Abstract
In transgenic experiments, we often face fundamental requirements such as overexpressing a certain gene, developing organelle markers, testing promoter activities, introducing large genomic fragments, and combinations of them. To fulfill these multiple requirements in rice, we developed simple binary vectors with or without maize ubiquitin (UBQ) promoter, Gateway cassette and fluorescent proteins. First, we compared stabilities of cauliflower mosaic virus 35S and maize UBQ promoters for constitutive gene expression in transgenic rice. We show that the 35S promoter was frequently silenced after shoot regeneration, whereas maize UBQ promoter achieved stable expression in various young tissues. Binary vectors with Gateway cassettes under the control of the UBQ promoter allowed us to develop stable organelle markers for nuclei, microtubules and P-bodies in rice. The maize UBQ promoter can be easily replaced with any promoters of interest as exemplified by reporters of mitotic cells and provascular bundles. Finally, by introducing two genomic fluorescent reporters, we showed utilities of the Gateway cassette and two selection markers in large DNA fragment transfer and sequential transformations, respectively. Thus, these binary vectors provide useful choices of transgenic experiments in rice.
Keywords: binary vector, fluorescent reporter, Gateway, maize ubiquitin promoter, rice
Introduction
Transgenic approaches provide wide range of experimental choices to test biological hypotheses. These include testing gene function by constitutive expression, visualizing protein subcellular localization or promoter activities, and introducing genomic DNA fragments to complement causal genes in mutants. We often encounter a situation that requires these approaches in combination, in which sequential transformation with different selection markers facilitates experimental progress. Thus, there is a demand to develop binary vectors that achieve a desirable plasmid construction promptly and certainly.
Cloning DNA fragments into binary vectors is an essential step to produce transformation constructs. Conventional methods using restriction enzymes and ligases are time consuming and are often limited by the availability of suitable restriction sites. Cloning methods using Gibson assembly allows seamless construction of desired plasmids (Gibson et al. 2009). This method is actually powerful and used also in this study. However, insert DNA and linearized vectors need to have homologous overhangs which are usually introduced by PCR amplification. Therefore, it is important to confirm the final product by Sanger sequencing to avoid undesired mutations during PCR, which is laborious in case that the size of amplified DNA is large or that the same fragment is transferred to various vectors. Gateway technology is a convenient way to transfer a DNA fragment from a vector to another. It allows reliable transfer of DNA of interest from entry to destination vectors without use of restriction enzymes and therefore has been utilized in wide range of transformation vectors. Once cloned into entry vectors, various experimental designs become feasible with binary vectors available for downstream applications.
To realize functional analyses of proteins of interest or to develop a series of organelle markers, constitutive promoters are important to drive transgene expression. Cauliflower mosaic virus 35S promoter is the most widely used constitutive promoter in plants (Benfey and Chua 1990). Several subsequent studies incorporated Gateway technology into 35S-based overexpression vectors and provided quick and reliable construction systems (Earley et al. 2006; Nakagawa et al. 2007a, b). It has been reported that the 35S promoter is active in rice, however, there are also other studies reporting a low activity of 35S promoter in monocot species (Battraw and Hall 1990; Christensen et al. 1992; Cornejo et al. 1993; Gupta et al. 2001; Terada and Shimamoto 1990). Thus, it is unclear how the 35S promoter activity is stable in rice. Previous studies identified several other promoters which drive constitutive expression in rice (Li et al. 2014; Park et al. 2010). Especially, the maize UBQ promoter has been characterized in detail and confirmed its activity in various tissues and organs such as leaf blade, spikelet, root, embryo and endosperm (Cornejo et al. 1993; Park et al. 2010). However, Gateway-based vectors with the promoter have not been reported.
We tested stabilities of 35S and maize UBQ promoters during rice transformation and transformant growth, and found that maize UBQ promoter achieved stable and constitutive expression in all rice tissues examined. Based on this result, we adopted maize UBQ promoter in a series of binary vectors with fluorescent proteins and Gateway cassettes for fluorescent organelle marker production in rice. Additional properties including exchangeability of the UBQ promoter, capacity to clone large DNA fragments and two choices of selection markers, further extend utilities of these vectors and facilitate transgenic experiments in rice.
Materials and methods
All DNA fragments amplified by PCR for construction were confirmed by Sanger sequencing to avoid undesired mutations. Primers used in PCR reactions were listed in Supplementary Table S1. Vector maps and sequences are available upon request.
Vector and plasmid construction
To construct GFP under the control of the 35S promoter (pro35S:GFP), the GFP coding sequence (CDS) was inserted between XhoI and XbaI sites in pBCH1 (Ito and Kurata 2008).
For pPUB construction, maize UBQ promoter was amplified by PCR from pBUH4 (Nigorikawa et al. 2012) and cloned between HindIII and KpnI sites in pPZP2H-lac (Fuse et al. 2001).
To construct pPUG1, pPUG2 and pPUG3, Gateway cassettes without GFP, with N-terminal GFP and with C-terminal GFP were PCR amplified from pGWB2, pGWB6 and pGWB4 (Nakagawa et al. 2007a), cloned between HindIII and XbaI sites in pBluescript SK- (Stratagene) and designated as pBS_PUG1, pBS_PUG2 and pBS_PUG3, respectively. These Gateway cassettes were then cloned into HindIII and SpeI sites in pPUB and named as pPUG1 (Gateway only), pPUG2 (N-terminal GFP and Gateway) and pPUG3 (Gateway and C-terminal GFP).
To construct mCherry binary vectors, pBS_PUG2 and pBS_PUG3 without GFP were amplified by inverse PCR using primers with overhangs to mCherry. These linearized pBS_PUG2 and pBS_PUG3 were circularized with mCherry CDS by NEBuilder HiFi DNA assembly (New England Biolabs) and designated as pBS_PUG4 and pBS_PUG5, respectively. Then, Gateway cassettes with mCherry were cloned into HindIII and SpeI sites in pPUB and resulting plasmids were designated as pPUG4 (N-terminal mCherry fusion) and pPUG5 (C-terminal mCherry fusion).
pPG was constructed by introducing a 1.7 kb SacI-HindIII fragment from pPUG1, which contained a Gateway cassette without any tags and terminators, into SacI-HindIII sites of pPZP2H-lac.
To construct pPUG1n–pPUG5n, we first replaced hpt in pPUB with nptII by introducing a DNA fragment containing OsAct1 promoter, nptII and AtPAB5 terminator amplified from pPN/hyPBase (Nishizawa-Yokoi et al. 2014) into MluI-StuI sites. This vector was designated as pPUBn. Gateway cassettes from pBS_PUG1–pBS_PUG5 (HindIII-XbaI fragments) were then introduced into HindIII-SpeI sites of pPUBn and these binary vectors were named as pPUG1n–pPUG5n, respectively.
For pPGn, the same nptII containing fragment amplified from pPN/hyPBase was cloned into PshAI-StuI sites in pPG using by NEBuilder.
Organelle marker and tissue specific reporter construction
CDSs of histone H2B, OsDCP2 and TOB3 were PCR amplified from rice cDNA from shoot apices and cloned into pENTR-D/TOPO (Thermo Fisher). The GUS CDS was amplified from pBGH1 (Ito and Kurata 2008). These CDSs were amplified without stop codons to allow C-terminal fusions. H2B, GUS and TOB3 CDSs were transferred to pPUG3 for C-terminal GFP fusion using LR clonase II (Thermo Fisher). OsDCP2 CDS was transferred to pPUG5n for C-terminal mCherry fusion. GFP- AtTUB6 in pENTR was a gift from Dr. Oda (Oda et al. 2010), and transferred to pPUG1.
To construct tissue specific reporters, CyclinB1;1 or Oshox1 promoter fragments were amplified from Nipponbare genomic DNA. These fragments were cloned into HindIII-KpnI sites in place of the UBQ promoter in pPUG3_GUS and pPUG3_TOB3, respectively, by NEBuilder HiFi assembly.
For genomic reporters, OSH15 and TOB3 genomic DNA fragments were cloned from BAC clones OSJNBa0031J04 and OSJNBa0007D12, respectively. To clone these genomic DNA fragments, pENTR2.2 was constructed by digesting pENTR_H2B with NotI and AscI and by replacing histone H2B CDS with double stranded oligo nucleotides listed in Supplementary Table S1. For OSH15, the BAC clone was digested with BmgBI and a 13.5 kb fragment containing 5.6 kb upstream, entire OSH15 gene and 2.3 kb downstream regions was purified from a gel slice after electrophoresis. In NEBuilder HiFi assembly, this genomic DNA fragment was cloned into pENTR2.2 vector, which was amplified by inverse PCR to introduce overhangs homologous to both ends of the OSH15 fragment. This plasmid was designated as pENTR2.2_gOSH15. The N-terminal region of OSH15 between AccIII and BstEII sites was PCR amplified and cloned into pCR_Blunt (Thermo Fisher). A coding sequence of GFP with 10x Alanine linker was amplified from gGFP-OSH1 (Tsuda et al. 2011) and inserted into N-terminus of OSH15 in this plasmid. This N-terminal GFP fusion of OSH15 was introduced into pENTR2.2_gOSH15 in place of the original OSH15 N-terminus between AccIII and BstEII sites using NEBuilder, and designated as pENTR2.2_gGFP-OSH15. gGFP-OSH15 was transferred to pPG by LR recombination.
TOB3 genomic reporter was constructed similarly. The BAC clone was digested PshAI and the 16 kb DNA fragment containing 9.6 kb upstream, 4.0 kb TOB3 gene and 2.4 kb downstream regions was cloned into pENTR2.2 using NEBuilder HiFi assembly. This plasmid was designated as pENTR2.2_gTOB3. The C-terminal region of TOB3 between two AccIII sites was PCR amplified and cloned into pCR_Blunt. A coding sequence of mCherry with 6x Alanine linker was inserted into C-terminus of TOB3 in this plasmid by NEBuilder HiFi assembly. This C-terminal mCherry fusion of TOB3 was introduced into pENTR2.2_gTOB3 in place of the original TOB3 C-terminus between two AccIII sites and the resultant plasmid was designated as pENTR2.2_gTOB3-mCherry. gTOB3-mCherry was transferred to pPGn by LB recombination.
Plant materials and transformation
Transgenes were introduced into Nipponbare using Agrobacterium mediated transformation as described previously (Hiei et al. 1994). pPG_gGFP-OSH15 was introduced into d6 homozygotes mutant and transgenic calli were selected on selection media with hygromycin (50 mg l−1). pPGn_gTOB3-mCherry was introduced into gGFP-OSH15 d6 transgenic calli and the double transgenic lines were selected on selection media containing G418 (35 mg l−1). We note that when using G418, agar (8.0 g l−1) was used instead of gellan gum to allow solubilization of G418 (Hiei and Komari 2008).
RNA extraction, cDNA synthesis and quantitative PCR
Total RNA was extracted from shoots and calli using Nucleospin RNA plant (Macherey-Nagel). After treating 400 ng of total RNA with recombinant DNase I (TaKaRa), cDNA was synthesized using SuperscriptIII Reverse Transcriptase (Thermo Fisher) in 20 µl reaction and diluted with 80 µl of Tris-EDTA buffer. Quantitative PCR was carried out using Thermal Cycler Dice Real Time System (TaKaRa) and KAPA SYBR Fast qPCR Kit (KAPA Biosysmtems) with 1 µl of cDNA as template in 10 µl reaction. Three technical replicates were performed for each sample. Relative expression levels were calculated using 2−∆∆Ct method and Rice Actin1 (RAc1) as an internal standard.
Fluorescent reporter observation
Samples were fixed in 4% paraformaldehyde overnight at 4°C. Anthers and root tips were directly mounted in VECTASHIELD (Vector Laboratories) on slide glasses. To observe fluorescence in shoot apices and young stems, fixed samples were embedded in 7% of Certified Low Range Ultra Agarose (Bio-Rad), and sliced into 50 µm sections using a microslicer DTK-1000 (DOSAKA). Sections were mounted in VECTASHIELD on slide glasses. Fluorescent images were captured under Fluoview FV300 CLSM system (Olympus). Photographs for larger samples were taken under the dissecting microscope (SZX16, Olympus) or using a digital camera (TG-6, Olympus) with a GFP filter.
Beta-glucuronidase (GUS) staining
GUS staining was conducted as described previously (Jefferson et al. 1987). Samples were dissected out to expose tissues of interest and incubated in 90% acetone at 4°C for 1 h. After removal of acetone, samples were rinsed twice in GUS buffer (100 mM sodium phosphate buffer [pH 7.2], 10 mM EDTA, 5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 0.1% Triton X-100), vacuumed for 1 h in GUS-staining buffer (1 mM X-gluc in GUS buffer) and incubated at room temperature for overnight. The staining reaction was terminated by replacing GUS-staining buffer with 70% ethanol and samples were observed under the dissecting microscope (SZX16, Olympus).
Results
Comparison of promoter activities between 35S and UBQ
Rice calli transformed with the pro35S:GFP via Agrobacterium infection showed a strong GFP signal (Figure 1B). However, downregulation of GFP fluorescence was frequently observed in plants regenerated from the pro35S:GFP calli (Figure 1C). Nine out of 10 independent lines showed severely reduced GFP transcript levels in shoots at one month after regeneration when compared to their derived calli (Figure 1D, E). Thus, 35S promoter was frequently silenced in rice after shoot regeneration.
Figure 1. Comparison of activities between 35S and maize UBQ promoters. (A) GFP fluorescence in proUBQ:GFP callus. (B) GFP fluorescence in pro35S:GFP callus. (C) GFP fluorescence in stems of proUBQ:GFP (left) and pro35S:GFP (right). pro35S:GFP plants were outlined with dashed lines for clarity. (D) Quantification of GFP transcript levels in calli. (E) Quantification of GFP transcript levels in regenerated shoots. GFP transcript levels were normalized relative to those of rice actin gene RAc1. Scale bars are 1 mm in (A) and (B), 1 cm in (C).
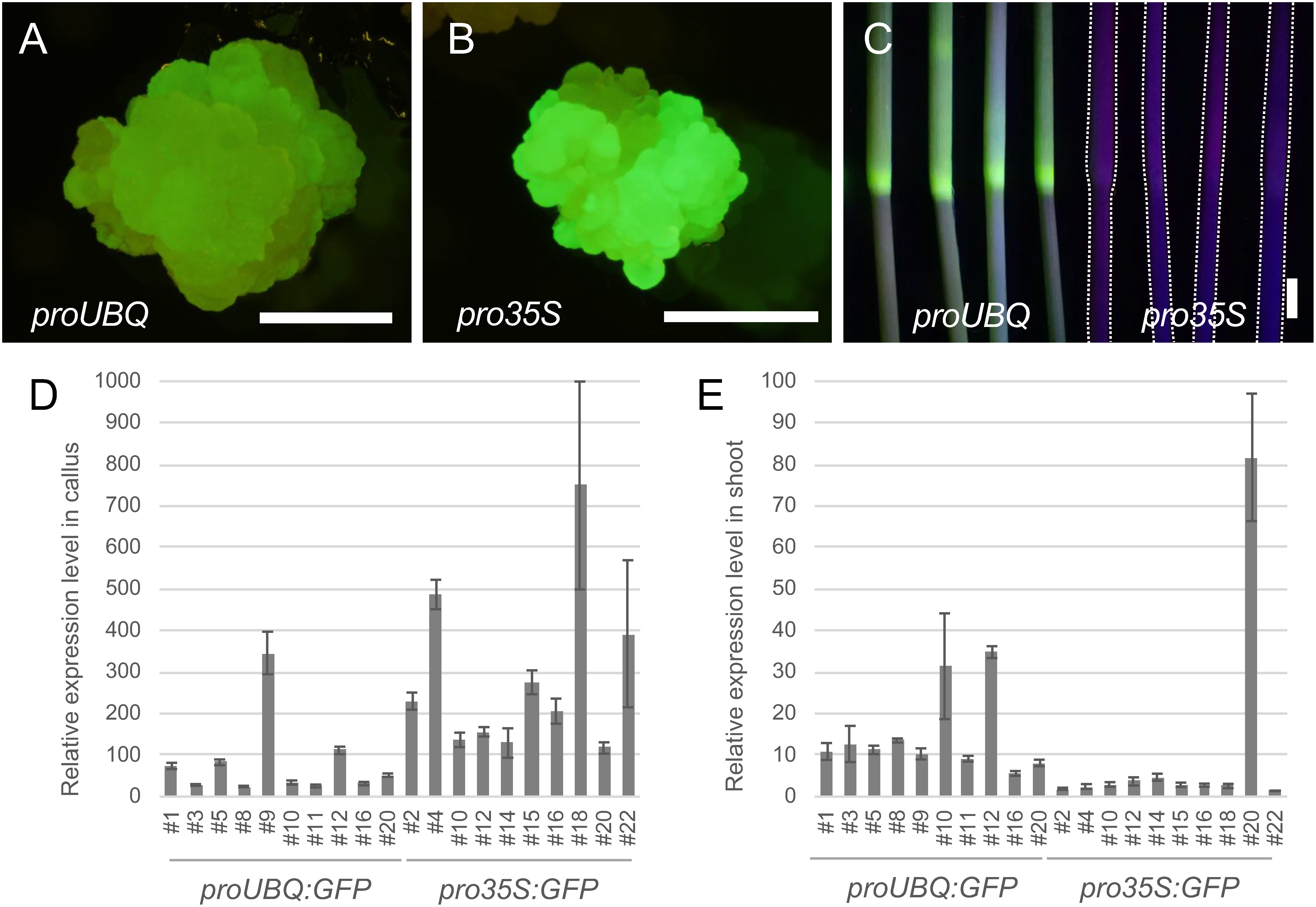
The maize UBQ promoter is known as a constitutive promoter in monocot species especially in young tissues (Christensen et al. 1992; Cornejo et al. 1993; Park et al. 2010). We next observed GFP expression under the control of the UBQ promoter in transgenic rice. In proUBQ:GFP calli on the selecting medium, clear GFP fluorescence was observed, although it was weaker than that in pro35S:GFP (Figure 1A). However, proUBQ:GFP kept showing high GFP fluorescence after shoot regeneration (Figure 1C). Compared to pro35S:GFP, GFP transcript levels in proUBQ:GFP were lower in callus but higher in shoots (Figure 1D, E).
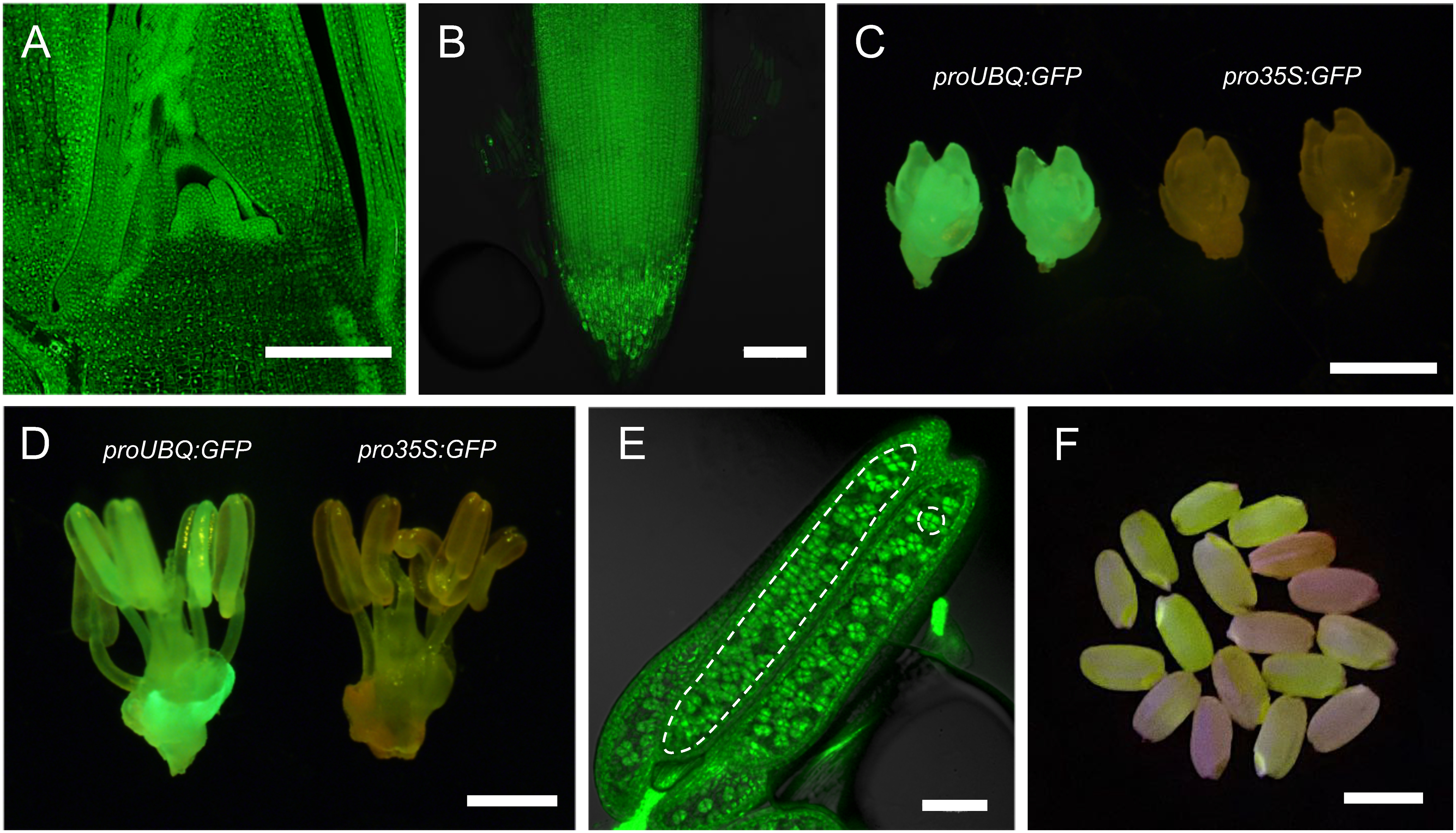
In proUBQ:GFP shoot and root apices, the GFP expression was stable and ubiquitous (Figure 2A, B). The constitutive expression was maintained after reproductive transition (Figure 2C, D, E). In anthers, the GFP fluorescence was clearly observed in tetrad microspores in addition to somatic wall cells, indicating this promoter was active in germline cells (Figure 2E). GFP expression was also maintained in T1 generation seeds (Figure 2F). Importantly, these stable and ubiquitous expression was observed in multiple transgenic lines independently. In contrast, once silenced, GFP fluorescence in pro35S:GFP was no longer recovered in all tissue and organs examined. These results highlighted the suitability of the maize UBQ promoter as constitutive promoter in rice.
Figure 2. Observation of GFP fluorescence in proUBQ:GFP in various organs. (A, B and E) Confocal images of GFP fluorescence in the shoot apex (A), in the root tip (B) and in the anther (E). In (E), an anther locule was encircled with a dashed line on the left to distinguish it from somatic anther walls. A unit of tetrad microspores was also indicated with a dashed circle on the right. (C, D and F) GFP fluorescence in young spikelet (C), in anther and pistils (D) and in T1 seeds (F). Note that GFP positive and negative seeds are segregating (F). In C and D, pro35S:GFP samples were shown on the right as comparison. Scale bars are 100 µm in A, B and E, 1 mm in C and D, and 5 mm in F.
Development of Gateway-based binary vectors
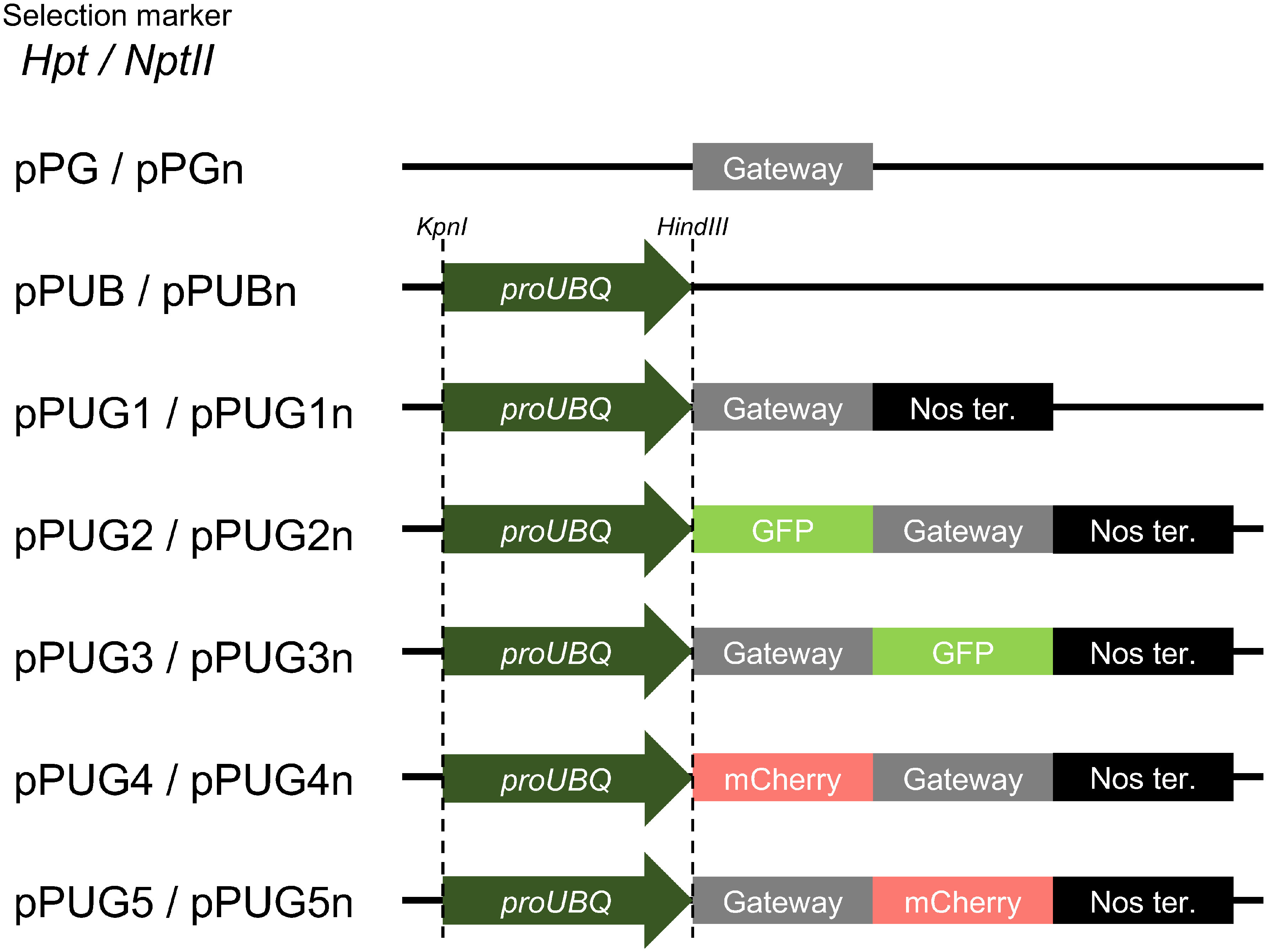
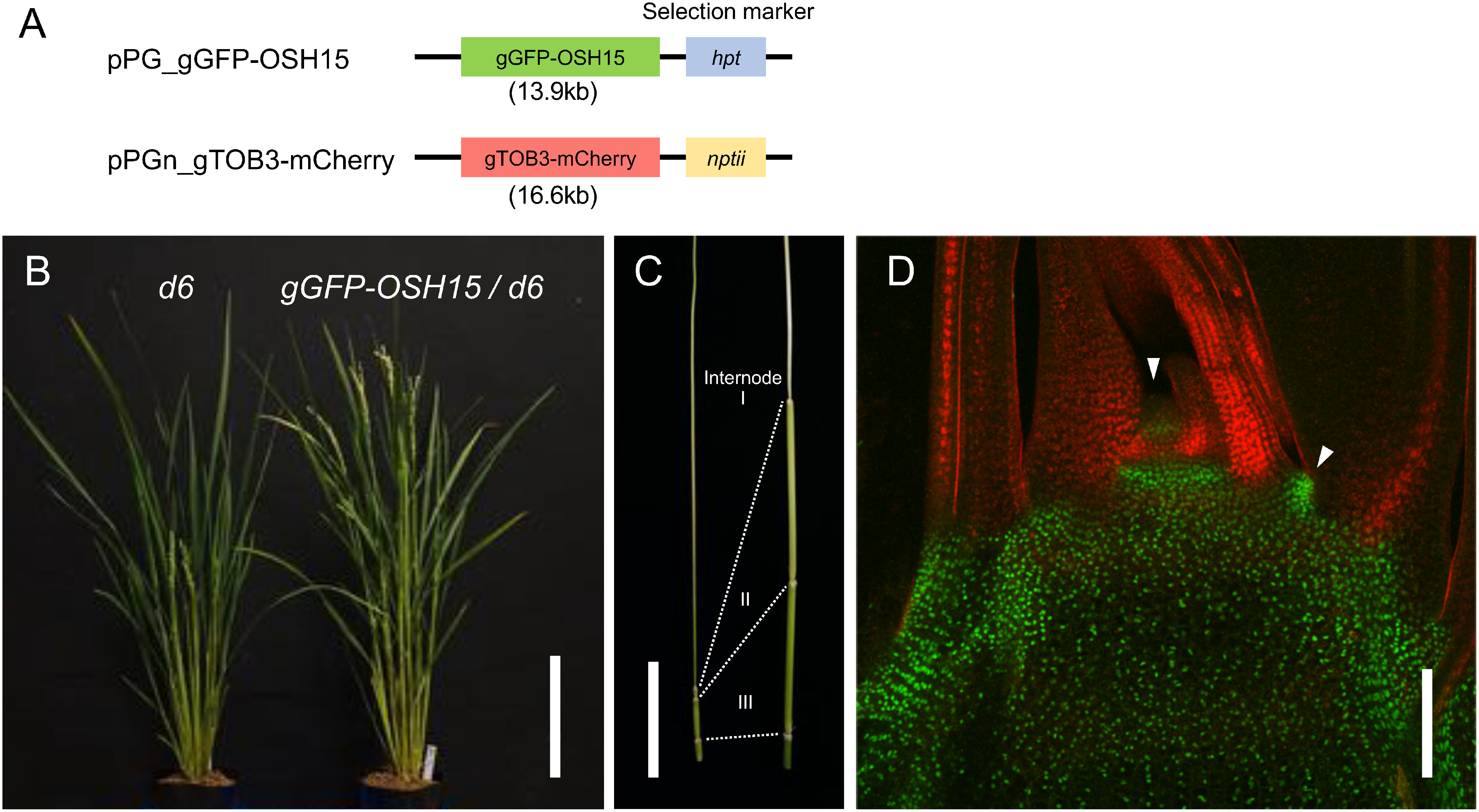
To facilitate the maize UBQ promoter-based plasmid construction, we developed a series of binary vectors that consists of combinations of Gateway cassettes, the UBQ promoter, fluorescent proteins and selection markers (Figure 3). First, the UBQ promoter was inserted into the multi-cloning site of pPZP2H-lac, a binary vector with the hygromycin-resistant gene htp (Fuse et al. 2001), and named as pPUB. Next, we produced five types of pPUB-based Gateway cassette-containing vectors for constitutive expression; pPUG1 carrying the UBQ promoter simply fused with a Gateway cassette, pPUG2 and pPUG3 carrying Gateway cassettes under the UBQ promoter with GFP at N- or C-termini, respectively, and pPUG4 and pPUG5, in which GFP of pPUG2 and pPUG3 were replaced with mCherry, respectively. All above Gateway cassettes with or without fluorescent proteins were followed by nopaline synthase (Nos) terminator. In addition, we prepared pPG, which harbors only a Gateway cassette in pPZP2H-lac to solely transfer a DNA fragment for experiments such as complementation tests. We also made pPUG1n–pPUG5n and pPGn, the htp-replaced version of pPUG1–pPUG5 and pPG with nptII, the kanamycin resistant gene reported to work in rice cells (Nishizawa-Yokoi et al. 2014) .
Figure 3. Gateway-based binary vectors for rice developed in this study.
Production of organelle markers and tissue specific reporters
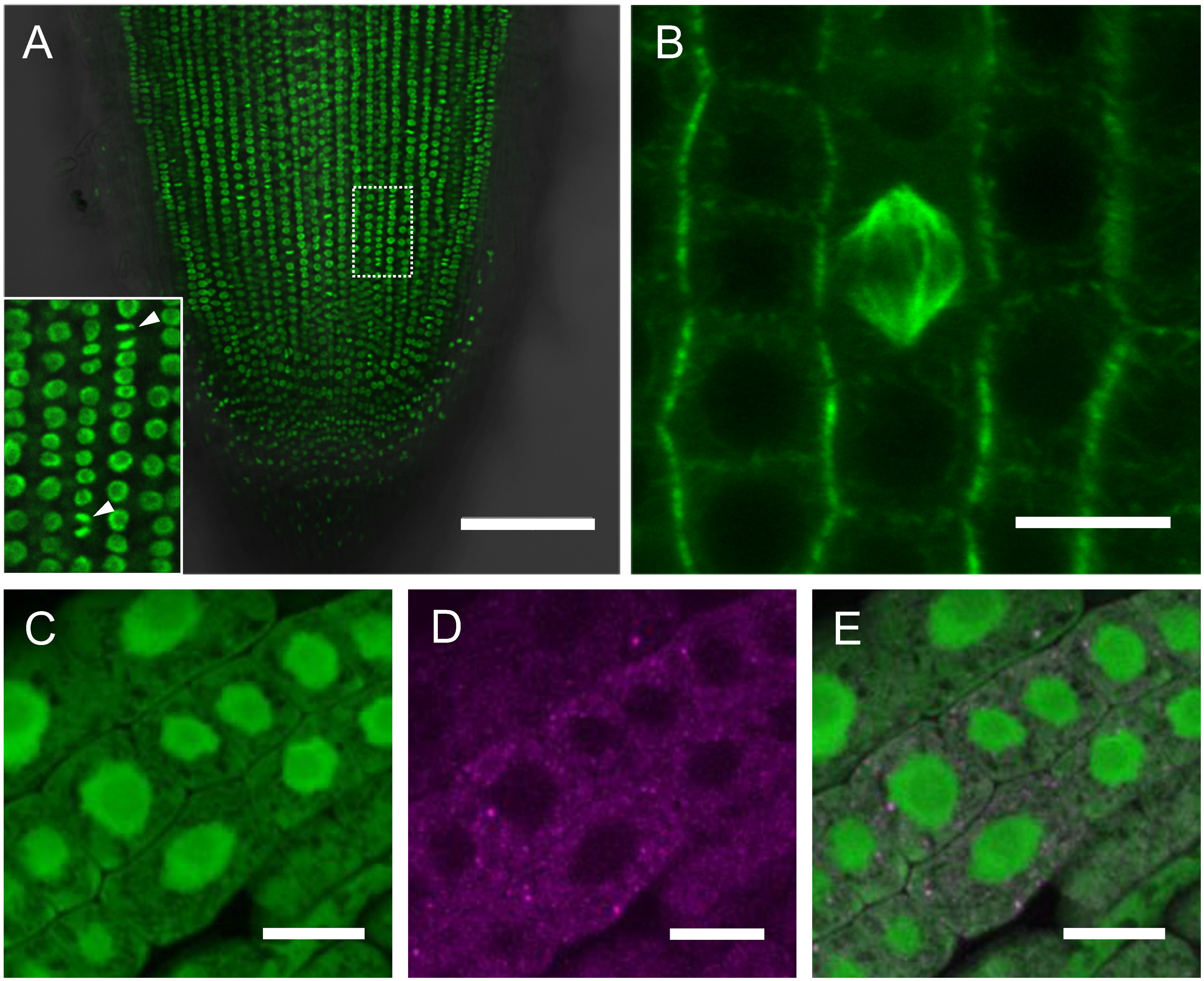
To verify the utility of above vectors for subcellular marker construction in rice, we selected three rice and Arabidopsis genes, encoding proteins whose subcellular localizations are previously reported. All gene CDSs were once cloned into the pENTR vector, and then transferred to respective pPUG vectors via LR recombination. As a nuclear marker, a rice histone H2B gene was transferred into pPUG3 and introduced into rice calli. As expected, the proUBQ:H2B-GFP plants showed clear nuclear signals constitutively in roots (Figure 4A). For microtubules, Tubulin6 from Arabidopsis fused to GFP (GFP- AtTUB6) (Oda et al. 2010) was introduced into pPUG1. In mitotic cells, GFP signals were clearly observed alongside with spindle fibers in roots (Figure 4B). As a third example, the rice gene encoding a decapping enzyme, OsDCP2 (Guo et al. 2016) was inserted into pPUG5n. OsDCP2-mCherry showed a punctate dotted pattern with faint diffused signals in cytosol (Figure 4C, D, E). DCP proteins are known as components of the processing body (P-body), a ribonucleoprotein granules required for mRNA processing or decay (Decker and Parker 2012). Thus, these markers showed patterns of subcellular localizations as expected, verifying the utility of these vectors for organelle marker production in rice.
Figure 4. Examples of fluorescent protein-tagged organelle markers. (A) Nuclei marked by proUBQ:H2B-GFP in a root apex. Arrowheads in the insets indicate nuclei at mitosis. A dashed box is the area magnified in the inset. (B) Microtubules in proUBQ: GFP-AtTUB6 root epidermis. Center is a spindle structure at metaphase in mitosis. (C–D) P-bodies in root marked by proUBQ:OsDCP2-mCherry introduced in proUBQ:GFP background. Fluorescence channels are for GFP in (C), for OsDCP2-mCherry in (D). (E) is a merged image of (C) and (D). Scale bars are 100 µm in (A), 10 µm in (B) to (E).
pPUG1–pPUG5 vectors harbor unique HindIII and KpnI sites at both ends of the UBQ promoter. These restriction sites enable us to easily replace the UBQ promoter with any promoter of interest using seamless cloning methods. We first constructed proUBQ:GUS-GFP by cloning GUS CDS into pPUG3 and replaced its UBQ promoter with a 2.7 kb regulatory sequence of rice CyclinB1;1. It included 1.6 kb promoter region, 5’ UTR and first four exons to place GUS-GFP under the control of both transcription and proteolysis during cell cycle. proCYCB1;1-GUS-GFP transgenic plants showed a patchy pattern of GUS staining at the base of leaf sheath, confirming its cell cycle-dependent reporter activity in mitotic cells (Figure 5A). As another example, we constructed a vascular bundle reporter. Oshox1 promoter was previously reported to drive vascular bundle-specific expression (Scarpella et al. 2000). Here, a CDS from YABBY transcription factor TONGARI BOUSHI3 (TOB3) was cloned in pPUG3 for C-terminal GFP fusion and the UBQ promoter was replaced with the Oshox1 promoter (Tanaka et al. 2017). In the stem of proOshox1:TOB3-GFP, we detected vascular bundle-enriched GFP signals in nuclei, although parenchymatous cells surrounding veins also showed weak expression (Figure 5B). These results exemplified compatibilities of our binary vectors with any promoters of interest.
Figure 5. Examples of cell-cycle or tissue specific reporters. (A) A proCYCB1;1-GUS seedling showing a band of mitotically active cells with GUS activity at the base of leaf sheath. (B) A cross section of the stem in proOshox1:TOB3-GFP. Confocal images of GFP (green) and cell wall stained with calcofluor white (gray) were merged. A few vascular bundles were indicated with dashed ovals to distinguish veins from surrounding cells. Scale bars are 2 mm in (A) and 100 µm in (B).
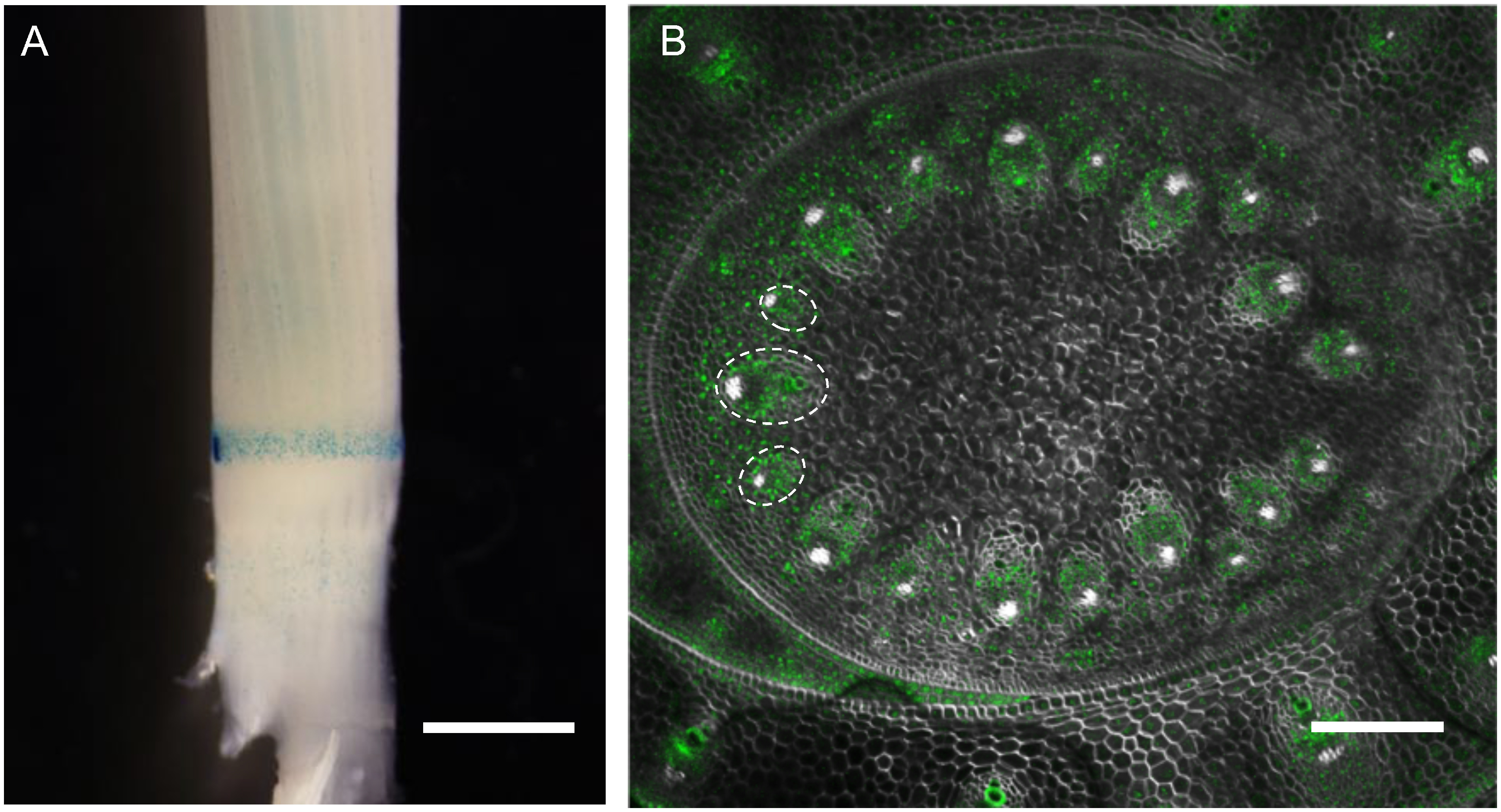
Introduction of GFP and mCherry genomic reporters through sequential transformation
In case that promoter regions are not sufficient to reproduce endogenous gene expression patterns, it is an effective approach to construct reporters using a genomic DNA fragment containing the entire gene structure. In addition, we often need to introduce genomic fragments to rescue mutants in complementation tests. For these purposes, we made pPG and pPGn that contain a Gateway cassette and hpt or nptII as selection markers, respectively (Figure 3). We constructed genomic fluorescent reporters of a rice knotted1-like homeobox (KNOX) gene Oryza sativa homeobox 15 (OSH15) with N-terminally fused GFP (gGFP-OSH15) and that of TOB3 with C-terminal mCherry (gTOB3-mCherry) in pPG and pPGn, respectively (Figure 6A). OSH15 is important for internode development and is specifically expressed in shoot meristems and young stems (Sato et al. 1999). TOB3 is expressed in incipient lateral organs and promote its differentiation (Tanaka et al. 2017). These genomic reporters were introduced into d6 mutant, a loss-of-function mutant of OSH15, through sequential transformations. To test if gGFP-OSH15 is functional, we regenerated shoots from a part of gGFP-OSH15 transgenic calli without gTOB3-mCherry introduction. The resultant transgenic plants recovered internode elongation compared to d6 mutants, confirming functional complementation of OSH15 (Figure 6B, C). In confocal imaging at the shoot apex of the gGFP-OSH15 gTOB3-mCherry plants, GFP-OSH15 was specifically expressed in the shoot meristems and young stems. On the other hand, TOB3-mCherry accumulated in leaf primordia (Figure 6D). These complementary expression patterns are consistent with previous reports (Sato et al. 1999; Tanaka et al. 2017), validating the utility of these binary vectors to introduce different genomic reporters in sequential transformation.
Figure 6. An example of double fluorescent genomic reporters. (A) Genomic reporter constructs introduced into d6 mutants. These two genomic reporter fragments were cloned in pPG and pPGn, which had hpt and nptII for selection, respectively. (B) Mature plants at seed setting showing that gGFP-OSH15 transgene recovered plant height in d6 mutant. Note that the gGFP-OSH15/d6 plant was regenerated without gTOB3-mCherry introduction to confirm the complementation. (C) Comparison of internode length between d6 (left) and gGFP-OSH15/d6 (right) at the heading stage. (D) A confocal image of the shoot apex in gGFP-OSH15 gTOB3-mCherry/d6 double genomic reporter line. GFP-OSH15 (green) accumulates in shoot meristems (arrowheads) and young stem, whereas TOB3-mCherry (red) is expressed in leaves. Scale bars are 20 cm in (B), 5 cm in (C) and 100 µm in (D).
Discussion
It has been reported that 35S promoter showed constitutive activities in various tissues in rice (Battraw and Hall 1990; Terada and Shimamoto 1990). In addition, it is the most widely used promoter in binary vectors to drive hpt expression during selection of transgenic rice calli using hygromycin. However, there are several studies reporting that 35S promoter activity was weak in monocot species including rice (Christensen et al. 1992; Cornejo et al. 1993; Gupta et al. 2001). In consistent with the latter reports, our trial to develop subcellular markers under the control of 35S promoter was not successful because the reporters were silenced in most transgenic plants. This prompted us to test the stability of 35S promoter activity in comparison with maize UBQ promoter, which was also known to drive constitutive expression in rice (Cornejo et al. 1993).
We observed very strong activity of the 35S promoter in callus in most independent lines, however, GFP fluorescence frequently became undetectable in regenerated shoots. Because GFP fluorescence was kept high as long as the calli were cultured on selection medium, it is likely that this silencing occurs and becomes evident after shoot regeneration or transplanting to soil with no antibiotic. This observation suggests that 35S promoter can be maintained active under certain selective pressures (such as hygromycin) but otherwise tends to be silenced. Thus, 35S promoter might be used as a constitutive promoter, but careful monitoring of transgene expression is especially important in use. From a perspective in genome editing, this property of 35S promoter in rice is rather advantageous, because its strong activity in callus enables high editing efficiency whereas frequent silencing after regeneration can stabilize mono-allelic mutations, which is valuable in case bi-allelic mutation results in lethality.
This study confirmed that as a constitutive promoter in rice, the maize UBQ promoter is more suitable than 35S promoter and its stable expression throughout the life cycle is especially important to obtain reporter production. Therefore, we propose that maize UBQ promoter- and Gateway-based binary vectors developed in this study are useful tools to make rice transgenic experiments more reproducible and handier than the binary vectors reported so far. We verified the utility of our vectors for reliable production of organelle markers. Two unique restriction sites (KpnI and HindIII) in these vectors allows to replace the maize UBQ promoter with any promoters of interest. In addition, our vectors were developed on pPZP2H-lac, which is a relatively high copy vector in Escherichia coli (E. coli) compared to pBI-based vectors (Jefferson et al. 1987). This is helpful when high yield of plasmid is required in experiments such as plasmid transfection assay into rice protoplasts (Zhang et al. 2011). Thus, our observation bore simple but versatile binary vectors in multiple types of experiments in rice.
We also showed that, by Gateway LR recombination, large genomic fragments can be reliably transferred from entry to binary vectors. In our case, gGFP-OSH15 and gTOB3-mCherry were 13.9 and 16.6 kb, respectively. We have confirmed successful transfer of insert DNA ranging from 0.4 to 18.3 kb through LR recombination in our experiments. Larger fragments might be cloned although we have not tested yet. This facilitates development of genomic reporters, which are often hampered by lack of available restriction sites in conventional cloning vectors. We note that, in case the size of DNA insert in pENTR was larger than 2 kb, linearizing binary vectors with XmaI located in Gateway cassette greatly improved efficiency of LR recombination. Another point to note is that lowering temperature from 37°C to 30°C or lower in E. coli culture often helped successful cloning especially when the insert size was large.
Taking together, by unveiling properties of two widely used constitutive promoters and developing a series of binary vectors, we paved several gaps which hampered transgenic experiments in rice. Addition of other protein tags, promoters and selection markers will widen the utility of these vectors.
Acknowledgments
We thank Kae Kato for technical assistance. We also thank Nishizawa-Yokoi Ayako for providing pPN/hyPBase plasmid and for advices on transgenic calli selection using G418, Yoshihisa Oda for GFP-AtTUB6 plasmid, Tsuyoshi Nakagawa for pGWB vectors, and Yukihiro Ito for pBCH1 and pBGH1 vectors. This work was supported by JSPS KAKENHI 19K05980 and 20H04891 to K. T. and by JSPS KAKENHI 21H04729 and JSPS Bilateral Program JPJSBP120213510 to K.-I. N..
Abbreviations
- GFP
green fluorescent protein
- UBQ
ubiquitin
Author contributions
K.T. designed the research, performed most experiments and wrote the manuscript with the help of K.-I. N.. M. M. and T. S. produced and observed OsDCP2-mCherry and GFP-AtTUB6, respectively.
Supplementary Data
References
- Battraw MJ, Hall TC (1990) Histochemical analysis of CaMV 35S promoter-beta-glucuronidase gene expression in transgenic rice plants. Plant Mol Biol 15: 527–538 [DOI] [PubMed] [Google Scholar]
- Benfey PN, Chua NH (1990) The cauliflower mosaic virus 35S promoter: Combinatorial regulation of transcription in plants. Science 250: 959–966 [DOI] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18: 675–689 [DOI] [PubMed] [Google Scholar]
- Cornejo MJ, Luth D, Blankenship KM, Anderson OD, Blechl AE (1993) Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol Biol 23: 567–581 [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R (2012) P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4: a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fuse T, Sakaki T, Yano M (2001) Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnol 18: 219–222 [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6: 343–345 [DOI] [PubMed] [Google Scholar]
- Guo C, Luo C, Guo L, Li M, Guo X, Zhang Y, Wang L, Chen L (2016) OsSIDP366, a DUF1644 gene, positively regulates responses to drought and salt stresses in rice. J Integr Plant Biol 58: 492–502 [DOI] [PubMed] [Google Scholar]
- Gupta P, Raghuvanshi S, Tyagi AK (2001) Assessment of the efficiency of various gene promoters via biolistics in leaf and regenerating seed callus of millets, Elusine coracana and Echinochloa crusgalli. Plant Biotechnol 18: 275–282 [Google Scholar]
- Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc 3: 824–834 [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Ito Y, Kurata N (2008) Disruption of KNOX gene suppression in leaf by introducing its cDNA in rice. Plant Sci 174: 357–365 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucoronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Xu RF, Qin RY, Ma H, Li H, Zhang YP, Li L, Wei PC, Yang JB (2014) Isolation and functional characterization of a novel rice constitutive promoter. Plant Cell Rep 33: 1651–1660 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007a) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamur S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007b) Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nigorikawa M, Watanabe A, Furukawa K, Sonoki T, Ito Y (2012) Enhanced saccharification of rice straw by overexpression of rice exo-glucanase. Rice (N Y) 5: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Endo M, Osakabe K, Saika H, Toki S (2014) Precise marker excision system using an animal-derived piggyBac transposon in plants. Plant J 77: 454–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Iida Y, Kondo Y, Fukuda H (2010) Wood cell-wall structure requires local 2D-microtubule disassembly by a novel plasma membrane-anchored protein. Curr Biol 20: 1197–1202 [DOI] [PubMed] [Google Scholar]
- Park SH, Yi N, Kim YS, Jeong MH, Bang SW, Choi YD, Kim JK (2010) Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J Exp Bot 61: 2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Sentoku N, Miura Y, Hirochika H, Kitano H, Matsuoka M (1999) Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J 18: 992–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Rueb S, Boot KJ, Hoge JH, Meijer AH (2000) A role for the rice homeobox gene Oshox1 in provascular cell fate commitment. Development 127: 3655–3669 [DOI] [PubMed] [Google Scholar]
- Tanaka W, Toriba T, Hirano HY (2017) Three TOB1-related YABBY genes are required to maintain proper function of the spikelet and branch meristems in rice. New Phytol 215: 825–839 [DOI] [PubMed] [Google Scholar]
- Terada R, Shimamoto K (1990) Expression of CaMV35S-GUS gene in transgenic rice plants. Mol Gen Genet 220: 389–392 [Google Scholar]
- Tsuda K, Ito Y, Sato Y, Kurata N (2011) Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23: 4368–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


