Abstract
Balance between sleep, wakefulness and arousal is important for survival of organisms and species as a whole. While, the benefits of sleep both in terms of quantity and quality is widely recognized across species, sleep has a cost for organismal survival and reproduction.
Here we focus on how sleep duration, sleep depth and sleep pressure affect the ability of animals to engage in courtship and egg-laying behaviors critical for reproductive success. Using isogenic lines from the Drosophila Genetic Reference Panel with variable sleep phenotypes we investigated the relationship between sleep and reproductive behaviors, courtship and oviposition. We found that three out of five lines with decreased sleep and increased arousal phenotypes, showed increased courtship and decreased latency to court as compared to normal and long sleeping lines. However, the male courtship phenotype is dependent on context and genotype as some but not all long sleeping-low courting lines elevate their courtship in the presence of short sleeping-high courting flies. We also find that unlike courtship, sleep phenotypes were less variable and minimally susceptible to social experience.
In addition to male courtship, we also investigated egg-laying phenotype, a readout of female reproductive output and find oviposition to be less sensitive to sleep length and parameters that are indicative of switch between sleep and wake states. Taken together our extensive behavioral analysis here shows complex bidirectional interactions between genotype and environment and add to the growing evidence linking sleep duration and sleep-wake switch parameters to behavioral decision making critical to reproductive output.
Keywords: Drosophila, Sleep, Mating, Oviposition, Behavior
1. Introduction
Sleep is an evolutionarily conserved neurobehavioral state that controls multiple physiological processes including development, metabolism, immune response, cognition and behavioral outputs critical for survival (Garbe et al., 2015; Harbison and Sehgal, 2008; Kayser et al., 2015; Keene and Duboue, 2018; Lesku et al., 2006; Sehgal and Mignot, 2011; Zielinski et al., 2016). Although, sleep has been observed and studied in mammals, birds, fish, reptiles, amphibians, and invertebrates, its effect on biological functions can be highly variable between and within species. In mammals, sleep deprivation and deficits have been associated with memory, attention and decision-making processes (Farahani et al., 2019; Krause et al., 2017; Sare et al., 2016) in addition to effects on metabolism, development and immune function which seem more generalizable across species (Besedovsky et al., 2012; Kayser and Biron, 2016; Sharma and Kavuru, 2010). Although, current research in human and animal models have demonstrated the importance of sleep on neural and non-neural processes, the precise function of sleep and the impacts of quality and quantity of sleep on an organism’s fitness and survival remain elusive.
Fruit flies have emerged as powerful systems in investigating the behavioral, physiological, cellular and molecular implications of sleep deficits and sleep-wake transitions. Across species, sleep is thought to be controlled by two processes, known as process S and process C, where Process S is a homeostatic process by which sleep pressure increases during wakefulness and decreases during sleep, and process C is a circadian process that is controlled by the circadian pacemaker circuits of the brain which determines the propensity to sleep during specific times of the day and night (Borbely, 1998; Borbely and Achermann, 1999; Borbely et al., 2016; Tobler et al., 1992). The process S and C model has provided a critical framework, but regulation of sleep and arousal goes beyond these processes. While sleep provides many benefits, it limits the ability of the organism to engage in other behaviors (e. g., foraging, mating, escaping predators etc.) critical for survival. Therefore, sleep states have to be actively balanced with cognitive and motivational processes critical for survival and rely on several external factors like availability of food sources, engagement in group interactions, temperature and humidity (Beckwith et al., 2017; Brown et al., 2018; Chen et al., 2017; Duhart et al., 2020; Keene et al., 2010; Machado et al., 2017; Masek et al., 2014; Yurgel et al., 2015).
Here we explore the relationship between sleep and the ability of the organisms to make behavioral decisions critical for reproductive success. Social interactions between male and female flies have been extensively studied in Drosophila and involves a complex series of stereotyped male-specific behaviors leading to copulation which is understood at the level of single genes and neurons (Yamamoto and Koganezawa, 2013; Yamamoto et al., 2014). Male flies suppress sleep in the presence of females or female cuticular pheromones. Conversely, male courtship behavior, female-induced arousal and egg-laying is attenuated by elevated sleep drive (Beckwith et al., 2017; Chen et al., 2017; Duhart et al., 2020; Machado et al., 2017; Potdar et al., 2018). Based on the current evidence pointing to a bidirectional relationship between sleep and reproductive behaviors (male courtship, copulation and egg laying) we probed how sleep parameters like sleep duration, sleep depth and sleep pressure affect male courtship and female oviposition behaviors.
Both sleep and reproductive behaviors are complex, variable, and highly dependent on genetic backgrounds making it difficult to study the subtle quantitative effects that result from naturally occurring alleles and SNPs. To circumvent this, we used a community resource, DGRP (Drosophila Genomic Resource Panel), a set of fully sequenced, inbred fly lines that were created by systematic mating of wild-caught isofemale full siblings for at least 20 generations. The isogenic genetic background and natural allelic variations make the DGRP an unparalleled resource to understand the evolutionary significance of sleep on reproductive behaviors important for organismal fitness. A previous screen of 168 DGRP lines reveals 10 inbred lines that had variable sleep duration which were classified as long and short sleepers. Remarkably, these lines with variable sleep phenotypes have similar longevity and egg to adult viability to normal sleeping lines (Harbison et al., 2013; Harbison and Sehgal, 2008; Harbison et al., 2017).
Using this collection, we first investigated the relationship between sleep length (the basis for classifying these lines as long-, short- and normal sleepers) and the complex sleep parameters sleep depth and pressure which have been linked to reproductive behaviors like courtship and egg-laying. Next, we asked if the DGRP lines with variable sleep phenotypes correlate with male courtship behavior and conversely if social experience alters sleep. We also investigated the effect of sleep length and sleep-wake switch parameters on female egg laying behavior. Lastly, we assayed for phenotypic plasticity in courtship behavior to test if the relationship between sleep parameters and reproductive behaviors is hard-wired or adaptable to changing environments.
2. Materials and methods
2.1. Fly strains and maintenance
Short-, long- and normal/moderate-sleeping flies from Drosophila Genome Research Panel were obtained from Bloomington Drosophila Stock Center (Harbison et al., 2013; Mackay et al., 2012). Stocks were maintained at 18οC and ~50% relative humidity (RH) under a 12:12 h light/dark (LD) cycle in standard fly food vials containing standard cornmeal-molasses media. Flies used in experiments were sub-cultured and maintained at 25 ◦C. All behavioral experiments were conducted at room temperature (21–25 ◦C) and included at least three independent trials. The stock number of flies used in the study is as follows (Table 1).
Table 1.
List of genotypes used in this study.
| DGRP Strain | Stock # (Bloomington Drosophila Resource Center) |
|---|---|
| DGRP-21 | 28122 |
| DGRP-38 | 28125 |
| DGRP-235 | 28275 |
| DGRP-301 | 25175 |
| DGRP-307 | 25179 |
| DGRP-310 | 28276 |
| DGRP-313 | 25180 |
| DGRP-335 | 25183 |
| DGRP-338 | 28173 |
| DGRP-365 | 25445 |
| DGRP-379 | 25189 |
| DGRP-808 | 28238 |
| DGRP-832 | 28245 |
| DGRP-859 | 25210 |
| CSMH line (originally from Heisenberg lab) | Gift from Janelia Research Campus, HHMI |
| W1118 (iso31) | 5905 |
2.2. Measurement of sleep and wakefulness by Drosophila activity monitoring system
To measure sleep, we used 3–10-day-old male and female flies. Individual flies were loaded into 5 mm × 65 mm glass tubes (Drosophila Activity Monitoring System, TriKinetics Inc) containing 5% sucrose 1% agarose solution. Tubes were loaded into Trikinetics DAM2 and placed in an incubator at 50% (RH) under a 12:12 h LD cycle at 25οC for 5 days as described in (Driscoll et al., 2019). The activity data were collected in one-minute bins for further processing using MATLAB (MathWorks)-based software SCAMP (Donelson et al., 2012). If no activity was detected by the Drosophila Activity Monitoring system for more than 5 min, the fly was considered to be in a sleep state (Garbe et al., 2015; Harbison and Sehgal, 2008; Kayser et al., 2015; Sehgal and Mignot, 2011; Shaw et al., 2000).
2.3. Video recording-based sleep measurement
The behavioral setup for video recording system was adapted from (Guo et al., 2016). Flies were briefly anesthetized and loaded into 96 well plates (Falcon™ 96-Well, Non-Treated, Fisher Scientific Inc) containing 5% sucrose and 1% agarose. The bottom of the plate was placed on and constantly illuminated using 850 nm LED board (Smart Vision Lights Inc). The 96-well plates with flies were imaged from top using a FLIR Flea 3 camera (Edmund Optics Inc) and the behavioral set up was placed in an incubator to control light/dark conditions and temperature. Fly movement was tracked using freely available Pysolo Video tracker and processed for sleep duration and sleep parameters using SCAMP 3 developed by Christopher Vecsey at Skidmore College and can be downloaded from https://academics.skidmore.edu/blogs/cvecsey/. The MATLAB scripts for analysis of P(Wake)/P(Doze) using locomotor date and hidden Markov model functions were accessed from https://github.com/Griffith-Lab/Fly_Sleep_Probability and were described in (Wiggin et al., 2020).
2.4. Courtship
Male virgin flies of each genotype were collected and housed in a separate vial for 2–3 days to mature. To minimize movement and rejection, female flies were decapitated using a razor blade (Spieth, 1966) and placed in courtship arena and allowed to recover prior to the behavioral assay. The use of decapitated female flies as a courtship target has been validated for genetic comparisons (Cheriyamkunnel et al., 2021; Ganter et al., 2011; Laturney and Billeter, 2014; Laturney et al., 2018; Pan and Baker, 2014; Pan et al., 2012; Pan et al., 2011; Rezaval et al., 2014; Rezaval et al., 2016; Zhang et al., 2018).
Male and female flies were placed into separate chambers of a 16-chamber bi-layer courtship wheel (diameter: 1 cm; height: 2.5 mm per layer) (Chen et al., 2017) backlit by a white LED panel (A2 Light Box Light Pad, Amazon Inc) in a 25οC incubator. Once flies recovered (~15–20 min) from anesthesia the layers of the courtship wheel were combined to place the male and female in the same chamber and flies were videotaped for 20 min. Courtship videos were analyzed single-blind and courtship index was measured as time spent by the male fly engaging in courtship behavior during the 20-min recording. Although, there are several behaviors like approach, licking and tapping associated with courtship these behaviors are also exhibited by flies when they are not courting. Hence, we used a more conservative approach and only coded unilateral wing extension to produce a courtship specific song, a behavior only exhibited in the context of male-female courtship (Cheriyamkunnel et al., 2021; Koganezawa et al., 2010; Massey et al., 2019; O’Sullivan et al., 2018; Pan and Baker, 2014; Pan et al., 2012; Pan et al., 2011).
2.5. Social isolation and enrichment for sleep measurements
3–7 days old flies were separated and grouped under socially enriched or isolated groups. DGRP lines that were enriched were housed in vials containing 20–25 females and 10–15 males from the time of eclosion to activity monitoring. Isolated flies were collected as pupae with a wet brush, sexed based on presence of sex combs and placed in 5 ml round-bottom polystyrene Test Tubes (352008, Falcon, Fisher Scientific) containing 1.5 ml of cornmeal molasses media. Both groups were collected and maintained 5–6 days before being placed in 65 mm glass tubes for sleep measurement using the DAM2 system described above.
2.6. Mate-competition assay
Male virgin flies of each genotype were collected and housed in a separate vial for 2–3 days to mature. Flies were then transferred onto standard cornmeal with red or blue food coloring (McCormick Inc) (Verspoor et al., 2015). To minimize movement and rejection, female flies were decapitated using a razor blade (Spieth, 1966) prior to the behavioral assay. Female flies and two males were placed into separate chambers of a 16-chamber bi-layer courtship wheel (diameter: 1 cm; height: 2.5 mm per layer) (Chen et al., 2017) backlit by a white LED panel (A2 Light Box Light Pad, Amazon Inc) in a 25οC incubator. Once flies recovered (~15–20 min) from anesthesia the layers of the courtship wheel were aligned to place the two males and decapitated female in the same chamber and flies were videotaped for 20 min. Courtship videos were analyzed single-blind and courtship index was measured as time spent by a male fly engaging in courtship behavior during the 20-min recording.
2.7. Oviposition
Age matched male and female flies (1–3 days) of each genotype were crossed for three days on standard cornmeal media. Mated females were then loaded on 24-well tissue culture plates (Cell treat Scientific 229123) containing apple-juice agar media (3 g sucrose, 125 ml apple juice, 3 g agarose, 125 ml water and 2.5 ml Tegosept). Female flies were allowed to lay eggs for 72 h and eggs were counted manually under single blind conditions.
2.8. Statistics
All sleep parameters (sleep amount, P (Wake), P(Doze), bout length and number of bouts) are presented as bar graphs and represent mean ± SEM. A one-way ANOVA was used for comparisons between group means and post hoc analysis was performed using Dunnett’s correction. For data sets that did not follow a gaussian/normal distribution (bout numbers and bout length) we used non-parametric analysis (one-way ANOVA of ranks and Kruskal Wallis Statistic). For comparisons of two genotypes or treatments we used t-tests (two-tailed). For correlation analysis, we conducted Spearman nonparametric correlation to avoid making assumption about the distribution of the values and also calculated two-tailed p values. Sample sizes for each experiment are presented in the figure legend. All statistical analyses and graphing were performed using Prism software (GraphPad Software 7.04; San Diego, California).
3. Results
3.1. Characterization of sleep duration and structure of DGRP lines
We first characterized the sleep phenotypes of the previously published DGRP lines that show altered sleep and wake patterns. Using the Drosophila activity monitoring system, we measured activity of 14 DGRP lines which were identified to have altered sleep phenotypes from a screen of 168 DGRP lines (Harbison et al., 2013). The DGRP is a panel of inbred lines created by crossing siblings of wild-caught female lines for over 20 generations and are in isogenic backgrounds (Mackay et al., 2012) allowing for more direct comparisons of parameters associated with quality and quantity of sleep.
We tested both males and females flies that were age matched (3–10 day old) and maintained in standard cornmeal media. We measured sleep duration over a 24-h period, mean length of sleep bout and number of sleep bouts for short sleepers (DGRP 38, 310, 365, 808 and 832), normal or moderate sleepers (DGRP 21, 301, 307, 859) and long sleepers (DGRP 235, 313, 335, 338 and 379). Data was analyzed by one-way ANOVA followed by pair-wise comparisons with DGRP 859, a moderate sleeper. To compare and contrast long and short sleeping lines we identified DGRP lines with moderate sleep phenotype as described in (Kumar et al., 2019). We did this by pooling the sleep data from several (~25 DGRP lines) and calculated mean sleep duration. Mean sleep duration over a 24-h period for DGRP 859 (~986 ± 8.28 mins in males and ~ 842 ± 10.11) was closest to the pooled data mean (~992 ± 6.12 mins in males and 839 ± 4.22). We also compared DGRP 859 with 2 strains commonly used as control in Drosophila sleep studies (w1118 (iso 31(Shi et al., 2014)) and CS) and have now added that data as Supplementary fig. 1.
In males, we found that short sleepers had sleep duration ranging from (~500–800 min) but only DGRP 38 (p < 0.0001) was statistically different from DGRP 859. Sleep duration of four normal sleepers ranged from ~800–1000 min, similar to other non-DGRP laboratory control strains CS and w1118 flies (Fig. S1). Four of the five long sleepers (~1000–1200 min) DGRP 235 (p < 0.0001), DGRP 335 (p < 0.0001), DGRP338 (p < 0.0001) and DGRP 379 (p < 0.0001) had significantly increased sleep duration (Fig. 1A).
Fig. 1. Sleep phenotypes of long and short sleeper DGRP male flies.
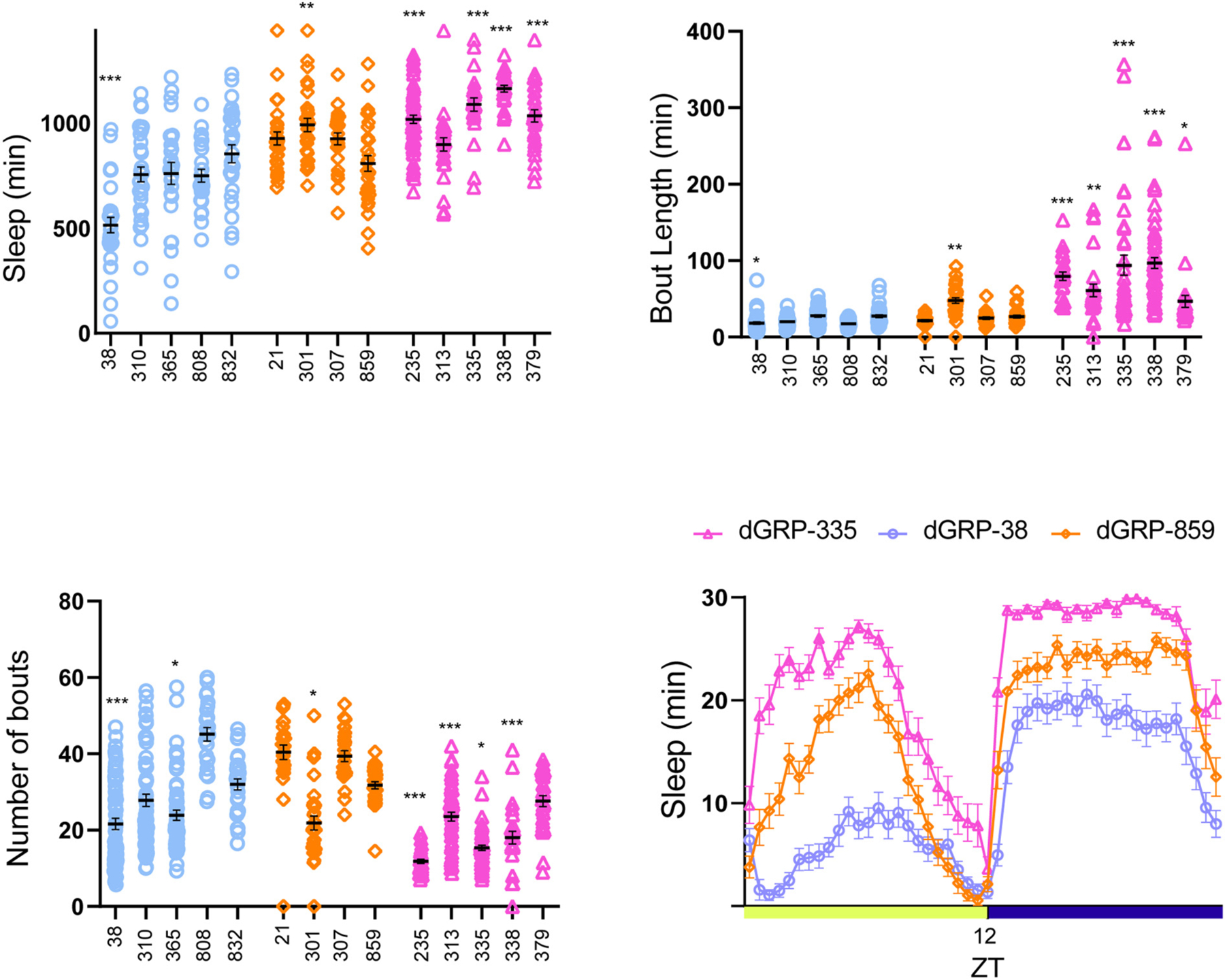
(A) 3-day average of sleep duration in males during a 24-h period.
(B) 3-day average of sleep bout length in males during a 24-h period.
(C)3-day average of sleep bout number in dGRP males during a 24-h period.
(D) 3-day average sleep (min/30 min) during a 24-h period. Sleep phenotype during daytime and nighttime of DGRP 38, 859 and 335 (each data point represents 30-min bin). ZT refers to zeitgeber time, ZT = 12 represents lights off.
21–63 flies of each genotype were assessed using Drosophila Activity Monitors with a 12:12 light/dark cycle for three days. Short sleepers (blue circles), normal sleepers (orange diamonds), and long sleepers (pink triangles). In this and all subsequent figures data represents mean and SEM *indicates p < 0.05, **indicates p < ***indicates p < 0.001.
Number of male flies for each genotype were: Short sleepers (dGRP-38: 31, dGRP-310: 32, dGRP-365: 26, dGRP-808: 25, dGRP-832: 29), Normal sleepers (dGRP-21: 28, dGRP-301: 29, dGRP-307: 25, dGRP-859: 30), Long Sleepers (dGRP-235: 63, dGRP-313: 27, dGRP-335: 24, dGRP-338: 31, dGRP-379: 29).
We saw a similar trend in female flies where short sleepers DGRP 38 (p < 0.0001), DGRP 310 (p < 0.00001), DGRP 365 (p = 0.018) and long sleepers DGRP 235 (p < 0.0001), DGRP 335 (p = 0.0002) and DGRP 379 (p = 0.0033) were significantly different from normal sleepers (Fig. 2A). Taken together, we found that the 10 DGRP lines have variable sleep duration both in males and females as previously reported (Harbison et al., 2013). In addition to the sleep duration, we assessed the sleep structure characterized by the average number of sleep bouts and length of sleep bouts.
Fig. 2. Sleep phenotypes of long and short sleeper DGRP female flies.
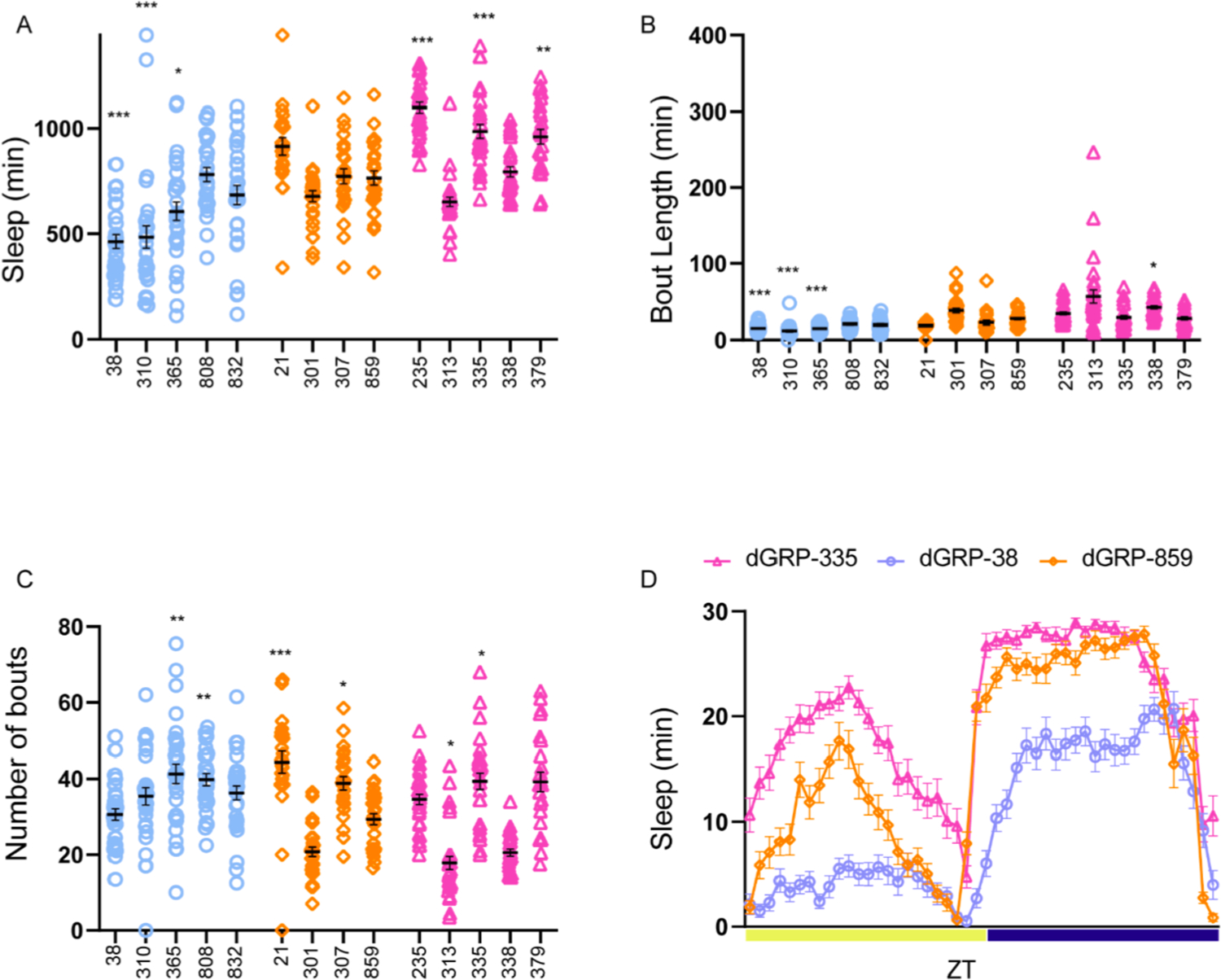
(A) 3-day average of sleep duration in female flies during a 24-h period.
(B) 3-day average of sleep bout length in female flies during a 24-h period.
(C) 3-day average of sleep bout number in female flies during a 24-h period.
(D) 3-day average sleep (min/30 min) during a 24-h period. Sleep phenotype during daytime and night-time of DGRP 38, 859 and 335 (each data point represents 30-min bin). ZT refers to zeitgeber time, ZT = 12 represents lights off.
24–31 flies of each genotype were assessed using Drosophila Activity Monitors and 12:12 light/dark cycle for three days. Short sleepers (blue circles), normal sleepers (orange diamonds), and long sleepers (pink triangles).
Number of female flies for each genotype were: Short sleepers (dGRP-38: 29, dGRP-310: 30, dGRP-365: 31, dGRP-808: 28, dGRP-832: 31), Normal sleepers (dGRP-21: 23, dGRP-301: 29, dGRP-307: 25, dGRP-859: 27), Long Sleepers (dGRP-235: 31, dGRP-313: 30, dGRP-335: 29, dGRP-338: 28, dGRP-379: 24) Statistical analysis was by one-way ANOVA using Dunnett multiple comparisons with normal sleeper dGRP-859 as control. In this and all subsequent figures data represents mean and SEM *indicates p < 0.05, **indicates p < 0.01. ***indicates p < 0.001.
In males, we found that short sleepers (DGRP 38, p = 0.017) had reduced bout length and all 5 tested long sleepers (DGRP 235 p < 0.0001, 313 p = 0.002, 335 p < 0.0001, 338 p < 0.0001 and 379 p = 0.04) had longer sleep bouts as compared to the normal sleeper DGRP 859 (Fig. 1B). Conversely, long sleepers (DGRP 235 p < 0.0001, 313 p < 0.0001, 335 p = 0.017, and 338 p < 0.0001) had fewer bouts as compared to normal sleepers consistent with increased sleep consolidation (Fig. 1C).
In females, we found that short sleepers DGRP 38 p < 0.0001, DGRP 310 p < 0.0001 and DGRP 365 p < 0.0001 had significantly shorter bouts as compared to normal sleepers, while only one long sleeper (DGRP 338 p = 0.03) had a modest increase in bout length (Fig. 2B). Number of bouts was less variable in females as compared to males; this could potentially be a result of reduced sleep duration in males as compared to females. Long sleeper DGRP 335 (p = 0.03) and short sleepers DGRP 365 (p = 0.007) and 808 (p = 0.01) had an increased number of bouts as compared to DGRP 859, while DGRP 313 (p = 0.02) had reduced bouts (Fig. 2C). We also saw that normal sleeper lines had a variable bout number phenotype even though sleep duration was not significantly different. Fig. 1D and 2D shows representative sleep profiles of a short- (DGRP 38), long- (DGRP 235) and normal sleeper (DGRP 859).
Taken together, our data shows that male and female long sleeper DGRP lines exhibited increases in sleep duration accompanied with fewer sleep bouts and longer bout duration suggesting that long sleepers have more consolidated sleep as compared to short sleepers. In contrast, short sleepers had significant reduction in sleep duration but bout structure was largely unaffected. These data suggest that the widely used metric of sleep phenotypes, sleep amount (24-h duration) is a good measure to identify short sleepers that exhibit prolonged wakefulness but don’t reliably define long sleepers as flies with even small increases in sleep amounts had more consolidated sleep as compared to normal and short sleepers.
The activity data and sleep measurement are essentially based on two states (movement or no movement). Conditional probability that defines the switch between these states have been recently used and validated as measures of sleep drive and arousal (Wiggin et al., 2020). In this approach Hidden Markov Model is applied to the locomotor data to reveal four states: deep sleep, light sleep, early wake, and full wake. Based on this model locomotor data can be used to calculate transition to sleep state and exit from sleep states or more specifically P(Wake) and P (Doze). P(Wake) quantifies the probability that a sleeping fly would wake up or move (sleep depth) and P(Doze) quantifies the probability that a waking fly would fall asleep (sleep pressure) (Wiggin et al., 2020). These probabilities depend on changes in response to sleep pressure and arousal and the algorithm for computing these probabilities using activity/inactivity instead of 5 in inactivity criteria is described in (Wiggin et al., 2020).
To get a better understanding of how the DGRP lines switch between sleep, wake and arousal states we mined the activity data to measure the probability of state transitions by calculating P (Wake) and P(Doze). Analysis of P(Wake) showed a clear trend related to sleep depth and propensity to switch to wakeful state in males and females. Short Sleepers ((DGRP 38, 310, 365, 808) p < 0.00001) had significantly higher P(Wake) as compared to control (DGRP 859) indicative of decreased sleep depth (Fig. 3A). Conversely, long sleepers ((DGRP 235, 313, 335, 338) p < 0.00001) had significantly lower P(Wake) as compared to control (DGRP 859) indicative of increased sleep depth. The normal sleepers DGRP 21, 301, 307 and 859 had similar P(Wake) and were not significantly different from each other (Fig. 3A).
Fig. 3. P(Wake), a measure of sleep depth and arousal in male and female DGRP lines.
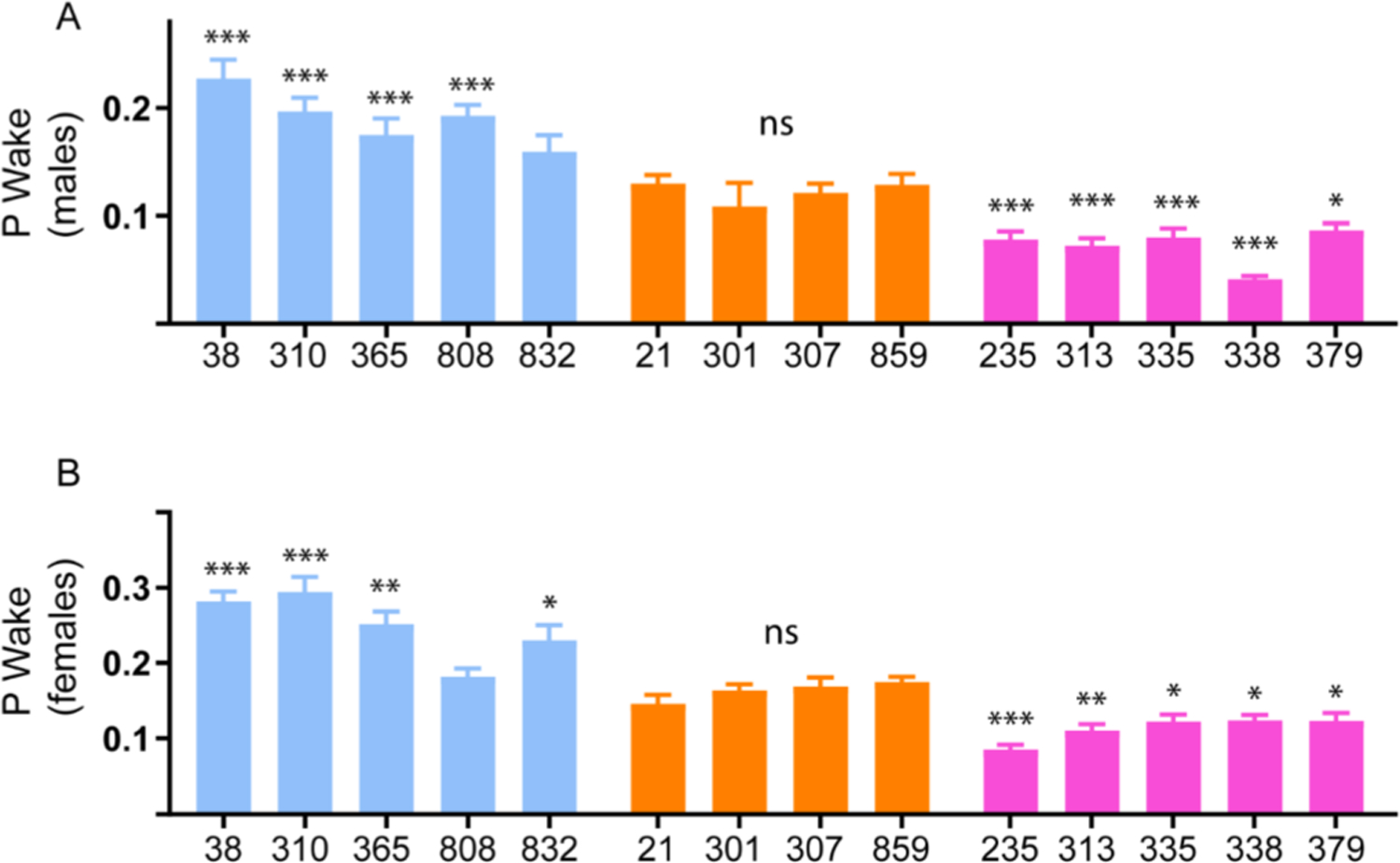
Average P(Wake) or probability of sleep to wake transitions/min in (A) Male and (B) female flies. 21–63 flies of each genotype were assessed using Drosophila Activity Monitors with a 12:12 light/dark cycle for three days.
Short sleepers (blue circles), normal sleepers (orange diamonds), and long sleepers (pink triangles). In this and all subsequent figures data represents mean and SEM *indicates p < 0.05, **indicates p < 0.01. ***indicates p < 0.001.
Number of male flies for each genotype were: Short sleepers (dGRP-38: 31, dGRP-310: 32, dGRP-365: 26, dGRP-808: 25, dGRP-832: 29), Normal sleepers (dGRP-21: 28, dGRP-301: 29, dGRP-307: 25, dGRP-859: 30), Long Sleepers (dGRP-235: 63, dGRP-313: 27, dGRP-335: 24, dGRP-338: 31, dGRP-379: 29).
Number of female flies for each genotype were: Short sleepers (dGRP-38: 29, dGRP-310: 30, dGRP-365: 31, dGRP-808: 28, dGRP-832: 31), Normal sleepers (dGRP-21: 23, dGRP-301: 29, dGRP-307: 25, dGRP-859: 27), Long Sleepers (dGRP-235: 31, dGRP-313: 30, dGRP-335: 29, dGRP-338: 28, dGRP-379: 24).
In females we observed a similar trend such that short sleepers (DGRP 38, 310 p < 0.00001, 365 p = 0.007 and 808 p = 0.009) had higher P(Wake) and long sleepers (DGRP 235 p < 0.0001, 335 p = 0.02, 313 p = 0.0020, 379 p = 0.04 and 338 p = 0.03) had smaller P(Wake) as compared to normal sleepers (Fig. 3B). Taken together, we conclude that short and long sleepers in our screen had decreased and increased sleep depth respectively, suggesting differences in their ability to transition from sleep to a wakeful/arousal state.
The transition from wake to sleep state, indicative of sleep pressure or sleep drive did not follow a specific pattern in males and females. Interestingly, three of the short sleepers (DGRP 310, 808 and 332 p < 0.00001) and two of long sleepers (DGRP 235 and 335, p < 0.00001) had increased P(Doze) in males and females suggesting higher probability of wake to sleep transition (Fig. S2A and 2B). Conversely, short sleeper DGRP 38 and 365 had comparable P(Doze) to control flies. Long sleeper DGRP 313 and 338 had significantly reduced P(Doze) as compared to controls. These data suggest that although long sleepers have increased sleep duration and low arousability, they have variable sleep pressure. Long sleepers DGRP 235 and 335 were specifically interesting as they had increased sleep duration, low P(Wake) and high P (Doze) in both males and females (Fig. S2A and 2B).
The Drosophila Activity monitoring (DAM) system used in the sleep measurements above detects movement based on IR beam breaks located in the midline of the tube and has limited spatial resolution. Hence, sleep measurements made by the widely used activity monitoring system is incapable of taking into account finer movements that represent a wakeful state. By detecting movement only at midline, the DAM system also overestimates sleep and limits the ability to accurately measure sleep duration, P(Wake) and P(Doze) (Garbe et al., 2015; Geissmann et al., 2019; Geissmann et al., 2017; Gilestro and Cirelli, 2009).
To address this, we tested 9 DGRP lines (3 short sleepers, 2 normal sleepers and 4 long sleepers) that have strong sleep phenotypes in the DAM system in a 96 well plate set-up using a video recording-based sleep measurement (Guo et al., 2016). The flies were entrained (12 h L:12 h D condition) and movement of flies were recorded for 2 days. Video files acquired at 1 frame/s were then tracked using an image analysis method based on frame subtraction (Guo et al., 2016).
Sleep duration during a 24 h period between DGRP lines were analyzed by one-sample t-test where mean of each genotypes was compared to mean of the population. We found that short and long sleepers had significantly reduced and increased sleep duration respectively consistent with the activity montoring system. Normal sleepers were not significantly different from the population mean (Fig. 4A and B). These phenotypes were consistent between male and female flies except for short sleepers DGRP 38 and 310 which had reduced sleep in males but not females. P(Wake) in male and female flies, a measure of sleep depth and sleep to wake transition was high for short sleepers DGRP 310 and 808. All long sleepers (DGRP 235, 335, 338 and 313) had lower P(Wake) as compared to normal sleepers supporting the finding that long sleepers are less arousable and short sleepers are more arousable (Fig. 4C and D).
Fig. 4. Sleep phenotypes of long and short sleeper DGRP male and female flies using video recordings and pixel movement-based measurement.
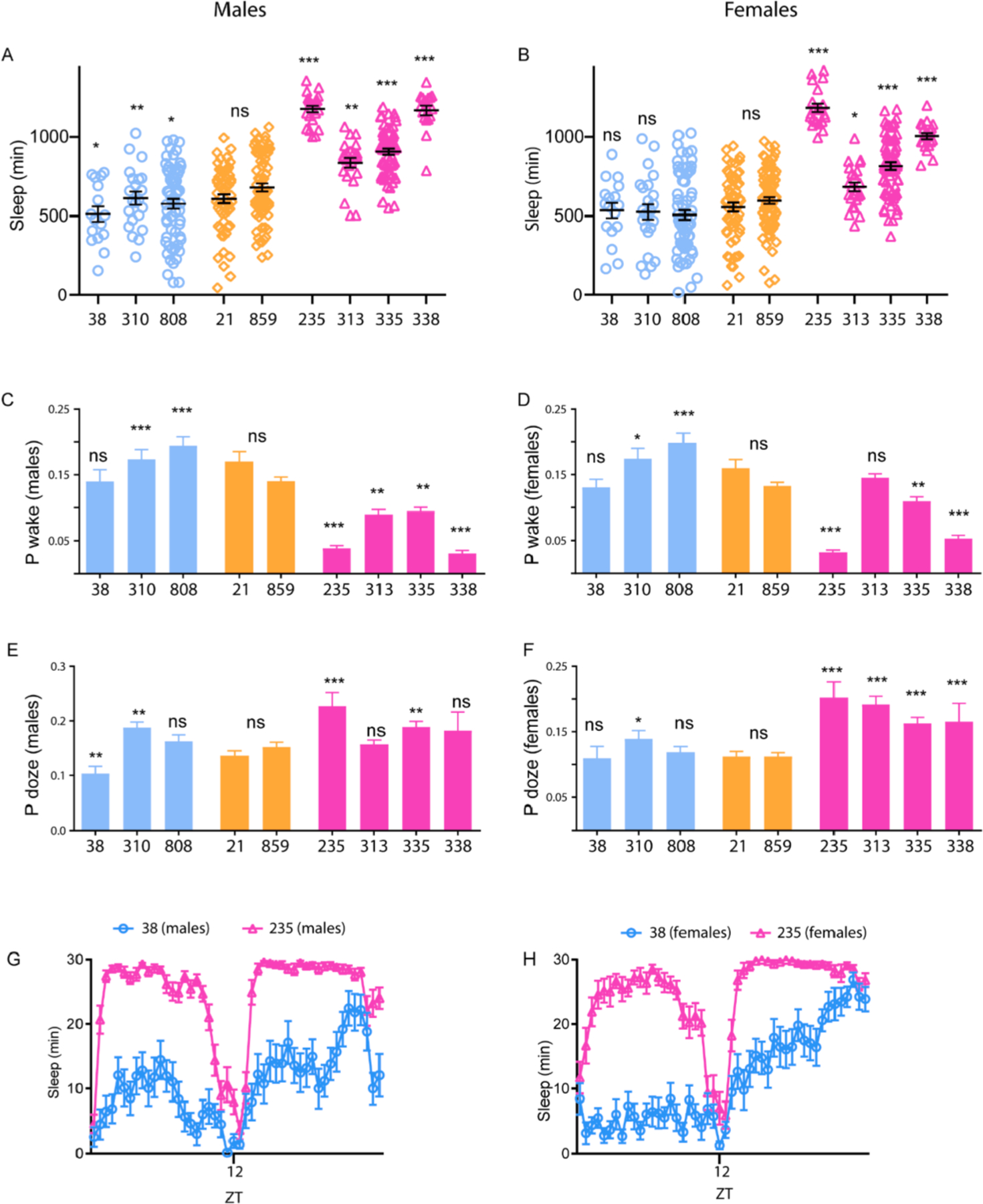
(A and B) Sleep duration (minutes) in male and female flies during a 24-h period.
(C and D) Average P(Wake) or probability of sleep to wake transitions/ min in male and female flies.
(E and F) Average P(Doze) or probability of wake to sleep transitions/ min in male and female flies.
(G and H) Sleep phenotype during daytime and nighttime of DGRP 38 and 235 (each data point represents 30-min bins). ZT refers to zeitgeber time, ZT = 12 represents lights off.
Short sleepers (blue circles), normal sleepers (orange diamonds), and long sleepers (pink triangles). In this and all subsequent figures data represents mean and SEM *indicates p < 0.05, **indicates p < 0.01. ***indicates p < 0.001, ***indicates p < 0.0001.
Number of male flies for each genotype were: Short sleepers (dGRP-38: 16, dGRP-310: 22, dGRP-808: 39), Normal sleepers (dGRP-21: 39, dGRP-859: 30), Long Sleepers (dGRP-235: 23, dGRP-313: 23, dGRP-335: 39, dGRP-338: 17).
Number of female flies for each genotype were: Short sleepers (dGRP-38: 17, dGRP-310: 24, dGRP-808: 45), Normal sleepers (dGRP-21: 40, dGRP-859: 36), Long Sleepers (dGRP-235: 21, dGRP-313: 23, dGRP-335: 42, dGRP-338: 20).
Although, probability of sleep to wake transition is a good measure of arousal and sleep depth, we also looked at probability of wake to sleep transition (P(Doze)). P(Doze) in male and female flies was consistently higher for long sleepers consistent with the idea that long sleepers have higher sleep pressure as compared to short and normal sleepers (Fig. 4E and F). Overall, the video based sleep measurements were consistent with the DAM data and is a more accurate measure of sleep depth and sleep pressure.
3.2. Male Courtship Phenotypes of short-, normal- and long-sleeper DGRP lines
Since, arousability measured by P(Wake) affects the ability of flies to switch states and engage in key behaviors critical for survival, we tested the ability of short-, long- and normal-sleepers to engage in courtship behavior. We specifically tested short sleepers (DGRP 38, 310, 365, 808, 832), normal sleepers (307 and 859) and long sleepers (235, 335 and 338). We selected these lines as they had P(wake), P(Doze) values that were significantly different from DGRP 859. Further, we only tested male long sleeper lines in courtship if they had increased sleep (p < 0.001) as compared to moderate sleeping controls.
We placed the DGRP male flies with wild type females in a courtship arena as previously described (Chen et al., 2017). We chose a 20-min courtship duration for these experiments as flies generally copulate within 12–20 min, flies that did not initiate courtship during the 20-min recording do not copulate at later time points (Luu et al., 2016).
We first measured courtship duration as the amount of time (in seconds) spent by male flies exhibiting courtship associated behaviors during the 20-min recording (Fig. 5A). We specifically quantified unilateral wing extensions exhibited by male flies. We found that short sleepers (DGRP 38 p < 0.0001, 310 p = 0.0075 and 365 p = 0.00012) showed significantly higher courtship index as compared to normal sleeper DGRP 859 (Fig. 5B). Indeed, three of these short sleepers initiated and sustained courtship through the entire duration of the recording (Supplementary video 1, representative courtship video of DGRP 38).
Fig. 5. Courtship Index and latency in long-, short- and normal- sleeping DGRP males.
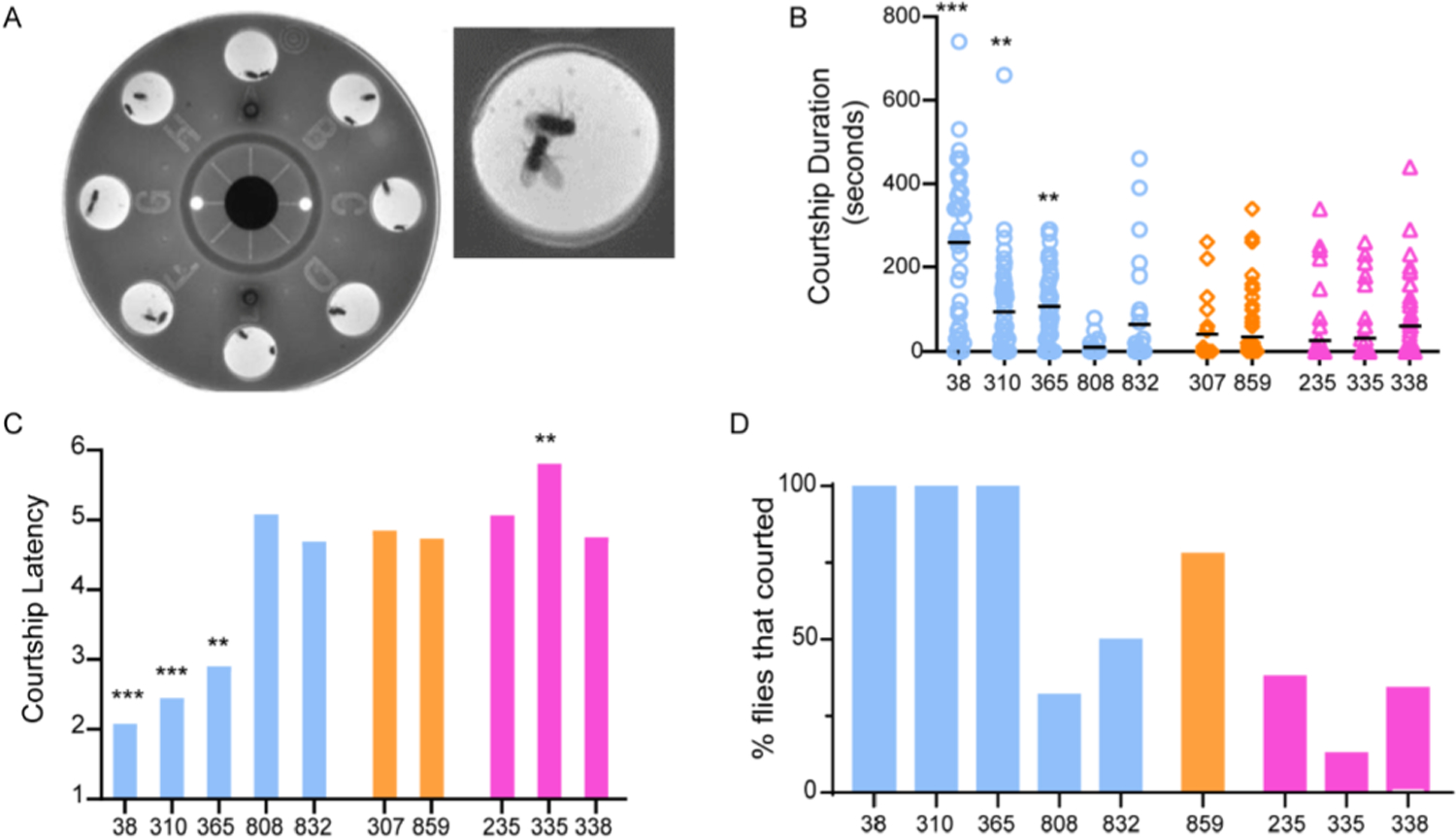
(A) Courtship wheel and arena used for assaying male courtship phenotypes. A pair of flies in an arena with male exhibiting unilateral wing extension.
(B) Courtship index measured as time spent (in seconds) by a male fly exhibiting unilateral wing extension (a courtship specific behavior) during a 20-min trial. Courtship index of short sleepers (blue circles), normal sleepers (orange diamonds), and long sleepers (pink triangles).
(C) Courtship latency measured as the time between start of the trial and initiation of the first bout of unilateral wing extension. Latency scores were coded such that 1 = 1–19 s 2 = 20–39 s, 3 = 40–59 s, 4 = 60–79 s, 5 = 80 s and above.
(D) Percentage of flies that exhibited at least one courtship bout.
23–63 flies of each genotype were recorded in courtship wheel arenas and unilateral wing extensions were manually counted. Males were separated from beheaded females until video recording began. Latency was measured as time to first unilateral wing extension and data was binned. Short sleeping dGRP-38 were found to court significantly more than all other genotypes. Short sleepers (blue circles), normal sleepers (orange diamonds), and long sleepers (pink triangles). Data represents mean and SEM *indicates p < 0.05, **indicates p < 0.01. ***indicates p < 0.001, ***indicates p < 0.0001.
In addition to courtship index, we also quantified courtship latency indicative of amount of time between start of recording and initiation of the first courtship bout (Fig. 5C). Like courtship index, short sleepers (DGRP 38 p < 0.0001, 310 p < 0.0001 and 365 p = 0.0002) had shorter latency and long sleeper DGRP 335 p = 0.0010 had longer latency as compared to normal sleeper DGRP 859. All long sleepers DGRP 335, 235 and 338 had very minimal courtship and high latency (Supplementary video 2, representative courtship video of DGRP 335).
In addition to latency and courtship index we also quantified percentage of flies that showed at least one courtship bout of unilateral wing extension. These values ranged from 13 to 38% for long sleepers and 32–100% for short sleepers (Fig. 5D) suggesting that sleep duration and arousability are correlated with the ability to engage in courtship behaviors critical for reproductive output.
3.3. Correlation between sleep and courtship parameters of DGRP lines
Based on our behavioral data we reasoned that altered propensity to switch between sleep and wake states are correlated with the ability to engage in courtship behaviors critical for reproductive output. To test the strength of this correlation we specifically focused on P(Wake) and P (Doze) as sleep measures, and courtship index and latency as courtship measures.
We computed correlation coefficients between P(Doze) and P(Wake) for all tested DGRP lines (Fig. 6A R = ‒0.2 and p = 0.002) and did not find a significant correlation between these parameters. Although, long sleepers exhibit longer sleep durations, they consistently show reduced sleep to wake transitions. On the other hand, short sleepers exhibited shorter sleep durations, increased sleep to wake transitions and high sleep pressure indicated by increased wake to sleep transitions.
Fig. 6. Correlational analysis within and between sleep and courtship parameters.
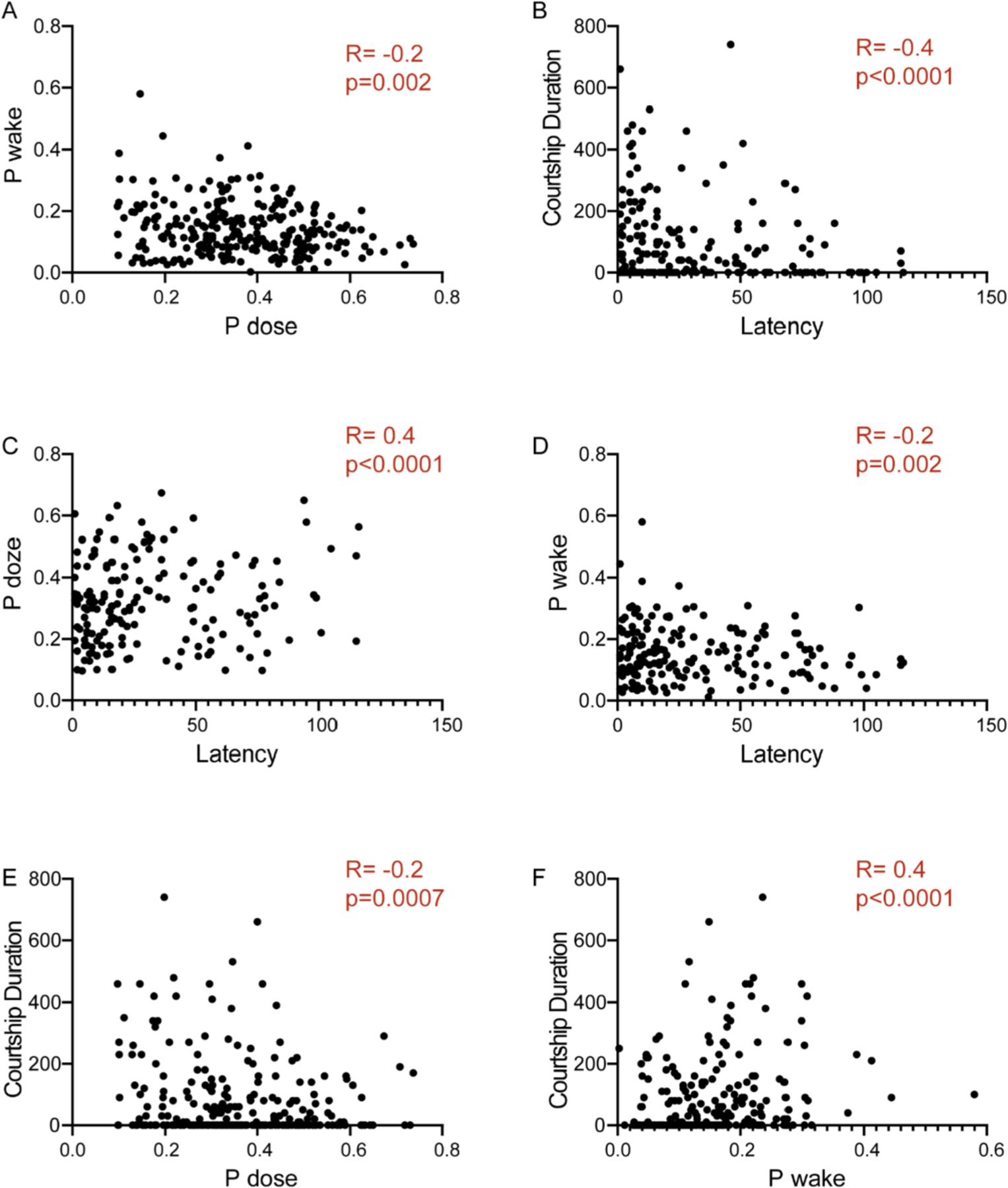
(A) Correlation analysis of P(Wake) and P(Doze) of male DGRP flies (short-, normal-, and long-sleeper lines).
(B) Correlation analysis of courtship duration and latency of male DGRP flies (short-, normal-, and long-sleeper lines).
(C and D) Correlation analysis of P(Doze) and P (Wake) vs courtship latency.
(E and F) Correlation analysis of P(Doze) and P(Wake) vs courtship duration.
We computed Spearman’s coefficient of correlation R between 2 groups at a time and two-tailed p values are indicated in all plots. Flies that did not court for the entirety of the 20-min duration are not included in the plots. For each of the genotypes we had 21–63 pairs. These genotypes included DGRP 38, 310, 365, 808, 832, 307, 859, 235, 335 and 338.
We next looked at how courtship parameters are correlated with each other and with P(Wake) and P(Doze). We did find a moderate negative correlation between courtship index (amount of time spent in courting/unilateral wing extension) and courtship latency (R = ‒0.4 and p < 0.0001) (Fig. 6B).
We asked if sleep parameters P(Doze) and P(Wake) are correlated with courtship index and latency. P Doze, indicative of sleep pressure, shows a moderate positive correlation with courtship latency (R = 0.4, p < 0.0001) and very weak negative correlation with courtship duration (R = ‒0.2, p = 0.0007) (Fig. 6C and E). On the other hand, P(Wake) indicative of arousal, shows moderate positive correlation (R = 0.4, p < 0.0001) with courtship duration and weak negative correlation (R = ‒0.2, p < 0.002) with latency (Fig. 6D and F). Taken together, we find a moderate correlation between P(Doze) and latency and P(Wake) and courtship duration within our DGRP population suggesting that transition between states is correlated with engagement in social behaviors like mating that are critical for reproductive output.
3.4. Socialization does not alter sleep duration
Based on our findings that short sleepers court more as compared to long and normal sleepers we asked if the relationship between sleep and courtship is bidirectional. Previous studies suggest that social isolation leads to increased male courtship drive and reduction in sleep duration (Ganguly-Fitzgerald et al., 2006; Liu et al., 2019; Ueda and Wu, 2009).
All the sleep results reported above were conducted on flies that were group housed in a socially enriched environment with specific sex ratio (20–30 females and 10–15 males). We compared sleep duration between socially isolated and enriched flies to test if increased courtship drive produced by social isolation alters sleep duration.
To test this, we focused on short-sleeping/high courting lines (DGRP 38, 310 and 365) and long-sleeping/low courting lines (DGRP 235 and 335). The socially isolated and enriched group for each DGRP line were compared using unpaired t-test and Welch’s correction for each of the 7 tested DGRP lines.
We found that other than normal sleeper DGRP 859 (p = 0.04) none of the short-sleepers and long-sleepers altered their sleep duration in conditions of social isolation (Fig. 7). Intriguingly, the long sleepers did not alter their sleep duration when socially isolated, a condition associated with increase courtship drive. Since, DGRP 859 (normal sleeper control) showed a decrease in sleep under isolated condition as compared to group-housed social condition we suspect that the genetic architecture of long and short sleepers is related to housing condition and sleep duration.
Fig. 7. Social isolation does not affect sleep duration in short- and long-sleepers.
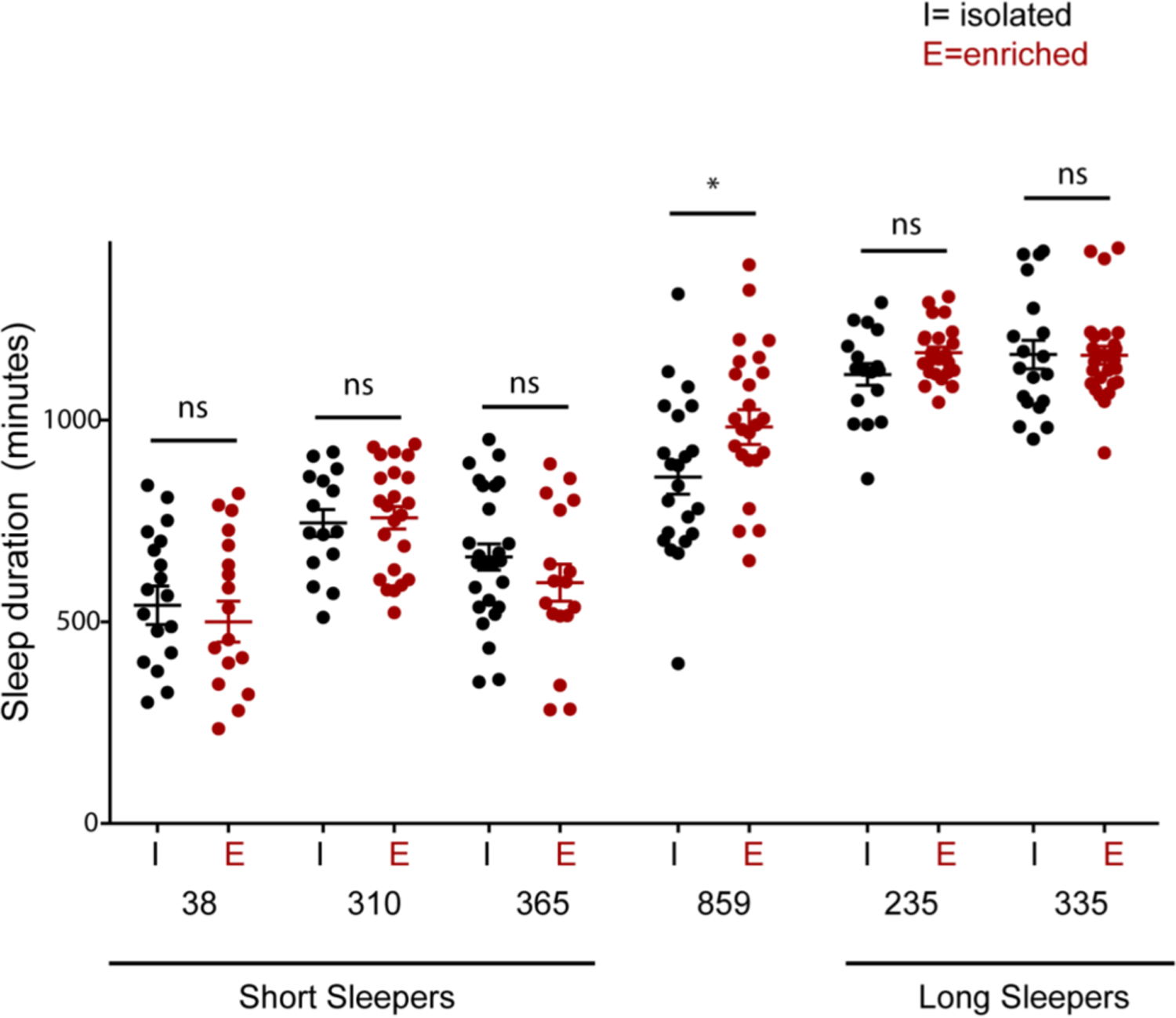
Sleep duration of socially isolated (I) and enriched (E) DGRP lines: short-sleepers with high courting phenotypes (38, 310 and 365), normal sleeper (859) and long sleepers with low courtship (235 and 335) in beam break-based DAM sleep assay.
Isolated and enriched groups for each of the tested lines were compared using unpaired t-test with Welch’s correction. Data represents mean and SEM *indicates p < 0.05.
Number of male flies for each genotype were between 18 and 26 and data were obtained from 2 independent trials.
3.5. Mating competition assay reveals phenotypic plasticity in long and short sleepers
Single male and female courtship assays in a controlled environment as described above have been widely used as a test for male courtship decision making and reproductive success. These assays are robust in measuring stereotypical pattern of context specific behaviors unilateral wing extension and following that are essential for copulation. However, male flies can modify their mating behavior when they face competition from other stimuli including other conspecific males. To address phenotypic plasticity in short and long sleepers we conducted a matecompetition assay. We paired high courting short sleeper DGRP 38 with long sleeper-low courters DGRP 235 and 335 male flies to test behavioral plasticity in mating duration towards wild type CS females (schematic of experimental design in Fig. 8A).
Fig. 8. Mate competition behavioral assays reveal that courtship phenotypes are plastic for longer sleeper DGRP 335 but not DGRP 235.
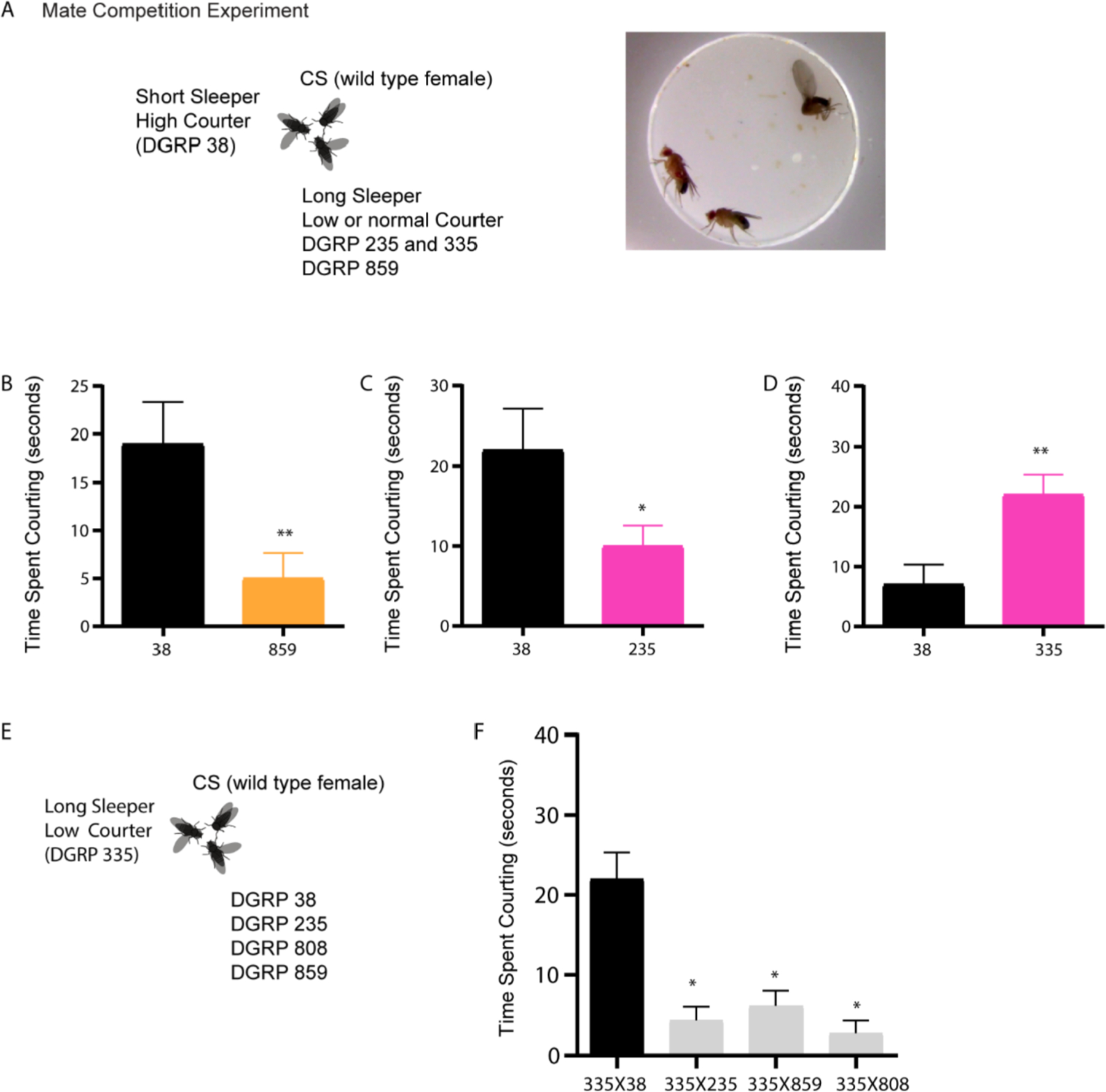
(A) Schematic of the mate competition assay showing a female CS (wild type fly) and two distinct DGRP males with variable sleep phenotype. One of the DGRP males is a short sleeper with high courtship phenotype. The other DGRP lines were long sleepers, low courters (DGRP 235 and 335) and normal sleeper (DGRP 859).
(B, C and D) Courtship phenotype (time spent engaging in unilateral wing extension) of DGRP 38 (black bar) with long (335 and 235) or normal sleepers (859). Pink bars represent courtship phenotypes of long sleepers DGRP 235 and 335 (pink bar) and yellow bar indicates courtship phenotypes of normal sleeper DGRP 859.
(E) Schematic of the mate competition assay showing a female CS (wild type fly) and two distinct DGRP males with variable sleep phenotype. One of the DGRP 38 male is a long sleeper with low courtship phenotype. The other DGRP lines were long sleepers-low courters (DGRP 235), short sleeper-high courter (DGRP 38), short sleeper-normal courter (DGRP 808) and moderate sleeper-normal courter (DGRP 859).
(F) Courtship phenotype (time spent engaging in unilateral wing extension) of DGRP 335 in mate competition assay with DGRP 38 (black bar) and DGRP 235, 808 and 859 (grey bar).
Data represents mean and SEM *indicates p < 0.05, **indicates p < 0.01. ***indicates p < 0.001, ***indicates p < 0.0001. Statistical analysis was conducted between two-samples by Unpaired t-test with Welch’s correction (B, C and D). For F, analysis was conducted between 4 samples by non-parametric Kruskal-Wallis test followed by Dunns’ correction.
As observed in single male-female courtship assays DGRP 38 courts the wild type female more as compared to long sleeper DGRP 235 and normal sleeper DGRP 859 in the same arena (Fig. 8B and C). However, DGRP 335 that exhibits increased sleep and reduced courtship in single male-female assay courts more than DGRP 38 when placed in the same arena. These results show that long sleeping-low courting DGRP 335 males increase their mating duration when presented with a high courting-short sleeping DGRP 38 male (Fig. 8D). In the mate competition with DGRP 335, we also found that DGRP 38 males adapted and reduced their mating duration by almost 50% indicating social experience dependent modification of mating duration.
We next asked if DGRP 335 male courtship is altered only in the presence of high courting male (DGRP 38) or any other male. To test this, we set up a modified mate competition protocol (schematic in Fig. 8E). In this second protocol we paired DGRP 335 with long sleeper-low courters DGRP 235, normal sleeper-normal courter DGRP 859 and short sleeper-normal courter DGRP 808. Increased male courtship in DGRP 335 occurs only in competition with DGRP 38 and not with the other lines (DGRP 235, 808 and 859) (Fig. 8F).
In summary, although there is a correlation between sleep parameters and courtship phenotypes based on a single male-single female assays, long sleeper males can increase courtship and short sleeper can exhibit reduced courtship and this phenotypic plasticity likelyalters reproductive output. However, these effects are dependent on genotype and not generalizable for all long sleeping-low courting lines since DGRP 235 and 859 do not exhibit the same plasticity.
3.6. Oviposition phenotypes as a measure of female reproductive output in short-, normal- and long-sleeper
Although increased courtship impacts the probability of mating and critical to reproductive success in male flies, it is an incomplete measure of reproductive fitness. We next tested the ability of DGRP female lines to mate and lay eggs. Like courtship, oviposition in Drosophila follows a series of processes including ovulation of eggs in females, mating, sperm storage, fertilization and extrusion by mated females (Aranha and Vasconcelos, 2018; Bracker et al., 2019). We mated DGRP lines for 3 days and placed the mated females in chambers containing apple juice agar, a preferable substrate for egg laying (Fig. 9A and B). We found that egg laying phenotypes of females were largely consistent and did not necessarily depend on sleep duration, sleep-depth or sleep-pressure.
Fig. 9. Oviposition behavior in short-, normal- and long-sleepers.
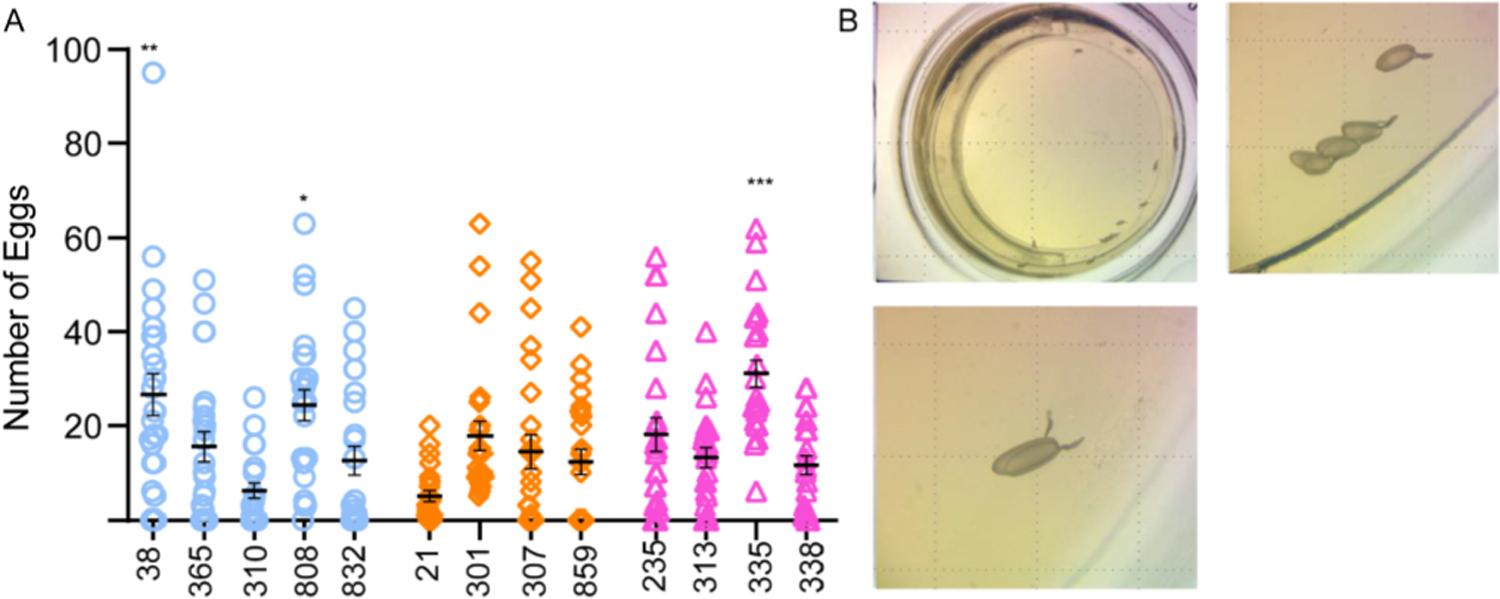
(A) Number of eggs laid per chamber of a 24-chamber arena during a 3-day period. Each chamber housed two age-matched, mated female flies and eggs were counted manually. Short sleepers (blue circles), normal sleepers (orange diamonds), and long sleepers (pink triangles).
(B) Single chamber of a 24 well plate arena containing apple juice agar media. Eggs were counted under the microscope.
Data represents mean and SEM *indicates p < 0.5, **indicates p < 0.01. ***indicates p < 0.001, ***indicates p < 0.0001. We tested 48 flies/genotype (2 flies/chamber). Statistical analysis was by one-way ANOVA using Dunnett multiple comparisons with normal sleeper dGRP-859 as control. Data represents mean and SEM *indicates p < 0.5, **indicates p < 0.01. ***indicates p < 0.001.
Among the short sleepers DGRP 38, a line that shows increased male courtship, increased sleep-depth and reduced sleep-pressure showed a significantly higher number of eggs (p = 0.008, Fig. 9A) as compared to normal sleeper DGRP 859. Another short sleeper, DGRP 808, that shows high sleep to wake transition and arousability also produced a significantly higher number of eggs (p = 0.04, Fig. 9A).
Lastly, long sleeper DGRP 335 that shows increased courtship latency, decreased sleep-depth and increased sleep-pressure also laid more eggs (p = 0.0001, Fig. 9A). These data reveal that although sleep duration and ability to switch between sleep-wake states are variable in female DGRP lines, they have a variable effect on female mating and oviposition.
3.7. Reproductive output in short sleeper DGRP 38 is contributed by males
Since oviposition was higher in 3 DGRP lines (2 short- 38 and 808 and 1 long sleeper- 335) that have different sleep parameters we probed specific male and female factors that contributed to increased eggs. Specifically, we asked if increased male courtship phenotypes associated with DGRP 38 and DGRP 335 identified in our study by single male-female courtship assay and mate-competition assay led to increased mating and eggs.
To address this, we mated males from 4 DGRP lines (38, 808, 859 and 335) with wild type females (CS) to control for the differential and variable reproductive contributions of DGRP females (Fig. S3A). We found that CS females mated with short sleeper DGRP 38 p < 0.0001 and long sleeper DGRP 335 p = 0.013 produced a significantly greater number of eggs as compared to DGRP 859 and 808.
Conversely, we mated female flies from 4 DGRP lines (38, 808, 859 and 335) with wild type males (CS) to control for the differential reproductive contributions of DGRP male. We found that under these conditions there was no significant difference between the short-, long- and normal sleepers suggesting that male courtship vigor is a critical regulator of reproductive fitness and that female flies don’t necessarily compensate for reduced mating vigor in males (Fig. S3B).
4. Discussion
Balance between sleep, wakefulness and arousal is important for survival of organisms and species as a whole. While, the benefits of sleep both in terms of quantity and quality is widely recognized across species, sleep has a cost for organismal survival. Sleep can make an animal vulnerable to it predators and also limit engagement in social behaviors (like mating or aggression) critical for survival. Here we focus on how sleep duration, sleep depth and sleep pressure correlate with the ability of animals to engage in behaviors critical for reproductive output. Previous studies show that sleep deprivation negatively impacts male courtship behaviors and female oviposition (Chen et al., 2017; Machado et al., 2017; Potdar et al., 2018). However, in both these cases sleep deficit was severe and induced by mechanical deprivation, dopamine dysregulation or caffeine. The ethological relevance of how sleep and arousal states are balanced and impact reproductive success is unexplored.
Genetic analysis of short sleeping mutants identified from wide-spread unbiased genetic screen of sleep duration reveals a strong genetic component underlying sleep regulation (Bringmann, 2019). Genetic manipulations including gene expression knockdowns by RNA interference, P-element knockdowns and large-scale mutagenesis are often conducted in variable genetic backgrounds so comparisons don’t always yield clear, reproducible results. In light of these technical challenges, we used the short and long sleeping natural variant lines from the DGRP panel to study the relationship between sleep and behavioral outputs specifically, courtship and egg laying that determine reproductive success. The 14 DGRP lines were selected from 168 lines and included flies with short-, long- and normal sleep duration. In addition to sleep duration, we characterized several additional parameters that define sleep quality as sleep is complex behavioral trait. First, we found minimal to no sexual dimorphism in sleep parameters except for sleep duration which is consistent with reduced sleep in females as compared to males. Second, sleep duration and parameters of variable sleeping lines were consistent between two independent methods of measuring sleep (video recording-tracking and activity monitoring). Third, we found that even between lines with consistent sleep duration, a parameter widely used to define sleep there were significant differences in ability to transition between sleep and wake states. In addition to measuring the probability of switching between states, we probed how complex sleep parameters are related to behavioral decisions that lead to increased reproductive output in flies.
We argued that sleep depth P(Wake) and sleep pressure P(Doze) are likely to be detrimental to the motivation of the animal to switch from sleep to arousal states to engage in social behaviors like mating. To directly address this, we assayed male courtship phenotypes of 3 of the longest sleepers, 5 short sleepers and 2 normal sleepers to test the correlation between sleep and courtship parameters. We found that 3 out of the 5 of the short sleepers showed increased courtship duration and reduced latency. We also found a moderate correlation between P (Wake) (sleep to wake transition) and courtship duration suggesting flies with reduced depth are more arousable when presented with a courtship target.
Conversely, we also found a moderate correlation between P(Doze) (wake to sleep transition) and latency suggesting that flies with greater sleep pressure tend to take longer to initiate courtship bout. Our data reveal that classification of lines based on sleep length alone has limited significance in understanding how animals balance sleep with other behaviors critical for survival. While our data support a relationship between prolonged waking and engagement in social behaviors like mating, they don’t necessarily mean that sleeping too much or too little alters reproductive fitness.
To address the effect of courtship drive on sleep duration we measured sleep in DGRP lines with increased sleep-low courtship or prolonged wakefulness-high courtship phenotype under socially enriched or isolation conditions. Previous studies show that social isolation increases male-male aggression, male courtship towards female target and reduce sleep duration (Ganguly-Fitzgerald et al., 2006; Liu et al., 2019; Ueda and Wu, 2009). Although, normal sleeper showed a decrease in sleep under social isolation, the long and short sleeper did not alter sleep duration in isolated condition. The short-sleepers exhibit prolonged wakefulness and it is likely that sleep drive is strong and unaffected by housing conditions. However, it is intriguing that long sleepers do not change their sleep duration when socially-isolated. Future studies are required to investigate gene and environment interactions that underlie how social experience like isolation, enrichment, modified group housing, starvation and sex ratios modifies sleep need and if short and long-sleepers investigated here have variable responses to these environmental conditions.
Mating behavior is highly plastic and dynamic and known to be altered by rival males, sexratio, nutritional status, age and female mating status (Bretman et al., 2009; Bretman et al., 2013; Fricke et al., 2008; Ruhmann et al., 2018). In our experiments, we controlled for several factors by using age-matched flies, reared in the same nutritional media and placed in vials with identical sex ratio. However, the courtship assays are often conducted in pairs (1 male and female) and don’t account for rival males. To address this, we conducted the mate competition assays.
Using this assay, we asked if high courting-short sleepers and low courting-long sleepers can modify their mating duration by placing them in the same arena with a wild type female target. Interestingly, we found an almost 50% reduction in mating duration of high courting-short sleeper DGRP 38 in the presence of long-sleeper DGRP 335 but no other lines. Conversely, DGRP 335 elevated its courtship duration in the presence of DGRP 38 but not with other lines. These data show that mating duration of short- and long-sleeper lines, specifically DGRP 38 and DGRP 335 exhibits adult behavioral plasticity but this is not generalizable for other long sleeper-low courting line DGRP 235. Hence, the effect of courtship success on reproductive output is correlated to sleep-wake transitions but can be modified by environmental factors like presence of rival males.
We also measured egg laying phenotypes for all lines, a more direct readout of reproductive output and intriguingly high courting-short sleepers (DGRP 38 and 808) and long sleeper (DGRP 335) produce higher number of eggs as compared to other lines.
We also find that sleep duration, sleep depth and sleep pressure in female flies may be less detrimental to female fecundity as compared to male courtship. This is consistent with previous findings that sleep deprivation has minimal impact on female mating behaviors (Chen et al., 2017). However, since female reproductive behaviors are poorly understood and characterized in flies as compared to males, we cannot unequivocally conclude that altered sleep does not impact female reproductive success (Aranha and Vasconcelos, 2018). Taken together, the relationship between sleep, behavioral decision making and reproductive output is more complex and context dependent than previously predicted and described (Beckwith et al., 2017; Chen et al., 2017; Duhart et al., 2020; Kayser et al., 2015; Machado et al., 2017).
In addition to isogenic background, highly reproducible behavioral phenotypes, and presence of naturally occurring genetic variations, DGRP lines used here are fully sequenced. Additionally, deep RNA sequencing of DGRP panel was recently published and mapped eQTLs (expression quantitative trait loci) for annotated genes, novel transcribed region and transposable elements for future analysis of gene transcript–trait associations (Everett et al., 2020; Mackay et al., 2012). Previous studies have identified genetic loci using GWAS (genome wide association study) that are distinct between DGRP lines used in this study and their potential role in sleep regulation. Although, several of these loci are novel, 19 genes were previously implicated in fly and/or human sleep (Harbison et al., 2013; Harbison et al., 2017). Several of these genes egfr, brk, fz, Hey, scrib, tkv, and Ubx have been tested in the context of development and their knockdown also produces sleep duration phenotypes (Harbison et al., 2013). However, the role of these genes in behaviors associated with reproductive output remain unexplored. Future studies involving analysis of these genes using RNAi knockdown and P-element disruptions can provide additional clues about the precise genetic relationship between sleep and courtship regulation.
Our extensive behavioral analysis shows that even between flies that have similar sleep duration, several sleep parameters like P(Wake) and P (Doze) are variable. Further, P(Wake) and P(Doze) are correlated with courtship duration and latency. Courtship duration is highly dependent on environmental context like presence of other males and affects DGRP lines differentially. Conversely, sleep duration in these lines were less variable and minimally susceptible to social isolation conditions that have been previously shown to reduce sleep duration, increase male-male aggression and male courtship. Unlike males, the mating and egg-laying phenotypes of female are minimally correlated to sleep duration and other state transition parameters.
These data support the growing literature that points to relationship between sleep and reproductive output but it is not clear if decreased male courtship performance is a consequence of long-sleeping pheno-type (high sleep pressure and sleep depth) or indicates a shared genetic architecture between sleep and male courtship duration. The behavioral analysis presented here paves way for a more detailed and mechanistic analysis of how genes and transcripts co-regulate complex sleep traits with behavioral choices important for reproductive output.
Supplementary Material
Acknowledgments
We would like to thank Lilian Mworia, Catherine Shorb, Zani Moore, and Austin Pavin for help with the experiments and aspects of fly rearing and behavioral analysis. The project was supported by College of Science Start-up funds (CSUEB), NIH grant 1R15GM125073–01, 7R15GM125073–02 and NSF grant IOS 2042873 awarded to Dr. Divya Sitaraman and research awards from Center for Student Research (CSUEB) and Office of Undergraduate Research (USD) to Steven Buchert, Aashaka Kalavadia, Martin Reyes and Pomai Murakami.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cbpa.2021.111114.
References
- Aranha MM, Vasconcelos ML, 2018. Deciphering Drosophila female innate behaviors. Curr. Opin. Neurobiol 52, 139–148. [DOI] [PubMed] [Google Scholar]
- Beckwith EJ, Geissmann Q, French AS, Gilestro GF, 2017. Regulation of sleep homeostasis by sexual arousal. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besedovsky L, Lange T, Born J, 2012. Sleep and immune function. Pflugers Arch 463, 121–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbely AA, 1998. Processes underlying sleep regulation. Horm. Res 49, 114–117. [DOI] [PubMed] [Google Scholar]
- Borbely AA, Achermann P, 1999. Sleep homeostasis and models of sleep regulation. J. Biol. Rhythm 14, 557–568. [DOI] [PubMed] [Google Scholar]
- Borbely AA, Daan S, Wirz-Justice A, Deboer T, 2016. The two-process model of sleep regulation: a reappraisal. J. Sleep Res 25, 131–143. [DOI] [PubMed] [Google Scholar]
- Bracker LB, Schmid CA, Bolini VA, Holz CA, Prud’homme B, Sirota A, Gompel N, 2019. Quantitative and discrete evolutionary changes in the egg-laying behavior of single Drosophila females. Front. Behav. Neurosci 13, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretman A, Fricke C, Chapman T, 2009. Plastic responses of male Drosophila melanogaster to the level of sperm competition increase male reproductive fitness. Proc. Biol. Sci 276, 1705–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretman A, Westmancoat JD, Chapman T, 2013. Male control of mating duration following exposure to rivals in fruitflies. J. Insect Physiol 59, 824–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H, 2019. Genetic sleep deprivation: using sleep mutants to study sleep functions. EMBO Rep 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EB, Torres J, Bennick RA, Rozzo V, Kerbs A, DiAngelo JR, Keene AC, 2018. Variation in sleep and metabolic function is associated with latitude and average temperature in Drosophila melanogaster. Ecol. Evol 8, 4084–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Sitaraman D, Chen N, Jin X, Han C, Chen J, Sun M, Baker BS, Nitabach MN, Pan Y, 2017. Genetic and neuronal mechanisms governing the sex-specific interaction between sleep and sexual behaviors in Drosophila. Nat. Commun 8, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheriyamkunnel SJ, Rose S, Jacob PF, Blackburn LA, Glasgow S, Moorse J, Winstanley M, Moynihan PJ, Waddell S, Rezaval C, 2021. A neuronal mechanism controlling the choice between feeding and sexual behaviors in Drosophila. Curr. Biol 31 (4231–4245), e4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson NC, Kim EZ, Slawson JB, Vecsey CG, Huber R, Griffith LC, 2012. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS One 7, e37250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll ME, Hyland C, Sitaraman D, 2019. Measurement of sleep and arousal in Drosophila. Bio. Protoc 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhart JM, Baccini V, Zhang Y, Machado DR, Koh K, 2020. Modulation of sleep-courtship balance by nutritional status in Drosophila. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett LJ, Huang W, Zhou S, Carbone MA, Lyman RF, Arya GH, Geisz MS, Ma J, Morgante F, St Armour G, et al. , 2020. Gene expression networks in the Drosophila genetic reference panel. Genome Res 30, 485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farahani FV, Fafrowicz M, Karwowski W, Douglas PK, Domagalik A, Beldzik E, Oginska H, Marek T, 2019. Effects of chronic sleep restriction on the brain functional network, as revealed by graph theory. Front. Neurosci 13, 1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke C, Bretman A, Chapman T, 2008. Adult male nutrition and reproductive success in Drosophila melanogaster. Evolution 62, 3170–3177. [DOI] [PubMed] [Google Scholar]
- Ganguly-Fitzgerald I, Donlea J, Shaw PJ, 2006. Waking experience affects sleep need in Drosophila. Science 313, 1775–1781. [DOI] [PubMed] [Google Scholar]
- Ganter GK, Panaitiu AE, Desilets JB, Davis-Heim JA, Fisher EA, Tan LC, Heinrich R, Buchanan EB, Brooks KM, Kenney MT, Verde MG, Downey J, Adams AM, Grenier JS, Maddula S, Shah P, Kincaid KM, O’Brien JR, 2011. Drosophila male courtship behavior is modulated by ecdysteroids. Journal of insect physiology 57 (9), 1179–1184. 10.1016/j.jinsphys.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe DS, Bollinger WL, Vigderman A, Masek P, Gertowski J, Sehgal A, Keene AC, 2015. Context-specific comparison of sleep acquisition systems in Drosophila. Biol. Open 4, 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann Q, Garcia Rodriguez L, Beckwith EJ, French AS, Jamasb AR, Gilestro GF, 2017. Ethoscopes: an open platform for high-throughput ethomics. PLoS Biol 15, e2003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann Q, Beckwith EJ, Gilestro GF, 2019. Most sleep does not serve a vital function: evidence from Drosophila melanogaster. Sci. Adv 5, eaau9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilestro GF, Cirelli C, 2009. pySolo: a complete suite for sleep analysis in Drosophila. Bioinformatics 25, 1466–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, Rosbash M, 2016. Circadian neuron feedback controls the Drosophila sleep—activity profile. Nature 536, 292–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, Sehgal A, 2008. Quantitative genetic analysis of sleep in Drosophila melanogaster. Genetics 178, 2341–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, McCoy LJ, Mackay TF, 2013. Genome-wide association study of sleep in Drosophila melanogaster. BMC Genomics 14, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison ST, Serrano Negron YL, Hansen NF, Lobell AS, 2017. Selection for long and short sleep duration in Drosophila melanogaster reveals the complex genetic network underlying natural variation in sleep. PLoS Genet 13, e1007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser MS, Biron D, 2016. Sleep and development in genetically tractable model organisms. Genetics 203, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser MS, Mainwaring B, Yue Z, Sehgal A, 2015. Sleep deprivation suppresses aggression in Drosophila. Elife 4, e07643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, Duboue ER, 2018. The origins and evolution of sleep. J. Exp. Biol 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene AC, Duboue ER, McDonald DM, Dus M, Suh GS, Waddell S, Blau J, 2010. Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr. Biol 20, 1209–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koganezawa M, Haba D, Matsuo T, Yamamoto D, 2010. The shaping of male courtship posture by lateralized gustatory inputs to male-specific interneurons. Curr. Biol 20, 1–8. [DOI] [PubMed] [Google Scholar]
- Krause AJ, Simon EB, Mander BA, Greer SM, Saletin JM, Goldstein-Piekarski AN, Walker MP, 2017. The sleep-deprived human brain. Nat. Rev. Neurosci 18, 404–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Smith KR, Serrano Negron YL, Harbison ST, 2019. Short-term memory deficits in the SLEEP inbred panel. Clocks Sleep 1, 471–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laturney M, Billeter JC, 2014. Neurogenetics of female reproductive behaviors in Drosophila melanogaster. Adv Genet 85, 1–108. 10.1016/B978-0-12-800271-1.00001-9.24880733. [DOI] [PubMed] [Google Scholar]
- Lesku JA, Roth TC 2nd, Amlaner CJ, Lima SL, 2006. A phylogenetic analysis of sleep architecture in mammals: the integration of anatomy, physiology, and ecology. Am. Nat 168, 441–453. [DOI] [PubMed] [Google Scholar]
- Laturney M, van Eijk R, Billeter JC, 2018. Last male sperm precedence is modulated by female remating rate in Drosophila melanogaster. Evolution letters 2 (3), 180–189. 10.1002/evl3.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Ganguly A, Huang J, Wang Y, Ni JD, Gurav AS, Aguilar MA, Montell C, 2019. Neuropeptide F regulates courtship in Drosophila through a male-specific neuronal circuit. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu P, Zaki SA, Tran DH, French RL, 2016. A novel gene controlling the timing of courtship initiation in Drosophila melanogaster. Genetics 202, 1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado DR, Afonso DJ, Kenny AR, Oztu Rk-Colak A, Moscato EH, Mainwaring B, Kayser M, Koh K, 2017. Identification of octopaminergic neurons that modulate sleep suppression by male sex drive. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482, 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masek P, Reynolds LA, Bollinger WL, Moody C, Mehta A, Murakami K, Yoshizawa M, Gibbs AG, Keene AC, 2014. Altered regulation of sleep and feeding contributes to starvation resistance in Drosophila melanogaster. J. Exp. Biol 217, 3122–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey JH, Chung D, Siwanowicz I, Stern DL, Wittkopp PJ, 2019. The yellow gene influences Drosophila male mating success through sex comb melanization. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan A, Lindsay T, Prudnikova A, Erdi B, Dickinson M, von Philipsborn AC, 2018. Multifunctional wing motor control of song and flight. Curr. Biol 28, 2705–2717 e2704. [DOI] [PubMed] [Google Scholar]
- Pan Y, Baker BS, 2014. Genetic identification and separation of innate and experience-dependent courtship behaviors in Drosophila. Cell 156, 236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Robinett CC, Baker BS, 2011. Turning males on: activation of male courtship behavior in Drosophila melanogaster. PLoS One 6, e21144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Meissner GW, Baker BS, 2012. Joint control of Drosophila male courtship behavior by motion cues and activation of male-specific P1 neurons. Proc. Natl. Acad. Sci. U. S. A 109, 10065–10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potdar S, Daniel DK, Thomas FA, Lall S, Sheeba V, 2018. Sleep deprivation negatively impacts reproductive output in Drosophila melanogaster. J. Exp. Biol 221. [DOI] [PubMed] [Google Scholar]
- Rezával C, Nojima T, Neville MC, Lin AC, Goodwin SF, 2014. Mar 31. Sexually dimorphic octopaminergic neurons modulate female postmating behaviors in Drosophila. Curr Biol 24 (7), 725–730. 10.1016/j.cub.2013.12.051. Epub 2014 Mar 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezával C, Pattnaik S, Pavlou HJ, Nojima T, Brüggemeier B, D’Souza LAD, Dweck HKM, Goodwin SF, 2016. Sep 26. Activation of Latent Courtship Circuitry in the Brain of Drosophila Females Induces Male-like Behaviors. Curr Biol 26 (18), 2508–2515. 10.1016/j.cub.2016.07.021. Epub 2016 Aug 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhmann H, Koppik M, Wolfner MF, Fricke C, 2018. The impact of ageing on male reproductive success in Drosophila melanogaster. Exp. Gerontol 103, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sare RM, Levine M, Hildreth C, Picchioni D, Smith CB, 2016. Chronic sleep restriction during development can lead to long-lasting behavioral effects. Physiol. Behav 155, 208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Mignot E, 2011. Genetics of sleep and sleep disorders. Cell 146, 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kavuru M, 2010. Sleep and metabolism: an overview. Int. J. Endocrinol 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G, 2000. Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837. [DOI] [PubMed] [Google Scholar]
- Shi M, Yue Z, Kuryatov A, Lindstrom JM, Sehgal A, 2014. Identification of Redeye, a new sleep-regulating protein whose expression is modulated by sleep amount. Elife 3, e01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth HT, 1966. Drosophilid mating behavior: the behaviour of decapitated females. Anim. Behav 14, 226–235. [DOI] [PubMed] [Google Scholar]
- Tobler II, Franken P, Trachsel L, Borbely AA, 1992. Models of sleep regulation in mammals. J. Sleep Res 1, 125–127. [DOI] [PubMed] [Google Scholar]
- Ueda A, Wu CF, 2009. Effects of social isolation on neuromuscular excitability and aggressive behaviors in Drosophila: altered responses by Hk and gsts1, two mutations implicated in redox regulation. J. Neurogenet 23, 378–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspoor RL, Heys C, Price TA, 2015. Dyeing insects for behavioral assays: the mating behavior of anesthetized Drosophila. J. Vis. Exp 98, 52645 10.3791/52645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggin TD, Goodwin PR, Donelson NC, Liu C, Trinh K, Sanyal S, Griffith LC, 2020. Covert sleep-related biological processes are revealed by probabilistic analysis in Drosophila. P. Natl. Acad. Sci. USA 117, 10024–10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto D, Koganezawa M, 2013. Genes and circuits of courtship behaviour in Drosophila males. Nat. Rev. Neurosci 14, 681–692. [DOI] [PubMed] [Google Scholar]
- Yamamoto D, Sato K, Koganezawa M, 2014. Neuroethology of male courtship in Drosophila: from the gene to behavior. J. Comp. Physiol. A 200, 251–264. [DOI] [PubMed] [Google Scholar]
- Yurgel ME, Masek P, DiAngelo J, Keene AC, 2015. Genetic dissection of sleep-metabolism interactions in the fruit fly. J. Comp. Physiol. A Neuroethol. Sens. Neural. Behav. Physiol 201, 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Guo C, Chen D, Peng Q, Pan Y, 2018. Hierarchical Control of Drosophila Sleep, Courtship, and Feeding Behaviors by Male-Specific P1 Neurons. Neurosci Bull 34, 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski MR, McKenna JT, McCarley RW, 2016. Functions and mechanisms of sleep. AIMS Neurosci 3, 67–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


