. When appropriate, antibiotics were added at the following concentrations: kanamycin 20 to 40 μg/ml; ampicillin, 100 μg/ml; tetracycline, 15 μg/ml; and streptomycin, 100 to 500 μg/ml. The molecular and genetic procedures used were standard techniques (50, 51). Transformation, conjugation, and complementation studies of Acinetobacter mutants and Southern hybridization experiments were performed as described previously (47). For triparental matings helper plasmid pRK2013 (13) was used. T. thermophilus HB27 was transformed by using a modified protocol described by Koyama et al. (31).
Transformation studies.
Transformation studies with Acinetobacter strains were performed as described previously (47). For transformation studies with T. thermophilus HB27 spontaneous streptomycin-resistant mutants were selected by plating 108 cells on TM medium plates containing streptomycin (500 μg/ml). The genomic DNA of the spontaneous streptomycin-resistant mutants were used as donor DNA for transformation studies as described previously (31).
Transposon mutagenesis in Acinetobacter.
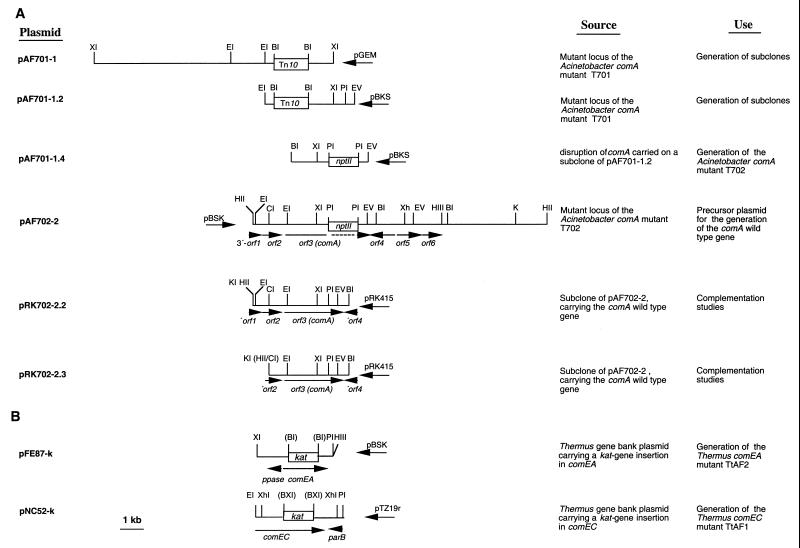
To identify competence loci in Acinetobacter sp. strain BD413, a genomic library of this strain was subjected to transposon mutagenesis by using a genetically engineered derivative of Tn10, mini-Tn10pLOF/Km, as described previously (20, 21). The BD413 library, which contained 6,000 clones harboring 2.5- to 6-kb XbaI DNA fragments, was generated in E. coli DH5α by using the mobilizable broad-host-range vector pRK415. Matings between the donor strain and pooled recipients were performed by using the filter mating technique as described recently (47). Transconjugants were selected on LB medium plates containing kanamycin and tetracycline and subjected to plasmid preparation. Plasmids were purified and digested with XbaI, and the XbaI fragments spanning the kanamycin resistance gene plus flanking BD413 DNA were transformed into Acinetobacter sp. strain ADP239. Selection of Acinetobacter sp. strain ADP239 transformants on LB medium plates containing kanamycin resulted in detection of 1,000 transformants that had acquired the kanamycin marker gene via homologous recombination of the marker-flanking DNA with ADP239 genomic DNA. Analysis of the transformants led to identification of one noncompetent mutant, T701. The proficiency of homologous recombination of this mutant was tested as described previously (16). The gene disrupted by mini-Tn10pLOF/Km was identified as comA. The mutant locus of T701 was recovered on a 10.5-kb XbaI fragment (pAF701-1) and an overlapping 3.9-kb EcoRI-EcoRV fragment (pAF701-1.2), which were cloned by using the vectors pGEM-7Zf(+) and pBluescriptII KS, respectively (Fig. 1A).
FIG. 1.
Physical maps of plasmids used in this study for Acinetobacter (A) and Thermus (B) gene disruption and complementation studies. pBKS, pBluescriptII KS; pBSK, pBluescriptII SK; pGEM, pGEM-7Zf(+). The arrows indicate directions of transcription. B, BamHI; BX, BstXI; CI, ClaI; EI, EcoRI; EV, EcoRV; HII, HincII; HIII, HindIII; KI, KpnI; XI, XbaI; Xh, XhoI; PI, PstI.
Generation of the wild-type comA gene.
To clearly confirm the essential role of comA in natural transformation, complementation studies with the wild-type comA gene were performed. Isolation or generation of the comA wild-type gene was a prerequisite for these studies. Mutant T701 could not be used to generate the comA wild-type allele. Therefore, a second mutant had to be generated. To generate the second comA mutant, a 1.9-kb BamHI-EcoRV DNA fragment that was located downstream of the mini-Tn10 insertion in pAF701-1.2 and included 5′-terminal truncated comA was subcloned from pAF701-1.2. The comA allele in the resulting plasmid, pAF701-1.3 (Table 1), was disrupted by a kanamycin marker (nptII), which gave rise to plasmid pAF701-1.4 (Fig. 1A). This construct containing part of comA, disrupted by the nptII marker digested with BamHI plus EcoRV, and the 3.2-kb BamHI-EcoRV fragment carrying the disrupted comA gene was introduced into Acinetobacter sp. strain ADP239 by natural transformation. Transformants were selected on LB medium containing kanamycin. Several of the kanamycin-resistant transformants were analyzed to determine their transformation phenotypes and, as expected, were found to be noncompetent. One such mutant, designated T702, was used for further studies. Correct allelic replacement of chromosomal wild-type comA in mutant T702 by the disrupted ORF was verified by Southern hybridization.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| Acinetobacter strains | ||
| Acinetobacter sp. strain BD413 | Wild type | 25 |
| Acinetobacter sp. strain ADP239 | Spontaneous pobA mutant of BD413 | 19 |
| Acinetobacter sp. strain ADP197 | recA mutant (rec100::Tn5) of BD413 | 16 |
| T701 | pobA comA::Tn10, noncompetent | This study |
| T702 | pobA comA::nptII, noncompetent | This study |
| T703 | pobA orf4::nptII | This study |
| Thermus strains | ||
| T. thermophilus HB27 | Wild type | 42 |
| TtAF1 | comEC::kat, noncompetent mutant | This study |
| TtAF2 | comEA::kat, noncompetent mutant | This study |
| E. coli strains | ||
| DH5α | F lacZΔM15 recA1 π− hsdR17 supE44 Δ(lacZYA argF) | 18 |
| S17-1(λpir) | trp SmrrecA thi pro hsdM+ RP4-Tc::Mu::Km Tn7 λpir, hsdR mutant | 54 |
| Plasmids | ||
| pBluescriptII KS/SK | Apr | Stratagene |
| pRK415 | TcrlacPOZ′ | 28 |
| pTZ19r | Apr | MBI, Fermentas |
| pGEM-7Zf(+) | Apr | Promega, Serva GmbH |
| pUC4K | Apr Kmr | Pharmacia |
| pLOF/Km | Apr, Tn10-based delivery plasmid with Kmr | 20 |
| pMK18 | Kmr | 6 |
| pKOK6.1 | Apr Kmr Cmr | 30 |
| pAF701-1 | 10.5-kb XbaI fragment of mutant T701 in pGEM-7Zf(+) (comA::Tn10) | This study |
| pAF701-1.2 | 3.9-kb EcoRI-EcoRV fragment of mutant T701 in pBIIKS (comA::Tn10) | This study |
| pAF701-1.3 | 1.9-kb BamHI-EcoRV fragment of pAF701-1.2 in pBIIKS (′comA′) | This study |
| pAF701-1.4 | nptII gene inserted into PstI site of pAF701-1.3 (comA::nptII) | This study |
| pAF702-2 | 12.5-kb HincII fragment of mutant T702 in pBIISK (comA::nptII) | This study |
| pAF702-2.1 | 5.3-kb HincII-BamHI fragment of pAF702-2 in pBIISK (comA::nptII) | This study |
| pAF702-2.2 | 4-kb HincII-BamII fragment (′orf1 orf2 comA ′orf4) | This study |
| pAF702-2.3 | 3.3-kb insert of pAF702-2.2 in pBIISK (′orf2 comA ′orf4) | This study |
| pRK702-2.2 | 4-kb BamHI-KpnI insert of pAF702-2.2 in pRK415 | This study |
| pRK702-2.3 | 3.3-kb BamHI-KpnI insert of pAF702-2.3 in pRK415 | This study |
| pAF702-3.2 | lacZ gene inserted into PstI site of pAF702-2.3 (comA::lacZ) | This study |
| pRK702-3.2 | 8-kb BamHI-KpnI insert of pAF702-3.2 in pRK415 (comA::lacZ) | This study |
| pAF702-4.4 | 3.4-kb PstI-HindIII insert of pAF702-2 in pBIIKS | This study |
| pRK702-4.2 | 3.4-kb insert of pAF702-4.4 in pRK415 | This study |
| pRK702-4.3 | nptII gene inserted into BamHI site of pRK702-4.2 (orf4::nptII) | This study |
| pNC52 | Thermus gene bank plasmid carrying a 2.6-kb fragment in pTZ19r (comEC) | This study |
| pFE87-HX | Thermus gene bank plasmid cloned as 2.1-kb HindIII-XbaI fragment in pBIIKS (comEA) | This study |
| pNC52-k | kat gene inserted into BstXI site of pNC52 (comEC::kat) | This study |
| pFE87-k | kat gene inserted into BamHI site of pFE87-HX (comEA::kat) | This study |
The T702 mutant locus was recovered as a 12.5-kb HincII fragment (pAF702-2) (Table 1); this fragment contained the entire chromosomal copy of comA. To generate the comA wild-type gene, a 5.3-kb HincII-BamHI DNA fragment was subcloned from pAF702-2 into pBluescriptII SK, resulting in plasmid pAF702-2.1 (Table 1). The nptII marker was eliminated by digestion with PstI and subsequent religation, resulting in plasmid pAF702-2.2 (Table 1). The sequence of the analogous comA region in plasmid pAF702-2.2 (spanning the PstI site) was found to be identical to the corresponding region in the 5′- and 3′-terminally truncated comA gene in pAF701-1.2 (Fig. 1A). This result confirmed that deletion of the nptII marker gene from pAF702-2.1 resulted in generation of the comA wild-type allele.
Complementation studies.
To perform complementation studies, the insert of pAF702-2.2 was cloned into pRK415 (20). This was done by cloning the 4-kb insert of pAF702-2.2 as a KpnI-BamHI fragment into linearized pRK415, which resulted in pRK702-2.2 (Fig. 1A). This fragment contained the 3′ end of orf1, orf2, comA, and the 3′ end of orf4. For further complementation studies a second plasmid containing only the complete comA gene was constructed. To delete the comA-flanking orf1 and orf2 regions, plasmid pAF702-2.2 was digested with HincII plus ClaI, incubated in the presence of deoxynucleoside triphosphates (dNTPs) and the Klenow enzyme, and ligated. The 3.3-kb insert in the resulting plasmid, pAF702-2.3 (Table 1), included the 3′ end of orf2, comA, and the 3′ end of orf4. The insert of pAF702-2.3 was cloned as a 3.3-kb KpnI-BamHI DNA fragment into pRK415, which resulted in pRK702-2.3 (Fig. 1A). Recombinant plasmids pRK702-2.2 and pRK702-2.3 were conjugatively transferred into comA mutants T701 and T702, respectively. Transconjugants were selected for growth on LB medium plates containing tetracycline and analyzed for the presence of recombinant plasmids and transformation phenotype.
Disruption of comA flanking ORFs.
To determine whether orf4 has a potential function in natural transformation, orf4 was disrupted by the kanamycin resistance gene, nptII. A 3.4-kb PstI-HindIII fragment of plasmid pAF702-2 spanning the 3′ end of comA, orf4, orf5, and orf6 was first subcloned into pBluescriptII KS (pAF702-4.4)(Table 1) and then was cloned as a 3.4-kb PstI-KpnI fragment into pRK415, which resulted in plasmid pRK702-4.2 (Table 1). The nptII marker was cloned into the unique BamHI site of orf4, which resulted in plasmid pRK702-4.3. Plasmid pRK702-4.3 was digested with EcoRV, and the resulting 3.3-kb EcoRV fragment carrying the disrupted orf4 DNA plus flanking DNA was introduced into Acinetobacter sp. strain ADP239 by natural transformation in order to disrupt chromosomal orf4 by homologous recombination. Transformants were selected for growth on LB medium plates containing kanamycin. Correct allelic replacement of chromosomal wild-type orf4 was verified by Southern hybridization. Several attempts were made to disrupt or to delete orf1 and orf2. For these experiments different subclones of pAF701-1 carrying orf1 and orf2 were generated and used to insert the nptII marker into orf1 and orf2, respectively. The resulting disrupted orf1 and orf2 DNA plus flanking wild-type DNA were transformed into Acinetobacter sp. strain ADP239, but despite several attempts with different constructs no replacement of wild-type orf1 and orf2 by the disrupted ORFs was observed.
Gene disruption in T. thermophilus HB27.
comEC and comEA of T. thermophilus HB27 were disrupted by a kanamycin resistance gene (kat) encoding a thermostable kanamycin nucleotidyltransferase (32, 36), which was derived from the shuttle vector pMK18. pMK18 confers kanamycin resistance in E. coli (37°C) and in Thermus (70°C) (6). Two plasmids from an HB27 gene bank were used for these disruption experiments. The gene bank was generated by cloning physically sheared, blunt-ended chromosomal DNA from T. thermophilus HB27 in HincII-digested vector pTZ19r. Based on the sequence information, two of the gene bank plasmids, designated pNC52 and pFE87, were determined to carry homologs of the B. subtilis comEC and comEA competence genes. These two plasmids were used for disruption of the HB27 comEC- and comEA-like genes via insertion of a thermostable kanamycin resistance marker gene (kat).
To insert the kanamycin resistance gene (kat) into the comEC-like ORF, plasmid pNC52 (Table 1) was digested with BstXI and incubated in the presence of dNTPs and the Klenow enzyme. pMK18 was digested with HindIII plus BamHI, which yielded a 1.2-kb fragment containing the kat gene. After incubation with the Klenow enzyme, the kat gene was ligated to BstXI-linearized pNC52, and the ligation mixture was transformed into E. coli DH5α. Transformants were selected on LB medium containing 100 μg of ampicillin per ml and 20 μg of kanamycin per ml. The resulting plasmid, pNC52-k (Fig. 1B), was digested with XhoI, and the resulting 3.2-kb fragment containing the disrupted comEC-like gene plus flanking DNA was introduced into T. thermophilus HB27 by natural transformation. Transformants were selected on TM medium containing 40 μg of kanamycin per ml.
To disrupt the comEA-like gene, the insert of pFE87 was cloned as an HindIII-XbaI fragment into pBluescriptII KS in order to delete restriction sites from the multiple cloning site. The resulting plasmid, pFE87-HX (Table 1), was digested with BamHI and incubated in the presence of dNTPs and the Klenow enzyme. The blunt-ended kat gene was ligated to blunt-ended, linearized plasmid pFE87-HX, and the ligation mixture was transformed into E. coli DH5α. Transformants were selected on LB medium containing 20 μg of kanamycin per ml and 100 μg of ampicillin per ml. The resulting recombinant plasmid, designated pFE87-k (Fig. 1B), was digested with PstI plus XbaI, and the 3.3-kb fragment containing the disrupted ORF plus flanking wild-type DNA was introduced into T. thermophilus HB27 via natural transformation. Transformants were selected on TM medium containing 40 μg of kanamycin per ml. Correct allelic replacement of chromosomal wild-type comEA- and comEC-like genes by the disrupted ORFs was verified by Southern hybridization.
Construction of comA::lacZ transcriptional fusions and assay for β-galactosidase activities.
To generate comA::lacZ transcriptional fusions, vector pKOK 6.1 (30) containing a promoter-free lacZ gene was digested with PstI. The 4.7-kb PstI fragment, carrying the promoter-free lacZ gene, was inserted into the PstI site in pAF702-2.3, which resulted in pAF702-3.2 (Table 1). Correct orientation of the insert with respect to the lacZ gene was confirmed by restriction analysis. The insert of pAF702-3.2 was subcloned as a 8-kb KpnI-BamHI fragment in the opposite orientation with respect to the lac promoter into the broad-host-range vector pRK415, resulting in plasmid pRK702-3.2 (Table 1). pRK702-3.2 was transferred into Acinetobacter sp. strain ADP239 via triparental mating. As a control Acinetobacter sp. strain ADP239 carrying the lacZ gene of pKOK6.1 cloned in the opposite direction with respect to the lac promoter in vector pRK415 was used. The β-galactosidase activities were determined as described by Miller (37) and were expressed in Miller units, which are proportional to the increase in absorbance of free o-nitrophenol per minute and cell density.
Nucleotide sequence accession number.
The sequence data have been deposited in the GenBank database under accession no. AF320001 and AF319938.
RESULTS
Isolation and characterization of a noncompetent Acinetobacter mutant.
Transposon mutagenesis of an Acinetobacter sp. strain BD413 genomic library resulted in 200 tetracycline- and kanamycin-resistant E. coli DH5α clones. Retransfer of the mutant loci into Acinetobacter sp. strain ADP239 led to 1,000 kanamycin-resistant transformants which were analyzed to determine their transformation phenotypes. These studies led to detection of one completely noncompetent mutant, designated T701 (Fig. 2A).
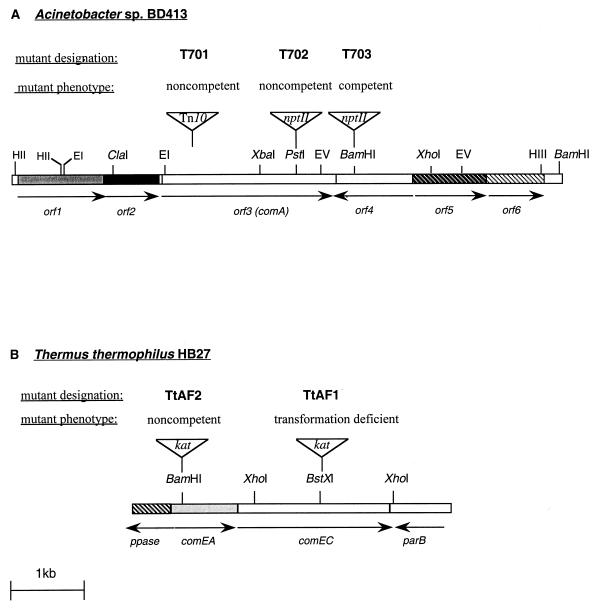
FIG. 2.
(A) Chromosomal region of Acinetobacter sp. strain BD413 containing orf1, orf2, orf3 (comA), orf4, orf5, and orf6 and physical map of the mutant loci. (B) Chromosomal region of T. thermophilus HB27 containing ppase, comEA, comEC, and parB. The arrows indicate directions of transcription. nptII, insertion site of the Kmr gene; Tn10, insertion site of mini-Tn10; kat, insertion site of the kat gene; EI, EcoRI; EV, EcoRV; HII, HincII; HIII, HindIII.
During previous transformation studies of mutants of Acinetobacter sp. strain ADP239, a deletion the pca operon, which encodes the key enzymes used for further breakdown of the p-hydroxybenzoate hydroxylase (PobA) reaction product (protocatechuate), was observed. A spontaneous large deletion in the pca operon would interfere with the screening system used for transformation-deficient mutants, which is based on the failure of transformation-deficient ADP239 mutants to grow on p-hydroxybenzoate in the presence of wild-type DNA since they cannot acquire the wild-type pobA allele. To exclude the possibility that there was a deletion in the pca operon, which could have resulted from integration of two noncontiguous DNA fragments into the ADP239 genome, mutant T701 was analyzed to determine its ability to use protocatechuate as a sole carbon and energy source. These studies revealed that mutant T701 was able to grow on protocatechuate, and so the possibility that there was a deletion in the pca operon was eliminated. Furthermore, mutant T701 showed a high proficiency for DNA repair during growth in the presence of the DNA-alkylating agent methyl methanesulfonate, which eliminated the possibility that the transformation defect was a result of an impaired RecA function. Complementation studies with recombinant plasmids carrying comP, comB, comC, comE, and/or comF revealed that the noncompetent phenotype of mutant T701 was not due to a defect in these recently identified competence genes. This was confirmed by hybridization studies (data not shown).
Cloning and nucleotide sequence analysis of the novel Acinetobacter competence locus.
The mutant locus of T701 was recovered on a 10.5-kb XbaI fragment (pAF701-1) and an overlapping 3.9-kb EcoRI-EcoRV fragment (pAF701-1.2) (Fig. 1A). Sequence analysis of the mini-Tn10-flanking genomic DNA of the mutant loci recovered led to identification of a 3′-end-truncated ORF, ′orf3, that was disrupted by transposon insertion. This finding suggests that orf3 plays an essential role in natural transformation of Acinetobacter sp. strain BD413. To allow cloning of the complete orf3 wild-type allele, an additional Acinetobacter orf3 mutant, designated T702, was generated by inserting a kanamycin marker gene (nptII) into the PstI site of orf3. Analysis of the transformation phenotype of the resulting mutant, T702 (Fig. 2A), revealed a completely noncompetent phenotype. The mutant locus of T702 was recovered on a 12.5-kb HincII fragment (pAF702-2) (Fig. 1A). To characterize the mutant loci of mutant T701 and T702, the inserts of plasmids pAF701-1 and pAF702-2 were subcloned into a series of overlapping clones, and a sequence analysis was performed. These studies led to identification of six ORFs, designated orf1, orf2, orf3, orf4, orf5, and orf6 (Fig. 2A).
Characterization of orf1, orf2, orf3, orf4, orf5, and orf6.
Each ORF starts with an ATG start codon and is preceded by a well-conserved and well-placed ribosome binding site (53). Two potential ATG start codons were found for orf3. Due to the well-placed conserved ribosome binding site, the second ATG is thought to represent the start codon. A conserved site for a presumptive ς70-dependent promoter was detected 68 bp upstream of this start codon. Additional conserved sites for presumptive ς70 -dependent promoters were found 25 bp upstream of orf1 and 31 bp upstream of orf5. A sequence with dyad symmetry that could form a stem-loop structure and that may act as a rho-independent terminator (46) was found 18 bp downstream of the stop codon of orf2 and 13 bp downstream of the stop codon of orf6. The ORFs are tightly clustered, and the stop codon of orf1 overlaps the start codon of orf2. This suggests that orf1 and orf2 may form an operon.
Similarities of the deduced proteins with proteins in databases.
The predicted proteins encoded by orf1, orf2, orf3, orf4, orf5, and orf6 comprise 411, 228, 792, 337, 338, and 268 amino acids, respectively, and have deduced molecular masses of 45, 25, 91, 39, 37, and 31 kDa, respectively. Orf1 and Orf2 are similar to components of ABC transporters. The protein product of orf1 is 22% identical to membrane protein HI1548 of H. influenzae RdKW20 (14) and 20% identical to DevC of Anabaena sp. strain PCC 7120, the integral membrane component of the DevBCA exporter (12). Hydropathy plots indicated that Orf1 is strongly hydrophobic and has at least four transmembrane helices. Orf2 is similar to ATP binding proteins of ABC transporters, exhibiting 41, 41, and 35% identity with UgpC (52) and GlnQ (40) of E. coli and DevA of Anabaena (12), respectively. ATP binding proteins of ABC transporters are characterized by an ATP binding motif (Walker sites A and B) which is essential for ATP binding and is highly conserved in Orf2 (11). The deduced protein encoded by orf3 is similar to proteins required for genetic transformation of different gram-negative and gram-positive bacteria; for example, Orf3 is 40% similar to gonococcal ComA (10), B. subtilis ComEC (18) and H. influenzae Rec-2 (5) and 37% similar to CelB of S. pneumoniae (45). Based on the significant similarities and the suggested function of Orf3 in natural transformation, Orf3 was designated ComA. Acinetobacter ComA also exhibits structural similarties with its homologs, including several hydrophobic domains in the central part of the protein. At least six hydrophobic potentially membrane-spanning domains were found in Acinetobacter ComA between residues 250 and 530, and two additional hydrophobic domains are present close to the N terminus.
The deduced protein encoded by orf4 is similar to HtrB proteins, exhibiting 35 and 33% similarity to HtrB of H. influenzae (33) and HtrB of E. coli (26), respectively. HtrB proteins are membrane-associated proteins that are involved in lipid A biosynthesis and are required for rapid growth of E. coli at temperatures above 33°C (3, 27).
The protein product encoded by orf5 is similar to proteases. The deduced protein is 36 and 41% similar to proteinase IV of E. coli (23) and PspA of Campylobacter jejuni (7), respectively. Proteinase IV of E. coli is a nonessential protease which digests cleaved signal peptides (23), and PspA of C. jejuni is thought to be involved in induction and/or processing of pilus subunits (7). Database searches with the deduced protein encoded by orf6 revealed no significant similarities to known proteins.
comA complementation studies and disruption of comA-flanking ORFs.
pRK415 derivatives pRK702-2.2 and pRK702-2.3 (Fig. 1A), both carrying the comA wild-type gene, were mobilized into noncompetent mutants T701 and T702, respectively, and were found to fully restore transformation competence. These results provide clear evidence that comA is essential for natural transformation in Acinetobacter sp. strain BD413. Complementation of the noncompetent mutants was independent of the orientation of the insert with respect to the lac promoter, indicating that expression of comA is under control of its native promoter.
To address the question whether orf1, orf2, and orf4 are also implicated in natural transformation, the ORFs were subjected to mutagenesis individually, and the resulting mutants were analyzed to determine their natural transformation phenotypes. Despite many different attempts, mutagenesis of orf1 and orf2 was not possible. This suggests that orf1 and orf2 have functions that are essential for cell viability. The nptII marker gene was successfully inserted into orf4 (Fig. 2A). The resulting mutant, T703, exhibited normal growth rates and was not affected in terms of natural transformation.
Growth phase-dependent transcription of comA in Acinetobacter sp. strain ADP239.
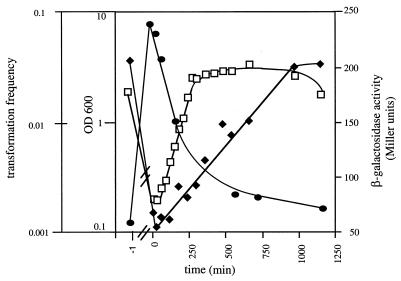
In order to gain insight into regulation of comA expression and to address the question of potential coregulation of comA with the recently identified pilin-like genes required for genetic transformation of Acinetobacter, we constructed transcriptional fusions of comA and a promoter-free lacZ gene and monitored β-galactosidase activities during growth. Upon transfer of the cells into fresh medium, comA transcription decreased rapidly, but then it recovered steadily during growth, reaching a maximum in the late stationary phase (Fig. 3). The course of growth phase-dependent regulation is correlated with growth phase-dependent transcriptional regulation of the pilin-like competence genes comB, comE, comF, and comP (21, 48).
FIG. 3.
Growth phase-dependent transcription of comA in Acinetobacter sp. strain ADP239. comA transcription was monitored by using comA::lacZ reporter fusions located on low-copy-number plasmid pRK415. β-Galactosidase activity was monitored as described by Miller (37). Optical densities at 600 nm (OD 600) (□), β-galactosidase activities (⧫), and transformation frequencies (●) during growth were determined at the times indicated.
Identification of comEC and comEA competence genes in T. thermophilus HB27.
Searches in the genome database for T. thermophilus HB27 with Acinetobacter sp. strain BD413 ComA led to identification of an ORF whose product shows significant similarities to the Acinetobacter sp. strain BD413 and N. gonorrhoeae ComA competence proteins and the B. subtilis ComEC competence protein. Analysis of the region upstream of this conserved ORF led to detection of an ORF encoding a putative protein with similarities to the B. subtilis protein ComEA, which is essential for genetic transformation of B. subtilis (17, 24). Based on the significant similarities and the organization of these two ORFs, which were analogous to the B. subtilis comEA-comEC cluster, they were designated comEA and comEC. The TGA stop codon of comEA in T. thermophilus HB27 overlaps the ATG start codon of comEC, indicating that putative cotranscription of the two ORFs occurs. Downstream of comEC an ORF nearly identical to the putative parB gene (accession no. AJ277593) of T. thermophilus HB8 was identified. The orientation of this ORF is opposite the orientation of comEA and comEC (Fig. 2B).
To examine the potential function of comEA and comEC in the transformation mechanism of the highly competent extreme thermophile T. thermophilus HB27, both ORFs were subjected to gene disruption by using the thermostable kanamycin gene kat. The resulting kanamycin-resistant T. thermophilus HB27 mutants, designated TtAF1 and TtAF2 (Table 1 and Fig. 2B), were analyzed to determine their transformation phenotypes by using the genomic DNA of a spontaneous streptomycin-resistant T. thermophilus HB27 mutant. In the comEA mutant natural transformation was completely absent, whereas in the comEC mutant natural transformation was significantly impaired (the natural transformation frequencies were 1,000-fold lower). Since the orientation of the downstream putative parB gene is opposite the orientation of comEA and comEC, the transformation defects of the comEA and comEC mutants cannot be due to a polar effect on the parB gene. The gene organization results together with the results of the mutant studies provide clear evidence that the Thermus comEA-comEC locus is involved in natural transformation. Since comEC is located downstream of comEA, the possibility that marker insertion in comEC has a polar effect on the preceding comEA gene can be excluded. The organization of comEA and comEC, together with the results of the mutant studies leads to the conclusion that comEC is important for natural transformation. Although the possibility that marker insertion in comEA has a polar effect on comEC cannot be excluded, the phenotypic differences of the comEA and comEC mutants indicate that analogous to B. subtilis comEA and comEC, both genes, comEA and comEC, are essential for T. thermophilus transformation.
Characterization of the comEC and comEA competence genes in T. thermophilus HB27.
The protein encoded by comEC consists of 677 amino acids and has a calculated mass of 72 kDa. Thermus ComEC is 40, 40, 40, 37, and 37% similar to ComA of Acinetobacter sp. strain BD413, ComEC of B. subtilis, Rec-2 of H. influenzae, ComA of N. gonorrhoeae, and CelB of S. pneumoniae, respectively. Like ComA of Acinetobacter sp. strain BD413, ComEC of T. thermophilus HB27 is predicted to be an inner membrane protein with six potentially membrane-spanning domains in the central part.
The ComEA protein of T. thermophilus HB27 consists of 298 amino acids, has a calculated mass of 30 kDa, and is similar to ComEA of B. subtilis and CelA of S. pneumoniae, which comprise 205 and 216 amino acids, respectively (17, 45). In a C-terminal 60-amino-acid overlap ComEA of T. thermophilus is 60% similar to the C-terminal domain of Bacillus ComEA and Streptococcus CelA (Fig. 4). This domain includes a helix-hairpin-helix motif which is responsible for DNA binding, as demonstrated in B. subtilis (49).
FIG. 4.
Alignment of ComEA proteins. Identical residues are indicated by gray boxes. B.s., B. subtilis; S.p., S. pneumoniae; T.t., T. thermophilus HB27.
DISCUSSION
Here we describe identification of novel competence genes in the mesophilic soil bacterium Acinetobacter sp. strain BD413 and the extremely thermophilic bacterium T. thermophilus HB27. The T. thermophilus HB27 competence genes are the first competence genes identified in an extremely thermophilic bacterium so far. Based on sequence similarities, the novel competence genes in Acinetobacter and Thermus were designated comA and comEC, respectively. Both comA of Acinetobacter sp. strain BD413 and comEC of T. thermophilus HB27 were found to be essential for natural transformation. Sequence analysis of the comA locus of Acinetobacter sp. strain BD413 revealed six tandemly arranged (ORFs), designated orf1, orf2, comA, orf4, orf5, and orf6. The comEC gene of T. thermophilus HB27 is preceded by an ORF that encodes a protein with significant similarities to the B. subtilis competence protein ComEA (17, 24). This ORF, designated comEA, was also found to be essential for natural transformation in Thermus. A comEA-like competence gene was not detected close to the comA gene in Acinetobacter sp. strain BD413.
Despite the fact that comA of Acinetobacter sp. strain BD413 is not closely associated with the recently identified Acinetobacter competence genes comP, comB, comE, and comF, growth phase-dependent transcriptional regulation this comA gene is similar to regulation of the pilin-like competence genes comP, comB, comE, and comF.
Although T. thermophilus, B. subtilis, and S. pneumoniae are not closely related bacteria, the genes of the conserved comEC locus of T. thermophilus and B. subtilis and the homologous celB locus of S. pneumoniae are organized analogously. In B. subtilis, T. thermophilus, and S. pneumoniae the competence gene comEC or celB is closely associated with a second competence gene that was first described in B. subtilis as comEA. However, in contrast to B. subtilis, a homolog of an additional gene, comEB, which is present in the Bacillus competence-specific operon, is not present T. thermophilus and S. pneumoniae (2, 17, 45).
Clues to the function of ComA in Acinetobacter sp. strain BD413 and the function of ComEC in T. thermophilus HB27 can be derived from the significant similarities of these proteins to ComA of N. gonorrhoeae, ComEC of B. subtilis, and Rec-2 of H. influenzae (8). comA mutants of N. gonorrhoeae have been shown to take up DNA in a DNase-resistant state (9). This finding, together with the prediction that there are several transmembrane α-helices, led to the proposal that the gonococcal ComA protein may be an integral membrane protein involved in DNA transfer across the cytoplasmic membrane (9, 10). The comEC product in B. subtilis was also predicted to be a polytopic integral membrane protein with 8 to 10 transmembrane segments and was found to be essential for DNA uptake but not for DNA binding (17, 24). An analogous function has been suggested for Rec-2 of H. influenzae (4). Based on secondary structure predictions for ComA of Acinetobacter sp. strain BD413 and ComEC of T. thermophilus HB27, six central and two N-terminal hydrophobic regions are present in ComA and ComEC, respectively. Furthermore, ComA of Acinetobacter sp. strain BD413 and ComEC of T. thermophilus both have a large, hydrophilic, positively charged domain near the N terminus, as well as a large hydrophilic, positively charged C-terminal domain which may be localized in the cytoplasm, based on the rule that positive charges of inner membrane proteins are oriented towards the cytoplasm (5). The N terminus of ComA of Acinetobacter sp. strain BD413 has structural features characteristic of signal peptides (56) including (i) a positively charged N terminus (Arg-4), (ii) a hydrophobic core (Ile-15 to Ile-23), and (iii) a small neutral C-terminal residue. Positions −1 and −3 are known to be particularly important for specifying the cleavage site, which could be between Ala-25 (position −1) and Ile-26 (position 0) according to the proposal of von Heinje (55). In contrast to ComA of Acinetobacter sp. strain BD413, the N terminus of ComEC of T. thermophilus does not exhibit structural features characteristic of signal peptides. Based on the structural characteristics and topology data, we propose that ComA and ComEC of Acinetobacter and Thermus, repectively, are channel-forming polytopic membrane proteins implicated in transport of DNA through the cytoplasmic membrane.
The broad distribution of ComA- and ComEC-like factors among very different bacteria independent of their phylogenetic relationships and their natural environments indicates that these proteins may play a central role in the transformation mechanisms of very different bacteria and that these components are highly conserved.
It has to be noted that except for the comEA gene of the comEC locus in T. thermophilus, no comEA-like genes have been found close to conserved comA/comEC-like genes in gram-negative bacteria, but interestingly, multiple copies of comEA-like genes have been identified in the genome of N. gonorrhoeae, and at least one of them is involved in natural transformation (I. Chen and E. C. Gotschlich, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. D/B-10, p. 211, 1999). Analogously, the possibility that there are genes in Acinetobacter sp. strain BD413 that exhibit similarities to the B. subtilis competence gene comEA also cannot be excluded, and this question remains an interesting topic for the future.
ACKNOWLEDGMENTS
This work was supported by grant Av 9/4-4 from the Deutsche Forschungsgemeinschaft. A. Friedrich was supported by the Fond der Chemischen Industrie.
REFERENCES
- 1.Busch S, Rosenplänter C, Averhoff B. Identification and characterization of ComE and ComF, two novel pilin-like competence factors involved in natural transformation of Acinetobacter sp. strain BD413. Appl Environ Microbiol. 1999;65:4568–4574. doi: 10.1128/aem.65.10.4568-4574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell E A, Choi S Y, Masure H R. A competence regulon in streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol. 1998;27:929–939. doi: 10.1046/j.1365-2958.1998.00737.x. [DOI] [PubMed] [Google Scholar]
- 3.Clementz T, Bednarski J J, Raetz C R. Function of the htrB high temperature requirement gene of Escherichia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J Biol Chem. 1996;271:12095–12102. doi: 10.1074/jbc.271.20.12095. [DOI] [PubMed] [Google Scholar]
- 4.Clifton S W, McCarthy D, Roe B A. Sequence of the rec-2 locus of Haemophilus influenzae: homologies to comE-ORF3 of Bacillus subtilis and msbA of Escherichia coli. Gene. 1994;146:95–100. doi: 10.1016/0378-1119(94)90840-0. [DOI] [PubMed] [Google Scholar]
- 5.Dalbey R E. Positively charged residues are important determinants of membrane protein topology. Trends Biochem Sci. 1990;15:253–257. doi: 10.1016/0968-0004(90)90047-f. [DOI] [PubMed] [Google Scholar]
- 6.de Grado M, Castan P, Berenguer J. A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid. 1999;42:241–245. doi: 10.1006/plas.1999.1427. [DOI] [PubMed] [Google Scholar]
- 7.Doig P, Yao R, Burr D H, Guerry P, Trust T J. An environmentally regulated pilus-like appendage involved in Campylobacter pathogenesis. Mol Microbiol. 1996;20:885–894. doi: 10.1111/j.1365-2958.1996.tb02526.x. [DOI] [PubMed] [Google Scholar]
- 8.Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 9.Facius D, Fussenegger M, Meyer T F. Sequential action of factors involved in natural competence for transformation of Neisseria gonorrhoeae. FEMS Microbiol Lett. 1996;137:159–164. doi: 10.1111/j.1574-6968.1996.tb08099.x. [DOI] [PubMed] [Google Scholar]
- 10.Facius D, Meyer T F. A novel determinant (comA) essential for natural transformation competence in Neisseria gonorrhoeae and the effect of comA defect on pilin variation. Mol Microbiol. 1993;10:699–712. doi: 10.1111/j.1365-2958.1993.tb00942.x. [DOI] [PubMed] [Google Scholar]
- 11.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiedler G, Arnold M, Hannus S, Maldener I. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol Microbiol. 1998;27:1193–1202. doi: 10.1046/j.1365-2958.1998.00762.x. [DOI] [PubMed] [Google Scholar]
- 13.Figurski D H, Helinski D R. Replication of an origin-containing derivate of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:495–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 15.Gebhard F, Smalla K. Transformation of Acinetobacter sp. strain BD413 by transgenic sugar beet DNA. Appl Environ Microbiol. 1998;64:1550–1554. doi: 10.1128/aem.64.4.1550-1554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregg-Jolly L A, Ornston L N. Properties of Acinetobacter calcoaceticus recA and its contribution to intracellular gene conversion. Mol Microbiol. 1994;12:985–992. doi: 10.1111/j.1365-2958.1994.tb01086.x. [DOI] [PubMed] [Google Scholar]
- 17.Hahn J, Inamine G, Kozlov Y, Dubnau D. Characterization of comE, a late competence operon of Bacillus subtilis required for the binding and uptake for transforming DNA. Mol Microbiol. 1993;10:99–111. doi: 10.1111/j.1365-2958.1993.tb00907.x. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D. Studies on transformation of Escherichia coli plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Hartnett G B, Averhoff B, Ornston L N. Selection of Acinetobacter calcoaceticus mutants deficient in the p-hydroxybenzoate hydroxylase gene (pobA), a member of a supraoperonic cluster. J Bacteriol. 1990;172:6160–6161. doi: 10.1128/jb.172.10.6160-6161.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herzberg C, Friedrich A, Averhoff B. comB, a novel competence gene required for natural transformation of Acinetobacter sp. BD413: identification, characterization and analysis of growth-phase-dependent regulation. Arch Microbiol. 2000;172:220–228. doi: 10.1007/s002039900134. [DOI] [PubMed] [Google Scholar]
- 22.Hidaka Y, Hasegawa M, Nakahara T, Hoshino T. The entire population of Thermus thermophilus cells is always competent at any growth phase. Biosci Biotechnol Biochem. 1994;58:1338–1339. doi: 10.1271/bbb.58.1338. [DOI] [PubMed] [Google Scholar]
- 23.Ichihara S, Suzuki T, Suzuki M, Mizushima S. Molecular cloning and sequencing of the sppA gene and characterization of the encoded protease IV, a signal peptidase, of Escherichia coli. J Biol Chem. 1986;261:9405–9411. [PubMed] [Google Scholar]
- 24.Inamine G S, Dubnau D. ComEA, a Bacillus subtilis integral membrane protein required for genetic transformation, is needed for both DNA binding and transport. J Bacteriol. 1995;177:3045–3051. doi: 10.1128/jb.177.11.3045-3051.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juni E, Janik A. Transformation of Acinetobacter calcoaceticus (Bacterium antitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karow M, Fayet O, Cegielska A, Ziegelhoffer T, Georgopoulos C. Isolation and characterization of the Escherichia coli htrB gene, whose product is essential for bacterial viability above 33°C in rich media. J Bacteriol. 1991;173:741–750. doi: 10.1128/jb.173.2.741-750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karow M, Georgopoulos C. The essential Escherichia coli msbA gene, a multicopy suppressor of null mutations in the htrB gene, is related to the universally conserved family of ATP-dependent translocators. Mol Microbiol. 1993;7:69–79. doi: 10.1111/j.1365-2958.1993.tb01098.x. [DOI] [PubMed] [Google Scholar]
- 28.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 29.Kloos D U, Strätz M, Guttler A, Steffan R J, Timmis K N. Inducible cell lysis system for the study of natural transformation and environmental fate of DNA released by cell death. J Bacteriol. 1994;176:7352–7361. doi: 10.1128/jb.176.23.7352-7361.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 31.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J Bacteriol. 1986;166:338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lasa I, Caston J R, Fernandez-Herrero L A, De Pedro M A, Berenguer J. Insertional mutagenesis in the extreme thermophile eubacterium Thermus thermophilus. Mol Microbiol. 1992;11:1555–1564. doi: 10.1111/j.1365-2958.1992.tb00877.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee N G, Sunshine M G, Engstrom J J, Gibson B W, Apicella M A. Mutation of the htrB locus of Haemophilus influenzae nontypable strain 2019 is associated with modifications of lipid A and phosphorylation of the lipo-oligosaccharide. J Biol Chem. 1995;270:27151–27159. [PubMed] [Google Scholar]
- 34.Link C, Eickernjäger S, Porstendörfer D, Averhoff B. Identification and characterization of a novel competence gene, comC, required for DNA binding and uptake in Acinetobacter sp. strain BD413. J Bacteriol. 1998;180:1592–1595. doi: 10.1128/jb.180.6.1592-1595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorenz M G, Wackernagel W. Bacterial gene transfer by genetic transformation in the environment. Microbiol Rev. 1994;58:563–603. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mather M W, Fee J A. Development of plasmid cloning vectors for Thermus thermophilus HB8: expression of a heterologous, plasmid-borne kanamycin nucleotidyltransferase gene. Appl Environ Microbiol. 1992;58:421–425. doi: 10.1128/aem.58.1.421-425.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller J H. Assay of β-galactosidase. In: Platt T, Miller-Hill B, Miller J H, editors. Experiments in molecular genetics Cold Spring Habor Laboratory Press. N.Y: Cold Spring Harbor; 1972. pp. 319–353. [Google Scholar]
- 38.Nielsen K M, van Elsas J D, Smalla K. Transformation of Acinetobacter sp. strain BD413 (pFG4ΔnptII) with transgenic plant DNA in soil microcosms and effects of kanamycin on selection of transformants. Appl Environ Microbiol. 2000;66:1237–1247. doi: 10.1128/aem.66.3.1237-1242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen K M, van Weerelt M D M, Berg T N, Bones A M, Hagler A N, van Elsas J D. Natural transformation and availability of transforming DNA to Acinetobacter calcoaceticus in soil microcosms. Appl Environ Microbiol. 1997;63:1945–1952. doi: 10.1128/aem.63.5.1945-1952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nohno T, Saito T, Hong J S. Cloning and complete nucleotide sequence of the Escherichia coli glutamine permease operon (glnHPQ) Mol Gen Genet. 1986;205:260–269. doi: 10.1007/BF00430437. [DOI] [PubMed] [Google Scholar]
- 41.Ornston L N, Stanier R Y. The conversion of catechol and protocatechuate to β-ketoadipate by Pseudomonas putida. J Biol Chem. 1966;241:3776–3786. [PubMed] [Google Scholar]
- 42.Oshima T, Imahori K. Description of Thermus thermophilus comb. nov., a nonsporulating thermophilic bacterium (Yoshida and Oshima) from a Japanese thermal spa. Int J Syst Bacteriol. 1974;24:102–112. [Google Scholar]
- 43.Palmen R, Buijsman P, Hellingwerf K J. Physiological regulation of competence induction for natural transformation in Acinetobacter calcoaceticus. Arch Microbiol. 1994;162:344–351. [Google Scholar]
- 44.Palmen R, Hellingwerf K J. Uptake and processing of DNA by Acinetobacter calcoaceticus. Gene. 1997;192:179–190. doi: 10.1016/s0378-1119(97)00042-5. [DOI] [PubMed] [Google Scholar]
- 45.Pestova E V, Morrison D A. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J Bacteriol. 1998;180:2701–2710. doi: 10.1128/jb.180.10.2701-2710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 47.Porstendörfer D, Drotschmann U, Averhoff B. A novel competence gene, comP, is essential for natural transformation of Acinetobacter sp. strain BD413. Appl Environ Microbiol. 1997;63:4150–4157. doi: 10.1128/aem.63.11.4150-4157.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Porstendörfer D, Gohl O, Mayer F, Averhoff B. ComP, a pilin-like protein essential for natural competence in Acinetobacter sp. strain BD413: regulation, modification, and cellular localization. J Bacteriol. 2000;182:3673–3680. doi: 10.1128/jb.182.13.3673-3680.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Provvedi R, Dubnau D. ComEA is a DNA receptor for transformation of competent Bacillus subtilis. Mol Microbiol. 1999;31:271–280. doi: 10.1046/j.1365-2958.1999.01170.x. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Habor, N.Y: Cold Spring Habor Laboratory Press; 1989. [Google Scholar]
- 51.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schweizer H, Boos W. Characterization of the ugp region containing the genes for the phoB dependent sn-glycerol-3-phosphate transport system of Escherichia coli. Mol Gen Genet. 1984;197:161–168. doi: 10.1007/BF00327937. [DOI] [PubMed] [Google Scholar]
- 53.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementary to nonsense triplets and ribosome-binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1345. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon R, Priefer U, Pühler A. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 55.von Heinje G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 56.von Heinje G. Signals for protein targeting into and across membranes. Subcell Biochem. 1994;22:1–19. doi: 10.1007/978-1-4615-2401-4_1. [DOI] [PubMed] [Google Scholar]