Abstract
Background:
Dietary sodium intake affects blood pressure (BP) and vascular function. Emerging studies in animal models suggest epigenetic changes such as DNA methylation might be involved. We hypothesized that dietary sodium restriction induces DNA methylation changes in T-cells and arterioles in humans.
Methods:
Fifty subjects (46% African American, 49% women) were placed on a 1200mg sodium/day diet for 2 weeks. All food was prepared and provided in accordance with subjects’ dietary preferences. BP and flow-mediated dilation (FMD) were evaluated. Methylation sequencing (pre- and post- low-sodium diet) was performed on DNA obtained from T-cells (n=50) and biopsied arterioles (n=10) using reduced representation bisulfite sequencing. RNA-seq was also performed on arterioles (n=11).
Results:
During the low-sodium diet period, subjects consumed an average of 946mg of sodium/day. The average systolic and diastolic BP reductions following the low-sodium intake were −8±13/−4±9 mmHg (p<0.001 for both). Brachial artery FMD improved (5.8±2.9 to 6.8±3.4%, p=0.03). T-cell DNA was broadly and substantially more methylated than arteriolar DNA. The differentially methylated regions (DMRs; false discovery rate <0.05) identified in T-cells and arterioles in response to the sodium restriction were located in intragenic or transcription start site regions of 117 and 71 genes, respectively. Four of these genes were identified in both T-cells and arterioles (p=0.009 for the overlap). The dietary effects on methylation in several DNA regions were associated with dietary effects on BP. Several differentially expressed genes in arterioles contained DMRs at the significance level of p<0.05. In addition, 46 genes contained DMRs in both human and SS/Mcw rat T-cells (p=0.03 for the overlap).
Conclusions:
Dietary sodium restriction has significant effects on DNA methylation in T-cells and arterioles, some of which are associated with BP. Methylation patterns and sodium effects on methylation are largely tissue-specific but also have overlaps between tissues and species. These findings may lead to better understanding of dietary sodium’s interactions with cellular processes and therefore novel therapeutic targets.
Keywords: salt intake, blood pressure, DNA methylation, gene expression, epigenetics
INTRODUCTION
Americans consume approximately 3400 mg of sodium (Na) per day, which is significantly above the USDA Dietary Guidelines recommendation of no more than 2,300 mg/day for most adults (DietaryGuidelines.gov, USDA). This recommendation has been endorsed by numerous professional societies, including the American Heart Association1. Hypertension, affecting over one billion people worldwide, is a major cardiovascular disease (CVD) risk factor and one of the most well-studied pathophysiologic phenomena in association with dietary sodium intake2-5. About 30-50% of individuals with hypertension are considered salt-sensitive based on blood pressure (BP) lowering on a low-sodium diet6. Another less well-known risk factor for CVD that is affected by dietary sodium is vascular endothelial function7-12. Several studies have shown that endothelial dysfunction is predictive of future cardiovascular events13, 14.
One of the proposed mechanisms by which dietary sodium may influence physiology such as BP and endothelial function is through epigenetic modifications of the DNA15. Epigenetic modifications are alterations to the genomic DNA without changes to its sequence; these modifications alter the expression of RNA encoded by genes16. Methylation marks are one of the many naturally occurring epigenetic modifications of genomic DNA. Animal studies have shown that factors such as dietary sodium changes induce methylation changes in genes that play a role in BP regulation 17, 18. In addition, animal studies have also shown that methylation pattern changes are associated with changes in expression of several genes implicated in BP regulation and/or hypertension development16. As an example, there were marked differences in epigenetic profiles of T cells isolated from blood and kidneys of Dahl salt sensitive rats on different diets. In response to high salt in the diet, the methylation marks in T cells isolated from the kidney of Dahl SS rates showed a significant increase in methylation levels among the differentially methylated regions 17. Using transcriptome and methylation analyses, a negative correlation was observed between gene expression and DNA methylation in T cells isolated from the same animals. It was also shown that de novo DNA methylation and demethylation can contribute the development of high blood pressure in response to sodium intake19.
While these animal studies make a robust case for implicating methylation changes as a mechanism for salt sensitive hypertension, few studies have been reported in humans. In the current study, we evaluated changes in DNA methylation patterns in T-lymphocytes (T-cells) and blood vessels and gene expression changes in blood vessels in subjects on a low sodium diet and their associations with BP levels and vascular function. We analyzed T-cells and arterioles because they have established physiological roles in the effect of sodium intake on BP and they are technically feasible, albeit still challenging, to obtain from human research participants. In addition, we compared the findings in humans to previously published data from salt-sensitive rats.
METHODS
Study participants
Post-menopausal women (50-65 years) and men (30-65 years) with baseline BP ≥ 130/80 mm Hg or history of hypertension being treated with medications were recruited by physician referrals and/or word of mouth. Subjects with known diabetes mellitus (glycosylated hemoglobin ≥6.5% or being treated with anti-diabetes medications), cardiovascular disease (including ischemic heart disease, congestive heart failure, peripheral vascular disease, cardiac arrhythmias, and stroke), chronic kidney disease (Glomerular Filtration Rate (GFR) ≤ 60 mL/min), gross proteinuria, and acute/chronic immunologic/hematologic conditions were excluded. Study visits were rescheduled in case of minor ailments such as upper respiratory tract infections or fever due to utilization of T-cells to assess methylation changes. Pre-menopausal women were excluded to avoid the effect of hormonal changes, during the menstrual cycle, on methylation marks. In addition, those with baseline (Visit 1) BPs ≥ 180/120 mm Hg were excluded for safety reasons. The protocol was approved by the Froedtert and Medical College of Wisconsin Institutional Review Board (IRB PRO00024645) and all subjects provided informed consent.
Study protocol
All volunteers were invited to the Translational Research Unit (TRU) for consenting and screening (Figure 1). During the Screening Visit (Visit 0), demographics, health history, anthropometrics (height, weight, and waist circumference), BP measurements, a blood sample (for basic metabolic panel and glycosylated hemoglobin), and a urine sample (for protein and electrolytes) were obtained. If a subject was on anti-hypertensive medications, he/she was evaluated by a physician to determine if antihypertensives could be held for three weeks. All subjects were off all BP medications one week prior to the start of the Study Visit 1 and during two-week duration of the study. Following consent and qualification for the study, subjects met with the TRU bionutritionist for an interview regarding food choices since all subjects were placed on a 2-week custom 1200 mg/day low-sodium diet. Subjects were also asked to keep a food log during the screening period (3-7 days) to assist in preparation of low sodium foods that subjects would consume. In addition, two questionnaires were administered to assess daily caloric, macronutrient, and sodium intake.
Figure 1: Synopsis of the study design.
Subjects were placed on 2 weeks of customized low-sodium diet; Blood pressure and vascular endothelial function studies were conducted. T cells were obtained to conduct DNA methylation analyses. Arterioles were obtained from a subset of the subjects for DNA methylation and gene expression analyses.
Subjects were given salt tablets (6000 mg of salt/day, given as 2000 mg salt [800 mg of Na] three times/day) for 3-days prior to start of a low sodium diet (Visit 1) in addition to their daily dietary sodium intake (Figure 1). Salt tablets were given to ensure that all subjects have a minimum and similar sodium intake prior to the start of the study due to significant variations in baseline intake (1372 −8624 mg/day). On the 3rd day of salt tablet use, subjects were asked to collect a 24-hour urine sample for electrolytes and creatinine and return it on the day of Study Visit 1. Urine creatinine was measured to assess completeness of the 24-hour urine collection. Subsequently, subjects were asked to consume a customized low sodium diet starting on the day of Study Visit 1.
All subjects were asked to come in fasting for 8-10 hour before noon for study visits and remain fasting until the conclusion of the Study Visit. During Study Visits 1 and 2 (Figure 1), repeat anthropometric and BP measurements were obtained. Blood was collected for methylation analyses before the start of low-sodium diet (see below) and urine was collected for spot urine studies. In addition, flow mediated dilation studies of brachial artery were performed. All of the above measurements and studies were repeated at the end of 2-weeks on the low-sodium diet. Subjects were asked to keep a food log during these 2 weeks to ensure the calorie count and macronutrient composition were not significantly altered and that they were compliant with the low-sodium diet. Subjects were instructed not to change their exercise or activity levels and not undertake any weight loss activities during the 2-week period. They were also asked not to use any new medications (prescribed or over the counter) during that period. Subjects were asked to check their BPs every day around the same time and keep a log of the values and inform the study team if they went above a threshold (≥160/110 mm Hg). BP cuffs and instructions were provided to all subjects while they were off BP medications. None of the subject’s BPs went above the threshold; and therefore, none of the studies had to be aborted.
Measurements
Anthropometrics
Anthropometric measures including weight, height, and waist circumference were measured using a calibrated scale (scale-tronix 5200, Welch Allen, Skaneateles Falls, NY, USA), a fixed wall-scale stadiometer (Harpenden stadiometer 602VR, Holtain, Wales, UK), and a tape measure (Gulick II, Country Technology, Inc., Gays Mills, WI), respectively. All participants were measured in scrubs and without shoes. Waist circumference was measured at the umbilicus. The average of three measures were used for each variable. Measurements were performed by the same bionutritionist for reproducibility, following the Center for Disease Control and Prevention’s National Health and Nutrition Examination Survey (NHANES) Anthropometry Procedures Manual (CDC Manual)20.
BP measurements
Standardized measurements of BP were obtained using an automated cuff according to the CDC Manual during the screening and study visits with subjects seated quietly, with arms and back supported, for at least 5 minutes with both feet flat on the ground20. Three measurements were obtained by a trained operator, 1 minute apart and averaged. BP logs kept by the subject after home measurements were used for safety when the participants were off of BP medications.
Diet plan and assessments
Low sodium menus were based on participant interviews for usual macronutrient intake, food intolerances, preferences, and meal patterns. Usual dietary intakes were assessed using a three-day food logs, two 24-hour dietary recalls and the Block Sodium Screener (BSS). BSS is a 25-item Block Food Frequency Questionnaire and takes 5-7 minutes to complete. Portion size is asked for 7 items. Salt added at the table is also included. A scoring key allows for scoring and determination of milligrams of sodium intake per day.21 Food logs and recalls were analyzed using Nutrition Data System for Research (NDSR)22-26. NDSR is a Windows-based dietary analysis program designed for the collection and analyses of 24-hour dietary recalls, food records, menus, and recipes. Calculation of nutrients occurs immediately providing data per ingredient, food, meal, and day in report and analysis file formats. The software includes a dietary supplement assessment module so that nutrient intake from both food and supplemental sources may be captured and quantified. We used both BSS and NDSR data to assess daily sodium intake during pre-study visit. Sodium intake during the study was assessed using daily food logs kept by the subject and entered into NDSR. Macronutrient and sodium totals for daily menus were calculated using NDSR. Meals were prepared in a metabolic kitchen on the premises of TRU. Food was either picked up or delivered to the participant every 48-72 hours in compliance with Wisconsin state laws for food preparation and delivery.
Dietary compliance was measured through self-report (food diaries, in-person interviews, and a survey). In addition, compliance with low sodium diet was objectively assessed by (1) measurement of 24 – hour urine sodium levels before and after low-sodium diet, (2) measurement of spot urine sodium (at screening visit and visits 1 and 2), and (3) BSS and NDSR data. While 24-hour urine measurement is the gold-standard, additional methods including BSS allowed us to be vigilant during the 2-week period. Results from the surveys and measurements were consistent with each other within each subject.
Biochemical Measurements
Basic metabolic panel was determined by a local clinical lab using the methodologies and reference intervals as noted - Glucose (Hexokinase, 65 - 99 mg/dL (fasting specimen)), Sodium (Ion Selective Electrode, Adults: 136 - 145 mmol/L), Potassium (Ion Selective Electrode, Adults: 3.4 - 5.1 mmol/L), Blood Urea Nitrogen (Kinetic test with urease and glutamate dehydrogenase, Adults: 6-23 mg/dL), and Creatinine (Modified Jaffe, Female: 0.5 −1.1 mg/dL, Male: 0.7 - 1.3 mg/dL). Glycohemoglobin was determined from whole blood specimen via a monoclonal antibody agglutination reaction using Siemens DCA Vantage HbA1c Analyzer® (CLIA Waived) and self-contained Immunoassay Cartridges. Determination of electrolyte levels in urine specimens were made by a local clinical lab using Roche Diagnostics cobas 8000 modular analyzer series and Ion-Selective Electrode (ISE) system module. The cobas ISE module measures the concentration of sodium (Na+), potassium (K+) and chloride (Cl−).
Vascular function assessments
Flow-mediated dilation (FMD) in conduit artery (brachial artery) was evaluated before and after the diet period to assess in vivo vascular function as described previously27 . The brachial artery was imaged using a high-resolution ultrasound at baseline and after a 5-minute occlusion of flow in the artery with a blood pressure cuff inflated to supra-systolic BP. Subsequent release of cuff pressure leads to a period of transient high blood flow, which serves as a stimulus for nitric oxide production by the endothelium. The brachial artery dilates as a result of increased flow within 1 minute of cuff release and its maximum diameter was recorded by ultrasound. All studies were performed in a 24°C temperature-controlled room before noon in a fasting state to limit the effects of temperature, meal, and diurnal variation on vascular function.
T-cell isolation
T-cells were isolated from peripheral blood with the process of separation starting within 30 minutes of blood collection. Peripheral blood mononuclear cells (PBMCs) were initially separated by centrifugation on Histopaque-1083. T-cells were specifically isolated from PBMCs utilizing CD3 human Microbeads (#130-050-101) and MACS Columns for magnetic cell isolation (Miltenyi Biotech).
T-cell genomic DNA library preparation for methylation sequencing
Genomic DNA was extracted from T-cells by Pure Link Genomic DNA kits (cat K1820, Invitrogen) following manufacturer’s protocol. DNA methylation profiles were analyzed at single-base resolution with reduced representation bisulfite sequencing (RRBS)28, with modifications to allow multiplexing18. The libraries underwent cluster generation using TruSeq PE Cluster Kit v4-cBot-HS and 100 cycles of paired-end sequencing using TruSeq SBS Kit v4-HS (Illumina). Fifteen libraries were sequenced in each lane with a control sample library prepared at the same time. The sequencing was done with a 350-Gb flowcell on an Illumina HiSeq 2500 sequencer.
Arteriole biopsy
Gluteal adipose tissue was used to isolate arterioles as we described27, 29. Adipose tissue was obtained by a 1-inch incision on the upper outer quadrant of the buttocks. Approximately 2-3 vessels (100-200 μm diameter) were separated from this adipose tissue for molecular studies.
Arteriolar DNA and RNA library preparation for methylation and RNA sequencing
RNA (n=11) and genomic DNA (n=10) were extracted from frozen arterioles using All Prep DNA/RNA/miRNA Universal Kit (Cat NO. 80224, Qiagen) following manufacturer’s protocol. Isolated gDNA were used for RRBS DNA library prep following the same protocol as DNA from above T-cells with modifications to accommodate the low amount of DNA input. Isolated RNA was used for RNAseq library preparation using the TruSeq Stranded mRNA Sample Preparation kit (A cat # FC-122-2101, B cat #FC-122-2102, Illumina) following Illumina TruSeq Stranded mRNA (LT) protocol. Both libraries prepared from frozen arterioles were sequenced with 150 bp paired end sequencing on HiSeq X Ten at BGI American Corporation (Cambridge, MA). Arteriolar RNA-seq data were obtained from eleven subjects and arteriolar RRBS data from 10 subjects, and 7 subjects had both arteriolar RNA-seq and RRBS data.
DNA methylation data analysis
RRBS data were analyzed as described previously18, 19, 30, 31. Raw sequences were processed, trimmed, and mapped to the human reference genome (hg19) using Bismark (v0.16.1) 18, 30, 31. Overall bisulfite conversion rates were estimated from the number of unconverted cytosines at Klenow filled-in 3’ MspI sites of sequencing reads that read through these sites. For each CpG site, the methylation rate was calculated as the percentage of unconverted cytosines in each library. Libraries were merged based on CpG coordinates. A modified software based on metilene (Version 0.23) and Wilcoxon Signed Rank Test, were applied to identify methylation regions (MRs) de novo in pre- and post-diet samples. Methylation rate of each region was calculated as the average methylation rate of each site across the region. Transcription start site (TSS) regions were defined as 1,000 bp upstream and downstream of a transcription start site.
RNA-Seq data analysis
An in-house pipeline was used to analyze RNA-seq data as previously described with modifications18. It included reads mapping and alignment using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml) and Tophat v2 (http://tophat.cbcb.umd.edu/)) as well as transcript construction using StringTie32. Transcript abundance was quantified as gene count data using Ballgown33. R package “DESeq2” with generalized linear mixed model was applied to identify the differentially expressed genes between pre- and post-diet samples34.
T-cell DNA methylation data from salt-sensitive rats
We previously published studies analyzing DNA methylation in circulating T-cells isolated from Dahl salt-sensitive (SS) rats fed a purified diet (AIN-76A) and 3 weeks of high sodium diet17, 35. Two lines of SS rats, SS/Mcw and SS/Crl, fed low sodium diet (0.4% NaCl AIN-76A Dyets, Inc for SS/Mcw rat and 0.75% NaCl Teklad 5L2F for SS/Crl rats) from weaning until 7 weeks of age were studied. Then rats were switched to the high sodium diet (4.0% NaCl AIN-76A Dyets, Inc) for 3 weeks. SS/Mcw has a greater level of BP salt-sensitivity than SS/Crl36. Metilene (Version 0.23) was applied to identify differentially methylated regions between high and low sodium diet in each line of SS rats.
Statistical analysis
RRBS and RNA-seq were analyzed as described above. Phenotypic and other data before and after the low sodium diet were compared using a paired t-test for normally distributed data and Wilcoxon Signed Rank test for data that are not normally distributed. Spearman correlation analysis was applied to examine if the changes in methylation were associated with changes in BP or other phenotypes. Permutation test by comparison with 100,000 random selections was performed to investigate whether any overlaps were greater than chance.
Benjamin-Hochberg procedure was used to control false discovery rates (FDR). FDR<0.05 was considered significant. All statistical analyses were performed using the R 3.5.1 statistical package.
RESULTS
Baseline characteristics
A total of 151 subjects were screened over telephone, 71 subjects signed consent, and 50 completed all study procedures. All subjects (25 [49%] women) were recruited in Milwaukee, Wisconsin between 2015 and 2019. Twenty-three (46%) subjects self-identified as African American, one subject (2%) was Hispanic, and the remaining 27 subjects (52%) were Caucasian. Anthropometric features are shown in Table 1. Their mean baseline systolic and diastolic BPs were 144±15 mm Hg (range: 116-178) and 85±12 mm Hg (range: 60-111). These BPs were obtained after the subjects were off BP medications for 1 week. Our inclusion criteria included baseline BP ≥ 130/80 mm Hg or history of hypertension being treated with medications. A few subjects (n=8), for inexplicable reasons, either had lower BPs or unchanged BPs when they came in for the Study Visit 1 (after being off of BP medications for 1 week). At that point, we continued the subjects in the study even though some had a BP that was below our threshold.
Table 1.
Demographic, phenotypic, and dietary information of the 50 study subjects.
| Measurement | Screening | Pre-diet | Post-diet | p-value# |
|---|---|---|---|---|
| Age (years) | 51±9 | |||
| BMI (kg/m2) | 31.8±6.7 | |||
| Waist circumference (cm) | 106±16 | |||
| Waist-to-hip ratio | 1.07±0.09 | |||
| Systolic blood pressure (mmHg) | 144±15* | 136±13 | <0.0001 | |
| Diastolic blood pressure (mm Hg) | 85±12* | 81 ±12 | 0.0007 | |
| Na intake (mmol/day) | 166±77 | Screen + salt tablets | 41±6 | 4.77E-16 |
| 24 h urine Na (mmol/day) | 211±90 | 51±53 | 1.59E-14 | |
| Spot urine Na (mmol/L) | 126±67 | 139±65 | 45±41 | 4.35E-13 |
| K intake (mmol/day) | 72.4±19.3 | Same as screen | 99.6±23.6 | 1.78E-15 |
| 24 h urine K (mmol/day) | 52±21 | 62±29 | 0.01 | |
| Spot urine K (mmol/L) | 61±29 | 53±26 | 64±28 | 0.03 |
| Body weight (kg) | 94.8±21.3 | 93.7±20.8 | 0.03 | |
| Daily caloric intake (kcal/day) | 2352±801 | 2392±773 | 0.61 | |
| Daily carbohydrate intake (g/day) | 244±97 | 261±115 | 0.11 | |
| Daily protein intake (g/day) | 103±39 | 108±32 | 0.22 | |
| Daily fat intake (g/day) | 102±38 | 101±31 | 0.86 |
Na: sodium; K: potassium.
obtained after holding BP medications for 1 week
from paired t-test comparing post-diet with pre-diet or, if pre-diet data were not available
Compliance and phenotypic effects of dietary sodium restriction
Average baseline sodium intake of subjects was 3807±1778 mg/day (166±77 mmol/day). During the diet period, subjects consumed an average of 946 ±141 mg of Na/day (41±6 mmol/day - 75% reduction from baseline sodium intake) (Table 1). Average 24-hour urine sodium excretion levels were 211±90 (pre-diet) and 51±53 (post-diet) mmol/24hr. Spot urine sodium levels were consistent with dietary sodium and 24-hour urinary sodium excretion data (Table 1). Twenty-four -hour urine sodium decreased in all subjects except 2, indicating compliance rate of 96% (Figure 2A). The two subjects were not excluded from subsequent analyses. Urine sodium data were obtained to assess compliance, but the analysis was based on intention to treat model and hence all subjects that entered the study were left to analyze. There were no statistically significant changes between baseline and Visit 2 in daily intakes of calories, carbohydrate, protein, and fat before and during the study period (Table 1). However, there was an increase in potassium consumption during the study period when compared to pre-study assessment. Potassium excretion also increased (Table 1).
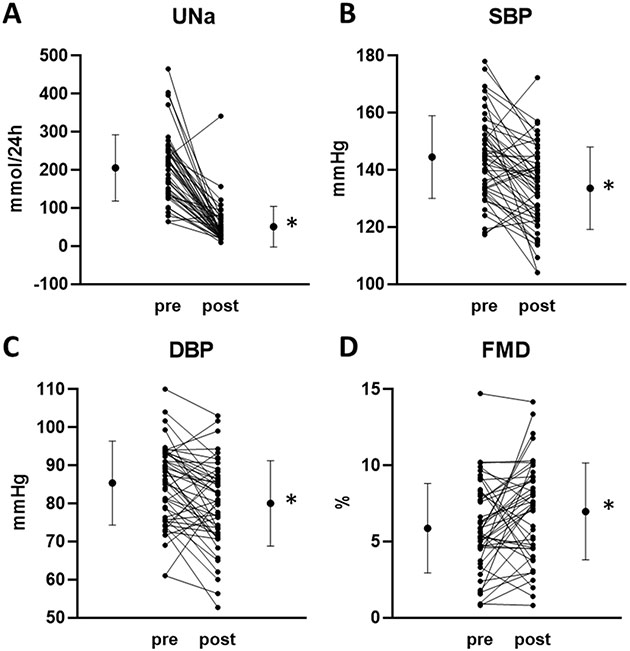
Figure 2. Urinary sodium excretion and phenotypes pre- and post-dietary restriction of sodium intake.
A. 24h urinary excretion of sodium (UNa). B. Systolic BP (SBP). C. Diastolic BP (DBP). D. Forearm blood flow mediated dilation (FMD). Individual subjects, means, and standard deviations were plotted. n=50, *, p<0.05.
The average systolic and diastolic BP reductions were −8±13/−4±9 mm Hg (p<0.001 for both) after 2 weeks of low sodium diet (Figure 2B, 2C). Approximately 75% of the subjects showed a decrease of systolic or diastolic BP. Brachial artery FMD improved from 5.8±2.9% to 6.8±3.4% (n=46, p=0.03) after 2 weeks (Figure 2D). Changes of systolic and diastolic BP in response to the dietary sodium restriction were significantly correlated (r=0.64, p<0.001). Changes of diastolic BP were correlated with changes in brachial artery FMD (r=0.30, p=0.04), but changes of systolic BP were not.
Effects of dietary sodium restriction on DNA methylation in T-cells
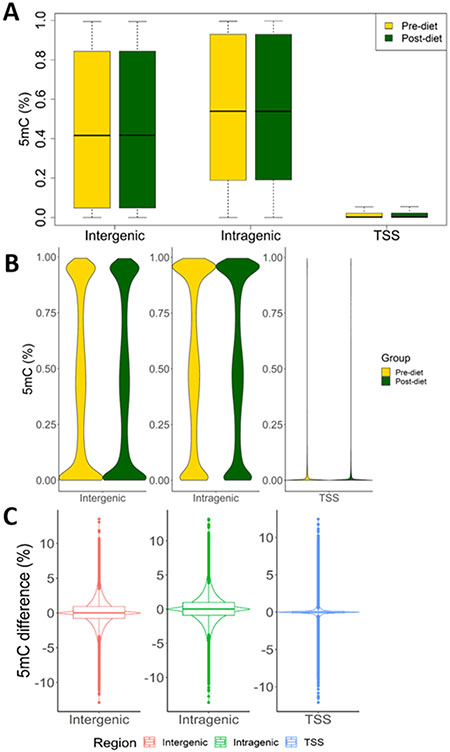
Circulating T-cells were isolated from all 50 subjects before and after two-week sodium restriction. DNA methylation profile data from 46 subjects (92 samples) passed quality control test and were used for analyses. On an average, 20.5 million sequencing reads per sample were obtained from the 92 samples (Supplemental Table S1). The average bisulfite conversion rate was 99.9% and 3,334,274 CpG sites were detected, 2,673,468 of which were detected with at least 5X coverage. The average methylation levels for CpG sites located in intergenic or intragenic regions were close to 50% but the distribution was wide (Figure 3A). Methylation levels at different sites varied from being fully methylated to fully unmethylated, with a group of sites showing methylation level of around 50% (Figure 3B). Methylation levels at TSS were much lower than intergenic and intragenic regions, as expected (Figure 3A, 3B). Methylation levels did not shift broadly from pre- to post- dietary sodium restriction but a large number of CpG sites showed a change of methylation level of greater than 5% (Figure 3C). For each site, methylation difference was defined as post-diet methylation level minus pre-diet methylation level34.
Figure 3. T-cell DNA methylation profiles and effect of dietary sodium restriction.
A. Box plots of methylation levels at CpG sites across all subjects (n=46). B. Violin plots of methylation levels at CpG sites. C. Violin plots with boxplots of differences in methylation levels at CpG sites between pre- and post-diet samples (n=46). Pre- and post-diet refer to before and after 2 weeks of dietary restriction of sodium intake. CpG sites are divided into intergenic, intragenic, and TSS (Transcription Start Site) regions, based on their locations relative to genes.
A modified Metilene (Version 0.23) was applied to identify differentially methylated regions (DMRs) de novo37. A CpG methylation region containing at least 5 CpG sites with at least 5X coverage and exhibiting an average 5mC difference of greater than 5% between paired samples with FDR < 0.05 was considered a DMR. A total of 133 DMRs were identified in T-cells (Supplemental Table S2). Of 133 DMRs, 41, 60 and 32 were located in intragenic, TSS, and intergenic regions. The result of DAVID pathway analysis of 117 genes containing intragenic and TSS DMRs is shown in Supplemental Figure 1. Of the 133 DMRs, 17 were located in known binding regions of CCCTC-binding factor (CTCF), a transcriptional factor and insulator protein important for the formation of chromatin loops.
We analyzed the association of methylation changes with changes in BP, BMI, FMD, weight, and several dietary components in response to the sodium restriction. Methylation changes in one intergenic CpG region in response to the sodium restriction was significantly (FDR <0.05) associated with changes in diastolic BP and one intragenic region was associated with baseline BMI (Table 2). The region associated with BMI was located in the gene MYO1D.
Table 2.
T-Cell and Arteriolar DNA regions with pre- and post-diet methylation differences associated with phenotypic differences.
| Chromosome | Start | Stop | Correlation | p-value | Adjusted p- value |
Location | Gene Name |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T-Cell DNA regions | ||||||||||||
| BMI | ||||||||||||
| 17 | 30821784 | 30821878 | 0.55 | 8.30E-05 | 0.01 | Intragenic | MYO1D | |||||
| Diastolic Blood Pressure | ||||||||||||
| 12 | 34499685 | 34499731 | 0.67 | 2.39E-06 | 0.01 | Intergenic | NA | |||||
| Arteriolar DNA regions | ||||||||||||
| BMI | ||||||||||||
| 1 | 33235530 | 33235594 | −0.98 | 4.27E-06 | 0.039 | Intragenic | KIAA1522 | |||||
| 19 | 2433330 | 2433532 | 0.96 | 7.50E-06 | 0.039 | TSS | MIR7108 | |||||
| Systolic Blood Pressure | ||||||||||||
| 7 | 130131947 | 130132002 | 0.930 | 9.60E-05 | 0.0067 | TSS | MESTIT1, MEST | |||||
| Diastolic Blood Pressure | ||||||||||||
| 17 | 26578249 | 26578315 | −0.979 | 4.27E-06 | 0.044 | NA | Intergenic | |||||
Effects of dietary sodium restriction on DNA methylation in arterioles
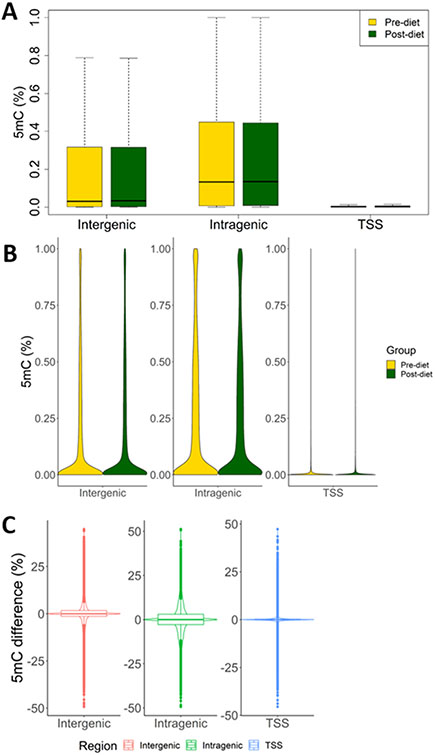
Arteriolar DNA was available for a subset of above subjects (n=10) to conduct methylation analyses. Clinical characteristics of this subset of subjects are shown in Supplemental Table S3. On average, 21.3 million sequencing reads per sample were obtained from 20 arteriole samples from 10 subjects that passed quality control (Supplemental Table S4). Bisulfite conversion rate was 99.8% and the number of CpG sites detected were 1,235,558, 995,140 of which were detected with at least 5X coverage. Global methylation patterns in arterioles were substantially different from T-cells. Average methylation levels of CpG sites, including intergenic and intragenic CpG sites, were much lower in arterioles than in T-cells (Figure 4A). While methylation levels of intergenic and intragenic CpG sites in arterioles also had a wide distribution, most of these CpG sites had low methylation levels and no large groups of CpG were found to have methylation levels around 50% or close to fully methylated as in T-cells (Figure 4B). A large number of CpG sites show large changes, up to 50%, in methylation level from before to after the dietary sodium restriction (Figure 4C), which was again unlike T-cells in which the methylation changes rarely exceeded 10%.
Figure 4. Arteriole DNA methylation profiles and effect of dietary sodium restriction.
A. Box plots of methylation levels at CpG sites across all subjects (n=10). B. Violin plots of methylation levels at CpG sites. C. Violin plots with boxplots of differences in methylation levels at CpG sites between pre- and post-diet samples (n=10). Pre- and post-diet refer to before and after 2 weeks of dietary restriction of sodium intake. CpG sites are divided into intergenic, intragenic, and TSS (Transcription Start Site) regions, based on their locations relative to genes.
A total of 70 DMRs were identified in arterioles (Supplemental Table S5). Of 70 DMRs, 27, 34, and 9 were located in intragenic, TSS, and intergenic regions. The result of DAVID pathway analysis of 71 genes containing intragenic or TSS DMRs is shown in Supplemental Figure S2. These genes were enriched for several pathways at unadjusted p<0.05, including protein targeting to Golgi and transcriptional regulation. Of the 70 DMRs, 6 were located in known CTCF binding regions.
Changes in several methylation regions in arterioles from before to after the dietary sodium restriction were significantly associated with changes in phenotypes (Table 2). These included one methylation region located in the TSS of MESTIT1 and MEST genes, which were associated with systolic BP, one intragenic methylation region in KIAA1522 and one TSS methylation region in MIR7108, which were associated with BMI, and two intergenic methylation regions associated with diastolic BP.
Association of DNA methylation changes with differential gene expression in arterioles
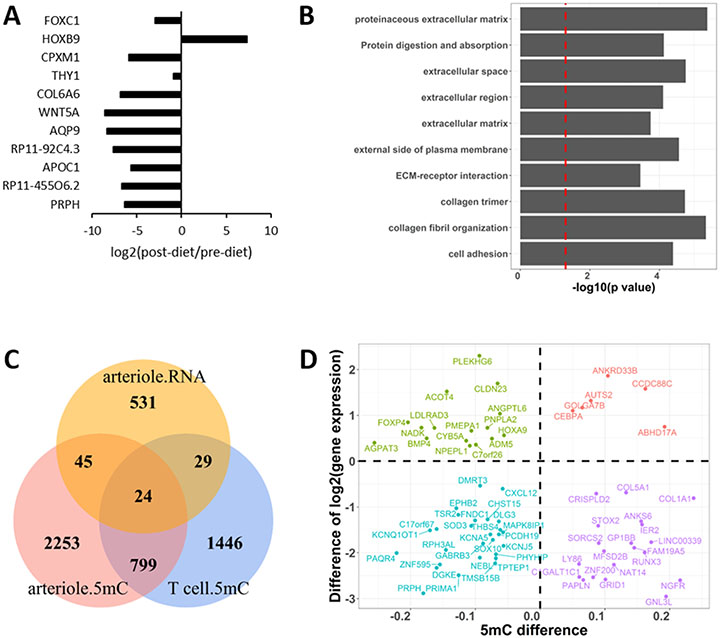
RNA-seq analysis was performed to examine gene expression profiles in arterioles before and after the sodium restriction (n=11). On average, 53 million sequencing reads were obtained from 22 RNA samples (Supplemental Table S6). Eleven genes were found to be significantly differentially expressed in arterioles between before and after the sodium restriction at the significance level of FDR<0.05 (Figure 5A & Supplemental Table S7). At the significance level of p<0.05, 629 genes were differentially expressed, which were enriched for 10 pathways at the significance level of FDR<0.05 for the enrichment (Figure 5B). Several of these pathways were related to extracellular matrix, suggesting the involvement of arteriole structural remodeling in response to the dietary sodium restriction. The 629 genes were involved in additional pathways, and those with p<0.01 for the enrichment are shown in Supplemental Figure S3.
Figure 5. Dietary sodium restriction induces gene expression changes in arterioles.
A. Genes differentially expressed in arterioles in response to 2 weeks of a low-sodium diet at the significance level of FDR<0.05. n=11. B. Biological pathways (FDR < 0.05) over-represented in 629 genes differentially expressed in arterioles at the significance level of p<0.05. C. Overlaps of genes differentially expressed in arterioles and genes containing differentially methylated regions (DMRs) in arterioles or T-cells, all at the significance level of p<0.05. The overlap between arteriole DMRs and T-cell DMRs was significantly greater than random chance (p<1x10−16). D. Expression and methylation differences in arterioles between pre- and post-dietary sodium restriction for the 69 overlapping genes shown in panel C.
Differentially expressed genes were cross-referenced with genes containing intragenic and TSS DMRs in arterioles. At the significance level of FDR < 0.05 for differential expression and methylation, no overlap was found between the two sets of genes. We analyzed the correlation between DMRs and their host genes or, for intergenic DMRs, nearest genes in seven subjects for whom both DNA methylation and RNA-seq data were available. In total, 78 genes and 70 DMRs were analyzed by Spearman correlation. The expression of 7 genes (CLYBL, NFATC1, C1GALT1C1, TPPP3, RP13-582O9.5, LRRFIP1, PDXDC1) was significantly correlated with methylation levels of corresponding DMRs (p < 0.05), four of which (CLYBL, LRRFIP1, PDXDC1, RP13-582O9.5) were inversely correlated.
At the significance level of p < 0.05, 629 genes were identified from the RNA-seq data and 3121 genes contained DMRs. Of these genes, 69 genes overlapped (Figure 5C). Of the 69 genes, 42 contain TSS DMRs. However, no specific directional relation was noted between methylation and gene expression differences regardless of whether all 69 genes (Figure 5D) or only the 42 genes were considered.
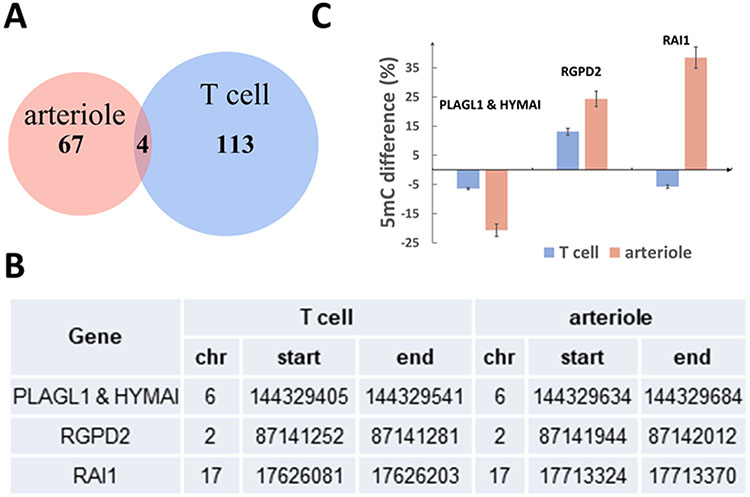
Comparing the effects of sodium restriction on DNA methylation in T-cells and arterioles
The DMRs identified in T-cells and arterioles were located in intragenic or TSS regions of 117 and 71 genes, respectively. Four of these genes were identified in both T-cells and arterioles (Figure 6A). Despite the apparently small number of overlapping genes, the overlap between T-cells and arterioles was statistically significant based on a permutation test (p=0.009). These genes were PLAGL1, HYMAI, RGPD2, and RAI1 (Figure 6B). The methylation level of the DMR in PLAGL1 and HYMAI was decreased in both T-cells and arterioles in response to the sodium restriction (Figure 6C). The methylation level of the DMR in RGPD2 was increased in both T-cells and arterioles. The methylation level of the DMR in RAI1 was decreased in T-cells and increased in arterioles.
Figure 6. Overlaps of differentially methylated regions between T-cells and arterioles.
A. Overlaps of genes containing intragenic or TSS (Transcription Start Site) regions differentially methylated, at the significance level of FDR<0.05, in T-cells and arterioles. The overlap was significantly greater than random chance (p=0.009). B. Genes containing regions differentially methylated (FDR<0.05) in both T-cells and arterioles. C. Changes of DNA methylation in T-cells and arterioles for the 4 genes shown in panel B.
At the significance level of p<0.05 for DMR, a significant overlap was also identified between genes containing DMRs in T-cells and arterioles. Of 2298 and 3121 genes containing DMRs with p<0.05 in T-cells and arterioles, respectively, 823 overlapped, which was significantly greater than what could be expected by chance alone (p<1 x10−16) (Figure 5C). These 823 genes, shown in Supplemental Table S8, were enriched for genes involved in transcriptional regulation, protein targeting to Golgi, intermediary metabolism, calcium signaling and other pathways (Supplemental Figure S4).
Comparing the effect of dietary salt intake on DNA methylation in T-cells in humans and rats
At the significance level of p<0.05 for differential methylation, 2298, 193, and 196 genes were found to contain DMRs in T-cells of human subjects, SS/Mcw and SS/Crl rats, respectively. Of these genes, 46 overlapped between human and SS/Mcw. The overlap was statistically significant (p=0.03). 38 genes overlapped between human and SS/Crl, which was not statistically significant. These overlapping genes are shown in Supplemental Table S9.
DISCUSSION
This is the first study to have evaluated DNA methylation in T-cells and arterioles in response to changes in dietary sodium intake in human subjects. We found that dietary sodium restriction resulted in T-cell and arteriolar DNA methylation changes and arteriolar gene expression changes along with improvements in BP and vascular function. Interestingly, the effect of dietary sodium restriction on DNA methylation is largely tissue-specific and yet significant overlaps exist between T-cells and arterioles. In addition, DNA methylation levels are broadly higher in T-cells than arterioles, especially in intergenic and intragenic regions. These findings are consistent with previous findings of a high degree of tissue specificity for DNA methylation in human38. Both T-cells and arterioles may sense sodium restriction-induced changes in circulating or humoral factors, leading to overlapping DNA methylation changes. Dietary sodium restriction may elicit tissue-specific DNA methylation responses because arterioles, but not T-cells, may sense mechanical and neural signals resulting from sodium restriction and decreased BP. Even the same circulating or humoral factors may elicit different DNA methylation changes because of tissue-specific signaling cascades, further contributing to the tissue-specific DNA methylation responses.
Changes in DNA methylation, a type of epigenetic modification, may underlie stable changes of DNA function and contribute to long-term effects of dietary sodium intake on BP and vascular disease. T-cells have an established role in the development of hypertension and cardiovascular disease39, 40. A high salt intake has been shown to stimulate the differentiation of T cells and other immune cells to a pro-inflammatory and pathogenic phenotype41, 42. DNA methylation changes in T-cells in response to low sodium diet occurred in genes enriched for pathways ranging from fatty acid oxidation to transcription regulation. It remains to be investigated whether the observed DNA methylation changes lead to changes in the biological activities of these pathways that, in turn, may contribute to the functional role of T-cells in the development of hypertension or vascular dysfunction. Methylation changes in an intragenic region in MYO1D gene in response to the sodium restriction were associated with BMI. Myosin ID (MYO1D) is a member of the class I myosin family43. Members of this class are single headed, bind calmodulin light chains and have lipid binding domains in their tails. The rat myo1d homologue has been implicated in endosome vesicle recycling in epithelial cells44. At this time, there is no explicit biological link between this gene and BMI. We found that the effect of dietary sodium intake on DNA methylation in T-cell in humans and Dahl SS/Mcw rats show significant overlaps, supporting the value of SS/Mcw rats as a model for studying mechanistic role of DNA methylation in the effect of sodium intake on BP.
Dietary sodium intake may influence arterial function, which may contribute to changes in BP or lead to vascular dysfunction independent of BP changes7. We observed a significant improvement in brachial artery FMD in response to the dietary sodium restriction. The arteriolar genes that we found to contain DMRs in response to the sodium restriction were enriched for pathways including protein targeting to Golgi and transcriptional regulation. Intragenic or TSS regions in several genes in arterioles showed methylation responses to the sodium restriction that are associated with BMI or BP responses. MESTIT1 and MEST code for non-protein coding antisense RNA that is imprinted, and preferentially expressed from the paternal allele. The antisense RNA is expressed predominantly in testis and mature motile spermatozoa, suggesting that it may be involved in development45. NXPH4 was reported to be a candidate gene in a BP quantitative trait locus in Dahl SS rats46. KIAA1522, gene with significant methylation changes associated with BMI, has been associated with oncogenic KRA pathway and is considered a biological marker for prognosis of non-small cell lung cancer47. Its biological function is unknown and no association has been previously identified with BMI. MIR7108 (microRNA 7108), associated with BMI, also has no known biological function or plausible relationship with BMI.
At the significance level of p<0.05, several dozen genes that are differentially expressed in arterioles in response to the sodium restriction contain differentially methylation regions. It remains to be investigated whether the differential methylation contributes to the differential expression of these genes. We did not observe a significant, inverse correlation between the methylation and expression changes of these genes, even if we only examined the genes containing differential methylation at their TSS regions. While DNA methylation is generally inversely correlated with gene expression when compared between genes or tissues, the relation between methylation and expression changes of a given gene in response to physiological stimuli is not monotonic18, 19. Methylation changes in response to physiological stimuli may play fine-tuning or compensatory roles in gene expression.
As with any dietary study, non-compliance with the dietary restriction of sodium can be a major problem48. Apart from recruiting subjects who volunteered willingly, we had excellent compliance due to (i) taking subject preferences into consideration while preparing the menu, (ii) providing prepared meals instead of leaving it to the subjects to follow sodium restriction, and (iii) moderate restriction in sodium intake (1200 mg/day), which may have allowed more subjects to be compliant in home environment than with severe sodium restriction, yet this degree of sodium restriction is still much lower than average daily consumption (3800 mg/day). In addition, we accommodated subject’s ability to come for follow-up visits by ± 2 days without compromising the study quality. We verified sodium restriction by multiple methods that were in congruence with each other including food diaries, BSS, spot urine, and two 24-hour urine analysis. Total caloric intake and macronutrient composition of the diet (protein, carbohydrate, and fat) did not change significantly before and during the low-sodium diet.
This study has several limitations. The duration of the low-sodium diet was determined after taking into consideration of compliance issues related to a long-term in-home dietary study, and the fact that sodium restriction for two weeks does result in a meaningful decrease in BP. We used research unit measured BPs instead of 24 hour measurements which may have been a better marker of improvements in various BP phenotypes. Sodium restriction over longer periods of time may result in additional changes in DNA methylation and gene expression. In addition, there was an increase in potassium consumption which may have had some impact on the BP levels and methylation marks. Subjects lost significant amount weight at the end of the study period despite same amount of calorie consumption and this may be secondary to decrease in plasma volume on low sodium diet. Our sample size is modest, especially arteriolar samples. However, even with the modest sample size, the study is one of the largest studies that have analyzed DNA methylation and RNA expression in human arterioles, and significant differences were identified. We did not further sort T-cells into its subtypes such as helper or suppressor T-cells for analysis. Nevertheless, majority of human DNA methylation studies were based on whole blood DNA. This study is one of the largest that have evaluated the effect of dietary sodium intake on human T-cell DNA methylation.
In summary, we have found that dietary sodium restriction has significant effects on DNA methylation in T-cells and arterioles, some of which are associated with change of BP. Methylation patterns and sodium effects on methylation are largely tissue-specific but also have significant overlaps between tissues and species. This study suggests methylation modification of DNA could be one of the mechanisms by which sodium intake may impact physiology and disease.
Supplementary Material
Sources of Funding
This work was supported by the American Heart Association (15SFRN23910002), the National Institutes of Health (HL149620), the Advancing a Healthier Wisconsin Endowment, and the Clinical and Translational Science Institute of Southeastern Wisconsin through the NIH Clinical and Translational Science Award UL1TR001436.
Footnotes
Disclosures
None.
References
- 1.Whelton PK, Carey RM, Aronow WS, Casey DE Jr., Collins KJ, Dennison Himmelfarb C, DePalma SM, Gidding S, Jamerson KA, Jones DW, MacLaughlin EJ, Muntner P, Ovbiagele B, Smith SC Jr., Spencer CC, Stafford RS, Taler SJ, Thomas RJ, Williams KA Sr., Williamson JD and Wright JT Jr. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol. 2018;71:1269–1324. [DOI] [PubMed] [Google Scholar]
- 2.James PA, Oparil S, Carter BL and et al. 2014 evidence-based guideline for the management of high blood pressure in adults: Report from the panel members appointed to the eighth joint national committee (jnc 8). JAMA. 2014;311:507–520. [DOI] [PubMed] [Google Scholar]
- 3.Weinberger MH. Salt sensitivity of blood pressure in humans. Hypertension. 1996;27:481–90. [DOI] [PubMed] [Google Scholar]
- 4.Cook NR, Cutler JA, Obarzanek E, Buring JE, Rexrode KM, Kumanyika SK, Appel LJ and Whelton PK. Long term effects of dietary sodium reduction on cardiovascular disease outcomes: observational follow-up of the trials of hypertension prevention (TOHP). BMJ. 2007;334:885–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saint-Remy A, Somja M, Gellner K, Weekers L, Bonvoisin C and Krzesinski JM. Urinary and dietary sodium and potassium associated with blood pressure control in treated hypertensive kidney transplant recipients: an observational study. BMC Nephrol. 2012;13:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinberger MH, Miller JZ, Luft FC, Grim CE and Fineberg NS. Definitions and characteristics of sodium sensitivity and blood pressure resistance. Hypertension. 1986;8:II127–34. [DOI] [PubMed] [Google Scholar]
- 7.Farquhar WB, Edwards DG, Jurkovitz CT and Weintraub WS. Dietary sodium and health: more than just blood pressure. J Am Coll Cardiol. 2015;65:1042–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor RS, Ashton KE, Moxham T, Hooper L and Ebrahim S. Reduced dietary salt for the prevention of cardiovascular disease: a meta-analysis of randomized controlled trials (Cochrane review). Am J Hypertens. 2011;24:843–53. [DOI] [PubMed] [Google Scholar]
- 9.He FJ and MacGregor GA. Salt reduction lowers cardiovascular risk: meta-analysis of outcome trials. Lancet. 2011;378:380–2. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Huang T and Lombard JH. Effect of high-salt diet on vascular relaxation and oxidative stress in mesenteric resistance arteries. J Vasc Res. 2007;44:382–90. [DOI] [PubMed] [Google Scholar]
- 11.Sanders PW. Vascular consequences of dietary salt intake. Am J Physiol Renal Physiol. 2009;297:F237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edwards DG and Farquhar WB. Vascular effects of dietary salt. Curr Opin Nephrol Hypertens. 2015;24:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitta Y, Obata JE, Nakamura T, Hirano M, Kodama Y, Fujioka D, Saito Y, Kawabata K, Sano K, Kobayashi T, Yano T, Nakamura K and Kugiyama K. Persistent impairment of endothelial vasomotor function has a negative impact on outcome in patients with coronary artery disease. J Am Coll Cardiol. 2009;53:323–30. [DOI] [PubMed] [Google Scholar]
- 14.Modena MG, Bonetti L, Coppi F, Bursi F and Rossi R. Prognostic role of reversible endothelial dysfunction in hypertensive postmenopausal women. J Am Coll Cardiol. 2002;40:505–10. [DOI] [PubMed] [Google Scholar]
- 15.Liang M, Cowley AW Jr., Mattson DL, Kotchen TA and Liu Y. Epigenomics of hypertension. Semin Nephrol. 2013;33:392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang M Epigenetic Mechanisms and Hypertension. Hypertension (Dallas, Tex : 1979). 2018;72:1244–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dasinger JH, Alsheikh AJ, Abais-Battad JM, Pan X, Fehrenbach DJ, Lund H, Roberts ML, Cowley AW Jr., Kidambi S, Kotchen TA, Liu P, Liang M and Mattson DL. Epigenetic Modifications in T Cells: The Role of DNA Methylation in Salt-Sensitive Hypertension. Hypertension. 2020;75:372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Liu P, Yang C, Cowley AW Jr. and Liang M. Base-resolution maps of 5-methylcytosine and 5-hydroxymethylcytosine in Dahl S rats: effect of salt and genomic sequence. Hypertension. 2014;63:827–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu P, Liu Y, Liu H, Pan X, Li Y, Usa K, Mishra MK, Nie J and Liang M. Role of DNA De Novo (De)Methylation in the Kidney in Salt-Induced Hypertension. Hypertension. 2018;72:1160–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CDC. NHANES Physical Examination Procedures Manual. 2007.
- 21.Quest N. Block Food Frequency Questionnaire. 2020.
- 22.Feskanich D, Sielaff BH, Chong K and Buzzard IM. Computerized collection and analysis of dietary intake information. Comput Methods Programs Biomed. 1989;30:47–57. [DOI] [PubMed] [Google Scholar]
- 23.Schakel SF, Sievert YA and Buzzard IM. Sources of data for developing and maintaining a nutrient database. J Am Diet Assoc. 1988;88:1268–71. [PubMed] [Google Scholar]
- 24.Sievert YA, Schakel SF and Buzzard IM. Maintenance of a nutrient database for clinical trials. Control Clin Trials. 1989;10:416–25. [DOI] [PubMed] [Google Scholar]
- 25.Harnack L, Stevens M, Van Heel N, Schakel S, Dwyer JT and Himes J. A computer-based approach for assessing dietary supplement use in conjunction with dietary recalls. J Food Compost Anal. 2008;21:S78–S82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson RK, Driscoll P and Goran MI. Comparison of multiple-pass 24-hour recall estimates of energy intake with total energy expenditure determined by the doubly labeled water method in young children. J Am Diet Assoc. 1996;96:1140–4. [DOI] [PubMed] [Google Scholar]
- 27.Widlansky ME, Jensen DM, Wang J, Liu Y, Geurts AM, Kriegel AJ, Liu P, Ying R, Zhang G, Casati M, Chu C, Malik M, Branum A, Tanner MJ, Tyagi S, Usa K and Liang M. miR-29 contributes to normal endothelial function and can restore it in cardiometabolic disorders. EMBO Mol Med. 2018:10:e8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu H, Smith ZD, Bock C, Boyle P, Gnirke A and Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 2011;6:468–81. [DOI] [PubMed] [Google Scholar]
- 29.Dass N, Kilakkathi S, Obi B, Moosreiner A, Krishnaswami S, Widlansky ME and Kidambi S. Effect of gender and adiposity on in vivo vascular function in young African Americans. J Am Soc Hypertens. 2017;11:246–257. [DOI] [PubMed] [Google Scholar]
- 30.Geurts AM, Mattson DL, Liu P, Cabacungan E, Skelton MM, Kurth TM, Yang C, Endres BT, Klotz J, Liang M and Cowley AW, Jr. Maternal diet during gestation and lactation modifies the severity of salt-induced hypertension and renal injury in Dahl salt-sensitive rats. Hypertension. 2015;65:447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krueger F and Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics (Oxford, England). 2011;27:1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT and Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frazee AC, Pertea G, Jaffe AE, Langmead B, Salzberg SL and Leek JT. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 2015;33:243–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abais-Battad JM, Alsheikh AJ, Pan X, Fehrenbach DJ, Dasinger JH, Lund H, Roberts ML, Kriegel AJ, Cowley AW Jr., Kidambi S, Kotchen TA, Liu P, Liang M, and Mattson DL. Dietary Effects on Dahl Salt-Sensitive Hypertension, Renal Damage, and the T Lymphocyte Transcriptome. Hypertension. 2019;74:854–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abais-Battad JM, Lund H, Fehrenbach DJ, Dasinger JH, Alsheikh AJ and Mattson DL. Parental Dietary Protein Source and the Role of CMKLR1 in Determining the Severity of Dahl Salt-Sensitive Hypertension. Hypertension. 2019;73:440–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juhling F, Kretzmer H, Bernhart SH, Otto C, Stadler PF and Hoffmann S. metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome research. 2016;26:256–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lokk K, Modhukur V, Rajashekar B, Martens K, Magi R, Kolde R, Koltsina M, Nilsson TK, Vilo J, Salumets A and Tonisson N. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014;15:r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drummond GR, Vinh A, Guzik TJ and Sobey CG. Immune mechanisms of hypertension. Nat Rev Immunol. 2019;19:517–532. [DOI] [PubMed] [Google Scholar]
- 40.Norlander AE, Madhur MS and Harrison DG. The immunology of hypertension. J Exp Med. 2018;215:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, Muller DN and Hafler DA. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature. 2013;496:518–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, Regev A and Kuchroo VK. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. 2013;496:513–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McAlpine W, Wang KW, Choi JH, San Miguel M, McAlpine SG, Russell J, Ludwig S, Li X, Tang M, Zhan X, Choi M, Wang T, Bu CH, Murray AR, Moresco EMY, Turer EE and Beutler B. The class I myosin MYO1D binds to lipid and protects against colitis. Dis Model Mech. 2018;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LeBlanc-Straceski JM, Sokac A, Bement W, Sobrado P and Lemoine L. Developmental expression of Xenopus myosin 1d and identification of a myo1d tail homology that overlaps TH1. Dev Growth Differ. 2009;51:443–51. [DOI] [PubMed] [Google Scholar]
- 45.Li T, Vu TH, Lee KO, Yang Y, Nguyen CV, Bui HQ, Zeng ZL, Nguyen BT, Hu JF, Murphy SK, Jirtle RL and Hoffman AR. An imprinted PEG1/MEST antisense expressed predominantly in human testis and in mature spermatozoa. J Biol Chem. 2002;277:13518–27. [DOI] [PubMed] [Google Scholar]
- 46.Crespo K, Chauvet C, Blain M, Menard A, Roy J and Deng AY. Normotension in Lewis and Dahl salt-resistant rats is governed by different genes. J Hypertens. 2011;29:460–5. [DOI] [PubMed] [Google Scholar]
- 47.Liu YZ, Yang H, Cao J, Jiang YY, Hao JJ, Xu X, Cai Y and Wang MR. KIAA1522 is a novel prognostic biomarker in patients with non-small cell lung cancer. Sci Rep. 2016;6:24786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luft FC, Morris CD and Weinberger MH. Compliance to a low-salt diet. Am J Clin Nutr. 1997;65:698S–703S [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.