ABSTRACT
N‐glycosylation is an important post‐translational modification of proteins that has been highly conserved during evolution and is found in Eukaryota, Bacteria and Archaea. In eukaryotes, N‐glycan processing is sequential, involving multiple specific steps within the secretory pathway as proteins travel through the endoplasmic reticulum and the Golgi apparatus. In this review, we first summarize the different steps of the N‐glycan processing and further describe recent findings regarding the diversity of N‐glycan structures in eukaryotic clades. This comparison allows us to explore the different regulation mechanisms of N‐glycan processing among eukaryotic clades. Recent findings regarding the regulation of protein N‐glycosylation are highlighted, especially the regulation of the biosynthesis of complex‐type N‐glycans through manganese and calcium homeostasis and the specific role of transmembrane protein 165 (TMEM165) for which homologous sequences have been identified in several eukaryotic clades. Further research will be required to characterize the function of TMEM165 homologous sequences in different eukaryotic clades.
Keywords: calcium homeostasis, endoplasmic reticulum, eukaryotes, glycosylation, glycosyltransferases, Golgi apparatus, manganese homeostasis, N ‐glycans, regulation, structural diversity
I. INTRODUCTION
N‐glycosylation is a co‐ and post‐translational modification (PTM) of proteins resulting in the attachment of an oligosaccharide to the nascent protein through the formation of an N‐glycosyl bond between the monosaccharide moiety and an asparagine residue belonging to the consensus sequence Asn‐X‐Ser/Thr/Cys (Gil, Velander & Van Cott, 2009; Zielinska et al., 2010; Matsui et al., 2011; Aebi, 2013). N‐glycosylation represents the most abundant PTM of proteins (Khoury, Baliban & Floudas, 2011) and influences their physicochemical properties and biological functions (Lingg et al., 2012; Varki, 2017a). Glycoproteomics studies have identified at least 2,000 glycoproteins in each of the following model organisms Caenorhabditis elegans, Arabidopsis thaliana, Drosophila melanogaster and Danio rerio (Zielinska et al., 2012), with this number a likely underestimate. This process is universal and is found in Eukaryota, Bacteria, and Archaea (Nothaft & Szymanski, 2010). In bacteria, N‐glycan processing starts in the cytoplasmic compartment and continues in the periplasm (Li et al., 2017). By contrast, in eukaryotes, the N‐glycosylation pathway is initiated in the endoplasmic reticulum (ER) and continues in the Golgi apparatus while glycoproteins travel along the secretory pathway (Colley, Varki & Kinoshita, 2015). In this review, we mainly focus on N‐glycosylation pathways in eukaryotes, describing the main steps leading to the extensive diversity of N‐glycan structures found in different organisms. We describe the more processed structures of N‐glycans isolated from the main groups Amorphea, Archaeplastida, Excavates and TSAR (Telonemia, Stramenopila, Alveolata and Rhizaria). We also highlight recent findings regarding the regulation of this process. It was recently reported in mammals and plants that the biosynthesis of complex‐type N‐glycans is regulated through manganese (Mn2+) and calcium (Ca2+) homeostasis involving the newly identified transmembrane protein 165 (TMEM165). Future directions for research on protein N‐glycosylation are also discussed.
II. N‐GLYCAN BIOSYNTHESIS IN THE ER: A CONSERVED PROCESS DURING EVOLUTION
In eukaryotes, protein glycosylation involves hundreds of different molecular actors, including enzymes involved in the biosynthesis and transport of nucleotide sugars as well as glycosyltransferases (GTs) and glycosylhydrolases (GHs), called glycoenzymes. In humans for example, more than 500 glycoenzymes take part in N‐ and O‐glycan processing including 114 enzymes and transporters involved in carbohydrate metabolism and transport, 76 glycosidases and more than 200 GTs (Neelamegham & Mahal, 2016). GTs involved in N‐glycan biosynthesis or processing predominantly use activated nucleotide monosaccharides (also called nucleotide sugars) as donor substrates and catalyse their transfer to an acceptor substrate that can either be a lipid‐linked oligosaccharide or an N‐glycan (Rini & Esko, 2017). They represent between 1 and 2% of eukaryotic genomes (Lairson et al., 2008). These actors are localised in different compartments, allowing efficient stepwise and well‐controlled N‐glycan processing. In eukaryotes, the synthesis of nucleotide sugars such as uridine diphosphate‐galactose (UDP‐Gal), guanidine diphosphate‐fucose (GDP‐Fuc), uridine diphosphate‐N‐acetylglucosamine (UDP‐GlcNAc), uridine diphosphate‐glucose (UDP‐Glc) or guanidine diphosphate‐mannose (GDP‐Man) starts in the cytoplasm. By contrast, cytidine monophosphate‐N‐acetylneuraminic acid (CMP‐Neu5Ac) synthesis occurs in the nucleus and uridine diphosphate‐xylose (UDP‐Xyl) in animals is formed in the Golgi apparatus from imported uridine diphosphate‐glucuronic acid (UDP‐GlcA). These nucleotide sugars are used as donor substrates in the different steps of N‐glycan processing (Bar‐Peled & O'Neill, 2011). N‐glycan processing is initiated in the ER with the synthesis of an oligosaccharide intermediate precursor linked to a dolichol (Dol) lipid, commonly known as lipid‐linked oligosaccharide (LLO), of diverse final structure ranging from Glc3Man9GlcNAc2‐P‐P‐Dol (Fig. 1) to a minimal form GlcNAc2‐P‐P‐Dol in protists (Samuelson & Robbins, 2015). LLO synthesis begins at the cytosolic face of the ER and requires the successive action of many different GTs called ALGs (asparagine‐linked glycosylation). These enzymes are embedded in the ER membrane and use activated nucleotide sugars and specific sugar acceptors to elongate the LLO (Aebi, 2013).
Fig. 1.
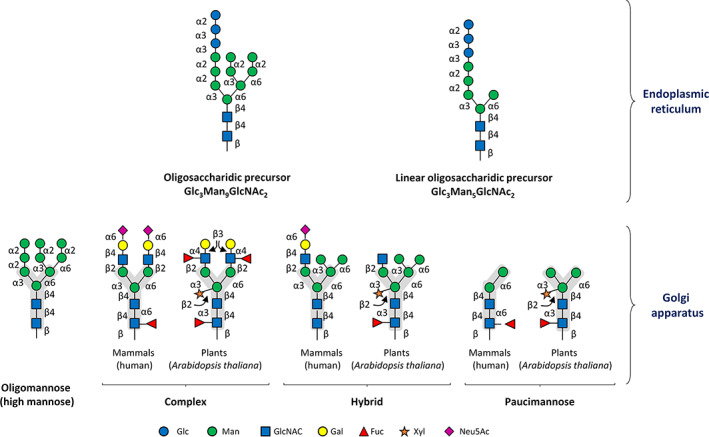
Structures of oligosaccharidic precursors and examples of mature N‐glycans. In the endoplasmic reticulum, the oligosaccharidic precursor is synthesized and then transferred en bloc to the nascent protein. This structure is found in almost all eukaryotes. However, in some protists and microalgae, the oligosaccharidic precursor can be linear and shortened. The oligosaccharide is then processed further in the Golgi apparatus to yield a completely different structure. As exemplified here, the four main types of human and plant N‐glycans found in mature N‐glycoproteins are oligomannose, complex, hybrid and paucimannose. The linkages between each monosaccharide are shown. N‐glycan structures drawn according to the recently updated Symbol Nomenclature for Glycans (Varki, 2017b). Note that all eukaryotic N‐glycans share the common core glycan structure Man2‐3GlcNAc2 (shaded in grey). Fuc, fucose; Gal, galactose; Glc, glucose; GlcNAc, N‐acetylglucosamine; Man, mannose; Neu5Ac, N‐acetylneuraminic acid; Xyl, xylose.
The first step during LLO biosynthesis is catalysed by dolichol‐phosphate GlcNAc phosphotransferase 1 (DPAGT1), also known as ALG7, and results in the transfer of a phospho‐GlcNAc group from UDP‐GlcNAc on the dolichol phosphate (Dol‐P). A GlcNAc and five Man residues then are sequentially added from UDP‐GlcNAc and GDP‐Man by the successive actions of multi‐enzymatic complexes ALG13/14 and the three mannosyltransferases ALG1 (β(1,4)‐mannosyltransferase), ALG2 (α(1,3)/(1,6)‐mannosyltransferase) and ALG11 (α(1,2)‐mannosyltransferase) (Gao, Nishikawa & Dean, 2004) to yield the dolichol pyrophosphate heptasaccharide Man5GlcNAc2‐P‐P‐Dol. At this stage, the LLO undergoes a trans bilayer translocation across the ER membrane, requiring the activity of a flippase (Alaimo et al., 2006; Rush, 2016; Verchère et al., 2021). In humans, deficiency of the proposed flippase RFT1 leads to a severe decrease in N‐glycosylation site occupancy on newly synthesized glycoproteins (Vleugels et al., 2009). After flipping of Man5GlcNAc2‐P‐P‐Dol, synthesis in most eukaryotes is completed on the luminal side of the ER (Breitling & Aebi, 2013) with four mannosylation steps respectively catalysed by ALG3, ALG9, ALG12 and ALG9. Then, three terminal Glc residues are consecutively added by ALG6, ALG8 and ALG10 to achieve the biosynthesis of the oligosaccharide precursor Glc3Man9GlcNAc2‐P‐P‐Dol that will be used as a donor for the N‐glycosylation of specific Asn residues (Fig. 1). Its en bloc transfer from the lipid‐linked precursor to newly synthesized proteins is catalysed by oligosaccharyltransferase (OST) (Silberstein & Gilmore, 1996; Lizak et al., 2011; Breitling & Aebi, 2013). An N‐glycosyl bond is then formed between the anomeric carbon of the proximal‐reducing GlcNAc of the oligosaccharide and the nitrogen atom of the lateral chain of the asparagine belonging to the consensus N‐glycosylation site of the protein (Kowarik et al., 2002). In most organisms, OST is an enzymatic complex composed of several different subunits (Ruiz‐Canada, Kelleher & Gilmore, 2009; Hamieh et al., 2017; Wild et al., 2018; Ramírez, Kowal & Locher, 2019). The oligosaccharide structure linked to the newly synthesized N‐glycoprotein will then be further trimmed in the ER lumen by the action of specific glycosidases: α‐glucosidase I, α‐glucosidase II and eventually an ER‐mannosidase leading to Man8‐9GlcNAc2 oligomannosides. Interaction of glucosylated glycan intermediates with two ER‐resident lectin‐like chaperones, calnexin and calreticulin, ensures ER quality control of N‐glycoproteins (Parodi, Cummings & Aebi, 2015). Once correctly folded, glycoproteins leave the ER to reach the Golgi apparatus.
III. N‐GLYCAN BIOSYNTHESIS IN THE GOLGI APPARATUS LEADS TO A RICH DIVERSITY OF N‐GLYCAN STRUCTURES
Oligosaccharides ranging from Man8‐9GlcNAc2 to the canonical Man5GlcNAc2 are present in most eukaryotes and are derived from the trimming of ER Man8‐9GlcNAc2 by α‐mannosidases. By contrast, a specific Man5GlcNAc2 structure has been recently identified in organisms synthesizing a truncated LLO Glc3Man5GlcNAc2‐P‐P‐Dol in the ER (Fig. 1), as reported for Trichomonas vaginalis, Tetrahymena thermophila, Chlamydomonas reinhardtii and Trypanosoma brucei (Jones et al., 2005; Manthri et al., 2008; Paschinger et al., 2012; Levy‐Ontman et al., 2014; Lucas et al., 2018). This non‐canonical oligomannoside will be called ‘linear’ Man5GlcNAc2 herein since it exhibits a Manα(1,2)Manα(1,2)Man sequence α(1,3)‐linked to the β‐Man residue of the core.
After leaving the ER, oligomannosidic N‐glycans (Fig. 1) are processed further in the Golgi apparatus. However, in contrast to the ER processing steps, Golgi N‐glycan maturation involves a large diversity of glycosidases and GTs giving rise to a diversity of organism‐specific complex, hybrid and paucimannosidic N‐glycans (Fig. 1). This N‐glycan diversity is discussed in the following subsections for Amorphea, Excavates, Archaeplastida and the TSAR supergroup.
(1). Amorphea
Amorphea are members of a taxonomic supergroup that includes fungi and animals and can be divided into two main subgroups: Amoebozoa and Obazoa. The latter includes Opisthokonta that can further be subdivided into two groups: Holomycota (fungi, yeasts) and Holozoa (animals and their closest single‐cell relatives) (Fig. 2).
Fig. 2.
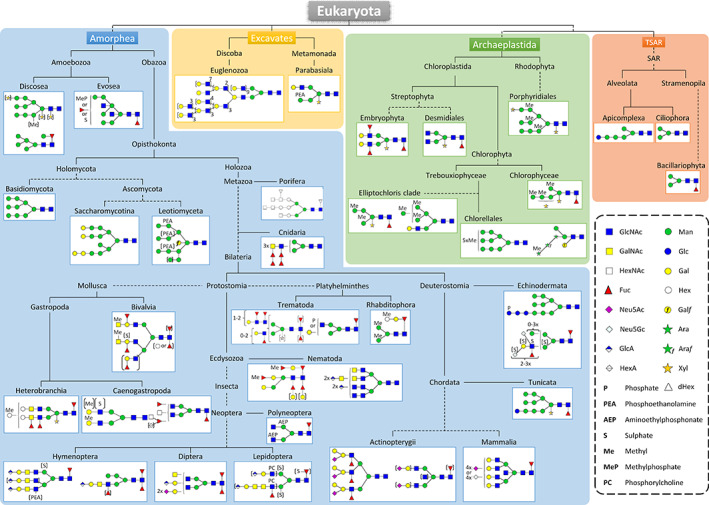
The structural diversity of N‐glycans synthesized in eukaryotes overlaid on the phylogenetic distribution of the different organisms. The most mature N‐glycan structures are shown for each clade. N‐glycan structures drawn according to the recently updated Symbol Nomenclature for Glycans (Varki, 2017b) using the Glycoworkbench tool v2.1. Phylogeny adapted from Burki et al. (2020) and http://lifemap.univ‐lyon1.fr/explore.html. Ara, arabinose; Araƒ, arabinofuranose; dHex, deoxyhexose; Fuc, fucose; Gal, galactose; Galƒ, galactofuranose; GalNAc, N‐acetylgalactosamine; Glc, glucose; GlcA, glucuronic acid; GlcNAc, N‐acetylglucosamine; Hex, hexose; HexA, Hexuronic acid; HexNAc, N‐acetylhexosamine; Man, mannose; Neu5Ac, N‐acetylneuraminic acid; Neu5Gc, N‐glycolylneuraminic acid; Xyl, xylose.
(a). Amoebozoa
To date, protein N‐glycan profiles have been reported for only three amoebozoan species: two human parasites, Acanthamoeba strains (clade Discosea) and Entamoeba histolytica (clade Evosea), and the non‐infectious slime mould Dictyostelium discoideum (clade Evosea). Amoebozoan species synthesize oligomannosides ranging from Man5GlcNAc2 to Man9GlcNAc2 with some additional substitutions (Fig. 2) (Magnelli et al., 2008; Schiller et al., 2009, 2012; Feasley et al., 2010; Feasley, van der Wel & West, 2015; Obregón et al., 2019). In Acanthamoeba strains, the major N‐glycan structures are oligomannosides with 7–10 hexoses (Hex) partially substituted with methyl (Me) and pentose residues for which the position is still ambiguous (Fig. 2) (Schiller et al., 2012). Further analyses have led to the biochemical characterization of a canonical Man5GlcNAc2 N‐glycan substituted by an α(1,6)‐Fuc residue capped with an α‐linked Man (Schiller et al., 2012). Similar substitutions of the core Fuc with a Hex have also been reported in nematodes (Gutternigg et al., 2007b; Yan et al., 2012; Paschinger & Wilson, 2015), cephalopods (Zhang et al., 1997), gastropods (Wuhrer et al., 2004) and platyhelminthes (Paschinger et al., 2011). However, in these organisms, the Fuc residue is capped with a Gal residue instead of a Man residue.
The most abundant N‐glycan found in E. histolytica is Man5GlcNAc2 (Magnelli et al., 2008). N‐glycome analysis of E. histolytica has also highlighted smaller oligomannosides from Man2GlcNAc2 to Man5GlcNAc2 carrying a Gal residue linked to the terminal mannoses. This Gal residue might eventually be substituted by an additional Glc (Magnelli et al., 2008). N‐glycans from D. discoideum are characterized by the presence of additional GlcNAc residues on oligomannoside N‐glycans (Couso et al., 1987; Schiller et al., 2009; Feasley et al., 2010; Nakagawa, Tojo & Fujii, 2011). Additional glycan modifications like fucosylation of the core proximal GlcNAc that bisects the GlcNAc have been reported, as well as substitutions on terminal monosaccharides with Fuc, sulfate or methylphosphate (Fig. 2) (Feasley et al., 2010; Nakagawa et al., 2011; Hykollari et al., 2013, 2017). Concerning core fucosylation, both α(1‐3)‐ and α(1‐6)‐ linkages have been reported (Schiller et al., 2009; Nakagawa et al., 2011).
(b). Obazoa
(i). Holomycota
Holomycota include yeasts and the large kingdom of Fungi. In this group, the main N‐glycan structures found on glycoproteins are oligomannosides (Fig. 2). For the phylum Basidiomycota, the two species Pseudozyma antarctica and Mallassezia furfur synthesize oligomannosides composed of 3–9 Man residues (Flores et al., 2019). Although oligomannosides are also observed in Saccharomycotina species (Gong et al., 2009), substitutions by Gal residues have been reported (Ballou, Ballou & Ball, 1994; Ziegler et al., 1999). Moreover, yeasts often extend their N‐glycan structures to form large polymannosidic structures (de Pourcq, de Schutter & Callewaert, 2010). Concerning Leotiomyceta, especially Penicillium species, similar N‐glycans have been identified with additional modifications such as the attachment of galactofuranose (Galf) residues, phosphoethanolamine and phosphorylcholine (Fig. 2) (Hykollari et al., 2016).
(ii). Holozoa (metazoans)
(A). Porifera
Sponges were long considered members of the Archaeplastida kingdom but are now classified as a branch of Metazoa. N‐glycosylation in Porifera seems not to be as complex as those of Bilateria described below (Fig. 2). The sponge Haliclona caerulea synthesizes protein N‐linked glycans based on a Man7GlcNAc2 structure substituted with N‐acetylhexosamine (HexNAc) and deoxyhexose (dHex) residues leading to a HexNAc7Man7dHex2 structure (Fig. 2). Larger N‐glycans with up to 13 additional Hex residues have also been detected but these still require structural characterization (Carneiro et al., 2013).
(B). Cnidaria
In Cnidaria, investigation of the protein N‐glycan profiles of the freshwater Hydra magnipapillata revealed polyfucosylated LacdiNAc (GalNAcβ(1,4)GlcNAc) structures that play a role in its regenerative process (Sahadevan et al., 2014).
(C). Bilateria
Bilateria is a very large group composed of Protostomia (arthropods, annelids, and molluscs) and Deuterostomia (echinoderms, chordates and hemichordates). The diversity of the N‐glycan structures within this phylum is large, ranging from paucimannosidic to di‐, tri‐ and tetra‐antennary complex N‐glycans (Fig. 2). In Protostomia, protein N‐glycosylation is more sophisticated.
For molluscs, data have been reported mainly for two subgroups: Bivalvia and Gastropoda which includes Heterobranchia and Caenogastropoda. N‐glycan analysis of these clades has revealed a large variety of structures, including oligomannosidic and complex N‐glycans with an immense variety of modifications including core xylose, methylated Hex, Fuc and N‐acetylgalactosamine (GalNAc) substitutions (Fig. 2) (Lommerse et al., 1997; Dolashka‐Angelova et al., 2003; Gutternigg et al., 2007a). The di‐ and tri‐antennary structures can also be mono‐ or di‐sulfated on the Hex residues or O‐methylated on the terminal Gal, Man or GalNAc (Dolashka‐Angelova et al., 2003; Kurz et al., 2013). A core di‐fucosylation has been observed in the bivalve mollusc Crassostrea virginica in addition to a repertoire of sulfated and blood group‐A epitopes (Kurz et al., 2013). Alternatively, the core α(1,3)‐Fuc residue can be replaced by a Hex residue on the proximal GlcNAc. Polyantennary N‐glycans containing 4‐O‐methyl GlcA(1,4)GlcNAc(1,3)Fuc(1,4)GlcNAc have been described in the mollusc Mytilus edulis (Zhou et al., 2013). Monofucosylation on the LacdiNAc motif associated with or without a core xylose has been reported in Biomphalaria glabrata (Lehr et al., 2007). Within Mollusca, xylosylation of the core seems to be specific to the phylum Heterobranchia (Gutternigg et al., 2007a). Other remarkable features are the presence in Rapana venosa of a HexNAc(HexA)Fuc motif linked to a terminal GlcNAc (Sandra et al., 2007). In Volvarina rubella, a carnivorous and scavenging marine gastropod, a complex N‐glycome encompasses a range of oligomannosidic, paucimannosidic, core‐modified and complex N‐glycans. The latter include highly modified N‐glycans bearing N‐methyl‐2‐aminoethylphosphonate, phosphorylcholine, methyl, sulfate, or aminoethylphosphonate substitutions and also a core α(1,6)‐Fuc capped with a Hex and an additional Fuc residue (Fucα(1,2)Galβ(1,4)Fucα(1,6)) (Eckmair et al., 2016).
N‐glycans of Platyhelminthes exhibit less‐sophisticated structures compared to other bilaterians (Fig. 2). They perform core α(1,6)‐fucosylation as well as galactosylation and phosphorylation of the terminal GlcNAc residues as reported for Fasciola hepatica N‐glycans (Ravidà et al., 2016). N‐glycan structures identified on proteins of Dugesia japonica more commonly have a Galβ(1,4)Fuc disaccharide on the proximal GlcNAc residue and an additional methyl Hex linked to this motif (Paschinger et al., 2011). More complex N‐glycan structures (not shown in Fig. 2) have been observed in trematodes, notably the presence of polygalactosylation (up to five Gal residues, which may be methylated) on the core fucose. Methylation was also observed on the Man residues leading to polymethylated glycans in Schmidtea mediterranea (Subramanian et al., 2018). In addition, LacdiNAc bearing three Fuc residues, Lewis x epitope (Fucα(1,3)‐Galβ(1,4)), core xylose or even core difucosylation were identified in Schistosoma species (Khoo et al., 1997; Wuhrer et al., 2006a,b; Jang‐Lee et al., 2007; Mickum et al., 2016).
Nematodes represent 80% of land animals (van den Hoogen et al., 2019). The structural diversity of the N‐glycans found in nematodes is very large, for example approximately 150 different N‐glycan structures have been identified in Dirofilaria immitis (Martini et al., 2019). A large complexity and diversity of N‐glycans exist in the model organism Caenorhabditis elegans, which shares similar N‐glycan features with other nematode species, such as Fuc residues or a Gal‐Fuc disaccharide linked to the core, and attachment of phosphorylcholine which can also be found in Ascaris suum, Oesophagostomum dentatum and Haemonchus contortus (van Die et al., 1999; Cipollo et al., 2005; Pöltl et al., 2007; Butschi et al., 2010; Yan et al., 2012, 2018; Paschinger & Wilson, 2015). However, C. elegans also has species‐specific N‐glycan modifications, such as the presence of a bisecting Gal motif that, to our knowledge, has never been reported in other organisms (Yan et al., 2015). A detailed list of the diversity of N‐glycan structures in C. elegans is provided by Paschinger, Yan & Wilson (2019) with the most processed structure being MeFuc5Gal5Man2GlcNAc2 (Fig. 2). Other structures such as poly‐fucosylated GlcNAc motifs and tetra‐antennary N‐glycans with terminal GlcA have been identified in D. immitis (Martini et al., 2019). Multi‐antennary LacdiNAc structures have also been described in D. immitis and Trichinella spiralis (Fig. 2; Martini et al., 2019; Morelle et al., 2000).
Insects are divided into four main phyla: Polyneoptera, Hymenoptera, Diptera and Lepidoptera (Fig. 2). As reported for nematodes, insects have the ability to attach GlcA to their N‐glycans. This residue has been found in all insect phyla investigated except Polyneoptera for which this substitution has not yet been described (Aoki & Tiemeyer, 2010; Stanton et al., 2017; Hykollari et al., 2018). The polyneopteran Locusta migratoria was the first example of an organism that can synthesise N‐glycans exhibiting an aminoethylphosphonate motif (Hård et al., 1993). This substitution was subsequently observed in glycoproteins from royal jelly and bee venom (Hykollari et al., 2018). LacdiNAc motifs similar to those described above for Heterobranchia also have been reported. Surprisingly, Lepidoptera can synthesise protein N‐glycans with phosphorylcholine substituents (Stanton et al., 2017). Sulfation of Man and Fuc residues of the core N‐glycan also have been observed in Lepidoptera. By contrast, in Hymenoptera, only Man sulfation has been reported (Stanton et al., 2017). Such sulfation of N‐glycans has also been reported in Crustacea, especially in the spiny lobster Panulirus interruptus (Van Kuik et al., 1986, 1987). The presence of sialic acids substitutions in Drosophila melanogaster has also been reported (Aoki & Tiemeyer, 2010).
The super‐phylum Deusterostomia is composed of echinoderms, chordates and hemichordates. Proteins from echinoderms carry oligomannosides up to Man9GlcNAc2 (Şahar & Deveci, 2017). Recently, mass spectrometry analyses demonstrated the presence of more complex N‐glycans in Holothuria atra. Indeed, Vanbeselaere et al. (2020) identified oligomannoside structures (72%), the most sophisticated being an unusual N‐glycan containing a phosphate (P) and three Glc residues (P1Glc3Man9GlcNAc2). Truncated forms containing only seven Man have also been identified. The remaining 28% of N‐glycans are complex and hybrid structures exhibiting core α(1,6)‐Fuc and a LacNAc (Galβ(1,4)GlcNAc) motif. This latter motif can also be fucosylated and substituted by two N‐glycolylneuraminic acids (Neu5Gc), the first being α(2,3)‐linked to the Gal residue and the second attached in α(2,6) to the GlcNAc of the antennae. The N‐glycome of the brittle star Ophiactis savignyi was recently described. In this species, core fucosylation, and various sulfate and Neu5Gc substitutions that can be methylated have been reported. Structures with polyLacNAc motifs or a terminal GlcNAc bearing a phosphate residue are also synthesized in O. savignyi (Eckmair et al., 2020).
Chordates include vertebrates (fish, amphibians, reptiles, birds, and mammals) and tunicates. The tunicate Ciona intestinalis synthesises mainly hybrid N‐glycans containing nine Man residues, one Glc and with core β(1,2)‐Xyl and core α(1,3)‐Fuc residues attached (Yagi et al., 2008). Species from the phylum Actinopterygii (ray‐finned fishes) synthesize very complex poly‐antennary N‐glycans substituted with core α(1,6)‐Fuc, bisecting GlcNAc, LacNAc motif with or without α(2,3)‐linked sialic acid and/or β(1,4)‐Gal and a Fuc on the outer GlcNAc residues (Taguchi et al., 1994; Hanzawa, Suzuki & Natsuka, 2017). In mammals, complex N‐glycans are di‐ to tetra‐antennary structures composed of LacNAc sequences carrying a core α(1,6)‐Fuc. These antennae are terminated by sialic acid residues, either Neu5Ac or Neu5Gc (Figs 1 and 2). Humans do not have Neu5Gc (Brinkman‐Van der Linden et al., 2000; Tangvoranuntakul et al., 2003; Bardor et al., 2005). Some mammalian species, but not humans, are able to synthesize the ‘alpha Gal’ epitope that consists of α(1,3)‐Gal linked to the LacNAc motif (Galili et al., 1988; Galili, 2013).
(2). Excavates
Excavates are diverse single‐cell flagellate organisms ranging from free‐living species such as the freshwater alga Euglena gracilis to human parasites like Trypanosoma brucei (clade Euglenozoa) or Trichomonas vaginalis (clade Metamonada).
(a). Euglenozoa
The freshwater alga E. gracilis is able to synthesise oligomannoside N‐glycans ranging from Man6GlcNAc2 to Man9GlcNAc2 (de la Canal & Parodi, 1985; O'Neill et al., 2017), a small proportion of which are substituted by a putative aminoethylphosphonate moiety (O'Neill et al., 2017). Oligomannosidic N‐glycans have also been observed on the variant surface glycoproteins of the human pathogen T. brucei (Zamze et al., 1990, 1991; Jones et al., 2005; Manthri et al., 2008; Damerow et al., 2016). However, in contrast to E. gracilis, T. brucei synthesizes complex N‐glycans with unusually large poly‐LacNAc‐containing structures (Fig. 2) (Zamze et al., 1991; Mehlert et al., 1998; Atrih et al., 2005; Manthri et al., 2008; Damerow et al., 2016). Such poly‐LacNAc‐containing structures have only been observed on proteins of the bloodstream form but not in the procyclic form of T. brucei (Mehlert et al., 1998; Nolan, Geuskens & Pays, 1999). Although rare, these poly‐LacNAc structures have also been found in cancerous tissue in humans (Ichikawa et al., 1999; Ishida et al., 2005; Holst, Wuhrer & Rombouts, 2015).
(b). Parabasiala
Protein N‐glycosylation in the clade Parabasiala has received little attention to date and is restricted to the sexually transmitted parasite T. vaginalis. The main N‐glycan of T. vaginalis is a linear Man5GlcNAc2 oligomannoside that results from the transfer in the ER of a truncated precursor Glc3Man5GlcNAc2‐P‐P‐Dol on the N‐glycosylation site of the protein (Samuelson et al., 2005; Paschinger et al., 2012; Lombard, 2016). This linear Man5GlcNAc2 oligomannoside is then trimmed into Man4GlcNAc2 and Man3GlcNAc2 that are substituted by a pentose (likely Xyl), phosphoethanolamine or LacNAc motif, thus generating hybrid N‐glycan structures (Fig. 2) (Paschinger et al., 2012).
(3). Archaeplastida
The Archaeplastida are a major group of autotrophic eukaryotes comprising Rhodophyta and Chloroplastida. This latter subgroup is subdivided into organisms of the clade Streptophyta, which includes land plants and desmids, and the Chlorophyta clade containing the green algae.
(a). Chloroplastida
(i). Streptophyta
In addition to oligomannosides, Embryophytes (land plants and mosses) and Desmidiales are able to synthesise complex‐type N‐glycans (Lerouge et al., 1998). To date, studies have mainly been carried out in land plants and mosses, but similar structures have been recently reported in the charophycean green alga Penium margaritaceum belonging to Desmidiales (Ruiz‐May et al., 2018). Complex N‐glycans of streptophytes are characterized by the presence of a core GlcNAc2Man3GlcNAc2 similar to that of mammals and insects that is substituted by a core α(1,3)‐Fuc and a core β(1,2)‐Xyl (Fig. 1). In land plants and mosses, additional modifications of N‐glycans could result in the synthesis of the Lewis a (Lea) epitope that results from the transfer of both an α(1,4)‐Fuc and a β(1,3)‐Gal to terminal GlcNAc residues (Figs 1 and 2). These complex N‐glycan epitopes are highly conserved in land plants (Lerouge et al., 1998) and also in mosses such as Physcomitrella patens (Fitchette et al., 1999; Viëtor et al., 2003; Mega, 2007). Proteins from land plants also carry truncated oligosaccharides consisting of mature N‐glycans lacking terminal GlcNAc residues (Lerouge et al., 1998). Such structures are called paucimannosidic N‐glycans (Fig. 1). These glycans result from the action of hexosaminidases in the secretory system, the extracellular matrix or the storage organelles (e.g. vacuole) (Strasser et al., 2007; Liebminger et al., 2011; Shin et al., 2017).
(ii). Chlorophyta
The clade Chlorophyta includes green algae such as Chlorella and Chlamydomonas species. Although phylogenetically close to Embryophytes, their protein N‐glycan profiles largely differ from those of land plants. Oligomannosides represent the major N‐glycans of proteins isolated from Chlorophytes. Man5GlcNAc2 oligomannoside found in Chlamydomonas reinhardtii exhibits a linear sequence (Vanier et al., 2017). This non‐canonical structure is derived from Glc3Man5GlcNAc2‐P‐P‐Dol occurring in the ER of this green microalga (Fig. 1) (Lucas et al., 2018). In addition, oligomannosides from chlorophytes are partially mono‐ or di‐ O‐methylated and substituted with deoxyhexoses and/or pentoses (Mathieu‐Rivet et al., 2013, 2014; Levy‐Ontman et al., 2014; Schulze et al., 2017; Vanier et al., 2017; Mócsai et al., 2019; Lucas et al., 2020; Oltmanns et al., 2020). In Chlamydomonas reinhardtii, N‐linked glycans are substituted by α(1,3)‐Fuc residues on the proximal GlcNAc and Xyl residues linked to α‐Man and β‐Man residues of Man4GlcNAc2 and Man5GlcNAc2 oligomannosides (Lucas et al., 2020). Two recent studies reporting the protein N‐glycomes of Chlorella species (clade Chlorellales) revealed an unsuspected variety of new N‐glycans composed of short oligomannosides substituted by various Gal, arabinose (Ara) and GlcNAc residues, some of which (Gal and Ara) exhibited either furanose or pyranose forms (Mócsai et al., 2019, 2020a,b). In addition, N‐glycans exhibiting GlcNAc at their non‐reducing end have also been reported in Botryococcus braunii (clade Elliptochloris) (Schulze et al., 2017). The presence of such GlcNAc‐terminated glycans suggests the action of an N‐acetylglucosaminyltransferase I in the N‐glycan processing of these microalgae, a transferase that is absent in C. reinhardtii (Vanier et al., 2017).
(b). Rhodophyta
In rhodophytes, protein N‐glycosylation profiles are mainly from Porphyridium sp. Proteins from this red alga carry O‐methylated oligomannosides substituted with one or two Xyl residues. Of particular interest, one of these two Xyl residues is described to be linked to the second GlcNAc of the core N‐glycan structure (Levy‐Ontman et al., 2011).
(4). TSAR
The TSAR supergroup includes the clades Telonemia, Stramenopila, Alveolata and Rhizaria. To date, data regarding N‐glycosylation have only been published for proteins isolated from alveolates and stramenopiles.
(a). Alveolata
The phylum Alveolata consists of unicellular organisms (protozoans) such as dinoflagellates, ciliates and apicomplexans (protozoan parasites including those responsible for the malaria and toxoplasmosis diseases). In contrast to dinoflagellates for which no information on protein N‐glycosylation is available, several studies have focused on protein N‐glycan structures in ciliates and apicomplexans. Short oligomannosides have been characterized in apicomplexans such as Toxoplasma gondii and Cryptosporidium parvum. For other apicomplexan species, such as the malaria parasite Plasmodium falciparum, only one or two GlcNAc residues are transferred to the asparagine of the N‐glycosylation consensus site (Fauquenoy et al., 2008, 2011; Garénaux et al., 2008; Luk, Johnson & Beckers, 2008; Haserick et al., 2017; Gas‐Pascual et al., 2019). Glucose‐terminated linear Man5GlcNAc2 N‐linked glycan has been identified in apicomplexan species (Taniguchi et al., 1985; Fauquenoy et al., 2008, 2011; Luk et al., 2008; Calow et al., 2016; Haserick et al., 2017). More recently, unexpected structures like Glc2Man6GlcNAc2 and Man6GlcNAc2 have been reported in T. gondii (Gas‐Pascual et al., 2019). These glycans derive from the trimming of a truncated Glc2‐3Man5‐6GlcNAc2‐P‐P‐Dol precursor synthesized in the ER (Taniguchi et al., 1985; Garénaux et al., 2008; Haserick et al., 2017). In ciliates, shorter oligomannosides (Man3GlcNAc2) have been characterized, especially in Tetrahymena thermophila and Tetrahymena pyriformis (Taniguchi et al., 1985; Becker & Rüsing, 2003; Weide et al., 2006; Calow et al., 2016).
(b). Stramenopila
Data on protein N‐glycans in stramenopiles are restricted to Phaeodactylum tricornutum. In contrast to apicomplexan species, this diatom is able to synthesize oligomannoside N‐glycans resulting from complete ER processing of the lipid‐linked precursor up to Glc2Man9GlcNAc2‐P‐P‐Dol (Lucas et al., 2018). After quality control events occurring in the ER, this glycan precursor is then trimmed into oligomannosides ranging from Man9GlcNAc2 to Man5GlcNAc2 N‐glycans (Baïet et al., 2011). These oligomannosides have recently been demonstrated to be identical to those of mammals and plants (Dumontier et al., 2021). With regards to Golgi maturation, expression of an N‐acetylglucosaminyltransferase gene [for α‐1,3‐mannosyl‐glycoprotein 2‐β‐GlcNAc transferase (GnT I)] from P. tricornutum was demonstrated to be able to restore complex N‐glycan maturation in the mammalian Chinese Hamster Ovary (CHO) cell line Lec1 mutant that lacks its endogenous GnT I (Baïet et al., 2011). However, although this diatom expresses a functional GnT I and other molecular actors of N‐glycan processing in the Golgi machinery (Zhang et al., 2019), only small amounts of paucimannosidic N‐glycans bearing a core α(1,3)‐Fuc have been detected in P. tricornutum proteins.
IV. REGULATION OF THE N ‐GLYCOSYLATION PATHWAY
At the evolutionary level, glycosylation provides eukaryotes with a rich combinatorial system, generating an incredible diversity of N‐glycan structures without prior genome modifications (Gagneux, Aebi & Varki, 2015; Varki & Gagneux, 2015). Fig. 2 shows the most common processed N‐glycans for the different eukaryotic clades. However, within a given organism, N‐glycoproteins exist as multiple glycosylation variants depending on their maturation level and number of N‐glycosylation sites. Regulation of the glycoproteome therefore is essential for numerous biological and physiological functions and hence represents an unprecedented level of complexity. An important aspect of this regulation is that there is no gene encoding N‐glycan structures per se, but 2% of the mammalian genome is estimated to encode genes involved in the synthesis of these N‐glycan structures. As noted in Section II, a prodigious enzymatic arsenal secures the biosynthesis of N‐glycan structures (Moremen, Tiemeyer & Nairn, 2012). Expression of these actors is a key parameter in the observed N‐glycan structural diversity, depending on many cellular parameters such as regulation at the transcriptional (transcription factors, epigenetic regulation), post‐transcriptional (microRNA, miRNA) (Nairn et al., 2008; Antony et al., 2014; Dewal et al., 2015; Lau et al., 2015), translational (speed of synthesis) and post‐translational (conformation, modifications, interaction with other proteins) levels (Rabouille et al., 1995; Tu & Banfield, 2010; Struwe & Reinhold, 2012; Gao et al., 2014; Neelamegham & Mahal, 2016; Blackburn et al., 2018; Liu, Doray & Kornfeld, 2018). Interestingly, the glycan structure itself, at least for the N‐glycan processing, governs the abundance of certain glycoproteins. Indeed, for N‐glycoproteins, quality control based on structures of the glycan chains takes place in the ER (Ruddock & Molinari, 2006; Xu & Ng, 2015). This quality control not only monitors N‐glycoprotein folding but also discriminates correctly folded glycoproteins from poorly folded ones destined to be degraded. Folding state can be considered as a key element in governing the N‐glycan structure generated through the Golgi apparatus. The accessibility of the oligosaccharide side chains to GTs and glycosidases in the Golgi apparatus determines whether a specific N‐glycan is processed into a complex‐type N‐glycan or not. This has been illustrated nicely for the well‐known phytohemagglutinin‐L (PHA‐L), the lectin of the common bean Phaseolus vulgaris, which possesses a non‐processed oligomannosidic N‐glycan at the asparagine‐12 and a complex‐type N‐glycan at asparagine‐60 (Sturm & Chrispeels, 1986; Sturm, Bergwerff & Vliegenthart, 1992; Bardor et al., 1999). Besides the notion of accessibility, the N‐glycan structures generated through the Golgi apparatus are also dictated by the strong substrate specificities and sequential action of different Golgi GTs and GHs. This led to the concept of “Go/No Go” pathways proposed by H. Schachter, the best example being the activity of the GnT I considered as a ‘Go’ signal for the activity of the Golgi mannosidase II (Schachter, 1991; Ünligil et al., 2000).
The spatiotemporal organization of the glycosylation actors in the secretory pathway is also crucial (Blackburn, D'Souza & Lupashin, 2019). The molecular mechanisms that govern this differential subcellular localization are far from being identified and understood. Nevertheless, about 10 years ago the importance of intra‐vesicular Golgi trafficking in the regulation of protein glycosylation was highlighted in human patients (Wu et al., 2004; Foulquier et al., 2006, 2007; D'Souza, Taher & Lupashin, 2020). Many mutations affecting seven of the eight constituent subunits of the conserved oligomeric Golgi (COG) complex (COG3 being the exception) were subsequently identified in patients with congenital disorders of glycosylation (CDG), a rare metabolic disease affecting glycosylation (Reynders et al., 2011; Péanne et al., 2018). The COG complex has been studied in many different organisms (Saccharomyces cerevisiae, A. thaliana, D. melanogaster, C. elegans) and in most studied organisms, COG defects were shown to be associated with Golgi morphological disruption, protein glycosylation and vesicular trafficking defects (Blackburn et al., 2019). The mechanisms by which glycosylation defects are generated have been the subject of numerous studies and have been partially elucidated. COG defects disturb the Golgi structure and the retrograde intra‐Golgi trafficking of vesicles containing important proteins for the glycosylation process, such as GTs and/or nucleotide‐sugar transporters (Reynders et al., 2009, 2011). Instability of some GTs, resulting from inadequate lysosomal targeting, in particular GlcNAc transferases I and II) (MGAT), α‐mannosidase class 2A member 1 (Man2A1), β‐1,4‐galactosyltransferase 1 (β4GalT1) and β‐galactoside α‐2,6‐sialyltransferase 1 (ST6Gal1) has been reported in COG‐depleted cells (Foulquier et al., 2006; Reynders et al., 2009; Pokrovskaya et al., 2011). Altogether, these processes account for the observed microheterogeneity of the N‐glycan structures on a specific glycoprotein. In addition, regulation of the Golgi N‐glycan process through Mn2+ and Ca2+ homeostasis has been investigated very recently. A specific discussion of this recent and original regulation is provided below.
(1). Overview of the different GTs and their Mn2+ dependency within N ‐glycan processing
Human Golgi GTs can be classified into three main groups according to their structural folds: GT‐A, GT‐B and GT‐C (Oriol et al., 2002; Lairson et al., 2008; Breton, Fournel‐Gigleux & Palcic, 2012; Gloster, 2014; Albuquerque‐Wendt et al., 2019). GT‐A and GT‐B share a close topology and use nucleotide sugars as donor substrates. Structurally, GT‐A possess two abutting secondary structures called Rossmann folds (alternative β‐sheets and α‐helices, β/α/β). By contrast, in GT‐B, the two β/α/β Rossmann domains face each other and are flexibly linked. One of the main differences between GT‐A and GT‐B is the presence of a highly conserved DXD motif (aspartic acid–any amino acid–aspartic acid) in the active site of GT‐A that is absent in GT‐B. This motif is crucial as it allows the carboxylates to coordinate a divalent cation and/or a ribose required for the stabilization of the nucleotide sugar. Note that for a specific GT, GnT I, this motif is comprised of three specific amino acids: glutamate, aspartic acid and aspartic acid (EDD) (Breton et al., 2012). GT‐As are thus considered metal‐ion‐dependent enzymes while GT‐Bs are generally metal‐ion‐independent. More recently, GT‐Cs have been identified as a new structural family of GTs possessing several hydrophobic transmembrane domains and using lipid phosphate‐linked sugar as the donor substrate (Oriol et al., 2002; Gloster, 2014; Albuquerque‐Wendt et al., 2019). These specific GTs act in the ER during the so‐called dolichol cycle. Like GT‐As, GT‐Cs possess a DXD or even a DD motif that is crucial for their enzymatic activity (Lommel et al., 2011; Albuquerque‐Wendt et al., 2019). While magnesium (Mg2+), zinc (Zn2+) or cobalt (Co2+) ions can be used as a cofactor by GT‐A enzymes, Mn2+ is considered the most prominent. This has been investigated by following the kinetics of catalytic activity of β4GalT1 (EC 2.4.1.38), a key enzyme of N‐glycan processing that catalyses, only in the presence of Mn2+, the transfer of Gal from UDP‐Gal to the non‐reducing end GlcNAc of complex N‐glycans (Ramakrishnan, Ramasamy & Qasba, 2006). β4GalT1 is a GT‐A folded enzyme containing a D248XD250 motif located in the cleft of its catalytic domain, which serves as a Mn2+ binding site that then allows the binding of UDP‐Gal (Arnold et al., 2000; Ramakrishnan et al., 2006). Fig. 3 summarizes the known and putative Mn2+‐dependent glycoenzymes that are involved in the N‐glycan‐processing pathway in mammals (human) and plants (Arabidopsis thaliana). The affinity of these enzymes for Mn2+ is not yet known and it could well be that according to their subcellular localization, a tight regulation of local Mn2+ homeostasis is required to sustain the corresponding glycosylation reactions. These Mn2+‐dependent enzymes are Golgi localized, highlighting the importance of Golgi Mn2+ homeostatic regulation in N‐glycan processing. Although surprising, the molecular mechanisms of Golgi Mn2+ homeostasis were unknown until recently.
Fig. 3.
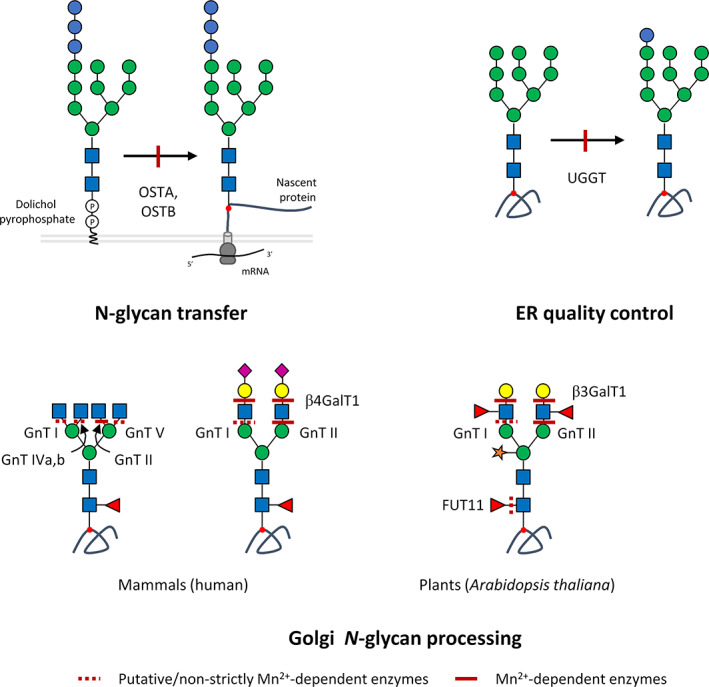
Mn2+‐dependent glycoenzymes of N‐glycosylation processing. During the biosynthesis of N‐linked glycans, the oligosaccharyltransferase (OST) complex ensures the transfer of the oligosaccharide precursor to the nascent protein. The catalytic activity of this complex is driven by subunits STT3A for co‐translational glycosylation and STT3B for co‐ and post‐translational glycosylation. Both subunits act as glycosyltransferases (GTs) and are Mn2+ dependent. Afterwards, during endoplasmic reticulum (ER) quality control of N‐glycoproteins, UGGT activity also requires Mn2+ ions as well as GnT I, GnT II, β4GalT1, GnTIVa,b and GnTVb involved in Golgi N‐glycan processing in mammals and GnT I, Gn T II, FUT11 and β3GalT1 in Arabidopsis thaliana. Symbol nomenclature of monosaccharides follows Varki (2017b), see Fig. 1 for key. The red dot indicates the asparagine residue from the Asn‐X‐Ser/Thr/Cys consensus sequence. β3GalT1, β(1,3)‐galactosyltransferase 1; β4GalT1, β(1,4)‐galactosyltransferase 1; FUT11, α(1,3)‐fucosyltransferase; GnT I, mannosyl α(1,3)‐glycoprotein β(1,2)‐N‐acetylglucosamine transferase; GnT II, mannosyl β‐(1,6)‐glycoprotein β(1,2)‐N‐acetylglucosamine transferase; GnT IVa,b, mannosyl α(1,3)‐glycoprotein β(1,4)‐N‐acetylglucosamine transferase 4a and 4b; GnT V, mannosyl α(1,6)‐glycoprotein β(1,6)‐N‐acetylglucosamine transferase 5; mRNA, messenger RNA; UGGT, UDP‐glucose glycoprotein glucosyltransferase 1.
(2). TMEM165, an essential protein in Golgi N ‐glycan processing and Mn2+ homeostasis
The discovery of TMEM165 as a gene related to human CDG (Houdou & Foulquier, 2020) began a new era in the understanding of the regulation of Golgi Mn2+ homeostasis (Foulquier et al., 2012). An understanding of the crucial role of TMEM165 in Golgi glycosylation maintenance arose unambiguously from the identification of CDG patients with mutations in TMEM165. All TMEM165‐CDG patients were diagnosed as type II CDG based on serum‐transferrin isoelectric focusing. In addition, mass spectrometry analysis of total N‐glycans from patients’ sera highlighted strong Golgi glycosylation defects with an increased level of abnormal N‐glycan structures lacking both Gal and sialic acid residues (Foulquier et al., 2012). This was confirmed in TMEM165 knock‐out cell lines (HEK cells) (Morelle et al., 2017) where synthesised N‐glycans provided evidence for a strong galactosylation defect, a mild GlcNAcylation defect and a slight sialylation defect. It is now clear that TMEM165 deficiency not only affects N‐glycan processing but also all other Golgi Mn2+‐dependent glycosylation pathways such as glycosaminoglycans and glycolipid synthesis (Bammens et al., 2015; Morelle et al., 2017; Haouari et al., 2020). TMEM165 was also recently found to be involved in lactose biosynthesis during milk production (Snyder et al., 2019).
The glycosylation defect is not restricted to TMEM165 but can be extended to all investigated uncharacterized protein family 0016 (UPF0016) members, a conserved family of transporter to which TMEM165 belongs (Demaegd et al., 2014; Thines, Stribny & Morsomme, 2020). One of the best examples is Gdt1p, the yeast ortholog of TMEM165. The phenotype of the yeast null mutant gdt1Δ is a strong growth defect when cultured in the presence of high external calcium chloride concentration that can be correlated to strong N‐ and O‐glycosylation abnormalities (Demaegd et al., 2013). In addition to yeast Gdt1p, Yang et al. (2021) recently characterized the function of PML3 (photosynthesis‐affected mutant 71 like 3; At5g36290), one of the five UPF0016 family members found in Arabidopsis thaliana, and demonstrated that PML3 played a critical role in Golgi glycosylation and cell wall biosynthesis under Mn2+‐deficient conditions. They also demonstrated interestingly a lower abundance of core α(1,3)‐Fuc and β(1,2)‐Xyl on the N‐glycan structures isolated from protein extracts of pml3 mutants. This defect is completely suppressed when the plant mutants are grown under Mn2+‐supplemented hydroponic nutrient solution (Yang et al., 2021).
(3). TMEM165 structure and functional domains
TMEM165 belongs to the UPF0016 family, a highly conserved family of membrane proteins. A phylogenetic analysis of TMEM165 homologous sequences among different eukaryotic clades shows strong sequence homologies (Fig. 4). The sequence alignments reveal the presence of two copies of the hydrophobic domain E‐Ø‐G‐D‐(KR)‐(ST) (where Ø indicates any hydrophobic residue) oriented in an antiparallel manner and highly conserved throughout species independently of the synthesis of complex‐type N‐glycans. This specific orientation could result from an ancient gene‐duplication event as proposed by Demaegd et al. (2014) based on bacterial UPF0016 members. Many UPF0016 homologous sequences can be found in databases but only a few have been characterized, including the eukaryotic TMEM165 (human, mouse, zebrafish), Gdt1p (Saccharomyces cerevisiae) (Foulquier et al., 2012; Demaegd et al., 2013, 2014; Reinhardt, Lippolis & Sacco, 2014; Bammens et al., 2015; Snyder et al., 2019) and PAM71 (photosynthesis‐affected mutant 71), CMT1 (chloroplast manganese transporter 1) and PML3–5 (plant A. thaliana) (Schneider et al., 2016; Hoecker, Leister & Schneider, 2017; Eisenhut et al., 2018; Hoecker et al., 2020). During the last decade, many studies have attempted to unravel the exact functions of these different members (Demaegd et al., 2013; Reinhardt et al., 2014; Bammens et al., 2015; Colinet et al., 2016; Potelle et al., 2017; Snyder et al., 2017, 2019; Thines et al., 2018; Stribny et al., 2020; Wang et al., 2020). So far, all these studies agree upon a function as a cation transporter required to maintain Ca2+, Mn2+ and H+ homeostasis in the Golgi apparatus. Site‐directed mutagenesis within the two consensus motifs E‐Ø‐G‐D‐(KR)‐(ST) demonstrated the crucial involvement of amino acid residues in these consensus motifs for Ca2+ and Mn2+ transport activities. The resulting altered transport function could be due to impaired cation affinity or pocket conformation changes, emphasizing that the amino acids of the two conserved motifs constitute the cation binding sites of the UPF0016 members.
Fig. 4.
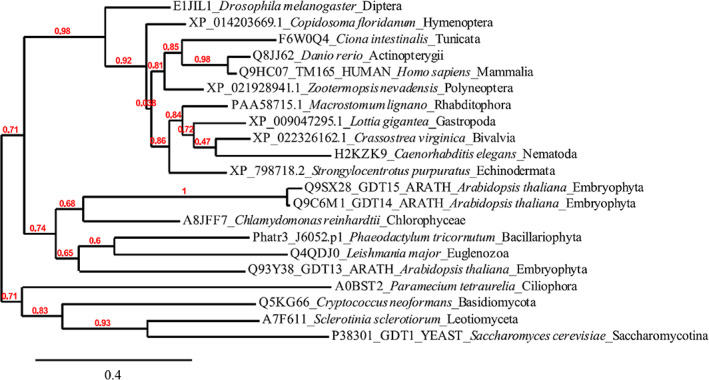
Phylogenetic tree showing a selection of transmembrane protein 165 (TMEM165) homologs in Eukaryotes. Putative TMEM165 protein sequences from organisms belonging to the clades depicted in Fig. 2 were retrieved in the subfamily PTHR12608:SF1 (TRANSMEMBRANE PROTEIN 165) of the PANTHER database version 16 (Mi et al., 2021; http://www.pantherdb.org/). A basic local alignment tool for proteins (BlastP) search with the human TMEM165 protein sequence (accession number Q9HC07) was also carried using the non‐redundant protein sequences (https://blast.ncbi.nlm.nih.gov/) or in the EnsemblProtists database (Phaeodactylum tricornutum), to identify additional similar sequences. Phylogeny created using the Phylogeny.fr platform (Dereeper et al., 2008; http://www.phylogeny.fr/index.cgi) with the selected 21 sequences. The ‘One Click’ mode was chosen to perform the analysis with the following steps: the protein sequences were aligned using MUSCLE 3.8.31 (Edgar, 2004), a curation step by Gblocks 0.91b (Castresana, 2000) was included and the phylogenetic tree was subsequently built using the PhyML 3.1/3.0 aLRT program (Guindon & Gascuel, 2003; Anisimova & Gascuel, 2006). Graphical representation and editing of the phylogenetic tree were done with TreeDyn 198.3 (Chevenet et al., 2006). The red numbers show the percentage of replicate trees in which the associated taxa clustered together, thus reflecting evolutionary distances based on sequence homologies.
(4). TMEM165 homologous sequences in eukaryotic clades and future research
For TMEM165 homologous sequences, the two consensus domains E‐Ø‐G‐D‐(KR)‐(ST) are highly conserved across the different eukaryotic clades (Fig. 4) although an extensive diversity of N‐glycan structures can be found. Some species clearly show evidence for Golgi glycosylation maturation while some only possess oligomannosidic‐type N‐glycans resulting from ER glycosylation. As shown for A. thaliana, TMEM165 orthologs are required for proper maturation of N‐glycan structures (Yang et al., 2021). The function of TMEM165 in the eukaryotic clades synthesizing only oligomannosidic‐type N‐glycans can be questioned. We noted from our phylogenetic analysis that the highest variability and heterogeneity of TMEM165 proteins is found in the variable length of the N‐terminal region which does not show any obvious conservation. According to the species, the N‐terminal extension of the eukaryotic UPF0016 members may thus have a role in targeting to reach their proper final destination. The observed diversity and variability in the N‐terminal part of the eukaryotic UPF0016 members could then explain their different subcellular localizations. Specific targeting systems have certainly evolved to ensure the proper localization of proteins and the inherent complexity of the N‐terminal region found in TMEM165 homologous sequences may coincide with the evolution of protein translocation systems used for membrane insertion and targeting. In terms of function, differential subcellular localization could allow the transport of Mn2+/Ca2+ from the cytoplasm to specific luminal compartments such as the ER/Golgi in order to enable specific enzymatic reactions such as glycosylation and/or to protect the cytoplasm from Mn2+excess and toxicity. Further studies are required to understand better the contribution of TMEM165 homologous sequences throughout the different eukaryotic clades and its possible involvement in regulation of N‐glycosylation.
V. CONCLUSIONS
The extensive diversity observed for most mature N‐glycan structures, summarized in Fig. 2, indicates that N‐glycan processing steps in the Golgi apparatus involve a large repertoire of organism‐specific glycosidases and GTs, as well as other enzymes required for specific sugar substitutions (e.g. methyl, sulfate, methylphosphate, phosphoethanolamine, phosphorylcholine, etc.).
The initial investigations of N‐glycosylation have mainly focused on mammalian proteins, attempting to unravel this essential protein PTM and to understand the functions of mature N‐glycans in human physiology and pathology. More recently, N‐glycosylation of proteins from other eukaryotes has been investigated. The emergence of sensitive and efficient analytical technologies for the structural identification of mixture of complex oligosaccharides have allowed in‐depth study of N‐glycan profiles from numerous organisms, even for low amounts of available proteins, highlighting the rich structural diversity of mature N‐glycans among eukaryotes.
The functions of such N‐glycan diversity of secreted proteins in eukaryotes are diverse. To date, numerous studies carried out on the functions of protein N‐glycosylation in mammals have demonstrated their importance in cell–cell interactions, cell–protein recognition and interaction, and in human pathology (Varki, 2017a). The diversity of mature N‐glycans in other eukaryotes suggests that N‐glycosylation of proteins in all of these organisms does not solely function to ensure their proper folding in the ER. Indeed, considering the energy cost for each step of the N‐glycan pathway, we can assume that the N‐glycans resulting from Golgi maturation play important biological functions in organisms belonging to the different clades.
Beyond the elucidation of the biological relevance of such N‐glycan diversity, the regulation of the Golgi processing steps is a key issue. Indeed, such regulation is multifactorial. Moreover, recent studies have demonstrated that in some organisms, complex N‐glycan processing steps are regulated through Golgi Mn2+ homeostasis, including molecular actors like TMEM165. More research is needed to understand the regulation of the N‐glycan processing steps in other eukaryotes and to establish whether regulation of N‐glycosylation through Mn2+ homeostasis is universal.
In this context, future studies should attempt to decipher the roles of TMEM165 in the regulation of N‐glycan processing in organisms that synthesise oligomannosides or mature N‐glycans.
VI. ACKNOWLEDGEMENTS
F.F. and M.B. are grateful to the CNRS for supporting the collaboration between the University of Lille, CNRS UMR UGSF8576, and the University of Rouen Normandie, Glyco‐MEV lab. This work was funded by the European Union's Horizon 2020 Research and Innovation Programme under Grant Agreement 774078 (Pharma‐Factory). The PhD work of M.‐L.W.‐B. was funded by the European Union's Horizon 2020 Research and Innovation Programme under Grant Agreement 774078 (Pharma‐Factory). The PhD work of C.T. was funded by the Region Normandie and the Carnot I2C. The funding bodies were not involved in the design of the study, collection and interpretation of data nor in the writing of the manuscript.
Contributor Information
François Foulquier, Email: francois.foulquier@univ-lille.fr.
Muriel Bardor, Email: muriel.bardor@univ-rouen.fr.
REFERENCES
- Aebi, M. (2013). N‐linked protein glycosylation in the ER. Biochimica et Biophysica Acta 1833, 2430–2437. [DOI] [PubMed] [Google Scholar]
- Alaimo, C. , Catrein, I. , Morf, L. , Marolda, C. L. , Callewaert, N. , Valvano, M. A. , Feldman, M. F. & Aebi, M. (2006). Two distinct but interchangeable mechanisms for flipping of lipid‐linked oligosaccharides. The EMBO Journal 25, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque‐Wendt, A. , Hütte, H. J. , Buettner, F. F. R. , Routier, F. H. & Bakker, H. (2019). Membrane topological model of glycosyltransferases of the GT‐C superfamily. International Journal of Molecular Sciences 20, 4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova, M. & Gascuel, O. (2006). Approximate likelihood‐ratio test for branches: A fast, accurate, and powerful alternative. Systematic Biology 55, 539–552. [DOI] [PubMed] [Google Scholar]
- Antony, P. , Rose, M. , Heidenreich, A. , Knüchel, R. , Gaisa, N. T. & Dahl, E. (2014). Epigenetic inactivation of ST6GAL1 in human bladder cancer. BMC Cancer 14, 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki, K. & Tiemeyer, M. (2010). The glycomics of glycan glucuronylation in Drosophila melanogaster . Methods in Enzymology 480, 297–321. [DOI] [PubMed] [Google Scholar]
- Arnold, S. M. , Fessler, L. I. , Fessler, J. H. & Kaufman, R. J. (2000). Two homologues encoding human UDP‐glucose: glycoprotein glucosyltransferase differ in mRNA expression and enzymatic activity. Biochemistry 39, 2149–2163. [DOI] [PubMed] [Google Scholar]
- Atrih, A. , Richardson, J. M. , Prescott, A. R. & Ferguson, M. A. J. (2005). Trypanosoma brucei glycoproteins contain novel giant poly‐N‐acetyllactosamine carbohydrate chains. The Journal of Biological Chemistry 280, 865–871. [DOI] [PubMed] [Google Scholar]
- Baïet, B. , Burel, C. , Saint‐Jean, B. , Louvet, R. , Menu‐Bouaouiche, L. , Kiefer‐Meyer, M.‐C. , Mathieu‐Rivet, E. , Lefebvre, T. , Castel, H. , Carlier, A. , Cadoret, J.‐P. , Lerouge, P. & Bardor, M. (2011). N‐Glycans of Phaeodactylum tricornutum diatom and functional characterization of its N‐acetylglucosaminyltransferase I enzyme. The Journal of Biological Chemistry 286, 6152–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou, C. E. , Ballou, L. & Ball, G. (1994). Schizosaccharomyces pombe glycosylation mutant with altered cell surface properties. Proceedings of the National Academy of Sciences of the United States of America 91, 9327–9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammens, R. , Mehta, N. , Race, V. , Foulquier, F. , Jaeken, J. , Tiemeyer, M. , Steet, R. , Matthijs, G. & Flanagan‐Steet, H. (2015). Abnormal cartilage development and altered N‐glycosylation in TMEM165‐deficient zebrafish mirrors the phenotypes associated with TMEM165‐CDG. Glycobiology 25, 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardor, M. , Loutelier‐Bourhis, C. , Marvin, L. , Cabanes‐Macheteau, M. , Lange, C. , Lerouge, P. & Faye, L. (1999). Analysis of plant glycoproteins by matrix‐assisted laser desorption ionisation mass spectrometry: application to the N‐glycosylation analysis of bean phytohemagglutinin. Plant Physiology and Biochemistry 37, 319–325. [Google Scholar]
- Bardor, M. , Nguyen, D. H. , Diaz, S. & Varki, A. (2005). Mechanism of uptake and incorporation of the non‐human sialic acid N‐glycolylneuraminic acid into human cells. The Journal of Biological Chemistry 280, 4228–4237. [DOI] [PubMed] [Google Scholar]
- Bar‐Peled, M. & O'Neill, M. A. (2011). Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annual Review of Plant Biology 62, 127–155. [DOI] [PubMed] [Google Scholar]
- Becker, B. & Rüsing, M. (2003). Structure of N‐glycosidic carbohydrates of secretory proteins of Tetrahymena thermophila . The Journal of Eukaryotic Microbiology 50, 235–239. [DOI] [PubMed] [Google Scholar]
- Blackburn, J. B. , D'Souza, Z. & Lupashin, V. V. (2019). Maintaining order: COG complex controls Golgi trafficking, processing, and sorting. FEBS Letters 593, 2466–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn, J. B. , Kudlyk, T. , Pokrovskaya, I. & Lupashin, V. V. (2018). More than just sugars: conserved oligomeric Golgi complex deficiency causes glycosylation‐independent cellular defects. Traffic 19, 463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling, J. & Aebi, M. (2013). N‐linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harbor Perspectives in Biology 5, a013359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton, C. , Fournel‐Gigleux, S. & Palcic, M. M. (2012). Recent structures, evolution and mechanisms of glycosyltransferases. Current Opinion in Structural Biology 22, 540–549. [DOI] [PubMed] [Google Scholar]
- Brinkman‐Van der Linden, E. C. , Sjoberg, E. R. , Juneja, L. R. , Crocker, P. R. , Varki, N. & Varki, A. (2000). Loss of N‐glycolylneuraminic acid in human evolution: implications for sialic acid recognition by siglecs. The Journal of Biological Chemistry 275, 8633–8640. [DOI] [PubMed] [Google Scholar]
- Burki, F. , Roger, A. J. , Brown, M. W. & Simpson, A. G. B. (2020). The new tree of eukaryotes. Trends in Ecology & Evolution 35, 43–55. [DOI] [PubMed] [Google Scholar]
- Butschi, A. , Titz, A. , Wälti, M. A. , Olieric, V. , Paschinger, K. , Nöbauer, K. , Guo, X. , Seeberger, P. H. , Wilson, I. B. H. , Aebi, M. , Hengartner, M. O. & Künzler, M. (2010). Caenorhabditis elegans N‐glycan core β‐galactoside confers sensitivity towards nematotoxic fungal galectin CGL2. PLOS Pathogens 6, e1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calow, J. , Behrens, A.‐J. , Mader, S. , Bockau, U. , Struwe, W. B. , Harvey, D. J. , Cormann, K. U. , Nowaczyk, M. M. , Loser, K. , Schinor, D. , Hartmann, M. W. W. & Crispin, M. (2016). Antibody production using a ciliate generates unusual antibody glycoforms displaying enhanced cell‐killing activity. mAbs 8, 1498–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro, R. F. , de Melo, A. A. , de Almeida, A. S. , da Moura, R. M. , Chaves, R. P. , de Sousa, B. L. , do Nascimento, K. S. , Sampaio, S. S. , Lima, J. P. M. S. , Cavada, B. S. , Nagano, C. S. & Sampaio, A. H. (2013). H‐3, a new lectin from the marine sponge Haliclona caerulea: purification and mass spectrometric characterization. The International Journal of Biochemistry & Cell Biology 45, 2864–2873. [DOI] [PubMed] [Google Scholar]
- Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 17, 540–552. [DOI] [PubMed] [Google Scholar]
- Chevenet, F. , Brun, C. , Bañuls, A.‐L. , Jacq, B. & Christen, R. (2006). TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollo, J. F. , Awad, A. M. , Costello, C. E. & Hirschberg, C. B. (2005). N‐glycans of Caenorhabditis elegans are specific to developmental stages. Journal of Biological Chemistry 280, 26063–26072. [DOI] [PubMed] [Google Scholar]
- Colinet, A.‐S. , Sengottaiyan, P. , Deschamps, A. , Colsoul, M.‐L. , Thines, L. , Demaegd, D. , Duchêne, M.‐C. , Foulquier, F. , Hols, P. & Morsomme, P. (2016). Yeast Gdt1 is a Golgi‐localized calcium transporter required for stress‐induced calcium signaling and protein glycosylation. Scientific Reports 6, 24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley, K. J. , Varki, A. & Kinoshita, T. (2015). Cellular organization of glycosylation. In Essentials of Glycobiology (eds Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L. and Seeberger P. H.), pp. 41–49, Third Edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor. [PubMed] [Google Scholar]
- Couso, R. , van Halbeek, H. , Reinhold, V. & Kornfeld, S. (1987). The high mannose oligosaccharides of Dictyostelium discoideum glycoproteins contain a novel intersecting N‐acetylglucosamine residue. Journal of Biological Chemistry 262, 4521–4527. [PubMed] [Google Scholar]
- Damerow, M. , Graalfs, F. , Güther, M. L. S. , Mehlert, A. , Izquierdo, L. & Ferguson, M. A. J. (2016). A gene of the β3‐glycosyltransferase family encodes N‐acetylglucosaminyltransferase II function in Trypanosoma brucei . Journal of Biological Chemistry 291, 13834–13845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Canal, L. & Parodi, A. J. (1985). Glycosylation of proteins in the protozoan Euglena gracilis . Comparative Biochemistry and Physiology Part B: Comparative Biochemistry 81, 803–805. [Google Scholar]
- de Pourcq, K. , de Schutter, K. & Callewaert, N. (2010). Engineering of glycosylation in yeast and other fungi: current state and perspectives. Applied Microbiology and Biotechnology 87, 1617–1631. [DOI] [PubMed] [Google Scholar]
- Demaegd, D. , Colinet, A.‐S. , Deschamps, A. & Morsomme, P. (2014). Molecular evolution of a novel family of putative calcium transporters. PLoS One 9, e100851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaegd, D. , Foulquier, F. , Colinet, A.‐S. , Gremillon, L. , Legrand, D. , Mariot, P. , Peiter, E. , van Schaftingen, E. , Matthijs, G. & Morsomme, P. (2013). Newly characterized Golgi‐localized family of proteins is involved in calcium and pH homeostasis in yeast and human cells. Proceedings of the National Academy of Sciences of the United States of America 110, 6859–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper, A. , Guignon, V. , Blanc, G. , Audic, S. , Buffet, S. , Chevenet, F. , Dufayard, J.‐F. , Guindon, S. , Lefort, V. , Lescot, M. , Claverie, J.‐M. & Gascuel, O. (2008). Phylogeny.fr: robust phylogenetic analysis for the non‐specialist. Nucleic Acids Research 36, W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewal, M. B. , DiChiara, A. S. , Antonopoulos, A. , Taylor, R. J. , Harmon, C. J. , Haslam, S. M. , Dell, A. & Shoulders, M. D. (2015). XBP1s links the unfolded protein response to the molecular architecture of mature N‐glycans. Chemistry & Biology 22, 1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolashka‐Angelova, P. , Beck, A. , Dolashki, A. , Beltramini, M. , Stevanovic, S. , Salvato, B. & Voelter, W. (2003). Characterization of the carbohydrate moieties of the functional unit RvH1‐a of Rapana venosa haemocyanin using HPLC/electrospray ionization MS and glycosidase digestion. Biochemical Journal 374, 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza, Z. , Taher, F. S. & Lupashin, V. V. (2020). Golgi inCOGnito: from vesicle tethering to human disease. Biochimica Et Biophysica Acta. General Subjects 1864, 129694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumontier, R. , Loutelier‐Bourhis, C. , Walet‐Balieu, M.‐L. , Burel, C. , Mareck, A. , Afonso, C. , Lerouge, P. & Bardor, M. (2021). Identification of N‐glycan oligomannoside isomers in the diatom Phaeodactylum tricornutum . Carbohydrate Polymers 259, 117660. [DOI] [PubMed] [Google Scholar]
- Eckmair, B. , Jin, C. , Abed‐Navandi, D. & Paschinger, K. (2016). Multistep fractionation and mass spectrometry reveal zwitterionic and anionic modifications of the N‐ and O‐glycans of a marine snail. Molecular & cellular proteomics: MCP 15, 573–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmair, B. , Jin, C. , Karlsson, N. G. , Abed‐Navandi, D. , Wilson, I. B. H. & Paschinger, K. (2020). Glycosylation at an evolutionary nexus: the brittle star Ophiactis savignyi expresses both vertebrate and invertebrate N‐glycomic features. The Journal of Biological Chemistry 295, 3173–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut, M. , Hoecker, N. , Schmidt, S. B. , Basgaran, R. M. , Flachbart, S. , Jahns, P. , Eser, T. , Geimer, S. , Husted, S. , Weber, A. P. M. , Leister, D. & Schneider, A. (2018). The Plastid Envelope chloroplast manganese transporter 1is essential for manganese homeostasis in Arabidopsis . Molecular Plant 11, 955–969. [DOI] [PubMed] [Google Scholar]
- Fauquenoy, S. , Hovasse, A. , Sloves, P.‐J. , Morelle, W. , Dilezitoko Alayi, T. , Dilezitoko Ayali, T. , Slomianny, C. , Werkmeister, E. , Schaeffer, C. , van Dorsselaer, A. & Tomavo, S. (2011). Unusual N‐glycan structures required for trafficking Toxoplasma gondii GAP50 to the inner membrane complex regulate host cell entry through parasite motility. Molecular & cellular proteomics: MCP 10, M111.008953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquenoy, S. , Morelle, W. , Hovasse, A. , Bednarczyk, A. , Slomianny, C. , Schaeffer, C. , Dorsselaer, A. V. & Tomavo, S. (2008). Proteomics and glycomics analyses of N‐glycosylated structures involved in Toxoplasma gondii‐host cell interactions. Molecular & Cellular Proteomics 7, 891–910. [DOI] [PubMed] [Google Scholar]
- Feasley, C. L. , Johnson, J. M. , West, C. M. & Chia, C. P. (2010). Glycopeptidome of a heavily N‐glycosylated cell surface glycoprotein of Dictyostelium implicated in cell adhesion. Journal of Proteome Research 9, 3495–3510. [DOI] [PubMed] [Google Scholar]
- Feasley, C. L. , van der Wel, H. & West, C. M. (2015). Evolutionary diversity of social amoebae N‐glycomes may support interspecific autonomy. Glycoconjugate Journal 32, 345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitchette, A.‐C. , Cabanes‐Macheteau, M. , Marvin, L. , Martin, B. , Satiat‐Jeunemaitre, B. , Gomord, V. , Crooks, K. , Lerouge, P. , Faye, L. & Hawes, C. (1999). Biosynthesis and immunolocalization of Lewis a‐containing N‐glycans in the plant cell. Plant Physiology 121, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, R. J. D. , Ohashi, T. , Sakai, K. , Gonoi, T. , Kawasaki, H. & Fujiyama, K. (2019). The neutral N‐linked glycans of the Basidiomycetous yeasts Pseudozyma antarctica and Malassezia furfur (Subphylum Ustilaginomycotina). The Journal of General and Applied Microbiology 65, 53–63. [DOI] [PubMed] [Google Scholar]
- Foulquier, F. , Amyere, M. , Jaeken, J. , Zeevaert, R. , Schollen, E. , Race, V. , Bammens, R. , Morelle, W. , Rosnoblet, C. , Legrand, D. , Demaegd, D. , Buist, N. , Cheillan, D. , Guffon, N. , Morsomme, P. , et al. (2012). TMEM165 deficiency causes a congenital disorder of glycosylation. The American Journal of Human Genetics 91, 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulquier, F. , Ungar, D. , Reynders, E. , Zeevaert, R. , Mills, P. , García‐Silva, M. T. , Briones, P. , Winchester, B. , Morelle, W. , Krieger, M. , Annaert, W. & Matthijs, G. (2007). A new inborn error of glycosylation due to a Cog8 deficiency reveals a critical role for the Cog1‐Cog8 interaction in COG complex formation. Human Molecular Genetics 16, 717–730. [DOI] [PubMed] [Google Scholar]
- Foulquier, F. , Vasile, E. , Schollen, E. , Callewaert, N. , Raemaekers, T. , Quelhas, D. , Jaeken, J. , Mills, P. , Winchester, B. , Krieger, M. , Annaert, W. & Matthijs, G. (2006). Conserved oligomeric Golgi complex subunit 1 deficiency reveals a previously uncharacterized congenital disorder of glycosylation type II. Proceedings of the National Academy of Sciences of the United States of America 103, 3764–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux, P. , Aebi, M. & Varki, A. (2015). Evolution of glycan diversity. In Essentials of Glycobiology (eds Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L. and Seeberger P. H.). Cold Spring Harbor Laboratory Press, Cold Spring Harbor.Third Edition [Google Scholar]
- Galili, U. (2013). Anti‐Gal: an abundant human natural antibody of multiple pathogeneses and clinical benefits. Immunology 140, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili, U. , Shohet, S. B. , Kobrin, E. , Stults, C. L. & Macher, B. A. (1988). Man, apes, and old world monkeys differ from other mammals in the expression of alpha‐galactosyl epitopes on nucleated cells. Journal of Biological Chemistry 263, 17755–17762. [PubMed] [Google Scholar]
- Gao, C. , Cai, Y. , Wang, Y. , Kang, B.‐H. , Aniento, F. , Robinson, D. G. & Jiang, L. (2014). Retention mechanisms for ER and Golgi membrane proteins. Trends in Plant Science 19, 508–515. [DOI] [PubMed] [Google Scholar]
- Gao, X.‐D. , Nishikawa, A. & Dean, N. (2004). Physical interactions between the Alg1, Alg2, and Alg11 mannosyltransferases of the endoplasmic reticulum. Glycobiology 14, 559–570. [DOI] [PubMed] [Google Scholar]
- Garénaux, E. , Shams‐Eldin, H. , Chirat, F. , Bieker, U. , Schmidt, J. , Michalski, J.‐C. , Cacan, R. , Guérardel, Y. & Schwarz, R. T. (2008). The dual origin of Toxoplasma gondii N‐glycans. Biochemistry 47, 12270–12276. [DOI] [PubMed] [Google Scholar]
- Gas‐Pascual, E. , Ichikawa, H. T. , Sheikh, M. O. , Serji, M. I. , Deng, B. , Mandalasi, M. , Bandini, G. , Samuelson, J. , Wells, L. & West, C. M. (2019). CRISPR/Cas9 and glycomics tools for Toxoplasma glycobiology. Journal of Biological Chemistry 294, 1104–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil, G.‐C. , Velander, W. H. & Van Cott, K. E. (2009). N‐glycosylation microheterogeneity and site occupancy of an Asn‐X‐Cys sequon in plasma‐derived and recombinant protein C. Proteomics 9, 2555–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloster, T. M. (2014). Advances in understanding glycosyltransferases from a structural perspective. Current Opinion in Structural Biology 28, 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, B. , Cukan, M. , Fisher, R. , Li, H. , Stadheim, T. A. & Gerngross, T. (2009). Characterization of N‐linked glycosylation on recombinant glycoproteins produced in Pichia pastoris using ESI‐MS and MALDI‐TOF. In Glycomics: Methods and Protocols (eds Packer N. H. and Karlsson N. G.), pp. Totowa, NJ: Humana Press, 213–223. [DOI] [PubMed] [Google Scholar]
- Guindon, S. & Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52, 696–704. [DOI] [PubMed] [Google Scholar]
- Gutternigg, M. , Bürgmayr, S. , Pöltl, G. , Rudolf, J. & Staudacher, E. (2007a). Neutral N‐glycan patterns of the gastropods Limax maximus, Cepaea hortensis, Planorbarius corneus, Arianta arbustorum and Achatina fulica . Glycoconjugate Journal 24, 475–489. [DOI] [PubMed] [Google Scholar]
- Gutternigg, M. , Kretschmer‐Lubich, D. , Paschinger, K. , Rendić, D. , Hader, J. , Geier, P. , Ranftl, R. , Jantsch, V. , Lochnit, G. & Wilson, I. B. H. (2007b). Biosynthesis of truncated N‐linked oligosaccharides results from non‐orthologous hexosaminidase‐mediated mechanisms in nematodes, plants, and insects. Journal of Biological Chemistry 282, 27825–27840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamieh, A. , Cartier, D. , Abid, H. , Calas, A. , Burel, C. , Bucharles, C. , Jehan, C. , Grumolato, L. , Landry, M. , Lerouge, P. , Anouar, Y. & Lihrmann, I. (2017). Selenoprotein T is a novel OST subunit that regulates UPR signaling and hormone secretion. EMBO Reports 18, 1935–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa, K. , Suzuki, N. & Natsuka, S. (2017). Structures and developmental alterations of N‐glycans of zebrafish embryos. Glycobiology 27, 228–245. [DOI] [PubMed] [Google Scholar]
- Haouari, W. , Dubail, J. , Lounis‐Ouaras, S. , Prada, P. , Bennani, R. , Huber, C. , Afenjar, A. , Colin, E. , Vuillaumier‐Barrot, S. , Foulquier, F. , Poüs, C. , Cormier‐Daire, V. & Bruneel, A. (2020). Serum bikunin isoforms in congenital disorders of glycosylation and linkeropathies. Journal of Inherited Metabolic Disease 43(6), 1349–1359. [DOI] [PubMed] [Google Scholar]
- Hård, K. , van Doorn, J. M. , Thomas‐Oates, J. E. , Kamerling, J. P. & van der Horst, D. J. (1993). Structure of the asn‐linked oligosaccharides of apolipophorin III from the insect Locusta migratoria. Carbohydrate‐linked 2‐aminoethylphosphonate as a constituent of a glycoprotein. Biochemistry 32, 766–775. [DOI] [PubMed] [Google Scholar]
- Haserick, J. R. , Leon, D. R. , Samuelson, J. & Costello, C. E. (2017). Asparagine‐linked glycans of Cryptosporidium parvum contain a single long arm, are barely processed in the endoplasmic reticulum (ER) or Golgi, and show a strong bias for sites with threonine. Molecular & Cellular Proteomics 16, S42–S53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker, N. , Honke, A. , Frey, K. , Leister, D. & Schneider, A. (2020). Homologous proteins of the manganese transporter PAM71 are localized in the Golgi apparatus and endoplasmic reticulum. Plants 9, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker, N. , Leister, D. & Schneider, A. (2017). Plants contain small families of UPF0016 proteins including the photosynthesis affected mutant71 transporter. Plant Signaling & Behavior 12, e1278101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst, S. , Wuhrer, M. & Rombouts, Y. (2015). Chapter six ‐ glycosylation characteristics of colorectal cancer. In Advances in Cancer Research (eds Drake R. R. and Ball L. E.), pp. 203–256. Cambridge, MA: Academic Press. [DOI] [PubMed] [Google Scholar]
- Houdou, M. & Foulquier, F. (2020). Anomalies congénitales de la glycosylation (CDG) ‐ 1980‐2020, 40 ans pour comprendre. Médecine/Sciences 36(8–9), 735–746. [DOI] [PubMed] [Google Scholar]
- Hykollari, A. , Balog, C. I. A. , Rendić, D. , Braulke, T. , Wilson, I. B. H. & Paschinger, K. (2013). Mass spectrometric analysis of neutral and anionic N‐glycans from a Dictyostelium discoideum model for human congenital disorder of glycosylation CDG IL. Journal of Proteome Research 12, 1173–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hykollari, A. , Eckmair, B. , Voglmeir, J. , Jin, C. , Yan, S. , Vanbeselaere, J. , Razzazi‐Fazeli, E. , Wilson, I. B. H. & Paschinger, K. (2016). More than just oligomannose: an N‐glycomic comparison of Penicillium Species. Molecular & Cellular Proteomics: MCP 15, 73–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hykollari, A. , Malzl, D. , Eckmair, B. , Vanbeselaere, J. , Scheidl, P. , Jin, C. , Karlsson, N. G. , Wilson, I. B. H. & Paschinger, K. (2018). Isomeric separation and recognition of anionic and zwitterionic N‐glycans from royal jelly glycoproteins. Molecular & Cellular Proteomics: MCP 17, 2177–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hykollari, A. , Malzl, D. , Yan, S. , Wilson, I. B. H. & Paschinger, K. (2017). Hydrophilic interaction anion exchange for separation of multiply modified neutral and anionic Dictyostelium N‐glycans. Electrophoresis 38, 2175–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa, T. , Nakayama, J. , Sakura, N. , Hashimoto, T. , Fukuda, M. , Fukuda, M. N. & Taki, T. (1999). Expression of N‐acetyllactosamine and β1,4‐galactosyltransferase (β4GalT‐I) during adenoma‐carcinoma sequence in the human colorectum. Journal of Histochemistry & Cytochemistry 47, 1593–1601. [DOI] [PubMed] [Google Scholar]
- Ishida, H. , Togayachi, A. , Sakai, T. , Iwai, T. , Hiruma, T. , Sato, T. , Okubo, R. , Inaba, N. , Kudo, T. , Gotoh, M. , Shoda, J. , Tanaka, N. & Narimatsu, H. (2005). A novel β1,3‐N‐acetylglucosaminyltransferase (β3Gn‐T8), which synthesizes poly‐N‐acetyllactosamine, is dramatically upregulated in colon cancer. FEBS Letters 579, 71–78. [DOI] [PubMed] [Google Scholar]
- Jang‐Lee, J. , Curwen, R. S. , Ashton, P. D. , Tissot, B. , Mathieson, W. , Panico, M. , Dell, A. , Wilson, R. A. & Haslam, S. M. (2007). Glycomics analysis of Schistosoma mansoni egg and cercarial secretions. Molecular & Cellular Proteomics 6, 1485–1499. [DOI] [PubMed] [Google Scholar]
- Jones, D. C. , Mehlert, A. , Güther, M. L. S. & Ferguson, M. A. J. (2005). Deletion of the glucosidase II gene in Trypanosoma brucei reveals novel N‐glycosylation mechanisms in the biosynthesis of variant surface glycoprotein. The Journal of Biological Chemistry 280, 35929–35942. [DOI] [PubMed] [Google Scholar]
- Khoo, K.‐H. , Chatterjee, D. , Caulfield, J. P. , Morris, H. R. & Dell, A. (1997). Structural mapping of the glycans from the egg glycoproteins of Schistosoma mansoni and Schistosoma japonicum: identification of novel core structures and terminal sequences. Glycobiology 7, 663–677. [DOI] [PubMed] [Google Scholar]
- Khoury, G. A. , Baliban, R. C. & Floudas, C. A. (2011). Proteome‐wide post‐translational modification statistics: frequency analysis and curation of the swiss‐prot database. Scientific Reports 1, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowarik, M. , Küng, S. , Martoglio, B. & Helenius, A. (2002). Protein folding during cotranslational translocation in the endoplasmic reticulum. Molecular Cell 10, 769–778. [DOI] [PubMed] [Google Scholar]
- Kurz, S. , Jin, C. , Hykollari, A. , Gregorich, D. , Giomarelli, B. , Vasta, G. R. , Wilson, I. B. H. & Paschinger, K. (2013). Hemocytes and plasma of the eastern oyster (Crassostrea virginica) display a diverse repertoire of sulfated and blood group A‐modified N‐glycans. The Journal of Biological Chemistry 288, 24410–24428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lairson, L. L. , Henrissat, B. , Davies, G. J. & Withers, S. G. (2008). Glycosyltransferases: structures, functions, and mechanisms. Annual Review of Biochemistry 77, 521–555. [DOI] [PubMed] [Google Scholar]
- Lau, E. , Feng, Y. , Claps, G. , Fukuda, M. N. , Perlina, A. , Donn, D. , Jilaveanu, L. , Kluger, H. , Freeze, H. H. & Ronai, Z. A. (2015). The transcription factor ATF2 promotes melanoma metastasis by suppressing protein fucosylation. Science Signaling 8, ra124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehr, T. , Geyer, H. , Maaß, K. , Doenhoff, M. J. & Geyer, R. (2007). Structural characterization of N‐glycans from the freshwater snail Biomphalaria glabrata cross‐reacting with Schistosoma mansoni glycoconjugates. Glycobiology 17, 82–103. [DOI] [PubMed] [Google Scholar]
- Lerouge, P. , Cabanes‐Macheteau, M. , Rayon, C. , Fischette‐Lainé, A.‐C. , Gomord, V. & Faye, L. (1998). N‐Glycoprotein biosynthesis in plants: recent developments and future trends. In Protein Trafficking in Plant Cells (ed. Soll J.), Dordrecht: Springer Netherlands, pp. 31–48. [PubMed] [Google Scholar]
- Levy‐Ontman, O. , Arad, S. , Harvey, D. J. , Parsons, T. B. , Fairbanks, A. & Tekoah, Y. (2011). Unique N‐glycan moieties of the 66‐kDa cell wall glycoprotein from the red microalga Porphyridium sp. The Journal of Biological Chemistry 286, 21340–21352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy‐Ontman, O. , Fisher, M. , Shotland, Y. , Weinstein, Y. , Tekoah, Y. & Arad, S. M. (2014). Genes involved in the endoplasmic reticulum N‐glycosylation pathway of the red microalga Porphyridium sp.: a bioinformatic study. International Journal of Molecular Sciences 15, 2305–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Debowski, A. W. , Liao, T. , Tang, H. , Nilsson, H.‐O. , Marshall, B. J. , Stubbs, K. A. & Benghezal, M. (2017). Understanding protein glycosylation pathways in bacteria. Future Microbiology 12, 59–72. [DOI] [PubMed] [Google Scholar]
- Liebminger, E. , Veit, C. , Pabst, M. , Batoux, M. , Zipfel, C. , Altmann, F. , Mach, L. & Strasser, R. (2011). β‐N‐acetylhexosaminidases HEXO1 and HEXO3 are responsible for the formation of paucimannosidic N‐Glycans in Arabidopsis thaliana . Journal of Biological Chemistry 286, 10793–10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingg, N. , Zhang, P. , Song, Z. & Bardor, M. (2012). The sweet tooth of biopharmaceuticals: importance of recombinant protein glycosylation analysis. Biotechnology Journal 7, 1462–1472. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Doray, B. & Kornfeld, S. (2018). Recycling of Golgi glycosyltransferases requires direct binding to coatomer. Proceedings of the National Academy of Sciences of the United States of America 115, 8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizak, C. , Gerber, S. , Numao, S. , Aebi, M. & Locher, K. P. (2011). X‐ray structure of a bacterial oligosaccharyltransferase. Nature 474, 350–355. [DOI] [PubMed] [Google Scholar]
- Lombard, J. (2016). The multiple evolutionary origins of the eukaryotic N‐glycosylation pathway. Biology Direct 11, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel, M. , Schott, A. , Jank, T. , Hofmann, V. & Strahl, S. (2011). A conserved acidic motif is crucial for enzymatic activity of protein O ‐mannosyltransferases. Journal of Biological Chemistry 286, 39768–39775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommerse, J. P. M. , Thomas‐Oates, J. E. , Gielens, C. , Préaux, G. , Kamerling, J. P. & Vliegenthart, J. F. G. (1997). Primary structure of 21 novel monoantennary and diantennary N‐linked carbohydrate chains from αD‐hemocyanin of Helix pomatia . European Journal of Biochemistry 249, 195–222. [DOI] [PubMed] [Google Scholar]
- Lucas, P.‐L. , Dumontier, R. , Loutelier‐Bourhis, C. , Mareck, A. , Afonso, C. , Lerouge, P. , Mati‐Baouche, N. & Bardor, M. (2018). User‐friendly extraction and multistage tandem mass spectrometry based analysis of lipid‐linked oligosaccharides in microalgae. Plant Methods 14, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, P.‐L. , Mathieu‐Rivet, E. , Song, P. C. T. , Oltmanns, A. , Loutelier‐Bourhis, C. , Plasson, C. , Afonso, C. , Hippler, M. , Lerouge, P. , Mati‐Baouche, N. & Bardor, M. (2020). Multiple xylosyltransferases heterogeneously xylosylate protein N‐linked glycans in Chlamydomonas reinhardtii . The Plant Journal 102, 230–245. [DOI] [PubMed] [Google Scholar]
- Luk, F. C. Y. , Johnson, T. M. & Beckers, C. J. (2008). N‐linked glycosylation of proteins in the protozoan parasite Toxoplasma gondii . Molecular and biochemical parasitology 157, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnelli, P. , Cipollo, J. F. , Ratner, D. M. , Cui, J. , Kelleher, D. , Gilmore, R. , Costello, C. E. , Robbins, P. W. & Samuelson, J. (2008). Unique Asn‐linked oligosaccharides of the human pathogen Entamoeba histolytica . The Journal of Biological Chemistry 283, 18355–18364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthri, S. , Güther, M. L. S. , Izquierdo, L. , Acosta‐Serrano, A. & Ferguson, M. A. J. (2008). Deletion of the TbALG3 gene demonstrates site‐specific N‐glycosylation and N‐glycan processing in Trypanosoma brucei . Glycobiology 18, 367–383. [DOI] [PubMed] [Google Scholar]
- Martini, F. , Eckmair, B. , Štefanić, S. , Jin, C. , Garg, M. , Yan, S. , Jiménez‐Castells, C. , Hykollari, A. , Neupert, C. , Venco, L. , Varón Silva, D. , Wilson, I. B. H. & Paschinger, K. (2019). Highly modified and immunoactive N‐glycans of the canine heartworm. Nature Communications 10, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu‐Rivet, E. , Kiefer‐Meyer, M.‐C. , Vanier, G. , Ovide, C. , Burel, C. , Lerouge, P. & Bardor, M. (2014). Protein N‐glycosylation in eukaryotic microalgae and its impact on the production of nuclear expressed biopharmaceuticals. Frontiers in Plant Science 5, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu‐Rivet, E. , Scholz, M. , Arias, C. , Dardelle, F. , Schulze, S. , Mauff, F. L. , Teo, G. , Hochmal, A. K. , Blanco‐Rivero, A. , Loutelier‐Bourhis, C. , Kiefer‐Meyer, M.‐C. , Fufezan, C. , Burel, C. , Lerouge, P. , Martinez, F. , et al. (2013). Exploring the N‐glycosylation pathway in Chlamydomonas reinhardtii unravels novel complex structures. Molecular & Cellular Proteomics 12, 3160–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, T. , Takita, E. , Sato, T. , Kinjo, S. , Aizawa, M. , Sugiura, Y. , Hamabata, T. , Sawada, K. & Kato, K. (2011). N‐glycosylation at noncanonical Asn‐X‐Cys sequences in plant cells. Glycobiology 21, 994–999. [DOI] [PubMed] [Google Scholar]
- Mega, T. (2007). Plant‐type N‐glycans containing fucose and xylose in Bryophyta (mosses) and Tracheophyta (ferns). Bioscience, Biotechnology, and Biochemistry 71, 2893–2904. [DOI] [PubMed] [Google Scholar]
- Mehlert, A. , Zitzmann, N. , Richardson, J. M. , Treumann, A. & Ferguson, M. A. (1998). The glycosylation of the variant surface glycoproteins and procyclic acidic repetitive proteins of Trypanosoma brucei . Molecular and Biochemical Parasitology 91, 145–152. [DOI] [PubMed] [Google Scholar]
- Mi, H. , Ebert, D. , Muruganujan, A. , Mills, C. , Albou, L.‐P. , Mushayamaha, T. & Thomas, P. D. (2021). PANTHER version 16: a revised family classification, tree‐based classification tool, enhancer regions and extensive API. Nucleic Acids Research 49, D394–D403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickum, M. L. , Prasanphanich, N. S. , Song, X. , Dorabawila, N. , Mandalasi, M. , Lasanajak, Y. , Luyai, A. , Secor, W. E. , Wilkins, P. P. , van Die, I. , Smith, D. F. , Nyame, A. K. , Cummings, R. D. & Rivera‐Marrero, C. A. (2016). Identification of antigenic glycans from Schistosoma mansoni by using a shotgun egg glycan microarray. Infection and Immunity 84, 1371–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mócsai, R. , Blaukopf, M. , Svehla, E. , Kosma, P. & Altmann, F. (2020a). The N‐glycans of Chlorella sorokiniana and a related strain contain arabinose but have strikingly different structures. Glycobiology 30, 663–676. [DOI] [PubMed] [Google Scholar]
- Mócsai, R. , Figl, R. , Sützl, L. , Fluch, S. & Altmann, F. (2020b). A first view on the unsuspected intragenus diversity of N‐glycans in Chlorella microalgae. The Plant Journal: For Cell and Molecular Biology 103, 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mócsai, R. , Figl, R. , Troschl, C. , Strasser, R. , Svehla, E. , Windwarder, M. , Thader, A. & Altmann, F. (2019). N‐glycans of the microalga Chlorella vulgaris are of the oligomannosidic type but highly methylated. Scientific Reports 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelle, W. , Haslam, S. M. , Olivier, V. , Appleton, J. A. , Morris, H. R. & Dell, A. (2000). Phosphorylcholine‐containing N‐glycans of Trichinella spiralis: identification of multiantennary lacdiNAc structures. Glycobiology 10, 941–950. [DOI] [PubMed] [Google Scholar]
- Morelle, W. , Potelle, S. , Witters, P. , Wong, S. , Climer, L. , Lupashin, V. , Matthijs, G. , Gadomski, T. , Jaeken, J. , Cassiman, D. , Morava, E. & Foulquier, F. (2017). Galactose supplementation in patients with TMEM165‐CDG rescues the glycosylation defects. The Journal of Clinical Endocrinology and Metabolism 102, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen, K. W. , Tiemeyer, M. & Nairn, A. V. (2012). Vertebrate protein glycosylation: diversity, synthesis and function. Nature Reviews. Molecular Cell Biology 13, 448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairn, A. V. , York, W. S. , Harris, K. , Hall, E. M. , Pierce, J. M. & Moremen, K. W. (2008). Regulation of glycan structures in animal tissues: transcript profiling of glycan‐related genes. The Journal of Biological Chemistry 283, 17298–17313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, M. , Tojo, H. & Fujii, S. (2011). A glycan of Ψ‐factor from Dictyostelium discoideum contains a bisecting‐GlcNAc, an intersecting‐GlcNAc, and a core α‐1,6‐fucose. Bioscience, Biotechnology, and Biochemistry 75, 1964–1970. [DOI] [PubMed] [Google Scholar]
- Neelamegham, S. & Mahal, L. K. (2016). Multi‐level regulation of cellular glycosylation: from genes to transcript to enzyme to structure. Current Opinion in Structural Biology 40, 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan, D. P. , Geuskens, M. & Pays, E. (1999). N‐linked glycans containing linear poly‐N‐acetyllactosamine as sorting signals in endocytosis in Trypanosoma brucei . Current Biology: CB 9, 1169–1172. [DOI] [PubMed] [Google Scholar]
- Nothaft, H. & Szymanski, C. M. (2010). Protein glycosylation in bacteria: sweeter than ever. Nature Reviews. Microbiology 8, 765–778. [DOI] [PubMed] [Google Scholar]
- Obregón, A. , Flores, M. S. , Rangel, R. , Arévalo, K. , Maldonado, G. , Quintero, I. & Galán, L. (2019). Characterization of N‐glycosylations in Entamoeba histolytica ubiquitin. Experimental Parasitology 196, 38–47. [DOI] [PubMed] [Google Scholar]
- Oltmanns, A. , Hoepfner, L. , Scholz, M. , Zinzius, K. , Schulze, S. & Hippler, M. (2020). Novel insights into N‐glycan fucosylation and core xylosylation in C. reinhardtii . Frontiers in Plant Science 10, 1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, E. C. , Kuhaudomlarp, S. , Rejzek, M. , Fangel, J. U. , Alagesan, K. , Kolarich, D. , Willats, W. G. T. & Field, R. A. (2017). Exploring the glycans of Euglena gracilis . Biology 6, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oriol, R. , Martinez‐Duncker, I. , Chantret, I. , Mollicone, R. & Codogno, P. (2002). Common origin and evolution of glycosyltransferases using Dol‐P‐monosaccharides as donor substrate. Molecular Biology and Evolution 19, 1451–1463. [DOI] [PubMed] [Google Scholar]
- Parodi, A. , Cummings, R. D. & Aebi, M. (2015). Glycans in glycoprotein quality control. In Essentials of Glycobiology (eds Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L. and Seeberger P. H.), pp. 503–511, Third Edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor. [Google Scholar]
- Paschinger, K. , Hykollari, A. , Razzazi‐Fazeli, E. , Greenwell, P. , Leitsch, D. , Walochnik, J. & Wilson, I. B. H. (2012). The N‐glycans of Trichomonas vaginalis contain variable core and antennal modifications. Glycobiology 22, 300–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschinger, K. , Razzazi‐Fazeli, E. , Furukawa, K. & Wilson, I. B. H. (2011). Presence of galactosylated core fucose on N‐glycans in the planaria Dugesia japonica . Journal of Mass Spectrometry 46, 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschinger, K. & Wilson, I. B. H. (2015). Two types of galactosylated fucose motifs are present on N‐glycans of Haemonchus contortus . Glycobiology 25, 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschinger, K. , Yan, S. & Wilson, I. B. H. (2019). N‐glycomic complexity in anatomical simplicity: Caenorhabditis elegans as a non‐model nematode? Frontiers in Molecular Biosciences 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péanne, R. , de Lonlay, P. , Foulquier, F. , Kornak, U. , Lefeber, D. J. , Morava, E. , Pérez, B. , Seta, N. , Thiel, C. , van Schaftingen, E. , Matthijs, G. & Jaeken, J. (2018). Congenital disorders of glycosylation (CDG): quo vadis? European Journal of Medical Genetics 61, 643–663. [DOI] [PubMed] [Google Scholar]
- Pokrovskaya, I. D. , Willett, R. , Smith, R. D. , Morelle, W. , Kudlyk, T. & Lupashin, V. V. (2011). Conserved oligomeric Golgi complex specifically regulates the maintenance of Golgi glycosylation machinery. Glycobiology 21, 1554–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöltl, G. , Kerner, D. , Paschinger, K. & Wilson, I. B. H. (2007). N‐glycans of the porcine nematode parasite Ascaris suum are modified with phosphorylcholine and core fucose residues. The FEBS Journal 274, 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potelle, S. , Dulary, E. , Climer, L. , Duvet, S. , Morelle, W. , Vicogne, D. , Lebredonchel, E. , Houdou, M. , Spriet, C. , Krzewinski‐Recchi, M.‐A. , Peanne, R. , Klein, A. , de Bettignies, G. , Morsomme, P. , Matthijs, G. , et al. (2017). Manganese‐induced turnover of TMEM165. Biochemical Journal 474, 1481–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille, C. , Hui, N. , Hunte, F. , Kieckbusch, R. , Berger, E. G. , Warren, G. & Nilsson, T. (1995). Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides. Journal of Cell Science 108(Pt 4), 1617–1627. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, B. , Ramasamy, V. & Qasba, P. K. (2006). Structural snapshots of β‐1,4‐galactosyltransferase‐I along the kinetic pathway. Journal of Molecular Biology 357, 1619–1633. [DOI] [PubMed] [Google Scholar]
- Ramírez, A. S. , Kowal, J. & Locher, K. P. (2019). Cryo–electron microscopy structures of human oligosaccharyltransferase complexes OST‐A and OST‐B. Science 366, 1372–1375. [DOI] [PubMed] [Google Scholar]
- Ravidà, A. , Aldridge, A. M. , Driessen, N. N. , Heus, F. A. H. , Hokke, C. H. & O'Neill, S. M. (2016). Fasciola hepatica surface coat glycoproteins contain mannosylated and phosphorylated N‐glycans and exhibit immune modulatory properties independent of the mannose receptor. PLoS Neglected Tropical Diseases 10, e0004601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, T. A. , Lippolis, J. D. & Sacco, R. E. (2014). The Ca2+/H+ antiporter TMEM165 expression, localization in the developing, lactating and involuting mammary gland parallels the secretory pathway Ca2+ ATPase (SPCA1). Biochemical and Biophysical Research Communications 445, 417–421. [DOI] [PubMed] [Google Scholar]
- Reynders, E. , Foulquier, F. , Annaert, W. & Matthijs, G. (2011). How Golgi glycosylation meets and needs trafficking: the case of the COG complex. Glycobiology 21, 853–863. [DOI] [PubMed] [Google Scholar]
- Reynders, E. , Foulquier, F. , Leão Teles, E. , Quelhas, D. , Morelle, W. , Rabouille, C. , Annaert, W. & Matthijs, G. (2009). Golgi function and dysfunction in the first COG4‐deficient CDG type II patient. Human Molecular Genetics 18, 3244–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rini, J. M. & Esko, J. D. (2017). Glycosyltransferases and glycan‐processing enzymes. In Essentials of Glycobiology (eds Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L. and Seeberger P. H.), pp. 65–75, Third Edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor. [Google Scholar]
- Ruddock, L. W. & Molinari, M. (2006). N‐glycan processing in ER quality control. Journal of Cell Science 119, 4373–4380. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Canada, C. , Kelleher, D. J. & Gilmore, R. (2009). Cotranslational and posttranslational N‐glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136, 272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐May, E. , Sørensen, I. , Fei, Z. , Zhang, S. , Domozych, D. S. & Rose, J. K. C. (2018). The secretome and N‐glycosylation profiles of the charophycean green alga, Penium margaritaceum, resemble those of embryophytes. Proteomes 6, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush, J. (2016). Role of flippases in protein glycosylation in the endoplasmic reticulum. Lipid Insights 8, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahadevan, S. , Antonopoulos, A. , Haslam, S. M. , Dell, A. , Ramaswamy, S. & Babu, P. (2014). Unique, polyfucosylated glycan‐receptor interactions are essential for regeneration of Hydra magnipapillata . ACS chemical Biology 9, 147–155. [DOI] [PubMed] [Google Scholar]
- Şahar, U. & Deveci, R. (2017). Profiling N‐glycans of the egg jelly coat of the sea urchin Paracentrotus lividus by MALDI‐TOF mass spectrometry and capillary liquid chromatography electrospray ionization‐ion trap tandem mass spectrometry systems. Molecular Reproduction and Development 84, 401–407. [DOI] [PubMed] [Google Scholar]
- Samuelson, J. , Banerjee, S. , Magnelli, P. , Cui, J. , Kelleher, D. J. , Gilmore, R. & Robbins, P. W. (2005). The diversity of dolichol‐linked precursors to Asn‐linked glycans likely results from secondary loss of sets of glycosyltransferases. Proceedings of the National Academy of Sciences of the United States of America 102, 1548–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson, J. & Robbins, P. W. (2015). Effects of N‐glycan precursor length diversity on quality control of protein folding and on protein glycosylation. Seminars in Cell & Developmental Biology 41, 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandra, K. , Dolashka‐Angelova, P. , Devreese, B. & van Beeumen, J. (2007). New insights in Rapana venosa hemocyanin N‐glycosylation resulting from on‐line mass spectrometric analyses. Glycobiology 17, 141–156. [DOI] [PubMed] [Google Scholar]
- Schachter, H. (1991). The ‘yellow brick road’ to branched complex N‐glycans. Glycobiology 1, 453–461. [DOI] [PubMed] [Google Scholar]
- Schiller, B. , Hykollari, A. , Voglmeir, J. , Pöltl, G. , Hummel, K. , Razzazi‐Fazeli, E. , Geyer, R. & Wilson, I. B. H. (2009). Development of Dictyostelium discoideum is associated with alteration of fucosylated N‐glycan structures. Biochemical Journal 423, 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller, B. , Makrypidi, G. , Razzazi‐Fazeli, E. , Paschinger, K. , Walochnik, J. & Wilson, I. B. H. (2012). Exploring the unique N‐glycome of the opportunistic human pathogen Acanthamoeba . The Journal of Biological Chemistry 287, 43191–43204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, A. , Steinberger, I. , Herdean, A. , Gandini, C. , Eisenhut, M. , Kurz, S. , Morper, A. , Hoecker, N. , Rühle, T. , Labs, M. , Flügge, U. I. , Geimer, S. , Schmidt, S. B. , Husted, S. , Weber, A. P. M. , et al. (2016). The evolutionarily conserved protein photosynthesis affected mutant71 is required for efficient manganese uptake at the thylakoid membrane in Arabidopsis . The Plant Cell 28(4), 892–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze, S. , Urzica, E. , Reijnders, M. J. M. F. , van de Geest, H. , Warris, S. , Bakker, L. V. , Fufezan, C. , dos Santos, V. A. P. M. , Schaap, P. J. , Peters, S. A. & Hippler, M. (2017). Identification of methylated GnT I‐dependent N‐glycans in Botryococcus brauni . New Phytologist 215, 1361–1369. [DOI] [PubMed] [Google Scholar]
- Shin, Y.‐J. , Castilho, A. , Dicker, M. , Sádio, F. , Vavra, U. , Grünwald‐Gruber, C. , Kwon, T.‐H. , Altmann, F. , Steinkellner, H. & Strasser, R. (2017). Reduced paucimannosidic N‐glycan formation by suppression of a specific β‐hexosaminidase from Nicotiana benthamiana . Plant Biotechnology Journal 15, 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein, S. & Gilmore, R. (1996). Biochemistry, molecular biology, and genetics of the oligosaccharyltransferase. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology 10, 849–858. [PubMed] [Google Scholar]
- Snyder, N. A. , Palmer, M. V. , Reinhardt, T. A. & Cunningham, K. W. (2019). Milk biosynthesis requires the Golgi cation exchanger TMEM165. Journal of Biological Chemistry 294, 3181–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder, N. A. , Stefan, C. P. , Soroudi, C. T. , Kim, A. , Evangelista, C. & Cunningham, K. W. (2017). H + and Pi byproducts of glycosylation affect Ca 2+ homeostasis and are retrieved from the Golgi complex by homologs of TMEM165 and XPR1. G3 (Bethesda) Genes|Genomes|Genetics 7, 3913–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton, R. , Hykollari, A. , Eckmair, B. , Malzl, D. , Dragosits, M. , Palmberger, D. , Wang, P. , Wilson, I. B. H. & Paschinger, K. (2017). The underestimated N‐glycomes of lepidopteran species. Biochimica et Biophysica Acta 1861, 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser, R. , Bondili, J. S. , Schoberer, J. , Svoboda, B. , Liebminger, E. , Glössl, J. , Altmann, F. , Steinkellner, H. & Mach, L. (2007). Enzymatic properties and subcellular localization of Arabidopsis beta‐N‐acetylhexosaminidases. Plant Physiology 145, 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribny, J. , Thines, L. , Deschamps, A. , Goffin, P. & Morsomme, P. (2020). The human Golgi protein TMEM165 transports calcium and manganese in yeast and bacterial cells. Journal of Biological Chemistry 295, 3865–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe, W. B. & Reinhold, V. N. (2012). The conserved oligomeric Golgi complex is required for fucosylation of N‐glycans in Caenorhabditis elegans . Glycobiology 22, 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm, A. , Bergwerff, A. A. & Vliegenthart, J. F. (1992). 1H‐NMR structural determination of the N‐linked carbohydrate chains on glycopeptides obtained from the bean lectin phytohemagglutinin. European Journal of Biochemistry 204, 313–316. [DOI] [PubMed] [Google Scholar]
- Sturm, A. & Chrispeels, M. J. (1986). The high mannose oligosaccharide of phytohemagglutinin is attached to asparagine 12 and the modified oligosaccharide to asparagine 60. Plant Physiology 81, 320–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, S. P. , Babu, P. , Palakodeti, D. & Subramanian, R. (2018). Identification of multiple isomeric core chitobiose–modified high‐mannose and paucimannose N‐glycans in the planarian Schmidtea mediterranea . The Journal of Biological Chemistry 293, 6707–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi, T. , Seko, A. , Kitajima, K. , Muto, Y. , Inoue, S. , Khoo, K. H. , Morris, H. R. , Dell, A. & Inoue, Y. (1994). Structural studies of a novel type of pentaantennary large glycan unit in the fertilization‐associated carbohydrate‐rich glycopeptide isolated from the fertilized eggs of Oryzias latipes . The Journal of Biological Chemistry 269, 8762–8771. [PubMed] [Google Scholar]
- Tangvoranuntakul, P. , Gagneux, P. , Diaz, S. , Bardor, M. , Varki, N. , Varki, A. & Muchmore, E. (2003). Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proceedings of the National Academy of Sciences of the United States of America 100, 12045–12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, T. , Mizuochi, T. , Banno, Y. , Nozawa, Y. & Kobata, A. (1985). Carbohydrates of lysosomal enzymes secreted by Tetrahymena pyriformis . Journal of Biological Chemistry 260, 13941–13946. [PubMed] [Google Scholar]
- Thines, L. , Deschamps, A. , Sengottaiyan, P. , Savel, O. , Stribny, J. & Morsomme, P. (2018). The yeast protein Gdt1p transports Mn 2+ ions and thereby regulates manganese homeostasis in the Golgi. Journal of Biological Chemistry 293, 8048–8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines, L. , Stribny, J. & Morsomme, P. (2020). From the uncharacterized protein family 0016 to the GDT1 family: molecular insights into a newly‐characterized family of cation secondary transporters. Microbial Cell 7, 202–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, L. & Banfield, D. K. (2010). Localization of Golgi‐resident glycosyltransferases. Cellular and Molecular Life Sciences: CMLS 67, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ünligil, U. M. , Zhou, S. , Yuwaraj, S. , Sarkar, M. , Schachter, H. & Rini, J. M. (2000). X‐ray crystal structure of rabbit N‐acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily. The EMBO Journal 19, 5269–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen, J. , Geisen, S. , Routh, D. , Ferris, H. , Traunspurger, W. , Wardle, D. A. , de Goede, R. G. M. , Adams, B. J. , Ahmad, W. , Andriuzzi, W. S. , Bardgett, R. D. , Bonkowski, M. , Campos‐Herrera, R. , Cares, J. E. , Caruso, T. , et al. (2019). Soil nematode abundance and functional group composition at a global scale. Nature 572, 194–198. [DOI] [PubMed] [Google Scholar]
- van Die, I. , Gomord, V. , Kooyman, F. N. J. , van den Berg, T. K. , Cummings, R. D. & Vervelde, L. (1999). Core α1→3‐fucose is a common modification of N‐glycans in parasitic helminths and constitutes an important epitope for IgE from Haemonchus contortus infected sheep. FEBS Letters 463, 189–193. [DOI] [PubMed] [Google Scholar]
- van Kuik, J. A. , Breg, J. , Kolsteeg, C. E. M. , Kamerling, J. P. & Vliegenthart, J. F. G. (1987). Primary structure of the acidic carbohydrate chain of hemocyanin from Panulirus interruptus . FEBS Letters 221, 150–154. [Google Scholar]
- van Kuik, J. A. , van Halbeek, H. , Kamerling, J. P. & Vliegenthart, J. F. (1986). Primary structure of the neutral carbohydrate chains of hemocyanin from Panulirus interruptus . European Journal of Biochemistry 159, 297–301. [DOI] [PubMed] [Google Scholar]
- Vanbeselaere, J. , Jin, C. , Eckmair, B. , Wilson, I. B. H. & Paschinger, K. (2020). Sulfated and sialylated N‐glycans in the echinoderm Holothuria atra reflect its marine habitat and phylogeny. The Journal of Biological Chemistry 295, 3159–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanier, G. , Lucas, P.‐L. , Loutelier‐Bourhis, C. , Vanier, J. , Plasson, C. , Walet‐Balieu, M.‐L. , Tchi‐Song, P. C. , Remy‐Jouet, I. , Richard, V. , Bernard, S. , Driouich, A. , Afonso, C. , Lerouge, P. , Mathieu‐Rivet, E. & Bardor, M. (2017). Heterologous expression of the N‐acetylglucosaminyltransferase I dictates a reinvestigation of the N‐glycosylation pathway in Chlamydomonas reinhardtii . Scientific Reports 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki, A. (2017a). Biological roles of glycans. Glycobiology 27, 3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki, A. (2017b). New and updated glycoscience‐related resources at NCBI. Glycobiology 27, 993. [DOI] [PubMed] [Google Scholar]
- Varki, A. & Gagneux, P. (2015). Biological functions of glycans. In Essentials of Glycobiology (eds Varki A., Cummings R. D., Esko J. D., Stanley P., Hart G. W., Aebi M., Darvill A. G., Kinoshita T., Packer N. H., Prestegard J. H., Schnaar R. L. and Seeberger P. H. ). Cold Spring Harbor Laboratory Press, Cold Spring Harbor.Third Edition [Google Scholar]
- Verchère, A. , Cowton, A. , Jenni, A. , Rauch, M. , Häner, R. , Graumann, J. , Bütikofer, P. & Menon, A. K. (2021). Complexity of the eukaryotic dolichol‐linked oligosaccharide scramblase suggested by activity correlation profiling mass spectrometry. Scientific Reports 11, 1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viëtor, R. , Loutelier‐Bourhis, C. , Fitchette, A.‐C. , Margerie, P. , Gonneau, M. , Faye, L. & Lerouge, P. (2003). Protein N‐glycosylation is similar in the moss Physcomitrella patens and in higher plants. Planta 218, 269–275. [DOI] [PubMed] [Google Scholar]
- Vleugels, W. , Haeuptle, M. A. , Ng, B. G. , Michalski, J.‐C. , Battini, R. , Dionisi‐Vici, C. , Ludman, M. D. , Jaeken, J. , Foulquier, F. , Freeze, H. H. , Matthijs, G. & Hennet, T. (2009). RFT1 deficiency in three novel CDG patients. Human Mutation 30, 1428–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Yang, Y. , Huang, F. , He, Z. , Li, P. , Zhang, W. , Zhang, W. & Tang, B. (2020). In situ fluorescent and photoacoustic imaging of Golgi pH to elucidate the function of transmembrane protein 165. Analytical Chemistry 92, 3103–3110. [DOI] [PubMed] [Google Scholar]
- Weide, T. , Herrmann, L. , Bockau, U. , Niebur, N. , Aldag, I. , Laroy, W. , Contreras, R. , Tiedtke, A. & Hartmann, M. W. (2006). Secretion of functional human enzymes by Tetrahymena thermophila . BMC Biotechnology 6, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild, R. , Kowal, J. , Eyring, J. , Ngwa, E. M. , Aebi, M. & Locher, K. P. (2018). Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N‐glycosylation. Science 359, 545–550. [DOI] [PubMed] [Google Scholar]
- Wu, X. , Steet, R. A. , Bohorov, O. , Bakker, J. , Newell, J. , Krieger, M. , Spaapen, L. , Kornfeld, S. & Freeze, H. H. (2004). Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nature Medicine 10, 518–523. [DOI] [PubMed] [Google Scholar]
- Wuhrer, M. , Koeleman, C. A. M. , Deelder, A. M. & Hokke, C. H. (2006a). Repeats of LacdiNAc and fucosylated LacdiNAc on N‐glycans of the human parasite Schistosoma mansoni . The FEBS Journal 273, 347–361. [DOI] [PubMed] [Google Scholar]
- Wuhrer, M. , Koeleman, C. A. M. , Fitzpatrick, J. M. , Hoffmann, K. F. , Deelder, A. M. & Hokke, C. H. (2006b). Gender‐specific expression of complex‐type N‐glycans in schistosomes. Glycobiology 16, 991–1006. [DOI] [PubMed] [Google Scholar]
- Wuhrer, M. , Robijn, M. L. M. , Koeleman, C. A. M. , Balog, C. I. A. , Geyer, R. , Deelder, A. M. & Hokke, C. H. (2004). A novel Gal(beta1‐4)Gal(beta1‐4)Fuc(alpha1‐6)‐core modification attached to the proximal N‐acetylglucosamine of keyhole limpet haemocyanin (KLH) N‐glycans. The Biochemical Journal 378, 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. & Ng, D. T. W. (2015). Glycosylation‐directed quality control of protein folding. Nature Reviews Molecular Cell Biology 16, 742–752. [DOI] [PubMed] [Google Scholar]
- Yagi, H. , Nakagawa, M. , Takahashi, N. , Kondo, S. , Matsubara, M. & Kato, K. (2008). Neural complex‐specific expression of xylosyl N‐glycan in Ciona intestinalis . Glycobiology 18, 145–151. [DOI] [PubMed] [Google Scholar]
- Yan, S. , Bleuler‐Martinez, S. , Plaza, D. F. , Künzler, M. , Aebi, M. , Joachim, A. , Razzazi‐Fazeli, E. , Jantsch, V. , Geyer, R. , Wilson, I. B. H. & Paschinger, K. (2012). Galactosylated fucose epitopes in nematodes increased expression in a Caenorhabditis mutant associated with altered lectin sensitivity and occurrence in parasitic species. Journal of Biological Chemistry 287, 28276–28290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, S. , Brecker, L. , Jin, C. , Titz, A. , Dragosits, M. , Karlsson, N. G. , Jantsch, V. , Wilson, I. B. H. & Paschinger, K. (2015). Bisecting galactose as a feature of N‐glycans of wild‐type and mutant Caenorhabditis elegans . Molecular & Cellular Proteomics: MCP 14, 2111–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, S. , Vanbeselaere, J. , Jin, C. , Blaukopf, M. , Wöls, F. , Wilson, I. B. H. & Paschinger, K. (2018). Core richness of N‐glycans of Caenorhabditis elegans: a case study on chemical and enzymatic release. Analytical Chemistry 90, 928–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.‐H. , Wang, C. , Singh, S. , Fan, N. , Liu, S. , Zhao, L. , Cao, H. , Xie, W. , Yang, C. & Huang, C.‐F. (2021). Golgi‐localised manganese transporter PML3 regulates Arabidopsis growth through modulating Golgi glycosylation and cell wall biosynthesis. New Phytologist 6, 2200–2214. [DOI] [PubMed] [Google Scholar]
- Zamze, S. E. , Ashford, D. A. , Wooten, E. W. , Rademacher, T. W. & Dwek, R. A. (1991). Structural characterization of the asparagine‐linked oligosaccharides from Trypanosoma brucei type II and type III variant surface glycoproteins. The Journal of Biological Chemistry 266, 20244–20261. [PubMed] [Google Scholar]
- Zamze, S. E. , Wooten, E. W. , Ashford, D. A. , Ferguson, M. A. , Dwek, R. A. & Rademacher, T. W. (1990). Characterisation of the asparagine‐linked oligosaccharides from Trypanosoma brucei type‐I variant surface glycoproteins. European Journal of Biochemistry 187, 657–663. [DOI] [PubMed] [Google Scholar]
- Zhang, P. , Burel, C. , Plasson, C. , Kiefer‐Meyer, M.‐C. , Ovide, C. , Gügi, B. , Wan, C. , Teo, G. , Mak, A. , Song, Z. , Driouich, A. , Lerouge, P. & Bardor, M. (2019). Characterization of a GDP‐fucose transporter and a fucosyltransferase involved in the fucosylation of glycoproteins in the diatom Phaeodactylum tricornutum . Frontiers in Plant Science 10, 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Iwasa, T. , Tsuda, M. , Kobata, A. & Takasaki, S. (1997). A novel monoantennary complex‐type sugar chain found in octopus rhodopsin: occurrence of the Galβ1→4Fuc group linked to the proximal N‐acetylglucosamine residue of the trimannosyl core. Glycobiology 7, 1153–1158. [DOI] [PubMed] [Google Scholar]
- Zhou, H. , Hanneman, A. J. , Chasteen, N. D. & Reinhold, V. N. (2013). Anomalous N‐glycan structures with an internal fucose branched to GlcA and GlcN residues isolated from a Mollusk shell‐forming fluid. Journal of Proteome Research 12, 4547–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, F. D. , Cavanagh, J. , Lubowski, C. & Trimble, R. B. (1999). Novel Schizosaccharomyces pombe N‐linked GalMan9GlcNAc isomers: role of the Golgi GMA12 galactosyltransferase in core glycan galactosylation. Glycobiology 9, 497–505. [DOI] [PubMed] [Google Scholar]
- Zielinska, D. F. , Gnad, F. , Schropp, K. , Wiśniewski, J. R. & Mann, M. (2012). Mapping N‐glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Molecular Cell 46, 542–548. [DOI] [PubMed] [Google Scholar]
- Zielinska, D. F. , Gnad, F. , Wiśniewski, J. R. & Mann, M. (2010). Precision mapping of an in vivo N‐glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907. [DOI] [PubMed] [Google Scholar]


