Abstract
Background and Purpose
Osteoarthritis, a major cause of disability in developed countries does not have effective treatment. Activation of TLR4 and innate immune response factors contribute to osteoarthritis progressive cartilage degradation. There are no clinically available TLR4 inhibitors. Interestingly, the antidepressant amitriptyline could block this receptor. Thus, we evaluated amitriptyline anti‐TLR4 effects on human osteoarthritis chondrocytes in order to repurpose it as an inhibitor of innate immune response in joint inflammatory pathologies.
Experimental Approach
Using in silico docking analysis, RT‐PCR, siRNA, elisa, proteomics and clinical data mining of drug consumption, we explored the clinical relevance of amitriptyline blockade of TLR4‐mediated innate immune responses in human osteoarthritis chondrocytes.
Key Results
Amitriptyline bound TLR4 but not IL‐1 receptor. Interestingly, amitriptyline binding to TLR4 inhibited TLR4‐ and IL‐1 receptor‐mediated innate immune responses in human osteoarthritis chondrocytes, synoviocytes and osteoblasts cells. Amitriptyline reduced basal innate immune responses and promoted anabolic effects in human osteoarthritis chondrocytes. Supporting its anti‐innate immune response effects, amitriptyline down‐regulated basal and induced expression of NLRP3, an inflammasome member from IL‐1 receptor signalling linked to osteoarthritis and gout pathologies. Accordingly, mining of dissociated and aggregated drug consumption data from 107,172 elderly patients (>65 years) revealed that amitriptyline consumption was significantly associated with lower colchicine consumption associated with inflammatory gout flare treatment.
Conclusion and Implications
Amitriptyline blocks TLR4‐, IL‐1 receptor and NLRP3‐dependent innate immune responses. This together with clinical data amitriptyline could be repurposed for systemic or local innate immune response management in diverse joint inflammatory pathologies.
Keywords: chondrocytes, cohort study, gout, innate immunity, osteoarthritis
Abbreviations
- 2‐ddCt
2‐delta–delta cycle threshold
- dCt
delta CT
- DDA
data‐dependent acquisition
- NAMPT
nicotinamide phosphoribosyl transferase
- SWATH
sequential window acquisition of all theoretical mass spectra
What is already known
TLR4 is a therapeutic target linked to the production of osteoarthritis innate immune factors.
Amitriptyline is an antidepressant with potential anti‐TLR4 properties.
What does this study add
Amitriptyline inhibits TLR4, IL‐1 receptor and NLRP3‐dependent innate immune responses in human chondrocytes, synoviocytes and osteoblasts.
Amitriptyline intake is associated with reduced gout inflammatory flares.
What is the clinical significance
Amitriptyline could be repurposed for systemic or local management of joint inflammatory pathologies like osteoarthritis.
Amitriptyline anti‐inflammatory effects should be considered when selecting the antidepressant treatment for inflammatory arthritis patients.
1. INTRODUCTION
The rising prevalence of musculoskeletal pathologies in modern societies is causing detrimental effects on our social welfare. Particularly alarming is the higher incidence of osteoporosis, gout, rheumatoid arthritis and osteoarthritis, the responsible for most disability cases (Gómez et al., 2014). Osteoarthritis pathogenesis is characterised by the progressive degradation of the articular cartilage that alters surrounding tissues including subchondral bone, synovium and tendons. Altogether, these disturbances lead to pain, disability and joint failure (Gómez et al., 2014). The absence of efficient osteoarthritis treatments reaching the clinic is a burden for the health system, which is a critical matter considering that novel drug discovery is a long unguaranteed process. Therefore, the search for new properties in currently approved safe drugs is a cost‐effective and time‐saving strategy (Parvathaneni et al., 2019).
Osteoarthritis is a heterogeneous musculoskeletal pathology, hence its development has been linked to risk factors such as female sex, specific genetic profiles, ageing, joint mechanical stress, certain metabolic disorders and also inflammation (Gómez et al., 2014). Interestingly, osteoarthritis‐associated inflammation has been linked to the promotion of innate immune response via different mechanisms including the activation of toll‐like receptors (TLRs). TLRs are members of a highly conserved family of receptors that recognise either pathogen or damage‐associated molecular patterns (DAMPs), which are host‐derived molecules released as a response to tissue stress and injury (Alonso‐Pérez et al., 2018; Gómez et al., 2014). Among TLRs, the toll‐like receptor 4 (TLR4) has been largely involved in neuroinflammation (Bruno et al., 2018), lung injury (Togbe et al., 2007) and osteoarthritis pathogenesis (Alonso‐Pérez et al., 2018; Gómez et al., 2014).
TLR4 is involved in the production of innate immune factors that boost synovitis, cartilage degradation and osteoarthritis joint inflammation (Gómez et al., 2014). Among others, these factors include NO (Clancy et al., 2004), cytokines like IL‐1ß and IL‐6 (Gómez et al., 2014), adipokines such as visfatin/nicotinamide phosphoribosyl transferase (NAMPT) (Franco‐Trepat et al., 2019) and lipocalin 2, and catabolic factors like MMP3 (Tong et al., 2017) and MMP13 (Wang et al., 2013). Interestingly, TLR4 downstream signalling is partially shared with the IL‐1 receptor type I (Loiarro et al., 2010), the innate immune response receptor of IL‐1ß, that is involved in osteoarthritis pathophysiology (Vincent, 2019). This receptor is involved in multiple arthritides, and its blockade is used to treat rheumatoid arthritis (Mertens & Singh, 2009), gout (Pascart & Richette, 2017) and crystal‐induced arthritis (Aouba et al., 2015). Inflammatory in vitro models have been successful to unveil potential TLR4 inhibitory agents such as resatorvid (TAK‐242; Hussey et al., 2012) or enexasogaol (96‐shogaol; Villalvilla et al., 2014). Nevertheless, a clinical viable TLR4 inhibitor for arthritides like osteoarthritis has yet to be found. Therefore, targeting TLR4 is an interesting approach for drug repurposing (Gómez et al., 2014).
Amitriptyline is a synthetic tricyclic compound prescribed to treat depression (Couch & Amitriptyline Versus Placebo Study Group, 2011) that was first approved by the Food and Drug Administration (FDA) in 1961 under the name Elavil. Later uses addressed the treatment of anxiety (Kelly, 1973), migraine (Couch & Amitriptyline Versus Placebo Study Group, 2011), insomnia (Saddichha, 2010), autism (Hellings et al., 2017) and fibromyalgia (Rico‐Villademoros et al., 2015). Interestingly, it was reported that amitriptyline bound and blocked the TLR4 receptor of mouse microglial cells (Hutchinson et al., 2010). Accordingly, amitriptyline repurposing could be useful to control TLR4‐mediated innate immune responses in joint inflammatory pathologies like osteoarthritis.
In the present study, we investigated whether amitriptyline blocked TLR4‐ and osteoarthritis‐associated innate immune responses. We demonstrated in different articular cells the ability of amitriptyline to block TLR4 and IL‐1 receptor signalling and their downstream innate immune responses. Moreover, amitriptyline anti‐innate immune response effects were confirmed by clinical data mining of gout patients treated with amitriptyline, suggesting that amitriptyline could be repurposed to treat innate immune response in diverse joint pathologies.
2. METHODS
2.1. Cell isolation and culture
Tissue samples were obtained from osteoarthritis patients who had undergone total knee replacement surgery, and synovial liquid was removed via joint aspiration under local anaesthetic from eight osteoarthritis and eight goutpatients. All patients signed an informed consent approved by the Santiago Hospital Ethics Committee (CAEIG‐2016/258). Primary human osteoarthritis chondrocytes were isolated from cartilage matrix using pronase and collagenase digestion (Merk Sigma‐Aldrich, Germany), whereas primary human osteoblasts were obtained from smiting the trabecular bone into flat (1 × 1 × 0.1 cm) pieces (Villalvilla et al., 2014). Synovial liquid was treated with type I‐S testicular bovine hyaluronidase (1500 IU·ml−1) from Merk Sigma‐Aldrich (Germany), incubated 30 min at 37°C and centrifuged for 10 min at 410 g. Supernatant was recollected and further centrifuged for 10 min at 1450 g, pellet was discarded and purified synovial liquid was obtained.
Cell culture of primary human osteoarthritis chondrocytes, primary human osteoblasts, murine chondrogenic ATDC5 (RCB Cat# RCB0565, RRID:CVCL_3894) and human synovial sarcoma SW982 (ATCC Cat# HTB‐93, RRID:CVCL_1734) was performed in P12 cell culture plates (105 cells per well) as previously described (Villalvilla et al., 2014). Except otherwise mentioned, all cell culture reagents were purchased from Merk Sigma‐Aldrich (Germany).
2.2. Cell culture treatments
After 18 h of starvation, if needed, a 3‐h pretreatment was done with the TLR4 inhibitor CLI‐095/TAK‐242 (InvivoGen, USA) and culture media were renewed. Cells were stimulated with TLR4 activator LPS (E. coli 026:B6; Merk Sigma‐Aldrich, Germany) and IL‐1 receptor activator IL‐1β (Merk Sigma‐Aldrich, Germany), or alternatively, synovial liquid (10% v/v) isolated from patients was used to activate inflammatory and catabolic pathways. Then, cell cultures were co‐treated with amitriptyline (tebu‐bio, France) or TriFECTa NLRP3 siRNA (IDT, USA) (Hannon & Rossi, 2004) for 24 h.
2.3. Biochemical assays
The culture media nitrite accumulation was measured by Griess assay (Merk Sigma‐Aldrich, Germany) to indirectly determine NO release, whereas an MTT assay (Merk Sigma‐Aldrich, Germany) was performed to determine cell viability as previously described (Villalvilla et al., 2014). Malachite green assay was used to determine total phosphoproteome following the manufacturer's instructions (Merk Sigma‐Aldrich, Germany).
2.4. Gene expression (mRNA) assays
To measure gene expression (mRNA), comparative RT‐PCR (500 μg) was performed as previously described (Villalvilla et al., 2014) using iTaq Universal SYBR Green Supermix (Bio‐Rad, USA) and KiCqStart SYBR Green primers (Merk Sigma‐Aldrich, Germany) in a QuantStudio3 thermocycler (Thermo Fisher, USA). TRIzol and EZNA Total RNA Kit I (Omega Bio‐Tek, USA) were used to isolate RNA, and High‐Capacity RNA‐To‐cDNA Kit (Thermo Fisher, USA) was used to retrotranscribe 500 μg of RNA.
2.5. RNA‐mediated gene expression silencing
To transiently silence NLRP3 mRNA, siRNA targeting NLRP3 was administered into 80% confluent murine ATDC5 chondrocytes or human SW982 synoviocytes using specific TriFECTa DsiRNA Kit (IDT, USA). Briefly, cells were plated at a density of 50,000 cells per well in P24‐well plates for 4 h. Cells were washed with Opti‐MEM and DMEN F12 (2% FBS) and incubated 60 min with DMEN F12 (2% FBS). Then NLRP3 siRNA (15 nM) composed of an equal mixture of three different oligos (sequence data not shown) was added to the cell culture. After 24 h of siRNA incubation, cells were treated with LPS or IL‐1β as required. Both HPRT‐S1 Positive Control DsiRNA and Negative Control DsiRNA were used, and depletion of gene‐specific mRNA levels was calculated by comparative RT‐PCR of NLRP3 and HPRT1 expression levels.
2.6. Protein expression assays
Proteins were isolated using RIPA 1× buffer (Merck, USA) with HALT protease and phosphatase inhibitors (Thermo Fisher, USA) and quantified using BSA Pierce Bradford assay (Thermo Fisher, USA). Human IL‐6 and IL‐1β were measured by elisa in cell culture supernatants (EliKine™ elisa Kit; Abbkine, USA). Absorbances were determined using a Multiskan EX (Helsinki, Finland).
Cellular proteome and secretome (supporting information) from human osteoarthritis chondrocytes co‐treated with amitriptyline and LPS (100 ng·ml−1) or IL‐1β (0.1 ng·ml−1) for 24 h were studied by micro‐LC–MS/MS using a hybrid quadrupole TOF/TripleTOF 6600 (Sciex, USA). Both proteomes were identified by the qualitative shotgun data‐dependent acquisition (DDA) method (Couselo‐Seijas et al., 2019; Shilov et al., 2007), and protein levels were measured by the quantitative sequential window acquisition of all theoretical mass spectra (SWATH) method (del Pilar Chantada‐Vázquez et al., 2020; Shilov et al., 2007). A 5% false discovery rate (FDR) and P ≤ 0.05 were used to filter the datasets (Shilov et al., 2007). FunRich software (Pathan et al., 2017) was used to determine the proteome enrichment in immune‐related categories.
2.7. Docking analysis
Ligand structures amitriptyline (TP0/3APV), LPS and IL‐1β were obtained from PubChem, and crystallographic docking analysis on TLR4 (3FXI.pdb) and IL‐1 receptor (1ITB.pdb) structures RCSB PDB bank (Burley et al., 2019) was performed with ‘1‐Click‐Docking’ (mCule Inc) and ‘AutoDock Vina’ (Trott & Olson, 2010). Potential docking positions or poses were identified within the extracellular active domains of TLR4 (B. S. Park et al., 2009) (grid box: x = 17, y = −27 and z = −35) and IL‐1 receptor (Thomas et al., 2012) (grid box: x = 56, y = 27 and z = 34). The highest binding affinity pose was determined by the lowest, most negative, docking score. Docking scores ≥−2 kcal·mol−1 were described as null docking.
2.8. Materials
Materials used in cell culture include DMEM F12 (Ref. D8437‐6X500ML; Merk Sigma‐Aldrich, Germany), FBS (Ref. F7524‐500ML; Merk Sigma‐Aldrich, Germany), trypsin (Ref. T3924‐100ML; Merk Sigma‐Aldrich, Germany), l‐glutamine (Ref. G7513‐100ML; Merk Sigma‐Aldrich, Germany) and penicillin–streptomycin (Ref. P4458‐100ML; Merk Sigma‐Aldrich, Germany). Furthermore, primary human osteoarthritis chondrocytes and osteoblasts (Clinical Hospital of Santiago [CHUS]; Santiago Ethics Committee—CAEIG‐2016/258), murine chondrogenic ATDC5 (RCB Cat# RCB0565, RRID:CVCL_3894) and human synovial sarcoma SW982 (ATCC Cat# HTB‐93, RRID:CVCL_1734) were used. Additionally, pronase (Ref. 14379324; Merk Sigma‐Aldrich, Germany), collagenase P (Ref. 34598926; MERK Sigma‐Aldrich, Germany), type I‐S testicular bovine hyaluronidase (H3506‐500MG; Merk Sigma‐Aldrich, Germany), CLI‐095 (Ref. tlrl‐cli95; InvivoGen, USA), LPS Escherichia coli 026:B6 (Ref. L2654‐1MG; Merk Sigma‐Aldrich, Germany), IL‐1β (Ref. H6291‐10UG; Merk Sigma‐Aldrich, Germany) and amitriptyline (Ref. BCBQ4634V; Merk Sigma‐Aldrich, Germany) were employed.
Materials used in biochemical assays include Griess assay (Ref. MAK367; Merk Sigma‐Aldrich, Germany), MTT assay (Ref. CGD1‐1KT; Merk Sigma‐Aldrich, Germany) and malachite green assay (Ref. M689; Thermo Fisher Scientific, USA).
Materials used in gene expression (mRNA) assays include iTaq Universal SYBR Green Supermix (Ref. 172‐5125; Bio‐Rad, USA), KiCqStart SYBR Green primers (Ref. KSPQ12012G; Merk Sigma‐Aldrich, Germany), TRIzol TRI Reagent (Ref. T9424‐200ML; Merk Sigma‐Aldrich, Germany), EZNA Total RNA Kit I (Ref. R6834‐02; Omega Bio‐Tek, USA) and High‐Capacity RNA‐To‐cDNA Kit (Ref. 773886; Thermo Fisher Scientific, USA).
Materials used in RNA‐mediated gene expression silencing include TriFECTa NLR family pyrin domain‐containing 3 (NLRP3) siRNA (mmRi NIrp3.13 and hsRi NIrp3.13; IDT, USA) and Opti‐MEM I (Ref. 212767; Thermo Fisher Scientific, USA).
Materials used in protein expression assays include RIPA 1× buffer (Ref. 20‐188; Merck, USA), HALT protease and phosphatase inhibitors (Ref. 78442; Thermo Fisher Scientific, USA), BSA Pierce Bradford assay (Ref. 23200; Thermo Fisher Scientific, USA), EliKine™ Human IL‐6 elisa Kit (Ref. 311KET6017‐96T; Abbkine, USA) and EliKine™ Human IL‐1β elisa Kit (Ref. 311KET6013‐96T; Abbkine, USA).
2.9. Experimental design and statistical analysis
The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology (Curtis et al., 2018). As such, it was determined as a four‐condition scheme:‐ control (nontreated and non‐stimulated cells), drug (amitriptyline [0.1–1 μM]), inflammatory stimulus (LPS [100 ng·ml−1] or IL‐1β [0.1 ng·ml−1]) and drug plus inflammatory stimulus (LPS [100 ng·ml−1] or IL‐1β [0.1 ng·ml−1] + amitriptyline [0.1–1 μM]). Randomisation and blinding were not applicable in this experimental set‐up.
In specific experiments, amitriptyline was substituted by CLI‐095 (1 μM) or NLRP3 siRNA (15 nM). Concentrations were selected for LPS and IL‐1β based on previous reports (Villalvilla et al., 2016), whereas amitriptyline concentrations overlapped the serum concentrations observed in amitriptyline‐treated patients (Shimoda et al., 1997).
In terms of sample size, we established a group size (n) of 6 independent biological replicas, each one with 3 dependent technical replicas to all determinations for all cell types. Remarkably, each independent biological replica for human osteoarthritis chondrocyte come from a different patient. Nonetheless, the availability of primary cells is scarce and proteomic‐based experiments such as MALDI/TOFF method, for either DDA or SWATH, required about ten times more cells to perform reliable determinations for each condition studied. Accordingly, we used primary cultures from cartilage samples that were big enough that allowed us to have the required number of cells from the same patient for all the treatments. Considering the scarce number of samples of this size proteomic experiments were limited to 3 patients. With the protein extracts from these primary cultures it was performed an accumulative signal approach that consists in the accumulation of thousands of technical replica runs into different independent output data.
Data were normalised by ‘control’ treatment, displayed as a scatter plot and expressed as the mean ± SEM. Gene expression (mRNA) was analysed by RT‐PCR, normalised by housekeeping gene (HPRT), compared using delta cycle threshold (dCt) method and displayed as 2‐delta–delta Ct (2‐ddCt). Likewise, heat maps and the volcano plot data were transformed into fold change (log2) in order to be properly displayed.
In terms of statistical significance and tests, we applied the New England Journal of Medicine (NEJM) statistical significance method of representation (* P <0.05) in the whole manuscript. Normality test Shapiro–Wilk and Kolmogorov–Smirnov was used to determine data distribution and decided whether to proceed with parametric or non‐parametric tests. Data originated from experiments using human samples such as primary human osteoarthritis chondrocytes, osteoblasts and synovial liquids were analysed by non‐parametric Kruskal–Wallis for multiple comparison or Mann–Whitney test for individual comparison if needed. Otherwise, data were analysed by parametric one‐way ANOVA for multiple comparison or unpaired t‐test Welch's correction for individual comparison if needed. In that sense, multiple comparison in non‐parametric Kruskal‐Wallis was performed by comparing the mean rank of each column with the mean rank of every other column and corrected using the recommended Dunn's hypothesis testing. Nontheless, we only proceeded to multiple comparion if Krustal Wallis statistic value presented statistical significance (P<0.05).
Similarly, multiple comparison in parametric One Way‐ANOVA was performed by comparing the mean of each column with the mean of every other column and corrected using the recommended Tukey hypothesis testing. Nontheless, we only proceeded to multiple comparion if F value was statistical significance (P<0.05).
Furthermore, statistical analysis of gene expression (mRNA) was done using the dCt values in order to consider the variability of all treatments including ‘control’. To analyse dissociated and aggregated clinical data of drug consumption for colchicine (0.5–1 mg) and amitriptyline (10–75 mg; 27.4 mg average dose) mined for 5 years from 107,172 elderly patients of Santiago de Compostela (Spain) healthcare area, Fisher contingency and Spearman correlation tests were used.
2.10. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in the IUPHAR/BPS Guide to PHARMACOLOGY (http://www.guidetopharmacology.org) and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander, Cidlowski, et al., 2019; Alexander, Fabbro, et al., 2019a, 2019b).
3. RESULTS
3.1. Amitriptyline has a high binding affinity towards TLR4
The TLR4 complex is a dimeric receptor coupled with accessory molecule myeloid differentiation factor 2 (lymphocyte antigen 96/MD‐2) that presents extracellular domain pockets for agonist LPS to dock and start the innate immune response signal transduction (B. S. Park et al., 2009). An in silico docking analysis was performed to assess whether amitriptyline could bind to the same extracellular domain pockets for LPS, as previously suggested (Hutchinson et al., 2010). Interestingly, amitriptyline exhibited (Figure 1a) a stronger binding affinity (−6.03 ± 0.06 kcal·mol−1) than LPS (−4.53 ± 0.01 kcal·mol−1) to the same pockets (Figure 1b).
FIGURE 1.
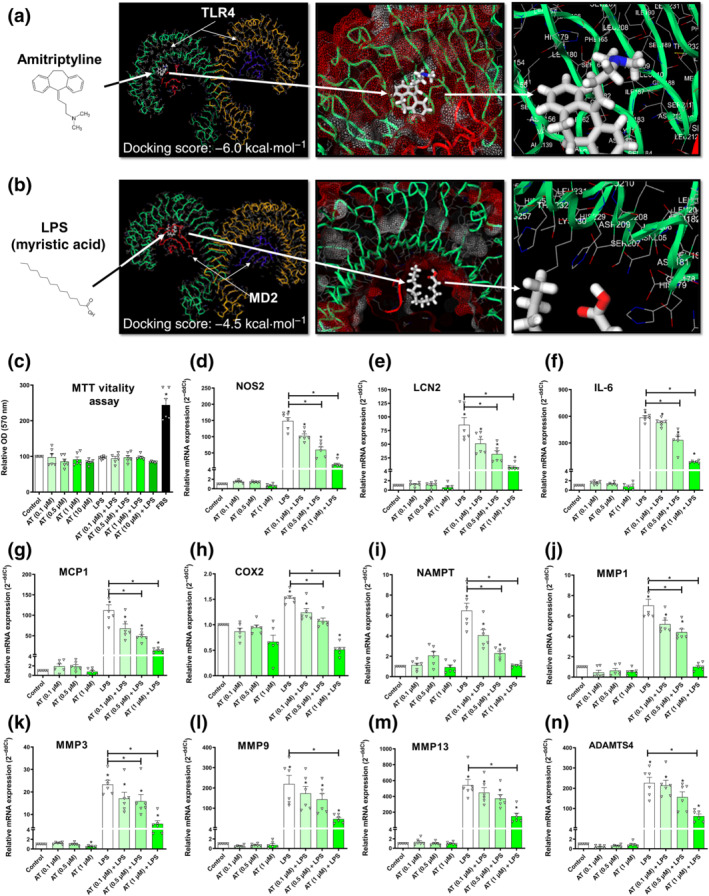
Amitriptyline (AT) effects on TLR4‐mediated innate immune response factors. All experiments were independent, normalised by control and expressed as the mean ± SEM and statistically analysed by non‐parametric Krustal‐Wallis test coupled with Dunn's post‐test (*P <0.05). (a) Best docking score (−6.0 kcal·mol−1) for amitriptyline (white) into the TLR4 extracellular domain. The potential pocket from TLR4 dimer (green and yellow) uses protein sequence positions (154th to 208th) next to accessory protein myeloid differentiation factor 2/lymphocyte antigen 96 (MD‐2; red and blue). (b) Best docking score (−4.5 kcal·mol−1) for LPS (white) into the TLR4 extracellular domain. The potential pocket from TLR4 dimer (green and yellow) uses protein sequence positions (154th to 208th) next to accessory protein MD‐2 (red and blue). (c–n) MTT cell viability assay and mRNA expression (RT‐PCR) innate immune response factors were determined in TLR4‐activated (LPS [100 ng·ml−1]) human osteoarthritis (OA) chondrocytes (hOCs) cotreated with AT (0.1, 0.5 and 1.0 μM) for 24 h (n = 6)
3.2. Amitriptyline blocked TLR4‐mediated innate immune response factors in human osteoarthritis chondrocytes
To validate the in silico analysis, we studied the effects of amitriptyline on TLR4‐induced innate immune response factors. We selected amitriptyline concentrations (0.1, 0.5 and 1 μM) to mimic its serum concentrations (0.033–1.130 μM) found in patients treated with amitriptyline (Shimoda et al., 1997). We used the TLR4 agonist LPS (100 ng·ml−1) to activate the TLR4 receptor in mouse chondrocytes as well as in human osteoarthritis chondrocytes. Interestingly, 24‐h cotreatment with amitriptyline reduced nitrite accumulation in the cell supernatant of mouse chondrocytes without any cytotoxic effect (Figure S1A,B). This was consistent with the reduced gene expression of the inducible NOS (iNOS) gene, NOS2 (Figure S1C). Similarly, amitriptyline inhibitory effects were observed in human osteoarthritis chondrocytes (Figure 1c,d). Moreover, in these cells, amitriptyline significantly reduced the gene expression of other TLR4‐induced factors (Figure 1e–n) such as IL‐6, lipocalin 2, monocyte chemoattractant protein 1 (MCP1/CCL2), COX2 and NAMPT, as well as osteoarthritis catabolic factors induced by TLR4 like MMP1, MMP3, MMP9, MMP13 and a disintegrin and metalloproteinase with thrombospondin motifs 4 (ADAMTS4). These effects were also observed on ATDC5 cells (Figure S1D–I).
3.3. Amitriptyline inhibits TLR4‐mediated proteomic changes in human osteoarthritis chondrocytes
Amitriptyline blockade of TLR4‐activation was further explored in human osteoarthritis chondrocyte cellular proteome and secretome (Figure 2a). Proteins from the qualitative proteomic analysis were clustered and categorised by molecular function (Figure 2b). Interestingly, amitriptyline depleted TLR4‐enriched categories such as ‘cytokine release’ and ‘metallopeptidase’ (Figure 2b). Quantitative cellular proteomic analysis revealed significant down‐regulation of multiple key proteins including vascular cell adhesion protein 1 (VCAM1), MMP3 and iNOS) (Figure 2c). Principal component analysis confirmed consistent differences between treatments (Figure S1K). Pathway enrichment analysis showed that amitriptyline significantly reverted TLR4‐enriched innate immune response pathways (Figure 2d), including TLR4‐related categories like ‘activated TLR4 signalling’, ‘cytokine signalling in immune system’ and ‘innate immune system’ (Figure 2d). Interestingly, amitriptyline also depleted TLR4‐enriched pathways like ‘IL‐1 signalling’ and ‘The NLRP3 inflammasome’ (Figure 2d), which are key pathways involved in diverse articular inflammatory processes, such as microcrystals‐induced IL‐1β secretion (Aouba et al., 2015; Mangan et al., 2018; Szekanecz et al., 2019; Zamyatina & Heine, 2020) (Figure 2d).
FIGURE 2.
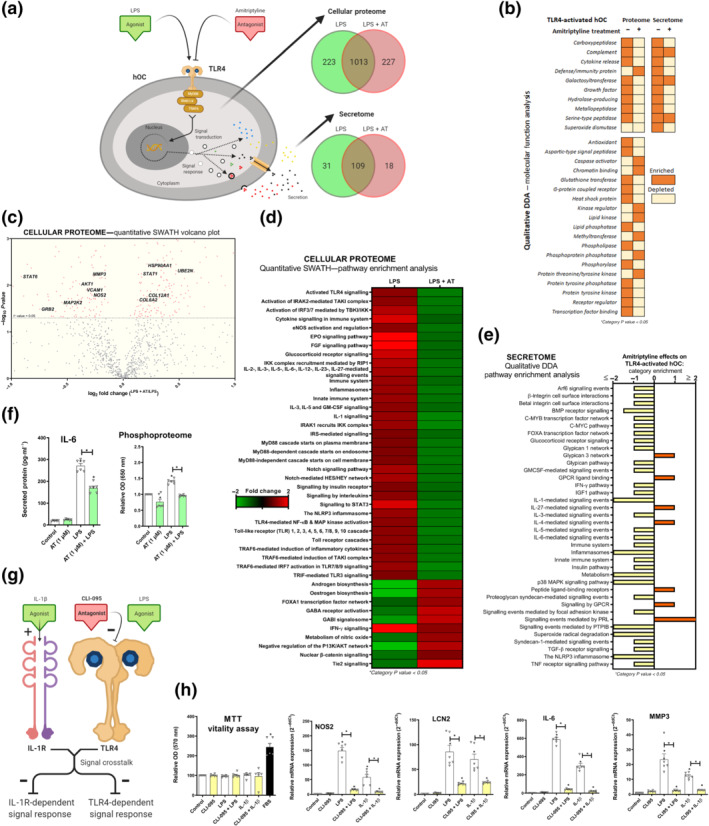
Amitriptyline (AT) reverts TLR4‐mediated proteomic changes in human osteoarthritis (OA) chondrocytes (hOCs). All experiments were independent, normalised by control and expressed as the mean ± SEM and statistically analysed by non‐parametric Krustal‐Wallis test coupled with Dunn's post‐test (* P<0.05). hOCs were stimulated by LPS (100 ng·ml−1) and cotreated with AT (1 μM) for 24 h. (a) Diagram of TLR4 signalling inhibition and proteome number identification in qualitative analysis (DDA) data (n = 3). (b) Qualitative data (DDA) clustering into molecular function categories from TLR4‐activated hOC proteome (n = 3). (c) Quantitative data (SWATH) shown as a volcano plot (P value vs. fold change) of protein expression changes by AT in TLR4‐activated hOCs (n = 3). (d) Quantitative data (SWATH) pathway enrichment analysis of AT effects on TLR4‐activated hOCs (n = 3). (e) Qualitative secretome data (DDA) pathway enrichment analysis of AT effects on TLR4‐activated hOCs (n = 3). (f) Secreted levels of protein IL‐6 (pg·ml−1) (elisa) and relative OD of malachite green total phosphoproteome (n = 3). (g) Diagram of TLR4 and IL‐1 receptor ® crosstalk in the context of CLI‐095 inhibition in hOCs. (h) MTT cell viability assay and innate immune response factor mRNA expression (RT‐PCR) were determined in hOCs pretreated with CLI‐095 (1 μM) for 3 h before TLR4 or IL‐1R (IL‐1β [0.1 ng·ml−1]) activation for 24 h (n = 6)
According to amitriptyline effects on the cellular proteome, pathway enrichment analysis on secretome data showed that amitriptyline also depleted TLR4‐enriched catabolic and inflammatory pathways (Figure 2e). In fact, protein expression of secreted IL‐6, a TLR4‐induced protein from these pathways, was reduced by amitriptyline in human osteoarthritis chondrocytes (Figure 2f). Amitriptyline also reverted the TLR4‐enriched ratio between phosphorylase and phosphatase categories (Figure 2b), which was confirmed by the cellular phosphoproteome of human osteoarthritis chondrocytes and mouse chondrocytes (Figures 2f and S1J).
3.4. TLR4 inhibition blocked TLR4‐ and IL‐1 receptor‐mediated innate immune responses in human osteoarthritis chondrocytes
TLR4 antagonism has shown to block IL‐1 receptor‐driven arthritis in animal models (Abdollahi‐Roodsaz et al., 2007). Thus, considering that the amitriptyline antagonism of TLR4 depleted IL‐1‐related pathways (Figure 2d,e), we determined whether TLR4 inhibition could block IL‐1 receptor‐mediated innate immune response (Figure 2g). To do so, we pretreated chondrocytes for 3 h with the specific TLR4 inhibitor CLI‐095 (1 μM) before the TLR4 activation by LPS (100 ng·ml−1) and IL‐1 receptor activation by IL‐1β (0.1 ng·ml−1) for 24 h (Figure 2g). Interestingly, specific TLR4 inhibition blocked both TLR4‐ and IL‐1 receptor ‐mediated innate immune responses in human osteoarthritis chondrocytes (Figure 2h) and mouse chondrocytes (Figure S1L–Q) without any toxic effect.
3.5. Amitriptyline does not bind IL‐1 receptor but blocks IL‐1 receptor‐mediated innate immune response factors in human osteoarthritis chondrocytes
To exclude a hypothetical binding of amitriptyline to IL‐1 receptor, we analysed their binding affinity (Figure 3a) and found it negligible (−0.1 kcal·mol−1) and far from the canonical agonist IL‐1β (−172 kcal·mol−1) (Figure 3b). To test whether amitriptyline might block IL‐1 receptor‐mediated innate immune response without binding to IL‐1 receptor, human osteoarthritis chondrocytes and ATDC5 cells were stimulated with IL‐1β (0.1 ng·ml−1) and amitriptyline. All the studied IL‐1β‐induced innate immune response factors, iNOS, IL‐6, lipocalin 2, MCP1, COX2, NAMPT, MMP1, MMP3, MMP9, MMP13 and ADAMTS4, were significantly decreased by amitriptyline without cytotoxic effects (Figures 3c–n and S1R–Z).
FIGURE 3.
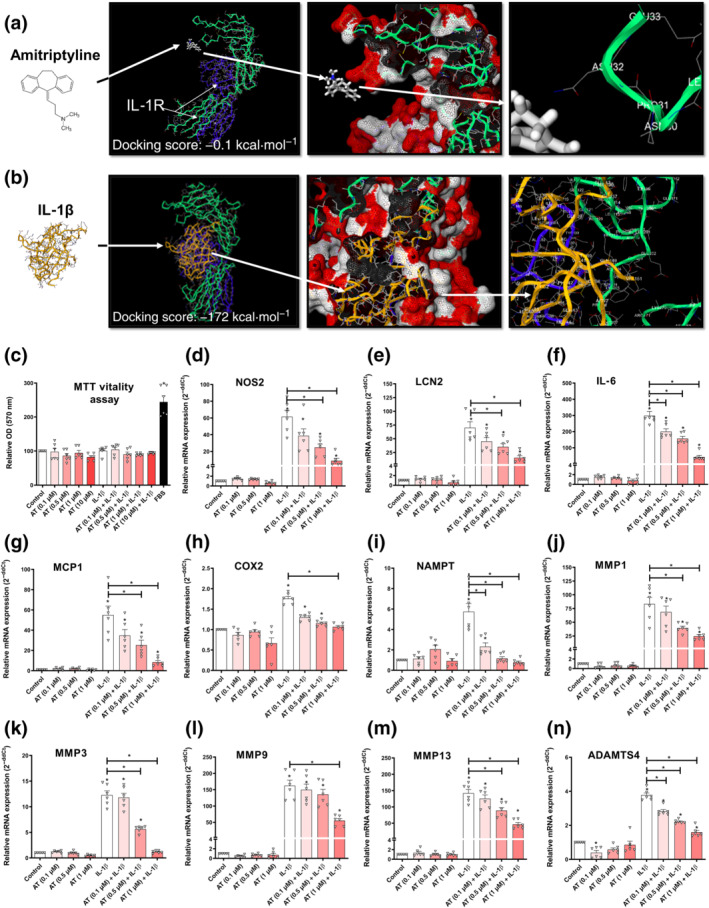
Amitriptyline (AT) effects on IL‐1 receptor (R)‐mediated innate immune response factors. All experiments were independent, normalised by control and expressed as the mean ± SEM.and statistically analysed by non‐parametric Krustal‐Wallis test coupled with Dunn's post‐test (* P<0.05). (a) Best docking score (−0.1 kcal·mol−1) for amitriptyline (white) into the IL‐1 receptor (R) extracellular domain. Amitriptyline does not bind to a pocket but to an energetically unfeasible radical from a protein sequence position (232nd) in IL‐1 receotor subunit IL‐1 receptor 1 (green) far from subunit IL‐1 receptor AP (blue). (b) Best docking score (−172 kcal·mol−1) for IL‐1β (yellow) into the IL‐1R extracellular domain. The pocket comprises the whole IL‐1 receptor subunit IL‐1 receptor 1 (green), which is next to subunit IL‐1RAP (blue). (c–n) MTT cell viability assay and innate immune response factor mRNA expression (RT‐PCR) were determined in IL‐1 receptor‐activated (IL‐1β [0.1 ng·ml−1]) human osteoarthritis (OA) chondrocytes (hOCs) cotreated with AT (0.1, 0.5 and 1.0 μM) for 24 h (n = 6)
3.6. Amitriptyline reduces IL‐1 receptor‐induced proteomic changes in human osteoarthritis chondrocytes
Amitriptyline blockade of IL‐1 receptor‐activation was further explored in human osteoarthritis chondrocyte cellular proteome and secretome (Figure 4a). Proteins from the qualitative proteomic analysis were clustered and categorised by molecular function (Figure 4b). Amitriptyline depleted IL‐1 eceptor ‐enriched categories such as ‘cytokine release’, ‘defence/immunity protein’ and ‘metallopeptidase’ in the cellular secretome, though effects on the cellular proteome were not that evident (Figure 4b). Nonetheless, quantitative cellular proteomic analysis revealed that amitriptyline significantly down‐regulated multiple key inflammatory and catabolic proteins including intercellular adhesion molecule 1 (ICAM1), MMP1 and MMP3 (Figure 4c). The principal component analysis confirmed consistent clustering among similar treatments (Figure S2A). Moreover, pathway enrichment analysis from cellular proteome and secretome data showed that amitriptyline significantly reverted IL‐1 receptor‐enriched innate immune response pathways (Figure 4d,e), including ‘activated TLR4 signalling’, ‘IL‐1 signalling’ and ‘the NLRP3 inflammasome’ (Figure 4d,e). To further confirm the innate immune response blockade, we found that secreted IL‐6 concentration was halved after amitriptyline co‐treatment with IL‐1β (Figure 4f). Consistent with IL‐1 receptor signalling blockade, amitriptyline reduced basal and IL‐1 receptor‐activated phosphorylation levels in human osteoarthritis chondrocytes and mouse chondrocytes (Figures 4f and S1AA).
FIGURE 4.
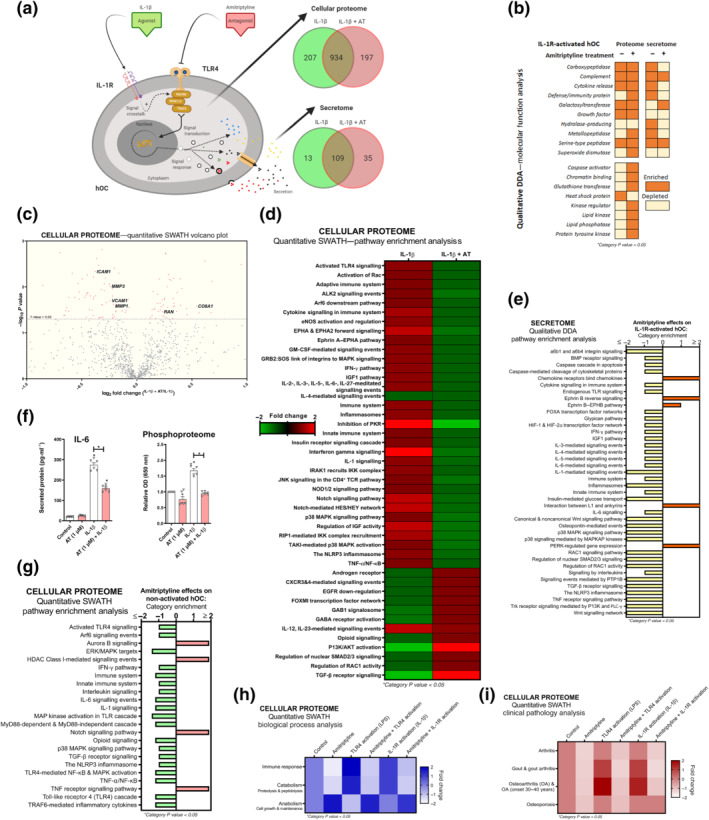
Amitriptyline (AT) reverts IL‐1 receptor (R)‐mediated proteomic changes in human OA chondrocytes (hOCs). All experiments were independent, (n=6) normalised by control and expressed as the mean ± SEM and statistically analysed by non‐parametric Krustal‐Wallis test coupled with Dunn's post‐test (* P<0.05). hOCs were stimulated with IL‐1β (0.1 ng·ml−1) and cotreated with AT (1 μM) for 24 h. (a) Diagram of TLR4 signalling inhibition and qualitative data (DDA) protein number identification from IL‐1 receptor (R)‐activated hOC proteome (n = 3). (b) Qualitative data (DDA) clustering into molecular function categories from IL‐1 receptor‐activated hOC proteome (n = 3). (c) Quantitative data (SWATH) of protein expression changes by AT shown as a volcano plot (P value vs. fold change) from IL‐1 receptor‐activated hOC proteome (n = 3). (d) Quantitative data (SWATH) pathway enrichment analysis of AT effects on IL‐1 receptor‐activated hOCs (n = 3). (e) Qualitative secretome data (DDA) pathway enrichment analysis of AT effects on IL‐1receptor‐activated hOCs (n = 3). (f) Secreted levels of protein IL‐6 (pg·ml−1) (elisa) and relative OD of malachite green total phosphoproteome (n = 3). (g) Amitriptyline effects on pathway enrichment of non‐activated hOCs using quantitative (SWATH) proteome data (n = 3). (h) Quantitative data (SWATH) clustering into biological processes categories from IL‐1 receptor‐activated hOC proteome (n = 3). (i) On clinical pathology analysis using quantitative data (SWATH) of AT effects on stimulated and non‐stimulated hOCs (n = 3)
Considering that osteoarthritis is a disease of the whole joint, we elucidated whether amitriptyline also exhibited similar anti‐inflammatory and anticatabolic effects in other articular cells. Amitriptyline blocked the gene expression of TLR4‐ and IL‐1 receptor‐mediated innate immune response factors in SW982 human synoviocytes (Figure S2B–G) and primary human osteoblasts‐like cells (Figure S2H–M).
3.7. Amitriptyline blocks basal innate immune response in human osteoarthritis chondrocytes
To expand amitriptyline clinical uses, we investigated whether amitriptyline blocked basal innate immune response in unstimulated human osteoarthritis chondrocytes (Figure S3A). Both TLR4 and NLRP3 pathways were specifically depleted after amitriptyline treatment of these cells (Figure 4g). In fact, biological process analysis of quantitative cellular proteomic data from amitriptyline‐treated human osteoarthritis chondrocytes showed that it promoted anabolism and blocked immune response and catabolism (Figure 4h).
These results widened the potential clinical use of amitriptyline in other articular pathologies. In fact, an algorithm that associates proteome signatures to known diseases (Pathan et al., 2017) suggested that amitriptyline might prevent or block processes linked to osteoarthritis, gout and/or osteoporosis (Figure 4i).
3.8. Amitriptyline blocks NLRP3 inflammasome signalling
Amitriptyline depleted basal and induced NLRP3 signalling, which is involved in osteoarthritis (McAllister et al., 2018) and gout (So & Martinon, 2017) pathogenesis. Accordingly, amitriptyline reduced basal and induced NLRP3 mRNA expression, as well as the downstream expression of IL‐1β in human osteoarthritis chondrocytes (Figure 5a–c) and ATDC5 cells (Figure 5d,e). This effect was associated with TLR4 inhibition because CLI‐095‐mediated TLR4 blockade mimicked these results. Moreover, silencing of NLRP3 mRNA expression was enough to block TLR4‐ and IL‐1 receptor‐mediated inflammatory responses without affecting cell vitality (Figures 5f,g and S3B,C). Consistent with this, similar results were obtained in SW982 human synoviocytes. Either basal or induced NLRP3 and IL‐1β mRNA expression were reduced by amitriptyline, NLRP3 siRNA and/or CLI‐095 with no cytotoxic effects (Figures 5h–k and S3D). Furthermore, in the same human synoviocyte cell line and without cytotoxic effects, treatment with amitriptyline also blocked basal and induced NLRP3 and IL‐1β mRNA expression elicited by synovial liquids from osteoarthritis and gout patients (Figure 5l–n).
FIGURE 5.
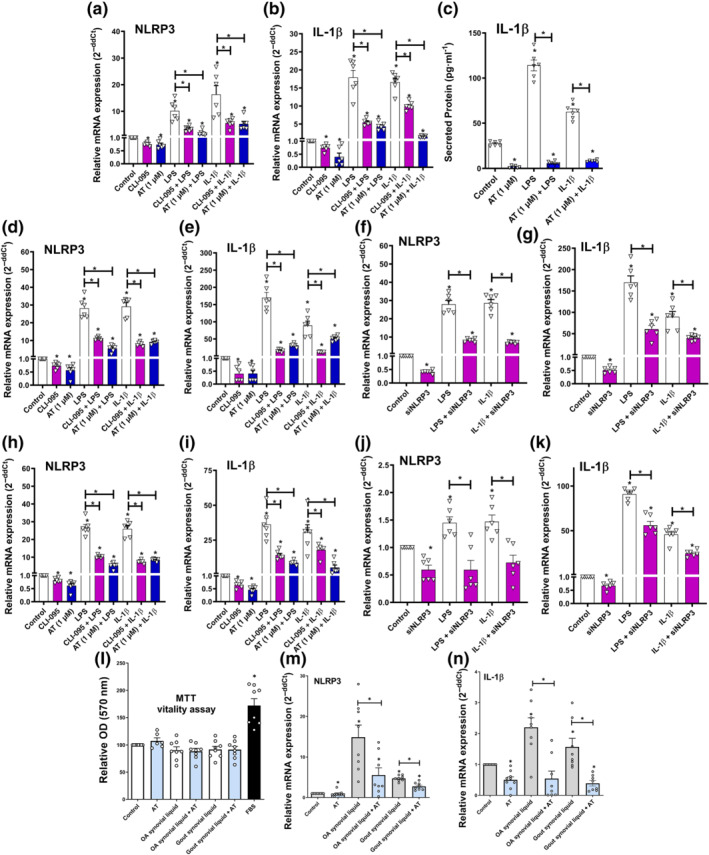
Amitriptyline (AT) blocks NLRP3 inflammasome. All experiments were independent, normalised by control and expressed as the mean ± SEM and statistically analysed by non‐parametric Krustal‐Wallis test coupled with Dunn's post‐test (* P<0.05). (a–c) Gene expression (RT‐PCR) of NLRP3 and IL‐1β mRNA (n = 6) and IL‐1β secreted levels (n = 3) were determined in human osteoarthritis (OA) chondrocytes (hOCs) pretreated with CLI‐095 (1 μM) for 3 h before TLR4 activation (LPS [100 ng·ml−1]) or IL‐1 receptor (R) activation (IL‐1β [0.1 ng·ml−1]) and cotreated with AT (1 μM) for 24 h. (d–g) Gene expression (RT‐PCR) of NLRP3 and IL‐1β mRNA were determined in ATDC5 chondrocytes pretreated with CLI‐095 (1 μM) for 3 h before TLR4 activation (LPS [100 ng·ml−1]) or IL‐1 receptor (R) activation (IL‐1β [0.1 ng·ml−1]) and co‐treated with AT (1 μM) or NLRP3 siRNA (15 nM) for 24 h (n = 6). (h–k) Gene expression (RT‐PCR) of NLRP3 and IL‐1β mRNA were determined in human SW982 synoviocytes pretreated with CLI‐095 (1 μM) for 3 h before TLR4 activation (LPS [100 ng·ml−1]) or IL‐1 receptor activation (IL‐1β [0.1 ng·ml−1]) and cotreated with AT (1 μM) or NLRP3 siRNA (15 nM) for 24 h (n = 6). (l–n) MTT cell viability assay and gene expression (RT‐PCR) of NLRP3 and IL‐1β mRNA were determined in human SW982 synoviocytes stimulated with OA (n = 6) or gout (n = 4) synovial liquid (10% v/v) and cotreated with AT (1 μM) for 24 h. Results from 5M were also statistically analysed by non‐parametric Mann‐Whitney test (* P<0.05)
3.9. Clinical data mining identifies evidence about amitriptyline anti‐innate immune response in gout patients
Mining clinical drug data to study the association between amitriptyline consumption and osteoarthritis innate immune process susceptibility is not possible. In short, osteoarthritis does not have a specific treatment that could be used to track down osteoarthritis patients to test their consumption of amitriptyline. Fortunately, gout patients can be tracked down using colchicine, a treatment for gout flares. Consequently, we analysed dissociated and aggregated clinical data from amitriptyline and colchicine consumption in our healthcare area for 5 years. Among the patients of our healthcare area, the elderly (>65 years) (n = 107,172) had a higher incidence (5%) in colchicine consumption than the rest of the patients (1%). In these elderly patients, the number of colchicine consumers was 37% lower in those taking amitriptyline (Figures 6a and S3E,F). Specifically, amitriptyline intake was negatively associated and significantly correlated to colchicine consumption in a gender‐independent manner (Figures 6b and S3G,H).
FIGURE 6.
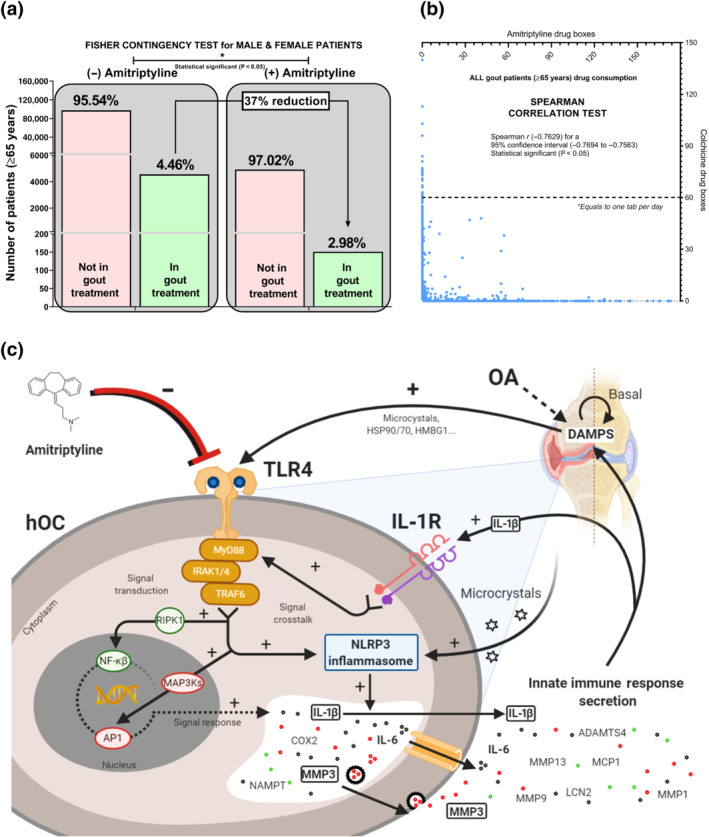
Amitriptyline reduces the need for colchicine gout treatment. Dissociated and aggregated clinical data of 107,172 elderly (>65 years) patients from Santiago de Compostela (Spain) healthcare area were mined for 5 years. (a) Fisher Contingency exact test (* P<0.05) comparing the number of elderly (>65 years) patients requiring or not colchicine and amitriptyline. (b) Spearman correlation test (* P<0.05) comparing the number of colchicine (dose: 0.5 mg most used; 0.66 mg exact mean) and amitriptyline (dose: 25 mg most used; 27.3 mg exact mean) drug boxes consumed by individual elderly (>65 years) patients. (c) Amitriptyline effects on TLR4 and IL‐1 receptor ® crosstalk signalling activation and inhibition including NLRP3/IL‐1β axis, NF‐κB and AP1 MyD88‐dependent pathways and in hOCs
4. DISCUSSION
In this work, we have identified that amitriptyline inhibits TLR4‐ and IL‐1 receptor‐mediated innate immune responses as a consequence of blocking TLR4 signalling and inhibiting NLRP3 expression in chondrocytes, synoviocytes and osteoblasts (Figure 6c). Furthermore, amitriptyline treatment was linked to lower colchicine consumption in gout patients, underpinning the repurpose to block articular innate immune responses in different musculoskeletal pathologies like osteoarthritis and gout.
The innate immune receptors TLR4 and IL‐1 receptor have a key role in several articular pathologies like osteoarthritis (Gómez et al., 2014). The blockade of IL‐1 receptor is used to treat rheumatoid arthritis (Mertens & Singh, 2009), gout (Pascart & Richette, 2017) and crystal‐induced arthritis (Aouba et al., 2015). Although TLR4 has been proposed as a therapeutic target for osteoarthritis and other arthritides (H. Park et al., 2020), a viable TLR4 inhibitor has yet to be found. Considering this, we determined whether amitriptyline, a drug that binds to TLR4 in mouse microglial cells (Hutchinson et al., 2010), could be repurposed as a TLR4 blocking agent to inhibit articular innate immune responses.
Considering that agonist TLR4 activation is species specific (Oblak & Jerala, 2015), we computationally explored the docking of amitriptyline on human TLR4 and found an even greater binding affinity than docking canonical agonist (LPS). Clinical trials testing novel pharmacological TLR4 inhibitors were not successful enough (Kuzmich et al., 2017), which suggests that amitriptyline could be useful to treat TLR4‐related diseases like rheumatic diseases, inflammatory bowel diseases (IBDs) (Dejban et al., 2020) or COVID‐19 (Sohn et al., 2020) among others.
To facilitate the translation into the clinic, we confirmed TLR4 inhibition in vitro using amitriptyline serum concentrations (0.033–1.130 μM) found in patients (Shimoda et al., 1997). Therefore, we selected three concentrations that represent the entire range of serum amitriptyline concentrations, 0.1 μM for lower range, 0.5 μM for medium range and 1 μM for highest range. In human osteoarthritis chondrocytes and mouse chondrocytes, amitriptyline in a dose–response manner blocked the gene expression of TLR4‐activated innate immune response factors broadly linked to osteoarthritis progression (Krasselt & Baerwald, 2019). This activity was devoid of cytotoxic effects, which further suggests that serum amitriptyline concentrations found in patients might exert relevant anti‐inflammatory effects in joint tissues. High‐throughput analysis of human osteoarthritis chondrocyte cellular proteome and secretome validated amitriptyline inhibition of TLR4 signalling and confirmed the down‐regulation of multiple proteins associated with innate immune‐related pathways (Pathan et al., 2017).
Interestingly, proteomic analysis revealed that IL‐1 signalling was also inhibited by amitriptyline in TLR4‐activated human osteoarthritis chondrocytes. Supporting this, TLR4 and IL‐1 receptor are known to be closely related because they share a part of their signalling pathway (Li & Qin, 2005; Martin & Wesche, 2002). In fact, IL‐1 receptor activation generates damage‐associated molecular patterns that activate TLR4 (Gómez et al., 2014), and TLR4 antagonism has shown to block IL‐1 receptor‐driven arthritis in animal models (Abdollahi‐Roodsaz et al., 2007). Consistent with this, we found that specific inhibition of TLR4 by CLI‐095 or amitriptyline in IL‐1 receptor‐activated human osteoarthritis chondrocytes blocked IL‐1 receptor‐induced innate immune response factors related to osteoarthritis progression (Krasselt & Baerwald, 2019). These data, together with the null binding affinity of amitriptyline towards IL‐1 receptor, revealed that TLR4 activation is required for full IL‐1 receptor signalling and suggest a mechanism connecting both receptors. Accordingly, amitriptyline inhibitory effects on TLR4‐ and IL‐1 receptor‐activated human osteoarthritis chondrocytes were akin.
It is noteworthy that amitriptyline dual effect to inhibit TLR4‐ and IL‐1 receptor‐activated innate immune responses was also identified in human synoviocytes and osteoblasts. This finding not only excludes a tissue‐specific connection between TLR4 and IL‐1 receptor but also stiffens the potential repurposing of amitriptyline to cope with innate immune responses across the whole joint.
To gain further insight into the potential connection between TLR4 and IL‐1 receptor, we investigated the effects of amitriptyline on unstimulated human osteoarthritis chondrocytes. Proteomic analysis revealed that amitriptyline promoted anabolic changes and blocked basal TLR4 and NLRP3 signalling. This suggests that amitriptyline repurposing might be used as a preventive and therapeutic tool against articular innate immune response. Interestingly, blockade of TLR4 and NLRP3 signalling was a common denominator of amitriptyline effects.
It is well known that NLRP3 signalling is linked to innate immunity activation (Demidowich et al., 2016), including TLR4 and IL‐1 receptor signalling in osteoarthritis (Qing et al., 2013) and gout (Mangan et al., 2018; Szekanecz et al., 2019). Consistent with this, we found that amitriptyline and/or CLI‐095 inhibition of TLR4 down‐regulated basal and inducible NLRP3 expression in osteoarthritis chondrocytes, osteoblasts and synoviocytes. Interestingly, this down‐regulation may be responsible for the anti‐inflammatory effects of TLR4 blockade because NLRP3 silencing was enough to reproduce amitriptyline inhibitory effects on TLR4‐ and IL‐1 receptor‐activated chondrocytes and synoviocytes. This idea was further underpinned in a more clinical context (osteoarthritis and gout) after amitriptyline reduced in synoviocytes the expression of NLRP3 and IL‐1β induced by the synovial liquids from osteoarthritis and gout and patients. In agreement with this, NLRP3 modulation has been considered a promising therapeutic approach for diseases like gout (Mangan et al., 2018; Szekanecz et al., 2019).
Evaluating amitriptyline effects in vitro could be considered a limitation of this study. However, to compensate for this, all the experiments were performed in primary human cells from osteoarthritis patients using amitriptyline concentrations within the serum concentrations observed in amitriptyline‐treated patients. In addition, logistic and ethical limitations blocked the clinical evaluation of amitriptyline effects in a big cohort of patients with joint inflammatory diseases. Nonetheless, these limitations were overcome, by the mining of dissociated and aggregated drug consumption data from our healthcare area, which allowed us to track down amitriptyline‐treated patients, as well as gout patients through their colchicine consumption. Underpinning the clinical relevance of amitriptyline blockade of TLR4/NLRP3 axis, amitriptyline intake significantly reduced colchicine consumption in a dose‐dependent manner. This suggests that amitriptyline action could reduce gout flares. Although these clinical data do not demonstrate causality, namely, the mechanism might be TLR4 independent, they strongly support amitriptyline repurposing to manage inflammation across diverse musculoskeletal pathologies like gout and osteoarthritis.
5. CONCLUSIONS
In this work, we present evidence supporting amitriptyline anti‐inflammatory and anticatabolic properties. We demonstrate that amitriptyline blockade of TLR4 signalling involves IL‐1 receptor and NLRP3 signalling inhibition. Interestingly, amitriptyline down‐regulation of NLRP3 expression could be responsible for amitriptyline anti‐inflammatory effects. Moreover, we show clinical evidence that amitriptyline consumption is associated with lower colchicine intake, which suggests an amitriptyline blockade of inflammatory gout flares.
Altogether, we present a solid amount of evidence supporting local or systemic amitriptyline repurposing to block articular innate immune responses.
CONFLICT OF INTEREST
The authors declare no conflicts of interest. Funders had no role in the study design; data collection, analysis, or interpretation; writing of the manuscript; or decision to publish the results.
AUTHOR CONTRIBUTIONS
All the authors meet the four British Journal of Pharmacology authorship criteria, which are experimental data design, acquisition, analysis and/or interpretation; involved in manuscript drafting or revising; take public responsibility of the final approved version; and account to the accuracy or integrity of any part of the work. We list the following as non‐author contributors: Veronica López for lab technician support, Ana Suárez Rodríguez for clinical pharmacological advice, Alberto Franco Estadella for dataset screening support and BioRender.com for science figure creation support.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for Design & Analysis and as recommended by funding agencies, publishers and other organisations engaged with supporting research.
Supporting information
Data S1. Supporting Information
Figure S1. Amitriptyline (AT) effects on ATDC5. All experiments were independent, normalised by control and expressed as the Mean ±SEM and statistically analysed by parametric One Way ANOVA coupled with Tukey post‐test (* P<0.05). A‐J: MTT cell viability assay, Griess nitrite accumulation assay, and mRNA expression (RT‐PCR) were determined in TLR4 (LPS [100 ng·kg‐1]) activated ATDC5 chondrocytes co‐treated with AT [0.1, 0.5, 1.0 μM] for 24 h (n = 6). K: Unsupervised multivariate square mean centre principal component analysis (PCA) of TLR4‐activated human osteoarthritis chondrocyte (hOC) co‐treated with AT [1 μM] for 24 h (n = 3). L‐Q: MTT cell viability assay, Griess nitrite accumulation assay, and innate immune response factors mRNA expression (RT‐PCR) were determined in hOC pre‐treated with CLI‐095 [1 μM] for 3 h before TLR4 or IL1 receptor (R) (IL1β [0.1 ng·ml‐1]) activation, and co‐treated with AT [0.1, 0.5, 1.0 μM] for 24 h (n = 6). R‐AA: MTT cell viability assay, Griess nitrite accumulation assay, and mRNA expression (RT‐PCR) were determined in IL1R activated ATDC5 chondrocytes co‐treated with AT [0.1, 0.5, 1.0 μM] for 24 h (n = 6).
Figure S2. Amitriptyline (AT) effects on TLR4 & IL1receptor (R)‐activated human osteoarthritis chondrocyte (hOC), synoviocytes and osteoblasts. All experiments were independent, normalised by control and expressed as the Mean ±SEM and statistically analysed by non‐parametric Krustal‐Wallis test coupled with Dunn's post‐test (* P<0.05). A: Unsupervised multivariate square mean centre principal component analysis (PCA) of IL1 receptor (R)‐activated (IL1β [0. ng·ml‐1]) human OA chondrocytes (hOC) co‐treated with AT [1 μM] for 24 h (n = 3). B‐G: MTT cell viability assay, Griess nitrite accumulation assay, and innate immune response factors mRNA expression (RT‐PCR) were determined in TLR4 activated (LPS [100 ng·ml‐1]) or IL1R activated human SW982 synoviocytes co‐treated with AT [0.1, 0.5, 1.0 μM] for 24 h (n = 6). H‐M: MTT cell viability assay, Griess nitrite accumulation assay, and innate immune response factors mRNA expression (RT‐PCR) were determined in TLR4 activated (LPS [100 ng/ml]) or IL1R activated primary human osteoblasts co‐treated with AT [0.1, 0.5, 1.0 μM] for 24 h (n = 6).
Figure S3. Amitriptyline effects on hOC, ATDC5, synoviocytes, and colchicine consumption. All experiments were independent (n=6), normalised by control and expressed as the Mean ±SEM. A: Quantitative data (SWATH) shown as a volcano plot (p‐value vs fold‐change) of protein expression changes by amitriptyline in non‐activated hOC (n = 3). B‐C: Griess nitrite accumulation assay and MTT cell viability assay were determined in ATDC5 chondrocytes pre‐treated with CLI‐095 [1 μM] for 3 h before TLR4 activation (LPS [100 ng·ml‐1]) or IL1 receptor (R) activation (IL1β [0.1 ng·ml‐1]) and co‐treated with NLRP3 siRNA [15 nM] for 24 h. Results were statistically analysed by parametric One Way ANOVA coupled with Tukey post‐test (* P<0.05). D: MTT cell viability assay was determined in human SW982 synoviocytes pre‐treated with CLI‐095 [1 μM] for 3 h before TLR4 or IL1R activation and co‐treated with NLRP3 siRNA [15 nM] for 24 h (n = 6). E‐F: Fisher Contingency exact test comparing the number of female or male patients (>65 years) requiring or not colchicine and amitriptyline (n = 107,172). G‐H: Spearman correlation test comparing the absolute amount of drug boxes of colchicine (0.5 mg most used dose; 0.66 mg exact mean) and amitriptyline (25 mg most used dose; 27.3 mg exact mean) consumed for 5 years in each individual female or male (>65 years) patient (n = 107,172).
ACKNOWLEDGEMENTS
This research was supported by grants from Fondo de Investigación Sanitaria, which is funded by the Instituto de Salud Carlos III (ISCIII) and co‐funded by the European Regional Development Fund (Fondo Europeo de Desarrollo Regional [FEDER]) (PI19/01446, PI16/01870, CP15/00007, PI14/00016 and PI17/00409). This work was also funded by the Spanish Ministry of Science (Ministerio de Ciencia e Innovación) and Fundación Española para la Ciencia y la Tecnologia (FECYT), through a Precipita crowdfunding (STOP ARTROSIS). R.G.B. is funded by ISCIII and Servizo Galego de Saúde (SERGAS), through a Miguel Servet II programme. A.J.‐M and O.G. are SERGAS personnel. This work was also supported by grants from Mutua Madrileña Foundation (Fundación Mutua Madrileña) (MMA 2018 and MMA 2020). E.F.‐T. and A.P.‐P. are funded by grants from Mutua Madrileña Foundation (MMA 2018). A.A.‐P. and S.B.B. are funded by the Instituto de Investigación Sanitaria de Santiago de Compostela (IDIS). M.G.‐F. is funded by the Ministry of Science and Innovation through an FPU grant (FPU17/01706). M.L.‐F. is funded by ISCIII (FI20/00210).
Franco‐Trepat, E. , Alonso‐Pérez, A. , Guillán‐Fresco, M. , Jorge‐Mora, A. , Crespo‐Golmar, A. , López‐Fagúndez, M. , Pazos‐Pérez, A. , Gualillo, O. , Belén Bravo, S. , & Gómez Bahamonde, R. (2022). Amitriptyline blocks innate immune responses mediated by toll‐like receptor 4 and IL‐1 receptor: Preclinical and clinical evidence in osteoarthritis and gout. British Journal of Pharmacology, 179(2), 270–286. 10.1111/bph.15707
Eloi Franco‐Trepat and Ana Alonso‐Pérez contributed equally to this work.
Funding information Fundación Mutua Madrileña, Grant/Award Numbers: MMA 2018, MMA 2020; Instituto de Investigación Sanitaria de Santiago de Compostela, Grant/Award Number: Fellowship; Servizo Galego de Saúde; Fundación Española para la Ciencia y la Tecnologia; Ministerio de Ciencia e Innovación, Grant/Award Numbers: FECYT ‐ PRECIPITA, FPU17/01706, STOP ARTROSIS ‐ PR250; European Regional Development Fund; Instituto de Salud Carlos III, Grant/Award Numbers: CP15/00007, FI20/00210, Miguel Servet II, PI14/00016, PI16/01870, PI17/00409, PI19/01446
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. Some data may not be made available because of privacy or ethical restrictions.
REFERENCES
- Abdollahi‐Roodsaz, S. , Joosten, L. A. B. , Roelofs, M. F. , Radstake, T. R. D. J. , Matera, G. , Popa, C. , van der Meer, J. W. M. , Netea, M. G. , & van den Berg, W. B. (2007). Inhibition of toll‐like receptor 4 breaks the inflammatory loop in autoimmune destructive arthritis. Arthritis and Rheumatism, 56(9), 2957–2967. 10.1002/art.22848 [DOI] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Cidlowski, J. A. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Armstrong, J. F. , Faccenda, E. , Harding, S. D. , Pawson, A. J. , Sharman, J. L. , Southan, C. , Davies, J. A. , & CGTP Collaborators (2019). THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Nuclear hormone receptors. British Journal of Pharmacology, 176, S229–S246. 10.1111/bph.14750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Armstrong, J. F. , Faccenda, E. , Harding, S. D. , Pawson, A. J. , Sharman, J. L. , Southan, C. , Davies, J. A. , & CGTP Collaborators (2019a). THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Catalytic receptors. British Journal of Pharmacology, 176, S247–S296. 10.1111/bph.14751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Armstrong, J. F. , Faccenda, E. , Harding, S. D. , Pawson, A. J. , Sharman, J. L. , Southan, C. , Davies, J. A. , & CGTP Collaborators (2019b). THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Enzymes. British Journal of Pharmacology, 176, S297–S396. 10.1111/bph.14752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso‐Pérez, A. , Franco‐Trepat, E. , Guillán‐Fresco, M. , Jorge‐Mora, A. , López, V. , Pino, J. , Gualillo, O. , & Gómez, R. (2018). Role of toll‐like receptor 4 on osteoblast metabolism and function. Frontiers in Physiology, 9, 504. 10.3389/fphys.2018.00504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouba, A. , Deshayes, S. , Frenzel, L. , Decottignies, A. , Pressiat, C. , Bienvenu, B. , Boue, F. , Damaj, G. , Hermine, O. , & Georgin‐Lavialle, S. (2015). Efficacy of anakinra for various types of crystal‐induced arthritis in complex hospitalized patients: A case series and review of the literature. Mediators of Inflammation, 2015, 1–7. 10.1155/2015/792173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, K. , Woller, S. A. , Miller, Y. I. , Yaksh, T. L. , Wallace, M. , Beaton, G. , & Chakravarthy, K. (2018). Targeting toll‐like receptor‐4 (TLR4)—An emerging therapeutic target for persistent pain states. Pain, 159(10), 1908–1915. 10.1097/j.pain.0000000000001306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley, S. K. , Berman, H. M. , Bhikadiya, C. , Bi, C. , Chen, L. , Di Costanzo, L. , Christie, C. , Dalenberg, K. , Duarte, J. M. , Dutta, S. , Feng, Z. , Ghosh, S. , Goodsell, D. S. , Green, R. K. , Guranović, V. , Guzenko, D. , Hudson, B. P. , Kalro, T. , Liang, Y. , … Zardecki, C. (2019). RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Research, 47(D1), D464–D474. 10.1093/nar/gky1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy, R. M. , Gomez, P. F. , & Abramson, S. B. (2004). Nitric oxide sustains nuclear factor kappaB activation in cytokine‐stimulated chondrocytes. Osteoarthritis and Cartilage, 12(7), 552–558. 10.1016/j.joca.2004.04.003 [DOI] [PubMed] [Google Scholar]
- Couch, J. R. , & Amitriptyline Versus Placebo Study Group . (2011). Amitriptyline in the prophylactic treatment of migraine and chronic daily headache. Headache, 51(1), 33–51. 10.1111/j.1526-4610.2010.01800.x [DOI] [PubMed] [Google Scholar]
- Couselo‐Seijas, M. , López‐Canoa, J. N. , Agra‐Bermejo, R. M. , Díaz‐Rodriguez, E. , Fernandez, A. L. , Martinez‐Cereijo, J. M. , Durán‐Muñoz, D. , Bravo, S. B. , Velo, A. , González‐Melchor, L. , Fernández‐López, X. A. , Martínez‐Sande, J. L. , García‐Seara, J. , González‐Juanatey, J. R. , Rodriguez‐Mañero, M. , & Eiras, S. (2019). Cholinergic activity regulates the secretome of epicardial adipose tissue: Association with atrial fibrillation. Journal of Cellular Physiology, 234(7), 10512–10522. 10.1002/jcp.27723 [DOI] [PubMed] [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, M. A. , Hoyer, D. , Insel, P. A. , Izzo, A. A. , Ji, Y. , MacEwan, D. J. , Sobey, C. G. , Stanford, S. C. , Teixeira, M. M. , Wonnacott, S. , & Ahluwalia, A. (2018). Experimental design and analysis and their reporting II: Updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 175(7), 987–993. 10.1111/bph.14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejban, P. , Nikravangolsefid, N. , Chamanara, M. , Dehpour, A. , & Rashidian, A. (2020). The role of medicinal products in the treatment of inflammatory bowel diseases (IBD) through inhibition of TLR4/NF‐kappaB pathway. Phytotherapy Research, 35, 835–845. 10.1002/ptr.6866 [DOI] [PubMed] [Google Scholar]
- del Pilar Chantada‐Vázquez, M. , López, A. C. , Vence, M. G. , Vázquez‐Estévez, S. , Acea‐Nebril, B. , Calatayud, D. G. , Jardiel, T. , Bravo, S. B. , & Núñez, C. (2020). Proteomic investigation on bio‐corona of Au, Ag and Fe nanoparticles for the discovery of triple negative breast cancer serum protein biomarkers. Journal of Proteomics, 212, 103581. 10.1016/j.jprot.2019.103581 [DOI] [PubMed] [Google Scholar]
- Demidowich, A. P. , Davis, A. I. , Dedhia, N. , & Yanovski, J. A. (2016). Colchicine to decrease NLRP3‐activated inflammation and improve obesity‐related metabolic dysregulation. Medical Hypotheses, 92, 67–73. 10.1016/j.mehy.2016.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco‐Trepat, E. , Guillán‐Fresco, M. , Alonso‐Pérez, A. , Jorge‐Mora, A. , Francisco, V. , Gualillo, O. , & Gómez, R. (2019). Visfatin connection: Present and future in osteoarthritis and osteoporosis. Journal of Clinical Medicine, 8(8), 1178. 10.3390/jcm8081178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez, R. , Villalvilla, A. , Largo, R. , Gualillo, O. , & Herrero‐Beaumont, G. (2014). TLR4 signalling in osteoarthritis—Finding targets for candidate DMOADs. Nature Reviews Rheumatology, 11(3), 1–12. 10.1038/nrrheum.2014.209 [DOI] [PubMed] [Google Scholar]
- Hannon, G. J. , & Rossi, J. J. (2004). Unlocking the potential of the human genome with RNA interference. Nature, 431(7006), 371–378. 10.1038/nature02870 [DOI] [PubMed] [Google Scholar]
- Hellings, J. A. , Arnold, L. E. , & Han, J. C. (2017). Dopamine antagonists for treatment resistance in autism spectrum disorders: Review and focus on BDNF stimulators loxapine and amitriptyline. Expert Opinion on Pharmacotherapy, 18(6), 581–588. 10.1080/14656566.2017.1308483 [DOI] [PubMed] [Google Scholar]
- Hussey, S. E. , Liang, H. , Costford, S. R. , Klip, A. , DeFronzo, R. A. , Sanchez‐Avila, A. , Ely, B. , & Musi, N. (2012). TAK‐242, a small‐molecule inhibitor of Toll‐like receptor 4 signalling, unveils similarities and differences in lipopolysaccharide‐ and lipid‐induced inflammation and insulin resistance in muscle cells. Bioscience Reports, 33(1), 37–47. 10.1042/BSR20120098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson, M. R. , Loram, L. C. , Zhang, Y. , Shridhar, M. , Rezvani, N. , Berkelhammer, D. , Phipps, S. , Foster, P. S. , Landgraf, K. , Falke, J. J. , Rice, K. C. , Maier, S. F. , Yin, H. , & Watkins, L. R. (2010). Evidence that tricyclic small molecules may possess toll‐like receptor and myeloid differentiation protein 2 activity. Neuroscience, 168(2), 551–563. 10.1016/j.neuroscience.2010.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, D. (1973). Anxiety and drug therapy. Proceedings of the Royal Society of Medicine, 66(3), 252–255. http://www.ncbi.nlm.nih.gov/pubmed/4572667. 10.1177/003591577306600325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasselt, M. , & Baerwald, C. (2019). Celecoxib for the treatment of musculoskeletal arthritis. Expert Opinion on Pharmacotherapy, 20(14), 1689–1702. 10.1080/14656566.2019.1645123 [DOI] [PubMed] [Google Scholar]
- Kuzmich, N. N. , Sivak, K. V. , Chubarev, V. N. , Porozov, Y. B. , Savateeva‐Lyubimova, T. N. , & Peri, F. (2017). TLR4 signaling pathway modulators as potential therapeutics in inflammation and sepsis. Vaccine, 5(4), 34. 10.3390/vaccines5040034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , & Qin, J. (2005). Modulation of toll–interleukin 1 receptor mediated signaling. Journal of Molecular Medicine, 83(4), 258–266. 10.1007/s00109-004-0622-4 [DOI] [PubMed] [Google Scholar]
- Loiarro, M. , Ruggiero, V. , & Sette, C. (2010). Targeting TLR/IL‐1R signalling in human diseases. Mediators of Inflammation, 2010, 12. 10.1155/2010/674363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan, M. S. J. , Olhava, E. J. , Roush, W. R. , Seidel, H. M. , Glick, G. D. , & Latz, E. (2018). Targeting the NLRP3 inflammasome in inflammatory diseases. Nature Reviews Drug Discovery, 17(8), 588–606. 10.1038/nrd.2018.97 [DOI] [PubMed] [Google Scholar]
- Martin, M. U. , & Wesche, H. (2002). Summary and comparison of the signaling mechanisms of the Toll/interleukin‐1 receptor family. Biochimica et Biophysica Acta‐Molecular Cell Research, 1592(3), 265–280. 10.1016/S0167-4889(02)00320-8 [DOI] [PubMed] [Google Scholar]
- McAllister, M. J. , Chemaly, M. , Eakin, A. J. , Gibson, D. S. , & McGilligan, V. E. (2018). NLRP3 as a potentially novel biomarker for the management of osteoarthritis. Osteoarthritis and Cartilage, 26(5), 612–619. 10.1016/j.joca.2018.02.901 [DOI] [PubMed] [Google Scholar]
- Mertens, M. , & Singh, J. A. (2009). Anakinra for rheumatoid arthritis. The Cochrane Database of Systematic Reviews, (1), CD005121. 10.1002/14651858.CD005121.pub3 [DOI] [PubMed] [Google Scholar]
- Oblak, A. , & Jerala, R. (2015). The molecular mechanism of species‐specific recognition of lipopolysaccharides by the MD‐2/TLR4 receptor complex. Molecular Immunology, 63(2), 134–142. 10.1016/j.molimm.2014.06.034 [DOI] [PubMed] [Google Scholar]
- Park, B. S. , Song, D. H. , Kim, H. M. , Choi, B. S. , Lee, H. , & Lee, J. O. (2009). The structural basis of lipopolysaccharide recognition by the TLR4–MD‐2 complex. Nature, 458(7242), 1191–1195. 10.1038/nature07830 [DOI] [PubMed] [Google Scholar]
- Park, H. , Hong, J. , Yin, Y. , Joo, Y. , Kim, Y. , Shin, J. , Kwon, H. H. , Shin, N. , Shin, H. J. , Beom, J. , Kim, D. W. , & Kim, J. (2020). TAP2, a peptide antagonist of Toll‐like receptor 4, attenuates pain and cartilage degradation in a monoiodoacetate‐induced arthritis rat model. Scientific Reports, 10(1), 17451. 10.1038/s41598-020-74544-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvathaneni, V. , Kulkarni, N. S. , Muth, A. , & Gupta, V. (2019). Drug repurposing: A promising tool to accelerate the drug discovery process. Drug Discovery Today, 24, 2076–2085. 10.1016/J.DRUDIS.2019.06.014 [DOI] [PubMed] [Google Scholar]
- Pascart, T. , & Richette, P. (2017). Current and future therapies for gout. Expert Opinion on Pharmacotherapy, 18(12), 1201–1211. 10.1080/14656566.2017.1351945 [DOI] [PubMed] [Google Scholar]
- Pathan, M. , Keerthikumar, S. , Chisanga, D. , Alessandro, R. , Ang, C. S. , Askenase, P. , Batagov, A. O. , Benito‐Martin, A. , Camussi, G. , Clayton, A. , Collino, F. , di Vizio, D. , Falcon‐Perez, J. M. , Fonseca, P. , Fonseka, P. , Fontana, S. , Gho, Y. S. , Hendrix, A. , Nolte‐'t Hoen, E. , … Mathivanan, S. (2017). A novel community driven software for functional enrichment analysis of extracellular vesicles data. Journal of Extracellular Vesicles, 6(1), 1321455. 10.1080/20013078.2017.1321455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing, Y. F. , Zhang, Q. B. , & Zhou, J. G. (2013). Innate immunity functional gene polymorphisms and gout susceptibility. Gene, 524(2), 412–414. 10.1016/j.gene.2013.04.039 [DOI] [PubMed] [Google Scholar]
- Rico‐Villademoros, F. , Slim, M. , & Calandre, E. P. (2015). Amitriptyline for the treatment of fibromyalgia: A comprehensive review. Expert Review of Neurotherapeutics, 15(10), 1123–1150. 10.1586/14737175.2015.1091726 [DOI] [PubMed] [Google Scholar]
- Saddichha, S. (2010). Diagnosis and treatment of chronic insomnia. Annals of Indian Academy of Neurology, 13(2), 94–102. 10.4103/0972-2327.64628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov, I. V. , Seymour, S. L. , Patel, A. A. , Loboda, A. , Tang, W. H. , Keating, S. P. , Hunter, C. L. , Nuwaysir, L. M. , & Schaeffer, D. A. (2007). The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Molecular & Cellular Proteomics, 6(9), 1638–1655. 10.1074/mcp.T600050-MCP200 [DOI] [PubMed] [Google Scholar]
- Shimoda, K. , Yasuda, S. , Morita, S. , Shibasaki, M. , Someya, T. , Bertilsson, L. , & Takahashi, S. (1997). Significance of monitoring plasma levels of amitriptyline, and its hydroxylated and desmethylated metabolites in prediction of the clinical outcome of depressive state. Psychiatry and Clinical Neurosciences, 51(1), 35–41. 10.1111/j.1440-1819.1997.tb02364.x [DOI] [PubMed] [Google Scholar]
- So, A. K. , & Martinon, F. (2017). Inflammation in gout: Mechanisms and therapeutic targets. Nature Reviews Rheumatology, 13(11), 639–647. 10.1038/nrrheum.2017.155 [DOI] [PubMed] [Google Scholar]
- Sohn, K. M. , Lee, S. G. , Kim, H. J. , Cheon, S. , Jeong, H. , Lee, J. , Kim, I. S. , Silwal, P. , Kim, Y. J. , Paik, S. , Chung, C. , Park, C. , Kim, Y. S. , & Jo, E. K. (2020). COVID‐19 patients upregulate toll‐like receptor 4‐mediated inflammatory signaling that mimics bacterial sepsis. Journal of Korean Medical Science, 35(38). e343–e343. 10.3346/JKMS.2020.35.E343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekanecz, Z. , Szamosi, S. , Kovács, G. E. , Kocsis, E. , & Benkő, S. (2019). The NLRP3 inflammasome–interleukin 1 pathway as a therapeutic target in gout. Archives of Biochemistry and Biophysics, 670, 82–93. 10.1016/j.abb.2019.01.031 [DOI] [PubMed] [Google Scholar]
- Thomas, C. , Bazan, J. F. , & Garcia, K. C. (2012). Structure of the activating IL‐1 receptor signaling complex. Nature Structural and Molecular Biology, 19(4), 455–457. 10.1038/nsmb.2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togbe, D. , Schnyder‐Candrian, S. , Schnyder, B. , Doz, E. , Noulin, N. , Janot, L. , Secher, T. , Gasse, P. , Lima, C. , Coelho, F. R. , Vasseur, V. , Erard, F. , Ryffel, B. , Couillin, I. , & Moser, R. (2007). Toll‐like receptor and tumour necrosis factor dependent endotoxin‐induced acute lung injury. International Journal of Experimental Pathology, 88(6), 387–391. 10.1111/j.1365-2613.2007.00566.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, Z. , Liu, Y. , Chen, B. , Yan, L. , & Hao, D. (2017). Association between MMP3 and TIMP3 polymorphisms and risk of osteoarthritis. Oncotarget, 8(48), 83563–83569. 10.18632/oncotarget.18745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott, O. , & Olson, A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalvilla, A. , García‐Martín, A. , Largo, R. , Gualillo, O. , Herrero‐Beaumont, G. , & Gómez, R. (2016). The adipokine lipocalin‐2 in the context of the osteoarthritic osteochondral junction. Scientific Reports, 6(1), 29243. 10.1038/srep29243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalvilla, A. , Silva, A. , Largo, R. , & Gualillo, O. (2014). 6‐Shogaol inhibits chondrocytes' innate immune responses and cathepsin‐K activity. Molecular Nutrition & Food, 58, 256–266. 10.1002/mnfr.201200833 [DOI] [PubMed] [Google Scholar]
- Vincent, T. L. (2019). IL‐1 in osteoarthritis: Time for a critical review of the literature. F1000Research, 8, 934. 10.12688/f1000research.18831.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Sampson, E. R. , Jin, H. , Li, J. , Ke, Q. H. , Im, H. J. , & Chen, D. (2013). MMP13 is a critical target gene during the progression of osteoarthritis. Arthritis Research and Therapy, 15(1), R5. 10.1186/ar4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamyatina, A. , & Heine, H. (2020). Lipopolysaccharide recognition in the crossroads of TLR4 and caspase‐4/11 mediated inflammatory pathways. Frontiers in Immunology, 11(585146), 1–22. 10.3389/fimmu.2020.585146 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supporting Information
Figure S1. Amitriptyline (AT) effects on ATDC5. All experiments were independent, normalised by control and expressed as the Mean ±SEM and statistically analysed by parametric One Way ANOVA coupled with Tukey post‐test (* P<0.05). A‐J: MTT cell viability assay, Griess nitrite accumulation assay, and mRNA expression (RT‐PCR) were determined in TLR4 (LPS [100 ng·kg‐1]) activated ATDC5 chondrocytes co‐treated with AT [0.1, 0.5, 1.0 μM] for 24 h (n = 6). K: Unsupervised multivariate square mean centre principal component analysis (PCA) of TLR4‐activated human osteoarthritis chondrocyte (hOC) co‐treated with AT [1 μM] for 24 h (n = 3). L‐Q: MTT cell viability assay, Griess nitrite accumulation assay, and innate immune response factors mRNA expression (RT‐PCR) were determined in hOC pre‐treated with CLI‐095 [1 μM] for 3 h before TLR4 or IL1 receptor (R) (IL1β [0.1 ng·ml‐1]) activation, and co‐treated with AT [0.1, 0.5, 1.0 μM] for 24 h (n = 6). R‐AA: MTT cell viability assay, Griess nitrite accumulation assay, and mRNA expression (RT‐PCR) were determined in IL1R activated ATDC5 chondrocytes co‐treated with AT [0.1, 0.5, 1.0 μM] for 24 h (n = 6).
Figure S2. Amitriptyline (AT) effects on TLR4 & IL1receptor (R)‐activated human osteoarthritis chondrocyte (hOC), synoviocytes and osteoblasts. All experiments were independent, normalised by control and expressed as the Mean ±SEM and statistically analysed by non‐parametric Krustal‐Wallis test coupled with Dunn's post‐test (* P<0.05). A: Unsupervised multivariate square mean centre principal component analysis (PCA) of IL1 receptor (R)‐activated (IL1β [0. ng·ml‐1]) human OA chondrocytes (hOC) co‐treated with AT [1 μM] for 24 h (n = 3). B‐G: MTT cell viability assay, Griess nitrite accumulation assay, and innate immune response factors mRNA expression (RT‐PCR) were determined in TLR4 activated (LPS [100 ng·ml‐1]) or IL1R activated human SW982 synoviocytes co‐treated with AT [0.1, 0.5, 1.0 μM] for 24 h (n = 6). H‐M: MTT cell viability assay, Griess nitrite accumulation assay, and innate immune response factors mRNA expression (RT‐PCR) were determined in TLR4 activated (LPS [100 ng/ml]) or IL1R activated primary human osteoblasts co‐treated with AT [0.1, 0.5, 1.0 μM] for 24 h (n = 6).
Figure S3. Amitriptyline effects on hOC, ATDC5, synoviocytes, and colchicine consumption. All experiments were independent (n=6), normalised by control and expressed as the Mean ±SEM. A: Quantitative data (SWATH) shown as a volcano plot (p‐value vs fold‐change) of protein expression changes by amitriptyline in non‐activated hOC (n = 3). B‐C: Griess nitrite accumulation assay and MTT cell viability assay were determined in ATDC5 chondrocytes pre‐treated with CLI‐095 [1 μM] for 3 h before TLR4 activation (LPS [100 ng·ml‐1]) or IL1 receptor (R) activation (IL1β [0.1 ng·ml‐1]) and co‐treated with NLRP3 siRNA [15 nM] for 24 h. Results were statistically analysed by parametric One Way ANOVA coupled with Tukey post‐test (* P<0.05). D: MTT cell viability assay was determined in human SW982 synoviocytes pre‐treated with CLI‐095 [1 μM] for 3 h before TLR4 or IL1R activation and co‐treated with NLRP3 siRNA [15 nM] for 24 h (n = 6). E‐F: Fisher Contingency exact test comparing the number of female or male patients (>65 years) requiring or not colchicine and amitriptyline (n = 107,172). G‐H: Spearman correlation test comparing the absolute amount of drug boxes of colchicine (0.5 mg most used dose; 0.66 mg exact mean) and amitriptyline (25 mg most used dose; 27.3 mg exact mean) consumed for 5 years in each individual female or male (>65 years) patient (n = 107,172).
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Some data may not be made available because of privacy or ethical restrictions.


