Abstract
ISAV is the causative agent of the infectious salmon anaemia (ISA), a disease listed by the OIE that has caused important economic losses to the Atlantic salmon (Salmo salar) industry. ISAV variants are identified as pathogenic or non‐pathogenic based on the presence or absence of a deletion in the highly polymorphic region (HPR) of segment 6 (S6). HPRΔ variants (pathogenic) are the only forms of the virus known to grow in cell culture. This is the first report of a HPR0 variant isolated in cell culture. The isolate is, however, atypical as it shows a marker of virulent variants on another segment (S5), which has never been reported for any other HPR0 variants. The significance of this finding remains unclear until more in‐depth work is carried out but does challenge current knowledge.
Keywords: ASK, HPR0, ISAV, Salmo salar, SHK‐1, virus isolation
The infectious salmon anaemia virus (ISAV) is the causative agent of the infectious Salmon Anaemia (ISA), a disease listed by the World Organisation for Animal Health (OIE) that has caused important economic losses to the Atlantic salmon (Salmo salar) aquaculture industry in the Northern Hemisphere and Chile (reviewed in OIE, 2021). In Canada, ISAV is endemic to the Atlantic coast and has caused severe disease outbreaks in the late 1990s, costing millions of CAD$ in fish loss over the next decade (Martin, 2007). Important changes in management practices were implemented, resulting in a decrease of the number and severity of outbreaks (Chang et al., 2014; Martin, 2007). To this day, the virus is still regularly detected and under close surveillance by provincial regulators and industry (CFIA, 2021).
ISAV is a virus that consists of eight RNA segments (Mjaaland et al., 1997), and variants can be grouped into two genetic clades: the European (EU) and the North American (NA) (Gagne & LeBlanc, 2018; Krossoy et al., 2001). Both NA and EU variants are regularly reported in Atlantic Canada (Gagne & LeBlanc, 2018). Not all variants cause disease, and two markers of virulence have been identified on segment 5 (S5) and 6 (S6), encoding two surface proteins: fusion and hemagglutinin esterase, respectively (Kibenge et al., 2007; Markussen et al., 2008; Rimstad & Markussen, 2020). On S6, a deletion in the highly polymorphic region (HPR) is considered to be a prerequisite of virulence. HPRΔ variants (deleted) can cause disease whereas HPR0 variants (no deletion) are not known to cause disease (Cook‐Versloot et al., 2004; Cunningham et al., 2002). With the exception of an artificial recombinant HPR0 variant (Cardenas et al., 2020), only HPRΔ variants are known to grow in cell culture (Aamelfot et al., 2016; OIE, 2021) and evidence suggests that HPRΔ originate from HPR0 variants, suggesting that transient variants with some but not all virulence markers may exist (Christiansen et al., 2017; Gagne & LeBlanc, 2018). On S5, a prerequisite of virulence seems to be a Q266→L266 substitution or Q266 with an insertion close to the putative cleavage site (Devold, 2006; Markussen et al., 2008). Association of these markers to virulent variants of ISAV has been supported over the years (Cárdenas et al., 2019; LeBlanc et al., 2018). This is the first report of the successful isolation in cell culture, supported with data, of a natural HPR0 variant, as defined by current genotyping, but is also the first report of a natural ISAV variant possessing a S6 typical of non‐virulent variants (no deletion in the HPR) and a S5 typical of virulent variants (Q266 with an insertion).
In late 2020, ISAV is detected by RT‐PCR in Atlantic salmon from a marine site in Atlantic Canada, through routine surveillance. The laboratory at Research Productivity Council (RPC), New Brunswick (N.B.), Canada, also detected ISAV by real‐time RT‐PCR (RT‐qPCR) in Atlantic salmon follow‐up samples. Genotyping identified a NA HPR0 sequence and isolation in cell culture was successful. The Canadian Food Inspection Agency (CFIA) submitted additional gill samples from the same apparently healthy population for testing at the National Aquatic Animal Health Laboratory in Winnipeg (NAAHLS‐FWI), Manitoba, Canada. Five detected samples were genotyped, verifying the initial laboratory report from RPC. The national reference laboratory for ISAV in Moncton (NAAHLS‐GFC), N.B., Canada, received infected tissues collected in 2020 and preserved at −80°C to corroborate isolation of this variant in cell culture.
Sample provided included a pool of gill, heart, kidney, spleen and pyloric caeca sampled from 5 adult Atlantic salmon. A homogenate of a subsample of tissues was used to inoculate ASK and SHK‐1 cells (see Supplemental File S1 for details). After 21 days post‐inoculation (dpi), cytopathic effects (CPE) were observed, and some wells displayed cytotoxicity. Cell lysates were filtered and used to inoculate new preparations of cells. CPE were observed after 6 and 11 days, for ASK and SHK‐1 cells, respectively, verifying the presence of a filterable replicating agent. The lysate was collected, filtered and immediately frozen at −80°C. Inoculum (0 dpi) and cell lysates at 21 dpi were also used to support viral growth by RT‐qPCR and to complete partial sequencing of S5 and S6. Partial sequencing of S5 was also done on samples received at NAAHLS‐FWI and RPC. Primers and probes used for RT‐qPCR and for sequencing are given in Table 1. Protocols used for the virology assay, RT‐qPCR and sequencing are available in Supplemental File S1.
TABLE 1.
Primers and probes used for real‐time RT‐PCR and partial sequencing analysis
| ISAV segment | Analysis | Primer/Probe name | Sequence 5’‐ 3’ |
|---|---|---|---|
| Segment 8 * | RT‐qPCR | ISAV‐S8−404F | TGGGCAATGGTGTATGGTATGA |
| ISAV‐S8−583R (RA3) | GAAGTCGATGAACTGCAGCGA | ||
| ISAV‐S8−491P | FAM‐CAG GAT GCA GAT GTA TGC‐MGB | ||
| Segment 6 | Sequencing |
ISAV‐S6−321F ISAV‐S6−1130R |
GGACCTGTACCTGGGAGCAT ACAGAGCAATCCCAAAACCTGC |
| Segment 5 | Sequencing |
ISAV S5−1F ISAV‐S5−311F ISAV‐S5−1476R |
AGTTAAAGATGGCTTTTCTAACAATT ATGGAGAACTGTGCAGTGAA TGGCTATTTATACAATTAATAATGCAT |
(Caraguel et al., 2012).
For both cell lines, CPE were as previously described for HPR‐deleted variants, that is rounding of cells and detachment of the monolayer observed for ASK cells (Devold et al., 2000) while vacuoles were visible on SHK‐1 cells (Dannevig et al., 1995) (Figure 1). At 14 dpi, the ASK monolayer was completely destroyed and some SHK‐1 cells were still attached (Figure 1). RT‐qPCR results supported viral growth with an increase in RNA copies at 21 compared with 0 dpi of approximately 4.4 and 4.5 logs for ASK and SHK‐1, respectively. Partial sequencing of S6 by NAAHLS‐GFC confirmed results from RPC and NAAHLS‐FWI, that is a NA HPR0 variant, with 100% identity between sequences obtained. An alignment of HPR0 sequences identified in Atlantic Canada since 2012 is presented in Figure 2. Within NA HPR0 sequences, more than 85% present a LEAQ motif, a typical signature of NA variants and 100% identity for the 113 aa available between them. The HPR0 variant reported here, CA/NS/2021‐31/2020 (NCBI: OK166559), was the most divergent with a PEAQ motif and 7/113 substitutions in total. In comparison with S6, a limited number of S5 sequences are available for NA variants. The alignment of available amino acids sequences is presented in Figure 3. All sequences showed the expected marker, that is Q266 for the HPR0 variants, with no insertion, and L266 or an insertion in proximity of Q266 for the HPRΔ variants. The isolate from this case study showed the expected Q266 but also a two amino acids (NF) insertion (NCBI: OK166560). At the nucleotide level, the 500‐bp S5 sequence is similar at 99.8% to an HPRΔ isolate previously found in Atlantic Canada in 2012 (CA/NS/G0008/2012), which translates into a 1 aa substitution for the 166 aa sequence examined.
FIGURE 1.
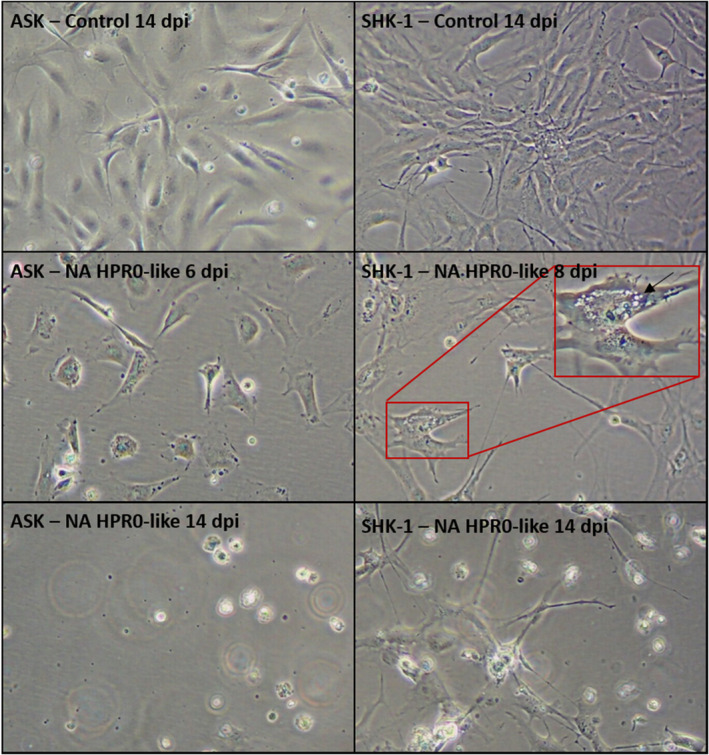
Cytopathic effects (CPE) on ASK and SHK‐1 cell lines inoculated with the isolated HPR0‐like variant (CA/NS/2021‐31/2020) at different time points in days post‐infection (dpi). Images taken at 100X (optical microscope). A close‐up at 200x is given for SHK‐1 cells at 8 dpi to show vacuoles (black arrow)
FIGURE 2.
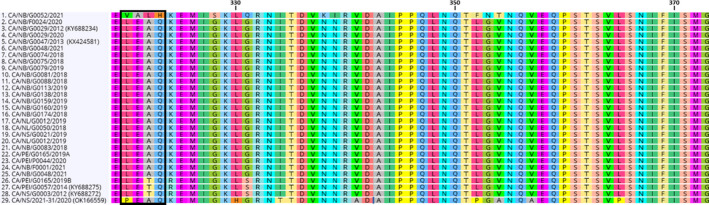
ISAV S6 partial amino acid sequences of the HPR0‐like under study (#29) and other HPR0 variants identified in Atlantic Canada between 2012 and 2021. All NA HPR0 sequences available were included (n = 27 independent cases). A EU HPR0 sequences (#1) were added as a point of comparison to NA variants. Amino acids motif used to distinguish NA from EU variants are boxed (VALH and LEAQ/LETQ/PEAQ for EU and NA variants, respectively). Accessions numbers of corresponding nucleic acid sequence, when available, are indicated between brackets
FIGURE 3.

ISAV S5 partial amino acid sequences of the HPR0‐like under study (#6) and other available variants in Atlantic Canada. Variant #1 is from a EU HPR0 variant. Variant #4 is from a NA HPR0 variant detected in a hatchery in New Brunswick in 2013. Isolates #2, #3 and #5 are from NA HPRΔ variants isolated in 2012 from disease outbreaks in open net pens in Newfoundland (#3) and Nova Scotia (#2 and #5). Accessions numbers of corresponding nucleic acid sequence, when available, are indicated between brackets
The significance of this finding remains unclear until more in‐depth work is completed. This challenges the current assumption that ISAV with no deletion in the HPR of S6 cannot grow in cell culture. To our knowledge, it is also the first report of a HPR0 variant with an insertion in S5. The possibility of a mixed infection was considered but judged unlikely; identical S5 and S6 sequences were repeatedly obtained independently by different laboratories, with clean sequences (i.e. no double peaks or unsatisfying portions). Recent work from Cárdenas and colleagues presented sequencing results for both segments 5 and 6 of 299 ISAV variants (Cárdenas et al., 2019). All HPR0 variants showed a Q266 with no insertion. Almost 98% of HPRΔ isolates presented either a L266 or a Q266 with insertion, but 2.3% had a Q266 with no insertion, suggesting that transition from a non‐virulent HPR0 to deleted HPR and virulence may require modifications of S6 prior or concomitantly to the modification in S5. Results presented here support alternative scenarios and that modification in S5 may be the only requirement to allow replication in cells; however, substitutions in S6 found in this unique isolate or in other un‐sequenced regions of the genome may contribute to the growth in cells. Characterization using controlled in vivo challenge will be necessary to determine whether this isolate can cause disease in Atlantic salmon.
Phylogenetic studies to determine how this variant relates to other known variants will require more S5 sequences, especially for the NA clade. Genotyping through surveillance and diagnostic activities in Canada focusses on S6. Partial sequencing of S5 is in progress on archived diagnostic samples available from 2009 to present. This work will allow phylogenetic studies based on both segments and may help determine how this novel HPR0 variant evolved or if transient variants with some but not both S5 and S6 virulence markers can be identified. Full sequencing will be done for selected variants, based on results. Until a nomenclature system can be revised to include both S5 and S6 information, isolate CA/NS/2021‐31/2020 will be identified as a HPR0‐like variant.
CONFLICT OF INTEREST
The authors declared no potential conflicts of interest with respect to the research, authorship and/or publication of this article.
Supporting information
Supplemental File S1
Supplementary Material
ACKNOWLEDGEMENTS
This work was financed by Fisheries and Oceans Canada and the National Aquatic Animal Health Program (NAAHP). We would like to acknowledge the Canadian Food Inspection Agency (CFIA) for authorizing sequencing of S5 on samples submitted to the NAAHLS, and Tamara Schroeder and the team at NAAHLS‐FWI for providing RNA.
Ditlecadet, D. , Gautreau, C. , Boston, L. , Liston, R. , Johnsen, E. , & Gagné, N. (2022). First report of successful isolation of a HPR0‐like variant of the infectious salmon anaemia virus (ISAV) using cell culture. Journal of Fish Diseases, 45, 479–483. 10.1111/jfd.13556
DATA AVAILABILITY STATEMENT
The data that support the finding of this study are available from the corresponding author upon request.
REFERENCES
- Aamelfot, M. , Christiansen, D. H. , Dale, O. B. , McBeath, A. , Benestad, S. L. , & Falk, K. (2016). Localised infection of atlantic salmon epithelial cells by hpr0 infectious salmon anaemia virus. PLoS One, 11(3), e0151723. 10.1371/journal.pone.0151723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraguel, C. , Stryhn, H. , Gagne, N. , Dohoo, I. , & Hammell, L. (2012). A modelling approach to predict the variation of repeatability and reproducibility of a RT‐PCR assay for infectious salmon anaemia virus across infection prevalences and infection stages. Preventive Veterinary Medicine, 103(1), 63–73. 10.1016/j.prevetmed.2011.08.012 [DOI] [PubMed] [Google Scholar]
- Cárdenas, C. , Ojeda, N. , Labra, Á. , & Marshall, S. H. (2019). Molecular features associated with the adaptive evolution of Infectious Salmon Anemia Virus (ISAV) in Chile. Infection, Genetics and Evolution, 68, 203–211. 10.1016/j.meegid.2018.12.028 [DOI] [PubMed] [Google Scholar]
- Cárdenas, M. , Galleguillos, C. , Acevedo, K. , Ananias, C. , Alarcón, J. , Michelson, S. , Toledo, J. , Montoya, M. , Meneses, C. , Castro‐Nallar, E. , Vásquez‐Martínez, Y. , & Cortez‐San Martin, M. (2020). Rapid sequence modification in the highly polymorphic region (HPR) of the hemagglutinin gene of the infectious salmon anaemia virus (ISAV) suggests intra‐segmental template switching recombination. Journal of Fish Diseases, 43(12), 1483–1496. 10.1111/jfd.13242 [DOI] [PubMed] [Google Scholar]
- CFIA (2021). Locations infected with infectious salmon anemia. Retrieved from https://inspection.canada.ca/animal‐health/aquatic‐animals/diseases/reportable‐diseases/isa/locations‐infected/eng/1549521878704/1549521878969 [Google Scholar]
- Chang, B. D. , Coombs, K. A. , & Page, F. H. (2014). The development of the salmon aquaculture industry in southwestern new brunswick, bay of fundy, including steps toward integrated coastal zone management. Aquaculture Economics & Management, 18(1), 1–27. 10.1080/13657305.2014.855952 [DOI] [Google Scholar]
- Christiansen, D. H. , McBeath, A. J. A. , Aamelfot, M. , Matejusova, I. , Fourrier, M. , White, P. , Petersen, P. E. , & Falk, K. (2017). First field evidence of the evolution from a non‐virulent HPR0 to a virulent HPR‐deleted infectious salmon anaemia virus. Journal of General Virology, 98(4), 595–606. 10.1099/jgv.0.000741 [DOI] [PubMed] [Google Scholar]
- Cook‐Versloot, M. , Griffiths, S. , Cusack, R. , McGeachy, S. , & Ritchie, R. (2004). Identification and characterisation of infectious salmon anaemia virus (ISAV) haemagglutinin gene highly polymorphic region (HPR) type 0 in North America. Bulletin of the European Association of Fish Pathologists, 24(4), 203–208. [Google Scholar]
- Cunningham, C. O. , Gregory, A. , Black, J. , Simpson, I. , & Raynard, R. S. (2002). A novel variant of the infectious salmon anaemia virus (ISAV) haemagglutinin gene suggests mechanisms for virus diversity. Bulletin of the European Association of Fish Pathologists, 22(6), 366–374. [Google Scholar]
- Dannevig, B. H. , Falk, K. , & Namork, E. (1995). Isolation of the causal virus of infectious salmon anemia (ISA) in a long‐term cell‐line from Atlantic Salmon Head Kidney. Journal of General Virology, 76, 1353–1359. 10.1099/0022-1317-76-6-1353 [DOI] [PubMed] [Google Scholar]
- Devold, M. , Karlsen, M. , & Nylund, A. (2006). Sequence analysis of the fusion protein gene from infectious salmon anemia virus isolates: Evidence of recombination and reassortment. Journal of General Virology, 87(7), 2031–2040. 10.1099/vir.0.81687-0 [DOI] [PubMed] [Google Scholar]
- Devold, M. , Krossøy, B. , Aspehaug, V. , & Nylund, A. (2000). Use of RT‐PCR for diagnosis of infectious salmon anaemia virus (ISAV) in carrier sea trout Salmo trutta after experimental infection. Diseases of Aquatic Organisms, 40, 9–18. 10.3354/dao040009 [DOI] [PubMed] [Google Scholar]
- Gagne, N. , & LeBlanc, F. (2018). Overview of infectious salmon anaemia virus (ISAV) in Atlantic Canada and first report of an ISAV North American‐HPR0 subtype. Journal of Fish Diseases, 41(3), 421–430. 10.1111/jfd.12670 [DOI] [PubMed] [Google Scholar]
- Kibenge, F. S. B. , Kibenge, M. J. T. , Wang, Y. , Qian, B. , Hariharan, S. , & McGeachy, S. (2007). Mapping of putative virulence motifs on infectious salmon anemia virus surface glycoprotein genes. Journal of General Virology, 88(11), 3100–3111. 10.1099/vir.0.83097-0 [DOI] [PubMed] [Google Scholar]
- Krossoy, B. , Nilsen, F. , Falk, K. , Endresen, C. , & Nylund, A. (2001). Phylogenetic analysis of infectious salmon anaemia virus isolates from Norway, Canada and Scotland. Diseases of Aquatic Organisms, 44(1), 1–6. 10.3354/dao044001 [DOI] [PubMed] [Google Scholar]
- LeBlanc, F. , Leadbeater, S. , Laflamme, M. , & Gagne, N. (2018). In vivo virulence and genomic comparison of infectious Salmon Anaemia Virus isolates from Atlantic Canada. Journal of Fish Diseases, 41(9), 1373–1384. 10.1111/jfd.12832 [DOI] [PubMed] [Google Scholar]
- Markussen, T. , Jonassen, C. M. , Numanovic, S. , Braaen, S. , Hjortaas, M. , Nilsen, H. , & Mjaaland, S. (2008). Evolutionary mechanisms involved in the virulence of infectious salmon anaemia virus (ISAV), a piscine orthomyxovirus. Virology, 374(2), 515–527. 10.1016/j.virol.2008.01.019 [DOI] [PubMed] [Google Scholar]
- Martin, W. (2007). The way forward: management of infectious Salmon Anemia in the bay of fundy (pp. 106). S.W. Martin Consulting Inc. Report. [Google Scholar]
- Mjaaland, S. , Rimstad, E. , Falk, K. , & Dannevig, B. H. (1997). Genomic characterization of the virus causing infectious salmon anemia in Atlantic salmon (Salmo salar L.): An orthomyxo‐like virus in a teleost. Journal of Virology, 71(10), 7681–7686. 10.1128/jvi.71.10.7681-7686.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE . (2021). Infection with HPR‐Deleted or HPR0 Infectious Salmon Anaemia Virus. In Manual of Diagnostic Tests for Aquatic Animals; (Vol. Chapter 2.3.4). Retrieved from https://www.oie.int/fileadmin/Home/eng/Health_standards/aahm/current/2.3.04_ISAV.pdf [Google Scholar]
- Rimstad, E. , & Markussen, T. (2020). Infectious salmon anaemia virus—molecular biology and pathogenesis of the infection. Journal of Applied Microbiology, jam. 129(1), 85–97. 10.1111/jam.14567 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental File S1
Supplementary Material
Data Availability Statement
The data that support the finding of this study are available from the corresponding author upon request.


