Abstract
Clostridium botulinum neurotoxin type A (BTx-A) is known to inhibit the release of acetylcholine at the neuromuscular junctions and synapses and to cause neuroparalysis and death. In this study, we have identified two monoclonal antibodies, BT57-1 and BT150-3, which protect ICR mice against lethal doses of BTx-A challenge. The neutralizing activities for BT57-1 and BT150-3 were 103 and 104 times the 50% lethal dose, respectively. Using immunoblotting analysis, BT57-1 was recognized as a light chain and BT150-3 was recognized as a heavy chain of BTx-A. Also, applying the phage display method, we investigated the antibodies' neutralizing B-cell epitopes. These immunopositive phage clones displayed consensus motifs, Asp-Pro-Leu for BT57-1 and Cys-X-Asp-Cys for BT150. The synthetic peptide P4M (KGTFDPLQEPRT) corresponded to the phage-displayed peptide selected by BT57-1 and was able to bind the antibodies specifically. This peptide was also shown by competitive inhibition assay to be able to inhibit phage clone binding to BT57-1. Aspartic acid (D5) in P4M was crucial to the binding of P4M to BT57-1, since its binding activity dramatically decreased when it was changed to lysine (K5). Finally, immunizing mice with the selected phage clones elicited a specific humoral response against BTx-A. These results suggest that phage-displayed random-peptide libraries are useful in identifying the neutralizing epitopes of monoclonal antibodies. In the future, the identification of the neutralizing epitopes of BTx-A may provide important information for the identification of the BTx-A receptor and the design of a BTx-A vaccine.
Clostridium botulinum neurotoxin A (BTx-A), produced by the anaerobic bacterium C. botulinum, is one of the most potent toxins known to humans (14, 15). The seven serologically different BTxs are highly potent protein toxins that inhibit neurotransmitter release from peripheral cholinergic synapses. BTxs are synthesized as single-chain polypeptides of approximately 150 kDa (10, 33, 35). These neurotoxins are structurally similar, with all consisting of a heavy chain (100 kDa) and a light chain (50 kDa) linked by a disulfide bridge (10, 35). The 50-kDa N terminus of the heavy chain has been postulated to form channels in membranes that induce the translocation of the light chain into the cytosol (6, 31). The light chains of neurotoxins act as zinc-dependent endoproteases, cleaving proteins that are essential for release of the neurotransmitter acetylcholine and leading to paralysis (5, 28, 33). Although there are several different routes through which the toxin can enter the body, most cases involve ingestion of toxin or ingestion of bacteria that produce the toxin. Because the identification of neutralizing epitopes for BTx might provide important information leading to the development of a safe, effective vaccine and might contribute to our understanding of the mechanism by which BTx binds to its receptor, we aimed in this study to generate neutralizing monoclonal antibodies (MAbs) against BTx-A and to use the phage display method to investigate their B-cell epitopes.
Phage display is a selection technique in which a peptide or protein is expressed as a fusion with a coat protein of bacteriophage, resulting in a display of the fused protein on the surface of the virion. Therefore, phage-displayed random-peptide libraries allow for the rapid identification the peptide ligands for a variety of target molecules (antibodies, enzymes, and cell surface receptors) through an in vitro selection process called biopanning. This method has been used for several purposes, including mapping B-cell epitopes (29, 40, 41), mapping protein-protein contacts (3, 23, 34), selecting bioactive peptides bound to receptors (19, 21, 39) or proteins (8, 20, 25), selecting disease-specific antigen mimics (13, 26), selecting cell (4, 22, 36)- and organ (1, 24, 27)-specific peptides, producing peptides that mimic the effect of neutralizing antibodies (17), and identifying peptides that mimic nonpeptide ligands (9).
In the present study, two neutralizing MAbs were generated that exhibited highly protective activity against BTx-A. The neutralizing epitopes of both antibodies were also identified by use of a phage-displayed random-peptide library. Using the hybrid phage system, we also demonstrated that the phage-displayed epitope could elicit antibody against BTx-A when the hybrid phage particles were used directly as antigens. This method proved useful in the identification of B-cell epitopes, and it might lead to the facile creation of antibodies against designated peptides.
MATERIALS AND METHODS
Antigen preparation and immunization.
The BTx-A was purified from cultures of C. botulinum type A by a previously described method (11). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and immunoblotting further identified the purified BTx-A. For immunization, BALB/c mice received an intraperitoneal (i.p.) injection of 25 μg of Formalin-inactivated BTx-A in 100 μl of phosphate-buffered saline (PBS) emulsified in an equal volume of complete Freund's adjuvant. After intervals of 2 and 4 weeks, booster injections were given as outlined above except that we used incomplete Freund's adjuvant instead. Three weeks after the third injection, final boosters containing 50 μg of antigen were administered via i.p. injection. Fusion with the spleen cells of the donor mouse was performed 5 days after the last injection.
Generation of MAbs.
Hybridomas secreting anti-BTx-A antibodies were generated by standard procedures (18). Briefly, the spleen of the immunized mouse was removed, and the splenocytes were fused with NSI/1-Ag4-1 (NS-1) myeloma cells. The splenocytes and the myeloma cells were washed twice with serum-free Dulbecco modified Eagle medium (DMEM). The final pellet was mixed in a 15-ml conical tube, and 1 ml of 50% (vol/vol) polyethylene glycol (GIBCO BRL) was added with gently stirring over a 1-min period. The mixture was diluted by the slow addition (over 1 min) of 1 ml of DMEM twice, followed by the slow addition (over 2 min) of 8 ml of DMEM without serum. The mixture was centrifuged at 400 × g for 5 min, and the fused cell pellet was resuspended in DMEM supplemented with 15% fetal bovine serum, hypoxanthine-aminopterin-thymidine medium, and hybridoma cloning factor (ICN, Aurora, Ohio) and distributed (150 μl per well) in 96-well tissue culture plates. Hybridoma colonies were screened by enzyme-linked immunosorbent assay (ELISA) for secretion of MAbs that would bind to BTx-A. Selected clones were subcloned by the limiting-dilution method. Immunoglobulin classes and subclasses were determined using a subtyping kit (Roche Diagnostics, Penzberg, Germany). Ascitic fluids were produced in pristane-primed BALB/c mice.
Screening of neutralizing antibodies against BTx-A.
Sixteen MAb-producing cell lines that could recognize BTx-A were generated. To screen for neutralizing antibodies against BTx-A, 101 to 106 times the 50% lethal dose (LD50) of BTx-A was mixed with anti-BTx-A or normal control ascites for 1 h prior to i.p. administration to ICR mice. Survivors were observed daily for 14 days following the challenge.
Selection of immunopositive phage clones by biopanning.
The ELISA plate was coated with a 100-μg/ml concentration of MAbs against BTx-A in 0.1 M NaHCO3 (pH 8.6) buffer at room temperature and gently agitated for 2 h. The plate was then incubated with blocking buffer (1% bovine serum albumin in PBS) at 4°C overnight and washed rapidly five times with PBS plus 0.5% (wt/vol) Tween 20 (PBST). The phage-displayed random-peptide libraries (Ph.D.-12; New England Biolabs, Inc., Beverly, Mass.) were diluted to 4 × 1010 phage particles, added to the coated plate, and rocked gently for 50 min at room temperature. The plate was then washed 10 times with PBST. The bound phage was eluted with 100 μl of 0.2 M glycine-HCl (pH 2.2)–1 mg of bovine serum albumin per ml and neutralized with 15 μl of 1 M Tris-HCl (pH 9.1). The eluted phage was amplified at 37°C in an Escherichia coli ER2537 culture, which was vigorously shaken for 4.5 h. The amplified phage was centrifuged for 20 min at 10,000 × g at 4°C, and the supernatant was removed to a fresh tube and respun. The upper 80% of the supernatant was removed to a fresh tube, and one-sixth volume of 20% (wt/vol) polyethylene glycol 8000–2.5 M NaCl was added to precipitate the phage particles at 4°C overnight. The phage particles were isolated by centrifugation, and the phage pellet was suspended in 1 ml of PBS. The supernatant was reprecipitated with one-sixth volume of 20% polyethylene glycol 8000–2.5 M NaCl, and the phage particles were resuspended in 200 μl of PBS containing 0.02% NaN3. The isolated phage particles were centrifuged for 1 min to precipitate any remaining insoluble matter, and the titer was determined on Luria-Bertani medium–IPTG (isopropyl-β-d-thiogalactopyranoside)–X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates. The biopanning protocol for the second and third rounds was identical to that for the first round. The titer of the unamplified third-round phage particles was determined on Luria-Bertani medium–IPTG–X-Gal plates, and the immunopositive phage clones were screened by ELISA.
Purification of phage DNA for sequencing.
The purified phage was suspended in 100 μl of iodide buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 4 M NaI), and then 250 μl of ethanol was added and incubated at room temperature for 10 min. Phage DNA was isolated by centrifugation at 12,000 × g for 10 min, the supernatant was discarded, and the pellet was washed in 70% ethanol and dried briefly. The phage DNA was sequenced by the dideoxynucleotide chain termination method using an automated DNA sequencer (ABI PRISM 377; Perkin-Elmer, Foster City, Calif.) or manually using a Sequenase kit 2.0 (United States Biomedical Corp., Cleveland, Ohio). The primer used for phage DNA sequencing was 5′-CCCTCATAGTTAGCGTAA-3′. This primer is located in the antisense strand of gene III of the M13 phage and has 96 nucleotides separated from the inserted DNA. The phage-displayed peptide sequences were aligned using MacDNASIS software (Hitachi Software Engineering Co., Ltd., Yokohama, Japan).
Identification of phage clones by ELISA.
The ELISA plate was coated with 100 μg of antibody per ml and blocked with blocking buffer at 4°C overnight. The serially diluted phage was added to the antibody-coated plate and incubated at room temperature for 1 h. The plate was incubated with horseradish peroxidase-conjugated antibacteriophage antibody (Pharmacia no. 27-9411-01) for 1 h with agitation and washed six times with PBST. The peroxidase substrate, o-phenylenediamine dihydrochloride (Sigma), was added to each well and incubated at room temperature. The o-phenylenediamine dihydrochloride reaction was stopped with 3 N HCl, and the absorbance at 490 nm was read with a microplate reader.
Immunoblotting analysis.
Proteins or antigens were mixed with an equal volume of the sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 0.1% bromophenol blue, 10% glycerol), separated by SDS-polyacrylamide gel electrophoresis under native or denaturing conditions, and transferred to a nitrocellulose membrane (Hybond-C Super; Amersham, Little Chalfont, United Kingdom). The nonspecific antibody-binding sites were blocked with 3% skim milk in PBS, and the filters were incubated with primary antibodies. The blot was then treated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (Jackson ImmunoResearch Laboratories, West Grove, Pa.) and developed with chemiluminescence reagents (ECL; Amersham).
Mouse immunization.
Immunizing phage was prepared from E. coli ER2537-infected cells and purified as described above. Phage particles were resuspended as a 0.9% NaCl suspension at a concentration of 1013 phage particles/ml. Four- to 6-week-old female ICR mice were immunized by i.p. injection of 100 μl of phage solution or 50 μg of immunogens with adjuvant at days 0, 14, 28, and 42 and were bled at day 52. Antibody titers were measured by ELISA and immunoblotting assay.
RESULTS
Reactivities of the anti-BTx-A neutralizing MAbs in ELISA and immunoblotting assay.
We generated 16 MAbs against BTx-A, 2 of which were identified as neutralizing MAbs against BTx-A: BT57-1 and BT150-3. The interaction of each MAb with BTx-A was analyzed by ELISA and immunoblotting. The ELISA titers were derived from curves represented graphically in Fig. 1A. The BT57-1 and BT150-3 MAbs recognized BTx-A antigens specifically and dose dependently. Both MAbs were found to have absorbances of >0.4 at a 105-fold dilution. Control ascites produced absorbances of <0.1 at a 100-fold dilution (Fig. 1A). To further investigate the specificities of the two MAbs in recognizing BTx-A, immunoblotting analysis was performed. The results showed that both MAbs recognized BTx-A in a native gel without β-mercaptoethanol (Fig. 1B, lanes 1 and 2). When the light and heavy chains of BTx-A were separated in a denatured gel with β-mercaptoethanol, we found that the BT57-1 and BT150-3 MAbs recognized the light chain and heavy chain of BTx-A, respectively (Fig. 1B, lanes 3 and 4).
FIG. 1.
(A) ELISA titers of MAbs against BTx-A. The reactivities of MAbs were determined with 10-fold serial dilutions (from 102 to 108) of ascites-incubated plates coated with BTx-A antigen as indicated in Materials and Methods. Error bars indicate standard deviations. NA, normal control ascites; OD490 nm, optical density at 490 nm. (B) Analysis of MAbs against BTx-A by immunoblotting. BTx-A and the heavy chain and light chain of BTx-A were size fractionated on SDS-polyacrylamide gels with or without β-mercaptoethanol (2ME) and blotted. The blot was incubated with anti-BTx-A MAbs. Lanes 1 and 2 show that both antibodies recognized the intact BTx-A when sample buffer without 2ME was used. Lanes 3 and 4 show that the MAbs BT57-1 and BT150-3 recognized the light (L) and heavy (H) chains of BTx-A, respectively. Numbers on the left are molecular masses in kilodaltons.
Measurement of neutralizing abilities of MAbs against BTx-A in vivo.
The neutralizing activities against BTx-A were demonstrated by challenging ICR mice with lethal doses of BTx-A. An LD50 of purified BTx-A was determined by i.p. injection into ICR mice. Three individual experiments determined that the LD50 was 10 pg of purified BTx-A (data not shown). All of the mice showed typical symptoms of BTx poisoning, namely, labored breathing, hind limb paralysis, and death in 2 days, when they were challenged with 102 times the LD50 of BTx-A. However, the mice were found to be protected from such poisoning when the same dose of BTx-A was preincubated with BT57-1 and BT150-3 ascites (data not shown). The protective abilities of the two MAbs were further determined by using them against different doses of BTx-A and comparing their activities. Our results revealed that BT57-1 and BT150-3 protected all of the mice from 103 (10 ng) and 104 (100 ng) times the LD50 of BTx-A, respectively (Table 1). In contrast, all of the mice died when doses of 102 and 103 times the LD50 of BTx-A incubated with control ascites were used (Table 1).
TABLE 1.
Characterization and determination of the neutralizing activities of anti-BTx-A MAbs
| Antibody | Immunoglobulin class | Subunit recognized by antibody | No. of survivors/total no. at challenge dose (LD50)a of:
|
||||
|---|---|---|---|---|---|---|---|
| 102 | 103 | 104 | 105 | 106 | |||
| BT57-1 | IgG1 | Light chain | 6/6 | 6/6 | 2/6 | 1/6 | 0/6 |
| BT150-3 | IgG1 | Heavy chain | 6/6 | 6/6 | 6/6 | 3/6 | 0/6 |
| Controlb | 0/8 | 0/6 | |||||
Neutralization results at 2 weeks following challenge.
NS-1 myeloma cell-induced ascites.
Screening of phage-displayed peptide libraries with neutralizing antibody.
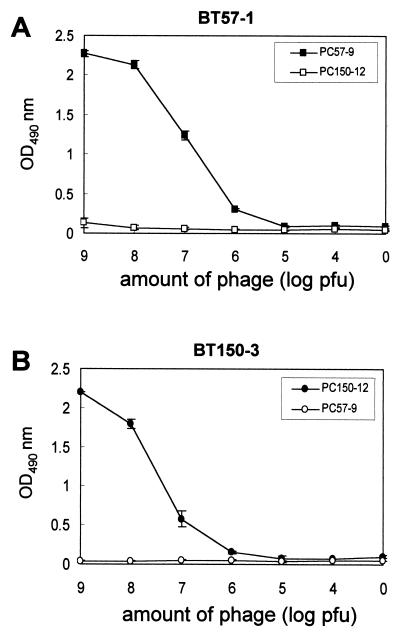
To study the B-cell epitopes of neutralizing MAbs against BTx-A, the phage display method was used. The affinity-purified antibodies were immobilized on the ELISA plate, and the bound phage clones were selected after three biopanning cycles. The selected phage clones produced by BT57-1 and BT150-3 showed significant increases in reactivity to their antibodies. The selected phage clones did not bind to normal mouse serum or immunoglobulin G (IgG) (data not shown). To further confirm that the immunopositive phage clone bound BT57-1 specifically, the antibody was incubated with a 10-fold serial dilution of selected (PC57-9) and control (PC150-12, selected by BT150-3) phage clones. The results showed that only the BT57-1-selected phage clone (PC57-9) bound its antibody specifically in a dose-responsive manner, whereas the control phage clone (PC150-12) did not react with the antibody (Fig. 2A). The other neutralizing antibody BT150-3-selected phage clone (PC150-12) bound its antibody specifically and did not react with BT57-selected phage clone PC57-9 (Fig. 2B).
FIG. 2.
Specific reactivities of selected phage clones with MAbs. (A) BT57-1 was incubated with 10-fold serial dilutions (from 109 to 104 PFU) of the selected phage clone (PC57-9) and control phage clone (PC150-12). (B) BT150-3 was incubated with 10-fold serial dilutions of the selected phage clone (PC150-12) and control phage clone (PC57-9). The selected phage clones reacted with their antibodies specifically. Error bars indicate standard deviations. OD490 nm, optical density at 490 nm.
Identification of neutralizing epitopes.
Eight (PC57-3, -4, -5, -6, -7, -8, -9, and -10) and six (PC150-2, -5, -12, -14, -15, and -23) immunopositive phage clones highly reactive with neutralizing antibodies BT57-1 and BT150-3, respectively, were further sequenced. The phage-displayed peptide sequences were aligned using MacDNASIS software to analyze the epitopes of the neutralizing antibodies. The binding motif D-P-L was found in phage clones PC57-3, -4, -9, and -10; the binding motif D-A-L was found in phage clones PC57-6 and -7; and the binding motif D-V-L/F was found in phage clones PC57-5 and -8 (Table 2). Through phage-displayed peptide sequence alignment using MacDNASIS software, the binding motif of the antibody BT150-3 was shown to be C-X-D-C. This motif was exhibited in all six immunopositive phage clones (Table 2). Alignment of the phage-displayed peptide sequences with the protein sequence of BTx-A revealed that there was no significant homology between them, and both epitopes may have belonged to mimotopes (mimic epitopes).
TABLE 2.
Alignment of phage-displayed peptide sequences selected by anti-BTx-A neutralizing antibodies
| Phage clone | Phage-displayed peptide sequencea |
|---|---|
| BT57-1 selected | |
| PC57-9 | K G T F D P L Q E P R T |
| PC 57-10 | Q D P L F P A M A T W A |
| PC 57-3 | W D P L L P P F P P P N |
| PC 57-4 | S P M D R F D P L H L P |
| PC 57-6 | N N P Y D A L Y N F P R |
| PC 57-7 | L S K H Q F D A L L P P |
| PC 57-8 | D S M D V F Q P P P I W |
| PC 57-5 | S L F D V L W S P P P K |
| BT150-3 selected | |
| PC150-2 | M C I D C Y L S T L G L |
| PC 150-5 | Y I K P P V S C Y D C Q |
| PC 150-12 | M C W D C T L H A R S D |
| PC 150-14 | Y I K P P V S C Y D C Q |
| PC 150-15 | M C I D C Y L S T L G L |
| PC 150-23 | C Y S M C L D C I M F G |
Phage-displayed consensus amino acids are shown in boldface.
Binding assay of synthetic peptide mimic.
In order to verify that the phage-displayed peptide sequences were indeed recognized by MAb BT57-1, synthetic peptide-binding assays were performed. The peptides were synthesized in multiple-antigen peptide form, because the ELISA sensitivity for this eight-chain multiple-antigen peptide was greater than that for single-chain peptides (37). As shown in Fig. 3A, the synthetic peptide KGTFDPLQEPRT (P4M), displayed by phage clone PC57-9, binds the antibody in a concentration-dependent manner. Two unrelated control peptides, SHRLHNTMPSES (P7M) and ELKYSWKS (14Mu), revealed no such reactivity. To further confirm that the phage-displayed peptide was the epitope of BT57-1, a peptide-competitive inhibition assay was performed to determine whether the synthetic peptide P4M and the selected phage clone (PC57-9) competed for the same antibody-binding site. The binding activity of BT57-1 with the phage clone (PC57-9) was inhibited by synthetic peptide P4M in a dose-dependent manner. The arbitrary control peptide SHRLHNTMPSES (P7M) had no effect on the ability of the phage particles to bind BT57-1 (Fig. 3B). A 1-μg/ml concentration of P4M peptide was able to inhibit 91% of the phage clone binding to BT57-1 antibody.
FIG. 3.
Characterization of the neutralizing epitope of BT57-1. (A) Assay of binding of synthetic peptide with BT57-1. The P4M peptide antigen reacted with BT57-1 specifically, but the control peptide antigens 14Mu and P7M did not. OD490 nm, optical density at 490 nm. (B) Competitive inhibition of phage clone binding to BT57-1 by synthetic peptide. The BT57-1-selected phage clone (PC57-9) binding to BT57-1 was inhibited by the P4M peptide, while a control peptide (P7M) had no effect. (C) Identification of the amino acid residue for binding to BT57-1. The reactivity of P4M with BT57-1 was markedly decreased when the Asp in the P4M peptide was changed to Lys in the P4M-m peptide. Two control peptides, P7M and P7M-m1, had no reactivity with BT57-1. Error bars indicate standard deviations.
Using the phage display method, we found that the binding motif for BT57-1 was Asp-Pro-Leu. The aspartic acid (Asp) was found in all of the immunopositive phage clones (Table 2). Therefore, this amino acid might play a crucial role in the binding to the antibody. To prove this hypothesis, we synthesized the peptide P4M-m (KGTFKPLQEPRT), which has only one amino acid residue difference from P4M (KGTFDPLQEPRT), and ELISA was performed. The binding activity was markedly weaker when the negatively charged Asp in the peptide P4M was changed to positively charged Lys (Fig. 3C). The control peptides P7M (SHRLHNTMPSES) and P7M-m1 (SLRLHNTMPSES) had no reactivity with BT57-1 (Fig. 3C). These experimental observations strongly suggested that Asp was an important amino acid residue for binding to MAb BT57-1.
Antigenicities of the phage-displayed peptides.
To test the antigenicities of the peptides as displayed in the bacteriophages, hybrid phages were purified as immunizing agents without the addition of any immunostimulants. PC57-9 phage particles were injected into five ICR mice, while five mice were immunized with wild-type phage particles. All sera from mice immunized with phage PC57-9 showed an anti-BTx-A response ranging from 2- to 10-fold the background signal observed using the sera from five mice immunized with wild-type phage (Fig. 4A). The difference in reactivity of immunized sera with BTx-A was also observed between sera from individual animals injected with the same phage particles. The specificity of the antibody induced by phage was further demonstrated by immunoblotting analysis. BTx-A antigens (0.1 μg/lane) were separated on denatured gels and transferred to nitrocellulose membranes. The membranes were immunostained with MAb BT57-1, PC57-9 phage-immunized mouse sera, and mixed control sera. The results showed that antibodies generated by phage clone PC57-9-immunized mice recognized the 50-kDa light chain of BTx-A (Fig. 4B).
FIG. 4.
The phage-displayed mimotope is an immunogenic mimic of BTx-A protein. The reactivities with BTx-A of five serum samples (M1, M2, M3, M4, and M5) from mice immunized with phage clone PC57-9 were assayed by ELISA (A) and immunoblotting (B). Five control serum samples (C1, C2, C3, C4, and C5) were obtained from mice immunized with wild-type phage. Error bars indicate standard deviations. Numbers on the left in panel B are molecular masses in kilodaltons. CM, mixed control sera (C1, C2, C3, C4, and C5); OD490 nm, optical density at 490 nm.
DISCUSSION
To study the biochemical and functional properties of BTx-A, we generated MAbs which bound BTx-A in several types of assays. While doing this, we characterized two neutralizing antibodies that exhibited highly protective activities against BTx-A. Our study also became the first to identify neutralizing B-cell epitopes of BTx-A. We further proved that phage-displayed mimotopes could induce antibodies that were able to recognize native BTx-A antigen.
Two out of 16 MAbs against BTx-A, BT57-1 and BT150-3, were shown to exhibit highly neutralizing effects on lethal doses of BTx-A by using the mouse lethality bioassay neutralization test, which has been the standard method of measuring antibodies to BTxs for many years. The neutralizing antibody BT57-1 was found by immunoblotting analysis to recognize the light chain of BTx-A (50 kDa) and was tested for its ability to protect against a dose of 103 time the LD50 of BTx-A. The light chain of BTx-A, which includes the HEXXH zinc-binding motif of metalloendopeptidase, exhibits proteolytic activity for the synaptic vesicle membrane proteins (5, 33). Cleavage of these proteins leads to a blockade of neurotransmitter release and paralysis. The BT57-1 protection of mice from our BTx-A challenge could have been due to the binding of the epitope, which might have inhibited the endopeptidase activity or inhibited penetration of the light chain into the cytosol. The other neutralizing antibody, BT150-3, recognized the heavy chain of BTx-A (100 kDa) as determined by immunoblotting analysis and was also tested for its ability to protect against our challenge with 104 times the LD50 of BTx-A. The high protective activity of BT150-3 could possibly be due to the binding of its epitope, which might involve the cell-binding site on the 50-kDa C terminus of the heavy chain (30) or membrane channel formation to induce translocation on the 50-kDa N terminus of the heavy chain (6, 31). However, this hypothesis needs further investigation.
BTx is the etiologic agent associated with the disease botulism and is the most toxic substance known to science. This disease is typically contracted by ingesting food contaminated with organisms that can manufacture this toxin in the gut. By generating neutralizing MAbs against BTx and studying their B-cell epitopes, we might be able to discover important information leading to the treatment or prevention of the disease botulism. Therefore, we also identified the B-cell epitopes of the neutralizing antibodies by the phage display method. The immunopositive phage clones with higher reactivity selected by BT57-1 contained the binding motif Asp-Pro-Leu. The synthetic peptide P4M, which mimics the phage-displayed peptide sequence KGTFDPLQEPRT, was shown to bind BT57-1 specifically. Such reactivity was dramatically decreased when the negatively charged Asp in the P4M peptide was changed to positively charged Lys. Therefore, we concluded that P4M was the epitope of BT57-1 and that the negatively charged residue Asp played an important role in the interaction of the antigenic epitope with the antibody. Furthermore, we also demonstrated that P4M was able to compete for the same antibody-binding site with phage clone PC57-9 by competitive inhibition assay. However, when aligning the P4M peptide sequence with BTx-A, no significant homology was found between them, indicating that KGTFDPLQEPRT (P4M) was a mimotope of BT57-1. The epitope of BT150-3 was also identified by random-peptide libraries displayed on phage. All of the immunopositive phage clones selected by BT150-3 contained the Cys-Xxx-Asp-Cys binding motif. The synthetic peptide P5M, mimicking the phage (PC150-12)-displayed peptide sequence MCWDCTLHARSD, was not soluble in many solvents, and so we were not able to study its binding activity with BT150-3. Our results demonstrated that BT150-3 had a greater neutralizing activity than BT57-1 and that it recognized the heavy chain of BTx-A, making it a very interesting candidate for study of the receptor of BTx-A. Unfortunately, the synthetic peptide to this mimotope was insoluble. In fact, we changed some hydrophobic amino acid residues to hydrophilic amino acid residues to increase the solubility of the synthetic peptides, but these peptides lost reactivity and were not recognized by BT150-3. Phage clone PC150-12 selected by MAb BT150-3 reacted highly with the antibodies. Therefore, construction of recombinant fusion proteins to display this mimotope or expression of the mimotope on a region of the major coat protein of phage particles in the future may be helpful in the characterization of the B-cell epitope of BT150-3 and in the study of the possible receptor of BTx-A.
The use of phage-displayed random-peptide libraries has made it possible for us to identify B-cell epitopes using MAbs as selector molecules. Typically, B-cell epitopes identified by this method have an easily recognizable consensus sequence, often corresponding to the peptide sequence found in the natural antigen. In one of our recent studies, we identified the B-cell epitope of dengue virus type 1 from random-peptide libraries displayed on phage. The phage-displayed peptides had a consensus motif which corresponded to amino acid residues 111 to 116 of nonstructural protein 1 of the virus (40). However, sometimes the phage-displayed consensus sequence cannot be found in the sequence of the natural antigens. For example, our present study and unpublished data, as well as some other reports (12, 13), have shown that the phage-displayed epitopes interacted with the antigen-binding sites of antibodies by peptide-binding or competition assays. However, their consensus sequences did not show any similarity with the sequence of the natural antigen (12, 13). In these cases, it is possible that the epitopes mimic natural epitopes or conformational epitopes. The B-cell epitopes of BTx-A have also been identified by overlapping synthetic peptides for PEPSCAN. However, this approach, which requires many overlapping synthetic peptides, has difficulty identifying the conformational epitopes, and the amino acid-binding motif cannot be obtained (2). Random-peptide libraries displayed on phage, which mimic continuous or discontinuous epitopes, can be selected using purified antibodies or serum samples. This method is useful for analyzing conformational epitopes or mimotopes, which, as mentioned above, are generally difficult to characterize.
Phage clone PC57-9 displaying the identified mimotope mimics the binding properties of P4M common to the antibody BT57-1 (Fig. 3). However, their peptide sequences shared no homology with the natural BTx-A. It was therefore important to verify whether the phage-displayed mimotope mimicked the natural epitope to induce antibody that could recognize BTx-A. In this way, we demonstrated by ELISA and immunoblotting analysis that five mice immunized with phage clone PC57-9 generated a specific response against BTx-A (Fig. 4). The phage itself is an excellent immunogen. Some reports have demonstrated that injecting mice with phage without adjuvant can elicit a T-cell-dependent response and antibodies against the displayed epitope (13, 16, 26, 38). Our data also confirmed that the phage-displayed peptide could act as an immunogenic mimic and elicit an immune response when the hybrid phage particles were used as antigen without adjuvant (Fig. 4). We also challenged the phage-immunized mice with BTx-A to evaluate their protection activities, although no significant protection response was observed. The poor protective competence of phage-immunized mice might be due to the generation of lower titers of antibodies by the low copy numbers (five copies of each phage particle) of the minor coat protein in each phage particle. Expression of a peptide on a region of the major coat protein (approximately 2,700 copies, encoded by phage gene VIII) should induce a more powerful immune response (13, 16, 26). Further development of a neutralizing epitope displayed on major coat protein or an epitope-based peptide to improve the immune response is in progress.
Human botulism is most frequently caused by BTx-A, -B, and -E. In this study, we have generated neutralizing MAbs against BTx-A and identified their B-cell epitopes. Generation of neutralizing MAbs against BTx-B and -E is in progress. We also plan to identify the neutralizing epitopes of BTx-B and -E in the future. Such results will provide important information for the development of safe and effective botulism vaccines. In addition to the currently used toxoids, some of the potential botulism vaccine candidates, such as recombinant vaccines (7) and DNA vaccines (32), have been developed. Using overlapping 19-residue peptides from residue 855 to 1296 of BTx-A, Atassi et al. identified six synthetic peptides that could bind with human antitoxin antibodies and that may be useful as immunogens to induce immunity against BTx-A in the future (2). Random-peptide libraries displayed on phage could be applied to identify the epitopes that fit the antibodies and were shown to be good immunogens. Therefore, the preparation of disease-specific peptide vaccines by use of random-peptide libraries might dramatically simplify the identification, cloning, and expression of the recombinant immunogen (13).
In summary, the 16 MAbs against BTx-A, including two neutralizing antibodies, that we generated can be useful reagents in the diagnosis of food-borne botulism, in the study of the structure and function of BTx-A, or in the routine monitoring of BTx-A content in therapeutic preparations. Furthermore, the application of phage-displayed peptide libraries to identify the neutralizing epitopes of BTx-A may prove to be invaluable in the study of its receptor and in the creation of a vaccine for the disease.
ACKNOWLEDGMENTS
We thank H.-Y. Chao for his kind gift of BTx-A.
This work was supported by research grants NSC 89-2320-B-016-027 and NSC 89-2320-B-016-064 from the National Science Council, Republic of China, to H.-C.W. and by grant 89-0303 from the Institute of Preventive Medicine, National Defense Medical Center, Taipei, Republic of China, to H.-C.W.
REFERENCES
- 1.Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 2.Atassi M Z, Dolimbek B Z, Hayakari M, Middlebrook J L, Whitney B, Oshima M. Mapping of the antibody-binding regions on botulinum neurotoxin H-chain domain 855–1296 with antitoxin antibodies from three host species. J Protein Chem. 1996;15:691–700. doi: 10.1007/BF01886751. [DOI] [PubMed] [Google Scholar]
- 3.Atwell S, Ultsch M, De Vos A M, Wells J A. Structural plasticity in a remodeled protein-protein interface. Science. 1997;278:1125–1128. doi: 10.1126/science.278.5340.1125. [DOI] [PubMed] [Google Scholar]
- 4.Barry M A, Dower W J, Johnston S A. Toward cell-targeting gene therapy vectors: selection of cell-binding peptides from random peptide-presenting phage libraries. Nat Med. 1996;2:299–305. doi: 10.1038/nm0396-299. [DOI] [PubMed] [Google Scholar]
- 5.Binz T, Blasi J, Yamasaki S, Baumeister A, Link E, Sudhof T C, Jahn R, Niemann H. Proteolysis of SNAP-25 by types E and A botulinal neurotoxins. J Biol Chem. 1994;269:1617–1620. [PubMed] [Google Scholar]
- 6.Blaustein R O, Germann W J, Finkelstein A, DasGupta B R. The N-terminal half of the heavy chain of botulinum type A neurotoxin forms channels in plant phospholipid bilayers. FEBS Lett. 1987;226:115–120. doi: 10.1016/0014-5793(87)80562-8. [DOI] [PubMed] [Google Scholar]
- 7.Byrne M P, Titball R W, Holley J, Smith L A. Fermentation, purification, and efficacy of a recombinant vaccine candidate against botulinum neurotoxin type F from Pichia pastoris. Protein Expr Purif. 2000;18:327–337. doi: 10.1006/prep.2000.1200. [DOI] [PubMed] [Google Scholar]
- 8.Castano A R, Tangri S, Miller J E, Holcombe H R, Jackson M R, Huse W D, Kronenberg M, Peterson P A. Peptide binding and presentation by mouse CD1. Science. 1995;269:223–226. doi: 10.1126/science.7542403. [DOI] [PubMed] [Google Scholar]
- 9.Cortese R, Felici F, Galfre G, Luzzago A, Monaci P, Nicosia A. Epitope discovery using peptide libraries displayed on phage. Trends Biotechnol. 1994;12:262–267. doi: 10.1016/0167-7799(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 10.DasGupta B R, Sathyamoorthy V S. Purification and amino acid composition of type A botulinum neurotoxin. Toxicon. 1984;22:415–424. doi: 10.1016/0041-0101(84)90085-0. [DOI] [PubMed] [Google Scholar]
- 11.DasGupta B R, Sugiyama H. A common subunit structure in Clostridium botulinum type A, B and E toxins. Biochem Biophys Res Commun. 1972;48:108–112. doi: 10.1016/0006-291x(72)90350-6. [DOI] [PubMed] [Google Scholar]
- 12.Felici F, Luzzago A, Folgori A, Cortese R. Mimicking of discontinuous epitopes by phage-displayed peptides. II. Selection of clones recognized by a protective monoclonal antibody against the Bordetella pertussis toxin from phage peptide libraries. Gene. 1993;128:21–27. doi: 10.1016/0378-1119(93)90148-v. [DOI] [PubMed] [Google Scholar]
- 13.Folgori A, Tafi R, Meola A, Felici F, Galfre G, Cortese R, Monaci P, Alfredo N. A general strategy to identify mimotopes of pathological antigens using only random peptide libraries and human sera. EMBO J. 1994;13:2236–2243. doi: 10.1002/j.1460-2075.1994.tb06501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franz D R, Jahrling P B, Friedlander A M, McClain D J, Hoover D L, Bryne W R, Pavlin J A, Christopher G W, Eitzen E M. Clinical recognition and management of patients exposed to biological warfare agents. JAMA. 1997;278:399–411. doi: 10.1001/jama.278.5.399. [DOI] [PubMed] [Google Scholar]
- 15.Gill D M. Bacterial toxins: a table of lethal amounts. Microbiol Rev. 1982;46:86–94. doi: 10.1128/mr.46.1.86-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenwood J, Willis A E, Perham R N. Multiple display of foreign peptides on a filamentous bacteriophage. J Mol Biol. 1991;220:821–827. doi: 10.1016/0022-2836(91)90354-9. [DOI] [PubMed] [Google Scholar]
- 17.Heiskanen T, Lundkvist A, Vaheri A, Lankinen H. Phage-displayed peptide targeting on the Puumala hantavirus neutralization site. J Virol. 1997;71:3879–3885. doi: 10.1128/jvi.71.5.3879-3885.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 19.Koivunen E, Arap W, Rajotte D, Lahdenranta J, Pasqualini R. Identification of receptor ligands with phage display peptide libraries. J Nucl Med. 1999;40:883–888. [PubMed] [Google Scholar]
- 20.Kraft S, Diefenbach B, Mehta R, Jonczyk A, Luckenbach G A, Goodman S L. Definition of an unexpected ligand recognition motif for alphav beta6 integrin. J Biol Chem. 1999;274:1979–1985. doi: 10.1074/jbc.274.4.1979. [DOI] [PubMed] [Google Scholar]
- 21.Li B, Tom J Y, Oare D, Yen R, Fairbrother W J, Wells J A, Cunningham B C. Minimization of a polypeptide hormone. Science. 1995;270:1657–1660. doi: 10.1126/science.270.5242.1657. [DOI] [PubMed] [Google Scholar]
- 22.Mazzucchelli L, Burritt J B, Jesaitis A J, Nusrat A, Liang T W, Gewirtz A T, Schnell F J, Parkos C A. Cell-specific peptide binding by human neutrophils. Blood. 1999;93:1738–1748. [PubMed] [Google Scholar]
- 23.Nord K, Gunneriusson E, Ringdahl J, Stahl S, Uhlen M, Nygren P A. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat Biotechnol. 1997;15:772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- 24.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 25.Pasqualini R, Koivunen E, Ruoslahti E. A peptide isolated from phage display libraries is a structural and functional mimic of an RGD-binding site on integrins. J Cell Biol. 1995;130:1189–1196. doi: 10.1083/jcb.130.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prezzi C, Nuzzo M, Meola A, Delmastro P, Galfre G, Cortese R, Nicosia A, Monaci P. Selection of antigenic and immunogenic mimics of hepatitis C virus using sera from patients. J Immunol. 1996;156:4504–4513. [PubMed] [Google Scholar]
- 27.Rajotte D, Ruoslahti E. Membrane dipeptidase is the receptor for a lung-targeting peptide identified by in vivo phage display. J Biol Chem. 1999;274:11593–11598. doi: 10.1074/jbc.274.17.11593. [DOI] [PubMed] [Google Scholar]
- 28.Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino L P, DasGupta B R, Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- 29.Scott J K, Smith G P. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 30.Shone C C, Hambleton P, Melling J. Inactivation of Clostridium botulinum type A neurotoxin by trypsin and purification of two tryptic fragments. Proteolytic action near the COOH-terminus of the heavy subunit destroys toxin-binding activity. Eur J Biochem. 1985;151:75–82. doi: 10.1111/j.1432-1033.1985.tb09070.x. [DOI] [PubMed] [Google Scholar]
- 31.Shone C C, Hambleton P, Melling J. A 50-kDa fragment from the NH2-terminus of the heavy subunit of Clostridium botulinum type A neurotoxin forms channels in lipid vesicles. Eur J Biochem. 1987;167:175–180. doi: 10.1111/j.1432-1033.1987.tb13320.x. [DOI] [PubMed] [Google Scholar]
- 32.Shyu R H, Shaio M F, Tang S S, Shyu H F, Lee C F, Tsai M H, Smith J E, Huang H H, Wey J J, Huang J L, Chang H H. DNA vaccination using the fragment C of botulinum neurotoxin type A provided protective immunity in mice. J Biomed Sci. 2000;7:51–57. doi: 10.1007/BF02255918. [DOI] [PubMed] [Google Scholar]
- 33.Simpson L L. Molecular pharmacology of botulinum toxin and tetanus toxin. Annu Rev Pharmacol Toxicol. 1986;26:427–453. doi: 10.1146/annurev.pa.26.040186.002235. [DOI] [PubMed] [Google Scholar]
- 34.Smith W C, McDowell J H, Dugger D R, Miller R, Arendt A, Popp M P, Hargrave P A. Identification of regions of arrestin that bind to rhodopsin. Biochemistry. 1999;38:2752–2761. doi: 10.1021/bi982643l. [DOI] [PubMed] [Google Scholar]
- 35.Syuto B, Kubo S. Separation and characterization of heavy and light chains from Clostridium botulinum type C toxin and their reconstitution. J Biol Chem. 1981;256:3712–3717. [PubMed] [Google Scholar]
- 36.Szardenings M, Tornroth S, Mutulis F, Muceniece R, Keinanen K, Kuusinen A, Wikberg J E. Phage display selection on whole cells yields a peptide specific for melanocortin receptor 1. J Biol Chem. 1997;272:27943–27948. doi: 10.1074/jbc.272.44.27943. [DOI] [PubMed] [Google Scholar]
- 37.Tam J P, Zavala F. Multiple antigen peptide: a novel approach to increase detection sensitivity of synthetic peptides in solid-phase immunoassays. J Immunol Methods. 1989;124:53–61. doi: 10.1016/0022-1759(89)90185-3. [DOI] [PubMed] [Google Scholar]
- 38.Willis A E, Perham R N, Wraith D. Immunological properties of foreign peptides in multiple display on a filamentous bacteriophage. Gene. 1993;128:79–83. doi: 10.1016/0378-1119(93)90156-w. [DOI] [PubMed] [Google Scholar]
- 39.Wrighton N C, Farrell F X, Chang R, Kashyap A K, Barbone F P, Mulcahy L S, Johnson D L, Barrett R W, Jolliffe L K, Dower W J. Small peptides as potent mimetics of the protein hormone erythropoietin. Science. 1996;273:458–463. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 40.Wu H C, Huang Y L, Chao T T, Jan J T, Huang J L, Chiang H Y, King C C, Shaio M F. Identification of B-cell epitope of dengue virus type 1 and its application in diagnosis of patients. J Clin Microbiol. 2001;39:977–982. doi: 10.1128/JCM.39.3.977-982.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young A C, Valadon P, Casadevall A, Scharff M D, Sacchettini J C. The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library: implications for the identification of peptide mimotopes. J Mol Biol. 1997;274:622–634. doi: 10.1006/jmbi.1997.1407. [DOI] [PubMed] [Google Scholar]