Abstract
Fibroblastic reticular cells (FRCs) are specialized stromal cells of lymphoid organs that generate the structural foundation of the tissue and actively interact with immune cells. Distinct FRC subsets position lymphocytes and myeloid cells in specialized niches where they present processed or native antigen and provide essential growth factors and cytokines for immune cell activation and differentiation. Niche‐specific functions of FRC subpopulations have been defined using genetic targeting, high‐dimensional transcriptomic analyses, and advanced imaging methods. Here, we review recent findings on FRC‐immune cell interaction and the elaboration of FRC development and differentiation. We discuss how imaging approaches have not only shaped our understanding of FRC biology, but have critically advanced the niche concept of immune cell maintenance and control of immune reactivity.
Keywords: cell‐fate mapping, fibroblastic reticular cells, immune cell niches, lineage tracing, transgenic mouse models
1. THE FIBROBLASTIC RETICULAR CELL NETWORK—A BRIEF HISTORY
Fibroblasts of lymphoid organs, also known as fibroblastic reticular cells (FRCs), build and maintain microenvironmental niches that host and nurture innate and adaptive immune cells. 1 , 2 FRCs generate the three‐dimensional fiber and conduit channel networks that form the physical scaffold of secondary lymphoid organs (SLOs) and contribute to the fluid homeostasis of the tissue. 3 The regional expression of cytokines by FRCs guides the migration and interaction of different immune cell populations in SLOs. 4 Specialized FRC subsets provide growth factors to maintain distinct immune cell populations in dedicated niches and regulate immune cell interactions by on‐demand supply of differentiation factors. 5 FRC subset definition and functional characterization have been fostered through the application of single cells transcriptomics analyses of FRCs in murine SLOs. 6 , 7 , 8 , 9 , 10 , 11 In broad strokes, the conserved zonal segregation of SLOs is underpinned by four major FRC subsets: marginal reticular cells (MRCs) line epithelial or endothelial barriers and form antigen‐sampling zones; B‐cell zone reticular cell (BRC) networks generate environments for the generation of humoral immunity and include follicular dendritic cells (FDCs) that present native antigen to B cells; T‐cell zone reticular cells (TRCs) define areas for T cell activation and differentiation and foster the interaction of T cells with dendritic cells (DCs); perivascular reticular cell (PRCs) are connected to blood vessels and possess the potential to act as embryonic and adult FRC progenitors. Despite their phenotypical and functional diversity, all FRC subsets show a conserved pattern of extracellular matrix (ECM) deposition and spindle‐like morphology with the formation of dendritic protrusions and veil‐like extensions, which are required to provide the fibrous scaffold and to generate liquid‐ and ECM‐free spaces for direct membrane interactions with and between immune cells. 3 , 12 In this review, we focus on the immune‐anatomy underlying FRC subset definition and the elaboration of functional properties through the combination of imaging and genetic FRC targeting. Since the field of stromal (FRC)‐immune cell interaction has only recently gained substantial traction and the nomenclature still needs to be consolidated, a short overview on the history of key discoveries in FRC biology and the connection with the advancement of imaging techniques is warranted.
The first descriptions of “reticular cells” in B cell follicles of spleens and lymph nodes in rats have been published in the early 1950s. It had been noted that a network of channels and chords, termed the reticulo‐endothelial system, exists in lymphoid organs. The reticulo‐endothelial system could be highlighted by the deposition of intravenously injected proteins, suggesting that the involved cells had a function in the retention of antigens. 13 Such experimental approaches became feasible with the availability of fluorophores, such as fluorescein isothiocyanate, and the covalent coupling to antibodies directed against bovine albumin that enabled immunohistochemical detection of antigen deposition in lymphoid organs. 13 The recording of the antigen deposition in the reticulo‐endothelial system was facilitated with the concomitant development of innovative sectioning techniques, such as dry ice‐cooled microtomes for the production of native thin sections, and the first fluorescence microscopes. The initial interpretation drawn from the deposition of bacterial polysaccharides in subcapsular and follicular regions of murine lymph nodes was that the antigen is retained by macrophages and can transiently attach to lymphocytes. 13 , 14 More than 10 years later, tracing of Salmonella adelaide‐derived flagellar antigens in rat lymph nodes revealed a cellular reticulum in follicles, which trapped the antigen on reticulin fibers. 15 Further studies using radioactive Iodine‐labeled Salmonella‐derived flagellin as antigen revealed the connection between antigen retention on (“dendritic”) reticular cells in primary and secondary follicles/germinal centers that appear during B cell memory responses. 16 Additional seminal studies by Nossal and colleagues 17 revealed the discrete distribution patterns of radioactively labeled antigen on reticular networks in the center of primary follicles and more superficial (perifollicular) aspects of secondary follicles (Figure 1A). The authors speculated that the antigen‐retaining reticular networks were generated by “dendritic macrophages,” an interpretation that was further promoted in studies using the detection of radioactive iodine‐labeled flagellar antigen by transmission electron microscopy. 18 Antigen was identified near the surface of fine cell processes belonging to branches of “dendritic follicular reticular cells”. The authors also observed that bona fide “tingible body” macrophages of germinal centers did not participate in antigen retention. 18 The most commonly used term for antigen‐retaining FRCs in the germinal center still follows the nomenclature established in the 1980s, that is, follicular dendritic cells (FDCs) 19 (Figure 1A,B). Later studies revealed that FDCs express surface proteins shared with other lymphoid organ fibroblasts, 20 that they are resistant to radiation and not replaced by bone marrow‐derived cells, 21 and that they originate from perivascular myofibroblastic progenitors. 22 The initial (and still prominent) confusion of one of the key FRC subsets as a dendritic (myeloid) cell population is indicative for the eminent function of lymphoid organ fibroblasts in immunological processes.
FIGURE 1.
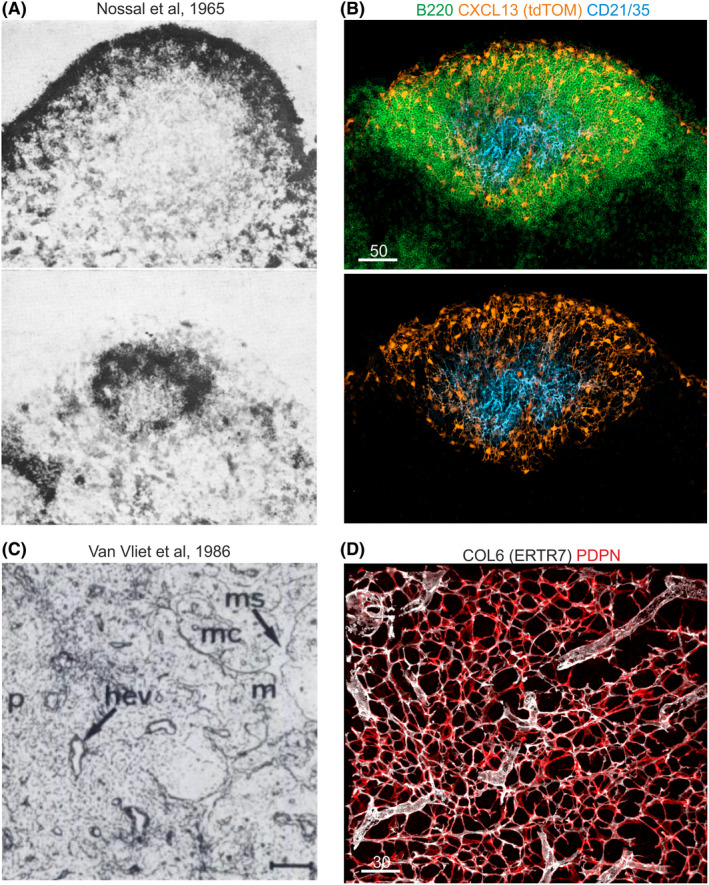
Fibroblastic reticular cell networks. (A) Nossal et al 17 have analyzed the retention of flagellar antigens labeled with radioactive Iodine (125I) in primary and secondary follicles of rat lymph nodes. Scintillation counting and autoradiography highlighted a reticular cell network, which retains antigen early in perifollicular (upper image) and later in the center of the follicle (lower image). Reprinted with the kind permission from Immunology (John Wiley & Sons). (B) Cxcl13 promoter activity revealed by the Cxcl13‐Cre/tdTomato model as fluorescent staining in B cell zone reticular cell networks that underpin subcapsular and perifollicular regions of mouse lymph nodes. Central follicles exhibit follicular dendritic cells (FDCs), which express the Cxcl13‐tdTomato transgene and complement receptors 1 and 2 (CD21/35). (C) Staining of mouse lymph node fibroblasts by Van Vliet et al 24 using the ER‐TR7 antibody that binds to the extracellular matrix component collagen type VI and stains the reticular fibroblast network in paracortical (p) and medullary (m) regions, surrounding medullary sinuses (ms) and within medullary chords (mc). Reprinted with the kind permission from The Journal of Histochemistry and Cytochemistry (SAGE Publications). (D) ER‐TR7 staining of the murine lymph node T cell zone marks the fibrillar core of the microfluidic conduit network, which is produced by podoplanin (PDPN)‐expressing T cell zone reticular cells
Research into the non‐hematopoietic cellular components that contribute to the spatial organization of lymphoid organs and interact with immune cells increased during the 1970s and precipitated the generation of antibodies that recognize proteins expressed by lymphoid organ fibroblasts (Table 1). For example, the antibody Erasmus University Rotterdam‐thymic reticulum antibody 7 (ER‐TR7) enabled specific staining of reticular networks in the splenic white pulp and the T cell zone of lymph nodes 23 , 24 (Figure 1C,D). The ER‐TR7 antibody recognizes collagen type VI 25 and facilitates hence the visualization of the extracellular matrix organization generated by the FRC network. Improved and refined immunohistochemistry protocols and the acquisition of high resolution images by laser scanning confocal microscopy and transmission electron microscopy revealed that the reticular fiber network, that is generated and ensheathed by FRCs, serves as a conduit for soluble factors and antigens. 26 , 27 , 28 FRCs have been further characterized through the expression of the chemokines CCL19 and CCL21 29 supporting the view that FRCs tightly interact with migrating CCR7‐positive T cells and dendritic cells in the T cell zone. CXCL13 was identified as B cell attractant, secreted by fibroblasts underpinning lymph node follicles (ie, FDCs). 30 Intravital two‐photon microscopy experiments revealed that T and B cells migrate on TRC and BRC networks of murine lymph nodes. 21 The importance of FRC‐lymphocyte interaction was demonstrated by PDPN‐positive FRC‐mediated improvement of T cell survival in murine lymph nodes, 31 FRC‐dependent coordination of lymph node remodeling following viral infection, 32 and the immune‐regulatory role of FRC‐derived nitric oxide. 33 , 34 The diverse functions of FRCs during immune homeostasis and activation indicate that such specialization depends on the generation of specialized microenvironmental niches for coordinated immune cell interaction and maintenance. The elaboration of FRC‐generated immune niches was supported by the establishment of genetic tools to visualize and functionally manipulate FRC subsets in vivo.
TABLE 1.
Markers, cellular targets, and basic methodological parameters for the imaging‐based characterization of murine lymphoid organ fibroblasts
| Marker a | Organs | FRC subset b | Methodological details c | References | ||
|---|---|---|---|---|---|---|
| Sectioning | Fixation | Detection | ||||
| ACTA2 | LN, SWP, PP | All, but FDC | C, V | PFA | IF | 9, 11, 31, 52 |
| CCL21 / Ccl21a | LN, SWP, PP | TRC, TBRC | C, V | PFA | IF, IHC | 9, 11, 117, 118 |
| LN | C, V | PFA | RNA | 88, 104 | ||
| CXCL13 / Cxcl13 | LN | BRC, MedRC | V | PFA | IF | 8, 119 |
| LN | C | PFA | RNA | 88 | ||
| SWP, PP | BRC | V | PFA, Ace | IF | 9, 120, 121 | |
| CD34 | LN | MedRC | C | PFA | IF | 6 |
| PP | TRC1 | V | PFA | IF | 11 | |
| CD35 (CR1) | LN, SWP, PP | FDC, LZ‐FDC | C, V | PFA, Ace | IF, IHC | 11, 22, 122 |
| CD157 | LN, SWP, PP | All, but MedRC, PRC | C | Ace | IF | 123 |
| Ch25h | LN | IFRC, TBRC | C | PFA/EtOH | RNA | 6, 100 |
| COL1 | LN, PP | All, but FDC | C, V | PFA | IF | 11, 88 |
| COL6 | LN, PP | All, but BRC | C, V | PFA | IF | 21, 52, 124 |
| SWP | TRC, MRC | C | PFA | IF | 125, 126 | |
| DESMIN | LN, SWP, PP | All subsets | C, V | PFA | IF | 21, 52, 127 |
| FcɛR2A | LN | LZ‐FDC | V | PFA | IF | 8 |
| ICAM1 | LN, SWP, PP | All, but PRC | C, V | Ace | IF | 123 |
| Inmt | LN | MedRC | C | PFA | RNA | 6 |
| MADCAM1 | LN, SWP | MRC | V | PFA | IF | 8, 9 |
| PP | MRC, FDC, TBRC | V | PFA | IF | 11 | |
| MYH11 | LN | LZ‐FDC | V | PFA | IF | 8 |
| LAMININ | SWP, PP | All subsets | C | PFA | IF | 124, 128 |
| LEPR | LN | All subsets | C | PFA | IF | 118 |
| LUMICAN | LN | MedRC | V | PFA | IF | 8 |
| PP | TRC, TBRC | V | PFA | IF | 11 | |
| PDGFRβ | LN | All, but BRC | C | PFA | IF | 129 |
| PDLIM3 | LN | DZ‐FDC | V | PFA | IF | 8 |
| PDPN | LN, PP | All, but PRC | C, V | PFA | IF | 52 |
| SWP | TRC | V | PFA | IF | 9 | |
| Pthlh | LN | FDC | C | PFA | RNA | 6 |
| TNFSF11 (RANKL) | LN | MRC, IFRC | C, V | PFA | IF | 6, 8 |
| SWP, PP | MRC | C, V | PFA, AF | IF | 11, 130, 131 | |
| TMEM119 | LN | FDC | C | PFA | IF | 6 |
| VCAM1 | LN, SWP, PP | All, but PRC | C, V | PFA, Ace | IF | 120, 123 |
Murine lymphoid organ fibroblasts can be detected in situ using either protein or mRNA expression.
FRC subsets in murine lymphoid organs include marginal zone reticular cells (MRC), interfollicular reticular cells (IFRC), light zone follicular dendritic cells (LZ‐FDC), dark zone follicular dendritic cells (DZ‐FDC), T‐B border reticular cells (TBRC), B cell zone reticular cells (BRC) comprising MRCs, FDCs, TBRCs and IFRCs, T cell zone reticular cells (TRC), perivascular reticular cells (PRC), and medullary reticular cells (MedRC).
Methodological details for the imaging of the respective FRC subset(s) in the indicated tissues according to the listed references include sectioning (cryo‐sectioning (C) or vibratome‐sectioning (V)), fixation (paraformaldehyde (PFA), paraformaldehyde and post‐fixation with ethanol (PFA/EtOH), acetone fixation (Ace), or Antigenfix (AF)), and the detection method (immunofluorescence (IF), immunohistochemistry (IHC), RNAscope in situ hybridization with nucleotide probes (RNA).
2. GENETIC TRACING OF LYMPHOID ORGAN FIBROBLASTS
The array of antibodies listed in Table 1 has enabled refined immunohistochemical definition of FRCs and has opened pathways for flow cytometric analyses and the selection of markers that permit genetic targeting of different FRC populations. The knowledge about FRC‐specific protein expression has helped to identify a number of promoters that drive the expression of real‐time reporters and/or facilitate Cre‐mediated genetic recombination, thereby paving the way for studies on origin, phenotype, and function of FRCs.
2.1. Visualization of FRCs in the tissue context
The elaboration of FRC biology requires the visualization of FRCs and interacting immune cells in situ. Microscopic analysis relies on the optimal preservation of tissue structure during sample preparation, which includes dissection of the native tissue, preservation, and fixation procedures as part of stringent workflows (Figure 2A). For optimal imaging of FRCs in different tissue contexts, the consecutive steps in the overall process should be optimized. The best‐suited tissue fixation strategy, that is, chemical or physical fixation, depends on the particular antigen‐antibody interaction because different fixatives alter the antibody‐binding site. Nevertheless, the use of chemical fixation with protein‐crosslinking agents such as paraformaldehyde or glutaraldehyde has become the standard for the 3‐dimensional assessment of FRC networks (Figure 2B,C). High‐purity paraformaldehyde generally preserves SLO tissue size, structure, and antigen‐binding sites for the majority of FRC markers (Table 1), although concentrations between 1% and 4% (w/v), incubation temperature and time have to be optimized empirically for different staining protocols. In addition, paraformaldehyde‐based fixation procedures allow for the production of thick tissue sections using standard sectioning procedures such as the vibratome. Agarose embedding of paraformaldehyde‐fixed tissues at room temperature followed by vibratome‐sectioning preserve tissue integrity and thereby improve the resolution of intracellular structures and FRC‐immune cell interactions (Figure 2D,E). The visualization and quantitative examination of 3‐dimensional FRC network topologies require the definition of imaging volumes before sectioning. The approximate size of a single murine FRC ranges between 10 and 20 µm, depending on the FRC subtype. Hence, the topological examination of FRC networks, for example in the T cell zone, requires at least 30 µm thick sections. Since human cells are bigger than their murine counterparts, the analysis of human lymphoid organ fibroblasts generally requires the acquisition of even larger volumes. Future studies will most likely focus on the 3‐dimensional positioning of multiple FRC communities to resolve the interconnection between different FRC subsets, comparable to the analysis of neuronal networks in the central nervous system. 35 Thus, an important step toward the goal of high‐resolution analyses of extended FRC networks in murine and human SLOs is the acquisition of the whole organ (Figure 2F). Different tissue clearing procedures have been developed to minimize light absorption and scattering within the tissue in order to make large tissue volumes accessible for optical imaging. 36 Most of these protocols, such as 3DISCO and BABB, include organic solvent‐based clearing and achieve optical clearance of murine organs. 37 Intact murine lymph nodes have been imaged and analyzed at single‐cell resolution by the clearing‐enhanced 3D‐imaging (Ce3D) pipeline, 38 which facilitates mainly antibody‐based labeling of hematopoietic cells. In a similar approach, B cell dynamics during germinal center reactions 39 and clonally expanding FRC networks in developing and intact Peyer's patches 11 could be recently imaged at a larger scale using aqueous clearing agents.
FIGURE 2.
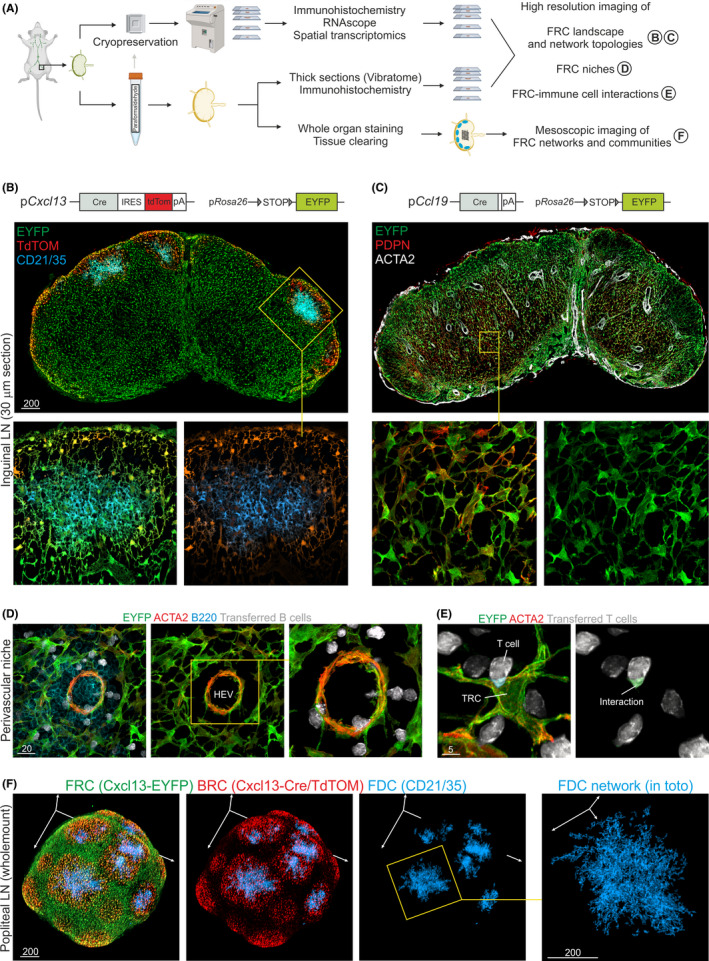
Multiscale imaging analyses of fibroblastic reticular cells (FRCs) in transgenic mouse models. (A) Dedicated workflows for lymphoid tissue preparation involve chemical and/or physical fixation strategies, which depend on the experimental question and the analyzed antigen combinations. The production of thin sections for immunohistochemical approaches or RNAscope analyses requires cryopreservation freezing of the tissue with subsequent sectioning using microtome cryostats. The assessment of the FRC landscape and network topologies can be achieved by producing thick sections from paraformaldehyde (or glutaraldehyde) fixed tissue, while global FRC networks and communities can only be imaged using stained and cleared whole‐mount tissues. (B) Analysis of FRC landscapes using thick vibratome sections produced from Cxcl13‐Cre/tdTomato R26R‐EYFP lymph nodes (upper image) shows the presence of tdTomato expressing B cell zone reticular networks in the cortex of the lymph node. Confocal microscopy imaging of 30 µm (or more) z‐stacks facilitates the analysis of BRC networks underpinning B cell follicles (boxed area and images below). (C) Analysis of FRC landscapes using thick vibratome sections produced from Ccl19‐Cre R26R‐EYFP lymph nodes (upper image) show the presence of EYFP expressing FRC networks in the T cell zone and other niches of the lymph node. Confocal microscopy imaging of 30 µm (or more) z‐stacks facilitates the analysis of coherent TRC networks within the paracortex (boxed area and images below). (D) Perivascular reticular cells situated around high endothelial venules (HEV) and interacting with migrating B cells (transferred 4 hours prior to analysis) in lymph nodes of Ccl19‐Cre R26R‐EYFP mice were imaged at high resolution by Airy‐scan microscopy. (E) TRC‐T cell interactions were imaged on thick vibratome sections of Ccl19‐Cre R26R‐EYFP lymph nodes (T cells were transferred 4 hours prior analysis) at high resolution by Airy‐scan microscopy. (F) Cxcl13‐Cre/tdTomato R26R‐EYFP lymph node stained in whole mount and cleared before imaging. The size of popliteal lymph nodes allows for adequate analysis of FRC networks in toto by mesoscopic imaging techniques such as selective plane illumination microscopy or conventional confocal microscopy
The continued improvement of confocal laser scanning microscopy (CLSM) has advanced the analysis of the 3‐dimensional structure of the FRC landscape at high resolution. In particular, topological properties of the FRC and conduit networks 12 , 40 have been elaborated using CLSM. The 3‐dimensional analysis of the TRC landscape at high resolution has resolved the small‐world property of FRC networks that determines immune cell migration and lymph node functionality. 12 , 40 The formation of chemokine gradients along FRC networks guides the migration of immune cells, as shown for CXCL13 gradients present on the BRC network 41 or CCL21 gradients between the cortical and paracortical regions in murine lymph nodes. 42 Three‐dimensional stromal cell network analysis by CLSM in combination with intravital two‐photon microscopy revealed the territorial migration of lymphocytes on FRC networks in greater detail. 21 , 40 , 43 More recently, mesoscopic imaging technologies have been developed together with the advent of optical clearing methods. For example, selective plane illumination microscopy 44 can be used to study dendritic cell‐T cell interactions in whole murine lymph nodes 45 , 46 or the development of FRC networks covering the subepithelial dome and germinal centers of murine Peyer's patches. 11 Further technical progress in CLSM, mesoscopic imaging, and high‐dimensional data analyses will facilitate the visualization and quantitative description of FRC network topology on a larger scale and will, at the same time, provide valuable information on cellular interactions and subcellular processes.
2.2. Assessing FRC functions in genetic mouse models
Genetic targeting of different cell types and elaboration of their phenotype and function can be achieved through cell type‐specific expression of the Cre recombinase and/or reporter genes. Cre recombinase expression cleaves genetically engineered Cre‐recognition (lox‐p) sites that flank transcriptional stop‐cassettes upstream of single color reporter genes (eg, R26R‐EYFP) 47 or stochastic multicolor reporter genes (eg, Brainbow transgenes) 48 to achieve permanent marker expression in the cells and their progeny. Moreover, the approach can generate conditional gain‐of‐function gene expression or conditional gene deficiency. A number of promoter constructs have been developed to drive Cre recombinase expression specifically in lymphoid organ fibroblasts (reviewed in 5 ). The finding that murine lymph node FRCs can be highlighted by flow cytometry through a combination of PDPN and the endothelial cell marker CD31 31 had motivated the generation of a bacterial artificial chromosome (BAC) transgenic mouse line with Cre recombinase expression driven by the murine Pdpn promoter (Pdpn‐Cre mice). 49 Cre recombinase‐mediated reporter gene expression in Pdpn‐Cre R26R‐EYFP mice showed that only a small fraction of FRCs and lymphatic endothelial cells is targeted by this transgene. 49 Likewise, Il7‐Cre mice 50 showed rather low transgene penetrance in murine lymph node FRCs 51 rendering these strains not suitable for studying the impact of conditional gene deletion in FRCs. It appears that the murine Ccl19 promoter used in BAC transgenic mice to drive Cre recombinase expression is well‐suited to functionally target lymphoid organ fibroblasts 9 , 52 (Figure 2C). The robust expression of Cre recombinase in Ccl19‐Cre mice along the differentiation trajectory of FRCs from their embryonic progenitors to all adult progeny has facilitated the visualization and molecular characterization FRC development in lymph nodes, 52 spleen, 9 , 53 and Peyer's patches. 11 Moreover, functions of lymphoid organ fibroblasts during viral infection 10 , 54 , 55 , 56 or inflammatory diseases 57 have been elaborated. Ccl19‐Cre activity can also be detected in FRCs underpinning non‐classical SLOs such a fat‐associated lymphoid clusters in the omentum (milky spots) 58 or tertiary lymphoid structures of the lung (ie, bronchus‐associated lymphoid tissues). 59 The possibility to genetically target FRCs, as demonstrated for the Ccl19‐Cre model, has advanced functional and phenotypical analyses of lymphoid organ FRCs and has paved the way to characterize FRC‐like cells that appear as immune‐interacting fibroblasts in inflamed tissues 60 , 61 and in the tumor microenvironment. 62
Ccl19‐Cre transgene activity is initialized in all embryonic SLO anlagen during the third trimester of gestation and serves as a reliable lineage tracer for adult FRCs. 9 , 11 , 63 However, to elaborate the FRC progenitor differentiation pathways and to study the formation of specific FRC subsets, a FRC fate‐mapping mouse model had to be developed. To this end, the BAC transgenesis strategy from Ccl19‐Cre mice was applied to place the tetracycline transactivator (tTA) in combination with a tandem‐repeated Tomato (tdTomato) reporter gene under the control of the Ccl19 promotor (Ccl19‐tTA/tdTomato). 9 Ccl19‐tTA/tdTomato mice were crossed to LC‐1 mice 64 to facilitate tetracyclin‐mediated regulation of Cre recombinase expression. This inducible system was further crossed to the R26R‐EYFP reporter (triple transgenic mice were named Ccl19‐iEYFP) and enabled a timed control of Cre recombinase activity and imprinting of EYFP expression in FRC progenitors. High‐resolution confocal microscopy of embryonic spleens in Ccl19‐iEYFP mice identified periarterial progenitors of splenic FRCs that give rise to all (known) FRC subsets of the splenic white pulp. 9 Differentiation trajectories of embryonic progenitors of Peyer's patch FRCs were studied in a combination of lineage‐tracing and fate‐mapping experiments using Ccl19‐iEYFP mice. 11 Imaging of whole Peyer's patch anlagen of Ccl19‐iEYFP mice during the perinatal period, when first immune cells populate the Peyer's patch primordia, identified perivascular progenitors that give rise to TRC and BRC subsets of adult Peyer's patches. Interestingly, early FRC progenitors clonally expand in the perivascular niche of the developing Peyer's patch, as demonstrated by imaging analysis of the fate of FRC progenitors in Peyer's patch primordia in Ccl19‐iBrainbow mice. 11 A similar differentiation and clonal expansion was demonstrated for MRC and FDC subsets in B cell follicles of murine lymph nodes. 65 Taken together, deciphering progenitor‐progeny relationships during FRC differentiation requires the combination of genetic fate‐mapping tools with powerful imaging technologies.
Physical depletion of Ccl19‐Cre‐expressing cells has a fundamental impact on B cell follicle integrity and B cell survival. 55 However, since Ccl19 mRNA expression is largely confined to TRCs and neighboring FRC subsets, certain fractions of BRCs are not targeted by this transgene, particularly in lymph nodes 6 , 10 and in Peyer's patches. 11 The notion that stromal expression of CXCL13 is restricted to B cell zones of adult SLOs 30 , 66 prompted the generation of Cxcl13‐Cre/tdTomato dual reporter mice, 63 which facilitate lineage tracing by Cre‐recombinase expression and detection of current Cxcl13 promoter activity as tdTomato expression (Figure 2B). The dual reporter system facilitated assessment of virus‐induced BRC remodeling and to dissect the molecular processes underlying FDC‐mediated governance of the germinal center reaction. 8 The possibility to enrich CXCL13‐expressing cells for single‐cell transcriptomic analyses combined with CLSM‐based in situ validation has led to the refined definition of B cell zone‐underpinning BRC subsets including light zone (LZ)‐ and dark zone (DZ)‐FDCs, MRCs, TBRCs, and interfollicular FRCs (IFRCs). 8 Furthermore, differentiation trajectory analyses of FRCs in Cxcl13‐Cre/tdTomato mice confirmed the presence of CXCL13‐expressing mesenchymal progenitors in early lymph node anlagen 63 that had been previously described by Mebius and colleagues. 67 Overall, the promoters of the chemokines CXCL13 and CCL19 appear to be particularly well suited to genetically target lymphoid organ FRCs. This approach facilitates the delineation of functional properties through genetic ablation of critical pathways and the definition of different immune cell niches that are generated by specialized FRC subsets.
In sum, the continued progress in imaging approaches has defined FRC‐generated immune cell niches as spatially confined microenvironments in lymphoid tissues that undergo remodeling during the course of immune responses. Furthermore, the detection of immunologically relevant molecules expressed by FRCs and the definition of molecular interfaces during the interaction with immune cells using high‐resolution imaging (Figure 2E) provide important correlative information on FRC functions during immunological processes. The confirmation of the functional importance of particular immune pathways steered by FRCs within microenvironmental niches can be obtained through genetic manipulating of the FRC‐immune cell crosstalk.
3. FRC NICHES IN SECONDARY LYMPHOID ORGANS
The maintenance of immune cells and their differentiation within tissues requires the establishment of a niche environment that facilitates both long‐lasting residence of a progenitor population and the regulated generation and differentiation of progeny. The niche concept has been developed by Schofield to explain the need for an environment that maintains “immortal” hematopoietic stem cells in the bone marrow. 68 The concept has been further extended to the presence of dedicated niches within tissues that support general tissue maintenance, 69 the evolution of cancer stem cells, 70 and the provision of niche environments for specific immune cell populations such as macrophages. 71 Recent evidence from single‐cell transcriptomics analyses has confirmed that fibroblastic stromal cells in perivascular spaces exhibit stemness/progenitor potential and support maintenance of different populations of lymphoid organ fibroblasts. 72 The FRC‐dependent immune cell niche concept (Figure 3) includes the ability of lymphoid organ fibroblasts to receive and integrate signals from dynamic immune cell interactions and to provide both maintenance and differentiation factors to different immune cells populations.
FIGURE 3.
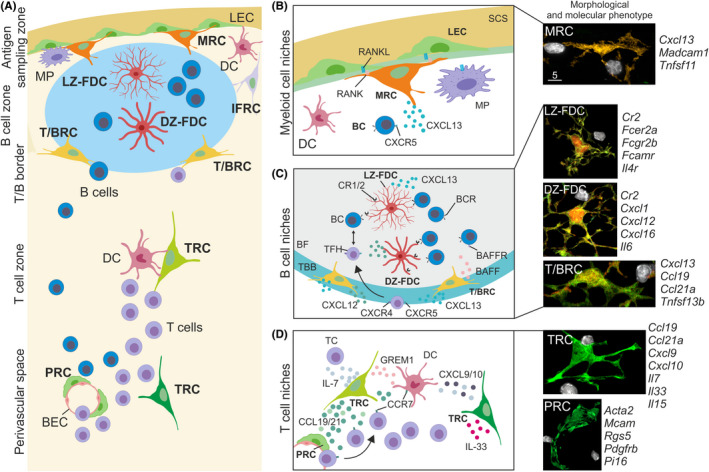
Fibroblastic reticular cell niches in murine lymph nodes. (A) Lymph node compartments are underpinned by different fibroblastic reticular cell subsets, such as marginal zone reticular cells (MRC) situated below the subcapsular sinus in close contact with lymphatic endothelial cells (LEC) and macrophages (MP). B cell follicles harbor light zone follicular dendritic cells (LZ‐FDC) and dark zone FDC (DZ‐FDC) forming germinal centers and the adjacent B cell follicle areas are populated by interfollicular fibroblastic reticular cells (IFRC) and T‐B border reticular cells (TBRC). T cell zone reticular cells (TRC) are situated in the T cell zone and close to perivascular regions, tightly interacting with T cells and dendritic cells (DC). Blood vessels of lymph nodes are constructed by blood endothelial cells (BEC) and surrounded by perivascular reticular cells (PRC) forming the perivascular space, which functions as an area of immune cell trafficking from blood vessels into the lymph node parenchyma. (B) The myeloid cell niche is found in the antigen sampling zone of lymph nodes and is characterized by a multicellular crosstalk between MRCs, LECs, and MPs. MRCs are situated near the B cell follicle and closely interact with B cells, as shown in the right high‐resolution micrograph with an MRC (orange) interacting with B cells (white). (C) B cell follicles harbor distinct B cell niches, such as the T‐B border (TBB) containing TBRCs expressing chemoattractants to guide T follicular helper (TFH) and B cells to the B cell follicle. Central B cell follicles (BF) contain LZ‐ and DZ‐FDC subsets, which present native antigen and instruct B cell differentiation in germinal centers. High‐resolution images on the right show morphological features of BRC subsets (green, red) forming B cell niches and interacting with single B cells (white). (D) The T cell zone provides several niches for the activation and differentiation of T cells by the provision of cytokines and chemokines. Blood vessels traverse through the T cell zone and PRCs are involved in the recruitment of immune cells from the blood circulation. High‐resolution images on the right show the TRC and PRCs morphology (green) of forming T cell and perivascular niches, respectively, and during the interaction with T cells (white). The adjacent molecular marker genes for FRC subsets have been derived from single‐cell RNA‐sequencing studies mentioned in the text. This figure was created using elements from BioRender.com
3.1. The myeloid cell niche in antigen‐sampling zones
FRC networks are highly heterogeneous in their topology depending on immune cell composition in the compartment that they underpin (Figure 3A). Antigens and inflammatory components including pathogens, ligands of innate immune recognition receptors, chemokines, and cytokines, first encounter a layer of macrophages and dendritic cells in the antigen‐sampling zone of the particular SLO. The antigen‐sampling zone is underpinned by MRCs, which provide key signals for the sustenance and differentiation of myeloid cells. For example, lymph‐borne antigens drain and accumulate in the subcapsular sinus of lymph nodes and are captured by subcapsular sinus macrophages 73 , 74 and dendritic cells of interfollicular regions. 75 The subcapsular sinus floor is built by lymphatic endothelial cells, which separate the lymphatic sinus from the cortex of the lymph node, 76 for example, B cell follicles and interfollicular regions. Lymphatic endothelial cells of the subcapsular sinus floor express atypical chemokine receptor 4 to scavenge CCL21 and to generate a CCL21 chemokine gradient that guides CCR7‐expressing cells into the lymph node parenchyma during inflammation. 42 Lymphatic endothelial cells and MRCs form a niche that fosters the development of subcapsular sinus macrophages through the interaction between receptor activator of nuclear factor‐κB (RANK, TNFRSF11A) and RANK‐ligand (TNFSF11). 77 The dependence of subcapsular sinus floor lymphatic endothelial cells on the provision of RANK‐ligand by MRCs 77 indicates that MRCs are part of a multicellular niche environment in the antigen‐sampling zone of lymph nodes (Figure 3B). In addition, the subcapsular sinus floor macrophage niche is supported by lymphatic endothelial cell‐mediated production of colony‐stimulating factor 1 (CSF‐1, macrophage colony‐stimulating factor) 78 highlighting the importance of a multi‐layered regulation within the niche environment.
In the splenic white pulp, blood‐borne antigens are captured by marginal zone macrophages and specialized marginal zone B cells that reside in areas adjacent to the marginal sinus, which is formed by specialized blood endothelial cells. Splenic fibroblasts situated on both sides of the marginal sinus have been shown to support marginal zone B cell and red pulp macrophage survival. 79 , 80 Moreover, the sustenance and differentiation of marginal zone B cells require the expression of the Notch‐2 ligand delta‐like 1 by Ccl19‐Cre‐positive splenic FRCs. 81 Red pulp fibroblasts maintain red pulp macrophage populations through the production of CSF1 and replenish the red pulp myeloid cell compartment through the secretion of the chemokines CCL2 and CCL7, 80 indicating that the cellular dynamics within the antigen‐sampling zone of the splenic white pulp is controlled by growth factors and the adaptive generation of migration and differentiation factors.
Peyer's patches are situated in the intestinal wall and have direct access to the content of the gut lumen. The antigen‐sampling zone in Peyer's patches is located in a compartment known as the subepithelial dome, which is separated from the gut lumen by a single layer of follicle‐associated epithelium. Antigens and inflammatory components from the gut lumen can traverse into the Peyer's patch parenchyma through specialized microfold epithelial cell (M cells) and are captured by myeloid cells for the presentation to B cells to promote IgA production. 82 Targeted ablation of RANK‐ligand in Peyer's patch MRCs revealed that M cell differentiation depends on functional MRC‐M cell interaction. 83 A more recent study by Prados et al 11 showed that activation of Peyer's patch MRCs via the tumor necrosis factor receptor‐1 is critical for M cell maintenance. In sum, the MRC niche in SLOs maintains and steers the activation of myeloid cells and forms critical interfaces with non‐hematopoietic cells such as lymphatic endothelial cells in lymph nodes or epithelial cells in Peyer's patches.
3.2. Dedicated B cell niches for immune cell positioning and activation
B cell follicles are situated directly adjacent to antigen‐sampling zones in all SLOs and are underpinned by a network of CXCL13‐expressing BRCs. 6 , 8 The BRC subset that forms the border between the T cell zone and B cell follicles (TBRCs) is a major source for the B cell survival factor BAFF. 8 , 55 Counter gradients of the chemokines CXCL12 and CXCL13 regulate B cell migration in primary B cell follicles and during the germinal center reaction. 8 , 41 , 84 The germinal center reaction triggers the reorganization of the B cell follicle into light zone (LZ) and dark zone (DZ) areas underpinned by phenotypically distinct FDC subsets. 84 , 85 Transcriptomic analysis of CXCL13‐expressing BRCs further showed that LZ‐ and DZ‐FDC subsets are present in the primary follicle and are poised to generate the respective microenvironments that steer the germinal center reaction through CXCL12‐mediated positioning of follicular helper T cells and B cells 8 (Figure 3C). Furthermore, FDCs have been shown to promote the transition of LZ to DZ germinal center B cells through the activation of TGFβ signaling 86 and scavenge IL‐4 at later stages of the germinal center reaction to foster the generation of memory B cells. 87 Interestingly, FRCs situated in the lymph node medulla (medullary reticular cells, MedRC) share transcriptomic similarities with BRCs. 8 , 10 It is most likely that the close interaction with plasma cells in the medullary chords stimulates MedRCs to provide IL‐6 and BAFF to the interacting B cells. 88 It is noteworthy that while the BRC niche in murine lymph nodes has been studied extensively, the knowledge about the identity and activation of BRC subsets in Peyer's patches and the splenic white pulp is still limited. Clearly, the niche formed by BRCs is complex and conclusions concerning key niche factors need to take into account the proximity and partial overlap with the adjacent antigen‐sampling zone and the T cell area.
3.3. Dedicated niches control T cell activation and differentiation
Lymphoid organ fibroblasts occupying the T cell zone had been considered the major, or even only, FRC population, mainly due to the association of PDPN expression, IL‐7 production and the effect of IL‐7 on T cell homeostasis and T cell memory maintenance. 31 , 89 Recent single‐cell transcriptomics analyses have indicated that the TRC landscape in murine SLOs is complex and consists of several TCR subsets. 6 , 10 , 11 The overarching function of TRCs is, in addition to the formation of the reticular fiber and conduit network, the control of T cell and dendritic cell migration and their interaction 21 , 40 , 90 (Figure 3D). In addition, TRCs in Peyer's patches are essential to maintain the pool of type 1 innate lymphoid cells and conventional NK cells through the provision of the cytokine IL‐15. 54 Parafollicular TBRCs expressing the bone morphogenic protein inhibitor GREM1 support the survival of conventional dendritic cells and/or their precursors. 91 Likewise, parafollicular FRCs express cholesterol‐25‐hydroxylase (CH25H) to produce hydroxycholesterol, a ligand for G protein‐coupled receptor 183 (GPR183, also known as EBI2), 6 which is most likely a key step to direct the positioning of CD4+ T cells in close contact with dendritic cells in parafollicular areas to foster T cell immunity. 92 It still remains unknown whether the expression of Cxcl9 and Cxcl10 by different TRC populations 6 , 10 , 11 warrants the assignment as a distinct subset. During viral infection, both dendritic cells and lymphoid organ fibroblasts acquire an activated cell status to coordinate the distribution and differentiation of effector and central memory T cells, 93 suggesting that T cell niches formed by TRC subsets can dynamically adapt during immune activation. Overall, specialized niches generated by different FRC subsets maintain immune cell homeostasis and facilitate dynamic and regional adaptation of particular microenvironments to secure immunocompetence during infection and the development of cancer.
4. MOLECULAR MECHANISMS GOVERNING FRC‐IMMUNE CELL INTERACTION
The FRC network in SLOs accommodates immune cells to facilitate their positioning, activation, and differentiation. Importantly, FRC niches rapidly adapt to changing immune cell dynamics during the course of immune responses. The visualization and quantitative assessment of these processes represent a substantial experimental challenge. The major approach to resolve the processes underpinning FRC‐immune cell interactions over time has been—and still is—the sequential visualization, mainly by CLSM, of static snapshots during immune responses. In addition, real‐time imaging by intravital two‐photon microscopy of immune cell migration in SLOs has revealed the importance of the FRC network for the guidance and activation of lymphocytes. The perturbation of FRC‐immune cell interaction by infection, cancer, or autoinflammatory triggers in combination with advanced imaging methods has provided key insight into the mechanisms underlying FRC‐immune cell interaction.
4.1. Immune cell migration is guided by FRCs
The compartmentalization of SLO structures is determined by FRC subsets that produce distinct sets of chemokines and generate gradients of cholesterol derivatives to orchestrate immune cell migration and positioning. Seminal work by the Germain group has shown by intravital two‐photon microscopy that naive T and B cells enter lymph nodes through high endothelial venules and migrate along the FRC network toward the T or B cell zones. 21 The entry of lymphocytes into lymph nodes via the high endothelial venule route depends mainly on the expression of CCR7 and the production of CCL19 and CCL21 by TRCs. 29 Moreover, assessment of the migration of lymph‐derived T cells into the lymph node parenchyma by a combination of CLSM and intravital imaging revealed that T cells utilize CCR7‐dependent routes through medullary sinuses, whereas dendritic cells transmigrate through the floor of the subcapsular sinus. 94 It remains to be established which additional FRC‐derived chemokine signals are required for the establishment of cell type‐specific intranodal migration. Similar challenges remain to be mastered for the full elucidation of T cell migratory pathways in the spleen, where lymphocytes gain passively access to the red pulp via arterioles that end in the sinuses of the red pulp. Real‐time imaging in combination with genetically modified T cells and genetically highlighted PRCs and TRCs in the white pulp has revealed that naive T cells migrate from the red pulp into the white pulp via perivascular tracks. 43 CCR7‐dependent T cell entry into the splenic white pulp occurs through FRC‐underpinned bridging channels in the marginal zone. 90 T cells then follow a CCL19 chemokine gradient into the T cell zone that is formed by PRCs. 43 It appears that naive B cells follow the same migration route to transit from the red pulp into the white pulp. 90 However, it is still unclear which FRC‐derived migratory cues, that is, chemokine gradients, guide B cells along the TRC scaffold into the B cell follicles.
Antigen‐presenting cells such as dendritic cells secure surveillance of peripheral tissues through transport of immunologically relevant information, that is, pathogens, antigens, and inflammatory substances, to lymph nodes. Migratory dendritic cells enter lymph nodes via afferent lymphatics and transmigrate through the subcapsular sinus toward the T cell zone guided by CCL19/CCL21 gradients that are generated by TRCs. 94 Intravital two‐photon microscopy has shown that the CCR7‐dependent initial migration of activated dendritic cells from skin through lymphatic vessels to draining lymph nodes is fostered by immune complexes. 95 Immune activation, for example following influenza vaccination, also activates intranodal DC migration as revealed by live‐imaging of T cell zone‐resident DCs that move toward the medulla to capture antigen and to initiate the immune response. 96 Such inflammation‐associated intranodal migration is most likely enhanced by the expression of the chemokines CXCL9 and CXCL10 by multiple immune cell populations including dendritic cells and T cells, 96 and by several FRC subsets. 6 , 10 DCs situated in medullary sinuses can directly capture lymph‐borne antigens and migrate to the T‐B border to steer activation of humoral immune responses. 97 Positioning of particular dendritic cell subsets at the T‐B border is directed by the chemoattractant receptor EBI2/GPR183 92 that senses oxysterol concentration gradients and determines the positioning of immune cells in SLOs. 98 The enzyme CH25H that synthesizes oxysterol derivatives is expressed by MRCs, TBRCs, and IFRCs in the parafollicular areas, 6 whereas the oxysterol metabolizing enzyme HSD3B7 is enriched in stromal cells of the T cell zone. 99 Quantitative immunohistochemistry has shown that the differential expression of these enzymes regulates oxysterol gradients in the spleen and directs the migration of CD11b+ conventional type 2 dendritic cells to the T‐B border to stimulate B cell activation. 100 In sum, classical and advanced imaging approaches have been shown to be important to elaborate how the FRC network generates pathways for immune cell migration to and within SLOs and to characterize chemokine and cholesterol metabolite gradients that foster immune cell interaction.
4.2. FRC‐tailored microenvironments for immune cell activation and differentiation
Immune activation leads to fundamental remodeling processes in SLOs, mainly due to rapid changes in immune cell influx or efflux and shuffling of immune cells between different niche environments. The influx of antigen‐presenting cells and other immune cells increases overall cell numbers and FRC networks physically adapt to these changing cell densities. 3 Analysis of lymph node swelling reactions by high‐resolution CLSM deconvoluted the physical stretching behavior of the FRC cell body and the FRC network to compensate for the changing space restrictions during the early phase of a response when immune cell influx and proliferation increase. 101 Stretching of FRC networks requires the inhibition of PDPN‐mediated contraction through interaction with C‐type lectin receptor on dendritic cells. 101 , 102 Importantly, dendritic cells sustain FRC differentiation during this highly dynamic process through the induction of lymphotoxin beta receptor signaling and thereby foster FRC proliferation in swelling lymph nodes. 103 , 104 FRCs undergo profound phenotypical changes during immune activation including enhanced production of effector cytokines such as IL‐4, IL‐33, and IL‐6, 105 , 106 and increased expression of surface molecules that modulate T cell activation such as inducible T cell costimulator ligand, 106 major histocompatibility complex molecules 58 or co‐inhibitory molecules. 10 The fine‐tuned expression of cytokines and other stimulatory factors by FRCs are decisive for the differentiation of T cells in effector or helper subsets and establish setpoints for ongoing immune response. Conversely, FRCs receive important signals from immune cells, for example through the type 1 interferon receptor alpha, which is necessary to control local lymphocytic choriomeningitis virus infection. 10 Interestingly, the lack of IFNAR signaling in FRCs led to altered LCMV infection patterns in the lymph node FRC network as revealed by imaging of viral nucleoproteins in lymph nodes of acutely infected mice, 10 indicating that sensing of innate immunological signals is important for the control of infectious agents. During later phases of an acute immune response, FRCs contribute to the contraction of lymphocyte populations through the activation of the COX2 pathway, 107 expression of programmed death ligand 1, 10 or through the restriction of IL‐15 availability. 54 Noteworthy, the important role of lymphoid organ fibroblasts in the activation and regulation of T cell responses as outlined in these studies could only be elaborated through the combination of functional approaches with advanced imaging methods that facilitate the localization of FRC‐immune cell interaction.
The FRC niche concept predicts that the dynamic regulation of FRC “catering,” that is, provision of migration, growth, and differentiation factors is key for the regulation of immune responsiveness. Hence, one major challenge for the assessment FRC‐immune cell interaction is the quantification of effector molecules and the determination of their distribution patterns. The understanding of FRC niche furnishment is particularly important during a multicellular crosstalk, for example during T follicular helper cell‐B cell interaction in the course of antiviral B cell response when the expression of the chemotactic receptor CXCR5 is changing rapidly. 108 Indeed, the expression of the CXCR5 ligand CXCL13 by BRCs forms both soluble and immobilized gradients express complex gradients of these chemokines. 41 Quantitative assessment of such gradients on FRC small‐world networks became feasible through the combination of high‐resolution microscopy and computer simulations leading to the conclusion that mainly immobilized CXCL13 gradients promote B cell trafficking. 41 High‐resolution confocal imaging has been most helpful to disentangle how and to what extent CXCL12 expression by TBRCs and DZ‐FDCs supports T follicular helper cell‐B cell interactions in the germinal center. 8 Clearly, further advances in physical, chemical, and biological system will generate tools to quantify the molecular and cellular mechanisms underlying the processes in FRC‐generated niches with high fidelity. Improved and more accurate data acquisition will be suitable for the analysis in mathematical and computational models of immune function as has been recently achieved for the analysis of single‐cell transcriptomics or high‐dimensional flow cytometry data. 109
5. CONCLUSIONS AND OUTLOOK
The advancement of imaging approaches has been key for the initial description and the subsequent characterization of specialized lymphoid organ fibroblasts. Antibody‐based staining and visualization are still the main methodological approach to provide insight into FRC morphology and phenotype (Table 1). The localization and quantification of protein expression by FRCs and the morphological characterization of different FRC subsets in tissue sections by high‐resolution imaging have become particularly important for the validation of gene expression profiles that can be generated by single‐cell transcriptomics. Genetic targeting of murine FRCs with expression of fluorescent markers has further advanced our understanding of FRC biology, mainly through the combination with Cre‐mediated gain‐of‐function or gene ablation approaches. However, the imaging of immune dynamics has not yet fully exploited the sophisticated methods for in vivo FRC visualization and functional manipulation offered by the available genetic models. In sum, imaging approaches have guided and constantly improve the elaboration of the specialized immune cell niches in SLOs that are generated by FRCs.
One of the main challenges for future studies that aim at the resolution of FRC‐immune cell interaction is the 3‐dimensional positioning of multiple FRC communities, the interconnection between different FRC subsets, and the changes in FRC phenotype and function that are associated with lymphoid organ remodeling during infection, inflammatory diseases or cancer. The size of human lymphoid organs represents a similar challenge for the imaging‐based characterization of FRC‐immune cell interaction during disease processes. First efforts have been made to isolate and characterize human FRCs from tonsils, 110 lymph nodes, 111 or spleen 112 revealing the expression of the chemokines CCL21 or CXCL13 in situ. The remodeling of lymphoid organ structure and the associated phenotypic changes in the human FRC landscape has been highlighted during HIV infection 113 or in patients with follicular lymphoma. 114 Recent single‐cell transcriptomic analyses of human tissues have revealed the cellular complexity of fibroblasts in lymphoid organs. 72 However, FRC community structures and the interaction patterns with immune cells are still elusive and need to be resolved using high‐resolution and large‐scale imaging methods. While conventional CLSM facilitates high‐resolution imaging to assess morphological and subcellular detail, larger volumes cannot be acquired. In contrast, mesoscopic imaging is able to scan more than 50 mm3 tissue volumes in human sections, 115 , 116 but does not permit in situ assessment of molecular detail. Hence, the combination of confocal, high‐resolution, and mesoscopic imaging will be required for the full characterization of the FRC landscape in human lymphoid organs. This combined approach needs to be supported by mathematical and bioinformatics‐based integration to define critical processes in FRC‐generated immune cell niches that could be therapeutically targeted in infection, cancer, or autoimmune diseases.
CONFLICT OF INTEREST
LO, H.‐WC, and BL are co‐founders and shareholders in Stromal Therapeutics AG, St. Gallen, Switzerland.
ACKNOWLEDGEMENTS
This study received financial support from the Swiss National Science Foundation (grants 177208 and 182583 to BL). The funder had no role in preparation of the manuscript.
Onder L, Cheng H‐W, Ludewig B. Visualization and functional characterization of lymphoid organ fibroblasts. Immunol Rev.2022;306:108–122. doi: 10.1111/imr.13051
This article is part of a series of reviews covering Insights into immune function from imaging appearing in Volume 306 of Immunological Reviews.
REFERENCES
- 1. Perez‐Shibayama C, Gil‐Cruz C, Ludewig B. Fibroblastic reticular cells at the nexus of innate and adaptive immune responses. Immunol Rev. 2019;289(1):31‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Krishnamurty AT, Turley SJ. Lymph node stromal cells: cartographers of the immune system. Nat Immunol. 2020;21(4):369‐380. [DOI] [PubMed] [Google Scholar]
- 3. Acton SE, Onder L, Novkovic M, Martinez VG, Ludewig B. Communication, construction, and fluid control: lymphoid organ fibroblastic reticular cell and conduit networks. Trends Immunol. 2021;42(9):782‐794. [DOI] [PubMed] [Google Scholar]
- 4. Schulz O, Hammerschmidt SI, Moschovakis GL, Forster R. Chemokines and chemokine receptors in lymphoid tissue dynamics. Annu Rev Immunol. 2016;34:203‐242. [DOI] [PubMed] [Google Scholar]
- 5. Lütge M, Pikor N, Ludewig B. Differentiation and activation of fibroblastic reticular cells. Immunol Rev. 2021;302(1):32‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodda LB, Lu E, Bennett ML, et al. Single‐cell RNA sequencing of lymph node stromal cells reveals niche‐associated heterogeneity. Immunity. 2018;48(5):1014‐1028 e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pezoldt J, Pasztoi M, Zou M, et al. Neonatally imprinted stromal cell subsets induce tolerogenic dendritic cells in mesenteric lymph nodes. Nat Commun. 2018;9(1):3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pikor NB, Mörbe U, Lütge M, et al. Remodeling of light and dark zone follicular dendritic cells governs germinal center responses. Nat Immunol. 2020;21(6):649‐659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng HW, Onder L, Novkovic M, et al. Origin and differentiation trajectories of fibroblastic reticular cells in the splenic white pulp. Nat Commun. 2019;10(1):1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perez‐Shibayama C, Islander U, Lütge M, et al. Type I interferon signaling in fibroblastic reticular cells prevents exhaustive activation of antiviral CD8(+) T cells. Sci Immunol. 2020;5(51):eabb7066. doi: 10.1126/sciimmunol.abb7066 [DOI] [PubMed] [Google Scholar]
- 11. Prados A, Onder L, Cheng HW, et al. Fibroblastic reticular cell lineage convergence in Peyer's patches governs intestinal immunity. Nat Immunol. 2021;22(4):510‐519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Novkovic M, Onder L, Bocharov G, Ludewig B. Topological structure and robustness of the lymph node conduit system. Cell Rep. 2020;30(3):893‐904 e896. [DOI] [PubMed] [Google Scholar]
- 13. Coons AH, Leduc EH, Kaplan MH. Localization of antigen in tissue cells. VI. The fate of injected foreign proteins in the mouse. J Exp Med. 1951;93(2):173‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaplan ME, Coons AH, Deane HW. Localization of antigen in tissue cells; cellular distribution of pneumococcal polysaccharides types II and III in the mouse. J Exp Med. 1950;91(1):15‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller JJ 3rd, Nossal GJ. Antigens in immunity. Vi. The phagocytic reticulum of lymph node follicles. J Exp Med. 1964;120(6):1075‐1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nossal GJ, Austin CM, Ada GL. Antigens in immunity. VII. Analysis of immunological memory. Immunology. 1965;9(4):333‐348. [PMC free article] [PubMed] [Google Scholar]
- 17. Nossal GJ, Ada GL, Austin CM, Pye J. Antigens in immunity. 8. Localization of 125‐I‐labelled antigens in the secondary response. Immunology. 1965;9(4):349‐357. [PMC free article] [PubMed] [Google Scholar]
- 18. Nossal GJ, Abbot A, Mitchell J, Lummus Z. Antigens in immunity. XV. Ultrastructural features of antigen capture in primary and secondary lymphoid follicles. J Exp Med. 1968;127(2):277‐290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mandel TE, Phipps RP, Abbot AP, Tew JG. Long‐term antigen retention by dendritic cells in the popliteal lymph node of immunized mice. Immunology. 1981;43(2):353‐362. [PMC free article] [PubMed] [Google Scholar]
- 20. Bofill M, Akbar AN, Amlot PL. Follicular dendritic cells share a membrane‐bound protein with fibroblasts. J Pathol. 2000;191(2):217‐226. [DOI] [PubMed] [Google Scholar]
- 21. Bajenoff M, Egen JG, Koo LY, et al. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25(6):989‐1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krautler NJ, Kana V, Kranich J, et al. Follicular dendritic cells emerge from ubiquitous perivascular precursors. Cell. 2012;150(1):194‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Vliet E, Melis M, Van Ewijk W. Monoclonal antibodies to stromal cell types of the mouse thymus. Eur J Immunol. 1984;14(6):524‐529. [DOI] [PubMed] [Google Scholar]
- 24. Van Vliet E, Melis M, Foidart JM, Van Ewijk W. Reticular fibroblasts in peripheral lymphoid organs identified by a monoclonal antibody. J Histochem Cytochem. 1986;34(7):883‐890. [DOI] [PubMed] [Google Scholar]
- 25. Schiavinato A, Przyklenk M, Kobbe B, Paulsson M, Wagener R. Collagen type VI is the antigen recognized by the ER‐TR7 antibody. Eur J Immunol. 2021;51(9):2345‐2347. [DOI] [PubMed] [Google Scholar]
- 26. Gretz JE, Kaldjian EP, Anderson AO, Shaw S. Sophisticated strategies for information encounter in the lymph node: the reticular network as a conduit of soluble information and a highway for cell traffic. J Immunol. 1996;157(2):495‐499. [PubMed] [Google Scholar]
- 27. Gretz JE, Anderson AO, Shaw S. Cords, channels, corridors and conduits: critical architectural elements facilitating cell interactions in the lymph node cortex. Immunol Rev. 1997;156:11‐24. [DOI] [PubMed] [Google Scholar]
- 28. Sixt M, Kanazawa N, Selg M, et al. The conduit system transports soluble antigens from the afferent lymph to resident dendritic cells in the T cell area of the lymph node. Immunity. 2005;22(1):19‐29. [DOI] [PubMed] [Google Scholar]
- 29. Luther SA, Tang HL, Hyman PL, Farr AG, Cyster JG. Coexpression of the chemokines ELC and SLC by T zone stromal cells and deletion of the ELC gene in the plt/plt mouse. Proc Natl Acad Sci USA. 2000;97(23):12694‐12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gunn MD, Ngo VN, Ansel KM, Ekland EH, Cyster JG, Williams LT. A B‐cell‐homing chemokine made in lymphoid follicles activates Burkitt's lymphoma receptor‐1. Nature. 1998;391(6669):799‐803. [DOI] [PubMed] [Google Scholar]
- 31. Link A, Vogt TK, Favre S, et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat Immunol. 2007;8(11):1255‐1265. [DOI] [PubMed] [Google Scholar]
- 32. Scandella E, Bolinger B, Lattmann E, et al. Restoration of lymphoid organ integrity through the interaction of lymphoid tissue‐inducer cells with stroma of the T cell zone. Nat Immunol. 2008;9(6):667‐675. [DOI] [PubMed] [Google Scholar]
- 33. Lukacs‐Kornek V, Malhotra D, Fletcher AL, et al. Regulated release of nitric oxide by nonhematopoietic stroma controls expansion of the activated T cell pool in lymph nodes. Nat Immunol. 2011;12(11):1096‐1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Siegert S, Huang HY, Yang CY, et al. Fibroblastic reticular cells from lymph nodes attenuate T cell expansion by producing nitric oxide. PLoS One. 2011;6(11):e27618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Susaki EA, Tainaka K, Perrin D, et al. Whole‐brain imaging with single‐cell resolution using chemical cocktails and computational analysis. Cell. 2014;157(3):726‐739. [DOI] [PubMed] [Google Scholar]
- 36. Tian T, Yang Z, Li X. Tissue clearing technique: Recent progress and biomedical applications. J Anat. 2021;238(2):489‐507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Susaki EA, Ueda HR. Whole‐body and whole‐organ clearing and imaging techniques with single‐cell resolution: toward organism‐level systems biology in mammals. Cell Chem Biol. 2016;23(1):137‐157. [DOI] [PubMed] [Google Scholar]
- 38. Li W, Germain RN, Gerner MY. Multiplex, quantitative cellular analysis in large tissue volumes with clearing‐enhanced 3D microscopy (C(e)3D). Proc Natl Acad Sci USA. 2017;114(35):E7321‐E7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Biram A, Strömberg A, Winter E, et al. BCR affinity differentially regulates colonization of the subepithelial dome and infiltration into germinal centers within Peyer's patches. Nat Immunol. 2019;20(4):482‐492. [DOI] [PubMed] [Google Scholar]
- 40. Novkovic M, Onder L, Cupovic J, et al. Topological small‐world organization of the fibroblastic reticular cell network determines lymph node functionality. PLoS Biol. 2016;14(7):e1002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cosgrove J, Novkovic M, Albrecht S, et al. B cell zone reticular cell microenvironments shape CXCL13 gradient formation. Nat Commun. 2020;11(1):3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ulvmar MH, Werth K, Braun A, et al. The atypical chemokine receptor CCRL1 shapes functional CCL21 gradients in lymph nodes. Nat Immunol. 2014;15(7):623‐630. [DOI] [PubMed] [Google Scholar]
- 43. Chauveau A, Pirgova G, Cheng HW, et al. Visualization of T cell migration in the spleen reveals a network of perivascular pathways that guide entry into T zones. Immunity. 2020;52(5):794‐807 e797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Verveer PJ, Swoger J, Pampaloni F, Greger K, Marcello M, Stelzer EH. High‐resolution three‐dimensional imaging of large specimens with light sheet‐based microscopy. Nat Methods. 2007;4(4):311‐313. [DOI] [PubMed] [Google Scholar]
- 45. Abe J, Ozga AJ, Swoger J, Sharpe J, Ripoll J, Stein JV. Light sheet fluorescence microscopy for in situ cell interaction analysis in mouse lymph nodes. J Immunol Methods. 2016;431:1‐10. [DOI] [PubMed] [Google Scholar]
- 46. Ozga AJ, Moalli F, Abe J, et al. pMHC affinity controls duration of CD8+ T cell‐DC interactions and imprints timing of effector differentiation versus expansion. J Exp Med. 2016;213(12):2811‐2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Srinivas S, Watanabe T, Lin CS, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Livet J, Weissman TA, Kang H, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450(7166):56‐62. [DOI] [PubMed] [Google Scholar]
- 49. Onder L, Scandella E, Chai Q, et al. A novel bacterial artificial chromosome‐transgenic podoplanin‐cre mouse targets lymphoid organ stromal cells in vivo. Front Immunol. 2011;2:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Repass JF, Laurent MN, Carter C, et al. IL7‐hCD25 and IL7‐Cre BAC transgenic mouse lines: new tools for analysis of IL‐7 expressing cells. Genesis. 2009;47(4):281‐287. [DOI] [PubMed] [Google Scholar]
- 51. Onder L, Narang P, Scandella E, et al. IL‐7‐producing stromal cells are critical for lymph node remodeling. Blood. 2012;120(24):4675‐4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chai Q, Onder L, Scandella E, et al. Maturation of lymph node fibroblastic reticular cells from myofibroblastic precursors is critical for antiviral immunity. Immunity. 2013;38(5):1013‐1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schaeuble K, Britschgi MR, Scarpellino L, et al. Perivascular fibroblasts of the developing spleen act as LTalpha1beta2‐dependent precursors of both T and B zone organizer cells. Cell Rep. 2017;21(9):2500‐2514. [DOI] [PubMed] [Google Scholar]
- 54. Gil‐Cruz C, Perez‐Shibayama C, Onder L, et al. Fibroblastic reticular cells regulate intestinal inflammation via IL‐15‐mediated control of group 1 ILCs. Nat Immunol. 2016;17(12):1388‐1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cremasco V, Woodruff MC, Onder L, et al. B cell homeostasis and follicle confines are governed by fibroblastic reticular cells. Nat Immunol. 2014;15(10):973‐981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aparicio‐Domingo P, Cannelle H, Buechler MB, et al. Fibroblast‐derived IL‐33 is dispensable for lymph node homeostasis but critical for CD8 T‐cell responses to acute and chronic viral infection. Eur J Immunol. 2021;51(1):76‐90. [DOI] [PubMed] [Google Scholar]
- 57. Majumder S, Amatya N, Revu S, et al. IL‐17 metabolically reprograms activated fibroblastic reticular cells for proliferation and survival. Nat Immunol. 2019;20(5):534‐545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Perez‐Shibayama C, Gil‐Cruz C, Cheng HW, et al. Fibroblastic reticular cells initiate immune responses in visceral adipose tissues and secure peritoneal immunity. Sci Immunol. 2018;3(26):eaar4539. [DOI] [PubMed] [Google Scholar]
- 59. Cupovic J, Ring SS, Onder L, et al. Adenovirus vector vaccination reprograms pulmonary fibroblastic niches to support protective inflating memory CD8(+) T cells. Nat Immunol. 2021;22(8):1042‐1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cupovic J, Onder L, Gil‐Cruz C, et al. Central nervous system stromal cells control local CD8(+) T cell responses during virus‐induced neuroinflammation. Immunity. 2016;44(3):622‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kinchen J, Chen HH, Parikh K, et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell. 2018;175(2):372‐386 e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cheng HW, Onder L, Cupovic J, et al. CCL19‐producing fibroblastic stromal cells restrain lung carcinoma growth by promoting local antitumor T‐cell responses. J Allergy Clin Immunol. 2018;142(4):1257‐1271.e4. [DOI] [PubMed] [Google Scholar]
- 63. Onder L, Morbe U, Pikor N, et al. Lymphatic endothelial cells control initiation of lymph node organogenesis. Immunity. 2017;47(1):80‐92. [DOI] [PubMed] [Google Scholar]
- 64. Schonig K, Schwenk F, Rajewsky K, Bujard H. Stringent doxycycline dependent control of CRE recombinase in vivo. Nucleic Acids Res. 2002;30(23):e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jarjour M, Jorquera A, Mondor I, et al. Fate mapping reveals origin and dynamics of lymph node follicular dendritic cells. J Exp Med. 2014;211(6):1109‐1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ansel KM, Ngo VN, Hyman PL, et al. A chemokine‐driven positive feedback loop organizes lymphoid follicles. Nature. 2000;406(6793):309‐314. [DOI] [PubMed] [Google Scholar]
- 67. van de Pavert SA, Olivier BJ, Goverse G, et al. Chemokine CXCL13 is essential for lymph node initiation and is induced by retinoic acid and neuronal stimulation. Nat Immunol. 2009;10(11):1193‐1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schofield R. The relationship between the spleen colony‐forming cell and the haemopoietic stem cell. Blood Cells. 1978;4(1–2):7‐25. [PubMed] [Google Scholar]
- 69. Voog J, Jones DL. Stem cells and the niche: a dynamic duo. Cell Stem Cell. 2010;6(2):103‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Boesch M, Sopper S, Zeimet AG, et al. Heterogeneity of cancer stem cells: rationale for targeting the stem cell niche. Biochim Biophys Acta. 2016;1866(2):276‐289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Guilliams M, Thierry GR, Bonnardel J, Bajenoff M. Establishment and maintenance of the macrophage niche. Immunity. 2020;52(3):434‐451. [DOI] [PubMed] [Google Scholar]
- 72. Buechler MB, Pradhan RN, Krishnamurty AT, et al. Cross‐tissue organization of the fibroblast lineage. Nature. 2021;593(7860):575‐579. [DOI] [PubMed] [Google Scholar]
- 73. Junt T, Moseman EA, Iannacone M, et al. Subcapsular sinus macrophages in lymph nodes clear lymph‐borne viruses and present them to antiviral B cells. Nature. 2007;450(7166):110‐114. [DOI] [PubMed] [Google Scholar]
- 74. Iannacone M, Moseman EA, Tonti E, et al. Subcapsular sinus macrophages prevent CNS invasion on peripheral infection with a neurotropic virus. Nature. 2010;465(7301):1079‐1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gerner MY, Torabi‐Parizi P, Germain RN. Strategically localized dendritic cells promote rapid T cell responses to lymph‐borne particulate antigens. Immunity. 2015;42(1):172‐185. [DOI] [PubMed] [Google Scholar]
- 76. Rantakari P, Auvinen K, Jäppinen N, et al. The endothelial protein PLVAP in lymphatics controls the entry of lymphocytes and antigens into lymph nodes. Nat Immunol. 2015;16(4):386‐396. [DOI] [PubMed] [Google Scholar]
- 77. Camara A, Cordeiro OG, Alloush F, et al. Lymph node mesenchymal and endothelial stromal cells cooperate via the RANK‐RANKL cytokine axis to shape the sinusoidal macrophage niche. Immunity. 2019;50(6):1467‐1481 e1466. [DOI] [PubMed] [Google Scholar]
- 78. Mondor I, Baratin M, Lagueyrie M, et al. Lymphatic endothelial cells are essential components of the subcapsular sinus macrophage niche. Immunity. 2019;50(6):1453‐1466 e1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Magri G, Miyajima M, Bascones S, et al. Innate lymphoid cells integrate stromal and immunological signals to enhance antibody production by splenic marginal zone B cells. Nat Immunol. 2014;15(4):354‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bellomo A, Mondor I, Spinelli L, et al. Reticular fibroblasts expressing the transcription factor WT1 define a stromal niche that maintains and replenishes splenic red pulp macrophages. Immunity. 2020;53(1):127‐142 e127. [DOI] [PubMed] [Google Scholar]
- 81. Fasnacht N, Huang HY, Koch U, et al. Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch‐regulated immune responses. J Exp Med. 2014;211(11):2265‐2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Reboldi A, Cyster JG. Peyer's patches: organizing B‐cell responses at the intestinal frontier. Immunol Rev. 2016;271(1):230‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nagashima K, Sawa S, Nitta T, et al. Targeted deletion of RANKL in M cell inducer cells by the Col6a1‐Cre driver. Biochem Biophys Res Commun. 2017;493(1):437‐443. [DOI] [PubMed] [Google Scholar]
- 84. Rodda LB, Bannard O, Ludewig B, Nagasawa T, Cyster JG. Phenotypic and morphological properties of germinal center dark zone Cxcl12‐expressing reticular cells. J Immunol. 2015;195(10):4781‐4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bannard O, Horton RM, Allen CD, An J, Nagasawa T, Cyster JG. Germinal center centroblasts transition to a centrocyte phenotype according to a timed program and depend on the dark zone for effective selection. Immunity. 2013;39(5):912‐924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Suzuki K, Maruya M, Kawamoto S, et al. The sensing of environmental stimuli by follicular dendritic cells promotes immunoglobulin A generation in the gut. Immunity. 2010;33(1):71‐83. [DOI] [PubMed] [Google Scholar]
- 87. Duan L, Liu D, Chen H, et al. Follicular dendritic cells restrict interleukin‐4 availability in germinal centers and foster memory B cell generation. Immunity. 2021;54(10):2256‐2272 e2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang HY, Rivas‐Caicedo A, Renevey F, et al. Identification of a new subset of lymph node stromal cells involved in regulating plasma cell homeostasis. Proc Natl Acad Sci USA. 2018;115(29):E6826‐E6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Raeber ME, Zurbuchen Y, Impellizzieri D, Boyman O. The role of cytokines in T‐cell memory in health and disease. Immunol Rev. 2018;283(1):176‐193. [DOI] [PubMed] [Google Scholar]
- 90. Bajénoff M, Glaichenhaus N, Germain RN. Fibroblastic reticular cells guide T lymphocyte entry into and migration within the splenic T cell zone. J Immunol. 2008;181(6):3947‐3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kapoor VN, Müller S, Keerthivasan S, et al. Gremlin 1(+) fibroblastic niche maintains dendritic cell homeostasis in lymphoid tissues. Nat Immunol. 2021;22(5):571‐585. [DOI] [PubMed] [Google Scholar]
- 92. Baptista AP, Gola A, Huang Y, et al. The chemoattractant receptor Ebi2 drives intranodal naive CD4(+) T cell peripheralization to promote effective adaptive immunity. Immunity. 2019;50(5):1188‐1201 e1186. [DOI] [PubMed] [Google Scholar]
- 93. Duckworth BC, Lafouresse F, Wimmer VC, et al. Effector and stem‐like memory cell fates are imprinted in distinct lymph node niches directed by CXCR3 ligands. Nat Immunol. 2021;22(4):434‐448. [DOI] [PubMed] [Google Scholar]
- 94. Braun A, Worbs T, Moschovakis GL, et al. Afferent lymph‐derived T cells and DCs use different chemokine receptor CCR7‐dependent routes for entry into the lymph node and intranodal migration. Nat Immunol. 2011;12(9):879‐887. [DOI] [PubMed] [Google Scholar]
- 95. Clatworthy MR, Aronin CE, Mathews RJ, Morgan NY, Smith KG, Germain RN. Immune complexes stimulate CCR7‐dependent dendritic cell migration to lymph nodes. Nat Med. 2014;20(12):1458‐1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Woodruff MC, Heesters BA, Herndon CN, et al. Trans‐nodal migration of resident dendritic cells into medullary interfollicular regions initiates immunity to influenza vaccine. J Exp Med. 2014;211(8):1611‐1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gonzalez SF, Lukacs‐Kornek V, Kuligowski MP, et al. Capture of influenza by medullary dendritic cells via SIGN‐R1 is essential for humoral immunity in draining lymph nodes. Nat Immunol. 2010;11(5):427‐434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hannedouche S, Zhang J, Yi T, et al. Oxysterols direct immune cell migration via EBI2. Nature. 2011;475(7357):524‐527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yi T, Wang X, Kelly LM, et al. Oxysterol gradient generation by lymphoid stromal cells guides activated B cell movement during humoral responses. Immunity. 2012;37(3):535‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lu E, Dang EV, McDonald JG, Cyster JG. Distinct oxysterol requirements for positioning naïve and activated dendritic cells in the spleen. Sci Immunol. 2017;2(10):eaal5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Acton SE, Farrugia AJ, Astarita JL, et al. Dendritic cells control fibroblastic reticular network tension and lymph node expansion. Nature. 2014;514(7523):498‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Astarita JL, Cremasco V, Fu J, et al. The CLEC‐2‐podoplanin axis controls the contractility of fibroblastic reticular cells and lymph node microarchitecture. Nat Immunol. 2015;16(1):75‐84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kumar V, Dasoveanu DC, Chyou S, et al. A dendritic‐cell‐stromal axis maintains immune responses in lymph nodes. Immunity. 2015;42(4):719‐730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Yang CY, Vogt TK, Favre S, et al. Trapping of naive lymphocytes triggers rapid growth and remodeling of the fibroblast network in reactive murine lymph nodes. Proc Natl Acad Sci USA. 2014;111(1):E109‐118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kallert SM, Darbre S, Bonilla WV, et al. Replicating viral vector platform exploits alarmin signals for potent CD8+ T cell‐mediated tumour immunotherapy. Nat Commun. 2017;8:15327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Brown FD, Sen DR, LaFleur MW, et al. Fibroblastic reticular cells enhance T cell metabolism and survival via epigenetic remodeling. Nat Immunol. 2019;20(12):1668‐1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Yu M, Guo G, Zhang X, et al. Fibroblastic reticular cells of the lymphoid tissues modulate T cell activation threshold during homeostasis via hyperactive cyclooxygenase‐2/prostaglandin E(2) axis. Sci Rep. 2017;7(1):3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular helper T cells. Annu Rev Immunol. 2016;34:335‐368. [DOI] [PubMed] [Google Scholar]
- 109. Efremova M, Vento‐Tormo R, Park JE, Teichmann SA, James KR. Immunology in the era of single‐cell technologies. Annu Rev Immunol. 2020;38:727‐757. [DOI] [PubMed] [Google Scholar]
- 110. Bar‐Ephraim YE, Konijn T, Gonultas M, Mebius RE, Reijmers RM. A reproducible method for isolation and in vitro culture of functional human lymphoid stromal cells from tonsils. PLoS One. 2016;11(12):e0167555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Link A, Hardie DL, Favre S, et al. Association of T‐zone reticular networks and conduits with ectopic lymphoid tissues in mice and humans. Am J Pathol. 2011;178(4):1662‐1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Steiniger BS, Wilhelmi V, Seiler A, Lampp K, Stachniss V. Heterogeneity of stromal cells in the human splenic white pulp. Fibroblastic reticulum cells, follicular dendritic cells and a third superficial stromal cell type. Immunology. 2014;143(3):462‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zeng M, Southern PJ, Reilly CS, et al. Lymphoid tissue damage in HIV‐1 infection depletes naïve T cells and limits T cell reconstitution after antiretroviral therapy. PLoS Pathog. 2012;8(1):e1002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Mourcin F, Verdière L, Roulois D, et al. Follicular lymphoma triggers phenotypic and functional remodeling of the human lymphoid stromal cell landscape. Immunity. 2021;54(8):1788‐1806 e1787. [DOI] [PubMed] [Google Scholar]
- 115. Kong C, Bobe S, Pilger C, et al. Multiscale and multimodal optical imaging of the ultrastructure of human liver biopsies. Front Physiol. 2021;12:637136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Migliori B, Datta MS, Dupre C, et al. Light sheet theta microscopy for rapid high‐resolution imaging of large biological samples. BMC Biol. 2018;16(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Okada T, Miller MJ, Parker I, et al. Antigen‐engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 2005;3(6):e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Takeuchi A, Ozawa M, Kanda Y, et al. A distinct subset of fibroblastic stromal cells constitutes the cortex‐medulla boundary subcompartment of the lymph node. Front Immunol. 2018;9:2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mionnet C, Mondor I, Jorquera A, et al. Identification of a new stromal cell type involved in the regulation of inflamed B cell follicles. PLoS Biol. 2013;11(10):e1001672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Glanville SH, Bekiaris V, Jenkinson EJ, Lane PJ, Anderson G, Withers DR. Transplantation of embryonic spleen tissue reveals a role for adult non‐lymphoid cells in initiating lymphoid tissue organization. Eur J Immunol. 2009;39(1):280‐289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Nakagawa R, Togawa A, Nagasawa T, Nishikawa S. Peyer's patch inducer cells play a leading role in the formation of B and T cell zone architecture. J Immunol. 2013;190(7):3309‐3318. [DOI] [PubMed] [Google Scholar]
- 122. Schneider P, Takatsuka H, Wilson A, et al. Maturation of marginal zone and follicular B cells requires B cell activating factor of the tumor necrosis factor family and is independent of B cell maturation antigen. J Exp Med. 2001;194(11):1691‐1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Katakai T, Suto H, Sugai M, et al. Organizer‐like reticular stromal cell layer common to adult secondary lymphoid organs. J Immunol. 2008;181(9):6189‐6200. [DOI] [PubMed] [Google Scholar]
- 124. Katakai T, Hara T, Sugai M, Gonda H, Shimizu A. Lymph node fibroblastic reticular cells construct the stromal reticulum via contact with lymphocytes. J Exp Med. 2004;200(6):783‐795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Mueller SN, Matloubian M, Clemens DM, et al. Viral targeting of fibroblastic reticular cells contributes to immunosuppression and persistence during chronic infection. Proc Natl Acad Sci USA. 2007;104(39):15430‐15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lokmic Z, Lämmermann T, Sixt M, Cardell S, Hallmann R, Sorokin L. The extracellular matrix of the spleen as a potential organizer of immune cell compartments. Semin Immunol. 2008;20(1):4‐13. [DOI] [PubMed] [Google Scholar]
- 127. Bogdanova D, Takeuchi A, Ozawa M, et al. Essential role of canonical NF‐kappaB activity in the development of stromal cell subsets in secondary lymphoid organs. J Immunol. 2018;201(12):3580‐3586. [DOI] [PubMed] [Google Scholar]
- 128. Inra CN, Zhou BO, Acar M, et al. A perisinusoidal niche for extramedullary haematopoiesis in the spleen. Nature. 2015;527(7579):466‐471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Choi SY, Bae H, Jeong SH, et al. YAP/TAZ direct commitment and maturation of lymph node fibroblastic reticular cells. Nat Commun. 2020;11(1):519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Knoop KA, Kumar N, Butler BR, et al. RANKL is necessary and sufficient to initiate development of antigen‐sampling M cells in the intestinal epithelium. J Immunol. 2009;183(9):5738‐5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Habbeddine M, Verthuy C, Rastoin O, et al. Receptor activator of NF‐kappaB orchestrates activation of antiviral memory CD8 T cells in the spleen marginal zone. Cell Rep. 2017;21(9):2515‐2527. [DOI] [PMC free article] [PubMed] [Google Scholar]


