Abstract
Fluorescent in situ hybridization (FISH) using 16S and 23S rRNA-targeted probes together with construction of an archaeal 16S ribosomal DNA (rDNA) clone library was used to characterize the microbial populations of an anaerobic baffled reactor successfully treating industrial dye waste. Wastewater produced during the manufacture of food dyes containing several different azo and other dye compounds was decolorized and degraded under sulfidogenic and methanogenic conditions. Use of molecular methods to describe microbial populations showed that a diverse group of Bacteria and Archaea was involved in this treatment process. FISH enumeration showed that members of the gamma subclass of the class Proteobacteria and bacteria in the Cytophaga-Flexibacter-Bacteroides phylum, together with sulfate-reducing bacteria, were prominent members of a mixed bacterial population. A combination of FISH probing and analysis of 98 archaeal 16S rDNA clone inserts revealed that together with the bacterial population, a methanogenic population dominated by Methanosaeta species and containing species of Methanobacterium and Methanospirillum and a relatively unstudied methanogen, Methanomethylovorans hollandica, contributed to successful anaerobic treatment of the industrial waste. We suggest that sulfate reducers, or more accurately sulfidogenic bacteria, together with M. hollandica contribute considerably to the treatment process through metabolism of dye-associated sulfonate groups and subsequent conversion of sulfur compounds to carbon dioxide and methane.
Synthetic organic colorants (e.g., azo dyes) are used universally in manufacturing processes ranging from food and textile production to the printing and pharmaceutical industries (9, 58). By design, the majority of these dyes are recalcitrant so that they can confer color on various raw materials (21); moreover, certain dyes and dye precursors and some aromatic amines produced through biotransformation of dye compounds have been shown to be carcinogenic (28, 34). In addition, very low concentrations of dyes (less than 1 mg/liter) can be highly visible in solution and are therefore aesthetically unacceptable. Some 10,000 dyes are currently manufactured; many of these are azo dyes due to their —N⩵N— bond structure, and it is estimated that at least 15% of them are released into the environment (58), predominantly by textile and dye manufacturing industries (41). Wastewaters produced by dye manufacturers typically comprise mixtures of the various dyes produced by the manufacturers and their intermediate precursors. Appropriate treatment of these wastewaters to remove both color and synthetic dye compounds is clearly an important issue for dye manufacturers.
It has been reported that biodegradation of dye compounds can occur in both aerobic and anaerobic environments (57), although certain azo dyes are known to be resistant to degradation by aerobic bacteria due to the strong electron-withdrawing property of the azo group thought to protect against attack by oxygenases (17, 25, 44). Seshadri and coworkers proposed that biotransformation of azo dyes is a two-step process, in which the azo bond is reduced under anaerobic conditions, producing two aromatic amines, which are then mineralized by aerobic microorganisms (51). In contrast to this proposed process, subsequent research showed that in the presence of readily utilizable cosubstrates, two azo dyes were reduced and decolorized under methanogenic conditions (48), and breakdown products from one azo dye were further mineralized. Due to comparatively low operation costs, use of anaerobic digestion to treat dye wastewater is a cost-effective alternative to the physical and chemical methods commonly used to do this.
An anaerobic baffled reactor (ABR) is a high-rate reactor that contains between three and eight compartments in which the liquid flow is alternately upward and downward between compartment partitions (for a review see reference 4). One of the advantages of the ABR design is the potential for spatial separation of acidogenic and methanogenic populations in the reactor compartments. This design characteristic enables separation of more sensitive anaerobic populations, such as methanogens, from the front of the reactor, where exposure to toxic or unfavorable growth conditions may occur. Successful treatment of an industrial dye waste containing potentially toxic synthetic dye compounds using an ABR has been demonstrated (5).
For accurately describing microbial populations, rRNA-based approaches utilizing the techniques of fluorescent in situ hybridization (FISH) with nucleic acid probes, together with other genetic analyses, have dramatically increased our knowledge of many ecosystems and have yielded a clearer overall picture of microbial diversity (for reviews see references 2, 18, and 20). Research into anaerobic digestion using rRNA-based molecular techniques has provided detailed descriptions of the complex bacterial and archaeal populations present, obviating the need for anaerobic culture techniques (14, 16, 40, 47). An obvious advantage of using FISH with rRNA-targeted nucleic acid probes is that metabolically active cells are detected, so descriptions of the physiologically important population members can be obtained (45). Genetic analysis of 16S rRNA gene sequences extracted directly from microbial ecosystems provides the means to accurately identify population members for which specific nucleic acid probes do not exist and provides additional information for the development of new probes.
Use of anaerobic digesters to decolorize and treat industrial dye waste is a promising alternative to current treatment approaches. By studying the microbial populations involved in this process, we aim to describe the significant microorganisms present in an ABR. Here we describe the use of molecular techniques to characterize the microbial populations in an ABR in order to understand this treatment process. We hope that a more detailed understanding of the microbiology in an ABR will provide information useful for its optimization.
MATERIALS AND METHODS
Food dye manufacturer waste.
The industrial effluent used in this research was obtained from a food dye manufacturer in England. The precise chemical composition of the effluent was not determined and varied according to the production schedule of the manufacturer. The effluent typically contains the intermediate precursors and final synthesis products of up to 15 different azo dyes and other synthetic colorants, most of which are sulfonated compounds (Table 1). The concentration of dye compounds in the effluent is not known; however, an estimate of the total dye concentration based on observations made with the azo dye tartrazine was obtained. Tartrazine typically makes up at least 50% of the manufacturer's production and therefore is one of the most abundant dyes present in the effluent. A solution containing 0.5 mg of tartrazine per liter is almost colorless (as determined by absorbance at 400 nm). The industrial effluent had to be diluted at least 1 in 1,000 to obtain a comparable colorless solution. This suggests that tartrazine was probably present at a concentration of at least 0.5 g/liter. If the presence of other dyes and colorless intermediate precursor compounds is also considered, then the concentration of dye and dye-related compounds was probably at least 1 to 9 g/liter or possibly more than 10 g/liter. Each of the dye compounds has been assayed to determine its anaerobic biodegradability and toxicity for methanogenesis (5), and the compounds range from biodegradable nontoxic compounds to recalcitrant toxic compounds. The pH of the dye effluent was 8 ± 0.2, and analysis of samples with an ion chromatograph (Dionex, United Kingdom) revealed levels of SO42− up to 1,770 mg/liter. The color of the effluent, obtained every 3 to 4 weeks, varied between dark red-brown and dark green according to the production schedule of the manufacturer.
TABLE 1.
Dye compounds routinely produced by the dye manufacturer that are typically present in the industrial wastewater
| Commercial dye | Color index classification | Dye class |
|---|---|---|
| Sunset Yellow Supra | CI Yellow 3 | Monoazo |
| Amaranth Supra | CI Red 9 | Monoazo |
| Carmoisine Supra | CI Red 3 | Monoazo |
| Brown FK Standard | CI Brown 1 | Monoazo |
| Allura Red AC Supra | CI Red 17 | Monoazo |
| Red 2G Supra | CI Red 10 | Monoazo |
| Tartrazine Supra | CI Yellow 4 | Monoazo |
| Ponceau 4R Supra | CI Red 7 | Monoazo |
| Black PN Extra | CI Black 1 | Diazo |
| Green S Supra | CI Green 4 | Triarylmethane |
| Patent Blue V Supra | CI Blue 5 | Triarylmethane |
| Brilliant Blue Supra | CI Blue 2 | Triarylmethane |
| Quinoline Yellow | CI Yellow 13 | Quinoline |
| Erythrosine Supraa | CI Red 14 | Xanthene |
| Indigo Carmine Supra | CI Blue 1 | Indigoid |
Erythrosine Supra is the only nonsulfonated dye compound.
Operation of the ABR.
Operation of the ABR has been described previously (5). Briefly, a 10-liter eight-compartment ABR was seeded with screened digested sludge (18 g of total suspended solids per liter and 12 g of volatile suspended solids per liter) and maintained at 35°C in a water bath. A synthetic feed containing (per liter) 0.67 g of sugar (standard white sugar), 0.2 g of peptone, 0.07 g of meat extract, 0.02 g of K2HPO4, 0.81 g of NaHCO3, and the trace minerals CoCl2 · 6H2O, FeCl2 · 4H2O, MnCl2 · 4H2O, Na2MoO4 · 2H2O, and NiCl2 · 6H2O was added at an organic loading rate of 1.2 g of chemical oxygen demand (COD) per liter with a hydraulic retention time (HRT) of 40 h. Over time, the HRT was gradually reduced to 20 h until the reactor reached a steady state. On day 68 of operation, effluent obtained from a food dye-manufacturing plant was added to the reactor influent at a concentration of 5% (vol/vol) without any change in the HRT or the total organic loading rate. On day 95 of operation, the dye waste concentration was increased to 10% (vol/vol).
Reactor chemical analyses.
The following parameters were measured in each ABR compartment: pH, COD (determined by the closed reflux colorimetric method [American Public Health Association, 1989]), total organic carbon (TOC) (used instead of COD due to interference from the dye waste; determined with a Shimadzu TOC-5050 apparatus), volatile fatty acids (VFAs; determined with a Shimadzu SCL-10A high-performance liquid chromatograph by using an Aminex column, 0.01 M H2SO4 carrier at a flow rate of 0.5 ml/min, and an oven temperature of 35°C), compartment headspace gas composition (determined by gas chromatography with a Shimadzu GC-TCD gas chromatograph and a Poropak N column), and color at the maximum absorbance of 500 nm (Shimadzu 1201 spectrophotometer).
Collection of samples for microbial analyses.
On days 60, 80, and 100 samples of sludge from each compartment of the ABR were collected through a sampling port at the top of each compartment by using a needle and a syringe. An aliquot of each of the samples was fixed for in situ hybridization by using 4% paraformaldehyde and ethanol as previously described (55), and these aliquots were stored at −20°C before they were used. The remaining unfixed portions of the samples were stored at −20°C for later use. Immediately prior to hybridization, smears of fixed sludge samples were dehydrated in an ascending ethanol series (50, 80, and 96% ethanol for 3 min each).
Oligonucleotide probes and in situ hybridization.
Oligonucleotide probes (Table 2) were synthesized with a C6-trifluoroacetyl amino linker at the 5′ end and labelled with either FLUOS, TAMRA, or CY3 (MWG Biotech, Ebersberg, Germany, or Interactiva, Ulm, Germany). In situ hybridization of fixed sludge samples was performed on Teflon-coated multiwell glass slides (Merck) in a buffer containing 0.9 M NaCl, 20 mM Tris-HCl (pH 7.4), 0.01% sodium dodecyl sulfate (SDS), 50 ng of the oligonucleotide probe per ml, and appropriate amounts of formamide (Table 2). Slides containing hybridization buffer were incubated for 2 h at 46°C inside an equilibrated humidity chamber. Following this, the slides were rinsed with and then immersed in a wash buffer containing 20 mM Tris-HCl (pH 7.4), 0.01% SDS, and between 0.9 M and 7 mM NaCl according to the formula of Lathe (26) for 10 to 15 min at 48°C. After washing, the slides were rinsed in distilled water, air dried, and mounted with Vectashield mounting medium containing DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories, Peterborough, United Kingdom). Probes BET42a and GAM42a were used with competitor probes as described previously (36).
TABLE 2.
Oligonucleotide probes used in this study
| Probe | Specificity (rRNA target, position)a | Sequence | % Formamide | Reference |
|---|---|---|---|---|
| EUB338 | Bacteria (16S, 338–355) | GCTGCCTCCCGTAGGAGT | 20 | 1 |
| ARC915 | Archaea (16S, 915–934) | GTGCTCCCCCGCCAATTCCT | 20 | 52 |
| ALF1b | α-Proteobacteria (16S, 19–35) | CGTTCGYTCTGAGCCAG | 20 | 36 |
| BET42a | β-Proteobacteria (23S, 1027–1043) | GCCTTCCCACTTCGTTT | 35 | 36 |
| GAM42a | γ-Proteobacteria (23S, 1027–1043) | GCCTTCCCACATCGTTT | 35 | 36 |
| SRB385 | δ-Proteobacteria (16S, 385–402) | CGGCGTCGCTGCGTCAGG | 20 | 1 |
| BAC303 | Bacteroides-Prevotella (16S, 303–319) | CCAATGTGGGGGACCTT | 0 | 37 |
| CF319a | Cytophaga-Flavobacterium (16S, 319–336) | TGGTCCGTGTCTCAGTAC | 35 | 37 |
| HGC69a | Actinobacteria (23S, 1901–1918) | TATAGTTACCACCGCCGT | 25 | 49 |
| LGC354a | Firmicutes with low G+C content (16S, 354–371) | TGGAAGATTCCCTACTGC | 20 | 39 |
| LGC354b | Firmicutes with low G+C content (16S, 354–371) | CGGAAGATTCCCTACTGC | 20 | 39 |
| LGC354c | Firmicutes with low G+C content (16S, 354–371) | CCGAAGATTCCCTACTGC | 20 | 39 |
| DSV698 | Desulfovibrionaceae (16S, 698–717) | GTTCCTCCAGATATCTACGG | 20 | 38 |
| DSB985 | Desulfobacteriaceae (16S, 985–1004) | CACAGGATGTCAAACCCAG | 0 | 38 |
| MX825 | Methanosaeta (16S, 825–847) | TCGCACCGTGGCCGACACCTAGC | 20 | 47 |
| MS821 | Methanosarcina (16S, 821–844) | CGCCATGCCTGACACCTAGCGAGC | 20 | 47 |
E. coli numbering (8).
Cells were visualized with a Zeiss (Jena, Germany) Axioskop epifluorescence microscope equipped with a 50-W high-pressure bulb or with a Zeiss LSM 510 confocal laser scanning microscope using argon and helium-neon lasers for imaging. Images were obtained with the confocal laser scanning microscope and were prepared further by using Adobe Photoshop. Cells were counted by sampling at least 20 randomly selected fields, and usually at least 1,500 cells were counted for each probing event.
Extraction and purification of total sample DNA.
A sample of frozen sludge collected from compartment 1 on day 100 of ABR operation was thawed, and 500 μl of the sample sludge was combined with an equal volume of buffer containing 200 mM NaCl, 200 mM Tris-HCl, 2 mM sodium citrate, and 10 mM CaCl2 adjusted to pH 8 with HCl. To this, lysozyme was added to a final concentration of ca. 5 mg/ml, and this was followed by gentle mixing and incubation at 37°C for 40 min. SDS and proteinase K were added to final concentrations of 0.3% and 2 mg/ml, respectively, and after further gentle mixing the samples were incubated at 50°C for 30 min. Then physical lysis was performed by adding SDS to a final concentration of 5% together with an equal volume of phenol-chloroform-isoamylalcohol (24:24:1) and 0.33 volume of acid-washed 0.1-mm-diameter zirconia glass beads (Stratech, Bedfordshire, United Kingdom). Samples were shaken on a Mini Bead-Beater (BioSpec Products, Bartlesville, Tenn.) for 2 min at the low setting. Samples were then centrifuged for 3 min at 12,000 × g and 4°C to pellet the beads, and each supernatant was transferred to a new tube. Samples were then extracted with an equal volume of phenol-chloroform-isoamyl alcohol (24:24:1) by thorough mixing and centrifugation for 3 min at 12,000 × g. Nucleic acids were precipitated by adding an equal volume of isopropanol and ca. 0.1 volume of 3 M sodium acetate (pH 5.2), and samples were placed on ice for 30 min and then centrifuged for 20 min at 12,000 × g and 4°C. The pellets were rinsed with 500 μl of chilled 70% ethanol and then air dried for 30 min before being resuspended in 50 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8). The extracted DNA was purified with the Wizard DNA Clean-Up system (Promega, Madison, Wis.) and eluted in 50 μl of TE buffer.
Amplification, cloning, and sequencing of archaeal 16S rDNA.
PCR amplification of archaeal 16S ribosomal DNA (rDNA) was performed by using archaea-specific primer 1Af (forward); (5′-TCYGKTTGATCCYGSCRGAG-3′) (33) and universal primer 1492r (5′-TACGGYTACCTTGTTACGACTT-3′). PCR mixtures (total volume, 100 μl) contained 10 μl of 10× PCR buffer, 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 0.2 μg of each primer, 0.1 to 0.2 μg of DNA template, and 2 U of BioTaq polymerase (Bioline, London, United Kingdom). Thermal cycling was performed with a hot start at 96°C for 5 min before the polymerase was added, followed by 28 cycles of 52°C for 1 min, 72°C for 2 min, and 94°C for 1 min and a final extension at 72°C for 5 min in a thermal cycler (Hybaid, Teddington, United Kingdom). The reaction products were visualized by 1% agarose gel electrophoresis and then purified with the Wizard PCR-Prep DNA purification system (Promega). A purified PCR product was inserted into the TA cloning vector (pCR2.1) by using a TOPO-TA cloning kit (Invitrogen Corporation, San Diego, Calif.) and then transformed into the competent Escherichia coli cells provided. Plasmid inserts were amplified by PCR by using the M13 primer set (Invitrogen), and full-sized-insert clones were screened by using two restriction endonucleases, HhaI and HaeIII (New England Biolabs), and grouped into operational taxonomic units (OTUs) on the basis of their restriction profiles. Representative clones from different OTU groups were selected for sequence analysis. Plasmids were prepared from selected clones for DNA sequencing by using a Flexi-prep kit (Pharmacia). Automated DNA sequencing was performed with an ABI model 377 sequencer (Applied Biosystems) by using the M13 primer set.
Sequence analysis.
Initially, sequences were compared with sequences in publicly accessible sequence databases by using the basic local alignment search tool (BLAST) (15) to determine the approximate phylogeny. Sequence data were aligned and analyzed with the ARB program package (available at http://www.mikro.biologie.tu-muenchen.de/) by comparing clone sequences with sequences in the database released with ARB and the RDP_SSU database (35). The program Check_Chimera was used to screen clone sequence data for the presence of chimeras (35). Other sequences used in the analyses were obtained from GenBank. Phylogenetic trees were constructed with distance matrix calculations by using the Jukes-Cantor correction (22) and neighbor-joining functions available in the ARB program. Bootstrapping (1,000 samples) was used to test the stability of the branching patterns in the phylogenetic trees.
Nucleotide sequence accession numbers.
The partial nucleotide sequences obtained in this study have been deposited in the EMBL database under accession numbers AJ288310 to AJ288324.
RESULTS
Reactor performance.
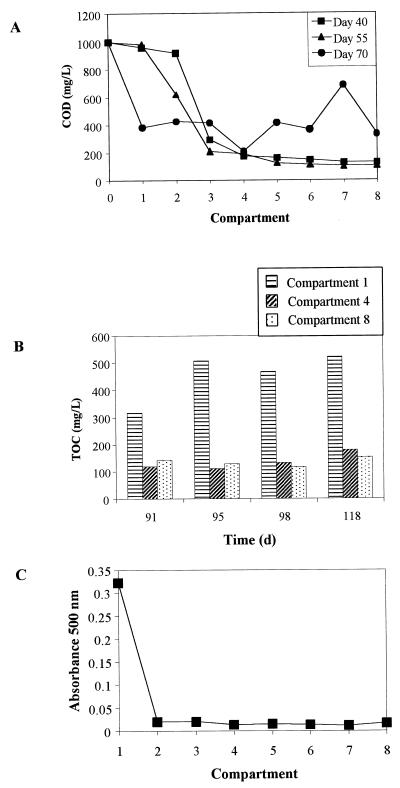
During operation of the ABR before the dye waste was added, COD removal values of around 90% were achieved, and following addition of the dye waste, TOC removal values between 70 and 80% were consistently achieved. Figure 1 shows COD and TOC values for reactor compartments at selected times. A profile of color removal in the reactor for samples taken on day 100 (Fig. 1C) shows that color removal was almost complete by compartment 2.
FIG. 1.
ABR performance. (A) Compartment COD levels before addition of the dye waste (days 40 and 55) and just after addition (day 70). (B) TOC levels in compartments 1, 4, and 8 after addition of the dye waste. (C) Profile of color removal in the ABR compartments measured by using absorbance at 500 nm for samples taken on day 100.
Population analysis.
Initial hybridizations performed with universal bacterial and archaeal probes revealed an abundance of members of both of these phyla in the first three compartments on each sampling date. Comparisons in which DAPI staining was used to quantify the relative proportion of each of the population groups were attempted; however, due to the extensive filamentous or tightly clustered microcolonies of the archaea present, accurate counting of individual cells by visual means was not possible. However, it was estimated that at least 80% of all DAPI-stained cells hybridized with either the universal bacterial or archaeal probes in compartments 1, 2, and 3. The signal intensity of cells in the first three compartments when they were hybridized to the bacterial or archaeal probes was relatively strong compared with the signal intensity of cells present in compartments 4 to 8, indicating that there was decreased metabolic activity in the latter compartments. The metabolic activity profiles for different compartments correlated with the measured COD-TOC removal profiles in the ABR (Fig. 1A). Estimates of the relative ratios of the numbers of bacteria and archaea in compartments 1 to 3 ranged from ca. 70:30 in compartment 1 to 50:50 in compartment 3 for each sampling date. Analyses of samples taken from compartments 4 to 8 revealed that the bacterial populations declined relative to the archaeal populations, and we estimated that 90% of the DAPI-stained cells were detected by primer ARC915 by compartments 7 and 8. The characteristic morphology of Methanosaeta cells (long sheathed filaments) was visualized by using the universal archaeal probe, and the identity of these filaments was confirmed by using genus-specific probe MX825. Other morphotypes observed hybridizing to ARC915 included Methanospirillum-like shorter filaments, single rods, and sarcina-like clusters of irregular cocci often found in microcolonies consisting of up to 50 or more cells. On the basis of the intense probe-conferred fluorescence, these cocci appeared to be very active. These cocci, either in a sarcina-like arrangement or as microcolonies, did not hybridize with Methanosarcina-specific probe MS821.
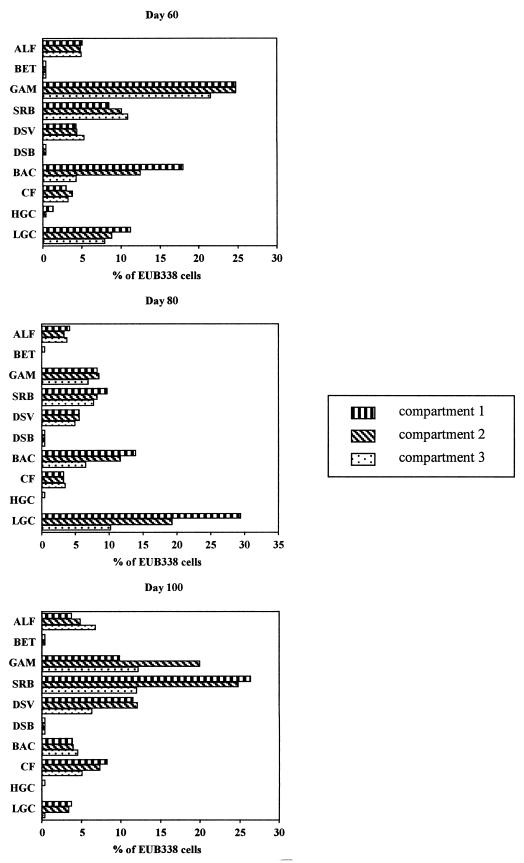
Further probing of the samples with group-specific probes targeting various bacterial phyla (Table 2) revealed the presence of considerable diversity. Cell counts obtained with EUB338 for the first three compartments for each of the sampling dates are shown in Figure 2. The sum of the percentages obtained with the group-specific probes was less than 100% for each of the samples analyzed, indicating that bacteria which were not detected with these probes were present. The bacterial populations present prior to addition of the dye waste (day 60) were dominated by cells detected with the GAM42a probe (between 20 and 25% in each compartment). Considerable numbers of cells (usually >5%) were detected with probes BAC303 and SRB385 and also with the LGC probe set in the day 60 samples. With probes CF319a, ALF1b, BET42a, and HGC69a, cells were detected at levels of ca. 5% or less in each compartment except compartment 3, in which no cells were detected with the HGC69a probe. In general, the percentage of cells detected with each group-specific probe remained fairly constant from compartment 1 to compartment 3; the exceptions were probe HGC69a, with which no cells were detected in compartment 3, and probe BAC303, with which the level varied from 18% in compartment 1 to 4% in compartment 3. Throughout operation of the ABR, especially after addition of the dye waste, considerable blackening of the inside of the first compartment and the influent tube due to production of sulfide was observed. Two probes, DSV698 and DSB985, were used in hybridizations to provide further descriptions of sulfate-reducing bacteria. The cell counts with the DSV698 probe were between 4 and 6% for each compartment, whereas with probe DSB985 the counts were very low in compartments 1 and 2 and no cells were detected in compartment 3.
FIG. 2.
Bacterial community analysis for ABR compartments 1, 2, and 3 sampled on days 60, 80, and 100, showing counts obtained with 10 different group-specific probes expressed as percentages of the total bacterial counts obtained with probe EUB338. ALF, probe ALF1b; BET, probe BET42a; GAM, probe GAM42a; SRB, probe SRB385; DSV, probe DSV698; DSB, probe DSB985; BAC, probe BAC303; CF, probe CF319a; HGC, probe HGC69a; LGC, LGC probe set.
In the day 80 samples (after addition of the dye waste), cells detected with the LGC probe set were numerically dominant in compartments 1 and 2 (ca. 29 and 19%, respectively). Also, cells detected with the BAC303 probe accounted for more than 10% of the bacteria in compartments 1 and 2. With probes SRB385, DSV698, and GAM42a, between 5 and 10% of the bacterial cells were detected in all three compartments. With probes CF319a and ALF1b, between 3 and 5% of all bacteria were detected in each compartment. Other probes were used, but only in compartment 1 were cells detected with either HGC69a or BET42a, and probe DSB985 detected less than 1% of the bacterial cells in each compartment. Analysis of the day 100 samples revealed a different population structure. Cells detected with the SRB385 and GAM42a probes were present at higher levels in each compartment (10% or more), and ca. 25% of all bacterial cells detected with EUB338 in compartments 1 and 2 were also detected with SRB385. In the first two compartments, the counts obtained with probe DSV698 were ca. 12% of the bacterial cells. Figure 3 shows an example of the cells detected with probe DSV698. Probe CF319a detected between 5 and 10% of the bacterial cells in each compartment, whereas the counts obtained with BAC303 and the LGC probe set were less than 5% of the bacterial cells. The counts obtained with ALF1b varied from less than 5% of the bacterial cells in compartments 1 and 2 to almost 7% in compartment 3. Low numbers of cells were detected with probes HGC69a and BET42a in some compartments, and low numbers of cells were detected with probe DSB985 in all compartments. In order to rationalize the counts obtained with probes SRB385 and DSV698, simultaneous hybridizations using these probes with the day 100 compartment 1 sample revealed that 44.1% of the cells detected with SRB385 also hybridized with DSV698. Cells with a vibrio morphology comprised the majority of the cells detected with DSV698, compared with the mixed cell morphotypes exhibited by cells detected with SRB385.
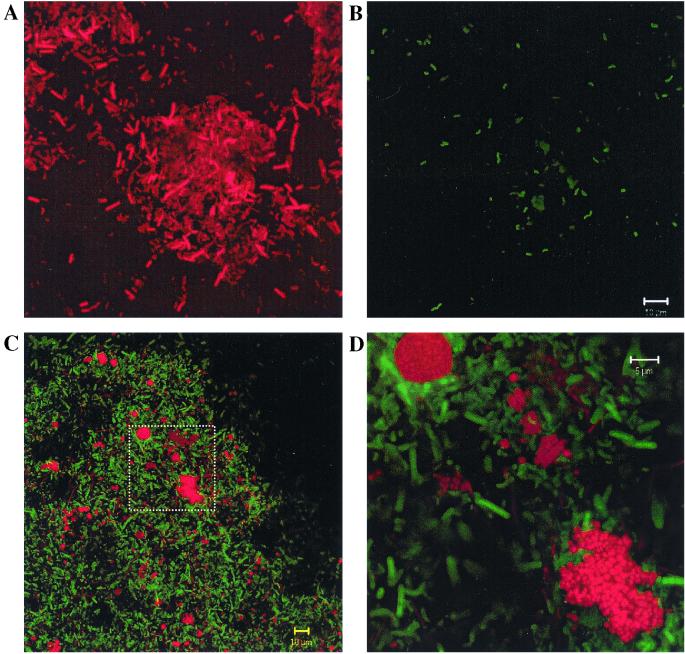
FIG. 3.
Confocal laser scanning micrographs of FISH-probed ABR samples. (A and B) Cells in the same field. Cells were detected with probe EUB338 (A) and with probe DSV698 (B). The vibrio morphology typical of Desulfovibrio spp. is exhibited by several cells in panel B. (C and D) Cells in a sludge floc detected with probes ARC915 (red) and EUB338 (green). The rectangle in panel C is magnified in panel D. In this image, the cells detected with the ARC915 probe appear more clearly as microcolonies of coccoid cells.
A noticeable trend with respect to the archaeal population members in the compartments sampled was the increase in the relative proportion of sarcina or tightly clustered microcolonies of cells detected with the ARC915 probe. Although some sarcina cells were present in the day 60 sample, the number appeared to increase by day 80, and these cells were very numerous in the day 100 sample. The sarcina cells and tight clusters of irregular cocci detected with ARC915 (Fig. 3) were abundant in compartment 1 and virtually nonexistent in compartments 2 to 8, and the archaeal morphotypes were dominated by filaments typical of Methanosaeta and, to a lesser degree, Methanospirillum-like morphotypes with single rods.
rDNA clone analysis.
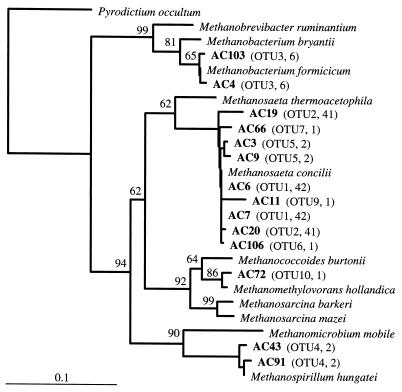
An effort to identify the archaea observed when FISH was performed with ARC915, particularly the clusters of irregular colonies present in the first compartment, was made by constructing a 16S rDNA clone library. Total community DNA was extracted from a day 100 compartment 1 sample. A total of 98 clones (designated with the prefix AC) with full-size inserts were obtained for analysis after PCR amplification of archaeal 16S rDNA. Screening of these 98 clones by HhaI and HaeIII restriction enzyme analysis grouped the clones into 10 OTUs. OTUs 1 and 2 were the largest groups, containing 42 and 41 clones, respectively. OTU 3 contained six clones, OTUs 4 and 5 contained two clones, and the remaining OTUs contained one clone each. When possible, at least two representatives of each OTU were sequenced. Phylogenetic analysis of partial sequence data (ca. 650 nucleotides) for clones representing OTUs was used to determine the phylogenetic position of each clone (Fig. 4). According to these analyses, all but one of the clones were affiliated with the domain Archaea. All clones affiliated with members of the Archaea were most closely related to methanogenic members of the Euryarchaeota. Sequence data for OTU 1 and 2 representative clones grouped closely with sequence data for Methanosaeta concilii. Clones in OTUs 5, 6, 7, and 9 grouped with the representatives of OTUs 1 and 2; assuming that unsequenced members of OTUs 1 and 2 have similar sequences, then ca. 90% of the clones obtained were affiliated with the genus Methanosaeta. Clones that grouped in OTU 3 were closely related to Methanobacterium formicicum and accounted for ca. 6% of the library. The only two clones in OTU 4 grouped closely with the sequence of Methanospirillum hungatei. The single clone in OTU 10 (AC72) was closely affiliated with the sequence of Methanomethylovorans hollandica, a member of the Methanosarcinaceae.
FIG. 4.
Phylogenetic tree based on partial 16S rRNA sequences, showing the phylogenetic positions of archaeal clones obtained from compartment 1 of the ABR on day 100. The tree was constructed by using distance matrix methods with the Jukes-Cantor correction and neighbor joining. Bootstrap values greater than 50% indicate areas of stable tree topology based on 1,000 bootstrap resamplings. Scale bar = 0.1 substitution per sequence position. The information in parentheses indicates the OTU for each clone and the total number of clones in each OTU. The accession numbers for the reference sequences are as follows: Pyrodictium occultum, M21087; Methanobrevibacter ruminantium, RDP Locus Mbb. rumina; Methanobacterium bryantii, M59124; Methanobacterium formicicum, M36508; Methanosaeta thermoacetophila, M59141; Methanosaeta concilii, M59146; Methanococcoides burtonii, X65537; Methanomethylovorans hollandica, AF120163; Methanosarcina barkeri, M59144; Methanosarcina mazei, AJ012095; Methanomicrobium mobile, M59142; and Methanospirillum hungatei, M60880.
DISCUSSION
Reduction of azo dyes has been shown to occur under anaerobic conditions, particularly in association with intestinal microorganisms (7, 48). Several different mechanisms have been proposed for reduction or degradation of azo dyes and similar compounds. A description of a nonspecific azo reductase system involved in azo dye reduction has been provided for selected bacterial species, and it has been shown that the relevant gene is relatively conserved in various anaerobic and facultative bacteria (24, 46). Redox mediator compounds, such as flavins, have been shown to enhance degradation of azo dyes by acting as electron shuttles that facilitate reduction of the azo dye (13). In this research it was also hypothesized that coenzyme reducing equivalents involved in normal electron transport through oxidation of organic substrates may act as electron donors for reduction of azo dyes. This would likely explain the observation that azo dye reduction or degradation occurs more readily as a cometabolic event when additional readily degradable substrates are provided. When a readily utilizable synthetic feed was provided together with the dye waste, it was hoped that metabolism of the dye compounds in the waste would be enhanced because of the greater metabolic activity due to degradation of the synthetic feed components. Recent research has shown not only that sulfide is able to chemically decolor azo dyes, but also that azo reduction in living sulfidogenic anaerobic sludge is three times more rapid than the chemical reaction due to sulfide alone (54).
One key to successful degradation of azo dyes and nitroaromatic compounds is the toxicity of the compounds for the microorganisms. In anaerobic digestion, mineralization of organic compounds to carbon dioxide and methane relies heavily on the methanogenic Archaea for the final phase. Previous research has shown that azo dyes are often more toxic to methanogens than their breakdown products are (10). As determined by FISH and the rRNA clone library constructed from day 100 compartment 1 sample material, methanogenic species of Archaea were present in compartment 1. These organisms can be considered metabolically active based on their detection by FISH, and therefore the data may indicate that the potential toxicity of dye compounds for methanogens is negligible at the dye loading rate used. In this study, it was not possible to discern any inhibition of methanogenesis in the reactor compartments. Analysis of the volatile fatty acids in compartment 1, however, showed that there was an increase in the level of acetate from <100 to ca. 800 mg/liter after the dye waste was added to the ABR, that the level of acetate returned to ca. 100 mg/liter after several days, and that there was another slightly smaller increase in the level of acetate when the load of dye waste was increased to 10% (vol/vol) (J. Bell, unpublished data). It appears that at first the dye waste may have slightly inhibited the acetotrophic methanogen populations before these populations acclimated to the changed environment and resumed more complete acetate utilization.
Separation of microbial trophic groups in the compartments of the ABR (4) was observed to some extent when FISH probing was used. The results obtained with universal bacterial and archaeal probes revealed an aspect of this separation; there was a higher percentage of bacteria relative to archaea in the front of the reactor than in compartments 7 and 8, in which comparatively few bacteria were detected. Detailed descriptions of microbial populations obtained with fluorescently labelled probes provided insight into the population dynamics in the front compartments of the ABR. The use of group-specific probes targeting different bacterial phyla revealed different bacterial population structures at the three sampling times examined. Before the dye waste was added at day 60, the numerical dominance of members of the gamma subclass of the class Proteobacteria (gamma-Proteobacteria) was clear. These bacteria, particularly the organisms which are fermentative, were expected to be present in the front of the reactor. The relative decline in this population of bacteria seen in the day 80 counts was a noticeable change in population structure, as was the increase in the number of gram-positive bacteria detected with the LGC probe set. It was not possible to determine whether these changes in population structure were related to the dye effluent or were simply due to background population fluctuations. The degree of microbial population change that occurs while steady-state conditions are maintained has been studied previously (11). Although this research employed PCR-based methods subject to several biases, which limited the accuracy of the quantitative descriptions, the authors claim that an extremely dynamic community can maintain a stable ecosystem function. Further efforts to study these community dynamics using more accurate enumerative techniques, such as FISH with rRNA-targeted probes, could be made. The increase in the dye waste level from 5 to 10% (vol/vol) may have favored growth of other microorganisms, such as gamma-Proteobacteria and cells detected with the SRB385 probe. The specificity of probe SRB385 is not phylogenetically consistent, as the probe target sequence is present not only in members of the delta-Proteobacteria (some sulfate-reducing bacteria) but also in some Actinobacteria (e.g., Frankia species, some clostridia, at least one species of Nitrospira, and many other phylogenetically diverse organisms). At times, SRB385 has been used as a general probe for detecting sulfate-reducing bacteria; however, Manz et al. highlighted the limitations of this probe for studying sulfate reducers and designed and tested several other more specific probes for these organisms (38). Use of two of these probes in this study, DSV698 and DSB985 (Table 2), provided more accurate enumeration of sulfate-reducing bacteria. The counts obtained with DSV698 show that species of Desulfovibrio comprised a considerable proportion of the community in each of the compartments on day 100. The results of the simultaneous hybridizations performed with SRB385 and DSV698 probably demonstrate the relatively broad specificity of the SRB385 probe. The proportions of cells that hybridized to SRB385 but not to DSV698 are unknown, but such cells accounted for considerable proportions of the populations in the ABR. PCR amplification of community 16S rDNA using SRB385 as a primer followed by cloning and analysis of clone inserts could be used to characterize this diversity.
The precise role of sulfate-reducing bacteria in the samples studied is not clearly defined, although as mentioned above, sulfide is capable of chemically reducing azo dyes. Sulfate-reducing bacteria are commonly detected in anaerobic reactors, and their abundance has been shown to vary depending on the sulfate level (14, 40). The sources of the sulfate used as a terminal electron acceptor in the ABR are likely to be compounds in the synthetic feed (ca. 50 mg/liter [L. H. Freese and J. J. Plumb, unpublished data]) and the dye waste (up to 1,770 mg/liter). The 1 in 20 dilution and later 1 in 10 dilution of the dye waste in the influent therefore resulted in a combined influent sulfate level of less than 250 mg/liter. Although this sulfate concentration is greater than 100 mg/liter, a limiting concentration for sulfate reduction (43), it seems unlikely that it is high enough to explain the sulfide production observed in the ABR and the 2.5-fold increase in sulfate-reducing bacteria. The cell counts in compartments 1 and 2 increased from <5% on day 60 to ca. 12% on day 100 with probe DSV698, and the cell counts obtained with the less specific probe SRB385 showed a similar increase. Based on these increases, it appears that there were favorable conditions for sulfidogenesis in the first two compartments of the ABR.
The range of compounds known to be used as terminal electron acceptors by sulfidogenic bacteria has been extended. In addition to sulfate and sulfite, other inorganic ions, such as nitrate, nitrite, and chromate, and organic molecules, such as fumarate and the sulfonic acid taurine, can serve as terminal electron acceptors (27, 29, 30). The reduction of sulfonates by sulfate-reducing bacteria suggests that this metabolism comprises a significant part of the global sulfur cycle particularly in ecosystems such as sediments and forest soils which contain up to 50% of their sulfur in the form of sulfonates (23). As Table 1 shows, all but one of the dyes typically found in the waste stream are sulfonated dyes. Dye waste therefore provides a considerable source of sulfonates that can be used as terminal electron acceptors. Members of the genus Desulfovibrio and Bilophila wadsworthia are among the growing list of bacteria that have been shown to reduce sulfonates (27, 31). Counts obtained with probe DSV698, which is specific for B. wadsworthia as well as the genus Desulfovibrio, probably indicate the levels of sulfonate-reducing bacteria. It is also likely that counts obtained with the less specific probe SRB385 represent other species of sulfate-reducing bacteria that also reduce sulfonates. Lie et al. (29) have suggested that cleavage of the carbon-sulfur bond of sulfonates is an energy-yielding step which results in accumulation of thiol compounds before final conversion into sulfide. Although metabolism of sulfonates under anaerobic conditions is not completely understood (for a review see reference 8), it appears that sulfate-reducing bacteria may both directly (sulfonate reduction) and indirectly (sulfide production) aid in overall degradation of dye waste.
Members of the Cytophaga-Flexibacter-Bacteroides phylum (cells detected with probes CF319a and BAC303) comprised a considerable proportion (usually at least 10%) of the bacterial community in each of the samples analyzed. Members of the Cytophaga-Flexibacter group within this phylum are ubiquitous microorganisms and have diverse physiologies. Members of the Bacteroides group are obligate anaerobes which commonly comprise a considerable proportion of the intestinal or ruminal microflora (19, 53). The possibility that the intestinal microflora is involved in degradation of ingested food dyes has been considered for a long time, and experimental evidence has shown that predominant intestinal microorganisms are capable of azo dye reduction (7).
Analysis of clones obtained with an archaea-specific primer and a universal primer after PCR amplification of 16S rDNA from the day 100 compartment 1 sample revealed some of the diversity of the methanogens present. The dominance of Methanosaeta phylotypes in the clone library was not unexpected as the numerous Methanosaeta-like sheathed filaments that hybridized to the ARC915 probe also hybridized to Methanosaeta-specific probe MX825. Strains of Methanosaeta concilii are known to be important members of anaerobic methanogenic communities due to their ability to metabolize acetate into carbon dioxide and methane, and their numerical dominance compared to other methanogens in anaerobic reactors has been reported previously (12, 40, 50). The presence of phylotypes closely related to the well-studied methanogenic species Methanobacterium formicicum and Methanospirillum hungatei also correlates with microscopic observations. These two species utilize hydrogen and formate for methanogenesis and are commonly found in anaerobic reactors. The presence of many tightly clustered sarcina-like irregular cocci detected with the ARC915 probe suggested that Methanosarcina species would be represented in the clone library, although these cells did not hybridize with Methanosarcina-specific probe MS821. This was not the case despite the suitability of the 1Af primer for amplifying 16S rDNA from Methanosarcina species based on sequence database comparisons. One phylotype, whose sequence was very similar (99% similarity) to sequence data for the recently described organism Methanomethylovorans hollandica (33), was the only other representative of the Methanosarcinaceae in the clone library. Unlike Methanosarcina species, this organism does not utilize hydrogen, carbon dioxide, or acetate and was isolated from a freshwater sediment by using dimethyl sulfide as the sole source of carbon and energy. The morphology of this species has been described as being between the morphology of Methanosarcina cell clusters and the morphology of the irregular cocci typical of Methanolobus and Methanococcoides species (33). This accurately describes the morphotypes seen in this study, and aggregation of small clusters into larger microcolonies has also been observed for this species. It has been claimed that as an obligate methylotroph, Methanomethylovorans hollandica is a key consumer of dimethyl sulfide and methanethiol in anaerobic environments, although this organism is also able to utilize methanol and methylamines. It seems possible that compounds such as methanethiol are formed during reduction of sulfonate from dye compounds. The methanethiol could then be utilized by Methanomethylovorans hollandica for methanogenesis. Alternatively, it has been shown that formation of methanethiol and dimethyl sulfide occurs readily under anaerobic conditions when sulfide concentrations are high (32). It seems likely that conditions in the front of the ABR approximate these conditions, resulting in production of these two key substrates for growth of Methanomethylovorans hollandica. Although not as numerous as Methanosaeta cells, these methanogenic cocci account for a considerable proportion of the archaeal biomass, possibly as much as 15%, and appear to be highly active on the basis of the very intense fluorescence observed when FISH is used. Together with sulfidogenic bacteria, Methanomethylovorans hollandica appears to play an important role in the overall degradation of dye waste.
Concluding remarks.
The molecular approaches used in this study provided useful descriptions of the microorganisms actively involved in treatment of an industrial dye waste. FISH performed with group-specific probes together with rRNA clone library analysis showed that a diverse community of microorganisms combined to successfully treat an industrial dye effluent in the ABR. While it is not clear what the roles of individual members of the population are in the metabolism of the dye waste, we suggest that sulfidogenic bacteria, such as Desulfovibrio spp., may be involved in reduction of dye-associated sulfonate groups and production of sulfide, which can reduce azo dyes chemically. The role of the organosulfur-utilizing organism Methanomethylovorans hollandica present in the first compartment of the reactor remains unclear, but this organism may be associated with sulfonate reduction. Also, this study is the first study in which rRNA-based molecular approaches were used to study the population dynamics of an ABR.
ACKNOWLEDGMENTS
We acknowledge the financial support of the Engineering and Physical Sciences Research Council, the South African Water Research Commission, the National Research Foundation, and the British Council.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber W P, Stuckey D C. The use of the anaerobic baffled reactor (ABR) for wastewater treatment: a review. Water Res. 1999;33:1559–1578. [Google Scholar]
- 5.Bell J, Plumb J J, Buckley C A, Stuckey D C. Treatment and decolorization of dyes in an anaerobic baffled reactor. J Environ Eng. 2000;126:1026–1032. [Google Scholar]
- 6.Brosius J, Dull T L, Steeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 7.Chung K-T, Fulk G E, Egan M. Reduction of azo dyes by intestinal anaerobes. Appl Environ Microbiol. 1978;35:558–562. doi: 10.1128/aem.35.3.558-562.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook A M, Laue H, Junker F. Microbial desulfonation. FEMS Microbiol Rev. 1999;22:399–419. doi: 10.1111/j.1574-6976.1998.tb00378.x. [DOI] [PubMed] [Google Scholar]
- 9.Cooper P. Colour in dyehouse effluent. Society of Dyers and Colourists. London, United Kingdom: Alden Press; 1995. [Google Scholar]
- 10.Donlon B, Razo-Flores E, Luijten M, Swarts H, Lettinga G, Field J. Detoxification and partial mineralization of the azo dye Mordant Orange 1 in a continuous anaerobic sludge-blanket reactor. Appl Microbiol Biotechnol. 1997;47:83–90. [Google Scholar]
- 11.Fernández A, Huang S, Seston S, Xing J, Hickey R, Criddle C, Tiedje J. How stable is stable? Function versus community composition. Appl Environ Microbiol. 1999;65:3697–3704. doi: 10.1128/aem.65.8.3697-3704.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ficker M, Krastel K, Orlicky S, Edwards E. Molecular characterization of a toluene-degrading methanogenic consortium. Appl Environ Microbiol. 1999;65:5576–5585. doi: 10.1128/aem.65.12.5576-5585.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gingell R, Walker R. Mechanisms of azo reduction by Streptococcus faecalis. II. The role of soluble flavins. Xenobiotica. 1971;1:231–239. doi: 10.3109/00498257109033172. [DOI] [PubMed] [Google Scholar]
- 14.Godon J J, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenberg A E, Clesceri L S, Eaton A D, editors. Standard methods for the examination of water and wastewater. 18th ed. Washington, D.C.: American Public Health Association; 1992. [Google Scholar]
- 16.Harmsen H J M, Kengen H M P, Akkermans A D L, Stams A J M, de Vos W M. Detection and localization of syntrophic propionate-oxidizing bacteria in granular sludge by in situ hybridization using 16S rRNA-based oligonucleotide probes. Appl Environ Microbiol. 1996;62:1656–1663. doi: 10.1128/aem.62.5.1656-1663.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haug W, Schmid A, Nörtemann B, Hempel D C, Stolz A, Knackmuss H-J. Mineralization of the sulfonated azo dye Mordant Yellow 3 by a 6-aminonaphthalene-2-sulfonate-degrading bacterial consortium. Appl Environ Microbiol. 1991;57:3144–3149. doi: 10.1128/aem.57.11.3144-3149.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology. A decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 19.Holdeman L V, Good I J, Moore W E C. Human fecal flora: variation in bacterial composition within individuals and a possible effect on emotional stress. Appl Environ Microbiol. 1976;31:359–375. doi: 10.1128/aem.31.3.359-375.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Idaka E, Ogawa T, Yatome C, Horitsu H. Behavior of activated sludge with dyes. Bull Environ Contam Toxicol. 1985;35:729–734. doi: 10.1007/BF01636580. [DOI] [PubMed] [Google Scholar]
- 22.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 23.Kertesz M A. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in gram-negative bacteria. FEMS Microbiol Rev. 1999;24:135–175. doi: 10.1016/S0168-6445(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 24.Kudlich M, Keck A, Klein J, Stolz A. Localization of the enzyme system involved in anaerobic reduction of azo dyes by Sphingomonas sp. strain BN6 and effect of artificial redox mediators on the rate of azo dye reduction. Appl Environ Microbiol. 1997;63:3691–3694. doi: 10.1128/aem.63.9.3691-3694.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulla H G, Klausener F, Meyer U, Lüdeke B, Leisinger T. Interference of aromatic sulfo groups in microbial degradation of azo dye Orange I and Orange II. Arch Microbiol. 1983;135:1–7. [Google Scholar]
- 26.Lathe R. Synthetic oligonucleotide probes deduced from amino acid sequence data. Theoretical and practical considerations. J Mol Biol. 1985;183:1–12. doi: 10.1016/0022-2836(85)90276-1. [DOI] [PubMed] [Google Scholar]
- 27.Laue H, Denger K, Cook A M. Taurine reduction in anaerobic respiration of Bilophila wadsworthia RZATAU. Appl Environ Microbiol. 1997;63:2016–2021. doi: 10.1128/aem.63.5.2016-2021.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine W G. Metabolism of azo dyes: implication for detoxification and activation. Drug Metab Rev. 1991;23:253–309. doi: 10.3109/03602539109029761. [DOI] [PubMed] [Google Scholar]
- 29.Lie T J, Leadbetter J R, Leadbetter E R. Metabolism of sulfonic acids and other organosulfur compounds by sulfate-reducing bacteria. Geomicrobiol J. 1998;15:135–149. [Google Scholar]
- 30.Lie T J, Clawson M J, Godchaux W, Leadbetter E R. Sulfidogenesis from 2-aminoethanesulfonate (taurine) fermentation by a morphologically unusual sulfate-reducing bacterium, Desulforhopalus singaporensis sp. nov. Appl Environ Microbiol. 1999;65:3328–3334. doi: 10.1128/aem.65.8.3328-3334.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lie T J, Godchaux W, Leadbetter E R. Sulfonates as terminal electron acceptors for growth of sulfite-reducing bacteria (Desulfitobacterium spp.) and sulfate-reducing bacteria: effects of inhibitors of sulfidogenesis. Appl Environ Microbiol. 1999;65:4611–4617. doi: 10.1128/aem.65.10.4611-4617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lomans B P, Smolders A, Intven L, Pol A, Op den Camp H J M, van der Drift C. Formation of dimethyl sulfide and methanthiol in anoxic freshwater sediments. Appl Environ Microbiol. 1997;63:4741–4747. doi: 10.1128/aem.63.12.4741-4747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomans B P, Maas R, Luderer R, Op den Camp H J M, Pol A, Van Der Drift C, Vogels G D. Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol. Appl Environ Microbiol. 1999;65:3641–3650. doi: 10.1128/aem.65.8.3641-3650.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Longstaff E. An assessment and categorization of the animal carcinogenicity data on selected dyestuffs and an extrapolation of those data on the relative carcinogenic risk to man. Dyes Pigments. 1983;4:243–304. [Google Scholar]
- 35.Maidak B L, Cole J R, Parker C T, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligonucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 37.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 38.Manz W, Eisenbrecher M, Neu T R, Szewzyk U. Abundance and spatial organization of gram-negative sulfate reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol. 1998;25:43–61. [Google Scholar]
- 39.Meier H, Amann R, Ludwig W, Schleifer K-H. Specific oligonucleotide probes for in situ detection of a major group of Gram-positive bacteria with low DNA G+C content. Syst Appl Microbiol. 1999;22:186–196. doi: 10.1016/S0723-2020(99)80065-4. [DOI] [PubMed] [Google Scholar]
- 40.Merkel W, Manz W, Szewzyk U, Krauth K. Population dynamics in anaerobic wastewater reactors: modelling and in situ characterization. Water Res. 1999;33:2392–2402. [Google Scholar]
- 41.Meyer U. Biodegradation of synthetic organic colorants. FEMS Symp. 1981;12:371–385. [Google Scholar]
- 42.Munson M A, Nedwell D B, Embley T M. Phylogenetic analysis of Archaea in sediment samples from a coastal salt marsh. Appl Environ Microbiol. 1997;63:4729–4733. doi: 10.1128/aem.63.12.4729-4733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Overmeire A, Lens P, Verstraete W. Mass transfer limitation of sulphate in methanogenic aggregates. Biotechnol Bioeng. 1994;44:387–391. doi: 10.1002/bit.260440318. [DOI] [PubMed] [Google Scholar]
- 44.Pagga U, Brown D. The degradation of dyestuffs: part II, behaviour of dyestuffs in aerobic biodegradation test. Chemosphere. 1986;15:479–491. [Google Scholar]
- 45.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rafii F, Coleman T. Cloning and expression in Escherichia coli of an azoreductase gene from Clostridium perfringens and comparison with azoreductase genes from other bacteria. J Basic Microbiol. 1999;39:29–35. [PubMed] [Google Scholar]
- 47.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Razo-Flores E, Luijten M, Donlon B, Lettinga G, Field J. Biodegradation of selected azo dyes under methanogenic conditions. Water Sci Technol. 1997;36:65–72. [Google Scholar]
- 49.Roller C, Wagner M, Amann R, Ludwig W, Schleifer K-H. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 50.Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl Environ Microbiol. 1999;65:1280–1288. doi: 10.1128/aem.65.3.1280-1288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seshadri S, Bishop P L, Agha A M. Anaerobic/aerobic treatment of selected azo dyes in wastewater. Waste Manag. 1994;15:127–137. [Google Scholar]
- 52.Stahl D A, Amann R. Development and application of nucleic acid probes in bacterial systematics. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons Ltd.; 1991. pp. 205–248. [Google Scholar]
- 53.Stewart C S, Bryant M P. The rumen bacteria. In: Hobson P N, Stewart C S, editors. The rumen microbial ecosystem. London, United Kingdom: Blackie; 1988. pp. 21–75. [Google Scholar]
- 54.van der Zee F P, Lettinga G, Field J A. The role of (auto)catalysis in the mechanism of an anaerobic azo reduction. Water Sci Technol. 2000;42:301–308. [Google Scholar]
- 55.Wagner M, Erhart R, Manz W, Amann R, Lemmer H, Wedi D, Schleifer K-H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Widdel F. Microbiology and ecology of sulphate- and sulphur-reducing bacteria. In: Zehnder A J B, editor. Biology of anaerobic microorganisms. New York, N.Y: Wiley-Liss, John Wiley & Sons Inc.; 1988. pp. 469–586. [Google Scholar]
- 57.Zissi U, Lyberatos G. Azo-dye biodegradation under anoxic conditions. Water Sci Technol. 1996;34:495–500. [Google Scholar]
- 58.Zollinger H. Colour chemistry—synthesis, properties and applications for organic dyes and pigments. New York, N.Y: VCH Publishers, Inc.; 1987. pp. 92–102. [Google Scholar]