Abstract
Strychnine is a natural product that, through isolation, structural elucidation and synthetic efforts, shaped the field of organic chemistry. Currently, strychnine is used as a pesticide to control rodents1 because of its potent neurotoxicity2,3. The polycyclic architecture of strychnine has inspired chemists to develop new synthetic transformations and strategies to access this molecular scaffold4, yet it is still unknown how plants create this complex structure. Here we report the biosynthetic pathway of strychnine, along with the related molecules brucine and diaboline. Moreover, we successfully recapitulate strychnine, brucine and diaboline biosynthesis in Nicotiana benthamiana from an upstream intermediate, thus demonstrating that this complex, pharmacologically active class of compounds can now be harnessed through metabolic engineering approaches.
Subject terms: Biochemistry, Plant sciences
The biosynthetic pathway of strychnine, brucine and diaboline is described, and the biosynthesis of these complex, pharmacologically active compounds has been successfully recapitulated in Nicotiana benthamiana from an upstream intermediate.
Main
Strychnine—a complex monoterpene indole alkaloid—was isolated in 1818 from the seeds of Strychnos nux-vomica (poison nuts)5, which were used in traditional medicine in China and South Asia. Currently, strychnine is used as a pesticide1 because of its neurotoxicity, which is mediated by high-affinity binding to the glycine receptor2,3. Approximately 130 years after its isolation, the structure of strychnine was independently elucidated by Robinson in 1946 (refs. 6,7) and Woodward in 1947 (ref. 8). Robinson noted that ‘for its molecular size, it is the most complex substance known’9. For centuries, strychnine had a large role in the field of chemistry through its isolation, structural elucidation and synthesis (Supplementary Fig. 1). Its polycyclic architecture inspired chemists to develop new synthetic transformations and strategies, and ultimately led to a number of total syntheses4 since the first seminal total synthesis in 1954 (ref. 10). Surprisingly, it is still unknown how plants create this complex structure. Here we report the biosynthetic pathways of strychnine, brucine and diaboline.
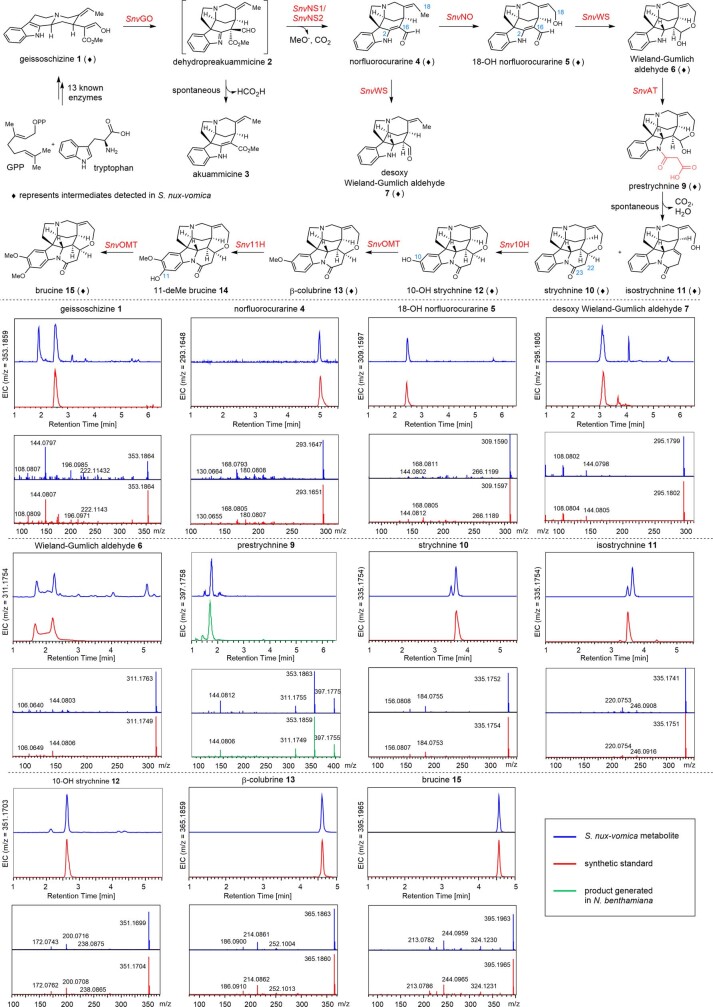
A partial biosynthetic hypothesis of strychnine was proposed in 1948 (ref. 11), which was substantiated by feeding studies of radioisotope-labelled substrates in S. nux-vomica12–15. These labelling studies demonstrated that, like all monoterpene indole alkaloids, strychnine 10 originates from tryptophan and geranyl pyrophosphate13. These starting materials are converted to two central intermediates, first geissoschizine 1 and then, through a series of unknown steps, to Wieland–Gumlich aldehyde 6 (refs. 14,15 and Fig. 1; see Supplementary Fig. 2 for full biosynthetic hypothesis). Wieland–Gumlich aldehyde 6 has been proposed to be converted to strychnine 10 through the incorporation of acetate to form the piperidone moiety, although the mechanism of acetate incorporation and ring cyclization has remained unclear12,13(ring G in Fig. 1; see Supplementary Fig. 3 for carbon and ring annotations). Subsequent hydroxylations and methylations of strychnine 10 would yield brucine 15 (ref. 16 and Fig. 1).
Fig. 1. The proposed biosynthesis pathway for strychnine and brucine.
The partial biosynthetic pathway was predicted on the basis of previous radioisotopic feeding experiments. OPP, pyrophosphate; GPP, geranyl pyrophosphate.
To identify strychnine biosynthetic genes, we selected two members of the Strychnos genus (family: Loganiaceae), one known producer of strychnine 10, S. nux-vomica17 and one non-producer, Strychnos sp.18, to investigate this biosynthetic pathway. Metabolic analysis of S. nux-vomica revealed the presence of several strychnos alkaloids, including strychnine 10, isostrychnine 11, β-colubrine 13 and brucine 15, all of which accumulate in the roots (Supplementary Fig. 4). These alkaloids were absent in the non-producer, although a biosynthetically related compound, strychnos alkaloid diaboline 8, was detected in its roots and stems (Supplementary Fig. 5). We generated tissue-specific RNA-sequencing data from these two plants to enable gene discovery.
The biosynthetic pathway of geissoschizine 1 from tryptophan and geranyl pyrophosphate has been completely elucidated in the phylogenetically related plant Catharanthus roseus (family: Apocynaceae) (see Supplementary Fig. 6 for the phylogenetic relationship of C. roseus and S. nux-vomica). C. roseus produces monoterpene indole alkaloids unrelated to strychnine19. A homologue for each biosynthetic gene in the geissoschizine 1 pathway was readily identified in the S. nux-vomica transcriptome, suggesting that the biosynthetic pathway of geissoschizine 1 is conserved in C. roseus and S. nux-vomica. These genes are all expressed preferentially in S. nux-vomica roots (Supplementary Fig. 7), consistent with previous feeding studies that suggest strychnine 10 biosynthesis occurs primarily in the roots12,13. Candidate genes for subsequent steps were selected according to three criteria: (1) high expression in the roots of S. nux-vomica (fragments per kilobase of transcript per million mapped reads (FPKM) ≥ 20); (2) co-expression with putative upstream genes; and (3) genes that could encode proteins with catalytic functions that are consistent with the chemical logic of our hypothesized biosynthetic pathway (Fig. 2).
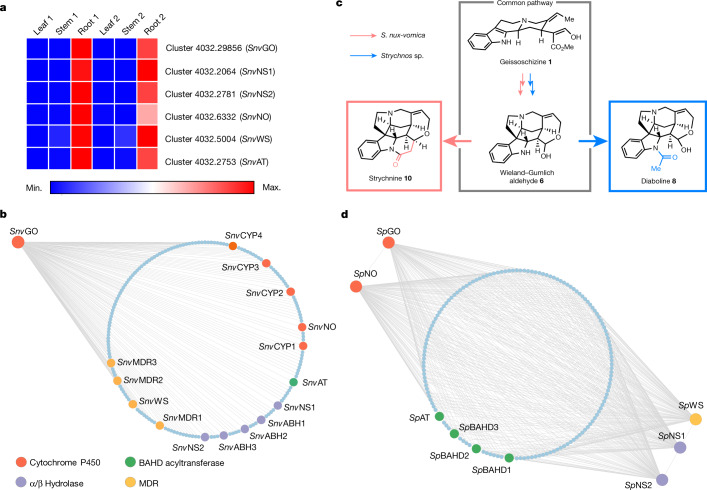
Fig. 2. Expression analysis of candidate genes in S. nux-vomica (strychnine producer) and Strychnos sp. (diaboline producer).
Both strychnine and diaboline are derived from the same biosynthetic intermediate, the Wieland–Gumlich aldehyde. a, Expression profiles of identified genes in S. nux-vomica. The expression of each identified gene is represented as the FPKM of S. nux-vomica transcriptomes. Sample sets 1 and 2 represent two biological replicates. b, Co-expression analysis using SnvGO as bait in S. nux-vomica. The circle of dots represents genes co-expressed with SnvGO (Pearson’s r ≥ 0.95; 470 genes in total). c, S. nux-vomica and Strychnos sp. share a common pathway from geissoszhizine 1 to Wieland–Gumlich aldehyde 6. d, Co-expression analysis in Strychnos sp. SpGO, SpNS1, SpNS2, SpNO and SpWS were used as baits. The circle depicts genes co-expressed with all the baits (r > 0.6; 3,999 genes in total). Enlarged and annotated dots in (b and d) represent genes tested in N. benthamiana.
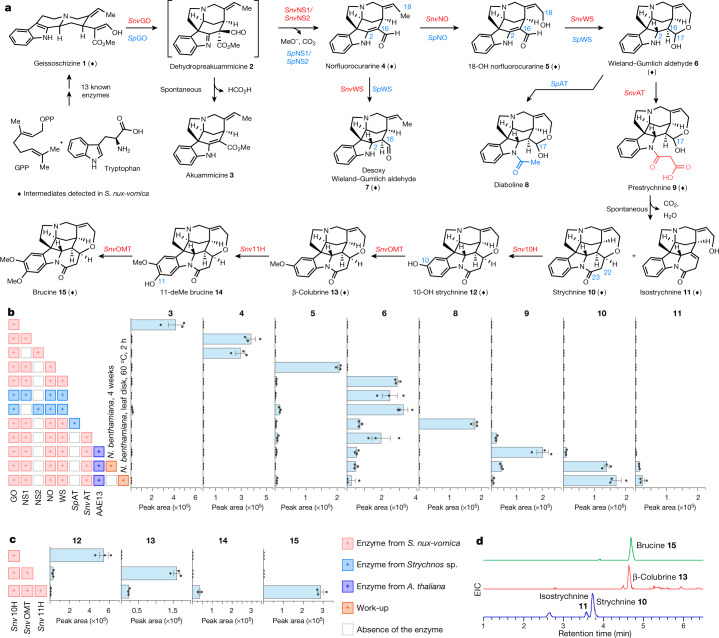
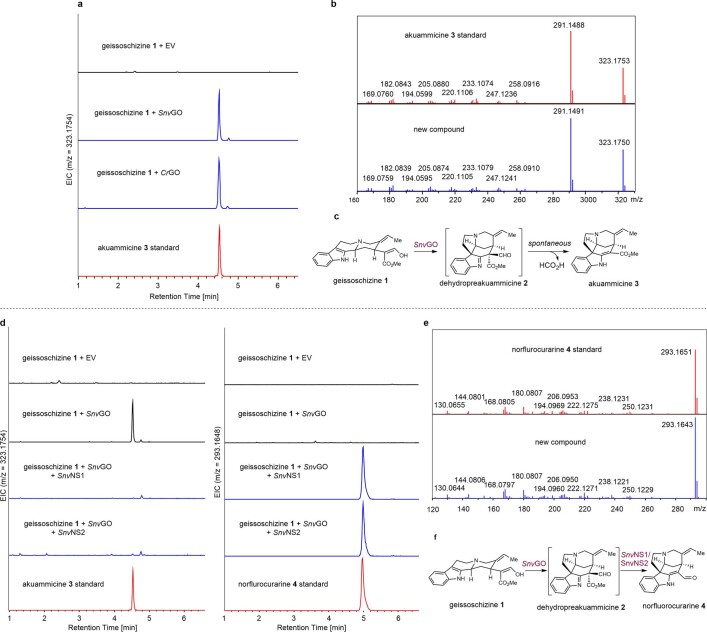
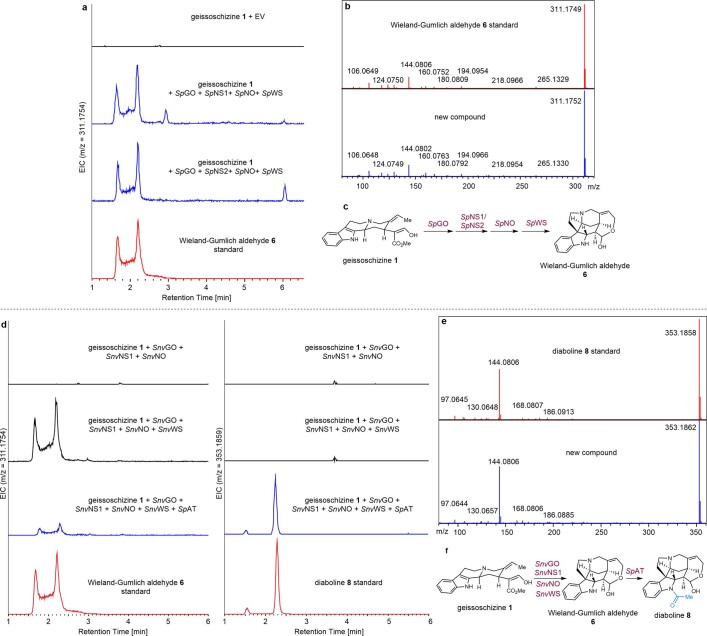
The chemical steps for transformation of geissoschizine 1 to Wieland–Gumlich aldehyde 6 are not known. However, given the structural similarity between the Wieland–Gumlich aldehyde 6 and the known early alkaloid intermediate dehydropreakuammicine 2 (ref. 19 and Fig. 3a), chemical logic suggests that Wieland–Gumlich aldehyde 6 could form from dehydropreakuammicine 2 through ester hydrolysis, decarboxylation, oxidation and reduction (Supplementary Fig. 2). If this hypothesis is correct, S. nux-vomica should contain a homologue of geissoschizine oxidase, which has also been isolated from C. roseus (CrGO). In vitro, CrGO converts geissoschizine 1 to akuammicine 3, presumably through the spontaneous deformylation of dehydropreakuammicine 2 (ref. 19, Fig. 3a and Supplementary Fig. 2). A BLAST search using CrGO as the query against the S. nux-vomica transcriptome identified one hit (transcript cluster 4032.29856; CYP71AY6) with 46% amino-acid sequence identity (Supplementary Fig. 8) that showed similar expression profiles with upstream biosynthetic gene candidates (Fig. 2a). We expressed this gene in N. benthamiana leaves through Agrobacterium tumefaciens-mediated transient expression followed by infiltration of geissoschizine 1. Liquid chromatography–mass spectrometry analysis of leaf extracts revealed the deformylation product of dehydropreakuammicine, akuammicine 3 (Fig. 3b and Extended Data Fig. 1). Therefore, cluster 4032.29856 was named SnvGO.
Fig. 3. Discovery of a diaboline, strychnine and brucine biosynthesis pathway.
a, The complete biosynthetic pathway leading to the production of diaboline 8, strychnine 10 and brucine 15. Diamonds represent intermediates detected in S. nux-vomica (Extended Data Fig. 10). b, The liquid chromatography–mass spectrometry peak area of products produced in N. benthamiana after expression of the indicated enzymes and geissoschizine 1 infiltration. Date are mean ± s.e.m.; n = 3 biological replicates. c, The liquid chromatography–mass spectrometry peak area of products produced in N. benthamiana after expression of the indicated enzymes and strychnine 10 infiltration. Date are mean ± s.e.m.; n = 3 biological replicates. Work-up: manipulation after expression of the indicated enzymes and substrate infiltration. d, Extracted ion chromatograms (EIC) for strychnine 10, isostrychnine 11, β-colubrine 13 and brucine 15 in N. benthamiana leaves expressing all nine enzymes with the infiltration of geissoschizine 1 and disodium malonate. All intermediates were validated by comparison to synthetic authentic standards (see Supplementary Information for synthetic procedures).
Extended Data Fig. 1. Functional characterization of SnvGO, SnvNS1 and SnvNS2.
a. Transient expression of SnvGO in N. benthamiana with co-infiltration of geissoschizine 1. Extracted ion chromatograms for akuammicine 3 (m/z [M+H]+ = 323.1754 ± 0.05). CrGO was used as a positive control. This experiment was repeated three times with similar results. b. MS/MS (20 to 50 eV) spectra of akuammicine 3 produced in N. benthamiana (blue) compared to standard (red). c. Reaction catalyzed by SnvGO. d. Transient expression of SnvGO, SnvNS1, and SnvNS2 in N. benthamiana with co-infiltration of geissoschizine 1. Extracted ion chromatograms for akuammicine 3 (m/z [M+H]+ = 323.1754 ± 0.05, left) and norflurocurarine 4 (m/z [M+H]+ = 293.1648 ± 0.05, right). This experiment was repeated three times with similar results. e. MS/MS (20 to 50 eV) spectra of norflurocurarine 4 produced in N. benthamiana (blue) compared to standard (red). f. Reaction catalyzed by SnvNS1 and SnvNS2. EV, empty vector.
Because it is known that decarboxylation of a methyl ester can be triggered by ester hydrolysis20, we speculated that an α/β hydrolase20,21 would hydrolyse the ester moiety of dehydropreakuammicine 2 and therefore lead to decarboxylation before spontaneous deformylation to akuammicine 3 occurs. This would result in the formation of the strychnos alkaloid norfluorocurarine 4 (Fig. 3a and Supplementary Fig. 2). On the basis of a co-expression analysis using SnvGO as bait, we initially selected five α/β hydrolases (r ≥ 0.95, Pearson correlation coefficient) for functional characterization (Fig. 2b). Each was tested in N. benthamiana along with SnvGO and geissoschizine 1 as substrate. Two of these candidates (clusters 4032.2064 and 4032.2781) led to the production of norfluorocurarine 4, along with substantially decreased levels of the deformylation product akuammicine 3 (Fig. 3b and Extended Data Fig. 1). Therefore, we named these two α/β hydrolases norfluorocurarine synthase 1 and 2 (SnvNS1 and SnvNS2). SnvNS1 and SnvNS2 share 74% identity at the protein level and showed the same reactivity in the N. benthamiana transient-expression system. We used SnvNS1 in all subsequent experiments.
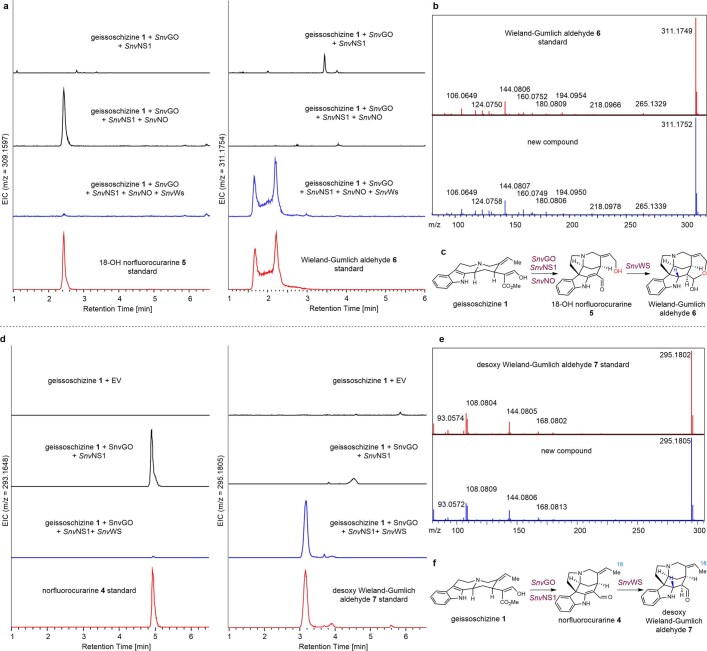
To convert norfluorocurarine 4 to Wieland–Gumlich aldehyde 6, a hydroxylase and a reductase are required to install the C18 hydroxyl group and reduce the 2,16 double bond, respectively (Fig. 3a). A total of five cytochrome P450 proteins22 and four medium-chain dehydrogenase/reductases (MDRs)23 that were co-expressed (r ≥ 0.95) (Fig. 2b) with SnvGO were initially considered, because these two protein families are often involved in alkaloid biosynthesis. Because the order of hydroxylation and reduction is unknown, combinatorial transient-expression experiments in N. benthamiana24–26 were adopted. Simultaneous expression of all candidate cytochrome P450 proteins and MDRs in N. benthamiana leaves combined with SnvGO and SnvNS1 indeed resulted in the consumption of norfluorocurarine 4 and production of Wieland–Gumlich aldehyde 6 (Fig. 3a). Co-infiltration of one cytochrome P450 (cluster 4032.6332; CYP71A144) along with SnvGO, SnvNS1 and geissoschizine 1 in N. benthamiana leaves produced a hydroxylated product 18-OH norfluorocurarine 5 that co-eluted with the synthetic standard (Fig. 3b and Extended Data Fig. 2). Intermediate 5 is consumed after one candidate MDR (cluster 4032.5004) is added to the co-infiltration experiments and the accumulation of Wieland–Gumlich aldehyde 6 is observed (Fig. 3b and Extended Data Fig. 3). Therefore, we named this cytochrome P450 norfluorocurarine oxidase (SnvNO) and the MDR Wieland–Gumlich aldehyde synthase (SnvWS). Notably, in planta and in vitro assays showed that SnvWS could reduce the 2,16 double bond in both norfluorocurarine 4 and 18-OH norfluorocurarine 5 (Fig. 3a, Extended Data Fig. 3 and Supplementary Fig. 9). Stereoselective reduction by SnvWS is probably initiated by the tautomerization of the enamine moiety in 4 and 5 through protonation at the α face, followed by NADPH reduction at the β face. The subsequent spontaneous cyclization between the C18-OH and C16 aldehyde, possibly facilitated by the conformational flexibility of the reduced substrate, forms the hemiacetal in 6 (Supplementary Fig. 10). In vitro steady-state kinetics indicated that SnvWS had a higher catalytic efficiency with 5 than with 4 (kcat/Km = 0.297 min−1 μM−1 for 5 compared with 0.068 min−1 μM−1 for 4) (Supplementary Fig. 11). A model of SnvWS docked with 18-OH norfluorocurarine 5 suggests that Thr95 and Ser309 in SnvWS may hydrogen bond with the C18 hydroxyl group in 18-OH norfluorocurarine 5, providing an explanation for the differences in catalytic efficiency between norfluorocurarine 4 and 18-OH norfluorocurarine 5 (Supplementary Fig. 10). No cytochrome P450, including SnvNO, could hydroxylate desoxy Wieland–Gumlich aldehyde 7, suggesting that the order of the reactions is first oxidation to form 18-OH norfluorocurarine 5, followed by reduction.
Extended Data Fig. 2. Functional characterization of SnvNO.
a. Transient expression of SnvGO, SnvNS1, and SnvNO in N. benthamiana with co-infiltration of geissoschizine 1. Extracted ion chromatograms for norflurocurarine 4 (m/z [M+H]+ = 293.1648 ± 0.05, left) and 18-OH norflurocurarine 5 (m/z [M+H]+ = 309.1567 ± 0.05, right). This experiment was repeated three times with similar results. b. MS/MS (20 to 50 eV) spectra of 18-OH norflurocurarine 5 produced in N. benthamiana (blue) compared to standard (red). c. Reaction catalyzed by SnvNO.
Extended Data Fig. 3. Functional characterization of SnvWS.
a. Transient expression of SnvGO, SnvNS1, SnvNO and SnvWS in N. benthamiana with co-infiltration of geissoschizine 1. Extracted ion chromatograms for 18-OH norflurocurarine 5 (m/z [M+H]+ = 309.1567 ± 0.05, left) and Wieland-Gumlich aldehyde 6 (m/z [M+H]+ = 311.1754 ± 0.05, right). The broad two peaks of Wieland-Gumlich aldehyde 6 in chromatogram due to the hemiacetal diastereomers. This experiment was repeated three times with similar results. b. MS/MS (20 to 50 eV) spectra of Wieland-Gumlich aldehyde 6 produced in N. benthamiana (blue) compared to synthetic standard (red). c. Reaction catalyzed by SnvGO, SnvNS1, SnvNO and SnvWS. d. Transient expression of SnvGO, SnvNS1, and SnvWS in N. benthamiana with co-infiltration of geissoschizine 1. Extracted ion chromatograms for norflurocurarine 4 (m/z [M+H]+ = 293.1648 ± 0.05, left) and desoxy Wieland-Gumlich aldehyde 7 (m/z [M+H]+ = 295.1805 ± 0.05, right). This experiment was repeated three times with similar results. e. MS/MS (20 to 50 eV) spectra of desoxy Wieland-Gumlich aldehyde 7 produced in N. benthamiana (blue) compared to synthetic standard (red). f. Reaction catalyzed by SnvGO, SnvNS1, and SnvWS.
To complete the biosynthesis of strychnine 10 from Wieland–Gumlich aldehyde 6, a new piperidone ring containing two additional carbon atoms must be installed (ring G in Fig. 1). However, the intermediates or the reaction steps for this ring construction are not known; the only clue is that the additional two-carbon unit (C22 and C23) originates from [14C]acetate12,13. To facilitate the discovery of these cryptic late biosynthetic steps, we compared the strychnine producing and non-producing Strychnos plants. Metabolic analysis showed that the major alkaloid in the non-strychnine producer Strychnos sp. is diaboline 8 (Supplementary Fig. 5), a compound that is most likely derived from N-acetylation of Wieland–Gumlich aldehyde 6 (Fig. 3a). Therefore, we hypothesized that S. nux-vomica and Strychnos sp. should share the same biosynthetic pathway from geissoschizine 1 to Wieland–Gumlich aldehyde 6 (Fig. 2c). Indeed, a BLAST search against the non-producer transcriptome identified orthologues SpGO (CYP71AY7, 92% amino-acid identity to SnvGO), SpNS1 (92% amino-acid identity to SnvNS1), SpNS2 (88% amino-acid identity to SnvNS2), SpNO (CYP71A145, 91% amino-acid identity to SnvNO) and SpWS (93% amino-acid identity to SnvWS). To validate the function of these genes, we expressed them in two combinations (SpGo, SpNS1, SpNO and SpWS; and SpGo, SpNS2, SpNO and SpWS) in N. benthamiana leaves with co-infiltration of geissoschizine 1. Both combinations led to the formation of Wieland–Gumlich aldehyde 6 (Fig. 3b and Extended Data Fig. 4). The only remaining step for the biosynthesis of diaboline 8 is the acetylation of the indole amine (Fig. 3a), which in alkaloid biosynthesis is often catalysed by a BAHD acyltransferase using acetyl-CoA as an acyl donor27. Four BAHD acyltransferase candidates were co-expressed with all five genes (r > 0.6) (Fig. 2d). Transient expression of one candidate (SpAT) with upstream genes generated diaboline 8 in N. benthamiana (Fig. 3b and Extended Data Fig. 4).
Extended Data Fig. 4. Functional characterization of SpGO, SpNS1, SpNS2, SpNO, SpWS and SpAT.
a. Transient expression of SpGO, SpNS1, SpNS2, SpNO, and SpWS in N. benthamiana with co-infiltration of geissoschizine 1. Extracted ion chromatograms for Wieland-Gumlich aldehyde 6 (m/z [M+H]+ = 311.1754 ± 0.05). This experiment was repeated three times with similar results. b. MS/MS (20 to 50 eV) spectra of Wieland-Gumlich aldehyde 6 produced in N. benthamiana (blue) compared to synthetic standard (red). c. Reaction catalyzed by SpGO, SpNS1, SpNS2, SpNO, and SpWS. d. Transient expression of SnvGO, SnvNS1, SnvNO, SnvWS and SpAT in N. benthamiana with co-infiltration of geissoschizine 1. Extracted ion chromatograms for Wieland-Gumlich aldehyde 6 (m/z [M+H]+ = 311.1754 ± 0.05, left) and diaboline 8 (m/z [M+H]+ = 353.1859 ± 0.05, right). The two peaks of diaboline 8 in chromatogram due to the hemiacetal diastereomers. This experiment was repeated three times with similar results. ‑e. MS/MS (20 to 50 eV) spectra of diaboline 8 produced in N. benthamiana (blue) compared to synthetic standard (red). f. Reaction catalyzed by SpAT.
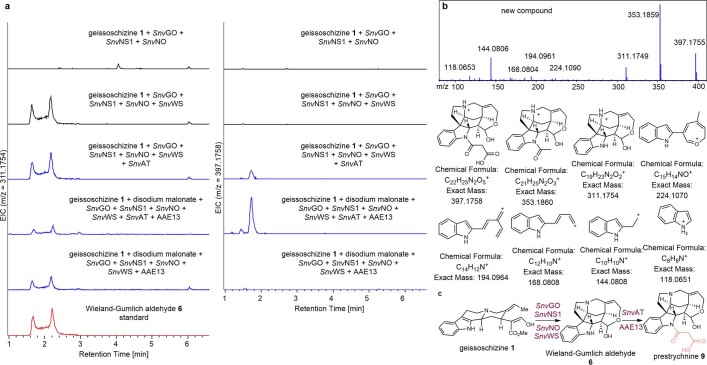
S. nux-vomica contains an orthologue (cluster 4032.2753; SnvAT) of SpAT (85% amino-acid identity to SpAT) that is highly expressed in the roots and showed high expression correlation with previously identified genes (r ≥ 0.99 with each gene) (Fig. 2a,b). However, S. nux-vomica does not produce diaboline 8, and previous feeding studies demonstrated that diaboline 8 is not a biosynthetic precursor of strychnine 10 (ref. 14). We surmised that SnvAT and SpAT may have distinct enzymatic activities, and indeed, simultaneous expression of SnvAT and SnvGO, SnvNS1, SnvNO, SnvWS and geissoschizine 1 in N. benthamiana led to only trace levels of diaboline 8. However, a new compound with a mass corresponding to a malonylated product was detected in the leaf extracts, which suggested that SnvAT is a BAHD acyltransferase with predominantly malonyltransferase activity (Fig. 3b and Extended Data Fig. 5). Although the expression of this enzyme in N. benthamiana resulted in only the partial consumption of Wieland–Gumlich aldehyde 6, we hypothesized that the conversion might be limited by the low concentration of malonyl-CoA in N. benthamiana leaves. Therefore, we expressed these enzymes along with AAE13 (Arabidopsis thaliana), a cytosolic enzyme that produces malonyl-CoA accessible to cytosolic SnvAT28 (Supplementary Fig. 12). The addition of AAE13 and co-infiltration of the co-substrate disodium malonate to the transient-expression system resulted in a tenfold increase in the production of malonylated product (Fig. 3b and Extended Data Fig. 5). During purification, this product rapidly decomposed, so we treated the crude methanolic extracts of N. benthamiana leaves with trimethylsilyldiazomethane to methylate the carboxylic acid, followed by aldehyde reduction with sodium borohydride. The derivatized products were confirmed by comparison to synthetic standards (Supplementary Fig. 13), indicating that the SnvAT product was N-malonyl Wieland–Gumlich aldehyde 9 (Fig. 3a). Therefore, although SnvAT and SpAT share 85% amino acid identity, they have distinct catalytic activities. Phylogenetic analysis showed that SnvAT clusters with SpAT in an acetyltransferase clade, which is evolutionarily distinct from the canonical malonyltransferase clade (Supplementary Fig. 14). Homology models of SnvAT and SpAT29 (Supplementary Fig. 15) were used to identify one amino acid (SnvAT(R424F) and SpAT(F421R)) that controls the selectivity between acetyl and malonyl transferase activity (Supplementary Figs. 16 and 17). These models suggest that the arginine residue is responsible for the malonyl-CoA selectivity by forming a bidentate salt bridge with the carboxylate of malonyl-CoA30,31 (Supplementary Fig. 18), providing a straightforward mechanistic explanation for the difference in alkaloid accumulation in these two plants. Notably, the 17-O-acylation product was predominant in in vitro assays at physiological pH (Supplementary Fig. 19), which may be because of changes in the protein activity in a non-cellular environment or differences in the equilibration of the open and closed forms of the Wieland–Gumlich aldehyde substrate.
Extended Data Fig. 5. Functional characterization of SnvAT.
a. Transient expression of SnvGO, SnvNS1, SnvNO, SnvWS, SnvAT, and AAE13 in N. benthamiana with co-infiltration of geissoschizine 1 and disodium malonate. Extracted ion chromatograms for for Wieland-Gumlich aldehyde 6 (m/z [M+H]+ = 311.1754 ± 0.05, left) and prestrychnine 9 (m/z [M+H]+ = 397.1758 ± 0.05, right). The two peaks of 9 in chromatogram due to the hemiacetal diastereomers. This experiment was repeated three times with similar results. b. MS/MS (20 to 50 eV) spectra and putative ion fragments of generated m/z [M+H]+ 397.1758. c. Reaction catalyzed by SnvAT.
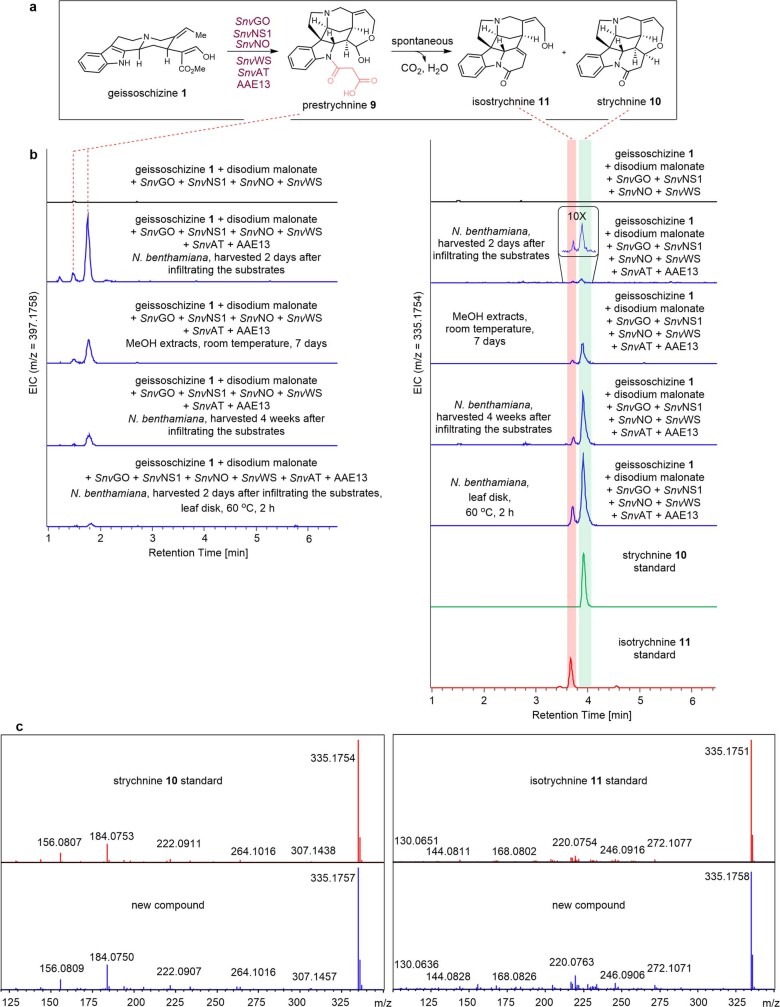
Notably, a trace amount of strychnine 10 and isostrychnine 11 could be detected in the methanolic extracts of N. benthamiana leaves that produce malonylated Wieland–Gumlich aldehyde 9 (Fig. 3b and Extended Data Fig. 6). These two alkaloids accumulated and 9 decreased over time when stored at room temperature (Supplementary Fig. 22). Indeed, most of 9 was converted to strychnine 10 and isostrychnine 11 in N. benthamiana leaves that were harvested 4 weeks after infiltrating the substrates (Fig. 3b and Extended Data Fig. 6). Incubating 9 with recombinant SnvAT or N. benthamiana crude protein extracts did not accelerate the conversion of 9 to 10 (Supplementary Fig. 23). These experiments suggest that conversion of 9 to strychnine 10 and isostrychnine 11 could occur spontaneously both in vitro and under physiological conditions. Alternatively, heating N. benthamiana leaves at 60 °C for 2 h substantially accelerated the conversion (Fig. 3b and Extended Data Fig. 6). We think that 10 and 11 are formed through the decarboxylation of the β-keto acid moiety in 9 to form an α,β-unsaturated amide. Subsequent oxa-Michael addition by C18 hydroxyl group would generate strychnine 10. The α,β-unsaturated amide can also tautomerize to the β,γ-unsaturated amide to form isostrychnine 11 (Supplementary Fig. 24).
Extended Data Fig. 6. Conversion of prestrychnine 9 to strychnine 10 and isostrychnine 11.
a. Reaction of the conversion. b. Extracted ion chromatograms for prestrychnine 9 (m/z [M+H]+ = 397.1758 ± 0.05, left) and strychnine 10 and isostrychnine 11 (m/z [M+H]+ = 335.1754 ± 0.05, right). These experiments were repeated three times with similar results. c. MS/MS (20 to 50 eV) spectra of generated m/z [M+H]+ 335.1754 (strychnine 10 and isostrychnine 11, blue) compared to standards (red).
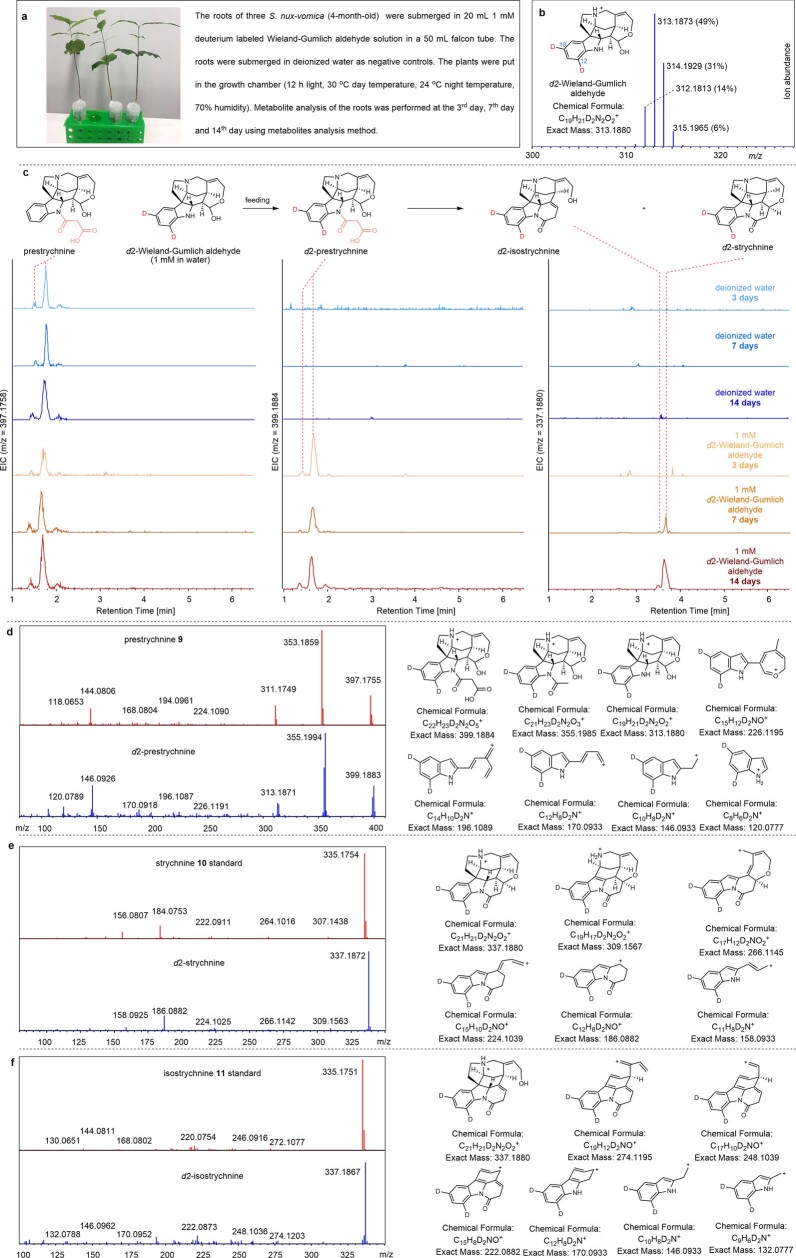
Previous radioisotopic labelling studies indicated that a structurally uncharacterized biosynthetic intermediate could be converted to strychnine by warming the acid extracts from S. nux-vomica roots14,15. The reported chemical properties of this intermediate14,15, which was called prestrychnine (see Supplementary Fig. 2 for the previously proposed structure), are similar to 9. Therefore, we suggest that the proposed structure of prestrychnine be revised to 9. Notably, in this feeding study the levels of radioisotope-labelled prestrychnine was 9 times higher than strychnine 10 after 3 days of feeding of S. nux-vomica with 14C-tryptophan14, suggesting that the conversion of prestrychnine to strychnine 10 is a slow process in S. nux-vomica. Indeed, we screened numerous α/β hydrolases21,32 and polyketide synthases33, as well as members of these two families that are known to catalyse decarboxylation of β-keto acid functionalities, and we also screened numerous transporters that could transfer prestrychnine to the vacuole where the acidic environment might accelerate the decarboxylation. However, none of these gene candidates accelerated the formation of strychnine 10 and isostrychnine 11. To establish whether conversion of prestrychnine to strychnine is a slow, non-enzymatic process in S. nux-vomica, we performed hydroponic feeding of deuterium-labelled Wieland–Gumlich aldehyde 6 to the roots of S. nux-vomica. Labelled prestrychnine 9 could be detected after 3 days, but trace amounts of strychnine 10 and isostrychnine 11 appeared only after 7 days (Extended Data Fig. 7). Collectively, these data are consistent with the previously published experiments14,15 and with the rate of strychnine formation in our heterologous expression system. The fact that prestrychnine 9 is converted to strychnine 10 slowly in S. nux-vomica is consistent with a non-enzymatic process, although the involvement of an enzyme with only modest rate acceleration cannot be definitively ruled out.
Extended Data Fig. 7. Hydroponic feedings to the roots of 4-month-old S. nux-vomica seedlings with deuterium labelled Wieland-Gumlich aldehyde.
a. Hydroponic feedings of S. nux-vomica in 50 mL falcon tube with 20 mL 1 mM deuterium labelled Wieland-Gumlich aldehyde. b. HRMS (ESI) [M+H]+ chromatogram of synthetic deuterium labelled Wieland-Gumlich aldehyde mixture. The major component (49%) is d2-Wieland-Gumlich aldehyde. c. Extracted ion chromatograms for prestrychnine 9 (m/z [M+H]+ = 397.1758 ± 0.005), d2-prestrychnine (m/z [M+H]+ = 399.1884 ± 0.005), d2-isostrychnine (m/z [M+H]+ = 337.1880 ± 0.005), d2-strychnine (m/z [M+H]+ = 337.1880 ± 0.005). d. MS/MS (20 to 50 eV) spectrum and putative ion fragments of generated d2-prestrychnine (m/z [M+H]+ = 399.1884, blue) compared to prestrychnine (m/z [M+H]+ = 397.1758, red). e. MS/MS (20 to 50 eV) spectra and putative ion fragments of generated d2-strychnine (m/z [M+H]+ = 337.1880, blue) compared to strychnine standard (m/z [M+H]+ = 335.1754, red). f. MS/MS (20 to 50 eV) spectra and putative ion fragments of generated d2-isostrychnine (m/z [M+H]+ = 337.1880, blue) compared to isostrychnine standard (m/z [M+H]+ = 335.1754, red). This experiment was repeated three times with similar results.
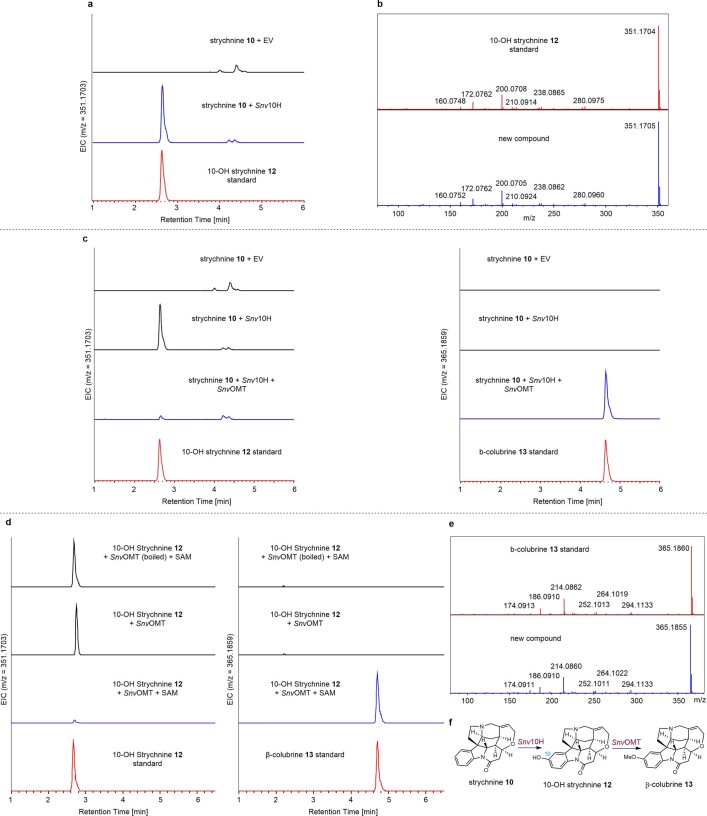
Brucine 15, which is a dimethoxylated derivative of strychnine 10, is also highly accumulated in the roots of S. nux-vomica (Fig. 3a and Supplementary Fig. 4). To identify the hydroxylase, 12 full-length cytochrome P450 proteins that shared a relatively high co-expression correlation with SnvGO (Pearson’s r > 0.7) were selected for subsequent tests (Supplementary Table 1). When one cytochrome P450 (cluster 4032.17050; CYP82D367) was expressed in the presence of strychnine 10 in N. benthamiana, 10-OH strychnine 12 was formed (strychnine-10-hydroxylase (Snv10H)) (Fig. 3c and Extended Data Fig. 8). The presence of β-colubrine 13 in S. nux-vomica suggests that the two methoxy groups are installed sequentially (Fig. 3a), so we next identified five methyltransferases34 that were highly expressed in the roots of S. nux-vomica (Supplementary Table 2). Expression of one of the methyltransferases (cluster 4032.16453; SnvOMT) with Snv10H in N. benthamiana resulted in the formation of a compound corresponding to synthetic β-colubrine 13 (Fig. 3c and Extended Data Fig. 8). None of the aforementioned 12 co-expressed cytochrome P450 proteins catalysed the hydroxylation of β-colubrine 13, but the high accumulation of the final product brucine 15 in roots led us to identify all 13 other cytochrome P450 proteins that were strongly expressed (FPKM ≥ 20) in roots (Supplementary Table 1). Of these 13 proteins, we initially targeted the 3 within the CYP71 clade (Supplementary Fig. 25). One of these cytochrome P450 proteins (cluster 4032.16581; CYP71AH44, Snv11H)—assayed in combination with strychnine, Snv10H and SnvOMT—produced brucine 15 as a major product along with trace amounts of the hydroxylated product 11-deMe brucine 14 (Fig. 3c and Extended Data Fig. 9). When we infiltrated synthetic β-colubrine 13 into tobacco leaves that express Snv11H alone only 11-deMe brucine 14 is formed; brucine 15 is formed only in the presence of SnvOMT (Extended Data Fig. 9). In vitro and in planta assays showed that SnvOMT could also methylate 11-OH strychnine 16 to α-colubrine 17 (Supplementary Fig. 26), and 10-deMe brucine 18 to brucine 15 (Supplementary Fig. 27), although with lower efficiency. Overall, these results highlight the promise for production of strychnos-type alkaloids using synthetic biology approaches, although substantial optimization of the heterologous host production system is required.
Extended Data Fig. 8. Functional characterization of Snv10H and SnvOMT.
a. Transient expression of Snv10H in N. benthamiana with co-infiltration of strychnine 10. Extracted ion chromatograms for 10-OH strychnine (m/z [M+H]+ = 351.1703 ± 0.05). This experiment was repeated three times with similar results. b. MS/MS (20 to 50 eV) spectra of 10-OH strychnine 12 produced in N. benthamiana (blue) compared to standard (red). c. Transient expression of Snv10H and SnvOMT in N. benthamiana with co-infiltration of strychnine 10. Extracted ion chromatograms for 10-OH strychnine (m/z [M+H]+ = 351.1703 ± 0.05, left) and β-colubrine 13 (m/z [M+H]+ = 365.1859 ± 0.05, right). This experiment was repeated three times with similar results. d. In vitro assays using purified SnvOMT from SoluBL21 E. coli with 10-OH strychnine 12. Extracted ion chromatograms for 10-OH strychnine (m/z [M+H]+ = 351.1703 ± 0.05, left) and β-colubrine 13 (m/z [M+H]+ = 365.1859 ± 0.05, right). This experiment was repeated more than three times with similar results. e. MS/MS (20 to 50 eV) spectra of β-colubrine 13 produced in N. benthamiana (blue) compared to standard (red). f. Reaction catalyzed by Snv10H and SnvOMT.
Extended Data Fig. 9. Functional characterization of Snv11H.
a. Reaction catalyzed by Snv11H and SnvOMT. b. Transient expression of Snv10H, SnvOMT, and Snv11H in N. benthamiana with co-infiltration of strychnine 10. Extracted ion chromatograms for β-colubrine 13 (m/z [M+H]+ = 365.1859 ± 0.05, left), 11-deMe brucine 14 (m/z [M+H]+ = 381.1808 ± 0.05, middle), and brucine 15 (m/z [M+H]+ = 395.1965 ± 0.05, right). This experiment was repeated three times with similar results. c. Transient expression of Snv11H in N. benthamiana with co-infiltration of β-colubrine 13. Extracted ion chromatograms for 11-deMe brucine 14 (m/z [M+H]+ = 381.1808 ± 0.05). This experiment was repeated three times with similar results. d. Transient expression of Snv11H and SnvOMT in N. benthamiana with co-infiltration of β-colubrine 13. Extracted ion chromatograms for 11-deMe brucine 14 (m/z [M+H]+ = 381.1808 ± 0.05, left) and brucine 15 (m/z [M+H]+ = 395.1965 ± 0.05, right). This experiment was repeated three times with similar results. e. Transient expression of SnvOMT in N. benthamiana with co-infiltration of 11-deMe brucine 14. Extracted ion chromatograms for brucine 15 (m/z [M+H]+ = 395.1965 ± 0.05). This experiment was repeated three times with similar results. f. In vitro assays using purified SnvOMT from SoluBL21 E. coli. Extracted ion chromatograms for 11-deMe brucine 14 (m/z [M+H]+ = 381.1808 ± 0.05, left) and brucine 15 (m/z [M+H]+ = 395.1965 ± 0.05, right). This experiment was repeated three times with similar results. g. MS/MS (20 to 50 eV) spectra of generated 11-deMe brucine 14 and brucine 15 (blue) compared to standards (red).
Having completed the pathway of brucine 15, we then reconstituted the pathway in N. benthamiana from geissoschizine 1. We transiently expressed all of the enzymes (SnvGO, SnvNS1, SnvNO, SnvWS and SnvAT, AAE13, Snv10H, SnvOMT and Snv11H) in tobacco leaves followed by infiltrating geissoschizine 1 and disodium malonate. If the tobacco leaves were harvested 1 week after infiltrating the substrates, the accumulation of strychnine 10, isostrychnine 11, β-colubrine 13 and brucine 15 was observed (Fig. 3d and Supplementary Fig. 28). Additionally, all of the intermediates in the pathway except for 11-deMe brucine 14 could be detected in the roots of S. nux-vomica (Fig. 3a and Extended Data Fig. 10), suggesting that the heterologously reconstituted pathway in N. benthamiana matches the physiologically relevant pathway in Strychnos plants.
Extended Data Fig. 10. Comparison of S. nux-vomica metabolites to standards or enzymatic product generated in N. benthamiana transient expression.
Intermediates detected in methanolic extracts of S. nux-vomica roots (blue) were compared to standards (red) or products produced in N. benthamiana transient expression experiment (green) by retention time and MS/MS spectra.
Here we report the discovery of nine enzymes that convert geissoschizine 1 to diaboline 8, strychnine 10 and brucine 11, using a combination of chemical logic, -omics datasets and enzymatic characterization. Pioneering studies of the structure and synthesis of strychnine provided the foundation for discovery of the enzymes of strychnine biosynthesis as it occurs in nature. These discoveries not only shed light on how plants produce these diverse alkaloids, but also provide a genetic basis for heterologous production of strychnos alkaloid derivatives to discover potent lead compounds through metabolic engineering approaches, providing a new challenge for synthetic biology.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-022-04950-4.
Supplementary information
This file contains Supplementary Methods, Supplementary Figs. 1–30, details regarding the synthesis of the compounds, NMR spectra data and Supplementary References.
Co-expression analysis of root highly expressed (FPKM ≥ 20) P450s in S. nux-vomica genes with SnvGO.
Root highly expressed methyltransferases (FPKM ≥ 20) in S. nux-vomica.
A list of primers used in the study.
Source Data for Supplementary Figs. 4, 7, 11, 16, 17, 23, 28 and 29.
Acknowledgements
We thank S. Arndt and C. Löhne for providing plant material; M. Florean for her help with feeding experiments; D. Ayled Serna Guerrero and M. Kunert for assistance with mass spectrometry; Y. Nakamura and C. Paetz for assistance with NMR analysis; E. Rothe and the greenhouse team for taking care of the plants; D. Nelson for his assistance in the systematic naming of the cytochrome P450 enzymes characterized in this study. This work was supported by grants from the European Research Council (788301) and the Max Planck Society. B.H. is grateful to the Alexander von Humboldt Foundation for a postdoctoral fellowship.
Extended data figures and tables
Source data
Author contributions
B.H. and S.E.O’C. designed the study and wrote the manuscript; B.H. performed all synthesis, cloning, protein-expression and protein-assay experiments; D.G. assisted with transcriptome preparation and analysis; L.C. assisted with enzyme cloning and data analysis; P.S. cloned AAE13 from A. thaliana cDNA and assisted with transient expression; C.E.R.L. helped with bioinformatic analyses; M.O.K. provided advice with cloning and protein purification; N.J.H.L. carried out protein homology modelling; and V.G. performed microscopy.
Peer review
Peer review information
Nature thanks Jing-Ke Weng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Funding
Open access funding provided by Max Planck Society.
Data availability
The sequence of genes characterized in this article are deposited in the National Center for Biotechnology (NCBI) GenBank under the following accession numbers: SnvGO (OM304290), SnvNS1 (OM304291), SnvNS2 (OM304292), SnvNO (OM304293), SnvWS (OM304294), SnvAT (OM304295), Snv10H (OM304296), SnvOMT (OM304297), Snv11H (OM304298), SpGO (OM304299), SpNS1 (OM304300), SpNS2 (OM304301), SpNO (OM304302), SpWS (OM304303) and SpAT (OM304304). The raw reads from the RNA-sequencing profiling analysis of S. nux-vomica and Strychnos sp. are deposited in the NCBI Sequence Read Archive (SRA) database under the BioProject accessions PRJNA825510 and PRJNA826736, respectively. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/5/2022
A Correction to this paper has been published: 10.1038/s41586-022-05177-z
Extended data
is available for this paper at 10.1038/s41586-022-04950-4.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-022-04950-4.
References
- 1.US EPA Strychnine. EPA-738-F-96-033 (1996); https://www3.epa.gov/pesticides/chem_search/reg_actions/reregistration/fs_PC-076901_1-Jul-96.pdf
- 2.Valdes F, Orrego F. Strychnine inhibits the binding of glycine to rat brain-cortex membrane. Nature. 1970;226:761–762. doi: 10.1038/226761a0. [DOI] [PubMed] [Google Scholar]
- 3.Huang X, Chen H, Michelsen K, Schneider S, Shaffer PL. Crystal structure of human glycine receptor-α3 bound to antagonist strychnine. Nature. 2015;526:277–280. doi: 10.1038/nature14972. [DOI] [PubMed] [Google Scholar]
- 4.Cannon JS, Overman LE. Is there no end to the total syntheses of strychnine? Lessons learned in strategy and tactics in total synthesis. Angew. Chem. Int. Edn. 2012;51:4288–4311. doi: 10.1002/anie.201107385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelletier PJ, Caventou JB. Note sur un nouvel alcali. Ann. Chim. Phys. 1818;8:323. [Google Scholar]
- 6.Openshaw HT, Robinson R. Constitution of strychnine and the biogenetic relationship of strychnine and quinine. Nature. 1946;157:438. doi: 10.1038/157438a0. [DOI] [PubMed] [Google Scholar]
- 7.Robinson, R. The constitution of strychnine. Experientia2, 28–29, (1946). [DOI] [PubMed]
- 8.Woodward RB, Brehm WJ, Nelson AL. The structure of strychnine. J. Am. Chem. Soc. 1947;69:2250. doi: 10.1021/ja01201a526. [DOI] [PubMed] [Google Scholar]
- 9.Robinson, R. Progress in Organic Chemistry (ed. Cook, J. W.) (Butterworths, 1952).
- 10.Woodward RB, et al. The total synthesis of (−)-strychnine. J. Am. Chem. Soc. 1954;76:4749–4751. doi: 10.1021/ja01647a088. [DOI] [Google Scholar]
- 11.Woodward RB. Biogenesis of the strychnos alkaloids. Nature. 1948;162:155–156. doi: 10.1038/162155a0. [DOI] [PubMed] [Google Scholar]
- 12.Schlatter C, et al. Über die Herkunft der C-Atome 22 und 23 im Strychnin. Helv. Chim. Acta. 1966;49:1714–1175. doi: 10.1002/hlca.19660490534. [DOI] [PubMed] [Google Scholar]
- 13.Schlatter C, Waldner EE, Schmid H, Maier W, Gröger D. Zur Biosynthese des Strychnins. 135. Mitteilung über Alkaloide. Helv. Chim. Acta. 1969;52:776–789. doi: 10.1002/hlca.19690520326. [DOI] [Google Scholar]
- 14.Heimberger, S. I. & Scott, I. A. Biosynthesis of strychnine. J. Chem. Soc. Chem. Commun.1973, 217–218 (1973).
- 15.Heimberg, S. I. The Biosynthesis of Strychnine. PhD Thesis, Yale Univ. (1973).
- 16.Maier W, Gröger D. Nichtverwertung von 5-Hydroxy-tryptophan für die Biosynthese von Strychnos-Alkaloiden. Z. Naturforsch. B. 1970;25:1192. doi: 10.1515/znb-1970-1032. [DOI] [PubMed] [Google Scholar]
- 17.Başer KHC, Bisset NG. Alkaloids of Sri Lankan Strychnos nux-vomica. Phytochemistry. 1982;21:1423–1429. doi: 10.1016/0031-9422(82)80155-6. [DOI] [Google Scholar]
- 18.Singh H, Kapoor VK, Phillipson JD, Bisset NG. Diaboline from Strychnos potatorum. Phytochemistry. 1975;14:587–588. doi: 10.1016/0031-9422(75)85141-7. [DOI] [Google Scholar]
- 19.Tatsis EC, et al. A three enzyme system to generate the Strychnos alkaloid scaffold from a central biosynthetic intermediate. Nat. Commun. 2017;8:316. doi: 10.1038/s41467-017-00154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mindrebo JT, Nartey CM, Seto Y, Burkart MD, Noel JP. Unveiling the functional diversity of the alpha/beta hydrolase superfamily in the plant kingdom. Curr. Opin. Struct. Biol. 2016;41:233–246. doi: 10.1016/j.sbi.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Dogru E, et al. The gene encoding polyneuridine aldehyde esterase of monoterpenoid indole alkaloid biosynthesis in plants is an ortholog of the α/β hydrolase super family. Eur. J. Biochem. 2000;267:1397–1406. doi: 10.1046/j.1432-1327.2000.01136.x. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen T-D, Dang T-TT. Cytochrome P450 enzymes as key drivers of alkaloid chemical diversification in plants. Front. Plant Sci. 2021;12:682181. doi: 10.3389/fpls.2021.682181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stavrinides A, et al. Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity. Nat. Commun. 2016;7:12116. doi: 10.1038/ncomms12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed J, Osbourn A. Engineering terpenoid production through transient expression in Nicotiana benthamiana. Plant Cell Rep. 2018;37:1431–1441. doi: 10.1007/s00299-018-2296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau W, Sattely ES. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science. 2015;349:1224–1228. doi: 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nett RS, Lau W, Sattely ES. Discovery and engineering of colchicine alkaloid biosynthesis. Nature. 2020;584:148–153. doi: 10.1038/s41586-020-2546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Auria JC. Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Kim HU, Weng H, Browse J. Malonyl-CoA synthetase, encoded by ACYL ACTIVATING ENZYME13, is essential for growth and development of Arabidopsis. Plant Cell. 2011;23:2247–2262. doi: 10.1105/tpc.111.086140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, et al. Structural and biochemical insights into two BAHD acyltransferases (AtSHT and AtSDT) involved in phenolamide biosynthesis. Front. Plant Sci. 2021;11:610118. doi: 10.3389/fpls.2020.610118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manjasetty BA, et al. Structural basis for modification of flavonol and naphthol glucoconjugates by Nicotiana tabacum malonyltransferase (NtMaT1) Planta. 2012;236:781–793. doi: 10.1007/s00425-012-1660-8. [DOI] [PubMed] [Google Scholar]
- 31.Oefner C, Schulz H, D’Arcy A, Dale GE. Mapping the active site of Escherichia coli malonyl-CoA-acyl carrier protein transacylase (FabD) by protein crystallography. Acta Crystallogr. D. 2006;D62:613–618. doi: 10.1107/S0907444906009474. [DOI] [PubMed] [Google Scholar]
- 32.Auldridge ME, et al. Emergent decarboxylase activity and attenuation of α/β-hydrolase activity during the evolution of methylketone biosynthesis in tomato. Plant Cell. 2012;24:1596–1607. doi: 10.1105/tpc.111.093997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Austin MB, Noel JP. The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 2003;20:79–110. doi: 10.1039/b100917f. [DOI] [PubMed] [Google Scholar]
- 34.Levac D, Murata J, Kim WS, de Luca V. Application of carborundum abrasion for investigating the leaf epidermis: molecular cloning of Catharanthus roseus 16-hydroxytabersonine-16-O-methyltransferase. Plant J. 2008;53:225–236. doi: 10.1111/j.1365-313X.2007.03337.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Methods, Supplementary Figs. 1–30, details regarding the synthesis of the compounds, NMR spectra data and Supplementary References.
Co-expression analysis of root highly expressed (FPKM ≥ 20) P450s in S. nux-vomica genes with SnvGO.
Root highly expressed methyltransferases (FPKM ≥ 20) in S. nux-vomica.
A list of primers used in the study.
Source Data for Supplementary Figs. 4, 7, 11, 16, 17, 23, 28 and 29.
Data Availability Statement
The sequence of genes characterized in this article are deposited in the National Center for Biotechnology (NCBI) GenBank under the following accession numbers: SnvGO (OM304290), SnvNS1 (OM304291), SnvNS2 (OM304292), SnvNO (OM304293), SnvWS (OM304294), SnvAT (OM304295), Snv10H (OM304296), SnvOMT (OM304297), Snv11H (OM304298), SpGO (OM304299), SpNS1 (OM304300), SpNS2 (OM304301), SpNO (OM304302), SpWS (OM304303) and SpAT (OM304304). The raw reads from the RNA-sequencing profiling analysis of S. nux-vomica and Strychnos sp. are deposited in the NCBI Sequence Read Archive (SRA) database under the BioProject accessions PRJNA825510 and PRJNA826736, respectively. Source data are provided with this paper.