Abstract
Although metals are thought to inhibit the ability of microorganisms to degrade organic pollutants, several microbial mechanisms of resistance to metal are known to exist. This study examined the potential of cadmium-resistant microorganisms to reduce soluble cadmium levels to enhance degradation of 2,4-dichlorophenoxyacetic acid (2,4-D) under conditions of cocontamination. Four cadmium-resistant soil microorganisms were examined in this study. Resistant up to a cadmium concentration of 275 μg ml−1, these isolates represented the common soil genera Arthrobacter, Bacillus, and Pseudomonas. Isolates Pseudomonas sp. strain H1 and Bacillus sp. strain H9 had a plasmid-dependent intracellular mechanism of cadmium detoxification, reducing soluble cadmium levels by 36%. Isolates Arthrobacter strain D9 and Pseudomonas strain I1a both produced an extracellular polymer layer that bound and reduced soluble cadmium levels by 22 and 11%, respectively. Although none of the cadmium-resistant isolates could degrade 2,4-D, results of dual-bioaugmentation studies conducted with both pure culture and laboratory soil microcosms showed that each of four cadmium-resistant isolates supported the degradation of 500-μg ml−1 2,4-D by the cadmium-sensitive 2,4-D degrader Ralstonia eutropha JMP134. Degradation occurred in the presence of up to 24 μg of cadmium ml−1 in pure culture and up to 60 μg of cadmium g−1 in amended soil microcosms. In a pilot field study conducted with 5-gallon soil bioreactors, the dual-bioaugmentation strategy was again evaluated. Here, the cadmium-resistant isolate Pseudomonas strain H1 enhanced degradation of 2,4-D in reactors inoculated with R. eutropha JMP134 in the presence of 60 μg of cadmium g−1. Overall, dual bioaugmentation appears to be a viable approach in the remediation of cocontaminated soils.
Cocontaminated soils, soils contaminated with both metals and organics, are considered difficult to remediate because of the mixed nature of the contaminants. A treatment alternative to expensive excavation and incineration (9) of metal-contaminated soils is bioaugmentation with metal-detoxifying and/or organic-degrading microorganisms (1, 3, 4, 6, 18). Many microorganisms are known to degrade a variety of organics, and likewise, a number of metal-resistant microorganisms are known to detoxify metals, such as selenium, mercury, and cadmium (23, 27). In cocontaminated sites, metal toxicity inhibits the activity of organic-degrading microorganisms (24). Consequently, bioremediation efforts focus on reducing metal toxicity in sites with mixed contaminants. Until recently, bioaugmentation studies focused on the introduction of a microorganism that was both metal resistant and capable of organic degradation. Under field conditions, such an approach is often unsuccessful. One reason may be that the energy requirements to maintain concurrent metal resistance and organic degradation are too high, and the introduced organism cannot perform both activities under environmental conditions. The issue of cocontamination is a serious one, since approximately 37% of all contaminated sites in the United States alone contain both metal and organic contaminants (20; W. W. Kovalich, Jr., keynote lecture, 4th World Congr. Chem. Eng., p. 281–295, 1991).
The approach used in this study was to coinoculate with a metal-detoxifying population and an organic-degrading population that cooperatively functioned to remediate both metal and organic pollutants in a cocontaminated system. We hypothesized that the metal-resistant population could protect the metal-sensitive organic-degrading population from metal toxicity. Stephen et al. (27) used metal-resistant bacteria to protect indigenous soil β-subgroup proteobacterium ammonia oxidizers.
Metals, including cadmium, lead, and mercury, are, in most cases, microcidal; however, some bacteria have developed the ability to resist and detoxify these metals. Metal detoxification strategies, including those for cadmium, may include metal sequestration and precipitation (2, 10, 14, 26), which reduce soluble metal concentrations. Unlike organics, metals cannot be degraded, and thus most biological metal remediation approaches rely on the detoxification and immobilization of the metal both to reduce the biological toxicity and to retard metal transport.
The objective of this study was to determine the efficacy of dual bioaugmentation with metal-detoxifying and organic-degrading bacteria to facilitate organic degradation within cocontaminated systems. This objective was examined in coamended solution studies, in cocontaminated soils in the laboratory, and in a pilot field experiment. Four different cadmium-resistant bacterial isolates that did not degrade 2,4-dichlorophenoxyacetic acid (2,4-D) were tested for the ability to allow 2,4-D degradation to occur in the presence of toxic levels of cadmium, using Ralstonia eutropha JMP134 as the 2,4-D degrader.
MATERIALS AND METHODS
Bacterial strains.
Four highly cadmium-resistant soil bacteria were chosen for this study: Arthrobacter sp. strain D9, Bacillus sp. strain H9, Pseudomonas sp. strain H1, and Pseudomonas sp. strain I1a (Table 1). The isolation and characterization of these isolates have been described by Roane and Pepper (22). Cadmium-resistant bacteria were cultured on a defined mineral salts medium (MSM) amended with soluble cadmium as CdCl2 in concentrations from 0 to 45 μg ml−1 to represent concentrations observed at contaminated sites. The MSM contained the following: 0.5 g of sodium citrate (C6H5Na3O7), 0.1 g of magnesium sulfate (MgSO4- · 7H2O), 1.0 g of ammonium sulfate [(NH4)2SO4], 1.0 g of glucose (C6H12O6), and 0.1 g of sodium pyrophosphate [Na4P2O7(H2O)10], buffered to pH 6.0 with potassium phthalate (KHC8H4O4). In the 2,4-D biodegradation studies, a modified MSM was used wherein the glucose was replaced with 500 μg of 2,4-D ml−1, and 2-[N-morpholino]ethanesulfonic acid (C6H13NO4S) replaced potassium phthalate, which interfered with the 2,4-D absorbance readings.
TABLE 1.
Cadmium-resistant isolates and examined mechanisms of cadmium resistance
| Isolate | Cd MRLa (μg ml−1) | Size of plasmid (bp) | Presence of gene
|
EPS production | Intracellular accumulation | Extracellular accumulation | |
|---|---|---|---|---|---|---|---|
| cadA | cadC | ||||||
| Pseudomonas sp. strain I1a | 20 | NDb | − | − | Yes | No | Yes |
| Arthrobacter sp. strain D9 | 50 | ND | − | − | Yes | No | NEc |
| Pseudomonas sp. strain H1 | 225 | 18,504 | − | − | No | Yes | No |
| Bacillus sp. strain H9 | 275 | 10,408 | − | − | No | Yes | No |
Cadmium maximum resistance level.
ND, not detected.
NE, not examined.
All subsequent culturing took place in 25 ml of MSM amended with cadmium and incubated at 28°C on a rotary shaker at 180 rpm.
The maximum resistance level (MRL) was defined as the highest concentration of cadmium at which at least 104 cells ml−1 remained culturable after 48 h from an initial inoculation of 106 cells ml−1. The MRL of cadmium reflected the degree of resistance to cadmium. Cadmium concentrations were determined using a flame atomic absorption spectrophotometer following centrifugation at 10,000 × g for 20 min and filtration of the sample with a 0.2-μm-pore-size filter.
Ralstonia eutropha JMP134 (previously Alcaligenes eutrophus JMP134) contains the 80-kb pJP4 plasmid that codes for the degradation of 2,4-D to 3-oxoadipate (13). The degradation to succinic acid is in part mediated by chromosomally encoded enzymes (19, 25, 28).
Cadmium fate experiment.
Following individual inoculation with each of the isolates, any reduction in the amount of soluble cadmium in the broth was measured using atomic absorption. Thus, any reduction in bioavailable cadmium due to specific microbial interactions could be evaluated. In replicate flasks, each isolate was grown in 25 ml of MSM broth for 48 h at 28°C at various cadmium levels up to the MRL. Precipitated and cell-associated cadmium was collected following centrifugation at 10,000 × g for 20 min and acidified with 1 N HCl to solubilize the cadmium. An initial microscopic assessment was performed to confirm cell lysis upon acidification. Both the supernatant and the acidified cell suspension were examined for cadmium.
Mechanism of resistance to cadmium. (i) Detection of the Cad operon.
Primers developed by Endo and Silver (7) were used to detect the Cad operon, coding for a cadmium efflux pump. Chromosomal DNA was extracted using direct cell lysis at 98°C. The alkaline lysis procedure of Kado and Liu (12) was used to isolate and purify plasmid DNA.
Touchdown PCR with step annealing temperatures ranging from 54 to 67°C was used to amplify target sequences in lysed cell extracts and from plasmid DNA. The primer concentration used was 7.7 pmol per reaction. PCR products were run on a Tris-borate-EDTA–1.2% agarose gel at 100 V cm−1, stained with ethidium bromide (1 μg ml−1), and viewed under UV light.
(ii) Production of extracellular polymers.
Many microorganisms have extracellular polymeric layers that confer metal resistance. These polymeric layers are anionic in nature and thus attract and sequester cationic metals. The rapid screening method developed by Liu et al. (17) was used to screen the cadmium-resistant isolates for the production of two bacterial exopolysaccharides (EPSs), succinoglycan or galactoglucon (EPS II). The method relies on the differential staining of polymer-producing versus nonpolymer-producing organisms.
(iii) TEM.
Transmission electron microscopy (TEM) was used to assess morphologic changes in response to cadmium exposure. Bacteria (1.5 ml) grown in MSM (pH 6.0) containing 20 μg of cadmium ml−1 for Pseudomonas strain I1a and Arthrobacter strain D9 and 125 μg of cadmium ml−1 for Pseudomonas strain H1 and Bacillus strain H9 were harvested during logarithmic growth by pelleting at 14,000 × g for 2 min. The cells were rinsed in sterile deionized water and fixed in 3% glutaraldehyde in 0.1 M cacodylate buffer, pH 6.0, saturated with oxine (Sigma Inc.), using microwave fixation (8, 16) to minimize the cadmium leaching associated with traditional fixation. Oxine reacts specifically with heavy metals, increasing contrast under the TEM (30) for visual affirmation of metal deposits. Cells were postfixed in 2% osmium in 0.1 M cacodylate buffer, pH 6.0. Following fixation, cells were dehydrated using a reagent-grade ethanol-H2O gradient at each concentration: 30, 50, 70, 95, 100, 100, and 100% (11). Cells were infiltrated with Spurr resin at concentrations of 50% and then 100% (Ted Pella Inc., Redding, Calif.). Samples were polymerized overnight at 70°C, thin sectioned with an RMC MT-7000 Microtome (Research Manufacturing Corp., Tucson, Ariz.), and viewed at 60 kV with a Philips 420 TEM (Philips Electron Optics Inc., Mahwah, N.J.). Cadmium accumulation was confirmed using Noran Series Voyager II X3 elemental dispersive spectroscopy (Noran Instruments, Inc., Middleton, Wisc.).
(iv) Plasmid profiles and curing.
Since metal resistance can be plasmid encoded, the four isolates were examined for the presence of plasmids ranging in size from 2.6 to 350 MDa following incubation in the presence of 3 or 12 μg of cadmium ml−1 depending on the resistance level of the isolate. Plasmid extractions used the alkaline lysis procedure of Kado and Liu (12).
Cadmium-resistant isolates containing plasmids were cured of their plasmids using increased temperature and a nonselective medium. Isolates underwent four successive transfers (0.1 ml of isolates into 25 ml of broth) into nutrient broth (Difco, Baltimore, Md.) that did not contain cadmium, thereby removing any possible selection pressure. Isolates were incubated at 37°C on a rotary shaker at 180 rpm. Plasmid extractions were performed at the end of the four transfers. “Cured” isolates were then reinoculated into MSM with and without cadmium stress to reevaluate the MRL of each isolate to cadmium.
Degradation studies.
R. eutropha JMP134 was cadmium sensitive in this study in that at levels greater than 3 μg of cadmium ml−1, no viable R. eutropha cells were detected (<102 cells ml−1 from an initial inoculum of 104 cells ml−1). Note, however, that cadmium sensitivity is inoculant concentration dependent, and very high cell concentrations of R. eutropha JMP134 are more cadmium tolerant (K. L. Josephson, personal communication). Since degradation is often inhibited in the presence of metal(s), the ability of cadmium-resistant isolates to support 2,4-D degradation by R. eutropha JMP134 was examined. The degradation of 2,4-D by R. eutropha JMP134 was monitored in the presence of various cadmium levels upon inoculation with one of the cadmium-resistant isolates. Concentrations of 2,4-D in culture extracts were measured every 24 h at 230 nm following centrifugation at 14,000 × g for 2 min to remove cell debris. The relationship between the concentration of 2,4-D and its absorbance at 230 nm was linear, with a y value of 0.03x − 0.07 (r2 = 0.991).
(i) Pure culture.
In replicate pure culture experiments, 25 ml of MSM buffered to pH 6.0 with 2-[morpholino]ethanesulfonic acid (Sigma Inc.) was amended with 500 μg of 2,4-D ml−1 and either 12 or 24 μg of cadmium ml−1 depending on the MRL of each individual isolate. Each culture flask was inoculated with 104 CFU of either cadmium-resistant isolate Arthrobacter strain D9, Pseudomonas strain H1, Bacillus strain H9, or Pseudomonas strain I1a ml−1 and incubated at 28°C for 48 h at 180 rpm.
(ii) Soil microcosms.
Once established in pure culture, the abilities of successful isolates to protect R. eutropha JMP134 from cadmium toxicity were examined in artificially metal-contaminated soil. To determine if 2,4-D degradation could be facilitated in cadmium-contaminated soil, 100 g of an uncontaminated Brazito sandy loam soil was amended with 1% (wt/wt) glucose, 500 μg of 2,4-D ml−1, and 60 μg of cadmium ml−1 (final concentrations). Glucose was used as a readily metabolizable carbon source to support the cadmium-resistant populations. Soil microcosms (500-ml wide-mouth polypropylene jars) were incubated at 28°C and kept at 14% (wt/wt) soil moisture (75% of field capacity). Similar to the pure culture experiments, each soil microcosm was inoculated with one of the cadmium-resistant isolates (104 CFU g−1) by including the isolate with the initial moisture amendment (to 75% field capacity) followed by vigorous soil mixing. Following a 48-h incubation, appropriate microcosms were inoculated with 104 CFU of R. eutropha JMP134 g−1, again in conjunction with moisture amendment. Control microcosms consisted of soil amended with glucose, 2,4-D, and cadmium without inocula and soil amended with glucose, 2,4-D, and cadmium inoculated with only a cadmium-resistant isolate or R. eutropha JMP134.
Concentrations of 2,4-D in soil extracts were measured daily for a total of 50 days. One-to-ten soil slurries were made using 0.1% (wt/vol) sodium pyrophosphate to neutralize soil particle charge, centrifuged at 14,000 × g for 10 min, and read spectrophotometrically at 230 nm. Samples were analyzed in duplicate, and the soil microcosm experiment was performed three times. Soil samples without 2,4-D were used as blanks to subtract background absorbance, which was <1% of the total absorbance.
(iii) Field bioreactors.
Laboratory soil microcosm studies with the isolate Pseudomonas strain H1 were repeated at an intermediate field scale level (other isolates were not examined at the field scale). Pseudomonas strain H1 was chosen because of its cadmium resistance (to a concentration of 225 μg ml−1) and culturability. The Bacillus strain H9 isolate was not examined even though it was also highly resistant (to a concentration of 275 μg ml−1), so as to avoid complications resulting from spore formation. Bioreactors were set up under field conditions to confirm laboratory microcosm results. The field study was initiated in June 1998 and concluded in September 1998.
Five-gallon polypropylene bioreactors (45.7 by 76.2 cm), located at the University of Arizona Campbell Avenue Agricultural Station, Tucson, were placed under a constructed shaded area so as to preclude direct sunlight, since daytime temperatures were routinely in excess of 37.8°C (100°F). Each reactor contained approximately 27 kg of Brazito sandy loam at 14% (wt/wt) moisture content amended with 500 μg of 2,4-D g−1 and/or 60 μg of cadmium g−1. The Pseudomonas sp. strain H1 isolate and the 2,4-D degrader R. eutropha JMP134 were inoculated at 104 CFU g (dry weight) of soil−1.
Inoculants and soil amendments were thoroughly mixed into the soil using a cement mixer (rinsed with 70% ethanol between treatments) prior to the start of the experiment while providing a 48-h incubation period between inoculation with Pseudomonas strain H1 and addition of R. eutropha JMP134. Treatments were set up so as to minimize cross-contamination between the amendments, e.g., 2,4-D-only treatments followed by 2,4-D-plus-cadmium treatments followed by 2,4-D-plus-R. eutropha followed by Pseudomonas strain H1 and so forth. There were 7 treatments (see Table 4), each replicated twice for a total of 14 bioreactors. Soil moisture was maintained at 14% (wt/wt) throughout the experiment. Ambient air temperature ranged from 16.7°C (62°F) to 48.9°C (120°F). Soil cores (45 by 2.5 cm) were collected weekly and analyzed for 2,4-D concentrations. Background absorbance at 230 nm was monitored in reactors without 2,4-D amendment and was subtracted from each sample 2,4-D reading. To eliminate cross-contamination, the soil corer was disinfected with 10% bleach after each sample collection.
TABLE 4.
Comparison of 2,4-D degradation levels in field study treatments based on analysis of variation followed by Fisher's protected least-significant-difference pairwise comparison
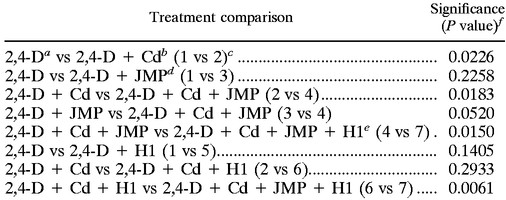 |
500 μg g of soil−1 (dry weight).
60 μg g of soil−1 (dry weight).
Treatment numbers are in parentheses.
JMP, R. eutropha JMP134 (2,4-D degrader).
H1, Pseudomonas strain (cadmium-resistant).
P values of ≤0.05 indicate 95% significance.
Culturable 2,4-D-degrading microorganisms were enumerated on an eosin methylene blue (EMB) medium developed by DiGiovanni et al. (5). The acidity produced during 2,4-D degradation caused the eosin blue to stain the colony dark purple. Previous studies in our laboratory have confirmed that the purple colony appearance is indicative of 2,4-D degradation (5). Some of the isolates from the EMB medium were further screened to identify possible transconjugants. The screening process included performing enterobacterial intragenic consensus PCR for genomic fingerprints (29) and PCR to amplify the tfdB gene found on pJP4 (5), a plasmid profile to detect the 80-kb pJP4 (12), and analysis of 2,4-D degradation using either high-pressure liquid chromatography or spectroscopy at 230 nm. Comparison of results from the screening process to those with R. eutropha JMP134 allowed transconjugant enumeration.
RESULTS
Resistance to cadmium.
As summarized in Table 1, the four isolates were resistant to a wide range of cadmium concentrations, from 20 to 275 μg ml−1. Only Pseudomonas strain H1 and Bacillus strain H9 had plasmids, of 18.5 and 10.4 kb, respectively. Upon plasmid curing, neither H1 nor H9 remained cadmium resistant, and they were unable to grow in the presence of 125 μg of cadmium ml−1 (levels approximately half of the MRLs), respectively. The Cad operon was not identified in any of the isolates.
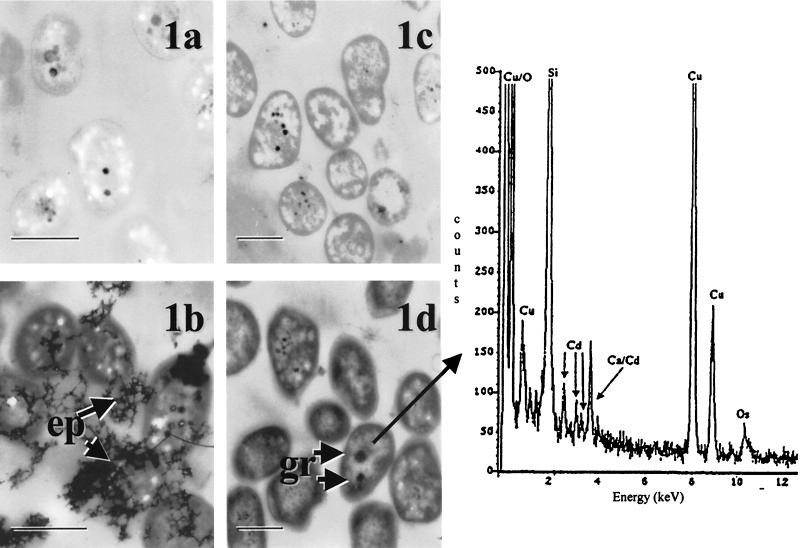
Observed mechanisms of cadmium resistance included extracellular and intracellular sequestration. Extracellular binding of cadmium was observed with Arthrobacter strain D9 and Pseudomonas strain I1a (data for I1a shown; Fig. 1a and b). After EPS production in isolate Pseudomonas strain I1a was confirmed with staining, the response of the isolate to 20 μg of cadmium ml−1 was observed under the TEM. Cadmium sequestration by the EPS layer was evident as a dark precipitate surrounding the cells (Fig. 1b).
FIG. 1.
TEM micrographs of Pseudomonas strain I1a in the absence of cadmium (a) and when exposed to 20 μg of cadmium ml−1 (b). Note the dark precipitate (ep) associated with the surrounding EPS layer (b), confirmed as cadmium by elemental X-ray analysis. TEM micrographs of Pseudomonas strain H1 in the absence of cadmium (c) and when exposed to 125 μg of cadmium ml−1 (d) are shown. Note the dense accumulations (gr), confirmed to be cadmium with elemental analysis. In both panels b and d, overall cellular density increased due to nonspecific metal binding. Bars equal 0.5 μm. In the spectrum provided, the copper (Cu) peaks were from the copper grid, the silicon (Si) peak was from the embedding medium Spurrs, and the osmium (Os) peak was from staining with OsO4. The cadmium (Cd) peaks confirmed the presence of cadmium.
In contrast, intracellular accumulation of cadmium in Pseudomonas strain H1 and Bacillus strain H9 cultures was observed (data for H1 shown; Fig. 1c and d). For Pseudomonas strain H1, under TEM and in the presence of 125 μg of cadmium ml−1, large intracellular accumulations of cadmium were confirmed with EDS (Fig. 1d). There was also an increase in cellular density indicative of nonspecific cadmium binding to the cells. Negative controls included cells grown in the absence of cadmium.
The effect of bacterial growth and metal resistance on cadmium solubility was also examined in a cadmium fate experiment (Table 2). The amount of cadmium present in solution decreased with isolates I1a, D9, H1, and H9 with growth from 104 to 108 CFU ml−1. The most dramatic decreases in levels of soluble cadmium were seen with Pseudomonas strain H1 and Bacillus strain H9, such that an average 36% was lost with growth. Growth of Arthrobacter strain D9 and Pseudomonas strain I1a resulted in 22 and 11% decreases in soluble cadmium. Based on controls with metal and no inocula, >99% of the total cadmium remained soluble.
TABLE 2.
The influence of microbial growth from 104 to 108 cells ml−1 on the solubility of cadmium in MSM broth, pH 6.0
| Total cadmium (μg/25 ml) | % Total cadmium remaining soluble (48 h postinoculation) with:
|
||||
|---|---|---|---|---|---|
| No inoculum | Pseudomonas strain H1 | Bacillus strain H9 | Arthrobacter strain D9 | Pseudomonas strain Ila | |
| 300 | 99.9 | 79 ± 0.4 | 85 ± 4.6 | ||
| 625 | 99.9 | 76 ± 2.5 | 58 ± 10.7 | ||
| 1,250 | 98.8 | 67 ± 4.0 | 75 ± 29.3 | ||
| 1,575 | 98.2 | 40 ± 0.0 | 35 ± 5.5 | ||
| 3,125 | 98.5 | 56 ± 5.0 | 68 ± 0.1 | ||
| 75 | 99.9 | NDa | 86 ± 1.9 | ||
| 150 | 99.9 | 82 ± 6.6 | 89 ± 4.2 | ||
| 300 | 99.9 | 86 ± 2.3 | 95 ± 2.0 | ||
| 625 | 99.9 | 84 ± 1.6 | 85 ± 6.5 | ||
| 950 | 99.9 | 62 ± 1.4 | ND | ||
ND, not determined.
Degradation studies.
Several experiments were conducted to determine the method of coinoculation. It was found that if a cadmium-resistant population and R. eutropha JMP134 were coinoculated at the same time, no degradation occurred and R. eutropha JMP134 was not recoverable. The same result was found if R. eutropha JMP134 was inoculated 24 h after the cadmium-resistant population was added to the cadmium–2,4-D culture medium. However, following a 48-h postinoculation with a cadmium-resistant population, the culture flasks inoculated with 104 CFU of R. eutropha JMP134 ml−1 did show degradation. A small inoculating biomass was used in both the pure culture and the soil experiments to assess the abilities of the inoculated populations to grow and perform under contaminated and, in the soil, nonsterile conditions. Augmentation with a smaller biomass in field scenarios can be desirable. It was also found that >105 cells ml−1 “artificially” decreased levels of soluble metal due to increased cell binding.
In order for the cadmium-sensitive 2,4-D degradation to occur in the presence of cadmium, bioavailable cadmium concentrations had to be detoxified. The abilities of the four cadmium-resistant soil isolates, Arthrobacter strain D9, Pseudomonas strain H1, Bacillus strain H9, and Pseudomonas strain I1a (Table 1), to detoxify cadmium such that R. eutropha JMP134 could degrade 500-μg ml−1 2,4-D was determined first in broth (Fig. 2). Experiments showed that 104 CFU of R. eutropha JMP134 ml−1 alone in the presence of >3 μg of cadmium ml−1 did not degrade 2,4-D, presumably because of cadmium toxicity. Additionally, none of the cadmium-resistant isolates could degrade 2,4-D (data not shown). Consequently, the dual-bioaugmentation approach was initially examined with MSM broth amended with 500 μg of 2,4-D and cadmium ml−1, wherein the cocontaminated broth was inoculated with 104 CFU of a cadmium-resistant isolate ml−1, incubated for 48 h at 28°C, and then reinoculated with 104 CFU of R. eutropha JMP134 ml−1. Complete 2,4-D degradation by R. eutropha JMP134 occurred in the presence of 12 μg of cadmium ml−1 with the cadmium-resistant isolate Pseudomonas strain I1a and 24 μg of cadmium ml−1 with the cadmium-resistant isolates Arthrobacter strain D9, Bacillus strain H9, and Pseudomonas strain H1. Interestingly, when the levels of soluble cadmium following growth of each of the cadmium-resistant isolates analyzed in Table 2 were examined, it seemed that the levels of cadmium solubility remained too high to support degradation of 2,4-D by R. eutropha JMP134. There may have been additional unidentified mechanisms of cadmium detoxification that did not reduce soluble cadmium concentrations but did render the cadmium nontoxic, as seen with chelation.
FIG. 2.
In broth, cadmium detoxification by isolates Arthrobacter strain D9, Bacillus strain H9, Pseudomonas strain H1, and Pseudomonas strain I1a allowed 2,4-D degradation by cadmium-sensitive R. eutropha JMP134 in the presence of 12 μg of cadmium ml−1 for isolate I1a and 24 μg of cadmium ml−1 for isolates D9, H1, and H9. Within 120 h, all isolates allowed the degradation of 500-μg ml−1 2,4-D to undetectable levels. Times as indicated were 48 h following inoculation with the cadmium-resistant isolate.
The uncontaminated soil used in the laboratory soil microcosms was a Brazito sandy loam with 12% clay, 0.21% organic matter, pH 8.2, and no known previous metal exposure. Indigenous microbial numbers in the soil were 3.2 × 107 ± 9.1 × 106 culturable CFU g−1 of dry weight on R2A medium (Difco, Baltimore, Md.) and 7.2 × 107 ± 3.2 × 106 total cells g−1 of dry weight as determined by acridine orange direct microscopic counts.
In laboratory soil microcosms, the cadmium-resistant isolates Pseudomonas strain H1, Bacillus strain H9, Arthrobacter strain D9, and Pseudomonas strain I1a appeared to detoxify cadmium, thereby protecting R. eutropha JMP134 from cadmium toxicity as 2,4-D degradation occurred in the presence of 60 μg of cadmium g−1. Soluble cadmium was not detectable in the amended soils; however, cadmium toxicity effects were observed in the contaminated soils as R. eutropha JMP134 2,4-D degradation was inhibited. Table 3 summarizes the specific rates of degradation for each isolate. Within 50 days, the cadmium-resistant Pseudomonas strains H1 and I1a allowed the complete degradation of 500-μg of 2,4-D ml−1. Upon addition of cadmium-resistant Bacillus strain H9 and Arthrobacter strain D9, degradation occurred within 35 days. Interestingly, neither the indigenous microbial flora nor the cadmium-resistant isolates could degrade 2,4-D in the cadmium-contaminated soil system within the 50-day time frame. Under the conditions of this experiment, R. eutropha JMP134 also did not degrade 2,4-D when cadmium was present. In Brazito soil amended only with 2,4-D, complete 2,4-D degradation by R. eutropha JMP134 occurred within 5 days. It should be noted that the Brazito soil used in this study was not sterile and consequently presented competitive challenges for the introduced organisms, and yet degradation still occurred in the cocontaminated soils upon inoculation with the cadmium-resistant isolates.
TABLE 3.
Degradation of 500-μg g−1 2,4-D by R. eutropha JMP134a in laboratory soil microcosms to undetectable levels with 60-μg g−1 cadmium and a cadmium-detoxifying isolate present
| Day | 2,4-D concn (μg g−1) with:
|
|||
|---|---|---|---|---|
| Pseudomonas strain I1a | Pseudomonas strain H1 | Bacillus strain H9 | Arthrobacter strain D9 | |
| 7 | 412 ± 123 | 400 ± 143 | 550 ± 19 | 500 ± 118 |
| 14 | 505 ± 7 | 550 ± 50 | 555 ± 59 | 538 ± 48 |
| 21 | 550 ± 0 | 500 ± 0 | 472 ± 39 | 158 ± 0 |
| 28 | 433 ± 10 | 495 ± 10 | 113 ± 0 | 146 ± 30 |
| 35 | 300 ± 50 | 289 ± 28 | 10 ± 0 | 10 ± 2 |
| 42 | 300 ± 35 | 238 ± 14 | NDb | ND |
| 50 | 10 ± 5 | 10 ± 12 | ND | ND |
R. eutropha JMP134 alone degraded the 500 μg of 2,4-D g−1 in the absence of cadmium to an undetectable level in 5 days.
ND, not detected.
We also tested the dual-bioaugmentation strategy in an intermediate field scale experiment. At the intermediate field scale, more variability was evident than in the bench-scale studies (Fig. 3). However, several conclusions can still be drawn. When 2,4-D was added to the Brazito soil without cadmium, slow rates of degradation ultimately occurred without bioaugmentation with R. eutropha JMP134 (Fig. 3a), as indigenous microbial populations acclimated to the 2,4-D. However, even after 10 weeks, degradation was incomplete, and 2,4-D levels did not decrease between weeks 5 and 10. The apparent degradation observed in the absence of inoculation in the field indicates the presence of a native population of 2,4-D-degrading microorganisms that were not detected in the soil microcosm experiments. In the presence of 60 μg of cadmium g−1, indigenous degradation appeared to be inhibited in the absence of bioaugmentation (Fig. 3b) even though 2,4-D degraders were culturable within the soil after 8 weeks. Under all treatment conditions, in the absence of R. eutropha JMP134 inoculation, 2,4-D-degrading organisms did not appear until week 8. The occurrence of these degraders may have been due to microsite variation in cadmium concentrations in soil or the use of alternate carbon sources available in the soil. In the laboratory, we have observed bacterial populations that were cadmium resistant and could degrade 2,4-D that were unable to resist cadmium and degrade 2,4-D concurrently, possibly due to the energy demand placed on the organism. Consequently, the indigenous 2,4-D degraders may not have been able to degrade 2,4-D in the presence of the cadmium, since the EMB medium used to select for 2,4-D-degrading organisms did not contain cadmium. In the reactors inoculated with R. eutropha JMP134, the number of 2,4-D-degrading organisms fell from the inoculated 104 CFU of R. eutropha JMP134 g−1 to <102 CFU of 2,4-D-degrading organisms g−1 during weeks 1 through 3. By week 4 or 5, however, the number of 2,4-D degraders increased dramatically, to 107 CFU g−1 (Fig. 3c to e).
FIG. 3.
The degradation of 500-μg g−1 2,4-D and the appearance of 2,4-D-degrading microbial populations with time in weeks, as detected in pilot field scale bioreactors containing Brazito soil amended with 2,4-D only (Treatment 1) (a); 2,4-D and 60 μg of cadmium g−1 (Treatment 2) (b); 2,4-D and 104 CFU of R. eutropha JMP134 g−1 (Treatment 3) (c); 2,4-D, 60 μg of cadmium g−1, and 104 CFU of R. eutropha JMP134 g−1 (Treatment 4) (d); 2,4-D, 60 μg of cadmium g−1, 104 CFU of R. eutropha JMP134 g−1, and 104 CFU of cadmium-resistant Pseudomonas strain H1 g−1 (Treatment 7) (e). The original number of 2,4-D degraders was fewer than 102 CFU g−1 of soil prior to bioaugmentation. Treatment 5 (2,4-D and Pseudomonas strain H1) and Treatment 6 (2,4-D, 60 μg of cadmium g−1, and Pseudomonas strain H1) are not shown.
In Treatment 3 (Fig. 3c), with 2,4-D and R. eutropha JMP134, as observed in all the reactors inoculated with R. eutropha JMP134, a 2,4-D-degrading population was evident by week 5 (105 CFU g−1). The concentration of 2,4-D in Treatment 3 was reduced to 300 mg kg−1 in the first 4 weeks and then remained stable, indicative of incomplete degradation or slow rates of degradation. Interestingly, degradation occurred prior to the appearance of 2,4-D degraders, probably due in part to some nonculturable degraders on the EMB medium. In Treatment 4 reactors (cadmium and R. eutropha JMP134; Fig. 3d), degradation was less apparent, and even though by week 6, 107 2,4-D degraders g−1 could be recovered, cadmium appeared to have an effect on 2,4-D degradation. The variation in 2,4-D degradation observed in Treatment 4 and Treatment 2 correlated with the cadmium amendment that resulted in sporadic degradation due to microscale toxicity effects. As expected from both the pure culture and soil microcosm experiments, cadmium-resistant Pseudomonas strain H1 (Treatment 5 and Treatment 6) did not degrade or facilitate the increased degradation (above background levels seen in Treatment 1) of 2,4-D within the 70-day time frame of the field study, regardless of whether cadmium was present or not.
In Treatment 7 reactors (2,4-D, cadmium; R. eutropha JMP134 and Pseudomonas strain H1), the extent of degradation was noticeably enhanced upon addition of the coinoculants (Fig. 3e). As observed in the soil microcosms, the reactors with 2,4-D and cadmium, coinoculated with cadmium-resistant Pseudomonas strain H1 and R. eutropha JMP134, exhibited substantial degradation in conjunction with the appearance of >106 CFU of 2,4-D-degrading organisms g−1 of dry weight, suggesting that Pseudomonas strain H1 conferred a protective effect. Degradation to 100 mg of 2,4-D kg−1 occurred by week 6 in conjunction with the appearance of 107 2,4-D degraders g−1. Thus, it appears that initial inoculation with the cadmium-resistant isolate Pseudomonas strain H1 detoxified the cadmium. Interestingly, an estimated 90% of the recovered 2,4-D-degrading organisms were strains other than R. eutropha JMP134.
Mean values for duplicate 2,4-D readings at each time point for weeks 5 through 10 where 2,4-D degradation occurred were used for analysis of variation followed by Fisher's protected least-significant-difference pairwise comparison between treatments (Table 4). The finding of no significant difference between the 2,4-D (Treatment 1) and 2,4-D plus R. eutropha JMP134 (Treatment 3) treatments indicated that indigenous microbial populations were capable of some 2,4-D degradation. This was surprising, since no indigenous degradation was observed in the soil microcosms. However, in both soil microcosms and field bioreactors, cadmium inhibited both indigenous degradation and that by R. eutropha JMP134 (Treatment 2 and Treatment 4, respectively). With a P value of ≤0.05 taken to be 95% confidence, there was not a significant difference in degradation rates between the R. eutropha JMP134 augmented reactors with and without cadmium (P = 0.052). However, cadmium did affect the degradation by increasing the variability of the readings, and in Treatment 7 (Fig. 3d), the addition of Pseudomonas strain H1 significantly increased the rate of degradation of 2,4-D (P = 0.015). Significant degradation was observed in treatments with both Pseudomonas strain H1 and R. eutropha JMP134 (Treatment 4 versus Treatment 7 and Treatment 6 versus Treatment 7). No degradation was observed in treatments with Pseudomonas strain H1 alone (Treatment 1 versus Treatment 5 and Treatment 2 versus Treatment 6).
DISCUSSION
This study has demonstrated the use of a dual-bioaugmentation strategy in the remediation of cocontaminated systems. This strategy involved the coinoculation of a metal-resistant microbial population with an organic-degrading population, the primary mode of action being metal detoxification, such that organic degradation was no longer inhibited. Based on promising results in laboratory experiments with both pure culture and soil microcosms, we examined this strategy in a field trial. The rates of 2,4-D degradation by R. eutropha JMP134 in the presence of Pseudomonas strain H1 were surprisingly similar in both the laboratory soil microcosms and in the field study (approximately 50 days), though 2,4-D degradation was not complete in the field study. Also interesting was the observation that many of the 2,4-D-degrading isolates preliminarily examined for the pJP4 plasmid recovered in the field experiment were not R. eutropha JMP134. This and the observation that indigenous 2,4-D degradation was not significant with cadmium suggested that transfer of the pJP4 plasmid to indigenous populations occurred. The field study did demonstrate that bioaugmentation using coinoculants is a viable option for the remediation of metal- and organically contaminated soils.
While the Cad operon was not found in any of the isolates, this does not exclude the presence of a cadmium-efflux system; however, the isolates examined reduced soluble cadmium concentrations, indicating the use of an alternative mechanism of resistance. Since soluble metal is thought to be more toxic than bound or precipitated metal, each of the four isolates effectively reduced cadmium toxicity. Of the four isolates, Pseudomonas strain H1 and Bacillus strain H9 appeared to use an intracellular mechanism of cadmium sequestration. While intracellular microbial cadmium accumulation has not been well documented, the observed increase in outer membrane density upon exposure to cadmium indicated binding of metal to lipopolysaccharides, as found by Landley and Beveridge (15). Metallothionein production and polyphosphate precipitation represent two possible explanations wherein cadmium is sequestered intracellularly. However, the precise mechanism of intracellular accumulation of cadmium merits further investigation.
The two other cadmium-resistant isolates, Pseudomonas strain I1a and Arthrobacter strain D9, showed evidence of EPS production upon staining and under the TEM showed cadmium accumulation external to the cells. Similarly, a study by Roane (21) found that EPS production resulted in extracellular lead sequestration. Metal binding to exopolymers is known to reduce metal toxicity. While generally associated with adhesion and protection against desiccation, exopolymers act as strong ionic attractants and, thus, readily bind metals.
It was interesting that the two most cadmium-resistant isolates were Pseudomonas strain H1 (resistant up to 225 μg ml−1) and Bacillus strain H9 (resistant up to 275 μg ml−1), which both exhibited intracellular cadmium accumulation. The resistance mechanisms of these two organisms were also plasmid encoded. Finally, the most dramatic decreases in soluble cadmium upon growth were seen with Pseudomonas strain H1 and Bacillus strain H9, in that there was a 36% loss in soluble cadmium with growth. Arthrobacter strain D9 and Pseudomonas strain I1a showed less detoxification, with a resulting decrease of 22 and 11% in soluble cadmium with growth.
This study found that while dual bioaugmentation with metal-detoxifying and organic-degrading microbial populations is effective at cocontaminant remediation, time must be allowed for metal detoxification to occur before organic degradation is observed. Evidence of this was observed in the 48 h needed between inoculation with the metal-detoxifying population and inoculation with the organic-degrading population. We found that the viability of the organic-degrading population decreased when it was added to the system prior to metal detoxification. Only using this staggered approach to bioaugmentation was remediation of a cocontaminated soil successful.
ACKNOWLEDGMENTS
This work was supported in part by grant no. 5 P42 ESO4940-07 from the National Institute of Environmental Health Sciences, Superfund Program, grant no. DE-FG03-97-ER62470 from the U.S. Department of Energy, Joint Program on Bioremediation, and by a graduate fellowship from the U.S. Environmental Protection Agency STAR Program.
We thank David Bentley of the University of Arizona Imaging Facility for his assistance with the transmission electron microscopy and Scot Dowd for his assistance with primer development. Special thanks to Christine Stauber and Miriam Eaton for their assistance during the field study.
REFERENCES
- 1.Crowley D E, Brennerova M V, Irwin C, Brenner V, Focht D D. Rhizosphere effects on biodegradation of 2,5-dichlorobenzoate by a bioluminescent strain of root-colonizing Pseudomonas fluorescens. FEMS Microbiol Ecol. 1996;20:79–89. [Google Scholar]
- 2.Cunningham D P, Lundie L L., Jr Precipitation of cadmium by Clostridium thermoaceticum. Appl Environ Microbiol. 1993;59:7–14. doi: 10.1128/aem.59.1.7-14.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daane L L, Haggblom M M. Earthworm egg capsules as vectors for the environmental introduction of biodegradative bacteria. Appl Environ Microbiol. 1999;65:2376–2381. doi: 10.1128/aem.65.6.2376-2381.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dejonghe W, Goris J, El Fantroussi S, Hofte M, De Vos P, Verstraete W, Top E M. Effect of dissemination of 2,4-dichlorophenoxyacetic acid (2,4-D) degradation plasmids on 2,4-D degradation and on bacterial community structure in two different soil horizons. Appl Environ Microbiol. 2000;66:3297–3304. doi: 10.1128/aem.66.8.3297-3304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiGiovanni G D, Neilson J W, Pepper I L, Sinclair N A. Gene transfer of Alcaligenes eutrophus JMP134 plasmid pJP4 to indigenous soil recipients. Appl Environ Microbiol. 1996;62:2521–2526. doi: 10.1128/aem.62.7.2521-2526.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Fantroussi S, Belkacemi M, Top E M, Mahillon J, Naveau H, Agathos S N. Bioaugmentation of a soil bioreactor designed for pilot-scale anaerobic bioremediation studies. Environ Sci Technol. 1999;33:2992–3001. [Google Scholar]
- 7.Endo G, Silver S. CadC, the transcriptional regulatory protein of the cadmium resistance system of Staphylococcus aureus plasmid pI258. J Bacteriol. 1995;177:4437–4441. doi: 10.1128/jb.177.15.4437-4441.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibberson R, Demare R S, Jr, Nordhausen R W. Four-hour processing of clinical/diagnostic specimens for electron microscopy using a microwave technique. J Vet Diagn Investig. 1997;9:61–67. doi: 10.1177/104063879700900111. [DOI] [PubMed] [Google Scholar]
- 9.Gieger G, Federer P, Sticher H. Reclamation of heavy metal-contaminated soils: field studies and germination experiments. J Environ Qual. 1993;22:201–207. [Google Scholar]
- 10.Gupta A, Whitton B A, Morby A P, Huckle J W, Robinson N J. Amplification and rearrangement of a prokaryotic metallothionein locus SMT in Synechococcus PCC-6301 selected for tolerance to cadmium. Proc R Soc Lond Ser B Biol Sci. 1992;248:273–281. doi: 10.1098/rspb.1992.0072. [DOI] [PubMed] [Google Scholar]
- 11.Hayat M A. Electron microscopy: biological applications. Boca Raton, Fla: CRC Press; 1989. [Google Scholar]
- 12.Kado C I, Liu S-T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasberg T, Daubaras D L, Chakrabarty A M, Kinzelt D, Reineke W. Evidence that operons tcb, tfd, and clc encode maleylacetate reductase, the fourth enzyme of the modified ortho pathway. J Bacteriol. 1995;177:3885–3889. doi: 10.1128/jb.177.13.3885-3889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurek E, Francis A J, Bollag J-M. Immobilization of cadmium by microbial extracellular products. Arch Environ Contam Toxicol. 1991;20:106–111. [Google Scholar]
- 15.Landley S, Beveridge T J. Effect of O-side-chain-lipopolysaccharide chemistry on metal binding. Appl Environ Microbiol. 1999;65:489–498. doi: 10.1128/aem.65.2.489-498.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindley V A. A new procedure for handling impervious biological specimens. Microsc Res Tech. 1992;21:355–360. doi: 10.1002/jemt.1070210411. [DOI] [PubMed] [Google Scholar]
- 17.Liu M, Gonzalez J E, Willis L B, Walker G C. A novel screening method for isolating exopolysaccharide-deficient mutants. Appl Environ Microbiol. 1998;64:4600–4602. doi: 10.1128/aem.64.11.4600-4602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newby D T, Gentry T J, Pepper I L. Comparison of 2,4-dichlorophenoxyacetic acid degradation and plasmid transfer in soil resulting from bioaugmentation with two different pJP4 donors. Appl Environ Microbiol. 2000;66:3399–3407. doi: 10.1128/aem.66.8.3399-3407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perkins E J, Gordon M P, Caceres O, Lurquin P F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990;172:2351–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riley R G, Zachara J M, Wobber F J. Chemical contaminants on DOE lands and selection of contaminant mixtures for subsurface science research. U.S. Department of Energy publication no. DOE/ER-0547T. U.S. Washington, D.C.: Department of Energy; 1992. [Google Scholar]
- 21.Roane T M. Lead resistance in two bacterial isolates from heavy metal-contaminated soils. Microb Ecol. 1999;37:218–224. doi: 10.1007/s002489900145. [DOI] [PubMed] [Google Scholar]
- 22.Roane T M, Pepper I L. Microbial responses to environmentally toxic cadmium. Microb Ecol. 2000;38:358–364. doi: 10.1007/s002489901001. [DOI] [PubMed] [Google Scholar]
- 23.Roane T M, Pepper I L, Miller R M. Microbial remediation of metals. In: Crawford R L, Crawford D L, editors. Bioremediation: principles and applications. Cambridge, United Kingdom: Cambridge University Press; 1996. pp. 312–340. [Google Scholar]
- 24.Said W A, Lewis D A. Quantitative assessment of the effects of metals on microbial degradation of organic chemicals. Appl Environ Microbiol. 1991;57:1498–1503. doi: 10.1128/aem.57.5.1498-1503.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Short K A, Doyle J D, King R J, Seidler R J, Stotzky G, Olsen R H. Effects of 2,4-dichlorophenol, a metabolite of a genetically engineered bacterium, and 2,4-dichlorophenoxyacetate on some microorganism-mediated ecological processes in soil. Appl Environ Microbiol. 1991;57:412–418. doi: 10.1128/aem.57.2.412-418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silver S, Phung L T. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 27.Stephen J R, Chang Y J, Macnaughton S J, Lowalchuk G A, Leung K T, Flemming C A, White D C. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl Environ Microbiol. 1999;65:95–101. doi: 10.1128/aem.65.1.95-101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Top E M, Holben W E, Forney L J. Characterization of diverse 2,4-dichlorophenoxyacetic acid-degradative plasmids isolated from soil by complementation. Appl Environ Microbiol. 1995;61:1691–1698. doi: 10.1128/aem.61.5.1691-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Versalovic J, Schneider M, de Bruijn F J, Lupski J R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol. 1994;5:25–40. [Google Scholar]
- 30.Yoshizuka M, McCarthy K J, Kaye G I, Fujimoto S. Cadmium toxicity to the cornea of pregnant rats: electron microscopy and X-ray microanalysis. Anat Rec. 1990;227:138. doi: 10.1002/ar.1092270116. [DOI] [PubMed] [Google Scholar]