Abstract
Ryanodine receptor type 2 (RyR2) mutations have been linked to an inherited form of exercise-induced sudden cardiac death called catecholaminergic polymorphic ventricular tachycardia (CPVT). CPVT results from stress-induced sarcoplasmic reticular Ca2+ leak via the mutant RyR2 channels during diastole. We present atomic models of human wild-type (WT) RyR2 and the CPVT mutant RyR2-R2474S determined by cryo–electron microscopy with overall resolutions in the range of 2.6 to 3.6 Å, and reaching local resolutions of 2.25 Å, unprecedented for RyR2 channels. Under nonactivating conditions, the RyR2-R2474S channel is in a “primed” state between the closed and open states of WT RyR2, rendering it more sensitive to activation that results in stress-induced Ca2+ leak. The Rycal drug ARM210 binds to RyR2-R2474S, reverting the primed state toward the closed state. Together, these studies provide a mechanism for CPVT and for the therapeutic actions of ARM210.
Cryo-EM structures of human RyR2, pathological primed RyR2-R2474S, and RyR2-R2474S reveal a mechanism for Rycal ARM210 treatment.
INTRODUCTION
Ryanodine receptors mediate the rapid release of Ca2+ from the intracellular stores of the endo/sarcoplasmic reticulum (ER/SR) (1). Among the three mammalian isoforms, ryanodine receptor type 2 (RyR2) is the major form expressed in cardiac muscle, where it mediates excitation-contraction coupling via a mechanism known as Ca2+-induced Ca2+ release (CICR) (2–4). During CICR, RyR2 is activated by a small influx of Ca2+ through voltage-gated L-type Ca2+ channels of the opposing plasma membrane, initiating a larger release of Ca2+ from the intracellular SR/ER stores. Dysfunctional RyR2 has been linked to cardiovascular diseases, including heart failure (5), atrial fibrillation (6), and catecholaminergic polymorphic ventricular tachycardia (CPVT) (7, 8). CPVT is an inherited form of exercise-induced arrhythmia, and sudden cardiac death linked to single amino acid mutations in RyR2 (7, 9, 10). Intense exercise and emotional stress induce the release of catecholamines, which activate the β-adrenergic signaling pathway. In CPVT patients, the β-adrenergic response culminates in pathological SR Ca2+ release during diastole, leading to delayed afterdepolarizations and ventricular arrhythmia (11). The β-adrenergic signaling pathway activates protein kinase A (PKA), which phosphorylates RyR2 predominantly at residue RyR2-S2808 (5). This has been shown to play a vital role in CPVT and other RyR2 pathologies (7, 12–15). Among the more than 300 familial CPVT variants identified to date, RyR2-R2474S is one of the most severe and most studied CPVT mutations (10, 16, 17). RyR2-R2474S has been directly linked to stress-induced syncope and juvenile sudden death (18). Exercise-induced sudden cardiac death has been demonstrated in knock-in mice expressing RyR2 mutations linked to CPVT in patients (10). Treatment with RyR2 stabilizing drugs known as Rycals (19), the inhibitor dantrolene (20), and enhanced binding of calmodulin (CaM) have been shown to prevent fatal arrhythmias in murine models of CPVT (10, 21, 22).
Elucidating structure-function relationships of RyRs is essential to understanding the mechanism of the SR Ca2+ leak that causes fatal cardiac arrhythmias and for determining the mechanism of action of drugs that fix this leak. Structural analyses of RyRs have been challenging because of their size (>2 MDa), dynamic behavior, and membrane-bound nature. Nevertheless, the resolution revolution of cryo-electron microscopy (cryo-EM) has enabled elucidation of the structures of skeletal muscle RyR1 by our group and others (23–26). However, RyR2 structures have been less well resolved compared to RyR1, as purification from heart tissue provides low yields of RyR2 with a variety of different posttranslational modifications (27, 28). Recently, recombinant purification of mouse RyR2 overexpressed in human embryonic kidney (HEK) 293 cells resulted in a greatly improved structure, reaching a resolution of 3.27 Å (29). Here, we provide structures of the human wild-type (WT) RyR2 and CPVT mutant RyR2-R2474S, the latter in the presence of the Rycal ARM210 and the modulator protein CaM, at reproducible excellent global resolutions (2.6 to 3.1 Å for the closed states and 2.9 to 3.6 Å for the open states). To our knowledge, these are the most complete RyR2 structures and the first showing the Rycal binding site and its mechanisms of action in disease-related RyR2 channels. Our models of RyR2 include previously unresolved peripheral structural elements, including the RY3&4 phosphorylation domain, the secondary adenosine triphosphate (ATP) binding site in the RY1&2 domain, the elusive BSol domain, and the previously unidentified S-1/S0 intramembrane helices. Last, we provide insights into the structure-function relationship and the mechanism underlying the gain-of-function mutation in the PKA-phosphorylated RyR2-R2474S as well as the mechanism of stabilization of the mutant CPVT RyR2 by the Rycal drug ARM210 and by CaM. We show that the PKA-phosphorylated RyR2-R2474S adopts a “primed” state that implies that less energy is required to rearrange the cytoplasmic shell, allowing gating of the channel at lower ligand concentrations. We show how binding of ARM210 to the cleft of the RY1&2 domain reverses this conformation and provides more evidence to support the function of the RY1&2 domain as a metabolic sensor—previously reported to bind ATP—[Protein Data Bank (PDB): 6UHH] (30, 31).
RESULTS
The CPVT-linked mutation RyR2-R2474S induces a primed state, while ARM210 and CaM stabilize the closed state
To obtain high-resolution structures of WT and mutant human RyR2, we optimized the purification of channels expressed recombinantly in HEK293 cells (fig. S1). For cryo-EM experiments, we used conditions that resemble those found during the resting phase of the cardiac cycle known as diastole (150 nM free Ca2+ and 10 mM ATP) when the RyR2 channels are tightly closed, but CPVT RyR2 variants are leaky (8, 19). We pretreated WT and RyR2-R2474S with PKA to induce hyperphosphorylation and mimic exercise-induced β-adrenergic signaling, which is the trigger for fatal cardiac arrhythmias in patients with CPVT (fig. S1A). We added xanthine (500 μM), which is a physiologically relevant analog of the RyR2 agonist caffeine. Xanthine activates RyRs (32) and increases to micromolar range during exercise (33–35), suggesting that it may be an endogenous activator of RyR2 that becomes more important during exercise. Under these conditions, we obtained 10 structures of human RyR2, including the closed and open states of PKA-phosphorylated RyR2 (PKA RyR2), PKA-phosphorylated RyR2-R2474S (PKA RyR2-R2474S), and PKA-phosphorylated RyR2-R2474S with the addition of ARM210 or CaM (fig. S2 and table S1). We have been able to consistently obtain ~3-Å resolution for most structures and, hence, to build more complete models of human RyR2 (fig. S3). Our highest-resolution cryo-EM coulombic potential maps (cryo-EM maps) show well-defined densities of most domains, including the NTD, SPRY1-3, JSol, BSol1, and activation core and channel domains, where we were able to observe near-atomic details including “holes” in aromatic residues (fig. S4). For the remaining dynamic domains usually missing in previously published structures (RY1&2, RY3&4, and BSol2), the local resolution is reduced but sufficient to follow the backbone and detect bulky side chains. The cryo-EM map quality was sufficient to build atomic models with a high level of confidence for the previously less well-resolved domains of RyR2 and the accessory proteins calstabin-2 (known as PPIase FKBP1B or, alternatively, as FKBP12.6) and CaM. Using this high-resolution model of the human RyR2 isoform and all known domains are resolved, we updated the residue span and nomenclature of domains and subdomains for human RyR2 (table S2).
To separate the particles in the closed state from those in the open states in the same dataset, we implemented local three-dimensional (3D) variability, clustering, and heterogeneous refinement, which rendered better results than 3D classification or heterogeneous refinement alone (fig. S2). We attribute this to the dynamic cytosolic shell of RyR2, which, being almost 90% of the mass of the protein, can predominate and result in mixed populations. Comparison of the pore region from all the structures showed no differences in the closed states and no differences in the open states, suggesting that the methodology implemented is reproducible (fig. S5). Of the three activators present in the sample, only ATP was detected in the closed state structures (fig. S6A). On the other hand, all three activators Ca2+, ATP, and xanthine were present in the open state structures (fig. S6B), suggesting that the binding of the additional activators, Ca2+ and xanthine, promotes opening of the channel. Xanthine occupied the previously identified caffeine site formed by W4645, I4926, and Y4944 (36). To confirm the functional role of xanthine, we performed single-channel experiments showing that, at presumed physiological concentrations during exercise (10 μM), there was a ~25-fold increase in the open probability of RyR2 in the presence of xanthine (fig. S6, C and D).
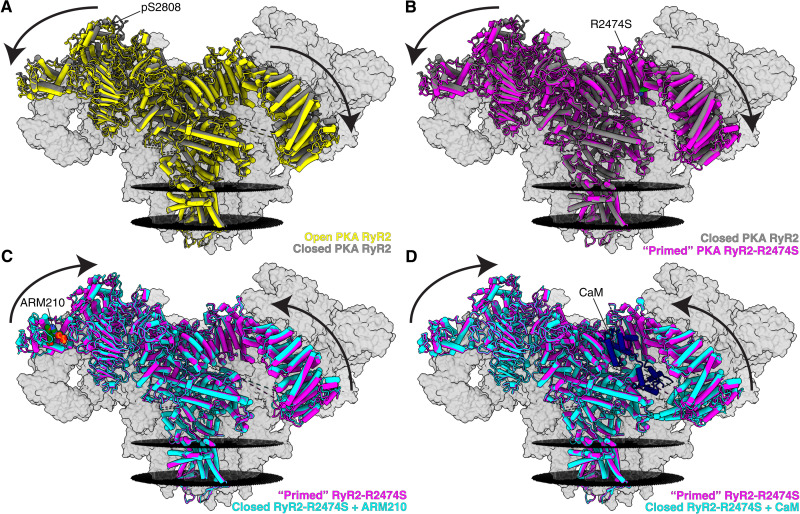
To better understand the mechanism underlying CPVT, we compared the structures of the PKA-phosphorylated RyR2 to the PKA-phosphorylated RyR2-R2474S. As seen for other RyRs, the cytosolic shell of the PKA-phosphorylated RyR2 was shifted downward and outward when going from the closed state (PDB: 7U9Q) to the open state (PDB: 7U9R) (Fig. 1A and fig. S7B). The cytosolic shell of PKA-phosphorylated RyR2-R2474S (PDB: 7U9X) was shifted downward and outward compared to closed PKA-phosphorylated RyR2, suggesting that the CPVT mutant RyR2-R2474S is in a primed state (Fig. 1B and fig. S7F). This primed state presents a structure that is approximately halfway between the closed and open states of PKA-phosphorylated RyR2. This is also the case for the open state; the cytosolic shell of open PKA-phosphorylated RyR2-R2474S (PDB: 7U9Z) was also shifted downward and outward compared to open PKA-phosphorylated RyR2 (fig. S7G), suggesting that the CPVT-related cytosolic shell destabilization is independent of the state of the pore.
Fig. 1. Cryo-EM reconstructions of human RyR2 showing that the CPVT mutant RyR2-R2474S puts the channel into the primed state, and treatment with the Rycal ARM210 and CaM puts the channel back toward the closed state.
(A) Overlapped models of open PKA-phosphorylated RyR2 (PDB: 7U9R, yellow) and closed PKA-phosphorylated RyR2 (PDB: 7U9Q, gray). The arrows show that the cytosolic shell of the PKA-phosphorylated RyR2 shifts downward and outward when going from the closed to the open state. To facilitate visualization, only the front protomer is shown in colors, while the other three protomers are shown as gray transparent volumes. The positions of the sarcoplasmic reticular membranes are shown as black discs. Conditions include 10 mM ATP, 150 nM free Ca2+, and 500 μM xanthine. (B) Overlapped models of closed PKA-phosphorylated RyR2 (PDB: 7U9Q, gray) and primed PKA-phosphorylated RyR2-R2474S (PDB: 7U9X, magenta). The arrows show that the cytosolic shell of RyR2-R2474S shifts downward and outward compared to closed PKA-phosphorylated RyR2, similar to the structural changes observed for PKA-phosphorylated RyR2 going from the closed state to the open state. We define this intermediate between closed and open states as the primed state. (C) Overlapped models of primed PKA-phosphorylated RyR2-R2474S (PDB: 7U9X, magenta) and closed PKA-phosphorylated RyR2-R2474S + ARM210 (PDB: 7UA1, cyan). The arrows show that the cytosolic shell of PKA-phosphorylated RyR2-R2474S + ARM210 cytosolic domain shifts upward and inward compared to the RyR2-R2474S reversing the primed state back toward the closed state. (D) Overlapped models of primed PKA-phosphorylated RyR2-R2474S (PDB: 7U9X, magenta) and closed PKA-phosphorylated RyR2-R2474S + CaM (PDB: 7UA3, cyan). Similar to the effects of the Rycal ARM210, CaM reverses the primed state back toward the closed state.
The PKA-phosphorylated RyR2-R2474S in the presence of the Rycal drug ARM210 (PKA-phosphorylated RyR2-R2474S + ARM210; PDB: 7UA1)—a benzothiazepine derivative closely related to S107, which we have shown to effectively prevent ventricular tachycardiac and sudden cardiac death in murine models of CPVT (10)—exhibited an upward and inward shift of the cytosolic shell compared to primed PKA-phosphorylated RyR2-R2474S, thus reversing the primed state back toward the closed state of the channel (Fig. 1C and fig. S7H). The PKA-phosphorylated RyR2-R2474S in the presence of CaM (PKA RyR2-R2474S + CaM; PDB: 7UA3) exhibits a similar but less pronounced shift of the cytosolic shell upward and inward compared to primed PKA-phosphorylated RyR2-R2474S, also reversing the primed state back toward the closed state of the channel similar to the effects of the Rycal ARM210 (Fig. 1D and fig. S7J).
ARM210 stabilizes the closed state of the PKA-phosphorylated RyR2-R2474S CPVT variant
We have previously reported that heterozygous Ryr2R2474S/WT mice undergo sustained ventricular tachycardia and sudden cardiac death after exercise and epinephrine (10). We show here that treatment with the Rycal ARM210 reduced the frequency of ventricular tachycardia and prevented sudden cardiac death (fig. S8A). ARM210 treatment also prevented SR Ca2+ leak via RyR2-R2474S channels (fig. S8B). Together, these in vivo and in vitro functional data show that the pathological state of RyR2-R2474S is effectively reversed by ARM210, in agreement with our single-particle cryo-EM data.
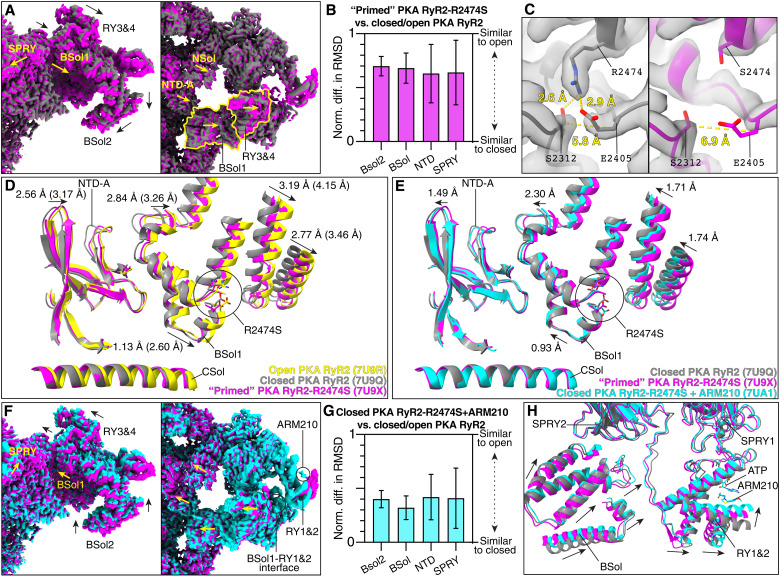
Visual inspection of the cryo-EM maps shows clear conformation changes that suggest a primed state of the PKA-phosphorylated RyR2-R2474S channels (Fig. 2A). To quantify the structural changes in the primed PKA-phosphorylated RyR2-R2474S channels, we measured the normalized differences in root mean square deviation (RMSD) between the primed PKA-phosphorylated RyR2-R2474S (PDB: 7U9X) and closed (PDB: 7U9Q) and open (PDB: 7U9R) PKA-phosphorylated RyR2 (Fig. 2B and table S3A). Values close to 0 indicate that the conformation is similar to the closed state, whereas values close to 1 indicate that the conformation is similar to the open state. Normalization allows the direct comparison between different domains. Our RMSD analysis showed an average value of ~0.7, indicating that the cytosolic domains of the primed PKA-phosphorylated RyR2-R2474S are in nearly open positions, reducing the energetic barrier of the primed state to adopt a fully open state. Thus, the primed PKA-phosphorylated RyR2-R2474S is more readily activated and promotes an SR Ca2+ leak during diastole when the activating [Ca2+]cyt is very low (~150 nM) and the WT channels are tightly closed.
Fig. 2. Stabilization of RyR2-R2474S by the Rycal ARM210.
(A) Cryo-EM maps of closed PKA-phosphorylated RyR2 (gray) and primed PKA-phosphorylated RyR2-R2474S (magenta) from the side (left) and top (right) views. Conformation changes are shown with arrows. (B) Normalized differences in RMSD of the primed PKA-phosphorylated RyR2-R2474S. (C) Close-up view of the region around residue 2474 of closed PKA-phosphorylated RyR2 (left) and primed PKA-phosphorylated RyR2-R2474S (right). Distances are measured between Cβ atoms. (D) Aligned models of closed PKA-phosphorylated RyR2 (PDB: 7U9Q, gray), open PKA-phosphorylated RyR2 (PDB: 7U9R, yellow), and primed PKA-phosphorylated RyR2-R2474S (PDB: 7U9X, magenta). Conformational changes are shown with arrows. Distances between closed PKA-phosphorylated RyR2 and primed PKA-phosphorylated RyR2-R2474S, and between closed and open PKA-phosphorylated RyR2 (in parentheses) are labeled. (E to G) Same as (A), (B), and (D) but including closed PKA-phosphorylated RyR2-R2474S + ARM210 (PDB: 7UA1, cyan). Distances between primed PKA-phosphorylated RyR2-R2474S and closed PKA-phosphorylated RyR2-R2474S + ARM210 are labeled (E). Changes introduced by the R2474S mutation are partially reversed by the addition of ARM210. The densities of ARM210 and BSol1-RY1&2 interface are highlighted (F, right). (H) Aligned models of closed PKA-phosphorylated RyR2 (PDB: 7U9Q, gray), primed PKA-phosphorylated RyR2-R2474S (PDB: 7U9X, magenta), and closed PKA-phosphorylated RyR2-R2474S + ARM210 (PDB: 7UA1, cyan). Conformational changes of the RY1&2 and BSol domains are shown with arrows.
It is surprising that a single point mutation can have such large effect over the cytosolic shell. Analyzing in detail the region of the mutation, the Arg to Ser mutation was readily identified because of the shortened side-chain density for the mutant residue S2474 compared to the WT residue R2474 (Fig. 2C). According to our models, R2474 stabilizes the surrounding structure via interactions with S2312 and E2405. In the mutant channel, this network is disrupted and the distance between residues S2312 and E2405 increases from 5.8 to 6.9 Å (Fig. 2C). The increased distances observed in the RyR2-R2474S mutant channel affects the helix-loop-helix motif formed by residues 2406 to 2418, which propagates to the adjacent NTD-A domain (Fig. 2D). Therefore, a single mutation destabilizes the interaction between the NTD and the BSol, which leads to destabilization of the entire cytosolic shell (NTD, SPRY, JSol, and BSol domains).
Incubation with the Rycal drug ARM210 partially reversed the primed state of PKA-phosphorylated RyR2-R2474S, placing the channel in a state closer to that of the closed PKA-phosphorylated RyR2 (Fig. 2, E and F, and movie S1). In this case, the RMSD analysis of closed PKA-phosphorylated RyR2-R2474S + ARM210 (PDB: 7UA1) showed a reduction of more than 50% toward the closed state in some domains compared to primed PKA-phosphorylated RyR2-R2474S (Fig. 2G and table S3B). Analyzing in detail the cryo-EM map of closed PKA-phosphorylated RyR2-R2474S in the presence of ARM210, we detected a clear density in the cleft of the RY1&2 domain (Fig. 2F and fig. S9, A and B). This agrees with our recent publication showing the presence of ARM210 in the cleft of the RY1&2 domain of RyR1 at a local resolution of 3.10 Å (31). Analysis of the rest of the cryo-EM map showed no additional densities that could correspond to ARM210. Not surprisingly, the ARM210 density was absent in the particles in the open state, in agreement with previous observations that Rycal compounds act by stabilizing the closed state of RyR2 (6, 37). In the absence of ARM210, the RY1&2 domain of RyR2 showed a weak density that we attribute to one ATP molecule as suggested by a previous report (fig. S9, A and C) (PDB: 6UHH) (30). In the presence of ARM210, this density is stronger and larger than one ATP molecule, suggesting that at least two molecules are present (ATP and ARM210; fig. S9D). However, the low local resolution limits our ability to model the precise position of both molecules. In addition, the RY1&2 domain adopts a conformation that closes around both ATP and ARM210 (Fig. 2H). Comparing PKA-phosphorylated RyR2-R2474S atomic models in the absence and presence of ARM210 (PDB: 7U9X versus PDB: 7UA1), we observed that the biggest changes were not in the contiguous SPRY1 domain, but instead in the adjacent BSol domain (Fig. 2H). The interface between the BSol1 and the RY1&2 domains showed enhanced density, suggesting that the transduction of the signal from the ARM210-loaded RY1&2 domain is through the stabilization of the BSol1 domains (Fig. 2F and fig. S9, B and E). On the basis of the model, the interface between the BSol1 and RY1&2 domains would be stabilized by His2995 in the BSol1 domain and Asp1070 in the RY1&2 domain; however, since the local resolution of this region is limited, Arg2988 (BSol1) and His1071 (RY1&2) may also be involved in strengthening the BSol1-RY1&2 interaction (fig. S9, E and F).
CaM stabilizes the closed PKA-phosphorylated RyR2-R2474S CPVT variant
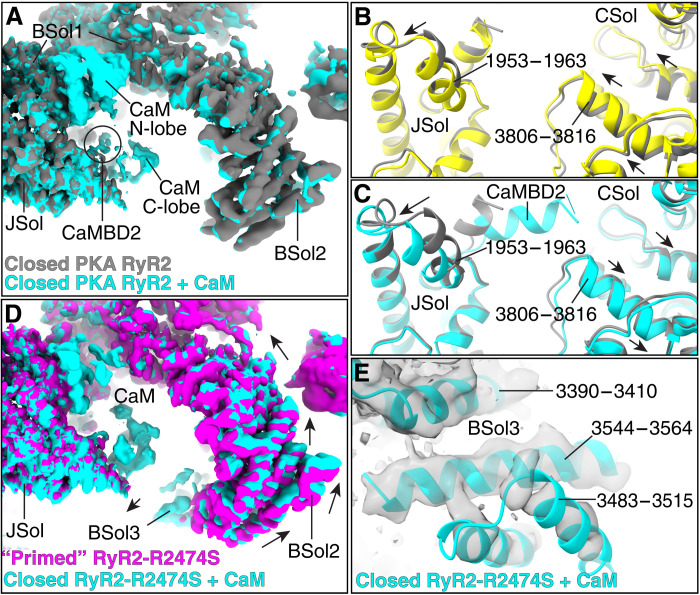
We analyzed the structure of closed PKA-phosphorylated RyR2 + CaM (PDB: 7U9T) and closed PKA-phosphorylated RyR2-R2474S + CaM (PDB: 7UA3) to better understand the role of CaM in CPVT. We found that, at a free Ca2+ concentration of 150 nM, CaM binds to PKA-phosphorylated RyR2 and PKA-phosphorylated RyR2-R2474S in the extended conformation that corresponds to the previously reported apo-CaM state (Fig. 3A and fig. S7E) (38). In the closed PKA-phosphorylated RyR2 + CaM (PDB: 7U9T), the apo-CaM N-lobe binds to the BSol1 domain but does not substantially affect the BSol1 conformation, suggesting an exclusive binding role for the N-lobe. The C-lobe binds to CaMBD2 or helix α-1 (3594 to 3604) and pushes the JSol domain outward, although this change is not propagated to the rest of the domain. CaM-bound helix α-1 also interacts with the α9 helix of the CSol (3806 to 3816), as previously reported (38), inducing small changes to the CSol domain in the opposite direction of the open state (Fig. 3, B and C). Although small, the changes observed in the CSol domain show that apo-CaM may counteract signals from the cytosolic domains, such as the phosphorylation of the RY3&4 domain and the changes resulting from CPVT-causing mutations found in the NTD and BSol domains, as previously described (39, 40). Not surprisingly, no particles in the open state showed substantial density of bound apo-CaM, confirming that binding of apo-CaM stabilizes the closed state.
Fig. 3. Stabilization of PKA-phosphorylated RyR2 and RyR2-R2474S by CaM.
(A) Aligned cryo-EM maps of closed PKA-phosphorylated RyR2 (gray) and closed PKA-phosphorylated RyR2 + CaM (cyan). (B) JSol and CSol models of closed PKA-phosphorylated RyR2 (PDB: 7U9Q, gray) and open PKA-phosphorylated RyR2 (PDB: 7U9R, yellow). Conformation changes are shown with arrows. (C) JSol and CSol models of closed PKA-phosphorylated RyR2 (PDB: 7U9Q, gray) and closed PKA-phosphorylated RyR2 + CaM (PDB: 7U9T, cyan). Conformation changes are shown with arrows. (D) Aligned cryo-EM maps of primed PKA-phosphorylated RyR2-R2474S (magenta) and closed PKA-phosphorylated RyR2-R2474S + CaM (cyan). Conformation changes are shown with arrows. CaM reverses the changes in the BSol2 domain introduced by the mutation by stabilizing the BSol3 domain. (E) Model with cryo-EM map of closed PKA-phosphorylated RyR2-R2474S + CaM (PDB: 7UA3) centered on the BSol3 domain that is stabilized by CaM.
In the case of closed PKA-phosphorylated RyR2-R2474S + CaM (PDB: 7UA3), apo-CaM reverses the changes in the BSol2 domain of the closed state introduced by the RyR2-R2474S mutation (Fig. 3D and fig. S7J). We suggest that this is also a result of apo-CaM stabilizing the BSol3 domain (Fig. 3, D and E), which interacts with the SPRY2, JSol, and CSol domains stabilizing the whole BSol domain. This is a surprising finding because the BSol3 domain was not detected in any previous RyR2 structures. Unlike PKA-phosphorylated RyR2 + CaM, we found that apo-CaM bound to the open PKA-phosphorylated RyR2-R2474S + CaM (PDB: 7UA4), where the cytosolic domains were further shifted downward and outward enhancing the open state (fig. S7K). This suggests that, in the presence of the activator xanthine, apo-CaM could also act as an activator of PKA-phosphorylated RyR2-R2474S, in agreement with an increased number of particles found for the open state in this dataset (60% compared to 10 to 20% for the other conditions). Looking in detail, we observed that the JSol of open PKA-phosphorylated RyR2-R2474S + CaM (fig. S7K), which is in direct contact with apo-CaM, is shifted downward, enhancing and stabilizing the open state.
PKA phosphorylation of RyR2 and the structure-function of the RY3&4 domain
As stated above, PKA treatment of the samples was performed to obtain hyperphosphorylated channels. We confirmed that residue RyR2-S2808 was targeted by PKA by specific immunoblots (fig. S1A). In agreement with previous reports (41), we found that RyR2-S2808 was already partially phosphorylated in HEK293 cells (approximately 30% according to quantification of immunoblots; fig. S1A). We showed by mass spectroscopic analysis that RyR2-S2808 was the major and only significantly PKA-phosphorylated site in RyR2 treated with PKA (coverage of 75%; fig. S10). We detected a secondary site with one order of magnitude lower intensity at RyR2-S1869 and other phosphorylation sites with even lower intensity at RyR2-T16, RyR2-S1856, RyR2-S2363, RyR2-S2814, and RyR2-S2822 (fig. S10A). Although being covered by two detected peptides (GRLLSLVEK and LLSLVEKVTYLKK), we found no phosphorylation of RyR2-S2031 (fig. S10B), a controversial PKA phosphorylation site detected by some investigators (42), and not detected by others (43, 44).
To determine the effect that PKA treatment has on the structures of RyR2 and the phosphorylation RY3&4 domain, we also pretreated WT RyR2 with phosphatase λ to obtain a completely dephosphorylated control (fig. S1B). We compared the structures of the dephosphorylated (deP RyR2) and PKA-phosphorylated (PKA RyR2) RyR2 channels. Analysis of the global structure showed that the cytosolic shell of the closed PKA-phosphorylated RyR2 (PDB: 7U9Q) showed a small shift outward and downward compared to the closed dephosphorylated RyR2 (PDB: 7UA5; fig. S7, A to D). RMSD analysis showed values of ~0.2 mainly in the BSol and SPRY domains, which are adjacent to the RY3&4 phosphorylation domain where the residue S2808 is located (fig. S11A, table S3C), suggesting that PKA phosphorylation has a “priming” effect. These changes would reduce the global energetic barrier for reaching the open state, thus sensitizing the channel to CICR. This conformational change is smaller than the one introduced by the CPVT mutation (~0.2 versus ~0.7). The cytosolic shell conformation remains closer to the closed state, in agreement with the physiological role of PKA phosphorylation and the pathological role of the CPVT mutation.
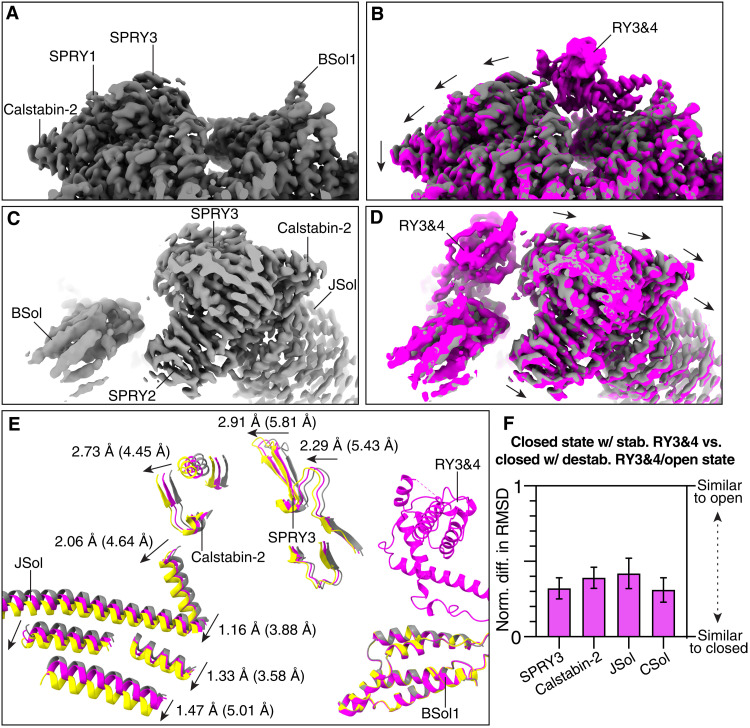
The RY3&4 phosphorylation domain had not been previously resolved in the cryo-EM structures of any RyRs due to its intrinsic dynamic behavior. We hypothesized that this dynamic nature of the RY3&4 domain would be necessary for interacting with modulator enzymes such as PKA and protein phosphatase 1 (PP1) (45). Therefore, we performed 3D variability analysis centered on the BSol containing the RY3&4 domain. This 3D variability analysis revealed two distinct populations: one where the RY3&4 domain was stabilized and could be resolved, and one where there was no substantial density, suggesting that RY3&4 is detached from the BSol domain (Fig. 4, A to D). In other words, in the same RyR2 particle, which has four RY3&4 domains, some of the RY3&4 domains are stabilized and some are destabilized. This behavior was observed in all the structures, suggesting that it is independent of the phosphorylation state, closed/open state, and WT/CPVT mutation (fig. S11B). The distribution of stabilized versus destabilized RY3&4 domains is roughly 50/50, but the limitations of this methodology prevent a precise measurement of the particle distribution.
Fig. 4. Structure-function relationship of the RY3&4 phosphorylation domain.
(A and B) Cryo-EM maps of the closed particles with destabilized (gray) and stabilized (magenta) RY3&4 domain. A downward shift in surrounding domains can be observed. Individual domains are labeled. (C and D) Same as (A) and (B) from a different point of view. (E) Aligned models of the closed state with destabilized (gray) and stabilized (magenta) RY3&4 domain, and open state (yellow). Models were aligned at the BSol1 domains to facilitate interpretation of conformational changes. When the RY3&4 domain is stabilized, the changes are in the same direction as the open state. Shown with arrows are the distance between the closed state with destabilized RY3&4 domain and the closed state with stabilized RY3&4 domain, and between the closed state with destabilized RY3&4 domain and the open state (in parentheses). (F) RMSD analysis of the closed state with stabilized RY3&4 domain. This analysis was performed on primed PKA-phosphorylated RyR2-R2474S because this dataset presents the most particles and best local refinement resolution.
To improve the local cryo-EM map of the RY3&4 domain, we clustered and analyzed the symmetry expanded particles with the stabilized RY3&4 domain. The best resulting cryo-EM map with a local resolution of 2.90 Å allowed us to build an atomic model of this domain with high confidence (fig. S11C). On the basis of the model, we observed weak interactions between the RY3&4 and BSol1 domains and almost no interaction between the RY3&4 and SPRY3 domains (fig. S11D). Our first hypothesis is that phosphorylation of RyR2-S2808 would strengthen the interaction between the RY3&4 and SPRY3 domains due to the introduction of a salt bridge between the negatively charged RyR2-pS2808 and either the positively charged RyR2-R1500 or RyR2-K1525 in SPRY3 (fig. S11E). The same conclusion can be obtained when analyzing the residue RyR2-S2814, which is preferentially phosphorylated by CaM-dependent protein kinase II (CaMKII) (46, 47). Hence, we suggest that the phosphorylation of either RyR2-S2808 or RyR2-S2814 stabilizes this conformation of the RY3&4 phosphorylation domain. However, we found no extra densities around these residues, suggesting that the interaction is still very dynamic.
Furthermore, we analyzed how the stabilization of the RY3&4 domain affects the surrounding RyR2 structure. Comparison to the destabilized RY3&4 domain in the closed state shows that stabilization of the RY3&4 domain increases the distance between the adjacent BSol1 and SPRY3 domains, adopting a conformation that is closer to the open conformation (Fig. 4, fig. S11F, and movie S2). This movement is propagated from the SPRY3 domain to the adjacent JSol and CSol domains. To quantify those changes, we again compared the normalized difference in RMSD between whole domains (Fig. 4, E and F, and table S3D). The RMSD analysis shows values in the range of 0.3 to 0.4, confirming that the stabilization of the RY3&4 domain has a priming effect (28, 36). In the case of the open channels, stabilization of the RY3&4 domain shows no effect on open dephosphorylated RyR2 but shows a small effect on open PKA-phosphorylated RyR2 and open PKA-phosphorylated RyR2-R2474S, confirming that the adopted conformation is similar to the open state and that PKA phosphorylation still has a priming effect on the open states (fig. S11G). Last, we cannot discard that binding of accessory proteins to the phosphorylation loop could shift the stabilization distribution of the RY3&4 domain to either completely destabilized or completely stabilized. It has been shown that phosphomimic at RyR2-S2814 induces the formation of an α helix (45), and this could be a substrate for protein-protein interaction that could further stabilize or destabilize the RY3&4 domain. Additional experiments to detect protein partners of the phosphorylated RY3&4 domain are necessary.
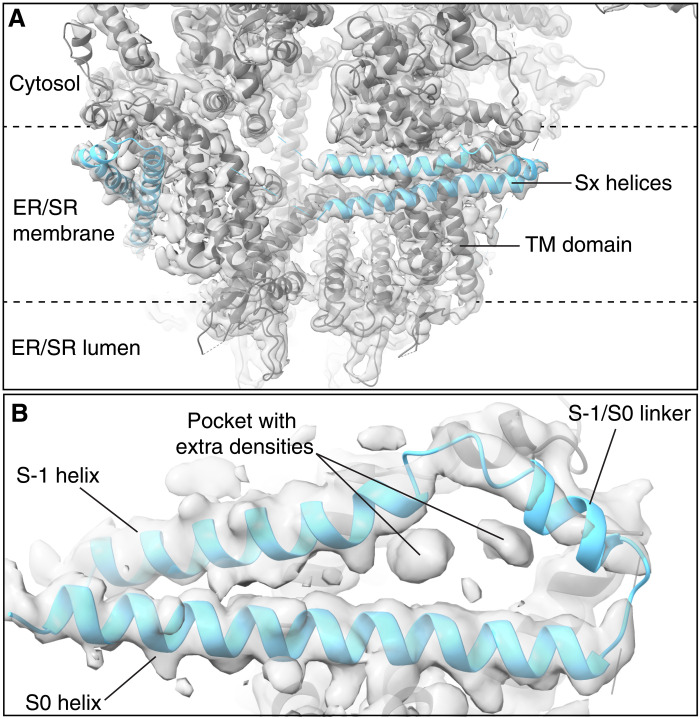
Auxiliary intramembrane helices
We observed two intramembrane helices laterally positioned and encircling the TM domain (Fig. 5A). While these densities were previously observed, they were attributed to detergent and lipids (48). These auxiliary helices (Sx) were predicted to exist in the RyR2 sequence upstream of the six transmembrane helices that form the pore (S1 to S6) (49). We used the Jpred4 algorithm (50) to predict the secondary structure of the fragment, which was previously reported to encompass these helices (4150 to 4350; fig. S12A). We modeled the predicted Sx helices because of the presence of well-resolved densities for large side chains, including W4288 as well as several tyrosine and phenylalanine residues (fig. S12B). Since the helices are upstream of the S1 to S6 helices, we named them S-1 (4238 to 4259), S-1/S0 linker (4262 to 4271), and S0 (4277 to 4309), in keeping with the current nomenclature. In addition, we detected a pocket formed by the interactions with helices S1, S2, and S3, where we found two densities that could correspond to detergent molecules or protein fragments (Fig. 5B). The S-1/S0 linker contains several positively charged lysine residues and is positioned at the cytosolic surface of the SR membrane, where these lysine residues would be able to interact with the negatively charged phospholipid head groups or other transmembrane proteins (fig. S12D). The Sx density is strong in the closed states but weaker in the open states (fig. S12E), suggesting that they may play a role in stabilizing the RyR2 closed state, as previously reported (49). The CaM binding motif CaMBD3 is also contained in this sequence, consistent with the recently solved crystal structure (51). In the presence of CaM, the Sx density was absent only in the open state. This suggests that, in the closed state of RyR2, the CaMBD3 motif would be inaccessible to CaM, but in the open state of RyR2, it would interact with CaM and completely destabilize the Sx helices.
Fig. 5. Resolution of auxiliary intramembrane helices in RyR2.
Model with overlapped cryo-EM map of PKA-phosphorylated RyR2 (PDB: 7U9Q) highlighting the Sx helices from the side view (A) and bottom view (B). Auxiliary helices and pocket with extra densities are labeled.
DISCUSSION
Mechanism of RyR2 leak in CPVT
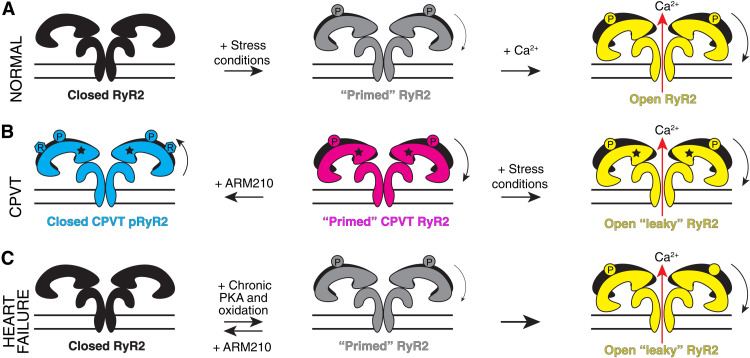
In CPVT patients, SR Ca2+ leak occurs via mutant RyR2 channels during diastole when the heart is supposed to be electrically silent, resulting in afterdepolarizations, arrhythmias, and eventually sudden cardiac death. As a mechanism underlying RyR2-associated CPVT, we propose that intense exercise and adrenergic stimulation cause two independent but synergistic events that affect RyR2. The β-adrenergic response to exercise (i) results in PKA phosphorylation of RyR2 mainly at S2808 and (ii) activates SR Ca2+ uptake via SERCA2a, thus increasing the SR Ca2+ load, and the driving force for Ca2+ leak out of the membrane via RyR2 channels. By virtue of being in a primed state, the CPVT variant RyR2-R2474S is likely more sensitive to channel-activating posttranslational modifications, resulting in a diastolic SR Ca2+ leak that can trigger fatal cardiac arrhythmias during intense exercise (Fig. 6, A and B).
Fig. 6. Proposed mechanism of CPVT-related RyR2 variants, other gain-of-function mutants, and heart failure.
(A) Schematic representation of the normal function of RyR2. (B) Schematic representation of the CPVT-related Ca2+ leak during diastole under intense exercise or stress conditions. In the case of CPVT variants, the resting state is already in a primed state. This correlates with the higher open probability during exercise or stress conditions, which results in opening during diastole, afterdepolarizations, arrhythmias, and sudden cardiac death. This pathological state can be reversed by treatment with the Rycal ARM210 (R). This basal primed state scenario could be a shared mechanism among other RyR1 and RyR2 gain-of-function mutants. (C) Schematic representation of the heart failure–related primed state and Ca2+ leak. The heart failure–related primed state remains hypothetical as no structure has been solved yet.
We have previously shown that binding of activators (Ca2+, ATP, and caffeine) at the TaF-CTD interfaces of RyR1 initiates a rearrangement of the activation core, which primes these domains to adopt a conformation and orientation poised for pore dilation: the primed state (36). In the present study, we show that the CPVT mutation R2474S puts the channel into a primed state that is independent of the binding of the activators including Ca2+. This explains why the mutant CPVT channel RyR2-R2474S is able to be inappropriately activated during diastole when the [Ca2+]cyt is too low to activate the WT RyR2 channel. This inappropriate activation of the mutant channel results in diastolic SR Ca2+ leak and triggers fatal cardiac arrhythmias (8).
We demonstrate the stabilizing effects of the Rycal ARM210 and CaM through two different mechanisms. One involves the stabilization of the BSol domain through the interaction with the ARM210-bound RY1&2 domain, and the other occurs via the stabilization of the CSol and BSol3 domains through the binding of CaM to CAMBD2 and the BSol domain. These findings provide a better understanding of RyR2-related pathological cardiac disorders and the mechanisms underlying the potential therapeutic actions of the novel Rycal class of drugs.
Mechanism of action of drug-induced stabilization of leaky RyR2 channels
Together, the data in the present study show that the CPVT mutation disrupts local interactions that destabilize the BSol domain and induces a primed state. This primed state leads to inappropriate opening of RyR2 channels that can be reversed by treatment with the RyR2 stabilizer ARM210. ARM210 binds to a cleft in the RY1&2 domain where it stabilizes interactions between key residues that are required to reduce flexibility between domains, particularly with the BSol1 domain, of the cytosolic shell. The net effect of ARM210 binding is to stabilize the overall channel structure closer to the closed state. This renders the channel less likely to be inappropriately activated during diastole when the conditions favor the closed state of the channel (e.g., low nonactivating [Ca2+]cyt). Melville et al. (31) showed high-resolution binding of ARM210 to the cleft of the RY1&2 domain of RyR1 and presented functional data of single point mutations in the cleft of the RY1&2 domain, confirming that this specific ARM210 binding site is the bona fide drug binding site. High homology between RY1&2 domains of RyR1 and RyR2, and the absence of any extra density that could correspond to ARM210 in our cryo-EM maps suggest that the same is true for RyR2.
Calstabin-2 depletion in cardiac pathologies
We had previously proposed that leaky RyR2 is related to calstabin-2 depletion in CPVT and other cardiac pathologies (6). In this case, we detected no differences in calstabin-2 binding in our cryo-EM structures between WT versus CPVT mutant channels, open versus closed states, or PKA-phosphorylated versus dephosphorylated states. We attribute this finding to the purification methodology: Since we add an excess of glutathione S-transferase (GST)–calstabin-2 to purify RyR2, RyR2 structures are saturated with calstabin-2. On the other hand, we have previously shown that Rycal drugs revert this depletion, although we detected no conformational changes in the vicinity of calstabin-2 in the presence of ARM210. To address the effect of ARM210 on the calstabin-2 binding site, we would need to resolve the structure of RyR2 in the absence of calstabin-2. On the other hand, RyR affinity for calstabin have been shown to be dependent on the conformational state of RyRs, having higher affinity in the closed state [dissociation constant (Kd) = 0.1 to 0.01 pM] than in the open state (Kd = 1 nM) (52, 53). The primed state would likely have a reduced affinity for calstabin-2, as we previously showed in the case of RyR2-R2474S and other RyR2 CPVT variants (8). Moreover, RyR2 affinity for calstabin-2 could be further reduced because of the effects of posttranslational modifications (e.g., phosphorylation, oxidation, or nitrosylation), as part of a vicious cycle that amplifies SR Ca2+ leak, which in turn further oxidizes RyR2 and increases the leak (6). Binding of ARM210 would stabilize the closed state over the primed state, which would increase the affinity for calstabin-2 and indirectly reduce the oxidative modifications by diminishing the Ca2+ leak.
A role for xanthine
Exercise up-regulates the purine catabolism pathway, temporarily increasing the levels of xanthine through the activation of xanthine oxidoreductase (XOR) (35, 54). XOR is present in the SR of cardiomyocytes (55) and colocalizes with RyR2 (56). Hence, the increased XOR activity may produce locally increased xanthine concentrations that bind to RyR2 and contribute to activation of the channel. Xanthine activation of RyRs has been reported with a concentration threshold lower than the commonly used activator caffeine (32). Structurally, xanthine and caffeine are very similar, having the same scaffold structure but the latter being trimethylated (fig. S6E). Xanthine is predicted to be more polar because of the lack of the methyl groups, but solubility in water is much lower: 0.069 versus 21.6 mg/ml (fig. S6E). This is explained in part by the absolute planarity of xanthine, which boosts the π-π self-stacking association and lowers the water solubility, as seen for other purines (57). We suggest that these properties would increase the polar interaction with residue RyR2-Y4944 and the π-π interaction with residue RyR2-W4645, increasing the affinity of this binding site for xanthine compared to caffeine.
CaM and CPVT
CaM has been closely linked to CPVT (58) not only because CaM is essential to protecting against CPVT (40) but also because mutations in CaM are also linked to CPVT in patients (59). CaM is a key modulator in Ca2+ signaling, binding to RyR2 and other related proteins, and activating CaMKII in the presence of Ca2+ (60). CaM is a physiological inhibitor of RyR2 and binds to the BSol domain, where residue RyR2-R2474 is located (38). CaM has also been reported to preferentially inhibit PKA-phosphorylated RyR2 channels (39).
In resting conditions, when xanthine and Ca2+ are not present, CaM also reverses the conformation changes in the CPVT-linked RyR2-R2474S mutant channel by stabilizing the BSol3 domain. However, impaired binding between RyR2 and CaM has been reported for the PKA-phosphorylated RyR2-R2474S mutant channel (61), preventing physiological CaM from executing its inhibitory role. In agreement, enhancing binding between RyR2-R2474S and CaM reverses the pathological effects in cardiomyocytes and mice models (22, 40). In a sense, together, the CPVT mutation, PKA phosphorylation, and reduced CaM binding likely play a role in destabilizing the closed state of the mutant channel and promoting leak that triggers fatal cardiac arrhythmias.
Comparison to other gain-of-function RyR mutants
Few RyR mutant cryo-EM structures have been published so far (29, 62). The CPVT-related mouse RyR2-R176Q structure was found to have a similar conformation as the WT mouse RyR2, showing no structural evidence of a primed state (29). However, some technical differences, such as the absence of the activator ATP, the absence of PKA phosphorylation, and the presence of nanodiscs only in the mutant channel, might obscure differences between WT and CPVT structures. The other possible explanation is that R176Q is related to a weak CPVT phenotype, leading to imperceptible changes in the resting state, and the need for stress conditions to achieve a primed state.
In the case of RyR1, mutant rabbit RyR1-R164C (29) and porcine RyR1-R615C (62) structures have been solved. Although these mutants are related to human malignant hyperthermia, which is an RyR1-related Ca2+ leak pathology induced by inhalation of volatile anesthetics, the structures of these mutant channels showed a similar primed or “pathological intermediate” state in the absence of activators. This presents an opportunity to generate a hypothesis that pathological gain-of-function RyR mutants induce a basal primed state, which can be opened under stress conditions in the absence of an activating signal, leading to Ca2+ leak and the onset of the pathology (Fig. 6B). This could also be the case in heart failure, where chronic hyperphosphorylation and oxidation have been detected and could lead to a persistent primed state and Ca2+ leak (Fig. 6C).
The study with porcine RyR1-R615C also included the analysis of the interaction between the mutant channels and CaM (62). Apo-CaM interaction with porcine RyR1-R615C disrupts the original “intermediate” conformation, leading to a closed state and an open state that looks more open, especially at the level of the BSol. This is surprisingly similar to what we have reported here with PKA-phosphorylated RyR2-R2474S in the presence of CaM. Apo-CaM disrupts the RyR2-R2474S primed state to achieve a closed state and an open state, which looks more open. Unlike RyR2, CaM acts as an activator of RyR1 at low Ca2+ concentration. Although there is high homology between RyR1 and RyR2, there are some differences, especially in the BSol domain. While in RyR1 the BSol is mostly stable and all residues can be detected, in RyR2 the BSol is more flexible: RyR2 BSol1 is always detected, BSol2 is only detected after the constant presence of calstabin-2, and BSol3 is always absent, except in our PKA-phosphorylated RyR2-R2474S + CaM. We hypothesize that the increased flexibility of the BSol domain due to the RyR2-R2474S CPVT mutation leads to a BSol behavior intermediate between RyR1 and RyR2, and therefore, CaM binding has a similar effect to porcine RyR1-R615C. Even more, in the presence of xanthine, CaM could act as an activator of PKA-phosphorylated RyR2-R2474S as seen for RyR1, adding another level of pathogenicity to the concurrence of CPVT mutation, PKA phosphorylation, CaM, and exercise-induced xanthine production.
MATERIALS AND METHODS
Generation of stable cell lines
Complementary DNA (cDNA) for human RyR2 was subcloned into the A1.2 vector in two steps, using the Nhe I–Xho I fragment and then the Not I–Nhe I fragment of human RYR2. Human RYR2 was inserted 3′ of a cytomegalovirus (CMV) promoter and followed in 5′ by internal ribosomal entry site (IRES)–green fluorescent protein (GFP). The sequence of the construct was confirmed by Sanger sequencing and restriction digestion with Afl III enzyme. The resulting vector (G418-resistant) was cotransfected into HEK293T cells with a plasmid carrying a puromycin resistance gene using calcium chloride. Cells were maintained in a G418- and puromycin-containing medium for approximately 3 weeks and then underwent two cycles of clonal selection, where the top 0.1% of the most highly fluorescent cells were propagated (63).
Generation of human RYR2-R2474S DNA and HEK293 transfection
Constructs expressing RyR2-R2474S were formed by introducing the respective mutation into fragments of human RYR2 using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent). Three nucleotide changes were introduced (WT sequence: AGGGTCTAT to R2474S mutant: AGCGTATAC). The first is the mutation R2474S, and the other two are silent mutations introducing BstZ17I restriction site (GTATAC) to facilitate screening for mutant clones. Each fragment was subcloned into a full-length human RYR2 construct in pCMV5 vector and confirmed by sequencing and expressed in 293T/17 cells using Lipofectamine 2000 (Thermo Fisher Scientific). For final expression, HEK293 cells grown in 150-mm dishes with Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (Invitrogen), penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM l-glutamine were cotransfected with 25 mg per dish of human RYR2-R2474S cDNA using PEI MAX at a 1:5 ratio (Polysciences) (64). Cells were collected 48 hours after transfection.
Purification and treatment of recombinant human RyR2
All purification steps were performed on ice unless otherwise stated. HEK293 cells (25 to 50 dishes) expressing human RyR2 or RyR2-R2474S were harvested by centrifugation for 10 min at 1500g. The pellet fraction was resuspended in tris malate buffer [10 mM tris malate (pH 6.8), 1 mM EGTA, 1 mM dithiothreitol (DTT), 1 mM menzamidine, 0.5 mM 4-benzenesulfonyl fluoride hydrochloride (AEBSF), and protease inhibitor cocktail] and was sonicated with six pulses of 20 s at 35% amplitude. The membrane fraction was precipitated by centrifugation at 100,000g for 30 min and was resuspended with a glass homogenizer in CHAPS buffer [10 mM Hepes (pH 7.4), 1 M NaCl, 1.5% CHAPS, 0.5% phosphatidylcholine (PC), 1 mM EGTA, 2 mM DTT, 0.5 mM AEBSF, 1 mM benzamidine, and protease inhibitor cocktail]. The sample was diluted 1:4 with the same buffer without NaCl. To achieve stabilization of peripherical domains, we added 100 nmol of GST–calstabin-2 to both tris malate buffer and CHAPS buffer. The remaining insoluble material was separated with a second centrifugation at 100,000g for 30 min. The supernatant—containing detergent-solubilized human RyR2—was filtered and loaded into a 5-ml HiTrap Q HP column (Cytiva) previously equilibrated with buffer A [10 mM Hepes (pH 7.4), 0.4% CHAPS, 1 mM EGTA, 0.001% dioleoylphosphatidylcholine (DOPC), 250 mM NaCl, and 0.5 mM TCEP (tris(2-carboxyethyl)phosphine)]. The HiTrap Q HP column was eluted with a linear gradient between 300 and 600 mM NaCl. The fractions containing human RyR2 (300 to 350 mM NaCl) were pooled, and 100 nmol of GST–calstabin-2 was added. The pooled fractions were loaded into a 1-ml GSTrap HP column (Cytiva), which was left recirculating overnight. The GSTrap HP column was washed with buffer A and eluted with glutathione buffer {10 mM Hepes (pH 8), 0.4% CHAPS, 1 mM EGTA, 0.001% DOPC, 200 mM NaCl, 10 mM GSH [glutathione (reduced form)], and 1 mM DTT}. Immediately after, a second 1-ml HiTrap Q HP column (Cytiva) binding/elution step was applied to separate human RyR2 from the excess of unbound GST–calstabin-2 and GSH. Simultaneous cleavage of GST tag and PKA phosphorylation was performed by addition of 50 U of thrombin and 100 U of PKA (+ 10 mM EGTA, 8 mM MgCl2, and 100 μM ATP for activity), respectively, for 30 min on ice. For samples requiring dephosphorylation treatment, PKA was replaced by 2000 U of phosphatase lambda (Pλ from NEB, + 1× Protein MetalloPhosphatases (PMP) buffer, and 1 mM MnCl2). The sample was concentrated to 0.5 ml, and a gel filtration step was run with TSKgel G4SWXL (TOSOH Biosciences) with buffer A. RyR2 fractions were pooled and concentrated to a concentration of 4 to 8 mg/ml (with centrifugal filters of 100-kDa cutoff) and were filtered (with centrifugal filters of 0.22-μm cutoff) to eliminate aggregates. To resemble exercise diastolic conditions, 10 mM NaATP, 500 μM xanthine, 150 nM Ca2+ free (650 μM total Ca2+), and 200 μM cAMP were added to all samples. For conditions with CaM or ARM210: 20 μM CaM was added to PKA-phosphorylated RyR2, 40 μM CaM was added to PKA-phosphorylated RyR2-R2474S, and 500 μM ARM210 was added to PKA-phosphorylated RyR2-R2474S. MaxChelator webserver was used to calculate total/free Ca2+ concentrations (65). Quality control was assessed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblots using anti–RyR-5029 and anti–RyR2-pS2809 antibodies for total and phosphorylated RyR2, respectively (66).
Purification of recombinant GST–calstabin-2
Recombinant human GST–calstabin-2 was expressed in BL21 (DE3) Escherichia coli cells with a thrombin protease cleavage site between GST and calstabin-2. Protein expression was induced with 0.8 mM isopropyl-β-d-thiogalactopyranoside (IPTG) added to E. coli at an OD600 (optical density at 600 nm) of 0.8 with overnight incubation at 18°C before centrifugation for 10 min at 6500g. The pellets were resuspended in buffer A (phosphate-buffered saline + 0.5 mM AEBSF) and lysed using an emulsiflex (Avestin EmulsiFlex-C3). The lysate was pelleted by centrifugation for 10 min at 100,000g. The supernatant was then loaded into a 5-ml GSTrap HP column (Cytiva) and washed with 5 column volume (CV) of buffer A to remove contaminants before elution with buffer B [tris (pH 8), 2 mM DTT, and 20 mM glutathione]. Fractions containing GST–calstabin-2 were pooled, concentrated, and dialyzed overnight at 4°C into buffer A. final concentration was determined by spectroscopy using NanoDrop 1000 (Thermo Fisher Scientific) with absorbance at 280 nm and an extinction coefficient of 46,200 M−1 cm−1. GST–calstabin-2 was stored at −80°C.
Purification of recombinant CaM and TEV protease
Recombinant human CaM was expressed in BL21 (DE3) E. coli cells with an N-terminal 6-histidine tag and a tobacco etch virus (TEV) protease cleavage site. Protein expression was induced with 0.8 mM IPTG added to E. coli at an OD600 of 0.8 with overnight incubation at 18°C before centrifugation for 10 min at 6500g and storage at −80°C. CaM was purified using a two-step 5-ml HisTrap HP column (Cytiva) purification. In brief, the pellets were resuspended in buffer A [20 mM Hepes (pH 7.5), 150 mM NaCl, 20 mM imidazole, 5 mM 2-Mercaptoethanol, and 0.5 mM AEBSF] and lysed using an emulsiflex (Avestin EmulsiFlex-C3). The lysate was pelleted by centrifugation for 10 min at 100,000g. The supernatant was then loaded over a HisTrap column and washed with 5 CV of buffer A to remove contaminants before elution using a linear gradient from buffer A to buffer B (buffer A containing 500 mM imidazole). Fractions containing CaM were pooled, 1 to 2 mg of purified TEV protease was added, and the mixture was dialyzed overnight at 4°C into buffer C (buffer A with no imidazole). CaM was then loaded onto a HisTrap column with the flowthrough collected and the wash fractionated to retain fractions containing CaM before elution of TEV and any remaining contaminants with a linear gradient from buffer C to buffer B. The flowthrough and any fractions containing CaM were pooled, concentrated to >2 mM, and determined by spectroscopy using NanoDrop 1000 (Thermo Fisher Scientific) with absorbance at 280 nm and the extinction coefficient of CaM (3000 M−1 cm−1). CaM was stored at −20°C. TEV protease was purified in the same manner except for using an uncleavable his-tag and thus ending after the first HisTrap column wherein the purified protease was stored at −80°C in buffer C with 10% glycerol.
Cryo-EM sample preparation and data collection
The final sample (3 μl) was applied to UltrAuFoil holey gold grids (Quantifoil R 0.6/1.0, Au 300) previously cleaned with easiGlow (PELCO). Grids were blotted with ashless filter paper (Whatman) using blot force 10 and blot time 8 s before vitrification by plunge-freezing into liquid ethane chilled with liquid nitrogen using Vitrobot Mark IV (Thermo Fisher Scientific) operated at 4°C with 100% relative humidity.
Prepared grids were screened in-house on a Glacios Cryo-TEM (Thermo Fisher Scientific) microscope with a 200-kV x-FEG source and a Falcon 3EC direct electron detector (Thermo Fisher Scientific). Microscope operations and data collection were carried out using EPU software (Thermo Fisher Scientific). High-resolution data collection was performed at Columbia University on a Titan Krios 300-kV (Thermo Fisher Scientific) microscope equipped with an energy filter (slit width 20 eV) and a K3 direct electron detector (Gatan). Data were collected using Leginon (67) and at a nominal magnification of ×105,000 in electron counting mode, corresponding to a pixel size of 0.83 Å. The electron dose rate was set to 16 e−/pixel per second with 2.5-s exposures for a total dose of 50 to 60 e/Å2.
Cryo-EM data processing and model building
Cryo-EM data processing was performed in cryoSPARC (68) with image stacks aligned using Patch motion, defocus value estimation by Patch CTF estimation. Particle picking was performed using the template picker with templates created from preexisting cryo-EM maps. Particles were subjected to 2D classification in cryoSPARC with 100 classes. Particles from the highest-resolution classes were pooled for ab initio 3D reconstruction with a single class followed by homogeneous refinement with C4 symmetry imposed. 3D variability and further clustering were performed using a mask comprising the TM domain to separate those particles in the closed and open states followed by heterogeneous refinement with four classes to further select the best particles. C4 symmetry expansion was performed before local refinements. The masks used were TaF + TM + CTD domains (residues 4131 to 4967), calstabin-2 + NTD + SPRY domains (residues 1 to 1646), JSol + CSol domains (residues 1700 to 2476, 3590 to 4130), and BSol domain (residues 2400 to 3344). Only the TaF + TM + CTD mask used C4 symmetry. Smaller masks, for a second round of 3D variability analysis, clustering, and local refinement, were RY1&2 (residues 862 to 1076), RY3&4 (residues 2685 to 2909), and BSol2 (residues 3042 to 3344). Local refinements used a dynamical mask with a far distance of 10, 50, and 150 Å for the initial masks, small masks, and expanded RY3&4 mask for comparing the effect of RY3&4 stabilization, respectively. The resulting maps were combined in ChimeraX (69) to generate a composite map before calibration of the pixel size using correlation coefficients with a map generated from the crystal structure of the NTD of RyR2 (4JKQ) (70). The pixel size was altered by 0.001 Å per step, up to 20 steps in each direction.
Initial model of RYR2 was generated with Phenix tool sculptor (71) from the 2.45-Å structure of RyR1 (PDB: 7TZC). Truncated residues were manually corrected to obtain the full side chains. Only domains RY1&2 and RY3&4, which show weak cryo-EM density and confidence in structure, were based on crystallographic structures. The initial model of RyR2 RY1&2 domain was generated with Phenix tool sculptor from the RY1&2 domain of RyR3 with ATP (PDB: 6UHH). The model of RyR2 RY3&4 domain was obtained as it is (PDB: 4ETV) (72). Calstabin-2 and CaM models were obtained as they are (PDB: 6JI8) (38). Model building was performed in Coot (73) and refined with Phenix tool RealSpaceRefine (71). Figures of the final structure were created using ChimeraX (69). Cryo-EM statistics are summarized in table S1.
Normalized difference in RMSD analyses: Pairwise 1-domain comparison
The analysis performed here aims to determine the conformation of a certain query domain from a model X relative to the reference closed and open states. To this end, the RMSD of Cα of the query domain is measured between model X and the closed state (RMSDX-closed), and between model X and the open state (RMSDX-open). The normalized difference in RMSD is calculated as RMSDX-closed/(RMSDX-closed + RMSDX-open). A value of 0 means that the conformation of the query domain is identical to the same domain in the closed state. A value of 1 means that the conformation of the query domain is identical to the same domain in the open state. A value of 0.5 means that the conformation of the query domain is equally distant to the closed and open states. The error, considered as the intrinsic variability between atomic models, was calculated as the RMSD between aligned domains of the respective models (i.e., CSol for analysis in table S3, A to C, and BSol1 for analysis in table S3D). Propagation of error was calculated using the webserver uncertaintycalculator.com. Bar graphs were made with GraphPad Prism software. RMSD was measured in ChimeraX (69). When needed, previous alignment of the atomic models centered on the control domains was performed.
Telemetric electrocardiogram recordings
Mice were implanted with radio telemetry transmitters (Data Sciences International) as described in detail elsewhere (74). Briefly, the transmitter (PhysioTel, ETA-F10 transmitter) was inserted in mice subcutaneously along the back under general anesthesia (5% inhaled isoflurane). Two electrocardiogram (ECG) electrodes were placed hypodermically in the region of the right shoulder (negative pole) and toward the lower left chest (positive pole) to approximate lead II of the Einthoven surface ECG. During the procedure, respiratory and cardiac rhythm, adequacy of anesthetic depth, muscle relaxation, body temperature, and analgesia were monitored to avoid anesthesia-related complications. Postoperating pain was considered during a 1-week postimplantation period, and carprofen (5 mg/kg, subcutaneously) was given. A minimum period of 2 weeks was allowed for recovery from the surgery. Animals were housed in individual stainless steel cages for telemetry recordings. Environmental parameters were recorded continuously and maintained within a fixed range: room temperature at 15° to 21°C and 45 to 65% relative humidity. The artificial day/night cycle was 12-hour light/12-hour dark with light on at 0700 hours. Drinking water was provided ad libitum. Solid diet (300 g) was given daily in the morning. ECG waveforms were continuously recorded at a sampling rate of 2000 Hz using a signal transmitter-receiver (RPC-1) connected to a data acquisition system (Ponemah system, Data Sciences International). ARM210 (50 mg/kg per day) was given in drinking water for 2 weeks before ECG recordings. Mice were recorded for 1 hour at baseline and then given epinephrine injection (1 mg/kg, intraperitoneally) and recorded for another 2 hours. The animal used in the study were maintained and studied according to protocols approved by the Institutional Animal Care and Use Committee of Columbia University (reference no. AC-AABP1551).
SR Ca2+ leak assay
Cardiac muscle SR microsomes were prepared by homogenizing heart samples on ice using a Teflon glass homogenizer (50 times) with 2 volumes of 20 mM tris-maleate (pH 7.4), 1 mM EDTA, 1 mM DTT, and protease inhibitors (Roche). Homogenate was then centrifuged at 4000g for 15 min at 4°C, and the following supernatant was centrifuged at 50,000g for 45 min at 4°C. Pellets were resuspended in lysis buffer containing 300 mM sucrose. Microsomes (5 μg/ml) were diluted into a buffer (pH 7.2) containing 8 mM K-phosphocreatine, and creatine kinase (2 U/ml), mixed with 3 μM Fluo-4 and added to multiple wells of a 96-well plate. Ca2+ loading of the microsomes was initiated by adding 1 mM ATP. After Ca2+ uptake (50 s), 3 μM thapsigargin was added to inhibit the Ca2+ reuptake by SERCA. SR Ca2+ leak was measured by the increase in intensity of the Fluo-4 signal (measured in a Tecan fluorescence plate reader). Ca2+ leak was quantified as the difference between the average Fluo-4 signal before and after addition of thapsigargin. Graphs were plotted with GraphPad Prism software.
Single-channel recordings
ER vesicles from HEK293 cells expressing RyR2 were prepared by homogenizing cell pellets on ice using a Teflon glass homogenizer with two volumes of solution containing 20 mM tris-maleate (pH 7.4), 1 mM EDTA, 1 mM DTT, and protease inhibitors (Roche). Homogenate was then centrifuged at 4000g for 15 min at 4°C, and the resulting supernatant was centrifuged at 40,000g for 30 min at 4°C. The final pellet, containing the ER fractions, was resuspended and aliquoted in 250 mM sucrose, 10 mM Mops (pH 7.4), 1 mM EDTA, 1 mM DTT, and protease inhibitors. Samples were frozen in liquid nitrogen and stored at −80°C. ER vesicles were fused to planar lipid bilayers formed by painting a lipid mixture of phosphatidylethanolamine and PC (Avanti Polar Lipids) in a 5:3 ratio in decane across a 200-μm hole in polysulfonate cups (Warner Instruments) separating two chambers. The trans chamber (1.0 ml), representing the intra-SR (luminal) compartment, was connected to the head stage input of a bilayer voltage clamp amplifier. The cis chamber (1.0 ml), representing the cytoplasmic compartment, was held at virtual ground. The following asymmetrical solutions were used: for the cis solution, 1 mM EGTA, 250/125 mM Hepes/tris, 50 mM KCl (pH 7.35); for the trans solution, 53 mM Ca(OH)2, 50 mM KCl, 250 mM Hepes (pH 7.35). The concentration of free Ca2+ in the cis chamber was calculated as previously described. ER vesicles were added to the cis side, and fusion with the lipid bilayer was induced by making the cis side hyperosmotic by the addition of 400 to 500 mM KCl. After the appearance of potassium and chloride channels, the cis side was perfused with the cis solution. At the end of each experiment, 10 μM ryanodine was added to block the RyR2 channel. Single-channel currents were recorded at 0 mV using Bilayer Clamp BC-525D (Warner Instruments), filtered at 1 kHz using Low-Pass Bessel Filter 8 Pole (Warner Instruments), and digitized at 4 kHz. All experiments were performed at room temperature (23°C). Data acquisition was performed by using Digidata 1322A and Axoscope 10.1 software (Axon Instruments). The recordings were analyzed using Clampfit 10.1 (Molecular Devices) and GraphPad Prism software.
Mass spectrometry analyses
ER vesicles from HEK293 cells expressing RyR2 were separated on 4 to 12% gradient SDS-PAGE and sent for mass spectrometry analysis to in-house Columbia Proteomics Shared Resource (HICCC). Protein gel slices were excised, and in-gel digestion was performed as previously described (75), with minor modifications. Digested peptides were collected and further extracted from gel slices in extraction buffer (1:2 ratio by volume of 5% formic acid:acetonitrile) at high speed, shaking in an air thermostat. Peptides were separated within 80 min at a flow rate of 400 nl/min on a reversed-phase C18 column with an integrated CaptiveSpray Emitter (25 cm × 75 μm, 1.6 μm, IonOpticks). Mobile phases A and B were with 0.1% formic acid in water and 0.1% formic acid in acetonitrile. The fraction of B was linearly increased from 2 to 23% within 70 min, followed by an increase to 35% within 10 min and a further increase to 80% before reequilibration. The timsTOF Pro was operated in parallel accumulation–serial fragmentation (PASEF) mode (76) with the following settings: mass range, 100 to 1700 mass/charge ratio (m/z); 1/K0 start, 0.6 V·s/cm2; end, 1.6 V·s/cm2; ramp time, 100 ms; lock duty cycle to 100%; capillary voltage, 1600 V; dry gas, 3 l/min; and dry temperature, 200°C. PASEF settings: 10 Tandem Mass Spectrometry frames (1.16 s duty cycle); charge range, 0 to 5; active exclusion for 0.4 min; target intensity, 20,000; intensity threshold, 2500; and collision-induced dissociation collision energy, 59 eV. A polygon filter was applied to the m/z and ion mobility plane to select features most likely representing peptide precursors rather than singly charged background ions. Acquired PASEF raw files were analyzed using the MaxQuant environment v.2.0.1.0 and Andromeda for database searches at default settings with a few modifications (76). MaxQuant was set up to search with the reference human proteome database downloaded from UniProt. The following modifications were used for protein identification and quantification: Carbamidomethylation of cysteine residues (+57.021 Da) was set as static modifications, while the oxidation of methionine residues (+15.995 Da), deamidation (+0.984) on asparagine and glutamine, and phosphorylation (+79.966) on serine, threonine, and tyrosine were set as a variable modification. Results obtained from MaxQuant, Phospho (STY) sites table was used for RyR2 phospho-site quantification. Sequence coverage was obtained with the software Scaffold 5.
Acknowledgments
Some of this work was performed at the Columbia University Cryo-Electron Microscopy Center and New York Structural Biology Center. We would also like to acknowledge the molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from the National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases. We would like to acknowledge O. Clarke, F. Mancia, and members of their laboratories for helpful discussions and input, and R. Grassucci and Z. Zhang from the cryo-EM facility and R. Soni from the proteomics facility for their essential services.
Funding: These studies were supported by NIH grants R01HL145473, R01DK118240, R01HL142903, R01HL140934, R01AR070194, R25HL156002R25, R25NS076445, and T32 HL120826 (to A.R.M.).
Author contributions: Conceptualization: A.R.M. Methodology: M.C.M., G.W., Z.M., S.R., H.D., and A.R.M. Investigations: M.C.M., G.W., H.D., Q.Y., Y.L., A.W., L.S., and S.R. Validation: M.C.M., G.W., Z.M., Q.Y., and S.R. Formal analysis: M.C.M., A.R.M., G.W., Z.M., Q.Y., S.R., and A.W. Resources: A.R.M. Data curation: M.C.M., Q.Y., and S.R. Writing—original draft: M.C.M. Writing—review and editing: M.C.M., G.W., Z.M., and A.R.M. Visualization: M.C.M., Z.M., Q.Y., and S.R. Supervision: A.R.M. Project administration: A.R.M. Funding acquisition: A.R.M.
Competing interests: A.R.M. is a member of the scientific advisory board and board of directors and an equity owner at ARMGO Inc. Columbia University also owns equity in ARMGO Inc. A.R.M. has patent applications or pending or awarded patents, including U.S. 2014/0378437 and U.S. 7,718,644. The authors declare no other competing interests.
Data and materials availability: Atomic models and cryo-EM maps are publicly available at wwPDB and EMDB under the following accession codes, respectively: 7U9Q and EMD-26405 (closed state of PKA-phosphorylated hRyR2), 7U9R and EMD-26407 (open state of PKA-phosphorylated hRyR2), 7U9T and EMD-26408 (closed state of PKA-phosphorylated hRyR2 + CaM), 7U9X and EMD-26409 (primed state of PKA-phosphorylated hRyR2-R2474S), 7U9Z and EMD-26410 (open state of PKA-phosphorylated hRyR2-R2474S), 7UA1 and EMD-26412 (closed state of PKA-phosphorylated hRyR2-R2474S + ARM210), 7UA3 and EMD-26413 (closed state of PKA-phosphorylated hRyR2-R2474S + CaM), 7UA4 and EMD-26414 (open state of PKA-phosphorylated hRyR2-R2474S + CaM), 7UA5 and EMD-26415 (closed state of dephosphorylated hRyR2), and 7UA9 and EMD-26416 (open state of dephosphorylated hRyR2).
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Tables S1 to S3
Other Supplementary Material for this manuscript includes the following:
Movies S1 and S2
REFERENCES AND NOTES
- 1.Flucher B. E., Andrews S. B., Fleischer S., Marks A. R., Caswell A., Powell J. A., Triad formation: Organization and function of the sarcoplasmic reticulum calcium release channel and triadin in normal and dysgenic muscle in vitro. J. Cell Biol. 123, 1161–1174 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Otsu K., Willard H. F., Khanna V. K., Zorzato F., Green N. M., MacLennan D. H., Molecular cloning of cDNA encoding the Ca2+ release channel (ryanodine receptor) of rabbit cardiac muscle sarcoplasmic reticulum. J. Biol. Chem. 265, 13472–13483 (1990). [PubMed] [Google Scholar]
- 3.Nakai J., Imagawa T., Hakamat Y., Shigekawa M., Takeshima H., Numa S., Primary structure and functional expression from cDNA of the cardiac ryanodine receptor/calcium release channel. FEBS Lett. 271, 169–177 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Fabiato A., Calcium-induced release of calcium from the cardiac sarcoplasmic reticulum. Am. J. Physiol. 245, C1–C14 (1983). [DOI] [PubMed] [Google Scholar]
- 5.Marx S. O., Reiken S., Hisamatsu Y., Jayaraman T., Burkhoff D., Rosemblit N., Marks A. R., PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): Defective regulation in failing hearts. Cell 101, 365–376 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Shan J., Xie W., Betzenhauser M., Reiken S., Chen B. X., Wronska A., Marks A. R., Calcium leak through ryanodine receptors leads to atrial fibrillation in 3 mouse models of catecholaminergic polymorphic ventricular tachycardia. Circ. Res. 111, 708–717 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehnart S. E., Wehrens X. H., Laitinen P. J., Reiken S. R., Deng S. X., Cheng Z., Landry D. W., Kontula K., Swan H., Marks A. R., Sudden death in familial polymorphic ventricular tachycardia associated with calcium release channel (ryanodine receptor) leak. Circulation 109, 3208–3214 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Wehrens X. H., Lehnart S. E., Huang F., Vest J. A., Reiken S. R., Mohler P. J., Sun J., Guatimosim S., Song L. S., Rosemblit N., D’Armiento J. M., Napolitano C., Memmi M., Priori S. G., Lederer W. J., Marks A. R., FKBP12.6 deficiency and defective calcium release channel (ryanodine receptor) function linked to exercise-induced sudden cardiac death. Cell 113, 829–840 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Hayashi M., Denjoy I., Extramiana F., Maltret A., Buisson N. R., Lupoglazoff J. M., Klug D., Hayashi M., Takatsuki S., Villain E., Kamblock J., Messali A., Guicheney P., Lunardi J., Leenhardt A., Incidence and risk factors of arrhythmic events in catecholaminergic polymorphic ventricular tachycardia. Circulation 119, 2426–2434 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Lehnart S. E., Mongillo M., Bellinger A., Lindegger N., Chen B. X., Hsueh W., Reiken S., Wronska A., Drew L. J., Ward C. W., Lederer W. J., Kass R. S., Morley G., Marks A. R., Leaky Ca2+ release channel/ryanodine receptor 2 causes seizures and sudden cardiac death in mice. J. Clin. Invest. 118, 2230–2245 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Priori S. G., Corr P. B., Mechanisms underlying early and delayed afterdepolarizations induced by catecholamines. Am. J. Physiol. 258, H1796–H1805 (1990). [DOI] [PubMed] [Google Scholar]
- 12.Dobrev D., Wehrens X. H., Role of RyR2 phosphorylation in heart failure and arrhythmias: Controversies around ryanodine receptor phosphorylation in cardiac disease. Circ. Res. 114, 1311–1319; discussion 1319 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kontula K., Laitinen P. J., Lehtonen A., Toivonen L., Viitasalo M., Swan H., Catecholaminergic polymorphic ventricular tachycardia: Recent mechanistic insights. Cardiovasc. Res. 67, 379–387 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Shan J., Betzenhauser M. J., Kushnir A., Reiken S., Meli A. C., Wronska A., Dura M., Chen B. X., Marks A. R., Role of chronic ryanodine receptor phosphorylation in heart failure and β-adrenergic receptor blockade in mice. J. Clin. Invest. 120, 4375–4387 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shan J., Kushnir A., Betzenhauser M. J., Reiken S., Li J., Lehnart S. E., Lindegger N., Mongillo M., Mohler P. J., Marks A. R., Phosphorylation of the ryanodine receptor mediates the cardiac fight or flight response in mice. J. Clin. Invest. 120, 4388–4398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wehrens X. H., Marks A. R., Altered function and regulation of cardiac ryanodine receptors in cardiac disease. Trends Biochem. Sci. 28, 671–678 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Uchinoumi H., Yano M., Suetomi T., Ono M., Xu X., Tateishi H., Oda T., Okuda S., Doi M., Kobayashi S., Yamamoto T., Ikeda Y., Ohkusa T., Ikemoto N., Matsuzaki M., Catecholaminergic polymorphic ventricular tachycardia is caused by mutation-linked defective conformational regulation of the ryanodine receptor. Circ. Res. 106, 1413–1424 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Priori S. G., Napolitano C., Tiso N., Memmi M., Vignati G., Bloise R., Sorrentino V., Danieli G. A., Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation 103, 196–200 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Wehrens X. H., Lehnart S. E., Reiken S. R., Deng S. X., Vest J. A., Cervantes D., Coromilas J., Landry D. W., Marks A. R., Protection from cardiac arrhythmia through ryanodine receptor-stabilizing protein calstabin2. Science 304, 292–296 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Penttinen K., Swan H., Vanninen S., Paavola J., Lahtinen A. M., Kontula K., Aalto-Setala K., Antiarrhythmic effects of dantrolene in patients with catecholaminergic polymorphic ventricular tachycardia and replication of the responses using iPSC models. PLOS ONE 10, e0125366 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi S., Yano M., Uchinoumi H., Suetomi T., Susa T., Ono M., Xu X., Tateishi H., Oda T., Okuda S., Doi M., Yamamoto T., Matsuzaki M., Dantrolene, a therapeutic agent for malignant hyperthermia, inhibits catecholaminergic polymorphic ventricular tachycardia in a RyR2(R2474S/+) knock-in mouse model. Circ. J. 74, 2579–2584 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Fukuda M., Yamamoto T., Nishimura S., Kato T., Murakami W., Hino A., Ono M., Tateishi H., Oda T., Okuda S., Kobayashi S., Koseki N., Kyushiki H., Yano M., Enhanced binding of calmodulin to RyR2 corrects arrhythmogenic channel disorder in CPVT-associated myocytes. Biochem. Biophys. Res. Commun. 448, 1–7 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Zalk R., Clarke O. B., des Georges A., Grassucci R. A., Reiken S., Mancia F., Hendrickson W. A., Frank J., Marks A. R., Structure of a mammalian ryanodine receptor. Nature 517, 44–49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zalk R., Marks A. R., Ca2+ release channels join the ‘Resolution Revolution’. Trends Biochem. Sci. 42, 543–555 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Efremov R. G., Leitner A., Aebersold R., Raunser S., Architecture and conformational switch mechanism of the ryanodine receptor. Nature 517, 39–43 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Yan Z., Bai X., Yan C., Wu J., Li Z., Xie T., Peng W., Yin C., Li X., Scheres S. H. W., Shi Y., Yan N., Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature 517, 50–55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng W., Shen H., Wu J., Guo W., Pan X., Wang R., Chen S. R., Yan N., Structural basis for the gating mechanism of the type 2 ryanodine receptor RyR2. Science 354, aah5324 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Chi X., Gong D., Ren K., Zhou G., Huang G., Lei J., Zhou Q., Yan N., Molecular basis for allosteric regulation of the type 2 ryanodine receptor channel gating by key modulators. Proc. Natl. Acad. Sci. U.S.A. 116, 25575–25582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer K. A., Hu Y., Nayak A. R., Kurebayashi N., Murayama T., Samso M., Structural mechanism of two gain-of-function cardiac and skeletal RyR mutations at an equivalent site by cryo-EM. Sci. Adv. 6, eabb2964 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popova O. B., Baker M. R., Tran T. P., Le T., Serysheva I. I., Identification of ATP-binding regions in the RyR1 Ca2+ release channel. PLOS ONE 7, e48725 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melville Z., Dridi H., Yuan Q., Reiken S., Wronska A., Liu Y., Clarke O. B., Marks A. R., A drug and ATP binding site in type 1 ryanodine receptor. Structure, (2022). [DOI] [PubMed] [Google Scholar]
- 32.Butanda-Ochoa A., Hojer G., Diaz-Munoz M., Modulation of the skeletal muscle Ca2+ release channel/ryanodine receptor by adenosine and its metabolites: A structure-activity approach. Bioorg. Med. Chem. 11, 3029–3037 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Yisireyili M., Hayashi M., Wu H., Uchida Y., Yamamoto K., Kikuchi R., Shoaib Hamrah M., Nakayama T., Cheng X. W., Matsushita T., Nakamura S., Niwa T., Murohara T., Takeshita K., Xanthine oxidase inhibition by febuxostat attenuates stress-induced hyperuricemia, glucose dysmetabolism, and prothrombotic state in mice. Sci. Rep. 7, 1266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maimaiti Y., Aikebaier A., Maimaitiaili A., Wubulikasimu W., Li Y. L., Aziguli A., Yan J., Kelimu A., Effects of psychological stress on xanthine oxidase expression, activity and related markers in adipose tissue of mice. Zhongguo Ying Yong Sheng Li Xue Za Zhi 35, 537–542 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Zielinski J., Slominska E. M., Krol-Zielinska M., Krasinski Z., Kusy K., Purine metabolism in sprint- vs endurance-trained athletes aged 20–90 years. Sci. Rep. 9, 12075 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.des Georges A., Clarke O. B., Zalk R., Yuan Q., Condon K. J., Grassucci R. A., Hendrickson W. A., Marks A. R., Frank J., Structural basis for gating and activation of RyR1. Cell 167, 145–157.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lehnart S. E., Terrenoire C., Reiken S., Wehrens X. H., Song L. S., Tillman E. J., Mancarella S., Coromilas J., Lederer W. J., Kass R. S., Marks A. R., Stabilization of cardiac ryanodine receptor prevents intracellular calcium leak and arrhythmias. Proc. Natl. Acad. Sci. U.S.A. 103, 7906–7910 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong D., Chi X., Wei J., Zhou G., Huang G., Zhang L., Wang R., Lei J., Chen S. R. W., Yan N., Modulation of cardiac ryanodine receptor 2 by calmodulin. Nature 572, 347–351 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Walweel K., Gomez-Hurtado N., Rebbeck R. T., Oo Y. W., Beard N. A., Molenaar P., Dos Remedios C., van Helden D. F., Cornea R. L., Knollmann B. C., Laver D. R., Calmodulin inhibition of human RyR2 channels requires phosphorylation of RyR2-S2808 or RyR2-S2814. J. Mol. Cell. Cardiol. 130, 96–106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura Y., Yamamoto T., Kobayashi S., Tamitani M., Hamada Y., Fukui G., Xu X., Nishimura S., Kato T., Uchinoumi H., Oda T., Okuda S., Yano M., Ryanodine receptor-bound calmodulin is essential to protect against catecholaminergic polymorphic ventricular tachycardia. JCI Insight 4, e126112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao B., Zhong G., Obayashi M., Yang D., Chen K., Walsh M. P., Shimoni Y., Cheng H., Ter Keurs H., Chen S. R., Ser-2030, but not Ser-2808, is the major phosphorylation site in cardiac ryanodine receptors responding to protein kinase A activation upon beta-adrenergic stimulation in normal and failing hearts. Biochem. J. 396, 7–16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao B., Jiang M. T., Zhao M., Yang D., Sutherland C., Lai F. A., Walsh M. P., Warltier D. C., Cheng H., Chen S. R., Characterization of a novel PKA phosphorylation site, serine-2030, reveals no PKA hyperphosphorylation of the cardiac ryanodine receptor in canine heart failure. Circ. Res. 96, 847–855 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Campbell H. M., Quick A. P., Abu-Taha I., Chiang D. Y., Kramm C. F., Word T. A., Brandenburg S., Hulsurkar M., Alsina K. M., Liu H. B., Martin B., Uhlenkamp D., Moore O. M., Lahiri S. K., Corradini E., Kamler M., Heck A. J. R., Lehnart S. E., Dobrev D., Wehrens X. H. T., Loss of SPEG inhibitory phosphorylation of ryanodine receptor type-2 promotes atrial fibrillation. Circulation 142, 1159–1172 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J., Imtiaz M. S., Beard N. A., Dulhunty A. F., Thorne R., vanHelden D. F., Laver D. R., ß-Adrenergic stimulation increases RyR2 activity via intracellular Ca2+ and Mg2+ regulation. PLOS ONE 8, e58334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haji-Ghassemi O., Yuchi Z., Van Petegem F., The cardiac ryanodine receptor phosphorylation hotspot embraces PKA in a phosphorylation-dependent manner. Mol. Cell 75, 39–52.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Wehrens X. H., Lehnart S. E., Reiken S. R., Marks A. R., Ca2+/calmodulin-dependent protein kinase II phosphorylation regulates the cardiac ryanodine receptor. Circ. Res. 94, e61–e70 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Kushnir A., Shan J., Betzenhauser M. J., Reiken S., Marks A. R., Role of CaMKIIdelta phosphorylation of the cardiac ryanodine receptor in the force frequency relationship and heart failure. Proc. Natl. Acad. Sci. U.S.A. 107, 10274–10279 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dhindwal S., Lobo J., Cabra V., Santiago D. J., Nayak A. R., Dryden K., Samso M., A cryo-EM-based model of phosphorylation- and FKBP12.6-mediated allosterism of the cardiac ryanodine receptor. Sci. Signal. 10, eaai8842 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Du G. G., Avila G., Sharma P., Khanna V. K., Dirksen R. T., MacLennan D. H., Role of the sequence surrounding predicted transmembrane helix M4 in membrane association and function of the Ca(2+) release channel of skeletal muscle sarcoplasmic reticulum (ryanodine receptor isoform 1). J. Biol. Chem. 279, 37566–37574 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Drozdetskiy A., Cole C., Procter J., Barton G. J., JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Q., Anderson D. E., Kaur R., Fisher A. J., Ames J. B., The crystal structure of calmodulin bound to the cardiac ryanodine receptor (RyR2) at residues Phe4246-Val4271 reveals a fifth calcium binding site. Biochemistry 60, 1088–1096 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones J. L., Reynolds D. F., Lai F. A., Blayney L. M., Ryanodine receptor binding to FKBP12 is modulated by channel activation state. J. Cell Sci. 118, 4613–4619 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Blayney L. M., Jones J. L., Griffiths J., Lai F. A., A mechanism of ryanodine receptor modulation by FKBP12/12.6, protein kinase A, and K201. Cardiovasc. Res. 85, 68–78 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Dudzinska W., Suska M., Lubkowska A., Jakubowska K., Olszewska M., Safranow K., Chlubek D., Comparison of human erythrocyte purine nucleotide metabolism and blood purine and pyrimidine degradation product concentrations before and after acute exercise in trained and sedentary subjects. J. Physiol. Sci. 68, 293–305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan S. A., Lee K., Minhas K. M., Gonzalez D. R., Raju S. V., Tejani A. D., Li D., Berkowitz D. E., Hare J. M., Neuronal nitric oxide synthase negatively regulates xanthine oxidoreductase inhibition of cardiac excitation-contraction coupling. Proc. Natl. Acad. Sci. U.S.A. 101, 15944–15948 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonzalez D. R., Treuer A. V., Castellanos J., Dulce R. A., Hare J. M., Impaired S-nitrosylation of the ryanodine receptor caused by xanthine oxidase activity contributes to calcium leak in heart failure. J. Biol. Chem. 285, 28938–28945 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tavagnacco L., Schnupf U., Mason P. E., Saboungi M. L., Cesaro A., Brady J. W., Molecular dynamics simulation studies of caffeine aggregation in aqueous solution. J. Phys. Chem. B 115, 10957–10966 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wleklinski M. J., Kannankeril P. J., Knollmann B. C., Molecular and tissue mechanisms of catecholaminergic polymorphic ventricular tachycardia. J. Physiol. 598, 2817–2834 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crotti L., Spazzolini C., Tester D. J., Ghidoni A., Baruteau A. E., Beckmann B. M., Behr E. R., Bennett J. S., Bezzina C. R., Bhuiyan Z. A., Celiker A., Cerrone M., Dagradi F., De Ferrari G. M., Etheridge S. P., Fatah M., Garcia-Pavia P., Al-Ghamdi S., Hamilton R. M., Al-Hassnan Z. N., Horie M., Jimenez-Jaimez J., Kanter R. J., Kaski J. P., Kotta M. C., Lahrouchi N., Makita N., Norrish G., Odland H. H., Ohno S., Papagiannis J., Parati G., Sekarski N., Tveten K., Vatta M., Webster G., Wilde A. A. M., Wojciak J., George A. L., Ackerman M. J., Schwartz P. J., Calmodulin mutations and life-threatening cardiac arrhythmias: Insights from the International Calmodulinopathy Registry. Eur. Heart J. 40, 2964–2975 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoeflich K. P., Ikura M., Calmodulin in action: Diversity in target recognition and activation mechanisms. Cell 108, 739–742 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Xu X., Yano M., Uchinoumi H., Hino A., Suetomi T., Ono M., Tateishi H., Oda T., Okuda S., Doi M., Kobayashi S., Yamamoto T., Ikeda Y., Ikemoto N., Matsuzaki M., Defective calmodulin binding to the cardiac ryanodine receptor plays a key role in CPVT-associated channel dysfunction. Biochem. Biophys. Res. Commun. 394, 660–666 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woll K. A., Haji-Ghassemi O., Van Petegem F., Pathological conformations of disease mutant ryanodine receptors revealed by cryo-EM. Nat. Commun. 12, 807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mancia F., Patel S. D., Rajala M. W., Scherer P. E., Nemes A., Schieren I., Hendrickson W. A., Shapiro L., Optimization of protein production in mammalian cells with a coexpressed fluorescent marker. Structure 12, 1355–1360 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Longo P. A., Kavran J. M., Kim M. S., Leahy D. J., Transient mammalian cell transfection with polyethylenimine (PEI). Methods Enzymol. 529, 227–240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bers D. M., Patton C. W., Nuccitelli R., A practical guide to the preparation of Ca2+ buffers. Methods Cell Biol. 99, 1–26 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Reiken S., Wehrens X. H., Vest J. A., Barbone A., Klotz S., Mancini D., Burkhoff D., Marks A. R., Beta-blockers restore calcium release channel function and improve cardiac muscle performance in human heart failure. Circulation 107, 2459–2466 (2003). [DOI] [PubMed] [Google Scholar]
- 67.Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C. S., Carragher B., Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., Ferrin T. E., UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borko L., Bauerova-Hlinkova V., Hostinova E., Gasperik J., Beck K., Lai F. A., Zahradnikova A., Sevcik J., Structural insights into the human RyR2 N-terminal region involved in cardiac arrhythmias. Acta Crystallogr. D Biol. Crystallogr. 70, 2897–2912 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liebschner D., Afonine P. V., Baker M. L., Bunkoczi G., Chen V. B., Croll T. I., Hintze B., Hung L. W., Jain S., McCoy A. J., Moriarty N. W., Oeffner R. D., Poon B. K., Prisant M. G., Read R. J., Richardson J. S., Richardson D. C., Sammito M. D., Sobolev O. V., Stockwell D. H., Terwilliger T. C., Urzhumtsev A. G., Videau L. L., Williams C. J., Adams P. D., Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuchi Z., Lau K., Van Petegem F., Disease mutations in the ryanodine receptor central region: Crystal structures of a phosphorylation hot spot domain. Structure 20, 1201–1211 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Dridi H., Liu X., Yuan Q., Reiken S., Yehia M., Sittenfeld L., Apostolou P., Buron J., Sicard P., Matecki S., Thireau J., Menuet C., Lacampagne A., Marks A. R., Role of defective calcium regulation in cardiorespiratory dysfunction in Huntington’s disease. JCI Insight 5, e140614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M., In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Meier F., Brunner A. D., Koch S., Koch H., Lubeck M., Krause M., Goedecke N., Decker J., Kosinski T., Park M. A., Bache N., Hoerning O., Cox J., Rather O., Mann M., Online parallel accumulation–serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics 17, 2534–2545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S1 to S3
Movies S1 and S2