Abstract
Purpose
There has been converging evidence of reliable detections of blood oxygenation level dependent (BOLD) signals evoked by neural stimulation and in a resting state in white matter (WM), within which few studies examined the relationship between BOLD functional signals and tissue metabolism. The purpose of the present study was to explore whether such relationship exists using combined functional MRI and positron emission tomography (PET) measurements of glucose uptake.
Methods
Functional and metabolic imaging data from 25 right‐handed healthy human adults (aged 18–23 years, 18 females) were analyzed. Measures, including average resting state functional connectivity (FC) with respect to 82 Brodmann areas, fractional amplitude of low‐frequency fluctuations (FALFF), and average fluorodeoxyglucose (FDG) uptake by PET, were computed for 48 predefined WM bundles. Pearson correlations across the bundles and 25 subjects studied were calculated among these measures. Linear mixed effects models were used to estimate the variance explainable by a predictor variable in the absence of inter‐subject variations.
Results
Analysis of six separate imaging intervals found that average FC the bundles was significantly correlated with local FDG uptake (r = 0.25, p < 0.001), and the FC also covaried significantly with FALFF (r = 0.41, p < 0.001). When random effects from inter‐subject variations were controlled, these correlations appeared to be medium to strong (r = 0.41 for FC vs. FDG uptake, and r = 0.65 for FALFF vs. FC).
Conclusion
This study indicates that BOLD signals in WM are directly related to variations in metabolic demand and engagement with cortical processing and suggests they should be incorporated into more complete models of brain function.
Keywords: BOLD, fALFF, FDG, fMRI, functional connectivity, white matter
Abbreviations & Acronyms
- BAs
Brodmann areas
- FALFF
fractional amplitude of low frequency fluctuations
- FC
function connectivity
- FDG
fluorodeoxyglucose
- GM
gray matter
- Montreal Neurological Institute
MNI
- regions of interest
ROIs
- white matter
WM
1. INTRODUCTION
Since the discovery of correlations between spontaneous low frequency fluctuations in blood oxygenation level dependent (BOLD) signals around the mid‐90s, 1 resting state functional connectivity (FC) has been extensively studied, leading to the identification of several resting state networks in the human brain. 2 Whereas the vast majority of these studies have hitherto focused on cortical gray matter (GM), there has been growing interest in the evaluation of functional networks in white matter (WM). 3 , 4 In particular, it has been recently demonstrated that spontaneous low frequency fluctuations in WM BOLD signals are robustly detectable and reflect specific neural activities, 5 , 6 which suggests the potential of analyzing and characterizing FC in WM.
Notwithstanding compelling evidence provided by experimental studies 7 and supportive clinical data that have recently emerged, 7 , 8 , 9 , 10 , 11 , 12 , 13 the interpretation of the observed fluctuations in WM signals remains unclear. 14 Physiologically, the vascular density of WM is approximately one‐fourth that of GM, so hemodynamic responses to increases in energy demand in WM are expected to be proportionally reduced. Thus, BOLD signals are weaker and may fall below the sensitivity of conventional acquisitions and analyses. Moreover, it is not clear what processes within WM modulate local metabolic needs or regulate flow and oxygenation to couple neural activity and vascular hemodynamics. The observed signal fluctuations in WM could plausibly originate from venous draining effects from upstream GM or other non‐neural confounds that impact BOLD signals throughout the brain parenchyma.
Although previous studies have shown WM BOLD signals are affected concomitantly with changes in neural activity in cortex, there is a residual need to clarify whether those changes reflect an intrinsic metabolic demand within WM itself. The primary energy substrate of brain tissues is glucose, and variations in glucose uptake reflect variations in baseline metabolic rates. We hypothesized the engagement of regions of WM in brain functions in a resting state is reflected in the magnitudes of the spontaneous fluctuations in local BOLD signals and in the strengths of the correlations of BOLD signals across time with other areas, which is interpreted as FC. We analyzed PET and MRI data previously acquired and reported by Jamadar et al. 15 We demonstrate that, by analyzing simultaneous recordings of the uptake of fluorodeoxyglucose (FDG) by dynamic positron emission tomography (PET) 16 and BOLD signals by functional MRI (fMRI), there are strong and significant spatial correlations between FDG uptakes and FC in WM, and FC is associated with the fractional amplitude of low frequency fluctuations (FALFF) in the BOLD signals. These observations lend strong support to the notion that BOLD signal fluctuations in WM are linked to neural activities through local variations in glucose metabolism.
2. METHODS
A schematic diagram of data analysis for this study is shown in Figure 1. Procedures of data collections were reviewed by the Monash University Human Research Ethics Committee, following the Australian National Statement of Ethical Conduct in Human Research (2007). Participants provided informed consent to participate in the study. Administration of ionizing radiation was approved by the Monash Health Principal Medical Physicist, following the Australian Radiation Protection and Nuclear Safety Agency Code of Practice (2005).
FIGURE 1.
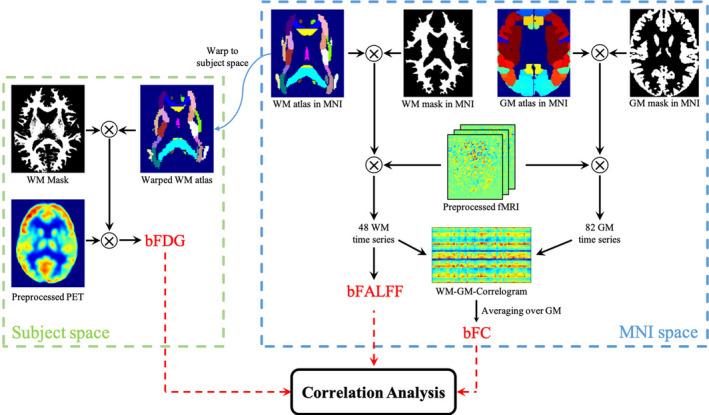
Schematic diagram of analysis framework. PET recordings are processed in the native space of each individual subject, from which bFDG is derived for each WM bundle by referencing the JHU‐ICBM WM atlas that is warped into the subject space. Meanwhile, BOLD signals are processed in the MNI space. To compute bFC, the brain is parcellated into 48 WM bundles and 82 GM regions, using the JHU‐ICBM atlas and BAs definitions, respectively. The coefficient of Pearson correlation in BOLD time series between each pair of WM bundle and GM region is calculated to obtain a WM‐GM correlogram, from which mean bFC of each WM bundle is derived by averaging over the 82 GM regions. A second measure of WM function, bFALFF, is derived for each WM bundle directly from the BOLD time series by referencing the JHU‐ICBM atlas. Finally, pair‐wise correlations are sought among the three measures derived
2.1. Participants
The subjects in the current study included 25 right‐handed human adults (aged 18–23 years, 18 F and 7 M) with no diagnosed mental illness, diabetes, or cardiovascular illness. Other inclusion and exclusion criteria are provided in previous reports. 15 , 17
2.2. MR‐PET imaging
Detailed imaging parameters and procedures can be found elsewhere. 17 Briefly, each participant underwent a 95‐min simultaneous MRI‐PET scan in a supine position in a Siemens (Erlangen) 3T Biograph molecular MR (mMR) scanner (Syngo VB20 P). The infusion of [18F] FDG (36 mL/h) was synchronized with the start of PET acquisitions. In the initial 30 min, while the PET signal rose to a detectable level, only non‐functional MRI scans were acquired, including T1 3D MPRAGE, and some other scans that are not reported in this study. This procedure was followed by six intervals of resting‐state PET‐fMRI, wherein each lasted for 10 min and subsequently went through a series of processing procedures, as described below.
2.3. Preprocessing of PET
For each individual, motion correction was performed on 225 PET volumes using the realign module in SPM (https://www.fil.ion.ucl.ac.uk/spm/software/spm12/) where the first volume was regarded as reference. Each of the realigned PET volumes was then coregistered to the T 1 weighted image of the same individual. Summing these coregistered volumes produced a static PET image, from which measurements of FDG uptakes were extracted.
2.4. Preprocessing of resting state fMRI data
Resting state data preprocessing involved a few steps as follows. First, the fMRI images were corrected for slice timing and head motion. Second, T 1 weighted images were segmented into GM, WM, and cerebrospinal fluid (CSF) using SPM, and all these images were registered to the fMRI data space of each individual. Third, mean signals from the CSF mask were regressed out as nuisance covariates from the fMRI time series. Fourth, the fMRI data, along with the coregistered T 1 weighted images as well as the GM and WM segments, were normalized into the Montreal Neurological Institute (MNI) space. Fifth, linear trends from the BOLD images were removed to correct for signal drift.
2.5. Computation of FALFF
To compute FALFF, we defined 48 WM bundle templates based on the Johns Hopkins University‐International Consortium for Brain Mapping (JHU‐ICBM) WM atlas. These WM templates were multiplied by the WM segment obtained in the preceding step, which was thresholded at 0.5 and eroded for 3 mm to eliminate potential partial volume effects from GM (see Supporting Information Figure S4, which is available online). The time‐series in each bundle template were averaged, and the average was then normalized to unit variance and Fourier transformed to derive the power spectrum. The FALFF was defined to be the average of the square root of the low‐frequency power spectrum (0.01–0.05 Hz) divided by that from the full frequency range, similarly to a previous study. 18
2.6. Computation of FC
Following the procedure of a previous study, 6 a set of time series was extracted from 130 regions of interest (ROIs), including 82 Brodmann areas (BAs) in GM and 48 WM bundles (based on the eroded WM mask as above) for each subject. Each time series was temporally filtered using a bandpass filter (0.01–0.1 Hz). Pearson correlations were calculated for the time series of each pair of WM and GM regions. This resulted in an 82 × 48 FC matrix, from which BA‐averaged FC was computed for each of the WM bundles to represent its overall FC profile.
2.7. Computation of FDG uptake
The FDG uptake was computed by first normalizing the image intensity of each static PET voxel with the global mean of the entire brain, so that the mean value of the entire brain is 1. Then WM bundle templates defined above were warped back to each individual space of the PET data, from which averaged FDG uptake values from each bundle were obtained.
3. RESULTS
3.1. Measurements of FC, FDG uptake, and FALFF
FC, FDG uptake, and FALFF measures were computed on the basis of individual WM bundles, which here are denoted as bFC, bFDG and bFALFF. As shown in Table 1, average bFC, bFDG, and bFALFF values across all the bundles and subjects studied and all six intervals ranged from −0.06 to 0.47 (0.27 ± 0.11), 0.47 to 1.00 (0.72 ± 0.15), and 0.20 to 0.25 (0.23 ± 0.01), respectively. Spatial distributions of bFC and bFDG (see left and middle panels in Figure 2A) were highly symmetrical, with a Pearson correlation coefficient of 0.966 and 0.995, respectively, comparing the bilateral WM bundles. By comparison, the inter‐hemispheric similarity of bFALFF distributions (see the right panel in Figure 2A) was somewhat reduced (r = 0.744).
TABLE 1.
Summary of Pearson correlations between the three measure pairs for each of the six fMRI sessions
| Session | r | p‐value |
|---|---|---|
| bFC vs. bFDG | ||
| 1 | 0.25 | <0.001 |
| 2 | 0.26 | <0.001 |
| 3 | 0.21 | <0.001 |
| 4 | 0.26 | <0.001 |
| 5 | 0.23 | <0.001 |
| 6 | 0.28 | <0.001 |
| r value (mean ± SD) | 0.25 ± 0.02 | – |
| bFALFF vs. bFC | ||
| 1 | 0.40 | <0.001 |
| 2 | 0.43 | <0.001 |
| 3 | 0.45 | <0.001 |
| 4 | 0.34 | <0.001 |
| 5 | 0.46 | <0.001 |
| 6 | 0.40 | <0.001 |
| r value (mean ± SD) | 0.41 ± 0.04 | – |
| bFALFF vs. bFDG | ||
| 1 | 0.12 | 0.002 |
| 2 | 0.17 | <0.001 |
| 3 | 0.14 | <0.001 |
| 4 | 0.07 | 0.08 |
| 5 | 0.10 | 0.01 |
| 6 | 0.11 | 0.006 |
| r value (mean ± SD) | 0.12 ± 0.03 | – |
FIGURE 2.
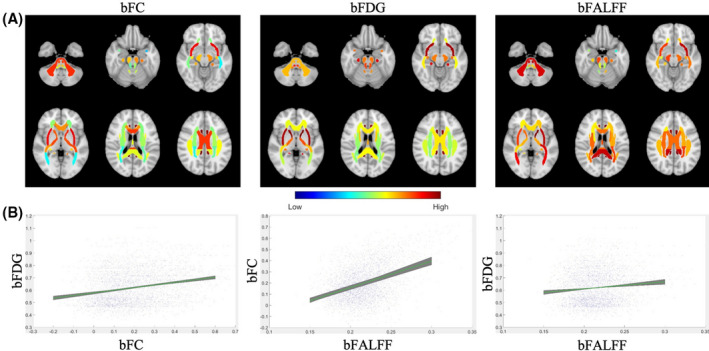
Distributions of bFC, bFDG and bFALFF measures and pairwise correlations between them. A, Distributions of average bFC (left), bFDG (middle), and bFALFF (right) in selected axial slices (see Supporting Information Figures S1–S3 for full brain distributions). Note that for visualization purposes, average values of these measures are mapped to the original atlas with no WM mask erosions. Note that the values of bFALFF have been rescaled from the original range of [0.20, 0.25] to [0.40, 1] for enhanced visualization. B, Scatter plots of linear relationships between bFC and bFDG, bFALFF and bFC, and bFALFF and bFDG. Tight regions shaded in green illustrate highly consistent linear fitting of the three measure pairs across six fMRI sessions
3.2. Correlations between bFC and bFDG
Pearson correlations between bFC and bFDG pooled over all the WM bundles and subjects studied are shown in Figure 2B (left). As seen, bFC exhibited significant correlation with bFDG, which was highly consistent across the six fMRI intervals (r = 0.25 ± 0.02, all p < 0.001). bFC was also found to correlate significantly with bFALFF for each session (r = 0.41 ± 0.04, all p < 0.001) (Figure 2B middle), likely because measures of FC are larger when BOLD FALFF increases compared to physiological noise. For completeness, Pearson correlation between bFALFF and bFDG was also computed (Figure 2B right), which appeared to be weaker and had only five of the six sessions reaching significance (r = 0.12 ± 0.03, all p < 0.01 except for session 4).
To assess the amount of variance in a dependent variable explainable by a predictor variable in the absence of inter‐subject variations, a linear mixed effects model was evaluated by treating the subject as a random effect in each of the three paired comparisons. The p‐values for all the three models were <0.001, and the R 2‐adjusted = 17%, 42% and 8%, respectively, for bFC vs. bFDG, bFALFF vs. bFC, and bFALFF vs. bFDG correlations (see Table 2). The corresponding coefficients for the three pairs of correlations were 0.41, 0.65 and 0.28, respectively, indicating the existence of moderate to strong correlations between these measures when the random effects introduced by inter‐subject variations were controlled.
TABLE 2.
Summary statistics of mixed effect models
| Model parameters | Response | ||
|---|---|---|---|
| bFDG | bFC | bFDG | |
| Fixed effect | bFC | bFALFF | bFALFF |
| Random effect | Subjects | Subjects | Subjects |
| R 2‐adjusted | 0.17 | 0.42 | 0.08 |
| p‐value | <0.001 | <0.001 | <0.001 |
See text for explanations.
3.3. Assessments of partial volume effects in WM
To examine whether the observed correlation between bFC and bFDG in WM was corrupted by the effects of partial volume averaging with GM, the correlations derived with WM masks eroded from 0 to 4 mm were compared (see Figure 3). It can be seen in Table 3, with the level of erosions increasing from 0 to 3 mm, the correlation varied from r = 0.37 ± 0.03 to 0.25 ± 0.02. However, the correlation tended to stabilize with further erosions of WM masks. This trend indicated that with WM masks eroded at 3 mm, the effects of partial volume averaging were quite minimal if any. Also note that the gradual decrease in the correlation coefficient from WM mask erosions of 1–3 mm may be due, at least in part, to the density gradient of interstitial neurons in WM, which tend to be more abundant toward the superficial WM zone. 19
FIGURE 3.
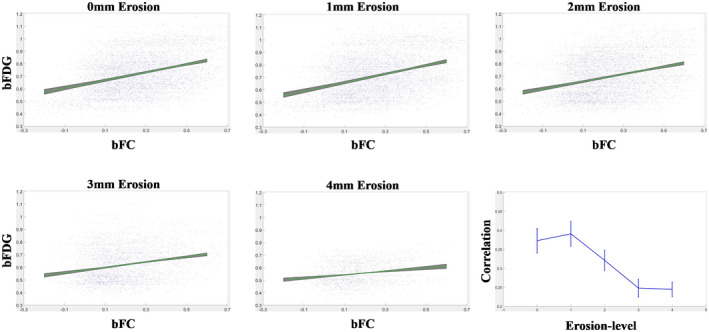
Effects of WM mask erosions on correlations between bFC and bFDG. Linear fittings between bFC and bFDG and correlation coefficients at the levels of WM mask erosions from 0 to 4 mm
TABLE 3.
Mean and SD of correlation coefficient across six imaging sessions at each level of WM mask erosion
| Level of erosion (mm) | Pearson correlation (mean ± SD) |
|---|---|
| 0 | 0.37 ± 0.03 |
| 1 | 0.39 ± 0.03 |
| 2 | 0.32 ± 0.03 |
| 3 | 0.25 ± 0.02 |
| 4 | 0.24 ± 0.02 |
4. DISCUSSION
WM constitutes nearly half the volume of the human brain, in which axonal fibers serve as information conduits that transmit neural activities between cortical regions. A complete understanding of brain functional architecture, therefore, requires both GM and WM functions be taken into consideration, which have important implications to FC modeling of the human brain. To date, such modeling is almost exclusively based on FC among GM regions without regard to the functional pathways that subserve GM function. 20 Physiologically, the diversity of brain functionality derives from the diverse patterns of axonal connections in WM, with each specific function engaging a distinct set of connection pathways underneath the cortex. Peering into the function of connection pathways in WM and linking them to those of GM will undoubtedly foster a more complete understanding of brain functional organization and how it evolves with development, aging, and pathology. This essentially shifts the paradigm of FC modeling of the brain from the current partial view on the sole basis of GM function to a full brain functional connectomics. Perhaps more provocatively, a WM‐centric brain connectome may be constructed with WM function wherein GM regions serve to relay and transfer information that flows in WM pathways; this could not only yield unique perspectives on brain functions unavailable from existing GM‐centric analyses, but also reveal more mechanistic insights into brain disease of WM origin.
As mentioned previously, there have been several reports of successful detections of functional signals in WM using fMRI, 21 but their interpretation remains unclear. It has been recognized that BOLD signals in GM are correlated with local field potentials, 22 and much of the cortical energy consumption is accounted for by neural activity of a type not found in WM. Specifically in GM, 23 it was estimated that 55% of the total ATP used on action potentials, synaptic transmission, and the resting potentials of neurons and glia, was consumed by the pre‐ and postsynaptic mechanisms involving in mediating synaptic transmission. 24 While in WM far fewer synapses exist, 25 oligodendrocyte activities make up a significant portion of the metabolic demands. 26 Thus, there have been concerns as to whether the observed fluctuations in WM reflect BOLD effects consequent on transient variations in metabolic demand. Interestingly, subsequent studies have found that BOLD signals are also correlated with postsynaptic spiking activity, which increases oxygen consumption as well. 27 , 28 Direct measurements of metabolism promise to clarify the origins of MRI signal changes. It was found earlier that, although WM has about one‐fourth of the vascular density of GM, the oxygen extraction fraction is quite uniform throughout the brain parenchyma. 29 A combined PET and fMRI study found that, during a hypocapnia challenge, BOLD signals increased in WM but with reduced magnitudes compared to GM, an effect that was largely attributable to reduced blood flow and volume in WM. 30 Moreover, it was recently observed that global glucose metabolic activities in WM, measured by FDG‐PET, vary with the functional state of the brain. 31 These findings suggest that the blood volume and oxygenation level in WM could also fluctuate with neural activities, thereby producing BOLD effects similar but smaller than those found in GM. We therefore analyzed regional glucose metabolism using dynamic PET data and explored their relations with BOLD signals. It was found that FC in WM bundles is significantly correlated with local metabolism and that the FC is associated with the power density of low frequency fluctuations of BOLD signals. These findings support the notion that BOLD signals in WM are modulated by cortical activities and reflect local metabolic variations and are robustly detectable.
In this work, seemingly low adjusted‐R 2 values of 17% for bFDG ~bFC and 8% for bFDG ~ bFALFF correlation were observed (In our companion experiments for GM with the same experiment and analysis procedures, the adjusted‐R 2 values for these correlations were found to be 13% and 16%, not included in this brief communication). These corresponded to Pearson correlation R values of 0.41 and 0.28, respectively, which are reasonably good given the existence of many confounds that could potentially reduce the underlying correlations, including, e.g., different levels of instrumental imperfection and sensitivity between fMRI and PET, inadequacy of proposed measures to fully capture the bona fide neural activity and metabolism, deviations of defined bundles from their actual anatomy, interferences from non‐neural or autonomic physiological activities, and limited signal to noise ratio of acquired signals, to name a few. Practically, this correlation level is very common in many human studies, e.g., correlation between the extent of coronary atherosclerosis and lipid profile, 32 and the correlation between biomarkers of Alzheimer disease and executive functioning scores in brain studyies. 33
It should be emphasized that both PET and fMRI are highly susceptible to noise, 34 , 35 particularly in WM where metabolic rates and hemodynamic responses are much reduced as compared to GM. Thus, it can be anticipated that correlations across the two imaging modalities with different profiles of noise sources (and signal sensitivity) are not as strong as those within the same imaging modality (e.g., between bFC and bFALFF), and those involving only WM regions (e.g., between bFDG and bFALFF) tend to be even weaker. It is also worth noting that bFALFF may be inversely related to WM transmission efficiency. 36 While this phenomenon has energetics benefits, the lower WM BOLD fluctuations associated with increased transmission efficiency also decreases the SNR that tends to add uncertainties to measured activity signals, thereby leading to a weaker correlation between bFDG and bFALFF as observed. By itself, bFALFF does not directly reflect the magnitudes of BOLD fluctuations.
Finally, the purpose of this study was to demonstrate the relationship between BOLD signal fluctuations and local metabolism in WM. Admittedly, the simple linear model of mixed effects with empirical fMRI and PET measures employed in this work is not able to fully capture the complicated relation between WM hemodynamic responses and metabolic demands. More sophisticated non‐linear models, such as that proposed by Tomasi et al. 18 for analysis of energy budget for FC in GM, could better reveal such relation, which is warranted for our future work.
CONFLICT OF INTEREST
The authors declare no competing interests.
Supporting information
FIGURE S1 Distributions of bFC. Bundle values are averaged across all the subjects studied and all six imaging sessions, and are superimposed onto the anatomical images in the MNI space
FIGURE S2 Distributions of bFDG. Bundle values are averaged across all the subjects studied and all six imaging sessions, and are superimposed onto the anatomical images in the MNI space
FIGURE S3 Distributions of bFALFF. Bundle values are averaged across all the subjects studied and all six imaging sessions, and are superimposed onto the anatomical images in the MNI space. Note that the values of bFALFF have been rescaled from the original range of [0.20, 0.25] to [0.40, 1] for enhanced visualization
FIGURE S4 Illustrations of WM masks before erosion (A), after erosion of 1 mm (B), 2 mm (C), and 3 mm (D). 3D renderings are shown in the right three columns for sagittal, coronal and axial views
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (NIH) grants R01 NS093669 (JCG) and R01 NS113832 (JCG), the National Key R&D Program of China Grants 2018YFA0704100 and 2018YFA0704101, and the National Natural Science Foundation of China Grant 61601012. We wish to sincerely thank Dr. Hakmook Kang (Department of Biostatistics, Vanderbilt University School of Medicine) for his valuable advice on our statistical analysis.
Guo B, Zhou F, Li M, Gore JC, Ding Z. Correlated functional connectivity and glucose metabolism in brain white matter revealed by simultaneous MRI/positron emission tomography. Magn Reson Med.2022;87:1507–1514. doi: 10.1002/mrm.29107
REFERENCES
- 1. Biswal B, Zerrin Yetkin F, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo‐planar MRI. Magn Reson Med. 1995;34:537‐541. [DOI] [PubMed] [Google Scholar]
- 2. Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8:700‐711. [DOI] [PubMed] [Google Scholar]
- 3. Li J, Biswal BB, Wang P, et al. Exploring the functional connectome in white matter. Hum Brain Mapp. 2019;40:4331‐4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peer M, Nitzan M, Bick AS, Levin N, Arzy S. Evidence for functional networks within the human brain's white matter. J Neurosci. 2017;37:6394‐6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ding Z, Newton AT, Xu R, Anderson AW, Morgan VL, Gore J. C Spatiotemporal correlation tensors reveal functional structure in human brain. PLoS One. 2013;8:e82107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ding Z, Huang Y, Bailey SK, et al. Detection of synchronous brain activity in white matter tracts at rest and under functional loading. Proc Natl Acad Sci U S A. 2018;115:595‐600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sarma MK, Pal A, Keller MA, et al. White matter of perinatally HIV infected older youths shows low frequency fluctuations that may reflect glial cycling. Sci Rep. 2021;11:3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cui W, Shang K, Qiu B, et al. White matter network disorder in mesial temporal epilepsy: an fMRI study. Epilepsy Res. 2021;172:106590. [DOI] [PubMed] [Google Scholar]
- 9. Faragó P, Tóth E, Kocsis K, et al. Altered resting state functional activity and microstructure of the white matter in migraine with aura. Front Neurol. 2019;10:1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frizzell TO, Grajauskas LA, Liu CC, Hajra SG, Song X, D’Arcy RC. White matter neuroplasticity: motor learning activates the internal capsule and reduces hemodynamic response variability. Front Hum Neurosci. 2020;14:509258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gore JC, Li M, Gao Y, et al. Functional MRI and resting state connectivity in white matter‐a mini‐review. Magn Reson Imaging. 2019;63:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu N, Lencer R, Yang Z, et al. Altered functional synchrony between gray and white matter as a novel indicator of brain system dysconnectivity in schizophrenia. Psychol Med. 2021:1‐9. 10.1017/S0033291720004420 [DOI] [PubMed] [Google Scholar]
- 13. Zhang Y, Kong Y, Liu X, et al. Desynchronized functional activities between brain white and gray matter in major depression disorder. J Magn Reson Imaging. 2021;53:1375‐1386. [DOI] [PubMed] [Google Scholar]
- 14. Gawryluk JR, Mazerolle EL, D'Arcy RC. Does functional MRI detect activation in white matter? A review of emerging evidence, issues, and future directions. Front Neurosci. 2014;8:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jamadar SD, Ward PGD, Close TG, et al. Simultaneous BOLD‐fMRI and constant infusion FDG‐PET data of the resting human brain. Sci Data. 2020;7:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lameka K, Farwell MD, Ichise M. Positron emission tomography. Handb Clin Neurol. 2016;135:209‐227. [DOI] [PubMed] [Google Scholar]
- 17. Jamadar SD, Ward PG, Li S, et al. Simultaneous task‐based BOLD‐fMRI and 18‐F FDG functional PET for measurement of neuronal metabolism in the human visual cortex. NeuroImage. 2019;189:258‐266. [DOI] [PubMed] [Google Scholar]
- 18. Tomasi D, Wang GJ, Volkow ND. Energetic cost of brain functional connectivity. Proc Natl Acad Sci U S A. 2013;110:13642‐13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sedmak G, Judaš M. The total number of white matter interstitial neurons in the human brain. J Anat. 2019;235:626‐636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horn A, Ostwald D, Reisert M, Blankenburg F. The structural–functional connectome and the default mode network of the human brain. NeuroImage. 2014;1:142‐151. [DOI] [PubMed] [Google Scholar]
- 21. Grajauskas LA, Frizzell T, Song X, D’Arcy RC. White matter fMRI activation cannot be treated as a nuisance regressor: overcoming a historical blind spot. Front Neurosci. 2019;13:1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature. 2001;412:150‐157. [DOI] [PubMed] [Google Scholar]
- 23. Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab. 2001;21:1133‐1145. [DOI] [PubMed] [Google Scholar]
- 24. Harris JJ, Jolivet R, Attwell D. Synaptic energy use and supply. Neuron. 2012;75:762‐777. [DOI] [PubMed] [Google Scholar]
- 25. Royeck M, Nishiyama A, Dietrich D. Persistance of neuronal transmitter release onto oligodendrocyte precursor cells during progressive myelination in the mouse optic tract. Soc Neurosci Abstr. 2010;36:554.11. [Google Scholar]
- 26. Harris JJ, Attwell D. The energetics of CNS white matter. J Neurosci. 2012;32:356‐371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heeger DJ, Ress D. What does fMRI tell us about neuronal activity? Nat Rev Neurosci. 2002;3:142‐151. [DOI] [PubMed] [Google Scholar]
- 28. Mukamel R, Gelbard H, Arieli A, Hasson U, Fried I, Malach R. Coupling between neuronal firing, field potentials, and FMRI in human auditory cortex. Science. 2005;309:951‐954. [DOI] [PubMed] [Google Scholar]
- 29. Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98:676‐682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rostrup E, Law I, Blinkenberg M, et al. Regional differences in the CBF and BOLD responses to hypercapnia: a combined PET and fMRI study. NeuroImage. 2000;11:87‐97. [DOI] [PubMed] [Google Scholar]
- 31. Thompson GJ, Riedl V, Grimmer T, Drzezga A, Herman P, Hyder F. The whole‐brain “global” signal from resting state fMRI as a potential biomarker of quantitative state changes in glucose metabolism. Brain Connect. 2016;6:435‐447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tarchalski J, Guzik P, Wysocki H. Correlation Between the Extent of Coronary Atherosclerosis and Lipid Profile. Vascular Biochemistry. Springer; 2003:25‐30. [PubMed] [Google Scholar]
- 33. Hammond TC, Xing X, Wang C, et al. β‐amyloid and tau drive early Alzheimer’s disease decline while glucose hypometabolism drives late decline. Commun Biol. 2020;3:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teymurazyan A, Riauka T, Jans H‐S, et al. Properties of noise in positron emission tomography images reconstructed with filtered‐backprojection and row‐action maximum likelihood algorithm. J Digit Imaging. 2013;26:447‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu TT. Noise contributions to the fMRI signal: an overview. NeuroImage. 2016;143:141‐151. [DOI] [PubMed] [Google Scholar]
- 36. Frizzell TO, Phull E, Khan M, et al. Imaging functional neuroplasticity in human white matter tracts. Brain Struct Funct. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Distributions of bFC. Bundle values are averaged across all the subjects studied and all six imaging sessions, and are superimposed onto the anatomical images in the MNI space
FIGURE S2 Distributions of bFDG. Bundle values are averaged across all the subjects studied and all six imaging sessions, and are superimposed onto the anatomical images in the MNI space
FIGURE S3 Distributions of bFALFF. Bundle values are averaged across all the subjects studied and all six imaging sessions, and are superimposed onto the anatomical images in the MNI space. Note that the values of bFALFF have been rescaled from the original range of [0.20, 0.25] to [0.40, 1] for enhanced visualization
FIGURE S4 Illustrations of WM masks before erosion (A), after erosion of 1 mm (B), 2 mm (C), and 3 mm (D). 3D renderings are shown in the right three columns for sagittal, coronal and axial views


