Abstract
The scope of non‐hydrogenative parahydrogen hyperpolarization (nhPHIP) techniques has been expanding over the last years, with the continuous addition of important classes of substrates. For example, pyruvate can now be hyperpolarized using the Signal Amplification By Reversible Exchange (SABRE) technique, offering a fast, efficient and low‐cost PHIP alternative to Dynamic Nuclear Polarization for metabolic imaging studies. Still, important biomolecules such as amino acids have so far resisted PHIP, unless properly functionalized. Here, we report on an approach to nhPHIP for unmodified α‐amino acids that allows their detection and quantification in complex mixtures at sub‐micromolar concentrations. This method was tested on human urine, in which natural α‐amino acids could be measured after dilution with methanol without any additional sample treatment.
Keywords: amino acids, NMR spectroscopy, PHIP, SABRE, urine
Non‐hydrogenative parahydrogen hyperpolarization allows the detection and quantification of α‐amino acids down to sub‐micromolar concentrations. This method can also be applied to aqueous mixtures after dilution with methanol without any additional sample treatment. This direct approach was tested on human urine, allowing a well‐resolved detection of amino acids despite the complexity of the mixture.
Introduction
Nuclear Magnetic Resonance (NMR) and Magnetic Resonance Imaging (MRI) are indispensable tools in chemical analysis and medical research. Magnetic resonance techniques suffer, however, from a low sensitivity, limiting their applications to relatively concentrated solutions (e.g. at least micromolar for NMR). Nuclear spin hyperpolarization techniques such as dynamic nuclear polarization (DNP)[ 1 , 2 ] and parahydrogen‐induced hyperpolarization (PHIP)[ 3 , 4 , 5 ] allow to overcome this sensitivity limit. They open, thereby, doors to biomedical applications such as real‐time metabolic imaging and biomarkers discovery.[ 6 , 7 , 8 , 9 ]
In this respect amino acids have attracted special attention due to their crucial roles in metabolism. [10] For instance, high concentrations of hyperpolarized alanine have been generated with DNP and utilized for in vivo study of enzyme activity. [11] Similarly, DNP has allowed a sensitive detection of amino acids present in complex (bio)mixtures.[ 12 , 13 ] Despite these achievements with DNP, cheaper and instrumentally less demanding hyperpolarization techniques would be of great interest to transform such proof of principles into real‐case implementations. This has motivated a large number of recent studies utilizing PHIP to hyperpolarize amino acids. [14]
Amino acids have mainly been hyperpolarized with hydrogenative PHIP.[ 3 , 4 ] Since this technique relies on the hydrogenation of a substrate with parahydrogen (p‐H2), only unsaturated molecules can be hyperpolarized. Two different routes were, therefore, followed for amino acids: either unsaturated amino acid precursors were used,[ 15 , 16 , 17 , 18 ] or an unsaturated side‐arm was attached to the amino acid.[ 19 , 20 ]
Alternatively, the non‐hydrogenative Signal Amplification By Reversible Exchange (SABRE) technique can be employed.[ 5 , 21 , 22 , 23 ] SABRE relies on the reversible association of substrates and p‐H2 to a catalyst, which mediates the transfer of hyperpolarization from p‐H2 to the substrate nuclei. Due to the high levels of enhancements reached for pyridine derivatives, such molecules were chemically attached to amino acids and oligopeptides, allowing the enhancement of the resulting constructs via SABRE.[ 24 , 25 , 26 ]
Non‐hydrogenative PHIP (nhPHIP) of unmodified amino acids for analytical applications is, on the other hand, largely unexplored. This may be due to the fact that SABRE/nhPHIP on amino acids was only reported at very low magnetic fields (3.9 mT), [27] lacking the spectral resolution to discriminate the hyperpolarized signals of different amino acids, orthohydrogen and hydrides, therefore hampering chemical analysis. A direct PHIP analysis of amino acids would, however, be highly desirable over the above mentioned derivatization techniques, not only in terms of costs and time, but also to prevent a potential bias for quantification.
Here, we demonstrate that α‐amino acids can be detected with a nhPHIP‐NMR chemosensing method. By shedding light onto their interaction with the nhPHIP catalyst, we provide an approach to detect, resolve and quantify (dilute) α‐amino acids in complex mixtures. Moreover, we show that α‐amino acids can be detected from aqueous samples containing high concentrations of competing ligands, with practically no sample pre‐treatment.
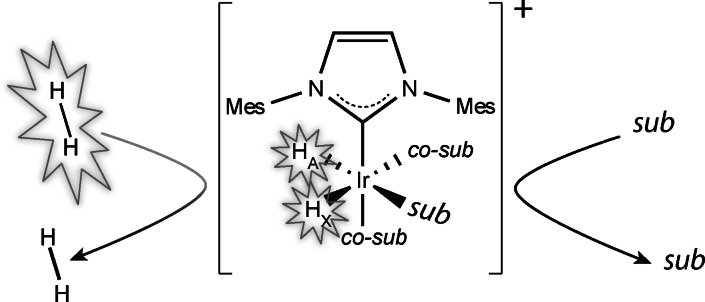
NhPHIP‐NMR chemosensing is based on the reversible binding of substrates to a complex of iridium with an N‐heterocyclic carbene ligand, such as the Ir‐IMes catalyst, together with p‐H2 and a suitable co‐substrate, as illustrated in Figure 1.[ 28 , 29 , 30 ] Formation of these transient asymmetric complexes at high magnetic field allows the conversion of the singlet state originating from p‐H2 to hydrides magnetization that can be detected via NMR with enhanced sensitivity. We have previously shown that the hydrides chemical shifts are highly sensitive to the structure of the analyte associating to the iridium complex and, therefore, can act as hyperpolarized probes to reveal the presence of specific substrates in the sample. [29]
Figure 1.
Schematic representation of the nhPHIP‐NMR method: substrates (sub) and p‐H2 bind reversibly to an organometallic catalyst, allowing the signalling of the substrates by a pair of hydrides that can be detected in an NMR spectrum with enhanced sensitivity. The Ir‐IMes complex is formed after hydrogenation of the [Ir(IMes)(COD)Cl] pre‐catalyst in the presence of an excess of a co‐substrate (co‐sub), where IMes stands for 1,3‐bis(2,4,6‐trimethylphenyl)imidazole‐2‐ylidene and COD for cyclooctadiene.
The same approach can be applied to detect most α‐amino acids with enhanced NMR sensitivity. It was previously demonstrated that these compounds can bind to iridium catalysts as bidentate ligands, via their amino‐ and carboxy groups. [31] Similarly, we will show that they can associate to the Ir‐IMes catalyst in the presence of the co‐substrate pyridine.
Results and Discussion
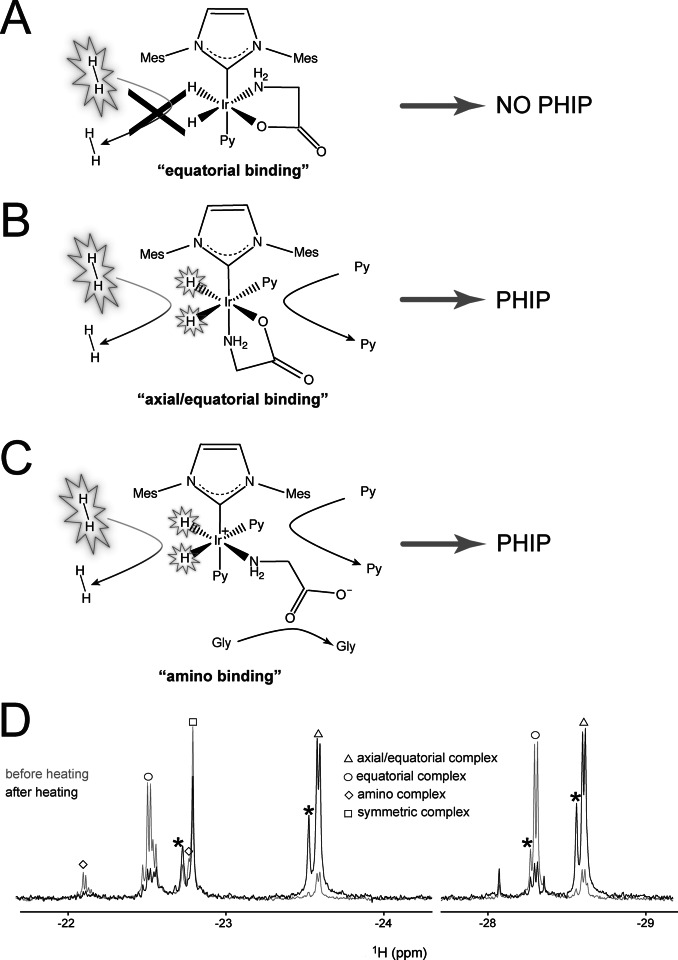
Upon activation of the catalyst precursor in the presence of glycine, an excess of pyridine as co‐substrate[ 32 , 33 , 34 ] and H2, a pair of hydrides signals at −22.5 ppm and −28.2 ppm appears in the thermal NMR spectrum, indicative of the formation of an asymmetric complex. However, in the presence of p‐H2, these signals are not enhanced by nhPHIP, similar as for other chelating ligands, previously reported in the literature.[ 35 , 36 ] This loss of nhPHIP performance can result from the tight binding of a bidentate ligand in the equatorial plane of the iridium complex. Consequently, p‐H2 refreshment, which is an associative process that requires (co‐)substrate dissociation, [37] cannot take place, thereby inactivating nhPHIP. Also glycine can act as a chelating ligand, binding via the amino‐ and carboxy group in the equatorial plane, as sketched in Figure 2 A. Note that the tight bidentate binding of the amino acids in these Ir‐IMes‐pyridine complexes also prevents their NMR signal enhancement via SABRE.
Figure 2.
A–C) Schematic representation of the main conformation geometries of glycine upon binding the Ir‐IMes complex in the presence of an excess of the co‐substrate pyridine. A) “Equatorial” binding. No ligand dissociation occurs on the NMR time scale, and accordingly p‐H2 refreshment cannot take place, abolishing nhPHIP for this complex. B) “Axial/equatorial” binding. Fast pyridine exchange is allowed and, consequently, reversible binding of p‐H2 and nhPHIP on the hydrides. C) “Amino” binding. Fast ligands exchange and, consequently, reversible binding of p‐H2 allows nhPHIP on the hydrides. Please note that a bidentate binding of the amino acid involving also the carboxy group in the axial position is not excluded (see Supporting Information). D) 1D NMR thermal spectra of the hydrides acquired in a 95 vol % [D4]MeOH, 5 vol % H2O solution of 0.66 mM glycine in the presence of 0.82 mM Ir‐IMes catalyst, 15.2 mM pyridine and 5 bar H2 at 278 K. The spectra were recorded at 500 MHz 1H resonance frequency, before and after sample heating, as described in the main text. A tentative assignment of the hydrides signals corresponding to the complexes in solution is indicated (see Supporting Information). Note that the structure observed for the signals at −22.15 and −22.5 results from the H/D isotope effect due to the amino group in the equatorial plane of the “amino” and “equatorial” complexes assigned to these signals, respectively. The signals marked with an asterisk originate from deuteration of one of the two hydrides in the complex.
A small fraction of the amino acid is, however, bound to the Ir‐IMes complex via the equatorial and axial binding sites (see Figure 2 B), giving rise to a pair of hydride signals at −23.6 and −28.6 ppm. Fast pyridine dissociation in the equatorial plane of this “axial/equatorial” complex allows quick exchange of the hydrides with p‐H2 and, thereby, their hyperpolarization via nhPHIP. Although the “equatorial” conformation (Figure 2 A) is kinetically favored, the “axial/equatorial” mode of binding (Figure 2 B) is energetically preferred. Reaching thermodynamic equilibrium is a slow process that takes several hours at room temperature but it can be significantly accelerated by warming up the sample. After 7–8 minutes at 50 °C, most of the amino acid complexes have adopted an “axial/equatorial” conformation. Note that this situation is exactly opposite to what was previously reported for the carboxyimine complexes with primary amines as co‐substrate, in which the equatorial binding mode appears to be both kinetically and thermodynamically favored. [35] In addition, at equilibrium a third complex conformation is present (revealed by a pair of hydride signals at −22.15 and −22.8 ppm), resulting from the association of the amino group in the equatorial plane, as displayed in Figure 2 C.
Figure 2 D displays the thermal NMR hydrides spectra acquired for a solution of glycine in the presence of the iridium catalyst, pyridine and H2, before and after heating the solution to 50 °C. The changes of the hydrides integrals clearly reflect the population redistribution of the amino acid complexes upon sample heating.
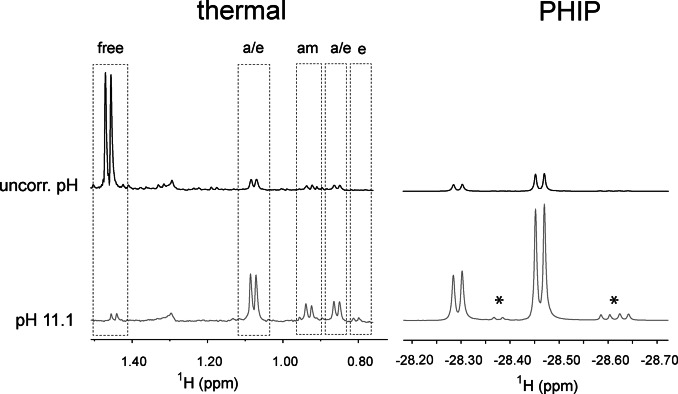
Not surprisingly, the interaction of amino acids with the iridium catalyst is strongly influenced by the pH of the solution. In Figure 3 (left) the binding of alanine to the complex at thermodynamic equilibrium is compared for two solutions, with and without pH correction. Depending on the pH, the thermal 1H spectra display a large difference in the concentration of amino acid complexes. Accordingly, a 6‐fold increase of the nhPHIP hydrides signals of the “axial/equatorial” complexes is observed for the solution at pH 11.1 (Figure 3, right). In order to afford a reproducible and almost complete binding of amino acids to the Ir‐IMes catalyst, the nhPHIP experiments presented in this work were acquired on samples in which the pH was set to ≈11, by adding either KOH/HCl or a piperidine/piperidinium buffer to the solution. Under these conditions, a 300‐fold enhancement of the hydrides signals with respect to thermal equilibrium was typically found using 51 %‐enriched p‐H2.
Figure 3.
Effect of pH on the association of alanine to the iridium complex (left) and on the nhPHIP‐NMR signals of the corresponding hydrides (right). The spectra at the top were acquired without pH correction, while the spectra at the bottom were measured in the presence of a 20 mM buffer of piperidine/piperidinium at pH 11.1. Left: NMR signals of the methyl protons of alanine after catalyst activation at 50 °C for 7.5 minutes. The signal assignment for the amino acid complexes is indicated as “a/e” (axial/equatorial), “am” (amino) and “e” (equatorial). Right: nhPHIP‐NMR signals of the high‐field hydrides for the diastereomeric “axial/equatorial” complexes formed by alanine. The signals marked with asterisks originate from impurities. The spectra were acquired under identical conditions (100 μM 13C Ala, 0.83 mM Ir‐IMes catalyst, 15 mM pyridine, 5 bar 51 %‐enriched p‐H2 and 0.5 vol‐% water) at 278 K.
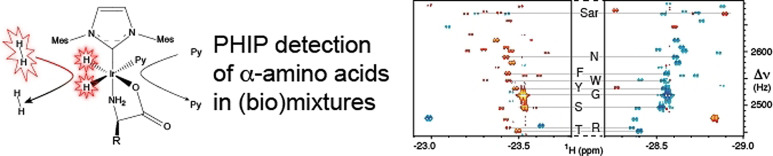
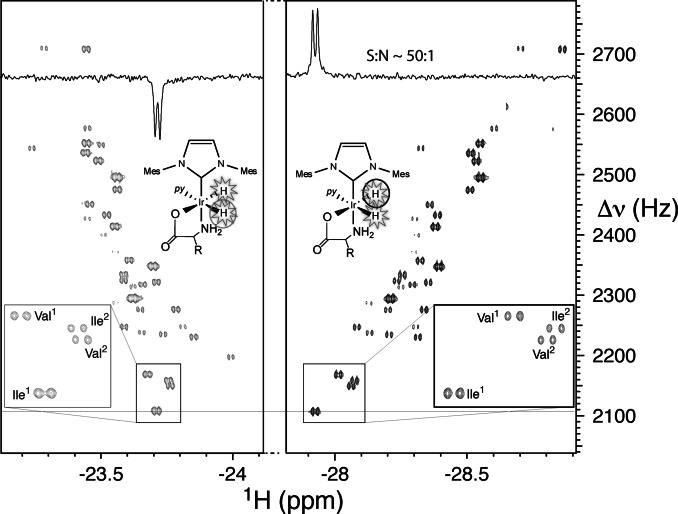
The “axial/equatorial” complexes can be employed as sensitive nhPHIP‐NMR chemosensing probes for amino acids detection in solution. We tested this approach on a mixture of the twenty natural α‐amino acids and sarcosine (N‐methyl glycine), each at 1 μM concentration, using the recently proposed 2D nhPHIP‐Zero‐Quantum (ZQ) experiment. [29] The hydrides resonances prove highly sensitive to the structure of the amino acids side chains, providing sufficient dispersion to resolve the nhPHIP‐NMR signals of all complexes, as illustrated in Figure 4. Note that, due to the chirality at the Cα, two diastereomeric nhPHIP‐active “axial/equatorial” complexes are formed for most α‐amino acids, as revealed by the doubling of the number of hydrides signals in the NMR spectrum (see inset Figure 4).
Figure 4.
2D nhPHIP‐ZQ hydrides’ spectrum of a mixture of 21 α‐amino acids at 1 μM concentration in [D4]MeOH, acquired at 500 MHz 1H resonance frequency, in the presence of 0.88 mM Ir‐IMes catalyst, 15.7 mM pyridine as co‐substrate and 5 bar 51 %‐enriched p‐H2. The experiment was acquired at 278 K in ca. 40 minutes. The low‐field and high‐field hydrides are indicated by gray and black circles, respectively. The inset displays the doubling of the hydrides signals for isoleucine and valine, due to the formation of diastereomeric complexes.
Dissociation/association of pyridine causes an interconversion between these two diastereoisomers that can be probed by inserting an exchange delay in the pulse scheme of the 2D nhPHIP‐ZQ experiment (see Supporting Information). The dissociation rate of pyridine determines also the lifetime of the amino acids complexes and, consequently, the nhPHIP enhancement as well as the hydrides signals line widths. Optimal spectral resolution and nhPHIP enhancement were obtained by reducing the measurement temperature to 5 °C (see Supporting Information), despite a lower refreshment rate of p‐H2. As indicated in Figure 4, nhPHIP‐NMR acquisition at this temperature affords a signal‐to‐noise ratio of ≈50:1 at an amino acid concentration of 1 μM, indicating a sub‐micromolar detection limit.
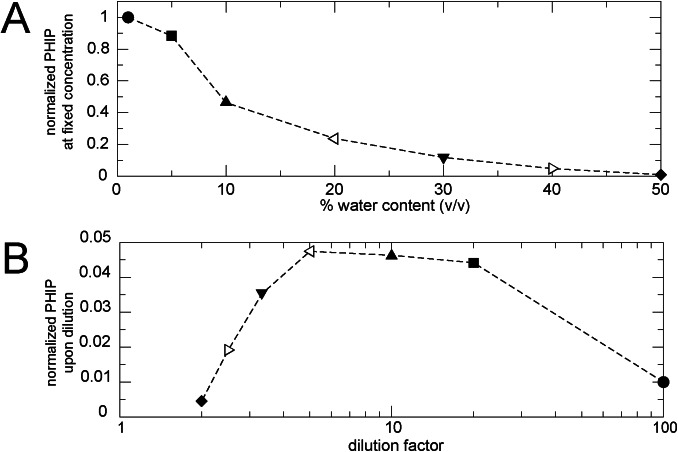
Despite the well‐known loss of PHIP/SABRE performance of the Ir‐IMes catalyst in an aqueous environment,[ 38 , 39 ] we demonstrate that α‐amino acid detection in water–methanol mixtures is possible, which avoids the previously reported Solid Phase Extraction pre‐treatment of the sample. [40] The nhPHIP response to a progressive increase of water content is shown in Figure 5 A for a solution of alanine at fixed concentration, taken as model system.
Figure 5.
A) Dependence of the nhPHIP hydrides signal integrals on the water content for a 200 μM solution of alanine in H2O/CH3OH, in the presence of Ir‐IMes catalyst (0.84 mM), pyridine (15.1 mM), 5 bar 51 %‐enriched p‐H2 and piperidine/piperidinium (20 mM). The integrals are normalized with respect to the value measured for the lowest water content (1 %). The data were acquired at 278 K at 500 MHz 1H resonance frequency. B) Dependence of nhPHIP on the dilution factor derived from the plot in (A). Identical symbols are used to indicate solutions with the same solvent composition.
These data allow estimating the dependence of nhPHIP upon diluting an aqueous α‐amino acid sample in methanol, as displayed in Figure 5 B, by dividing each (normalized) signal integral by the corresponding dilution factor. It appears that nhPHIP of α‐amino acids can tolerate water, with an optimal performance found for 80 vol %/20 vol % methanol/water mixture. This observation suggests that nhPHIP measurements of amino acids in natural extracts/biofluids should be feasible by simply diluting the aqueous sample with a solution of the Ir‐IMes catalyst and pyridine in methanol.
Human urine represents a challenging aqueous mixture for this direct nhPHIP approach, due to the large number of potential Ir‐IMes catalyst ligands, their concentration range (at least 5 orders of magnitude), and the high salt content. Particularly, the presence of concentrated ligands (e.g. urea (ca. 250 mM), ammonia (ca. 20–30 mM) and creatinine (ca. 15 mM) [41] ) should be carefully considered, as these species might compete with the pyridine co‐substrate. This would result in scrambling of each amino acid among different complexes, further increasing the spectral crowding, and attenuating the nhPHIP‐NMR signals. In order to minimize the chances of such interference, it was decided to perform a 20‐fold rather than a 5‐fold dilution of urine. Indeed, when recording nhPHIP spectra in the presence of urea, ammonia and creatinine (at concentrations corresponding to 20‐fold diluted urine), no additional signals were detected that could be assigned to amino acid complexes. Because of the relatively high levels of natural α‐amino acids in urine (between 20 μM for isoleucine and 2 mM for glycine [41] ), the concentrations attained after a 20‐fold dilution with methanol are well within the detection limits of nhPHIP‐NMR.
It should be noted that, when following this direct approach, urine and, in general, aqueous samples should be diluted in protonated rather than deuterated methanol to prevent isotope effects. In fact, when working with a mixed H2O/[D4]MeOH solvent, partial deuteration quickly occurs at the ortho positions of pyridine [42] and at the amino group of the amino acids, which results in the scrambling of the hydrides signals for the amino acid complexes, with the consequent degradation of both spectral resolution and sensitivity (see Supporting Information).
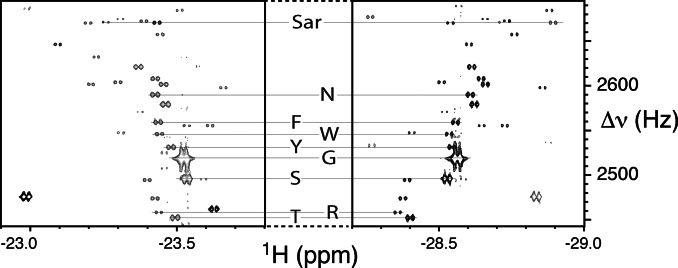
A region of the 2D nhPHIP‐ZQ NMR spectrum of human urine diluted in methanol is shown in Figure 6, demonstrating the high quality of the measurement. The hydride assignment for some natural α‐amino acids is indicated. Note the presence of several unassigned signals, possibly corresponding to urinary metabolites containing α‐amino acid motifs.
Figure 6.
Regions of the 2D nhPHIP‐ZQ NMR spectrum of a sample of human urine diluted 20 times in methanol. The spectrum was acquired at 500 MHz 1H resonance frequency, in the presence of 0.88 mM Ir‐IMes catalyst, 15.8 mM pyridine as co‐substrate and 5 bar of 51 %‐enriched p‐H2. The experiment was recorded at 278 K in ca. 40 minutes. The assignment of the hydride resonances of sarcosine (Sar) and some natural α‐amino acids (using the one‐letter code) is indicated. The complete amino acid regions of this spectrum are presented in the Supporting Information.
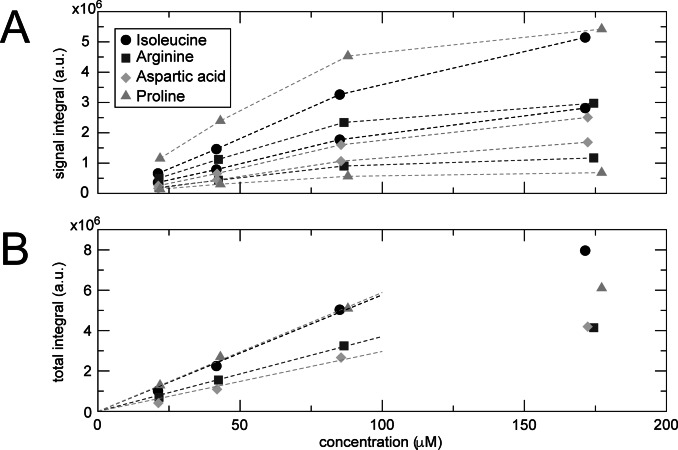
Analytes’ quantification from nhPHIP‐NMR spectra is in general not straightforward due to nhPHIP's reliance on a multitude of competing binding equilibria. [43] As previously demonstrated, however, a linear relation between substrate concentration and nhPHIP signal integral is obtained when an excess of catalyst and of a suitable co‐substrate are present.[ 44 , 45 ] As shown in Figure 7 A, this linear concentration dependence holds also for nhPHIP of amino acids, which allows their quantification via calibration techniques such as standard addition. [43] Note that a clear deviation from the linear trend is observed at high amino acids concentration due to the saturation of the Ir‐IMes catalyst (c=0.43 mM).
Figure 7.
nhPHIP dilution series for a mixture of isoleucine, arginine, aspartic acid and proline. A) Plot of the nhPHIP signal integrals of the high‐field hydrides for the Ir‐IMes complexes of the four amino acids as a function of their analytical concentration. B) Linear dependence of the sum of the integrals of the two diastereomeric complexes as a function of amino acid analytical concentration. The series was acquired at 278 K in the presence of Ir‐IMes catalyst (0.43 mM), pyridine (15 mM) and 5 bar 51 %‐enriched p‐H2 in 5 vol % D2O, 95 vol % [D4]MeOH. The most concentrated sample contained also 2.3 mM NH4Cl and 11.2 mM NaCl to mimic a 20‐fold diluted urine environment. These compounds were diluted in the same fashion as the amino acids.
In Figure 7 A, two sets of signal integrals are displayed for each amino acid, corresponding to the two diastereomeric “axial/equatorial” complexes formed in solution. Note that the signal integrals for the two complexes are generally different. This reflects the concentration ratio of the two diastereomers, as their nhPHIP enhancement is expected to be identical. Notably, the two diastereomeric complexes formed by proline display a different behavior, as they show nhPHIP enhancements that differ by almost an order of magnitude while their relative populations in solution are comparable. Interestingly, no interconversion between these two diastereomeric complexes by pyridine dissociation/association was observed (see Supporting Information).
The sum of the contributions of the two diastereomers, displayed in Figure 7 B as a function of concentration, is a measure of the efficiency of nhPHIP for each amino acid. The different linear dependencies indicate that nhPHIP enhancement can vary among amino acids. However, it appears that similar enhancements are observed for homologous side chains (see Supporting Information). This offers the possibility to obtain an estimate of amino acids concentrations from nhPHIP spectra, based on integrals comparison with an internal homologous standard. However, for a more accurate quantification, standard addition should be used, as previously demonstrated.[ 29 , 45 ]
Conclusion
We have shown that α‐amino acids can interact with the Ir‐IMes catalyst in a fashion that is compatible with nhPHIP. p‐H2 reversible association to the catalyst can provide continuous hydrides hyperpolarization for α‐amino acids complexes, which allows their detection in complex mixtures at sub‐micromolar concentrations. This nhPHIP‐NMR chemosensing method can be directly applied to biofluids or aqueous extracts with no sample manipulation, provided the water content is reduced to ca. 20 %. This can be achieved simply by diluting the aqueous sample with a solution of the Ir‐IMes catalyst and pyridine in methanol. This direct nhPHIP approach sets obviously higher concentration requirements and is, therefore, not universally applicable. However, when these requirements are met, as for urine, this approach offers a fast route to α‐amino acids detection and quantification, minimizing the potential bias deriving from sample treatment.
Our results expand in the direction of nhPHIP‐chemosensing for NMR detection of α‐amino acids in complex (bio)mixtures. However, the issue of p‐H2 hyperpolarization of free amino acids in solution (e.g. via SABRE), crucial for in vivo studies of metabolic processes, remains to be addressed. Still, our work clearly identifies the tight binding with the Ir‐IMes catalyst as the bottleneck in the attainment of free α‐amino acids hyperpolarization. Possible routes to enhance amino acids dissociation rate, involving a modification of the experimental parameters (co‐substrate, solvent or pH) and of the N‐heterocyclic carbene ligand of the iridium catalyst, are currently being explored in our lab.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
L. Sellies, R. L. E. G. Aspers, M. C. Feiters, F. P. J. T. Rutjes, M. Tessari, Angew. Chem. Int. Ed. 2021, 60, 26954.
Data Availability Statement
Sample preparation, experimental conditions and NMR parameters are detailed in the Supporting Information. Approval for the study was obtained from the Research Ethics Committee of the Faculty of Science of the Radboud University (Approval number: REC21111).
References
- 1. Ardenkjær-Larsen J. H., Fridlund B., Gram A., Hansson G., Hansson L., Lerche M. H., Servin R., Thaning M., Golman K., Proc. Natl. Acad. Sci. USA 2003, 100, 10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abragam A., Goldman M., Rep. Prog. Phys. 1978, 41, 395–467. [Google Scholar]
- 3. Bowers C. R., Weitekamp D. P., J. Am. Chem. Soc. 1987, 109, 5541–5542. [Google Scholar]
- 4. Pravica M. G., Weitekamp D. P., Chem. Phys. Lett. 1988, 145, 255–258. [Google Scholar]
- 5. Adams R. W., Aguilar J. A., Atkinson K. D., Cowley M. J., Elliott P. I. P., Duckett S. B., Green G. G. R., Khazal I. G., López-Serrano J., Williamson D. C., Science 2009, 323, 1708–1711. [DOI] [PubMed] [Google Scholar]
- 6. Golman K., in't Zandt R., Thaning M., Proc. Natl. Acad. Sci. USA 2006, 103, 11270–11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nelson S. J., Kurhanewicz J., Vigneron D. B., Larson P. E. Z., Harzstark A. L., Ferrone M., van Criekinge M., Chang J. W., Bok R., Park I., Reed G., Carvajal L., Small E. J., Munster P., Weinberg V. K., Ardenkjaer-larsen J. H., Chen A. P., Hurd R. E., Odegardstuen L.-I., Robb F. J., Tropp J., Murray J. A., Sci Transl Med. 2013, 5, 198ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Olsson L. E., Chai C.-M., Axelsson O., Karlsson M., Golman K., Petersson J. S., Magn. Reson. Med. 2006, 55, 731–737. [DOI] [PubMed] [Google Scholar]
- 9. Cavallari E., Carrera C., Sorge M., Bonne G., Muchir A., Aime S., Reineri F., Sci. Rep. 2018, 8, 8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simińska E., Koba M., Amino Acids 2016, 48, 1339–1345. [DOI] [PubMed] [Google Scholar]
- 11. Jensen P. R., Karlsson M., Meier S., Duus J. Ø., Lerche M. H., Chem. Eur. J. 2009, 15, 10010–10012. [DOI] [PubMed] [Google Scholar]
- 12. Wilson D. M., Hurd R. E., Keshari K., Van Criekinge M., Chen A. P., Nelson S. J., Vigneron D. B., Kurhanewicz J., Proc. Natl. Acad. Sci. USA 2009, 106, 5503–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katsikis S., Marin-Montesinos I., Ludwig C., Günther U. L., J. Magn. Reson. 2019, 305, 175–179. [DOI] [PubMed] [Google Scholar]
- 14. Pravdivtsev A. N., Buntkowsky G., Duckett S. B., Koptyug I. V., Hövener J.-B., Angew. Chem. Int. Ed. 2021, 60, 23496; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2021, 133, 23688. [Google Scholar]
- 15. Soon P. C., Xu X., Zhang B., Gruppi F., Canary J. W., Jerschow A., Chem. Commun. 2013, 49, 5304–5306. [DOI] [PubMed] [Google Scholar]
- 16. Trantzschel T., Plaumann M., Bernarding J., Lego D., Ratajczyk T., Dillenberger S., Buntkowsky G., Bargon J., Bommerich U., Appl. Magn. Reson. 2013, 44, 267–278. [Google Scholar]
- 17. Tang J. A., Gruppi F., Fleysher R., Sodickson D. K., Canary J. W., Jerschow A., Chem. Commun. 2011, 47, 958–960. [DOI] [PubMed] [Google Scholar]
- 18. McCormick J., Grunfeld A. M., Ertas Y. N., Biswas A. N., Marsh K. L., Wagner S., Glöggler S., Bouchard L.-S., Anal. Chem. 2017, 89, 7190–7194. [DOI] [PubMed] [Google Scholar]
- 19. Kaltschnee L., Jagtap A. P., McCormick J., Wagner S., Bouchard L.-S., Utz M., Griesinger C., Glöggler S., Chem. Eur. J. 2019, 25, 11031–11035. [DOI] [PubMed] [Google Scholar]
- 20. Glöggler S., Wagner S., Bouchard L.-S., Chem. Sci. 2015, 6, 4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iali W., Roy S. S., Tickner B. J., Ahwal F., Kennerley A. J., Duckett S. B., Angew. Chem. Int. Ed. 2019, 58, 10271–10275; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2019, 131, 10377–10381. [Google Scholar]
- 22. Iali W., Rayner P. J., Duckett S. B., Sci. Adv. 2018, 4, eaao6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Theis T., Truong M. L., Coffey A. M., Shchepin R. V., Waddell K. W., Shi F., Goodson B. M., Warren W. S., Chekmenev E. Y., J. Am. Chem. Soc. 2015, 137, 1404–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ratajczyk T., Gutmann T., Bernatowicz P., Buntkowsky G., Frydel J., Fedorczyk B., Chem. Eur. J. 2015, 21, 12616–12619. [DOI] [PubMed] [Google Scholar]
- 25. Ratajczyk T., Buntkowsky G., Gutmann T., Fedorczyk B., Mames A., Pietrzak M., Puzio Z., Szkudlarek P. G., ChemBioChem 2021, 22, 855–860. [DOI] [PubMed] [Google Scholar]
- 26. Chae H., Min S., Jeong H. J., Namgoong S. K., Oh S., Kim K., Jeong K., Anal. Chem. 2020, 92, 10902–10907. [DOI] [PubMed] [Google Scholar]
- 27. Glöggler S., Müller R., Colell J., Emondts M., Dabrowski M., Blümich B., Appelt S., Phys. Chem. Chem. Phys. 2011, 13, 13759–13764. [DOI] [PubMed] [Google Scholar]
- 28. Hermkens N. K. J., Eshuis N., van Weerdenburg B. J. A., Feiters M. C., Rutjes F. P. J. T., Wijmenga S. S., Tessari M., Anal. Chem. 2016, 88, 3406–3412. [DOI] [PubMed] [Google Scholar]
- 29. Sellies L., Reile I., Aspers R. L. E. G., Feiters M. C., Rutjes F. P. J. T., Tessari M., Chem. Commun. 2019, 55, 7235–7238. [DOI] [PubMed] [Google Scholar]
- 30. Reimets N., Ausmees K., Vija S., Reile I., Anal. Chem. 2021, 93, 9480–9485. [DOI] [PubMed] [Google Scholar]
- 31. Roy C. P., Huff L. A., Barker N. A., Berg M. A. G., Merola J. S., J. Organomet. Chem. 2006, 691, 2270–2276. [Google Scholar]
- 32. Shchepin R. V., Barskiy D. A., Mikhaylov D. M., Chekmenev E. Y., Bioconjugate Chem. 2016, 27, 878–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tickner B. J., John R. O., Roy S. S., Hart S. J., Whitwood A. C., Duckett S. B., Chem. Sci. 2019, 10, 5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mewis R. E., Green R. A., Cockett M. C. R., Cowley M. J., Duckett S. B., Green G. G. R., John R. O., Rayner P. J., Williamson D. C., J. Phys. Chem. B 2015, 119, 1416–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tickner B. J., Iali W., Roy S. S., Whitwood A. C., Duckett S. B., ChemPhysChem 2019, 20, 241–245. [DOI] [PubMed] [Google Scholar]
- 36. Mewis R. E., Fekete M., Green G. G. R., Whitwood A. C., Duckett S. B., Chem. Commun. 2015, 51, 9857–9859. [DOI] [PubMed] [Google Scholar]
- 37. Cowley M. J., Adams R. W., Atkinson K. D., Cockett M. C. R., Duckett S. B., Green G. G. R., Lohman J. A. B., Kerssebaum R., Kilgour D., Mewis R. E., J. Am. Chem. Soc. 2011, 133, 6134–6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fekete M., Gibard C., Dear G. J., Green G. G. R., Hooper A. J. J., Roberts A. D., Cisnetti F., Duckett S. B., Dalton Trans. 2015, 44, 7870–7880. [DOI] [PubMed] [Google Scholar]
- 39. Hermkens N. K. J., Aspers R. L. E. G., Feiters M. C., Rutjes F. P. J. T., Tessari M., Magn. Reson. Chem. 2018, 56, 633–640. [DOI] [PubMed] [Google Scholar]
- 40. Reile I., Eshuis N., Hermkens N. K. J., van Weerdenburg B. J. A., Feiters M. C., Rutjes F. P. J. T., Tessari M., Analyst 2016, 141, 4001–4005. [DOI] [PubMed] [Google Scholar]
- 41. Bouatra S., Aziat F., Mandal R., Guo A. C., Wilson M. R., Knox C., Bjorndahl T. C., Krishnamurthy R., Saleem F., Liu P., Dame Z. T., Poelzer J., Huynh J., Yallou F. S., Psychogios N., Dong E., Bogumil R., Roehring C., Wishart D. S., PloS one 2013, 8, e73076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barskiy D. A., Kovtunov K. V., Koptyug I. V., He P., Groome K. A., Best Q. A., Shi F., Goodson B. M., Shchepin R. V., Coffey A. M., Waddell K. W., Chekmenev E. Y., J. Am. Chem. Soc. 2014, 136, 3322–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eshuis N., van Weerdenburg B. J. A., Feiters M. C., Rutjes F. P. J. T., Wijmenga S. S., Tessari M., Angew. Chem. Int. Ed. 2015, 54, 1481–1484; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 1501–1504. [Google Scholar]
- 44. Eshuis N., Hermkens N., van Weerdenburg B. J. A., Feiters M. C., Rutjes F. P. J. T., Wijmenga S. S., Tessari M., J. Am. Chem. Soc. 2014, 136, 2695–2698. [DOI] [PubMed] [Google Scholar]
- 45. Eshuis N., Aspers R. L. E. G., van Weerdenburg B. J. A., Feiters M. C., Rutjes F. P. J. T., Wijmenga S. S., Tessari M., Angew. Chem. Int. Ed. 2015, 54, 14527–14530; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 14735–14738. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
Sample preparation, experimental conditions and NMR parameters are detailed in the Supporting Information. Approval for the study was obtained from the Research Ethics Committee of the Faculty of Science of the Radboud University (Approval number: REC21111).