Abstract
Tardigrades are remarkable for their ability to survive harsh stress conditions as diverse as extreme temperature and desiccation. The molecular mechanisms that confer this unusual resistance to physical stress remain unknown. Recently, tardigrade‐unique intrinsically disordered proteins have been shown to play an essential role in tardigrade anhydrobiosis. Here, we characterize the conformational and physical behaviour of CAHS‐8 from Hypsibius exemplaris. NMR spectroscopy reveals that the protein comprises an extended central helical domain flanked by disordered termini. Upon concentration, the protein is shown to successively form oligomers, long fibres, and finally gels constituted of fibres in a strongly temperature‐dependent manner. The helical domain forms the core of the fibrillar structure, with the disordered termini remaining highly dynamic within the gel. Soluble proteins can be encapsulated within cavities in the gel, maintaining their functional form. The ability to reversibly form fibrous gels may be associated with the enhanced protective properties of these proteins.
Keywords: atomic force microscopy, gels, intrinsically disordered proteins, NMR spectroscopy, tardigrades
The conformational behaviour of an intrinsically disordered protein responsible for protecting tardigrades against extreme stress has been characterized. The protein assembles into fibres and fibrillar gels as a function of temperature and concentration. The disordered tails remain flexible within the gel cavities, which can encapsulate client proteins in their native state.
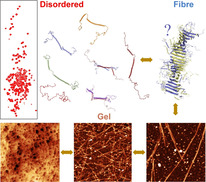
Tardigrades are microscopic aquatic animals [1] that exhibit remarkable resistance to environmental stresses such as extreme temperature, radiation, high pressure, or absence of oxygen or water, a characteristic that has remained mysterious for centuries.[ 2 , 3 , 4 ] In response to such stress, tardigrades transform into a characteristic tun state that can survive for long periods of time. [5] Although the molecular mechanisms that confer this protection against extreme physiological stress remains poorly understood, [6] recent studies have identified the importance of two families of tardigrade‐unique intrinsically disordered proteins (cytosolic abundant heat‐soluble (CAHS) proteins and secreted abundant heat‐soluble (SAHS) proteins).[ 7 , 8 , 9 , 10 ] These proteins are collectively known as tardigrade‐specific disordered proteins (TDPs). [9] RNA interference demonstrated that TDPs were required for desiccation tolerance in tardigrades, while heterologous expression of certain TDPs demonstrated their ability to increase desiccation tolerance in both yeast and E. coli. [9]
The protective role of TDPs remains under discussion in the literature.[ 11 , 12 , 13 ] The observation of the glass transition temperature of TDPs by differential scanning calorimetry was interpreted as evidence that TDPs can reversibly vitrify the cellular environment. [9] Similar phenomena were observed for the intrinsically disordered late embryogenesis abundant (LEA) proteins, [14] through the stabilization and apparent vitrification of co‐solutes such as trehalose. This sugar is known to play an important role in anhydrobiosis in a wide range of organisms, [15] although trehalose is not generally abundant in tardigrades. [16]
To better understand how TDPs alone can confer protective properties in response to desiccation and other environmental stresses, we investigated the conformational behaviour of a CAHS from Hypsibius exemplaris at atomic resolution. By using high‐field NMR spectroscopy, we reveal that the protein comprises an extended central helical domain, with apparent coiled‐coil propensity, flanked by long, highly disordered N‐ and C‐terminal domains. Small‐angle X‐ray scattering (SAXS), atomic force microscopy (AFM), and dynamic light scattering (DLS) were used to follow the successive formation of oligomers, long fibres, and finally gels constituted of fibres. We were able to probe the behaviour of soluble proteins that are encapsulated within the gel matrix, and propose possible mechanisms by which the formation of such fibrous gels can enable anhydrobiosis.
The 15N‐1H HSQC spectrum of CAHS‐8 (Figure 1 A) is typical of an intrinsically disordered protein, with low dispersion in the 1HN dimension. Nevertheless, >98 % of the backbone resonances could be assigned (see Methods in the Supporting Information), revealing the presence of a helical domain between residues 95 and 195 (Figure 1 B). This region is divided into two segments, α1 (E95–F150) and α2 (V167–L194), with α2 showing a lower level of helical propensity (Figure 1 B,C; estimated from 13C secondary shifts to be around 60 and 30 %, respectively). The linker between α1 and α2 (12 consecutive sites from 152–163, with 1H‐15N nOe effects <0.2 and 13C shifts showing a secondary structure population of <0.25) exhibits reduced helical propensity (Figure 1 B,C), and enhanced flexibility (Figure 1 D,E).
Figure 1.
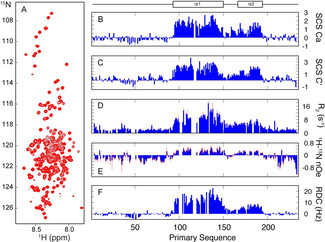
Characterisation of the structural and dynamic properties of CAHS‐8 in solution. A) The 15N‐1H correlation spectrum is characteristic of an intrinsically disordered protein. B, C) Secondary 13Ca and 13C′ chemical shifts showing strong α‐helical propensities from 95–195, divided into two distinct helices, α1 (E95‐F150) and α2 (V167‐L194). 15N relaxation (D; 850 MHz, E; 600 MHz), 15N‐1H RDCs (F), and PREs (see Figure S1) suggest the protein acts as a disordered chain comprising two highly populated α‐helices (303 K) and no persistent tertiary structure. The NMR spectrum was recorded at 350 μm (9.3 mg mL−1). The NMR assignments are deposited in BMRB (accession code 51115).
The tertiary folding of CAHS‐8 was probed using paramagnetic relaxation enhancements, with TEMPO‐maleimide labelling of positions A134C and A185C. The absence of significant enhancements beyond immediate neighbours indicates an absence of persistent long‐range structure (see Figure S1).
The presence of two helices connected by a more flexible linker is supported by the measurement of 15N‐1H residual dipolar couplings (RDCs; see Methods in the Supporting Information). The RDCs show a profile typical of IDPs with helical elements that do not specifically interact with each other, as predicted by simulation and experimentally validated in numerous systems. The RDCs exhibit positive values in the helical regions and lower or negative values in the unstructured parts of the chain (Figure 1 f).[ 17 , 18 ] A model of CAHS‐8, comprising a flexible chain with two consecutive helices connected by a short linking peptide and flanked by intrinsically disordered termini was calculated using the disordered protein ensemble generator flexible‐meccano[ 19 , 20 ] and compared to SAXS measurements. Ensemble selection using the ASTEROIDS approach suggests that the protein samples a largely extended conformation in the monomeric state with an average radius of gyration of 58±7 Å (Figure 2 A). The average radius of gyration R g,av of an ensemble of fully disordered conformers is 47±10 Å, while an ensemble of conformers containing the helical elements (R g,av =59±11 Å; see Figure S2) coincides with the experimental radius of gyration, thereby supporting the nature of the ensemble of unfolded chains comprising significant populations of helices α1 and α2.
Figure 2.
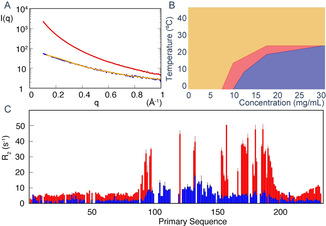
Monomeric CAHS‐8 forms higher order oligomers as a function of concentration and temperature. A) SAXS measured at 2 mg mL−1 (blue) and 5 mg mL−1 (red). At the higher concentration, the curve shows characteristics associated with fibril formation. The orange curve shows the predicted SAXS curve from an ASTEROIDS‐selected ensemble of 7 disordered conformers calculated using flexible‐meccano (average Rg of best‐fitting ensembles=58±7 Å). B) Phase diagram derived from DLS. Orange represents the isotropic phase, red represents increased oligomerization associated with slight gel formation, and blue represents gel formation. C) Transverse 15N relaxation (R2) measured at 303 K (blue) and 278 K (red) at 850 MHz, showing a very large increase in molecular tumbling at the lower temperature.
CAHS‐8 shows a strongly concentration‐dependent propensity to oligomerize, as illustrated by SAXS, where scattering curves that are characteristic of fibril formation are observed at higher concentrations (Figure 2 A). The 13C chemical shifts show strong temperature dependence, revealing a significantly higher helical content at 278 K for helix α2 (see Figure S3). The 15N relaxation is also temperature dependent (Figure 2 C and see also Figure S4), notably higher than that normally observed in intrinsically disordered proteins containing helical elements[ 21 , 22 ] (see Figure S2), thus supporting the formation of higher order oligomers at lower temperature. Dynamic light scattering (DLS) measurements were used to map the phase diagram of CAHS‐8 by observing the dependence of the decay in the autocorrelation of scattered intensity as a function of temperature and concentration (Figure 2 B). We observe the formation of an apparent physical hydrogel phase at low temperatures and higher concentration, in qualitative agreement with observations from the NMR and SAXS studies.
We used atomic force microscopy (AFM) in liquid and air modes to observe the apparent formation of fibrils that eventually form a dense mesh that resembles a gel and to investigate the nature of these gels. Extension often follows a twisted helix structure comprising paired filaments, exhibiting a pitch of approximately 600 nm (Figure 3 A). AFM was used to follow gel formation, and revealed the generation of fibrillar structures as a function of time (Figure 3 B, see also Figure S3), eventually leading to a dense mesh of fibres (Figure 3 C). Circular fibrillar structures with a radius of about 150 nm were also formed (see Figure S5). Application of a Laplacian‐weight method[ 23 , 24 ] highlights the twist, and again suggests the presence of paired strands forming single fibrils (see Figure S6). Placing concentrated CAHS‐8 on glass and cooling resulted in the formation of a porous structure comprising large cavities with diameters of 10–30 nm (Figure 3 D).
Figure 3.
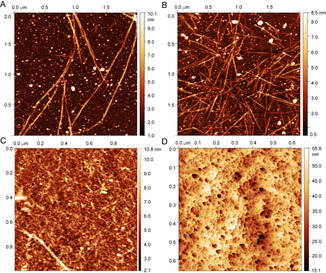
Different states of CAHS‐8 imaged by AFM in the liquid (A,B) and air mode (C,D). A) Dissolving the gel on mica revealed fibrils and allowed a kinetic series to be recorded (see Figure 3). B) Allowing fibrillation to continue whilst the sample is drying shows an increasingly dense matrix of fibrils. C) After the kinetic series from (A) was recorded and the sample fully dehydrated, air imaging displays the entanglement of the fibrils forming a gel. D) Placing a 30 mg mL−1 gel on glass reveals porous structures, comprising cavities with diameters of tens of nanometers.
NMR spectroscopic analysis of the gels formed from 15N‐labelled CAHS‐8 demonstrates that the intrinsically disordered domains (excluding helix α1) remain flexible in the context of the fibrillar gel (Figure 4 A). No signals from helix α1 were detected in the 15N‐1H HSQC spectrum of a gel formed from 15N‐labelled CAHS‐8, and 15N spin relaxation rates reveal an overall similar dynamic behaviour of the N‐terminal intrinsically disordered region at 303 K as the protein in free solution. The 62–72 strand, comprising both hydrophobic and acidic residues, shows elevated R2 values, which suggests increased correlation times, possibly arising from interaction with more rigid elements of the gel. The C‐terminal domain shows a similar dynamic behaviour to the monomeric phase, with the exception of the terminus, where the non‐native TEV‐cleavage site (228–233) also shows enhanced R2 values. Signals from helix α2 are visible in the spectrum, but show considerably higher relaxation rates than those in free solution (25 s−1 compared to 10 s−1), which suggests that this domain is tethered to a much larger, or immobile, object. These observations are compatible with the involvement of α1 in the formation of fibrils that constitute the gel, with α2 and the disordered domains remaining flexible within the fibril cavities.
Figure 4.
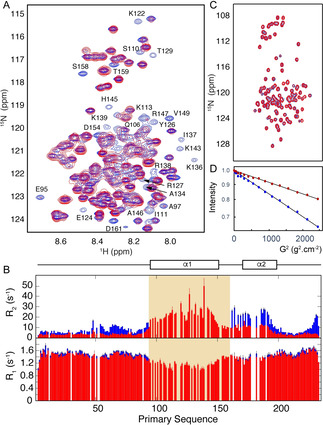
CAHS‐8 disordered domains remain flexible in gels that confine soluble proteins. A) Comparison of the 15N‐1H correlation spectra (293 K, 850 MHz) of CAHS‐8 in monomeric (blue) and gel (red) forms. Residues in helix α1 that disappear in the gel spectrum are annotated. B) Transverse (R2) and longitudinal (R1) 15N relaxation (293 K, 850 MHz) of monomeric (red) and gel (blue) CAHS‐8. Signals from the shaded region were not visible in the spectrum. C) 15N‐1H correlation spectra of the disordered domain of transcription factor ANP32a in buffer (blue) and gel (red) reveal that conformational sampling of ANP32a is unperturbed in gel. D) DOSY measurements of ANP32a in buffer (blue) and gel (red) reveal that translational diffusion of ANP32a is significantly reduced (4.8 compared to 9.6 10−11 m2s−1) in the gel.
To investigate the possible role of gel formation in the establishment of protective properties of CAHS‐8, gels were formed in mixtures of the TDP and soluble proteins that were isotopically labelled to allow observation by NMR spectroscopy. The results (illustrated in Figure 4 C) demonstrate that the 15N‐1H HSQC spectrum of the intrinsically disordered domain of ANP32a, [25] a transcription factor, is essentially unchanged in the gel. The translational diffusion of the protein is reduced by approximately a factor of two compared to standard buffer conditions (Figure 4 D). This level of restriction is similar to that measured for a small, folded protein in 6 % polyacrylamide gels. [26] Despite this slow diffusion, the conformational sampling of the sequestered protein is maintained in the CAHS‐8 gel cavity. Similar results were measured on the intrinsically disordered C‐terminal domain of Sendai virus nucleoprotein (NTAIL). [27] Similarly, a folded SH3 domain [28] exhibited an essentially identical HSQC spectrum in gel and isotropic solution (see Figure S7). The intensity of the spectrum is very weak, which suggests a significant fraction is partially immobilised in the gel.
Comparison of the sequence of α1 with related CAHS sequences reveals highest conservation in the helical domain, which exhibits an irregular coiled coil propensity comprising both canonical heptads and stutters,[ 29 , 30 ] as illustrated for the segment A133–R152 (Figure 5 A,B, see also Figure S8). Formation of a coiled‐coil conformation has also been associated with desiccation‐protectant LEA proteins upon dehydration.[ 14 , 31 , 32 ]
Figure 5.
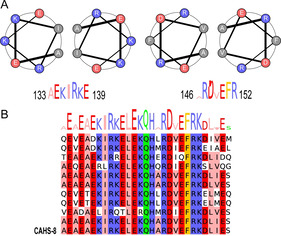
A) CAHS‐8 comprises coiled‐coil‐like sequences located specifically in helix α1. Two sequences are shown: A133–E139 and A146–R152, which exhibit a typical heptad repeat. B) This propensity is conserved in related sequences (see Figure S8).
On the basis of these observations, we propose that the long helical domains of the partially disordered protein associate through coiled‐coil‐like interactions under certain environmental conditions (low temperature, high concentration) to form oligomers, then fibrils (Figure 5 C), which eventually form a gel‐like matrix. Both charged and hydrophobic amino acids in this region are strongly conserved over CAHS proteins from different species. NMR relaxation data also show strong evidence that this region is central to the intermolecular interaction, with signals in this region disappearing at lower temperatures, where higher order oligomerization is favoured, and relaxation rates increasing more in this region than elsewhere. Although the molecular structure of the twisted fibril is not known, helix α1 likely contributes to the structured core, possibly through a super‐helical [33] or alternatively a cross‐α [34] configuration, while the remainder of the molecule remains flexible, as shown by NMR spectroscopy.
The exact mechanism by which the formation of such gels protects the organism remains unknown, although the total water volume required to maintain biomolecules in their functional state is clearly reduced in such a matrix. The disordered termini of CAHS‐8 remain flexible in the gel and may contribute to the solvation of sequestered biomolecules. In this respect, it is interesting to note that the C‐terminal IDR, in particular, is rich in polar amino acids. Cryoprotection may also be facilitated by a reduction in the volume of bulk water, thereby reducing the formation of ice crystals. [35] Under our conditions, we see no evidence of vitrification during the formation of fibrillar hydrogels, as has been observed for trehalose, sucrose, and LEA proteins and proposed as a mechanism of TDP protection.[ 12 , 13 ]
In conclusion, we present a detailed characterization of the transformation of the physical properties of CAHS‐8 as a function of environmental conditions. We describe the monomeric behaviour of the protein as partially disordered, with a long two‐helical domain flanked by long flexible tails. We propose that this domain mediates homo‐dimerization through a coiled‐coil interaction, and have experimentally observed the successive formation of oligomers, fibrils, and eventually gels in response to concentration and temperature change. Importantly the gels can sequester soluble proteins in their native state, maintaining their functional form. We propose that the observed transformation of this important protein from soluble monomer to intracellular matrix may be responsible for its protective role in the phenomenal response of tardigrades to environmental stress.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This work used the platforms of the Grenoble Instruct‐ERIC center (ISBG; UAR 3518 CNRS‐CEA‐UGA‐EMBL) within the Partnership for Structural Biology (PSB), supported by FRISBI (ANR‐10‐INBS‐05‐02) and GRAL, financed within the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH‐EUR‐GS (ANR‐17‐EURE‐0003). We thank Caroline Mas for assistance and access to the Biophysical platform. IBS acknowledges integration into the Interdisciplinary Research Institute of Grenoble (IRIG CEA). This work acknowledges the AFM platform at the IBS and the ESRF. M.B. acknowledges support from ERC advanced grant DynamicAssemblies.
A. Malki, J.-M. Teulon, A. R. Camacho-Zarco, S.-w. W. Chen, W. Adamski, D. Maurin, N. Salvi, J.-L. Pellequer, M. Blackledge, Angew. Chem. Int. Ed. 2022, 61, e202109961.
References
- 1. Greven H., in Water Bears: The Biology of Tardigrades (Ed.: Schill R. O.), Springer International Publishing, 2018, pp. 1–55. [Google Scholar]
- 2. Spallanzani L., Dalyell J. G., Baillie M., Tracts on the Nature of Animals and Vegetables., William Creech, Edinburgh, 1804. [Google Scholar]
- 3. Wełnicz W., Grohme M. A., Kaczmarek L., Schill R. O., Frohme M., J. Insect Physiol. 2011, 57, 577–583. [DOI] [PubMed] [Google Scholar]
- 4. Somme L., EJE 2013, 93, 349–357. [Google Scholar]
- 5. Bertolani R., Guidetti R., Jönsson I. K., Altiero T., Boschini D., Rebecchi L., J. Limnol. 2004, 63, 16–25. [Google Scholar]
- 6. Schill R. O., Mali B., Dandekar T., Schnölzer M., Reuter D., Frohme M., Biotechnol. Adv. 2009, 27, 348–352. [DOI] [PubMed] [Google Scholar]
- 7. Yamaguchi A., Tanaka S., Yamaguchi S., Kuwahara H., Takamura C., Imajoh-Ohmi S., Horikawa D. D., Toyoda A., Katayama T., Arakawa K., Fujiyama A., Kubo T., Kunieda T., PLoS One 2012, 7, e44209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hashimoto T., Horikawa D. D., Saito Y., Kuwahara H., Kozuka-Hata H., Shin-I T., Minakuchi Y., Ohishi K., Motoyama A., Aizu T., Enomoto A., Kondo K., Tanaka S., Hara Y., Koshikawa S., Sagara H., Miura T., Yokobori S.-I., Miyagawa K., Suzuki Y., Kubo T., Oyama M., Kohara Y., Fujiyama A., Arakawa K., Katayama T., Toyoda A., Kunieda T., Nat. Commun. 2016, 7, 12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boothby T. C., Tapia H., Brozena A. H., Piszkiewicz S., Smith A. E., Giovannini I., Rebecchi L., Pielak G. J., Koshland D., Goldstein B., Mol. Cell 2017, 65, 975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Piszkiewicz S., Gunn K. H., Warmuth O., Propst A., Mehta A., Nguyen K. H., Kuhlman E., Guseman A. J., Stadmiller S. S., Boothby T. C., Neher S. B., Pielak G. J., Protein Sci. 2019, 28, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boothby T. C., Pielak G. J., BioEssays 2017, 39, 1700119. [DOI] [PubMed] [Google Scholar]
- 12. Arakawa K., Numata K., Mol. Cell 2021, 81, 409–410. [DOI] [PubMed] [Google Scholar]
- 13. Boothby T. C., Mol. Cell 2021, 81, 411–413. [DOI] [PubMed] [Google Scholar]
- 14. Ingram J., Bartels D., Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 377–403. [DOI] [PubMed] [Google Scholar]
- 15. Crowe L. M., Comp. Biochem. Physiol. Part A 2002, 131, 505–513. [DOI] [PubMed] [Google Scholar]
- 16. Hengherr S., Heyer A. G., Köhler H.-R., Schill R. O., FEBS J. 2008, 275, 281–288. [DOI] [PubMed] [Google Scholar]
- 17. Salmon L., Nodet G., Ozenne V., Yin G., Jensen M. R., Zweckstetter M., Blackledge M., J. Am. Chem. Soc. 2010, 132, 8407–8418. [DOI] [PubMed] [Google Scholar]
- 18. Jensen M. R., Zweckstetter M., Huang J., Backledge M., Chem. Rev. 2014, 114, 6632–6660. [DOI] [PubMed] [Google Scholar]
- 19. Bernado P., Blanchard L., Timmins P., Marion D., Ruigrok R. W. H., Blackledge M., Proc. Natl. Acad. Sci. USA 2005, 102, 17002–17007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ozenne V., Bauer F., Salmon L., Huang J.-R., Jensen M. R., Segard S., Bernadó P., Charavay C., Blackledge M., Bioinformatics 2012, 28, 1463–1470. [DOI] [PubMed] [Google Scholar]
- 21. Abyzov A., Salvi N., Schneider R., Maurin D., Ruigrok R. W. H., Jensen M. R., Blackledge M., J. Am. Chem. Soc. 2016, 138, 6240–6251. [DOI] [PubMed] [Google Scholar]
- 22. Adamski W., Salvi N., Maurin D., Magnat J., Milles S., Jensen M. R., Abyzov A., Moreau C. J., Blackledge M., J. Am. Chem. Soc. 2019, 141, 17817–17829. [DOI] [PubMed] [Google Scholar]
- 23. Chen S.-W. W., Teulon J.-M., Godon C., Pellequer J.-L., J. Mol. Recognit. 2016, 29, 51–55. [DOI] [PubMed] [Google Scholar]
- 24. Chen S.-W. W., Banneville A.-S., Teulon J.-M., Timmins J., Pellequer J.-L., Nanoscale 2020, 12, 22628–22638. [DOI] [PubMed] [Google Scholar]
- 25. Camacho-Zarco A. R., Kalayil S., Maurin D., Salvi N., Delaforge E., Milles S., Jensen M. R., Hart D. J., Cusack S., Blackledge M., Nat. Commun. 2020, 11, 3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sass H.-J., Musco G., Stahl S. J., Wingfield P. T., Grzesiek S., J. Biomol. NMR 2000, 18, 303–309. [DOI] [PubMed] [Google Scholar]
- 27. Jensen M. R., Houben K., Lescop E., Blanchard L., Ruigrok R. W. H., Blackledge M., J. Am. Chem. Soc. 2008, 130, 8055–8061. [DOI] [PubMed] [Google Scholar]
- 28. Roldan J. L. O., Blackledge M., van Nuland N. A. J., Azuaga A. I., J. Biomol. NMR 2011, 50, 103–117. [DOI] [PubMed] [Google Scholar]
- 29. Brown J. H., Cohen C., Parry D. A., Proteins 1996, 26, 134–145. [DOI] [PubMed] [Google Scholar]
- 30. Lupas A. N., Gruber M., Adv. Protein Chem. 2005, 70, 37–38. [DOI] [PubMed] [Google Scholar]
- 31. Browne J., Tunnacliffe A., Burnell A., Nature 2002, 416, 38. [DOI] [PubMed] [Google Scholar]
- 32. Goyal K., Tisi L., Basran A., Browne J., Burnell A., Zurdo J., Tunnacliffe A., J. Biol. Chem. 2003, 278, 12977–12984. [DOI] [PubMed] [Google Scholar]
- 33. Mondal S., Adler-Abramovich L., Lampel A., Bram Y., Lipstman S., Gazit E., Nat. Commun. 2015, 6, 8615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tayeb-Fligelman E., Tabachnikov O., Moshe A., Goldshmidt-Tran O., Sawaya M. R., Coquelle N., Colletier J.-P., Landau M., Science 2017, 355, 831–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mazur P., Am. J. Physiol. Cell Physiol. 1984, 247, C125–C142. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information


