Abstract
Telomere length regulation is essential for cell viability in eukaryotes. While many pathways that affect telomere length are known, we do not yet have a complete understanding of the mechanism of length regulation. To identify new pathways that might regulate telomere length, we carried out a genetic screen in yeast and identified the cyclin‐dependent kinase complex Bur1/2 as a regulator of telomere length. Mutations in either BUR1 cyclin‐dependent kinase or the associated BUR2 cyclin resulted in short telomeres. This regulation did not function through the known role of BUR1 in regulating histone modification as bur1∆ set2∆ and bur2∆ set2∆ double mutants rescued cell growth but did not rescue the telomere shortening effects. We found that both bur1∆ and bur2∆ set2∆ were also defective in de novo telomere addition, and deletion of SET2 did also not rescue this elongation defect. The Bur1/2 cyclin‐dependent kinase regulates transcription of many genes. We found that TLC1 RNA levels were reduced in bur2∆ set2∆ mutants; however, overexpression of TLC1 restored the transcript levels but did not restore de novo telomere elongation or telomere length. These data suggest that the Bur1/2 kinase plays a role in telomere elongation separate from its role in transcription of telomerase components. Dissecting the role of the Bur1/2 kinase pathway at telomeres will help complete our understanding of the complex network of telomere length regulation.
Keywords: Bur1, Bur2, kinase, telomere length equilibrium
Bur1/2 is a cyclin‐dependent kinase that phosphorylates many substrates within the cell. Telomere binding complexes regulating telomere length equilibrium are regulated through protein modification. Phosphorylation by this kinase either directly or indirectly regulates telomere length elongation.
Take Away
Loss of Bur1/2 cyclin‐dependent kinase activity causes short telomeres.
Short telomere phenotype is not due to the role of Bur1/2 in histone modification.
Short telomeres are not due to decreased levels of telomerase components Est1, Est2, Est3, or Tlc1.
In absence of Bur1/2 activity, TLC1 deleted cells do not form survivors.
Bur1/2 kinase directly or indirectly regulates telomere length.
1. INTRODUCTION
Telomere length is maintained around a species‐specific equilibrium. Loss of equilibrium length maintenance can result in critically short telomeres that signal cell death, cell cycle arrest, or cellular senescence (di Fagagna et al., 2003; Harley et al., 1990; IJpma & Greider, 2003; Lundblad & Szostak, 1989). In humans, loss of telomere length maintenance and subsequent telomere shortening leads to specific age‐associated degenerative diseases such as bone marrow failure and pulmonary fibrosis (Armanios, 2009). Multiple factors contribute to telomere length equilibrium maintenance. First, the components of telomerase, required for telomere elongation, are limiting in cells (Armanios et al., 2005; Mozdy & Cech, 2006; Vulliamy et al., 2004). Having reduced levels of telomerase leads to progressive telomere shortening over many generations. Second, telomere length is regulated by telomere binding proteins through multiple independent mechanisms. In yeast, Tel1 and the Mre11/Rad50/Xrs2 (MRX) complex mediate telomere elongation (Greenwell et al., 1995; Keener et al., 2019; Ritchie & Petes, 2000) and the Rap1/Rif1/Rif2 proteins and the Cdc13/Stn1/Ten1(CST) complex bind telomeres and regulate telomere length (Gao et al., 2007; Marcand et al., 1997; Price et al., 2010). In human cells, the shelterin complex of telomere binding proteins also regulates the ability of telomerase to elongate telomeres (Palm & de Lange, 2008; Smogorzewska & de Lange, 2004).
The different telomere binding complexes that determine telomere length homeostasis are regulated through protein modification. The ATM/ATR (Tel1/Mec1 in yeast) kinases regulate telomere elongation; deletion of both TEL1 and MEC1 in yeast leads to short stable telomeres (Ritchie et al., 1999). The cyclin‐dependent kinase Cdk1 regulates the telomere elongation during S phase in the cell cycle (Frank et al., 2006; Li et al., 2009; Liu et al., 2014; Vodenicharov & Wellinger, 2006). Here, we describe a role for the Bur1/2 cyclin‐dependent kinase in the regulation of telomeres in the yeast Saccharomyces cerevisiae.
Bur1 is a kinase with homology to cyclin‐dependent kinases, and its activity is regulated by the Bur2 cyclin component (Yao et al., 2000). BUR1 is an essential gene, while BUR2 deletion mutants grow slowly, presumably because another cyclin can substitute, though poorly, for Bur2. The Bur1/2 kinase complex is involved in regulating transcriptional elongation at several levels in yeast. First, Bur1/2 regulates Set2, which is a histone methylase specific for H3K36 trimethylation (Chu et al., 2006). Deletion of SET2 rescues the lethality of a BUR1 deletion and improves the growth of BUR2 deletion (Chu et al., 2006), likely by decreasing the methylation of H3K36, since overexpression of histone demethylases also rescues bur1∆ growth (Kim & Buratowski, 2007). In addition to regulation of Set2 H3K36 trimethylation, the Bur1/2 kinase complex regulates transcriptional elongation. The kinase regulates the ubiquitination of histone H2B (Laribee et al., 2005), and it phosphorylates serine 2 on the C‐terminal domain (CTD) of RNA polymerase II (Liu et al., 2009; Murray et al., 2001). In addition, the Bur1/2 kinase complex promotes the recruitment of the PAF1 complex through its role as a CTD kinase and by phosphorylation of Spt5 (Qiu et al., 2012). Through these multiple levels of transcriptional regulation, the Bur1/2 complex regulates the mRNA levels of a large number of genes (Laribee et al., 2005).
We identified bur2Δ as having short telomeres from a collection of yeast deletion mutants. We demonstrated that the short telomeres in both bur1 and bur2 mutants were not rescued by SET2 deletion, which rescues the growth defect in these mutants. Telomere elongation in a de novo telomere addition assay was blocked by both bur1 and bur2 mutations. We tested whether the short telomere effect and loss of de novo elongation were due to low levels of TLC1, as previously reported in paf1 mutants (Mozdy et al., 2008), but found instead that overexpression of TLC1 did not rescue telomere elongation. Finally, bur1 mutants were defective in telomere recombination to generate survivors, which occurs in the absence telomerase. Taken together, our data suggest that the Bur1/2 kinase affects telomere elongation independent of its effects on transcription of telomerase components.
2. MATERIALS AND METHODS
2.1. Yeast strains and plasmids
Yeast strains and plasmids are listed in Tables 1 and 2, respectively. Oligonucleotides (primers) for polymerase chain reaction (PCR) and quantitative reverse transcription PCR (qRT‐PCR) are listed in Table 3. The haploid MAT a deletion collection of S. cerevisiae BY4741 and the yeast strains containing the histone H3 mutants were gifts from J. Boeke and E. Hyland. Haploid mutants, bur1–1, bur1–8, and a plasmid containing BUR1, were gifts from G. Prelich (Murray et al., 2001). Strains for the de novo telomere addition assay were generated using JHUY877 (Ma & Greider, 2009) by direct transformation followed by tetrad analysis. JHUY887 is the haploid strain derived from dissection of JHUY877. The bur1–8 allele was first backcrossed to YPH500 (Sikorski & Hieter, 1989), followed by mating to JHUY887. The diploid was sporulated to generate wild‐type and bur1–8 mutant strains for analysis. Gene replacement at the endogenous locus (Brachmann et al., 1998) was used to generate all deletion mutants in diploid strains, followed by tetrad analysis. Overexpression of TLC1 was done from the inducible galactose promoter with the plasmid pGalTLC1 that was derived from pESC‐TRP‐TLC1/HAT‐EST2, a generous gift from V. Zakian. This plasmid was modified by cutting with EcoRI to remove the HAT tag and 1.3 Kb of the coding region of EST2; after release of this fragment, the plasmid was reclosed at EcoRI. Passaging of this plasmid construct in cells was done in casamino acid supplemented media without tryptophan (CAA). The construction of pBUR1∆C was a derivative of pBUR1 where, by Gibson assembly (Gibson 2011), only the first 365aa were contained in the open reading frame of BUR1. All restriction enzymes, T4 DNA ligase, and Gibson assembly mix used in these experiments were from New England Biolabs (NEB).
Table 1.
Yeast strains used in this study
| Strain ID | Genotype | Source | Figure |
|---|---|---|---|
| OY249 (JHUY890) | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 bur1–8::kanMX4 | Prelich gift | 1a and 7b |
| GY114 (JHUy891) | MATa his4‐912δ lys2‐128δ suc2∆UAS(‐1900/‐390) ura‐352 ade8 bur1–1 | Prelich & Winston, 1993 | 1a and 7b |
| JHUY761 | MATa/MATα his3∆1/his3∆1 leu2∆0/leu2∆0 lys2∆0/lys2∆0 met15∆0/met15∆0 trp1∆63/trp1∆63 ura3∆0/ura3∆0 | Ma & Greider, 2009 | |
| YCC212, 213 | JHUY761 BUR2/bur2∆::LEU2 | This study | 1a |
| YCC221 | MATa his3∆1 leu2∆0 met15∆0 ura3∆0 bur2∆::kanMX4 | Winzeler et al., 1999 | 1a and 3 |
| JHUY896 | MATa/MATα his3∆1/his3∆1 leu2∆0/leu2∆ LYS2/lys2∆0 met15∆0/MET15 ura3∆0/ura3∆0 BUR1/bur1–8::kanMX4 | This study | 1b |
| YCC294 | MATa/MATa his3∆1/his3∆1 leu2∆0/leu2∆ LYS2/lys2∆0 met15∆0/MET15 ura3∆0/ura3∆0 BUR1/bur1–8::kanMX4 SET2/set2∆::LEU2 | This study | 1c |
| YCC241 | MATα his3∆1 leu2∆0 lys2∆0 ura3∆0 bur1∆::kanMX4 + pRS426BUR1 | This study | 1d |
| EMHy201 (YCC237) | MATa his3∆200 leu2∆1 lys2∆0 met15∆0 ura3–167 trp1∆63 ade2::his hht1‐hhf1hhf1::natMX hht2‐hhf2::hygMX4 RDN::mURA3/HIS3 + pBS4 (HHT2:K36A‐HHF2) | Gift, Hyland et al., 2005 | 1d |
| JHUY877 | MATa‐inc/MATα ade2‐101/ade2‐101 his3∆200/his3∆200 lys2‐801/lys2‐801 trp1∆63/trp1∆63 ura3‐52/ura3‐52 leu2∆1/leu2∆1:LEU2‐GAL‐HO VII‐L::ADE2‐TG(1‐3)‐HOsite‐LYS2 rad52∆::hphMX4/RAD52 | Ma & Greider, 2009 | |
| JHUY887 | MATa‐inc ade2‐101 his3∆200 lys2‐801 trp1∆63 ura3‐52 leu2∆1:LEU2‐GAL‐HO VII‐L::ADE2‐TG(1‐3)‐HOsite‐LYS2 rad52∆::hphMX4 | This study | |
| YCC243 | MATα his3∆0 leu2∆0 lys2∆0 met15∆0 bur1–8:KanMX | This study | |
| YCC265, 266 | MATa‐inc bur1–8:kanMX4 ade2‐101 lys2‐801 leu2 his3 ura3 trp1 leu2∆1:GAL1‐HO‐LEU2 VII‐L::ADE2‐TG(1‐3)‐HO SITE‐LYS2 rad52∆::hphMX4 | This study | 2a,b |
| YCC267 | MATa‐inc BUR1 ade2‐101 lys2–801 leu2 his3 ura3 trp1 leu2∆1:GAL1‐HO‐LEU2 VII‐L::ADE2‐TG(1‐3)‐HO SITE‐LYS2 rad52::hphMX4 | This study | 2a,b |
| YCC282 | MATa‐inc BUR1 ade2‐101 lys2‐801 leu2 his3 ura3 trp1 leu2∆1:GAL1‐HO‐LEU2 VII‐L::ADE2‐TG(1‐3)‐HO SITE‐LYS2 rad52::hphMX4 set2∆::URA3 | This study | 2c |
| YCC303, 304 | MATa‐inc bur1–8::kanMX4 ade2‐101 lys2‐801 leu2 his3 ura3 trp1 leu2∆1:GAL1‐HO‐LEU2 VII‐L::ADE2‐TG(1‐3)‐HO SITE‐LYS2 rad52::hphMX4 | This study | 2c |
| YCC306 | MATa‐inc BUR1 ade2‐101 lys2‐801 leu2 his3 ura3 trp1 leu2∆1:GAL1‐HO‐LEU2 VII‐L::ADE2‐TG(1‐3)‐HO SITE‐LYS2 rad52::hphMX4 | This study | 2c |
| YCC328 | MATa his3∆200 leu2∆1 lys2∆0 met15∆0 trp1∆63 ura3–167 ade2::hisG hht1‐hhf1::natMX4 hht2‐hhf2‐HHTS‐HHFS RDN1::Ty1‐MET15 TelV::ADE2 | Gift, Hyland et al., 2005 | 2c |
| YCC336 | MATa his3∆200 leu2∆1 lys2∆0 met15∆0 trp1∆63 ura3–167 ade2::hisG hht1‐hhf1::natMX4 hht2‐hhf2‐HHTS:K36A‐HHFS RDN1::Ty1‐MET15 TelV::ADE2 | Gift, Hyland et al., 2005 | 2c |
| YCC459, 460 | MATa‐inc/MATα ade2‐101/ade2‐101 his3∆200/his3∆200 lys2‐801/lys2‐801 trp1∆63/trp1∆63 ura3‐52/ura3‐52 leu2∆1/leu2∆1:LEU2‐GAL‐HO VII‐L::ADE2‐TG(1‐3)‐HOsite‐LYS2 rad52∆::hphMX4/RAD52 BUR2/bur2∆::kanMX4 SET2/set2∆::URA3 | This study | |
| YCC468, 469 | MATa‐inc ade2‐101 his3∆200 lys2‐801 trp1∆63 ura3‐52 leu2∆1:LEU2‐GAL‐HO VII‐L::ADE2‐TG(1‐3)‐HOsite‐LYS2 rad52∆::hphMX4 bur2∆::kanMX4 set2∆::URA3 | This study | 2d |
| YCC466 | MATa‐inc ade2‐101 his3∆200 lys2‐801 trp1∆63 ura3‐52 leu2∆1:LEU2‐GAL‐HO VII‐L::ADE2‐TG(1‐3)‐HOsite‐LYS2 rad52∆::hphMX4 set2∆::URA3 | This study | 2d |
| YCC379 | MATa ura3∆0 lys2∆0 leu2∆0 his3∆1 set2∆::kanMX4 | This study | 3 |
| YCC380 | MATα ura3∆0 leu2∆0 his3∆1 set2∆::kanMX4 bur1∆::LEU2 | This study | 3 |
| YCC381 | MATa ura3∆0 leu2∆0 his3∆1 SET2 BUR1 | This study | 3 |
| YCC498, 499, 500, 501 | MATa‐inc ade2‐101 his3∆200 lys2‐801 trp1∆63 ura3‐52 leu2∆1:LEU2‐GAL‐HO VII‐L::ADE2‐TG(1‐3)‐HOsite‐LYS2 rad52∆::hphMX4 bur2∆::kanMX4 set2∆::URA3 (lost plasmid pGal1TLC1) | This study | 4 and 5 |
| YCC502, 503, 504, 505 | MATa‐inc ade2‐101 his3∆200 lys2‐801 trp1∆63 ura3‐52 leu2∆1:LEU2‐GAL‐HO VII‐L::ADE2‐TG(1‐3)‐HOsite‐LYS2 rad52∆::hphMX4 bur2∆::kanMX4 set2∆::URA3 + pGal1TLC1 | This study | 4 and 5 |
| YCC506, 507 | MATa‐inc ade2‐101 his3∆200 lys2‐801 trp1∆63 ura3‐52 leu2∆1:LEU2‐GAL‐HO VII‐L::ADE2‐TG(1‐3)‐HOsite‐LYS2 rad52∆::hphMX4 BUR2 SET2 (lost plasmid pGal1TLC1) | This study | 4 and 5 |
| YCC508, 509 | MATa‐inc ade2‐101 his3∆200 lys2‐801 trp1∆63 ura3‐52 leu2∆1:LEU2‐GAL‐HO VII‐L::ADE2‐TG(1‐3)‐HOsite‐LYS2 rad52∆::hphMX4 BUR2 set2∆::URA3 (lost plasmid pGal1TLC1) | This study | 4 and 5 |
| YCC510, 511 | MATa‐inc ade2‐101 his3∆200 lys2‐801 trp1∆63 ura3‐52 leu2∆1:LEU2‐GAL‐HO VII‐L::ADE2‐TG(1‐3)‐HOsite‐LYS2 rad52∆::hphMX4 BUR2 SET2 + pGal1TLC1 | This study | 4 and 5 |
| YCC512, 513 | MATa‐inc ade2‐101 his3∆200 lys2‐801 trp1∆63 ura3‐52 leu2∆1:LEU2‐GAL‐HO VII‐L::ADE2‐TG(1‐3)‐HOsite‐LYS2 rad52∆::hphMX4 BUR2 set2∆::URA3 + pGal1TLC1 | This study | 4 and 5 |
| YCC562, 563 | MATa/MATα his4‐912δ/HIS4 his3∆1/HIS3 lys2‐128δ/lys2∆0 ura3‐52/ura3∆0 ade8/ADE8 suc2∆UAS(‐1900/‐390)/SUC2 bur1–1(ts)/BUR1 | This study | 6 |
| YCC214, 215 | JHUY761 TEL1/tel1∆::hphMX4 MEC1/mec1∆::kanMX SML1/sml1∆::TRP1 BUR2/bur2∆::LEU2 | This study | 7a |
| YCC222, 223 | JHUY761 MRE11/mrell∆::kanMX4 BUR2/bur2∆::LEU2 | This study | 7a |
| YCC226, 227 | JHUY761 KU80/ku80∆::kanMX4 BUR2/bur2∆::LEU2 | This study | 7a,b |
| YCC228, 229 | JHUY761 RIF1/rif1∆::kanMX4 BUR2/bur2∆::LEU2 | This study | 7a |
| YCC230, 231 | JHUY761 TLC1/tlc1∆::kanMX4 BUR2/bur2∆::LEU2 | This study | 7a |
| YCC252 | MATa/MATa his3∆1/his3∆1 leu2∆0/leu2∆ LYS2/lys2∆0 met15∆0/MET15 ura3∆0/ura3∆0 BUR1/bur1–8::kanMX4 TLC1/tlc1∆::LEU2 | This study | 8 |
| BY4742 (YCC36) | MATα his3∆1 leu2∆0 lys2∆0 ura3∆0 | Brachmann et al., 1998 | S1 |
| YPH500 (YCC20) | MATα ura3‐52 lys2‐801 ade2‐101 his3∆200 trp1∆63 leu2∆1 | Sikorski & Hieter, 1989 | S1 |
| W303 (OAy1003) | MATα ade2–1 trp1–1 ura3–1 leu2–3,112 his3–11,15 can1–100 RAD5 | Viggiani & Aparicio, 2006 | S1 |
| YCC205 | MATa/MATα ura3∆0/ura3∆0 LYS2/lys2∆0 his3∆1/his3∆1 leu2∆0/leu2∆0 met15∆0/MET15 TLC1/tlc1∆::kanMX | This study | S2 |
| JHUY895 | MATa/MATα ura3∆0/ura3∆0 LYS2/lys2∆0 his3∆1/his3∆1 leu2∆0/leu2∆0 met15∆0/MET15 BUR1/bur1::kanMX | This study | S3 |
Table 2.
Plasmids used in this study
| Plasmid name | Brief description | Source |
|---|---|---|
| pRS316 | Control plasmid for experiments | Sikorski & Hieter, 1989 |
| pBUR1 | Contains BUR1 coding region in pRS316 (GP111, JHU1169) | G. Prelich gift |
| pRS426BUR1 | Contains BUR1 coding region in pRS426 (JHU1171) | This study |
| pBUR1∆C | Contains BUR1 1–365aa only in pRS316 (JHU1255) | This study |
| pBS4 (pH3:K36A) | Plasmid in EMHy201 (HHT2:K36A HHF2/TRP/CEN/ARS/AmpR) | Hyland et al., 2005 |
| pESC‐TRP‐TLC1/HAT EST2 | Overexpression plasmid containing TLC1 and EST2 (JHU1054) | V. Zakian gift |
| pGal1TLC1 | Overexpression plasmid containing TLC1 derived from pesc‐TRP‐TLC1/HAT EST2 (JHU1238) | This study |
| SCR1/topo PCR2.1 | Control plasmid for SCR1 expression (BCC41) | This study |
| ARN1/topo PCR2.1 | Control plasmid for ARN1 expression (BCC43) | This study |
| pVL1091 (JHU995) | Control plasmid for EST1 expression (Cdc13‐Est1 fusion/pRS415) | Evans & Lundblad, 1999 |
| pVL369 (JHU612) | Control plasmid for EST2 expression (2μ ADH‐EST2 integrating) | Lingner et al., 1997 |
| pAY30 (JHU1166) | Control plasmid for TLC1 expression (pRS316 TLC1) | A. IJpma collection |
Table 3.
Primers used in this study
| Primer ID | 5′‐3′ sequence | Function | Source |
|---|---|---|---|
| OCC8 | CGAATATTTAGAGAGAATCCGTCAC | A primer for YLR226W BUR2 | Winzeler et al., 1999 |
| OCC16 | TCAGTTATGGCTGTAGGTATTCCAT | B primer for YLR226W BUR2 | Winzeler et al., 1999 |
| OCC162 | TTTTGAATCATATTGAAACAAGGGT | C primer for YLR226W BUR2 | Winzeler et al., 1999 |
| OCC163 | TCGAAAATATTATTGATGCTTGTGA | D primer for YLR226W BUR2 | Winzeler et al., 1999 |
| OCC164 | CGTAGTATTTTCGTTTAAAATATATTACAGTAAGATAATGAGATTGTACTGAGAGTGCAC | Upstream primer for KO of YLR226W BUR2 | Winzeler et al., 1999 |
| OCC165 | CTGATCCCTCCAATTAAACATAACTTGTACTCTATTTTTACTGTGCGGTATTTCACACCG | Downstream primer for KO of YLR266W BUR2 | Winzeler et al., 1999 |
| OCC166 | GACCTAGGTCTCATTGTGACT | A1 primer for YLR226W BUR2, 170 bp from ATG | This study |
| OCC167 | AAGGTACTGTTGACTGCTAT | D1 primer for YLR226W BUR2, 304 bp down from TAA | This study |
| OCC176 | GGTAGCAACTCTGATATTCCACTGT | A confirmation primer for YPR161C BUR1, 258 bp up from ATG | Winzeler et al., 1999 |
| OCC177 | CTGTACACCCGTAAACTTTCTCACT | B confirmation primer for YPR161C BUR1 | Winzeler et al., 1999 |
| OCC178 | AGGAGTTAATAGATACGGACCCAAC | C confirmation primer for YPR161C BUR1 | Winzeler et al., 1999 |
| OCC179 | TTTTTGGCACTCTTTTAAATGGTAT | D confirmation primer for YPR161C BUR1 | Winzeler et al., 1999 |
| OCC182 | CAGATGCAGATCATTCTTCAGGAAT | A1 confirmation primer for YPR161C BUR1 | This study |
| OCC183 | TTGAACCAGTGACTTAGCTGGGAGT | D1 confirmation primer for YPR161C BUR1 426 bp down from TAA | This study |
| OCC224 | GAGAAGAAGCTGACTTCGACTATTG | A confirmation primer for YJL168C SET2 | Winzeler et al., 1999 |
| OCC174 | TTCAGTATTTCTTTTTCATCTTCCG | B confirmation primer for YJL168C SET2 | Winzeler et al., 1999 |
| OCC175 | GCTAAAGACATCGTGAAAATCCTAA | C confirmation primer for YJL168C SET2 | Winzeler et al., 1999 |
| OCC225 | AAAAATAAAGACACTTGAAACGCAC | D confirmation primer for YJL168C SET2 | Winzeler et al., 1999 |
| OCC228 | GTGGGATGGGATACGTTGAG | SCR1(pol lll transcript) forward primer. Anneal to nt 20–39 (YM) | This study |
| OCC229 | TTTACGACGGAGGAAAGACG | SCR1(pol lll transcript) reverse primer. Anneals to nt 144–163 (YM) | This study |
| OCC230 | ACCGATCCTCTTCTCGACCT | TLC1 forward primer. Anneals to nt 417–436 (YM) | This study |
| OCC231 | TAAACAGCGAACTCGTGCAA | TLC1 reverse primer. Anneals to nt 516–535 (YM) | This study |
| OCC232 | GAGATGAACAACAGCGCAAA | EST1_F forward primer (SVC) | This study |
| OCC233 | GAAACGCCATCTTTTTCTGG | EST1_R reverse primer (SVC) | This study |
| OCC234 | AAAACTGGCTGACGATTTCC | EST2_F forward primer (SVC) | This study |
| OCC235 | TTGGGAGCTTACGGCTAAAA | EST2_R reverse primer (SVC) | This study |
| OCC236 | ATCATTGGCGATGCTGACTT | EST3_F forward primer (SVC) | This study |
| OCC237 | CAAATATCGTGGCCTGGTTT | EST3_R reverse primer (SVC) | This study |
| OCC238 | AATGGAGGGCCAGAAAGACT | ARN_F forward primer (SVC) | This study |
| OCC239 | TCAAAAGGACACCAACGACA | ARN_R reverse primer (SVC) | This study |
| OCC248 | TGACGGGCGAATTATACAAC | EST2_F new forward primer | This study |
| OCC249 | ATTTTCCAAGCAGCGCCTTT | EST2_R new reverse primer | This study |
| OCC274 | atccttggtttaaagaggactagGTTATACTATTCTCTCTTCCTTTCTGGC | GP111fwd Gibson deletion of BUR1 for BUR1∆C | This study |
| 0CC275 | AGAGAGAATAGTATAACctagtcctctttaaaccaaggatggtgt | GP111rev Gibson deletion of BUR1 for BUR1∆C | This study |
2.2. Telomere length screen in Saccharomyces non‐essential gene deletion collection
To minimize the generation of suppressors in the haploid yeast deletion collection (Winzeler et al., 1999), we obtained a very early passage of the entire collection. To further minimize any growth advantage of revertants, strains were amplified by growing on yeast extract‐peptone‐dextrose plates (YPD) directly from the 96‐well format plates. Six strains were propagated on one YPD plate in single strips. After overnight growth at 30°C, each strip of cells was scraped into water and genomic DNA was prepared from each mutant, cut with Xho1, and fractionated by 1% agarose electrophoresis as detailed in Kaizer et al. (2015) and Ritchie et al. (1999). Southern blot telomere length analysis was performed using a radiolabeled subtelomeric Y′ fragment and, in most cases, a radiolabeled 2‐log ladder (NEB). Images were captured on XAR‐5 film (Kodak) or Storage Phosphor screens (GE Healthcare). Bulk telomere lengths of the mutants were compared to those of wild type. The telomere lengths in the mutants were categorized based on a 5‐point scale as either very short, short, wild type, long, or very long. Candidate genes were then re‐tested by direct transformation to delete a given gene in a wild‐type diploid, and tetrad analysis was performed to determine linkage. A germinated spore from a tetrad was propagated by one passage in liquid culture overnight for Southern blot analysis. Southern blots shown in this study contain, in most cases, numbers on the y‐axis that indicate the sizes of the 2‐log ladder in kilobases. The numbers on the x‐axis describe the lane for samples analyzed on an agarose gel.
2.3. De novo telomere elongation assay
The de novo telomere elongation assay was performed as previously described (Diede & Gottschling, 1999). During the assay, cells were also collected for Southern, western, and qRT‐PCR. For the bur1–8 experiments, the temperature was shifted when the cells were resuspended in galactose at T = 0 h.
2.4. Western blot analysis
Samples for western blot analysis were prepared and analyzed as described (Kaizer et al., 2015). Inactivation of H3 K36 trimethylation was monitored by immunoblot using the rabbit polyclonal antibody to histone H3 (trimethyl K36) at 1:2000 dilution (Abcam ab9050). A mouse monoclonal antibody to phosphoglycerate kinase (PGK1) at 1:10,000 dilution (Invitrogen, 459250) was used as a loading control. Immunoblots were analyzed on an Odyssey Infrared Imaging System (LI‐COR Biosystems) using the quantification software provided. Band intensity of the samples were normalized to the loading control and expressed as a ratio of signal intensity.
2.5. Quantitative PCR analysis
To determine mRNA levels, we used qRT‐PCR. Total RNA was extracted according to the protocol recommended by the RNeasy Mini Kit (Qiagen, 74104) and quantitated using a NanoDrop ND‐1000 Spectrophotometer. One microgram of RNA was reverse‐transcribed according to the protocol using a Superscript III First‐Strand Synthesis System for RT‐PCR (Invitrogen, 18080‐051). Transcript levels were quantitated in triplicate with target gene‐specific primer pairs using IQ Cyber Green Supermix (Bio‐Rad) and detected in a CFX96 RT‐PCR machine (Bio‐Rad). Plasmids containing each target gene were serially diluted as standard controls. CFX Manager Software (Bio‐Rad) was used to obtain the relative expression of the target gene normalized to expression in wild type, and triplicate variation was quantified by standard error of the mean (SEM). During the de novo telomere elongation assay, samples of each strain were collected at the T = 0 h and T = 4 h for RNA preparation.
2.6. Detection of single‐stranded DNA
We followed the modified protocol for in‐gel native hybridization documented by the Wellinger lab (Dionne & Wellinger, 1996). Single‐stranded DNA signals were detected on a Typhoon 9410 scanner (GE Healthcare).
3. RESULTS
3.1. bur1 and bur2 mutants have short telomeres
To identify genes that regulate telomere length, we screened the yeast deletion collection of ~4800 non‐essential genes (Winzeler et al., 1999) using Southern blots to examine telomere length in individual knockout strains. Other groups also employed a similar strategy to look at genome wide regulators of telomere length and characterized a network of telomere length regulators revealing mutants we also found (Askree et al., 2004; Gatbonton et al., 2006; Ungar et al., 2009). However, in our screen, we identified a novel mutant, bur2, that was not reported in the other screens. bur2∆ cells are viable but grow very slowly (Yao et al., 2000), and the deletion strain can quickly pick up suppressors, which is likely why it was not identified in the other deletion collection screens. Our Southern analysis utilized a very early replicate of this collection that had not been grown extensively (a gift from J. Boeke), allowing us to avoid the overgrowth of suppressors (see Section 2).
To verify that bur2Δ mutants have short telomeres, we deleted BUR2 in a diploid strain and sporulated it to obtain wild‐type and bur2Δ haploid strains. The bur2Δ strain grew very slowly compared to wild type as expected. To examine telomere length in the mutant, we prepared DNA from patches of cells made immediately after sporulation to avoid enriching for suppressors. We found that bur2Δ cells have short telomeres compared to the isogenic wild‐type cells from the same tetrad. The original isolate from the deletion collection had even shorter telomeres, likely due to the increased number of cell divisions (Figure 1a).
Figure 1.
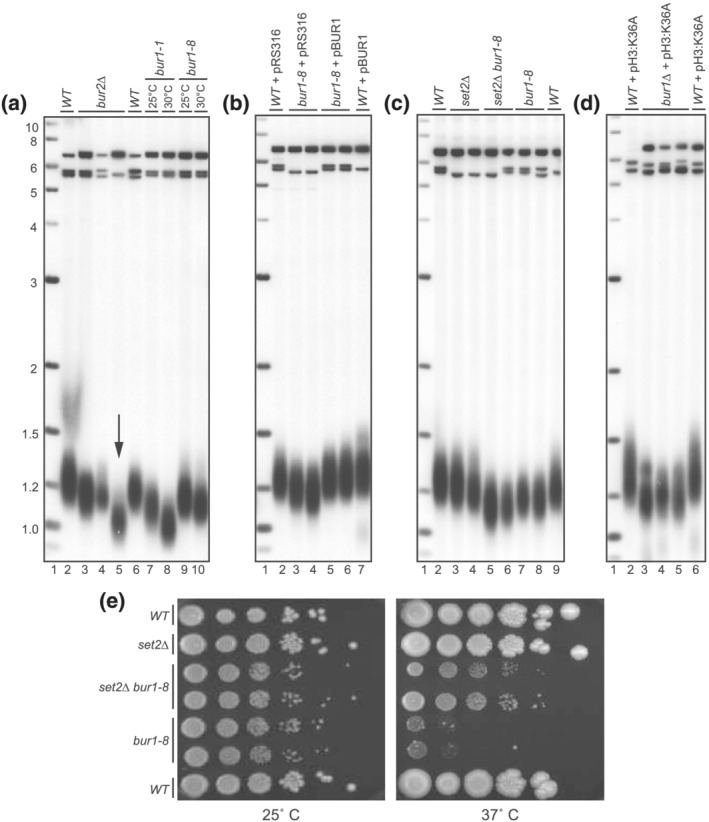
Short telomeres in bur1 and bur2 mutants. (a) Southern blot analysis of telomeres from WT, bur2∆, bur1–1, and bur1–8 mutants. bur2∆ isolated from the haploid screen of yeast deletion mutants is indicated with an arrow (Lane 5), in addition two independent segregants of bur2∆ haploids from BUR2/bur2∆ heterozygous diploids are shown (Lanes 3, 4). Telomere lengths of bur1–1 and bur1–8 temperature‐sensitive alleles of BUR1 grown at 25°C and 30°C, respectively, are shorter than WT (Lanes 7, 8 and 9, 10). (b) The short telomere length in bur1–8 is rescued by a centric plasmid expressing BUR1 (Lanes 5, 6). (c) Telomere length in bur1–8 and bur1–8 set2∆ mutants is shorter than WT and set2∆ (Lanes 5, 6). (d) Lethality of bur1∆ is rescued by a plasmid expressing histone mutant H3:K36A; however, telomere length is shorter than WT (Lanes 3, 4, 5). (e) Growth rate of bur1–8, set2∆, bur1–8 set2∆ mutants at permissive (25°C) and restrictive (37°C) temperature. Serial 10‐fold dilution of cells plated at different temperatures indicated bur1–8 set2∆ mutants grow better than bur1–8 mutants alone at 37°C
Since Bur2 is the cyclin component of a Cdk/cyclin complex (Yao et al., 2000), we wanted to test the role of its partner, Bur1, the catalytic Cdk kinase, in telomere length regulation. BUR1 is an essential gene and thus a knockout of this gene is not represented in the haploid deletion collection (Winzeler et al., 1999). To examine telomere length in bur1 mutants, we obtained two temperature‐sensitive alleles, bur1–1 and bur1–8 (gift from G. Prelich; see Murray et al., 2001), and measured telomere length by Southern blot. Both bur1–1 and bur1–8 showed short telomeres, even when grown at the permissive temperature (Figure 1a). Given the strong growth defect in bur1–1, it was difficult to characterize this mutant and we chose to further characterize bur1–8 which had a less severe growth defect and would thus not pick up suppressors as readily. To confirm that the short telomeres in the bur1–8 cells were due to the mutations in BUR1 and not a suppressor in this strain, we introduced a plasmid expressing BUR1, pBUR1, and found that this rescued the short telomere phenotype back to wild‐type levels, while the empty vector pRS316 did not (Figure 1b).
3.2. Short telomeres in bur1 and bur2 are independent of histone modification pathway
The Bur1/2 kinase complex plays several roles in transcriptional regulation (Chu et al., 2006). One major role is regulation of histone H3 modification by Set2. BUR1 deletion lethality is suppressed by deletion of the SET2 histone methyltransferase (Chu et al., 2006). SET2 deletion relieves the transcriptional repression caused by H3K36 trimethylation, thus allowing for growth in the absence of Bur1 kinase activity. To determine whether telomere length changes were mediated by this transcriptional repression pathway, we examined the growth potential and telomere length in set2Δ and bur1–8 set2Δ double mutants. SET2 deletion partially rescued the temperature‐sensitive growth defect of bur1–8 (Figure 1e) but did not rescue the short telomeres (Figure 1c), suggesting that the short telomeres in bur1–8 cells are independent of the histone methylation pathway of Set2.
To further examine the role of histone methylation on the viability of Bur1 mutants and on telomere length, we investigated a strain expressing only a mutant version of H3, H3:K36A, which cannot be methylated by Set2. Expression of this histone mutant has been shown to rescue the growth of bur1Δ mutants (Chu et al., 2006). By mating a bur1∆ haploid covered by pBUR1 to cells deleted for the H3 locus with a plasmid expressing H3:K36A (Dai et al., 2008), and selecting diploids for dissection, we show expression of H3:K36A rescued lethality of bur1Δ, but these cells still showed short telomeres (Figure 1d). The fact that rescuing the histone modification defects of bur1∆ rescued growth but not telomere length further suggests that the short telomeres in the bur1–8 cells are independent of changes in H3K36me3 in transcriptional regulation.
3.3. bur1 and bur2 mutants are defective in de novo telomere elongation
To more directly examine the role of the Bur1/2 cyclin kinase complex in telomere elongation, we use the de novo telomere elongation assay (Diede & Gottschling, 1999). In this assay, a short telomere seed is exposed by cutting with a galactose inducible HO endonuclease, and elongation of the seed sequence is followed over 7 h while cells are held in nocodazole to allow S phase to finish but then block cell cycle progression into the next cell cycle. We grew two independent isolates of bur1–8 at the permissive temperature, arrested them in nocodazole for 4 h, added galactose to induce HO cutting and kept half of the culture at the permissive temperature while the other half was shifted to the restrictive temperature of 38°C and time points were collected. The bur1–8 mutants showed some telomere elongation at the permissive temperature of 30°C (Figure 2a); however, at the restrictive temperature of 38°C, elongation was abolished (Figure 2b). Telomere elongation was seen in wild‐type cells at both temperatures (Figure 2a,b). To verify that bur1–8 function was compromised at the restrictive temperature, we examined the H3K36 modification with a trimethylation‐specific antibody and found that trimethylation was compromised in this mutant at 38°C (Figure 2c) as expected from previous work (Chu et al., 2006). The specificity of the antibody was shown by the absence of the H3K36 trimethylation band in the set2Δ mutants and in cells expressing the mutant H3:K36A, which cannot be methylated. These results indicate that loss of Bur1 function, over the course of a 4‐h temperature shift, blocks telomere elongation.
Figure 2.
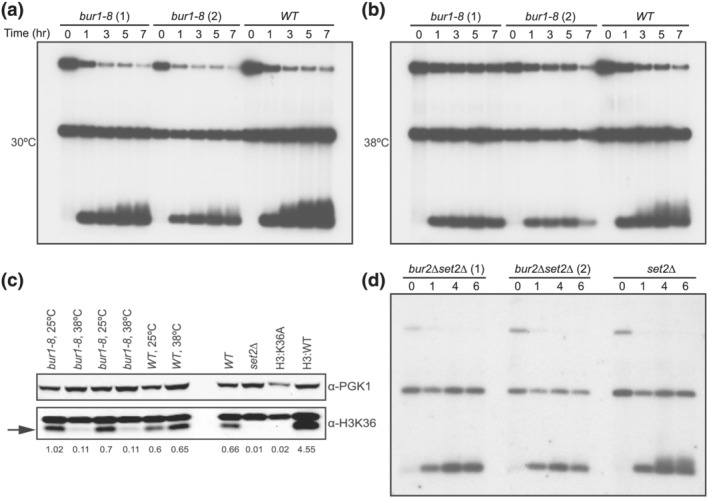
De novo telomere addition in bur1–8 and bur2∆ set2∆ mutants. (a) De novo telomere elongation assays were carried out for 0, 1, 3, 5, and 7 h after galactose induced cutting to expose a telomere seed. bur1–8 cells showed reduced elongation compared to WT cells at 30°C. (b) De novo telomere elongation assays performed in bur1–8 cells at 38°C showed no elongation when compared to WT cells. (c) Temperature sensitivity of histone trimethylation by western blot analysis of trimethylated H3:K36A in two independent bur1–8 mutants grown at permissive (25°C) or restrictive (38°C) temperatures. The upper panel shows the PGK1 protein loading control; the lower panel shows a non‐specific band just above the trimethylated H3K36 band (arrow). The four lanes on the right serve as controls for the H3K36 antibody specificity. (d) De novo telomere elongation assays performed in two independent bur2∆ set2∆ mutants compared to set2∆ alone
Several studies suggested that temperature shifts can affect telomere length (Paschini et al., 2012; Romano et al., 2013). To determine whether temperature affected telomere length in our experiments, we grew wild‐type cells in liquid culture at 37°C by continuous passaging over 4 days, collected cells each day for genomic DNA, and screened for telomere length by Southern blot (see Section 2). Telomere shortening occurred with progressive growth at 37°C as previously reported (Paschini et al., 2012; Romano et al., 2013). The degree of shortening was remarkably strain dependent with large changes in telomere length in the W303 strain which showed short telomeres when grown at 37°C that were comparable in length to a Ku‐deficient strain (Figure S1). This suggests that telomere length changes using temperature‐sensitive alleles must be interpreted with caution.
To avoid these effects of temperature shifts on telomere length (Paschini et al., 2012; Romano et al., 2013), we also examined the role of the Bur1/2 kinase complex without using temperature shifts in the de novo telomere elongation assay. We were unable to generate bur1Δ set2Δ double mutants in this genetic background; thus, we generated BUR2/bur2Δ, SET2/set2Δ double heterozygous diploids and analyzed fresh haploid spores to measure de novo telomere elongation. De novo telomere elongation was defective in two independent isolates of bur2Δ set2Δ, but elongation was still seen in the set2Δ mutant similar to wild‐type cells (Figure 2d). These data provide further evidence that the Bur1/2 cyclin‐dependent kinase complex is required for de novo telomere elongation.
3.4. Bur1/2 kinase regulates the mRNA levels of telomerase components
Telomere length is regulated in part by the very low expression levels of the protein component, Telomerase Reverse Transcriptase (TERT), or the RNA component (Cristofari & Lingner, 2006; Greider, 2006; Mozdy & Cech, 2006). Both TERT and the telomerase RNA are haploinsufficient in human and mouse cells (Armanios et al., 2005; Hao et al., 2005; Marrone et al., 2004; Strong et al., 2011). In yeast, mutations in transcription factors, including the PAF complex, result in a reduction in the levels of the telomerase RNA, TLC1, and telomere shortening (Mozdy et al., 2008). Given that the Bur1/2 kinase complex is also a transcriptional regulator, we wanted to determine whether changes in the abundance of telomerase components were causing the short telomeres in bur1 and bur2 mutants. Microarray experiments previously identified a large set of genes that are either upregulated or downregulated in bur2Δ mutants (Laribee et al., 2005). In their study, EST2 (yeast TERT component) showed a small increase in expression.
We examined the levels of mRNA for each of the four yeast telomerase components: EST1, EST2, EST3, and TLC1 by qRT‐PCR (see Section 2). As a control, we examined ARN1, which was previously shown to be upregulated in a bur2Δ mutant (Laribee et al., 2005). RNA was prepared from wild‐type, bur2Δ, bur1Δ set2Δ, and set2Δ mutants for qRT‐PCR, and the relative mRNA levels were normalized to wild type for each gene (Figure 3). The mRNA levels for EST1, EST2, and EST3 were elevated in the bur2Δ mutant, and the mRNA levels of EST1 and EST2 were elevated in the bur1Δ set2Δ double mutant. In contrast, TLC1 levels were significantly decreased in both bur2Δ and the bur1Δ set2Δ mutants. These gene expression data indicate that transcription of telomerase components is regulated by the Bur1/2 kinase complex, and in particular, TLC1 expression levels are decreased.
Figure 3.
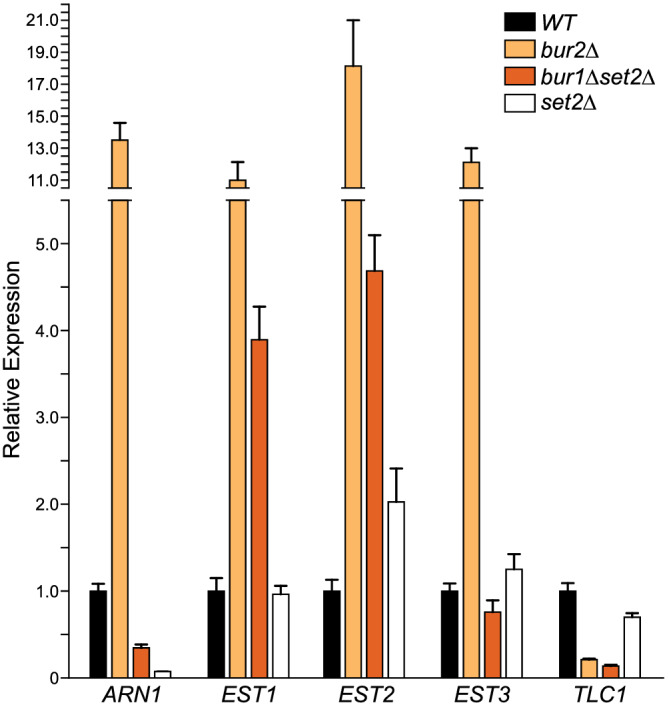
Expression of telomerase components in Bur1/2 kinase mutant cells. Quantitative PCR (qRT‐PCR) of telomerase components EST1, 2, 3, and TLC1 and a control ARN1 were analyzed in WT, bur2∆, bur1∆ set2∆, and set2∆ cells. Error bars represent SEM (N = 3 technical replicates) [Colour figure can be viewed at wileyonlinelibrary.com]
3.5. bur2Δ set2Δ mutants overexpressing TLC1 are still deficient in de novo telomere elongation
To examine whether deficient TLC1 levels were responsible for the short telomeres in Bur1/2 kinase complex mutants, as seen previously for the PAF complex mutations (Mozdy et al., 2008), we set out to overexpress TLC1. We modified a plasmid in which TLC1 is driven by the Gal promoter (Boule et al., 2005) and tested whether this construct, pGalTLC1, could rescue a null mutant in TLC1. We transformed TLC1/tlc1Δ heterozygous diploids with pGalTLC1, sporulated, and found that pGalTLC1 rescued telomere shortening in tlc1Δ cells (Figure S2). We next examined whether expression of pGalTLC1 could rescue de novo telomere addition in the bur2Δ set2Δ mutants. Two independent bur2Δ/BUR2, set2Δ/SET2 heterozygous diploids containing pGalTLC1 were sporulated to generate haploid bur2Δ set2Δ or set2Δ mutants, and de novo telomere elongation assays were carried out. The cells were first grown in raffinose minimal media without uracil to maintain plasmid selection and with nocodazole to allow S phase progression without allowing additional cell cycles. Galactose was added at time zero to induce both the cleavage of the chromosome and TLC1 transcription. No telomere elongation was seen in the two independent bur2Δ set2Δ haploids, while elongation was seen in the isogenic wild‐type and set2Δ mutants (Figure 4a). qRT‐PCR confirmed that Tlc1 RNA was overexpressed in the bur2Δ set2Δ cells that contained the plasmid (Figure 4b). These results indicate that the lack of telomere elongation in the bur2Δ set2Δ strains is not due to low levels of Tlc1.
Figure 4.
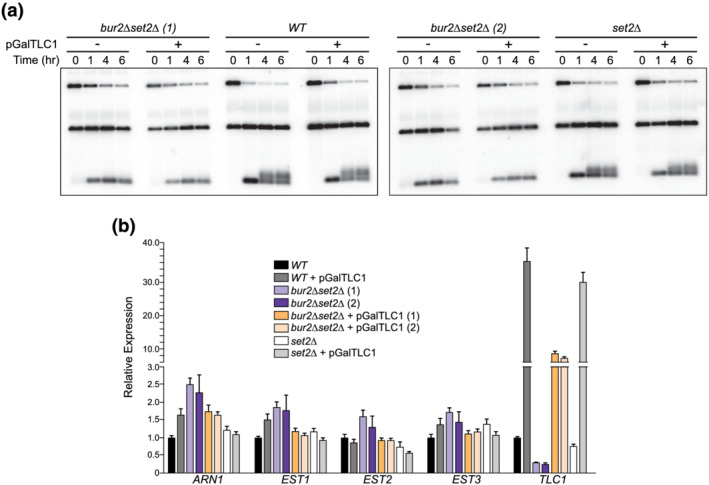
Overexpression of Tlc1 does not rescue de novo telomere elongation. (a) De novo telomere elongation in bur2∆ set2∆ cells with or without pGalTLC1. Two independent isolates of bur2∆ set2∆ were examined and compared to WT cells or set2∆ mutants alone. (b) qRT‐PCR of EST1, 2, 3, and TLC1 and the control ARN1 from RNA isolated during the de novo elongation assay at T = 4 h shown in part (a). Tlc1 is the only component downregulated in bur2∆ set2∆ cells, and the levels are increased over 10‐fold in cells containing the pGalTLC1 plasmid. Error bars represent SEM (N = 3 technical replicates [Colour figure can be viewed at wileyonlinelibrary.com]
3.6. bur2Δ set2Δ mutants overexpressing Tlc1 RNA have short telomeres
The lack of telomere elongation in the de novo assay suggests that Bur1/2 regulates telomere elongation in vivo over a short time frame. To determine whether overexpression of Tlc1 RNA would rescue telomere length given multiple cell divisions, we examined endogenous telomeres in cells expressing the pGalTLC1 construct for four successive passages. Overexpression of Tlc1 did not rescue the short telomere phenotype of the bur2Δ set2Δ mutants (Figure 5). However, as shown previously, long‐term overexpression lead to some telomere shortening, due to titration of the Ku proteins from telomeres (Singer & Gottschling, 1994; Stellwagen et al., 2003). These data, taken together with the absence of de novo telomere elongation, further suggest that the low level of Tlc1 is not responsible for the short telomeres in cells lacking Bur1/2 kinase complex activity.
Figure 5.
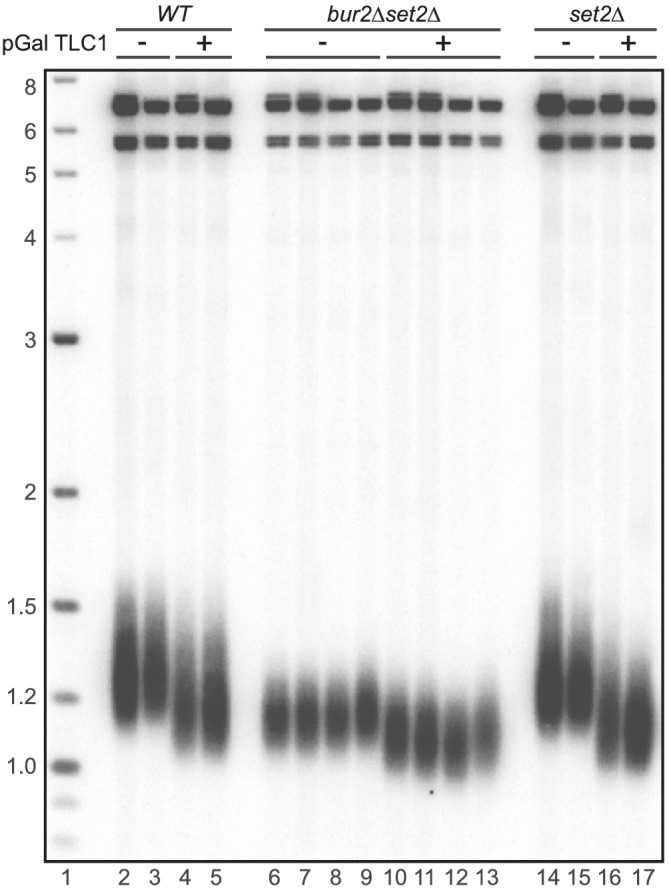
Overexpression of Tlc1 does not rescue telomere length in bur1∆ set2∆ cells. Telomere length was not increased by pGalTlc1 overexpression in bur1∆ set2∆ cells (Lanes 10, 11, 12, 13). Two of the haploids from independent diploid dissections are the same strains used for de novo telomere addition in Figure 4. The other two haploids are isogenic from the same dissections. These selectively grown haploids were propagated after the second passage in CAA media containing galactose and no tryptophan for plasmid selection. Cultures were harvested after two additional passages for analysis, for a total of four passages. Shortening in cells containing the overexpressed Tlc1 RNA is most likely due to titration of the Ku proteins from telomeres
3.7. The C‐terminal domain of Bur1 is not necessary for telomere maintenance
The C‐terminal domain of Bur1 was shown to interact with RPA and play a role in the DNA damage response (Clausing et al., 2010). To determine if the function of Bur1 at telomeres is mediated by this RPA interaction, we generated a C‐terminal truncation of Bur1 similar to that used by Clausing et al. and asked whether it would rescue the short telomeres in the bur1–1 mutant. A plasmid expressing full length BUR1 rescued the short telomeres in the bur1–1 mutant (Figure 6), as we saw previously (Figure 1a). In addition, the C‐terminal truncation of Bur1, BUR1ΔC (1–365aa), rescued telomere length to the same extent as full length Bur1, indicating that the C‐terminal region is not required for telomere maintenance. pBUR1∆C also rescued bur1∆ lethality and did not change telomere length after four passages (Figure S3).
Figure 6.
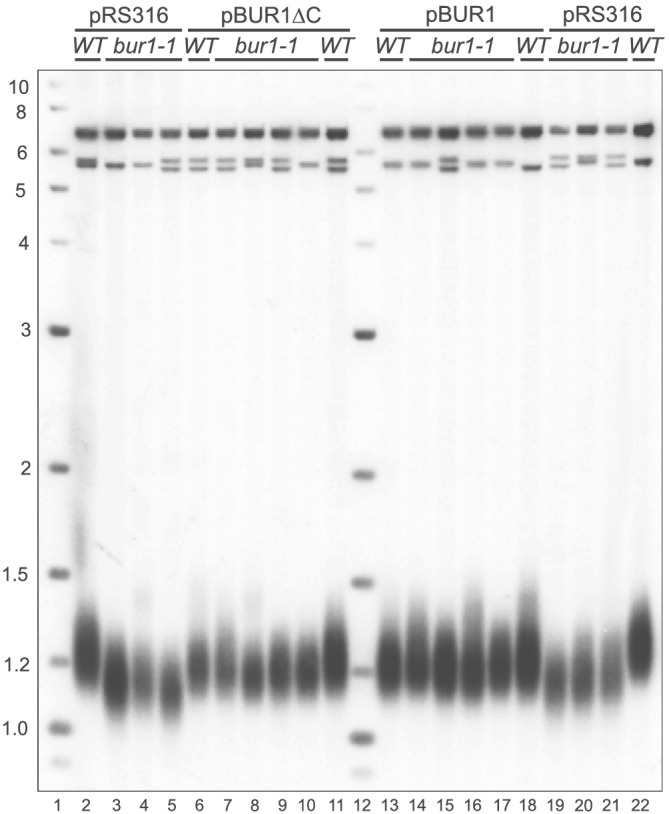
Telomere length is rescued by pBUR1∆C in bur1–1 cells. The short telomere length phenotype in the bur1–1 mutants was rescued by plasmids expressing full length BUR1 (Lanes 14–17) and truncated BUR1∆C (Lanes 7–10) as compared to vector alone (pRS316, Lanes 3–5 and 19–21). These haploids were only passaged once before growth for Southern analysis and may explain why there is a slight shortening of the telomere length in bur1–1 with both BUR clones
3.8. Epistasis analysis of BUR2 with genes involved in telomere maintenance
To determine whether Bur1/2 activity plays a role in a known pathway of telomere length regulation, we made double mutants of bur2Δ with genes known to affect telomere length, including tel1Δ, mre11Δ, rif1Δ, ku80Δ, and tlc1Δ (Figure 7a). All double mutants were constructed in a heterozygous diploid that was sporulated to generate isogenic single and double mutant combinations. Both bur2Δ tel1Δ and bur2Δ mre11Δ had telomeres that were shorter than either of the mutants alone. This suggested that BUR2 functions in a pathway separate from Tel1 and the MRX complex. rif1Δ mutants have significantly longer telomeres than wild type (Hardy et al., 1992). The double mutant bur2Δ rif1Δ had telomeres that were slightly shorter than rif1Δ but still longer than wild type, indicating that deletion of bur2Δ in a rif1Δ background partially blocks the excessive telomere elongation in this strain and that the two genes may be in different telomere length regulation pathways. The bur2Δ yku80Δ double mutants had shorter telomeres than both single mutants, suggesting that Bur2 is in a different pathway than Ku. Consistent with this, we saw a synthetic growth defect in the double mutants compared to either single mutant. Only the double mutants of bur2Δ tlc1Δ showed no additional shortening of telomeres compared to that seen in tlc1Δ, further suggesting that the Bur1/2 kinase complex functions in the regulation of telomerase.
Figure 7.
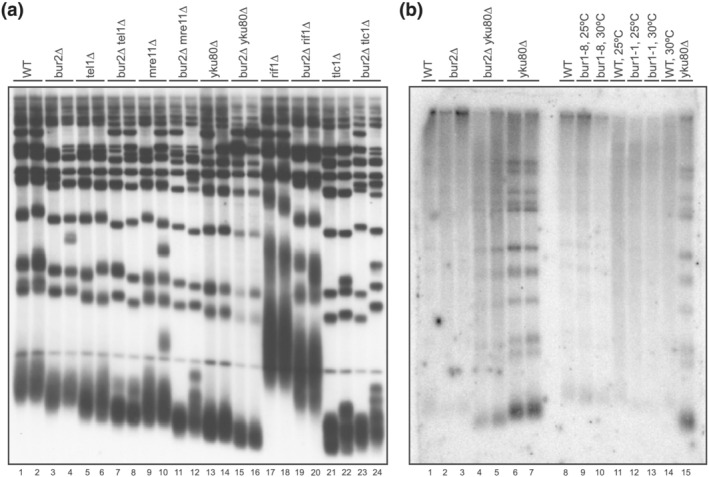
Epistasis analysis of bur2∆ mutants. (a) Telomere lengths of single and double mutant haploid spores were compared by Southern blot analysis. Heterozygous diploids were generated as double mutants with bur2∆ and several genes that affect different pathways of telomere maintenance including tel1∆ (Lanes 7, 8), mre11∆ (Lanes 11, 12), yku80∆ (Lanes 15, 16), rif1∆ (Lanes 19, 20), and tlc1∆ (Lanes 23, 24). In this Southern, as compared to the others in this study using detection of the Y′ region, the telomere G1‐3T repeats are detected using a poly GT probe. (b) Non‐denaturing in‐gel hybridization to examine single‐stranded DNA at telomeres. yku80∆ was used as a control to verify single‐stranded telomeric DNA (Lanes 6, 7, 15). Neither bur2∆, bur1–1, nor bur1–8 showed evidence of increased single‐stranded DNA. Note: A radiolabeled DNA ladder was not included with the probe in the hybridization mix
Because yku80Δ mutants have excessive single‐stranded DNA at their telomeres, we examined bur1–8 and bur2Δ mutants on non‐denaturing Southern blots to look at single‐stranded telomeric DNA. yku80Δ mutants produced the expected signal in non‐denaturing gels; however, we did not detect single‐stranded telomeric DNA in bur2Δ or bur1–8 and bur1–1 at either 25°C or 30°C (Figure 7b). We conclude that the Bur1/2 kinase complex does not regulate telomere integrity in the same pathway as Ku.
3.9. bur1–8 mutants block telomeric recombination in tlc1Δ survivors
To further study the interaction of Bur1/2 and telomerase, we examined the progressive telomere shortening in bur1–8 tlc1Δ cells passaged for successive days in liquid culture. The initial generation of a haploid bur1–8 tlc1Δ from a heterozygous diploid showed telomere lengths similar to tlc1Δ. When telomerase mutants are grown for 6 or more days in culture, progressive telomere shortening occurs, and ultimately, survivors arise via a Rad52‐mediated recombination process (Lundblad & Blackburn, 1993). We followed telomere length during progressive days of growth in bur1–8 tlc1Δ double mutants. With progressive growth, the rate of telomere shortening was similar in bur1–8 tlc1Δ double mutants and tlc1Δ mutants grown at semi‐permissive temperatures of 30°C or 34°C. However, while the tlc1Δ cells showed evidence of telomere recombination after 6 and 7 days in culture, strikingly, the bur1–8 tlc1Δ mutants showed no telomere recombination (Figure 8). Previous work linked RAD52 and DNA damage response with Bur1/2 (Clausing et al., 2010), and Bur1/2 regulation of histone ubiquitination has been shown to regulate recombination (Uckelmann & Sixma, 2017). Our data further indicate that Bur1/2 is required for generation of survivors in telomerase mutants through telomere recombination.
Figure 8.
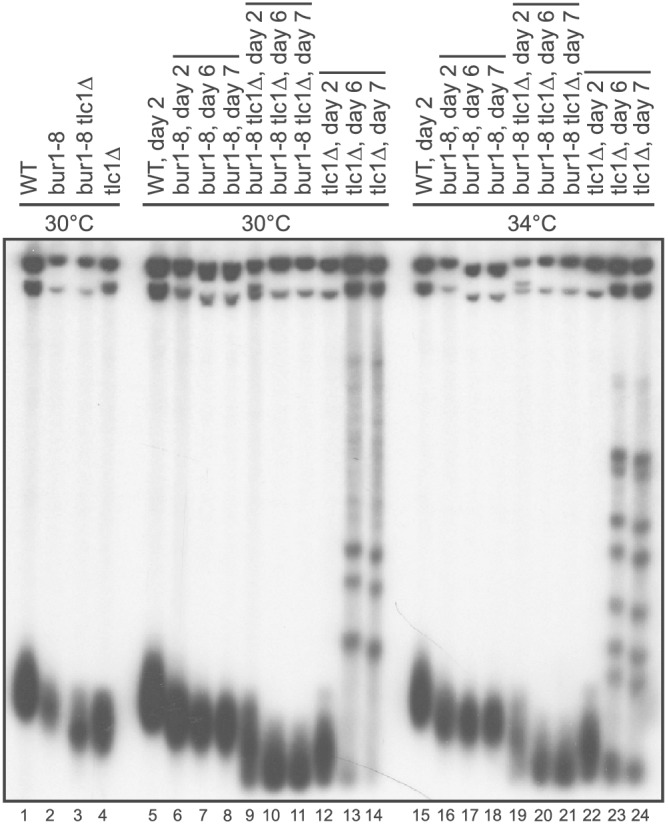
Survivors are not observed in bur1–8 tlc1∆ double mutants. Successive passages of bur1–8, tlc1∆, and bur1–8 tlc1∆ double mutants at either 30°C or 34°C. Cells were kept in continuous daily culture and DNA prepared at Day 2, 6, or 7. Survivors were generated at Days 6 and 7 in the tlc1∆ strains at both temperatures, as indicated by the higher molecular weight telomere bands in this Southern analysis (Lanes 13, 14 and 23, 24). Survivors were not seen in the bur1–8 tlc1∆ double mutants at this time period (Lanes 10, 11 and 20, 21)
4. DISCUSSION
The Bur1/2 kinase complex is well characterized for its multiple roles in regulating transcriptional elongation (Qiu et al., 2012). Besides regulating trimethylation of H3K36 through Set2, this kinase complex also phosphorylates the CTD of RNA polymerase II and directly phosphorylates the transcriptional regulator Spt5. Recent studies suggest that there are also other targets of the kinase (Qiu et al., 2012). Here, we describe a role for the Bur1/2 kinase complex in the regulation of telomere elongation.
4.1. The Bur1/2 kinase complex regulates telomere elongation
We found that bur1 and bur2 mutants have short telomeres and fail to elongate telomeres in a short‐term de novo telomere elongation assay. While several components of telomerase are transcriptionally regulated by Bur1/2, only Tlc1 showed decreased expression in the bur1 or bur2 mutants. Overexpression of Tlc1 in bur1 and bur2 mutants did not rescue the short telomeres or the telomere elongation defect.
Several different approaches suggest that the Bur1/2 kinase complex may play a role in telomere elongation that is separate from its role in transcriptional regulation. First, while deletion of SET2 rescues the growth defect of both bur1 and bur2 mutants, it did not rescue the telomere length defects. Second, while Tlc1 mRNA levels were reduced in the absence of bur1 and bur2, reintroduction of Tlc1 did not rescue the telomere length phenotype. This is in contrast to what has been found for mutations in the PAF transcription factor components, where reintroduction of Tlc1 did rescue the short telomeres (Mozdy et al., 2008). Third, telomere elongation in the de novo telomere length assay was blocked in both bur1–8 and bur2Δ set2Δ mutants even when Tlc1 RNA was overexpressed. This loss of telomere elongation during a single cell cycle suggests a critical role of the Bur1/2 kinase in telomere elongation.
4.2. Epistasis analysis suggests Bur1/2 regulates telomerase activity
To understand what role the Bur1/2 kinase complex might play in telomere function, we generated a series of double mutants to examine epistasis. Tel1 and the MRX complex are known to be in the same pathway of telomere length maintenance (Ritchie & Petes, 2000). Consistent with this, both bur2Δ tel1Δ and bur2Δ mre11Δ double mutants had shorter telomeres than bur2Δ, suggesting that Bur2 may be in a different pathway than Tel1 and the MRX complex. The double mutant bur2Δ yku80Δ also had shorter telomeres than either single mutant alone, again suggesting that the Bur1/2 kinase complex is in a different pathway than Ku at telomeres. The long telomeres in a rif1Δ mutant were shorter in bur2Δ rif1Δ double mutant but were still longer than wild type, suggesting the activity of the Bur1/2 kinase complex may be partially required for excessive elongation of rif1Δ at telomeres; however, other pathways also play a role. The telomere length of bur2Δ tlc1Δ double mutants was very similar to tlc1Δ alone, which suggests that the Bur1/2 kinase complex affects telomere length through telomerase activity. However, the lack of rescue of telomere length in strains overexpressing Tlc1 suggests that it is unlikely the transcriptional effect of bur1 on Tlc1 that meditates this effect.
There are several models for how Bur1/2 may regulate telomere length. One model suggests that the effects may be mediated through telomeric chromatin since Bur1/2 regulates histone modification. However, loss of SET2 did not rescue telomere length, and therefore, if there is a chromatin effect, it does not act through H3K36 trimethylation. A second model is that Bur1/2 directly phosphorylates a protein involved in telomere homeostasis. Our epistasis analysis suggests that the most likely target would be one of the telomerase protein components because the telomere lengths of the double and single mutants were similar. We cannot rule out the possibility that the Bur1/2 short telomere phenotype is due to transcriptional regulation of genes other than the telomerase components.
4.3. Bur1/2 regulates telomere recombination and the generation of survivors
The lack of survivors in bur2Δ tlc1Δ double mutants suggests a role for the Bur1/2 kinase complex in the break induced replication that elongates telomeres (Bosco & Haber, 1998). The Bur1 human homolog, Cdk9, is required for recruitment of BRCA1 and BARD1 to double‐strand breaks to regulate homologous recombination (Nepomuceno et al., 2017). In yeast, Bur1 regulates the monoubiquitination of histone H2B (Wood et al., 2005), which in turn regulates homologous recombination (Uckelmann & Sixma, 2017). The absence of survivors in bur2Δ tlc1Δ double mutants are very similar to what is seen in a tel1∆ mec1∆ double mutant, in which telomeres shorten and are maintained as short, but survivors are not generated (Ma & Greider, 2009). Strikingly, human ATM also regulates histone H2B monoubiquitination (Moyal et al., 2011), suggesting a common pathway of chromatin modification by which both the Bur1and Tel1/Mec1 kinases mediate telomere recombination and generate survivors in tlc1Δ yeast. Determining the mechanism of Bur1/2 on telomeres will be important for understanding telomere homeostasis.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Figure S1. Telomere shortening at 37°C in three different yeast strains. To determine whether growth of WT cells at 37°C might contribute to telomere shortening, we passaged three different common laboratory strains for 4 days in liquid culture at 30°C and 37°C. (A) Southern blot analysis of telomere length is shown for strains BY4742 and YPH500 grown under these conditions. (B) Southern blot analysis of strain W303 grown under these conditions
Figure S2. Overexpression construct pGalTLC1 rescues tlc1∆ mutants. pGalTLC1 overexpression rescues the tlc1∆ short telomere length indicating the telomerase RNA expressed from this plasmid is functional (lanes 6,7). Independent heterozygous TLC1/tlc1∆ diploid transformants (BY background) with pGal1TLC1 were dissected. Haploids were passaged two times on CAA media containing galactose and no tryptophan for plasmid selection. Background differences and passage number would explain the lack of telomere shortening in WT and mutants with overexpressed TLC as observed in Figure 5.
Figure S3. Rescue of bur1∆ lethality does not change telomere length. The lethality of bur1∆ mutants were rescued by plasmids expressing full length BUR1 (lanes 3–6) and truncated BUR1∆C (lanes 11–14). These rescued haploids from two independent diploid dissections did not change telomere length after four passages on selective media.
ACKNOWLEDGMENTS
We thank Greg Prelich for generously providing the bur1–1 and bur1–8 temperature‐sensitive strains and plasmids with BUR1. We thank V. Zakian for the Gal1 regulated TLC1 construct. We thank Courtney O'Farrell for her work on the bur1–8 tlc1Δ double mutant growth analysis, Molly Hyde for doing the bur2Δ ku80Δ single‐stranded telomere analysis, and Yunmei Ma Karanjawala for her help with the Southern blot for the epistasis analysis. We would also like to thank Catherine Connelly for her assistance in designing the figures for this study.
Connelly, C. J. , Vidal‐Cardenas, S. , Goldsmith, S. , & Greider, C. W. (2022). The Bur1 cyclin‐dependent kinase regulates telomere length in Saccharomyces cerevisiae . Yeast, 39, 177–192. 10.1002/yea.3680
REFERENCES
- Armanios, M. (2009). Syndromes of telomere shortening. Annual Review of Genomics and Human Genetics, 10, 45–61. 10.1146/annurev-genom-082908-150046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios, M. , Chen, J. L. , Chang, Y. P. , Brodsky, R. A. , Hawkins, A. , Griffin, C. A. , Eshleman, J. R. , Cohen, A. R. , Chakravarti, A. , Hamosh, A. , & Greider, C. W. (2005). Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proceedings of the National Academy of Sciences of the United States of America, 102, 15960–15964. 10.1073/pnas.0508124102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askree, S. H. , Yehuda, T. , Smolikov, S. , Gurevich, R. , Hawk, J. , Coker, C. , Krauskopf, A. , Kupiec, M. , & McEachern, M. J. (2004). A genome‐wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proceedings of the National Academy of Sciences of the United States of America, 101, 8658–8663. 10.1073/pnas.0401263101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco, G. , & Haber, J. E. (1998). Chromosome break‐induced DNA replication leads to non‐reciprocal translocations and telomere capture. Genetics, 150, 1037–1047. 10.1093/genetics/150.3.1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boule, J. B. , Vega, L. R. , & Zakian, V. A. (2005). The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature, 438(7064), 57–61. 10.1038/nature04091 [DOI] [PubMed] [Google Scholar]
- Brachmann, C. B. , Davies, A. , Cost, G. J. , Caputo, E. , Li, J. , Hieter, P. , & Boeke, J. D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR‐mediated gene disruption and other applications. Yeast, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Chu, Y. , Sutton, A. , Sternglanz, R. , & Prelich, G. (2006). The BUR1 cyclin‐dependent protein kinase is required for the normal pattern of histone methylation by SET2. Molecular and Cellular Biology, 26, 3029–3038. 10.1128/MCB.26.8.3029-3038.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausing, E. , Mayer, A. , Chanarat, S. , Muller, B. , Germann, S. M. , Cramer, P. , Lisby, M. , & Strasser, K. (2010). The transcription elongation factor Bur1‐Bur2 interacts with replication protein A and maintains genome stability during replication stress. The Journal of Biological Chemistry, 285, 41665–41674. 10.1074/jbc.M110.193292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari, G. , & Lingner, J. (2006). Telomere length homeostasis requires that telomerase levels are limiting. The EMBO Journal, 25, 565–574. 10.1038/sj.emboj.7600952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, J. , Hyland, E. M. , Yuan, D. S. , Huang, H. , Bader, J. S. , & Boeke, J. D. (2008). Probing nucleosome function: A highly versatile library of synthetic histone H3 and H4 mutants. Cell, 134, 1066–1078. 10.1016/j.cell.2008.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Fagagna, F. D. A. , Reaper, P. M. , Clay‐Farrace, L. , Fiegler, H. , Carr, P. , Von Zglinicki, T. , Saretzki, G. , Carter, N. P. , & Jackson, S. P. (2003). A DNA damage checkpoint response in telomere‐initiated senescence. Nature, 426, 194–198. 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- Diede, S. J. , & Gottschling, D. E. (1999). Telomerase‐mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell, 99, 723–733. 10.1016/S0092-8674(00)81670-0 [DOI] [PubMed] [Google Scholar]
- Dionne, I. , & Wellinger, R. J. (1996). Cell cycle‐regulated generation of single‐stranded G‐rich DNA in the absence of telomerase. Proceedings of the National Academy of Sciences of the United States of America, 93, 13902–13907. 10.1073/pnas.93.24.13902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, S. K. , & Lundblad, V. (1999). Est1 and Cdc13 as comediators of telomerase access. Science, 286, 117–120. 10.1126/science.286.5437.117 [DOI] [PubMed] [Google Scholar]
- Frank, C. J. , Hyde, M. , & Greider, C. W. (2006). Regulation of telomere elongation by the cyclin‐dependent kinase CDK1. Molecular Cell, 24, 423–432. 10.1016/j.molcel.2006.10.020 [DOI] [PubMed] [Google Scholar]
- Gao, H. , Cervantes, R. B. , Mandell, E. K. , Otero, J. H. , & Lundblad, V. (2007). RPA‐like proteins mediate yeast telomere function. Nature Structural & Molecular Biology, 14, 208–214. 10.1038/nsmb1205 [DOI] [PubMed] [Google Scholar]
- Gatbonton, T. , Imbesi, M. , Nelson, M. , Akey, J. M. , Ruderfer, D. M. , Kruglyak, L. , Simon, J. A. , & Bedalov, A. (2006). Telomere length as a quantitative trait: Genome‐wide survey and genetic mapping of telomere length‐control genes in yeast. PLoS Genetics, 2, e35. 10.1371/journal.pgen.0020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell, P. W. , Kronmal, S. L. , Porter, S. E. , Gassenhuber, J. , Obermaier, B. , & Petes, T. D. (1995). TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell, 82, 823–829. 10.1016/0092-8674(95)90479-4 [DOI] [PubMed] [Google Scholar]
- Greider, C. W. (2006). Telomerase RNA levels limit the telomere length equilibrium. Cold Spring Harbor Symposia on Quantitative Biology, 71, 225–229. 10.1101/sqb.2006.71.063 [DOI] [PubMed] [Google Scholar]
- Hao, L. Y. , Armanios, M. , Strong, M. A. , Karim, B. , Feldser, D. M. , Huso, D. , & Greider, C. W. (2005). Short telomeres, even in the presence of telomerase, limit tissue renewal capacity. Cell, 123, 1121–1131. 10.1016/j.cell.2005.11.020 [DOI] [PubMed] [Google Scholar]
- Hardy, C. F. , Sussel, L. , & Shore, D. (1992). A RAP1‐interacting protein involved in transcriptional silencing and telomere length regulation. Genes & Development, 6, 801–814. 10.1101/gad.6.5.801 [DOI] [PubMed] [Google Scholar]
- Harley, C. B. , Futcher, A. B. , & Greider, C. W. (1990). Telomeres shorten during ageing of human fibroblasts. Nature, 345, 458–460. 10.1038/345458a0 [DOI] [PubMed] [Google Scholar]
- Hyland, E. M. , Cosgrove, M. S. , Molina, H. , Wang, D. , Pandey, A. , Cottee, R. J. , & Boeke, J. D. (2005). Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Molecular and Cellular Biology, 25, 10060–10070. 10.1128/MCB.25.22.10060-10070.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJpma, A. , & Greider, C. W. (2003). Short telomeres induce a DNA damage response in Saccharomyces cerevisiae . Molecular Biology of the Cell, 14, 987–1001. 10.1091/mbc.02-04-0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizer, H. , Connelly, C. J. , Bettridge, K. , Viggiani, C. , & Greider, C. W. (2015). Regulation of telomere length requires a conserved N‐terminal domain of Rif2 in Saccharomyces cerevisiae . Genetics, 201, 573–586. 10.1534/genetics.115.177899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener, R. , Connelly, C. J. , & Greider, C. W. (2019). Tel1 activation by the MRX complex is sufficient for telomere length regulation but not for the DNA damage response in Saccharomyces cerevisiae . Genetics, 213, 1271–1288. 10.1534/genetics.119.302713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T. , & Buratowski, S. (2007). Two Saccharomyces cerevisiae JmjC domain proteins demethylate histone H3 Lys36 in transcribed regions to promote elongation. The Journal of Biological Chemistry, 282, 20827–20835. 10.1074/jbc.M703034200 [DOI] [PubMed] [Google Scholar]
- Laribee, R. N. , Krogan, N. J. , Xiao, T. , Shibata, Y. , Hughes, T. R. , Greenblatt, J. F. , & Strahl, B. D. (2005). BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Current Biology, 15, 1487–1493. 10.1016/j.cub.2005.07.028 [DOI] [PubMed] [Google Scholar]
- Li, S. , Makovets, S. , Matsuguchi, T. , Blethrow, J. D. , Shokat, K. M. , & Blackburn, E. H. (2009). Cdk1‐dependent phosphorylation of Cdc13 coordinates telomere elongation during cell‐cycle progression. Cell, 136, 50–61. 10.1016/j.cell.2008.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner, J. , Cech, T. R. , Hughes, T. R. , & Lundblad, V. (1997). Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proceedings of the National Academy of Sciences of the United States of America, 94, 11190–11195. 10.1073/pnas.94.21.11190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. C. , Gopalakrishnan, V. , Poon, L. F. , Yan, T. , & Li, S. (2014). Cdk1 regulates the temporal recruitment of telomerase and Cdc13‐Stn1‐Ten1 complex for telomere replication. Molecular and Cellular Biology, 34, 57–70. 10.1128/MCB.01235-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Warfield, L. , Zhang, C. , Luo, J. , Allen, J. , Lang, W. H. , Ranish, J. , Shokat, K. M. , & Hahn, S. (2009). Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Molecular and Cellular Biology, 29, 4852–4863. 10.1128/MCB.00609-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad, V. , & Blackburn, E. H. (1993). An alternative pathway for yeast telomere maintenance rescues est1 − senescence. Cell, 73, 347–360. 10.1016/0092-8674(93)90234-H [DOI] [PubMed] [Google Scholar]
- Lundblad, V. , & Szostak, J. W. (1989). A mutant with a defect in telomere elongation leads to senescence in yeast. Cell, 57, 633–643. 10.1016/0092-8674(89)90132-3 [DOI] [PubMed] [Google Scholar]
- Ma, Y. , & Greider, C. W. (2009). Kinase‐independent functions of TEL1 in telomere maintenance. Molecular and Cellular Biology, 29, 5193–5202. 10.1128/MCB.01896-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand, S. , Gilson, E. , & Shore, D. (1997). A protein‐counting mechanism for telomere length regulation in yeast. Science, 275, 986–990. 10.1126/science.275.5302.986 [DOI] [PubMed] [Google Scholar]
- Marrone, A. , Stevens, D. , Vulliamy, T. , Dokal, I. , & Mason, P. J. (2004). Heterozygous telomerase RNA mutations found in dyskeratosis congenita and aplastic anemia reduce telomerase activity via haploinsufficiency. Blood, 104, 3936–3942. 10.1182/blood-2004-05-1829 [DOI] [PubMed] [Google Scholar]
- Moyal, L. , Lerenthal, Y. , Gana‐Weisz, M. , Mass, G. , So, S. , Wang, S. Y. , Eppink, B. , Chung, Y. M. , Shalev, G. , Shema, E. , Shkedy, D. , Smorodinsky, N. I. , van Vliet, N. , Kuster, B. , Mann, M. , Ciechanover, A. , Dahm‐Daphi, J. , Kanaar, R. , Hu, M. C. T. , … Shiloh, Y. (2011). Requirement of ATM‐dependent monoubiquitylation of histone H2B for timely repair of DNA double‐strand breaks. Molecular Cell, 41, 529–542. 10.1016/j.molcel.2011.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy, A. D. , & Cech, T. R. (2006). Low abundance of telomerase in yeast: Implications for telomerase haploinsufficiency. RNA, 12, 1721–1737. 10.1261/rna.134706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy, A. D. , Podell, E. R. , & Cech, T. R. (2008). Multiple yeast genes, including Paf1 complex genes, affect telomere length via telomerase RNA abundance. Molecular and Cellular Biology, 28, 4152–4161. 10.1128/MCB.00512-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, S. , Udupa, R. , Yao, S. , Hartzog, G. , & Prelich, G. (2001). Phosphorylation of the RNA polymerase II carboxy‐terminal domain by the Bur1 cyclin‐dependent kinase. Molecular and Cellular Biology, 21, 4089–4096. 10.1128/MCB.21.13.4089-4096.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepomuceno, T. C. , Fernandes, V. C. , Gomes, T. T. , Carvalho, R. S. , Suarez‐Kurtz, G. , Monteiro, A. N. , & Carvalho, M. A. (2017). BRCA1 recruitment to damaged DNA sites is dependent on CDK9. Cell Cycle, 16, 665–672. 10.1080/15384101.2017.1295177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm, W. , & de Lange, T. (2008). How shelterin protects mammalian telomeres. Annual Review of Genetics, 42, 301–334. 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- Paschini, M. , Toro, T. B. , Lubin, J. W. , Braunstein‐Ballew, B. , Morris, D. K. , & Lundblad, V. (2012). A naturally thermolabile activity compromises genetic analysis of telomere function in Saccharomyces cerevisiae . Genetics, 191, 79–93. 10.1534/genetics.111.137869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich, G. , & Winston, F. (1993). Mutations that suppress the deletion of an upstream activating sequence in yeast: Involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics, 135, 665–676. 10.1093/genetics/135.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, C. M. , Boltz, K. A. , Chaiken, M. F. , Stewart, J. A. , Beilstein, M. A. , & Shippen, D. E. (2010). Evolution of CST function in telomere maintenance. Cell Cycle, 9, 3157–3165. 10.4161/cc.9.16.12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, H. , Hu, C. , Gaur, N. A. , & Hinnebusch, A. G. (2012). Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR‐independent recruitment of Paf1 complex. The EMBO Journal, 31, 3494–3505. 10.1038/emboj.2012.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, K. B. , Mallory, J. C. , & Petes, T. D. (1999). Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae . Molecular and Cellular Biology, 19, 6065–6075. 10.1128/MCB.19.9.6065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, K. B. , & Petes, T. D. (2000). The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics, 155, 475–479. 10.1093/genetics/155.1.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano, G. H. , Harari, Y. , Yehuda, T. , Podhorzer, A. , Rubinstein, L. , Shamir, R. , Gottlieb, A. , Silberberg, Y. , Pe'er, D. , Ruppin, E. , Sharan, R. , & Kupiec, M. (2013). Environmental stresses disrupt telomere length homeostasis. PLoS Genetics, 9, e1003721. 10.1371/journal.pgen.1003721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S. , & Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics, 122, 19–27. 10.1093/genetics/122.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, M. S. , & Gottschling, D. E. (1994). TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science, 266, 404–409. 10.1126/science.7545955 [DOI] [PubMed] [Google Scholar]
- Smogorzewska, A. , & de Lange, T. (2004). Regulation of telomerase by telomeric proteins. Annual Review of Biochemistry, 73, 177–208. 10.1146/annurev.biochem.73.071403.160049 [DOI] [PubMed] [Google Scholar]
- Stellwagen, A. E. , Haimberger, Z. W. , Veatch, J. R. , & Gottschling, D. E. (2003). Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes & Development, 17, 2384–2395. 10.1101/gad.1125903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong, M. A. , Vidal‐Cardenas, S. L. , Karim, B. , Yu, H. , Guo, N. , & Greider, C. W. (2011). Phenotypes in mTERT +/− and mTERT −/− mice are due to short telomeres, not telomere‐independent functions of telomerase reverse transcriptase. Molecular and Cellular Biology, 31, 2369–2379. https://doi/10.1128/MCB.05312-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckelmann, M. , & Sixma, T. K. (2017). Histone ubiquitination in the DNA damage response. DNA Repair (Amst), 56, 92–101. 10.1016/j.dnarep.2017.06.011 [DOI] [PubMed] [Google Scholar]
- Ungar, L. , Yosef, N. , Sela, Y. , Sharan, R. , Ruppin, E. , & Kupiec, M. (2009). A genome‐wide screen for essential yeast genes that affect telomere length maintenance. Nucleic Acids Research, 37, 3840–3849. 10.1093/nar/gkp259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viggiani, C. J. , & Aparicio, O. M. (2006). New vectors for simplified construction of BrdU‐Incorporating strains of Saccharomyces cerevisiae. Yeast, 23, 1045–1051. 10.1002/yea.1406 [DOI] [PubMed] [Google Scholar]
- Vodenicharov, M. D. , & Wellinger, R. J. (2006). DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell‐cycle kinase. Molecular Cell, 24, 127–137. 10.1016/j.molcel.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Vulliamy, T. , Marrone, A. , Szydlo, R. , Walne, A. , Mason, P. J. , & Dokal, I. (2004). Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nature Genetics, 36, 447–449. 10.1038/ng1346 [DOI] [PubMed] [Google Scholar]
- Winzeler, E. A. , Shoemaker, D. D. , Astromoff, A. , Liang, H. , Anderson, K. , Andre, B. , Bangham, R. , Benito, R. , Boeke, J. D. , Bussey, H. , Chu, A. M. , Connelly, C. , Davis, K. , Dietrich, F. , Dow, S. W. , el Bakkoury, M. , Foury, F. , Friend, S. H. , Gentalen, E. , … Davis, R. W. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- Wood, A. , Schneider, J. , Dover, J. , Johnston, M. , & Shilatifard, A. (2005). The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Molecular Cell, 20, 589–599. 10.1016/j.molcel.2005.09.010 [DOI] [PubMed] [Google Scholar]
- Yao, S. , Neiman, A. , & Prelich, G. (2000). BUR1 and BUR2 encode a divergent cyclin‐dependent kinase‐cyclin complex important for transcription in vivo. Molecular and Cellular Biology, 20, 7080–7087. 10.1128/MCB.20.19.7080-7087.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Telomere shortening at 37°C in three different yeast strains. To determine whether growth of WT cells at 37°C might contribute to telomere shortening, we passaged three different common laboratory strains for 4 days in liquid culture at 30°C and 37°C. (A) Southern blot analysis of telomere length is shown for strains BY4742 and YPH500 grown under these conditions. (B) Southern blot analysis of strain W303 grown under these conditions
Figure S2. Overexpression construct pGalTLC1 rescues tlc1∆ mutants. pGalTLC1 overexpression rescues the tlc1∆ short telomere length indicating the telomerase RNA expressed from this plasmid is functional (lanes 6,7). Independent heterozygous TLC1/tlc1∆ diploid transformants (BY background) with pGal1TLC1 were dissected. Haploids were passaged two times on CAA media containing galactose and no tryptophan for plasmid selection. Background differences and passage number would explain the lack of telomere shortening in WT and mutants with overexpressed TLC as observed in Figure 5.
Figure S3. Rescue of bur1∆ lethality does not change telomere length. The lethality of bur1∆ mutants were rescued by plasmids expressing full length BUR1 (lanes 3–6) and truncated BUR1∆C (lanes 11–14). These rescued haploids from two independent diploid dissections did not change telomere length after four passages on selective media.


