Abstract
Microbially mediated reduction and immobilization of U(VI) to U(IV) plays a role in both natural attenuation and accelerated bioremediation of uranium-contaminated sites. To realize bioremediation potential and accurately predict natural attenuation, it is important to first understand the microbial diversity of such sites. In this paper, the distribution of sulfate-reducing bacteria (SRB) in contaminated groundwater associated with a uranium mill tailings disposal site at Shiprock, N.Mex., was investigated. Two culture-independent analyses were employed: sequencing of clone libraries of PCR-amplified dissimilatory sulfite reductase (DSR) gene fragments and phospholipid fatty acid (PLFA) biomarker analysis. A remarkable diversity among the DSR sequences was revealed, including sequences from δ-Proteobacteria, gram-positive organisms, and the Nitrospira division. PLFA analysis detected at least 52 different mid-chain-branched saturate PLFA and included a high proportion of 10me16:0. Desulfotomaculum and Desulfotomaculum-like sequences were the most dominant DSR genes detected. Those belonging to SRB within δ-Proteobacteria were mainly recovered from low-uranium (≤302 ppb) samples. One Desulfotomaculum-like sequence cluster overwhelmingly dominated high-U (>1,500 ppb) sites. Logistic regression showed a significant influence of uranium concentration over the dominance of this cluster of sequences (P = 0.0001). This strong association indicates that Desulfotomaculum has remarkable tolerance and adaptation to high levels of uranium and suggests the organism's possible involvement in natural attenuation of uranium. The in situ activity level of Desulfotomaculum in uranium-contaminated environments and its comparison to the activities of other SRB and other functional groups should be an important area for future research.
Microbially mediated reduction of redox-sensitive metals offers the potential to remediate metal-contaminated groundwater in situ. Sulfate-reducing bacteria (SRB) are important members of microbial communities involved in such metal reduction and occur in a variety of environments, including oil- and gas-bearing formations, soils, and domestic, industrial, and mining wastewaters (39). Although traditionally considered obligate anaerobes, observations of sulfate reduction occurring in the aerobic environment reported in the last 15 years have demonstrated a much larger ecological range of the SRB than previously thought (5, 6, 13). The dissimilatory sulfate-reducing bacteria in particular are environmentally ubiquitous, are found over an extensive range of pH and salt concentrations, and exhibit a superior ability to reduce and accumulate metals (16, 30, 53). Additionally they can tolerate a variety of heavy metals and dissolved sulfide. Some of these organisms can use U(VI) as a terminal electron acceptor, reducing the toxic and soluble U(VI) to insoluble U(IV), and also generate insoluble uranium sulfides in the presence of H2S.
Uranium has no known biological function and is toxic to cells at low concentrations: 20 to 40 times more toxic than copper or nickel (20). The toxicity of uranium is primarily derived from its chemical properties rather than from its radioactivity (12). It has been reported that bacteria capable of reducing U(VI) to U(IV) are ubiquitous in nature (1, 2). The uranium reducers are also primarily sulfate reducers, and their growth can be stimulated by adding nutrients to the groundwater (1).
It has been well documented that Desulfovibrio species can reduce the soluble oxidized form of uranium, U(VI), to insoluble U(IV) (22, 23). A recent study demonstrated that a Desulfotomaculum strain isolated from heavy metal-contaminated sediment can grow with U(VI) as the sole electron acceptor (44). Overall the SRB play an important role in uranium geochemistry and may be a useful tool for removing uranium from contaminated environments by using ex situ treatments and stabilizing uranium in situ. However, little is known about the diversity of these bacteria, both in terms of community structure (the different SRB present in a single environmental community at a specific site) and community composition (the numbers or percentages of different SRB at a particular site). There is also little information available on variations in the SRB community structure and composition in response to changing environmental conditions.
The objective of this research is to establish the diversity of SRB at a heavy metal-contaminated site in relation to geochemical measurements, particularly uranium concentration. This work is part of a broader effort to study the dominant terminal electron accepting processes and biotransformation occurring in the subsurface at such sites. The Shiprock site was chosen because of its wide range of uranium concentrations in groundwater and a wide range of cocontaminant concentrations, particularly sulfate and nitrate. The site has been subject to uranium contamination since the 1950s, providing a significant length of time for microbial communities to adapt to elevated levels of uranium. Groundwater samples with a range of contaminant concentrations were used as the means of accessing and interpreting the subsurface microbial communities. Two culture-independent analyses were used. (i) The first was a molecular method based on PCR, restriction enzyme digestion, and sequence analysis of dissimilatory sulfite reductase (DSR) gene fragments (17, 29, 49), which provided detailed information on the major taxonomic groups of sulfate-reducers present in these samples. The presence of Desulfovibrio sp. (δ-Proteobacteria) was also directly assessed by specific PCR targeting the NiFe hydrogenase indicative of this genus (48, 50). (ii) The second method, signature lipid analysis, was used to quantify viable sulfate/metal reducers by measuring mid-chain branched saturates and branched monounsaturates in the community phospholipid fatty acids (PLFA) (51, 52). These techniques taken together and compared to measured groundwater geochemical parameters provide new information on SRB diversity. As similar studies are conducted at other sites, we anticipate insight into community structure and composition that will enable effective in situ bioremediation of uranium-contaminated sites.
MATERIALS AND METHODS
Site description.
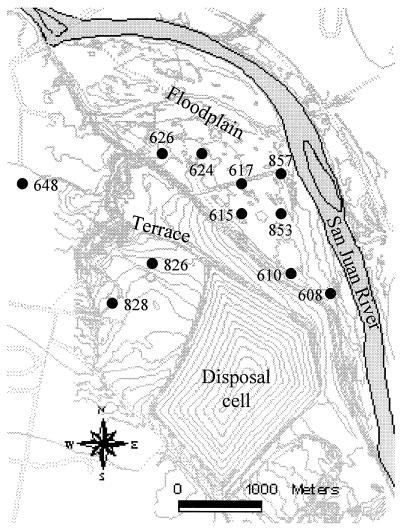
The U.S. Department of Energy is responsible for uranium mill tailings under the Uranium Mill Tailings Radiation Control Act of 1978. The Shiprock UMTRA site is on Navajo Nation land in San Juan County, N.Mex., located adjacent to and partly within the town of Shiprock, along the south side of the San Juan River on an elevated terrace about 21 m above the river (samples 828 and 826; Fig. 1). Bob Lee Wash flows northward on the terrace along the west side of the site and flows down onto the floodplain of the river. This wash would contain flowing water ephemerally, but the lower 200 m of the wash receives a constant discharge of about 230 liters min−1 from a potable-water artesian well (well 648). This water has created wetlands within Bob Lee Wash and at the mouth of the wash, where it discharges onto the river's floodplain (proximal to wells 608, 610, 615, 617, 624, 626, 853, and 857; Fig. 1). Several drainage ditches in the floodplain contain water year-round (7). An uncontaminated control sample was taken from well 648 (Fig. 1).
FIG. 1.
Map of the UMTRA Shiprock mill tailing site.
The Shiprock disposal cell is on unconsolidated alluvial terrace deposits underlain by Mancos Shale. Groundwater occurs at the contact between the terrace alluvium and the upper portion of the Mancos Shale, where it has been weathered. Uranium contamination occurs in the alluvium and upper Mancos Shale on the terrace and in the floodplain alluvium. The contaminated groundwater in the terrace alluvium and upper Mancos Shale beneath the site and in the floodplain alluvium along the river have exceeded the maximum concentration limits established by the Environmental Protection Agency EPA for nitrate and uranium. The volume of contaminated groundwater is estimated to be 610,000 m3 (160 million gallons).
Sample collection.
Prior to sample collection, all glassware used was washed in a 10% (vol/vol) Micro cleaning solution (VWR Scientific), rinsed 10 times in tap water, and then rinsed 10 times in deionized water. The glassware was then heated at 450°C for 4 h in a muffle furnace prior to use. All groundwater samples were collected in March 1999 with a downhole peristaltic or impeller pump. A minimum of 3 well volumes was purged from the well before sampling. Between sampling events, the pump and associated tubing were decontaminated with a dilute detergent and rinsed with deionized water. Samples (2 to 4 liters each) were filtered through sterile (methanol rinsed) Anodisc filters (Whatman International, Ltd., Maidstone, England), 47 mm diameter, 0.2 μm pore size. Filters were stored in muffle-sterilized glass petri dishes, preserved on dry ice, and shipped overnight to the University of Tennessee, Knoxville. The filtration method was designed to ensure that all suspended particles, including both sediment grains (with microorganisms attached) and individual microorganisms, are retained for analysis. A significant proportion of the microbial populations analyzed likely is attached to sediment particles.
Measurement of relevant geochemical properties.
Sulfate and sulfide were determined as components of a suite of anions analyzed by ion chromatography (Dionex Model DX-300; AS-4a column, chemical suppression, and conductivity detection) according to McKinley et al. (28). Samples were quantified against commercial standards that ranged from 0.1 to 100 mg liter−1. Uranium (U(VI)) concentrations were determined with a kinetic phosphorescence analyzer (model KPA-11, Chemchek Instruments, Inc.) according to McKinley et al. (27). The detection limit was 0.3 μg of uranium liter−1. Quantitation was against NIST-traceable standards over the standard concentration range of 0.25 μg of uranium liter−1 to 50 μg of uranium liter−1 in 11 steps. Samples were treated only by the addition of a phosphorescent complexant and were run in batch using an autosampler. When necessary, samples were diluted and rerun so that raw results fell within the standard concentration range and yielded acceptable counting statistics. A full suite of standards was run at the beginning and end of each analytical sample set as an internal check on accuracy and precision. Dissolved oxygen (DO) was measured with a flow cell during well purging. Stable (invariant) DO values typically occurred prior to completion of well purging; the minimum observed concentration was taken as the in situ value. The pH was measured by electrode against commercial standards.
Lipid analysis.
All solvents used were of GC grade (Fisher Scientific, Pittsburgh, Pa.). Glassware was washed in 10% (vol/vol) micro cleaner solution (VWR Scientific), rinsed 10 times in tap water and 5 times in deionized water, and then heated for 4 h in a muffle furnace at 450°C. Lipids were extracted from filters by the modified procedure of Bligh and Dyer (52). Total lipid fractions were then fractionated into glyco-, neutral, and polar lipids (14). The phospholipid-containing polar lipid was then subjected to a mild alkaline methanolysis, transesterifying the fatty acids into methyl esters (FAME) and recovered with hexane (14). The PLFA were separated, quantified, and identified by GC-MS (37). Fatty acids were identified by relative retention times, comparison with authentic standards (Matreya, Inc.) and by mass spectra (collected at an electron energy of 70 mV) (38). Fatty acid nomenclature is in the form of “A:Bω C,” where “A” designates the total number of carbons, “B” designates the number of double bonds, and “C” designates the distance of the closest unsaturation from the aliphatic end of the molecule. The suffixes “c” for cis and “t” for trans refer to geometric isomers. The prefixes “i,” “a,” and “me” refer to iso and anteiso methyl branching and mid-chain methyl branching, respectively. Cyclopropyl rings are indicated by “cy” (18).
DNA extraction and PCR from pure cultures and filters.
Pure cultures of the following Desulfotomaculum strains were kindly provided by David Boone, Portland State University; D. acetoxidans strain DSM771 (type strain), D. aeronauticum strain 9, D. luciae strain SLT, D. nigrificans strain Delft 74T, D. orientis strain DSM765 (type strain), D. putei strain TH-12. Desulfovibrio desulfuricans (ATCC 29577) was purchased from the American Type Culture Collection. The organisms were grown anaerobically to mid-log phase in MS enrichment medium (pH 7) (http://caddis.esr.pdx.edu/smccw/; for Desulfovibrio desulfuricans, ATCC medium 1250 was used as recommended), and nucleic acids were extracted from cell pellets by a bead-beating procedure (41). Anodisk filters were broken into shards by hand with solvent-sterilized forceps and placed into 2-ml screw-cap microcentrifuge tubes. DNA was extracted directly from filters by mechanical disruption as described above. PCR amplifications were performed with two sets of primers, one targeting the [NiFe] hydrogenase of Desulfovibrio sp. as described by Wawer et al. (50). The second employed general SRB-specific primers targeting the DSR gene (dsr1F, 5′-ACSCACTGGAAGCACG-3′; dsr4R, 5′-GTGTAGCAGTTACCGCA-3′) described by Wagner et al. (49). Briefly, thermocycling for DSR consisted of 30 cycles of 94°C for 45 s, 54°C for 30 s, and 68°C for 90s (10 min on final cycle) with 1.25 U of Expand HF polymerase (Boehringer, Indianapolis, Ind.) and 10 pmol each of the primers in a total volume of 25 μl to produce a ca. 1.9-kb DNA fragment encoding most of the α- and β- subunits of the gene (49). Thermocycling was performed with a “Robocycler” PCR block (Stratagene, La Jolla, Calif.). For the hydrogenase gene, a touchdown PCR from 66°C to 55°C (50) was performed to reduce the formation of spurious by-products. The primer targets the [NiFe] hydrogenase gene, which encodes an enzyme playing an important role in hydrogen metabolism of SRB (47) and in dissimilatory metal reduction by SRB (24, 25). This enzyme is present in all Desulfovibrio species (48), with a PCR product length around 450 bp using the primer described above.
Cloning and restriction digestion.
PCR products of the DSR gene fragment were gel purified and extracted with a Gene-Clean kit (BIO-101, Vista, Calif.). Purified fragments were cloned using the vector PCR2.1 TOPO and Escherichia coli TOP10F′ competent cells according to the manufacturer's instructions (Invitrogen, Carlsbad, Calif.). From each of those 11 libraries, 21 to 34 white colonies were randomly selected and the cloned inserts were reamplified with the vector primers M13 reverse and T7 (30 cycles of 94°C for30 s, 55°C for 30 s, and 72°C for 90 s). A portion (5 μl) of the resulting amplification product was digested at 37°C with the restriction endonuclease MspI according to the manufacturer's instructions (Boehringer) and analyzed by separation of fragments on a 2% agarose TAE gel.
Sequence analysis.
Representative plasmids from each digestion pattern were selected for sequencing. Clones with MspI digestion patterns that appeared more than once were sequenced from both the 3′ and 5′ ends of each insert with vector primers M13 reverse/T7, while those with unique MspI digestion patterns were sequenced with the DSR1F primer to obtain a partial α-subunit sequence of the gene. The M13 reverse/T7 amplification product was gel purified, extracted with a Gene Clean kit (BIO-101), and subjected to sequencing on an Applied Biosystems automated sequencer (model 310) with Prism Big-Dye terminators. The sequences were assembled and aligned by using D. G. Gilbert's SeqPup sequence alignment editor, version 0.6 (available from the author at ftp.bio.indiana.edu). Sequence identification was performed by use of the BLASTN facility of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Phylogenetic trees were constructed by the neighbor-joining method, with the ARB software environment (42). Cloned sequences were screened for possible chimeric origin by independent neighbor-joining analysis of the 5′ and 3′ halves of sequences within an alignment of all published DSR sequences, including DSR sequences from pure cultures generated in this study. Sequences from pure cultures were derived from direct analysis of amplification products from genomic DNA templates.
Statistical analysis.
Pearson r linear correlations between geochemical variables were performed using the basic stats module of Statistica, version 5.1 for Windows (Statsoft, Inc., Tulsa, Okla.). Logistic regression was used to test whether concentrations of environmental chemicals could explain occurrence frequencies for specific clades of DSR sequences, where sufficient samples for statistical analysis were detected (SAS Institute, version 8.1, Cary, N.C.).
Nucleotide sequence accession number.
Partial cloned DSR sequences recovered from Shiprock groundwater samples were submitted to GenBank under accession no. AY015500 to AY015569 (α-subunits) and AY015582 to AY015615 (β-subunits). Partial DSR sequences recovered from cultured reference strains were submitted as AY015493 to AY015495 and AY015496 to AY015499 for the α-subunits and AY015577 to AY015581 for the β-subunits.
RESULTS
Physical and chemical characteristics of UMTRA groundwater wells.
Samples collected on the terrace (wells 828 and 826) were collected from the top 3.0 m of the water table, averaging a minimal depth of 4.9 m below the ground surface. Samples collected on the floodplain (wells 608, 610, 615, 617, 624, 626, 853, and 857) were taken from the top 1.3 m of the water table, averaging a minimal depth of 2.1 m below the ground surface. The pH measured at all groundwater sites was nearly neutral and ranged from 6.53 to 7.12, except that for the control well 648, which was slightly alkaline (pH 7.8 to 8.0). (See Fig. 1 for well locations and Table 1 for geochemical data.) Well 648 is an artesian well with the water produced from the Morrison Formation (Jurassic age) through perforated casing from a depth of approximately 450 to 540 m. Well 648 was also warmer (30°C) than other wells (from 8.4°C to 15.5°C), due to its source depth.
TABLE 1.
Geochemical measurements of Shiprock groundwater
| Well | Concn of:
|
pH | Temp (°C) | ||||
|---|---|---|---|---|---|---|---|
| U(VI) (ppb) | DO (mg liter−1) | Nitrate (mg liter−1) | Sulfate (mg liter−1) | Sulfide (μg ml−1) | |||
| 826 | 2,848 | 0.41 | 148 | 13,396 | 0.0027 | 6.53 | 15.5 |
| 615 | 2,458 | 0.54 | 3,907 | 13,539 | 0.0027 | 6.84 | 10.9 |
| 608 | 2,089 | 0.31 | 2,688 | 11,715 | 0.0026 | 6.75 | 8.4 |
| 610 | 1,687 | 0.48 | 3,090 | 9,772 | 0.0014 | 7.06 | 9.6 |
| 624 | 1,205 | 0.18 | 189 | 8,673 | 0.0004 | 7.04 | 14.0 |
| 617 | 544 | 0.32 | 793 | 5,579 | 0.0024 | 7.00 | 11.5 |
| 828 | 343 | 3.05 | 168 | 3,294 | 0.0059 | 6.89 | 13.0 |
| 857 | 302 | 0.23 | <1 | 3,529 | 0.0088 | 7.08 | 13.3 |
| 853 | 227 | 0.24 | <1 | 2,021 | 0.0048 | 7.12 | 12.5 |
| 626 | 90 | 0.21 | 1 | 2,831 | 0.0027 | 7.31 | 9.4 |
| 648 | 0 | 0.00 | <1 | 2,000 | 0.0202 | 8.0 | 30.0 |
The highest uranium concentration (2,848 ppb) was in well 826, and the highest sulfate concentration (13,539 mg liter−1) was in well 615. Pearson r linear correlation analysis showed a significant correlation between the concentrations of uranium and sulfate (R = 0.98, P < 0.05).
PLFA profile of SRB.
PLFA biomarkers indicative of bacterial sulfate reducers have been identified in previous studies. The lipid marker Br17:1 (especially i17:1ω7) has been associated with Desulfovibrio (11, 46, 50, 51); 10me16:0 and 17:1 (especially 17:1ω6) were recognized as a major fatty acid component for Desulfobacter (10, 11) and Desulfobulbus (32), respectively. These biomarkers were determined for a small subset of isolates and may not be present in, or exclusive to, all members of the groups they are reported to represent. Within the genus Desulfotomaculum, fatty acid composition was only determined for strains of D. acetoxidans, D. orientis, D. ruminis, and D. nigrificans (19, 31). Unclassified components were predominant in all four of the species mentioned above, except for D. acetoxidans; other major fatty acids were i17:0, i15:0, 10me16:0, and i17:1ω7, etc. (19, 31).
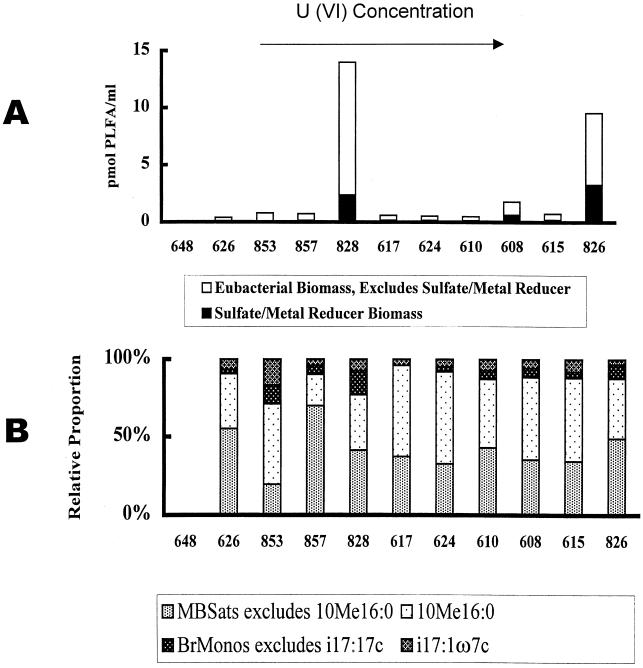
The highest biomass detected in groundwater samples was from well 828 (14.00 pmol ml−1), and the lowest was from well 648 (0.02 pmol ml−1) (Fig. 2A). In order to determine bacterial biomass, PLFA generally taken to be indicative of eukaryotes (normal saturates over 18 carbons in length, polynoics) and the trace quantities of PLFA of unknown structure were excluded. PLFA analysis showed that all samples contained biomarkers for SRB and metal-reducing bacteria (specifically 10me16:0). Of these, 10me16:0 comprised >10% of thePLFA of the SRB and metal-reducing bacteria. Well 853 has the highest proportion of i17:1ω7c PLFA (16.77%) compared with the other wells (3.53 to 8.00% of SRB and metal-reducing bacterial PLFA) (Fig. 2B).
FIG. 2.
PLFA profile of UMTRA Shiprock groundwater samples. (A) Community abundance as measured by total bacterial biomass level. ▪, SRB-metal reducer biomass measured as picomoles per milliliter of mid-chain branched saturates (MBSats) and branched monounsaturates (BrMonos). □, the remaining bacterial biomass measured as total PLFA, with normal saturates over 18 carbons in length, polynoics (eukaryote PLFA), SRB-metal reducer PLFA, and trace quantities of PLFA of unknown structure excluded. (B) Community composition as categorized via SRB-metal reducer PLFA structural group.
Based on PLFA analysis, these samples most likely contain complex SRB microbial communities with a wide range of biomass content. Community complexity was estimated from the number of different mid-chain branched saturates detected (in this case over 52), nearly half of them with unknown branch structures.
DNA extraction and PCR amplification of DSR genes, [NiFe] Hydrogenase genes.
Genomic DNA was extracted from a total of 13 samples with uranium concentrations varying from 0 to 2848 ppb. Genes encoding DSR were successfully detected from 11 sites. The amplicons were approximately the size generated in control amplifications of the dissimilatory sulfite reductase gene of D. vulgaris (1.9 kb). For [NiFe] hydrogenase gene amplification, only sample 853 produced a positive band of the expected size as the control organism Desulfovibrio desulfuricans (about 450 bp).
Clone library characterization.
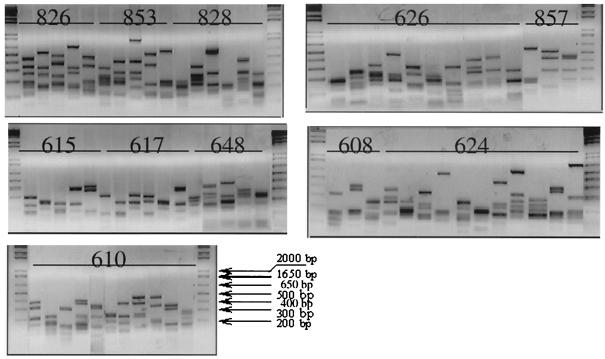
A clone library of DSR PCR products from each sample well was used to explore the diversity of DSR genes of the bacteria from this contaminated groundwater. A total of 305 clones were assembled into 70 clone families based on MspI restriction fragment banding patterns (Fig. 3). Some samples yielded as many as 11 different types of digestion patterns (e.g., well 610), while others were less diverse and produced 3 different restriction patterns (e.g., wells 857 and 608). Identical sequences were recognized between different samples mostly from floodplain wells 610, 615, 608, 624, 626, and 617.
FIG. 3.
Restriction digestion analysis of DSR clones with MspI. Cloned DSR gene fragments were amplified from the cloning vector by using primers directed at the T7 and M13 reverse RNA polymerase binding sites, producing a fragment with approximately 70 bp of vector sequence on each end. The vector sequence contained no MspI recognition sites. Products were digested with a twofold excess of MspI for 1 h at 37°C, analyzed by electrophoresis on a 2% agarose gel with TAE buffer, and visualized by ethidium bromide fluorescence.
Phylogenetic analysis of Shiprock DSR genes.
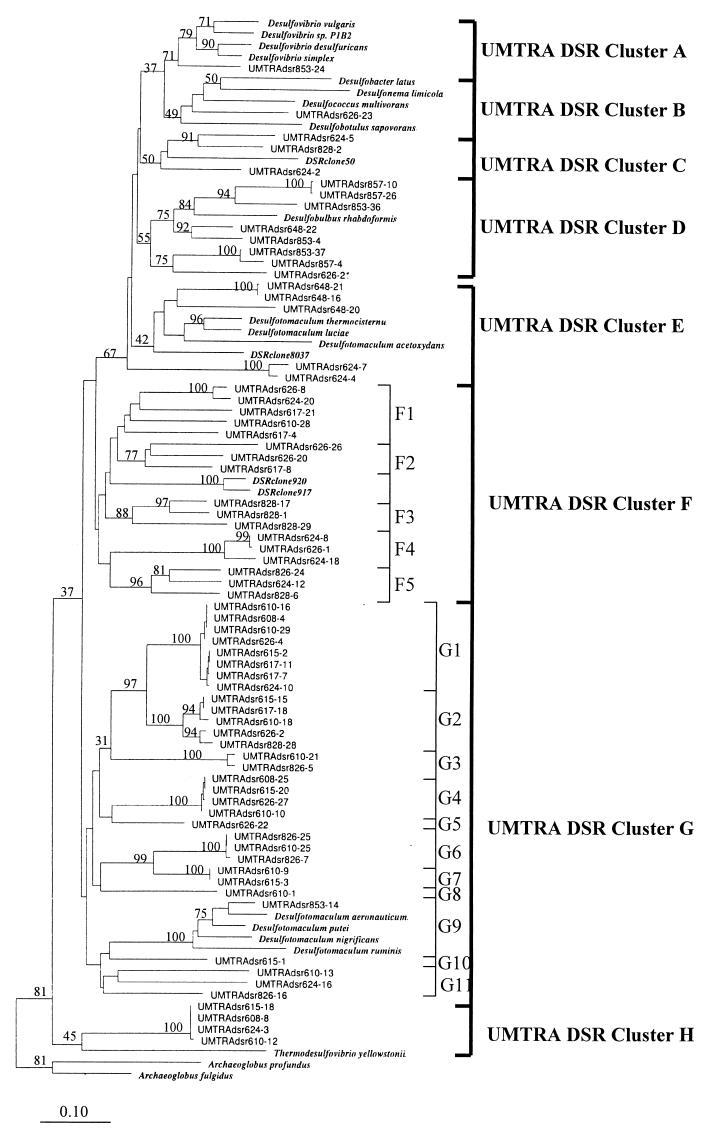
The α-subunits of recovered DSR gene fragments and of a variety of cultured Desulfotomaculum strains were sequenced. On average, 500 nucleotides were determined. For the groundwater clones, the sequence of representatives for each library and restriction pattern (total, 70 clones) was determined. Potential chimeric artifacts (one artifact) and non-DSR sequences (five sequences) were recognized in some of the clone library and were excluded from further analysis. In order to obtain an accurate description of the phylogenetic relationships of the SRB population in these groundwaters, we included in our analysis most environmental DSR clone sequences available from the database, as well as the newly characterized sequences of pure SRB reference cultures. Neighbor-joining analysis revealed the presence of eight well-resolved lineages of DSR sequences, designated clusters A to H (Fig. 4). Clusters F and G were further subdivided into 5 and 11 subclusters, respectively, most of which are well separated by similarity values between 70 and 85% (Fig. 4). Within subclusters, the similarity values were greater than 90%. The Desulfotomaculum pure culture sequences showed a division of the available organisms into two clusters, with group 1 containing D. thermocistern, D. luciae, and D. acetoxydans (in cluster E) and group 2 containing D. aeronauticum, D. putei, D. nigrificans, and D. ruminis (in cluster G-9).
FIG. 4.
Neighbor-joining analysis of DSR α-subunit fragments selected from clone libraries as frequently occurring MspI digestion patterns. Sequences prefixed “UMTRAdsr” were generated during this study. The prefix is followed by the sample well number, which is followed by the clone number. Clones were selected from libraries on the basis of MspI patterns to provide a preliminary survey of the most commonly recovered DSR gene sequences from each sample. Nucleotide sequence accession numbers are given in parentheses for the following organisms: Desulfobulbus rhabdoformis (AJ250473), Desulfovibrio desulfuricans (AJ289157), Desulfococcus multivorans (AJ277107), Desulfotomaculum thermocisternum (F074396), Archaeoglobus profundus (AF071499), Thermodesulfovibrio yellowstonii (U58122), Desulfobotulus sapovorans (U58120), Desulfovibrio sp. strain P1B2 (U58116), Desulfotomaculum ruminis (U58118), Desulfococcus multivorans (U58126), Desulfonema limicola (U58128), Desulfobacter latus (U58124), Desulfovibrio vulgaris (U16723), Archaeoglobus fulgidus (M95624), and Desulfovibrio simplex (U78738). Gene sequences from Desulfotomaculum luciae, D. acetoxidans, D. putei, D. aeronauticum, D. nigrificans, and Desulfovibrio desulfuricans were generated in this study under accession no. AY015493 to AY015499. The numbers on the tree refer to bootstrap values on 100 replicates; only values above 30 are given. Scale bar represents 10% estimated change.
Cluster A.
Cluster A contained all available DSR α-subunit sequences from cultured Desulfovibrio (δ-subclass of the class Proteobacteria) strains and one clone recovered from well 853, in which both uranium contamination and and the sulfate concentration were relatively low.
Cluster B.
The cluster B sequence also contained sequences derived from δ-subclass Proteobacteria, Desulfobacter latus, Desulfonema limicola, Desulfococcus multivorans, and Desulfobotulus sapovorans. A single clone recovered from well 626 grouped with these strains. This site was a low-uranium sample, in which sulfate was measured at 2,831 mg liter−1.
Cluster C.
Cluster C contains no cultured representatives. It is comprised of three unique sequences, from high-uranium to medium-uranium samples from wells 624 and 828. They are closely related to an environmental DSR clone (accession no. AF179329) (29) generated from a microbial mat at Solar Lake and are peripherally related to the genus Desulfobulbus.
Cluster D.
Cluster D contains the DSR α-subunit sequence of Desulfobulbus rhabdoformis, a δ-proteobacterium. Eight clones, all from low-uranium samples, fell into this group.
Cluster E.
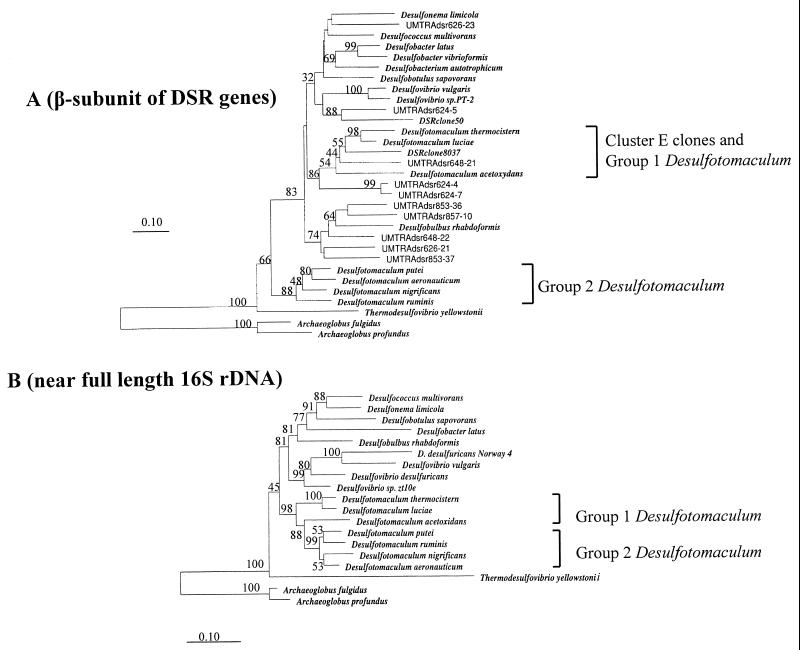
Cluster E contained the three cultured strains, D. thermocistern, D. luciae, and D. acetoxydans, referred to here as Desulfotomaculum group 1. Three clone sequences fell into this group; all had been recovered from well 648. Another two clones, generated from well 624 are loosely associated with this group. Although this group does not appear monophyletic, based on DSR α-subunit sequencing, phylogenetic analysis of the DSR β-subunits encoded in these clones showed that they branch together (Fig. 5A).
FIG. 5.
(A) Neighbor-joining analysis of some DSR β-subunit sequences from UMTRA Shiprock site clones and reference pure culture organisms. Sequences prefixed “UMTRAdsr” were generated during this study. Nucleotide sequence accession numbers are given in parentheses for the following organisms: Desulfobulbus rhabdoformis (AJ250473), Desulfococcus multivorans (U58127), Desulfotomaculum thermocisternum (AF074396), Archaeoglobus profundus (AF071499), Thermodesulfovibrio yellowstonii (U58123), Desulfobotulus sapovorans (U58121), Desulfovibrio sp. strain PT-2. (U58115), Desulfotomaculum ruminis (U58119), Desulfonema limicola (U58129), Desulfobacter latus (U58125), Desulfovibrio vulgaris (U16723), Archaeoglobus fulgidus (M95624), Desulfobacter vibrioformis (AJ250472), Desulfobacterium autotrophicum (Y15478), DSR clone 50 (AF179329), and DSR clone 8037 (AF179336). Gene sequences from Desulfotomaculum luciae, D. acetoxidans, D. putei, D. aeronauticum, and D. nigrificans were generated in this study under accession no. AY015577 to AY015581. (B) Neighbor-joining analysis of near full-length 16S rDNA from pure cultures. Nucleotide sequence accession numbers are given in parentheses for the following organisms: Desulfotomaculum ruminis (Y11572), D. putei strain SMCCW 459 (AF053929), D. aeronauticum (X98407), D. acetoxidans (Y11566), D. thermocisternum (U33455), D. nigrificans (X62176), D. luciae (AF069293), Desulfococcus multivorans ATCC 33890 (M34405), Desulfonema limicola (U45990), Desulfobacter latus (M34414), Desulfobotulus sapovorans (M34402), Desulfovibrio vulgaris (M34399), D. desulfuricans (AF098671), Archaeoglobus profundus. (AF272841), Desulfovibrio sp. strain zt10e (AF109469), Archaeoglobus fulgidus (Y00275), and D. desulfuricans sp. strain Norway 4 (M37312). The numbers on the trees refer to bootstrap values on 100 replicates; only values above 30 are given. The scale bar represents 10% estimated change.
Cluster F.
Cluster F contains no DSR α-subunit sequences from cultured organisms. This was the second most abundant group of clone sequences recovered (17 in total). Ten of them were recovered from high-uranium samples, 4 of them are from low-uranium well 626, and 3 are from well 828 (medium uranium). Although not closely related to any available pure culture sequences, this cluster showed close relationship with two DSR clones recovered from the Solar Lake microbial mat (DSR clone 917 and 920, accession no. AF179334 and AF179339) (29). Each subcluster contains three to five distinct sequences, except subcluster F4, which contained only closely related sequences.
Cluster G.
Cluster G includes the Desulfotomaculum strains designated as group 2. This is the largest single cluster of cloned sequences, all but one of which was associated basally with Desulfotomaculum group 2. Among 31 clones in this cluster, 25 originated from high-uranium samples. The remaining six clones were recovered from medium- to low-uranium wells, including the single sequence within Desulfotomaculum group 2 (well 853). The remaining four sequences were recovered from well 626. The phylogenetic depths of the individual subclusters are different: subclusters G1, G2, G3, G4, G6, and G7 contain clones sharing 95 to 100% sequence similarity, and subclusters G9 and G11 contain sequences that are more deeply diverged. Subclusters G5, G8, and G10 are represented by single clones.
Cluster H.
Cluster H is defined by four clone sequences, all generated from high-uranium samples, and shows a relationship to Thermodesulfovibrio yellowstonii, in the Nitrospira group.
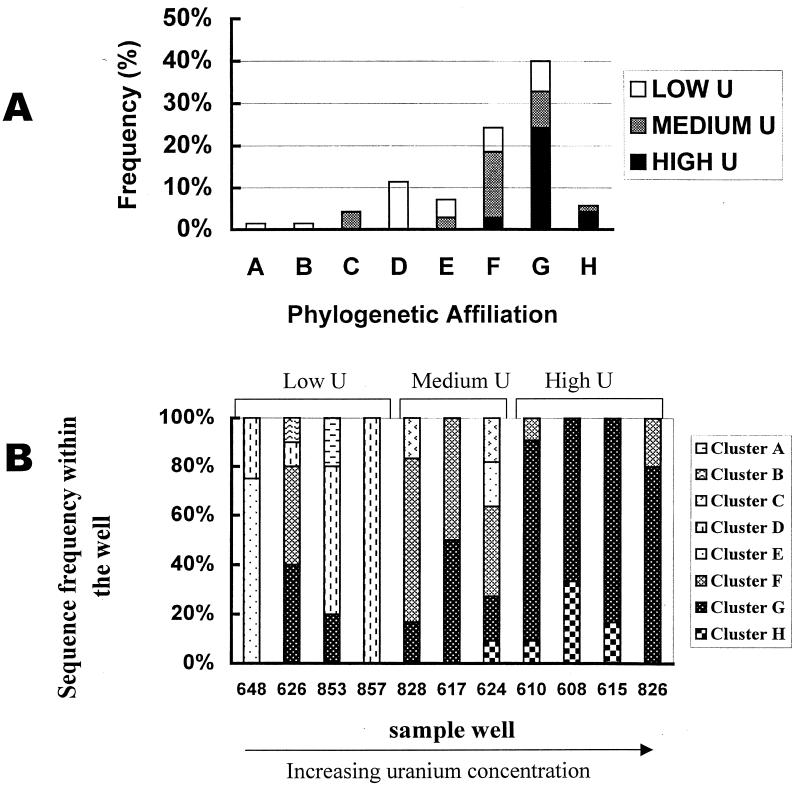
Population structure in relation to uranium concentration.
Of the sequenced clones, 40% grouped with lineage G, which includes the gram-positive, thermophilic Desulfotomaculum species D. nigrificans, D. putei, D. aeronauticum, and D. ruminis. Sequences recovered from high-uranium (>1,500 ppb) samples were dominated by this lineage, although cluster F and the less frequently recovered cluster H were also detected. DSR α-subunit sequences recovered from low-uranium (≤302 ppb) samples were more diverse, including representatives of clusters A, B, D, E, F, and G (Fig. 6A). Figure 6B shows the distribution of different DSR sequences detected among different sample wells. Cluster D, belonging to the δ-Proteobacteria, was recovered exclusively from low-uranium (≤302 ppb) samples. Together with sequences closely related to Desulfotomaculum group 1, they constitute the recovered SRB community of well 648, which had the lowest uranium concentration. Notably, sequences related to δ-Proteobacteria and Desulfotomaculum group 1 were not recovered from high-uranium samples. Lineage G dominated in all high-uranium samples. Cluster H, related to Thermodesulfovibrio yellowstonii, was recovered from samples 615, 608, 610, and 624, in which the uranium concentration is relatively high (1,205 to 2,458 ppb of uranium).
FIG. 6.
(A) Sequence distribution for 70 DSR clone sequences (α-subunit) obtained from high-uranium [samples 610, 608, 615, and 826; U(VI) from 2,847.5 to 1,687 ppb], medium-uranium (samples 624, 617, and 828; U(VI) from 1,205 to 342.7 ppb), and low-uranium samples (857, 853, 626, and 648; U(VI) from 301 to 0.001 ppb). (B) SRB population structure in different sample wells as measured by DSR sequences recovered and their affiliation with different cluster of SRB.
Logistic regression suggested a significant association between sequence cluster G and uranium concentration (slope = 0.00111, P = 0.0005; Table 2). With only 11 samples, many combinations of chemical concentrations were not observed, creating difficulty in separating the influences of the various chemical constituents. With that caution, however, important relationships between chemical concentrations were found in cluster F, but not for cluster G, although the overall model was significant (P = 0.0007, full model). DO had the largest effect, as measured by the slope. A model with only uranium showed that as uranium increased from 0 to 3,000 ppb, clade G frequencies were predicted to increase from 18.5% to 90% (Table 2).
TABLE 2.
Parameter estimates from two logistic regression models used to explain Cluster F and G frequencies in relation to selected geochemical measurements
| Parametera | Cluster F
|
Cluster G
|
||
|---|---|---|---|---|
| Estimate | P > Chi square | Estimate | P > Chi square | |
| Full model | ||||
| Intercept | −3.6181 | 0.009 | −0.6515 | 0.554 |
| Sulfate | 0.000969 | 0.038 | −0.00029 | 0.453 |
| DO | 0.8999 | 0.015 | −0.1827 | 0.647 |
| Uranium | −0.00416 | 0.047 | 0.00183 | 0.258 |
| Nitrate | −0.00066 | 0.088 | 0.000532 | 0.049 |
| Model with U(VI) as sole variable | ||||
| Intercept | −0.5934 | 0.1421 | −1.4794 | 0.0008 |
| Uranium | −0.00055 | 0.1086 | 0.00111 | 0.0005 |
P values of the full model are 0.0074 for cluster F and 0.0007 for cluster G. The P values of the model with U(VI) as the sole variable are 0.0902 for cluster F and 0.0001 for cluster G.
DISCUSSION
Genetic diversity and metabolic activity.
Until now, the abundance and diversity of SRB have been analyzed mostly through cultivation techniques and through hybridizations with rRNA-targeted oligonucleotide probes (8, 9, 15, 21, 26, 34–36). 16S ribosomal DNA (rDNA) sequences are currently popular as a useful criterion for the definition of taxa at several levels. However, 16S rRNA sequences by themselves provide no information about potential physiological differences between closely related bacteria. Here we present a field-scale study using an alternative PCR-based approach, targeting the dissimilatory sulfite reductase gene along with the [NiFe] hydrogenase gene. Although it is well established that PCR-based methods in microbial ecology have intrinsic biases (33, 43), these biases can be assumed to be equal for each of the groundwater samples described here. Furthermore, as a complement to this molecular diversity study, analysis of PLFA biomarkers for SRB groups provides quantitative confirmation of the presence of viable SRB.
Based on near full-length 16S rDNA sequence analysis, Desulfotomaculum spp. form a distinct cluster of related sequences (Fig. 5B). Comparison of partial α- and β-subunit DSR gene sequences revealed a greater genetic diversity, as expected, but suggested a similar grouping of Desulfotomaculum strains (Fig. 4 and 5A): i.e., a division of the Desulfotomaculum genus into at least two clusters, which we have designated as groups 1 and 2. This is consistent with recent findings about SRB taxonomy described by Stackebrandt et al. (40) and Hristova et al. (15).
MspI restriction enzyme digestion analysis revealed substantial diversity of the DSR gene sequence. A total of 70 DSR sequences (61 of them unique) were identified, and all were affiliated with the bacterial domain. Comparison of phylogenetic trees constructed with different portions of the DSR genes revealed, in general, consistent topologies for both α- and β-subunits of DSR, although slight variance was observed (Fig. 4 and 5A). Eight well-resolved lineages of DSR sequences are represented by the cloned sequences (A to H, Fig. 4). Some sequence differences within the subclusters (Fig. 4) involved only several base changes. It is entirely possible that this microheterogeneity may reflect PCR point errors. The finding of the same partial DSR sequences in different gene libraries suggests that most of the differences are real. The [NiFe] hydrogenase Desulfovibrio-specific PCR detected positive amplification only from well 853, and Desulfovibrio-like DSR fragments were indeed recovered from only this well. The PLFA profile of well 853 also showed highest proportion of i17:1ω7, a biomarker associated with Desulfovibrio spp. (11). The consistent occurrence of Desulfovibrio-like signals in well 853 may be significant, since it is a location at the Shiprock site, where active microbial reduction of U(VI) may be responsible for low uranium concentrations in groundwater (D. Elias, D. Wong, J. Senko, P. E. Long, J. P. McKinley, J. M. Suflita, and L. R. Krumholz, EOS, Trans. Am. Geophys. Union Fall Meet., vol. 81, no. 48, p. F214, 2000).
Since DNA recovered from environmental samples may be derived in part from dead or inactive cells, the recovery of DSR sequences alone does not provide direct evidence for an active sulfate-respiring population. However, a significant amount of lipids known to be markers for sulfate or metal reducers were found in 10 samples (all except background well 648) tested, which supported the presence of a viable SRB microbiota in this groundwater environment. An unusually high number of distinct mid-chain branched saturates (more than 50, constituting 7 to 25% of total bacteria biomass) suggested a diverse SRB community as well as a high relative abundance within the domain Bacteria.
Dominance of Desulfotomaculum and Desulfotomaculum-like sequences.
As dissimilatory SRB, the genus Desulfotomaculum and Desulfovibrio spp. were reported to exhibit a superior ability, over assimilatory organisms, to extract large amounts of metals from culture media (16). Tolerance and adaptation to heavy metals by Desulfovibrio and Desulfotomaculum strains of different origins have been investigated in enrichment cultures under a range of sulfate concentrations (4).This study revealed an overwhelming dominance of Desulfotomaculum and Desulfotomaculum-like sequences, particularly in those wells with high uranium (> 1,500 ppb) concentration. As many as 50 different DSR sequences associated with the genus Desulfotomaculum were recovered. The majority of them form subclusters representing sequences basal to the established Desulfotomaculum genus and are, as yet, to be characterized.
Previous work indicates that the abundance of Desulfotomaculum spp. in various environments is probably related to sulfate availability and exposure to adverse environmental conditions, such as regular exposure to oxygen (53). This study suggests that at the Shiprock site, uranium contamination is another factor influencing the Desulfotomaculum population. To test this hypothesis, we used logistic regression, a statistical technique that is widely used in medical research, but is rarely used in microbial ecology (3, 45). It is a variation of ordinary regression, useful when the observed outcome variable represents the occurrence or nonoccurrence of some outcome event (such as the occurrence or nonoccurrence of sequence cluster G or F). It produces a formula that predicts the probability of the occurrence as a function of the independent variables (such as uranium concentration, DO, and sulfate and nitrate concentration). Logistic regression predicted an increase in the frequency of the presence of cluster G from 18.5% to 90% as the uranium concentration increased from 0 to 3,000 ppb, clearly suggesting that this genus holds a selective advantage over other SRB populations at high U(VI) concentrations. The present work is the first report of Desulfotomaculum dominance among SRB in a mesophilic natural environment setting. Desulfotomaculum strains isolated from thermophilic environments have been reported more often than those from mesophilic environments; however, this frequency may be attributable to intensified research of extreme environments rather than to a preference of most Desulfotomaculum spp. for thermophilic conditions. While the correlation of Desulfotomaculum with uranium concentration is clear, we cannot entirely rule out the possibility of another factor related to uranium contamination. However, a full suite of groundwater geochemical parameters, including total organic carbon, were analyzed and included in a preliminary statistical analysis, and no parameters other than uranium and sulfate showed a strong correlation. Since sulfate is present in concentrations ranging from 2 to 3 orders of magnitude greater than that of uranium, sulfate clearly plays the dominant role in Desulfotomaculum metabolism. However, the more significant statistical linkage between Desulfotomaculum and uranium concentration suggests a competitive advantage for Desulfotomaculum conferred by the presence of uranium. This competitive advantage may result from uranium tolerance or from the ability of Desulfotomaculum to use U(VI) as a terminal electron acceptor or both.
Conclusions.
This study demonstrates a remarkable diversity of DSR sequences associated with bacteria from the δ-Proteobacteria, gram-positive, and Nitrospira divisions. Since strains with different functional genomic fingerprints also differ considerably in their physiological capabilities, this result suggests strongly that the high diversity detected at the Shiprock site is very likely of ecological significance. These data also showed that the genus Desulfotomaculum and Desulfotomaculum-like organisms dominated the SRB population of this uranium-contaminated environment. The overall level of SRB activity relative to those of other functional metabolic groups and the specific role that Desulfotomaculum may play in uranium reduction are not addressed by this study, nor is the role of sulfate. However, the strong association between DSR cluster G and uranium concentration indicates that Desulfotomaculum has remarkable tolerance and adaptation to high levels of uranium. In addition to confirming the results of this study at other sites, future research might well focus on the in situ activity level of Desulfotomaculum relative to uranium concentration and relative to those of other SRB and other functional groups. Desulfotomaculum apparently could play a role in both intrinsic and accelerated bioremediation of U(VI)-contaminated environments. For accelerated bioremediation of U(VI), it may be important to either suppress Desulfotomaculum to avoid production of toxic H2S and allow iron reducers to reduce U(VI) or to stimulate Desulfotomaculum to intentionally produce insoluble sulfide minerals such as FeS2 to stabilize U(IV) precipitates. Either case will require advances in understanding of SRB in uranium-contaminated environments.
ACKNOWLEDGMENTS
This work was supported by the U.S. Department of Energy, Office of Science, grant no. DE-FC02–96ER62278 to D. C. White as part of the Assessment Component of the Natural and Accelerated Bioremediation Research Program (NABIR), administered by Anna Palmisano. Support for sample collection and geochemistry was also provided by NABIR to the Pacific Northwest National Laboratory. D. C. White also received support from National Science Foundation grant DEB-9814813. The cooperation of the Navajo Tribal Nation and the U.S. Department of Energy, Uranium Mill Tailings Remedial Action (UMTRA) Program is gratefully acknowledged. Pacific Northwest National Laboratory is operated by Battelle Memorial Institute for the U.S. Department of Energy.
Don Metzler, Craig Goodknight, Stan Morrison, and Mark Kautsky of the UMTRA Program were particularly helpful in the successful conduct of fieldwork to obtain samples critical to this research. We are grateful to David Boone (Portland State University, Portland, Oreg.), who helped this project by kindly providing pure culture strains of SRB.
REFERENCES
- 1.Abdelouas A, Lutze W, Gong W, Nuttall E H, Strietelmeier B A, Travis B J. Biological reduction of uranium in groundwater and subsurface soil. Sci Total Environ. 2000;250:21–35. doi: 10.1016/s0048-9697(99)00549-5. [DOI] [PubMed] [Google Scholar]
- 2.Abdelouas A, Leutz W, Nuttall E H. Uranium contamination in the subsurface: characterization and remediation. In: Burns P C, Finch R, editors. Uranium: mineralogy, geochemistry, and the environment. Reviews in mineralogy. Vol. 38. Washington, D.C.: Mineralogical Society of America; 1999. pp. 433–473. [Google Scholar]
- 3.Agresti A. An introduction to categorical data analysis. New York, N.Y: John Wiley and Sons, Inc.; 1996. [Google Scholar]
- 4.Brecklinghaus J, Schwartz W, Naveke R. Geomicrobiological studies. XIV. Heavy metal tolerance of desulfurizing bacteria under various ecological conditions. Z Allg Mikrobiol. 1981;21:65–76. [PubMed] [Google Scholar]
- 5.Canfield D E, DeMarias D J. Aerobic sulfate reduction in microbial mats. Science. 1991;251:1471–1473. doi: 10.1126/science.11538266. [DOI] [PubMed] [Google Scholar]
- 6.Cypionka H. Oxygen respiration by Desulfovibrio species. Annu Rev Microbiol. 2000;54:827–848. doi: 10.1146/annurev.micro.54.1.827. [DOI] [PubMed] [Google Scholar]
- 7.Department of Energy. Final site observational work plan for the Shiprock, New Mexico, UMTRA Project site. GJO-2000–169-TAR, MAC-GWSHP 1:1. Rev 2. U.S. Grand Junction, Colo: Department of Energy; 2000. [Google Scholar]
- 8.Devereux R, Stahl D. Phylogeny of sulfate-reducing bacteria and a perspective for analyzing their natural communities. In: Odom J M, Singleton R Jr, editors. The sulfate-reducing bacteria: contemporary perspectives. Berlin, Germany: Springer; 1993. pp. 131–160. [Google Scholar]
- 9.Devereux R, Winfrey M R, Winfrey J, Stahl D A. Depth profile of sulfate-reducing bacterial ribosomal RNA and mercury methylation in an estuarine sediment. FEMS Microbiol Ecol. 1996;20:23–31. [Google Scholar]
- 10.Dowling N J E, Widdle F, White D C. Phospholipid ester-linked fatty acid biomarkers of acetate-oxidizing sulfate-reducers and other sulfide-forming bacteria. J Gen Microbiol. 1986;132:1815–1826. [Google Scholar]
- 11.Edlund A, Nichols P D, Roffey R, White D C. Extractable and lipopolysaccharide fatty acid and hydroxy fatty acid profiles from Desulfovibrio species. J Lipid Res. 1985;26:982–988. [PubMed] [Google Scholar]
- 12.Ehrlich H L. Geomicrobiology. New York, N.Y: Marcel Dekker; 1996. [Google Scholar]
- 13.Fründ C, Cohen Y. Diurnal cycles of sulfate reduction under oxic conditions in cyanobacterial mats. Appl Environ Microbiol. 1992;58:70–77. doi: 10.1128/aem.58.1.70-77.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guckert J B, Antworth C P, Nichols P D, White D C. Phospholipid ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol Ecol. 1985;31:147–158. [Google Scholar]
- 15.Hristova K R, Mau M, Zheng D, Aminov R I, Mackie R I, Gaskins H R, Raskin L. Desulfotomaculum genus- and subgenus-specific 16S rRNA hybridization probes for environmental studies. Environ Microbiol. 2000;2:143–159. doi: 10.1046/j.1462-2920.2000.00085.x. [DOI] [PubMed] [Google Scholar]
- 16.Jones H E, Trudinger P A, Chambers L A, Pyliotis N A. Metal accumulation by bacteria with particular reference to dissimilatory sulphate-reducing bacteria. Z Allg Mikrobiol. 1976;16:425–435. doi: 10.1002/jobm.3630160603. [DOI] [PubMed] [Google Scholar]
- 17.Karkhoff-Schweizer R R, Huber D P W, Voordouw G. Conservation of the genes for dissimilatory sulfite reductase from Desulfovibrio vulgaris and Archaeoglobus fulgidus allows their detection by PCR. Appl Environ Microbiol. 1995;61:290–296. doi: 10.1128/aem.61.1.290-296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kates M. Techniques in lipidology: isolation, analysis and identification of lipids. 2nd ed. Amsterdam, The Netherlands: Elsevier Press; 1986. [Google Scholar]
- 19.Kohring L L, Ringelberg D B, Devereux R, Stahl D A, Mittelman M W, White D C. Comparison of phylogenetic relationships based on phospholipid fatty acid profiles and ribosomal RNA sequence similarities among dissimilatory sulfate-reducing bacteria. FEMS Microbiol Lett. 1994;119:303–308. doi: 10.1111/j.1574-6968.1994.tb06905.x. [DOI] [PubMed] [Google Scholar]
- 20.LeDuc L G, Feroni G D, Trevors J T. Resistance to heavy metals in different strains of Thiobacillus ferooxidans. World J Microbiol Biotechnol. 1997;13:453–455. [Google Scholar]
- 21.Llobet-Brossa E, Rosello-Mora R, Amann R. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovley D R, Phillips E J P. Reduction of uranium by Desulfovibrio desulfuricans. Appl Environ Microbiol. 1992;58:850–856. doi: 10.1128/aem.58.3.850-856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovley D R. Dissimilatory metal reduction. Annu Rev Microbiol. 1993;47:263–290. doi: 10.1146/annurev.mi.47.100193.001403. [DOI] [PubMed] [Google Scholar]
- 24.Lovley D R, Roden E E, Phillips E J P, Woodward J C. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Mar Geol. 1993;113:41–53. [Google Scholar]
- 25.Lovley D R, Widman P K, Woodward J C, Phillips E J P. Reduction of uranium by cytochrome c3 of Desulfovibrio vulgaris. Appl Environ Microbiol. 1993;59:3572–3576. doi: 10.1128/aem.59.11.3572-3576.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manz W, Eisenbrecher M, Neu T R, Szewzyk U. Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol. 1998;25:43–61. [Google Scholar]
- 27.McKinley J P, Zachara J M, Smith S C, Turner G D. The influence of uranyl hydrolysis and multiple site-binding reactions on adsorption of U(VI) to montmorillonite. Clays Clay Minerals. 1995;43:586–598. [Google Scholar]
- 28.McKinley J P, Stevens T O, Fredrickson J K, Zachara J M, Colwell F S, Wagnon K B, Smith S C, Rawson S A, Bjornstad B N. Biogeochemistry of anaerobic lacustrine and paleosol sediments within an aerobic unconfined aquifer. Geomicrobiol J. 1997;14:23–39. [Google Scholar]
- 29.Minz D, Flax J L, Green S J, Muyzer G, Cohen Y, Wagner M, Rittmann B E, Stahl D A. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl Environ Microbiol. 1999;65:4666–4671. doi: 10.1128/aem.65.10.4666-4671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odom J M. Industrial and environmental activities of sulfate-reducing bacteria. In: Odom J M, Singelton R Jr, editors. The sulfate-reducing bacteria: contemporary perspectives. New York, N.Y: Springer-Verlag, Inc.; 1993. pp. 189–210. [Google Scholar]
- 31.O'Leary W M, Wilkinson S G . : Microbial lipids vol.1, edited by C. Ratledge and S.G. Wilkinson, Academic Press, Harcourt Brace Jovanovich, Publishers, p172–173. 1988. [Google Scholar]
- 32.Parkes R J, Calder A G. The cellular fatty acids of three strains of Desulfobulbus, a propionate-utilizing sulfate-reducing bacterium. FEMS Microbiol Ecol. 1985;31:361–363. [Google Scholar]
- 33.Polz M F, Cavanaugh C M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rabus R, Fukui M, Wilkes H, Widdle F. Degradative capacities and 16S rRNA-targeted whole-cell hybridization of sulfate-reducing bacteria in an anaerobic enrichment culture utilizing alkylbenzenes from crude oil. Appl Environ Microbiol. 1996;62:3605–3613. doi: 10.1128/aem.62.10.3605-3613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramsing N B, Kühl M, Jørgensen B B. Distribution of sulfate-reducing bacteria, O2 and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrode. Appl Environ Microbiol. 1993;59:3840–3849. doi: 10.1128/aem.59.11.3840-3849.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramsing N B, Fossing H, Ferdelman T, Anderson F, Thamdrup B. Distribution of bacterial populations in a stratified fjord (Mariager Fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol. 1996;62:1391–1404. doi: 10.1128/aem.62.4.1391-1404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ringelberg D B, Townsend G T, DeWeerd K A, Sulita J M, White D C. Detection of the anaerobic dechlorinating microorganism Desulfomonile tiedjei in environmental matrices by its signature lipopolysaccharide branch-long-chain hydroxy fatty acids. FEMS Microbiol Ecol. 1994;14:9–18. [Google Scholar]
- 38.Ringelberg D B, Davis J D, Smith G A, Pfiffner S M, Nichols P D, Nickels J S, Henson J M, Wilson J T, Yates M, Kampbell D H, Read H W, Stocksdale T T, White D C. Validation of signature polar lipid fatty acid biomarkers for alkane-utilizing bacteria in soils and subsurface aquifer materials. FEMS Microbiol Ecol. 1989;62:39–50. [Google Scholar]
- 39.Singleton R., Jr . The sulfate-reducing bacteria: an overview. In: Odom JM, Singleton R Jr, editors. The sulfate-reducing bacteria: contemporary perspectives. New York, N.Y: Springer Verlag, Inc.; 1993. pp. 1–20. [Google Scholar]
- 40.Stackebrandt E, Sproer C, Rainey F A, Burghardt J, Pauker O, Hippe H. Phylogenetic analysis of the genus Desulfotomaculum: evidence for the misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. Int J Syst Bacteriol. 1997;47:1134–1139. doi: 10.1099/00207713-47-4-1134. [DOI] [PubMed] [Google Scholar]
- 41.Stephen J R, Chang Y J, Macnaughton S J, Kowalchuk G A, Leung K T, Flemming C A, White D C. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl Environ Microbiol. 1999;65:95–101. doi: 10.1128/aem.65.1.95-101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strunk O, Ludwig W. ARB: a software environment for sequence data. Munich, Germany: Technische Universität München; 1996. [Google Scholar]
- 43.Suzuki M, Rappé M S, Giovannoni S J. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tebo B M, Obraztsova A Y. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe (III) as electron acceptors. FEMS Microbiol Lett. 1998;162:193–198. [Google Scholar]
- 45.Trexler J C, Travis J. Nontraditional regression analysis. Ecology. 1993;74:1629–1637. [Google Scholar]
- 46.Vainshtein M, Hippe H, Kroppenstedt R M. Cellular fatty acid composition of Desulfovibrio species and its use in classification of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:554–566. [Google Scholar]
- 47.Voordouw G. Hydrogenase genes in Desulfovibrio. In: Belaich J P, Bruschi M, Garcia J L, editors. Microbiology and biochemistry of strict anaerobes involved in interspecies hydrogen transfer. New York, N.Y: Plenum; 1990. pp. 37–51. [Google Scholar]
- 48.Voordouw G, Niviere V, Ferris F G, Fedorak P M, Westlake D W S. Distribution of hydrogenase genes in Desulfovibrio spp. and their use in identification of species from the oil field environment. Appl Environ Microbiol. 1990;56:3748–3754. doi: 10.1128/aem.56.12.3748-3754.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner M, Roger A J, Flax J L, Brusseau G A, Stahl D A. Phylogeny of dissimilatory sulfite reductase supports an early origin of sulfate respiration. J Bacteriol. 1998;180:2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wawer C, Jetten M S M, Muyzer G. Genetic diversity and expression of the [NiFe] hydrogenase large-subunit gene of Desulfovibrio spp. in environmental samples. Appl Environ Microbiol. 1995;63:4360–4369. doi: 10.1128/aem.63.11.4360-4369.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White D C, Leung K, Macnaughton S J, Flemming C, Wimpee M, Davis G. Lipid/DNA biomarker analysis for assessment of in situ bioremediation effectiveness. In: Alleman B C, Leeson A, editors. In situ and on-site bioremediation. Vol. 5. Columbus, Ohio: Battelle Press; 1997. pp. 319–324. [Google Scholar]
- 52.White D C, Davis W M, Nickels J S, King J D, Bobbie R J. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia. 1979;40:51–62. doi: 10.1007/BF00388810. [DOI] [PubMed] [Google Scholar]
- 53.Widdle F. Microbiology and ecology of sulfate- and sulfur-reducing bacteria. In: Zebnder A J B, editor. Biology of anaerobic microorganisms. New York, N.Y: John Wiley and Sons, Inc.; 1988. pp. 469–585. [Google Scholar]