Abstract
Within the overlapping geographical ranges of P. knowlesi monkey hosts and vectors in Southeast Asia, an estimated 1.5 billion people are considered at risk of infection. P. knowlesi can cause severe disease and death, the latter associated with delayed treatment occurring from misdiagnosis. Although microscopy is a sufficiently sensitive first-line tool for P. knowlesi detection for most low-level symptomatic infections, misdiagnosis as other Plasmodium species is common, and the majority of asymptomatic infections remain undetected. Current point-of-care rapid diagnostic tests demonstrate insufficient sensitivity and poor specificity for differentiating P. knowlesi from other Plasmodium species. Molecular tools including nested, real-time, and single-step PCR, and loop-mediated isothermal amplification (LAMP), are sensitive for P. knowlesi detection. However, higher cost and inability to provide the timely point-of-care diagnosis needed to guide appropriate clinical management has limited their routine use in most endemic clinical settings. P. knowlesi is likely underdiagnosed across the region, and improved diagnostic and surveillance tools are required. Reference laboratory molecular testing of malaria cases for both zoonotic and non-zoonotic Plasmodium species needs to be more widely implemented by National Malaria Control Programs across Southeast Asia to accurately identify the burden of zoonotic malaria and more precisely monitor the success of human-only malaria elimination programs. The implementation of specific serological tools for P. knowlesi would assist in determining the prevalence and distribution of asymptomatic and submicroscopic infections, the absence of transmission in certain areas, and associations with underlying land use change for future spatially targeted interventions.
1. Introduction
The recent increase in zoonotic transmission of the Cercopithecinae Old World monkey malaria parasite Plasmodium knowlesi, endemic to Southeast Asia, exemplifies how intensive anthropogenic land-use change is driving emerging disease spill-over to humans (Brock et al., 2019). Although a single presumed anomalous human P. knowlesi infection was first reported in the 1960s in Peninsular Malaysia (Chin et al., 1965), it was not until 2004 that Singh and colleagues using molecular detection methods identified a large number of naturally acquired human knowlesi malaria cases in Sarawak, East Malaysia (Singh et al., 2004). Molecular analysis of archival samples in Sarawak (Lee et al., 2009a) and Thailand (Jongwutiwes et al., 2011) demonstrated that in this region P. knowlesi had been misreported on routine microscopy at least as far back as the mid-1990s. Regional under-reporting of P. knowlesi infections is likely occurring, with PCR confirmed infections now documented from all countries in Southeast Asia where its major natural macaque hosts (Macaca fascicularis and M. nemestrina) and Anopheles Leucosphyrus group vectors are present (Herdiana et al., 2016; Iwagami et al., 2018; Lubis et al., 2017; Moyes et al., 2014; Singh and Daneshvar, 2013).
Changes in human land use are thought to play a major role in driving the increase in P. knowlesi exposure to humans, with the relative influence of associated environmental factors such as forest fragmentation varying at different spatial scales (Brock et al., 2019; Fornace et al., 2019b). Increased interaction between humans, vectors and the monkey parasite reservoir results from adaptive behaviours in response to these changing landscapes (Fornace et al., 2019a; Stark et al., 2019). Differences in monkey and human host biting preferences between An. leucosphyrus group species may also influence spatial variation in P. knowlesi transmission patterns (Vythilingam, 2010). Vector adaptation includes features such as earlier outside peak biting times in the evening for An. balabacensis, an incriminated P. knowlesi vector in Sabah (Wong et al., 2015). Adaptation to human-to-human transmission as a potential underlying driver of the emergence of P. knowlesi has not been evident in studies to date comparing parasite genetic lineages between macaque and human hosts (Divis et al., 2015; Jeslyn et al., 2011; Jongwutiwes et al., 2011; Lee et al., 2011). However, vector competence for this transmission mode has been experimentally proven (Chin et al., 1968), with incriminated P. knowlesi vectors such as An. dirus and An. balabacensis also being the primary vector for non-zoonotic Plasmodium species in certain areas (Marchand et al., 2011; Vythilingam, 2010). A decrease in cross-protective human immunity from the reduction of malaria transmission due to P. vivax may also be contributing to the prevalence of symptomatic disease or patent P. knowlesi infections detectable by microscopy in endemic areas (Muh et al., 2020).
Technological advances in malaria detection methods, specifically the advent of highly sensitive and specific molecular tools, have allowed accurate confirmation of P. knowlesi human infections across Southeast Asia (Singh and Daneshvar, 2013). However, first-line point-of-care testing for suspected malaria in most co-endemic settings in Southeast Asia primarily involves microscopy, which is inherently unreliable for diagnosing P. knowlesi due to morphological similarities with P. malariae and P. falciparum. Further complicating microscopic diagnosis, P. knowlesi co-infections particularly with P. vivax are common in endemic areas (Singh and Daneshvar, 2013). Current lateral flow malaria rapid diagnostic assays designed for non-zoonotic malaria species have shown poor sensitivity and specificity for P. knowlesi detection, particularly at the low parasite counts often seen even in symptomatic infections (Barber et al., 2013b; Grigg et al., 2014). Molecular tools including nested, real-time, and single-step PCR, and loop-mediated isothermal amplification (LAMP), have now been designed for P. knowlesi detection (Singh and Daneshvar, 2013). However, higher cost and inability to provide the timely point-of-care diagnosis needed to guide appropriate clinical management has limited their routine use in most endemic clinical settings.
Molecular tools used for research surveillance purposes, including in monkey hosts and mosquito vectors, have highlighted key knowledge gaps in estimating regional variation in the transmission and disease burden of P. knowlesi (Shearer et al., 2016). This includes the detection of underreported asymptomatic or submicroscopic infections (Fornace et al., 2016b; Grignard et al., 2019; Imwong et al., 2019; Jongwutiwes et al., 2011; Lubis et al., 2017; Marchand et al., 2011). In this context, understanding of P. knowlesi transmission has been further enhanced by the recent development of serological surveillance tools for P. knowlesi indicating past exposure. These multiplex sero-surveillance platforms also offer the potential for more detailed understanding of factors associated with exposure to P. knowlesi and other malaria species over time (Herman et al., 2018). Use of these sero-surveillance tools has already provided important insights into P. knowlesi transmission and specific environmental associations, including identifying infection exposure in demographic groups such as children and women thought to be at lower risk (Fornace et al., 2019b).
The emergence of P. knowlesi in Southeast Asia, with an intractable monkey parasite reservoir and ongoing misidentification with other Plasmodium species, threatens regional malaria elimination goals (Chin et al., 2020; Cox-Singh and Singh, 2008). In Malaysia, implementation of routine PCR for species confirmation of all malaria cases since 2012 has enabled public health authorities to demonstrate the near-elimination of P. falciparum and P. vivax malaria, alongside a concurrent increase in P. knowlesi cases in high-risk groups such as adult males with a history of forest exposure or agricultural work (Chin et al., 2020; Hussin et al., 2020; WHO, 2020). Molecular tools have also allowed accurate descriptions of the disease spectrum of P. knowlesi, ranging from asymptomatic infections, to severe malaria in 6%–9% (Grigg et al., 2018a; Singh and Daneshvar, 2013), and fatalities in 2.5 per 1000 cases (Rajahram et al., 2019). However, routine molecular confirmation of cases is not possible in many resource constrained settings, and WHO do not specifically include PCR-confirmed P. knowlesi case notifications in global yearly reporting (Barber et al., 2017b; WHO, 2020). Thus, the true regional prevalence and disease burden of P. knowlesi infections remains poorly understood, with limited use of accurate detection methods outside research settings.
In 2017 the WHO convened an evidence review group to guide research priorities in response to the public health threat posed by the emergence of P. knowlesi, which included a major recommendation to develop and improve laboratory detection methods (WHO, 2017). The following sections review current detection methods for both point-of-care diagnosis and surveillance of P. knowlesi human infections.
2. Point-of-care diagnosis
2.1. Microscopy
Microscopy remains the gold standard for malaria diagnosis in most endemic countries, as it is inexpensive, fast, sensitive and allows parasite quantification. Microscopy does however require trained laboratory technicians and equipment, an electricity supply and ongoing quality assurance procedures (WHO, 2015a). Older malaria microscopy teaching or reference tools generally in routine use in Southeast Asia do not include descriptions of P. knowlesi or comment on similarities with P. malariae or P. falciparum. Formal public health malaria reporting by microscopy in most countries outside Malaysia have not yet added provisions for suspected P. knowlesi. P. malariae remains the conventional reporting default, further limiting accurate regional estimates on the true burden of disease (Barber et al., 2013; Cox-Singh et al., 2008). Morphological similarities between P. knowlesi and P. malariae have allowed the existence of human P. knowlesi cases to previously remain undiagnosed. Misdiagnosis of P. knowlesi as the more benign P. malariae has also contributed to a lack of recognition of severe disease, with important adverse clinical and treatment outcomes (Rajahram et al., 2012; William et al., 2011), including case-fatalities (Rajahram et al., 2019). In contrast to P. knowlesi, P. malariae preferentially infects older senescent red blood cells (RBCs), which in addition to a slow 72-h replication cycle and host immunity results in chronic infections with low parasite counts (Collins and Jeffery, 2007). In P. knowlesi endemic areas, infections appearing morphologically similar to P. malariae, particularly with higher parasite counts, should be reported and treated as P. knowlesi with PCR confirmation for final reporting purposes where possible (Barber et al., 2017a,b).
2.1.1. P. knowlesi morphology in human infections
The first descriptions of the morphology of P. knowlesi in experimental inoculated human infections by Knowles and Das Gupta in 1932 commented on the similarity with P. malariae, including little or no amoeboid activity, and normal size of infected red blood cells (Knowles and Gupta, 1932). Sinton and Mulligan more extensively detailed the 24-h life-cycle of the parasite, the distinctive stippling occasionally observed in schizont-infected cells (different from that seen in P. vivax) and the presence of an accessory chromatin dot (Sinton and Mulligan, 1933), originally thought to be diagnostic for P. knowlesi but subsequently found to be present in other simian Plasmodium species (Coatney et al., 1971).
More recently, Lee et al. in 2009 described detailed microscopy findings in 10 human P. knowlesi cases from Sarawak, Malaysia (as in Fig. 1). Early trophozoites resembled those of P. falciparum, with a ring-like cytoplasm, and double chromatin dots, multiply infected red blood cells and applique forms were all seen. Late trophozoites were almost indistinguishable from P. malariae, with dense irregular cytoplasm and band-like forms often present. Schizonts were noted on average to contain 10 merozoites, and up to 16, in contrast to P. malariae, which has a maximum of 12 and are more regularly arranged (Lee et al., 2009b). Stippling was not observed in this study, although has been described in other case reports (Figtree et al., 2010; Jongwutiwes et al., 2004). Gametocytes have been described as being rare, spherical, and develop over 48h. Resembling P. malariae sexual stages, the mature female macrogametocyte stains blue and contains a small eccentrically placed nucleus, compared to the pink staining male microga-metocyte with a nucleus making up about half of the body of the parasite (Coatney et al., 1971).
Fig. 1.
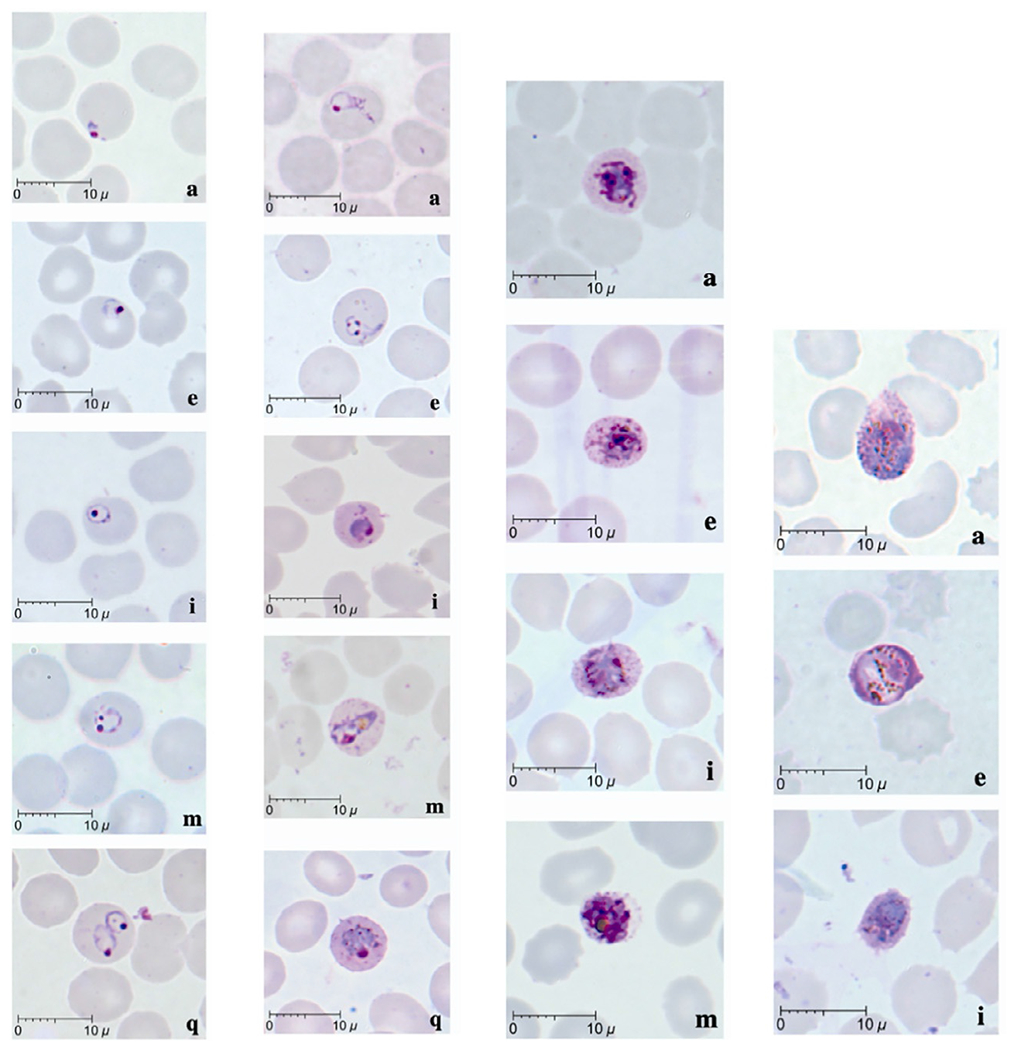
Morphology of P. knowlesi life-stages. L-R columns: 1. Early trophozoites (rings): note normal sized RBC, multiply infected RBCs, double chromatin dots; 2. Late trophozoites (including band forms); 3. Schizonts; 4. Gametocytes (a) macrogametocyte, (e) microgametocyte, (i) young gametocyte. Adapted from Lee, K.S., Cox-Singh, J., Singh, B., 2009b. Morphological features and differential counts of Plasmodium knowlesi parasites in naturally acquired human infections. Malaria J. 8, 73. https://doi.org/10.1186/1475-2875-8-73.
2.1.2. P. knowlesi parasite count measurements
The majority of symptomatic P. knowlesi human infections consist of patent low-level asexual parasites when quantified on peripheral blood films. A large prospective study of 481 knowlesi malaria cases presenting to primary health facilities in Sabah, Malaysia demonstrated a median parasite count of 2480/μL (IQR 538–8481/μL; range 20–263,772/μL). Median parasitemia was lower than that seen in falciparum or vivax malaria in both adults and children. Children less than 12 years of age with P. knowlesi infections had a lower median parasitemia of 1722/μL (IQR 386–4830/μL) compared to adults (Grigg et al., 2018a).
Extremely high parasite counts of greater than 500,000parasite/μL (10% of infected red blood cells) can occur, in part attributed to the fast 24-h parasite life-cycle, although more definitely linked to alternative red blood cell invasion pathways such as the presence of P. knowlesi normocyte binding proteins (Ahmed et al., 2014). A lower parasite count threshold of 100,000/μL to define hyperparasitemia compared to falciparum malaria was determined from severe knowlesi malaria cases in Malaysia (Barber et al., 2013a) and subsequently adopted as the WHO epidemiological and definition for hyperparasitemia defining severe malaria for P. knowlesi (WHO, 2014). These guidelines defined a parasitemia >20,000/μL as the clinical definition requiring treatment as severe malaria in those in whom other laboratory measures of severe malaria cannot be measured (WHO, 2014).
2.1.3. P. knowlesi life stages in human infections
Similar to P. malariae and P. vivax, the majority of P. knowlesi infections (86%) are asynchronous in data from Sabah, with a mean proportion of early trophozoite ring stages relative to other parasite life stages of 44%, compared to late trophozoites at 54% and schizonts comprising on average 2% (Grigg et al., 2018a), consistent with other prospective tertiary hospital data (Barber et al., 2013a). In contrast to P. falciparum infections, which due to microvascular cytoadherence demonstrate almost exclusively synchronous ring stage infections in peripheral blood films, P. knowlesi synchronous ring stage-only infections were seen in a small proportion (5%) of cases (Grigg et al., 2018a). P. knowlesi asynchronous infections had a higher median parasite count of 2177/μL, compared to synchronous ring-stage infections of 461/μL. Schizonts were present in 38% (95% CI 33%–42%) of P. knowlesi infections in this study overall, with a schizont proportion >10% relative to other life-stages found to be associated with higher parasite counts and severe disease (Barber et al., 2013a; Grigg et al., 2018a). Gametocytes were noted to be present by microscopy in 14% (95% CI 11%—18%) of patients (Grigg et al., 2018a), lower than in Lee et al. where detailed research microscopy demonstrated four (40%) patients had gametocytes, although also at very low levels of up to ~3% of infected red blood cells (Lee et al., 2009b).
2.1.4. Diagnostic performance of routine malaria microscopy for P. knowlesi infections
Studies involving reference PCR diagnostics have consistently demonstrated the poor utility of thick blood film microscopy to correctly identify symptomatic P. knowlesi infections (Mahittikorn et al., 2021). In addition to misidentification with P. malariae, P. knowlesi is often misreported as both P. falciparum and P. vivax, and vice versa, despite the latter having distinct morphological differences including enlargement of infected red blood cells (Barber et al., 2013; Coutrier et al., 2018). In countries north of Malaysia, P. knowlesi has been most commonly detected as co-infections with P. falciparum or P. vivax, creating further challenges for the use of microscopy for accurate diagnosis and reporting (Jongwutiwes et al., 2011; Marchand et al., 2011). Co-infections also highlight the shared mosquito vectors between zoonotic and human-only Plasmodium species in co-endemic areas (Marchand et al., 2011). Implications of these findings for co-endemic settings in Southeast Asia include the lack of anti-relapse therapy for P. vivax and, historically, the inappropriate treatment of P. falciparum with chloroquine.
In Malaysia, Barber et al. described the limitations of microscopy to reliably distinguish between Plasmodium species in a co-endemic region of Malaysia in a prospective evaluation of PCR-confirmed Plasmodium infections (Barber et al., 2013a). In routine hospital-based microscopy of 130 malaria patients, only 72% of PCR confirmed P. knowlesi monoinfections were identified as ‘P. malariae/P. knowlesi’, 13% as P. falciparum and 10% as P. vivax (Barber et al., 2013a). More recently in state-wide malaria notifications from Sabah over the period 2015–2017, despite nearing elimination of P. falciparum and P. vivax, the specificity of microscopy to correctly identify P. knowlesi remained low for at 68% (95% CI 62%–73%) against reference malaria PCR (Cooper et al., 2020).
In other countries where research studies have used appropriate PCR methods to detect P. knowlesi infections, routine microscopy has been shown to perform poorly in terms of differentiating all Plasmodium species. In Aceh Province, Indonesia, in 2014–2015, a small study of 41 microscopy positive malaria cases, including 3 cases reported as P. malariae, found 19 P. knowlesi monoinfections using PCR, with routine microscopy misidentifying 56% of all infections (Coutrier et al., 2018). In a study from North Sumatra, Indonesia, of 1169 participants with PCR confirmed Plasmodium species infections, 377 were found to be positive for P. knowlesi (including around half with mixed P. vivax infections) on PCR, with none of the 34 with patent microscopic infections reported as P. knowlesi (Lubis et al., 2017). In Thailand in a study conducted in 2008–2009, there were 3300 febrile patients with PCR confirmed malaria, nearly all of whom were diagnosed with either P. falciparum (52%) or P. vivax (48%) by microscopy. Of these patients 23 were subsequently confirmed as P. knowlesi (8 monoinfections, and 15 mixed infections) (Jongwutiwes et al., 2011). The monkey malaria species P. cynomolgi, also known to infect humans in Southeast Asia albeit more rarely, resembles P. vivax morphologically on microscopy, further complicating accurate diagnosis and reporting (Grignard et al., 2019; Imwong et al., 2019).
2.1.5. Automated visualisation of blood films for detecting P. knowlesi infections
Novel diagnostic devices based on automated visualization of malaria microscopy blood films using machine learning approaches are increasingly being validated for detection of Plasmodium species infections (Torres et al., 2018). These approaches hold promise, with a recent malaria microscopy scanner device evaluating laboratory cultured P. knowlesi and P. falciparum parasites demonstrating higher precision than microscopy for parasitemia at a lower limit of 0.1% of infected RBCs, although further development to allow differentiation between species on clinical samples using thick blood films is required (Firdaus et al., 2021).
2.2. Rapid diagnostic tests
Point-of-care diagnostics such as immunochromatographic rapid diagnostic tests (RDTs) play an important role in parasite-based malaria diagnosis world-wide, as they can be performed by people with minimal training in areas where good quality microscopy cannot be maintained (WHO, 2021). Other advantages, particularly compared to molecular detection tools, include the limited amount of time needed to conduct the RDT and receive a result (usually less than 20 min) and relatively low cost. Commercially available sensitive RDTs to detect Plasmodium genus and/or species-specific tests for P. falciparum and P. vivax are widely used, including in returned travellers from endemic settings (WHO, 2020). Of the most commonly used RDTs, many utilise antigen capture for parasite lactate dehydrogenase (pLDH), with a variety of monoclonal antibodies targeting different epitopes associated with P. falciparum and P. vivax pLDH isoforms allowing species specific identification (McCutchan et al., 2008). Although no specific P. knowlesi RDTs have been designed to date, cross-reactivity between certain pLDH epitopes for P. falciparum and P. vivax with a subset of P. knowlesi parasites also expressing these epitopes allows their detection, with sensitivity associated with the degree of binding affinity. In addition, tests targeting non-specific Plasmodium species pLDH or aldolase enzyme are able to detect P. knowlesi (McCutchan et al., 2008); Table 1.
Table 1.
Sensitivity of individual antigen/antibody targets in commercially available RDTs evaluated to date for detecting P. knowlesi.
| RDT name (manufacturer; product code) | WHO RDT testing round/product code | Target antibody (epitope) | Target Plasmodium species | Sensitivity for P. knowlesi, n/N, % (95% CI) | Parasitaemia Median parasites/μL, (IQR) [range] | Lowest parasite count/μL detected | Location | Year | References |
|---|---|---|---|---|---|---|---|---|---|
| First Response Malaria Antigen pLDH/HRP2 Combo Card Test Premier Medical Corporation Ltd., Mumbai, India | 1, 2 I16FRC25 |
Pan-pLDH | P. genus | 30/34, 88 (73–95) | 7493 [907–584,018] | 907 | Kota Kinabalu, Sabah, Malaysia | 2013 | Barber et al., (2013) |
| Pf-HRP2 | P. falciparum | 0/34, 0 (0) | |||||||
| ParaHIT Total Dipstick SPAN Diagnostics, Ltd., Surat, India | 1, 2 55IC204-10 |
Pan-aldolase | P. genus | 22/96, 23 (16–32) | 50,794 [395–340,954] | 4412 | Kota Kinabalu, Sabah, Malaysia | 2013 | Barber et al., (2013) |
| Pf-HRP2 | P. falciparum | 0/96, 0 (0) | |||||||
| BinaxNOW Malaria Inverness Medical Innovations, Inc., FL, USA | 1, 2 IN660050 |
Pan-aldolase | P. genus | 8/28, 29 (12–26) | 9131 [159–911,616] | 1/12 for <500/μL | Sarikei, Sarawak, Malaysia | 2014 | Foster et al., (2014) |
| Pf-HRP2 | P. falciparum | 0/28, 0 (0) | 9131 [159–911,616] | NA | |||||
| Paramax-3 Rapid Test for Malaria Pan/Pv/Pf Zephyr Biomedicals, India | 1, 2 50320025 |
Pan-pLDH | P. genus | 10/25, 40 (21–59) | 9131 [159–911,616] | 2/6 for <5000/μL | Sarikei, Sarawak, Malaysia | 2014 | Foster et al., (2014) |
| Pv-pLDH | P. vivax | 8/25, 32 (15–54) | 9131 [159–911,616] | 1/8 for <500/μL | |||||
| Pf-HRP2 | P. falciparum | 1/25a, 4 (0–20) | 9131 [159–911,616] | — | |||||
| OptiMAL-IT DiaMed, CA, USA | 1, 3 710024 |
Pf-pLDH (17E4) | P. falciparum | 18/28, 64 (44–81) | 9131 [159–911,616] | 1/3 for <500/μL | Sarikei, Sarawak, Malaysia | 2014 | Foster et al., (2014) |
| 55/190, 29 (22–35) | 2301 [30–641,464] | 30parasite/μL | Kota Kinabalu, Sabah, Malaysia | 2014 | Grigg et al., (2014) | ||||
| Pan-pLDH (19G4) | P. genus | 20/28, 71 (54–88) | 9131 [159–911,616] | 1/3 for <500/μL | Sarikei, Sarawak, Malaysia | 2014 | Foster et al., (2014) | ||
| 57/189, 30 (24–37) | 2301 [30–641,464] | 30parasite/μL | Kota Kinabalu, Sabah, Malaysia | 2014 | Grigg et al., (2014) | ||||
| CareStart™ Malaria HRP2/pLDH (Pf/VOM) Combo Access Bio, Inc. NJ, USA | 2, 4 G0171 |
VOM-pLDH | P. vivax, P. ovale, P. malariae | 77/178, 43 (36–51) | 2301 [30–641,464] | 50 parasites/μL | Kota Kinabalu, Sabah, Malaysia | 2014 | Grigg et al., (2014) |
| Pf-HRP2 | P. falciparum | 4/183a, 2 (0–4) | 2301 [30–641,464] | — |
False positive Pf-HRP2 result.
Systematic evaluations, pre-treatment, fresh whole blood samples only.
Importantly, P. knowlesi does not appear to cross-react with the highly specific P. falciparum histidine-rich protein-2 (HRP2), used in the most sensitive P. falciparum-based RDTs. The extensive use of P. falciparum-HRP2 based RDTs has resulted in selection of parasites with deletions in the pfhrp2 and pfhrp3 genes, rendering P. falciparum infections with these deletions undetectable. However, pfhrp2/3 deletions have been reported only in countries outside Southeast Asia, and non-expression of pLDH or aldolase targets has not been documented. WHO recommends ongoing use of P. falciparum-HRP2 based tests in areas where <5% false negative rates for P. falciparum detection are present (WHO, 2021).
2.2.1. Pan-pLDH based RDTs
The best performing RDT published to date for detecting P. knowlesi was the First Response Malaria pLDH/HRP2 Combo™ test (Premier Medical Ltd., India). Barber et al. demonstrated a sensitivity of the pan-pLDH component of 74% (95% CI 65%–80%) for detecting P. knowlesi infections, with a positive result in 95/129 patients. For samples tested prior to antimalarial treatment the sensitivity increased to 88% (95% CI 73%–95%), although with a higher median parasite count of 7493/μL (IQR 3823–19,895/μL) among those tested. Within this group, for those with parasite counts greater than 1000/μL sensitivity further increased to 97% (95% CI 83%–99%), although had a corresponding low sensitivity of 25% below this parasite count cut-off. Sensitivity in severe malaria with higher parasitemias was 95% (36/38; 95% CI, 83–99), however the pLDH target failed to detect two moderately high individual parasite counts of 3486 and 48,833 parasite/μL. All P. knowlesi samples tested negative for the concurrent P. falciparum HRP2 target (Barber et al., 2013b).
The OptiMAL-IT™ malaria RDT (DiaMed, USA) includes a pan-pLDH based component and was evaluated by Foster et al., with positive tests reported in 18 freshly collected P. knowlesi samples, resulting in a sensitivity of 64% (95% CI 44%–81%). The median parasite count of samples tested was 9131/μL, however sensitivity was markedly reduced in those with lower parasitemias, with a single positive result in the small number of three total samples tested with parasite counts below 500/μL (Foster et al., 2014). The poor performance of the pan-pLDH component in the OptiMAL-IT™ test was consistent with a separate larger study of 190 P. knowlesi clinical samples, which comprised a lower median parasite count of 2301/μL among those tested. A poorer sensitivity of 29% (95% CI 22%–35%) for the OptiMAL-IT™ pan-pLDH target was demonstrated, despite a low minimum detectable parasite count of 30 parasites/μL (Grigg et al., 2014).
The Paramax-3 Rapid Test for Malaria Pan/Pv/Pf™ (Zephyr Biomedicals, India) was evaluated by Foster et al., who reported a poor sensitivity of the pan-pLDH test component of 32% (95%CI 17%–46%) in fresh P. knowlesi infected samples (Foster et al., 2014). Grigg et al. evaluated the CareStart™ Malaria HRP2/pLDH (Pf/VOM) Combo RDT (Access Bio Inc., USA), with the VOM pLDH component targeting non-falciparum species (P. vivax, P. ovale and P. malariae). This pLDH based target was also relatively poor for detecting P. knowlesi, with 77/178 clinical samples positive, resulting in a low sensitivity of 43% (95% CI 36–51) (Grigg et al., 2014). For the studies evaluating the Paramax-3™ and CareStart™ RDTs which also contain a P. falciparum-HRP2 test, a small number of false positive results, less than 0.04%, were recorded for P. knowlesi samples read by research staff blinded to the underlying PCR-confirmed Plasmodium species result (Foster et al., 2014; Grigg et al., 2014).
2.2.2. P. falciparum and P. vivax–pLDH antibody cross-reactivity with P. knowlesi
The known P. knowlesi cross-reactivity with certain P. falciparum and P. vivax-pLDH epitopes has been exploited to evaluate RDTs containing these test components for detection of P. knowlesi infections. For P. knowlesi a high homology of 96.8% has been found with the pLDH ortholog for P. vivax, and over 90% for P. falciparum, P. malariae and P. ovale (Singh et al., 2012). Despite the inherent lack of specificity in this approach, cross-reactivity against P. knowlesi in clinical samples has been demonstrated. A P. falciparum-pLDH (rather than HRP2) target in the OptiMAL-IT™ RDT demonstrated positive test results in 18/28 P. knowlesi samples in Sarawak, Malaysia with a sensitivity of 64% (95% CI 44%–81%) (Foster et al., 2014). Positive results for P. falciparum-pLDH were also recorded among 190 patients with comparatively lower overall P. knowlesi parasite counts, resulting in a sensitivity of 29% (95% CI 22%–35%) in Sabah, Malaysia (Grigg et al., 2014). A P. vivax-pLDH target was evaluated in the Paramax-3™ RDT, with a lower sensitivity of 32% (95 %CI 15%–54%) for detecting P. knowlesi compared to the pan-pLDH component (Foster et al., 2014). The Core Malaria Pan/Pv/Pf™ malaria RDT (Core Diagnostics Ltd., USA) demonstrated a positive P. vivax-pLDH result for a single returned traveller with PCR confirmed knowlesi malaria and a parasite count of 40,000/μL (Berry et al., 2011). The addition of the P. falciparum-pLDH or P. vivax-pLDH tests in the OptiMAL-IT™ and Paramax-3™ RDTs respectively did not confer any improvement in overall sensitivity when results were combined with the pan-pLDH test components for P. knowlesi diagnosis.
2.2.3. Pan-aldolase based RDTs
The BinaxNOW Malaria RDT™ (Inverness Medical Innovations Inc., USA) contains a pan-aldolase target validated for conventional major human Plasmodium species (P. falciparum, P. vivax, P. malariae and P. ovale), in addition to a P. falciparum HRP2 test component. The pan-aldolase test performed poorly when conducted on P. knowlesi samples, with a reported sensitivity of 29% (95% CI 12%–26%), including negative results for all 10 P. knowlesi samples tested with a parasite count of less than 5000/μL (Foster et al., 2014). The ParaHIT Total Dipstick™ RDT was evaluated by Barber et al, which also contains a pan-aldolase target and P. falciparum HRP2 target. P. knowlesi detection was also poor, with the pan-aldolase target demonstrating a sensitivity of 23% (95% CI 16%–32%) and the lowest parasite count detected was 4412/μL (Barber et al., 2013b).
2.2.4. RDT use in returned travellers from endemic countries
Published case reports from returned travellers with P. knowlesi malaria who visited endemic countries in Southeast Asia including from the Netherlands, Sweden, Spain, Australia, France, Japan, Germany, and Italy most commonly described the use of the BinaxNOW™ RDT as a primary point-of-care diagnostic tool. The pan-aldolase test strip for Plasmodium species detection was positive in thtree case reports in patients with moderate to high parasite counts of 84,000/μL (van Hellemond et al., 2009), 40,000/μL (Berry et al., 2011) and 7700/μL (Ong et al., 2009), with the remainder negative. A number of P. knowlesi case reports from various non-endemic countries after the BinaxNOW™ evaluation was published in 2014 highlights the ongoing use of this RDT as a first line point of care malaria diagnostic tool in returned travellers. This is despite the poor performance demonstrated for detecting P. knowlesi, in addition to that seen for P. vivax at lower parasite counts in previous WHO malaria RDT product testing (WHO, 2015b). The OptiMAL-IT™ RDT has also been reported as having positive P. falciparum-pLDH results when conducted in Netherlands (van Hellemond et al., 2009) and Singapore (Ong et al., 2009) for P. knowlesi confirmed symptomatic infections. The Core Malaria Pan/Pv/Pf™ malaria RDT used in a single P. knowlesi case report demonstrated a positive pan-pLDH and negative P. falciparum-HRP2 test in addition to the positive P. vivax-pLDH result previously mentioned (Berry et al., 2011).
2.2.5. RDT detection for Plasmodium species in monkey hosts
Kawai et al. evaluated two malaria RDTs for the detection of different simian malaria species infections in non-natural Japanese macaque hosts. The study macaques were separately inoculated with P. knowlesi Hackeri strain, P. gonderi, P. cynomolgi B strain, and P. coatneyi CDC strain and monitored over the ensuing course of infection (Kawai et al., 2009). The Entebe Malaria Casette™ (Laboritorium Hepatika, Indonesia) was evaluated, which contains a P. vivax-pLDH target found to be initially negative for the P. knowlesi infected macaque at a parasitemia of 0.04%, and subsequently positive at later higher parasitemias of 11.2% and 28.8% of infected red blood cells. The OptiMAL-IT™ RDT was also concurrently evaluated with positive P. falciparum-pLDH and pan-pLDH results at the same parasite count levels. Serial dilutions of the quantified P. knowlesi sample were used to demonstrate weakly positive results remaining present at a minimum of 100parasites/μL for both RDTs. Both the P. vivax-pLDH target for the Entebe Malaria Cassette™ and the OptiMAL-IT™ pan-pLDH test demonstrated positivity when tested on other Old World non-human primate malaria species infections including P. gonderi, P. cynomolgi and P. coatneyi. The P. falciparum-pLDH test component for OptiMAL-IT™ was positive for P. gonderi, however negative for P. cynomolgi and P. coatneyi (Kawai et al., 2009).
2.2.6. Summary and recommendations for RDT use
Commercially available RDTs evaluated to date have limited utility in accurately detecting P. knowlesi clinical infections and should not replace microscopy as an existing first line malaria diagnostic tool in endemic areas, or PCR detection methods for accurate final Plasmodium species reporting. Neither pan-pLDH or pan-aldolase based tests have demonstrated sufficient sensitivity for detecting P. knowlesi, particularly with low parasite counts of less than 1000/μL which are seen in around 33% (95% CI 32%–34%) of over 8000 P. knowlesi malaria cases in endemic areas such as Sabah, Malaysia over recent years (unpublished data, Sabah Ministry of Health). Cross-reactivity of P. knowlesi with P. falciparum-pLDH and P. vivax-pLDH test components are also insufficiently sensitive to diagnose P. knowlesi reliably. In co-endemic areas specificity for the original P. falciparum and P. vivax pLDH targets is potentially less than manufacturer’s report (Grigg et al., 2014). Point-of-care malaria RDT use in returned travellers from P. knowlesi endemic areas requires the concurrent use of microscopy for initial diagnosis and clinical management, and validated PCR detection methods for accurate final Plasmodium species reporting.
RDTs remain a major point-of-care malaria diagnostic tool in Southeast Asia, with over 12.7 million distributed in the WHO regions encompassing P. knowlesi endemic areas in 2019 (WHO, 2020). Consistent with current WHO recommendations, in malaria endemic areas in Southeast Asia (or in returned travellers) where RDTs are used as a first line tool, a P. falciparum-HRP2 based test should be preferentially used to distinguish falciparum versus non-falciparum malaria. An additional pan-pLDH and P. falciparum-pLDH test component which cross-reacts with P. knowlesi, or a P. vivax-pLDH test utilising a specific epitope which has been definitively shown to lack any cross-reactivity with P. knowlesi (McCutchan et al., 2008), may theoretically assist in identifying P. vivax infections. However, in general current RDTs remain unable to distinguish mixed P. vivax/P. knowlesi infections, and the poor sensitivities demonstrated to date limit the possible use of any current combination of target antigens to accurately diagnose P. knowlesi monoinfections (Grigg et al., 2014).
Commercial appeal for developing and including P. knowlesi specific targets in malaria RDTs is constrained in part by the relatively low reported regional incidence of human infections, which in turn is underestimated by the lack of accurate point-of-care diagnostic tools. Current epitopes particularly for pLDH based tests are usually undisclosed by commercial manufacturers, with reported specificity for the major P. falciparum and P. vivax RDT targets not known to be determined against P. knowlesi or other simian malaria species able to cause human infections, such as P. cynomolgi or P. simium, for which cross-reactivity may also exist. WHO malaria RDT product testing also report rigorously conducted panel detection scores only against P. falciparum and P. vivax (WHO, 2018), and ideally would expand validation protocols to encompass P. knowlesi and other clinically relevant human or simian malaria species. Future novel RDTs such as magneto-optical detection of hemozoin, an intrinsic by product of Plasmodium species haemoglobin digestion, hold promise for sensitive P. knowlesi detection at low parasite counts, although also need to be able to reliably differentiate major Plasmodium species (Arndt, 2021) (Box 1).
BOX 1. Point-of-care summary and diagnostic reporting recommendations.
These summary guidelines refer to P. knowlesi endemic areas in Southeast Asia.
Microscopy
Microscopy remains a sufficiently sensitive first-line tool for P. knowlesi detection for the majority of low-level symptomatic infections.
Routine microscopy is unable to differentiate P. knowlesi infections from P. malariae, or early ring stage infections with P. falciparum, and misidentification with P. vivax also occurs.
In knowlesi-endemic areas where microscopy is used as a first-line tool, blood films in which parasite morphology appears similar to P. malariae and the parasite count is less than 5000/μL should be reported as ‘P. knowlesi/P. malariae’ (or ‘suspected P. knowlesi’ in relevant national guidelines).
Blood films in which parasite morphology appears similar to P. malariae and the parasite count is greater than 5000/μL should be reported as P. knowlesi, and for any infections with a parasite count greater than 15,000/μL consider early administration of intravenous artesunate due to the high risk of severe disease.
Rapid diagnostic tests
Current RDTs (pLDH or aldolase-based) remain insufficiently sensitive to detect low-level P. knowlesi infections and should not replace microscopy as a first-line diagnostic tool where possible.
Cross-reactivity of P. knowlesi with P. falciparum and P. vivax-pLDH based test components occurs, however is insufficiently sensitive and specific for diagnostic purposes.
Where RDTs are used as a first-line diagnostic tool, they should include a P. falciparum-specific HRP2 test component.
RDTs should additionally include the highest performing pan-pLDH test possible to detect non-falciparum malaria.
RDTs containing a pan-aldolase test component should not preferentially be used.
Positive RDT results should where possible have subsequent PCR conducted for final Plasmodium species confirmation.
3. Molecular detection
3.1. PCR assays
3.1.1. Background
The development of molecular detection assays for P. knowlesi has delivered enhanced sensitivity and specificity compared to microscopy. A number of assays utilizing different gene targets and methods have been developed, including the use of nested PCR, real-time PCR, single-step PCR; summarized in Table 2.
Table 2.
Molecular detection methods for P. knowlesi.
| Type of assay | Gene target | Primers or probe(s) sequence | Limit of detection | Advantages | Disadvantages | References |
|---|---|---|---|---|---|---|
| Nested PCR | SSU rRNA | Pmk8 Pmkr9 |
1–6 parasites/μL | Highly conserved; can be combined with nested non-zoonotic malaria | Can spuriously amplify P. vivax | Singh et al. (2004) |
| PKF1160 PKR1150 PKF1140 |
1–10 parasite genomes/sample | Sensitive/specific; can be combined with nested non-zoonotic malaria | Lengthy amplification rounds (105 mins each nest) | Imwong et al. (2009) | ||
| Kn1f Kn3r |
1–6 parasite/μL | Sensitive/specific; can be combined with nested non-zoonotic malaria | Lee et al. (2011) | |||
| PK18SF PK18SRc |
NR | Jongwutiwes et al. (2011) | ||||
| DHFR-thymidylate synthase | Pk-Lin-F Pk-Lin-R |
Not reported | 1 μL of genomic DNA; 0.4 U Taq; heminested; amplification efficiency and specificity less prone to genetic variations within species or cross-hybridization between species | Lengthy amplification rounds (90 and 105 mins) | Tanomsing et al. (2010) | |
| Mitochondrial cytochrome B | PKCBF PKCFR |
1–10 copies/μL | Detected on saliva and urine; highly sensitive - more copies in the mtDNA than 18S rRNA; highly conserved with limited variation; Cytb saliva detection better than microscopy | Secondary PCR has to be performed separately for each species | Putaporntip et al. (2011) | |
| PKCBF PkCBR-ed |
Not reported | Tanizaki et al. (2013) | ||||
| PKCBF PkCBR-ed |
Not reported | 1μL DNA; Pk DNA from urine and faeces is detected for a longer period than from blood | P. knowlesi only | Kawai et al. (2014) | ||
| SICAvar | SICAf1 SICAf2 SICAr1 |
0.1 parasites/μL | Sensitive | P. knowlesi only; specificity not cross-checked using P. vivax PCR on clinical samples | Lubis et al. (2017) | |
| Real-time PCR | SSU rRNA | Pk1 probe Pk2 probe |
10 copies/μL | Simultaneous detection of all Plasmodium species; no cross-reactivity with other pathogens | Cannot differentiate mixed P. vivax and P. knowlesi; four probes; expensive | Babady et al. (2009) |
| NVPK-F NVPK-R NVPK-Probe |
5 copies/reaction (<1 parasites/μL) | Sensitive | P. knowlesi only | Link et al. (2012) | ||
| Pke’F Pkg’R |
100 copies/μL | Detects all species; low cost compared to other RT-PCR | Still expensive; two probes; different melting temperatures | Oddoux et al. (2011) | ||
| Pk probe | 10 copies/μL (<3 parasites/μL on clinical samples) | Rapid results; low contamination risk; high throughput; quantification of parasites No cross-reactivity-including P. cynomolgi and P. inui; needs Plasmo1 and 2 primers |
Standard 5 μL DNA used | Divis et al. (2010) | ||
| Plasmepsin | Pk (probe) | 1–6 parasites/μL | Five species High reported sensitivity and specificity 4.5 h from DNA preparation to PCR amplification 96-well (for 5 species) | Limited to BioRad iQ5 multicolor RT PCR; no lower limit of detection tested with clinical samples | Reller et al. (2013) | |
| Single step PCR | SSU rRNA | 0.25 parasites/μL | 1.5 μL DNA; faster, less labour intensive; Detects all species; very sensitive; can detect co-infections; less expensive | Inconsistent results due to primers and DNA competition effects; minimal samples of P. ovale and P. malariae | Chew et al. (2012) | |
| aConsensus repeat sequences | Pkr140 | 1 parasite/μL | Non-nested; 1 μL DNA; sensitive | P. knowlesi only; specificity not well validated with lack of clinical samples tested; bioinformatics approach to sequence target selection | Lucchi et al. (2012) | |
| Cytochrome c oxidase subunit 1 | PlasmoCOI (FDB+RDB) | Not reported | Validated on a single P. knowlesi sample | Setiadi et al. (2016) | ||
| LAMP | β-tubulin | F3, B3, FIP, BIP, FLP, BLP | 100 plasmid copies/sample | Iseki et al. (2010) | ||
| AMA-1 | F3, B3, FIP, BIP, FLP, BLP | 10 plasmid copies/sample | Lau et al. (2011) | |||
| 18S rRNA | F3, B3, FIP, BIP, FLP, BLP | 10plasmid copies/sample | Lau et al. (2016) | |||
| Mitochondrial | Pk 101 | 0.2 parasites/μL | Britton et al. (2016) |
Consensus repeat sequences: chromosome ends and interiors; not protein-encoding.
Adapted from Singh, B., Daneshvar, C., 2013. Human infections and detection of Plasmodium knowlesi. Clin. Microbiol. Rev. 26, 165–184. https://doi.org/10.1128/cmr.00079-12.
SSU rRNA has been used as the most common target for all Plasmodium species identification as most Plasmodium parasites have two to three distinct genes that are differentially expressed during the parasite’s life cycle (Waters, 1994). The first P. knowlesi oligonucleotide primers, Pmk8-Pmkr9, developed by Singh et al. targeted the ssu rRNA-S gene expressed during the sexual stages (Singh et al., 2004). This PCR assay enables simultaneous examination for other Plasmodium infections using initial well-established genus-specific primers (rPLU6 with either rPLU1 or rPLU5) in the primary PCR followed by species-specific detection (Snounou and Singh, 2002). In conjunction with initial confirmatory sequencing of part of the P. knowlesi ssu rRNA and csp genes, this PCR assay was instrumental in demonstrating that the majority of malaria cases in the Kapit division of Sarawak, Malaysia, were actually single-species or mixed P. knowlesi infections (Singh et al., 2004).
Further study in Thailand however subsequently confirmed the possibility of cross-reactivity of these primers with P. vivax isolates. When the P. knowlesi and P. vivax ssu rRNA-S gene sequences were aligned, they showed identical sequences corresponding to the Pmkr9 primer and also similarities in the first 19 bases of the Pmk8 primer despite the differences at the 3′ end. Therefore, Pmk8-Pmkr9 primers may cause false amplification of P. vivax ssu rRNA under certain amplification conditions (Imwong et al., 2009). In addition, these 18S rRNA gene primers were also reported to cross-react with four other malaria parasites present in forest macaques of SE Asia: P. inui, P. hylobati, P. cynomolgi, and P. coatneyi (Lucchi et al., 2012). New primers using a fragment of the P. knowlesi ssu rRNA gene expressed during the asexual stages were subsequently designed. The PkF1160, PkF1140 and PkR1150 primers were shown to be specific for P. knowlesi with a level of detection of 1 to 10 parasite genomes per sample (Imwong et al., 2009). Updated ssu rRNA gene primers (Kn1f and Kn3r) were also developed in Sarawak, with specificity validated against other endemic simian Plasmodium species (Lee et al., 2011).
The Pmk8 and Pmk9 primers initially developed for P. knowlesi have now been updated or superseded with alternative primer pairs in routine practice in Malaysia at state reference laboratories (Divis et al., 2010; Nuin et al., 2020). Cross-reactivity of P. knowlesi infections with older P. vivax-designed primers amplifying parts of the ssu rRNA gene (Kimura et al., 1997) and the mitochondrial Cox1 gene (Tham et al., 1999) have also been demonstrated in returned travellers (Berry et al., 2011; Tanizaki et al., 2013). Other P. vivax ssu rRNA gene primers (Perandin et al., 2004) have also been demonstrated to show cross-reactivity with the closely genetically related zoonotic monkey parasite P. cynomolgi in Cambodia (Imwong et al., 2019). The routine use of newer PCR assays where specificity against P. knowlesi and other simian Plasmodium species has been tested is highly recommended.
3.1.2. Alternative target genes for P. knowlesi identification
Target genes of Plasmodium species that provide an alternative to the ssu rRNA gene include circumsporozoite surface protein (csp), a nuclear gene encoding a cysteine protease, cytochrome b (Conway, 2007), and dihydrofolate reductase (Tanomsing et al., 2010). More recently, the high copy number gene family encoding SICAvar antigens has been used as an alternative target (Lubis et al., 2017).
The use of mitochondrial-encoded cytochrome b is well established for diagnostics and species-discrimination in Plasmodium (Conway, 2007). It is highly conserved and possesses multiple copy numbers (20–150) in each parasite (Feagin, 1992; Mercereau-Puijalon et al., 2002), compared to 4–8 copies of the 18S rRNA (Vaidya and Mather, 2005). It has also been used to determine the evolutionary history of parasites (Escalante et al., 1998).
Dihydrofolate reductase (DHFR) and thymidylate synthase (TS) of Plasmodium species are enzymes with an essential role in the intra-erythrocytic life-cycle. They are relatively conserved between different isolates of the same species. Amplification of these genes has been used to investigate point mutations implicated in parasite antifolate resistance (Grigg et al., 2016a,b). The sensitivity and specificity to detect P. knowlesi has been reported to be similar to those detected by ssu rRNA assays. Other advantages are the ability to detect mixed-species infections, as these loci are less prone to genetic variation than the non-protein coding ribosomal genes (Tanomsing et al., 2010).
The schizont-infected cell agglutination variant antigens (SICavar) genes encode an antigen family unique to P. knowlesi, with an estimated number of more than 100 members, including both multiexon and truncated forms randomly distributed across all 14 chromosomes (Pain et al., 2008). SICAvar was designed to address potential cross-reactivity between P. vivax and P. knowlesi, with higher sensitivity also reported in one study compared to ssu rRNA and cytb amplification (Lubis et al., 2017).
3.1.3. Comparative performance of PCR assays
PCR based amplification assays for identification of malaria parasites to the species level have been developed in various formats, including nested PCR, heminested, multiplex PCR, and real-time PCR. The nested PCR is time-consuming, labour-intensive, and requires a large number of reagents and disposable consumables. It also has high risk of cross-contamination, although this can be minimised by training of laboratory staff and well-designed laboratory layout. Real-time PCR has the advantage of a rapid result, minimal contamination risk (as it is a sealed system) and also allows parasite quantification. However, it is more expensive due to the costs of fluorescent reagents and specialized assay platforms. A single-step multiplex PCR reduces the time needed for PCR amplification and reactions, therefore saving on costs. It allows simultaneous identification of all human-infecting Plasmodium species, however the primer design and optimisation steps are challenging to ensure a highly sensitive and specific assay (Chew et al., 2012).
Since its introduction in the 1990s, nested PCR targeting the ssu rRNA gene has been considered the gold standard method for malaria detection (Snounou et al., 1993). The nested Kn1f and Kn3r (Lee et al., 2011) and the heminested PkF1160, PkF1140 and PkR1150 (Imwong et al., 2009) PCR assays were developed with improved specificity against other zoonotic and non-zoonotic species, targeting the sexual and asexual stages of P. knowlesi ssu rRNA, respectively. Either primer set can be used as a nested reaction following primary amplification with broad genus-specific Plasmodium primers (Snounou et al., 1993), and have a reported level of detection of 1–6 parasites/μL. However, it takes six PCR amplifications to enable the identification of P. knowlesi and the four major human-only Plasmodium species, with further PCR amplifications required if necessary to distinguish other co-endemic macaque Plasmodium species such as P. cynomolgi, P. coatneyi, P. inui and P. fieldi (Lee et al., 2011). The latter macaque Plasmodium species have recently been described to naturally infect humans in Malaysia (Yap et al., 2021), with P. inui, P. inui-like, P. coatneyi, and also P. simiovale able to be described with the use of nested PCR (Lee et al., 2011) and additional sequencing of the ssu rRNA and COX1 genes (Yap et al., 2021).
In contrast to nested PCR, a significant advantage of qPCR is the straight-forward Plasmodium species detection analysis through the use of fluorophores, without the need for gel electrophoresis. Several assays for P. knowlesi detection using qPCR targeting either S-type (Babady et al., 2009; Divis et al., 2010; Link et al., 2012) or A-type (Oddoux et al., 2011) ssu loci have also been developed. Two of these incorporated a P. knowlesi-specific probe (Divis et al., 2010; Link et al., 2012), with the remaining assays using a multiplexed P. knowlesi-probe with the human-only Plasmodium probes to allow detection of all Plasmodium species in a single reaction (Babady et al., 2009; Oddoux et al., 2011). Babady et al. used four FRET probes to enable simultaneous detection of five Plasmodium species at two reading channels. Although this assay has the advantages of a qPCR assay and maintains the sensitivity and specificity for all Plasmodium species identification, the four probes used increased the cost of the reaction and remains of limited value for field deployment (Babady et al., 2009). Another approach to minimise the cost of all Plasmodium species identification by qPCR was the modification of a SYBR Green qPCR protocol to include a P. knowlesi-specific probe. However, the sensitivity of this assay is much poorer (100 copies/μL) than that offered by Babady (Oddoux et al., 2011).
These limitations led to the development of a single-step hexaplex PCR assay (Chew et al., 2012). This assay is comprised of a single reverse Plasmodium genus-specific and five forward species-specific primers of human malaria parasites (P. knowlesi, P. falciparum, P. vivax, P. malariae, P. ovale curtisi and P. ovale wallikeri). The single-step hexaplex approach allows detection of single or mixed malaria infections within a shorter laboratory timeframe (~3 h). In addition, the limit of detection for P. knowlesi was reported to be lower than previous published PCR assays at 0.25 parasites/μL. Although primers were robustly designed for Plasmodium species specificity, when testing specificity against 171 clinical malaria isolates the reference nested PCR assay used to confirm P. knowlesi included the original Pmk8 and Pmk9 primers known to falsely amplify a proportion of P. vivax monoinfections. Specificity was also not reported against the epidemiologically relevant P. cynomolgi. Despite this, with further validation this assay has the potential to provide an ideal and more affordable molecular detection method for malaria diagnosis (Chew et al., 2012).
The cytochrome b assay allows the identification of all Plasmodium species with high sensitivity and has been used with samples of body fluids other than blood. The first nested PCR developed targeting cytochrome b was designed by Putaporntip et al. The cytb PCR detected malarial DNA from a higher proportion of blood samples compared to the reference 18S rRNA assay, resulting in an improved sensitivity by 16%, with a reported limit of detection of 10 parasites/μL. When using saliva and urine samples, cytb amplification demonstrated marked superiority compared to the ssu rRNA assay, which was not able to identify P. knowlesi (Putaporntip et al., 2011). Cytb-targeted primers were further adapted to detect P. knowlesi in urine and additional faecal samples. After an inoculation of P. knowlesi infected RBCs into a macaque, P. knowlesi DNA in urine samples was detected early at day 2 similar to the detection of DNA in blood samples, while the DNA in faecal samples were only detected on day 7. However, the parasite DNA in faecal samples was detected up to 37 days after blood samples were PCR-negative. This may be due to the accumulation of parasite DNA in the gallbladder, from where it is gradually excreted to the faeces. This offers an attractive approach for studies in macaque hosts (Kawai et al., 2014; Putaporntip et al., 2011; Tanizaki et al., 2013).
The SICAvar gene assay was developed as a heminested PCR amplification protocol and was tested against human and simian malaria parasites. It was reported to be specific for P. knowlesi, with the limit of detection of 0.2 parasites/μL, lower than most of other reported P. knowlesi PCR assays. This assay identified a significant number of sub-microscopic and asymptomatic P. knowlesi cases in a malaria endemic area of Indonesia, of which nearly half were co-infected with other Plasmodium species, most commonly P. vivax. However, the apparent presence of the many P. knowlesi-P.vivax co-infections in the study could not be confirmed by performing head-to-head PCR assays using the other target genes due to the limited material available from the samples, which had been collected as dried blood spots (Lubis et al., 2017).
3.1.4. Detection of submicroscopic or asymptomatic infections
The use of highly sensitive PCR assays in research settings has increasingly demonstrated the extent of submicroscopic or asymptomatic P. knowlesi infections in population-based surveys across Southeast Asia. This includes a proportion of documented P. knowlesi infections from molecular surveys in Thailand (Jongwutiwes et al., 2011), Vietnam (Marchand et al., 2011), East Malaysia (Fornace et al., 2016b; Grignard et al., 2019; Siner et al., 2017), West Malaysia (Jiram et al., 2016; Noordin et al., 2020), Indonesia (Lubis et al., 2017), Myanmar (Ghinai et al., 2017) and Cambodia (Imwong et al., 2019). The potential role of asymptomatic or submicroscopic human P. knowlesi infections contributing to ongoing transmission in human or monkey host populations is not established, although these transmission modes have been experimentally demonstrated (Chin et al., 1968). It remains unclear what proportion of these low-level infections either spontaneously resolve, persist at levels sufficient to be transmitted, develop into symptomatic disease resulting in seeking treatment and detection, or progress further to high parasitemia and severe disease.
Different approaches to maximizing the sensitivity of molecular methods to detect very low-level Plasmodium species infections have been used. In a cross-sectional community malaria survey conducted in Cambodia, Imwong et al. initially used a previously validated high-volume P. genus quantitative assay (uPCR) able to detect to down to 22genomes/mL of blood (Imwong et al., 2014). Samples positive for malarial DNA on this screening assay were then evaluated using a novel nested PCR amplifying 18S ssu rRNA genes (both A and S type), which encompasses 14 zoonotic malaria species including the endemic macaque parasites P. knowlesi, P. cynomolgi, P. coatneyi and P. inui. The PCR products (233–298bp) were then sequenced and aligned against corresponding individual reference ssu rRNA sequences for final diagnosis. This method was able to detect 8 P. knowlesi monoinfections, and 11 P. cynomolgi infections (including 2 co-infections with P. vivax). The very low-level parasitemia of asymptomatic P. knowlesi or P. cynomolgi infections seen in this study (geometric mean 52 and 4parasite/μL respectively) means a large proportion of infections in this area are likely to be below the threshold of ~40 parasites/μL able to be detected using routine microscopy or pan-pLDH based RDTs (Imwong et al., 2019).
A study of household and village members of confirmed P. knowlesi cases in Sabah revealed a high proportion of submicroscopic P. knowlesi carriage, with an estimated prevalence of 6.9% using latent class analysis of multiple P. knowlesi species-specific PCR assays (Fornace et al., 2016a). Findings differed depending on the sensitivity of the molecular P. knowlesi-specific assay, with the ssu rRNA nested assay expected to identify 15% of these infections, compared to Cytb nested PCR (59.1%), ssRNA qPCR (87.9%), and Plasmepsin qPCR (81.3%) (Fornace et al., 2016a). A subsequent rigorously designed community cross-sectional survey of 10,100 participants conducted in the same area in Sabah used a pooled 10 × 10 matrix of samples for DNA extraction and PCR detection for submicroscopic Plasmodium species infections (Fornace et al., 2019b). A well-established nested P. genus and non-zoonotic species-specific PCR assay targeting the ssu rRNA was initially conducted (Snounou et al., 1993), followed by the SICAvar assay for P. knowlesi detection (Lubis et al., 2017), with 9 infections found: 2 P. knowlesi, 1 P. knowles/P. vivax, with the remainder non-zoonotic Plasmodium species. A further analysis using 876 selected high-risk samples from the same survey conducted the nested zoonotic malaria PCR and sequencing method developed previously (Imwong et al., 2019), demonstrating improved sensitivity with 54 (6.2%) submicroscopic Plasmodium species infections reported. This included 3 P. knowlesi infections, and 2 P. cynomolgi infections (previously misdiagnosed as P. vivax and mixed P. knowlesi/P. vivax), with the remainder non-zoonotic species infections (Grignard et al., 2019).
In a longitudinal molecular malaria survey in Sarawak, Siner et al. used a pooled sample of four individuals dried blood spots for initial nested ssu rRNA PCR P. genus screening in 2118 participants, before DNA re-extraction and individual participant screening on those positive with Pmk8-Pmkr9 primers for P. knowlesi (Singh et al., 2004), other zoonotic monkey Plasmodium species (Lee et al., 2011) and non-zoonotic malaria. A further 884 individuals underwent the same PCR assay protocol without initial pooling of four samples. There were seven P. knowlesi infections in total detected, including a single asymptomatic submicroscopic infection, and five asymptomatic infections with parasite counts <50/μL, with no apparent difference in detection between samples initially pooled vs non-pooled (Siner et al., 2017).
3.1.5. Molecular detection of P. knowlesi gametocytes
The 24-h replication cycle of P. knowlesi in human host RBCs is known to rapidly produce mature gametocytes within around 48h (Knowles and Gupta, 1932). A short period of gametocyte infectivity then follows, with the timing hypothesised to increase transmission efficiency by occurring mainly at night during peak blood-feeding of mosquitoes (Hawking et al., 1968), although other mechanisms related to circadian within-host immunity and RBC physiology have been proposed (Mideo et al., 2013). The ability to accurately detect and quantify the level of gametocyte production in human P. knowlesi infections would assist in exploring fundamental parasite biology questions related to adaptation and replication in human hosts. This includes more accurately defining the timing and degree of gametocytogenesis relative to asexual parasitemia, infectivity to mosquitoes, and the risk of sustained human-human transmission. Other major applications of sensitive gametocyte detection include the ability to identify or evaluate the effect of future transmission blocking vaccines or treatments.
Molecular methods to detect P. knowlesi sexual life-stage stages hold significant advantages over microscopy, with most P. knowlesi infections likely to have gametocyte densities below the threshold of microscopic detection (Grigg et al., 2018a). Microscopy is insensitive due to the scarcity of gametocytes present in peripheral blood films (Lee et al., 2009b), and the limited number of fields screened usually in thick films when concurrently quantifying asexual parasite infected RBCs by counting the presence of up to 200 white blood cells (WHO, 2015a). Targets for the design of molecular assays to detect gametocytes are based on the underlying sexual-stage parasite biology. In preparation for gametocyte activation after mosquito ingestion in the midgut, mRNA transcripts encoding stage-specific proteins are produced by gametocytes in the human host in order to repress translation for the subsequent rapid genome replication and nuclear division. For P. falciparum this includes pfs16 mRNA, present from the earliest gametocyte stage onwards, pfpeg3 and pfpeg4 present in stage II gametocytes, and pfs25 mRNA for which transcription only begins to occur in mature stage IV gametocytes (Bousema and Drakeley, 2011). Other female specific gametocyte surface proteins include pfs47, thought to be involved in immune evasion in the mosquito midgut, and their male gametocyte paralogs pfs48/45 (Molina-Cruz et al., 2020).
Molecular detection assays for P. falciparum gametocyte protein orthologs have been designed and evaluated for P. knowlesi, with various primers developed to detect pks25 mRNA from mature gametocytes (Armistead et al., 2018; Grigg et al., 2016a,b; Maeno et al., 2017). In a clinical trial of antimalarial treatment for patients with uncomplicated knowlesi malaria in Sabah, Malaysia, Grigg et al. demonstrated the presence of P. knowlesi pks25 mRNA transcripts in 85/100 (85%; 95%CI 76%–91%) of participants tested at enrolment (Grigg et al., 2016a,b). Of those positive for pks25 mRNA only 16% had gametocytes detected by microscopy (Grigg et al., 2016a,b), consistent with a separate clinical trial where only microscopy was performed and 32/123 (26%) patent infections had gametocytes present (Grigg et al., 2018b). There was an association with higher parasitemia and detectable with a median parasite count of 2140 vs 704 parasite/μL respectively; P = 0.019. At day 7, there were 5/97 (5%) patients persistently positive for pks25 mRNA: 3 p25 mRNA: 3 patients initially treated with artesunate-mefloquine, and 2 with chloroquine (Grigg et al., 2016a,b).
Maeno et al. conducted a comparative study of gametocyte detection for four species in Khan Hoa Province, Vietnam in both humans and mosquitoes. A nested real-time PCR assay was developed targeting pks25 mRNA to detect P. knowlesi gametocytes, with 15/32 (47%) of those with P. knowlesi confirmed asexual stage infections also positive for pks25 transcripts. All P. knowlesi samples were co-infections with one or more non-zoonotic Plasmodium species. The proportion of P. knowlesi infections with detectable pks25 mRNA was comparable to that seen for the P. falciparum and P. vivax gametocyte specific targets. Serial dilution of gametocytes from a P. knowlesi infected monkey was used to demonstrate a lower limit of detection of 1 gametocyt/μL for the RT-PCR pks25 assay. In this study, 70% of mosquitoes with P. knowlesi in their salivary glands also carried human malaria parasites, supporting the possibility that mosquitoes are infected with P. knowlesi from human infections (Maeno et al., 2017).
Armistead et al. designed multiple P. knowlesi RT-qPCR detection assays across gametocyte developmental stages, including targets for pks16, pks25, and pks47 mRNA. Using P. knowlesi H-strain cultured samples, they demonstrated infectivity of An. dirus through membrane infected RBC feeders, however none of the quantified transcripts of the 3 targets correlated well with infectivity, and peaks in pks25 and pks47 transcription were not associated with gametocyte maturation as may have been expected (Armistead et al., 2018).
3.2. Loop-mediated isothermal amplification (LAMP)
More recently LAMP has been evaluated as a potentially simple and sensitive diagnostic tool compared to microscopy as an alternative molecular diagnostic option for P. knowlesi infections in resource limited areas without specialized facilities for PCR (Britton et al., 2016). First described by Notomi, briefly, this molecular approach relies on a Bacillus stearothermophilus (Bst) polymerase to perform DNA strand separation and amplification of target sequences of 300 base pairs, at a single temperature of 65°C for 1 h using a set of four primers that form a stem-and-loop structure that promotes ongoing DNA amplification (Notomi et al., 2000). Magnesium pyrophosphate, a by-product of the LAMP DNA amplification process, forms a white precipitate following successful target amplification that can then be visualized by eye, by turbidimetry (Mori et al., 2001) or colourimetrically using hydroxynaphthol blue (Britton et al., 2016), malachite green (Lucchi et al., 2016b), phenol red (Lai et al., 2020) or SYBR green (Lai et al., 2021) to objectively confirm a positive result.
3.2.1. LAMP platforms
There are two commercially available LAMP platforms. The first, the Eiken Loopamp™ MALARIA detection kit (Eiken chemical company, Japan) offers a Plasmodium genus specific assay (panLAMP) for identifying parasites that cause human infection and one able to specifically detect Plasmodium falciparum (PfLAMP) infection. When used together to identify non-falciparum infections, this panLAMP platform was able to identify P. knowlesi from 50 patients with uncomplicated P. knowlesi infections, at a genus level, with 100% sensitivity and specificity for infections with mean parasitemia of 1197 parasites/μL (IQR 300-3920) and a limit of detection of 2parasite/μL (Piera et al., 2017). However, this platform does not have a P. knowlesi species-specific assay currently available. The second commercial platform, Illumigene Malaria LAMP (Meridian Biosciences Inc., Cincinnati, OH) has demonstrated good sensitivity and specificity for identifying Plasmodium species using undisclosed LAMP primers (Lucchi et al., 2016a; Rypien et al., 2017) but has not been validated using P. knowlesi clinical samples. Several non-commercial LAMP platforms have also been published, such as HTP-LAMP (Perera et al., 2017) and RealAMP (Patel et al., 2013) one of which, HtLAMP, has been evaluated in P. knowlesi samples (Britton et al., 2016).
3.2.2. P. knowlesi specific LAMP primers
Since its first use in identifying P. falciparum (Poon et al., 2006), several LAMP primers specific for detecting P. knowlesi have been developed (summarised in Table 2), each with different targets, advantages and drawbacks. The first P. knowlesi-specific LAMP primers targeting the beta-tubulin gene demonstrated good species specificity and limit of detection of 102 copies/μL of target DNA but was only validated in samples from infected macaque monkeys and not human clinical samples (Iseki et al., 2010). Lau et al. published P. knowlesi specific LAMP primers targeting the apical membrane antigen 1 (AMA-1) (Lau et al., 2011) followed by genus and species-specific primers targeting the 18S rRNA gene for each of the Plasmodium parasites that cause human infection (Lau et al., 2016). These LAMP primers demonstrated a reported 100% sensitivity and specificity on a limited number of clinical samples, n = 13 (Lau et al., 2011), n = 57 (Lau et al., 2016), and n = 71 (Lai et al., 2021), including validation against P. vivax, however lacked detail on the limit of detection for these primers. In keeping with other LAMP studies demonstrating superior analytical sensitivity of primers targeting mitochondrial genes (Polley et al., 2010), P. knowlesi-specific mitochondrial target LAMP primers have been shown to have a limit of detection of 0.2 parasites/μL (Britton et al., 2016). However, given the 97% sequence homology between P. vivax and P. knowlesi at the target gene, this assay demonstrated 96% sensitivity, 30% specificity overall, but 100% specificity only if P. vivax samples were excluded (Britton et al., 2016).
3.2.3. Challenges for LAMP as a diagnostic tool
The advantages of LAMP include less need for equipment and technical expertise compared with PCR, potential for high specificity and rapid turnaround time. However, the requirement for parasite DNA extraction processes, high potential risk of contamination given the large amount of DNA amplification and cost per sample remain limitations to the widespread use of the LAMP for diagnosis of malaria in primary care settings compared with microscopy.
Much effort has been made to simplify the preparation of DNA template for LAMP. LAMP has been performed on DNA templates generated directly from heat-treated blood samples (Poon et al., 2006), boil and spin method (Hopkins et al., 2013) and closed-circuit PURE extraction technology (Eiken chemical company) (Polley et al., 2013) with the latter two processes showing 97.5% sensitivity compared with nested PCR (Hopkins et al., 2013). A comparison of different volumes of whole blood with chelex or chelex-saponin extraction protocol found lower limit of detection with LAMP with a 20 μL blood volume with chelex-saponin compared with 5 or 10μL (Britton et al., 2015) confirming that, similar to high volume ultrasensitive PCR (Imwong et al., 2014), blood volume impacts LAMP performance particularly at low parasitemia. Filter paper dried blood spots (DBS) have also been extracted using a multistep Chelex-based process (Han et al., 2007) from symptomatic and asymptomatic patients (Aydin-Schmidt et al., 2017) or sodium dodecyle sulphate (SDS) (Polley et al., 2010). LAMP performed on whole blood extracted by SDS was able to detect 2–5 parasites/μL compared with 10 parasites/μL from DBS extracted with SDS (Polley et al., 2010). Commercial DNA extraction kits with closed systems such as PURE (Eiken Chemical company, Japan) for either small number of samples (Polley et al., 2013) or high throughput (Perera et al., 2017) minimize the risk of potential contamination found in the multistep DNA extraction protocols but with associated increased cost. LAMP performed on blood samples appeared to be more sensitive than when performed on saliva (Singh et al., 2013) or urine (Najafabadi et al., 2014) for identifying P. falciparum and P. vivax. However, no direct comparison of DBS with different volumes of whole blood or on different sample types has been performed using LAMP in the detection of P. knowlesi.
3.2.4. The future of LAMP for P. knowlesi diagnosis
Overall, the optimal role for LAMP, as either a point of care diagnostic for case detection or a molecular tool for surveillance, remains unresolved. Platforms such as non-instrumented nucleic acid amplification by LAMP (NINA-LAMP) (Sema et al., 2015), lateral-flow device LAMP (Mallepaddi et al., 2018) and LAMP MinION nanopore sequencing (Imai et al., 2017) combined with appropriately sensitive species-specific primers may improve its applicability as a point-of-care diagnostic test. However, these technologies remain constrained by need for DNA extraction, risk of contamination, low sample throughput capacity and cost. LAMP also has been incorporated into several lab-on-a-chip platforms such as multiplex microfluidic arrays (Mao et al., 2018) and microchambers on cell microarrays (Ido et al., 2019) which hold much promise for improved field applicability of LAMP technology. However, except for LFD-LAMP, all of these platforms are yet to be fully validated in large numbers of P. knowlesi clinical samples.
In terms of surveillance, two high throughput platforms have been published to overcome the limited number of samples and prohibitive costs associated with existing commercial platforms for this application. The colourimetric high-throughput LAMP (HtLAMP) platform based on a 96 well plate required minimal infrastructure, was low cost and was applied in a resource limited setting (Britton et al., 2015). However, this assay required a separate DNA extraction process, whereas an alternative high-throughput LAMP (HTP LAMP) was able to combine a 96 well plate platform with a closed DNA extraction process (Perera et al., 2017) albeit at likely high cost. However, only the HtLAMP platform has been evaluated with P. knowlesi specific primers (Britton et al., 2016).
Therefore, the applicability of LAMP for P. knowlesi diagnosis remains constrained by two significant challenges: analytical sensitivity to detect sub-microscopic parasitemia without compromising species specificity, and cost-effective applicability in resource limited settings as an alternative to PCR either as a point of care diagnostic tool or for purposes of P. knowlesi malaria surveillance.
4. Serology
4.1. Importance of serology for malaria surveillance
The use of serology as a tool to survey and monitor transmission and exposure to infection, in addition to measuring the success of interventions has been well documented for P. falciparum (Plucinski et al., 2018; Wu et al., 2020) and P. vivax (Edwards et al., 2021; Longley et al., 2020). Measuring antibodies provides a fast and simple means for evaluating transmission intensity (Stewart et al., 2009) and in combination with parasite prevalence data can be used to monitor changes in malaria transmission over time (Drakeley et al., 2005). In this context serological tools have a major advantage compared to molecular methods, as they can capture exposure from asymptomatic or transient symptomatic infections in those not seeking treatment (Corran et al., 2007). However, to date less focus has been afforded to understanding transmission for the other causes of human malaria including P. knowlesi using serological methods, meaning neglected infections from species such as P. knowlesi remain excluded from the elimination narrative (Barber et al., 2017b; Chin et al., 2020).
4.2. Development of serologic assays for P. knowlesi detection
To complement existing approaches for assessing exposure to infection, species-specific serological reagents are an essential part of the toolbox. The ability to accurately assess the prevalence of each infecting species and monitor the impact of control and intervention approaches on each could help to provide better guidance for country-level programmatic decision-making. Serology has a number of obvious advantages, specifically the ability to accurately measure recent and historical exposure to infection (Edwards et al., 2021; Helb et al., 2015). Increasingly serology assays are also being designed using multiplex platforms (Chan et al., 2021). These platforms have allowed for high-throughput biomarker discovery and evaluation which in turn has led to an expansion in both the number and utility of the protein targets available (Tran and Crompton, 2020). As more serology tools are discovered and developed, the utility of the individual, or panel of, targets need to be properly assessed. Early seroepidemiological studies aimed at assessing non-specific malaria exposure through cross-reactivity to P. knowlesi antigens were conducted using indirect haemaglutination tests (Kagan et al., 1969). This approach was based on crude parasite preparations generated from lysed infected erythrocytes taken from splenectomised rhesus monkeys. The unrefined nature of the antigen preparation would suggest there were antigens conserved across the Plasmodium spp. present in the assay, highlighted by the wide geographical distribution of seropositive responses in non-P. knowlesi endemic settings seen in military personnel when using this technique (Kagan et al., 1969).
Designing recombinant proteins provides a more focused means for developing specific targets for use in serology assays. However, previous approaches have focused on targeting proteins such as MSP1 and AMA1 that shared moderate to high levels of sequence homology, and or exhibited cross-reactive responses with other Plasmodium spp. (Muh et al., 2020; Narum and Thomas, 1994; Waters et al., 1990). This includes other previous P. knowlesi antigens designed mainly to interrogate underlying parasite biology rather than for use as sero-surveillance tools in co-endemic areas such as PkCSP (Sharma and Godson, 1985), PkDPB (Singh et al., 2002), PkSPATR (Mahajan et al., 2005), PkLDH (Singh et al., 2012), Pk1-Cys peroxiredoxin (Hakimi et al., 2013), Pk knowpains (Prasad et al., 2012), PkMSP1-42 (Cheong et al., 2013a,b), PkMSP1-33 (Cheong et al., 2013a, 2013b), PkMSP1-19 (Lau et al., 2014), PkTrags (Tyagi et al., 2015), PkMSP3 (Silva et al., 2016) and PkSBP1 (Lucky et al., 2016). Such reagents likely would lack the required specificity to accurately define exposure to dissect P. knowlesi infections in regions co-endemic for other species, increasing the potential for false positive responses.
In order to facilitate P. knowlesi sero-surveillance in areas of Southeast with co-endemic Plasmodium species, Herman et al. used an in silico modelling approach to design four novel recombinant P. knowlesi antigen constructs from three genes: PkSERA3-antigen 1, PkSERA3-antigen 2, PkSSP2/TRAP, and PkTSERA2-antigen 1 (Herman et al., 2018). Initial target candidate genes were selected from known seroreactive P. falciparum orthologues, with protein sequences processed using analytical in silico tools aiming to maximise specificity for P. knowlesi against other Plasmodium species. This included iterative BlastP interrogation of target sequences against Plasmodium specific and other relevant databases, domain prediction, exclusion of signal peptides and transmembrane domains and glutathione-S-transferase tagging to aid expression solubility, and construction of phylogenetic trees to exclude high identity cross-species amino acid alignments. An Escherichia coli expression system was used to produce soluble, multimerised and stable antigen products with final yields from 11.9 to 20.5 mg/L. Blood-stage transcription of SERA3 and TSERA2 candidate genes was also confirmed, in contrast to the sporozoite stage SSP/TRAP gene. Finally, non-synonymous SNPs associated with the three known distinct P. knowlesi genetic subpopulations were characterized and shown to predominantly exist outside regions covered by the recombinant constructs, highlighting their potential utility for all P. knowlesi strains (Herman et al., 2018).
Validation of the four new recombinant antigens was subsequently conducted using ELISA to detect IgG responses for 97 individuals with acute P. knowlesi infections at three time points (day 0, day 7 and day 28) following treatment in a clinical trial in Sabah, Malaysia. Negative controls consisted of 26 Ethiopian non-P. knowlesi and 29 UK malaria-naïve sera, with cut-off values to define reactivity for P. knowlesi antigens taken as the mean OD (±3 SD) from the latter group. A single weakly positive sample was recorded for both SERA3 ag 1 and SSP2/TRAP in the Ethiopian negative controls. The best performing antigen was the PkSERA3 antigen 2, which was positive in 67% of all time-points in P. knowlesi cases, with maximum positivity at day 7 and up to 50-fold increases compared to day 0 recorded. Antibody prevalence for PkSERA3 antigen 1, PkSSP2 and PkTSERA2 antigen 1 also peaked at day 7, however remained relatively low at 18%, 33% and 43% respectively. Using data adaptive boosted tree regression models, the combined ability of all four antigens to classify P. knowlesi exposure was 89% (IQR 43%–61%), defined as the cross-validated area under the receiver operating curve, with the highest relative variable importance contributed by PkSERA3 antigen 2 at 50% (Herman et al., 2018).
A separate panel of 9 recombinant P. knowlesi antigens was generated by Müller-Sienerth and colleagues, based on the most highly expressed P. falciparum and P. vivax protein orthologues known to produce high antibody responses in endemic areas: CyRPA, GAMA MSP10, MSP4, MSP5, P12, P38, P41, and P92 (Müller-Sienerth et al., 2020). This study differed in approach by utilizing a mammalian expression system in order to improve the likelihood that appropriate post-translational modifications such as disulfide bonds are present (Crosnier et al., 2013). This in turn allows correct protein folding to recreate the original conformation of epitopes and therefore improve antibody recognition (Forsström et al., 2015). Malaysian sera from PCR confirmed P. knowlesi infections initially demonstrated the highest antibody responses in P12, MSP10 and P38 when quantified by ELISA. Cross-species seropositivity of these three P. knowlesi antigens was minimal when tested as part of a larger panel including 19 selected antigens for P. falciparum, P. vivax, P. malariae and P. ovale using sera from small numbers of European returned travellers with malaria. Cross-reactivity was not shown to be related to the degree of sequence identity between protein orthologues, including for the P12 antigen despite a high sequence identity of 72% between P. knowlesi and P. vivax. Logistic regression classification models combining the three P. knowlesi antigens were then used to demonstrate on a larger dataset of 50 P. knowlesi sera and 66 uninfected controls from Malaysia a high AUC of the median ROC curve of 0.91. Using a model score of greater than 0.5 was shown to accurately diagnose 82% of P. knowlesi infections with a 3% false positive rate.
4.3. Potential use case scenarios for P. knowlesi serologic tools
In 2017 a meeting was convened in Paris to review the state-of-theart with antibody-based assays in the context of malaria surveillance (Greenhouse et al., 2019). A key aim was to define potential use-case scenarios in terms of use and evaluation of serological biomarkers in control and elimination programmes. Although focused on P. falciparum and P. vivax, many of the use cases could be applied to P. knowlesi, including: (i) documenting the absence of transmission over a given geographic space; (ii) stratification of transmission level; (iii) measuring the impact of interventions and monitoring changes in malaria transmission; and (iv) decentralized immediate public health response. The summary provided a detailed understanding of what tests would be required for a given situation/population, establishing target product profiles for the future development of diagnostics and surveillance tools (Greenhouse et al., 2019).
Using recently developed P. knowlesi antigens designed as species-specific reagents (Herman et al., 2018). Fornace and colleagues conducted a survey of 2503 participants to determine the level of exposure and infection within case-study communities across Sabah, Malaysia and neighbouring Palawan, Philippines (Fornace et al., 2018). This was the first study to assess P. knowlesi infection and associated risk using antibodies, demonstrating a seroprevalence of 7.1% to PkSERA3 antigen 2, including comparable seroreactivity between adult men and women (despite passive case detection of acute infections mainly consisting of men), and 4.2% seroprevalence in children <15 years. Despite a very low acute infection prevalence of 0.2% using PCR, the use of serological tools was able to demonstrate marked spatial heterogeneity in transmission intensity between sites (i.e., higher intensity in Malaysia compared to Philippines), and associations with P. knowlesi exposure and agricultural work, high forest cover or clearing around houses (Fornace et al., 2018).
This was followed by a large population-based cross-sectional malaria survey of 10,100 individuals to assess exposure and environmental risk to P. knowlesi across Northern Sabah, Malaysia (Fornace et al., 2019b). In this study, a multiplex Luminex platform was used to concurrently evaluate IgG responses for P. knowlesi SERA3 ag 2 and SSP2, in addition to 8 other antigens for P. falciparum and 5 for P. vivax. Classification of seropositive individuals for P. knowlesi (exposure in the last year), P. falciparum, and P. vivax was conducted using a data adaptive meta-learning algorithm (Super Learner, van der Laan et al., 2007). The overall seroprevalence of P. knowlesi was 5.1% (95% CI 4.8–5.4), with high antibody responses seen across all age groups. However, individual-level associations with higher P. knowlesi exposure risk in the final Bayesian hierarchical model were seen with increasing age and male sex, in addition to macaque contact and forest activities, with personal insecticide use protective. Household-level associations with exposure included raised level house construction, lower geographical elevation and fragmented land types classified at varying spatial scales around the household (intact forest, irrigated farming land, pulpwood and oil-palm plantations). The latter are consistent with separate modelling demonstrating the relative importance of different environmental variables at small scales of <1 km around households (proportion of cleared land, aspect, slope), compared to larger spatial scales of 4-5 km (fragmentation of deforested areas) (Brock et al., 2019).
The implementation of serological tools in national malaria control programmes would result in several potential benefits. Using serological exposure to understand the mechanistic links between land use change and P. knowlesi transmission highlights their utility in the possible design of spatially-targeted interventions, particularly for low-resource settings with scarce or absent occurrence data. Future use of serological tools outside highly studied areas in Malaysia are also vital to understand regional differences, including demonstrating the potential absence of transmission in certain areas. However, implementation of serology for P. knowlesi will require guidance on validated and consistent data analysis methods to define seropositivity cut-offs (Chan et al., 2021). This is particularly the case in multiplex platforms, which ideally need to take into account known antibody titers and kinetics, regional variation in transmission, and population-level immunity for other human-only Plasmodium species that may be providing some cross-protective effect (Muh et al., 2020) (Box 2).
BOX 2. Molecular and serology methods summary and diagnostic reporting recommendations.
These summary guidelines refer to P. knowlesi endemic areas in Southeast Asia.
PCR and LAMP
A number of PCR assays have been developed which are highly sensitive and specific for P. knowlesi detection.
A validated P. knowlesi species-specific PCR assay should be included routinely as part of any PCR assay conducted for suspected or microscopy positive malaria from Southeast Asia.
Nested PCR assays targeting the 18S ssu rRNA gene are the most commonly used for P. knowlesi detection. Compared to real-time PCR they have lower sensitivity and are more time consuming, although are less expensive.
The most well-validated nested PCR assays include an initial P. genus amplification round, followed by species specific detection for P. falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi. Where possible, species-specific detection for P. cynomolgi should also be considered.
Early P. knowlesi designed nested primers, Pmk8-Pmkr9, can cross-react with P. vivax monoinfections to produce a false-positive mixed P. vivax/P. knowlesi result.
Updated and more specific nested ssu rRNA primers for P. knowlesi include PkF1160, PkF1140 and PkR1150, or Kn1f and Kn3r, which have been validated against zoonotic and non-zoonotic Plasmodium species.
Some older nested PCR assays contain P. vivax species-specific primers which have been shown to cross-react with P. knowlesi and P. cynomolgi. The well-established VIV1-VIV2 primers do not.
PCR has been successfully used in Malaysia, a pre-elimination setting for non-zoonotic malaria, for accurate species reporting of all zoonotic and non-zoonotic malaria cases, and for research surveillance purposes across Southeast Asia including the detection of submicroscopic or asymptomatic infections.
Reference laboratory molecular testing of malaria cases for both zoonotic and non-zoonotic Plasmodium species needs to be more widely implemented by National Malaria Control Programmes across Southeast Asia to accurately identify the burden of zoonotic malaria and to more accurately monitor the success of human-only malaria elimination programmes.
P. knowlesi mature gametocytes are able to be detected by real-time qPCR assays targeting pks25 mRNA, with validation and implementation in naturally infected human clinical studies demonstrating much higher sensitivity compared to microscopy.
LAMP based platforms for P. knowlesi diagnosis remain constrained by two main challenges: the ability to detect submicroscopic infections while retaining high species-specificity, and any significant cost benefit compared to PCR for diagnostic species-confirmation or surveillance.
Serology
Antigenic reagents have been identified that can distinguish exposure to P. knowlesi infection from human-only malaria with a reasonable degree of specificity.
Antibody responses to these antigens can be used at the population-level to assess the degree of exposure, including the absence of exposure.
Antibody responses can also be used to identify demographic and spatial risk factors which can support targeting of control approaches.
Population-level evaluation of novel targets would allow the identification of antigens for development of species-specific lateral flow devices to help with rapid case management.
5. Conclusion
P. knowlesi is an emergent public health threat across Southeast Asia. Implementation of improved detection methods and reporting practices for point-of-care diagnostics are vital to understanding the disease burden and distribution of symptomatic human infections and allowing the timely administration of appropriate treatment. Newer lateral flow or novel RDTs with the ability to accurately differentiate P. knowlesi and other non-P. falciparum species are ideally required for this to occur. Molecular methods incorporating validated PCR assays for P. knowlesi can improve accurate regional case reporting to monitor progress towards elimination of other human-only species, with additional utility in detection of low-level infections to understand broader transmission dynamics. However, molecular tools remain expensive in low-resource settings and public health programmes may require further input to understand the background prevalence or predicted transmission risk in order to guide their optimal targeted deployment. The implementation of multiplex serological tools with Plasmodium species specific recombinant proteins allows improved understanding of geographically heterogenous P. knowlesi transmission intensity and associated underlying environmental drivers, in addition to the ability to guide both the design and evaluation of future spatially targeted interventions.
Acknowledgements
M.J.G., I.N.L., and R.N. are supported by the ZOOMAL project (‘Evaluating zoonotic malaria and agricultural land use in Indonesia’; #LS-2019-116), Australian Centre for International Agricultural Research and Department of Foreign Affairs, Australian Government. K.K.A.T. was supported by a Bloomsbury SET Innovation Fellowship (‘Development of a suspension bead assay targeting the zoonotic malaria parasite Plasmodium knowlesi’; BSA14). This work was also supported by the Australian National Health and Medical Research Council (grant numbers 1037304 and 1045156; fellowships to N.M.A. [1042072], B.E.B. [1088738]; M.J.G. [1138860] and ‘Improving Health Outcomes in the Tropical North: A multidisciplinary collaboration “Hot North”’, [grant 1131932]). The Sabah Malaria Research Program is supported by the US NIH IRIDA program. We thank Sitti Saimah binti Sakam from the IDSKKS-Menzies Clinical Research Unit, Sabah, Malaysia for input into the microscopic diagnosis section.
References
- Ahmed AM, Pinheiro MM, Divis PC, Siner A, Zainudin R, Wong IT, Lu CW, Singh-Khaira SK, Millar SB, Lynch S, Willmann M, Singh B, Krishna S, Cox-Singh J, 2014. Disease progression in Plasmodium knowlesi malaria is linked to variation in invasion gene family members. PLoS Negl. Trop. Dis 8, e3086. 10.1371/journal.pntd.0003086.s017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armistead JS, Barros RRM, Gibson TJ, Kite WA, Mershon JP, Lambert LE, Orr-Gonzalez SE, Sá JM, Adams JH, Wellems TE, 2018. Infection of mosquitoes from in vitro cultivated Plasmodium knowlesi H strain. Int. J. Parasitol 48, 601–610. 10.1016/j.ijpara.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt L, et al. , 2021. Magneto-optical diagnosis of symptomatic malaria in Papua New Guinea. Nat. Commun 10.1038/s41467-021-21110-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin-Schmidt B, Morris U, Ding XC, Jovel I, Msellem MI, Bergman D, Islam A, Ali AS, Polley S, Gonzalez IJ, Mårtensson A, Björkman A, 2017. Field evaluation of a high throughput loop mediated isothermal amplification test for the detection of asymptomatic Plasmodium infections in Zanzibar. PLos One 12, e0169037. 10.1371/journal.pone.0169037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babady NE, Sloan LM, Rosenblatt JE, Pritt BS, 2009. Detection of Plasmodium knowlesi by real-time polymerase chain reaction. Am. J. Trop. Med. Hyg 81, 516–518. [PubMed] [Google Scholar]
- Barber BE, William T, Grigg MJ, Yeo TW, Anstey NM, 2013. Limitations of microscopy to differentiate Plasmodium species in a region co-endemic for Plasmodium falciparum, Plasmodium vivax and Plasmodium knowlesi. Malaria J. 12, 8. 10.1186/1475-2875-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber BE, William T, Grigg MJ, Menon J, Auburn S, Marfurt J, Anstey NM, Yeo TW, 2013a. A prospective comparative study of Knowlesi, Falciparum, and Vivax Malaria in Sabah, Malaysia: high proportion with severe disease from Plasmodium Knowlesi and Plasmodium Vivax but no mortality with early referral and artesunate therapy. Clin. Infect. Dis 56, 383–397. 10.1093/cid/cis902. [DOI] [PubMed] [Google Scholar]
- Barber BE, William T, Grigg MJ, Piera K, Yeo TW, Anstey NM, 2013b. Evaluation of the sensitivity of a pLDH-based and an aldolase-based rapid diagnostic test for diagnosis of uncomplicated and severe malaria caused by PCR-confirmed Plasmodium knowlesi, Plasmodium falciparum, and Plasmodium vivax. J. Clin. Microbiol 51, 1118–1123. 10.1128/jcm.03285-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber BE, Grigg MJ, William T, Yeo TW, Anstey NM, 2017a. The treatment of Plasmodium knowlesi malaria. Trends Parasitol. 33, 242–253. 10.1016/j.pt.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Barber BE, Rajahram GS, Grigg MJ, William T, Anstey NM, 2017b. World Malaria report: time to acknowledge Plasmodium knowlesi malaria. Malaria J. 16, 135. 10.1186/s12936-017-1787-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A, Iriart X, Wilhelm N, Valentin A, Cassaing S, Witkowski B, Benoit-Vical F, Menard S, Olagnier D, Fillaux J, Sire S, Coustumier AL, Magnaval JF, 2011. Imported Plasmodium knowlesi malaria in a French tourist returning from Thailand. Am. J. Trop. Med. Hyg 84, 535–538. 10.4269/ajtmh.2011.10-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousema T, Drakeley C, 2011. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin. Microbiol. Rev 24, 377–410. 10.1128/cmr.00051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton S, Cheng Q, Sutherland CJ, McCarthy JS, 2015. A simple, high-throughput, colourimetric, field applicable loop-mediated isothermal amplification (HtLAMP) assay for malaria elimination. Malaria J. 14, 335. 10.1186/s12936-015-0848-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton S, Cheng Q, Grigg MJ, William T, Anstey NM, McCarthy JS, 2016. A sensitive, colorimetric, high-throughput loop-mediated isothermal amplification assay for the detection of Plasmodium knowlesi. Am. J. Trop. Med. Hyg 95, 120–122. 10.4269/ajtmh.15-0670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock PM, Fornace KM, Grigg MJ, Anstey NM, William T, Cox J, Drakeley CJ, Ferguson HM, Kao RR, 2019. Predictive analysis across spatial scales links zoonotic malaria to deforestation. Proc. R. Soc. B 286, 20182351. 10.1098/rspb.2018.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y, Fornace K, Wu L, Arnold BF, Priest JW, Martin DL, Chang MA, Cook J, Stresman G, Drakeley C, 2021. Determining seropositivity—a review of approaches to define population seroprevalence when using multiplex bead assays to assess burden of tropical diseases. PLos Negl. Trop. D 15, e0009457. 10.1371/journal.pntd.0009457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong FW, Fong MY, Lau YL, Mahmud R, 2013a. Immunogenicity of bacterial-expressed recombinant Plasmodium knowlesi merozoite surface protein-142 (MSP-142). Malaria J. 12, 454. 10.1186/1475-2875-12-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong F-W, Lau Y-L, Fong M-Y, Mahmud R, 2013b. Evaluation of recombinant Plasmodium knowlesi merozoite surface protein-1(33) for detection of human malaria. Am. J. Trop. Med. Hyg 88, 835–840. 10.4269/ajtmh.12-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew CH, Lim YAL, Lee PC, Mahmud R, Chua KH, 2012. Hexaplex PCR detection system for identification of five human Plasmodium species with an internal control. J. Clin. Microbiol 50, 4012–4019. 10.1128/jcm.06454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin W, Contacos PG, Coatney GR, Kimball HR, 1965. A naturally acquired quotidian-type malaria in man transferable to monkeys. Science 149, 865. [DOI] [PubMed] [Google Scholar]
- Chin W, Contacos PG, Collins WE, Jeter MH, Alpert E, 1968. Experimental mosquito-transmission of Plasmodium knowlesi to man and monkey. Am. J. Trop. Med. Hyg 17, 355–358. [DOI] [PubMed] [Google Scholar]
- Chin AZ, Maluda MCM, Jelip J, Jeffree MSB, Culleton R, Ahmed K, 2020. Malaria elimination in Malaysia and the rising threat of Plasmodium knowlesi. J. Physiol. Anthropol 39, 36. 10.1186/s40101-020-00247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coatney GR, Collins WE, Warren M, Contacos PG, 1971. The Primate Malarias. US Government. [Google Scholar]
- Collins WE, Jeffery GM, 2007. Plasmodium malariae: parasite and disease. Clin. Microbiol. Rev 20, 579–592. 10.1128/cmr.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway DJ, 2007. Molecular epidemiology of malaria. Clin. Microbiol. Rev 20, 188–204. 10.1128/cmr.00021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DJ, Rajahram GS, William T, Jelip J, Mohammad R, Benedict J, Alaza DA, Malacova E, Yeo TW, Grigg MJ, Anstey NM, Barber BE, 2020. Plasmodium knowlesi malaria in Sabah, Malaysia, 2015–2017: ongoing increase in incidence despite near-elimination of the human-only Plasmodium species. Clin. Infect. Dis 70. ciz237 10.1093/cid/ciz237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corran P, Coleman P, Riley E, Drakeley C, 2007. Serology: a robust indicator of malaria transmission intensity? Trends Parasitol. 23, 575–582. 10.1016/j.pt.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Coutrier FN, Tirta YK, Cotter C, Zarlinda I, González IJ, Schwartz A, Maneh C, Marfurt J, Murphy M, Herdiana H, Anstey NM, Greenhouse B, Hsiang MS, Noviyanti R, 2018. Laboratory challenges of Plasmodium species identification in Aceh Province, Indonesia, a malaria elimination setting with newly discovered P. knowlesi. PLoS Negl. Trop. Dis 12, e0006924. 10.1371/journal.pntd.0006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Singh J, Singh B, 2008. Knowlesi malaria: newly emergent and of public health importance? Trends Parasitol. 24, 406–410. 10.1016/j.pt.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, Rahman HA, Conway DJ, Singh B, 2008. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin. Infect. Dis 46, 165–171. 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Wanaguru M, McDade B, Osier FH, Marsh K, Rayner JC, Wright GJ, 2013. A library of functional recombinant cell-surface and secreted P. falciparum merozoite proteins. Mol. Cell Proteomics 12, 3976–3986. 10.1074/mcp.o113.028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divis PC, Shokoples SE, Singh B, Yanow SK, 2010. A TaqMan real-time PCR assay for the detection and quantitation of Plasmodium knowlesi. Malaria J. 9, 344. 10.1186/1475-2875-9-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divis PCS, Singh B, Anderios F, Hisam S, Matusop A, Kocken CH, Assefa SA, Duffy CW, Conway DJ, 2015. Admixture in humans of two divergent Plasmodium knowlesi populations associated with different macaque host species. PLoS Pathog. 11, e1004888. 10.1371/journal.ppat.1004888.s014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakeley CJ, Corran PH, Coleman PG, Tongren JE, McDonald SLR, Carneiro I, Malima R, Lusingu J, Manjurano A, Nkya WMM, Lemnge MM, Cox J, Reyburn H, Riley EM, 2005. Estimating medium- and long-term trends in malaria transmission by using serological markers of malaria exposure. Proc. Natl. Acad. Sci. U. S. A 102, 5108–5113. 10.1073/pnas.0408725102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards HM, Dixon R, de Beyl CZ, Celhay O, Rahman M, Oo MM, Lwin T, Lin Z, San T, Han KT, Nyunt MM, Plowe C, Stresman G, Hall T, Drakeley C, Hamade P, Aryal S, Roca-Feltrer A, Hlaing T, Thi A, 2021. Prevalence and seroprevalence of Plasmodium infection in Myanmar reveals highly heterogeneous transmission and a large hidden reservoir of infection. PLos One 16, e0252957. 10.1371/journal.pone.0252957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Freeland DE, Collins WE, Lal AA, 1998. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc. Natl. Acad. Sci. U. S. A 95, 8124–8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagin JE, 1992. The 6-kb element of Plasmodium falciparum encodes mitochondrial cytochrome genes. Mol. Biochem. Parasitol 52, 145–148. 10.1016/0166-6851(92)90046-m. [DOI] [PubMed] [Google Scholar]
- Figtree M, Lee R, Bain L, Kennedy T, Mackertich S, Urban M, Cheng Q, Hudson BJ, 2010. Plasmodium knowlesi in human, Indonesian Borneo. Emerg. Infect. Dis 16, 672–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firdaus ER, Park J-H, Muh F, Lee S-K, Han J-H, Lim C-S, Na S-H, Park WS, Park J-H, Han E-T, 2021. Performance evaluation of biozentech malaria scanner in Plasmodium knowlesi and P. falciparum as a new diagnostic tool. Korean J. Parasitol 59, 113–119. 10.3347/kjp.2021.59.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace KM, Abidin TR, Alexander N, Brock P, Grigg MJ, Murphy A, William T, Menon J, Drakeley CJ, Cox J, 2016a. Association between landscape factors and spatial patterns of Plasmodium knowlesi infections in Sabah, Malaysia. Emerg. Infect. Dis 22, 201–209. 10.3201/eid2202.150656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace KM, Nuin NA, Betson M, Grigg MJ, William T, Anstey NM, Yeo TW, Cox J, Ying LT, Drakeley CJ, 2016b. Asymptomatic and submicroscopic carriage of Plasmodium knowlesi malaria in household and community members of clinical cases in Sabah, Malaysia. J. Infect. Dis 213, 784–787. 10.1093/infdis/jiv475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace KM, Herman LS, Abidin TR, Chua TH, Daim S, Lorenzo PJ, Grignard L, Nuin NA, Ying LT, Grigg MJ, William T, Espino F, Cox J, Tetteh KKA, Drakeley CJ, 2018. Exposure and infection to Plasmodium knowlesi in case study communities in Northern Sabah, Malaysia and Palawan, The Philippines. PLos Negl. Trop. D 12, e0006432. 10.1371/journal.pntd.0006432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace KM, Alexander N, Abidin TR, Brock PM, Chua TH, Vythilingam I, Ferguson HM, Manin BO, Wong ML, Ng SH, Cox J, Drakeley C, 2019a. Local human movement patterns and land use impact exposure to zoonotic malaria in Malaysian Borneo. eLife 8, 327. 10.7554/elife.47602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace KM, Brock PM, Abidin TR, Grignard L, Herman LS, Chua TH, Daim S, William T, Patterson CLEB, Hall T, Grigg MJ, Anstey NM, Tetteh KKA, Cox J, Drakeley CJ, 2019b. Environmental risk factors and exposure to the zoonotic malaria parasite Plasmodium knowlesi across northern Sabah, Malaysia: a population-based cross-sectional survey. Lancet Planet. Heal 3, e179–e186. 10.1016/s2542-5196(19)30045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsström B, Axnäs BB, Rockberg J, Danielsson H, Bohlin A, Uhlen M, 2015. Dissecting antibodies with regards to linear and conformational epitopes. PLos One 10, e0121673. 10.1371/journal.pone.0121673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster D, Cox-Singh J, Mohamad DS, Krishna S, Chin PP, Singh B, 2014. Evaluation of three rapid diagnostic tests for the detection of human infections with Plasmodium knowlesi. Malaria J. 13, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghinai I, Cook J, Hla TTW, Htet HMT, Hall T, Lubis IN, Ghinai R, Hesketh T, Naung Y, Lwin MM, Latt TS, Heymann DL, Sutherland CJ, Drakeley C, Field N, 2017. Malaria epidemiology in central Myanmar: identification of a multi-species asymptomatic reservoir of infection. Malaria J. 16, 16. 10.1186/s12936-016-1651-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhouse B, Daily J, Guinovart C, Goncalves B, Beeson J, Bell D, Chang MA, Cohen JM, Ding X, Domingo G, Eisele TP, Lammie PJ, Mayor A, Merienne N, Monteiro W, Painter J, Rodriguez I, White M, Drakeley C, Müeller I, Convening MS, 2019. Priority use cases for antibody-detecting assays of recent malaria exposure as tools to achieve and sustain malaria elimination. Gates Open Res. 3, 131. 10.12688/gatesopenres.12897.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg MJ, William T, Barber BE, Parameswaran U, Bird E, Piera K, Aziz A, Dhanaraj P, Yeo TW, Anstey NM, 2014. Combining parasite lactate dehydrogenase-based and histidine-rich protein 2-based rapid tests to improve specificity for diagnosis of malaria due to Plasmodium knowlesi and other Plasmodium species in Sabah, Malaysia. J. Clin. Microbiol 52, 2053–2060. 10.1128/jcm.00181-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg MJ, Barber BE, Marfurt J, Imwong M, William T, Bird E, Piera KA, Aziz A, Boonyuen U, Drakeley CJ, Cox J, White NJ, Cheng Q, Yeo TW, Auburn S, Anstey NM, 2016a. Dihydrofolate-reductase mutations in Plasmodium knowlesi appear unrelated to selective drug pressure from putative human-to-human transmission in Sabah, Malaysia. PLos One 11, e0149519. 10.1371/journal.pone.0149519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg MJ, William T, Menon J, Dhanaraj P, Barber BE, Wilkes CS, von Seidlein L, Rajahram GS, Pasay C, McCarthy JS, Price RN, Anstey NM, Yeo TW, 2016b. Artesunate–mefloquine versus chloroquine for treatment of uncomplicated Plasmodium knowlesi malaria in Malaysia (ACT KNOW): an open-label, randomised controlled trial. Lancet Infect. Dis 16, 180–188. 10.1016/s1473-3099(15)00415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg MJ, William T, Barber BE, Rajahram GS, Menon J, Schimann E, Piera K, Wilkes CS, Patel K, Chandna A, Drakeley CJ,Yeo TW, Anstey NM, 2018a. Age-related clinical spectrum of Plasmodium knowlesi Malaria and predictors of severity. Clin. Infect. Dis 67, 350–359. 10.1093/cid/ciy065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg MJ, William T, Barber BE, Rajahram GS, Menon J, Schimann E, Wilkes CS, Patel K, Chandna A, Price RN, Yeo TW, Anstey NM, 2018b. Artemether-lumefantrine versus chloroquine for the treatment of uncomplicated Plasmodium knowlesi Malaria: an open-label randomized controlled trial CAN KNOW. Clin. Infect. Dis 66, 229–236. 10.1093/cid/cix779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignard L, Shah S, Chua TH, William T, Drakeley CJ, Fornace KM, 2019. Natural human infections with Plasmodium cynomolgi and other malaria species in an elimination setting in Sabah, Malaysia. J. Infect. Dis 220, 1946–1949. 10.1093/infdis/jiz397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi H, Asada M, Angeles JMM, Kawai S, Inoue N, Kawazu S, 2013. Plasmodium vivax and Plasmodium knowlesi: cloning, expression and functional analysis of 1-Cys peroxiredoxin. Exp. Parasitol 133, 101–105. 10.1016/j.exppara.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Han E-T, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, Iriko H, Jin L, Takeo S, Tsuboi T, 2007. Detection of four plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J. Clin. Microbiol 45, 2521–2528. 10.1128/jcm.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawking F, Worms MJ, Gammage K, 1968. Host temperature and control of 24-hour and 48-hour cycles in malaria parasites. Lancet 291, 506–509. 10.1016/s0140-6736(68)91469-4. [DOI] [PubMed] [Google Scholar]
- Helb DA, Tetteh KKA, Felgner PL, Skinner J, Hubbard A, Arinaitwe E, Mayanja-Kizza H, Ssewanyana I, Kamya MR, Beeson JG, Tappero J, Smith DL, Crompton PD, Rosenthal PJ, Dorsey G, Drakeley CJ, Greenhouse B, 2015. Novel serologic biomarkers provide accurate estimates of recent Plasmodium falciparum exposure for individuals and communities. Proc. Natl. Acad. Sci. U. S. A 112, E4438–E4447. 10.1073/pnas.1501705112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdiana H, Cotter C, Coutrier FN, Zarlinda I, Zelman BW, Tirta YK, Greenhouse B, Gosling RD, Baker P, Whittaker M, Hsiang MS, 2016. Malaria risk factor assessment using active and passive surveillance data from Aceh Besar, Indonesia, a low endemic, malaria elimination setting with Plasmodium knowlesi, Plasmodium vivax, and Plasmodium falciparum. Malaria J. 15, 468. 10.1186/s12936-016-1523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman LS, Fornace K, Phelan J, Grigg MJ, Anstey NM, William T, Moon RW, Blackman MJ, Drakeley CJ, Tetteh KKA, 2018. Identification and validation of a novel panel of Plasmodium knowlesi biomarkers of serological exposure. PLos Negl. Trop. D 12, e0006457. 10.1371/journal.pntd.0006457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins H, González IJ, Polley SD, Angutoko P, Ategeka J, Asiimwe C, Agaba B, Kyabayinze DJ, Sutherland CJ, Perkins MD, Bell D, 2013. Highly sensitive detection of malaria parasitemia in a malaria-endemic setting: performance of a new loop-mediated isothermal amplification kit in a remote clinic in Uganda. J. Infect. Dis 208, 645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussin N, Lim YA-L, Goh PP, William T, Jelip J, Mudin RN, 2020. Updates on malaria incidence and profile in Malaysia from 2013 to 2017. Malaria J. 19. 55–14 10.1186/s12936-020-3135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ido Y, Hashimoto M, Yatsushiro S, Tanaka M, Yokota K, Kajimoto K, Kataoka M, 2019. Loop-mediated isothermal amplification in microchambers on a cell microarray chip for identification of Plasmodium species. J. Parasitol 105, 69–74. 10.1645/18-107. [DOI] [PubMed] [Google Scholar]
- Imai K, Tarumoto N, Misawa K, Runtuwene LR, Sakai J, Hayashida K, Eshita Y, Maeda R, Tuda J, Murakami T, Maesaki S, Suzuki Y, Yamagishi J, Maeda T, 2017. A novel diagnostic method for malaria using loop-mediated isothermal amplification (LAMP) and MinION™ nanopore sequencer. BMC Infect. Dis 17, 621. 10.1186/s12879-017-2718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Tanomsing N, Pukrittayakamee S, Day NPJ, White NJ, Snounou G, 2009. Spurious amplification of a Plasmodium vivax small-subunit RNA gene by use of primers currently used to detect P. knowlesi. J. Clin. Microbiol 47, 4173–4175. 10.1128/jcm.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Hanchana S, Malleret B, Renia L, Day NPJ, Dondorp A, Nosten F, Snounou G, White NJ, 2014. High-throughput ultrasensitive molecular techniques for quantifying low-density malaria parasitemias. J. Clin. Microbiol 52, 3303–3309. 10.1128/jcm.01057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Madmanee W, Suwannasin K, Kunasol C, Peto TJ, Tripura R, von Seidlein L, Nguon C, Davoeung C, Day NPJ, Dondorp AM, White NJ, 2019. Asymptomatic natural human infections with the simian malaria parasites Plasmodium cynomolgi and Plasmodium knowlesi. J Infect Dis 219, 695–702. 10.1093/infdis/jiy519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseki H, Kawai S, Takahashi N, Hirai M, Tanabe K, Yokoyama N, Igarashi I, 2010. Evaluation of a loop-mediated isothermal amplification method as a tool for diagnosis of infection by the zoonotic simian malaria parasite Plasmodium knowlesi. J. Clin. Microbiol 48, 2509–2514. 10.1128/jcm.00331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwagami M, Nakatsu M, Khattignavong P, Soundala P, Lorphachan L, Keomalaphet S, Xangsayalath P, Kawai S, Hongvanthong B, Brey PT, Kano S, 2018. First case of human infection with Plasmodium knowlesi in Laos. PLoS Negl. Trop. Dis 12, e0006244. 10.1371/journal.pntd.0006244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeslyn WPS, Huat TC, Vernon L, Irene LMZ, Sung LK, Jarrod LP, Singh B, Ching NL, 2011. Molecular epidemiological investigation of Plasmodium knowlesi in humans and macaques in Singapore. Vector Borne Zoon. Dis 11, 131–135. 10.1089/vbz.2010.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiram A, Hisam S, Reuben H, Husin SZ, Roslan A, Ismail WRW, 2016. Submicroscopic evidence of the simian malaria parasite, Plasmodium knowlesi, in an Orang Asli community. Southeast Asian J. Trop. Med. Public Health 47, 591–599. [Google Scholar]
- Jongwutiwes S, Putaporntip C, Iwasaki T, Sata T, Kanbara H, 2004. Naturally acquired Plasmodium knowlesi malaria in human, Thailand. Emerg. Infect. Dis 10, 2211–2213. 10.3201/eid1012.040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongwutiwes S, Buppan P, Kosuvin R, Seethamchai S, Pattanawong U, Sirichaisinthop J, Putaporntip C, 2011. Plasmodium knowlesi malaria in humans and macaques, Thailand. Emerg. Infect. Dis 17, 1799–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan IG, Mathews HM, Rogers WA, Fried J, 1969. Seroepidemiological studies by indirect haemagglutination tests for malaria. Military recruit collections from Argentina, Brazil, Colombia, and the United States of America. Bull. World Heal. Organ 41, 825–841. [PMC free article] [PubMed] [Google Scholar]
- Kawai S, Hirai M, Haruki K, Tanabe K, Chigusa Y, 2009. Cross-reactivity in rapid diagnostic tests between human malaria and zoonotic simian malaria parasite Plasmodium knowlesi infections. Parasitol. Int 58, 300–302. 10.1016/j.parint.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Kawai S, Sato M, Kato-Hayashi N, Kishi H, Huffman MA, Maeno Y, Culleton R, Nakazawa S, 2014. Detection of Plasmodium knowlesi DNA in the urine and faeces of a Japanese macaque (Macaca fuscata) over the course of an experimentally induced infection. Malaria J. 13, 1–9. 10.1186/1475-2875-13-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Kaneko O, Liu Q, Zhou M, Kawamoto F, Wataya Y, Otani S, Yamaguchi Y, Tanabe K, 1997. Identification of the four species of human malaria parasites by nested PCR that targets variant sequences in the small subunit rRNA gene. Parasitol. Int 46, 91–95. 10.1016/s1383-5769(97)00013-5. [DOI] [Google Scholar]
- Knowles R, Gupta BMD, 1932. A study of monkey-malaria and its experimental transmission to man. Indian Med. Gaz, 246–249. [PMC free article] [PubMed] [Google Scholar]
- Lai MY, Ooi CH, Jaimin JJ, Lau YL, 2020. Evaluation of warmstart colorimetric loop-mediated isothermal amplification assay for diagnosis of malaria. Am. J. Trop. Med. Hyg 102, 1370–1372. 10.4269/ajtmh.20-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MY, Ooi CH, Lau YL, 2021. Validation of SYBR green I based closed-tube loop-mediated isothermal amplification (LAMP) assay for diagnosis of knowlesi malaria. Malaria J. 20, 166. 10.1186/s12936-021-03707-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau Y-L, Fong M-Y, Mahmud R, Chang P-Y, Palaeya V, Cheong F-W, Chin L-C, Anthony CN, Al-Mekhlafi AM, Chen Y, 2011. Specific, sensitive and rapid detection of human plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malaria J. 10, 197. 10.1186/1475-2875-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau YL, Cheong FW, Chin LC, Mahmud R, Chen Y, Fong MY, 2014. Evaluation of codon optimized recombinant Plasmodium knowlesi merozoite surface protein-119 (pkMSP-119) expressed in Pichia pastoris. Trop. Biomed 31, 749–759. [PubMed] [Google Scholar]
- Lau Y-L, Lai M-Y, Fong M-Y, Jelip J, Mahmud R, 2016. Loop-mediated isothermal amplification assay for identification of five human Plasmodium species in Malaysia. Am. J. Trop. Med. Hyg 94, 336–339. 10.4269/ajtmh.15-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Cox-Singh J, Brooke G, Matusop A, Singh B, 2009a. Plasmodium knowlesi from archival blood films: further evidence that human infections are widely distributed and not newly emergent in Malaysian Borneo. Int. J. Parasitol 39, 1125–1128. 10.1016/j.ijpara.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Cox-Singh J, Singh B, 2009b. Morphological features and differential counts of Plasmodium knowlesi parasites in naturally acquired human infections. Malaria J. 8, 73. 10.1186/1475-2875-8-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Divis PCS, Zakaria SK, Matusop A, Julin RA, Conway DJ, Cox-Singh J, Singh B, 2011. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 7, e1002015. 10.1371/journal.ppat.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link L, Bart A, Verhaar N, van Gool T, Pronk M, Scharnhorst V, 2012. Molecular detection of Plasmodium knowlesi in a Dutch traveler by real-time PCR. J. Clin. Microbiol 50, 2523–2524. 10.1128/jcm.06859-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley RJ, White MT, Takashima E, Brewster J, Morita M, Harbers M, Obadia T, Robinson LJ, Matsuura F, Liu ZSJ, Li-Wai-Suen CSN, Tham W-H, Healer J, Huon C, Chitnis CE, Nguitragool W, Monteiro W, Proietti C, Doolan DL, Siqueira AM, Ding XC, González IJ, Kazura J, Lacerda M, Sattabongkot J, Tsuboi T, Müeller I, 2020. Development and validation of serological markers for detecting recent Plasmodium vivax infection. Nat. Med 26, 1–19. 10.1038/s41591-020-0841-4. [DOI] [PubMed] [Google Scholar]
- Lubis IND, Wijaya H, Lubis M, Lubis CP, Divis PCS, Beshir KB, Sutherland CJ, 2017. Contribution of Plasmodium knowlesi to multispecies human malaria infections in North Sumatera, Indonesia. J. Infect. Dis 215, 1148–1155. 10.1093/infdis/jix091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchi NW, Poorak M, Oberstaller J, DeBarry J, Srinivasamoorthy G, Goldman I, Xayavong M, da Silva AJ, Peterson DS, Barnwell JW, Kissinger J, Udhayakumar V, 2012. A new single-step PCR assay for the detection of the zoonotic malaria parasite Plasmodium knowlesi. PLoS One 7, e31848. 10.1371/journal.pone.0031848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchi NW, Gaye M, Diallo MA, Goldman IF, Ljolje D, Deme AB, Badiane A, Ndiaye YD, Barnwell JW, Udhayakumar V, Ndiaye D, 2016a. Evaluation of the Illumigene malaria LAMP: a robust molecular diagnostic tool for malaria parasites. Sci. Rep. UK 6, 36808. 10.1038/srep36808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchi NW, Ljolje D, Silva-Flannery L, Udhayakumar V, 2016b. Use of malachite green-loop mediated isothermal amplification for detection of Plasmodium spp. Parasites. PLos One 11, e0151437. 10.1371/journal.pone.0151437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucky AB, Sakaguchi M, Katakai Y, Kawai S, Yahata K, Templeton TJ, Kaneko O, 2016. Plasmodium knowlesi skeleton-binding protein 1 localizes to the ‘Sinton and Mulligan’ stipplings in the cytoplasm of monkey and human erythrocytes. PLos One 11, e0164272. 10.1371/journal.pone.0164272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeno Y, Culleton R, Quang NT, Kawai S, Marchand RP, Nakazawa S, 2017. Plasmodium knowlesi and human malaria parasites in Khan Phu, Vietnam: gametocyte production in humans and frequent co-infection of mosquitoes. Parasitology 144, 527–535. 10.1017/s0031182016002110. [DOI] [PubMed] [Google Scholar]
- Mahajan B, Jani D, Chattopadhyay R, Nagarkatti R, Zheng H, Majam V, Weiss W, Kumar S, Rathore D, 2005. Identification, cloning, expression, and characterization of the gene for Plasmodium knowlesi surface protein containing an altered thrombospondin repeat domain. Infect. Immun 73, 5402–5409. 10.1128/iai.73.9.5402-5409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahittikorn A, Masangkay FR, Kotepui KU, Milanez GDJ, Kotepui M, 2021. Quantification of the misidentification of Plasmodium knowlesi as Plasmodium malariae by microscopy: an analysis of 1569 P. knowlesi cases. Malaria J. 20, 179. 10.1186/s12936-021-03714-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallepaddi PC, Lai M-Y, Lau Y-L, Liew JW-K, Polavarapu R, Podha S, Ooi C-H, 2018. Development of loop-mediated isothermal amplification–based lateral flow device method for the detection of malaria. Am. J. Trop. Med. Hyg 99, 704–708. 10.4269/ajtmh.18-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao R, Ge G, Wang Z, Hao R, Zhang G, Yang Z, Lin B, Ma Y, Liu H, Du Y, 2018. A multiplex microfluidic loop-mediated isothermal amplification array for detection of malaria-related parasites and vectors. Acta Trop. 178, 86–92. 10.1016/j.actatropica.2017.10.025. [DOI] [PubMed] [Google Scholar]
- Marchand RP, Culleton R, Maeno Y, Quang NT, Nakazawa S, 2011. Co-infections of Plasmodium knowlesi, P. falciparum, and P. vivax among humans and anopheles dirus mosquitoes, Southern Vietnam. Emerg. Infect. Dis 17, 1232–1239. 10.3201/eid1707.101551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutchan TF, McCutchan TF, Piper RC, Piper RC, Makler MT, Makler MT, 2008. Use of malaria rapid diagnostic test to identify Plasmodium knowlesi infection. Emerg. Infect. Dis 14, 1750–1752. 10.3201/eid1411.080480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercereau-Puijalon O, Barale J-C, Bischoff E, 2002. Three multigene families in Plasmodium parasites: facts and questions. Int. J. Parasitol 32, 1323–1344. 10.1016/s0020-7519(02)00111-x. [DOI] [PubMed] [Google Scholar]
- Mideo N, Reece SE, Smith AL, Metcalf CJE, 2013. The Cinderella syndrome: why do malaria-infected cells burst at midnight? Trends Parasitol. 29, 10–16. 10.1016/j.pt.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Cruz A, Canepa GE, Silva TLAE, Williams AE, Nagyal S, Yenkoidiok-Douti L, Nagata BM, Calvo E, Andersen J, Boulanger MJ, Barillas-Mury C, 2020. Plasmodium falciparum evades immunity of anopheline mosquitoes by interacting with a Pfs47 midgut receptor. Proc. Natl. Acad. Sci. U. S. A 117, 2597–2605. 10.1073/pnas.1917042117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Nagamine K, Tomita N, Notomi T, 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun 289, 150–154. 10.1006/bbrc.2001.5921. [DOI] [PubMed] [Google Scholar]
- Moyes CL, Henry AJ, Golding N, Huang Z, Singh B, Baird JK, Newton PN, Huffman M, Duda KA, Drakeley CJ, Elyazar IRF, Anstey NM, Chen Q, Zommers Z, Bhatt S, Gething PW, Hay SI, 2014. Defining the geographical range of the Plasmodium knowlesi reservoir. PLoS Negl. Trop. Dis 8, e2780. 10.1371/journal.pntd.0002780.s004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muh F, Kim N, Nyunt MH, Firdaus ER, Han J-H, Hoque MR, Lee S-K, Park J-H, Moon RW, Lau Y-L, Kaneko O, Han ET, 2020. Cross-species reactivity of antibodies against Plasmodium vivax blood-stage antigens to Plasmodium knowlesi. PLoS Negl. Trop. Dis 14. e0008323–21 10.1371/journal.pntd.0008323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Sienerth N, Shilts J, Kadir KA, Yman V, Homann MV, Asghar M, Ngasala B, Singh B, Färnert A, Wright GJ, 2020. A panel of recombinant proteins from human-infective Plasmodium species for serological surveillance. Malaria J. 19, 31. 10.1186/s12936-020-3111-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi ZG, Oormazdi H, Akhlaghi L, Meamar AR, Nateghpour M, Farivar L, Razmjou E, 2014. Detection of Plasmodium vivax and Plasmodium falciparum DNA in human saliva and urine: loop-mediated isothermal amplification for malaria diagnosis. Acta Trop. 136, 44–49. 10.1016/j.actatropica.2014.03.029. [DOI] [PubMed] [Google Scholar]
- Narum DL, Thomas AW, 1994. Differential localization of full-length and processed forms of PF83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol 67, 59–68. 10.1016/0166-6851(94)90096-5. [DOI] [PubMed] [Google Scholar]
- Noordin NR, Lee PY, Bukhari FDM, Fong MY, Hamid MHA, Jelip J, Mudin RN, Lau YL, 2020. Prevalence of asymptomatic and/or low-density malaria infection among high-risk groups in Peninsular Malaysia. Am. J. Trop. Med. Hyg 103, 1107–1110. 10.4269/ajtmh.20-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T, 2000. Loop-mediated isothermal amplification of DNA. Nucl. Acids Res 28, e63. 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuin NA, Tan AF, Lew YL, Piera KA, William T, Rajahram GS, Jelip J, Dony JF, Mohammad R, Cooper DJ, Barber BE, Anstey NM, Chua TH, Grigg MJ, 2020. Comparative evaluation of two commercial real-time PCR kits (QuantiFast™ and abTES™) for the detection of Plasmodium knowlesi and other Plasmodium species in Sabah, Malaysia. Malaria J. 19, 306. 10.1186/s12936-020-03379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddoux O, Debourgogne A, Kantele A, Kocken CH, Jokiranta TS, Vedy S, Puyhardy JM, Machouart M, 2011. Identification of the five human Plasmodium species including P. knowlesi by real-time polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol 30, 597–601. 10.1007/s10096-010-1126-5. [DOI] [PubMed] [Google Scholar]
- Ong CWM, Lee SY, Koh WH, Ooi EE, Tambyah PA, 2009. Monkey malaria in humans: a diagnostic dilemma with conflicting laboratory data. Am.J. Trop. Med. Hyg 80, 927–928. [PubMed] [Google Scholar]
- Pain A, Bohme U, Berry AE, Mungall K, Finn RD, Jackson AP, Mourier T, Mistry J, Pasini EM, Aslett MA, Balasubrammaniam S, Borgwardt K, Brooks K, Carret C, Carver TJ, Cherevach I, Chillingworth T, Clark TG, Galinski MR, Hall N, Harper D, Harris D, Hauser H, Ivens A, Janssen CS, Keane T, Larke N, Lapp S, Marti M, Moule S, Meyer IM, Ormond D, Peters N, Sanders M, Sanders S, Sargeant TJ, Simmonds M, Smith F, Squares R, Thurston S, Tivey AR, Walker D, White B, Zuiderwijk E, Churcher C, Quail MA, Cowman AF, Turner CMR, Rajandream MA, Kocken CHM, Thomas AW, Newbold CI, Barrell BG, Berriman M, 2008. The genome of the simian and human malaria parasite Plasmodium knowlesi. Nature 455, 799–803. 10.1038/nature07306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JC, Oberstaller J, Xayavong M, Narayanan J, DeBarry JD, Srinivasamoorthy G, Villegas L, Escalante AA, DaSilva A, Peterson DS, Barnwell JW, Kissinger JC, Udhayakumar V, Lucchi NW, 2013. Real-time loop-mediated isothermal amplification (RealAmp) for the species-specific identification of Plasmodium vivax. PLoS One 8, e54986. 10.1371/journal.pone.0054986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perandin F, Manca N, Calderaro A, Piccolo G, Galati L, Ricci L, Medici MC, Arcangeletti MC, Snounou G, Dettori G, Chezzi C, 2004. Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J. Clin. Microbiol 42, 1214. 10.1128/jcm.42.3.1214-1219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera RS, Ding XC, Tully F, Oliver J, Bright N, Bell D, Chiodini PL, Gonzalez IJ, Polley SD, 2017. Development and clinical performance of high throughput loop-mediated isothermal amplification for detection of malaria. PLos One 12, e0171126. 10.1371/journal.pone.0171126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piera KA, Aziz A, William T, Bell D, González IJ, Barber BE, Anstey NM, Grigg MJ, 2017. Detection of Plasmodium knowlesi, Plasmodium falciparum and Plasmodium vivax using loop-mediated isothermal amplification (LAMP) in a co-endemic area in Malaysia. Malaria J. 16, 29. 10.1186/s12936-016-1676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plucinski MM, Candrinho B, Chambe G, Muchanga J, Muguande O, Matsinhe G, Mathe G, Rogier E, Doyle T, Zulliger R, Colborn J, Saifodine A, Lammie P, Priest JW, 2018. Multiplex serology for impact evaluation of bed net distribution on burden of lymphatic filariasis and four species of human malaria in northern Mozambique. PLos Negl. Trop. D 12, e0006278. 10.1371/journal.pntd.0006278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SD, Mori Y, Watson J, Perkins MD, Gonzalez IJ, Notomi T, Chiodini PL, Sutherland CJ, 2010. Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J. Clin. Microbiol 48, 2866–2871. 10.1128/jcm.00355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SD, González IJ, Mohamed D, Daly R, Bowers K, Watson J, Mewse E, Armstrong M, Gray C, Perkins MD, Bell D, Kanda H, Tomita N, Kubota Y, Mori Y, Chiodini PL, Sutherland CJ, 2013. Clinical evaluation of a loop-mediated amplification kit for diagnosis of imported malaria. J. Infect. Dis 208, 637–644. 10.1093/infdis/jit183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon LLM, Wong BWY, Ma EHT, Chan KH, Chow LMC, Abeyewickreme W, Tangpukdee N, Yuen KY, Guan Y, Looareesuwan S, Peiris JSM, 2006. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin. Chem 52, 303–306. 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- Prasad R, Atul, Soni A, Puri SK, Sijwali PS, 2012. Expression, characterization, and cellular localization of knowpains, papain-like cysteine proteases of the Plasmodium knowlesi malaria parasite. PLos One 7, e51619. 10.1371/journal.pone.0051619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putaporntip C, Buppan P, Jongwutiwes S, 2011. Improved performance with saliva and urine as alternative DNA sources for malaria diagnosis by mitochondrial DNA-based PCR assays. Clin. Microbiol. Infect 17, 1484–1491. 10.1111/j.1469-0691.2011.03507.x. [DOI] [PubMed] [Google Scholar]
- Rajahram GS, Barber BE, William T, Menon J, Anstey NM, Yeo TW, 2012. Deaths due to Plasmodium knowlesi malaria in Sabah, Malaysia: association with reporting as Plasmodium malariae and delayed parenteral artesunate. Malaria J. 11, 284. 10.1186/1475-2875-11-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajahram GS, Cooper DJ, William T, Grigg MJ, Anstey NM, Barber BE, 2019. Deaths from Plasmodium knowlesi malaria: case series and systematic review. Clin. Infect. Dis 69, ciz011. 10.1093/cid/ciz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reller ME, et al. 2013. Multiplex 5′ nuclease quantitative real-time PCR for clinical diagnosis of malaria and species-level identification and epidemiologic evaluation of malaria-causing parasites, including Plasmodium knowlesi. J. Clin. Microbiol 9 (51). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rypien C, Chow B, Chan WW, Church DL, Pillai DR, 2017. Detection of plasmodium infection by the illumigene malaria assay compared to reference microscopy and real-time PCR. J. Clin. Microbiol 55, 3037–3045. 10.1128/jcm.00806-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sema M, Alemu A, Bayih AG, Getie S, Getnet G, Guelig D, Burton R, LaBarre P, Pillai DR, 2015. Evaluation of non-instrumented nucleic acid amplification by loop-mediated isothermal amplification (NINA-LAMP) for the diagnosis of malaria in Northwest Ethiopia. Malaria J. 14, 44. 10.1186/s12936-015-0559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiadi W, et al. 2016. A zoonotic human infection with simian malaria, Plasmodium knowlesi, in Central Kalimantan, Indonesia. Malaria J. 15, 218. 10.1186/s12936-016-1272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Godson G, 1985. Expression of the major surface antigen of Plasmodium knowlesi sporozoites in yeast. Science 228, 879–882. 10.1126/science.3890178. [DOI] [PubMed] [Google Scholar]
- Shearer FM, Huang Z, Weiss DJ, Wiebe A, Gibson HS, Battle KE, Pigott DM, Brady OJ, Putaporntip C, Jongwutiwes S, Lau Y-L, Manske M, Amato R, Elyazar IRF, Vythilingam I, Bhatt S, Gething PW, Singh B, Golding N, Hay SI, Moyes CL, 2016. Estimating geographical variation in the risk of zoonotic Plasmodium knowlesi infection in countries eliminating malaria. PLoS Negl. Trop. Dis 10, e0004915. 10.1371/journal.pntd.0004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JRD, Lau Y-L, Fong M-Y, 2016. Expression and evaluation of recombinant Plasmodium knowlesi merozoite surface protein-3 (MSP-3) for detection of human malaria. PLoS One 11, e0158998. 10.1371/journal.pone.0158998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siner A, Liew S-T, Kadir KA, Mohamad DSA, Thomas FK, Zulkarnaen M, Singh B, 2017. Absence of Plasmodium inui and Plasmodium cynomolgi, but detection of Plasmodium knowlesi and Plasmodium vivax infections in asymptomatic humans in the Betong division of Sarawak, Malaysian Borneo. Malaria J. 16, 1–11. 10.1186/s12936-017-2064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Daneshvar C, 2013. Human infections and detection of Plasmodium knowlesi. Clin. Microbiol. Rev 26, 165–184. 10.1128/cmr.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Puri SK, Chitnis CE, 2002. Antibodies raised against receptor-binding domain of Plasmodium knowlesi Duffy binding protein inhibit erythrocyte invasion. Mol. Biochem. Parasitol 121, 21–31. 10.1016/s0166-6851(02)00017-8. [DOI] [PubMed] [Google Scholar]
- Singh B, Sung LK, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ, 2004. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363, 1017–1024. 10.1016/s0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- Singh V, Kaushal DC, Rathaur S, Kumar N, Kaushal NA, 2012. Cloning, overexpression, purification and characterization of Plasmodium knowlesi lactate dehydrogenase. Prot. Exp. Purif 84, 195–203. 10.1016/j.pep.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Singh R, Savargaonkar D, Bhatt R, Valecha N, 2013. Rapid detection of Plasmodium vivax in saliva and blood using loop mediated isothermal amplification (LAMP) assay. J. Infect 67, 245–247. 10.1016/jjinf.2013.04.016. [DOI] [PubMed] [Google Scholar]
- Sinton JA, Mulligan HW, 1933. A critical review of the literature relating to the identification of the malarial parasites recorded from monkeys of the families Cercopithecidae and Colobidae. Rec. Malaria Surv. Ind 3, 381–444. [Google Scholar]
- Snounou G, Singh B, 2002. Nested PCR analysis of Plasmodium parasites. Meth. Mol. Med 72, 189–203. 10.1385/1-59259-271-6:189. [DOI] [PubMed] [Google Scholar]
- Snounou G, Viriyakosol S,Jarra W, Thaithong S, Brown KN, 1993. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol. Biochem. Parasitol 58, 283–292. 10.1016/0166-6851(93)90050-8. [DOI] [PubMed] [Google Scholar]
- Stark DJ, Fornace KM, Brock PM, Abidin TR, Gilhooly L, Jalius C, Goossens B, Drakeley CJ, Salgado-Lynn M, 2019. Long-tailed macaque response to deforestation in a Plasmodium knowlesi-endemic area. EcoHealth 6, e14592–e14599. 10.1007/s10393-019-01403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart L, Gosling R, Griffin J, Gesase S, Campo J, Hashim R, Masika P, Mosha J, Bousema T, Shekalaghe S, Cook J, Corran P, Ghani A, Riley EM, Drakeley C, 2009. Rapid assessment of malaria transmission using age-specific sero-conversion rates. PLoS One 4, e6083. 10.1371/journal.pone.0006083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizaki R, Ujiie M, Kato Y, Iwagami M, Hashimoto A, Kutsuna S, Takeshita N, Hayakawa K, Kanagawa S, Kano S, Ohmagari N, 2013. First case of Plasmodium knowlesi infection in a Japanese traveller returning from Malaysia. Malaria J. 12, 128. 10.1186/1475-2875-12-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanomsing N, Imwong M, Theppabutr S, Pukrittayakamee S, Day NPJ, White NJ, Snounou G, 2010. Accurate and sensitive detection of plasmodium species in humans by use of the dihydrofolate reductase-thymidylate synthase linker region. J. Clin. Microbiol 48, 3735–3737. 10.1128/jcm.00898-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham JM, Lee SH, Tan TM, Ting RC, Kara UA, 1999. Detection and species determination of malaria parasites by PCR: comparison with microscopy and with ParaSight-F and ICT malaria Pf tests in a clinical environment. J. Clin. Microbiol 37, 1269–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres K, Bachman CM, Delahunt CB, Baldeon JA, Alava F, Vilela DG, Proux S, Mehanian C, McGuire SK, Thompson CM, Ostbye T, Hu L, Jaiswal MS, Hunt VM, Bell D, 2018. Automated microscopy for routine malaria diagnosis: a field comparison on Giemsa-stained blood films in Peru. Malaria J. 17, 339. 10.1186/s12936-018-2493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TM, Crompton PD, 2020. Decoding the complexities of human malaria through systems immunology. Immunol. Rev 293, 144–162. 10.1111/imr.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi K, Gupta D, Saini E, Choudhary S, Jamwal A, Alam MS, Zeeshan M, Tyagi RK, Sharma YD, 2015. Recognition of human erythrocyte receptors by the tryptophan-rich antigens of monkey malaria parasite Plasmodium knowlesi. PLoS One 10, e0138691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya AB, Mather MW, 2005. A post-genomic view of the mitochondrion in malaria parasites. Curr. Top. Microbiol 295, 233–250. 10.1007/3-540-29088-5_9. [DOI] [PubMed] [Google Scholar]
- van der Laan MJ, Polley EC, Hubbard AE, 2007. Super learner. Stat. Appl. Genet. Mol. Biol 6. Article 25 10.2202/1544-6115.1309. [DOI] [PubMed] [Google Scholar]
- van Hellemond JJ, Rutten M, Koelewijn R, Zeeman AM, Verweij JJ, Wismans PJ, Kocken CH, van Genderen PJJ, 2009. Human Plasmodium knowlesi infection detected by rapid diagnostic tests for malaria. Emerg. Infect. Dis 15, 1478–1480. 10.3201/eid1509.090358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vythilingam I, 2010. Plasmodium knowlesi in humans: a review on the role of its vectors in Malaysia. Trop. Biomed 27, 1–12. [PubMed] [Google Scholar]
- Waters AP, 1994. The ribosomal RNA genes of Plasmodium. Adv. Parasitol 34, 33–79. 10.1016/s0065-308x(08)60136-0. [DOI] [PubMed] [Google Scholar]
- Waters AP, Thomas AW, Deans JA, Mitchell GH, Hudson DE, Miller LH, McCutchan TF, Cohen S, 1990. A merozoite receptor protein from Plasmodium knowlesi is highly conserved and distributed throughout Plasmodium. J. Biol. Chem 265, 17974–17979. [PubMed] [Google Scholar]
- WHO, 2014. Severe malaria. Trop. Med. Int. Heal 19 (Suppl. 1), 7–131. 10.1111/tmi.12313_2. [DOI] [PubMed] [Google Scholar]
- WHO, 2015a. Microscopy for the Detection, Identification and Quantification of Malaria Parasites on Stained Thick and Thin Blood Films in Research Settings. World Health Organisation. [Google Scholar]
- WHO, 2015b. Summary results of WHO product testing of malaria RDTs: round 1-6 (2008–2015). http://apps.who.int/iris/bitstream/10665/204119/1/9789241510042_eng.pdf. (Accessed 3 August 2016).
- WHO, M.P.A.C., 2017. Outcomes From the Evidence Review Group on Plasmodium knowlesi.
- WHO, 2018. Malaria Rapid Diagnostic Test Performance (Round 8).
- WHO, 2020. WHO|World Malaria Report 2020.
- WHO, 2021. WHO Guidelines for Malaria.
- William T, Menon J, Rajahram G, Chan L, Ma G, Donaldson S, Khoo S, Frederick C, Jelip J, Anstey NM, Yeo TW, 2011. Severe Plasmodium knowlesi malaria in a tertiary care hospital, Sabah, Malaysia. Emerg. Infect. Dis 17, 1248–1255. 10.3201/eid1707.101017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ML, Chua TH, Leong CS, Khaw LT, Fornace K, Wan-Sulaiman W-Y, William T, Drakeley C, Ferguson HM, Vythilingam I, 2015. Seasonal and spatial dynamics of the primary vector of Plasmodium knowlesi within a major transmission focus in Sabah, Malaysia. PLoS Negl. Trop. Dis 9, e0004135. 10.1371/journal.pntd.0004135.t004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Mwesigwa J, Affara M, Bah M, Correa S, Hall T, Singh SK, Beeson JG, Tetteh KKA, Kleinschmidt I, D’Alessandro U, Drakeley C, 2020. Antibody responses to a suite of novel serological markers for malaria surveillance demonstrate strong correlation with clinical and parasitological infection across seasons and transmission settings in The Gambia. BMC Med. 18, 304. 10.1186/s12916-020-01724-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap NJ, Hossain H, Nada-Raja T, Ngui R, Muslim A, Hoh B-P, Khaw LT, Kadir KA, Divis PCS, Vythilingam I, Singh B, Lim YA-L, 2021. Natural human infections with Plasmodium cynomolgi, P. inui, and 4 other simian malaria parasites, Malaysia. Emerg. Infect. Dis 27, 2187–2191. 10.3201/eid2708.204502. [DOI] [PMC free article] [PubMed] [Google Scholar]


