SUMMARY
Plant response to drought stress includes systems for intracellular regulation of gene expression and signaling, as well as inter‐tissue and inter‐organ signaling, which helps entire plants acquire stress resistance. Plants sense water‐deficit conditions both via the stomata of leaves and roots, and transfer water‐deficit signals from roots to shoots via inter‐organ signaling. Abscisic acid is an important phytohormone involved in the drought stress response and adaptation, and is synthesized mainly in vascular tissues and guard cells of leaves. In leaves, stress‐induced abscisic acid is distributed to various tissues by transporters, which activates stomatal closure and expression of stress‐related genes to acquire drought stress resistance. Moreover, the stepwise stress response at the whole‐plant level is important for proper understanding of the physiological response to drought conditions. Drought stress is sensed by multiple types of sensors as molecular patterns of abiotic stress signals, which are transmitted via separate parallel signaling networks to induce downstream responses, including stomatal closure and synthesis of stress‐related proteins and metabolites. Peptide molecules play important roles in the inter‐organ signaling of dehydration from roots to shoots, as well as signaling of osmotic changes and reactive oxygen species/Ca2+. In this review, we have summarized recent advances in research on complex plant drought stress responses, focusing on inter‐tissue signaling in leaves and inter‐organ signaling from roots to shoots. We have discussed the mechanisms via which drought stress adaptations and resistance are acquired at the whole‐plant level, and have proposed the importance of quantitative phenotyping for measuring plant growth under drought conditions.
Keywords: drought stress, abscisic acid (ABA), inter‐tissue signaling, inter‐organ signaling, peptide signals, phenotyping
Significance Statement
This review focuses on inter‐tissue signaling, inter‐organ signaling, and stress sensing in complex drought stress responses and tolerance, in which molecular patterns of stress signals are sensed by different sensors and transmitted to other tissues and organs to induce stress responses in total. Recent progress in quantitative phenotyping under drought conditions is also discussed to understand the whole system of complex drought responses and tolerance.
INTRODUCTION
Environmental conditions change frequently, both rapidly and incrementally, which affect plant growth and productivity. For example, global warming and climate change can cause droughts, water deficit during which negatively affects plant growth and productivity. Therefore, plants must recognize and respond to environmental changes and adapt to water deficits to survive and grow. Plants have evolved sophisticated systems for adaptation to drought stress to maintain optimal growth under water‐deficit conditions (Gupta et al., 2020). Furthermore, they have developed unique and complex mechanisms connecting various organs and tissues to resist severe environmental stresses. The entire plant body is composed of organs, such as roots, leaves, and stems, and each organ consists of tissues such as epidermal or vascular tissue. The analysis of inter‐tissue and inter‐organ communication systems will provide insights regarding plant responses to water‐deficit conditions, in addition to the cellular mechanism underlying plant responses to dehydration stress (Figure 1).
Figure 1.
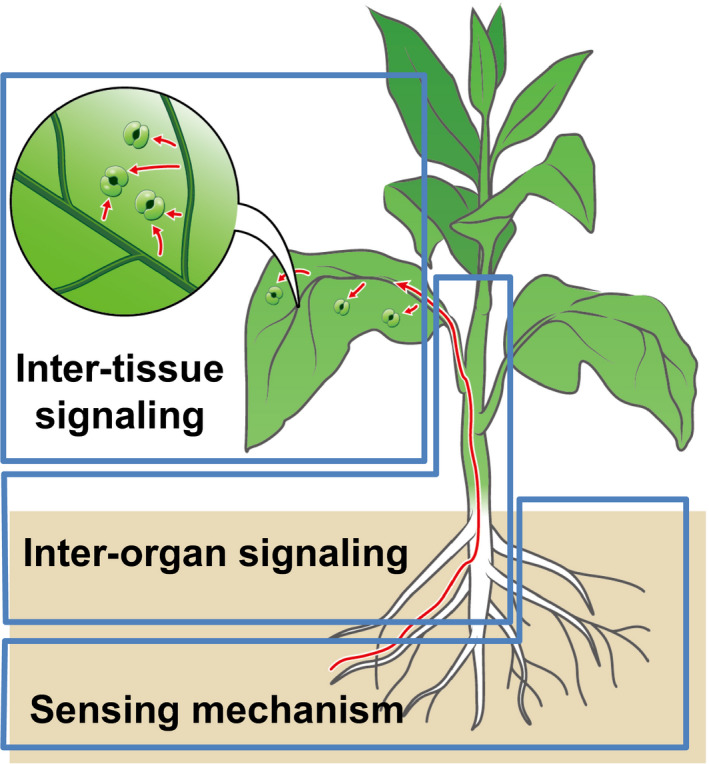
Hierarchy of drought stress responses in plants.
Under drought stress conditions, plants perceive water‐deficit via “sensing mechanisms” in roots. Several types of water‐deficit signals are transmitted from roots to leaves via “inter‐organ signaling.” Then, they are distributed between distal tissues in each organ via “inter‐tissue signaling” to adapt to drought stress; for example, abscisic acid functions as an inter‐tissue signal to close stomata and change gene expression in leaves.
Abscisic acid (ABA), a phytohormone, functions as an inter‐tissue signal in leaves, one of the above‐ground organs. ABA mediates drought stress responses and resistance by regulating stomatal closure and stress‐responsive gene expression. ABA accumulates mainly in the vasculature of leaves, as the enzymes involved in ABA biosynthesis are expressed in vascular tissues (Chen et al., 2020; Kuromori et al., 2018). In addition, several cellular membrane‐localized ABA transporters are predominantly expressed in vascular tissues. Drought‐induced ABA may be transported from the vasculature to tissues to mediate stomatal movements and gene expression in response to drought stress (Hsu et al., 2021; Kuromori et al., 2018; Munemasa et al., 2015). How tissue‐specific synthesis of ABA and ABA transporter networks control the level of ABA and stomatal closure under drought stress conditions have been discussed subsequently.
Water‐deficit signals are transmitted via inter‐organ signaling from roots to leaves to adapt to drought stress. The vascular system of plants connects the roots and shoots, and plays an important role in integrating stress information from underground organs to aerial organs. Hydraulic signals, Ca2+ waves, electric currents, and reactive oxygen species (ROS) mediate drought stress inter‐organ responses, as well as cellular responses (Hsu et al., 2021; Kollist et al., 2019; Takahashi et al., 2020; Zhu, 2016). Hormone‐like peptides act as signaling molecules that mediate inter‐organ stress responses (Gupta et al., 2020; Kim et al., 2021; Li et al., 2021; Takahashi et al., 2018b, 2019, 2020; Thomas and Frank, 2019). These findings suggest that peptides transported by the vasculature integrate water‐deficit stress signals for whole plant‐level communication. Here, we have reviewed the mechanism via which inter‐organ signaling mediates drought stress responses and resistance in plants and the signaling molecules involved.
Under drought stress conditions, plants sense changes in the water‐deficit status of their roots. Drought stress sensing systems are complex. They are stimulated by various stress signals, such as osmotic, ROS, and mechanical stresses, and involve numerous sensing molecules (Takahashi et al., 2020; Yoshida et al., 2021; Zhu, 2016). Environmental stresses are sensed by multiple sensing systems as molecular patterns of stress stimuli, which are transmitted to various tissues to induce specific and sequential stress responses for proper adaptation of plants to complex environmental stresses. We will subsequently discuss the importance of sensing factors of stress signals for correct responses not only at the cellular level, but also at the whole‐plant level. The sensors and signaling patterns mediate the physiological responses of plants to drought stress after sensing the water‐deficit status and downstream stress responses.
The analysis of plant growth under water‐deficit conditions is important for understanding plant responses to drought stress at the whole‐plant level. Toward this, imaging and information technologies have been developed based on artificial intelligence (Dhondt et al., 2013; Mochida et al., 2020; Singh et al., 2016, 2021). Furthermore, quantitative phenotyping enables the analysis of physiological plant responses and assessment of plant water‐use efficiency and drought resistance not only under controlled growth conditions, but also in the greenhouse and field (Singh et al., 2021). In this review, we have described recent advances in plant and crop phenotyping. These new technologies are expected to enhance our understanding of plant responses to drought stress.
SYNTHESIS AND TRANSPORT OF ABA AND ROLES OF OTHER HORMONES IN INTER‐TISSUE STRESS SIGNALING
Under drought conditions, ABA induces responses that help plants cope with water deficit. As an early drought stress factor, ABA induces guard cells to close the stomata and prevent water shortage. The enzymes involved in ABA biosynthesis are expressed in vascular tissues distant from the guard cells. Therefore, ABA must be transported from vascular tissues to guard cells. Biosynthesis, degradation, and modification of ABA have been investigated (Chen et al., 2020), and various types of ABA transporters have been reported (Takahashi et al., 2020). In response to dehydration, ABA is also rapidly perceived by the guard cells, where it triggers stomatal closure. However, the mechanisms underlying ABA synthesis, metabolism, and transport in drought stress responses are unclear. In this section, we have discussed the ABA synthesis sites, ABA‐mediated inter‐tissue signaling, and function of other hormones in leaves.
Sites of ABA synthesis in stress responses
Control of the water levels in plant bodies is an important environmental response. Being sessile, higher plants sense water deficiency via roots, which are underground organs. This information is transmitted to shoots, including the leaves, which are above‐ground organs, via root‐to‐shoot inter‐organ signals (Christmann et al., 2013; Schachtman and Goodger, 2008) (Figure 2).
Figure 2.
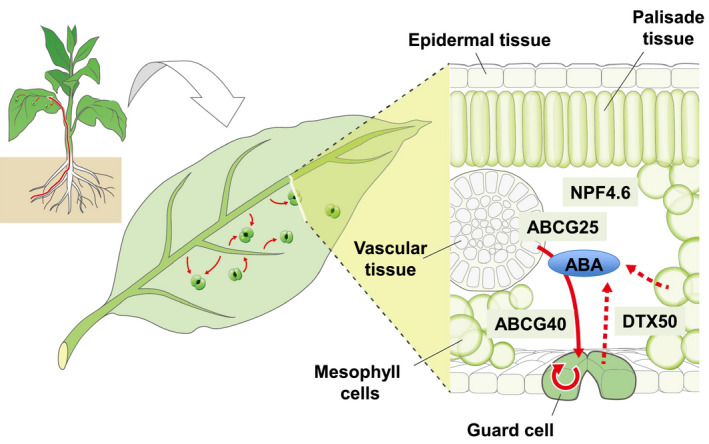
Abscisic acid (ABA) functions as an inter‐tissue signal in leaves.
Leaf cross‐section showing vascular tissues (sites of ABA biosynthesis) and guard cells (ABA action sites). In Arabidopsis, the ABA transporters ABCG25, ABCG40, NRF4.6, and DTX50, are expressed in vascular cells and/or guard cells. In the leaf section, the movement of ABA is indicated by solid and dashed arrows.
ABA is believed to be a major root‐to‐shoot inter‐organ signal. After plants sense water deficiency, ABA is synthesized in the roots and transported to the shoots via xylem flow (Wilkinson and Davies, 2002). However, shoots can also be a source of ABA under stress (Christmann et al., 2007). Foliage‐derived ABA affects drought stress considerably, with leaves being the predominant location of ABA biosynthesis (McAdam et al., 2016).
ABA is synthesized from precursors in plastids via sequential enzymatic reactions. The final three enzymes in the ABA biosynthesis pathway are encoded by NCED3, ABA2, and AAO3 (Finkelstein, 2013). NCED3 is strongly induced by drought stress, mainly in the vasculature of leaves (Endo et al., 2008), whereas ABA2 and AAO3 are expressed under both non‐stress and stress conditions (Cheng et al., 2002; Koiwai et al., 2004; Kuromori et al., 2014a). Although their expression patterns vary, these three genes are predominantly expressed in vascular tissues, which are the major sites of ABA biosynthesis. Drought‐inducible NCED3 is regulated by the NGA1 transcription factor, which contains a B3 domain (Sato et al., 2018). NCED3 induction contributes to ABA accumulation in leaves, inducing not only stomatal closure, but also the expression of stress genes.
Guard cells are also sites of ABA biosynthesis and may autonomously support the ABA responses required for stomatal closure (Bauer et al., 2013). They are sensitive to changes in aerial humidity because of their location in the leaf epidermis. In addition, mesophyll cells were found to be the sites of ABA biosynthesis in water‐stressed leaves (McAdam and Brodribb, 2018). In addition to vascular tissues, guard cells and other cells also produce ABA. However, the mechanism via which ABA biosynthesis is coordinated across various tissues in the leaves remains unclear.
In addition to de novo biosynthesis, ABA is produced via the hydrolysis of ABA glucose ester (ABA‐GE) by β‐glucosidase. ABA‐GE is a reversibly inactive form of ABA and is believed to act as a storage form of ABA (Lee et al., 2006). ABA‐GE hydrolysis occurs locally in the epidermis of leaf petioles during the early stages of drought stress (Han et al., 2020). Although the physiological effect of ABA‐GE on drought stress responses is unclear, it may contribute to ABA production in leaves (Hussain et al., 2020).
ABA is rapidly catabolized by CYP707As once stress is released. A member of this family, CYP707A3, is predominantly expressed in vascular tissues and regulates ABA accumulation in leaves (Umezawa et al., 2006). Another member, CYP707A1, is preferentially expressed in guard cells and contributes to leaf ABA content, with mutant analyses showing that it regulates the stomatal aperture similar to CYP707A3 (Okamoto et al., 2009). These results also indicated that site‐specific ABA metabolism might be related to ABA responses in leaves.
Inter‐tissue ABA transport under stress conditions
According to a theory regarding the importance of leaf‐derived ABA over root‐derived ABA, after sensing water depletion in roots, root‐derived signals other than ABA are transmitted to the shoots, including leaves (Kuromori et al., 2014b). Root‐to‐shoot signals induce ABA biosynthesis in leaves by activating NCED3 expression in vascular tissues, which is the rate‐determining step in ABA biosynthesis (Endo et al., 2008). This suggests that root‐to‐shoot signaling information is transferred via the vascular tissues.
ABA acts as a major inter‐tissue signaling factor in leaves under water stress. In leaves, ABA is transferred to guard cells to close the stomata for preventing water loss. This is followed by changes in gene expression patterns in other cells to cope with water‐deficit stress in tissues (Takahashi et al., 2018a; Yoshida et al., 2021). In general, the sites of action of hormones are distant from their sites of biosynthesis. To act as an inter‐tissue signaling molecule in leaves, ABA is transported to the target guard cells. Indeed, ABA membrane transporters have also been identified (Figure 2).
In Arabidopsis, four membrane proteins, AtABCG25, AtABCG40, AtNPF4.6, and AtDTX50, function as ABA transporters related to ABA inter‐tissue signaling in leaves (reviewed by Kuromori et al., 2018; Shimizu et al., 2021) and are localized to the plasma membrane, indicating that ABA can be exported out of the cells where it is synthesized and imported into cells where it is sensed via the ABA receptor. In addition, mutants of each gene exhibited ABA‐related phenotypes. ABA transporters are regulated at the post‐translational level; for example, the plasma membrane localization of AtABCG25 is regulated by abiotic stress, ABA (Park et al., 2016), and phosphorylated AtNPF4.6, which relocates from the plasma membrane to the membrane of the endoplasmic reticulum (Zhang et al., 2021). In addition, AtABCG22 and AtABCG21 may function in stomatal regulation, although whether they are directly associated with ABA transport across membranes is unclear (Kuromori et al., 2011, 2017).
These transporter genes are mainly expressed in vascular tissues and/or guard cells, corresponding to the sites of ABA synthesis and action, respectively. Furthermore, ABA membrane transporters are categorized as ABA exporters, which mediate ABA export, and ABA importers, which mediate ABA import. ABA is imported into cells for cytosolic proteins that function as ABA receptors responsible for stomatal closure (Ma et al., 2009; Park et al., 2009). Therefore, ABA transport is regulated by membrane transporters in leaf inter‐tissue networks (Kuromori et al., 2018).
Other ABA transporters also control seed germination (Kang et al., 2015). ABA membrane transporters have been identified in non‐model plant species, such as legumes, wheat, and rice (Takahashi et al., 2020). Many types of membrane transporters are involved in ABA transport.
Roles of other hormones in drought stress responses and tolerance
Hormones are inter‐tissue signals that regulate plant growth under various environmental conditions. ABA, as well as various other phytohormones, are implicated in drought stress tolerance. For example, similar to ABA, methyl jasmonate induces stomatal closure by elevating pH, and the levels of ROS, nitric oxide, and Ca2+, leading to the activation of anion channels (Bharath et al., 2021). Jasmonate is involved in the crosstalk between abiotic and biotic stress responses (Huang et al., 2017). Brassinosteroid and auxin responses are related to leaf and root growth under drought conditions (summarized by Gupta et al., 2020). Downstream components of brassinosteroid signaling act by activating ABA signaling. Independent of ABA, brassinosteroid receptors modulate hydrotropic responses in roots and coordinate plant growth and survival under drought stress by promoting the accumulation of osmoprotectant metabolites. In addition, non‐canonical auxin responses modulate root architecture patterning and depth to boost water absorption from the soil, thereby improving drought tolerance (Gupta et al., 2020). In Arabidopsis, auxin‐sensitive Aux/IAA proteins mediate drought tolerance by upregulating glucosinolate levels. The AUX/IAA repressors IAA5, IAA6, and IAA19 are involved in the maintenance of glucosinolate levels when plants are dehydrated. Glucosinolate may function in drought stress responses via ROS, suggesting links between auxin signaling, glucosinolate levels, and drought tolerance (Salehin et al., 2019).
Cytokinin is also involved in drought acclimation/adaptation and in stabilizing plant yield under drought conditions (Hai et al., 2020; Li et al., 2016; Nishiyama et al., 2011). Strigolactone and similar signaling molecules, such as karrikin, function in stomatal responses under conditions of dehydration. For example, shoot‐produced strigolactone induces SLAC1‐dependent stomatal closure by triggering the production of H2O2 and nitric oxide in guard cells. Moreover, these signaling molecules indirectly affect stomatal closure by positively regulating ABA sensitivity in guard cells (Cardinale et al., 2018). Karrikin and strigolactones function in hormone crosstalk related to drought stress resistance (Li et al., 2017; Mostofa et al., 2018).
The existence of multiple phytohormone functions is indicative of complex plant responses to drought stress in different plant tissues. The terminal phenotypes of drought resistance are achieved via various hormone‐regulatory systems. This reflects the involvement of brassinosteroids, cytokinins, auxin, strigolactones, and karrikin in drought stress resistance, in addition to ABA. Recent reviews have addressed this issue (Hai et al., 2020; Sirko et al., 2021).
INTER‐ORGAN SIGNALING IN DROUGHT RESPONSES
Higher plants have developed inter‐organ communication systems that involve various signaling molecules induced by environmental changes. These inter‐organ signals are transmitted via the vasculature to integrate stress responses at the whole‐plant level. Xylem and phloem are important vascular tissues that participate in inter‐organ signaling (Li et al., 2021) (Figure 3). Under drought stress, reduction in water potential in roots informs the plant of soil water deficit (Gupta et al., 2020; Li et al., 2021; Takahashi and Shinozaki, 2019). The water deficit signal is transmitted from the roots to the leaves via the vasculature. Ca2+ fluxes, turgor loss, and ROS production are involved in the perception and signaling of dehydration stress responses in the vasculature and guard cells (Chen et al., 2020; Kollist et al., 2019; Soma et al., 2021; Yoshida et al., 2021; Zhu, 2016). After long‐term dehydration, plants accumulate ABA to maintain stomatal closure and induce stress proteins and metabolites to protect organs from dehydration (Thomas and Frank, 2019). Inter‐organ signaling molecules, including peptides and metabolites, activate ABA production at different stages of drought stress response (Li et al., 2021; Takahashi and Shinozaki, 2019; Yoshida et al., 2021). Stepwise responses, including inter‐organ communication, are important for evaluating spatiotemporal drought responses at the whole‐plant level.
Figure 3.
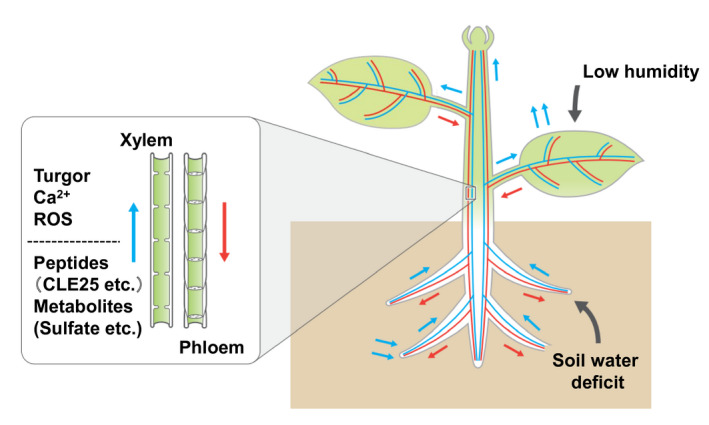
Inter‐organ signaling via the vasculature in drought stress responses.
Various inter‐organ signals are transmitted via the vasculature as part of drought stress response. Xylem and phloem are important for inter‐organ signaling. Soil water deficit and low humidity may induce drought stress responses in roots and guard cells, respectively. Osmotic changes, reactive oxygen species (ROS), and Ca2+ transients function in stress signaling from roots to leaves. Peptides and metabolites are synthesized in roots in response to drought stress and transported via the xylem to leaves. Among them, the roles of CLE25 peptide have been precisely analyzed (Takahashi et al., 2018b).
Signaling in early drought responses involving Ca2+ and ROS
Land plants sense water deficiency mainly in the roots by monitoring the water potential in root vasculature. The hydraulic stress signal from root cells is transmitted to the leaves to mediate stomatal closure and promote stress‐inducible gene expression to protect plants from dehydration (Christmann et al., 2013). Ca2+ flux across the plasma membrane is activated by various stress signals, including osmotic stress, and Ca2+ waves rapidly transmit stress information to distant tissues (Konrad et al., 2018). OSCA1 (REDUCED HYPEROSMOLALITY INDUCED Ca2+ INCREASE 1), which encodes a plasma membrane protein, functions as a Ca2+ influx channel during osmotic stress response (Choi et al., 2017; Murthy et al., 2018; Yuan et al., 2014) (Figures 3 and 4). osca1 mutant plants do not regulate stomatal closure and lose more water than wild‐type plants. However, stomatal closure is induced by ABA in osca1 mutant plants, suggesting a role for OSCA1 in stomatal closure induced by osmotic stress‐mediated Ca2+ influx into the stomata. OSCA1.2/CALCIUM‐PERMEABLE STRESS‐GATED CATION CHANNEL 1 (CSC1) functions as a hyperosmolality‐gated Ca2+‐permeable channel protein that mediates Ca2+ influx (Hou et al., 2014; Liu et al., 2018). Fifteen OSCA family genes have been identified across four clades in Arabidopsis (Yuan et al., 2014). Among them, OSCA1.3 and OSCA1.7 are involved in stomatal immunity downstream of the plasma membrane‐associated cytosolic kinase, BIK1, which is implicated in PAMP‐induced Ca2+ influx and stomatal closure (Thor et al., 2020). Therefore, OSCA family membrane proteins mediate Ca2+ influx in stress responses and are regulated downstream of receptor‐like kinases. Stretch‐activated Ca2+ channels are other candidate drought stress‐sensing factors. In plants, Ca2+‐permeable mechanosensitive channels (MCA) 1 and ‐2 sense osmotic changes in the plasma membrane to mediate Ca2+ influx and activate downstream cellular signaling (Nakagawa et al., 2007; Nishii et al., 2021). MCA1 is a homolog of yeast MID1, a Ca2+‐permeable stretch‐activated channel component. MCA1 and ‐2 are candidate osmosensors that respond to drought stress (Yoshimura et al., 2021).
Figure 4.
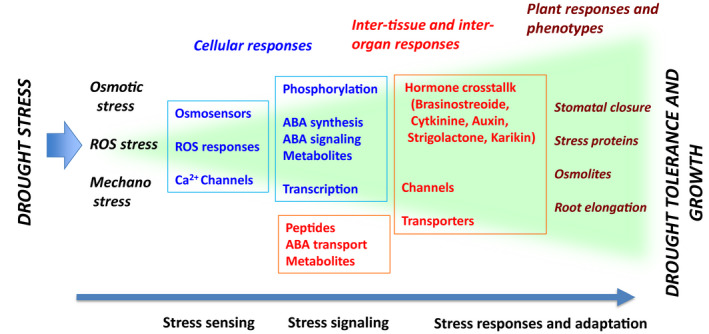
Drought stress signaling network from the perception of stress signals to cellular, inter‐organ, and whole‐plant responses and the acquisition of tolerance.
Drought imposes water‐deficit stress on plants at the cellular, organ, and whole‐plant levels. Water deficit induces osmotic stress, oxidative stress, and mechanical stress, which are sensed by osmosensors, reactive oxygen species (ROS) sensors, and Ca2+ channels. These intra‐ and inter‐tissue stress signals are mediated by phosphorylation, abscisic acid (ABA), and metabolites. Inter‐organ signaling molecules (peptides such as CLE2 and metabolites such as sulfate) are transported between tissues and organs. Stress signals regulate channels, transporters, transcription factors, and hormones to induce stomatal closure, stress‐responsive gene expression, and osmolyte/stress‐protein synthesis to prevent severe dehydration. After long‐term water‐deficit stress, phenotypic changes in plants such as drought tolerance and growth delay can be monitored precisely using quantitative phenotyping.
Oxidative stress signaling is also involved in the systemic response to various environmental stresses. In response to high light and/or heat stress, ROS function not only as local signals in cellular stress response, but also as systemic signals to induce stress responses in remote tissues. Under extreme drought conditions, heat, high light, and water deficit stresses can be severe, suggesting that oxidative stress signaling is induced by drought conditions (Figures 2 and 4). ROS waves are induced by respiratory burst oxidase D (RBOHD) and regulate stomatal closure in stressed leaves. Systemic ROS signaling coordinates responses in leaves to extreme abiotic stresses (Kollist et al., 2019; Zandalinas et al., 2020). HYDROGEN‐PEROXIDE‐INDUCED Ca2+ INCREASE (HPCA) is an H2O2 sensor in guard cells (Wu et al., 2020). HPCA1 is a leucine‐rich repeat receptor‐like kinase (LRR‐RLK), the extracellular domain of which is activated by H2O2 to induce Ca2+ influx into guard cells during stomatal closure. HPCA1 is identical to CANNOT RESPOND TO DMBQ 1 (CARD1), a receptor for quinone perception during haustorium formation in parasitic plants (Laohavisit et al., 2020). HPCA1/CARD1 of the LRR‐RLK subclass VIII‐1 may perform multiple functions in stress responses.
In early drought stress responses, several osmosensing systems recognize dehydration. Moreover, sensing systems in different organs, such as the roots, stomata, and vasculature, may recognize plant dehydration. Early drought stress signals can be recognized by multiple sensing systems based on Ca2+ influx and ROS production to activate stress signaling networks, including phosphorylation and downstream gene expression. Molecular patterns of drought stress conditions are important for the recognition of drought conditions to promote plant survival under changing environmental conditions (Figures 3 and 4).
Roles of peptides in inter‐organ and local dehydration stress signaling
Small peptides function as signaling molecules in plant stress responses. Many novel genes encoding small peptides and non‐coding RNAs have been identified (Takahashi et al., 2019). Proteomic analyses of Arabidopsis have revealed novel small peptides predicted to function as regulatory factors in plant development and environmental responses, which are classified as members of the families CLAVATA/EMBRYO‐ SURROUNDING REGION (CLE) and C‐TERMNAL ECODED PEPTIDES (CEP), as well as peptides containing sulfated tyrosine (PSY) (Oh et al., 2018; Ohkubo et al., 2017; Okamoto et al., 2015). CLE peptides are conserved in the plant kingdom (Boschiero et al., 2019; Fletcher, 2020; Goad et al., 2017; Takahashi and Shinozaki, 2019; Whitewoods, 2020). CLE peptides are synthesized as long propeptides, which are then processed into mature peptides of 12–14 amino acids. CLAVATA3 (CLV3) is a key regulator of shoot apical meristem development. In Arabidopsis, the CLE peptide family consists of 27 members that perform diverse functions in development and environmental responses. Among them, CLE25 mediates inter‐organ signaling from roots to shoots in drought stress responses. CLE25 is mainly expressed in the root vasculature and is upregulated in response to dehydration in root tissues (Figure 3). The cle25 mutant exhibits a drought‐sensitive phenotype and open stomata. In response to CLE25, NCED3 expression is induced in the roots and ABA accumulates in the leaves (Takahashi et al., 2018b). Grafting experiments revealed that CLE25 is transported from the roots to the shoots via the vasculature. Two LRR‐RLKs, BARELY ANY MERISTEM (BAM) 1 and 3, recognize CLE25 in leaves and induce NCED3 expression to close the stomata. Root‐derived CLE25 is an inter‐organ signaling factor that is transmitted via the xylem from roots to shoots and maintains the drought stress response in leaves (Figure 3) by maintaining high ABA levels in leaves. The processes upstream of CLE25 expression in the root vasculature and downstream of BAM1/BAM3 kinase activity have to be elucidated to understand long‐term responses to dehydration in roots. The NGA1 transcription factor, which has a B3 domain, regulates the dehydration‐induced expression of NCED3 (Sato et al., 2018). NGA1‐mediated induction of NCED3 expression may be activated by CLE25. These sequential processes are important for adaptation to long‐term dehydration and maintenance of stress resistance (Figure 3). Vasculature is important for inter‐organ signaling in the regulation of whole‐plant responses (Li et al., 2021). Xylem and phloem transmit mobile signals to distant organs, such as roots and leaves (Figure 3), and mediate upstream and downstream transport of signaling molecules, respectively. In xylem tissue, CLE25 is transported via a transport pattern that differs from that of CLE26 (Endo et al., 2019), indicating the existence of a CLE25‐specific xylem transporter in Arabidopsis. It is necessary to identify the mechanism via which CLE25 and other peptides are transported from the roots to the leaves in response to dehydration stress. Furthermore, the role of the vasculature in coordinating the response to water‐deficit stress status at the whole‐plant level should be investigated.
Other peptides have been shown to be involved in plant drought responses. CLE9 and CLE10 mediate dehydration stress responses in guard cells to regulate stomatal closure. Mitogen‐activated protein (MAP) kinases and SnRK2s are responsible for CLE9‐mediated stomatal closure (Zhang et al., 2019). In addition, CLE9 and CLE10 promote the proliferation of precursors of guard cells and xylem (Qian et al., 2018). Therefore, the tissue‐specific expression of CLE9/CLE10 is critical for their function in environmental responses and development. The genes encoding phytosulfokine precursor (proPSK) and subtilisin‐like protease (SBT) are upregulated in response to osmotic stress (Stuhrwohldt et al., 2021). Overexpression of proPSK and SBTs improved osmotic stress tolerance. SBT3.8 is involved in the posttranslational processing of proPSKs to bioactive PSKs, which also improved drought stress tolerance. Drought‐induced flower drop in tomatoes is regulated by PSK (Reichardt et al., 2020). Mature PSK is formed in response to drought stress by phytaspase 2, an SBT, which then acts in the abscission zone to induce cell wall hydrolases involved in abscission. In rice, OsDSSR1, which encodes a small peptide, has been shown to function in drought tolerance (Cui et al., 2018). Overexpression of OsDSSR1 enhances drought tolerance by inducing the accumulation of compatible osmolytes, superoxide dismutase, and ascorbate peroxidase activities. These recent reports suggest the involvement of different types of small peptides in drought stress response and tolerance.
The Arabidopsis genome contains >7000 small open reading frames (sORFs) and sequences for non‐coding RNAs. Transcriptome analyses have shown that the predicted sORFs are expressed under various environmental stresses, such as drought, heat, salinity, and cold stress, as summarized in the HanaDB database (Hanada et al., 2010, 2013; Takahashi et al., 2019). These stress‐responsive sORFs were analyzed by overexpressing them in transgenic Arabidopsis. One sORF, named Arabidopsis plant elicitor peptide3 (AtPep3), mediates salt‐stress resistance (Nakaminami et al., 2018). Further analyses of the sORFs regulated by environmental stress will provide insights regarding the functions of various peptides in Arabidopsis. Moreover, some non‐coding RNA genes have been reported to encode peptides (Lauressergues et al., 2015; Ren et al., 2021). Therefore, the functions of these predicted peptide‐encoding genes have to be analyzed. In maize, sORFs and small peptides (sPeptides) were systematically surveyed based on genomic and mass spectrometry data (Liang et al., 2021). Based on these systematic analyses, 9338 sORFs 3–300 nucleotides in length and 2695 sPeptides were identified in the maize genome. PlantPepDB is a manually curated plant peptide database (Das et al., 2020) consisting of 3848 peptides, of which 2821 are experimentally validated at the protein level, 458 at the transcript level, 530 at the predicted level, and 39 based on homology. PlantPepDB is a useful database for comprehensive information on plant peptides.
Signaling molecules involved in inter‐organ signaling in drought stress responses
Various inter‐organ signaling molecules are important for growth, development, and biotic and abiotic interactions (Li et al., 2021; Thomas and Frank, 2019). Grafting experiments have identified inter‐organ signaling molecules, including proteins, peptides, RNAs, and metabolites (Kurotani and Notaguchi, 2021; Thomas and Frank, 2019). Complex environmental stress signals are transmitted by multiple peptides, proteins, miRNAs, and mRNAs (Figure 3).
Soil drying activates the root‐to‐shoot transport of sulfate. Sulfate, a component of cysteine, induces ABA biosynthesis by activating NCED3 (Batool et al., 2018). The stomata of the nced3 mutant did not close after the application of sulfate or cysteine. Sulfate activates NADPH oxidases to induce ROS, which triggers stomatal closure. These findings suggested that sulfate uptake in roots is an inter‐organ signal that activates ABA biosynthesis and stomatal closure in leaves. Cysteine derived from sulfate is involved in ABA synthesis and stomatal closure. The role of sulfate and cysteine in response to soil drying warrants further investigation.
SENSING OF WATER‐DEFICIT STRESS IN ORGANS
Water‐deficit stress signals are sensed in the sensitive tissues of leaves and roots. In leaves, the dehydration status is sensed by stomata to control plant water status. Stomatal closure decreases water loss from leaves under drought stress conditions and significantly represses CO2 uptake to inhibit photosynthesis. In contrast, open stomata control leaf temperature by modulating evaporation. In roots, dry soil causes water deficiency and activates plant drought stress responses. Water deficiency is sensed by root cells, which then initiate drought stress responses (Christmann et al., 2013; Li et al., 2021). In this section, we will discuss the sensing of water‐deficit stress in the stomata and roots.
Signaling crosstalk in stomatal regulation
Stomata play important roles in CO2 and O2 exchange during photosynthesis. Moreover, stomata manage the water status of plants. Stomatal responses are regulated by various environmental stimuli, such as water status, light intensity and wavelength, humidity, CO2 level, and pathogen infection (Hsu et al., 2021; Yoshida et al., 2021). Furthermore, environmental stresses such as ABA, ROS, and Ca2+ were found to regulate stomatal responses. ABA induces rapid stomatal closure via the canonical ABA receptor signaling machinery in response to early stages of drought stress. This machinery includes the PYR/PYL/RCSR ABA receptor, protein phosphatase 2C (ABI homolog), and SnRK2 protein kinases, which activate downstream transporters to regulate stomatal responses. The ABA receptor‐signaling machinery has been reviewed elsewhere (Chen et al., 2020; Cutler et al., 2010). Ca2+/ROS signals also regulate stomatal closure by modulating the activity of channel proteins, including SLAC1 and KAT1, as reviewed by others (Hsu et al., 2021; Yoshida et al., 2021).
Sensing of water‐deficit status in the roots and vasculature
The water‐deficit status of soil is sensed in roots, of which vascular tissues respond to hydraulic stress and low water potential. Hydraulic stress signaling in roots is believed to be an early drought stress response (Figures 3 and 4). However, the mechanism via which hydraulic stress is sensed in the roots is unclear. Histidine kinases function as osmosensors in yeast and bacteria, including cyanobacteria. In yeast, Sln1p, an osmosensor, acts upstream of the HOG1 MAP kinase pathway. In Arabidopsis, histidine kinase 1 (ATHK1/AHK1) is a functional homolog of yeast Sln1p, which also functions as an osmosensor (Urao et al., 1999). Analyses of mutant and overexpression strains have revealed that ATHK1/AHK1 positively regulates drought stress tolerance (Tran et al., 2007; Wohlbach et al., 2008). However, ABA responses or stomatal closure was not affected in ahk1 mutant (Kumar et al., 2013; Sussmilch et al., 2017). These inconsistent data indicated the existence of complex osmosensing systems in plants, which include AHK1 and its downstream signaling pathway, along with MAP kinases. Plant MAP kinases are activated by abiotic and biotic stress signals to control downstream events, including gene expression and physiological responses (Lin et al., 2021). In contrast, AHK2, AHK3, and AHK4 (CRE1) are cytokinin receptors that function as negative regulators of drought response (Nishiyama et al., 2013). AHK4 (CRE1) complements yeast snl1 mutants in the presence of cytokinin, indicating its osmosensing ability (Reiser et al., 2003).
Root hydrotropic response is important for avoiding dry soil and obtaining water for growth. MIZU‐KUSSEI1 (MIZ1), involved in hydrotropic response, is expressed in the epidermis, cortex, and lateral root cap (Dietrich et al., 2017; Kobayashi et al., 2007; Moriwaki et al., 2012, 2013; Takahashi et al., 2002). MIZ1 is induced by light in the root cap, and its expression is significantly low in plants harboring the mutant HY5 transcription factor (Lee et al., 2007). HY5 functions as an inter‐organ signal from shoots to roots, mediates light‐responsive root growth (Chen et al., 2016), and stimulates the MIZ1‐mediated hydroponic response to increase water uptake. The role of HY5 in drought stress avoidance requires further analysis. MIZ1 also functions in the regulation of inter‐organ Ca2+ signaling in root tip cells (Shkolnik et al., 2018).
Root growth, morphogenesis, and architecture are genetically regulated for the efficient absorption of water and nutrients from soil. The long and deep phenotype of roots is regulated via quantitative traits, and the DEEPER ROOTING1 (DRO1) locus has been analyzed to identify the related genes in rice (Uga et al., 2013). Root branching is associated with water availability and seed production, a phenotype known as hydropatterning. Rice DRO1 encodes an unknown factor related to auxin signaling. In Arabidopsis, AUXIN RESPONSE FACTOR7 (ARF7) initiates lateral root growth. On the dry side of roots, ARF7 is modified by a small ubiquitin‐like modifier (SUMO) and inactivated by the repressor IAA3, triggering hydropatterning (Orosa‐Puente et al., 2018; Yoshida et al., 2021).
The vasculature transports nutrients and signals from the roots to the shoots. Water deficit is sensed in the vasculature with a reduction in the water potential (Figure 3). Xylem, phloem, phloem companion cells, and epidermal cells recognize the reduction in water potential caused by drought stress (Endo et al., 2019; Li et al., 2021; Lucas et al., 2013). However, it is not known which molecules in these cells of the vascular tissues that sense the reduction in water potential.
The time course of drought stress responses, their molecular networks in complex stress signaling, and drought tolerance phenotypes are schematically described in Figure 4. Spatiotemporal responses to drought stress are complex processes involving molecular patterns of different types of stress signals, including cellular and inter‐organ responses induced by osmotic, ROS, and mechanical stresses, which are integrated to induce drought stress responses and tolerance at the whole‐plant level.
HIGH‐THROUGHPUT PHENOTYPING OF DROUGHT‐STRESS RESPONSES AND TOLERANCE
In the previous sections, we have discussed inter‐tissue and inter‐organ signaling, and sensing in drought tolerance. To understand how stress resistance is acquired at the whole‐plant level via signal transduction at various spaces and scales, it is necessary to observe the whole‐plant phenotype over time in a system that allows precise and highly reproducible analysis. Therefore, this section introduces high‐throughput phenotyping technologies, which have developed at a rapid pace in recent years, to understand comprehensively how signaling at various levels in drought stress response affects the whole plant. In addition, high‐throughput phenotyping techniques in the laboratory under controlled conditions, as well as those in the field under highly uncertain conditions, will be discussed.
High‐throughput phenotyping of drought stress responses and tolerance in the laboratory
Environmental factors in the field are complex and unpredictable, necessitating laboratory studies under controlled environmental conditions. High‐throughput phenotyping has progressed rapidly, resulting in automation and non‐destructive analyses (Dhondt et al., 2013). In addition, non‐destructive analysis, such as imaging analysis, can be combined with machine and deep learning to improve accuracy and efficiency (Li et al., 2020; Singh et al., 2016, 2021). The analysis of the resulting big data has been enhanced by various innovations (Tardieu et al., 2017). In addition, multiomics analysis, which integrates data from multiple omics methods, is progressing (Mochida et al., 2020).
In this subsection, we have first focused on high‐throughput phenotyping of Arabidopsis thaliana, which is essential for the analysis of systems regulating drought stress responses and tolerance. Automated phenotyping platforms for A. thaliana, such as PHENOPSIS (Granier et al., 2006), WIWAM (Skirycz et al., 2011), Phenoscope (Tisne et al., 2013), Phenovator (Flood et al., 2016), and RIPPS (Fujita et al., 2018) have been developed.
The PHENOPSIS platform (INRA, France) has been used to identify loci involved in natural variation and water‐deficit responses in Arabidopsis accessions and inbred lines (Bac‐Molenaar et al., 2016; Ghandilyan et al., 2009; Rymaszewski et al., 2017; Schmalenbach et al., 2014; Tisne et al., 2010; Vasseur et al., 2014; Vile et al., 2012), and to evaluate the function of genes such as RD20 (Aubert et al., 2010) and SMR1 (Dubois et al., 2018) in drought stress. PHENOPSIS also enables the investigation of plant–microbe and abiotic–biotic stress interactions (Berges et al., 2018, 2020; Bresson et al., 2013).
WIWAM (VIB, Belgium) has been used to study the response of Arabidopsis to mild drought stress (Clauw et al., 2015, 2016; Dubois et al., 2017). The comparative analysis of Arabidopsis and its drought‐tolerant relatives revealed that important differences in drought responses among Brassica plants are likely to occur in downstream signaling and response networks rather than in initial water deficit‐sensing mechanisms (Marín‐de la Rosa et al., 2019).
The Phenovator is a benchtop high‐throughput photosynthesis phenotyping platform that has been used in studies on Arabidopsis. Unlike PHENOPSIS and WIWAM, rockwool blocks, instead of soil‐filled pots, are used for hydroponics, facilitating the analysis of regulatory systems responsible for drought stress responses in up to 1440 Arabidopsis plants (Flood et al., 2016). Benchtop phenotyping platforms may not be able to reproduce phenotypes due to uncontrolled noise sources, leading to micro‐ and macro‐environmental variability (Massonnet et al., 2010). To overcome these problems, conveyorized phenotyping platforms such as Phenoscope (France) and RIPPS (RIKEN, Japan) have been developed (Fujita et al., 2018; Tisne et al., 2010). Rotation of the pot significantly reduces the small environmental perturbations that exist even under well‐standardized conditions. Phenoscope analysis with Arabidopsis accessions showed that mild drought stress did not exert any epigenetic effects across generations (Van Dooren et al., 2020). The Phenoscope can analyze up to 735 plants, unlike 120 plants that can be analyzed using RIPPS. RIPPS, however, uses fewer root system constraints and larger pots, a system that minimizes water evaporation; furthermore, various state‐of‐the‐art equipment for drought analysis is used (Fujita et al., 2018). The RIPPS platform revealed that the ABA transporter, ABCG25, improves water‐use efficiency and drought tolerance (Kuromori et al., 2016) and that NCED3 (involved in ABA biosynthesis) and CYP707A (involved in ABA catabolism) are important for water‐use efficiency (Fujita et al., 2018). Thus, RIPPS enables the investigation of phenotypes of Arabidopsis mutants and stress‐exposed plants, and will promote the analysis of drought‐response mechanisms, particularly with the development of imaging systems and water/nutrient delivery systems. It is also expected to facilitate the mathematical analysis of drought‐response mechanisms.
The drought responses and tolerance of crops have been investigated using indoor high‐throughput phenotyping platforms. Scanalyzer 3D is a typical conveyor‐type high‐throughput phenotyping platform developed by LemnaTec GmbH (Aachen, Germany) and has been used to study drought responses and tolerance in barley (Chen et al., 2014; Neumann, 2015), sorghum (Neilson et al., 2015), Seteria (Fahlgren et al., 2015), rice (Campbell et al., 2020; Duan et al., 2018), and wheat (Bruning et al., 2019). As part of the Montpellier plant phenotyping platforms (Cabrera‐Bosquet et al., 2016; Tardieu et al., 2017), the PhenoArch platform was used to analyze the growth of maize ears and silique under drought conditions (Brichet et al., 2017), drought response in terms of leaf water potential and transpiration of grape vines (Coupel‐Ledru et al., 2014), and the water‐use efficiency of apple trees (Lopez et al., 2015). A high‐throughput rice phenotyping facility with image analysis pipeline at Huazhong Agricultural University in China (Yang et al., 2014) was used to quantify the dynamic response of rice to drought (Duan et al., 2018) and reveal the genetic architecture of drought resistance in rice (Guo et al., 2018) and cotton (Li and Shen, 2020). In addition, root phenotyping is being studied using X‐ray computed tomography (CT) and magnetic resonance imaging (Atkinson et al., 2019). A non‐destructive X‐ray CT method has been developed to analyze the effects of high temperature and drought stress on potato tubers over time (Van Harsselaar et al., 2021). OpenSimRoot, a functional structural three‐dimensional plant model, enables the mathematical description of root growth and function (Postma et al., 2017). This software can be applied to model three‐dimensional images of roots in soil using magnetic resonance imaging and X‐ray CT. Analysis with OpenSimRoot revealed that the metaxylem morphology interacts with root system depth to regulate water use under drought stress (Strock et al., 2021). A biological organic electrochemical transistor sensor‐based method (Bioristor) was developed to analyze the response of tomato plants to drought (Janni et al., 2019). An Internet of Things‐based pot system (iPOTs), in which the soil water condition can be adjusted via the application of optional treatments, was developed to monitor rice growth under drought stress conditions (Numajiri et al., 2021). The physiological state of the plant can be continuously monitored by embedding the Bioristor device into the tomato stem (Janni et al., 2019). Bioristors can detect drought stress‐induced changes in ion concentrations in the sap, enabling detection of the onset of drought stress immediately after the initiation of defense responses (Janni et al., 2019). In future, further developments in smart plant sensors based on nanobiotechnology (Giraldo et al., 2019) and in technologies for biomolecular detection based on wearable materials integrated with synthetic biology sensors (Nguyen et al., 2021) are expected to advance plant phenotyping considerably.
In pot‐based phenotyping of drought responses, the use of pots may limit root elongation and growth, and the high frequency of deficit irrigation may lead to uneven distribution of water in the soil, affecting plant growth, root distribution, water and nutrient uptake, and root–shoot interactions (Puertolas et al., 2017; Turner et al., 2019). The use of larger pots such as RhizoTubes (Jeudy et al., 2016), rooting columns (Gebre and Earl, 2020), and rhizotrons (Belachew et al., 2019; Canales et al., 2019) has been examined as possible solutions to these problems despite certain limitations. These problems associated with pot‐based phenotyping have not been observed in field phenotyping. However, field work is hampered by larger problems caused by uncontrollable environmental fluctuations. The weaknesses and strengths of laboratory and field phenotyping have to be recognized to identify drought‐response mechanisms and develop drought‐tolerant crop varieties.
Phenotyping to evaluate drought stress tolerance in the field
In the field, large experimental plots enable the collection of large amounts of data using unmanned aerial vehicles and remote sensing. In addition, data science is required to extract relevant information. Automated, non‐destructive, and image‐based high‐throughput phenotyping now allows acquisition of temporal data beyond the terminal phenotype with lesser effort than manual phenotyping (Li et al., 2020). Drought research using high‐throughput phenotyping is currently being developed, the progress of which is outlined below.
In wheat, Phenocart, a portable field phenotyping system (Crain et al., 2018), was used to assess simultaneously the normalized difference vegetation index (NDVI) and canopy temperature of 1170 lines grown under drought or high‐temperature conditions, and to evaluate several genomic selection models (Crain et al., 2018). Durum wheat accessions were grown and phenotyped under different irrigation conditions (Condorelli et al., 2018; Gomez‐Candon et al., 2021). A cost‐effective proximity‐sensing cart equipped with an infrared thermometer, ultrasonic transducer, multiple spectral reflectance sensors, weather station, and RGB camera was used to evaluate upland cotton (Thompson et al., 2018). A field phenotyping platform consisting of a high‐throughput phenotyping system with a gantry frame equipped with various sensors and integrated into a large‐scale automated rainfall shelter facility was constructed to investigate water and nitrogen stress responses in wheat (Beauchene et al., 2019). This environmental management system is promising, as it enables accurate comparative evaluation in the field. As environmental control other than that of soil moisture is dependent on environmental conditions and is expensive, the use of field systems in combination with laboratory studies will accelerate phenotypic analysis.
SUMMARY AND FUTURE PERSPECTIVES
Gene expression and signal transduction related to the plant drought stress response have been studied at the cellular and molecular levels in model plants and crops, and the functions of genes involved in stress tolerance have been analyzed in transgenic and mutant plants. In addition, the mechanisms underlying the induction of gene expression by drought stress and its regulation by ABA and protein phosphorylation have been analyzed. Stomatal closure due to water deficiency has been evaluated at the cellular and molecular levels, revealing complex regulatory systems mediated by protein phosphorylation. The molecular mechanisms underlying stomatal responses to dehydration, CO2, and light crosstalk, integrating complex responses to environmental changes.
The molecular transmission of stress signals in tissues and organs was analyzed. Research on plant responses to abiotic stresses has shifted from intra‐cellular to inter‐tissue or inter‐organ systemic regulation (Figure 1). To understand drought stress responses and tolerance in the whole plant, inter‐tissue transmission of stress signals has been analyzed with respect to ABA. For example, several types of transporters are involved in ABA transport between the vascular bundle, stomata, and the entire leaf (Figure 2). Furthermore, the induction of stress‐responsive gene expression and acquisition of stress tolerance via the systemic transport of ABA and stress signals have been investigated. Analyses of regulatory genes related to ABA transport have been performed, and research on inter‐tissue communication is underway.
Stress signals due to soil water loss are sensed by the roots and transmitted to the leaves. The roles of turgor pressure, Ca2+, and ROS have been analyzed in inter‐organ signal transduction from the roots to the leaves. Inter‐organ signal transduction from roots to leaves is mediated by the transport of peptides and metabolites via the vasculature, particularly the xylem, which involves various molecular mechanisms (Figure 3). The transport of these inter‐organ signaling molecules will be investigated based on functional analyses of the vascular transport systems. Novel peptide‐coding genes have been identified during the analysis of genomes and non‐coding RNAs. Predicted sORF and proteomics (PeptideAtlas/Arabidopsis; http://www.peptideatlas.org/builds/arabidopsis/) databases are publicly available, and additional functional analysis of predicted peptides will reveal novel regulatory systems.
Drought stress or water‐deficit stress causes complex physiological responses in different plant organs. Moreover, complex drought stress signals are sensed to induce complex molecular patterns of intra‐ and inter‐cellular stress signals, which are recognized by different cellular sensors to induce correct responses required for survival under drought stress conditions (Figure 4). Water‐deficit stress signals are also multilaterally sensed in the leaves and roots. In leaves, the dehydration status is sensed by stomata to control the water status. Stomatal closure decreases water loss from leaves under drought stress conditions. In roots, dry soil causes water deficiency, which is sensed mainly by the vascular tissues of roots to activate inter‐organ stress responses from roots to leaves. Water deficiency is sensed by root vascular cells, which initiates drought stress responses in all plant organs. Integration of these stress responses in different organs is necessary for proper responses of the entire plant (Figures 3 and 4).
Non‐destructive and quantitative analyses of plant phenotypes have been enabled by developments in imaging and data analyses. Plant development under stress conditions can be analyzed using automatic phenotyping systems under controlled environmental conditions. Imaging systems using high‐performance cameras allow monitoring of plant growth under various environmental conditions. For example, leaf temperature can be measured using an infrared camera, and plant water status can be measured using a near‐infrared camera to monitor dehydration. By continuously monitoring the water‐deficit status of plants, it is possible to measure quantitatively the response of plants to water loss in terms of plant growth, stomatal closure, and water status. Transcriptomic and metabolomic analyses facilitated the identification of novel gene sets using mutant plants and natural ecotypes. Furthermore, genome‐wide association studies of mutant strains can reveal novel genes involved in water‐use efficiency and dehydration resistance. In addition, the effects of drought stress on plants can be investigated in the reproductive stage, such as flower formation and seed maturation. The drought responses of plants in the field were predicted using the data obtained from quantitative phenotypic analysis. In the future, these data will be related to crop growth and the acquisition of tolerance during drought conditions, along with meteorological data. Further progress in information technology and data science will enable the use of big data in research on the environmental responses of plants. Interdisciplinary phenomics research on plant development, such as vegetative growth, flowering, and seed formation, will be facilitated by further technological developments and the anticipated availability of more plant and crop biological resources.
AUTHOR CONTRIBUTIONS
TK and KS wrote and edited a large part of this review article. MF mainly wrote the phenotyping section and FT wrote the inter‐organ signaling section. All the authors contributed to the edition and revision of this article.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ACKNOWLEDGEMENTS
The preparation of this review article was supported by RIKEN CSRS (to KS, TK), JSPS KAKENHI Grant Numbers 19H03255 and 20K21437 (to FT), Research Grant against Global Warming from the Ichimura Foundation for New Technology (to TK), and Bio‐oriented Technology Research Advancement Institution, grant number: JPJ009237 (to MF).
Contributor Information
Takashi Kuromori, Email: takashi.kuromori@riken.jp.
Kazuo Shinozaki, Email: kazuo.shinozaki@riken.jp.
DATA AVAILABILITY STATEMENT
This is a review article. All relevant data can be found within the manuscript and its supporting materials.
REFERENCES
- Atkinson, J.A. , Pound, M.P. , Bennett, M.J. & Wells, D.M. (2019) Uncovering the hidden half of plants using new advances in root phenotyping. Current Opinion in Biotechnology, 55, 1–8. 10.1016/j.copbio.2018.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert, Y. , Vile, D. , Pervent, M. , Aldon, D. , Ranty, B. , Simonneau, T. et al. (2010) RD20, a stress‐inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana . Plant and Cell Physiology, 51, 1975–1987. 10.1093/pcp/pcq155 [DOI] [PubMed] [Google Scholar]
- Bac‐Molenaar, J.A. , Granier, C. , Keurentjes, J.J. & Vreugdenhil, D. (2016) Genome‐wide association mapping of time‐dependent growth responses to moderate drought stress in Arabidopsis . Plant, Cell and Environment, 39, 88–102. 10.1111/pce.12595 [DOI] [PubMed] [Google Scholar]
- Batool, S. , Uslu, V.V. , Rajab, H. , Ahmad, N. , Waadt, R. , Geiger, D. et al. (2018) Sulfate is incorporated into cysteine to trigger ABA production and stomatal closure. The Plant Cell, 30, 2973–2987. 10.1105/tpc.18.00612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, H. , Ache, P. , Lautner, S. , Fromm, J. , Hartung, W. , Al‐Rasheid, K. et al. (2013) The stomatal response to reduced relative humidity requires guard cell‐autonomous ABA synthesis. Current Biology, 23, 53–57. 10.1016/j.cub.2012.11.022 [DOI] [PubMed] [Google Scholar]
- Beauchene, K. , Leroy, F. , Fournier, A. , Huet, C. , Bonnefoy, M. , Lorgeou, J. et al. (2019) Management and characterization of abiotic stress via PhenoField®, a high‐throughput field phenotyping platform. Frontiers in Plant Science, 10, 904. 10.3389/fpls.2019.00904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belachew, K.Y. , Nagel, K.A. , Poorter, H. & Stoddard, F.L. (2019) Association of shoot and root responses to water deficit in young Faba bean (Vicia faba L.) plants. Frontiers in Plant Science, 10, 1063. 10.3389/fpls.2019.01063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges, S.E. , Vasseur, F. , Bediee, A. , Rolland, G. , Masclef, D. , Dauzat, M. et al. (2020) Natural variation of Arabidopsis thaliana responses to cauliflower mosaic virus infection upon water deficit. PLoS Path, 16, e1008557. 10.1371/journal.ppat.1008557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges, S.E. , Vile, D. , Vazquez‐Rovere, C. , Blanc, S. , Yvon, M. , Bediee, A. et al. (2018) Interactions between drought and plant genotype change epidemiological traits of cauliflower mosaic virus. Frontiers in Plant Science, 9, 703. 10.3389/fpls.2018.00703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharath, R. , Gahir, S. & Raghaventra, A. (2021) Abscisic acid‐induced stomatal closure: an important component of plant defense against abiotic and biotic stress. Frontiers in Plant Science, 12, 615114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschiero, C. , Lundquist, P.K. , Roy, S. , Dai, X. , Zhao, P.X. & Scheible, W.R. (2019) Identification and functional investigation of genome‐encoded, small, secreted peptides in plants. Current Protocols in Plant Biology, 4, e20098. 10.3389/fpls.2021.615114 [DOI] [PubMed] [Google Scholar]
- Bresson, J. , Varoquaux, F. , Bontpart, T. , Touraine, B. & Vile, D. (2013) The PGPR strain Phyllobacterium brassicacearum STM196 induces a reproductive delay and physiological changes that result in improved drought tolerance in Arabidopsis . New Phytologist, 200, 558–569. 10.1111/nph.12383 [DOI] [PubMed] [Google Scholar]
- Brichet, N. , Fournier, C. , Turc, O. , Strauss, O. , Artzet, S. , Pradal, C. et al. (2017) A robot‐assisted imaging pipeline for tracking the growth of maize ear and silks in a high‐throughput phenotyping platform. Plant Methods, 13, 96. 10.1186/s13007-017-0246-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning, B. , Liu, H. , Brien, C. , Berger, B. , Lewis, M. & Garnett, T. (2019) The development of hyperspectral distribution maps to predict the content and distribution of nitrogen and water in wheat (Triticum aestivum). Frontiers in Plant Science, 10, 1380. 10.3389/fpls.2019.01380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera‐Bosquet, L. , Fournier, C. , Brichet, N. , Welcker, C. , Suard, B. & Tardieu, F. (2016) High‐throughput estimation of incident light, light interception, and radiation‐use efficiency of thousands of plants in a phenotyping platform. New Phytologist, 212, 269–281. 10.1111/nph.14027 [DOI] [PubMed] [Google Scholar]
- Campbell, M.T. , Grondin, A. , Walia, H. & Morota, G. (2020) Leveraging genome‐enabled growth models to study shoot growth responses to water deficit in rice. Journal of Experimental Botany, 71, 5669–5679. 10.1093/jxb/eraa280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales, F.J. , Nagel, K.A. , Muller, C. , Rispail, N. & Prats, E. (2019) Deciphering root architectural traits involved to cope with water deficit in oat. Frontiers in Plant Science, 10, 1558. 10.3389/fpls.2019.01558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale, F. , Korwin Krukowski, P. , Schubert, A. & Visentin, I. (2018) Strigolactones: mediators of osmotic stress responses with a potential for agrochemical manipulation of crop resilience. Journal of Experimental Botany, 69, 2291–2303. 10.1093/jxb/erx494 [DOI] [PubMed] [Google Scholar]
- Chen, D. , Neumann, K. , Friedel, S. , Kilian, B. , Chen, M. , Altmann, T. et al. (2014) Dissecting the phenotypic components of crop plant growth and drought responses based on high‐throughput image analysis. The Plant Cell, 26, 4636–4655. 10.1105/tpc.114.129601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. , Li, G.‐J. , Bresson, R.A. , Song, C.‐P. , Zhu, J.‐K. & Zao, Y. (2020) Abscisic acid dynamics, signaling, and functions in plants. Journal of Integrative Plant Biology, 62, 25–54. 10.1111/jipb.12899 [DOI] [PubMed] [Google Scholar]
- Chen, X. , Yao, Q. , Gao, X. , Jiang, C. , Harberd, N.P. & Fu, X. (2016) Shoot‐to‐root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Current Biology, 26, 640–646. 10.1016/j.cub.2015.12.066 [DOI] [PubMed] [Google Scholar]
- Cheng, W.H. , Endo, A. , Zhou, L. , Penney, J. , Chen, H.C. , Arroyo, A. et al. (2002) A unique short‐chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. The Plant Cell, 14, 2723–2743. 10.1105/tpc.006494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W.G. , Miller, G. , Wallace, I. , Harper, J. , Mittler, R. & Gilroy, S. (2017) Orchestrating rapid long‐distance signaling in plants with Ca2+, ROS and electrical signals. The Plant Journal, 90, 698–707. 10.1111/tpj.13492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann, A. , Grill, E. & Huang, J. (2013) Hydraulic signals in long‐distance signaling. Current Opinion in Plant Biology, 16, 293–300. 10.1016/j.pbi.2013.02.011 [DOI] [PubMed] [Google Scholar]
- Christmann, A. , Weiler, E.W. , Steudle, E. & Grill, E. (2007) A hydraulic signal in root‐to‐shoot signalling of water shortage. The Plant Journal, 52, 167–174. 10.1111/j.1365-313X.2007.03234.x [DOI] [PubMed] [Google Scholar]
- Clauw, P. , Coppens, F. , De Beuf, K. , Dhondt, S. , Van Daele, T. , Maleux, K. et al. (2015) Leaf responses to mild drought stress in natural variants of Arabidopsis . Plant Physiology, 167, 800–816. 10.1104/pp.114.254284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauw, P. , Coppens, F. , Korte, A. , Herman, D. , Slabbinck, B. , Dhondt, S. et al. (2016) Leaf growth response to mild drought: natural variation in Arabidopsis sheds light on trait architecture. The Plant Cell, 28, 2417–2434. 10.1105/tpc.16.00483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli, G.E. , Maccaferri, M. , Newcomb, M. , Andrade‐Sanchez, P. , White, J.W. , French, A.N. et al. (2018) Comparative aerial and ground based high throughput phenotyping for the genetic dissection of NDVI as a proxy for drought adaptive traits in Durum wheat. Frontiers in Plant Science, 9, 893. 10.3389/fpls.2018.00893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupel‐Ledru, A. , Lebon, E. , Christophe, A. , Doligez, A. , Cabrera‐Bosquet, L. , Pechier, P. et al. (2014) Genetic variation in a grapevine progeny (Vitis vinifera L. cvs GrenachexSyrah) reveals inconsistencies between maintenance of daytime leaf water potential and response of transpiration rate under drought. Journal of Experimental Botany, 65, 6205–6218. 10.1093/jxb/eru228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain, J. , Mondal, S. , Rutkoski, J. , Singh, R.P. & Poland, J. (2018) Combining high‐throughput phenotyping and genomic information to increase prediction and selection accuracy in wheat breeding. The Plant Genome, 11, 170043. 10.3835/plantgenome2017.05.0043 [DOI] [PubMed] [Google Scholar]
- Cui, Y. , Li, M. , Yin, X. , Song, S. , Xu, G. , Wang, M. et al. (2018) OsDSSR1, a novel small peptide, enhances drought tolerance transgenic rice. Plant Science, 270, 85−96. 10.1016/j.plantsci.2018.02.015 [DOI] [PubMed] [Google Scholar]
- Cutler, S. , Rodriquez, P.L. , Finkelstein, R.R. & Abrams, S.R. (2010) Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology, 61, 651–659. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- Das, D. , Jaiswal, M. , Khan, F.K. , Ahmad, S. & Kumar, S. (2020) PlantPepDB: a manually curated plant peptide database. Scientific Reports, 10, 2194. 10.1038/s41598-020-59165-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt, S. , Wuyts, N. & Inze, D. (2013) Cell to whole‐plant phenotyping: the best is yet to come. Trends in Plant Science, 18, 428–439. 10.1016/j.tplants.2013.04.008 [DOI] [PubMed] [Google Scholar]
- Dietrich, D. , Pang, L. , Kobayashi, A. , Fozard, J.A. , Boudolf, V. , Bhosale, R. et al. (2017) Root hydrotropism is controlled via a cortex‐specific growth mechanism. Nature Plants, 3, 17057. 10.1038/nplants.2017.57 [DOI] [PubMed] [Google Scholar]
- Duan, L. , Han, J. , Guo, Z. , Tu, H. , Yang, P. , Zhang, D. et al. (2018) Novel digital features discriminate between drought resistant and drought sensitive rice under controlled and field conditions. Frontiers in Plant Science, 9, 492. 10.3389/fpls.2018.00492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois, M. , Claeys, H. , Van den Broeck, L. & Inze, D. (2017) Time of day determines Arabidopsis transcriptome and growth dynamics under mild drought. Plant, Cell and Environment, 40, 180–189. 10.1111/pce.12809 [DOI] [PubMed] [Google Scholar]
- Dubois, M. , Selden, K. , Bediee, A. , Rolland, G. , Baumberger, N. , Noir, S. et al. (2018) SIAMESE‐RELATED1 is regulated posttranslationally and participates in repression of leaf growth under moderate drought. Plant Physiology, 176, 2834–2850. 10.1104/pp.17.01712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, A. , Sawada, Y. , Takahashi, H. , Okamoto, M. , Ikegami, K. , Koiwai, H. et al. (2008) Drought induction of Arabidopsis 9‐cis‐epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiology, 147, 1984–1993. 10.1104/pp.108.116632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, S. , Iwai, Y. & Fukuda, H. (2019) Cargo‐dependent and cell wall‐associated xylem transport in Arabidopsis . New Phytologist, 222, 159–170. 10.1111/nph.15540 [DOI] [PubMed] [Google Scholar]
- Fahlgren, N. , Feldman, M. , Gehan, M.A. , Wilson, M.S. , Shyu, C. , Bryant, D.W. et al. (2015) A versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in Setaria . Molecular Plant, 8, 1520–1535. 10.1016/j.molp.2015.06.005 [DOI] [PubMed] [Google Scholar]
- Finkelstein, R. (2013) Abscisic acid synthesis and response. The Arabidopsis Book, 11, e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, J.C. (2020) Recent advances in Arabidopsis CLE peptide signaling. Trends in Plant Science, 25, 1005–1016. 10.1016/j.tplants.2020.04.014 [DOI] [PubMed] [Google Scholar]
- Flood, P.J. , Kruijer, W. , Schnabel, S.K. , van der Schoor, R. , Jalink, H. , Snel, J.F. et al. (2016) Phenomics for photosynthesis, growth, and reflectance in Arabidopsis thaliana reveals circadian and long‐term fluctuations in heritability. Plant Methods, 12, 14. 10.1186/s13007-016-0113-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, M. , Tanabata, T. , Urano, K. , Kikuchi, S. & Shinozaki, K. (2018) RIPPS: A plant phenotyping system for quantitative evaluation of growth under controlled environmental stress conditions. Plant and Cell Physiology, 59, 2030–2038. 10.1093/pcp/pcy122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebre, M.G. & Earl, H.J. (2020) Effects of growth medium and water stress on soybean [Glycine max (L.) Merr.] growth, soil water extraction and rooting profiles by depth in 1 m rooting columns. Frontiers in Plant Science, 11, 487. 10.3389/fpls.2020.00487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandilyan, A. , Barboza, L. , Tisne, S. , Granier, C. , Reymond, M. , Koornneef, M. et al. (2009) Genetic analysis identifies quantitative trait loci controlling rosette mineral concentrations in Arabidopsis thaliana under drought. New Phytologist, 184, 180–192. 10.1111/j.1469-8137.2009.02953.x [DOI] [PubMed] [Google Scholar]
- Giraldo, J.P. , Wu, H. , Newkirk, G.M. & Kruss, S. (2019) Nanobiotechnology approaches for engineering smart plant sensors. Nature Nanotechnology, 14, 541–553. 10.1038/s41565-019-0470-6 [DOI] [PubMed] [Google Scholar]
- Goad, D.M. , Zhu, C. & Kellogg, E.A. (2017) Comprehensive identification and clustering of CLV3/ESR‐related (CLE) genes in plants finds groups with potentially shared function. New Phytologist, 216, 605–616. 10.1111/nph.14348 [DOI] [PubMed] [Google Scholar]
- Gomez‐Candon, D. , Bellvert, J. & Royo, C. (2021) Performance of the two‐source energy balance (TSEB) model as a tool for monitoring the response of Durum wheat to drought by high‐throughput field phenotyping. Frontiers in Plant Science, 12, 658357. 10.3389/fpls.2021.658357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier, C. , Aguirrezabal, L. , Chenu, K. , Cookson, S.J. , Dauzat, M. , Hamard, P. et al. (2006) PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytologist, 169, 623–635. 10.1111/j.1469-8137.2005.01609.x [DOI] [PubMed] [Google Scholar]
- Guo, Z. , Yang, W. , Chang, Y. , Ma, X. , Tu, H. , Xiong, F. et al. (2018) Genome‐wide association studies of image traits reveal genetic architecture of drought resistance in rice. Molecular Plant, 11, 789–805. 10.1016/j.molp.2018.03.018 [DOI] [PubMed] [Google Scholar]
- Gupta, A. , Rico‐Medina, A. & Caño‐Delgado, A.I. (2020) The physiology of plant responses to drought. Science, 368, 266–269. 10.1126/science.aaz7614 [DOI] [PubMed] [Google Scholar]
- Hai, N.N. , Chuong, N.N. , Tu, N.H.C. , Kisiala, A. , Hoang, X.L.T. & Thao, N.P. (2020) Role and regulation of cytokinins in plant response to drought stress. Plants, 9, 422. 10.3390/plants9040422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y. , Watanabe, S. , Shimada, H. & Sakamoto, A. (2020) Dynamics of the leaf endoplasmic reticulum modulate beta‐glucosidase‐mediated stress‐activated ABA production from glycosyl ester. Journal of Experimental Botany, 71, 2058–2071. 10.1093/jxb/erz528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada, K. , Akiyama, K. , Sakurai, T. , Toyoda, T. , Shinozaki, K. & Shiu, S.H. (2010) sORF finder: a program package to identify small open reading frames with high coding potential. Bioinformatics, 26, 399–400. 10.1093/bioinformatics/btp688 [DOI] [PubMed] [Google Scholar]
- Hanada, K. , Higuchi‐Takeuchi, M. , Okamoto, M. , Yoshizumi, T. , Shimizu, M. , Nakaminami, K. et al. (2013) Small open reading frames associated with morphogenesis are hidden in plant genomes. Proceedings of the National Academy of Sciences of the United States of America, 110, 2395–2400. 10.1073/pnas.1213958110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, C. , Tian, W. , Kleist, T. , He, K. , Garcia, V. , Bai, F. et al. (2014) DUF221 proteins are a family of osmosensitive calcium‐permeable cation channels conserved across eukaryotes. Cell Research, 24, 632–635. 10.1038/cr.2014.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, P.‐K. , Dubeaux, G. , Takahashi, Y. & Schroeder, J. (2021) Signaling mechanisms in abscisic acid‐mediated stomatal closure. The Plant Journal, 105, 307–321. 10.1111/tpj.15067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H. , Liu, B. , Liu, L. & Song, S. (2017) Jasmonate action in plant growth and development. Journal of Experimental Botany, 68, 1349–1359. 10.1093/jxb/erw495 [DOI] [PubMed] [Google Scholar]
- Hussain, S. , Brookbank, B.P. & Nambara, E. (2020) Hydrolysis of abscisic acid glucose ester occurs locally and quickly in response to dehydration. Journal of Experimental Botany, 71, 1753–1756. 10.1093/jxb/eraa026 [DOI] [PubMed] [Google Scholar]
- Janni, M. , Coppede, N. , Bettelli, M. , Briglia, N. , Petrozza, A. , Summerer, S. et al. (2019) In vivo phenotyping for the early detection of drought stress in tomato. Plant Phenomics, 2019, 6168209. 10.34133/2019/6168209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeudy, C. , Adrian, M. , Baussard, C. , Bernard, C. , Bernaud, E. , Bourion, V. et al. (2016) RhizoTubes as a new tool for high throughput imaging of plant root development and architecture: test, comparison with pot grown plants and validation. Plant Methods, 12, 31. 10.1186/s13007-016-0131-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J. , Yim, S. , Choi, H. , Kim, A. , Lee, K.P. , Lopez‐Molina, L. et al. (2015) Abscisic acid transporters cooperate to control seed germination. Nature Communications, 6, 8113. 10.1038/ncomms9113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.S. , Jeon, B.W. & Kim, J. (2021) Signaling peptides regulating abiotic stress responses in plants. Frontiers in Plant Science, 12, 704490. 10.3389/fpls.2021.704490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, A. , Takahashi, A. , Kakimoto, Y. , Miyazawa, Y. , Fujii, N. , Higashitani, A. et al. (2007) A gene essential for hydrotropism in roots. Proceedings of the National Academy of Sciences of the United States of America, 104, 4724–4729. 10.1073/pnas.0609929104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwai, H. , Nakaminami, K. , Seo, M. , Mitsuhashi, W. , Toyomasu, T. & Koshiba, T. (2004) Tissue‐specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis . Plant Physiology, 134, 1697–1707. 10.1104/pp.103.036970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollist, H. , Zandalinas, S. , Senqupta, S. , Nuhkat, M. , Kangasjaervi, J. & Mittler, R. (2019) Rapid responses to abiotic stress: Priming the landscape for the signal transduction network. Trends in Plant Science, 24, 25–37. 10.1016/j.tplants.2018.10.003 [DOI] [PubMed] [Google Scholar]
- Konrad, K.R. , Maierhofer, T. & Hedrich, R. (2018) Spatio‐temporal aspects of Ca2+ signalling: lessons from guard cells and pollen tubes. Journal of Experimental Botany, 69, 4195–4214. 10.1093/jxb/ery154 [DOI] [PubMed] [Google Scholar]
- Kumar, M.N. , Jane, W.N. & Verslues, P.E. (2013) Role of the putative osmosensor Arabidopsis histidine kinase1 in dehydration avoidance and low‐water‐potential response. Plant Physiology, 161, 942–953. 10.1104/pp.112.209791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori, T. , Fujita, M. , Urano, K. , Tanabata, T. , Sugimoto, E. & Shinozaki, K. (2016) Overexpression of AtABCG25 enhances the abscisic acid signal in guard cells and improves plant water use efficiency. Plant Science, 251, 75–81. 10.1016/j.plantsci.2016.02.019 [DOI] [PubMed] [Google Scholar]
- Kuromori, T. , Mizoi, J. , Umezawa, T. , Yamaguchi‐Shinozaki, K. & Shinozaki, K. (2014b) Stress signaling networks: drought stress. In Howell, S.H. (Ed.) Molecular biology. New York: Springer, pp. 383–409. [Google Scholar]
- Kuromori, T. , Seo, M. & Shinozaki, K. (2018) ABA transport and plant water stress responses. Trends in Plant Science, 23, 513–522. 10.1016/j.tplants.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Kuromori, T. , Sugimoto, E. , Ohiraki, H. , Yamaguchi‐Shinozaki, K. & Shinozaki, K. (2017) Functional relationship of AtABCG21 and AtABCG22 in stomatal regulation. Scientific Reports, 7, 12501. 10.1038/s41598-017-12643-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori, T. , Sugimoto, E. & Shinozaki, K. (2011) Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. The Plant Journal, 67, 885–894. 10.1111/j.1365-313X.2011.04641.x [DOI] [PubMed] [Google Scholar]
- Kuromori, T. , Sugimoto, E. & Shinozaki, K. (2014a) Intertissue signal transfer of abscisic acid from vascular cells to guard cells. Plant Physiology, 164, 1587–1592. 10.1104/pp.114.235556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotani, K.I. & Notaguchi, M. (2021) Cell‐to‐cell connection in plant grafting—molecular insights into symplasmic reconstruction. Plant and Cell Physiology, 62, 1362–1371. 10.1093/pcp/pcab109 [DOI] [PubMed] [Google Scholar]
- Laohavisit, A. , Wakatake, T. , Ishihama, N. , Mulvey, H. , Takizawa, K. , Suzuki, T. et al. (2020) Quinone perception in plants via leucine‐rich‐repeat receptor‐like kinases. Nature, 587, 92–97. 10.1038/s41586-020-2655-4 [DOI] [PubMed] [Google Scholar]
- Lauressergues, D. , Couzigou, J.‐M. , Clemente, H.S. , Martinez, Y. , Dunand, C. , Bécard, G. et al. (2015) Primary transcripts of microRNAs encode regulatory peptides. Nature, 520, 90–93. 10.1038/nature14346 [DOI] [PubMed] [Google Scholar]
- Lee, J. , He, K. , Stolc, V. , Lee, H. , Figueroa, P. , Gao, Y. et al. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. The Plant Cell, 19, 731–749. 10.1105/tpc.106.047688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.H. , Piao, H.L. , Kim, H.‐Y. , Choi, S.M. , Jiang, F. , Hartung, W. et al. (2006) Activation of glucosidase via stress‐induced polymerization rapidly increases active pools of abscisic acid. Cell, 126, 1109–1120. 10.1016/j.cell.2006.07.034 [DOI] [PubMed] [Google Scholar]
- Li, B. & Shen, X. (2020) Preliminary study on discrimination of transgenic cotton seeds using terahertz time‐domain spectroscopy. Food Science & Nutrition, 8, 5426–5433. 10.1002/fsn3.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, D. , Quan, C. , Song, Z. , Li, X. , Yu, G. , Li, C. et al. (2020) High‐throughput plant phenotyping platform (HT3P) as a novel tool for estimating agronomic traits from the lab to the field. Frontiers in Bioengineering and Biotechnology, 8, 623705. 10.3389/fbioe.2020.623705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Testerink, C. & Zhang, Y. (2021) How roots and shoots communicate through stressful times. Trends in Plant Science, 26, 940–952. 10.1016/j.tplants.2021.03.005 [DOI] [PubMed] [Google Scholar]
- Li, W. , Herrera‐Estrellla, L. & Tran, L.P. (2016) The yin‐yang of cytokinin homeostasis and drought acclimation/adaptation. Trends in Plant Science, 21, 548–550. 10.1016/j.tplants.2016.05.006 [DOI] [PubMed] [Google Scholar]
- Li, W. , Nguyen, K.H. , Chu, H.D. , Ha, C.V. , Watanabe, Y. , Osakabe, Y. et al. (2017) The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana . PLoS Genetics, 13, e1007076. 10.1371/journal.pgen.1007076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. , Zhu, W. , Chen, S. , Qian, J. & Li, L. (2021) Genome‐wide identification and characterization of small peptides in maize. Frontiers in Plant Science, 12, 695439. 10.3389/fpls.2021.695439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, L. , Wu, J. , Jian, M. & Wang, Y. (2021) Plant mitogen‐activated protein kinase cascades in environmental stresses. International Journal of Molecular Sciences, 22, 1543. 10.3390/ijms22041543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Wang, J. & Sun, L. (2018) Structure of the hyperosmolality‐gated calcium‐permeable channel OSCA1.2. Nature Communications, 9, 5060. 10.1038/s41467-018-07564-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, G. , Pallas, B. , Martinez, S. , Lauri, P.E. , Regnard, J.L. , Durel, C.E. et al. (2015) Genetic variation of morphological traits and transpiration in an apple core collection under well‐watered conditions: towards the identification of morphotypes with high water use efficiency. PLoS One, 10, e0145540. 10.1371/journal.pone.0145540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, W.J. , Groover, A. , Lichtenberger, R. , Furuta, K. , Yadav, S.R. , Helariutta, Y. et al. (2013) The plant vascular system: evolution, development. Journal of Integrative Plant Biology, 55, 294–388. 10.1111/jipb.12041 [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Szostkiewicz, I. , Korte, A. , Moes, D. , Yang, Y. , Christmann, A. et al. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science, 324, 1064–1068. 10.1126/science.1172408 [DOI] [PubMed] [Google Scholar]
- Marín‐de la Rosa, N. , Lin, C.‐W. , Kang, Y.J. , Dhondt, S. , Gonzalez, N. , Inzé, D. et al. (2019) Drought resistance is mediated by divergent strategies in closely related Brassicaceae. New Phytologist, 223, 783–797. 10.1111/nph.15841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet, C. , Vile, D. , Fabre, J. , Hannah, M.A. , Caldana, C. , Lisec, J. et al. (2010) Probing the reproducibility of leaf growth and molecular phenotypes: a comparison of three Arabidopsis accessions cultivated in ten laboratories. Plant Physiology, 152, 2142–2157. 10.1104/pp.109.148338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam, S.A.M. & Brodribb, T.J. (2018) Mesophyll cells are the main site of abscisic acid biosynthesis in water‐stressed leaves. Plant Physiology, 177, 911–917. 10.1104/pp.17.01829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam, S.A. , Brodribb, T.J. & Ross, J.J. (2016) Shoot‐derived abscisic acid promotes root growth. Plant, Cell and Environment, 39, 652–659. 10.1111/pce.12669 [DOI] [PubMed] [Google Scholar]
- Mochida, K. , Nishii, R. & Hirayama, T. (2020) Decoding plant–environment interactions that influence crop agronomic traits. Plant and Cell Physiology, 61, 1408–1418. 10.1093/pcp/pcaa064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriwaki, T. , Miyazawa, Y. , Fujii, N. & Takahashi, H. (2012) Light and abscisic acid signalling are integrated by MIZ1 gene expression and regulate hydrotropic response in roots of Arabidopsis thaliana . Plant, Cell and Environment, 35, 1359–1368. 10.1111/j.1365-3040.2012.02493.x [DOI] [PubMed] [Google Scholar]
- Moriwaki, T. , Miyazawa, Y. , Kobayashi, A. & Takahashi, H. (2013) Molecular mechanisms of hydrotropism in seedling roots of Arabidopsis thaliana (Brassicaceae). American Journal of Botany, 100, 25–34. 10.3732/ajb.1200419 [DOI] [PubMed] [Google Scholar]
- Mostofa, M.G. , Li, W. , Ngyuyen, K.H. , Fujita, M. & Tran, L.P. (2018) Strigolactones in plant adaptation to abiotic stresses: an emerging avenue of plant research. Plant, Cell and Environment, 41, 2227–2243. 10.1111/pce.13364 [DOI] [PubMed] [Google Scholar]
- Munemasa, S. , Hauser, F. , Park, J. , Waadt, R. , Brandt, B. & Schroeder, J.I. (2015) Mechanisms of abscisic acid‐mediated control of stomatal aperture. Current Opinion in Plant Biology, 28, 154–162. 10.1016/j.pbi.2015.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy, S.E. , Dubin, A. , Whitwam, T. , Jojoa‐Cruz, S. , Cahalan, S. , Mousavi, S.A.R. et al. (2018) OSCA/TMEM63 are an evolutionarily conserved family of mechanically activated ion channels. eLife, 7, e41844. 10.7554/eLife.41844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, Y. , Katagiri, T. , Shinozaki, K. , Qi, Z. , Tatsumi, H. , Furuichi, T. et al. (2007) Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proceedings of the National Academy of Sciences of the United States of America, 104, 3639–3644. 10.1073/pnas.0607703104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami, K. , Okamoto, M. , Higuchi‐Takeuchi, M. , Yoshizumi, T. , Yamaguchi, Y. , Fukao, Y. et al. (2018) AtPep3 is a hormone‐like peptide that plays a role in the salinity stress tolerance of plants. Proceedings of the National Academy of Sciences of the United States of America, 115, 5810–5815. 10.1073/pnas.1719491115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson, E.H. , Edwards, A.M. , Blomstedt, C.K. , Berger, B. , Moller, B.L. & Gleadow, R.M. (2015) Utilization of a high‐throughput shoot imaging system to examine the dynamic phenotypic responses of a C4 cereal crop plant to nitrogen and water deficiency over time. Journal of Experimental Botany, 66, 1817–1832. 10.1093/jxb/eru526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, G. (2015) The role of ethylene in plant adaptations for phosphate acquisition in soils ‐ a review. Frontiers in Plant Science, 6, 1224. 10.3389/fpls.2015.01224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, P.Q. , Soenksen, L.R. , Donghia, N.M. , Angenent‐Mari, N.M. , de Puig, H. , Huang, A. et al. (2021) Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nature Biotechnology, 39, 1366–1374. 10.1038/s41587-021-00950-3 [DOI] [PubMed] [Google Scholar]
- Nishii, K. , Moeller, M. & Iida, H. (2021) Mix and match: Patchwork domain evolution of the land plant‐specific Ca2+‐permeable mechanosensitive channel MCA. PLoS One, 16, e0249735. 10.1371/journal.pone.0249735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, R. , Watanabe, Y. , Fujita, Y. , Le, D.T. , Kojima, M. , Werner, T. et al. (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. The Plant Cell, 23, 2169–2183. 10.1105/tpc.111.087395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, R. , Watanabe, Y. , Leyva‐Gonzalez, M.A. , Ha, C.V. , Fujita, Y. , Tanaka, M. et al. (2013) Arabidopsis AFP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought‐stress response. Proceedings of the National Academy of Sciences of the United States of America, 110, 4840–4845. 10.1073/pnas.1302265110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numajiri, Y. , Yoshino, K. , Teramoto, S. , Hayashi, A. , Nishijima, R. , Tanaka, T. et al. (2021) iPOTs: internet of things‐based pot system controlling optional treatment of soil water conditions for plant phenotyping under drought stress. The Plant Journal, 107, 1569–1580. 10.1111/tpj.15400 [DOI] [PubMed] [Google Scholar]
- Oh, E. , Seo, P.J. & Kim, J. (2018) Signaling peptides and receptors coordinating plant root development. Trends in Plant Science, 23, 337–351. 10.1016/j.tplants.2017.12.007 [DOI] [PubMed] [Google Scholar]
- Ohkubo, Y. , Tanaka, M. , Tabata, R. , Ogawa‐Ohnishi, M. & Matsubayashi, Y. (2017) Shoot‐to‐root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nature Plants, 3, 17029. 10.1038/nplants.2017.29 [DOI] [PubMed] [Google Scholar]
- Okamoto, M. , Tanaka, Y. , Abrams, S.R. , Kamiya, Y. , Seki, M. & Nambara, E. (2009) High humidity induces ABA 8'‐hydroxylase in stomata and vasculature to regulate local and systemic ABA responses in Arabidopsis . Plant Physiology, 149, 825–834. 10.1104/pp.108.130823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, S. , Suzuki, T. , Kawaguchi, M. , Higashiyama, T. & Matsubayashi, Y. (2015) A comprehensive strategy for identifying long‐distance mobile peptides in xylem sap. The Plant Journal, 84, 611–620. 10.1111/tpj.13015 [DOI] [PubMed] [Google Scholar]
- Orosa‐Puente, B. , Leftley, N. , von Wangenheim, D. , Banda, J. , Srivastava, A.K. , Hill, K. et al. (2018) Root branching toward water involves posttranslational modification of transcription factor ARF7. Science, 362, 1407–1410. 10.1126/science.aau3956 [DOI] [PubMed] [Google Scholar]
- Park, S.Y. , Fung, P. , Nishimura, N. , Jensen, D.R. , Fujii, H. , Zhao, Y. et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science, 324, 1068–1071. 10.1126/science [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y. , Xu, Z.Y. , Kim, S.Y. , Lee, J. , Choi, B. , Lee, J. et al. (2016) Spatial regulation of ABCG25, an ABA exporter, is an important component of the mechanism controlling cellular ABA levels. The Plant Cell, 28, 2528–2544. 10.1105/tpc.16.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma, J.A. , Kuppe, C. , Owen, M.R. , Mellor, N. , Griffiths, M. , Bennett, M.J. et al. (2017) OpenSimRoot: widening the scope and application of root architectural models. New Phytologist, 215, 1274–1286. 10.1111/nph.14641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertolas, J. , Larsen, E.K. , Davies, W.J. & Dodd, I.C. (2017) Applying 'drought' to potted plants by maintaining suboptimal soil moisture improves plant water relations. Journal of Experimental Botany, 68, 2413–2424. 10.1093/jxb/erx116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, P. , Song, W. , Yokoo, T. , Minobe, A. , Wang, G. , Ishida, T. et al. (2018) The CLE9/10 secretory peptide regulates stomatal and vascular development through distinct receptors. Nature Plants, 4, 1071–1081. 10.1038/s41477-018-0317-4 [DOI] [PubMed] [Google Scholar]
- Reichardt, S. , Piepho, H.P. , Stintzi, A. & Schaller, A. (2020) Peptide signaling for drought‐induced tomato flower drop. Science, 367, 1482–1485. 10.1126/science.aaz5641 [DOI] [PubMed] [Google Scholar]
- Reiser, V. , Raitt, D.C. & Saito, H. (2003) Yeast Sln1 and plant cytokinin receptor Cre1 respond to changes in turgor pressure. Journal of Cell Biology, 161, 1035–1042. 10.1083/jcb.200301099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y. , Song, Y. , Zhang, L. , Guo, D. , He, J. , Wang, L. et al. (2021) Coding of non‐coding RNA: Insights into the regulatory functions of pre‐microRNA‐encoded peptides in plants. Frontiers in Plant Science, 12, 641351. 10.3389/fpls.2021.641351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymaszewski, W. , Vile, D. , Bediee, A. , Dauzat, M. , Luchaire, N. , Kamrowska, D. et al. (2017) Stress‐related gene expression reflects morphophysiological responses to water deficit. Plant Physiology, 174, 1913–1930. 10.1104/pp.17.00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin, M. , Li, B. , Tang, M. , Katz, E. , Song, L. , Ecker, J.R. et al. (2019) Auxin‐sensitive AUX/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nature Communications, 10, 4021. 10.1038/s41467-019-12002-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, H. , Takasaki, H. , Takahashi, F. , Suzuki, T. , Iuchi, S. , Mitsuda, N. et al. (2018) Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proceedings of the National Academy of Sciences of the United States of America, 115, 11178–11187. 10.1073/pnas.1811491115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman, D.P. & Goodger, J.Q. (2008) Chemical root to shoot signaling under drought. Trends in Plant Science, 13, 281–287. 10.1016/j.tplants.2008.04.003 [DOI] [PubMed] [Google Scholar]
- Schmalenbach, I. , Zhang, L. , Reymond, M. & Jiménez‐Gómez, J.M. (2014) The relationship between flowering time and growth responses to drought in the Arabidopsis Landsberg erecta x Antwerp‐1 population. Frontiers in Plant Science, 5, 609. 10.3389/fpls.2014.00609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, T. , Kanno, Y. , Suzuki, H. , Watanabe, S. & Seo, M. (2021) Arabidopsis NPF4.6 and NPF5.1 control leaf stomatal aperture by regulating abscisic acid transport. Genes, 12, 885. 10.3390/genes12060885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik, D. , Nuriel, R. , Bonza, M.C. , Costa, A. & From, H. (2018) MIZ1 regulates ECA1 to generate a slow, long‐distance phloem‐transmitted Ca2+ signal essential for root water tracking in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America, 115, 8031–8036. 10.1073/pnas.1804130115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. , Ganapathysubramanian, B. , Singh, A.K. & Sarkar, S. (2016) Machine learning for high‐throughput stress phenotyping in plants. Trends in Plant Science, 21, 110–124. 10.1016/j.tplants.2015.10.015 [DOI] [PubMed] [Google Scholar]
- Singh, A. , Jones, S. , Ganapathysubramanian, B. , Sarkar, S. , Mueller, D. , Sandhu, K. et al. (2021) Challenges and opportunities in machine‐augmented plant stress phenotyping. Trends in Plant Science, 26, 53–69. 10.1016/j.tplants.2020.07.010 [DOI] [PubMed] [Google Scholar]
- Sirko, A. , Wawrzyńska, A. , Brzywczy, J. & Sieńko, M. (2021) Control of ABA signaling and crosstalk with other hormones by the selective degradation of pathway components. International Journal of Molecular Sciences, 22, 4638. 10.3390/ijms22094638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz, A. , Vandenbroucke, K. , Clauw, P. , Maleux, K. , De Meyer, B. , Dhondt, S. et al. (2011) Survival and growth of Arabidopsis plants given limited water are not equal. Nature Biotechnology, 29, 212–214. 10.1038/nbt.1800 [DOI] [PubMed] [Google Scholar]
- Soma, F. , Takahashi, F. , Yamaguchi‐Shinozaki, K. & Shinozaki, K. (2021) Cellular phosphorylation signaling and gene expression in drought‐stress responses: ABA‐dependent and ABA‐independent regulatory systems. Plants, 10, 756. 10.3390/plants10040756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strock, C.F. , Burridge, J.D. , Niemiec, M.D. , Brown, K.M. & Lynch, J.P. (2021) Root metaxylem and architecture phenotypes integrate to regulate water use under drought stress. Plant, Cell and Environment, 44, 49–67. 10.1111/pce.13875 [DOI] [PubMed] [Google Scholar]
- Stuhrwohldt, N. , Bruehler, E. , Sauter, M. & Schaller, A. (2021) Phytosulfokine (PSK) precursor processing by subtilase SBT3.8 and PSK signaling improve drought stresss tolerance in Arabidopsis . Journal of Experimental Botany, 72, 3427–3440. 10.1093/jxb/erab017 [DOI] [PubMed] [Google Scholar]
- Sussmilch, F. , Brodribb, T.J. & McAdam, S. (2017) Upregulation of NCED3 and ABA biosynthesis occur within minutes of a decrease in leaf turgor but AHK1 is not required. Journal of Experimental Botany, 68, 2913–2918. 10.1093/jxb/erx124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, F. , Hanada, K. , Kondo, T. & Shinozaki, K. (2019) Hormone‐like peptides and small coding genes in plant stress signaling and development. Current Opinion in Plant Biology, 51, 88–95. 10.1016/j.pbi.2019.05.011 [DOI] [PubMed] [Google Scholar]
- Takahashi, F. , Kuromori, T. , Sato, H. & Shinozaki, K. (2018a) Regulatory gene networks in drought stress responses and resistance in plants. In: Iwaya‐Inoue, M. , Sakurai, M. & Uemura, M. (Eds.) Survival strategies in extreme cold and desiccation. Singapore: Springer Nature Singapore Pte Ltd., pp. 189–214. [DOI] [PubMed] [Google Scholar]
- Takahashi, F. , Kuromori, T. , Urano, K. , Yamaguchi‐Shinozaki, K. & Shinozaki, K. (2020) Drought‐stress responses and resistance in plants: from cellular responses to long‐distance intercellular communication. Frontiers in Plant Science, 11, 556972. 10.3389/fpls.2020.556972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, F. & Shinozaki, K. (2019) Long‐distance signaling in plant stress response. Current Opinion in Plant Biology, 47, 106–111. 10.1016/j.pbi.2018.10.006 [DOI] [PubMed] [Google Scholar]
- Takahashi, F. , Suzuki, T. , Osakabe, Y. , Betsuyaku, S. , Kondo, Y. , Dohmae, N. et al. (2018b) A small peptide modulates stomatal control via abscisic acid in long‐distance signaling. Nature, 556, 235–238. 10.1038/s41586-018-0009-2 [DOI] [PubMed] [Google Scholar]
- Takahashi, N. , Goto, N. , Okada, K. & Takahashi, H. (2002) Hydrotropism in abscisic acid, wavy, and gravitropic mutants of Arabidopsis thaliana . Planta, 216, 203–211. 10.1007/s00425-002-0840-3 [DOI] [PubMed] [Google Scholar]
- Tardieu, F. , Cabrera‐Bosquet, L. , Pridmore, T. & Bennett, M. (2017) Plant phenomics, from sensors to knowledge. Current Biology, 27, R770–R783. 10.1016/j.cub.2017.05.055 [DOI] [PubMed] [Google Scholar]
- Thomas, H.R. & Frank, M. (2019) Connecting the pieces: uncovering the molecular basis for long‐distance communication through plant grafting. New Phytologist, 223, 582–589. 10.1111/nph.15772 [DOI] [PubMed] [Google Scholar]
- Thompson, A.L. , Thorp, K.R. , Conley, M. , Andrade‐Sanchez, P. , Heun, J.T. , Dyer, J.M. et al. (2018) Deploying a proximal sensing cart to identify drought‐adaptive traits in upland cotton for high‐throughput phenotyping. Frontiers in Plant Science, 9, 507. 10.3389/fpls.2018.00507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor, K. , Jiang, S. , Michard, E. , George, J. , Scherzer, S. , Huang, S. et al. (2020) The calcium‐permeable channel OSCA1.3 regulates plant stomatal immunity. Nature, 585, 569–573. 10.1038/s41586-020-2702-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisne, S. , Schmalenbach, I. , Reymond, M. , Dauzat, M. , Pervent, M. , Vile, D. et al. (2010) Keep on growing under drought: Genetic and developmental bases of the response of rosette area using a recombinant inbred line population. Plant, Cell and Environment, 33, 1875–1887. 10.1111/j.1365-3040.2010.02191.x [DOI] [PubMed] [Google Scholar]
- Tisne, S. , Serrand, Y. , Bach, L. , Gilbault, E. , Ben Ameur, R. , Balasse, H. et al. (2013) Phenoscope: an automated large‐scale phenotyping platform offering high spatial homogeneity. The Plant Journal, 74, 534–544. 10.1111/tpj.12131 [DOI] [PubMed] [Google Scholar]
- Tran, L.P. , Urao, T. , Qin, F. , Maruyama, K. , Kakimoto, T. , Shinozaki, K. et al. (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America, 104, 20623–20628. 10.1073/pnas.0706547105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S. , Cotton, S.C. , Emele, C.D. , Thomas, R. , Fielding, S. , Gaillard, E.A. et al. (2019) Reducing asthma attacks in children using exhaled nitric oxide as a biomarker to inform treatment strategy: a randomized trial (RAACENO). Trials, 20, 573. 10.1186/s13063-019-3500-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga, Y. , Sugimoto, K. , Ogawa, S. , Rane, J. , Ishitani, M. , Hara, N. et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nature Genetics, 45, 1097–1102. 10.1038/ng.2725 [DOI] [PubMed] [Google Scholar]
- Umezawa, T. , Okamoto, M. , Kushiro, T. , Nambara, E. , Oono, Y. , Seki, M. et al. (2006) CYP7070A3, a major ABA 8’‐hydrooxylase involved in dehydration and rehydration response in Arabidopsis thaliana . The Plant Journal, 46, 171–182. 10.1111/j.1365-313X.2006.02683.x [DOI] [PubMed] [Google Scholar]
- Urao, T. , Yakubov, B. , Satoh, R. , Yamaguchi‐Shinozaki, K. , Seki, M. , Hirayama, T. et al. (1999) A transmembrane hybrid‐type histidine kinase in Arabidopsis functions as an osmosensor. The Plant Cell, 11, 1743–1754. 10.1105/tpc.11.9.1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dooren, T.J.M. , Silveira, A.B. , Gilbault, E. , Jimenez‐Gomez, J.M. , Martin, A. , Bach, L. et al. (2020) Mild drought in the vegetative stage induces phenotypic, gene expression, and DNA methylation plasticity in Arabidopsis but no transgenerational effects. Journal of Experimental Botany, 71, 3588–3602. 10.1093/jxb/eraa132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Harsselaar, J.K. , Claußen, J. , Lübeck, J. , Wörlein, N. , Uhlmann, N. , Sonnewald, U. et al. (2021) X‐ray CT phenotyping reveals bi‐phasic growth phases of potato tubers exposed to combined abiotic stress. Frontiers in Plant Science, 12, 613108. 10.3389/fpls.2021.613108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur, F. , Bontpart, T. , Dauzat, M. , Granier, C. & Vile, D. (2014) Multivariate genetic analysis of plant responses to water deficit and high temperature revealed contrasting adaptive strategies. Journal of Experimental Botany, 65, 6457–6469. 10.1093/jxb/eru364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vile, D. , Pervent, M. , Belluau, M. , Vasseur, F. , Bresson, J. , Muller, B. et al. (2012) Arabidopsis growth under prolonged high temperature and water deficit: independent or interactive effects? Plant, Cell and Environment, 35, 702–718. 10.1111/j.1365-3040.2011.02445.x [DOI] [PubMed] [Google Scholar]
- Whitewoods, C.D. (2020) Evolution of CLE peptide signalling. Seminars in Cell & Developmental Biology, 109, 12–19. 10.1016/j.semcdb.2020.04.022 [DOI] [PubMed] [Google Scholar]
- Wilkinson, S. & Davies, W.J. (2002) ABA‐based chemical signaling: the co‐ordination of responses to stress in plants. Plant, Cell and Environment, 25, 195–210. 10.1046/j.0016-8025.2001.00824.x [DOI] [PubMed] [Google Scholar]
- Wohlbach, D.J. , Quirino, B.F. & Sussman, M.R. (2008) Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. The Plant Cell, 20, 1101–1117. 10.1105/tpc.107.055871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Chi, Y. , Jiang, Z. , Xu, Y. , Xie, L. , Huang, F. et al. (2020) Hydrogen peroxide sensor HPCA1 is an LRR receptor kinase in Arabidopsis . Nature, 578, 577–581. 10.1038/s41586-020-2032-3 [DOI] [PubMed] [Google Scholar]
- Yang, W. , Guo, Z. , Huang, C. , Duan, L. , Chen, G. , Jiang, N. et al. (2014) Combining high‐throughput phenotyping and genome‐wide association studies to reveal natural genetic variation in rice. Nature Communications, 5, 5087. 10.1038/ncomms6087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, T. , Fernie, A.R. , Shinozaki, K. & Takahashi, F. (2021) Long‐distance stress and developmental signals associated with abscisic acid signaling in environmental responses. The Plant Journal, 105, 477–488. 10.1111/tpj.15101 [DOI] [PubMed] [Google Scholar]
- Yoshimura, K. , Iida, K. & Iida, H. (2021) MCAs in Arabidopsis are Ca2+‐permeable mechanosensitive channels inherently sensitive to membrane tension. Nature Communications, 12, 6074. 10.1038/s41467-021-26363-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, F. , Yang, H. , Xue, Y. , Kong, D. , Ye, R. , Li, C. et al. (2014) OSCA1 mediates osmotic‐stress‐evoked Ca2+ increases vital for osmosensing in Arabidopsis . Nature, 514, 367–371. 10.1038/nature13593 [DOI] [PubMed] [Google Scholar]
- Zandalinas, S. , Fichman, Y. , Devireddy, A. , Sengupta, S. , Azad, R.K. & Mittler, R. (2020) Systemic signaling during abiotic stress combination in plants. Proceedings of the National Academy of Sciences of the United States of America, 117, 13810–13820. 10.1073/pnas.2005077117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Shi, X. , Zhang, Y. , Wang, J. , Yang, J. , Ishida, T. et al. (2019) CLE9 peptide‐induced stomatal closure is mediated by abscisic acid, hydrogen peroxide, and nitric oxide in Arabidopsis thaliana . Plant, Cell and Environment, 42, 1033–1044. 10.1111/pce.13475 [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Yu, Z. , Xu, Y. , Yu, M. , Ren, Y. , Zhang, S. et al. (2021) Regulation of the stability and ABA import activity of NRT1.2/NPF4.6 by CEPR2‐mediated phosphorylation in Arabidopsis . Molecular Plant, 14, 1–14. 10.1016/j.molp.2021.01.009 [DOI] [PubMed] [Google Scholar]
- Zhu, J.‐K. (2016) Abiotic stress signaling and responses in plants. Cell, 167, 313–322. 10.1016/j.cell.2016.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is a review article. All relevant data can be found within the manuscript and its supporting materials.


