Abstract
Biomedical researchers routinely use a variety of biological models and resources, such as cultured cell lines, antibodies and laboratory animals. Unfortunately, these resources are not flawless: cell lines can be misidentified; for antibodies, problems with specificity, lot‐to‐lot consistency and sensitivity are common; and the reliability of animal models is questioned due to poor translation of animal studies to human clinical trials. In some cases, these problems can render the results of a study meaningless. As a response, some journals have implemented guidelines regarding the use and reporting of cell lines, antibodies and laboratory animals. In our study we use a portfolio of existing and newly created datasets to investigate identification and authentication information of cell lines, antibodies and organisms before and after guideline introduction, compared to journals without guidelines. We observed a general improvement of reporting quality over time, which the implementation of guidelines accelerated only in some cases. We therefore conclude that the effectiveness of journal guidelines is likely to be context dependent, affected by factors such as implementation conditions, research community support and monitoring and resource availability. Hence, journal reporting guidelines in themselves are not a quick fix to repair shortcomings in biomedical resource documentation, even though they can be part of the solution.
Keywords: biomedical resources, cell line authentication, journal guidelines, replication crisis, reporting standards
What's new?
Inadequately identified research materials and irreproducibility of results are significant issues in biomedical research. In response, some biomedical journals have added guidelines to help authors ensure the identity of research materials. Here, guideline compliance and effects concerning the reporting of materials were compared among journals before and after the implementation of guidelines for the authentication and validation of research materials. Reporting quality was found to have improved modestly over time, accelerated by authentication and validation guidelines only in certain instances. Hence, while journal guidelines can help ensure accurate resource reporting, additional measures are needed to address ongoing issues with misidentification.

Abbreviations
- ARRIVE
Animal Research: Reporting of In Vivo Experiments
- BMC
BioMed Central
- ICLAC
International Cell Line Authentication Committee
- IJC
International Journal of Cancer
- RRID
research resource identifier
- STR
short tandem repeats
1. INTRODUCTION
Biomedical researchers routinely use a variety of biological models and resources in their labs, including materials and specimens such as cultured cell lines, antibodies and laboratory animals. Unfortunately, these resources are not always flawless: trust in their stated provenance, identity and experimental suitability may not always be warranted. For example, cultured cell lines are frequently used as experimental models on the assumption that the cells correctly represent the tissue type from which they were originally derived. However, cell lines can become misidentified, potentially rendering study results meaningless. 1 Similar problems occur with antibodies: antibodies are used under the assumption that they bind exclusively to the protein of interest, but this may vary with experimental conditions. Problems with specificity, lot‐to‐lot consistency and sensitivity are common. 2 The validity and reliability of animal models used to study human physiology and anatomy 3 are likewise increasingly questioned, in response to poor translation of animal studies to human clinical trials 4 and irreproducibility, 5 at least partly due to similar problems of insufficient identification. 6
While some variability or imprecise identification in published research may be unavoidable and not even fatal to the published results, there is nevertheless cause for concern. Undocumented test organism identity, unaccounted sources of genetic variability, or even microbiome variation, can threaten correct interpretation and use of research results, or even invalidate results, spread error, lead to irreproducible research and jeopardise translation to clinical applications. 7 , 8 A Nature survey revealed that over 70% of researchers have tried and failed to reproduce another scientist's experiment in biomedical research, 9 in which inadequately identified research materials are a major factor of concern. 5 Even though the alarmist term ‘reproducibility crisis’ may wrongly suggest a recent outbreak, given that problems such as cell line misidentification have a long history, 10 extra attention for improved identification and validation does seem appropriate.
Some concerned scientists expect journals to play a central role in tackling unreliable, invalid or badly reported biomedical research resources. 11 , 12 In addition to the quality assurance provided by peer review, 13 journals' editorial policies could take a more directive role. Author guidelines have been suggested as a key instrument to encourage more complete identification and validation information, or even to mandate verification tests of key resource materials. In response, some prominent biomedical journals have devoted a section of their author guidelines to reporting standards for materials such as cell lines, antibodies and animals, including Nature Portfolio (previously Nature Research) and BMC journals.
While some studies have investigated the effectiveness of such guidelines, the analyses have typically focused on only one research resource or one journal, and remain overall inconclusive. 14 , 15 , 16 In our study, we present new data, and new analyses of existing data, about journal guideline compliance and effects concerning the reporting of cell lines, antibodies and animal models. By bringing together results assessing the effect of guidelines for a range of biomedical resources, we provide an overall assessment of journal guidelines' ability to improve resource identification and point out remaining uncertainties and avenues for further research.
2. METHODS AND DATA
2.1. Research design
To analyse whether journal guidelines improve the identification and authentication information of research resources we compared articles between journals with and without guidelines, and where possible also before and after guideline introduction. We assessed the reporting of cell line, antibody and organism identification information, that is, whether these resources can be uniquely identified by readers of the articles. In addition, we assessed levels of cell line authentication and articles' usage of misidentified cell lines. To control for the effect of changes over time, such as growing awareness in research communities, we selected journals with guidelines implemented in the same year and contrasted these with journals without guidelines in a before/after analysis with a control. However, this approach could only be used if it was clear when guidelines were precisely implemented or updated. We do know that BioMed Central (BMC) implemented guidelines regarding cell line authentication in 2015 (author's communication with Maria Hodges of BMC), and that Nature Portfolio implemented guidelines regarding cell line authentication in 2013 and these became stringent in 2015. 17 Thus, the journals of BMC and Nature Portfolio can be reliably contrasted with similar journals that lack guidelines regarding cell line authentication. Both BMC and Nature Portfolio also have guidelines regarding antibodies and refer to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines, which aim to improve the reporting of animal research. 18 However, precise implementation years for BMC's and Nature Portfolio's guidelines on antibodies and the ARRIVE guidelines could not be identified. In addition to the journal guidelines, Nature Portfolio implemented a checklist for authors with the aim to reduce irreproducibility. This checklist includes cell lines, antibodies and the ARRIVE guidelines, and was implemented in 2013. 19 However, it remains unclear whether Nature Portfolio's guidelines included antibodies and the ARRIVE guidelines previous to the introduction of the checklist. Therefore, for antibodies and the ARRIVE guidelines, we distinguished between journals of BMC, Nature Portfolio and other journals with guidelines on the one hand, and comparable journals without guidelines on the other, but could not establish a clear before and after (for the content of these guidelines see Supplementary material S1).
2.2. Data sources and analyses
We used data from a range of sources, including reanalysis of earlier studies. An overview is shown in Figure 1. Data analysis relied on coding articles and comparing rates of identification and validation information. We sometimes worked with relatively small sets of articles due to the splitting of the set into different journals and years. The number of articles analysed per category per year ranges from tens to thousands of articles. Generally, the number of articles included in our sample increased for later years (see supplementary tables for exact numbers in Appendix S1). For some of the data, we relied on expert readers, for most on automated searches. These methods have some disadvantages: they will not be able to identify all articles of interest and will contain some false positives. To avoid a misleading suggestion of certainty, we avoid advanced statistical analysis or significance testing.
FIGURE 1.
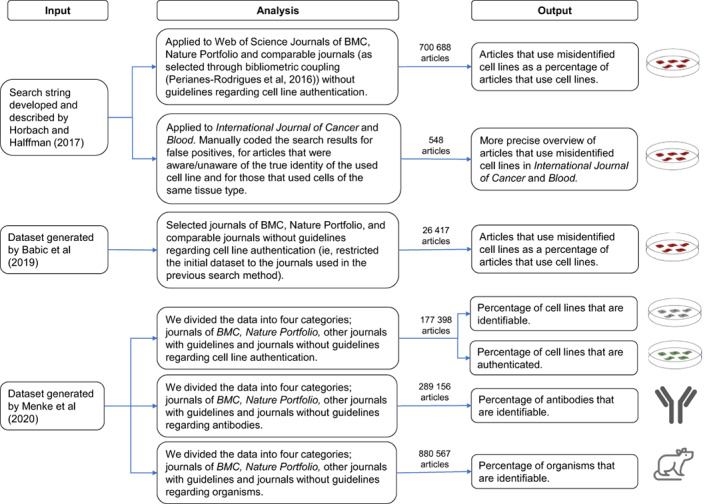
Overview of the input material, our analysis and the output [Color figure can be viewed at wileyonlinelibrary.com]
2.3. Horbach and Halffman string‐based search: Misidentified cell lines, before/after with control
To identify articles that used misidentified cell lines, we used a previously established search method based on version 8.0 of the list of misidentified cell lines of the International Cell Line Authentication Committee (ICLAC). 20 In short, this method searches Web of Science for articles with a misidentified cell line in the title, abstract or keywords and articles that cite the establishing paper of a cell line that later became misidentified. With this method, we searched journals of BMC and Nature Portfolio and comparable journals without guidelines regarding cell line authentication—comparable in the sense of bibliometric coupling 21 —for articles that used misidentified cell lines (for details see Supplementary material S2A,B). We calculated the number of articles that used misidentified cell lines as a percentage of articles that use cell lines in both sets of journals. Because cell lines will often be mentioned in the methods section rather than the title, abstract and keywords and because the establishing paper will not always be cited, this search cannot identify all articles that use misidentified cell lines.
2.4. Expert reader check: Misidentified cell lines, before/after with control
For two journals, we performed a more detailed study of the articles that used misidentified cell lines as identified by this search strategy. We chose the International Journal of Cancer (IJC) since this journal published multiple articles on the problem of misidentified cell lines and previously evaluated its guidelines as effective, 22 and Blood as a comparable journal without guidelines. We assessed the use of misidentified cell lines for all articles in the IJC (250) and Blood (initially 298, but 48 were meeting abstracts for which the full texts were unavailable, of which one was clearly a warning about misidentified cell lines; the other 47 articles were excluded from our analysis) that used misidentified cell lines as found by the search strategy of Horbach and Halffman 20 in the years 1995 to 2018. We distinguished two categories of false positives: papers warning about misidentified cell lines and papers in which no misidentified cell line was used. For the articles that did use a misidentified cell line, we distinguished three categories: misidentified cell line used but aware of problem (a), misidentified cell line used but of same tissue type (b) and misidentified cell line used and unaware of problem (c). For the last two categories we also distinguished between misidentified cell lines that were used before or after they were first reported as misidentified according to the ICLAC data (see Supplementary material S2C for details). We included in the analysis a small set of articles that did not use the misidentified cell line that triggered our search parameters (seemingly false positives), but that did use another misidentified cell line (3 articles in IJC and 11 in Blood; for details see Supplementary material S2C). The number of articles with cells was overall higher in Blood than in IJC (see Table S4 for precise numbers).
2.5. Algorithmic search Babic, Capes‐Davis, Martone, Bairoch, Ozyurt, Gillespie and Bandrowski: Misidentified cell lines, before/after with control
To further analyse the use of misidentified cell lines, we reanalysed part of a previously established dataset. 23 We restricted our analysis to the same journals as before (Nature Portfolio, BMC and comparable journals without guidelines), provided they were included in this dataset. In short, Babic, Capes‐Davis, Martone, Bairoch, Ozyurt, Gillespie and Bandrowski 23 used an algorithm to search for articles using cell lines in PubMed's openly accessible subset of articles and then matched these cell lines to the list of problematic cell lines. Besides misidentified cell lines, their problematic cell lines include those that are only partially misidentified. We excluded the partially misidentified cell lines, and continued only with the misidentified cell lines mentioned on the ICLAC list 1 for consistency with our own set (for details see Supplementary material S3).
2.6. Algorithmic search Menke, Roelandse, Ozyurt, Martone and Bandrowski: Positively identified and authenticated cell lines, before/after with control; positively identified antibodies and animals, guidelines vs control only
To analyse the authentication of cell lines and the identifiability of cell lines, antibodies and organisms, we reanalysed previously established datasets (cell lines supplementary data 8, antibodies supplementary data 7, organisms supplementary data 1 of Menke, Roelandse, Ozyurt, Martone and Bandrowski 24 ). In short, Menke, Roelandse, Ozyurt, Martone and Bandrowski 24 used an algorithm to search for articles that use cell lines, antibodies, or organisms in PubMed's subset of openly accessible articles. If enough information is supplied, namely name, supplier and catalogue number or a research resource identifier (RRID), the algorithm labels the cell line, antibody or organism as identifiable. In addition, the algorithm labelled cell lines as authenticated when there was information present for mycoplasma contamination or authentication assessment like STR (short tandem repeats) profiling (for details see Supplementary material S4).
3. RESULTS
3.1. Cell lines
We analysed whether guidelines improved cell line reporting, that is, whether cell lines are identifiable and misidentified cell lines are avoided. Figure 2 shows the percentage of articles that use misidentified cell lines as obtained through the search method described by Horbach and Halffman 20 (Figure 2A) and in the dataset of Babic, Capes‐Davis, Martone, Bairoch, Ozyurt, Gillespie and Bandrowski 23 (Figure 2B). Figure 2A shows a generally lower percentage of articles that use misidentified cell lines than Figure 2B. This is not surprising, because cell lines are usually mentioned in the methods section of an article, and the search method showed in Figure 2A is restricted to the title, keywords and abstract, as well as citation of an establishment article. Figure 2 shows that the use of misidentified cell lines—according to both methods—decreases both for journals with guidelines as well as for comparable journals without guidelines. This is in line with the recent decrease observed by Babic, Capes‐Davis, Martone, Bairoch, Ozyurt, Gillespie and Bandrowski. 23 Interestingly, the decrease seems to have started before the implementation of the guidelines in 2013 and 2015. Furthermore, the implementation of the guidelines in 2013 and 2015 does not seem to have accelerated the decrease. This suggests that the implementation of guidelines itself was not the prime driver of improved cell line usage. However, we have to be cautious in drawing this conclusion, since the implementation was relatively recent, and due to limitations of the search methods.
FIGURE 2.
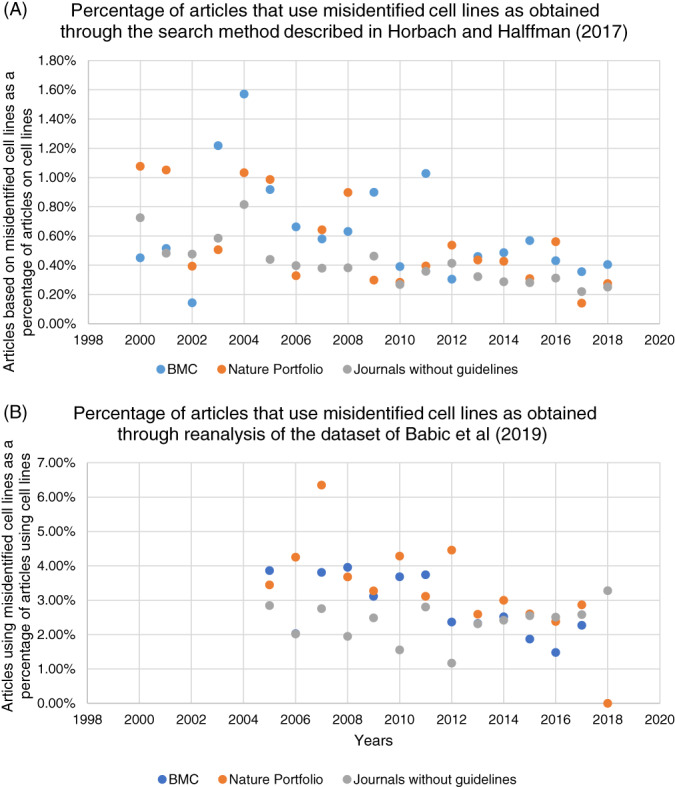
Distribution of articles in which the used cell lines were misidentified as obtained by search method described by Horbach and Halffman 20 (A) or Babic, Capes‐Davis, Martone, Bairoch, Ozyurt, Gillespie and Bandrowski 23 (B) in journals of BMC (guidelines since 2015) and Nature Portfolio (guidelines since 2013; stringent since 2015) and in comparable journals without guidelines [Color figure can be viewed at wileyonlinelibrary.com]
We also performed a more detailed study of the articles that used misidentified cell lines as obtained through the search method described by Horbach and Halffman 20 for the International Journal of Cancer (IJC). IJC implemented its guidelines in 2010—stringent since 2011—and evaluated its guidelines to be effective. 22 As a control, we used a comparable journal without guidelines: Blood. In our samples, we found a larger fraction of false positives than expected. Horbach and Halffman 20 estimated the share of articles falsely identified as using misidentified cell lines to be maximum 10%. However, in the sample used for our case study, we find that no misidentified cell line was used in 36% of IJC (90/250) and 55% of Blood (138/251) articles in this sample. Some of these articles—11 in IJC and 6 in Blood—were articles warning about (a) misidentified cell line(s). We used the remainder of the articles for our analysis.
Figure 3 shows the articles that used a misidentified cell line in IJC and Blood. In both IJC and Blood, the number of articles that use a misidentified cell line decreases over time, indicating an increase of awareness for the problem of misidentified cell lines. This general level of awareness might differ between research communities and their journals. We will revisit this issue in our discussion section. For IJC, this decrease seems to have accelerated after the implementation of guidelines (Figure 3A). However, there are still some articles that use a misidentified cell line after the implementation of the guidelines. Nevertheless, the majority of these articles (five out of six) did not use a misidentified cell line after it was first reported to be misidentified, which, in combination with the decrease in number of articles using misidentified cell lines without demonstrating awareness of their misidentification, constitutes a hopeful trend.
FIGURE 3.
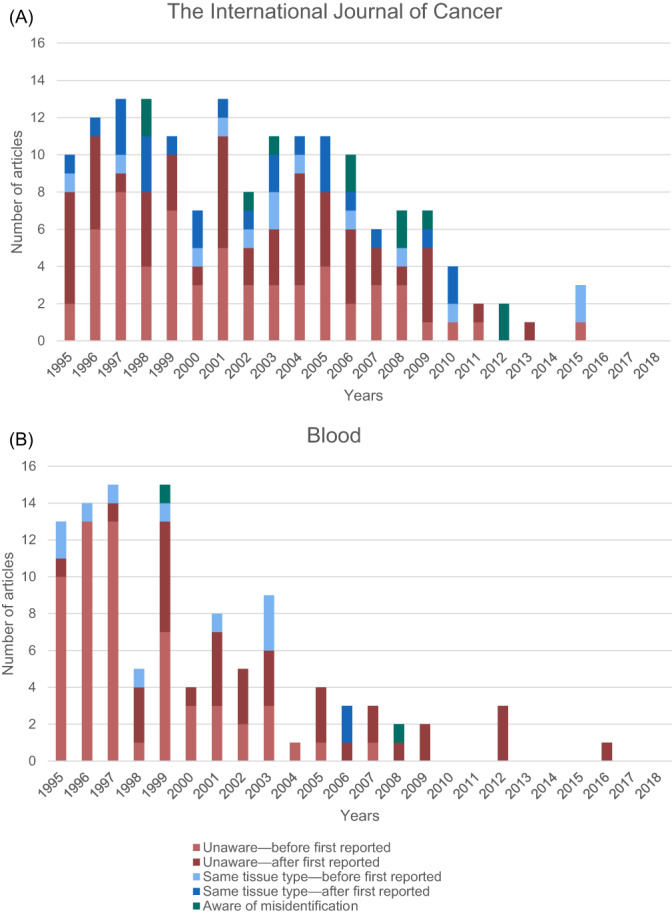
Assessment of how misidentified cell lines were used in 162 articles in The International Journal of Cancer (guidelines implemented in 2010, stringent since 2011) (A) and 107 articles in Blood (no guidelines) (B) as identified by applying the Web of Science search string of Horbach and Halffman. In which we distinguished articles in which a misidentified cell line was used while being aware of the misidentification (green), articles in which a misidentified cell line was used but the cell line was of the same tissue type (blue) and articles in which a misidentified cell line of a different tissue type was used while being unaware of it (red). For the last two categories, we distinguished between articles that used a misidentified cell line before it was first reported to be misidentified (light blue and light red) and after it was first reported to be misidentified (dark blue and dark red) [Color figure can be viewed at wileyonlinelibrary.com]
We then reanalysed the dataset of Menke, Roelandse, Ozyurt, Martone and Bandrowski 24 to show how the implementation of journal guidelines affected the percentage of cell lines that were authenticated or uniquely identifiable. Figure 4 shows the percentage of cell lines that were identifiable (Figure 4A) or authenticated (Figure 4B) in articles published in journals of BMC (with guidelines since 2015), Nature Portfolio (with guidelines since 2013; stringent since 2015), in other journals with guidelines, and in the control group consisting of journals without guidelines regarding cell line authentication.
FIGURE 4.
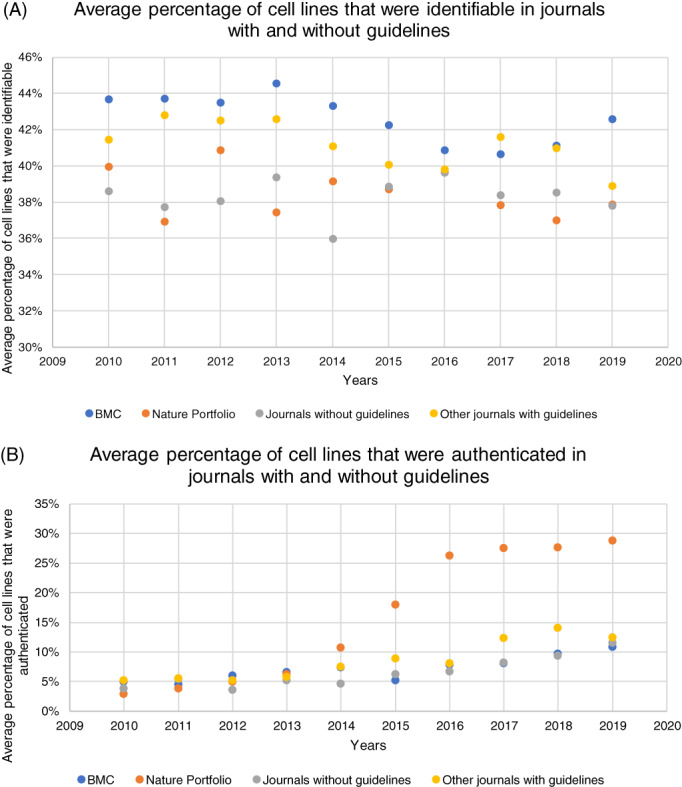
Distribution of articles in which the used cell lines were identifiable (A) or authenticated (B) in BMC (guidelines since 2015) and Nature Portfolio (guidelines since 2013, stringent since 2015) journals, in other journals with guidelines (implementation date unknown) and in journals without guidelines [Color figure can be viewed at wileyonlinelibrary.com]
The implementation of journal guidelines does not mark a general improvement in cell line identification (Figure 4A). In contrast, Figure 4B indicates a large increase of cell line authentication in Nature Portfolio journals, which does not occur in journals of BMC or journals without guidelines, suggesting this increase is caused by the specific conditions under which guidelines are implemented. In addition, Figure 4B shows a general, small but noticeable, increase in the percentage of authenticated cell lines over time. As this increase is also found in journals without guidelines, this suggests a general upsurge of awareness of the issue of cell line authentication. However, it is possible that this observed improvement mostly represents an increase in checking for mycoplasma contamination rather than cell line authentication, since the dataset of Menke, Roelandse, Ozyurt, Martone and Bandrowski 24 makes no distinction between these. Previous research indicated an overall decrease of mycoplasma‐contaminated cell lines, whereas the number of misidentified cell lines remained stable. 25
3.2. Antibodies
Similar to initiatives regarding cell lines, some journals have implemented guidelines to improve antibody validation and reporting. These journals require information on antibody validation or identification, such as via an RRID. Figure 5 shows the percentage of identifiable antibodies in articles of journals of BMC, Nature Portfolio, other journals with guidelines and in journals without guidelines as obtained through reanalysis of the dataset of Menke, Roelandse, Ozyurt, Martone and Bandrowski. 24 For all journals, the identifiability of antibodies seems to increase over time. This increase seems to be less steep for BMC journals and journals without guidelines than for journals of Nature Portfolio and other journals with guidelines. This again suggests that other factors beyond the presence of guidelines improve identification rates, a finding in line with previous research. 16
FIGURE 5.
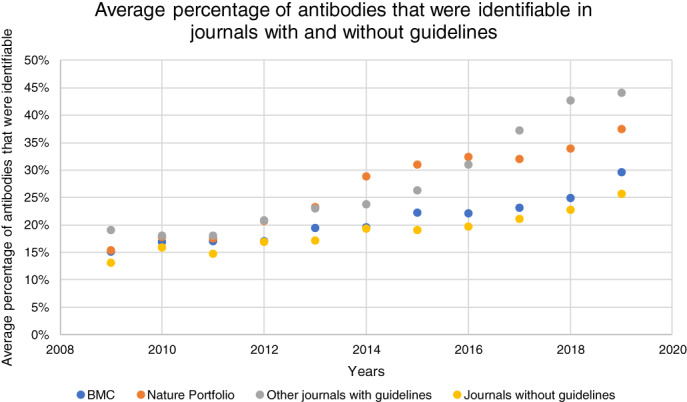
Distribution of articles in which the used antibodies were identifiable in journals of BMC (implementation year unknown), Nature Portfolio (checklist since 2013) and other journals with guidelines (implementation year unknown) and of journals without guidelines [Color figure can be viewed at wileyonlinelibrary.com]
3.3. Laboratory animals
The ARRIVE guidelines aim to improve the reporting of animal research, considering the reporting of the animal used in a uniquely identifiable way as a step in the right direction. We analysed how referring to the ARRIVE guidelines affected the number of articles with identifiable organisms. We must note that the Menke, Roelandse, Ozyurt, Martone and Bandrowski 24 dataset we could use here includes all organisms and not only animals, whereas the ARRIVE guidelines only concern animals. Figure 6 shows the percentage of identifiable organisms in articles in journals of BMC, Nature Portfolio, other journals that refer to the ARRIVE guidelines and in journals that do not refer to the ARRIVE guidelines. For all journals, the number of articles in which the organism used is identifiable does not seem to substantially increase or decrease over time, consistent with the generally low compliance with the ARRIVE guidelines as indicated in previous research. 14 , 26
FIGURE 6.
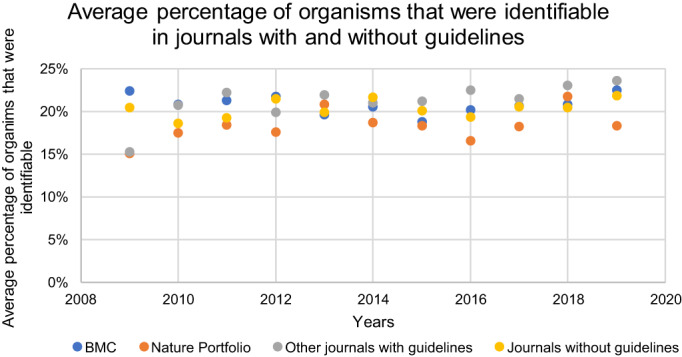
Distribution of articles in which the organisms used were identifiable in journals of BMC (implementation year unknown), Nature Portfolio (checklist since 2013), other journals that refer to the ARRIVE guidelines (implementation year unknown) and journals that do not refer to the ARRIVE guidelines [Color figure can be viewed at wileyonlinelibrary.com]
4. CONCLUSIONS AND DISCUSSION
Our study gathered data from a range of sources to assess the effect of journal guidelines on reported identification and validation information for biomedical experimental materials. While finding some variability, the general pattern presents a modest improvement of reporting adequacy over time. Overall, we observed an improvement in cell line and antibody reporting in both journals with and without guidelines which could be—partly—due to an increased awareness. Over the years, various efforts have been made to increase awareness for both problems with cell lines and antibodies. Many articles and editorials addressing problems with cell lines and antibodies have been published. 1 , 7 , 12 , 25 , 27 , 28 , 29 , 30 ICLAC was founded, 31 a standard for cell line authentication was set, 32 and the NIH now requires authentication of key biological resources, including cell lines and antibodies, for grant funding (NOT‐OD‐15‐103, NOT‐OD‐16‐011 and NOT‐OD‐16‐012). Antibody registries such as CiteAb 33 and the Antibody Registry 34 now exist, several workshops and international conferences have been hosted to tackle the problem of antibody validation, 35 , 36 and a series of webinars have been created to increase awareness. 37 Finally, journal guidelines—whether or not actively enforced—may also contribute to increased awareness.
However, while journal guidelines may constitute meaningful measures for increasing awareness, they only occasionally seem to cause drastic improvements in reporting quality. Therefore, we conclude that journal guidelines in themselves are not a quick fix to repair shortcomings in biomedical resource documentation, even though they can be part of the solution. Other, local and contextual factors most likely affect the efficacy of journal guidelines, such as the precise conditions of guideline implementation. For antibodies, similar factors were previously discussed regarding both identifiability 24 and validation information. 16
From a social science perspective, the variability of reporting guidelines' efficacy should not come as a surprise. Rules require legitimacy, enforcement and visibility among the regulated. Requirements must be doable under practical constraints, and general statements should be translatable to research practices in all their rich variation. 38 , 39 Rules that are considered toothless or meaningless bureaucracy may simply be ignored, or even worse: lead to cynicism or token compliance. Among researchers, cynicism may in turn even erode commitment to research integrity. 40 Organisations may also issue regulations primarily as symbolic tokenism in times of moral indignation, rather than as effective action plans, or they may simply lack effective power or resources to implement regulations. 41
After familiarising ourselves with the different guidelines, journals, editorial policies and literature, and after analysing available data, we can now hypothesise the factors that might influence guidelines' effectiveness. Future studies could assess these hypotheses in more detail.
One of the factors potentially influencing guidelines' effectiveness relates to their accessibility. We encountered strong variability of guidelines visibility between journals. Browsing through many Author Instructions, Editorial Policies, Guide to Authors and more, we noticed that these guidelines are often long and sometimes hard to find. To illustrate, Nature Portfolio refers to the ARRIVE guidelines under the heading Ethics and Biosecurity and not under Reporting standards and availability of data, materials, code and protocols, which is peculiar since the ARRIVE guidelines are a reporting standard. In addition, many of the journals of Nature Portfolio besides the Nature Portfolio Policies also have their own Guide to Authors, Editorial Policies and in some cases also a different Reproducibility Checklist. It therefore seems likely that not all authors—and possibly not even all editors—read and remember the complete author instructions. This becomes even more probable when considering the length and complexity of the instructions in combination with the increasing pressure on researchers to publish, and the common practice that a manuscript is often sent to multiple journals before it is accepted. Moreover, authors may only become aware of the existence of these guidelines when explicitly asked to comply upon manuscript submission when the experiments are already conducted.
Another potentially important factor relates to the guidelines' content and wording. Also in this respect, we noticed several differences among the guidelines studied for our analyses. For example, some journals ask researchers to check whether employed cell lines are on ICLAC's list of known misidentified cell lines, 1 whereas other journals ask researchers to actively authenticate their cell lines, such as by comparing STR profiles against online databases. 32 In addition, differences in wording suggest variation in guideline strictness, such as: ‘we recommend …’, ‘[we] strongly encourage authors to …,’ ‘author must declare …’ or ‘details need to be provided for …’ (see Supplementary material S1). There is, therefore, significant variation in the specificity, directiveness as well as resources and efforts required to comply.
Thirdly, guidelines' effectiveness might depend on the way in which they are embedded in the editorial process and how they are supported by other initiatives. To enhance compliance with their guidelines, several journals use various kinds of checklists. These range from internal checklists used in the editorial process, to checklists authors are required to complete at submission, such as Nature Portfolio's. 19 In some cases, the desired effect was indeed observed after the incorporation of these checklists in the publishing process, 15 , 42 but other studies, including in the context of the ARRIVE guidelines, show little improvement. 14 , 43 , 44 Besides checklists, some journals provide editorial support for compliance, such as regarding RRIDs: RRIDs were barely used in journals with only instructions to authors. Compliance was higher when, in addition to author instructions, letters were sent to the authors or the staff suggested specific RRIDs to the authors. 45 Some journals try to motivate their authors to comply to their guidelines by rewarding authors that do so with a badge. However, in the case of data sharing, there was no noticeably effect of this incentive. 46 Nevertheless, these endeavours suggest that putting additional effort in improving guidelines' practicality from an author, or 'user', perspective, is likely to increase compliance. This support and active enforcing of the guidelines by the editors, is, however, time‐consuming and thus expensive. 22 In addition, it requires expertise among the editorial staff that may not always be available or too heavy a burden for a journal's business model.
Hence, while the expectations for journals to enforce guidelines may be high, we should keep in mind that the power of journals to do so is not absolute. Journals also depend on researchers to submit papers and if journal guidelines are considered unreasonable, submissions may migrate to other journals. While the top journals may be in a strong position, most journals may not be able to run ahead of the herd. Our communication with an editor has provided anecdotal evidence in which a neurology journal made their antibody validation requirements less stringent 16 years after implementing them because of a submission‐drop and concerns about the journal's future (email communication with editor). These are all interesting avenues for further investigation.
When interpreting our study's findings, one should be considerate of the following limitations. The method described by Horbach and Halffman, 20 used to identify articles with a misidentified cell line, seems to lead to more false positives than previously thought. All other datasets are limited to the open access subset of PubMed, which does not only result in a relatively small sample size, but might also not be representative for entire research literature, still primarily consisting of nonopen access articles. In addition, our indicators may not be perfect proxies for what we aim to measure. For example, organism identifiability includes all organisms and not only animals. Finally, not all precise practices and even formal processes of editorial offices are known to us—among which the years of implementing guidelines regarding antibody reporting and endorsement of the ARRIVE guidelines. Lastly, we used Blood as a comparable journal without guidelines for IJC. However, since the German cell bank DSMZ published extensively on haematological cell lines, 25 , 27 , 47 , 48 , 49 , 50 , 51 the awareness of potential issues relating to misidentified cells may be above average in the community of researchers publishing in Blood. Thus, in other journals without guidelines, outside of this community, the awareness may still be lower, and, consequently, the use of misidentified cell lines higher than in Blood.
All in all, we conclude that while journal guidelines can be part of the solution of resource misidentification, they are not a quick fix for the problem of failing resource identification. While we find that journal guidelines may contribute to improved reporting and authentication, they are but one factor in a broader development of research standards. Their effect appears to depend on the precise conditions of implementation, as well as support in the research community, and requires a consideration of resource availability. In line with previous analyses of reporting guidelines more generally, 39 issuing and announcing guidelines in itself has a limited effect and requires additional measures. Future research on the effectiveness of guidelines to improve biomedical research material identification and authentication should therefore take the context of their implementation into account.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Appendix S1 Supporting Information.
ACKNOWLEDGEMENTS
We would like to thank Anita Bandrowski (Centre for Research in Biological Systems, University of California) for sharing the SciScore text‐mining dataset with us. We would like to thank Nees Jan van Eck (Centre for Science and Technology Studies, Universiteit Leiden) for sharing the algorithm for bibliographic coupling, and Patrick Kooij (Centre for Science and Technology Studies, Universiteit Leiden) for his help to apply this algorithm in our study. We also want to thank the members of the Research Quality Team (Institute for Science in Society, Radboud University) for valuable feedback on earlier drafts of this manuscript and to Camila R. van Ham for help with graphics. Lastly, we would also like to thank two anonymous reviewers for their detailed and constructive comments.
Hepkema WM, Horbach SPJM, Hoek JM, Halffman W. Misidentified biomedical resources: Journal guidelines are not a quick fix. Int. J. Cancer. 2022;150(8):1233‐1243. doi: 10.1002/ijc.33882
DATA AVAILABILITY STATEMENT
The data that support the findings of our study are available:
For Horbach and Halffman (2017) set: search string at DANS repository at https://doi.org/10.17026/dans-2ap-7bnu, conditional on approval from the Radboud Faculty of Science ethics committee, applied to WoS (webofknowledge.com). As analysed: in our Appendix S1 tables.
For Babic et al. (2019): full set courtesy of Anita Bandrowski (abandrowski@ncmir.ucsd.edu), expanded set of Appendix S1 with original publication: https://doi.org/10.7554/eLife.41676. As reanalysed: in our Appendix S1 tables.
For Menke et al. (2020): Appendix S1 to their publication: https://doi.org/10.1016/j.isci.2020.101698. As reanalysed: in our Appendix S1 tables.
Further information is available from the corresponding author upon request.
REFERENCES
- 1. Capes‐Davis A, Theodosopoulos G, Atkin I, et al. Check your cultures! A list of cross‐contaminated or misidentified cell lines. Int J Cancer. 2010;127:1‐8. [DOI] [PubMed] [Google Scholar]
- 2. Baker M. Antibody anarchy: a call to order. Nature. 2015;527:545. [DOI] [PubMed] [Google Scholar]
- 3. Hau J. Animal models for human diseases. In: Conn PM, ed. Sourcebook of Models for Biomedical Researched. Totowa, NJ: Humana Press; 2008:3‐8. [Google Scholar]
- 4. Hackam DG, Redelmeier DA. Translation of research evidence from animals to humans. JAMA. 2006;296:1727‐1732. [DOI] [PubMed] [Google Scholar]
- 5. Freedman LP, Cockburn IM, Simcoe TS. The economics of reproducibility in preclinical research. PLoS Biol. 2015;13:e1002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Witjes VM, Boleij A, Halffman W. Reducing versus embracing variation as strategies for reproducibility: the microbiome of laboratory mice. Animals. 2020;10:2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baker M. Reproducibility crisis: blame it on the antibodies. Nature. 2015;521:274‐276. [DOI] [PubMed] [Google Scholar]
- 8. Freedman LP, Gibson MC, Ethier SP, Soule HR, Neve RM, Reid YA. Reproducibility: changing the policies and culture of cell line authentication. Nat Methods. 2015;12:493‐497. [DOI] [PubMed] [Google Scholar]
- 9. Baker M. Is there a reproducibility crisis? A nature survey lifts the lid on how researchers view the'crisis rocking science and what they think will help. Nature. 2016;533:452‐455.27225100 [Google Scholar]
- 10. Gold M. A Conspiracy of Cells: One woman's Immortal Legacy and the Medical Scandal it Causeded. Albany, NY: SUNY Press; 1986. [Google Scholar]
- 11. Chambers K, Collings A, Graf C, et al. Towards minimum reporting standards for life scientists. MetaArXiv. 2019:1‐1. 10.31222/osf.io/9sm4x [DOI] [Google Scholar]
- 12. Vaughan L, Glänzel W, Korch C, Capes‐Davis A. Widespread use of misidentified cell line KB (HeLa): incorrect attribution and its impact revealed through mining the scientific literature. Cancer Res. 2017;77:2784‐2788. [DOI] [PubMed] [Google Scholar]
- 13. Horbach SPJM, Halffman W. The changing forms and expectations of peer review. Res Integr Peer Rev. 2018;3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baker D, Lidster K, Sottomayor A, Amor S. Two years later: journals are not yet enforcing the ARRIVE guidelines on reporting standards for pre‐clinical animal studies. PLoS Biol. 2014;12:e1001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han S, Olonisakin TF, Pribis JP, et al. A checklist is associated with increased quality of reporting preclinical biomedical research: a systematic review. PLoS One. 2017;12:e0183591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoek JM, Hepkema WM, Halffman W. The effect of journal guidelines on the reporting of antibody validation. PeerJ. 2020;8:e9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anon J. Announcement: time to tackle cells' mistaken identity. Nature. 2015;520:264. [Google Scholar]
- 18. Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anon J. Announcement: reducing our irreproducibility. Nature. 2013;496:398. [Google Scholar]
- 20. Horbach SPJM, Halffman W. The ghosts of HeLa: how cell line misidentification contaminates the scientific literature. PloS One. 2017;12:e0186281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perianes‐Rodriguez A, Waltman L, Van Eck NJ. Constructing bibliometric networks: a comparison between full and fractional counting. J Informet. 2016;10:1178‐1195. [Google Scholar]
- 22. Fusenig NE, Capes‐Davis A, Bianchini F, Sundell S, Lichter P. The need for a worldwide consensus for cell line authentication: experience implementing a mandatory requirement at the international journal of cancer. PLoS Biol. 2017;15:e2001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Babic Z, Capes‐Davis A, Martone ME, et al. Meta‐research: incidences of problematic cell lines are lower in papers that use RRIDs to identify cell lines. Elife. 2019;8:e41676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Menke J, Roelandse M, Ozyurt B, Martone M, Bandrowski A. The rigor and transparency index quality metric for assessing biological and medical science methods. iScience. 2020;23:101698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Drexler HG, Dirks WG, MacLeod RA, Uphoff CC. False and mycoplasma‐contaminated leukemia–lymphoma cell lines: time for a reappraisal. Int J Cancer. 2017;140:1209‐1214. [DOI] [PubMed] [Google Scholar]
- 26. Hair K, Macleod MR, Sena ES. A randomised controlled trial of an intervention to improve compliance with the ARRIVE guidelines (IICARus). Res Integr Peer Rev. 2019;4:1‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Drexler H, Dirks W, Matsuo Y, MacLeod R. False leukemia–lymphoma cell lines: an update on over 500 cell lines. Leukemia. 2003;17:416‐426. [DOI] [PubMed] [Google Scholar]
- 28. Capes‐Davis A, Neve RM. Authentication: a standard problem or a problem of standards? PLoS Biol. 2016;14:e1002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uhlen M, Bandrowski A, Carr S, et al. A proposal for validation of antibodies. Nat Methods. 2016;13:823‐827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. MacLeod RA, Dirks WG, Matsuo Y, Kaufmann M, Milch H, Drexler HG. Widespread intraspecies cross‐contamination of human tumor cell lines arising at source. Int J Cancer. 1999;83:555‐563. [DOI] [PubMed] [Google Scholar]
- 31. Capes‐Davis A, Reid YA, Kline MC, et al. Match criteria for human cell line authentication: where do we draw the line? Int J Cancer. 2013;132:2510‐2519. [DOI] [PubMed] [Google Scholar]
- 32. Almeida JL, Cole KD, Plant AL. Standards for cell line authentication and beyond. PLoS Biol. 2016;14:e1002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. CiteAb . https://www.citeab.com/2014
- 34. Antibody Registry . https://antibodyregistry.org/2011
- 35. GBSI . GBSI workshop report—Antibody Validation: Standards, Policies, and Practices; 2016.
- 36. Goodman SL. The 3rd Antibody Validation Meeting: Bath UK 20–21 st September 2018. F1000Research. 2018;7:1‐4. 10.12688/f1000research.17645.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Voskuil JL, Bandrowski A, Begley CG, et al. The antibody Society's antibody validation webinar series. MAbs. 2020;12:1794421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huising R, Silbey SS. Surveillance and Regulation of Laboratory Practices: The Handbook of Science and Technology Studiesed. Cambridge, MA: MIT Press; 2017:793‐822. [Google Scholar]
- 39. Blanco D, Altman D, Moher D, Boutron I, Kirkham JJ, Cobo E. Scoping review on interventions to improve adherence to reporting guidelines in health research. BMJ Open. 2019;9:e026589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clair JA. Procedural injustice in the system of peer review and scientific misconduct. Acad Manag Learn Edu. 2015;14:159‐172. [Google Scholar]
- 41. Edelman LB. Legal ambiguity and symbolic structures: organizational mediation of civil rights law. Am J Sociol. 1992;97:1531‐1576. [Google Scholar]
- 42. Plint AC, Moher D, Morrison A, et al. Does the CONSORT checklist improve the quality of reports of randomised controlled trials? A systematic review. Med J Aust. 2006;185:263‐267. [DOI] [PubMed] [Google Scholar]
- 43. Leung V, Rousseau‐Blass F, Beauchamp G, Pang DS. ARRIVE has not ARRIVEd: support for the ARRIVE (animal research: reporting of in vivo experiments) guidelines does not improve the reporting quality of papers in animal welfare, analgesia or anesthesia. PLoS One. 2018;13:e0197882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tanriver‐Ayder E, Gray LJ, McCann SK, et al. A randomised controlled trial of an intervention to improve compliance with the ARRIVE guidelines. J Physiol. 2019;598:3793‐3801. [Google Scholar]
- 45. Bandrowski A, Brush M, Grethe JS, et al. The resource identification initiative: a cultural shift in publishing. J Comp Neurol. 2016;524:8‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rowhani‐Farid A, Aldcroft A, Barnett AG. Did awarding badges increase data sharing in BMJ open? A randomized controlled trial. R Soc Open Sci. 2020;7:191818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. MacLeod RA, Nagel S, Scherr M, et al. Human leukemia and lymphoma cell lines as models and resources. Curr Med Chem. 2008;15:339‐359. [DOI] [PubMed] [Google Scholar]
- 48. Drexler HG, MaCleod RA. History of leukemia‐lymphoma cell lines. Hum Cell. 2010;23:75‐82. [DOI] [PubMed] [Google Scholar]
- 49. Tiacci E, Pucciarini A, Bigerna B, et al. Absence of BRAF‐V600E in the human cell lines BONNA‐12, ESKOL, HAIR‐M, and HC‐1 questions their origin from hairy cell leukemia. Blood. 2012;119:5332‐5333. [DOI] [PubMed] [Google Scholar]
- 50. Drexler HG, Chen S, Macleod RA. Would the real Waldenström cell line please stand up? Leuk Lymphoma. 2013;54:224‐226. [DOI] [PubMed] [Google Scholar]
- 51. Drexler HG, Eberth S, Nagel S, Quentmeier H. There is a scientific need for the right leukemia‐lymphoma cell lines. HemaSphere. 2019;3:e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting Information.
Data Availability Statement
The data that support the findings of our study are available:
For Horbach and Halffman (2017) set: search string at DANS repository at https://doi.org/10.17026/dans-2ap-7bnu, conditional on approval from the Radboud Faculty of Science ethics committee, applied to WoS (webofknowledge.com). As analysed: in our Appendix S1 tables.
For Babic et al. (2019): full set courtesy of Anita Bandrowski (abandrowski@ncmir.ucsd.edu), expanded set of Appendix S1 with original publication: https://doi.org/10.7554/eLife.41676. As reanalysed: in our Appendix S1 tables.
For Menke et al. (2020): Appendix S1 to their publication: https://doi.org/10.1016/j.isci.2020.101698. As reanalysed: in our Appendix S1 tables.
Further information is available from the corresponding author upon request.


